Immunology3-
Antigens, Antibodies and their associated proteins:
http://biosiva.50webs.org/
https://www.studyblue.com
Antigens:
In 1899, Ladislas Deutsch (Laszlo Detre) (1874�1939) named the hypothetical substances halfway between bacterial constituents and antibodies "substances immunogens or antigens".
An antigen is a macromolecule that evokes the production of an Antibody or antibodies. Antigen is also considered as immunogen for it induces immune response. Antigen in simple terms inducer an antibody (antibody generator), most of the time an antigen is foreign to animal body; rarely self-cellular molecules can activate or induce antibody formation.� The antigen can also refer to a substance that is presented by Major Histocompatibility Complex (MHC) to T cells.� Concepts immunogenicity and antigenicity are subtly different (according to present concept). Immunogenicity induces humoral and/or cell mediated immune response. Antigenicity is that the substance binds to specific antibodies and/or surface T-cell receptors.
An antigen is molecule of certain dimension made up of proteins, glycoproteins, proteoglycans and polysaccharides and the combination of them.� Antigens have specific structural features, which binds to specific antibodies.� The binding to antibodies on B cells or free circulating Igs is more or less like Lock and Key for they have molecular complementary surfaces.� But Idiotypes are antibody variants bind to the same antigen have an epitope. On the contrary the T cell receptors recognize only the protein components as antigens.
 �
�
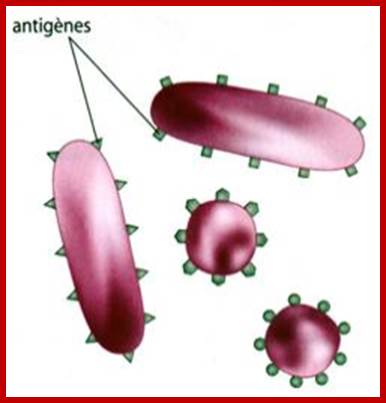
https://svt07.wordpress.com
The dark sites are antigenic sites; antigen determinants, colored for a kind of domains or motifs, which act as the binding surface for Abs.
Antigens mostly exogenous but they can be endogenous, generated from cells within the body due to viral or bacterial infection, that generates an antigen, which have to be presented by MHCI class proteins, if activated they are attacked by cytotoxic CD8T cells leading to lysis or apoptosis; or they can be auto antigens developed from cells within the body where immunological tolerance is lost. Many a time tumors generate specific surface proteins which are not the normal proteins/or such compounds.� They are if produced, often happens in cancer patients will be presented by MHC I and MHCII, such antigens are recognized by cytotoxic T-cells and destroy (if lucky) them.� A native antigen is a unprocessed by an APC to smaller parts. T cells cannot bind native antigens, but require that they be processed by APCs, whereas B cells can be activated by native ones.
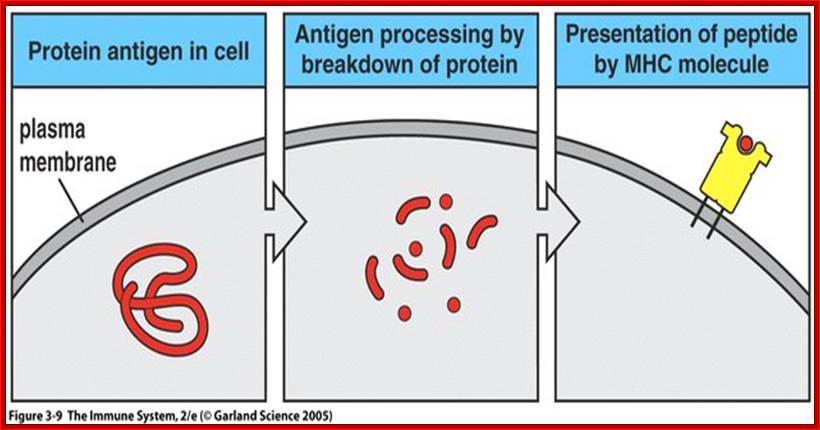
https://www.slideshare.net
An antigen or pathogen proteins released in the cell are broken down into pieces and then some are transferred to endoplasmic reticulum, from there the proteins are loaded on to MHC class of proteins which ultimately be found on the surface of plasma membranes.� There are two classes of MHC proteins MHCI and MHCII, which mature in ER.
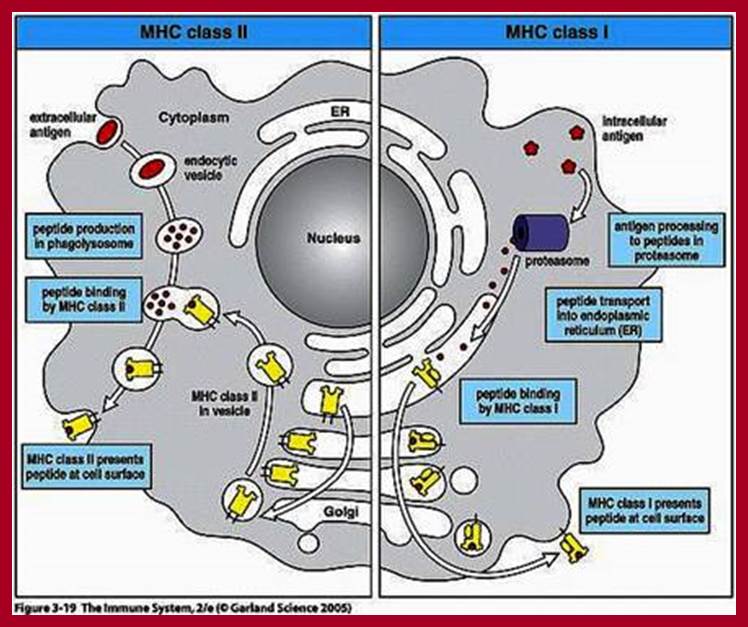
https://en.wikipedia.org
This diagram tells how a microbe ingested and its proteins are digested and passed though ER and assembled with MHC proteins, which are then transferred to membrane surface, thus the bacterial proteins (surface or otherwise) proteins are presented as antigens for the recognition by specific Abs. Viruses infected, and found in the cells are considered as internal proteins, which are also processed through ER and bind to MHCI and they are put on cell membranes.
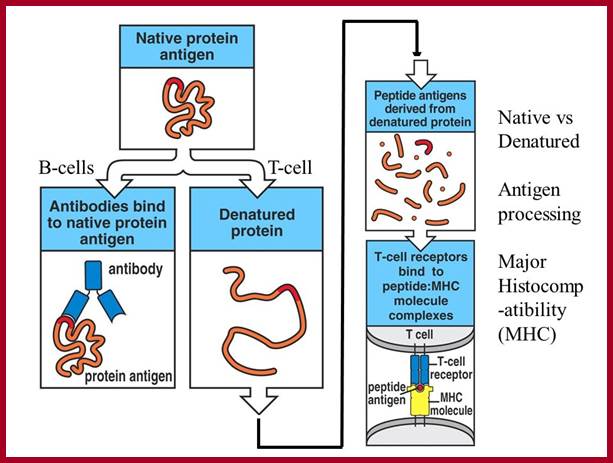
http://slideplayer.com
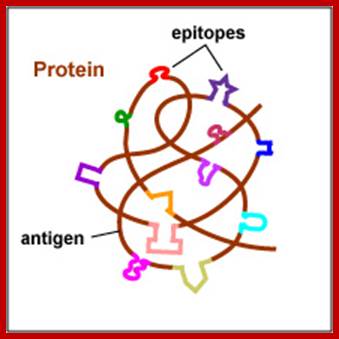
Proteins have many epitopes of different specificities; http://faculty.ccbcmd.edu
Epitope:� Epitope is an antigenic determinant. It is a stretch of amino acids with specific conformation consisting of 8-11 amino acids presented by MHCI and recognized by T-cells. The other epitope form of epitope consists of 16-18 a.a presented by MHC II.� Epitopes can be mapped by protein microarray or with ELLISPOT or ELISA techniques.� Paratope is the variant region of the antibody (15-20a.a) is the antigen binding region of the antibody called Fv region. The part of the antigen to which the paratope binds is called an epitope. This can be mimicked by a mimotope. For peptide antigens to elicit immune response the epitopic regions should contain at least 30% of immunogenic amino acids, they can be lysine, arginines, glutamic acid, aspartic acid, glutamine and asparagine.� Today it is possible to design a good peptide immunogen by recombinant biology.
Each of the antigens may contain a single epitope or determinants or multiples of them. Each of their specific structural domains or motifs acts as antigens. Linear adjacent amino acids can form an epitopes or folding can provide such determinants.
http://www.microbiologybook.org
Association and dissociation depends upon the kinetic constants (Kq). Polyvalent determinant antigens contain identical epitopes to which identical antibodies (Abs) bind. Abs can bind to two or in the case of IgM they can bind to ten identical epitopes, which leads to enhanced avidity of antigen and Ig interactions. An antigen can contain different epitopes depending upon the kind of antigen they present when infected.
Binding of antigens is more specific but it can happen to different determinant because of small differences in their structural sequences. So molecules in their specificity make mistakes in identifying the correct sequence based molecules.
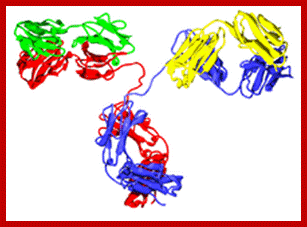
http://yang-sheng.com; http://www.bioon.com.cn; http://proteopedia.org; An Antibody or an Immunoglobulin.
Epitope Tags are generated by recombinant method so as to produce a protein with specific amino acid (a.a) sequence, so the proteins produced by the recombinant method can be easily purified or recognized (ex. Myc-Tag, FLAG-Tag, 6His Tag etc.)
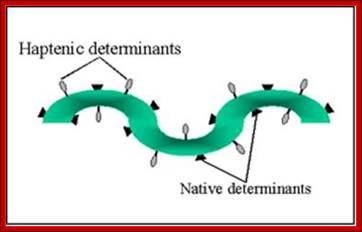
A hapten is a small molecule that can elicit an immune response only when attached to a large carrier molecule such as a protein; the carrier may be one that does not elicit an immune response by itself. (In general, only large molecules, infectious agents, or insoluble foreign matter can elicit an immune response in the body.) Once the body has generated antibodies to a hapten-carrier adduct, the small-molecule hapten may also be able to bind to the antibody, but it will usually not initiate an immune response; usually only the hapten-carrier adduct can do this. Sometimes the small-molecule hapten can even block immune response to the hapten-carrier adduct by preventing� the adduct from binding to the antibody, a process called hapten inhibition. The first haptens used were aniline and its carboxyl derivatives (o-, m-, and p-amino benzoic acid).
A well-known example of a hapten is Urushiol, which is a toxin found in poison ivy.� Haptens are small molecule act as adjuvants.�
Haptens vs. Epitopes;
Peptides and other small molecules that are used as antigens are referred to as haptens. They are able to act as recognition sites for production of specific antibodies but cannot by themselves stimulate the necessary immune response. Haptens can be made immunogenic by coupling them to a suitable carrier molecule.
Haptens can be made immunogenic by coupling them to a suitable carrier molecule.� Polyclonal antibodies are mixtures of serum Immunoglobulins and collectively are likely to bind to multiple epitopes on the antigen. Monoclonal antibodies by definition contain only a single antibody clone and have binding specificity for one particular epitope.
Carrier Proteins for Immunogen Preparation:
A carrier protein is any protein used for coupling with peptides or other haptens that are not sufficiently large or complex on their own to induce an immune response and produce antibodies. The two most commonly used carriers are keyhole limpet hemocyanin (KLH) and bovine serum albumin (BSA). To create the best immunogen, it may be beneficial to prepare the conjugates with several different carriers and with a range of hapten: carrier coupling ratios.
Keyhole Limpet Hemocyanin (KLH);
Keyhole limpet hemocyanin (KLH) is the most widely used carrier protein. The copper-containing polypeptide belongs to a group of non-heme proteins called hemocyanins, which are found in arthropods and mollusks. KLH is isolated from keyhole limpets (Megathura crenulata). Because KLH is from a class of proteins and a group of organisms that are evolutionarily distant from mammals, it is very "foreign" to the animal systems typically used to produce antibodies. The protein is also highly immunogenic because of its very large size and complex structure. The molecule is composed of 350kDa and 390kDa subunits that associate to form aggregates ranging from 0.5 to 8 million daltons.
Blue Carrier* Immunogenic Protein: Blue Carrier* Protein is a purified preparation of Concholepas concholepas hemocyanin (CCH). Bovine Serum Albumin as a Carrier Protein: Bovine serum albumin (BSA; 67kDa) belongs to the class of serum proteins called albumins.
Cationized BSA:
Cationized bovine serum albumin (cBSA) is prepared by modifying native BSA with excess ethylenediamine, essentially capping all negatively-charged carboxyl groups with positively-charged primary amines. The result is a highly positively-charged protein (pI > 11) that has significantly increased immunogenicity compared to native BSA. In addition, the increased number of primary amines provides for a greater number of antigen molecules to be conjugated with typical crosslinking methods.
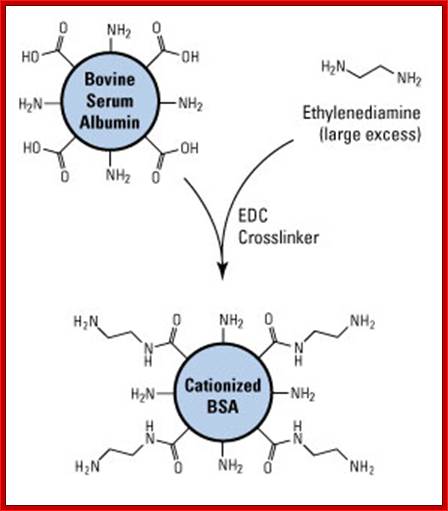
Prep of catonized BSA, it reacted with excess of ethylenediamine using EDC; Ovalbumin can be used as a Carrier Protein; Ovalbumin (OVA; 45kDa). https://www.thermofisher.com
Adjuvants:
An immunologic adjuvant is defined as any substance that acts to accelerate, prolong, or enhance antigen-specific immune responses when used in combination with specific vaccine antigens. �Adjuvants in immunology are often used to modify or augment the effects of a vaccine by stimulating the immune system to respond to the vaccine more vigorously, and thus providing increased immunity to a particular disease. Adjuvants accomplish this task by mimicking specific sets of evolutionarily conserved molecules, so called PAMPs, which include liposomes, lipopolysaccharide (LPS), molecular cages for antigen, components of bacterial cell walls, and endocytosed nucleic acids such as double-stranded RNA (dsRNA), single-stranded DNA (ssDNA), and unmethylated CpG dinucleotide-containing DNA. Because immune systems have evolved to recognize these specific antigenic moieties, the presence of an adjuvant in conjunction with the vaccine can greatly increase the innate immune response to the antigen by augmenting the activities of dendritic cells (DCs), lymphocytes, and macrophages by mimicking a natural infection.� Furthermore, because adjuvants are attenuated beyond any function of virulence, they pose little or no independent threat to a host organism.�
To enhance the immune response to an immunogen, various additives called adjuvants can be used. When mixed and injected with an immunogen, an adjuvant will enhance the immune response. An adjuvant is not a substitute for a carrier protein because it enhances the immune response to immunogens but cannot itself render haptens immunogenic. Adjuvants are nonspecific stimulators of the immune response, helping to deposit or sequester the injected material and causing a dramatic increase in the antibody response.
There are many popular adjuvants, including complete Freund's adjuvant (CFA or FCA). This reagent consists of a water-in-oil emulsion and killed Mycobacterium. The oil-and-water emulsion localizes the antigen for an extended period of time, and the Mycobacterium attracts macrophages and other appropriate cells to the injection site. Solutions of aluminum hydroxide (alum are convenient alternatives to Freund�s adjuvants.
Immunization Schedule for Mice:
- Day 0: Collect pre-immune serum from the mouse to use as a blank when performing ELISA screening after immunization. Store frozen. Inject 50 to 100�g of immunogen (equal to 100 to 200�L of antigen-adjuvant mixture) per mouse. Typical routes of injection include intraperitoneal (i.p.) or subcutaneous (s.c.). One or two such injections may be made per animal.
- Day 14: Boost with an equivalent amount of immunogen in adjuvant.
- Day 21: Test bleed and assay antibody response by ELISA. (Typically, mice are bled under anesthesia through the tail vein or the retro-orbital plexis).
- Day 28: Boost again if necessary. Continue with a similar schedule of alternating boosts and test bleeds until a satisfactory response is observed. For monoclonal antibody production, inject either i.p. or intravenously (i.v.) 4 to 5 days before fusion with the immunogen dissolved in saline (no adjuvant).
Immunization Schedule for Rabbits:
- Day 0: Collect pre-immune serum from the rabbit to use as a blank when performing ELISA after immunization. Store frozen. Inject 100�g of immunogen (equal to about 200�L of the antigen-adjuvant mixture) into each of 8 to 10 subcutaneous sites on the back of the rabbit. Other routes of injection may also be used, but this is by far the easiest with the rabbit.
- Day 14: Boost with an equivalent amount of adjuvant.
- Day 21: Test bleed and assay antibody response by ELISA. (Typically, rabbits are bled through the ear vein without anesthetic). It is not difficult to collect 5 to 10mL of blood, which is more than adequate for measuring antibody response.
Day 28: Boost again if necessary. Continue with a similar schedule of alternating boosts and test bleeds until a satisfactory response is observed.
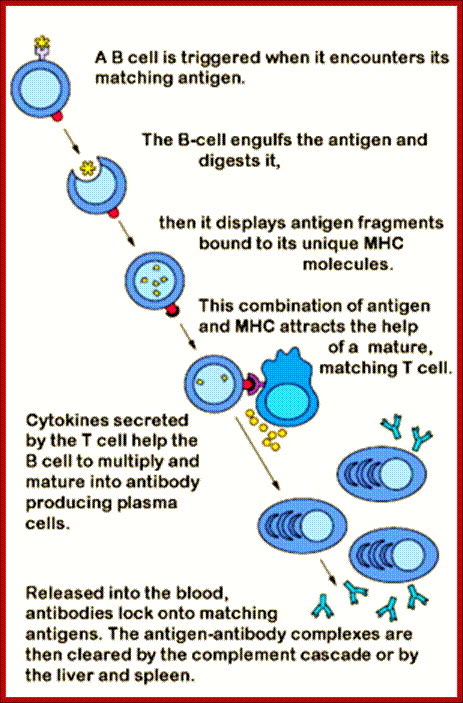
Binding of the antigen to the surface antibody molecule of virgin B cell is an absolute requirement for an antibody response.
In summary, a good immunogen has three chemical features:
- It must have an epitope that can be recognized by the cell-surface antibody found on B cells.
- It must have at least one site that can be recognized simultaneously by a class II protein and by a T-cell receptor.
- Usually, it must be degradable. These three properties are the only intrinsic chemical features needed for a molecule to elicit a strong antibody response.
Antibodies:
Antibodies are those proteins that recognize, non self molecules as outsiders, and respond to defend against them; so they are called antibodies to outside bodies which can harm the cells or animals.
The B lymphocytes are the producers of Immunoglobulins (Igs) of one clone (one specific antigen binding) can change in the course of immune response. To begin with it generates one specific antigenic variant as membrane bound protein, but activation leads to the production and secretion type of Igs of the same type and kind. Isotype switching is due to changes in the C region and without any changes in V region. Mutation in the V region can change to higher affinity or no affinity at all.
B Cells and Antibodies (NCBI);
B cells originate in bone marrow from pluripotent stem cells and move out and land in Bursa and then in spleen.� By the time they move out of bone marrow cells they are already endowed with surface B cell receptors.
The binding of antibodies to invading pathogens also recruits various types of white blood cells and a system of blood proteins, collectively called complement). The white blood cells and activated complement components work together to attack the invaders.
Development of B cells and Production of Immunoglobulins:
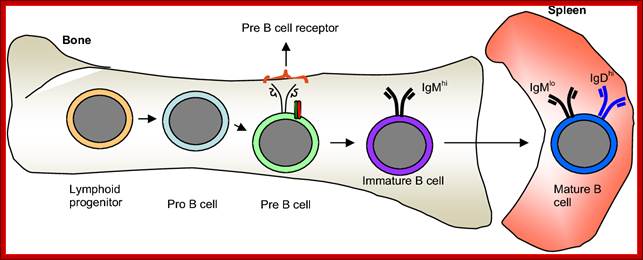
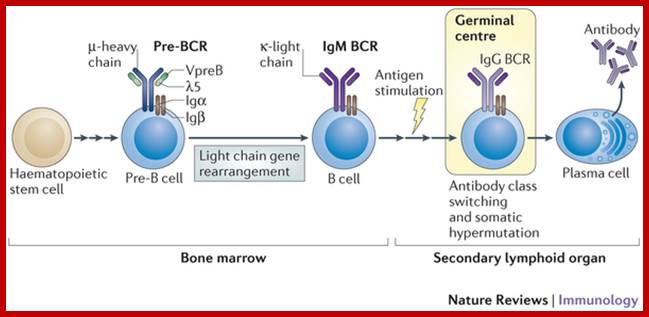
� Bone marrow has all the inputs for the development of Imunoglobulin producing cells and developing them; Max D. Cooper; �http://www.nature.com
B cells are produced in bone marrow by lymphoid progenitor cells, which develops into Pro B cells and then into Pre B cells with pre BCR that matures into what is called immature B cells with IgM.� These in turn are released into blood stream and reach spleen where they mature into mature B cells.� Synthesized exclusively by B cells, antibodies are produced in billions of forms, each with a different amino acid sequence for different antigen-binding site. Collectively called Immunoglobulins (abbreviated as Ig), they are among the most abundant protein components in the blood, constituting about 20% of the total protein in plasma by weight. Mammals make five classes of antibodies, each of which mediates a characteristic biological response following antigen binding.
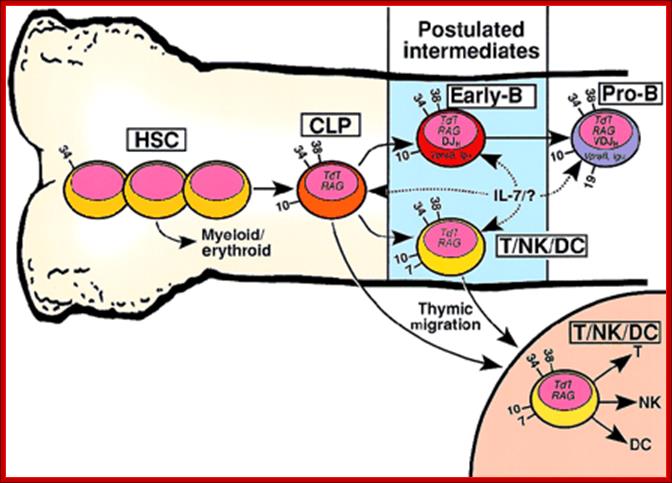
Developmental relationship between hematopoietic stem cells (HSCs), common lymphoid progenitors (CLPs), and putative early-B or T/NK/dendritic cell (DC) progenitors. Developmental relationship between hematopoietic stem cells (HSCs), common lymphoid progenitors (CLPs), and putative early-B or T/NK/dendritic cell (DC) progenitors http://www.bloodjournal.org
HSCs include all primitive CD34+/lineage−hematopoietic developmental stages prior to the CLP, shown schematically as 3 cells. Arrows with solid lines indicate developmental flow culminating in increased lineage restriction. Dashed arrows indicate possible cellular targets of IL-7 signaling or an unknown (?) ligand. Numbers on the cell surface indicate CD antigens useful in distinguishing the developmental compartments. Although not shown in this figure, the 3 reports that described the cell surface phenotype of CD19− lymphoid progenitors revealed considerable heterogeneity. �For example, CD7 and CD33 were detected on a minority of the lymphoid progenitors in each study. There is no known surface marker that distinguishes the CLP from the early-B cell. It is also likely that IL-7R expression and signaling vary both within and between the lymphoid progenitor compartments.
Development of B-Cells in Bone Marrow in Stages; https://www.slideshare.net
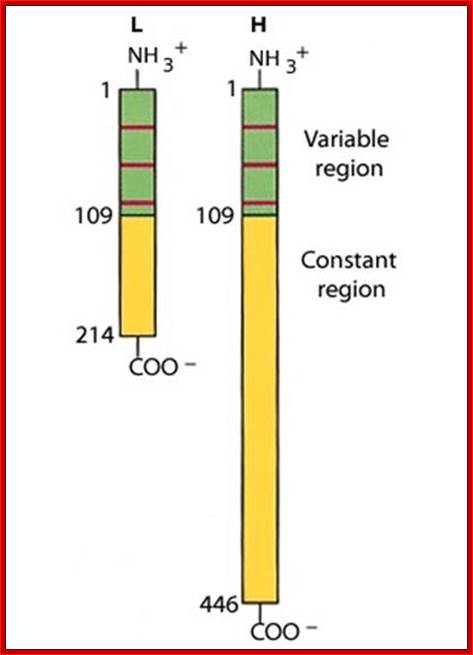
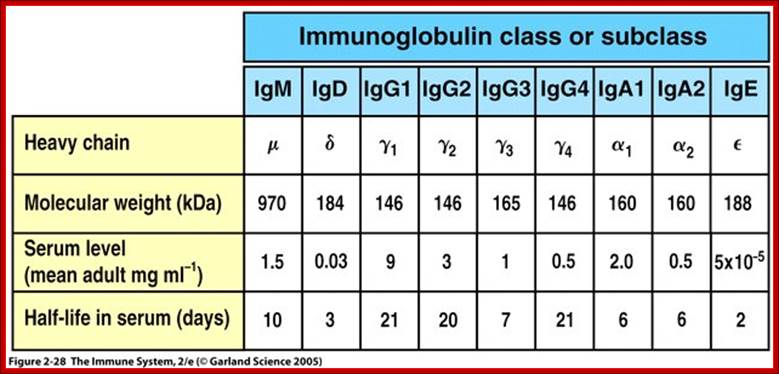
V/L and C/L contain 110 a.a, V/H 110 a.a and C/H 330-440 a.a Heavy and Light chains are made up of 446 (50-70kD) and 214 (23KD) amino acids respectively. www.suggest-keywords.com
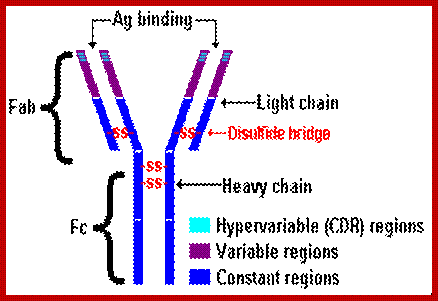
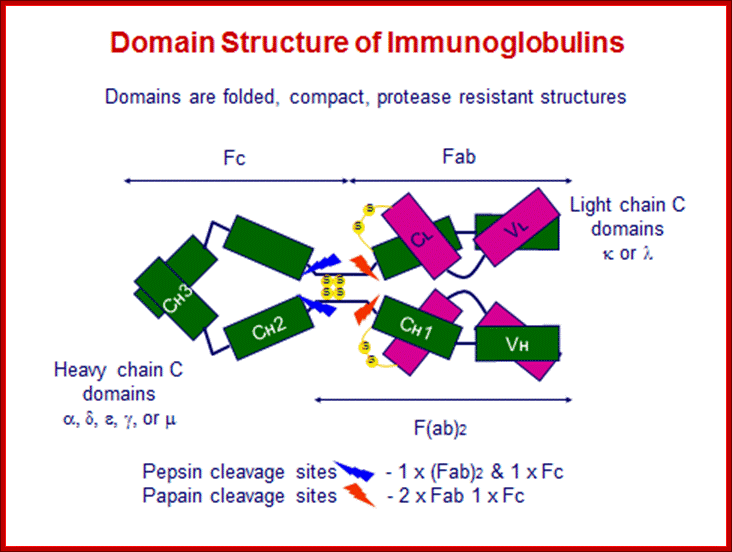
General features of an antibody.
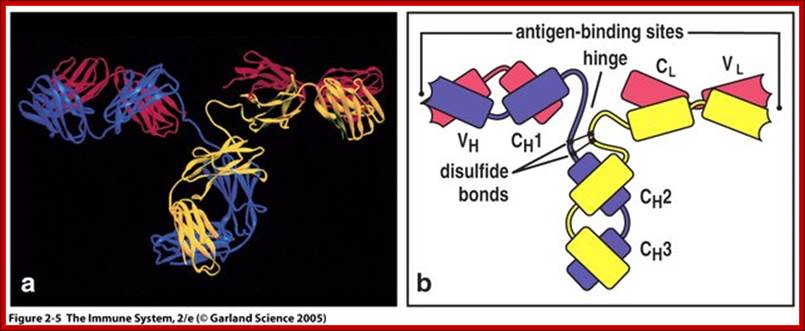
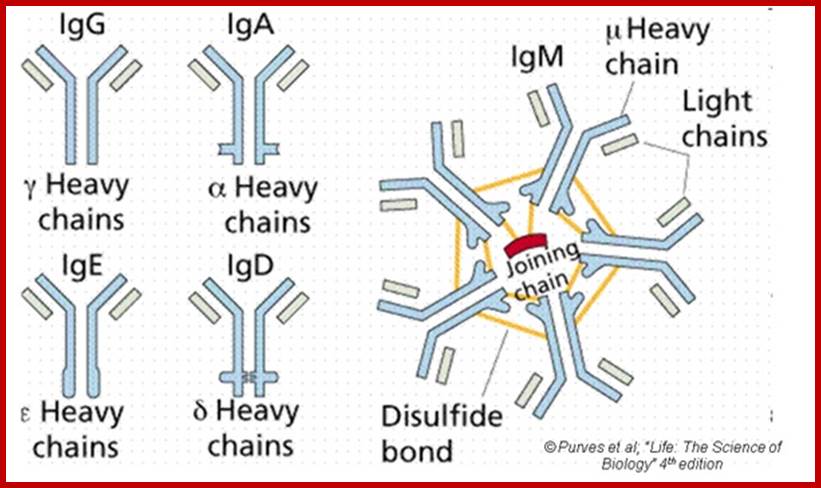
Basic structural features of Antibody proteins, with Heavy chain and Light chains having constant and variable regions. The Heavy and Light chains have inter strand S-S bonds, the number varies, they have non covalent interaction too.� They also have intrachain S-S bonds. Each of them has specific domains-VH, CH1, CH2 and CH3; CL and VL. https://forum.facmedicine.com
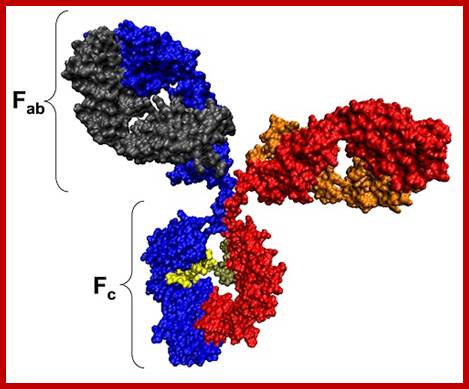
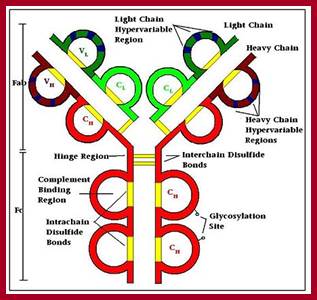
Model of an antibody general; http://www.azom.com
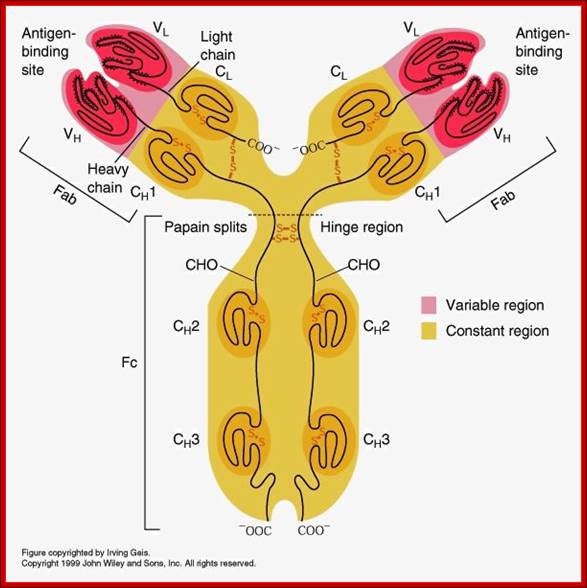
Detailed description of H and L chains; https://www.studyblue.com
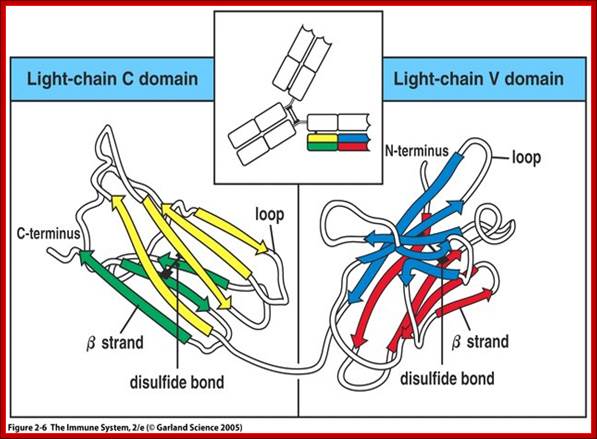
https://www.studyblue.com
Different classes of Antibodies:
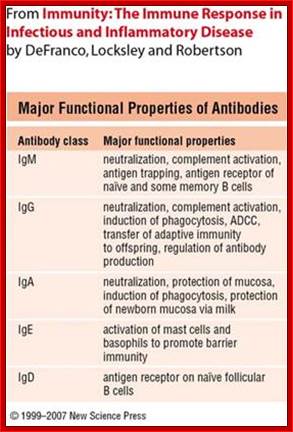
Antibodies are proteins and there are no parallels to such proteins in any systems. Antibodies are a family of glycoproteins produced by B-lymphocytes either in membrane bound or free forms. The membrane bound forms act as receptors to oligo-saccharides, poly-liposaccharides, glycoproteins, cell wall proteins, viral proteins (antigens in general) etc. all external to body cellular molecules. Binding triggers activation of B cells as a result it generates secreted Igs. These Abs act as mediators of specific humoral immunity by engaging various effector mechanisms that serves to eliminate the bound antigens.
http://www.edvotek.com
The antigen binding region of AB is highly variable and amazingly a single person can generate 10x^9 such variants of Abs, and the same number of them are also produced and stick to the B-lymphocytes as sentinels for invaders. Na�ve B cells have such variants one species in each of the lymphocytes.
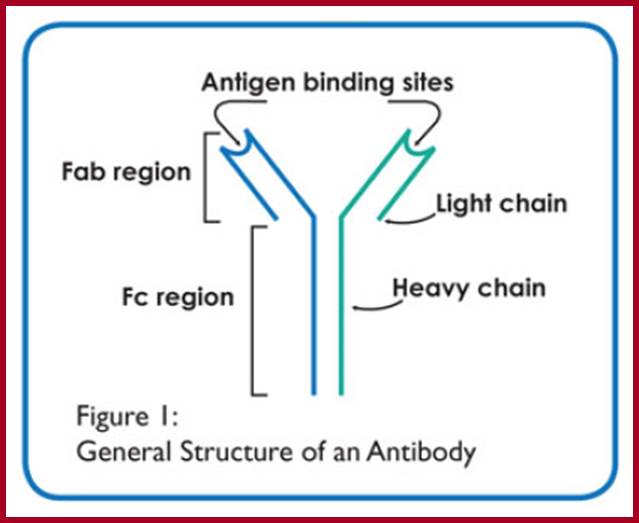
All Abs contain common symmetric core structural features of two identical covalently linked heavy chains and two identical light chains each linked to each other by S-S bonds. Each of the light chains consists of Variable V/L (110a.a) and Constant C/L(110a.a) containing folded Ig domains with conserved a.a sequences and interchain and intrachain S-S bonds. And the heavy chain consists of Variable V/H(110a.a) and constant C/H (330-440a.a). The light and heavy chains are held together by S-S bonds, the number of bonds vary. Each of the chains has a hinge region where they bend away from the axis to form �Y� shaped molecule. Both Light and heavy chains have domains which are folded into globular structures, they are named as V/L and C/L for light chains and V/H, C/H- C/H1, or C/H4.� Carbohydrates are added to C/H2 regions, but the position can vary.
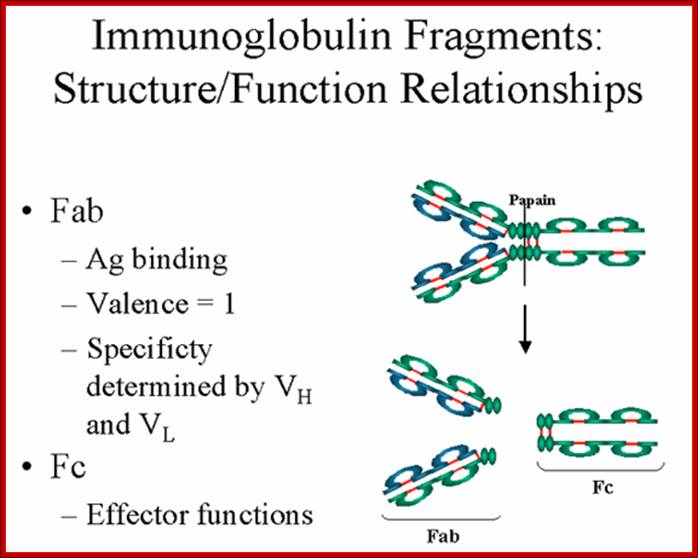
The N terminal sequences of both H and L chains contain variable regions called �V� which provides variability to Igs for they contain specific binding (complimentary) domains to specific antigens, the binding site called hyper variable regions (three blocks) of different complementary determining regions (CDR); it is this generates more than 10^9 variables and it is this region recognizes and binds to antigens in what famously called �enzyme-substrate binding� process; it is like lock and key process. Digestion of Igs with papain release two fragment called Fab fragments. The portion released is the hinge region above consisting S-S bonds and the lower region is called Fc.� The Fab consists of H portion and L portion.� This provides different combination of amino acids for recognizing and binding to antigens.� The Fc fragments are below consisting of heavy chain constant regions C/H2 and C/H3 domains.� The Fab regions can be further cleaved by pepsin protein.� These Igs are classified in to different classes based on a.a sequences in constant regions of heavy chains and subclasses based on heavy chains, such as IgG (gamma), IgM (Mu), IgA (alpha), IgD (delta) and IgE (epsilon),; the differences can be detected serologically too.
Each of the Ig class can be further divided into subclasses based on a.a sequences in their H/C regions. IgG has G1, G2, G3 and G4.� The IgA consists of IgA1 and IgA2.� Immunoglobulins are further classified into types and subtypes based on their light chains (serological differences). Kappa chains and Lambda chains; the lambda has subtypes such as Lambda 1, Lambda2, Lambda 3, and Lambda 4.� Often the terminologies used is interesting.
Abs are classified into different Isotypes and subtypes based on the H chain constant regions (Ig constant domain) with three to four Ig domains. They generate what is called Isotypes, Idiotypes and Allotypes. They have different functional properties; classified as IgM, IgD, IgG, IgE and IgA. In all these variants the light chain is either kappa or lambda type in their C-terminal region.
http;//www.slideshare.net
https://www.pinterest.com
Isotypes and Allotypes and Idiotypes of Immunoglobulins:

http://www.slideshare.net
An isotype usually refers to any related proteins/genes from a particular gene family. In immunology, the "immunoglobulin isotype" refers to the genetic variations or differences in the constant regions of the heavy and light chains. In humans, there are nine heavy chain isotypes and two light chain isotypes. Isotypes are distinct forms of light or heavy chains which are present in all members of a species, encoded at distinct genetic loci. Ex. Heavy chains-alpha IgA 1,2; delta IgD, gamma IgG1,2,3 and 4, epsilon IgE, micron-IgM, light chain Kappa and lambda
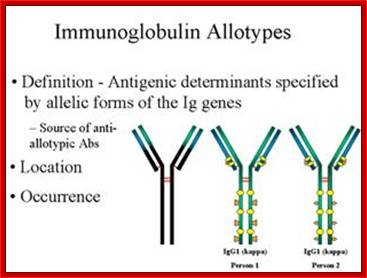

Igs Allotypes and Isotypes ; http://www.microbiologybook.org
In immunology, an immunoglobulin Allotypes is the allele of the antibody chains found in the individual. In zoological nomenclature, an Allotype is a specimen of the opposite sex to the Holotype
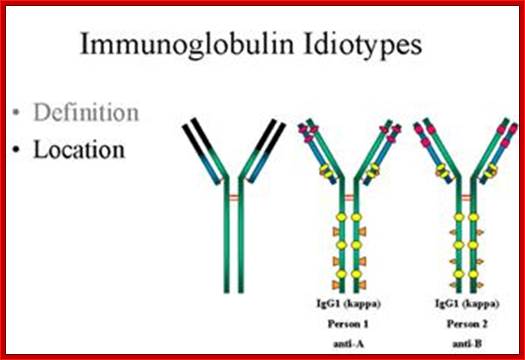
http://www.microbiologybook.org
In immunology, an Idiotype is a shared characteristic between a group of immunoglobulin or T cell receptor (TCR) molecules based upon the antigen binding specificity and therefore structure of their variable region. Antibody Idiotype is determined by Gene rearrangement, Junctional diversity, P-nucleotides (palindromic nucleotides at sites of single-strand breaks), N-nucleotides and Somatic hyper mutations.
Most of the effector functions of Igs is mediated by the C-regions of H-chain, but triggered by binding of antigens to spatially distant binding or combining site in the V region.
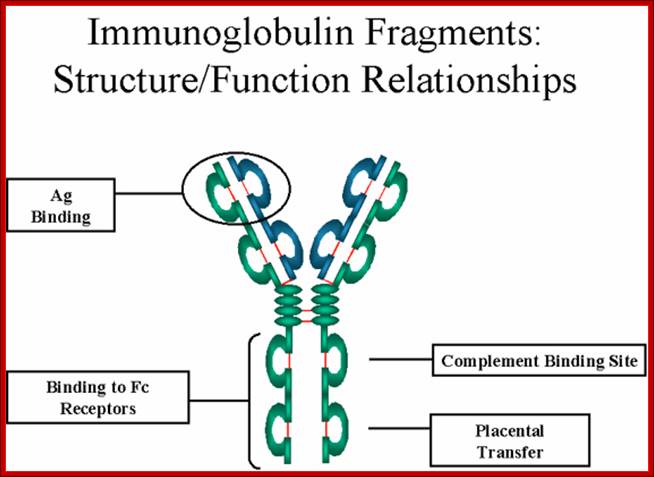
https://www.slideshare.net
http://www.microbiologybook.org
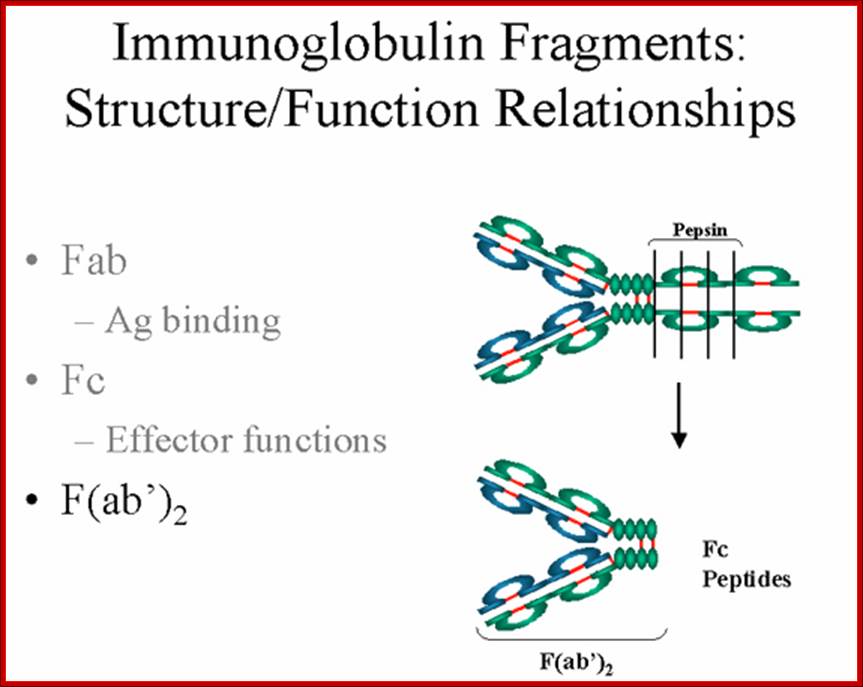
����������� Fab is papain digested fragments which includes basal S-S bonds; and Fc is Pepsin digested fragment, which provides the effector function to Igs. https://www.slideshare.net
Structure and Properties of Ig Classes of Abs: Immunoglobulins are classified into 5 different classes; By Gene Mayer , University of South Carolina
IgG
1.
Structure;
The structures of the IgG subclasses are presented in figure 7. All IgG's are
monomers (7S immunoglobulin). The subclasses differ in the number of disulfide
bonds and length of the hinge region.
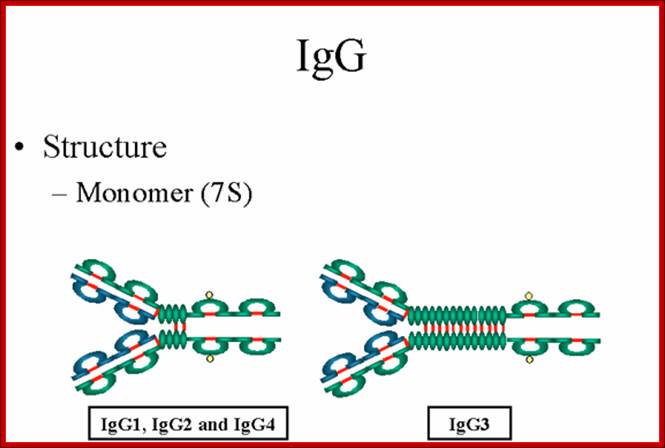
Gene Mayer,; http://www.microbiologybook.org
2.Properties;
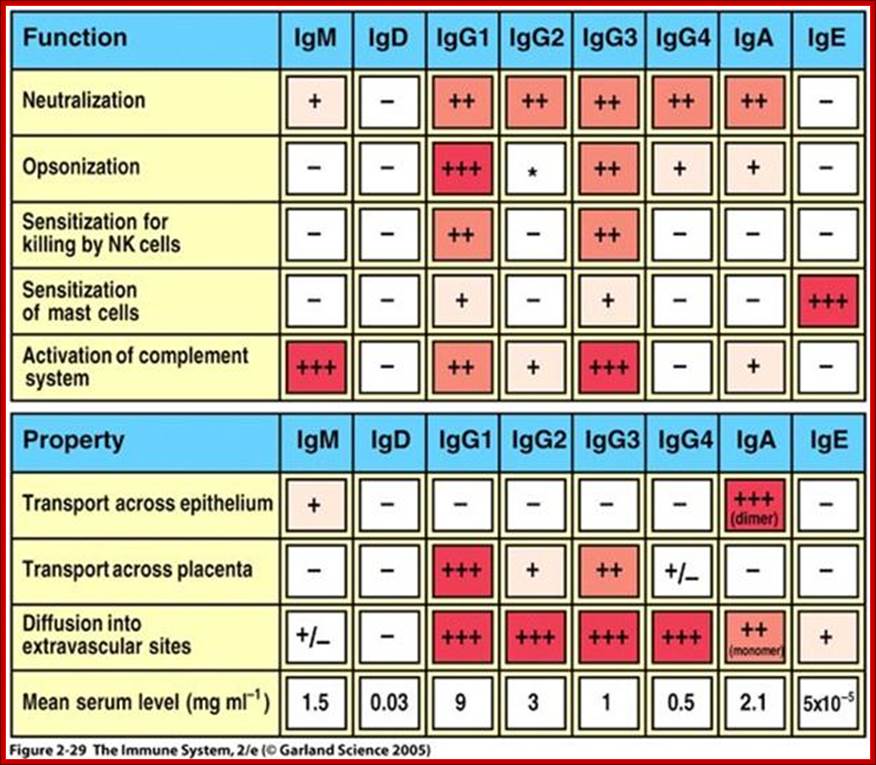
IgG is the most versatile immunoglobulin because it is capable of carrying out
all of the functions of immunoglobulin molecules.
a) IgG is the major Ig in serum - 75% of serum Ig is IgG
b) IgG is the major Ig in extra vascular spaces
c) Placental transfer - IgG is the only class of Ig that crosses the placenta. Transfer is mediated by a receptor on placental cells for the Fc region of IgG. Not all subclasses cross equally well; IgG2 does not cross well.
d) Fixes complement - Not all subclasses fix equally well; IgG4 does not fix complement
e) Binding to cells - Macrophages, monocytes, PMNs and some lymphocytes have Fc receptors for the Fc region of IgG. Not all subclasses bind equally well; IgG2 and IgG4 do not bind to Fc receptors. A consequence of binding to the Fc receptors on PMNs, monocytes and macrophages is that the cell can now internalize the antigen better. The antibody has prepared the antigen for eating by the phagocytic cells. The term opsonin is used to describe substances that enhance phagocytosis. IgG is a good opsonin. Binding of IgG to Fc receptors on other types of cells results in the activation of other functions.
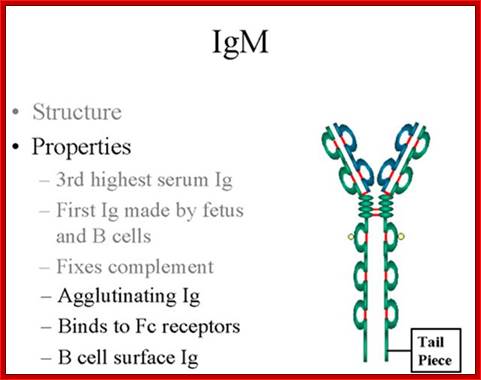
IgM:
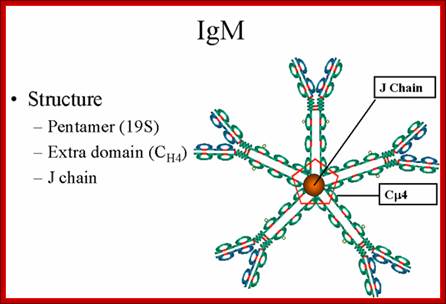
Structure-
The structure of IgM is presented in figure below. IgM normally exists as a
pentamer (19S immunoglobulin) but it can also exist as a monomer. In the
pentameric form all heavy chains are identical and all light chains are
identical. Thus, the valence is theoretically 10. IgM has an extra domain on
the mu chain (CH4) and it has another protein covalently bound via a
S-S bond called the J chain. This chain functions in polymerization of the
molecule into a Pentamer.
��� 
http://www.microbiologybook.org
�Properties-
a) IgM is the third most common serum Ig.
b) IgM is the first Ig to be made by the fetus and the first Ig to be made by a virgin B cells when it is stimulated by antigen.
c) As a consequence of its pentameric structure, IgM is a good complement fixing Ig. Thus, IgM antibodies are very efficient in leading to the lysis of microorganisms.
d) As a consequence of its structure, IgM is also a good agglutinating Ig . Thus, IgM antibodies are very good in clumping microorganisms for eventual elimination from the body.
e) IgM binds to some cells via Fc receptors.
f) B cell surface Ig- Surface of IgM exists as a monomer and lacks J chain but it has extra 20 amino acids at the C-terminus to anchor it into the membrane. �Cell surface IgM functions as a receptor for antigen on B cells. Surface IgM is noncovalently associated with two additional proteins in the membrane of the B cell called Ig-alpha and Ig-beta as indicated in figure. These additional proteins act as signal transducing molecules since the cytoplasmic tail of the Ig molecule itself is too short to transduce a signal. Contact between surface immunoglobulin and an antigen is required before a signal can be transduced by the Ig-alpha and Ig-beta chains. In the case of T-independent antigens, contact between the antigen and surface immunoglobulin is sufficient to activate B cells to differentiate into antibody secreting plasma cells. However, for T-dependent antigens, a second signal provided by helper T cells is required before B cells are activated.
IgA
1.Structure;
Serum IgA is a monomer but IgA found in secretions is a dimer as presented in
Figure below. When IgA exits as a dimer, a J chain is associated with it.
When IgA is found in secretions is also has another protein associated with it called the secretory piece or T piece; sIgA is sometimes referred to as 11S immunoglobulin. Unlike the remainder of the IgA which is made in the plasma cell, the secretory piece is made in epithelial cells and is added to the IgA as it passes into the secretions (Figure). The secretory piece helps IgA to be transported across mucosa and also protects it from degradation in the secretions.
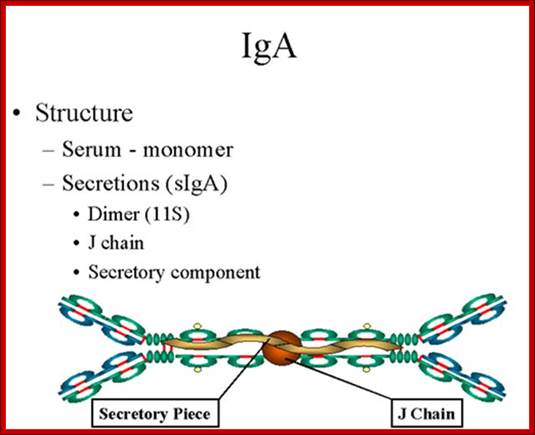
http://www.namrata.co
2. Properties- a) IgA is the 2nd most common serum Ig.
b) IgA is the major class of Ig in secretions - tears, saliva, colostrum, mucus. Since it is found in secretions secretory IgA is important in local (mucosal) immunity.
c) Normally IgA does not fix complement, unless aggregated.
d) IgA can binding to some cells - PMN's and some lymphocytes.
IgD
1. Structure-The structure of IgD is presented in the Figure. IgD exists only as a monomer.
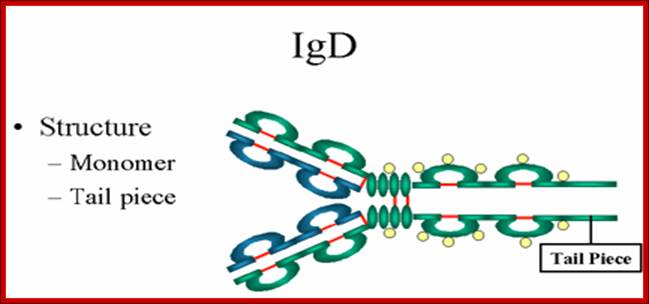
http://www.biologyexams4u.com
2. Properties- a) IgD is found in low levels in serum; its role in serum uncertain. b) IgD is primarily found on B cell surfaces where it functions as a receptor for antigen. IgD on the surface of B cells has extra amino acids at C-terminal end for anchoring to the membrane. It also associates with the Ig-alpha and Ig-beta chains. c) IgD does not bind complement. It is produced by Na�ve B cells when they still in bone marrow.
IgE
1. Structure- The structure of IgE is presented in Figure below. �IgE exists as a monomer and has an extra domain in the constant region.
2. Properties- a) IgE is the least common serum Ig since it binds very tightly to Fc receptors on basophils and mast cells even before interacting with antigen.
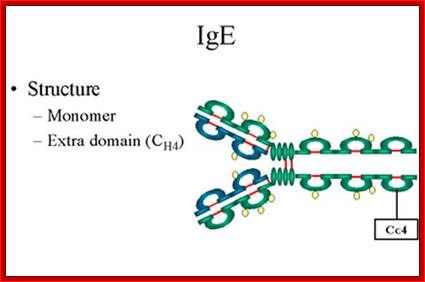
http://www.microbiologybook.org
b) Involved in allergic reactions - As a consequence of its binding to basophils and mast cells, IgE is involved in allergic reactions. Binding of an allergen to the IgE on cells results in the release of various pharmacological mediators that result in allergic symptoms.
c) IgE also plays a role in parasitic helminthes diseases. Since serum IgE levels rise in parasitic diseases, measuring IgE levels is helpful in diagnosing parasitic infections. Eosinophils have Fc receptors for IgE and binding of Eosinophils to IgE-coated helminthes results in killing of the parasite.
d) IgE does not fix complement.
Ig or Ab Genes:
Immunoglobulin protein coding genes are located on different chromosomes.� The number of each light chain and heavy chain genes vary.� Each of the Heavy chain genes and light chain genes has variable structural features such as constant region, diversity and variable regions; thus they can generate 10x10^14 types of IgGs and perform surveillance. Activated B cells by a specific antigen go through rearrangements by recombination (an unique process) in their gene loci DNA and generate specific antibody to that antigen.� Activated B cells get differentiated into large sized Plasma cells and produce IgG.� They also produce memory cells.� Plasma cells in bone marrow secrete 10,000 IgGs per second.� Remember B1 cells produce IgMs and B2 cells produce IgGs.� Isotype switching generate other forms of Igs.
https://www.pinterest.com/; https://dokuwiki.noctrl.edu
����������������������������������������������� Plasma cell; https://en.wikipedia.org
Naive B cells can be activated by T cell depended or T cell independent manner.
T cell independent Manner:
Many bacteria have repeating carbohydrate epitopes that stimulate B cells, through so called pattern recognition receptors, to respond with IgM synthesis in the absence of T cell help. Lipid A and dextran sulfate also can activate B cells.There are two types of T-cell independent activation; Type 1 T cell-independent (polyclonal) activation, and type 2 T cell-independent activation (in which macrophages present several of the same antigen in a way that causes cross-linking of antibodies on the surface of B cells).
T cell Dependent manner:
Bound pathogens� antigens (proteins, glycoproteins, proteoglycans) to their receptor IgM are ingested and their proteins are processed and transferred to ER where the bind to cavities in MHC II and the same are put on the surface of the B cells.� These B cells presents to Th form of T cells(T cells one form) and T cells bind, and get signaled and they secrete cytokines which activate B cells This leads to multiplication of B cells and by recombination of BCR genes especially the H constant regions, they start producing IgGs and the same are released free. Isotype class switching to IgG, IgA, and IgE and memory cell generation occur in response to T-dependent antigens. Class switching is due to ds Strand DNA breaking in a conserved region called S upstream of heavy chain constant region.� In this process DNA is nicked and broken at S site (by cytidine deminase and uracil DNA glycolase and apyrimidine/apurinic endonuclease) and rejoined by base pairing and gap filling. This Isotypes switching is known as Class Switch Recombination (CSR). Once this switch is on, that particular B-cell can no longer make the earlier isotypes, IgM or IgD.
B Cell Co-receptor Signal Transduction:
� The B cell possesses a co-receptor complex which can modulate BCR signal transduction.� The co-receptor complex is composed of CD21, CD19, and Tapa-1(CD81). CD21 binds opsonized antigenic particles, such as bacteria or enveloped viruses, via its affinity for the C3d complement fragment. CD19 is primarily responsible for signal transduction. The function of the Tetra-span protein Tapa-1 is unknown; but it may be involved in signal transduction leading to homotypic adhesion.
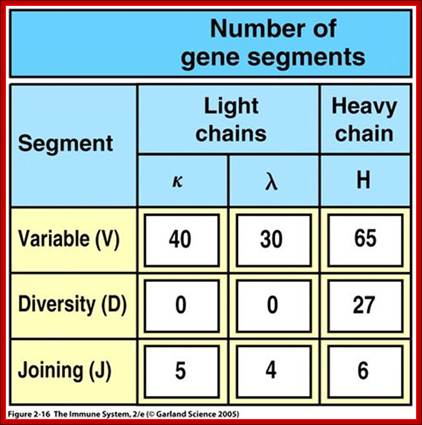
Multiple gene segments increase Ig diversity; http://slideplayer.com
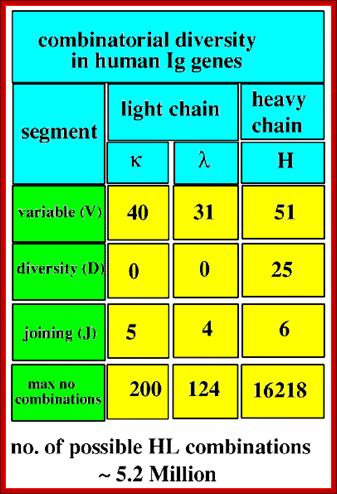
http://slideplayer.com
Human antibody molecules (and B cell receptors) comprise heavy and light chains with both constant (C) and variable (V) regions that are encoded by genes on three loci. Depending on the reference literature the above numbers given in the tables vary.
- Immunoglobulin heavy locus (IGH@) on chromosome 14, containing genes for the immunoglobulin heavy chain 14q32.33.
- Immunoglobulin kappa (κ) locus (IGK@) on chromosome 2, p11.2 (near CeN) containing genes for the immunoglobulin light chain.
- Immunoglobulin lambda (λ) locus (IGL@) on chromosome 22, containing genes for the immunoglobulin light chain. 22q11.2.
4. Multiple genes for the variable regions are encoded in the human genome that contains three distinct types of segments. For example, the immunoglobulin heavy chain region contains 44 Variable (V) genes plus 27 Diversity (D) genes and 6 Joining (J) genes. The light chains also possess numerous V and J genes, but do not have D genes. By the mechanism of DNA rearrangement of these regional genes it is possible to generate an enormous antibody repertoire; roughly 3�1011 combinations are possible, although some are removed due to self-reactivity.
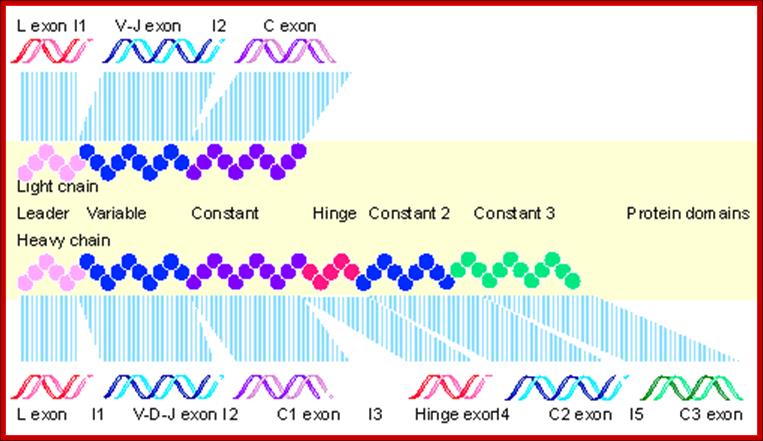
Immunoglobulin light chains and heavy chains are coded by genes whose structures (in their expressed forms) correspond with the distinct domains in the protein. Each protein domain corresponds to an exon; introns are numbered 1-5.� Each of the exons shown corresponds to specific functional domain of the immunoglobulin. http://genes.atspace.org
Variability of IgGs to various antigens, whose structural features complementary to each other like �lock and key� is generated because of existence of Igs segments.� Each of the domains are coded for by a specific exons. However exons are intervened by specific number of introns; this alternative slicing can generate more variability.
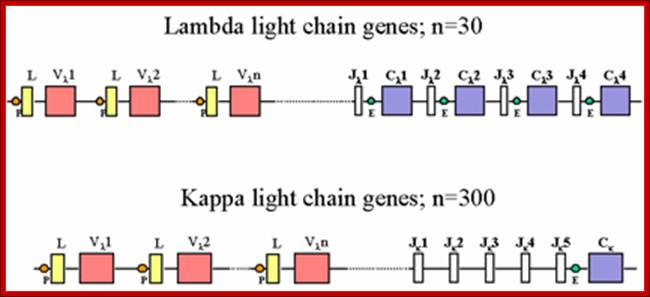
Light Chain has 250 V segments, 4 J and one Heavy C segment. �Each of the V segments can join with any one of the four J segments.
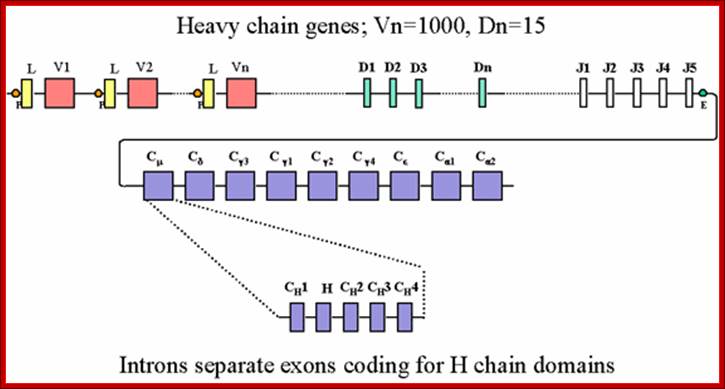
Heavy chains have 250 V segments, 15 D segments, 5 J segments followed by H constant segment.� One of the V segments can join with any one of the D segment and it can join with any one of the J segment.� Permutation combination of these can give extraordinary number of Igs (1 x 10^10 billion).
http://www.immunatorhoney.com
http://www.microbiologybook.org
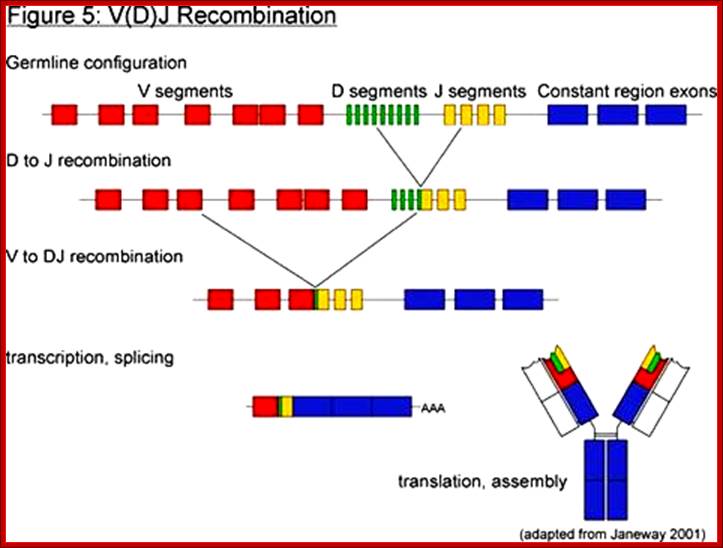
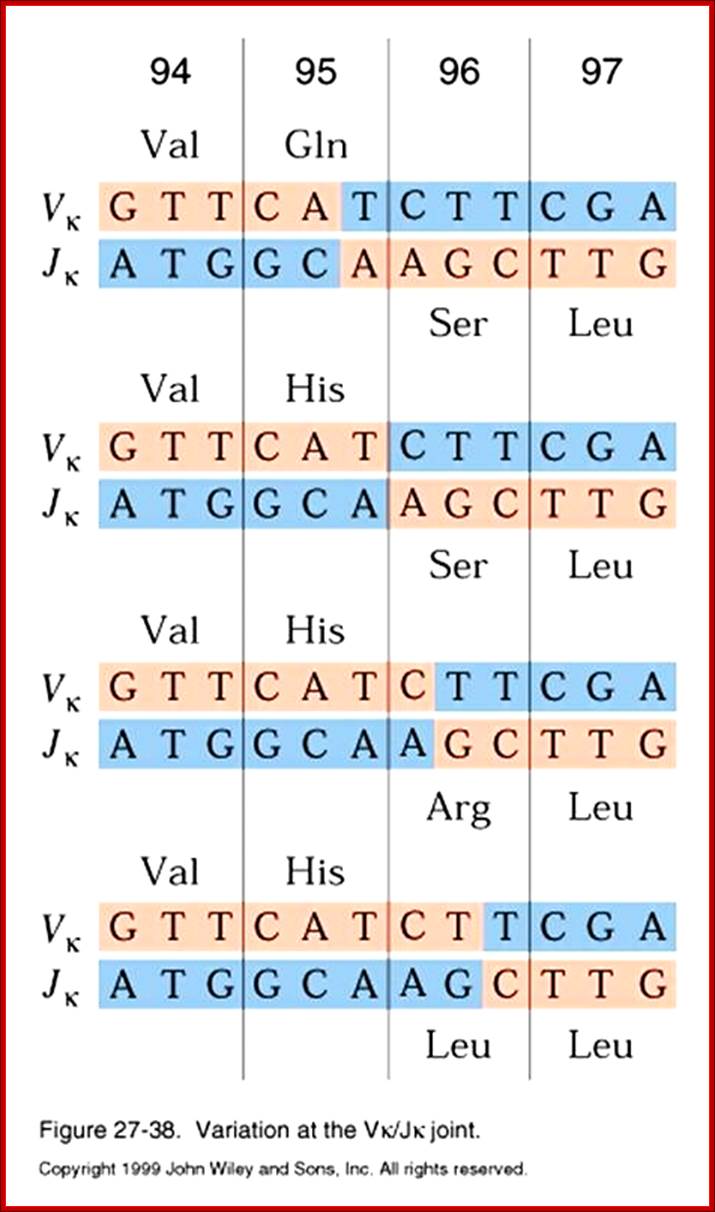
Process of random mode of joining of VDJ and C joining by simple site specific/ sequence specific DNA recombination method.� V= Variable, J = Joint, D = Diversity and C = Constant.
As the cell maturing heavy chains go through rearrangements; where one of the D is brought to one of the J; then one of the V is joined to DJ. This recombination leads to transcription where the promoter is associated V is brought close to E (enhancer) located in the intron between J and Cmu.� The pre mRNA contains sequences from L,V,D,J and Cmu and C delta plus intron between L and V, J and Cmu and Cmu and C delta.� The generated pre mRNA is spliced alternatively also.� This can generate mRNA containing VDJ next to Cmu; or VDJ next to Cdelta.� Similarly the light chains, such as kappa or lambda also go through such recombination and transcription and alternative splicing to produce an mRNA to be transported in to cytoplasm, where the translated protein is threaded through ER and finally, it is either placed on the B cell membrane (pre B cells) or secreted free from the B cell.
http://slideplayer.com/
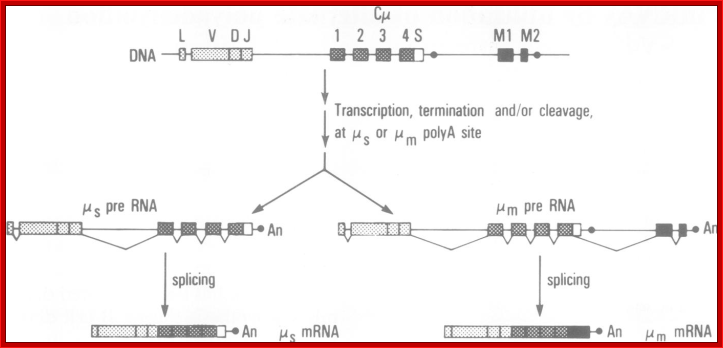
Possible RNA processing pathways for synthesis of ys and p, mRNAs; A productively rearranged Cu gene with VH, DH and JH segment joined already is shown in the top of the figure. The various boxes represent exons, and the line, introns and flanking sequences. As shown in the middle of the diagram, two precursor RNAs, one polyadenylated at the p poly(A) addition site (left) and the other at the,��� poly(A) addition site (right), are generated by separate transcription termination and/or cleavage near each poly(A) addition site, following polyadenylation at the 3' ends. The mature s (bottom left) and p, (bottom right) mRNAs are processed separately from these precursor RNAs. Poly(A) addition sites are shown as closed circles, and poly(A) as An. The schema was modified from the original figure by Early et al. (1980).
Transcriptional Regulation of Rearranged Ig Genes by Enhancers and Promoters:
� As a result of V-D-J recombination, promoter regions and enhancer regions come into proximity and the enhancers maximize transcriptional activity of the V gene promoters = high level transcription of V genes in lymphocytes
� In tumors of B and T lymphocytes, oncogenes are often translocated to Ig or TCR gene loci.
� Genes from other places, sometimes transcription factors, sometimes from viruses, are introduced into the rearranging Ag receptor genes��with bad results.
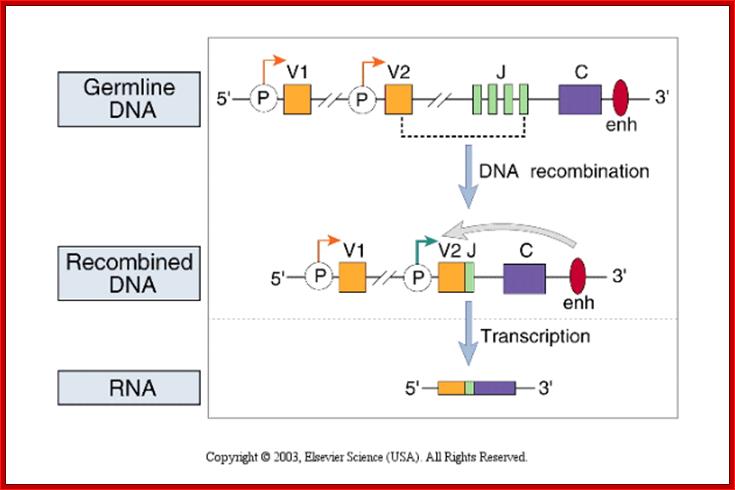
����������� Germline Ig Genome DNA recombination and Transcription; www.suggest-keywords.com
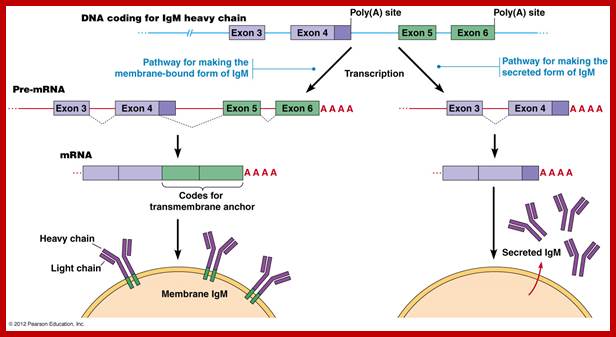
Transcription and poly A addition and translation to generate Igs. http://www.mun.ca
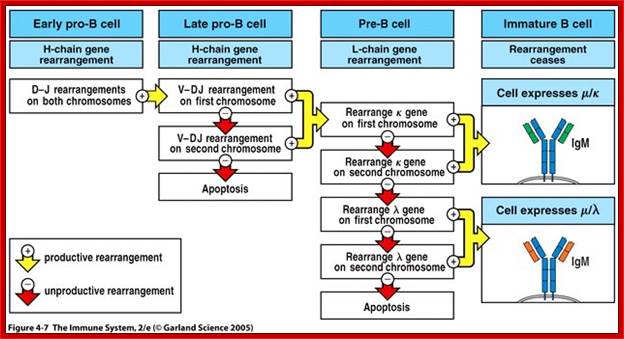
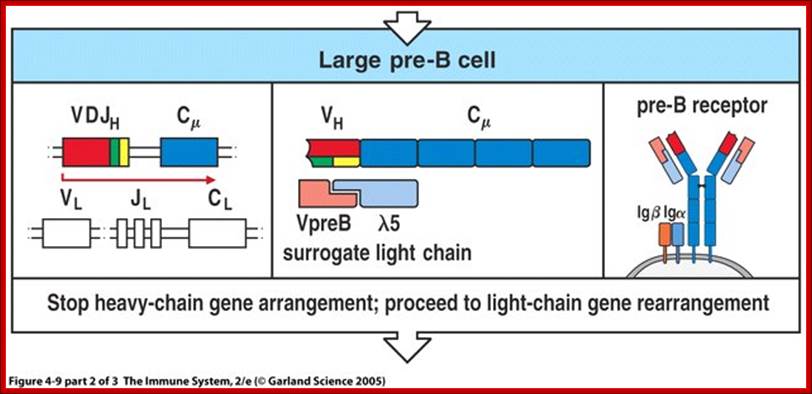
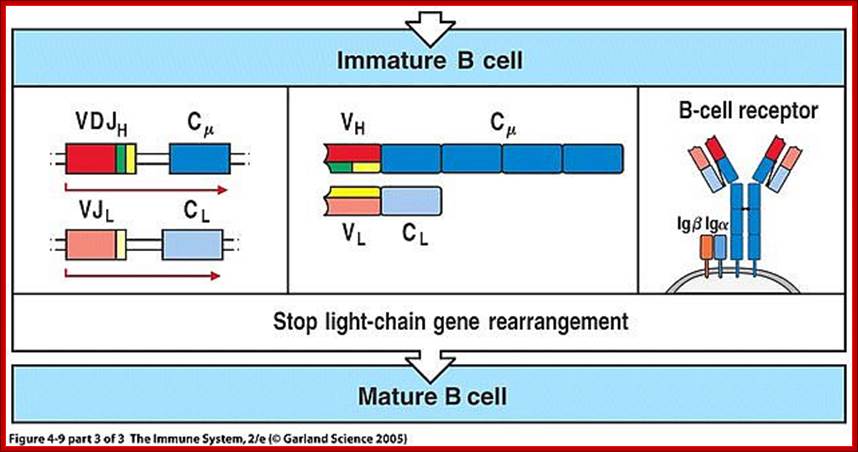
Summary of development of B cells:
- Progenitor B cells�Contains Germline H genes, Germline L genes
- Early Pro-B cells�undergoes D-J rearrangement on the H chains
- Late Pro-B cells�undergoes V-DJ rearrangement on the H chains
- Large Pre-B cells�the H chain is VDJ rearranged, Germline L genes
- Small Pre-B cells�undergoes V-J rearrangement on the L chains
- Immature B cells�VJ rearranged on L chains, VDJ rearranged on H chains. There is start of expression of IgM receptors.
- Mature B cells�There is start of expression of IgD
- Failed to develop get suppressed or anergy or subjected to apoptosis.
|
B Cell Receptor (Immunoglobulin) |
||
|
Heavy |
Kappa |
|
|
V gene segments |
1000 |
300 |
|
D gene segments |
15 |
- |
|
J gene segments |
4 |
4 |
|
N region insertion |
++ |
- |
|
Junctional diversity |
+++ |
+ |
|
Somatic mutation |
+ |
+ |
|
Combinatorial association |
V x D x J 1000 X 15 X 4 |
V x J 300 x 4 |
|
Total |
6 x 104 |
1.2 x 103 |
|
Combinatorial association |
7.2 x 107 |
|
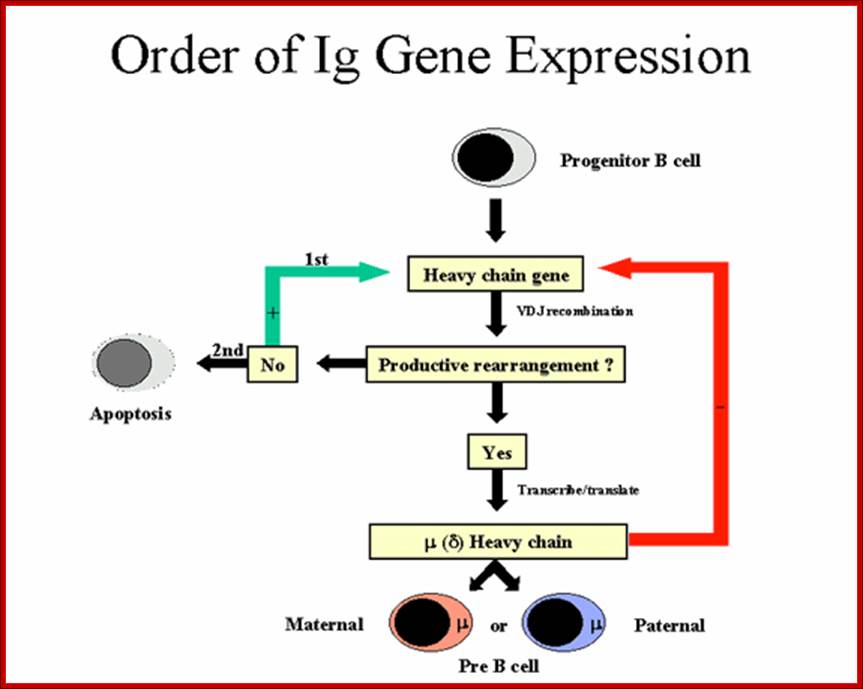
Order of Heavy chain gene expression; http://www.microbiologybook.org
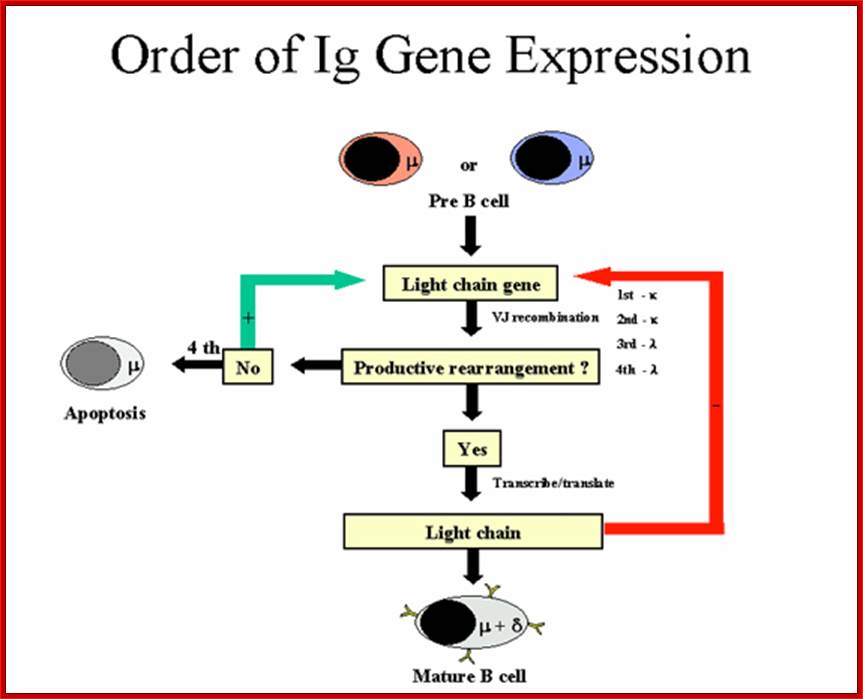
Order of Light chain gene expression; http://www.microbiologybook.org
Immunoglobulins recognize foreign antigens and initiate immune responses such as phagocytosis and the complement system. Each immunoglobulin molecule consists of two identical heavy chains and two identical light chains. There are two classes of light chains, kappa and lambda. This region represents the germ line organization of the lambda light chain locus. The locus includes V (variable), J (joining), and C (constant) segments. During B cell development, a recombination event at the DNA level joins a single V segment with a J segment; the C segment is later joined by splicing at the RNA level. Recombination of many different V segments with several J segments provides a wide range of antigen recognition. Additional diversity is attained by junctional diversity, resulting from the random additional of nucleotides by terminal deoxy nucleotidyl transferase, and by somatic hyper mutation, which occurs during B cell maturation in the spleen and lymph nodes. Several V segments and three C segments are known to be incapable of encoding a protein and are considered pseudo genes. The locus also includes several non-immunoglobulin genes, many of which are pseudo genes or are predicted by automated computational analysis or homology to other species.
http://www.keywordsking.com/
https://www.slideshare.net
Heavy chain recombination, transcription and translation generate B cell receptors.
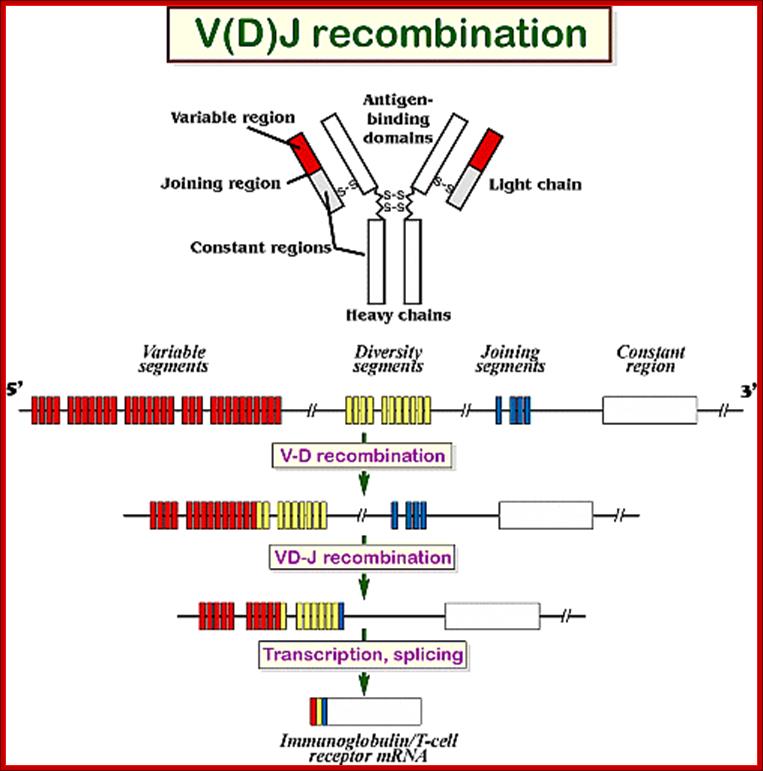
Recombination, transcription and translation-> produces IgM/IgD; http://www.alzakera.eu
Somatic Recombination and Antibody Diversity:
Bone marrow is the site for B and T cell origin. B cells mature in B/M and T cell progenitors migrate to Thymus and there they mature. Early maturation by cell to proliferation is activated by cytokines mainly by IL 7 leading to increase in immature lymphocytes.
B and T cell maturation involves somatic recombination of antigen receptor gene segments and the expression Ig molecules as BCR and TCR in B and T cells respectively. This expression provides a repertoire of useful antigen specificities.
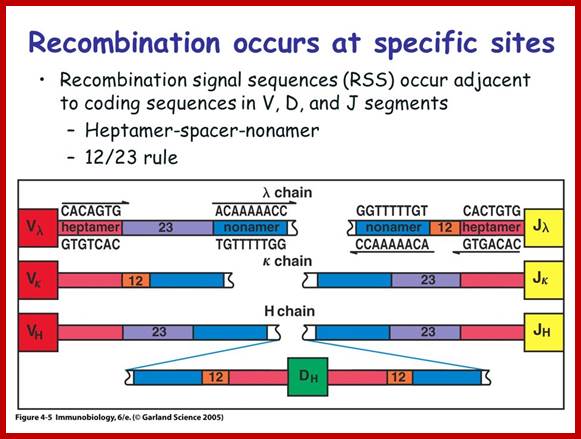
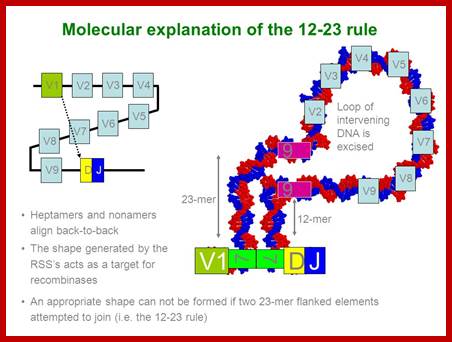
T and B cell receptor genes are spatially segregated in germ line antigen loci. They contain separate loci for Ig heavy and Ig kappa light, and Ig lambda chains, TCR beta and TCR alpha and TCR gamma and TCR delta. These loci contain V, J in Ig H chains and TCR beta and delta loci D gene segments. Somatic recombination of both Ig and TCR loci involves the joining of D and J segments in the loci that contains D segments, followed by the joining of V to recombined DJ in these loci or direct V to J joining in other loci. This recombination is facilitated by recombinase enzyme complex that includes lymphocyte-specific components RAG1 and RAG2.
Isotypes and class switching:
Starting from IgM recombination in Heavy chain constant regions can go through recombination; this leads to Isotypes.� The process is called as cxlass switching.
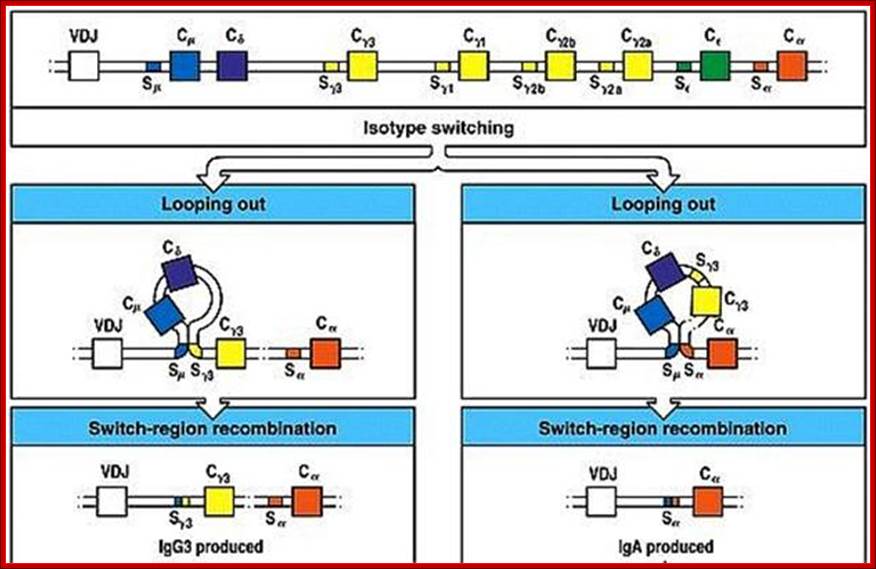
Isotype switching produces different Igs, this is due to somatic recombination at the level of DNA breaking and recombination in C loci. https://www.slideshare.net
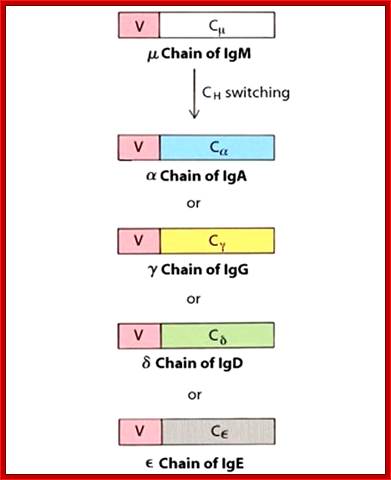
Class switching by somatic recombination; http://www.microbiologybook.org
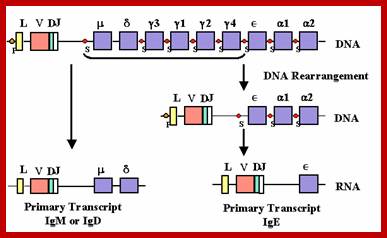
Dr Gene Mayer; http://www.microbiologybook.org
Class switching requires certain nucleotide sequences for recombination activity.Microbiology
In Immunoglobulin (Ig) class of genes switching recombination (CSR) involves the breakage and subsequent repair of two DNA sequences among the heavy chain constant regions, is known as switch (S) regions, which flank IgH constant region exons. The resolution of CSR-associated breaks is thought to require the nonhomologous end-joining (NHEJ) DNA repair pathway, but the role of the NHEJ factor DNA-dependent protein kinase catalytic subunit (DNA-PKcs) in this process has been unclear. A new study, in which broken IgH-containing chromosomes in switching B cells were visualized directly, clearly demonstrated that DNA-PKcs and, unexpectedly, the nuclease Artemis are involved in the resolution of switch breaks.
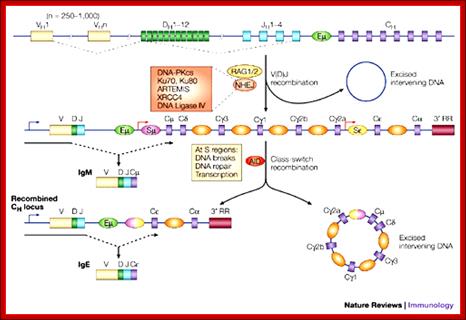
Class-switch recombination: interplay of transcription, DNA deamination and DNA repair; Class-switch recombination (CSR) of immunoglobulin heavy chains is the genetic process by which a B cell switches from the production of IgM to the production of IgG, IgE or IgA. Although the general characteristics of CSR have been known for some time, the detailed molecular mechanism of this process is only now emerging. CSR is unique, in that it seems to involve transcription-generated, higher-order RNA�DNA structures, specific DNA deamination and several DNA-repair pathways. In this review, we discuss our current knowledge of the mechanism of CSR and highlight the important unanswered questions. ; http://www.nature.com
https://www.boundless.com
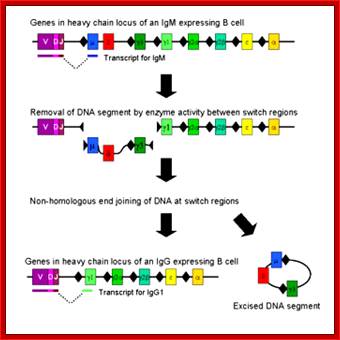
Immunoglobulin class switching (or isotype switching or isotypic commutation or class switch recombination (CSR)) is a biological mechanism that changes a B cell's production of antibody from one class to another, for example, from an isotype called IgM to an isotype called IgG. During this process, the constant region portion of the antibody heavy chain is changed, but the variable region of the heavy chain stays the same (the terms "constant" and "variable" refer to changes or lack thereof between antibodies that target different epitopes). Since the variable region does not change, class switching does not affect antigen specificity. Instead, the antibody retains affinity for the same antigens, but can interact with different effector molecules.
T cell cytokines are responsible for class switching in human (Table ). These cytokines may have suppressive effect on production of IgM.
|
Class switching in human Immunoglobulins |
||||||||
|
T cells |
Cytokines |
Immunoglobulin classes |
|
|||||
|
IgG1 |
IgG2 |
IgG3 |
IgG4 |
IgA |
IgE |
|
||
|
Th2 |
IL-4 |
↑ |
↑ |
|
||||
|
IL-5 |
↑ |
|
||||||
|
Th1 |
IFNγ |
|
||||||
|
Treg |
TGFβ |
↑ |
|
|||||
|
|
|
|
|
|
|
|
|
|
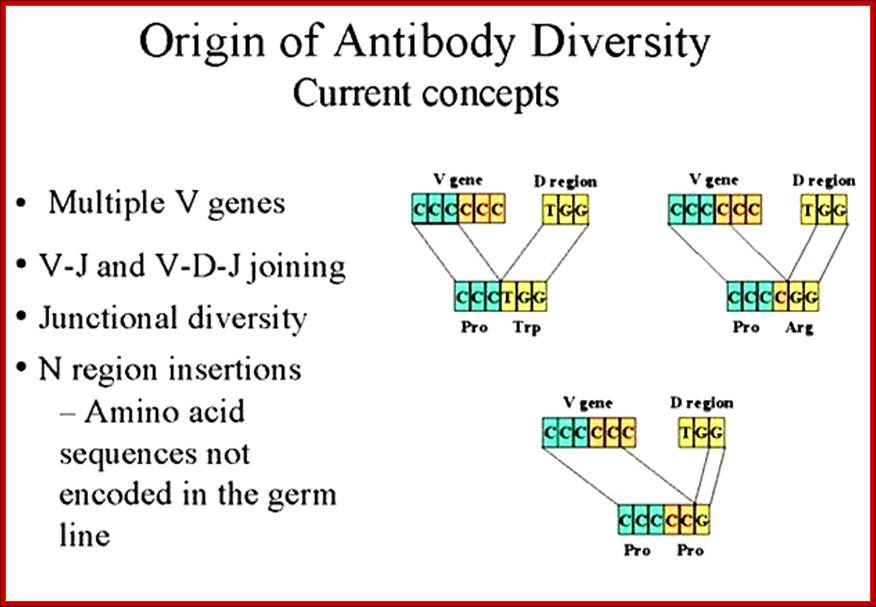
The diversity of Igs and TCR repertoire is generated by the combinatorial associations of multiple germ line V, D and J genes and junction diversity generated by the addition of random nucleotides to the sites of recombination These mechanisms generate maximum diversity at the junctions of V and C regions that form the third hyper variable region of both antibody and TCR polypeptides. http://www.microbiologybook.org
T Cell development:
Progenitors of T cells are produced in bone marrow and they are transported in to circulation, where they first lodge in Thymus.� It is in this tissue the cells undergo differentiation and maturation.
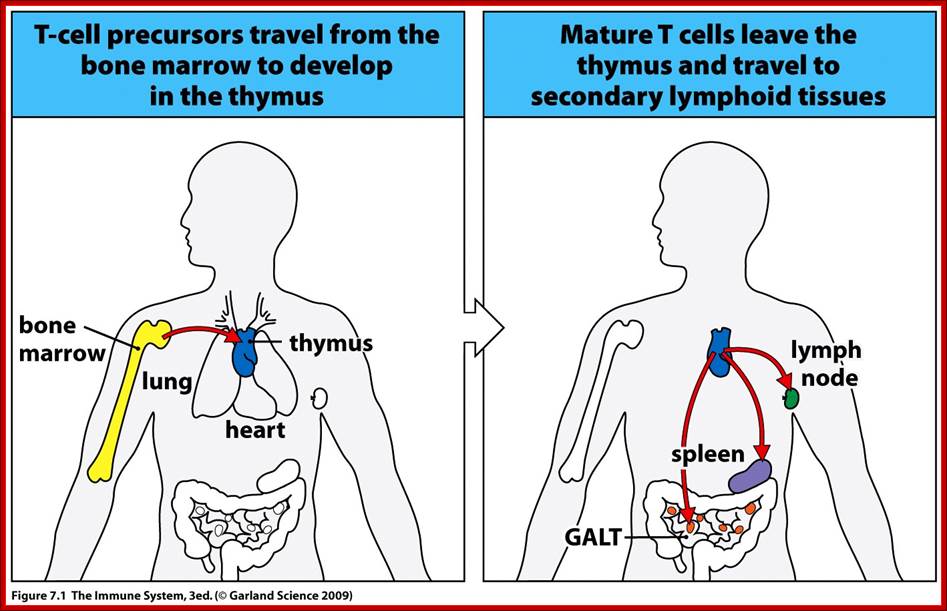
Precursor cells move into Thymus, there they mature and move to secondary lymphoid tissues such as lymph node and spleen. www.slideserve.com/
![T-cell development and the CD4|[ndash]|CD8 lineage decision](Cell_And_Molecular_Immunology3-Antigens_Antibodies_And_Its_Related_Proteins_files/image070.jpg)
T-cell development and the CD4�CD8 lineage decision; Ronald N. Germain; http://www.nature.com
Development in Thymus tissue;https://www.slideshare.net; https://www.studyblue.com
�
�Precursor cell to matured T cells. https://www.studyblue.com
Once developed they show important several surface molecules among them alpha and beta chains are important; they are anchored to the cell membrane.� In addition to these main receptor polypeptides there two others which act as co receptors/coactivators- they are CD4 and CD8 which are also anchored to membranes, they play important functions, they make T cells as T CD4 and TD8 cell types. Another group of proteins found are CD3.
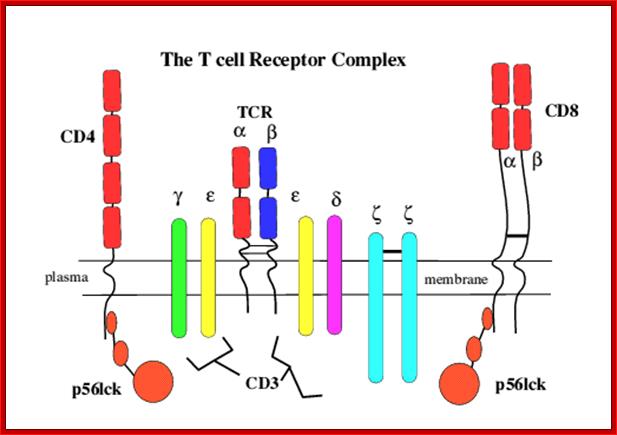
Engagement of the TCR by peptide antigen, in association with MHC gene products, leads to a series of intracellular biochemical events culminating in the transcription of new genes and cellular activation. The earliest identifiable intracellular change documented at present is the activation of one or more tyrosine kinases that phosphorylate first the CD3 chains themselves and subsequently other substrates. Subsequent to tyrosine kinase activation a series of secondary events have been observed to follow TCR engagement, including activation of serine/threonine kinases, activation of the GTP-binding protein p21ras and activation of transcription factors for receptors and growth factors such as the major T cell growth factor interleukin-2 (IL2). The CD4 and CD8 co-receptors bind a tyrosine kinase (p56lck) via their intra-cytoplasmic tail which plays a critical role in T cell signalling. https://www.slideshare.net
The intracellular portions of the CD3 g, d, e, and z subunits contain copies of a sequence motif termed ITAMs (immunoreceptor tyrosine-based activation motifs). ITAMs can serve as protein tyrosine kinase substrates and, after phosphorylation, as binding sites for SH2 domains of yet other kinases. The regulation and mechanism of the recruitment of protein kinases to the activated T cell receptor is being studied intensively; members of both the Syk family (ZAP-70) and Src family (Lck) of kinases are involved in this process.
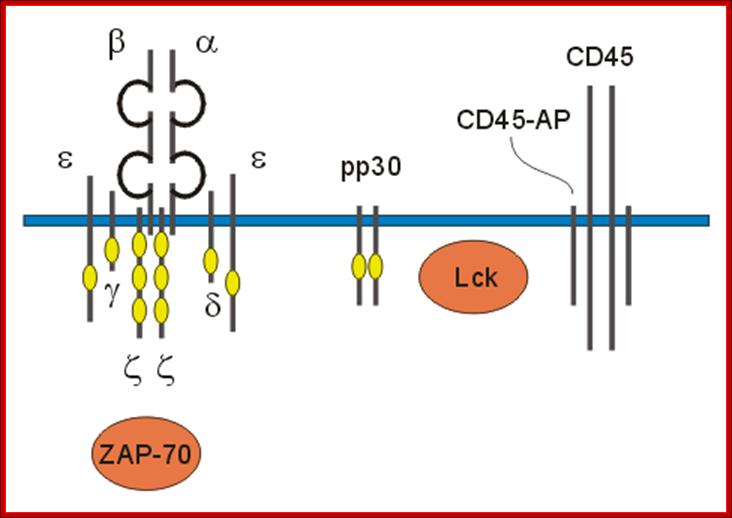
http://www.bioc.rice.edu/
Interactions between transmembrane domains are important for the function of the TCR. The TMs of TCR a and CD3 d appear to form an ion pair that is required for proper assembly and cell-surface expression of the TCR. Mutagenesis of the CD3 z TM shows that this domain mediates sequence-specific homodimerization as well as heteromeric interactions. Mutations to the TM domain of TCR b have been shown to abrogate signaling through certain downstream pathways without affecting receptor assembly.
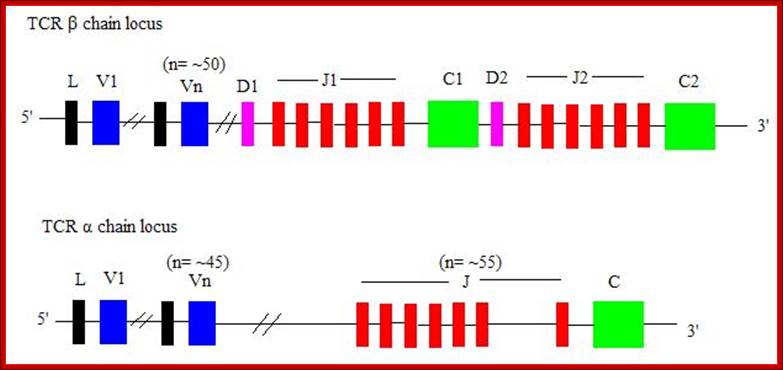
Genes for alpha and beta polypeptides of TCR;
Most T cell receptors are also composed of an alpha chain and a beta chain. The T cell receptor genes are similar to immunoglobulin genes in that they too contain multiple V, D and J genes in their beta chains (and V and J genes in their alpha chains) that are rearranged during the development of the lymphocyte to provide that cell with a unique antigen receptor.
Genes and their loci:
TRA and TRD 14q11.2 (127 genes in the locus);
TRB 7q34 (620 genes in the locus)
TRG 7p14 (19-22 total number of genes in the locus)
TRD 14q11.2 (A and 127 total number of genes in the locus);
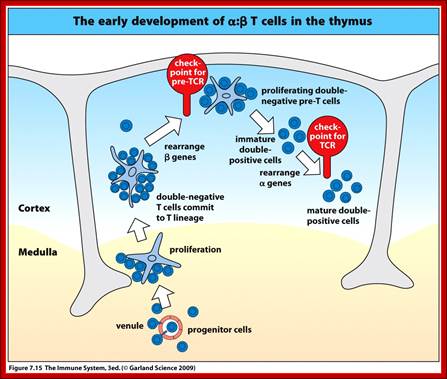
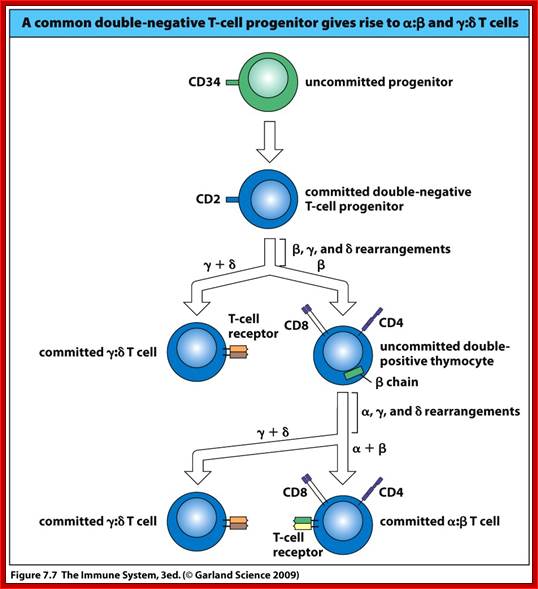
When T cells enter Thymus, they are normally unmatured thymocyte progenitor cells; they uncommitted, they double negative to CD8 and CD4.� It is at this stage, genes for alpha, beta and delta chains undergo somatic recombination to generate the main receptor proteins which will be anchored in the membrane.� There is competition between beta, gamma and delta for productive rearrangements. Those cells which fail to do so die by apoptosis. If both gamma and delta rearrange successfully before beta, T cell commits to being a gamma/delta T cell. If beta rearranges first, it will be expressed with a surrogate alpha chain as a pre-TCR. The successful Pre-T receptor with productive beta chain will clone itself and then become a double positive thymocyte (express CD4 and CD8).
Positive and Negative Selection of the T-cell repertoire; T-cells that can recognize self-MHC molecules are positively selected in the thymus . Positive selection determines expression of either CD4 or CD8 co-receptor. Alpha chain can rearrange and try again if the T cell does not recognize self-MHC. Negative selection of T cells remove self-recognizing cells while still in the thymus.� Thymic epithelial cells can express genes from other parts of the body, just so they can present these antigens to thymocytes.� Regulatory CD4 T cells comprise a distinct lineage of CD4 T cells.
https://dokuwiki.noctrl.edu
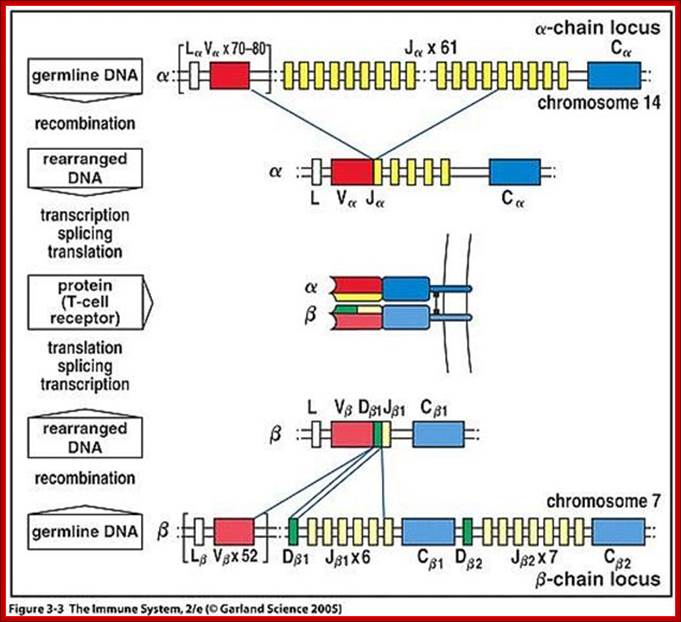
Heavy chain locus- alpha and beta gene components such as V, D, J and C loci recombination generates TCRs; www.slideshare.netbneta chain locus
Somatic Recombination:
Looking at the alpha and beta genes they have DNA segments similar to Igs, such as L,V,D, J and C regions.� The recombination process is more or less similar to Ig genes. After recombination, transcription leads to splicing and the same are transported into cytoplasm where they are translated and transported on to cell membrane.
V(D)J recombinase is a group of enzymes that mediate the somatic recombination of V and J, or V, D, and J gene segments and is only expressed in immature T-cells. V(D)J recombinase is made up of RAG-1 and RAG-2 proteins which recognized the DNA sequences that surround the V, D, and J, gene segments.
The recognized V, D, and J segments are brought close together by V(D)J recombinase which then cleaves the DNA at specific sites and the DNA is then repaired by ligase. This causes the V, D, and J segments to be rearranged to form a chain of TCR.
Combinatorial Diversity
Combinatorial diversity occurs by the different available number and combinations of V, D, and J gene segments and is therefore limited on the amount of variation. There are about 3�10^6 different combinations of V, J and V, D, and J genes.
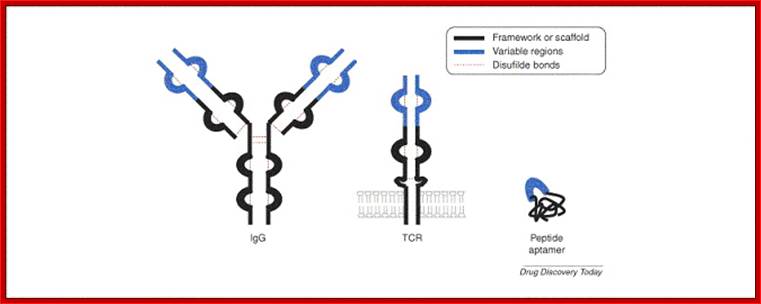
Peptide aptamers; Comparison between BCR and TCR proteins; http://aptamerstbc2013.wixsite.com/
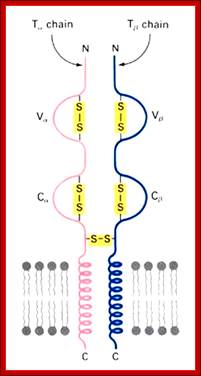
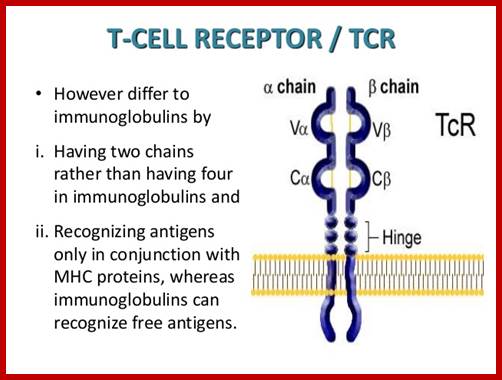
https://www.slideshare.net
T alpha and T beta chains anchored to cell membrane
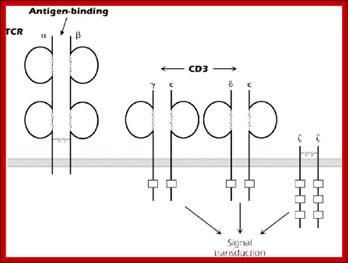
The T-cell receptor complex (TcR complex) is shown in figure 8.4. What is shown is the TcR and its associated protein molecules called CD3:;http://www2.hawaii.edu
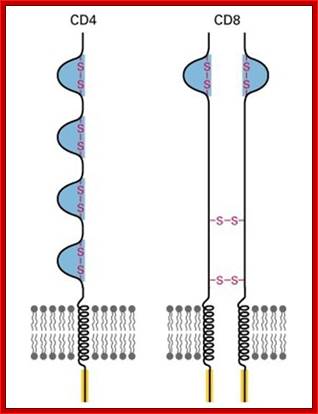
T cell receptors- CD4 with single chain and CD8 with two similar chains cross linked by S-S bonds
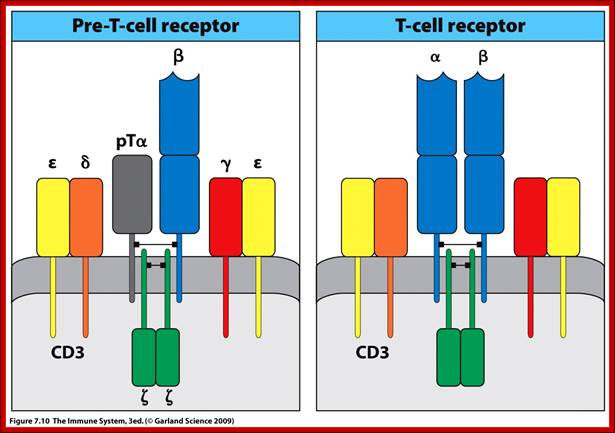
Diverse membrane protein populations are found on T lymphocytes. https://www.studyblue.com
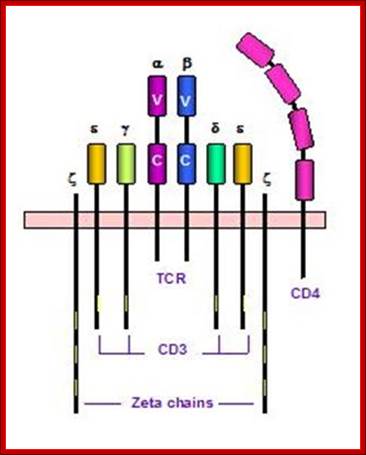
T cell receptors and associated co-receptors/activators; http:// allergycases.blogspot.com
![]()
Dendritic cells not only present the antigens but they should contain receptors such as B7 for na�ve T cell to be activated, and the T cell in turn should contain TCR plus CD4 and CD28 for their proper activity. The combination of an antigen specific signal and a co stimulatory signal is required to activate a na�ve T cell;https://www.studyblue.com
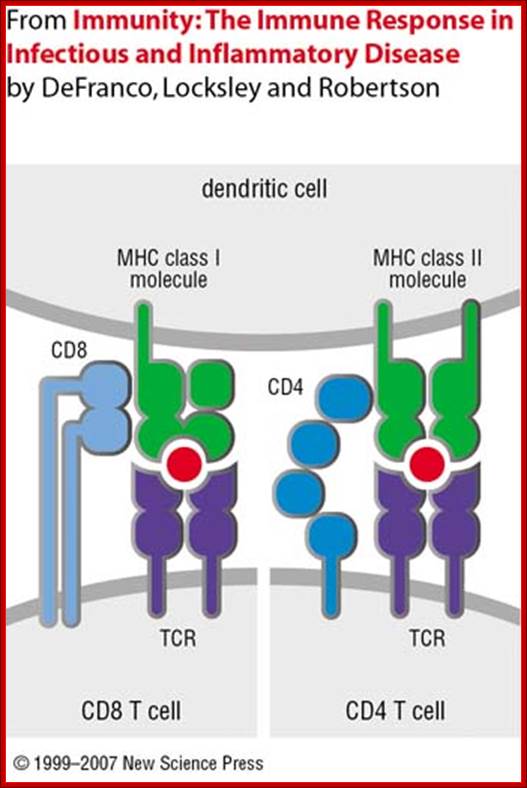
Dendrite cell with their MHCs containing antigens locked on to T cell receptors; https://www.slideshare.net
https://www.slideshare.net; T cell receptor with their additional helper proteins such as CD8 for MHCI and CD4 for MHCII molecules
T cell maturation in thymus also progresses in stages distinguished by expression of antigens receptors genes, CD4 and CD8 co receptor molecules and location in thymus. The earliest T cell lineage immigrants to thymus do not express TCRs or CD4 or CD8 molecules. Developing T cells within Thymus (thymocytes) initially populate the outer cortex, where they undergo proliferation, rearrangement of TCR genes and surface expression of TCR CD3, TCR CD4 and CD8. As cells mature they migrate from the cortex to the medulla.
The least mature thymocytes called pro-T cells are Cd4^- and CD8^- double negative and the TCR genes are in germ line configuration. In the pre-T stage, thymocytes remain double negative, but VDJ recombination occurs in TCR beta chain locus. Primary beta chain transcripts are expressed and processed to bring C beta segments adjacent to VDJ complex and beta chain polypeptides are produced. The beta chain associates with the invariant pre-T alpha protein to form a pre TCR. The pre TCR transduces signals that inhibit rearrangements on the other beta chain alleles (allelic exclusion) and promote CD4 and CD8 expression and further proliferation of immature thymocytes to the CD4+CD8+ (double positive) stage of T cell development, V-J recombination in the alpha locus , alpha chains are produced and low levels of TCR are expressed on cell surface.
Selection process drive maturation of TCR expressing double positive thymocytes and shape the T cell repertoire towards self MHC restriction and self tolerance. Positive selection of CD4+CD8+ TCR alpha/beta thymocytes require low avidity recognition of peptide �MHC complexes on thymic epithelial cells, leading to a rescue of the cells from PCD. Negative selection of CD4/CD8 plus TCR alpha/beta double positive thymocytes occur when these cells recognize with high avidity, antigens that are present at high concentration in thymus. This process is responsible for tolerance to many self antigens. Most of the cortical thymocytes do not survive these selection processes. As the surviving TCR alpha/beta cells mature they move into the medulla and become either CD4, CD8 plus or CD4/CD8 minus. Medullary thymocytes acquire the ability to differentiate into either helper or cytotoxic effector cells and finally emigrate to peripheral lymphoid tissues.
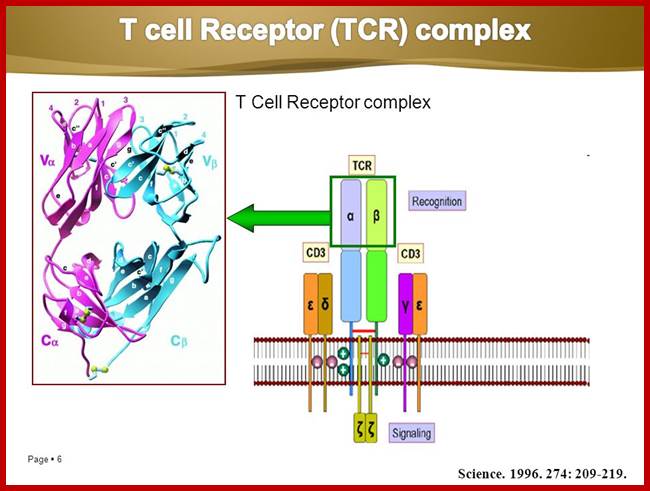
http://slideplayer.com
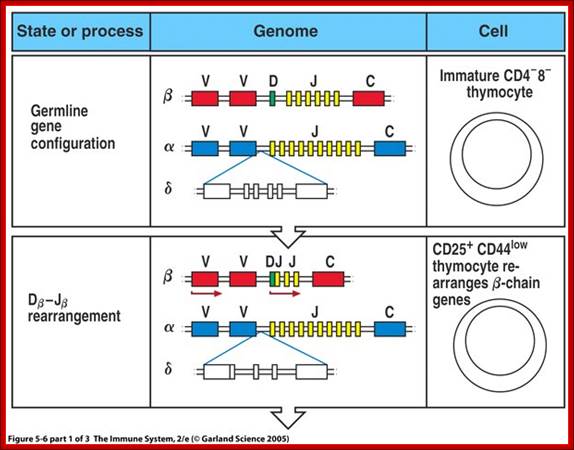
T cell receptors;
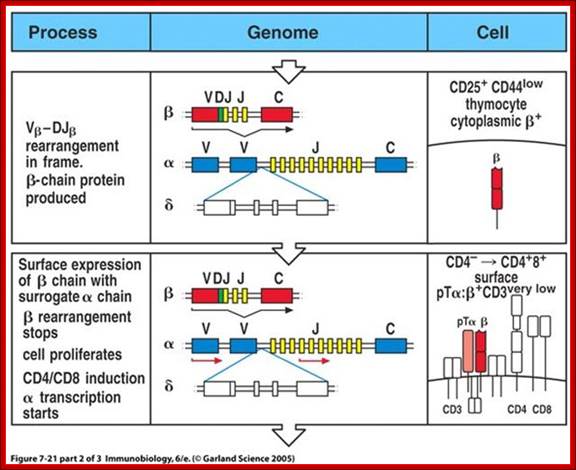
T cell Receptor gene loci and recombination to generate TCRs; slideplayer.com/slide/4225612/