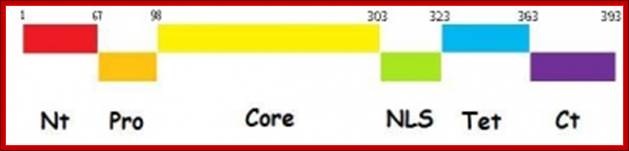
Cell Cycle and Its Regulation:
The Vision Of Jeff Johnson; SCIENCE PHOTO LIBRARY; Human Chromosomes; http://www.sciencephoto.com/
Saccharomyces cerevisiae; http://www.pha.jhu.edu/
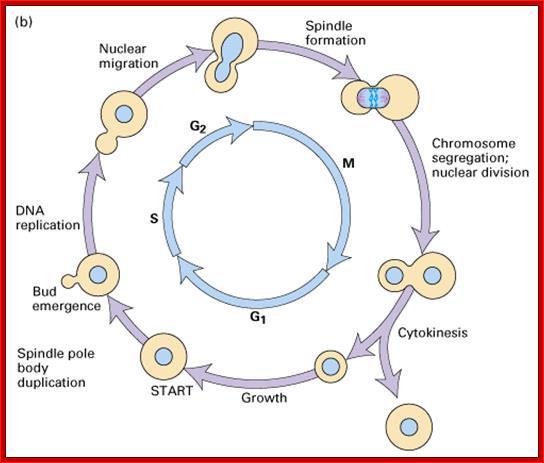
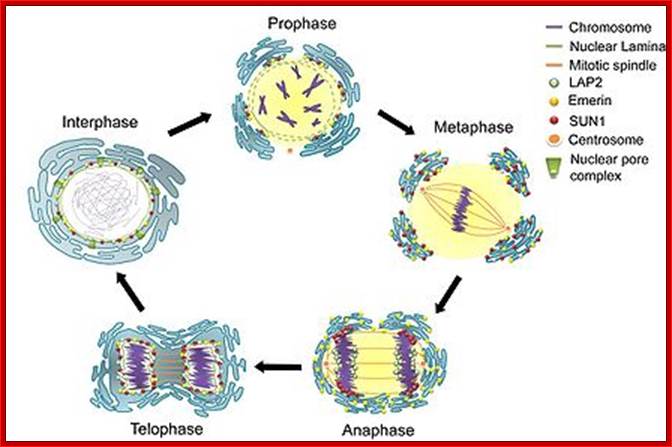
Cell cycle progression, nucleus divides without disintegration nuclear membrane; http://www.pha.jhu.edu/
Regulation of Cell Cycle:
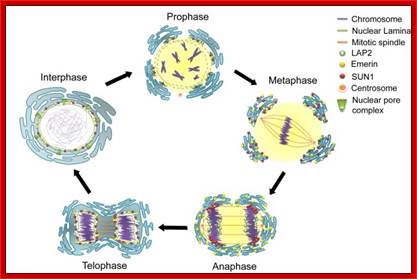
As in prokaryotes, Eukaryotic DNA replication is restricted to either Mitosis or Meiosis.� Mitosis is used for growth and development and in some lower forms it is one of the modes of reproduction.� But meiosis is mostly involved in reproductive stages.� Whether Mitosis or Meiosis, cell division is highly regulated and precise and exact.� Mitosis goes through several programmed phases such as Prophase, Metaphase, Anaphase, Telophase and Cytokinesis (not always) and then the cell enters Interphase, which is an intervening stage at which the cell prepares for the next cell division or goes into resting phase where the cells undergo differentiation to specific cell type.
Reversible chemical modifications that regulate the flow of genetic information:
In the central dogma, genetic information is passed from DNA to RNA and then to protein. Epigenetic DNA modifications (for example, the formation of 5-methylcytosine (m5C; also known as 5mC) and 5-hydroxymethylcytosine. Cellular RNAs carry diverse chemical modifications that used to be regarded as static and having minor roles in 'fine-tuning' structural and functional properties of RNAs. In this Review, we focus on reversible methylation through the most prevalent mammalian mRNA internal modification, N6-methyladenosine (m6A). Recent studies have discovered protein 'writers', 'erasers' and 'readers' of this RNA chemical mark, as well as its dynamic deposition on mRNA and other types of nuclear RNA. These findings strongly indicate dynamic regulatory roles that are analogous to the well-known reversible epigenetic modifications of DNA and histone proteins. This reversible RNA methylation adds a new dimension to the developing picture of post-transcriptional regulation of gene expression.
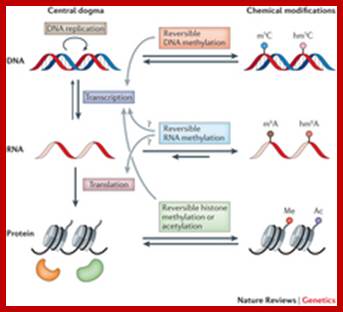
Gene expression regulation mediated through reversible m6A RNA methylation; Ye Fu, Dan Dominissini,Gideon Rechavi & ; He; http://www.nature.com
Rb/E2F protein regulation of the transition from G1 to S phase mammal cells� http://www.bio.miami.edu
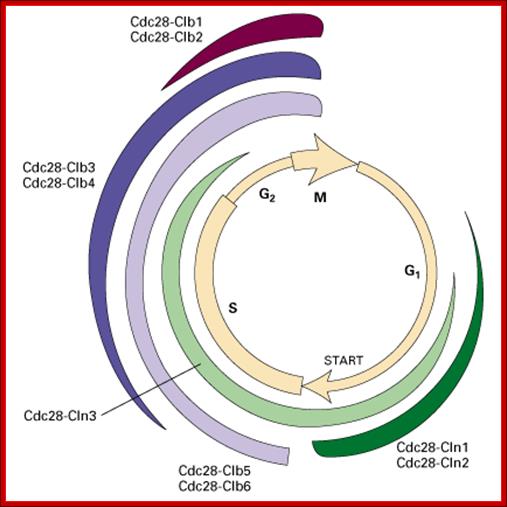
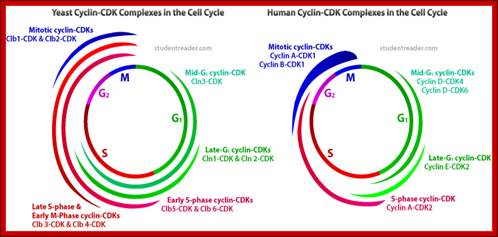
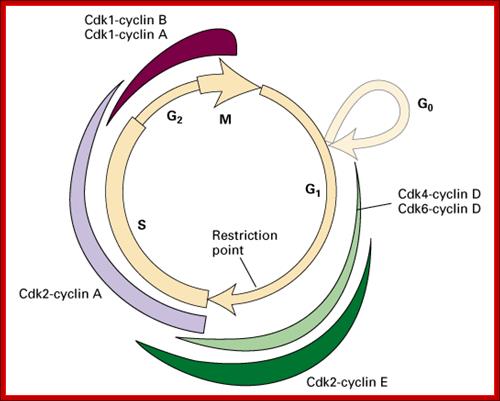
Role of CDK-Cyclins at different phases of the Cell cycle. Passage through G1, S and G2; http://www.pha.jhu.edu
In vertebrates, the G1 kinases are Cdk4/6 complexed with cyclin D. The late G1/S-phase kinase is Cdk2 complexed with cyclin E. Yeast G1 Cdk activity is regulated by Clns. The main job of the G1 kinases is to prepare the cell for S phase, so they stimulate the synthesis of the DNA replication enzymes and the S-phase kinase; Ghzheng http://www.pha.jhu.edu
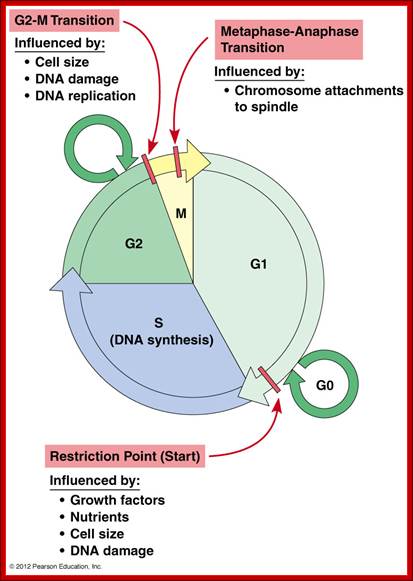
Cell cycle restriction Points; Hardin, Bertoni & Klein smith, 2012 http://www.mun.ca/
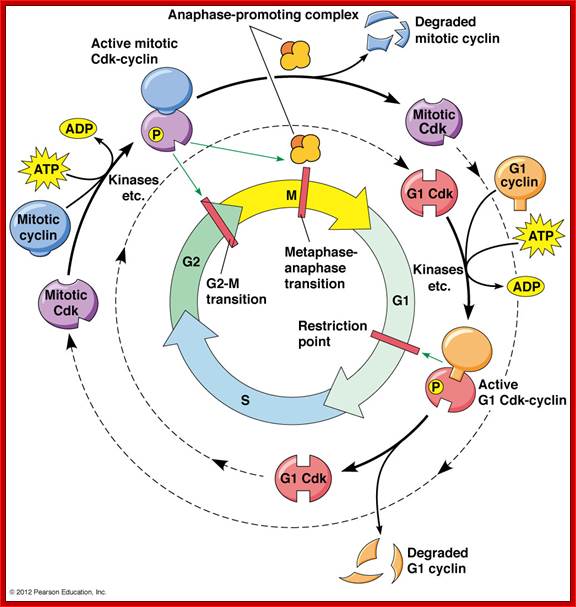
The cell cycle is regulated at the checkpoints by cyclin-Cdk complexes.; http://www.mun.ca/
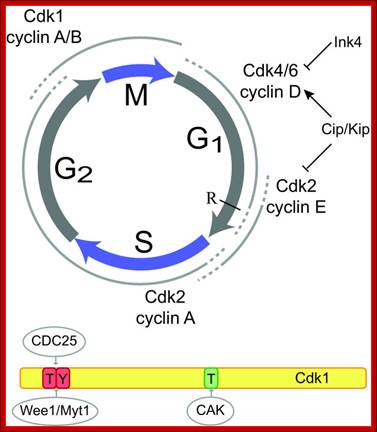
The Cell cycle regulators-Check points; The first includes cyclins, a regulator subunit. The second is that of cyclin-dependent kinases (CDKs), a catalytic subunit. Cyclins are synthesized in the cell cycle and have no catalytic activity. CDKs on the other hand, are simply found within the cell (not created during the cell cycle) and only become active once bound with cyclins. Although there are many types of CDKs, for simplicity�s sake, we will focus on a few. CDK-1 is commonly found/used in the G2 and M phases of the cell cycle; CDK-4 is found/used during the G1 phase. Paul Nurse and Tim Hunt received a Nobel Prize in Physiology or Medicine in 2001 for their efforts in this category. wordpress.com/cell-cycle-regulators/-
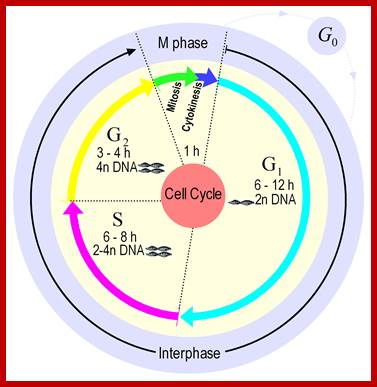
� Among the five stages in a ~24hrs cell cycle in animal cell cultures, interphase occupies longest period where as the other stage called M-phase lasts just about 0.30 min to 1 hr.� But embryonic cells go through the whole cell cycle is just ~15 �30 minutes or less; in this the cell after M-phase, directly enters to S-phase.� It also means all the inputs required for the next stage already exist for they are synthesized all the time in embryonic cells.���
� In Cerevisiae, G1 last for ~15 min, S-phase requires ~30 min, G2 takes 20-30 minutes and M-takes about ~75 min all together require ~150 min.� While S. pombe (fission yeast), G-1 requires ~20 min, S takes ~20 min, G2-takes ~35 min and M-phase lasts for ~45 min, in all ~120 min.�� In both the above-mentioned forms cell divides without dissolution of nuclear membrane.
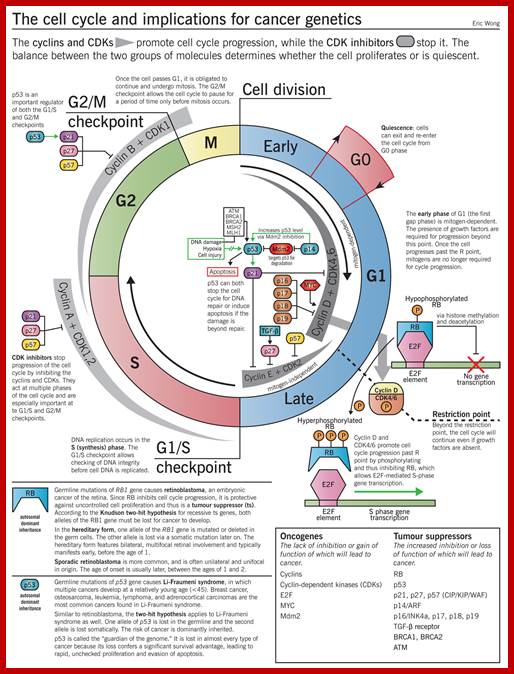
Image source: McMaster Pathophysiology Review (pathophys.org); http://epomedicine.com
http://epomedicine.com/medical-students/
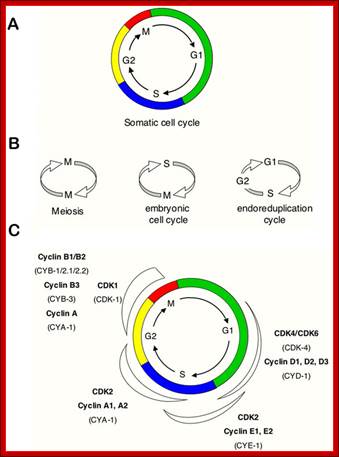
Simple representations of the cell cycle; (A) a typical (somatic) cell cycle, which can be divided in four sequential phases: G1, S, G2 and M. M phase consist of nuclear division (mitosis) and cytoplasmic division (cytokinesis) (B) variant cell cycles in which specific phases are omitted. (C) Approximate time of activity for different combinations of cyclins and CDKs, based on studies of mammalian cyclins and CDKs. C. elegans family members are indicated between brackets in the figure. Shapes outside the cycle indicate increase and reduction of corresponding CDK/cyclin activity. Sander van den Heuvei; http://www.wormbook.org
The cell cycle is controlled by a highly complex bio-chemical-molecular system. In this project authors investigate the mitotic transition control mechanisms. In order to capture their complexity on a systems level, we apply mathematical and computational methods. Time scale; http://users.minet.uni-jena.de
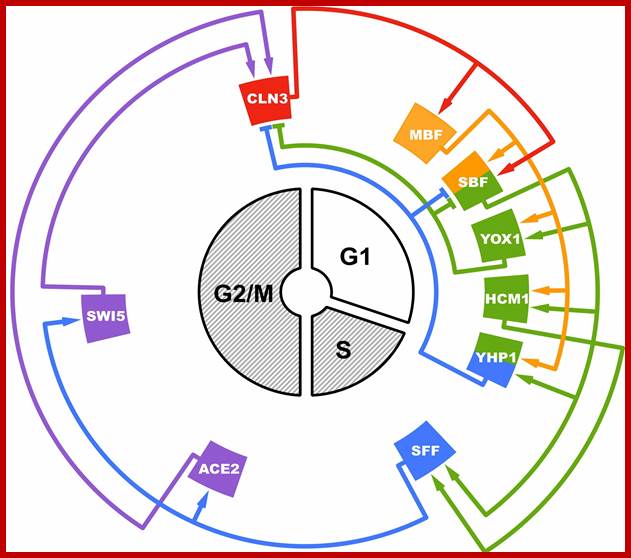
Haase group; http://sites.biology.duke.edu/
The Haase Lab is broadly interested in the structure/function of biological clocks. In 2008, the Haase group proposed a new clock model for the cell cycle in which a complex network of sequentially activated transcription factors (TFs) regulates the precise timing of gene expression during the cell cycle, and functions as a robust time-keeping oscillator. Greater than 1,000 genes are expressed at distinct phases of the cycle, and the control network itself consists of ~20 components. This dynamical system is far too complex to understand simply by biological intuition. The Haase Lab combined efforts with John Harer's group in the Department of Mathematics at Duke. Harer's expertise in the analysis of complex data, along with his understanding of dynamical systems, brought the two groups together to dissect the intricacies of the cell-cycle clock mechanism. Using a collection of tools, including molecular genetics, genomics, mathematical models, and statistical inference, the group aims to understand how the cell division clock works, how it might be perturbed in proliferative diseases such as cancer, and how the clock components might be targeted for new anti-tumor therapies. Cell-cycle models and network perturbation experiments are first conducted in the highly genetically tractable budding yeast, Saccharomyces cerevisiae. The oscillating TF network model proposed in 2008 is shown below: http://sites.biology.duke.edu/
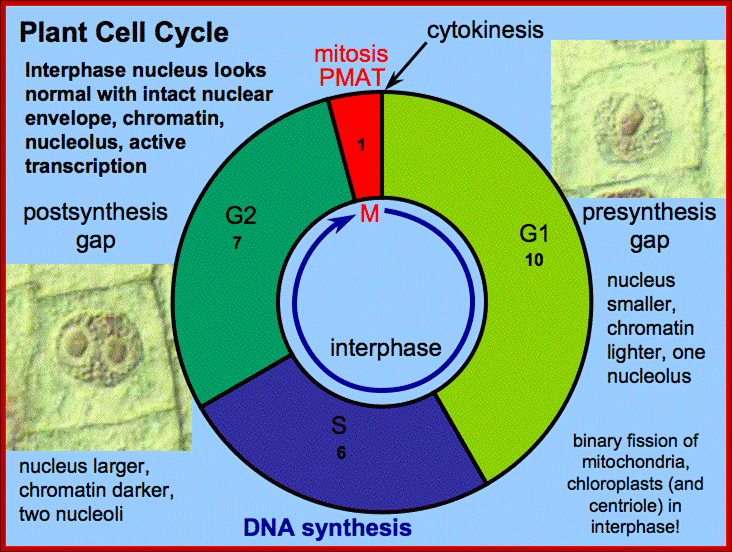
Interphase consists of sub-stages such as G1, S and G2; where, G stands for the gap in knowledge (in early years of cytology) of these stages so they are called G stages. G1 phase is further subdivided into early, mid and late phases; this view is based on the synthesis of cyclins such as early, mid and late G1 cyclins. In 24 hr cell cycle events G1 occupies ~10-12 hrs, S-stage about ~6-8 hr and G2 stage -4.5 hr.� The G1 phase is considered as preparatory phase for DNA replication, but the cells escape from G1 phase in terminally differentiating cells into what is called G� stage, where cells assume specific shape, structure and function. But some of the cells remain embryonic and such cells can be stimulated by some mitogens to become dividing cells, and they can be induced to differentiate depending upon the kind of stimulus they get, or stimulus provided.� In animal system they are called STEM cells such cells are found in most of the tissues. Such stem cells can be isolated from living beings and they can be induced to differentiate into their specific cell types and the same can be grafted to diseased humans.� But in plant tissues, all nuclei containing cells have greater potentiality to become mitotic by mitogens such as cytokinins. Plants too have stem cells I their living tissues except sieve tube cells.�
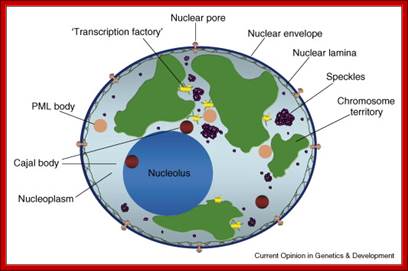
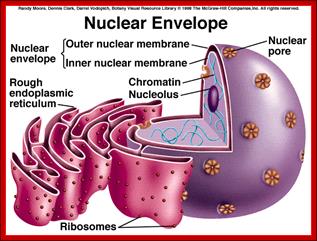
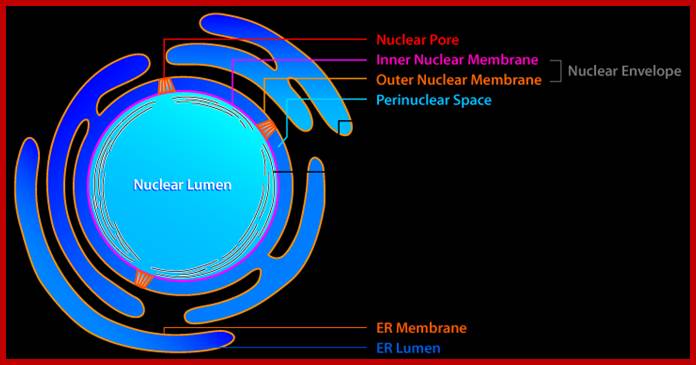
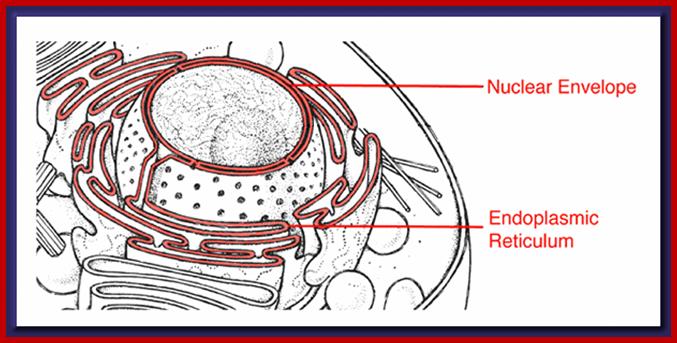
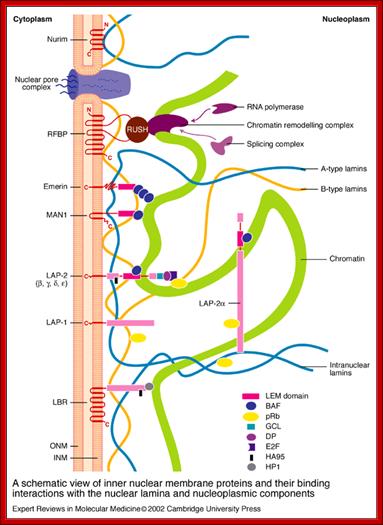
Compartments of Interphase Nucleus; Olga Pontes,� Craig S Pikaard; slideplayer.com; http://www.sciencedirect.com/
Compartments of the interphase cell nucleus. The nuclear envelope is a double membrane punctuated by nuclear pores through which molecules traffic to and from the cytoplasm. The nuclear lamina is a meshwork of proteins mediating nuclear envelope structure and chromatin attachment. In the nucleus, chromosomes occupy non-random positions known as chromosome territories. Genes transcribed simultaneously tend to cluster in �transcription factories. In the vicinity of transcription factories for Pol II-transcribed protein-coding genes, speckles serve as storage, recycling or assembly sites for snRNPs and other splicing proteins. The nucleolus is the site of ribosome biogenesis and numerous RNA-related functions. Cajal bodies are spherical structures involved in the biogenesis of ribonucleoprotein complexes including siRNA and miRNA RISC complexes in plants. PML bodies are linked to various aspects of transcriptional regulation, virus accumulation, tumor suppression, and DNA repair. siRNA and miRNA are involved in mRNA processing, including mRNA inactivation where Cajal bodies take part in the process. In certain plants Cajal bodies are the sites for the synthesis of mi and si RNAs. Olga Pontes,� Craig S Pikaard; http://www.cell.com/trends/genetics
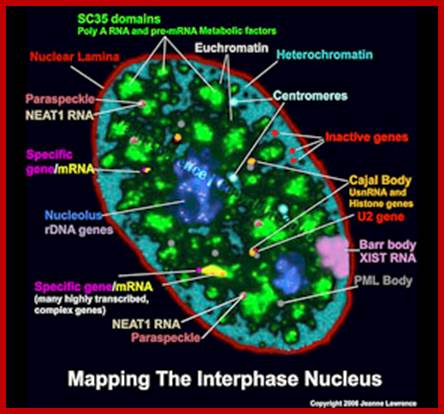
Pluripotent stem cell consists of unique nuclear organization lacking defined structural compartments; Lawrence lab; https://www.umassmed.edu
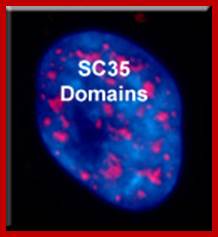
There are 25-30 sites where pre-mRNA is processed and the required components are located, suc structures are called nuclear Speckles SC35 domains; Splicing factor compartments (SFCs), they also contain numerous splicing factors and SR proteins, as well as poly(A) RNA, poly(A) RNA Binding Protein II, hyperphosphorylated RNA polymerase II, lamins, and factors implicated in RNA; they also contain active genes transcribing in the region, export.; http://labs.umassmed.edu/
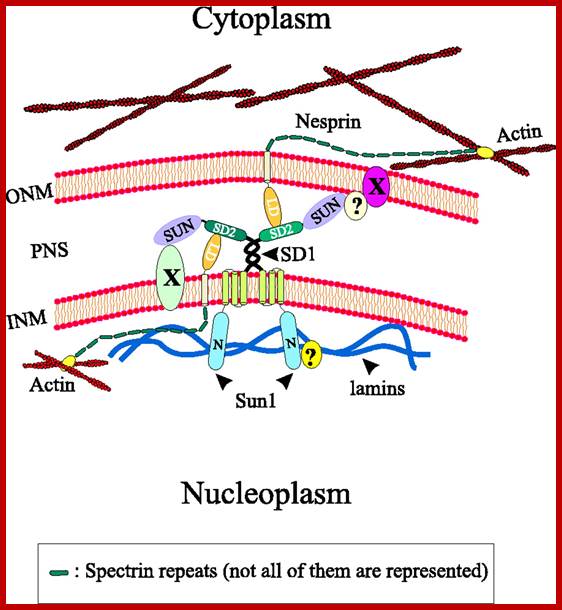
. Model illustrating the interactions of Sun1 with Nesprin at the nuclear envelope. Unknown nuclear envelope proteins and interactions are indicated by X and ? respectively. To reduce complexity a homotypic dimerization of Sun1 via the coiled-coil regions is postulated, although other coiled-coil-containing proteins might form heterotypic complexes with Sun1. INM, inner nuclear membrane; LD, luminal domain; N, N-terminal domain; ONM, outer nuclear membrane; PNS, perinuclear space; V.C. Padma Kumar et al; http://jcs.biologists.org
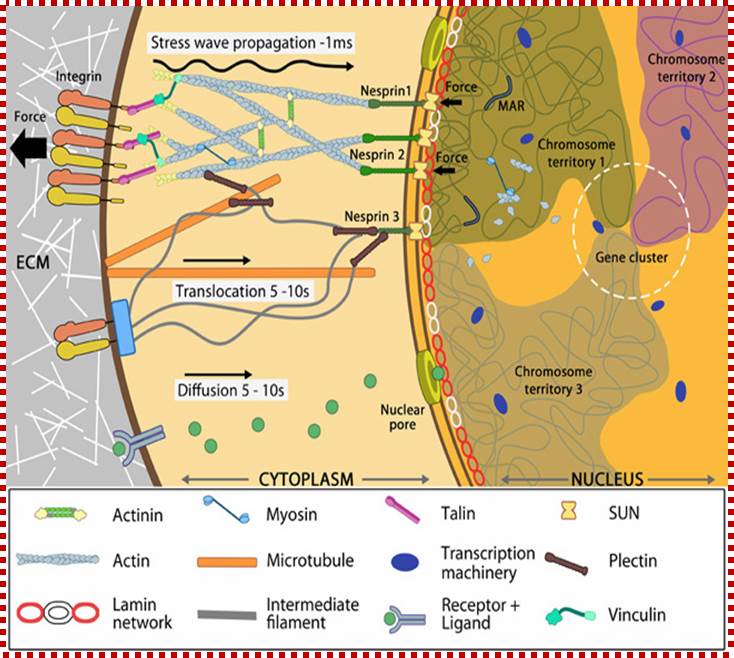
Nuclear connectivity and mechano-transduction; MB: Info, Assoc Prof G.V. Shiva Shankar, MBI, Singapore;
��Force experienced by integrins at the cell surface via mechanosensing structures like focal adhesions (integrin cluster linked to actin network), hemi desmosomes (blue rectangle) or cell-cell contact (not shown) is accumulated, channeled through SUN1/SUN2 form the LINC (linker of nucleoskeletal and cytoskeleton) complexes connecting further to the nuclear lamina (red and white lamin network) and hence the attached nuclear scaffold proteins (actin and myosin). Chromatin attaches directly to the lamina and to other scaffolding proteins through the matrix attachment regions (MARs). Upon sensing the force, the nuclear scaffold help repositioning the chromatin thus affecting nuclear pre-stress and activating genes within milliseconds. Spatial segregation of chromosomes with defined territories is represented as colored compartments inside the nucleus. The dotted circle highlights looping of genes from different chromosomes to form a cluster in 3D space and share transcription apparatus (navy ovals). On the contrary, chemical signaling mediated by motor-based translocation along cytoskeletal filaments or diffusion of activated regulatory factors takes few seconds. MB: Info, Assoc Prof G.V. Shivashankar, MBI, Singapore.
Cells can be stimulated by mitogens, to enter into cell division mode, where they enter again into G1 phase.� The S-phase is for DNA replication and repair if needed and G2 stage is a preparatory phase for M-phase, where the nucleus disassembles, chromatids are rendered inactive by histone methylation and deacetylation; and condensed by condensin proteins and sister chromatids are still held together by cohesins till mitotic apparatus assembles, the centromere splits (visually), sister chromatids are pulled to their respective poles; daughter nuclei reform and cytokinesis leads to division of cytoplasm into two cells.� This is a simplistic description of cell division.� In some cells mitosis continues without cytokinesis leading to multinucleate cells.
� Each of these phases is regulated by specific factors and molecular events, until each of the events in each of the phases, is completed, cell won�t enter the next stage.� Thus the entry and exit to and out of stages or phases is tightly regulated by a variety of factors; and the factors themselves undergo changes in their turn over (synthesis and degradation), activation and inactivation.
Nuclear pore complexes; http://reasonandscience.heavenforum.org
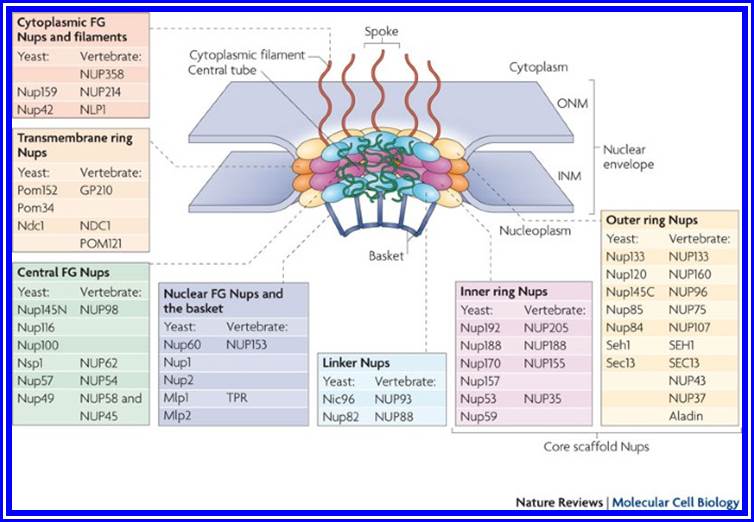
Nuclear pore complexes (NPCs) fuse the inner and outer nuclear membranes to form channels across the nuclear envelope. They are large macromolecular assemblies with a complex composition and diverse functions. Apart from facilitating nucleocytoplasmic transport, NPCs are involved in chromatin organization, the regulation of gene expression and DNA repair. Understanding the molecular mechanisms underlying these functions has been hampered by a lack of structural knowledge about the NPC. The recent convergence of crystallographic and biochemical in vitro analysis of nucleoporins (NUPs), the components of the NPC, with cryo-electron microscopic imaging of the entire NPC in situ has provided first pseudo-atomic view of its central core and revealed that an unexpected network of short linear motifs is an important spatial organization principle. These breakthroughs have transformed the way we understand NPC structure, and they provide an important base for functional investigations, including the elucidation of the molecular mechanisms underlying clinically manifested mutations of the nucleocytoplasmic transport system. https://www.nature.com/
����������������������������������������������� ��������� 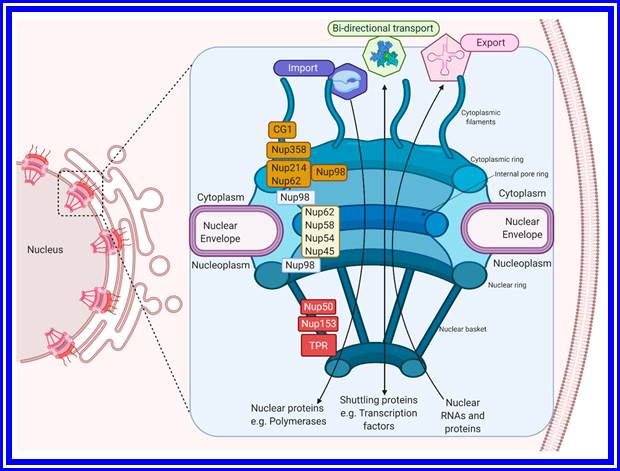
The machinery of the Nuclear Pore Complex. The NPC�s different components, which participate in the import and export of proteins and RNAs, and the FG-Nups, which participate in nuclear�cytoplasmic transport, are shown. https://www.mdpi.com/
Molecules smaller than approximately 40�50 kDa can pass freely through the nuclear envelope; however, higher molecular weight molecules such as proteins and RNAs from both cellular and viral origin are actively transported through the NPC, between the nucleoplasm and the cytoplasm. The nuclear import and export of molecules are regulated by NTR (nuclear transport receptors), such as importins, exportins, carriers, and small GTPases of the Ran family that regulate the activity of importins and exportins that transport cargo molecules (Figure 2) [36,37,38].
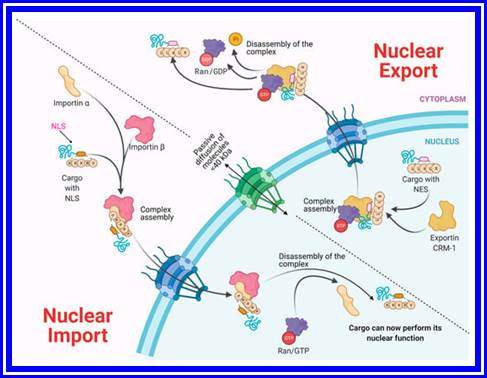
Bidirectional Nucleus�cytoplasm Transport. In the classical pathway of import (importin α/β), the importins and cargo complex are formed through the recognition of the NLS (nuclear location sequence). In the case of Export via CRM-1, the charge contains an NES (nuclear export sequence). In both cases, a GTP gradient is required. Molecules <40 kDa pass through passive diffusion towards the nucleus by the NPC. https://www.mdpi.com/
NTRs recognize specific sequences in cargo proteins that cross the nuclear membrane from the cytoplasm, such as nuclear location sequences (NLS) that contain repeated arginine (Arg or R) and lysine (Lys or K) amino acids. The classical NLS consists of five KKKRK amino acids. Moreover, some proteins possess bipartite NLS consisting of two groups of basic amino acids, separated by approximately ten amino acids. On the other hand, nuclear export sequences (NES) participate in the trafficking from the nucleus to the cytoplasm. They are composed of sequences rich in leucine or hydrophobic amino acids such as valine (Val), isoleucine (Ile), phenylalanine (Phe), or methionine (Met), which are found in motifs conserved in cargo proteins, such as some transcription or translation factors and mRNA transport proteins. �The nuclear localization of a given protein occurs by the recognition of the NLS by the NTRs; for example, importin α through its NTR binds to NLS-cargo, then importin β binds to importin α to form a trimeric complex and its cargo molecule. If the NLS is atypical, then importin β directly binds to its cargo molecule without the participation of importin α. The directionality of the cargo is given by the Ran-GTP gradient, regulated by the Ran-GTP/GDP cycle. Once the trimeric complex has entered the nucleus, the Ran-GTP activated by RCC1 (GEF) joins importin β and thereby is released from its cargo. Importin β is transported to the cytoplasm, and the Ran-GTP is deactivated to Ran-GDP by GAP (GTPase Activating Protein) to free itself from importin β for its next import cycle. https://www.mdpi.com/
On the other hand, the nuclear export is given by the recognition of the nuclear export sequences (NES). Nuclear export begins with Ran-GTP binding to exportin (e.g., CRM-1), which causes an increased affinity for the export cargo. Then, the complex moves to the nuclear pore and Ran-GTP hydrolyze (activated by RCC1), which forms the export complex. The complex crosses the NPC, and in the cytoplasm, GAP deactivates Ran-GTP (hydrolyses the GTP in GDP), causing the export protein to be released from its cargo
Nuclear fusion Experiments:
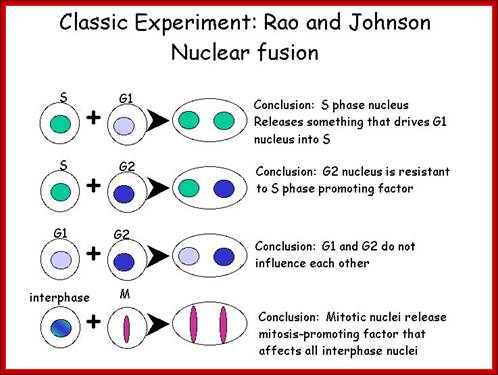
Cell cycle and its regulation-nuclear fusion; Rao and Johnson; http://www-bcf.usc.edu/
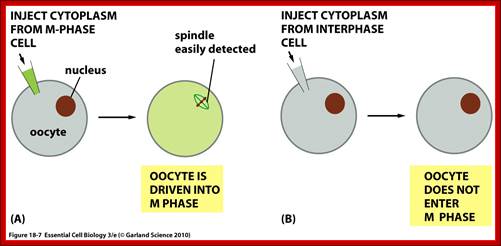
Essential Cell Biology (Garland Science (2010); http://oregonstate.edu/
In classical experiments, as shown above, that certain molecular events during each of the stages generate a set of factors and they are responsible for executing the stage and perhaps provide signals for the next stage.� For example, when a cell in S-stage is fused with G1 stage, the cell in G1 stage is stimulated to proceed into S-phase.� But if a cell in S-stage is fused with a cell at G2 stage, nothing happens, which means the components found in S-phase cells have no effect on G2, because the cells at G2 cells have already achieved what the S-phase components have provided. Fusion between G1 and G2 cells does not result in any changes in each of them.� But if an Interphase cell is fused with a cell at M stage the Interphase cells directly enter into M-phase with disastrous consequences.� This is because Interphase cell is not yet competent to enter into M phase, but M phase cells have all the components for nuclear disassembly and chromosomal separation.� So, there is a regulation at each of the entry points called check points, which is tightly regulated.
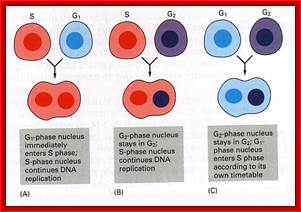
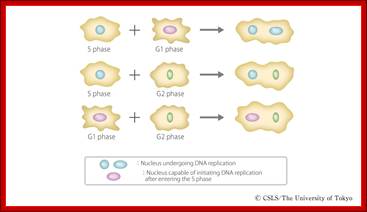
Cell fusion experiments show the existence of different stage specific regulators; http://personalpages.manchester.ac.uk; http://slideplayer.com ;http://csls-text3.c.u-tokyo.ac.jp/
The major checkpoints lie in between G1 and S phase and G2 and M-phase and another control point exists within the M-phase events at anaphase.� Not withstanding the said checkpoints, DNA damage can introduce its own checkpoint, where until the DNA damage is repaired, cell does not enter M-phase, this can happen at S-phase or at G2 phase; if the damage is beyond repair the cell is signaled for Apoptotic death.�
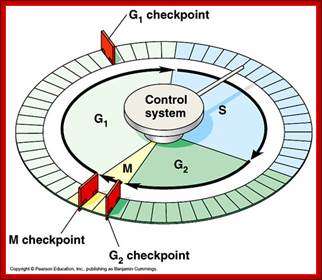
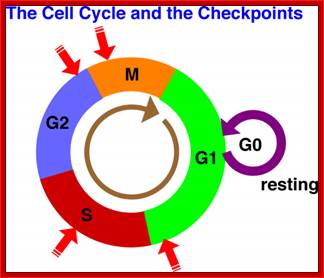
Red arrows indicate check points; G1 to S, with in S, G2 to M and within M phases; www.slideplayer.com;Eishi Noguchi; https://sharonap-cellrepro-p2
The cell cycle proceeds by a defined sequence of events where late events depend upon completion of early events 1. The aim of the dependency of events is to distribute complete and accurate replicas of the genome to daughter cells 2. To monitor this dependency, cells are equipped with the checkpoints that are set at various stages of the cell cycle. When cells have DNA damages that have to be repaired, cells activate DNA damage checkpoint that arrests cell cycle. According to the cell cycle stages, DNA damage checkpoints are classified into at least 3 checkpoints: G1/S (G1) checkpoint, intra-S phase checkpoint, and G2/M checkpoint. Upon perturbation of DNA replication by drugs that interfere with DNA synthesis, DNA lesions, or obstacles on DNA, cells activate DNA replication checkpoint that arrests cell cycle at G2/M transition until DNA replication is complete. There are more checkpoints such as Spindle checkpoint and Morphogenesis checkpoint. The spindle checkpoint arrests cell cycle at M phase until all chromosomes are aligned on spindle. This checkpoint is very important for equal distribution of chromosomes. Morphogenesis checkpoint detects abnormality in cytoskeleton and arrests cell cycle at G2/M transition.
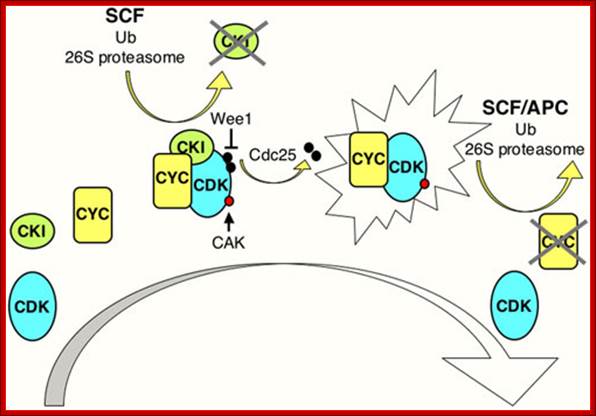
CDK Regulation: www.wormbook.org
Model illustrating general aspects of CDK regulation; CDK activation requires cyclin (CYC) expression and association. Cyclin/CDK complexes are kept inactive through association with CDK-inhibitory proteins (CKIs) and inhibitory phosphorylation by Wee1/Myt1 kinases (black circles). Activation requires ubiquitin-dependent proteolysis of the CKI, phosphorylation of the CDK by a CDK-activating kinase (CAK; red circle), and removal of the inhibitory phosphates by a Cdc25 phosphatase. Cyclin destruction leads to inactivation. Ubiquitin-dependent proteolysis of cell cycle regulators in late G1 and S involves cullin-based E3 ligases such as SCF, while in M phase and early G1 the anaphase-promoting complex (APC) is active. The exclamation figure denotes the active kinase complex, the large arrow indicates time. www.wormbook .org
Different classes of cyclins such as G1 cyclins and M phase cyclins are synthesized in temporal fashion; when their function is over, they are degraded, only to be synthesized in the next cell cycle.� This figure shows major check points (vertical Bars) such as G1-S and G2-M and one at M phase itself.
Checkpoints act at transition points or regulatory points where all the earlier events have to be completed before it progresses to the next stage.� They also act as surveillance systems.� It is a regulatory loop where initiation of one event depends on the completion of the earlier event, so progression through a checkpoint is strictly controlled.
Changes in cellular components:
Among the many cellular components involved in cell cycle, cyclin dependent kinases (Cdks) play a significant role.� Cells, on the whole, employ more than 1000 kinases and also employ equal number of phosphatases. The beauty of the interplay of these two components is that many proteins and other cellular components rendered active when they are phosphorylated at specific sites on them.� Some of them get inactivated when they are phosphorylated, but some, depending upon the individual component, become active when they are dephosphorylated.� Thus, specific kinases phosphorylate proteins or similar substrates at specific sites in temporal fashion; thereby they activate or inactivate cellular components.� Phosphatase in turn removes phosphate groups from specific sites in specific protein at specific time, so the substrate may be rendered active or inactive. ��But the cell cycle kinases are protein kinase and they are exclusively specific; hence they are called cyclin dependent protein kinases.� Similarly, there are a host of inhibitors especially Cyclin-Cdk kinase inhibitors, and they play a pivotal role.
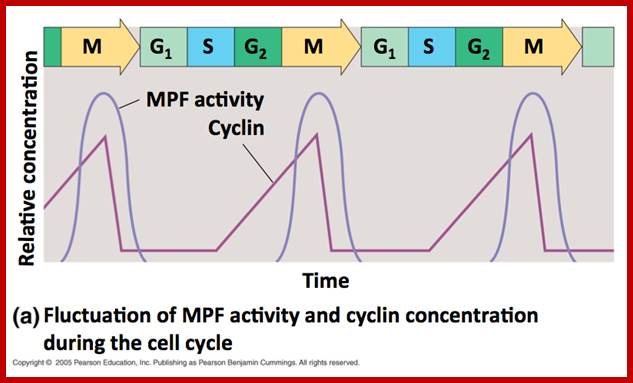
http://www.mun.ca/;https://www.google.co.in; Honors biology cell cyle; http://anitadotta.blogspot.com; http;//www.slideshare.net
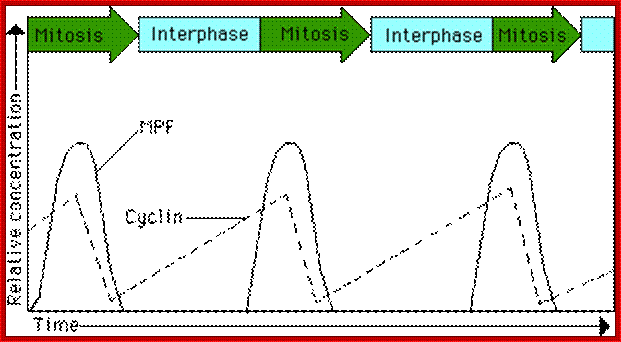
This diagram illustrates the levels of cyclins as mentioned above in graphic mode where mitosis promoting activity and cyclins peak in synchronic fashion at specific stages of the cell division. www.uic.edu/classes/bios/bios100/lecturesf04am
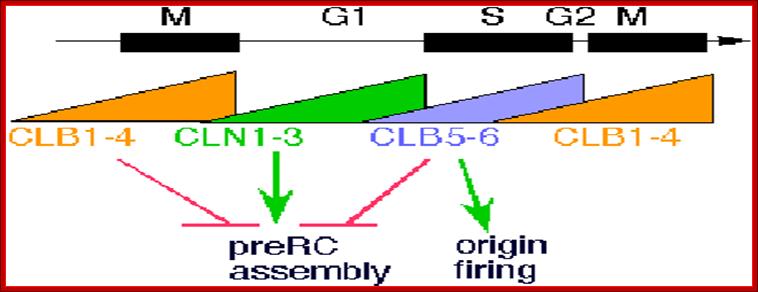
Dr Frostburg�s all-purpose cell Cylce-Notes; http://www-rcf.usc.edu/~forsburg/cclecture.html.
The diagram as shown above not only depicts levels of (rise and fall) of cell cycle dependent components such as CLN and CLBs but also check points at which they act, where they block the progression and some fire the origins into replication bubbles ex. S. cerevisiae.
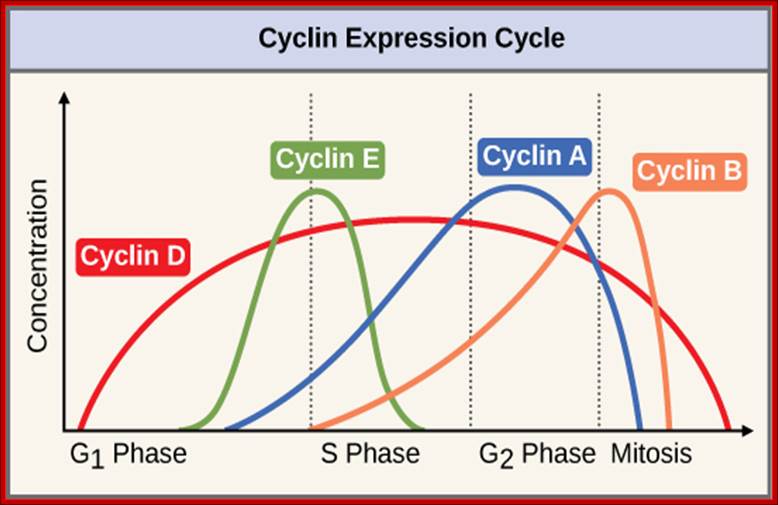
The concentrations of cyclin proteins change throughout the cell cycle. There is a direct correlation between cyclin accumulation and the three major cell cycle checkpoints. Also note the sharp decline of cyclin levels following each checkpoint (the transition between phases of the cell cycle), as cyclin is degraded by cytoplasmic enzymes. (Credit: modification of work by https://www.boundless.com "WikiMiMa"/Wikimedia Commons); OpenStax College.
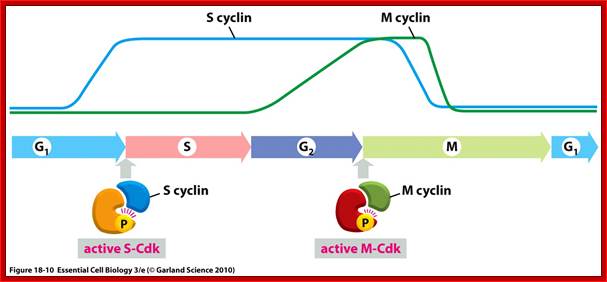
Stages at which cyclins produced as S cyclin and M cyclin, with binding to CDK they become active; Essential Cell Biology; http://oregonstate.edu/
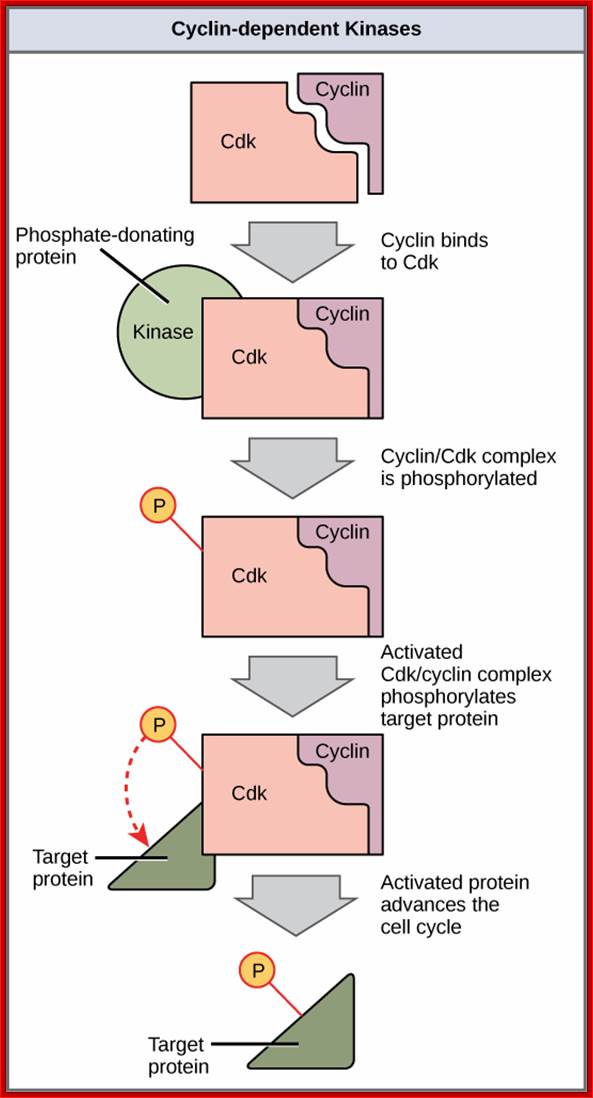
Cyclin-dependent kinases (Cdks) are protein kinases that, when fully activated, can phosphorylate and thus activate other proteins that advance the cell cycle past a checkpoint. To become fully activated, a Cdk must bind to a cyclin protein and then be phosphorylated by another kinase. OpenStax College. https://www.boundless.com
� In all this, the entry of cells into S phase is very crucial for the cell to divide and generate two individual cells with its genetic material equally duplicated for equal distribution without making a single mistake; it is very very important. If a mistake is made it has be corrected, but cells can over look such errors (in DNA) provided that segment of the DNA is not functionally important; ex. change in a nucleotide changes the meaning of a codon and so the amino acid; if the amino acid in the protein structure and function is non-significant, the mutational error can remain.
� Thus, DNA replication at S phase is critical.� For initiating DNA replication exactly at S-phase and completion of it in S phase requires an input of many qualitatively different factors. So DNA entry into replication mode, completion of replication and separation of replicated daughter DNA molecules in all its glory is controlled by the inputs of several factors.� The diagram below shows some of the crucial components required and events that trigger the initiation of replication at specific sites and at specific time is depicted.
� Reinitiation of S-phase depends on M-phase; till it is completed another round of DNA replication won�t be initiated.
The G1 stage, as said earlier, is the stage where cell prepares for DNA replication.� The cyclins required at this stage are cyclins D.� The required components for DNA replication, besides a large nucleotide and histone pools, and DNA polymerase, helicases, SSBs, and few others.
It is during late stage of G1 transcription of genes required for DNA replication is activated.� Transcription of the said genes requires transcription factors and their activation is sine quo non for the entry of the G1 to S-phase.� If there is a damaged DNA at G1 stage entry into S-phase is prevented by the mediation of p53 and its associated components.� If and only if all the required components for replication are provided then cell enters into S-phase.
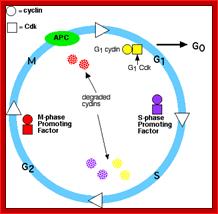
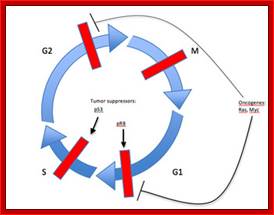
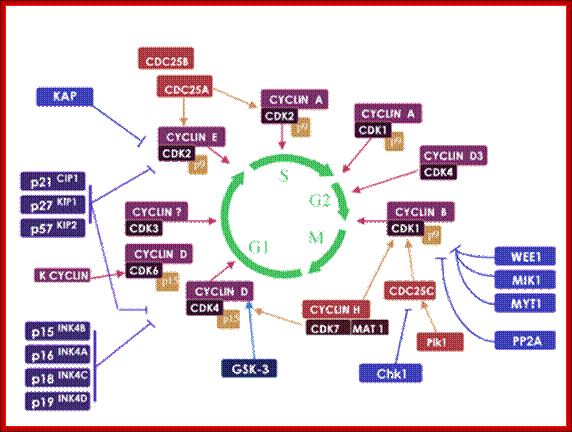
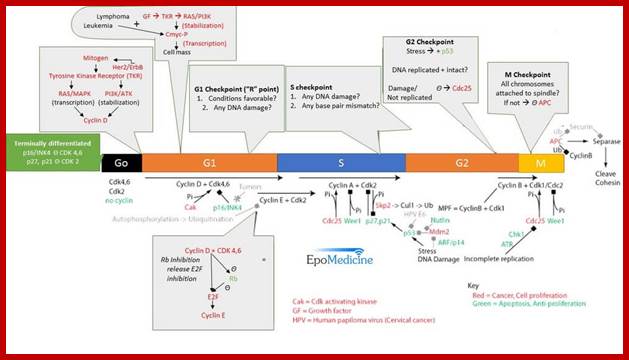
The above diagram shows the major checkpoints and the major cyclins and Cdks which act as SPF and MPF factors involvement at specific stages, which are required for the progression of the phases into the next stage; some cyclins get degraded at specific phases. Stage specific synthesis and degradation is important events in cell cycle. Right diagram check points are depicted as thick lines. �The stages of cell cycle G1 gap1, S, G2-Gap2 and M mitosis are indicated. Tumor suppressor�s act to maintain check points, whereas oncogenes allow checkpoints overcome (Konin 2000); http://www.nature.com/
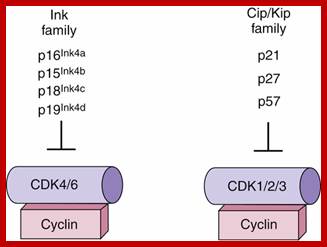
Cyclin and Cdk action at specific stages, CAK for phoshorylation and CDC25 removal phosphate group from Tyrosine 14/15; role of specific Cdks and Cyclins at different stages are shown. INKs stand for Cdk inhibitors, cip/kip are cyclin/Cdk inhibitors.
http://www2.technologyreview.com
https://clinicalgate.com
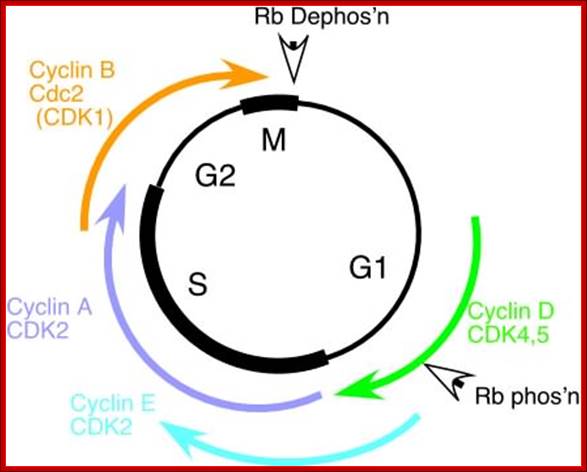
The diagram depict different stages of cell cycle regulated by different sets of cyclin-Cdks and RB proteins; temporal fashion of synthesis of cyclins and Cdks and RBs are found through out but their inactivation and activation to release E2Fs is very important. Dr. Forsburg; http://www-bcf.usc.edu/
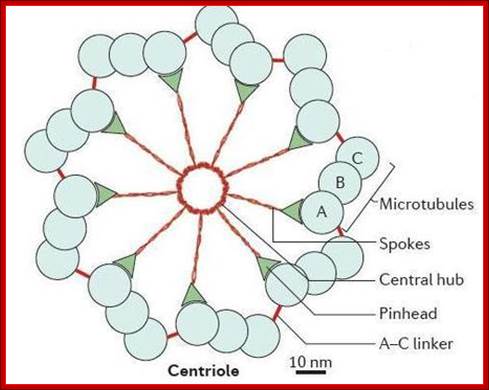
� The S-phase is critical for the single stranded chromosome becomes double stranded by means of DNA replication, yet they are held together all along the length of the chromosome and also at centromeric region which has an elaborate structural organization called kinetochore (Centrosome).� Each of the chromosomes contain one long DNA compacted by nucleosomal organization and the ends with telomeric structures.�
� Initiation of replication at origins (multiple origins) is governed by several factors.� Firing of replication is critical and it takes place only once in one cell cycle and second initiation is prevented before the M-phase is completed.� During replication if there are any errors; they are fixed, and if the damage is beyond repair, the cell is subjected to Apoptosis.�
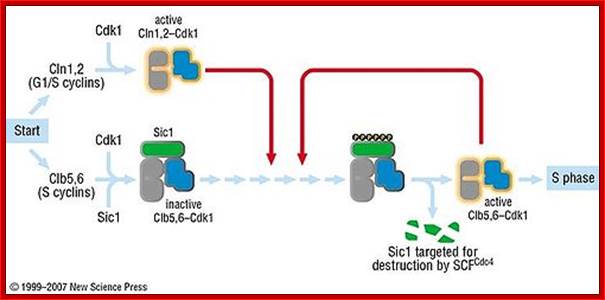
� The progression of S-phase requires cyclin-E, but inhibited by cyclin/Cdk inhibitors; and they have to be degraded by active SCF which acts as ubiquitin ligase system.� The SCF complex consists of Cdc 53, SKP1 and Cdc4; all together targets Sic-1 the inhibitor of S-phase Cdk-cyclins, thus the Cdk-cyclin (G1) complex is released from inhibition.
The G2 stage is again a preparatory stage for M-phase, which requires a whole set of proteins and organization of cellular components for chromosomal separation and cytoplasmic division.� If the DNA damage is not repaired in the S-phase and even in G2 phase the cell won�t enter into M-phase.� Cells have in built-in sensory system.
� The M phase, though short in duration, involves major physical changes; dismemberment of lamin cytoskeleton leads to dismemberment of nuclear membrane which retracts into endoplasmic reticulum and pore-complexes are released. �Disassembly of microtubules and actin filaments associated with the nucleus takes place, to their respective subunits such as Tubulins (alpha and beta) and actins (F and G actins), simultaneously mitotic apparatus assembles, including tractile fibers which bind to kinetochore complex.� Separation of chromatids and centromere movement to their respective poles, once completed cytoplasmic division ensues.� These events can be visualized. �
� Among the M-phase events, the MPF acts as progressive complex and Anaphase Promoting Complex (APC/C) called cyclosome acts as the regulatory complex.
Mutations that exert cell cycle control in yeast are:� Cdc 28 at G1-� S; Cdc 28 at S phase; cdc24 at G-2 and Cdc 34 at M > G1
Genetic analysis of single cell systems such as Saccharomyces cerevisiae (budding yeast) and Saccharomyces pombe (fission yeast) has yielded a wealth of information, including database.� Combined with genetic data, proteome search has provided information about the number of genes and gene products involved in cell cycle and its regulation. The database search to 95% accuracy shows that Homo sapiens have 99 gene products [28-proteins for G1/S, 28 proteins for -G2/M, 23 proteins for -M, 41-Sphase, 24-others]. Mus musculus contain 68 gene products and�� S. cerevisiae 87 to name only few.� Important factors that operate cell cycle control system are cell cycle specific kinase complexes and few cell cycle kinase inhibitors.� Among many of the cyclin- dependent kinases (CDK) are important; for their activity cyclins are required, hence the name cyclin dependent kinases.�
Kinases are effector molecules and cyclins are required for kinase effector function.� Perhaps the first such kinase discovered was from yeast and it was called Maturation Promoting Factor (MPF), later it turned out to be Mitotic Promoting Factor (MPF) or Mitosis Promoting Kinase (MPK).� This protein complex turned out to be a serine-threonine protein kinase but dependent on specific cyclin.� From a variety of sources many such cyclin dependent kinases and cyclins have been discovered their nomenclature has not been strictly adhered to the law of international nomenclature, so there is confusion in identifying, which is which.
Cyclin dependent kinase (Cdk):
The Cdk is protein kinase, but it requires cyclin as a factor for its activity. There are several Cdks, at least 11 or more and not all of them are involved cell cycle events. In general, Cdks have a molecular weight of ~34 kd, and they are monomers and function as kinase subunit i.e. as effectors.� The Cdks consist of N-terminal beta-sheet containing ATP binding site and an alpha helix with PSTAIRE sequence.� The C-terminal has helical domain.� When Cyclin binds, the PSTAIRE region fits into Cyclin structure (like hand in glove).� Cdks are constitutively synthesized and found in reasonably higher concentration. �When cyclin is not bound to Cdk, the C-terminal loop can fold back and mask the ATP binding site and block protein kinase site.� Cdk is a protein kinase and responsible for phosphorylating several target proteins, thus activate several components that leads to the progression of G1 to S-phase then to M-phase.
The N-terminal region of Cdk contains phosphorylating sites at tyrosine (Y) 15 (or threonine (T) 14); this depends upon the organism.� These sites are adjacent to the substrate-binding site.� Phosphorylated Y15/T14 prevents the binding of ATP to its site thus makes the Cdk inactive.� Another site for phoshorylation is Threonine 160 in (CDK2) and Threonine 161 in CDC2 (it is also called CDK in Schizosaccharomyces pombe).
 �
�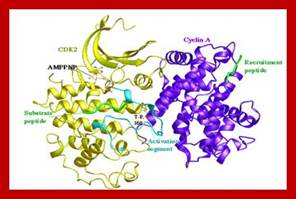

The diagrammatic representation of 3-D ribbon model of Cyclin-A and Cdk2 proteins; blue is Cdk(right) and purple cyclin(left); ATP binding site is located within the Cdk; Cdks PSTRAIRE domain contains cyclin binding site. http://folk.uio.no/ http://oregonstate.edu
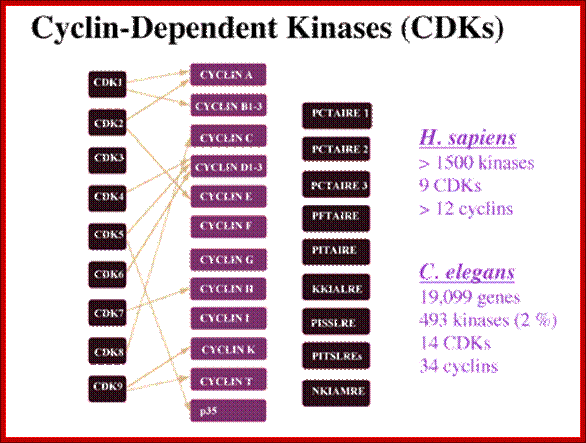
CDKs and their cyclin partners; http://www.biocristalografia.df.ibilce.unesp.br/
Cyclins:
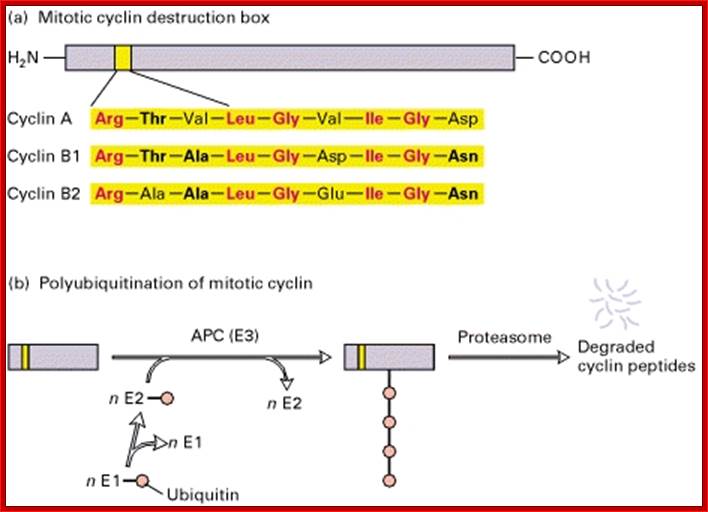
Cyclins are protein kinase activators. There are several cyclins at least 30 of them, which are synthesized and degraded (by proteasomes) in stage specific manner.� The molecular weight ranges form 35 to 90 KD.� Not all cyclins are involved in Cdk activation.� Cyclins are made up of helices (HTH and HLH), which provides a site into which CDK snugs in. �The 100aa long five-helix domain called cyclin box (shared by all cyclins).� The N region contains sequence of 9 a.a called destruction boxes, which is recognized by ubiquitination enzyme complex.�
Cyclin destruction box:
Cyclin-A:�� RTVLGVIGD,
Cyclin-B:��� RTVLGVIGN,
Cyclin-B2:� RAVLGVIGN.
http://2013.igem.org/Team:Shenzhen
Ubiquitin-APC/C mediated proteolysis of mitotic Cyclins is dependent on its 90 residues for ubiquitin mediated proteolysis.� The sequence used is called destruction box. When D-box exists in protein N-terminal, this protein is recognized more easily by E1 and then it is captured for degradation pathway. ttp://2013.igem.org/Team:Shenzhen
The said sequences of cyclins are targets for ubiquitination by E1, E2, and E3 by Anaphase Promoting Complex (APC/C) at the end of anaphase. However, cyclins-C, F, G and H have structural relationship but not involved in cell cycle regulation.� Example cyclin-H/Cdk 7 dimers are associated with eukaryotic TFII-H
Regulation of CDK activity:
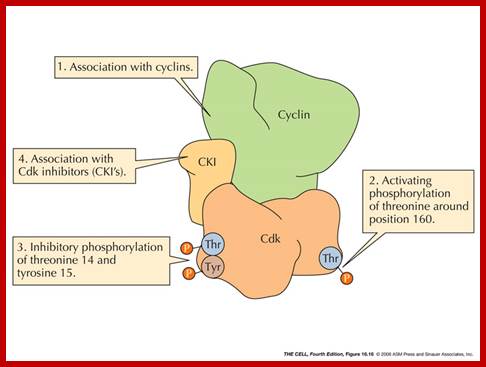
Cdk�s activity is regulated.� Depending upon the species, the Cdk has at its N-end has a Threonine 14 or Tyrosine 15.� Adjacent to kinase site there is another phosphorylating site i.e., Thr 160 or Thr 161. If these hydroxyl sites of Y/T amino acids are phosphorylated by Wee 1(a kinase); the enzyme remains inactive. �It will be active only when cyclin binds (binding of cyclin opens up Thr160 site) and dephosphorylation of Thr 14 or Tyr 15, but CDKs� �to be activated phosphorylation of Thr160 or Thr 161 is required.� Dephosphorylation of the same is performed by activated cdc25 (a phosphotase enzyme, becomes active when it is phosphorylated otherwise inactive).� Phoshorylation of Thr 160 (161) is a must.� This phoshorylation is believed to be by the enzyme called Cdk-activating kinase (CAK).� Phoshorylation of Thr160 (161) can also be achieved by autophosphorylation once the Cdk is active.� The cyclin/Cdk dimer protein is a serine and tyrosine protein kinase.
Note- the Wee 1 is active when it is unphosphorylated and become inactive if it is phosphorylated by nim1 enzyme.� Similarly Cdc25, a phosphotase becomes active if it is phosphorylated and remains inactive if it is not phosphorylated.
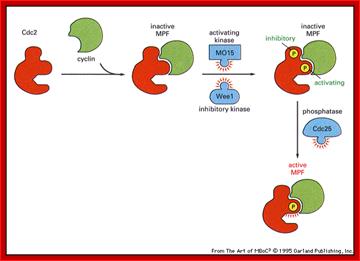
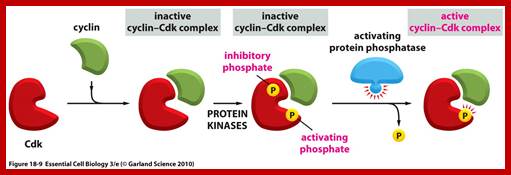
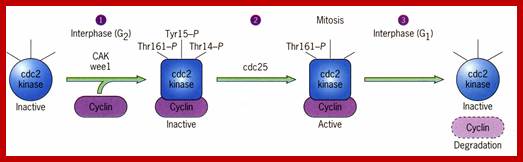
Activation by the binding of cyclin to Cdk; Phoshorylation of Cdk at Ty14/ Ty15 (Wee1) and Thr160/161 (CAK) renders inactive; but dephosphorylation of Ty14/Ty16 by cdc25 make Cyclin-Cdk active. http://greatcourse.cnu.edu.cn and www.oregonstate.edu
A list of Cdks and Cyclins:
G1/S Regulatory Components:
|
|
CDKs |
Cyclins |
|
Mammalian & Frog |
Cdk2,� 4 |
Cyclins D1, D2, D3,� E |
|
S. cerevisiae (budding) |
Cdc� 28 |
Clb B-1-4 (B-like) |
|
S. pombe (fission) |
Cdc 2 |
Cdc13 (B-like) |
G2 / M Regulatory Components:
|
|
Cdks |
Cyclins |
|
Mammals/frogs |
Cdk2, (cdc2,) |
Cyclin A, B1, B2 |
|
S. cerevisiae |
Cdk 28 (cdc28) |
Cyclin 1-4 (B-like) |
|
S. pombe |
Cdk2 (cdc2) |
Cyclin13 (cdc13) (A-like) |
Note-In S pombe- there is only one CDK (cdc2), and one mitotic cyclin (cdc13), in S. cerevisiae- there is only one CDK (cdc28), but there are mid G1 cyclin, late G1 cyclin, early S phase cyclins, late S phase cyclins, early Mitotic cyclins, and late mitotic cyclins.
In vertebrates there are mid G1 Cdks, (CDK4 and 6), late G1 and S phase Cdks (CDK2), Mitotic Cdks (CDK1, CDK2),� mid G1 cyclins ( cyclin D type), Late G1 cyclins and S-phase cyclins (Cyclin E), S-phase cyclins and mitotic cyclins� (cyclin A), mitotic cyclins (cyclin A and B).
*** Paul Nurse (UK), Thomas hunt (UK) and Leland Hartman (USA) were awarded Nobel prize for their work on Cdc cyclins.
Note that the cdc2 (Cdk2) of mammalian system is equivalent to cdc28 (Cdk 28) of S. cerevisiae, which is equivalent to Cdc 2 (Cdk 2) of S. pombe.� Here we use Cdk because each of them acts as Cyclin Dependent Kinase.�
The Cdk has a molecular weight of 34 KD. Cdc 13 is a homolog of cyclin-B.
Combination of Cyclin-Cdks; Their Functions at Different Stages:
S. cerevisiae:
|
|
cyclin |
Cdk |
|
|
|
G1>>S phase |
Cln 1,2,3 |
Cdc 28 |
|
|
|
S-phase |
Clb 5, 6 |
Cdc 28 |
|
|
|
Replication origin firing |
Dbf4 Clb 5 |
Cdc 7 Ccdc28 |
Firing replication origin |
|
|
M-phase entry |
Clb 3,4 |
Cdc28 |
|
|
|
M-phase progression |
Clb1,2 |
Cdc28 |
|
|
|
M-phase exit |
Clb destruction |
|
|
|
|
|
|
|
|
|
Human and Vertebrates:
|
Cyclins |
Cdk (protein kinase) |
Cyclin level |
Note |
|
Cyc-D1, D3 |
Cdk-4, 6 |
Increase |
START- G1 phase progression |
|
Cyc-E |
Cdk-2 |
E-Increase, D-decrease |
Onset of S phase, G1 >S |
|
Cyc-A |
Cdk-2 |
E-decrease, A-increase, |
S-phase progression |
|
Cyc-A |
Cdc-2 (cdk-1) |
A-decrease, |
S through G2 |
|
Cyc-B |
Cdc2 (cdk-1) |
B-increase |
M-phase progression |
|
Cyc 13(Mr 45-47KD) |
Cdc2 (Mr 34KD) |
|
Prevents S-phase before M-phase |
|
Cig 2 |
Cdc2 |
|
Prevent the start of M-phase before S-phase completion |
A list of CDKs with their regulator proteins-Cyclins or others in mammalian systems:
- CDK1; Cyclin A, Cyclin B
- CDK2; Cyclin A, Cyclin E
- CDK3: Cyclin A, Cyclin E,Cyclin C
- CDK4; Cyclin D1, Cyclin D2, Cyclin D3
- CDK5; CDK5R1, CDK5R2. See also CDKL5.
- CDK6; Cyclin D1, Cyclin D2, Cyclin D3
- CDK7; Cyclin H
- CDK8; Cyclin C
- CDK9; Cyclin T1, Cyclin T2a, Cyclin T2b, Cyclin K
- CDK10: Ets2-a TF binds,� G2-M phase;
- CDK11 (CDC2L2) ; Cyclin L
- CDK12 (CRKRS) ; Cyclin L
- CDK13 (CDC2L5) ; Cyclin L
Structural studies of cyclin dependent kinases; http://www.biocristalografia.df.ibilce.unesp.br/
Cyclin-dependent kinases (CDKs) play an essential role in the regulation of the cell division cycle. (Meijer, et al., 1995a; Morgan, 1995; Pines, 1995; Meijer et al, 1996b). A typical CDK consists of a catalytic subunit [CDK1 (=cdc2)-CDK9] and a regulatory subunit (cyclin A-cyclin H).
Each CDK/cyclin complex is believed to act at a specific stage of the cell cycle. Figure 1 shows the main CDKs involved in cell cycle control. CDKs are regulated by (1) transient transcription/translation of their subunits, (2) complex formation, (3) several posttranslational modifications (phosphorylation/dephosphorylation), (4) interaction with various protein inhibitors (p16INK4A, p21CIP1, p27KIP1, etc.) and interacting proteins (p9CKS), and (5) modifications of their cellular localization. The crystal structures of CDK2 in complex with ATP and various inhibitors, (De Bondt et al, 1993; Schulze-Gahmen, et al, 1995; Azevedo et al, 1996; Azevedo et al, 1997) CDK2/p9CKS, CDK2/cyclin A, and CDK2/cyclin A/kip1 have been determined, allowing a very precise understanding of the molecular mechanisms underlying CDK activation and inhibition http://www.biocristalografia.df.ibilce.unesp.br/
Kinases and Phosphatases:
|
CAK kinase |
Activates cyclin-Cdks by p-lating T160/161 |
|
Wee1 kinase |
�Inhibits cyclin-Cdks by p-latingh Y15/T14 |
|
Cdc25 phosphatase |
Activates cyclin-Cdks by dephophorylationY15/T14 |
|
Cdc14 phophotase |
Activates Cdh1 by dephosophorylating it, to inhibit cyclin-Cdks |
|
CDC 25A phosphatase |
Activates vertebrate M cyclin-Cdk |
|
CDC25C phosphatase |
Activates vertebrate Cyclin-Cdks |
|
ATM/ATR kinases |
Activates chK1/chk2 kinase check point control |
|
Chk1/chk2 kinases |
Inactivates cdc25C andcdc25A phosphotase |
|
|
|
Inhibitor Proteins:
|
Sic1 |
Binds and inhibits S phase cyclin-CDKs |
|
CDKIs-P27kip1, P57kp2, p21cip |
Inhibit cyclin-Cdks |
|
INK4 (p16) |
Inhibit mid G1 Cdks |
|
Mad2 |
Spindle assembly Chkp binds Cdc20 prevents onset of Anaphase and inactivation of B type cyclin-CDK |
|
RB (Retinoblastoma) |
BindsE2F-prevents cell cycle required transcription |
Ubiquitin-Protein ligases:
|
SCF |
Degradation of Sic1 or p27KIP1 to activates S phase cyclin-Cdks |
|
APC/C+cdc20 (specificity factor) |
Degrades Securin, initiates Anaphase, and partial degradation of B-cyclins |
|
APC/C+cdh1 (specificity factor) |
Induces Cyclin B degradation, allows pre replication complexes |
Check point Proteins:
|
Checkpoint |
Purpose |
Sensor |
Action |
|
Intra S-phase chkpt |
Ensure complete DNA replication before entry into M phase |
ATR detect Replication forks |
Inhibition CDC25C prevent activation of Mcycli-CDKs, prevent entry into M phase |
|
Spindle assembly chkp |
Ensures the binding of MTs to all Kinetochores |
Mad2 detects KC unattached to MTs |
Inhibition cdc20 prevet activation od seoparase and onset of anaphase |
|
Spindle position chkp |
Ensures chromosomes segregated before telophase |
Tem1 detects positioning of spindle pole body |
Prevents CDK14 activation and degradation M cyclins block late M events |
|
DNA damage check up |
Detects DNA Damage-all over |
ATM/ATR detectors |
Inhibit cdc25A,p21cipinhibits all cyclin-CDKs-cellcycle arrest |
Note:
S = S phase,
Chkp = Check point,
MT = Microtubules, Tem1 =
M = Mitotic phase, i.e., from prophase to Anaphase. Mad2 = Mitotic arrest defective 2,
A plethora of proteins involved in cell cycle in different systems:
|
CDK |
cyclin dependent kinase |
CDC28 |
Cdc2 |
Multiple CDKs: CDK1-6 |
|
G1 cyclin |
regulatory subunit of CDK for cell cycle entry |
CLN1,2 and 3 |
? |
Cdk4-cyclinD |
|
S phase cyclin |
regulatory subunit of CDK for S phase entry |
CLB5, 6 |
Cig2 |
Cdk2-cyclinE |
|
late S phase cyclin |
regulatory subunit of CDK for S phase progression |
CLB3, 4 |
? |
Cdk2-cyclinA |
|
M phase cyclin |
regulatory subunit of CDK for mitosis |
CLB1, 2 |
Cdc13 |
Cdc2<CDK1)-CYCLINB< TD> |
|
APC |
Multi-component ubiquitin ligase required for degradation of substrates in mitosis and G1 |
Many genes |
Many genes |
Many genes |
|
APC specificity factors |
target the APC towards different substrates |
CDC20 |
Slp1 |
Cdc20, fizzy |
|
Securin |
An APC target, inhibits sister chromatid separation |
PDS1 |
Cut2 |
securin |
|
|
|
|
|
|
|
checkpoint sensor |
Complex of proteins consisting of a clamp loader and a clamp that binds DNA and monitors damage |
RAD24 |
Rad17 |
Rad17 |
|
preRC |
Pre-replication complex, which marks a replication origin as ready to fire |
ORC1-6 |
Orp1-6 |
ORC1-6 |
�What the heck are all these
gene names anyway?
As any other fields of molecular biology, the cell cycle is complicated because
of the plethora of different gene names in different systems. One option in
lecture is to use just one generic name--but then you can't read any papers,
because everyone in the literature uses different gene names. Here is a table
that should help negotiate the different species and different nomenclatures in
this lecture.
It is important to note that the effector i.e. the protein kinase subunit, often called Cdc-28 in S. cerevisiae, or Cdc2 in S. pombe, and Cdk in other systems, are more or less same at all stages of cell cycle, but the cyclin, the partner varies from stage to stage, such as G1-cyclins (early, mid and late), S-cyclins, G2-cyclins, M-cyclins and so on.� When such cyclins combine with the specific phase dependent kinase subunits, Cdks become active, provided that the kinase subunit is phosphorylated at Thr.160 (or Thr 161 in other systems). The cyclin-Cdks act on different targets at different stages of the cell cycle.� Most of the cyclins are synthesized in temporal fashion, starting at the beginning of G1 and build up to M-phase and then they are degraded (in ubiquitination-proteasome mode), again in temporal fashion; and the timing of degradation is critical; so, cyclins act as regulators of the protein kinases, where kinase subunit is more or less same (not all) but cyclins are different.� Thus, the regulation of cell cycle is regulated by the synthesis and timely destruction of the said cyclins.
Accessory factors:
Though cyclins and Cdks are considered as the prime factors in controlling cell cycle events, there are other factors, which are as important as Cyclin-Cdks.� There are several kinases (some are cyclin dependent and some are cyclin independent) and several phosphotases.� They are- the following.
Nim 1(Never In Mitosis):� It is a signal mediated protein kinase Inhibits wee1 by phoshorylation.� Nim-1 pathway links to Cdc2/cyclin system to external signals.
Wee 1:� Wee1 is a kinase; inactivated by nim-1 by phoshorylation, Dephosphorylation makes it active when active it phosphorylates Cdk�s threonine 14 (or Tyrosine 15) and makes it inactive.� Wee-1 activity is determined by signal input and signal transduction across the cell membrane.
CDK kinase (CAK): Phosphorylates Cdk�s active site threonine 160 (or Threonine 161).
Cdc25: �It is a phosphatase, (counter part of this in the fly is �string� gene). Its Mr. is 80KD.� It is active when phosphorylated and inactive when dephosphorylated.� When Thr 14 (or Tyr 15) is phosphorylated the Cdk is inactive, but Cdc 25 dephosphorylates these sites; when this Dephosphorylation is coupled with the phoshorylation of Thr160 (Thr161) by CAK, Cdk-cyclin becomes active.� The level of cdc25 reaches a threshold at M-phase, perhaps marks the end of S-phase.
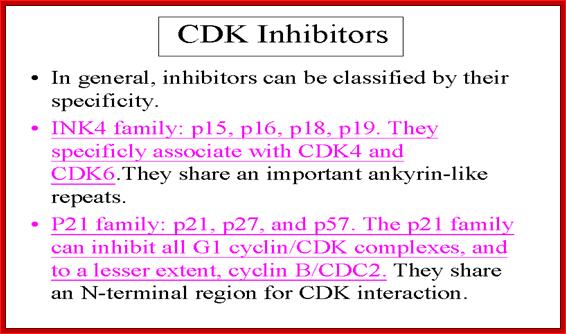
Cell cycle Cyclin/Cdk inhibitors:
Though Cyclin-Cdks play critical regulatory roles in cell cycle, there is another set of molecules that regulate the regulators; in yeasts they are Cdk inhibitors or generally they are cell cycle Cyclin/Cdk (kinase) inhibitors (CKIs).� There are different types of CKIs, such as far1p, Sic1p.� The inhibitors bind to Cyclin-Cdk complexes and prevent their activity.�
The CKIs are p21 (cip), p27(kip),p57(kip2).� Cki s inhibit late G1 Cyclin/Cdks, S-phase Cyclin/Cdks
In metazoans, there are inhibitors that bind to Cdk (Kinase subunits) called INK (Inhibitors of Kinases) family of inhibitors.� INKs interact with mid G1 cdks-Cdk4 and Cdk6 control G1 phase.
MP kinase inhibitors (in the form of dimers) bind to kinases to form inactive complexes.� Thus, they prevent phosphorylation of
Retinoblastoma proteins (RB).� So, the cell cycle is checked at G1 or Go stage.�
Mdlai /Handout/mitosis/sld016.htm
INKs (Inhibitor of Kinase):�
They are Cdk inhibitor proteins.� INK 4 family is specific to Cdk4 and Cdk 6.� Ink4 has four members- p15 (INK-4B), p16 (INK 4A), p18 (INK4C), and p19 (INK4D).� They contain ankyrin repeat sequences.� P16 and P19 bind next to ATP binding site, so prevent its catalytic activity.� It also induces conformational changes so cyclin cannot bind.� They act on cyclin D complexed either to Cdk4 or Cdk6.
Another class of inhibitors such as Sic I binds to Cdc28-clb2, in S. cerevisiae, inactivates the kinase at G1.� So, entry of cell cycle into S-phase requires the degradation of Sic-I by ubiquitination mode.� SCF acts as E3 ligase system.� The Skp1-Cullin factor (SCF complex) consists of Cdc53, Cdc4, Skp1 and Cdc34.� These are involved in G1 cyclin destruction.
Kips: Another class consists of P21, p27 and p57 and they are identified by their molecular weights.� They in general act on G1/S class Cdks.� P21 binds to all Cyclin/Cdks- Cdk2, 4 and 6, thus block progress through all stages of G1/S.� Increase in p21 concentration is inhibitory.� Many a times in cultured cells, one finds PCNA is also complexed with CDK-cyclin along with p21; so it controls G1/S stage progression.� P27 also binds to Cdk-cyclin and blocks progression into S-phase, but it�s over expression leads the cell to go into Go stage.
CyclinH/Cdk-7: it is associated with TF-II H and involved in phoshorylation of CTD tail of RNA polymerase II; TF-II B also contains cyclin like helix bundles.
Cdc7-cyc-Dbf4 kinase: It is serine/Thr protein kinase required for the onset of S-phase.� The cyclin Dbf4 is constitutively synthesized but rapidly degraded from late M to G1.� Activity peaks at the onset of DNA replication.� Human homolog is Hsk (Homolog of Cdk Seven Kinase, however this CDK lacks PSTAIRE sequence.�� The target of this complex is Mcm2.� Loading of Mcm2 on to ORE region is important in triggering the firing replication origin.
Cdk Activating Kinase (CAK):� Cdk 7-cyclin H has CAK activity.� Cyclin-A binding to cdc2 (homolog iscdc28) exposes active site and ATP binding site in Cdk protein, where Thr 160 (Thr 161) is made available for CAK to act upon.� CAK phosphorylates Thr 160 (161) of Cdk to make Cdk-cyclin A to be active.
Positive regulation of Cdk by cyclins is often counterbalanced by negative regulation by Inks, Cips and Kips.
Rum 1 protein:� Cdc2/cdc13 MPkinase is influenced by Rum-1 factor.� When rum-1 is over expressed cell does not enter M-phase, but s-phase goes through multiple cycles.� When rum1 is deleted, the cell enters M-phase prematurely.� This is expressed between G1 and G2 and keeps the MPK inactive.� So this is essential for the S-phase to proceed.
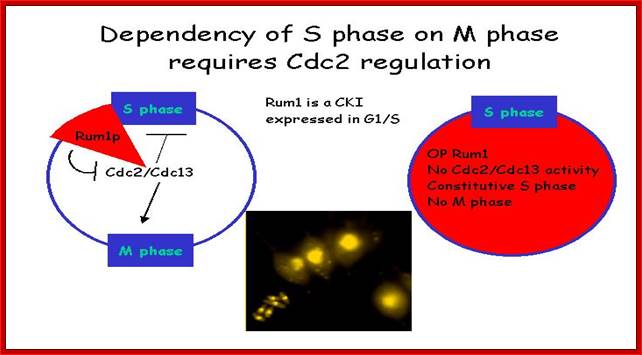
Dependence: S-phase on M-phase, which requires Cdc2 regulators.� Till S-phase is over M-phase will not start, and till M-phase is over another round of S-phase won�t begin. http://www-bcf.usc.edu/
Nucleophosmin:� It is a protein, in unphosphorylated form, binds to centrosomes at the end of M-phase prevents duplication of centrosome.� But Cyclin E /Cdk2 phosphorylate Nucleophosmin, at M-phase.� The phosphorylated Nucleophosmin then dissociates from centrosome.� This is further augmented by Calcium mediated calmodulin dependent kinase II activity at G1-S boundary and facilitates the duplication of centrosome.� Centrosome duplication is essential for the organization mitotic apparatus.
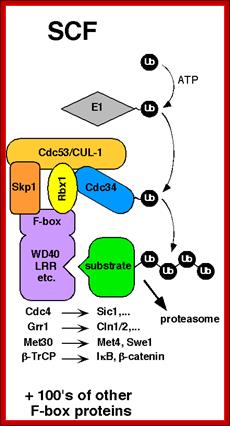
SCF complex:
It consists of at least 5-6 proteins acts as E3 ligase and ubiquitinates Sic1 protein (inhibitor of S-phase) and the same is subjected proteasome mediated decay.� The SCF complex consists of Cdc4, SKP Cullin1, Ring O and E2-Ub
![]()
Modular architecture of SCF and other cullin-RING ligases; Phosphorylated (P) Sic1 is recruited to SCF via the Cdc4 substrate receptor. Cdc4 is incorporated into SCF via its F-box domain (F), which binds Skp1. Skp1, in turn, binds the N-terminal domain of Cul1. The C-terminal domain of Cul1 binds the RING subunit, which recruits the E2 enzyme. Raymond J. Deshaies, Caltwech Division of Biological Engineering; http://www.deshaieslab.com/
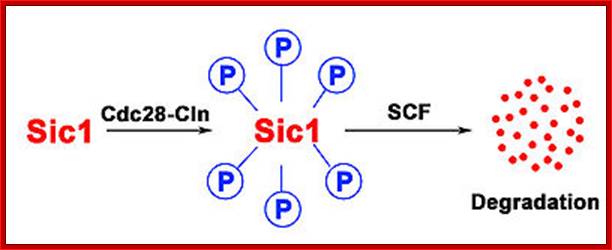
The first step of the degradation of Sic1 is its phosphorylation by Cdc28-Cln followed by the degradation through SCF; SCF complex; http://en.wikipedia.org/
The first step of the degradation of Sic1 is its phoshorylation by Cdc28-Cln followed by the degradation through SCF.
Sic1:� �Sic1 protein, is a stoichiometric inhibitor� of Cdk1-Clb (B-type cyclins) complexes in the budding yeast Saccharomyces cerevisiae. Because B-type cyclin-Cdk1 complexes are the drivers of S-phase initiation, Sic1 prevents premature S-phase entry. Multisite phoshorylation of Sic1 is thought to time Sic1 ubiquitination and destruction, and by extension, the timing of S-phase entry. When Sic1 is degraded, the Cdc28-Clb complex is no longer inhibited and the cell can enter the S/M-phase. Thus, Sic1 inactivation is essential for transition into S phase.
Sic1 has to be phosphorylated at least at 6 of the 9 Cdk sites. Sic1 can also be phosphorylated by other kinases, such as Pho85-Pc11, a kinase which becomes essential when Cln1 and Cln2 are absent. �Sic1 has also a role in the response to osmostress. The stress-activated protein kinase (SAPK) Hog1 phosphorylates Sic1 at a single residue at the carboxyl terminus. This leads to down regulation of cyclin expression and Sic1 stabilization which arrests the cell cycle�.
The diagram shows the role of Sic1 in Clb5,6-Cdk1 inhibition and its phoshorylation-mediated polyubiquitination and destruction. Destruction allows for Clb5,6-Cdk1 activity and S-phase entry; http://en.wikipedia.org/
Activated SCF acts on S-phase Sic1 protein complex and by ubiquitination it is degraded. https://www.ncbi.nlm.nih.gov/
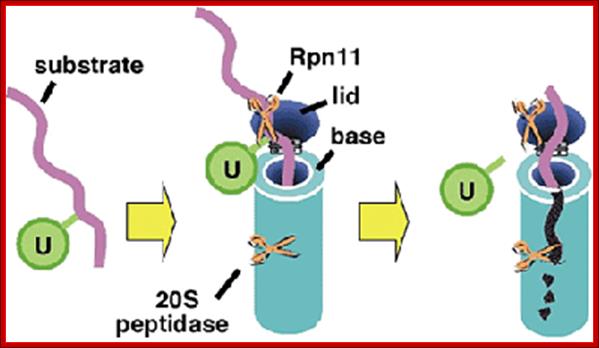
Whether it is SCF or APCs, ubiquitylated they are subjected to proteasome degradation; a simple model. http://www.deshaieslab.com/
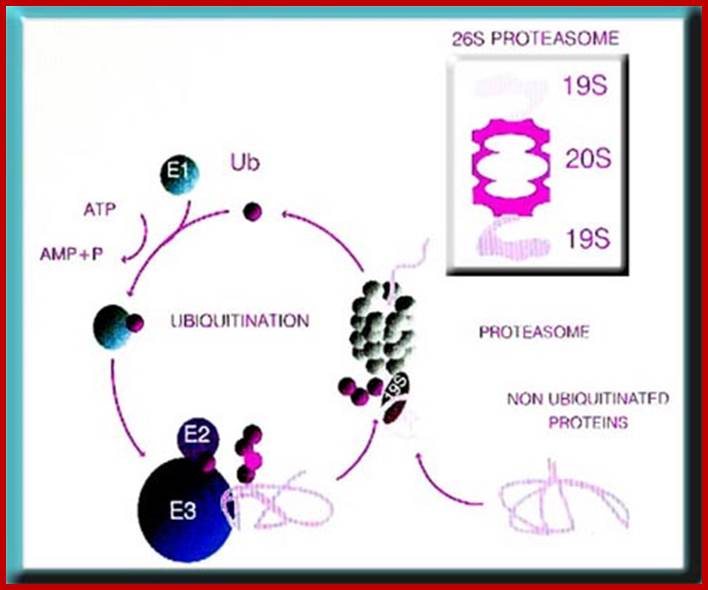
The ubiquitin-proteasome system. Most short-lived and abnormal proteins in the cell cytosol are degraded by the 26S proteasome, directly or after a covalent modification known as ubiquitination. The multiple enzymatic steps and several enzymes required for the ubiquitination of proteins are summarized in this figure. See the text for details. https://www.bioscience.org/
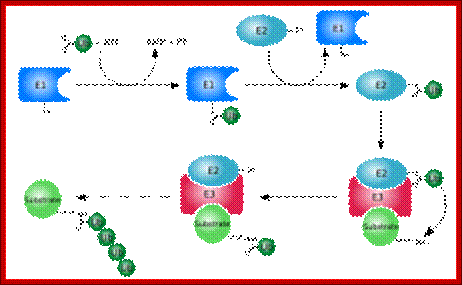
Ubiquitination of the ubiquitin to target substrates involves three enzymes, E1, E2 and E3.The ubiquitination system (showing a RING E3 ligase; http://en.wikipedia.org/
APC/C complexes:�
It is called Anaphase Promoting Complex/Cyclosome.� This is a multisubunit complex made up of eight proteins; the complex is also called Cyclosome.� Such complexes are found in yeast and animal tissues. �Activated APC/C complexes activate ubiquitinated proteasome mediated protein degradation. APC/C becomes active during M-phase.� It functions as E3-ligase in ubiquitinating target proteins for proteasome mediated protein degradation.� First the Cdc20 binds and then Cdh-1 displaces Cdc20 and activates its ubiquitination activity.� CDC/C cyclin is blocked till all kinetochores are attached by microtubulins.� Cdch1 is blocked till anaphase chromosomes reach poles.
APC/C-Cdc20: Cdc20 is inhibited by G1 cyclin/Cdk phoshorylation, till all kinetochores are attached with microtubules. Activated APC/C-cdc20 is essential for the degradation of Securin, an inhibitor of Separase that degrades cohesins that holds sister chromatids together.� Degradation of cohesins paves the way for separation of chromatids i.e. from Metaphase to Anaphase. Perhaps chromosomal arms are liberated from cohesin complexes first then centromeric region is freed from cohesins. APC?C-cdc2 20 though directs to cyclin B enough cyclin is made available for the condensation of chromosomes until late anaphase.
APC/C-cdh1: Phoshorylation of cdh1 by late G1 cyclin-Cdks inhibit its association with APC/C complex. Cdh1 is inhibited till chromosomes move to poles. It is at this point Cdc14 a phosphotase activates cdh1 by dephosphorylation; this allows cdh1 to bind to APC and activates APC/C-cdh1 complex. APC /Cdh1 is responsible for the degradation of mitotic cyclins such as Cyclin B at late M-phase; this renders MPF inactivation.
Two points of destruction: metaphase to anaphase transition, when chromosomes separate and mitotic exit, when cyclin degraded. These were distinguished because expression of a non-degradable cyclin did not prevent chromosome segregation.
![]()
Anaphase promoting complex also called cyclosome is activated by cdc20 to remove cohesins and after chromosomes move towards their poles it is activated cdc20 is displaced and cdh1 binds to make APC/C complex to be active and degrade cyclin A and cyclin B; which act prevents second round of replication initiation before the completion of once cycle of cell division.(?).
APC/C is active with cdh1 acts on cyclin A and cyclin B at late M stage; Jan-Michael Peters http://www.nature.com/
Model of how APC/C might recruit and ubiquitylate substrates:
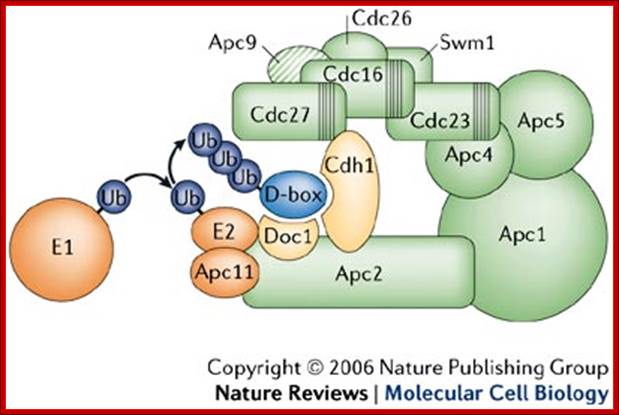
The anaphase promoting complex/cyclosome; a machine designed to destroy� specific proteins; Ubiquitin (Ub) is first activated and covalently bound through a thioester bond by the ubiquitin-activating (E1) enzyme and then transferred to the ubiquitin-conjugating (E2) enzyme with which the ubiquitin residue again forms a thioester bond. The ubiquitin-charged E2 enzyme interacts with anaphase promoting complex protein-11 (Apc11). This anaphase promoting complex/cyclosome (APC/C) subunit has ubiquitin ligase (E3) activity and promotes the transfer of the ubiquitin residue from the E2 enzyme to the substrate protein on which the C terminus of ubiquitin forms a covalent isopeptide bond with a lysine residue. In subsequent reactions, the attached ubiquitin can itself become ubiquitylated, resulting in the formation of a polyubiquitin chain. All proteins that are known to be involved in the catalysis of ubiquitylation reactions are shown in orange. Substrates are recruited to the APC/C if they contain a D-box or a KEN-box. Both of these sequences are recognized by an APC/C co-activator, such as Cdh1 or Cdc20. Cdh1 binds to APC/C by interacting with two subunits, Cdc27 and Apc2. Cdc27 is one of several TPR proteins that are present in the APC/C (TPR domains are shown as vertical stripes), and Apc2 is a scaffold subunit that binds to Apc11 via a Cullin domain. The small globular protein Doc1 is required for processive ubiquitylation of substrates and might also interact with the D-box of substrates, although direct evidence for such an interaction is lacking. The APC/C subunits that are implicated in substrate recognition are shown in yellow. The topology of subunits is based on biochemical data in Refs 13,29,30. Note that this model illustrates subunit interactions but does not represent a structural map of where subunits are located in the three dimensional models that are shown in Fig. Apc9 is hatched because so far it has only been detected in budding yeast APC/C. Human APC/C also contains another TPR subunit, APC7, and human DOC1 interacts not only with APC2 but also with another, unidentified subunit13 (not shown here). Swm, spore wall maturation; http://www.nature.com/
Degradation of poly-ubiquitinated proteins by the 26S-proteasome complex represents a crucial quantitative control mechanism.; APC/C activity and its targets change with time and their specific factors such as cdc1 cdch ; http://cardiovascres.oxfordjournals.org/
https://en.wikipedia.org/
�Two main classes of protein-ubiquitin ligase that target cell-cycle regulatory proteins are shown. Both the anaphase-promoting complex (APC/C) and the SCF (Skp1/Cullin/F-box protein) complex have a core catalytic domain consisting of a Cullin-like protein (APC/C2 in the APC/C, and Cul1/Cdc53 in SCF) and a ring-finger protein (APC/C11 in the APC/C and Roc1/Rbx1 in SCF). These two subunits interact with a ubiquitin-conjugating enzyme (Ubc), which provides activated ubiquitin for the reaction. a | For APC/C, there are two alternative substrate-binding-specificity factors, Cdc20 and Cdh1. The other subunits, shown in red, create a large hollow particle that functions as a scaffold for the catalytic core and is likely to regulate access of substrates or cofactors. b | For SCF ligases, substrate binding and specificity is provided by one of a number of F-box proteins. For SCF, the Skp1 subunit, which is shown in red, connects the F box protein to the catalytic core.
� Once activated, MPKs or MPF s initiate M-phase, the progress of it takes its own course and it does not require active other MP kinases any more, so to exit from M-phase, MPkinase has to be inactivated. �One way to inactivate is to block the catalytic site by inhibitors, or dissociate Cyclin from the kinase, or phosphorylate Thr14 (or Tyr15) or destroy cyclin the CDK partner. Actually, as the M-phase sets in, the first cyclin to be destroyed by ubiquitin mode is cyclin-A at Metaphase.� Then little later i.e., at Anaphase, Cyclin-B is degraded by ubiquitination mode, making MPkinase inactive.� This type of degradation mediated by Cdh1 activated APC complex (Cdh1 is essential for the degradation of Clb 2 which is B-like cyclins); this paves the way for the cell to exit from Mitosis�.
�When chromosomal DNA replicates, single stranded chromosome becomes double stranded, for reasons of stability, a protein complex called cohesins glue the two strands to each other.� But when they reach equatorial region or little earlier, the tightly held chromosomal strands release from one another by the activity of APC/C-cdc20 activated ligase-proteosome activity, yet they are still held at centromeric region.� For equal segregation of chromosomes, the kinetochore complex has to split and free chromosomal strands from one another, so the strands can move to their respective poles.� For the chromosomal strands to free from one another by chromatin glue called cohesin complex, that holds chromosomal strands, is degraded, so at Anaphase chromatin strands separate.� In some systems chromosomal strands are freed at the end of prophase itself, but centromere is still held together, in such cases kinetochore complex splits by the protease activity induced by APC complex�.
The APC/C complex that targets cohesion complex and kinetochore complex is activated by cdc20.� It is activated at M-phase or little earlier and performs destruction of securin (pds-1p) inhibitor of separase, which triggers the release of two chromosomal strands from one another.� This process is critical for the separation of sister chromatids at anaphase, so the complex is called Anaphase Promoting Complex.
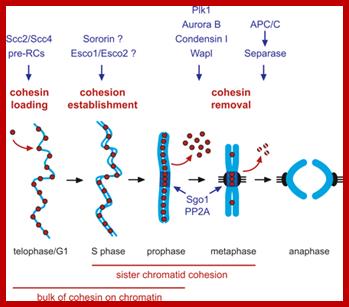
Regulation of sister chromatid cohesion during the vertebrate cell cycle. Loading of cohesin onto chromatin occurs during telophase and G1 and requires the cohesin loading factors Scc2 and Scc4. In Xenopus egg extracts, cohesin loading also depends on the assembly of pre-RCs on chromatin. During S phase, cohesion between sister chromatids is established, and this process may depend on sororin, Esco1, and Esco2. During prophase, the bulk of cohesin dissociates from chromatin, and this removal is regulated by Plk1, Aurora B kinase, condensin I, and Wapl. Cohesin at centromeres is protected by Sgo1 and PP2A. At the metaphase-to-anaphase transition, separase is activated by the APC/C and cleaves centromeric cohesin as well as residual cohesin on chromosome arms, enabling sister chromatid separation. See the text for details. http://genesdev.cshlp.org
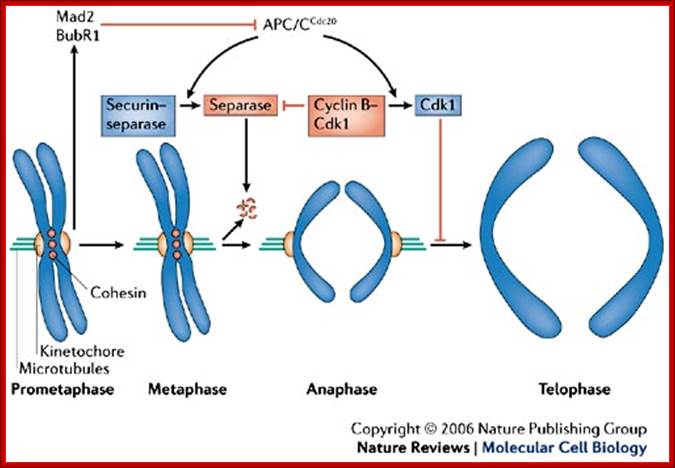
During prometaphase, spindle-assembly-checkpoint proteins such as Mad2 and BubR1 are activated at kinetochores that are not (or not fully) attached with microtubules (indicated in green). Activated Mad2 and BubR1 inhibit the capability of anaphase promoting complex/cyclosome Cdc20 (APC/CCdc20) to ubiquitylate securin and cyclin B and thereby prevent anaphase and mitotic exit. In metaphase, when all kinetochores are attached to microtubules, APC/CCdc20 ubiquitinates Securin and cyclin B and thereby activates the protease separase and inactivates the cyclin-dependent kinase-1 (Cdk1). Separase then cleaves cohesin complexes (shown as red circles) that are holding sister chromatids together and thereby initiates sister-chromatid separation. Cdk1 inactivation leads to the dephosphorylation of Cdk1 substrates by protein phosphatases, and thereby enables exit from mitosis. In vertebrates, CDK1 inactivation also contributes to separase activation.www.nature.com
DNA Chromosome dynamics; Rolf Jessberger;https://tu-dresden.de
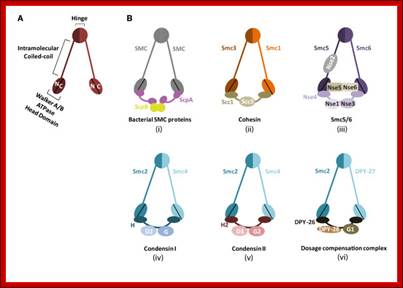
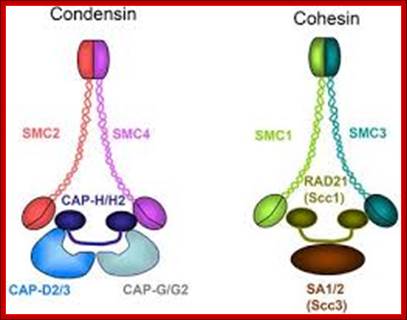
Cohesins and Condensins;� http://www.cellandbioscience.com/
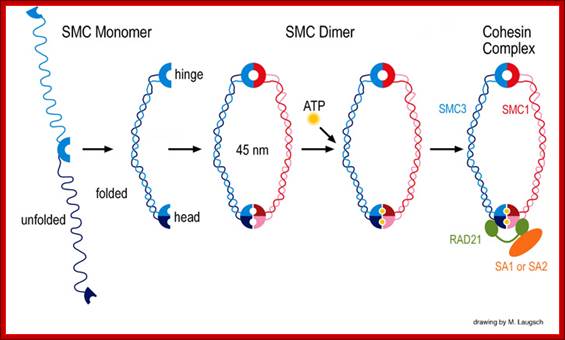
Architecture of SMC protein Complexes; (A) The core of each Smc complex is formed by two Smc proteins. Each Smc protein contains an ATPase head domain, a hinge domain, and an intramolecular antiparallel coiled coil that connects the two. The hinge domain mediates the dimerization of Smc proteins. (B) Various Smc complexes found in bacteria and eukaryotes. Each Smc complex is composed of a specific Smc dimer and several non-Smc subunits. (i) The bacterial Smc complex from Bacillus subtilis. ScpA connects the two ATPase heads of the Smc homodimer. (ii) The Smc1/3 cohesion complex. (iii) The Smc5/6 complex. (iv) The condensin I complex. H, D2, and G stand for CAP-H, CAP-D2, and CAP-G, respectively. (v) The condensin II complex. H2, D3, and G2 stand for CAP-H2, CAP-D3, and CAP-G2, respectively. (vi) The condensin-like dosage compensation complex in C. elegans. DPY-27 is an Smc4 variant.
The evolutionary highly conserved eukaryotic SMC protein family, with six members named SMC1 to SMC6, is involved in several key nuclear processes. The SMC family was defined as such in late 1994, and our publication in 1996 was the first on mammalian SMC proteins. Processes in which SMC proteins are involved are chromosome condensation, sister chromatid cohesion, DNA recombination and repair, in mitosis and meiosis. SMC proteins share a characteristic protein structure with coiled-coil-domains flanked by globular N- and C-terminal domains, and divided in the central region by a flexible hinge domain that allows movement of the arms. The six types of SMC proteins form three types of heterodimers: SMC1 & SMC3, SMC2 & SMC4, and SMC5 & SMC6. All the heterodimers constitute core components of larger multiprotein complexes that carry out specific, ATP-driven functions in chromosome dynamics.
Operation of Cell Cycle:
This description is general and applies to metazoan cells. Cells, irrespective of their ploidy divide either during growth or during reproduction.� During reproduction, a diploid gamete-producing cell undergoes reduction or meiotic division.� But a similar diploid or haploid cell during growth and development goes through a series of mitotic cell divisions.� Even in a fully grown organism, where most of the cells at all times are in resting or what is called Go state, occasionally undergo cell division in order to compensate cell loss in a given tissues.� Example, if the liver (a large organ in human body) is cut and removed, the cells in rest of the liver divide and redivide grow in numbers till the size of the liver reaches its original size. There is sensory mechanism to induce cell division and stop cell division. In culture condition cells also undergo cell division under mitogenic induction to multiply in numbers.
Recall what has been described earlier:
� In general cells in an environment provided with rich nutrients divide and redivide e.g., yeast, but cells under culture conditions initiate cell division when they are stimulated by mitogens.� Cells in a tissue require stimulation for division.� At that time cell size increases and when the cell mass reaches an optimum level to its volume, it initiates cell division, if it is somatic, it is called Mitosis.
Mitosis goes through several physical and biochemical changes in the form of stages or phases.� Mitosis has several stages such as M-phase and Interphase.� Between two M-phases there exist an intervening phase called Interphase.� The M-phase its self consists of sequential steps like Prophase, Metaphase, Anaphase and Telophase and finally Cytokinesis culminates in producing two daughter cells, which have inherited their genetic material equally.� Interphase, in general occupies longest period during cell division. �It can extend to 10-12 hrs in a 24 hr cell cycle.� But the M-phase takes just 30 minutes or 1 hr.
� Interphase when in resting phase exists in what is called Go stage, where all cell cycle processes are shut down.� But when the cell is stimulated, either by nutrient supply or by mitogens, they renter from Go stage and enter into G1 stage.� The G1 is a preparatory stage for the next phase called S-stage.� In the S-stage the chromosomal DNA replicates and generates two copies of them.� Then the cell enters into another stage called G2 stage; which is again another preparatory stage for M-phase.� G1, G2 are called so scientists did not know what exactly happen at these intervening stages, so they called it G1 and G2, which is a Gap in the knowledge about them.� Though these stages are sequential and temporal, they don�t enter to the next stage until and unless each of the stages is completed their requirements and functions.� To prevent any such early entry into the next stage, cells use checkpoints, which act as control loops where cellular events should be completed at the earlier stage to move to the next stage; other wise they remain in the same stage till all the requirement are met. Activated cells have surveillance system.
G1 Stage:
The G1 occupies approximately 10 to 12 hr where the cell prepares for S-phase.� Important check point here is called START point or Restriction point; once it passes through it there is no going back.
�
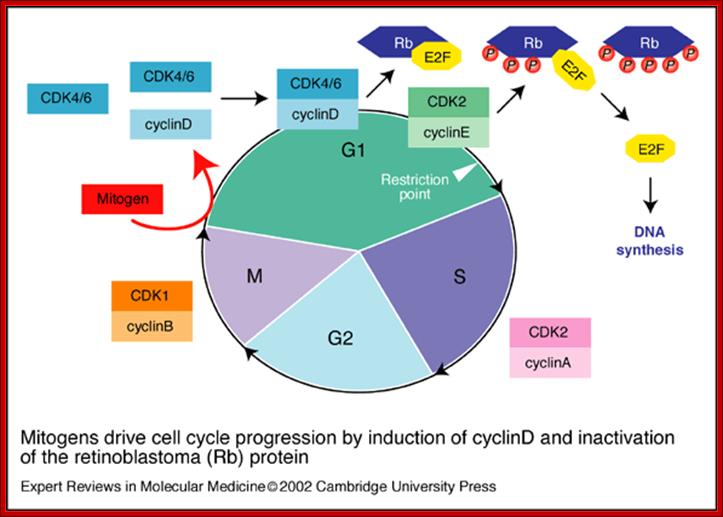
Mitogen driven Cell Cycle progression; Walter Kolch, Ashwin Kotwaliwale, Keith Vass and Petra Janosch; http://journals.cambridge.org/
Mitogens drive cell cycle progression by induction of cyclinD and inactivation of the retinoblastoma (Rb) protein: The cell cycle is driven by the co-ordinated activation of the cyclin-dependent kinases (CDKs). Whereas the CDKs are expressed throughout the cycle, their activating subunits � the cyclins � oscillate between rapid synthesis and degradation. The interface between mitogens and the cell cycle is cyclinD (and to a lesser extent cyclinE), whose expression is induced by mitogens. CyclinD- and cyclinE-dependent kinases phosphorylate (P) and thereby disable the Rb tumor suppressor protein, which is a principal checkpoint controlling the progression from G1 to S phase. Inactivation of the Rb protein marks the restriction point at which cell-cycle progression becomes independent of mitogens. Inactivated Rb releases E2F transcription factors, which stimulate the expression of downstream cyclins and other genes that are required for DNA synthesis, Walter Kolch, Ashwin Kotwaliwale, Keith Vass and Petra Janosch; http://journals.cambridge.org/
Once cells are stimulated by mitogenic signals they measure cell mass and cell volume.� There are genes, which do this function.� Once cell mass to cell volume is measured and full filled, it launches into a series of molecular events that sets the stage to next stage.�
In general at early stages of G1, inputs for DNA replication are shut off.� This is achieved by sequestering all those required Transcriptional Factors (TF-IIs) for activating genes that are essential as inputs for initiating and executing DNA replication to completion. �Some TFs have to be synthesized afresh. �One of the most important factors, besides Cyclin-Cdk inhibitors (specific), that blocks G1 transition to S-phase is RB protein, which was identified as a mutant gene causing Retinoblastoma disease (eye retinal cancer).� In its native state RBs sequester all transcriptional factors-E2Fs.� These are required for activating genes for cyclins, TFs and DNA synthesis enzymes.� Once cyclins are synthesized, they activate certain cyclin dependent kinases (Cdks).
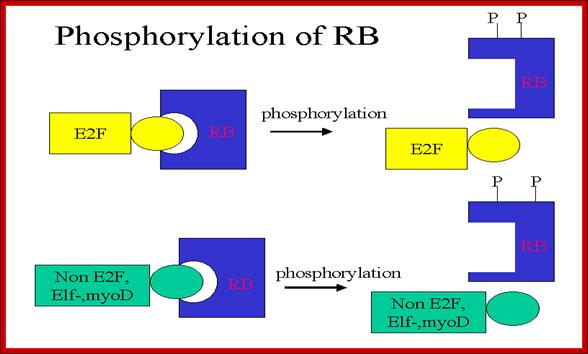
RB binds transcription factors like E2Fs and some non-E2Fs, which are sequestered when RBs are non-phosphorylated, but when, phosphorylated they are released for activating S-phase specific gene expression. www.keyword-suggestions.com
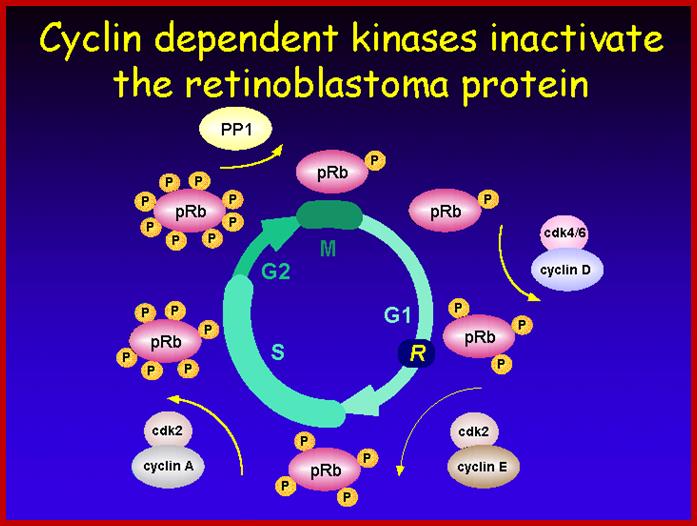
A simple diagram showing the possible phoshorylation of RBs by different combination of Cdc-Cdks at different stages.www.slideplayer.com
� Earlier signal transducing cellular events triggered by mitogens
or nutritional factors do cause specific phoshorylation and dephosphorylation reactions that have cascading effects on cellular activities.
Almost all cells have a battery of thousand or more Kinases and equal number or more phosphotases, which act specifically on their targets; thus they activate or inactivate certain substrates, which can be a protein or a carbohydrate or any other target cellular component.
Thus, RBs and similar proteins bind to E2F factors, thus transcription of genes required for DNA replication are kept inactive state.�
As the G1 Cdk-cyclin complex in its fully active state phosphorylate RBs and its associated proteins, thus make E2F factors released free of RBs.� There are several E2Fs such as E2F1, E2F2, E2F3, E2F4, and E2F5. These transcription factors with their associated DPs (Dimeric proteins) and pocket proteins such as p130 and p103 bind to their respective gene promoter elements and activate the transcription of genes whose products are required for additional cyclins and the other components for DNA replication.� Once the said factors are synthesized in sufficient amounts cell enters into S-stage. At this stage several cyclins are synthesized, such as cyclin E and others for M-phase activity.
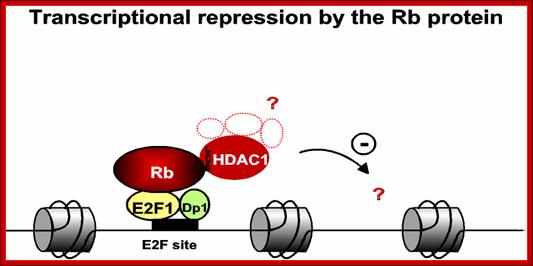
The figure just represents a model of how RB proteins repress gene expression even though E2F transcription factors are bound to their promoter; this inactivation is achieved perhaps through recruiting HDAC which deacetylate histones in chromatin.� Deacetylation of histone in chromatin makes the site of a gene inactive.
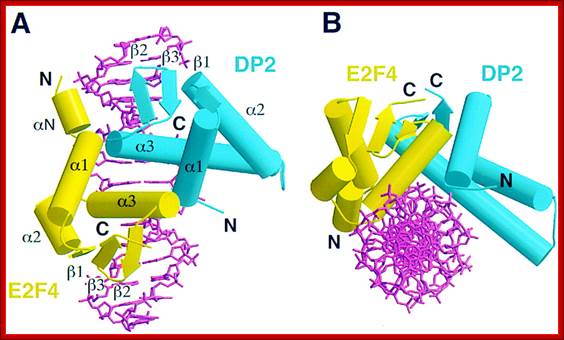
E2F4-DP2; https://www.bioscience.org

Pocket proteins interact with E2F/DP heterodimer. E2F family members seem to be able to heterodimerize with any DP protein. p130 associates with both E2F-4/DP and E2F-5/DP heterodimers. p107 associates primarily with E2F-4, *and has been found associated with E2F-5 in transfection assays. pRB associates with E2F-1 to 4/DP heterodimers.
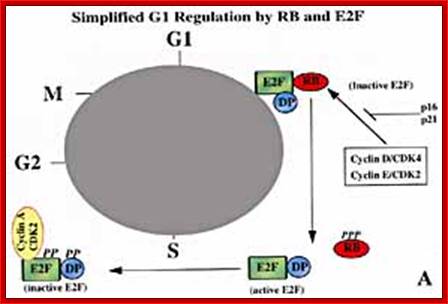
A simple representation of E2F cycle. https://www.bioscience.org
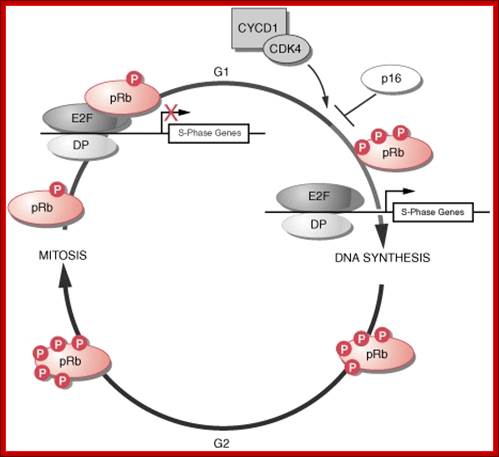
Phosphorylation regulates the function of pRb during the cell cycle. The pRb protein is hypo phosphorylated in the G1 phase of the cell cycle, and phosphorylation (P) of specific sites appears to increase during progression through the cell cycle. A protein complex that appears to phosphorylate pRb prior to DNA synthesis (S-phase) includes a cyclin (CYC) and a cyclin-dependent kinase (CDK) (probably, cyclin D1 and CDK4). The CYCD1/CDK4 complex is regulated by the p16 inhibitor protein, which is itself the product of a tumor-suppressor gene on chromosome 9p known as INK4a (see text). In its hypo phosphorylated state, pRb binds to E2F transcriptional regulatory proteins. E2F proteins dimerize with DP proteins and activate the transcription of genes, including those involved in DNA synthesis. However, when pRb is brought to the promoter regions of genes via its interaction with E2F proteins, pRb represses the expression of the E2F target genes. Phosphorylation of pRb releases it from the E2F/DP protein complex and results in gene activation. The figure also indicates that pRb phosphorylation increases in G2 with pRb dephosphorylated at or near anaphase. https://www.slideshare.net; https://bioinfo-btg10-grupo15
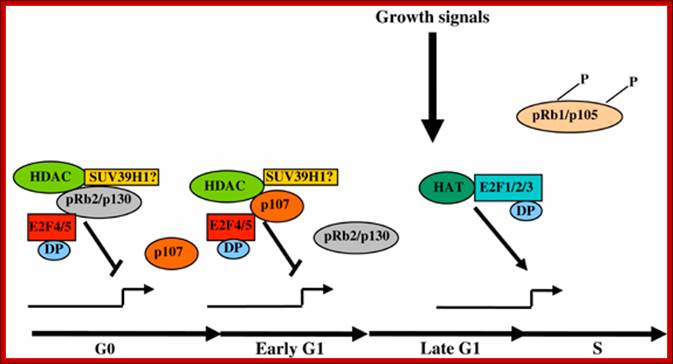
Rb family proteins as modulators of gene expression and new aspects regarding the interaction with chromatin remodeling enzymes; Model for the regulation of gene expression by pocket proteins, E2F transcription factors and chromatin modifiers: M Macaluso, M Montanari and A Giordano http://www.nature.com
E2Fs/pocket proteins/chromatin modifiers (histone deacetylase, HDAC, histone methyltransferase, SUV39H1) are present at cell cycle-regulated promoters in G0 and early G1 and function to repress transcription (Additional components of the complexes have been omitted for clarity). During G0 and early G1, the activity of E2Fs is mainly maintained by E2F4 and E2F5, which are bound preferentially to pRb2/p130 exerting an inhibitory effect on the E2F-responsive genes. pRb2/p130 is then replaced by p107 in mid to late G1 and then by pRb/p105 in the late G1- and S-phases. Upon mitogenic stimulation G1, CDKs phosphorylate pocket proteins and disrupt E2Fs/pocket proteins interaction. Switching from 'repressive' to 'activating' E2Fs and the recruitment of histone acetyltransferase activity allows G1- to S-phase transition.
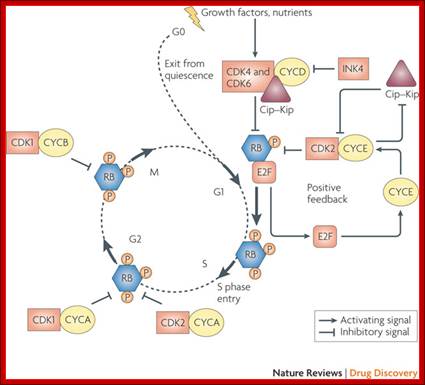
http://www.nature.com/
RB family members, or 'pocket proteins', play key parts in the control of cell proliferation. They negatively modulate the transition from the first gap (G1) phase to the DNA synthesis (S) phase (see figure), are growth-suppressive in a cell type-dependent manner, are implicated in various forms of differentiation and are crucial targets for inactivation by transforming oncoproteins of DNA tumor viruses; Silvia Lapenna & Antonio Giordano; http://www.nature.com/
� Members of the retinoblastoma (RB) protein family, comprising RB1 (also known as p105-RB), retinoblastoma-like protein 1 (RBL1; also known as p107) and RBL2 (also known as p130), share sequence homology in a bipartite domain known as the pocket domain, which folds into a globular pocket-like structure owing to the presence of a flexible 'spacer' region. The pocket domain mediates interactions with members of the E2F family of transcription factors and with proteins containing an LXCXE motif, such as D-type cyclins (CYCDs), histone deacetylases and viral oncoproteins. However, passage through restriction point requires early and mid cyclin-Cdk mediated phoshorylation of Retinoblastoma proteins (RBs).� RBs hitherto sequestered E2F factors are released.�� The RBs with E2Fs and DP (dimeric proteins) bind to specific gene promoter regions and recruit methylases and deacetylase; thereby chromatin is condensed and the genes get inactivated.� Release of E2Fs from RBs leads to activation of specific genes by recruiting histone acetylases, and also pockets proteins; there by activate genes for factors for DNA replication, late G1 cyclins, S-phase cyclins such as cyclin A and cyclin B and also activate its own transcription of E2F genes. S phase cyclin/Cdks initiate DNA replication. Progress of S-phase is greatly facilitated by S-phase Cyclins or cyclin-E, but once the S-phase is initiated S-Cyclins get degraded.� The next cyclin expressed is Cyclin-A from S to G2 stage.
S-Stage:� S-phase is of short duration of 6-8 hrs.� This is most precise and exact process and its execution should be error free.� S-phase initiation is again controlled by another set of factors.� Until and unless they are made available DNA replication is not initiated.
For the replication of DNA, replication origins have to be fired, that is the strands at origin have to open into replication bubbles.� In eukaryotes DNA is compacted by nucleosomal organization into higher order of compaction i.e., chromosome.� Chromosomes at this stage have to be relaxed and origins should be made available.�
DNA replication and Cell cycle:
Eukaryotic DNA is long (~100 million bp to 100 billion bp or so), and linear, unlike E. coli, which is circular.� The origins are located in what is called replication initiator zones, which contain several origins.� On the basis of yeast� ARS sites, most of the origins contain segments of 11 bp long ORE (Origin Recognition Elements) sequences.� Next to it there are DNA unwinding elements called DUE.� On either side of these two elements there can be auxiliary sequences.�
At the time of initiation of replication, the ORE eleven bp long sequence in S. cerevisiae, should be bound by a complex of proteins called ORC- origin recognition complex (hexamer > 400KD).� For firing the replication origin, it requires Cdc 6 and Cdt1 (licensing factors), which loads Mcms (mini chromosome maintenance proteins-Mcm2-Mcm7); they are hexamers; they are acquired when the nuclear membrane is dissolved.� Binding of ORC and cdc6 and cdt1 act as pre-replication complex (PRC); this happens only once in one cell cycle.�
The S-phase cyclin/Cdks phosphorylate pre-replication complex components. The origin is fired.� At the same time, two more factors join; they are Cdc45 and Geminin.� The Geminin prevents the loading Mcms second time before completing M-phase.� Joining of cdc45 leads to the release of cdc6/cdt1.
Loading of Mcms is crucial for they actually open the origin region into replication bubble.� Mcms act as ATP dependent helicases, located on either side of Origin they move in opposite direction.� Geminin also sequester factors required for replication, thus prevents second round of replication before the completion of mitosis.� The ORC complex remains bound to the origin.� Replication bubble provides scope for the assembly of RPA, RFC, PCNA and Pol alpha and Pol delta and replication ensues.
Propagation of genome replication:� Spatiotemporal changes of DNA replication were followed with a GFP-PCNA fusion protein by time lapse microscopy. The replication machinery starts at multiple sites (green) and over time it "spreads" in a domino-like manner activating adjacent sites (red) until all the genome is duplicated. Spatiotemporal changes of DNA replication were followed with a GFP-PCNA fusion protein by time lapse microscopy. The replication machinery starts at multiple sites (green) and over time it "spreads" in a domino-like manner activating adjacent sites (red) until all the genome is duplicated. Molecular and Cell Biology of the genome-Cardoso Lab � Research; http://www.cardoso-lab.org/
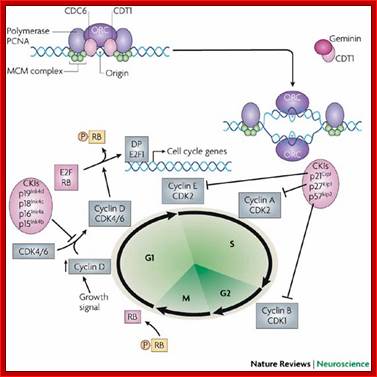
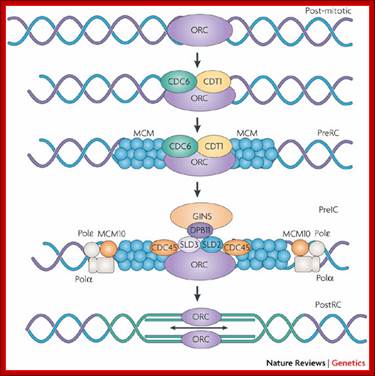
On activation of cell cycle genes ORC complex with the association of Cdc6/Cdt1 and MCM initiates opening of DNA as replication bubble.� This process is controlled and regulated by several factors. A six-subunit complex called the origin recognition complex (ORC) serves as a platform for the assembly of pre-replication complexes. ORC binds yeast replicators throughout the cell cycle; in metazoans, the binding of the large subunit of ORC, ORC1, to to chromatin is cell-cycle regulated. During the mitosis�G1-phase transition, chromatin-bound ORC recruits CDC6 and CDT1, which facilitate the loading of a helicase complex consisting of MCM (minichromosomal maintenance) proteins 2�7 (licensing). The resulting complex is termed the pre-replication complex (PreRC). The PreRC is activated to create the pre-initiation complex (PreIC) by recruitment of additional factors including CDC45, SLD2�3, DPB11, the GINS complex (SLD1 and PSF1�3) and MCM10. The initiation of DNA replication during S phase requires phosphorylation A detailed description of the formation and activation of PreRC and PreIC complexes is available in excellent recent reviews . Mirit I. Aladjem, http://www.nature.com/
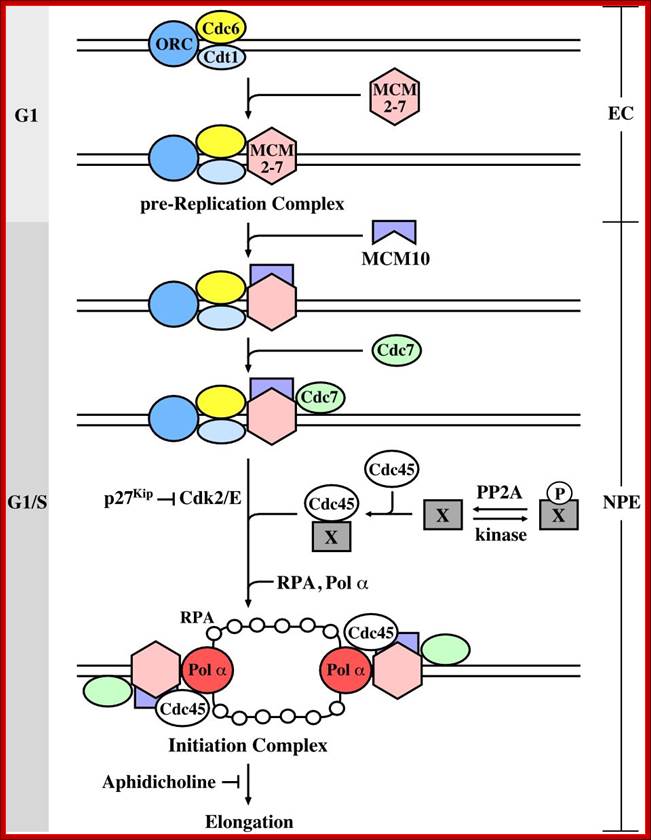
Cellular events and the components leading to activation of DNA replication at origin. https://superbpaper.com;salg365.no/mitosis-of-sythesis-phase; https://www.boundless.com/
Mitogens activate cells for mitotic activity through signal transduction process. As the cell enters into cell cycle mode mid G1 cyclin D and Cdk4 and Cdk6 are activated.� This leads to the activation of late G1 cyclin E and Cdk2.� This leads to the activation of S-phase cyclin/Cdk2 and then to cyclin/Cdk1 and cyclinB/Cdk2.
Late G1 cyclins/Cdks and S-phase cyclin/Cdks further phosphorylated RBs and keep them inactive till early M-phase.� Once the cell enters into secondGo/G1 state RBs get phosphorylated and E2Fs are sequestered.
� When all the inputs are made available DNA replication progresses to completion; then only the cell enters G 2 stage.� If during replication any error or damage is done to DNA or there is any pre-existing damaged DNA, cell cycle halts and starts repairing the damage.� Even the cell enters to G2, it still waits till it is repaired.� If the damage is beyond repair, the damage sensing system p53 protein, which acts as a sensor for DNA damage, activates specific genes for the synthesis of P21 and such proteins, which bind to Cdk-cyclin complexes and halt the cell cycle progress. As a last recourse p53 induces apoptosis. �
DNA damage induces intracellular signaling cascades that blocks DNA replication and cell cycle progression. Two parallel branches, the ATR-Chk1 and ATM-Chk2 pathways are activated by different forms of damage. Cdk activity is inhibited by degradation/inhibition of Cdc25 phosphotases and by the p53-dependent induction of the p21 Cdk inhibitor. Cdk-independent events also block origin firing through inhibition of Cdk and Cdc7 kinase activity. The Tlk1 kinase which promotes Asf1-dependent chromatin assembly is also inactivated in response to DNA damage.
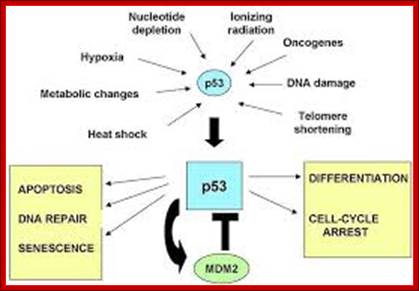
The p53 response. http://en.wikipedia.org/ Cancer biomarkers; http://www.cancer-biomarkers.com/
Damaged DNA is sensed by ATR; and ATM sensors and activate P53, which in turn turns on p21 and cell cycle inhibitor. This 'growth arrest' function of p53 is important in preventing cancer by suppressing tumours and p53 mutations or non-functioning p53 are observable in many human cancers. If the damage can be repaired, p53 starts a cascade of events that induces cell cycle arrest providing more time for DNA repair.
The role of p53 in apoptotic signaling primarily involves interaction with members of the ASPP protein family. The introduction of DNA damage or oncogene upregulation to a cell results in ASPP1 and ASPP2 expression. In humans the C-terminals of these proteins bind to p53. This activates the mitochondrial apoptotic pathway by activating PUMA (p53 upregulated modulator of apoptosis) and inhibiting activity of the ant-apoptotic Bcl-2 protein. Conversely, expression of the iASPP (inhibitory ASPP) competes with and prevents ASPP1 and ASPP2 binding to p53 and is therefore anti-apoptotic. http://en.wikipedia.org/; http://www.cancer-biomarkers.com; http://en.wikipedia.org/ Cancer biomarkers; http://www.cancer-biomarkers.com/
P 53 Structure:
The p53 protein is 400 amino acids long, and the human P53 is 393 amino acids long. <ref> Joerger AC, Fersht AR. It consists of several domains as shown below in the figure.
Schematic representation of p53 major domains; Agarwal ML, Taylor WR, et al; http://www.biomed-search.com/
Diagram showing the domain structure of the p53 protein; the p53 protein is a transcription factor that contains several well-defined domains. At the N-terminus are the transactivation domain and a proline-rich region, which is required for apoptotic function. Within the N-terminus are the interaction sites of p53 with components of the transcriptional machinery as well as ubiquitin ligase Mdm2. The central domain harbors the sequence specific DNA binding region, where most of the tumor associated mutations occur. This central region also contains binding sites for interaction with members of the Bcl2 protein family. The C-terminal region contains the oligomerization domain as well as nuclear localization and export signals. Several sites within the N-terminal region have been shown to be phosphorylated and the C-terminal region contains numerous sites of modification which influence stability, localization and activity of p53. Agarwal ML, Taylor WR, et al; http://www.biomed-search.com/
Schematic representation of p53 major domains; Proline-rich region (residues 67-98)
- Central core domain (residues 98-303)
- Nuclear localization signal containing region (residues 303-323)
- Oligomerisation domain (residues 323-363) and
- C-terminal basic domain (residues 363-393).
- N-terminal transcription activation domain (TAD), which can be further divided into two smaller sub-domains, TAD-I (residues 1-40) and TAD-II (residues 41-67). If the cellular damage cannot be repaired, it triggers gene activation to produce p21 which blocks progression of cell cycle.� If that fails, p53 induces apoptosis (cell death).� Even, if this fails the cell get transformed and continues to divide and redivide into cancerous cells.
MPF induces multiple nuclear and cytoplasmic changes at the onset of M phase, both by activating other protein kinases and by phosphorylating proteins such as condensins and the nuclear lamins.
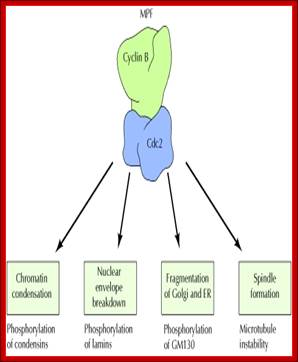
http://oregonstate.edu/
The nuclear lamina consists of a meshwork of lamin filaments. At mitosis, Cdc2 and other protein kinases phosphorylate the lamins, causing the filaments to dissociate into free lamin dimers.
A schematic view of inner nuclear membrane proteins and their binding interactions with the nuclear lamina and nucleoplasmic components. www.journals.cambridge.org
The outer and inner nuclear membranes (ONM and INM, respectively) are shown in cross-section, with a nuclear pore complex spanning the two membranes. The exact interactions and organization of the inner nuclear membrane, nuclear lamina and chromatin are unknown; and they are hypothetically depicted in the above figure. Twelve inner nuclear membrane proteins have been characterized in the mammalian nuclear envelope. These include: the multi-spanning membrane proteins nurim, lamin B receptor (LBR), ring-finger-binding protein (RFBP), the double-spanning membrane protein MAN1; and the single-spanning membrane protein emerin, lamina-associated protein 2 (LAP-2) isoforms and LAP-1 isoforms (A, B, C). All the LAP-2 isoforms, emerin and MAN1 share a homologous N-terminal domain called the LEM domain, which binds to BAF (�barrier to auto-integration factor�). Interactions occur between the inner nuclear membrane proteins and the A-type lamins (shown in blue) and B-type lamins (shown in orange), which are helical filamentous proteins. �Intranuclear lamina binds to the soluble LAP-2 isoform. Transcriptional regulators crosslink inner nuclear membrane proteins and chromatin; these include: the retinoblastoma protein pRB; the �germ-cell-less� protein GCL; the transcription factor E2F, RNA polymerase, RNA splicing complex and DP protein. Heterochromatin-binding proteins include HP1, BAF and HA95.
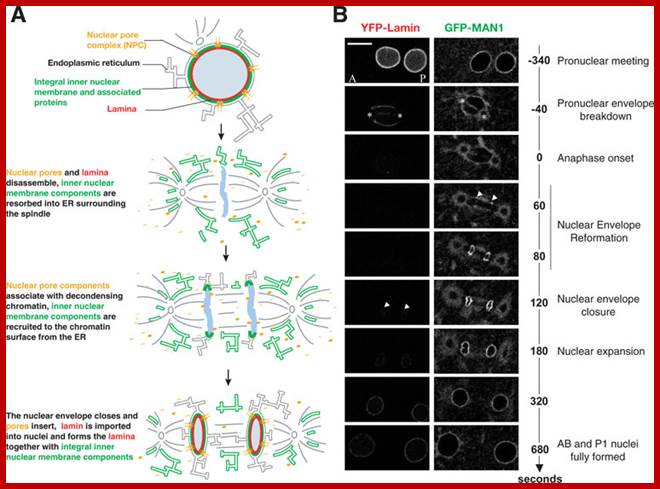
Karen Oegema and Anthony, A Hyman; http://www.wormbook.org/
Nuclear envelope dynamics in the C. elegans embryo; Figure courtesy of Vincent Galy. (A) Schematics illustrate the cycle of nuclear envelope breakdown and reassembly. (B) Still images of the first mitotic division of wild-type embryos expressing YFP-Lamin (left) and GFP-MAN1 (right). Times on the right are relative to first metaphase to anaphase transition. Arrowheads indicate the reappearance of GFP-MAN1 around the chromatin at t=60 sec and YFP-Lamin at t=120 sec. White stars mark the positions of the centrosomes. Note the persistence of membranes containing GFP-MAN1 around the mitotic spindle and centrosomes. Scale bar = 10μm.worm book.org
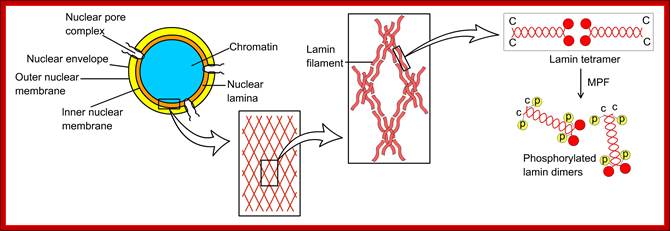
Nuclear membrane dismemberment due to disassembly of nuclear lamins. https://pt.slideshare.net
Chromosomal adhesion and condensing proteins:
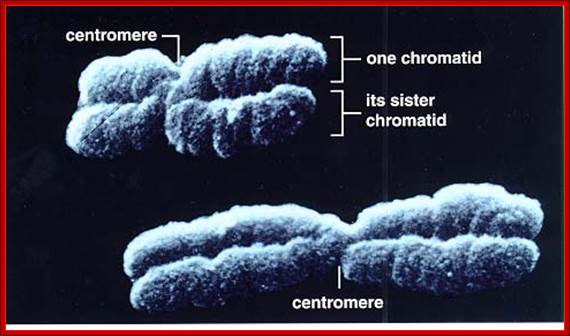
Resolution results from the decatenation of the sister DNAs, accompanied by the partial removal of cohesin molecules along the chromosome arms. http://reasonandscience.heavenforum.org
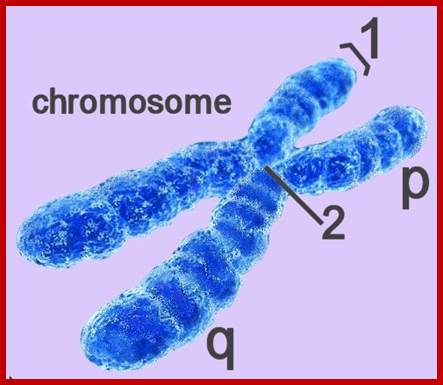
http://reasonandscience.heavenforum.org;http://genebiochem.blogspot.in
Chromosomes go through condensation and relaxation during cell cycles. When they replicate the two strands remain associated with each other for some time; it is at anaphase the strands separate. Among several proteins SMC and their associated proteins are important in condensation and cohesion of chromatids.
SMC proteins:
Regulation of genomic integrity by SMC complexes: The cases of the Nijmegen breakage syndrome and Smc5/6 complex.
Cancer cells can modify their genome by inducing structural defects in their genome, such as the fusion of DNA sequences from unrelated chromosomes. With this specific project, our laboratory is interested in understanding the molecular mechanisms responsible for preventing alterations in the basic structure of chromosomes.
We know that a family of proteins known as the Structural Maintenance of Chromosome (SMC) proteins plays an essential role in the maintenance of genome stability. This family of proteins is characterized by a unique structural organization. The four conserved eukaryotic SMC complexes play wide variety molecular functions including key roles in DNA repair and checkpoint signaling. In spite of recent advances in our knowledge, we still only have a partial mechanistic understanding of how SMC complexes promote DNA repair and genome integrity.
![]() Cohesins and
Condensins:
Cohesins and
Condensins:
Cohesins and Condensins are heteromeric proteins made up of Smc proteins (Structural Maintenance of Chromosomes) and non-Smc proteins.� Cohesins are made up of two Smc proteins, Smc-1p and Smc-3p and two non-Smc proteins such as Scc1p and Scc3p) Sec1p and Sec3p; whereas Condensins are made up of Smc2p and Smc4p and Sec2p and Sec4p.� Cohesins are responsible for adhesion of two sister strands together as parallel strands all along the length including kinetochore region.� But condensins are responsible for the condensation of chromatin DNA from long convoluted threads into short and stable threads at metaphase. They are also responsible for cohesion of homologous chromosomes during meiotic chromosomal pairing called synapses.
Cohesins:�� They are complex of proteins, made up of Smc1 & 3 and Scc1P & Scc3p.� Smc1 and Smc3 of cohesins are coiled coils with a flexible hinge in the middle and DNA binding and ATPase domain at either ends.� In Smc1 and Smc3 coiled coil protein pairs show V-shaped bending with 86 ^o apart.�
But the Smc2 and Smc 4 proteins too have helical arms with flexible hinge in the middle and ATPase domain at either ends when dimerizes the flexible angle is steep of 86^o. They function as dimers with opposite polarity.�
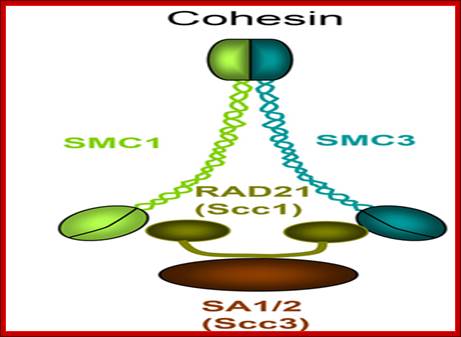
Cohesins are dimers of Smc1 and Smc2 they are helically coiled to each other in opposite orientation and the heads are associated with non Smc proteins.� The head are glued by kinesins. http://www.bioscience.org/
When Smc1 and Smc3 of cohesins dimerize parallel to each other oriented in antipolar fashion.��� The dimers of such Smc pairs with their heads can hold on to the same DNA at one site and the heads enclose the DNA as a ring and heads join with kinesin a screw that holds the heads together.� If two sets of Smc protein pairs, hold one strand and another hold the opposite strand or both together in as one ring.� When the free ends are paired and linked by kinesin (formerly called Scc1p and Scc-3p), two chromosomal strands will be held parallel to each other.� Several protein pairs all along the length of chromosomes provide such links, thus chromosomal strands are paired and glued.� Here the glue is kinesin (Scc1p and Scc3p), perhaps one more protein pds5 may also be present.� There are more non Smc proteins involved; in degradation of these makes chromosomal strands to separate from one another.
 �
�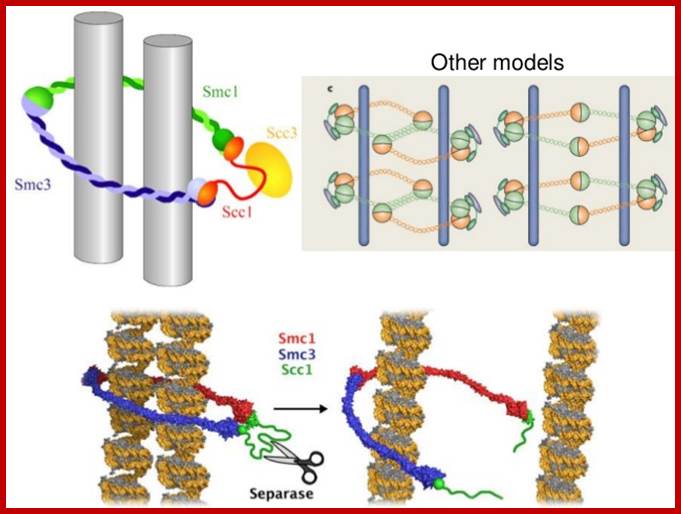
Structural model of the tripartite cohesin complex consisting of Smc1 (red), Smc3 (blue) and Scc1 (green) proteins encircling two 10nm sister chromatids, e.g. DNA (gold) wrapped around nucleosomes (grey); http://reasonandscience.heavenforum.org Kim Nasmyth; https://www.slideshare.net/
Condensins:� They are also made up of a complex of proteins, such as Smc 2 and Smc4 and two non-Smc proteins called Scc3 and Scc4.� Here the Smc proteins 2 & 4 coil to each other in antiparallel fashion.� The flexible portion can generate an 80^o angle.� These proteins bind to the same chromosomal DNA at different sites i.e., a pair at the base of one DNA loop rather binding leads to the formation of a loop and the other pair at another site, perhaps at a distance. When free heads glued by kinesins (Scc2 and Scc4) the DNA found in between is looped out.� This can lead to super coiling of DNA. If a free and long DNA is added with Smc2 and Smc4 proteins with kinesins one finds the clamps at the base of DNA loops.� Thus, chromatin gets condensed. Many such condensins all along the length of chromosomes act on at different sites and condense chromosome to a maximum at metaphase.� Whether or not there is any relation between Histone-1 phoshorylation and histone methylation induced condensation and condensin operated condensation, is not clear; sure, there should be a relationship.
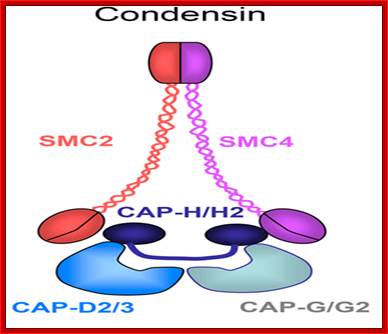
Condensins consists of heterodimer of smc2 and smc4 and they are associated with non Smc proteins such as CAPs (D2/3, G/G2 and H/H2, An SMC1-SMC3 heterodimer functions as the core of the cohesin complex, which contains two non-SMC subunits, Rad21/Scc1 and SA proteins/Scc3. http://www.bioscience.org/
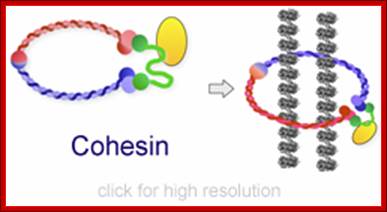
www.slideplayer.com
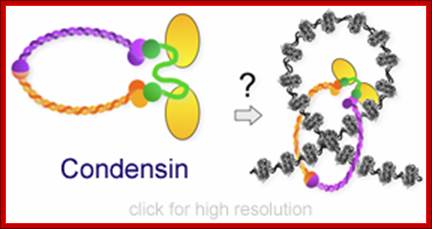
www.slideplayer.com
![]() �
�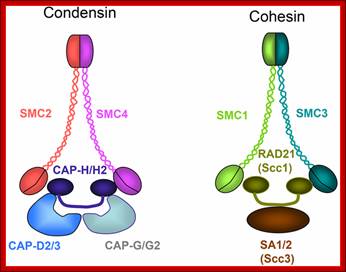
Figure. Condensin complex and cohesin complex in C. elegans; SMC-protein complexes, condensin, and cohesin, are conserved in eukaryotes. Their common structures are shown. An SMC2-SMC4 heterodimer functions as the core of condensin I and II. Condensin, I contain the non-SMC subunits CAP-D2, CAP-H, and CAP-G; Condensin II contains CAP-D3, CAP-H2, and CAP-G2. An SMC1-SMC3 heterodimer functions as the core of the cohesin complex, which contains two non-SMC subunits, Rad21/Scc1 and SA proteins/Scc3. �SMC2 and Smc4 condense chromatin and Smc1-Smc3 cohese chromatin strands. http://www.bioscience.org/
Structural maintenance of chromosomes (SMC) proteins are ubiquitous in organisms from bacteria to humans, and function as core components of the condensin and cohesin complexes in eukaryotes. SMC proteins adopt a V-shaped structure with two long arms, each of which has an ATP-binding head domain at the distal end. It is important to understand how these uniquely designed protein machines interact with DNA strands and how such interactions are modulated by the ATP-binding and -hydrolysis cycle. An emerging idea is that SMC proteins use a diverse array of intramolecular and intermolecular protein�protein interactions to actively fold, tether and manipulate DNA strands.
� A major controversy in the field of chromosome research has been whether condensin is required for achieving the highly compacted state of chromatin fibres in mitosis and meiosis. Through genetic experiments in mouse oocytes, condensin is now found to be indispensable for meiotic chromosome assembly by mediating chromosome compaction and disentanglement of sister chromatids and by conferring rigidity to chromosomes. Kota Nagasaka & Toru Hirota; https://www.nature.com
�
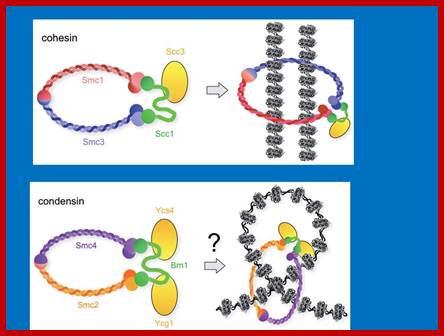
KA Hagstrom - 2003; http://slideplayer.it/www.nature.com;
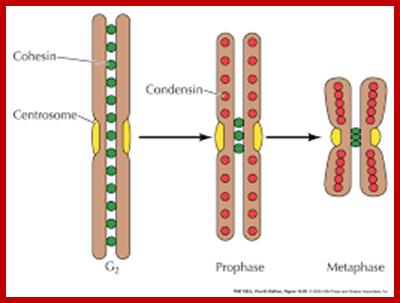
Cohesins brings homologous chromosomes pair with each other, Condensin contract the long nucleo-proteinaceous thread chromatin shorter; www.oregonstate.edu;
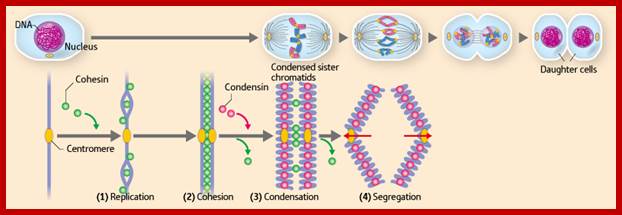
Chromosome condensation and segregation processes (lower) in cell division (upper). Upon DNA replication, cohesin works as a glue to bond together the pair of DNA sequences ((1) and (2)). The cohesin then detaches, except near the centromere, and condensin acts to condense the sister chromatids ((3)). The remaining cohesin then detaches and the sister chromatids are segregated, with one set distributed to each of two daughter cells ((4)). Stage (3) corresponds to the sequence shown in Fig. Copyright:RIKEN: https://phys.org/news/2010-12-chromosome.html#jCp;https://phys.org/news
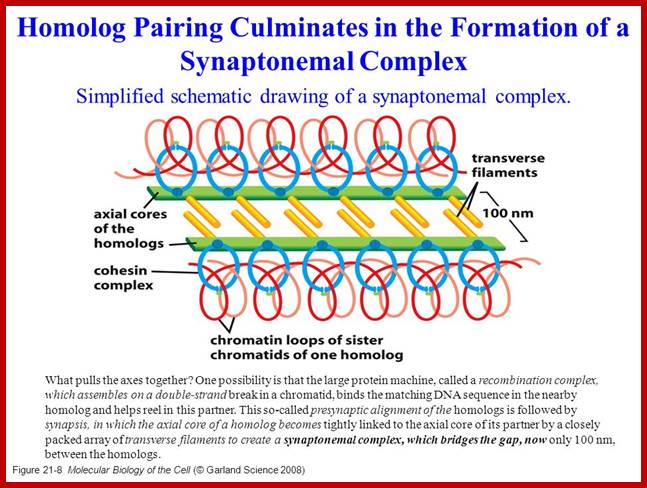
Molecular Biology of the Cell (� Garland Science 2008) Homolog Pairing Culminates in the Formation of a Synaptonemal Complex Simplified schematic drawing of a synaptonemal complex. What pulls the axes together? One possibility is that the large protein machine, called a recombination complex, which assembles on a double-strand break in a chromatid, binds the matching DNA sequence in the nearby homolog and helps reel in this partner. This so-called presynaptic alignment of the homologs is followed by synapsis, in which the axial core of a homolog becomes tightly linked to the axial core of its partner by a closely packed array of transverse filaments to create a synaptonemal complex, which bridges the gap, now only 100 nm, between the homologs.
Garland Science;http://slideplayer.com/
![]() The Smc5/6 complex contains eight subunits: Structural Maintenance
of Chromosomes (Smc)-5 and Smc6 plus six non-Smc element (Nse) subunits (Nse1�6
), (see figures). Nse1�4 are essential, whereas Nse5 and Nse6 are inessential
but are required for the DNA repair functions of Smc5/6. An additional loosely
associated subunit, Rad60, is required for both DNA repair and the essential
function of the Smc5/6 complex. The complex is closely related to cohesin and
condensin. The N- and C-terminal globular domains of each SMC subunit
self-associate to generate an ATPase. �Nse1 resembles a RING-finger ubiquitin
E3 ligase but no in vitro ubiquitylation activity has
been reported. Nse2 functions as an E3 small ubiquitin-related modifier (SUMO)
ligase in vitro. In vivo, several
proteins have been identified as potential Nse2 substrates. Nse3 contains a
MAGE (type II melanoma antigen) domain, and is the only MAGE-domain protein in
yeasts (by contrast, there are multiple MAGE-domain proteins in higher
eukaryotes). Nse4 resembles a kleisin and may bridge the heads of Smc5 and Smc6.
Nse5 and Nse6 also associate with the head domains, potentially forming a
second bridge.
The Smc5/6 complex contains eight subunits: Structural Maintenance
of Chromosomes (Smc)-5 and Smc6 plus six non-Smc element (Nse) subunits (Nse1�6
), (see figures). Nse1�4 are essential, whereas Nse5 and Nse6 are inessential
but are required for the DNA repair functions of Smc5/6. An additional loosely
associated subunit, Rad60, is required for both DNA repair and the essential
function of the Smc5/6 complex. The complex is closely related to cohesin and
condensin. The N- and C-terminal globular domains of each SMC subunit
self-associate to generate an ATPase. �Nse1 resembles a RING-finger ubiquitin
E3 ligase but no in vitro ubiquitylation activity has
been reported. Nse2 functions as an E3 small ubiquitin-related modifier (SUMO)
ligase in vitro. In vivo, several
proteins have been identified as potential Nse2 substrates. Nse3 contains a
MAGE (type II melanoma antigen) domain, and is the only MAGE-domain protein in
yeasts (by contrast, there are multiple MAGE-domain proteins in higher
eukaryotes). Nse4 resembles a kleisin and may bridge the heads of Smc5 and Smc6.
Nse5 and Nse6 also associate with the head domains, potentially forming a
second bridge.
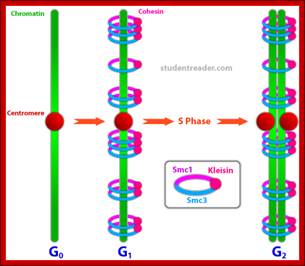
http://studentreader.com
Cohesin has two non-SMC subunits, sister chromatid cohesion protein-1 (Scc1) and Scc3. Scc1 is a member of the kleisin family and bridges the heads of Smc1 and Smc3 to form a ring-like structure. Cleavage of Scc1 at the metaphase�anaphase transition by a specialized protease opens the ring and allows the sister chromatids to segregate. It is likely that the Smc1�Smc3 ring encircles the two sister chromatids.
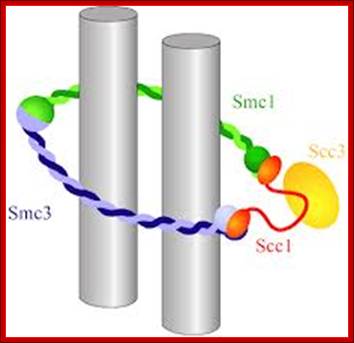
Cohesin embrace model. Sister chromatids are symbolized by gray cylinders; C. H. Haering; http://www.sciencemag.org/
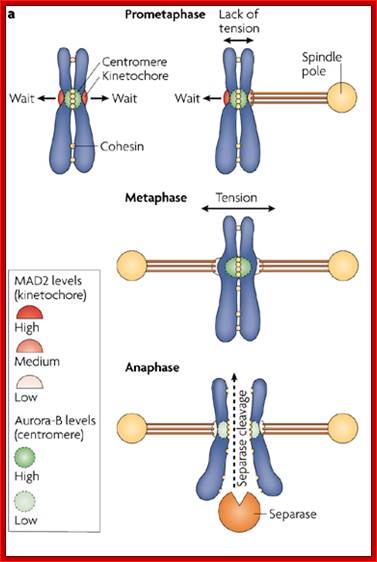
����������� As mitotic apparatus develops, tractile fibers attach to kinetochores; cohesins have to be degraded for anaphase movement. It is at this stage the MP-kinase activates Anaphase promoting complex/cyclosome (APC/C) which gets activated with binding of cdc20.� APC/C complex is ubiquitination (ubiquitylation) ligase complex that targets cohesins; first securin that gets degraded.� The Securin always keeps Separase sequestered.� Separin is an endopeptidase or call it endo-protease.� Degradation of Securin releases Separin, which now acts of Scc1 and Scc3 which are the components of Cohesin protein complex, thus the glue is dissolved and chromatin strand separate from one another; only chromosomal arms are freed but not kinetochore region.
Once the cdc20 is released from APC complex, it joins with cdh1. This complex leads to the degradation of cyclin A first and then cyclin B by the same ubiquitin-proteasome mediated decay.� It is at this point kinetochores held by cohesins are degraded and kinetochores separation facilitates anaphasic chromosomal movement. APC/C complex as described earlier is ligase system specific to M phase proteins for ubiquitinated proteosome mediated digestion.
Kinetochore
�Accurate segregation of chromosomes, essential for the stability of the genome, depends on 'bi-orientation'-simultaneous attachment of each individual chromosome to both poles of the mitotic spindle. On bi-oriented chromosomes, kinetochores (macromolecular complexes that attach the chromosome to the spindle) reside on the opposite sides of the chromosome's centromere. In contrast, sister kinetochores shift towards one side of the centromere on 'syntelic' chromosomes that erroneously attach to one spindle pole with both sister kinetochores. Syntelic attachments often arise during spindle assembly and must be corrected to prevent chromosome loss. It is assumed that restoration of proper centromere architecture occurs automatically owing to elastic properties of the centromere. Here authors test this assumption by combining laser microsurgery and chemical biology assays in cultured mammalian cells. Authors find that kinetochores of syntelic chromosomes remain juxtaposed on detachment from spindle microtubules. These findings reveal that correction of syntelic attachments involves an extra step that has previously been overlooked: external forces must be applied to move sister kinetochores to the opposite sides of the centromere. Furthermore, it is demonstrate that the shape of the centromere is important for spindle assembly, because bipolar spindles do not form in cells lacking centrosomes when multiple chromosomes with juxtaposed kinetochores are present. Thus, proper architecture of the centromere makes an important contribution to achieving high fidelity of chromosome segregation�.
DAIP fluorescent stained Centromere with H3-CENP-A Variant. http://www.zmbh.uni-heidelberg.de/Sylvia Erhardt/
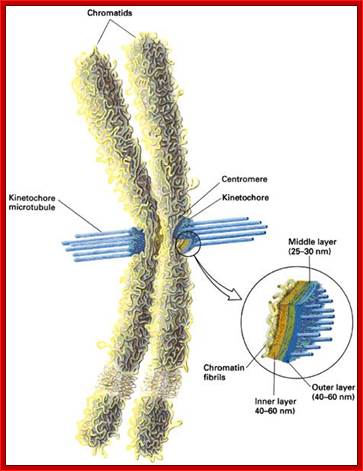
Structure and localization of Centromere and Kinetochore; the University of Texas at Austin (US); https://www.utexas.edu
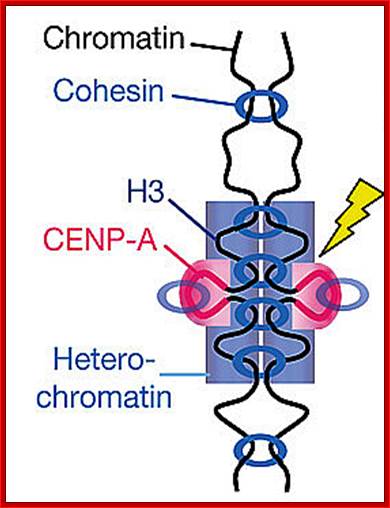
Centromere with special CEN-A nucleosomes are held by cohesin complexes. www.dianliwenmi.com
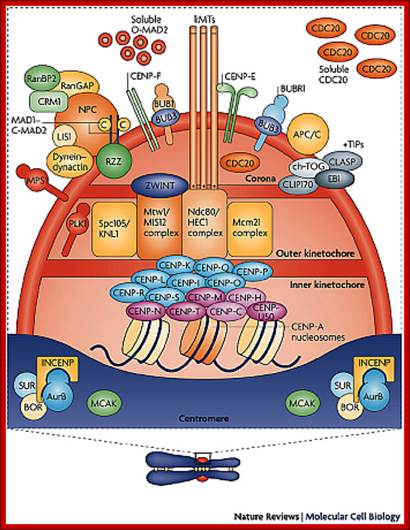
http://www.nature.com/
The centromere-kinetochore region: At the heart of the kinetochore is a specialized nucleosome that contains centromere protein (CENP)-A, a histone H3 homologue. Several inner kinetochore components (cyan and purple ovals) associate with kinetochores throughout the cell cycle. Many other proteins are recruited to the outer kinetochore specifically in mitosis. They provide a landing platform for the spindle-assembly checkpoint (SAC) proteins. The Ndc80/HEC1 complex seems to be directly involved in microtubule binding. Several microtubule-plus-end-binding proteins (+TIPs) are important for microtubule to kinetochore attachment. Most proteins indicated in this drawing are present at kinetochores in all metazoans.
Nature
Publishing Group Mustachio. et al. The spindle-assembly checkpoint in
space and time � 2007. Nature Reviews Molecular Cell Biology 8, 385 (2007). All rights reserved. ![]()
![Molecular architecture of the kinetochore|[ndash]|microtubule interface](Cell_Cycle_And_Its_Regulation_files/image122.jpg)
Vertebrate kinetochore ultrastructure;
a | A schematic of a mitotic chromosome with paired sister chromatids � the chromatid on the right is attached to microtubules and the chromatid on the left is unattached. The inner kinetochore, the outer kinetochore, the inner centromere and the fibrous corona, which is detectable on the unattached kinetochore, are highlighted. b | Electron micrograph of a human kinetochore (image courtesy of Y. Dong and B. McEwen, State University of New York at Albany, USA). The micrograph represents a single slice from a tomographic volume of a high-pressure frozen mitotic cell and has been labelled as in a to highlight the key structural features of the kinetochore. Scale bar, 100 nm. Iain M. Cheeseman & Arshad Desai http://www.nature.com/
Structural features of centromeric region, consists of layers of kinetochore complex proteins.� The complex consists of inner, middle and outer kinetochore complexes.� Microtubules are attached to outer kinetochore protein complex.� The kinetochore perse is associated CENP nucleosomal (HC) complex.
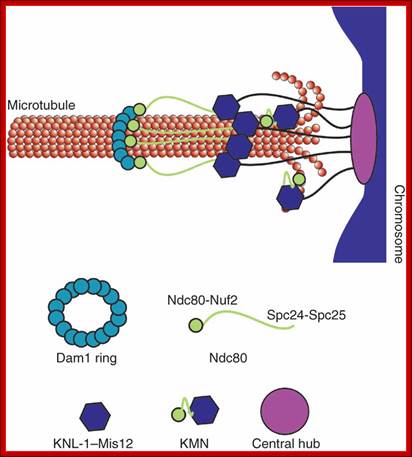
The structure of purified kinetochores reveals multiple microtubule-attachment sites; The central globular domain binds to the centromeric locus of the chromosome, and globular domains containing the KMN complex extend to attach to the microtubule. The Ndc80 sub complex makes an additional extension to contact a distal ring composed of the Dam1 sub complex. Shane Gonen, Bungo Akiyoshi, Matthew G Iadanza, Dan Shi, Nicole Duggan, Sue Biggins & Tamir Gonen; http://www.nature.com/
Schematic of the proposed model of kinetochore architecture. The structure of purified kinetochores reveals multiple microtubule-attachment sites:
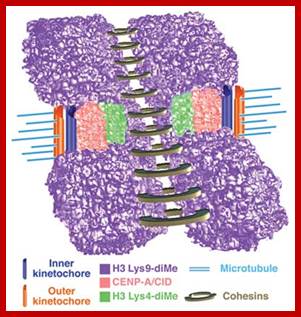
Model for three-dimensional organization of centromeric
(CEN) chromatin in D.
melanogaster and Centromeric chromatin
exhibits a histone modification pattern that is distinct from both euchromatin
and heterochromatin in Melanogaster and Humans; �
Beth A Sullivan & Gary H Karpen;
Incorporation of the two-dimensional and three-dimensional histone modification
patterns extends our understanding of the chromatin composition and
organization of the CEN region, and suggests that interspersed CENP-A/CID and H3 Lys4-dimethyl nucleosome blocks
comprise a unique chromatin state that is distinct from the flanking
heterochromatin. Associations between similarly modified nucleosome blocks are
proposed to contribute to the formation of distinct three-dimensional structures
in CEN and flanking chromatin. Interspersed CENP-A/CID and distinctly modified
H3 and H4 may mediate formation of the 'cylindrical' three-dimensional
structures observed in metaphase chromosomes15. H3 Lys9-diMe
chromatin, which recruits heterochromatin proteins such as HP1 and cohesion
proteins such as RAD21/SCC1, is present in the inner kinetochore space between
mitotic sister chromatids and in regions that flank CEN chromatin. This
arrangement may position CENP-A toward the poleward face of the mitotic
chromosome and facilitate recruitment of outer kinetochore proteins, and
promote HP1 self-interaction and proper chromosome condensation and cohesion.
Cohesins are presented as ringed structures, in
accord with recent model http://www.nature.com/
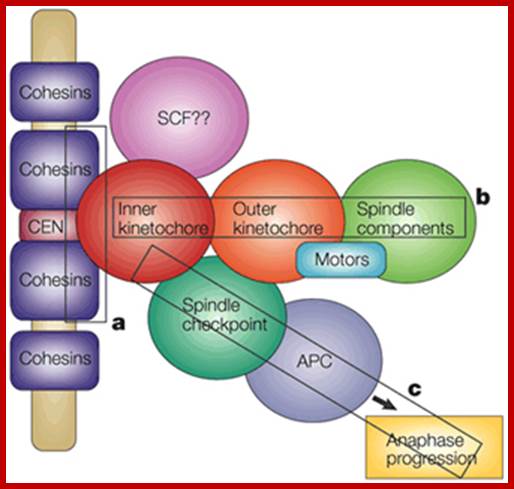
Chromosome Segregation in Mitosis: The Role of Centromeres; : Mitosis is the process by which a eukaryotic cell divides to produce two daughter cells that each contain the same number of chromosomes as the parent cell. As this definition suggests, the overall process of mitosis fails if the parent cell's chromosomes don't reach their correct destinations. One structure that plays a critical role in ensuring that this does not occur is the centromere. The centromere was first described by German biologist Walter Flehming in the 1880s as the "primary constriction" of the chromosome. Scientists now appreciate that the centromere is a region of specialized chromatin found within each constricted chromosome that provides the foundation for kinetochore assembly and serves as a site for sister chromatid attachment (Figure 1). Errors in centromere or kinetochore function are catastrophic for cells. Such errors can lead to aberrant division and chromosomal instability, both of which are often observed in cancerous cells. Clare O'Connor, Ph. D. http://www.nature.com
A brief view of kinetochore structure:
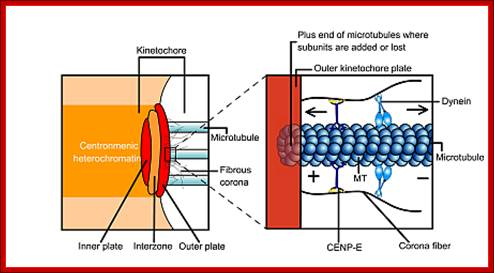
Attachment of microtubules to the kinetochore protein complex: Kinetochore is made up of three layers of proteins, emanating from special centromeric nucleosome- CeNP. http://www.cc.scu.edu.cn
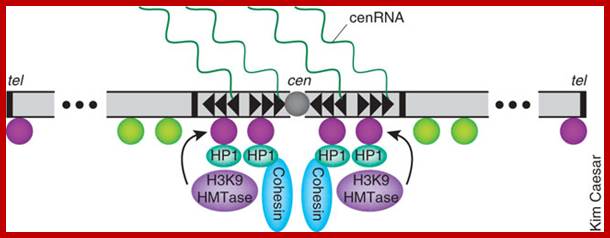
Kim Caesar
The centromere (cen) is surrounded by repetitive DNA elements, which are transcribed in S. pombe to produce noncoding cenRNAs. These sequences and the associated cenRNAs attract H3K9 HMTase, HP1 proteins and other complexes that mediate heterochromatin formation. HP1 recruits cohesin to promote sister-chromatid cohesion at centromeres. Nucleosomes associated with heterochromatic regions carry H3K9 methylation and are hypoacetylated (magenta circles), whereas nucleosomes in euchromatic regions carry H3K4 methylation and are hyperacetylated (green circles). Heterochromatic regions at telomeres (tel) and other chromosome regions (not shown) share many properties with centric heterochromatin. A role for noncoding cenRNAs in the assembly of centromeric heterochromatin has thus far been established only in S. pombe, although components of the RNAi pathway contribute to heterochromatic gene silencing at D. melanogaster pericentromeric regions. Transcription and RNAi in heterochromatic gene silencing; Marc B�hler & Danesh Moazed; http://www.nature.com
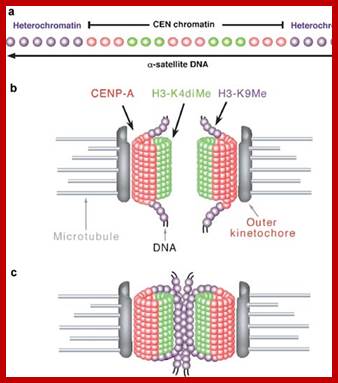
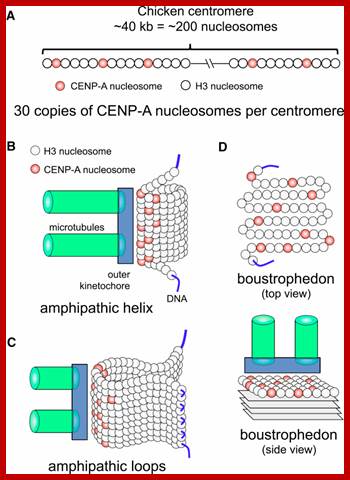
Unique organization of centromere regions in humans. a: On linear, two-dimensional chromatin fibers, subdomains of nucleosomes containing centromeric (CEN) histone CENP-A (red) are interspersed with H3 demethylated at lysine 4 (H3K4me2) (green) to form a domain of CEN chromatin on a fraction of the mega base regions of human α-satellite DNA. The remainder of the α-satellite DNA is assembled into heterochromatin (purple) that flanks one or both sides of CEN chromatin domain. b: At metaphase, when mitotic chromosomes condense, the interspersed domains promote coiling of the DNA so that stacks of CENP-A nucleosomes are presented to the poleward face of the chromosome where they can interact with other kinetochore proteins. H3-containing nucleosomes are oriented between sister kinetochores. c: Heterochromatin defined by nucleosomes containing H3-K9 methylation (purple) is assembled into a domain that is distinct from CEN chromatin. Higher-order packaging of heterochromatin between sister kinetochores may promote orientation of CENP-A, pushing it toward the outside of the chromosome. Heterochromatin in this region is also important for recruiting cohesion proteins that are sustained at the centromere until chromatid separation at anaphase. Reproduced from Schueler and Sullivan (2006). Journal of Cellular Biochemistry;
Organization of CENP-A and H3 Nucleosomes in Centromeres (A); �Based on Chip-seq analysis centromeres are ∼40 kb long in chicken, corresponding to 200 nucleosomes per centromere. Of these, 30 are predicted to contain CENP-A (roughly 1 in 6�8 centromeric nucleosomes). Thus, centromeric chromatin is largely composed of nucleosomes containing histone H3. (B and C) The CENP-A chromatin was originally suggested to form an amphipathic organization, with CENP-A on the exterior facing the kinetochore, and H3 largely on the interior. This chromatin was proposed to form either a helix or loop structure. The diagram in (B and C) is based on Blower et al. (2002), but modified to show the lower occupancy of CENP-A nucleosomes in the centromeric chromatin. (D) The boustrophedon model of centromeric CENP-A-containing chromatin was proposed based on super-resolution microscopy (Ribeiro et al., 2010). To account for the super resolution mapping data, it was suggested that CENP-A chromatin might be organized as a sinusoidally folded patch, or boustrophedon, at the surface of the centromeric chromatin (Ribeiro et al., 2010). The boustrophedon was proposed to be 4�5 layers deep (Figure 5), consistent with the 10 nm diameter of the nucleosome and the ∼60 nm thickness of the kinetochore plate typically observed in electron micrographs (Rieder, 1982). Thus, the architecture of CENP-A chromatin and the boustrophedon model remains questions in need of further experimentation. Bottom figure; http://www.researchgate.in
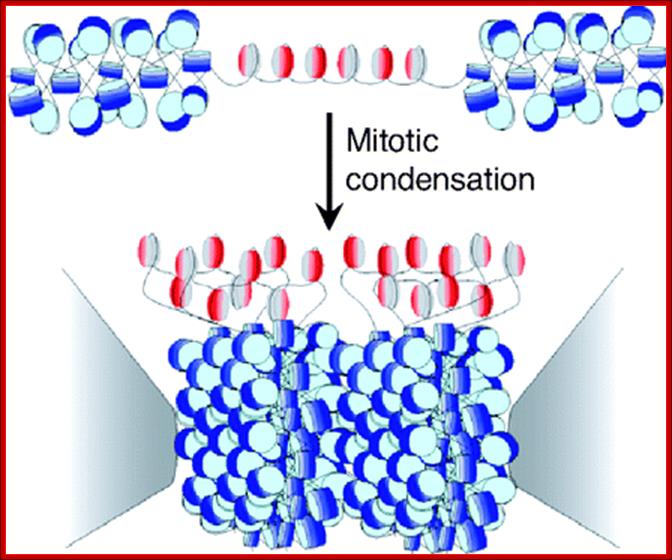
Nucleosomes at centromeric region; Structure, dynamics, and evolution of centromeric nucleosomes; Model for the kinetochore. CenH3 hemisomes (red/gray disks) are separated by extended linker DNAs and so are decondensed relative to surrounding heterochromatin (blue disks). Asymmetric CenH3 nucleosomes assemble in random orientations [CenH3/H4 (red) and H2A/H2B (gray)]. Only one unit of a CenH3-rich block is shown. During mitotic condensation, heterochromatin packs tightly as a result of its homogeneity. Intervening blocks of CenH3 chromatin cannot pack into this crystal-like structure because of its smaller size, long linkers, and heterogeneity in its relative orientation, resulting in extruded loops of uncondensed CenH3 nucleosomes that serve as the foundation for kinetochore formation. The flanking gray cones represent pericentric regions flanking the primary constriction. Yamini Dalal*,,Takehito Furuyama�, Danielle Vermaak* and Contributed by Steven Henikoff, August 13, 2007 (received for review July 27, 2007; http://www.pnas.org/content
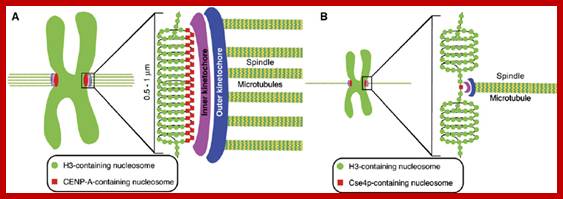
Centromeric Chromatin from Mammals to Budding Yeast Diagrams for the organization and mitotic kinetochore function of structurally distinct nucleosomes containing CENP-A found exclusively at active mammalian (A) and budding yeast (B) centromeres. https://www.researchgate
Model for the kinetochore: CenH3 hemisomes (red/gray disks) are separated by extended linker DNAs and so are decondensed relative to surrounding heterochromatin (blue disks). Asymmetric CenH3 nucleosomes assemble in random orientations [CenH3/H4 (red) and H2A/H2B (gray)]. Only one unit of a CenH3-rich block is shown. During mitotic condensation, heterochromatin packs tightly as a result of its homogeneity. Intervening blocks of CenH3 chromatin cannot pack into this crystal-like structure because of its smaller size, long linkers, and heterogeneity in its relative orientation, resulting in extruded loops of uncondensed CenH3 nucleosomes that serve as the foundation for kinetochore formation. The flanking gray cones represent pericentric regions flanking the primary constriction
Spindle check point:
During prometaphase, spindle-assembly-checkpoint proteins such as Mad2 and BubR1 are activated at kinetochores that are not (or not fully) attached with microtubules (indicated in green). Activated Mad2 and BubR1 inhibit the capability of anaphase promoting complex/cyclosome Cdc20 (APC/CCdc20) to ubiquitylate Securin and cyclin B and thereby prevent anaphase and mitotic exit. In metaphase, when all kinetochores are attached to microtubules, APC/CCdc20 ubiquitylate securin and cyclin B and thereby activates the protease separase and inactivates the cyclin-dependent kinase-1 (Cdk1). Separase then cleaves cohesin complexes (shown as red circles) that are holding sister chromatids together and thereby initiates sister-chromatid separation. Cdk1 inactivation leads to the dephosphorylation of Cdk1 substrates by protein phosphotases, and thereby enables exit from mitosis. In vertebrates, CDK1 inactivation also contributes to separase activation.
�
This anaphase promoting complex/cyclosome (APC/C) subunit has ubiquitin ligase (E3) activity and promotes the transfer of the ubiquitin residue from the E2 enzyme to the substrate protein on which the C terminus of ubiquitin forms a covalent isopeptide bond with a lysine residue. In subsequent reactions, the attached ubiquitin can itself become ubiquitylated, resulting in the formation of a polyubiquitin chains. All proteins that are known to be involved in the catalysis of ubiquitylation reactions are shown in orange. Substrates are recruited to the APC/C if they contain a D-box or a KEN-box. Both of these sequences are recognized by an APC/C co-activator, such as Cdh1 or Cdc20. Cdh1 binds to APC/C by interacting with two subunits, Cdc27 and Apc2. Cdc27 is one of several TPR proteins that are present in the APC/C (TPR domains are shown as vertical stripes), and Apc2 is a scaffold subunit that binds to Apc11 via a cullin domain. The small globular protein Doc1 is required for processive ubiquitylation of substrates and might also interact with the D-box of substrates, although direct evidence for such an interaction is lacking. The APC/C subunits that are implicated in substrate recognition are shown in yellow. The topology of subunits is based on biochemical data. Note that this model illustrates subunit interactions but does not represent a structural map of where subunits are located in the three-dimensional models that are shown in Fig. Apc9 is hatched because so far it has only been detected in budding yeast APC/C. Human APC/C also contains another TPR subunit, APC7, and human DOC1 interacts not only with APC2 but also with another, unidentified subunit (not shown here). Spore wall maturation. Modified from Ref. 30 � (2006) Cold Spring Harbor Laboratory Press.
Relationship-Centrosome duplication and Cell cycle: Nucleic acid research; volume 37 issue 19 2009.
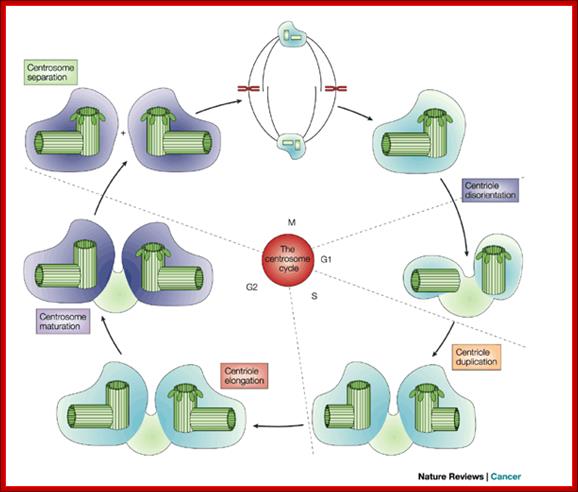
Erich A. Nigg; http://www.nature.com/
The centrosome duplication cycle can be subdivided
into several discrete steps (see figure). During mitosis, the centrosome at
each pole of the mitotic spindle contains a pair of centrioles. These two
centrioles usually display a conspicuous orthogonal orientation, indicating
that they are tightly connected. At the end of mitosis, this orthogonal
association is lost during a process that is referred to as centriole
disorientation. This step might relate to the final separation (abscission) of
the two incipient daughter cells95. In addition, it might be required
for the subsequent duplication step21, or for the re-establishment of a linker
structure between the two parental centrioles96. Centriole duplication then occurs
during S phase. At the morphological level, this event is characterized by the
formation of procentrioles at the proximal end of each parental centriole. So,
duplication is semi-conservative from the perspective of the whole centrosome,
but conservative from the perspective of the centriole97. How centriole duplication is brought
about remains a mystery, but the recent establishment of in vitro assays for centrosome duplication
might hopefully provide new opportunities for studying this fundamental problem.
Procentrioles then elongate until they reach their maximal length, but,
importantly, the two centriole doublets continue to function as a single
microtubule-organizing center until late G2. At the G2�M transition, centrosome
maturation occurs. This process involves the exchange of several PCM components
and culminates in the recruitment of additional ![]() -tubulin ring complexes � a prerequisite
for increased microtubule-nucleating activity. In response to the activation of
microtubule-dependent motor proteins, centrosomes then separate from each other
and instruct the formation of the two spindle poles. As a result, each
incipient daughter cell again inherits one centrosome. Erich A. Nigg; �http://www.nature.com/
-tubulin ring complexes � a prerequisite
for increased microtubule-nucleating activity. In response to the activation of
microtubule-dependent motor proteins, centrosomes then separate from each other
and instruct the formation of the two spindle poles. As a result, each
incipient daughter cell again inherits one centrosome. Erich A. Nigg; �http://www.nature.com/
The Centrosome Duplication Cycle; The centrosome consists of mother and daughter centrioles (green), that are interconnected by an intercentriolar linkage (red) and are embedded in the pericentriolar material (grey) which anchors the microtubules. The mother centriole can be distinguished by the presence of appendages (black lines). (a) During G1, cells lose their orthogonal arrangement. (b) As G1/S, a procentriole (blue) forms perpendicular to each centriole. (c) During S phase, the new centrioles elongate. (d) At G2, the two newly formed centriole pairs disconnect, and (e) by G2/M, the PCM is also divided between the centrioles. (f) At the end of the cycle, the daughter centrioles acquire appendages and behave as a mother centriole during the subsequent cycle.
The centrosome (centriole) duplication cycle in animal cells. At the end of mitosis, each daughter cell inherits only one centrosome. In the following cell cycle, the centrosome must duplicate, which is initiated at late G1/early S phase of the cell cycle by the splitting of the centriole pair. This is followed by the formation of daughter centrioles (procentrioles) in the vicinity of each preexisting centriole (shown in red). The procentrioles continue to elongate during S and G2; http://www.nature.com
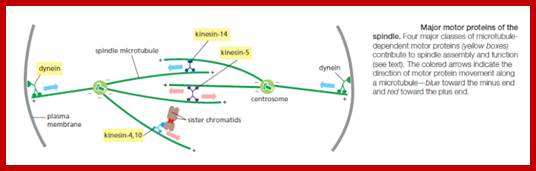
The astral microtubules, which elongate in all directions from the spindle poles during anaphase, are thought to play an additional role in anaphase B movement. In both fungi and animal spindles, severing the central spindle does not block the separation of the spindle poles but in fact accelerates it. This suggests that the astral microtubules that point away from the spindle generate a pulling force that assists the separation of poles during anaphase B, possibly by interacting with minus-end-directed motor proteins attached to the cell cortex or other cytoplasmic structures (Figure18-30B). Consistent with this suggestion, the injection of antibodies against the minus-end-directed motor protein dynein causes the spindle in cultured cells to collapse. This pulling force is believed to operate also during spindle assembly in prophase, and similar forces are thought to guide the specific positioning of spindles prior to asymmetric cell cleavages, as we discuss later. http://reasonandscience.heavenforum.org; http://reasonandscience.heavenforum.org
Chromosome movement in anaphase- A depends on a combination of
the two major poleward forces described earlier. The first is the force
generated by microtubule depolymerization at the kinetochore, which results in
the loss of tubulin subunits at the plus end as the kinetochore moves toward
the pole. The second is provided by microtubule flux, which is the poleward
movement of the microtubules toward the spindle pole, where minus-end
depolymerization occurs. The relative importance of these two forces during
anaphase varies in different cell types: in embryonic cells, chromosome
movement depends mainly on microtubule flux, for example, whereas movement in yeast
and vertebrate somatic cells results primarily from forces generated at the kinetochore.
Spindle-pole separation during anaphase B depends on motor driven mechanisms similar
to those that separate the two centrosomes in early mitosis. Plus, end directed
kinesin-5 motor proteins, which cross-link the overlapping plus ends of the
interpolar microtubules, push the poles apart. In addition, dynein motors that anchor astral microtubule plus
ends to the cell cortex pull the poles apart.
Although sister-chromatid separation initiates the chromosome movements of
anaphase A, other mechanisms also ensure correct chromosome movements in
anaphase A and spindle elongation in anaphase B. Most importantly, the
completion of a normal anaphase depends on the dephosphorylation of Cdk
substrates, which in most cells results from the APC/C-dependent destruction of
cyclins. If M-cyclin destruction is prevented�by the production of a mutant
form that is not recognized by the APC/C, for example�sister-chromatid separation generally occurs, but the chromosome
movements and microtubule behavior of anaphase are abnormal. The relative
contributions of anaphase A and anaphase B to chromosome segregation vary
greatly, depending on the cell type. In mammalian cells, anaphase B
begins shortly after anaphase A and stops when the spindle is about twice its
metaphase length; in contrast, the spindles of yeasts and certain protozoa
primarily use anaphase B to separate the chromosomes at anaphase, and their
spindles elongate to up to 15 times their metaphase length. http://reasonandscience.heavenforum.org.
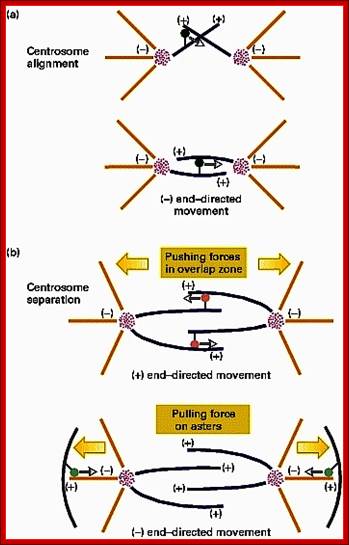
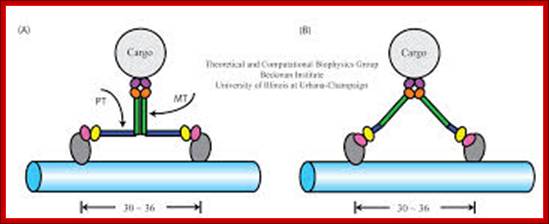
Alternative geometries suggested for myosin VI stepping. (A) Model assumes associated MT domains and extended PT domains. (B) Model assumes dissociated MT domains and compact PT domains. http://www.ks.uiuc.edu/
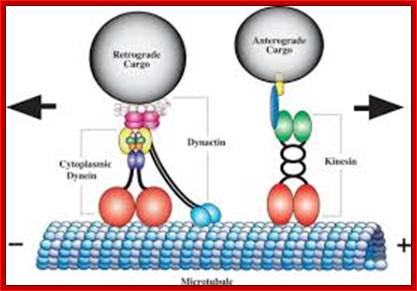
Duncan JE, Goldstein LS - PLoS Genet. (2006); http://openi.nlm.nih.gov/
Polarity of MTs + and � ends; MTs are associated with motor proteins that makes the MTs move on each other.� The � end of MTs is where MTs dissociate into monomers; the + ends are the sites at which monomeric Tubulins are added.
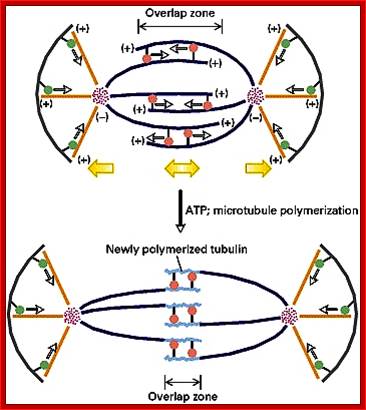
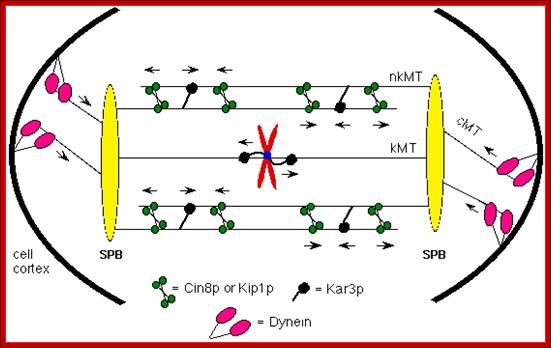
�Proposed roles for four mitotic spindle motor protein;. BimC-like Cin8p and Kip1p overlap in spindle assembling and elongating activities. They act by cross-linking and sliding midzone anti-parallel microtubules. Prior to the onset of anaphase, Cin8p an Kip1p are opposed by the actions of kinesin-related Kar3p, although the mechanism of this motor is not clear. (Two mechanisms are suggested here. There exists evidence to support both.) Dynein is proposed to contribute to pole separation from outside the poles at the cell cortex. Arrows indicate motor movement relative to microtubule substrate. SPB = Spindle Pole Body; cMT, kMT, and nkMT = cytoplasmic, kinetochore, and non-kinetochore microtubules�. M.Andrew Hoyt, Biology; http://www.bio.jhu.edu/

Highly dynamic, the mitotic spindle is composed of microtubules and associated proteins, which work together to form the mechanical framework needed for cellular division. During prophase, the important mitotic apparatus begins to form outside the nucleus at opposite ends of a cell, stretching from pole to pole. In metaphase, the mitotic spindle attaches to the centromere of each chromosome, moving them through the mitotic process. The chromatids of each chromosome are then pulled apart in anaphase and the spindle fibers disperse in telophase. Thomas J. Deerlick; http://www.microscopyu.com/
Mitotic apparatus with chromosomes at metaphasic plate. https://embryology.med.unsw.edu.au/ /Cell_Division
Cleavage and furrow formation; http://jcs.biologists.org
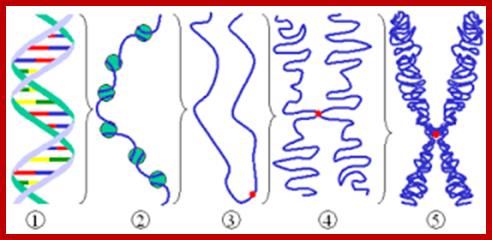
From Nucleosome 11nm to 30nm chromonemal thread; www.ck12.org
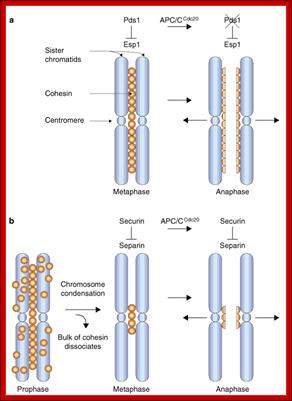
APC activates degradation of Cohesins; Control of sister-chromatid cohesion in yeast and vertebrates; Angelika Amon; http://www.nature.com/
a, In budding yeast, sister chromatids (blue) are held together by Cohesin complexes (orange) at metaphase. Degradation of Pds1 by APC/C Cdc20-dependent proteolysis releases Esp1, allowing it to cleave Scc1/Mcd1 and leading to the initiation of anaphase. b, In vertebrates, the bulk of Cohesin dissociates from chromosomes during prophase, perhaps as a result of chromosome condensation5, 8. A small amount of Cohesin remains on chromosomes, predominantly around centromeres. This pool of Cohesin is cleaved by separin at the metaphase�anaphase transition. Activation of separin at this cell-cycle transition is brought about by destruction of Securin by APC/C Cdc20. Angelika Amon.
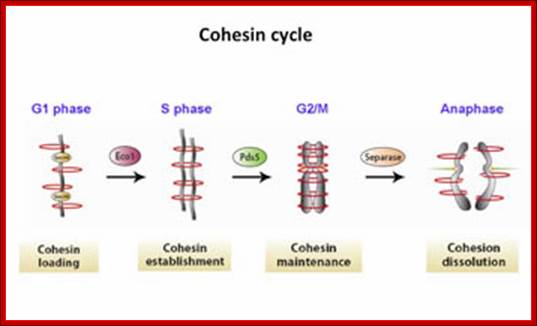
�������������������������� Dr. Jennifer Gerton; http://www.stowers.org/�
Molecular addresses of cohesion. Cohesin mutations cause human disorders known as cohesion-pathies, such as Cornelia de Lange syndrome, named for the doctor who discovered it.
Nuclear envelope vesicles, generated by the breakdown of the envelope during prophase, associate with decondensing chromosomes and then fuse. Sub pore complexes reassemble into nuclear pores, forming individual mininuclei called karyomeres. The enclosed chromosome further decondenses, and subsequent fusion of the nuclear envelopes of all the karyomeres at each spindle pole forms a single nucleus containing a full set of chromosomes. Reassembly of the nuclear lamina is not shown. [See G. P. Vigers and M. J. Lohka, 1991, J. Cell Biol. 112:545; adapted from A. Murray and T. Hunt, 1993, The Cell Cycle: An Introduction,W. H. Freeman and Company.]
Schematically depicts reassembly of the nuclear envelope, which occurs late in mitosis. Vesicles derived from the breakdown of the nuclear envelope during prophase associate with the surface of the decondensing chromosomes during telophase. These vesicles fuse to form continuous double membranes around each chromosome. Nuclear pore complexes, which disassemble into sub pore complexes during prophase, reassemble into the nuclear membrane around each chromosome, forming individual mininuclei called karyomeres. Subsequent fusion of the karyomeres associated with each spindle pole generates the two daughter-cell nuclei, each one containing a full set of chromosomes. Lamins A and C appear to be imported through the reassembled nuclear pore complexes during this period and reassemble into a new nuclear lamina. Reassembly of the nuclear lamina in the daughter nuclei probably is initiated on lamin B molecules, which remain associated with the nuclear-envelope vesicles throughout mitosis via the isoprenyl anchors covalently linked to the C-terminal region of lam
Assembly of Nuclear Pore Complex (NPC):
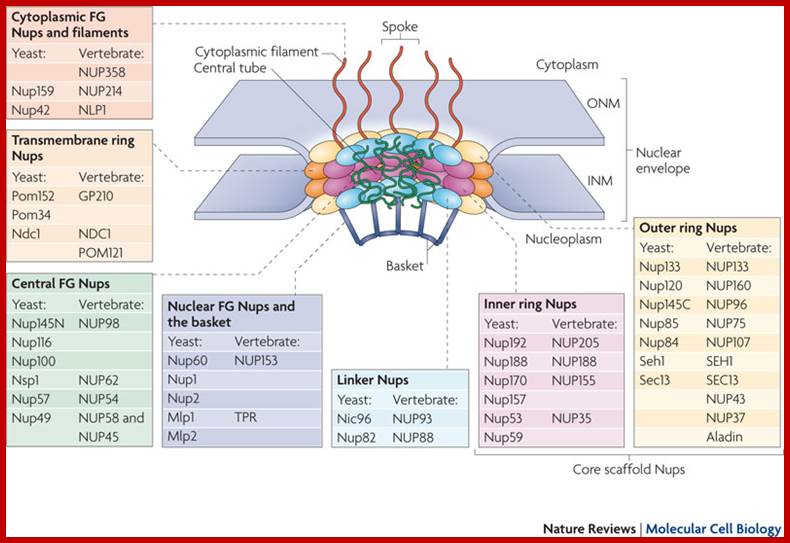
The nuclear pore complex: bridging nuclear transport and gene regulation; Each nuclear pore complex (NPC);� It is a cylindrical structure comprised of eight spokes surrounding a central tube that connects the nucleoplasm and cytoplasm. The outer and inner nuclear membranes (ONM and INM, respectively) of the nuclear envelope join to form grommets in which the NPC sits. The NPC is anchored to the nuclear envelope by a transmembrane ring structure that connects to the core scaffold and comprises inner ring and outer ring elements. Linker nucleoporins (Nups) help anchor the Phe-Gly (FG) Nups such that they line and fill the central tube. NPC-associated peripheral structures consist of cytoplasmic filaments, the basket and a distal ring. The Nups that are known to constitute each NPC substructure are listed, with yeast and vertebrate homologues indicated. Both inner and outer ring Nups are known to form biochemically stable NPC subcomplexes, which are thought to have a role in NPC biogenesis and nuclear envelope assembly. GP210, glycoprotein 210; Mlp, myosin-like protein; Ndc1, nuclear division cycle protein 1; Nic96, Nup-interacting component of 76 kDa; NLP1, Nup-like protein 1; Pom, pore membrane protein; Seh1, SEC13 homologue 1; TPR, translocated promoter region.� Nature Reviews Molecular Cell Biology
; Caterina Strambio-De-Castillia, Mario Niepel & Michael P. Rout; http://www.nature.com/
The nuclear
envelope is studded with pores or porters which allow regulated transport of
macromolecules into and out of the nucleus. Nuclear pores form through the
double membrane of the nuclear envelope, and NP complexes are assembled at the
site of the pore. Higher eukaryotes undergo an "open mitosis," in
which the nuclear envelope and NPCs break down during mitosis and later
reassemble. Yeast, however, undergo a "closed mitosis" in which the
nuclear envelope and NPCs remain intact during cell division. In each case,
however, new pores and pore complexes must continually form in the nuclear
envelope to maintain a consistent number of NPCs throughout the cell cycle. The
yeast Saccharomyces
cerevisiae NPC is composed of multiple copies of ~30 proteins.
Because of this complexity, NPC assembly is predicted to be a highly ordered
process. However, basic questions regarding membrane fusion, nucleation of
NPCs, and the ordered addition of factors or substructures have not been
answered.
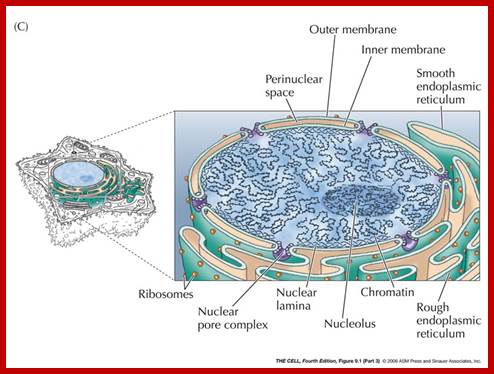
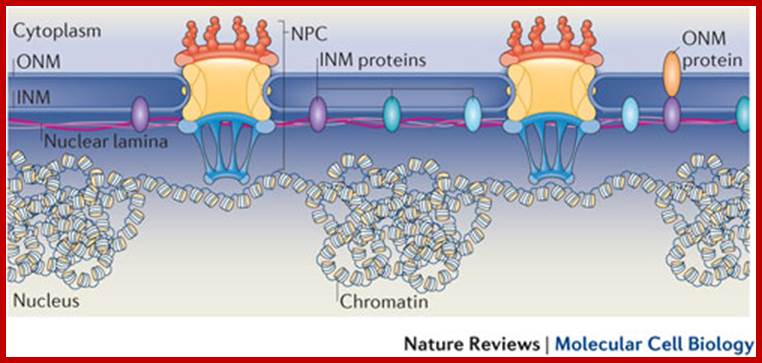
Since its first characterization3, our view of the nuclear envelope as a simplistic physical barrier that protects the genetic material from damage by the cytoskeleton has dramatically changed to the nuclear envelope being a highly elaborate, dynamic structure with key functions in gene expression regulation, genome organization, cell differentiation and disease. The nuclear envelope is composed of two concentric membranes: the outer nuclear membrane (ONM), which is contiguous with the endoplasmic reticulum; and the inner nuclear membrane (INM), which surrounds the nuclear interior. The INM and ONM associate at sites of nuclear pore complex (NPC) insertion (see the figure). The INM is decorated with specific transmembrane proteins that interact with the nuclear lamina and chromatin underneath the nuclear envelope. Some of these proteins also associate with ONM proteins to link the cytoskeleton with the nuclear lamina. To date, a large number of INM proteins have been identified by proteomic approaches, and many of these proteins show tissue-specific expression, suggesting a heterogeneity of the nuclear envelope composition in different cell types. Interestingly, a growing number of INM proteins are being linked to human diseases, which points to the nuclear envelope as a key regulator of cellular function. Consistent with their differential expression, most diseases associated with these proteins show tissue-specific phenotypes; http://www.nature.com
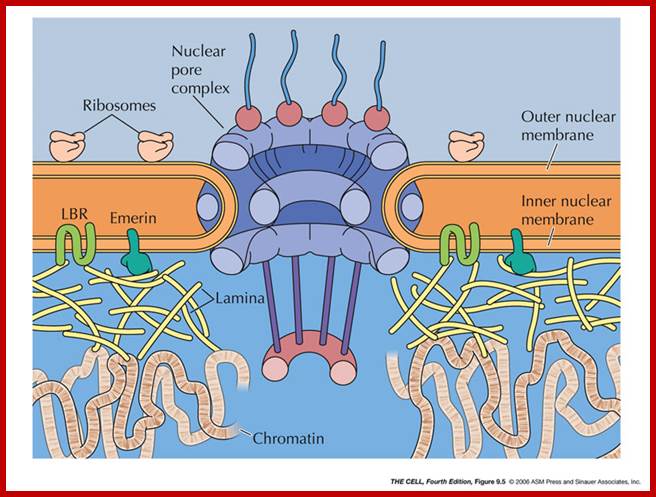
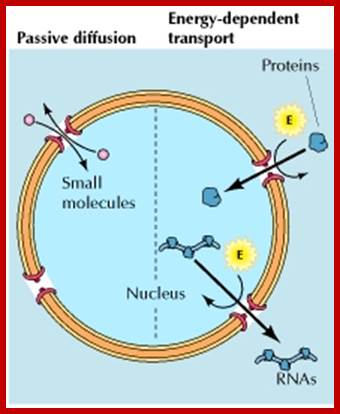
Molecular traffic through nuclear pore complexes: Small molecules are able to pass rapidly through open channels in the nuclear pore complex by passive diffusion. In contrast, macromolecules are transported by a selective, energy-dependent mechanism that acts predominantly to import proteins to the nucleus and export RNAs to the cytoplasm. https://www.ncbi.nlm.nih.gov
 �
�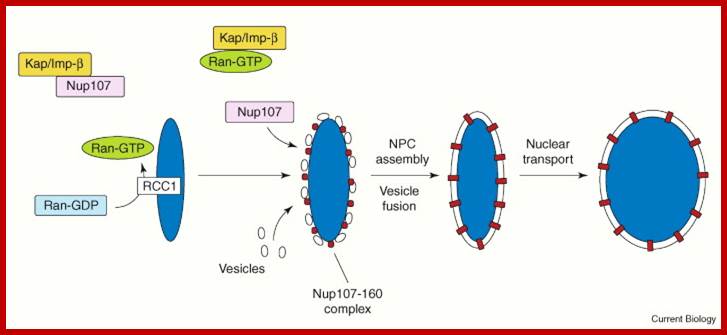
Nuclear envelope (NE) and nuclear pore complex (NPC) assembly.
In Xenopus egg extracts, Ran GTPase and its guanine nucleotide exchange factor RCC1 bind to chromatin. Local generation of Ran�GTP releases nucleoporins such as Nup107, Nup153 and Nup358 [6] from inhibitory complexes with karyopherin-β1/importin-β(Kap/Imp-β), allowing the assembly of the subcomplexes such as the Nup107�Nup160 complex which associates with chromatin and seeds NPC assembly. Membrane vesicles recruited by Ran-GTP through a distinct pathway fuse to form the double membrane that constitutes the NE. Vesicle fusion is also regulated by karyopherin-β1/importin-β; http://www.sciencedirect.com
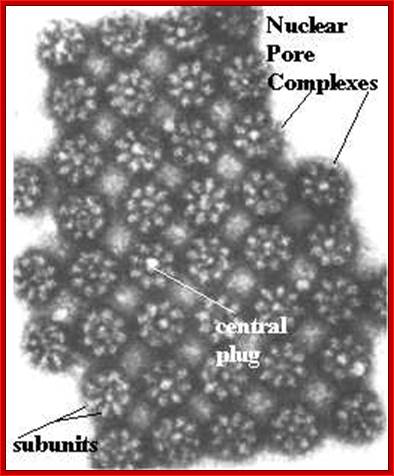
http://legiblyloco.blogspot.in/
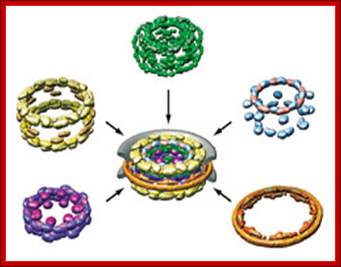
Different structural components of nuclear pore complex
Assembly Required Protein-based substructures-outer rings (yellow), inner rings (purple), membrane rings (brown), linker nucleoporins (green), and FG (phenylalanine-glycine) nucleoporins-make up the nuclear pore complex (center), analyzed structurally in better than ever detail this year. Pore membrane is grey.
Nuclear envelope dynamics in the C. elegans embryo; Figure courtesy of Vincent Gaily. (A) Schematics illustrate the cycle of nuclear envelope breakdown and reassembly. (B) Still images of the first mitotic division of wild-type embryos expressing YFP-Lamin (left) and GFP-MAN1 (right). Times on the right are relative to first metaphase to anaphase transition. Arrowheads indicate the reappearance of GFP-MAN1 around the chromatin at t=60 sec and YFP-Lamin at t=120 sec. White stars mark the positions of the centrosomes. Note the persistence of membranes containing GFP-MAN1 around the mitotic spindle and centrosomes. Scale bar = 10μm
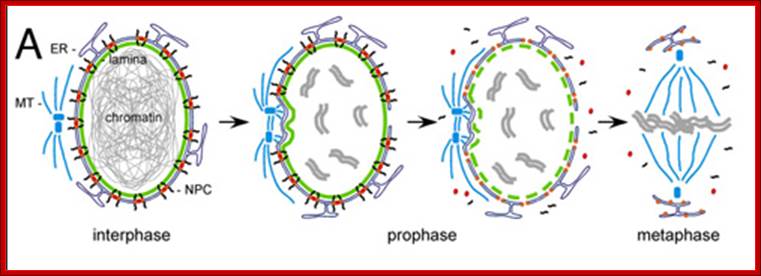
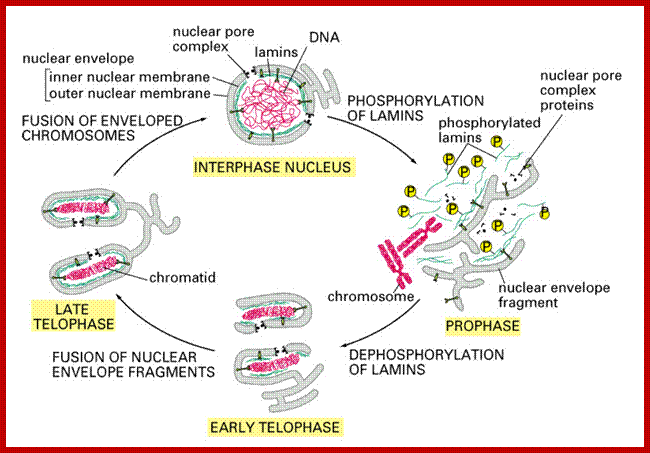
Cell Biol 1 Study guide; https://www.studyblue.com
Nuclear envelope dynamics in the C. elegans embryo:
Once the chromosomes have separated, the lamina begins to form again. Sometime at the beginning of mitosis, ends of the endoplasmic reticulum bound to the DNA, using lamin A receptor. (Duband-Goulet and Courvalin) Once the lamina begins to re-form, it forces tubules of ER to form a network across the surface of the chromatid. (Anderson and Hetzer) These tubes eventually are flattened out and merged, forming a solid nuclear membrane. It is not certain what mechanisms cause this to happen.
The ER and the LBR eventually detach themselves from the DNA. In a mature cell, the lamina forms a continuous layer just inside the nuclear envelope, and is attached to the nucleus by protein known as emerin.
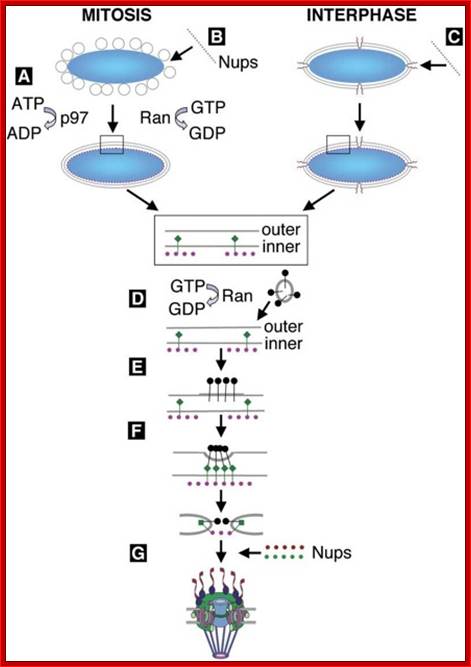
Adapted from Suntharalingam and Wente, 2003
After nine years of intense effort, a collaborative research team has succeeded in dissecting the structure of the nuclear pore complex (NPC), an assembly of 456 proteins that controls the flow of molecules between the DNA-storing nucleus and the rest of the cell. This figure highlights 30 different types of proteins found in the donut-shaped complex from yeast. The work�led by Andrej Sali of the University of California, San Francisco, and Michael P. Rout and Brian Chait of Rockefeller University�may shed light on the function and evolution of the NPC and other large protein assemblies. Video featured by permission from Macmillan Publishers Ltd: Nature 450:695-701, 2007.
Cytokinesis:
Microtubule organizing centers (aka centrosomes) are composed of asters at each end, with centrosomes spanning between them. The tubules that connect to the chromosome kinetochore are called kinetochore microtubules, while the tubules which interact with each other are polar microtubules (aka non-kinetochore microtubules). Microtubules are composed of α- and β-tubulin monomers polymerized to form hollow tubes. Depolymerization shortens the microtubules, while the chromosomes simultaneously migrate toward the asters, driving apart the spindle poles.
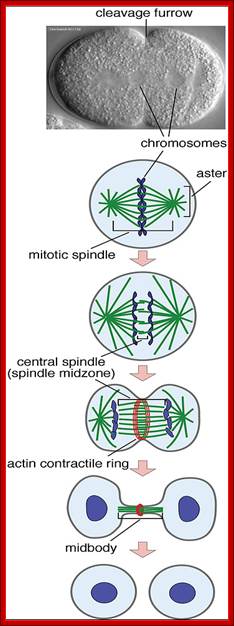
Cytokinesis by cleavage; Eukaryotic cytokinesis requires actomyosin-based contraction at the cleavage furrow to physically divide a mother cell into two daughter cells. The determination of the plane and position of the cleavage furrow during cytokinesis requires communication between microtubules and the actin cortex, which results in the assembly of F-actin (the filamentous form of actin) and non-muscle Myosin II (NM II) into contractile ring. The contractile ring is a highly dynamic structure with a rapid turnover of both F-actin and NM II. NM II is the major motor protein in cytokinesis and its movement along F-actin and F-actin depolymerization are required for furrow ingression. Ingression of the cleavage furrow proceeds by actomyosin-mediated force generation, but the underlying mechanism of actomyosin contraction remains largely unknown. They demonstrated that dioxygenase ALKBH4-mediated demethylation of a novel mono-methylated site in actin (K84me1) regulates actin-myosin interaction and actomyosin dependent processes including cytokinesis and cell migration. http://english.big.cas.cn/ ; http://www.nature.com���

How the cell membrane is pulled inwards by contractile proteins at the equatorial region of the cell. http://www.quia.com/
Plant cell cytokinesis:
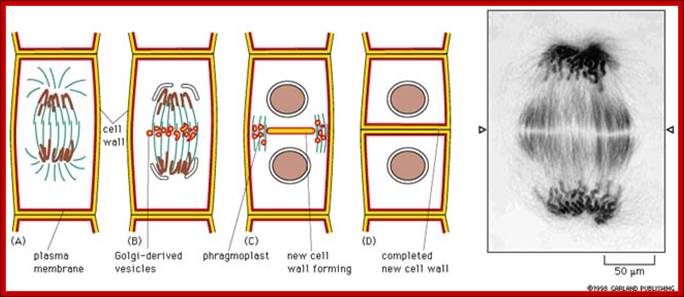 http://celldivisionandreproduction.weebly.com/
http://celldivisionandreproduction.weebly.com/
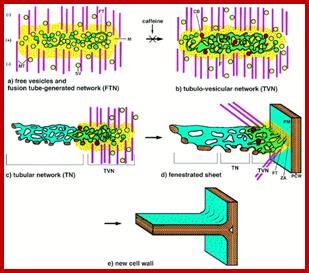
http://homeinsurancequotations.com/
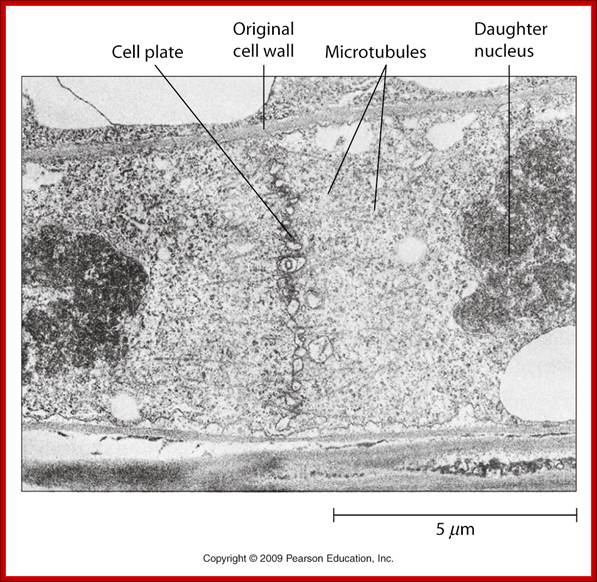
http://celldivisionandreproduction.weebly.com/;http://www.quia.com/
Comparison of cell cycle cyclin/Cdks of yeast and humans:
An obvious advantage of proteolysis for controlling passage through these critical points in the cell cycle is that protein degradation is an irreversible process, ensuring that cells proceed irreversibly in one direction through the cycle. http://studentreader.com/
A brief Review of Plant cell cycle: http://plantphys.info/
In this simplified model, the inactive CDK is shown at about 1 O'clock position as green with an empty binding site. This inactive CDK is produced at G1. As the cytokinesis is complete, the specific cyclin ( for DNA synthesis) is also made. Their concentration also rises; and cyclin binds to CDK. The combination alters the conformation of the CDK so that an active site is formed for a secondary messenger. This messenger protein binds and gets phosphorylated by ATP dependent kinase. The cell is now competent to go from G1 to S.
The cell begins replicating the DNA. As the DNA is synthesized, a cyclin ubiquitinating protein marks the s-cyclin for destruction and the 26S proteasome destroys it. This prevents the cell from attempting to duplicate its DNA again before the cell cycle is completely back to G1. The inactive cyclin dependent kinase is free for the next control point.
A new cyclin, the mitotic cyclin, is produced as the S-phase ends. This binds to CDK, altering its conformation to make an active site for a different messenger. The messenger is produced at G2 and binds to the active site, and gets phosphorylated to signal the cell to make the transition from G2 to M phase.
Later, during M-phase, the Anaphase Promoting Complex (APC) is produced and it marks the mitotic cyclins by ubiquitination. Then the 26S proteasome digests the marked mitotic-cyclins. This of course prevents the cell from going into mitosis without completing the cycle through to G2. The APC also includes enzymes that degrade the cohesion (cohesins) and condensation (cohesins) proteins permitting the separation of the sister chromatids at the centromere.
�
These points of control in the cell cycle can be stimulated or inhibited by plant growth substances (hormones). Auxins and cytokinins supplied with adequate sucrose stimulate the production of the synthesis cyclin, cytokinins stimulate the mitotic cyclin. Abscisic acid interferes with the synthesis CDK-cyclin complex, preventing the cell from cycling. These "effectors" can be manipulated by scientists to grow tissues in vitro, they become ingredients in the recipes for tissue culture. Such as those we use in our tissue culture project in lab.
�
To conclude, the observation that the cytoskeleton components are conserved is also reflected in the control elements for the cell cycle. In spite of the fact that the common ancestor of plants and humans is perhaps 1 billion years (exaggerated) in the past, the control elements in the cell cycle are shared. The protein kinases, cyclins, DNA replication enzymes, ubiquitin-tagging, proteosomes, and all of the substantive details of the mitosis event are shared. Thus, what we have only recently learned is in fact the product of some very ancient evolutionary events. Cell cycling is a fundamentally ancient process since eukaryotes developed.
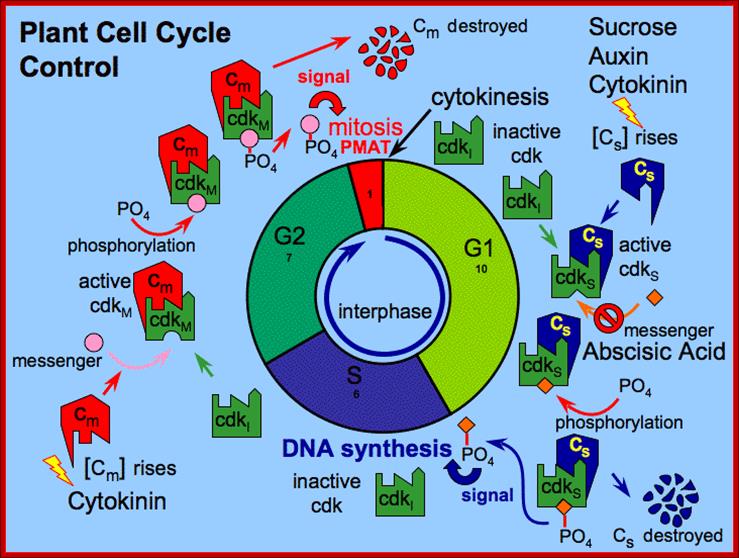
Cells use Cyclin-Dependent-Protein-Kinases (CDKs) to regulate the cell cycle; Ross Koning; http://plantphys.info/http://www. blc.arizona.edu/courses
One protein that is involved in cell cycle control is the CDK. This CDK protein, by itself, is useless but after combining with another protein called a cyclin and ATP, it can permit a cell to pass on to the next part of the cell cycle.
The cyclins are relatively labile proteins, and their availability is regulated in turn by other cell cycle control proteins. These cell cycle control proteins add a ubiquitin polymer to the cyclin, marking them for destruction by the 26S proteasome, a complex of proteases. By destroying the cyclins, the CDK is inactivated.
The CDK-cyclin complex also must combine with ATP to be active. This combination is accomplished by yet other cell-cycle control protein, a protein kinase. Protein kinases add a phosphate from ATP to a protein and phosphotases remove the phosphate from a protein. The relative balance of protein kinases to phosphotases also helps to determine whether the CDK-cyclin complex is active or inactive.
There are different kinds of protein kinases at work in the cell cycle. Some put the phosphate into one threonine of the CDK, other protein kinases put the phosphate on a different threonine of the CDK. Which threonine gets the phosphate determines whether the CDK-cyclin complex is active or inactive.
Now that we understand how a CDK can be active or inactive, we can go back to our cell cycle control figure and understand the rest of it.
New cells are made by a multistep process:
Cells that divide actively in plants are called meristematic. These cells divide repeatedly in what has been called the cell cycle. This process has two major parts: a long and rather interesting phase (called interphase) marked by typical biochemistry, and a short and quite dynamic phase (called mitosis) marked by nuclear changes and cellular changes culminating in cytokinesis.
The "typical" cell diagrams show cells in interphase. The nuclear envelope is intact, the chromatin is not condensed into chromosomes, but there may be one or more nucleoli visible in the nucleus. The usual biochemistry of cells occurs in this interphase. What might not be easily noticeable is that during the interphase the DNA in the nucleus. The replication process divides the interphase into three distinct time intervals: G1, S, and G2.
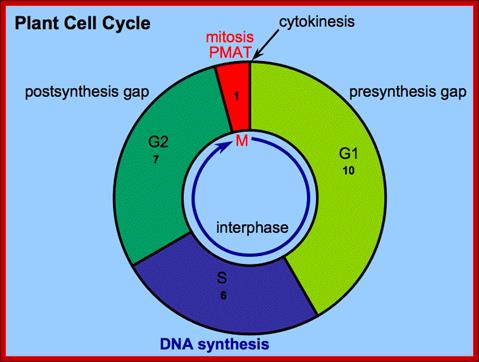
����������� ����������� ����������� Cell Cycle; http://plantphys.info/
During the first Gap (G1) the cell is doing its usual biochemistry, but this also includes preparing materials needed for the S phase. In the synthesis (S) interval, the cell is replicating its DNA. To accomplish that, proteins locate origins of replication in the genome. These occur at 66kb intervals on dicot and 47kb intervals on monocot chromosomes. DNA polymerase and ligase enzymes replicate the chromosomes.
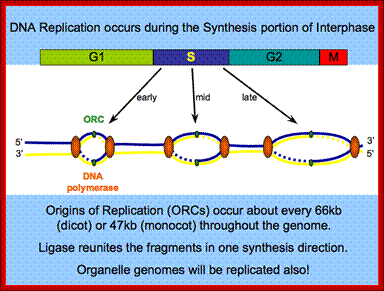
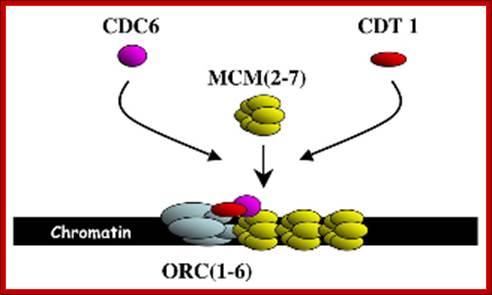
Plant Replication Group; http://bonaire.cshl.edu/
In the second Gap (G2), the cell returns to its usual biochemistry, but this also includes multiplication of the chloroplasts and mitochondria (by binary fission!) and production of components needed for mitosis (a nuclear! event).
The mitosis itself involves the condensation and separation of the replicated chromosomes. Mitosis has been subdivided into the phases: prophase (condensation), metaphase (alignment and attachment to microtubules), anaphase (centromere separation and chromatid migration), and telophase (recovery of nuclear envelope and decondensation).