Co and Post Translational Events
Precursor & Polyproteins
A majority of secretary and membrane proteins are synthesized longer than their final functional proteins, such proteins are called precursor proteins. They are also named as pre-pro or pre-proteins depending upon their N-terminal segments.
Proteins are synthesized in cytoplasm and their destination depends on the kind of protein synthesized.� Majority of the extracellular and membrane proteins start their synthesis in free cytoplasm as polysome and then they are threaded through ER using signal receptor protein (SRP) mediated process (it is delt in the chapter protein synthesis).� Some are synthesized in free-state and they are released into cytoplasm.� The other proteins that are destined to the nucleus, peroxisomes, mitochondria and chloroplast and lysosomes are synthesized on free polysomes and the released proteins are directed and transported to their respective destination by specific translocator proteins.� Almost all proteins destined to specific loci are synthesized as precursor proteins; by the time they reach their destination they are processed (cut and modified).
Pre-pro protein:
For example insulin is synthesized as 104 amino acids long pre-pro insulin polypeptide.� While it is synthesized it is transported as pre-pro protein into ER lumen using ER translocons. During translocation signal peptide sequences are removed.� This precursor moves from ER to Golgi complex where it is packed into proinsulin complex into vesicles.
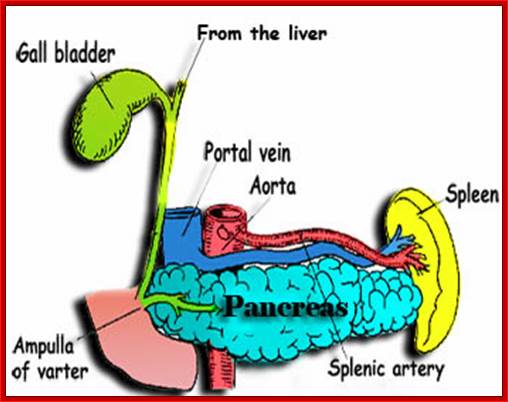
Pancreas with associated structural organs; http://www.sedico.net/
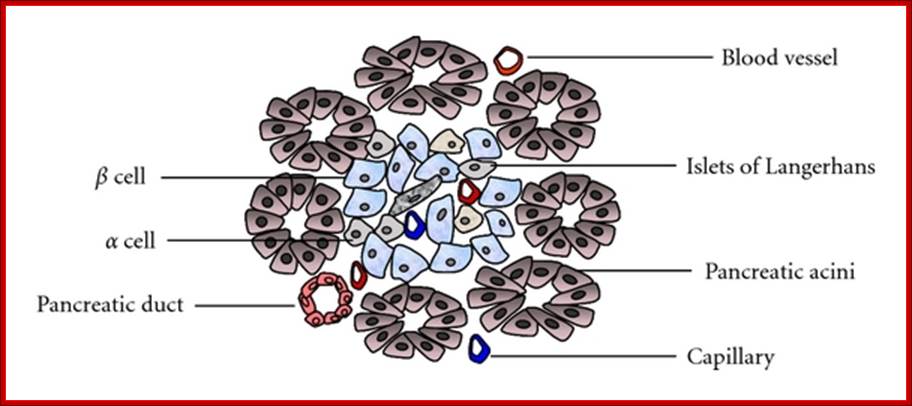
Structure of the pancreas; Pancreas tissue consists of both exocrine and endocrine glands. Islets of Langerhans mainly include α-, β-, and δ-cells. The islets are arranged in clusters associated with a dense network of capillaries. Most islet cells are directly in contact with blood vessels. http://www.hindawi.com/
The pancreas is a gland organ in the digestive and endocrine system of vertebrates. It is both exocrine (secreting pancreatic juice containing digestive enzymes) and endocrine (producing several important hormones, including insulin, glucagon, and somatostatin). It also produces digestive enzymes that pass into the small intestine. These enzymes help in the further breakdown of the carbohydrates, protein, and fat in the chyme www.sedico.net/ erhans, Alpha and beta cell
�����������������������������������
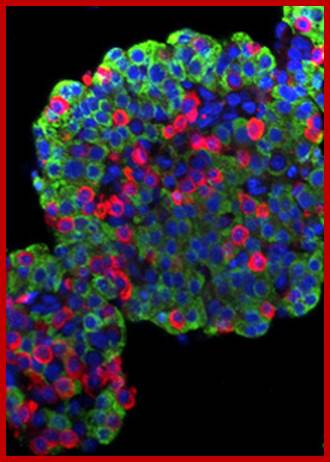
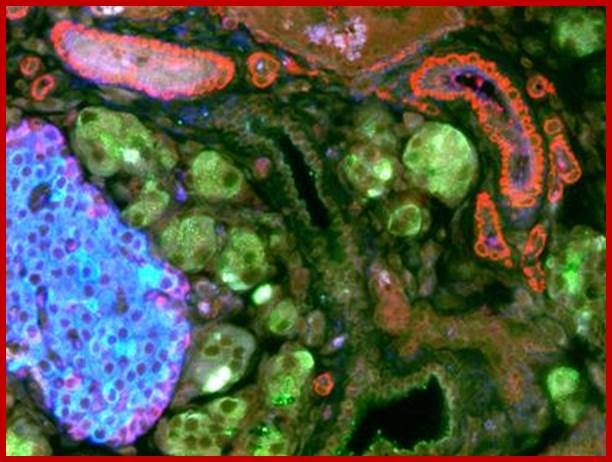
Islets of Langerhans are made of four different cell types. The majority are insulin-producing beta cells (shown in green) and glucagon-producing alpha cells (shown in red). The blue color is a marker used to locate the nucleus of the cells. Using Laser Scanning Cytometry (LSC), DRI scientists can study the physiology of the basic islet components to assess their viability before transplantation. http://www.diabetesresearch.org/
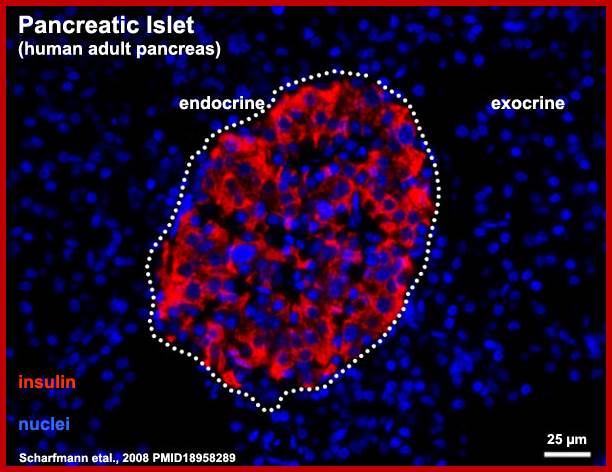
Human-Pancreatic Islets; http://embryology.med.unsw.edu.au/
�
During transportation, its N-end signal sequence is cleaved and removed. This precursor protein is packed as pro-insulin in Golgi complex into vesicles.
�
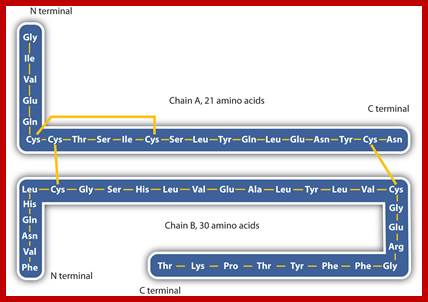
The pro-insulin is 84 amino acids long.� This is further cleaved in the vesicles, before it is released as compact two fragmented protein; A 21 aa and B 30aa, which are cross linked by two sulphydril bonds and one intra chain s-s bond within the A chain.� This is a functional protein. The functional proteins, if glycosylated, they will be effective, but often insulin in vivo and in vitro (synthetic insulin called Humulin) gets aggregated and become ineffective. Substantial number of proteins are synthesized like pre pro proteins and processed (cut and modified) as functional proteins. Insulin stimulates muscle and adipose cells to uptake glucose.

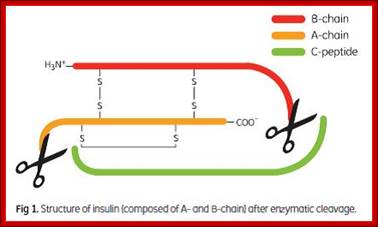
Insulin composed of A and B chains after cleavage; recombinant insulin from proinsulin; http://image.3sir.net/
����������� Signal B-chain��������� ������� dispensable������ �� A-chain
NH3---24aa----I---------------------I--------------------I-------------ICOO^-
����������������������������������������������� 30aa��������������� 33aa��� ��� 21aa
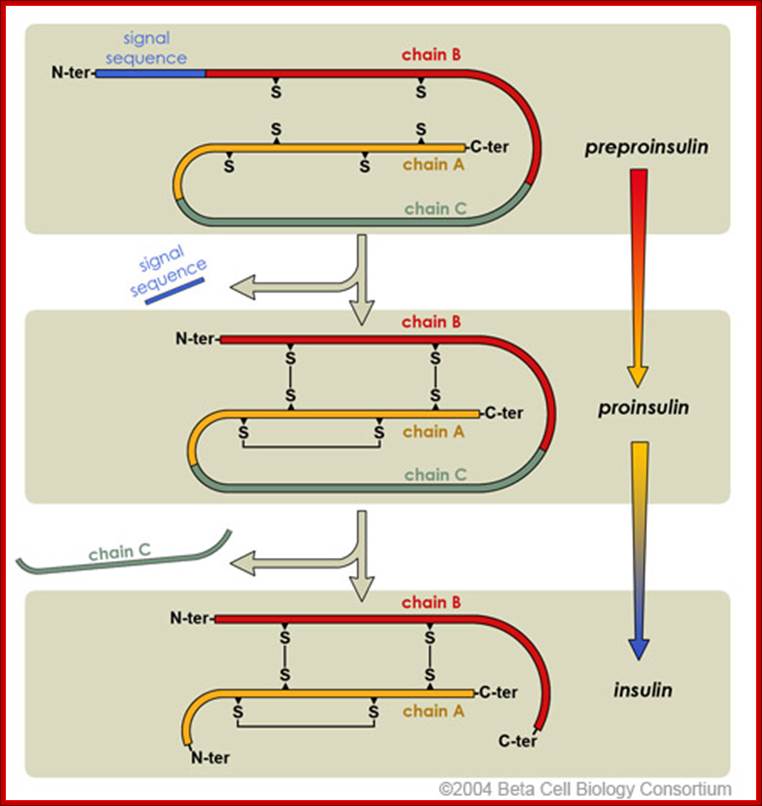
Insulin Synthesis
Insulin production involved intermediate steps.
Initially, preproinsulin is the inactive that is secreted into the endoplasmic
reticulum. Post-translational processing of proinsulin clips the N-terminal
signal sequence and forms the disulfide bridges. Lastly, the polypeptide is
clipped at two positions to release the intervening chain C. This and active
insulin are finally packaged into secretory granules for storage.
http://www.betacell.org/
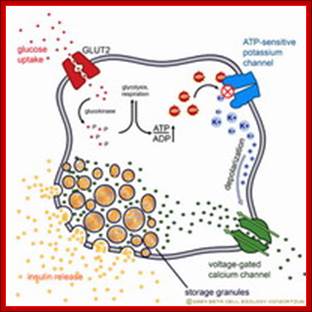
Insulin
secretion
Insulin secretion in beta cells is triggered by rising blood glucose levels.
Starting with the uptake of glucose by the GLUT2 transporter, the glycolytic
phosphorylation of glucose causes a rise in the ATP:ADP ratio. This rise
inactivates the potassium channel that depolarizes the membrane, causing the
calcium channel to open up allowing calcium ions to flow inward. The ensuing
rise in levels of calcium leads to the exocytotic release of insulin from their
storage granule. http://www.betacell.org/
http://saylordotorg.github.io
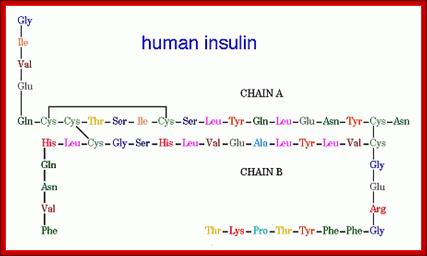
Insulin (Latin insula, "island", as it is produced in the Islets of Langerhans in the pancreas) is a polypeptide hormone that regulates carbohydrate metabolism. http://www.bio.davidson.edu/ Human Insulin; The amino acid diagram of human insulin, showing the A and B chains and 3 disulfide bonds. Permission pending, http://www.chem.uwec.edu/Chem406/Webpages/Ying/overview.htm;

3D model of Insulin-after folding; http://www.helsinki.fi; http://saylordotorg.github.io
Insulin �chemical structure
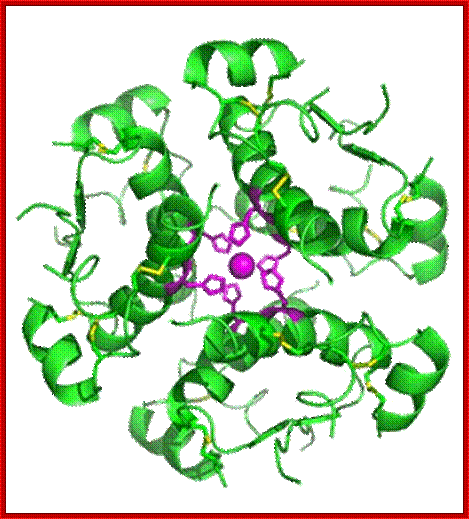
3D structure-Hexamer ; https://en.wikipedia.org
![[Insulin.jpg]](Co_and_Post_Translational_Events1-Precursor_&_Polyproteins_files/image014.jpg)
Diabetes
and abnormalities in glucose-stimulated insulin secretion; Glucose and other
nutrients regulate insulin secretion by the pancreatic beta cell. Glucose is
transported by the GLUT2 glucose transporter; subsequent glucose metabolism by
the beta cell alters ion channel activity, leading to insulin secretion. The
SUR receptor is the binding site for drugs that act as insulin secretagogues.
Mutations in the events or proteins underlined are a cause of maturity onset
diabetes of the young (MODY) or other forms of diabetes. SUR, sulfonylurea
receptor; ATP, adenosine triphosphate; ADP, adenosine diphosphate, cAMP, cyclic
adenosine monophosphate. (Adapted
from WL Lowe, in JL Jameson (ed): Principles of Molecular Medicine. Totowa, NJ, Humana, 1998.)The insulin secretion
mechanism in short. http://e-medicaltextbook.blogspot.com/
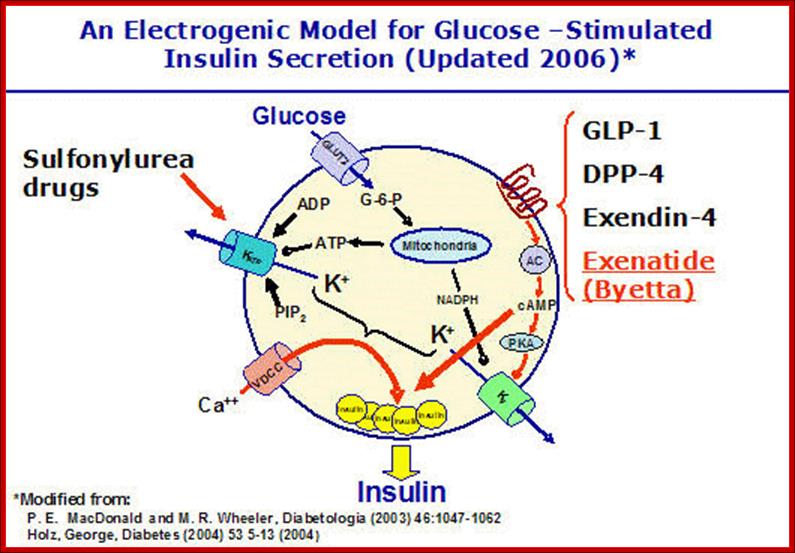
Activity of K=V channel is subject to control �Y� hormones acting through G-proteins, adenyl cyclase and protein kinase A. Phosphorylation of the Kv channel inhibits K+ transport. Kv is also inhibited by NADPH which is formed through glucose metabolism. Glucagon-like protein 1 (GLP-1), which stimulates insulin secretion, seems to act through Kv. The figure above, slightly modified from MacDonald�s review article, summarizes the potassium channel theory. http://www.medbio.info/
Intra-chain S-S bond is between a.a cyst 71 and cys 76. �Inter-chain S-S bonds are between a.a cys 7 and a.a cys 72 and another is between a.a cys 19 and a.a cys 85 found circulating in the blood. (Note- S-S bonds invariably between cysteines).� When the concentration of blood sugar is less than the optimum, responding to such situations, precursor protein is immediately processed to functional insulin.� This is not the only protein that undergoes such changes, there are several hundreds and thousands of proteins which are synthesized as large precursor proteins then they are processed in the sense they can be modified and can be cut into individual fragments, some of them act as individual proteins or associate with other proteins to form multimers and some are destined to certain locations and they are delivered to their respective sites.� It is a remarkable process that happens in every single cell.
�
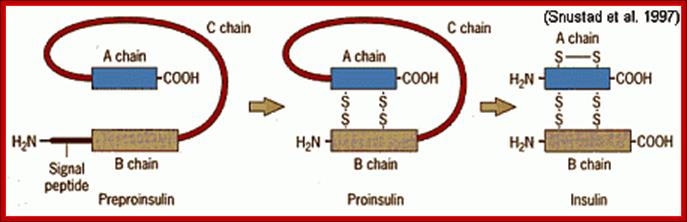
Pre-proinsulin is transcribed as 110 amino acid chain nad by removal of signal peptide, proinsulin is produced. Formation of S-S bonds between A and B chain are made and removal intervening C peptide chin produces biologically active insulin of 51 a.a; Insulin post translational modification. htm; http://www.bio.davidson.edu/
Synthetic Insulins:
International color code for human insulin preparations;
A universal color code means that similar preparations of insulin have the same color on the label regardless of manufacturer, and that this is the standardized worldwide. Such a color code helps to reduce confusion and uncertainty for people with diabetes who have to buy insulin abroad or from a different source.
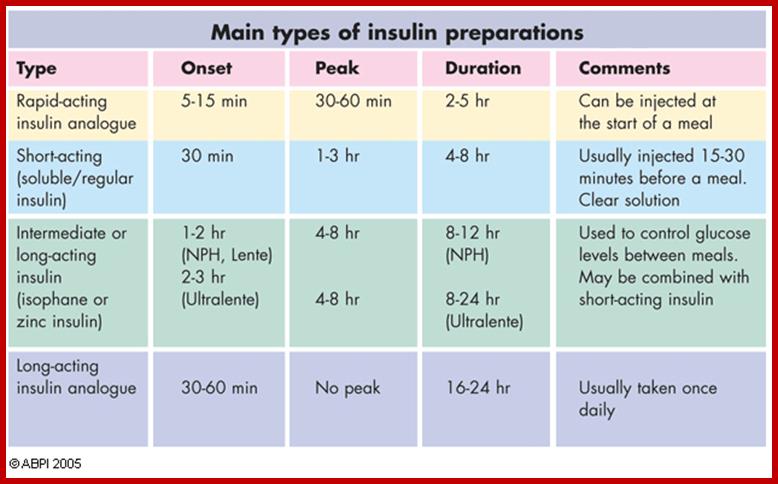
Range of insulin preparations, from short acting to long acting. https://235.stem.org.uk
Glucagon:
Two of the many gastrointestinal hormones have significant effects on insulin secretion and glucose regulation. These hormones are the glucagon-like peptides (GLP-1) and glucose-dependent insulinotropix peptide (GIP).� Both of these gut hormones constitute the class of molecules referred to as the gastro intestinal hormone have significant effects on insulin secretion and glucose regulation.� Incretins are molecules associated with stimulation of insulin secretion from the pancreas.
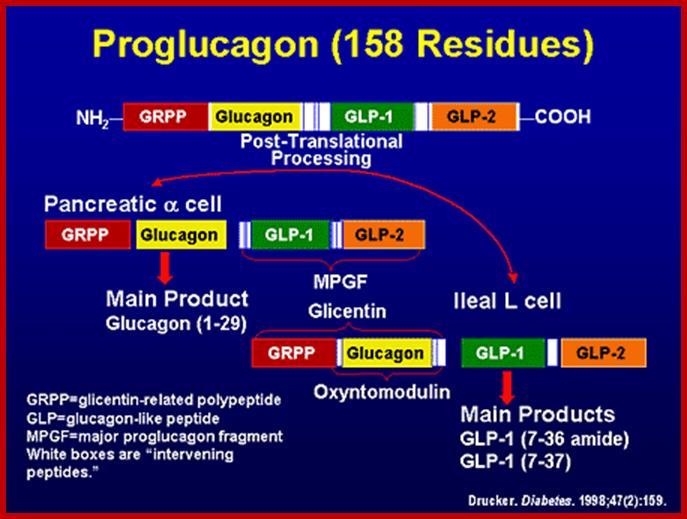
Preproglucagan:
Preproglucogon is 180 amioacids long and it is also cleaved in secretary vesicles by endo peptidases.� The cleaving is specific to sequences like Arg-Arg (R-R or R-K), Lys-Arg and performed by specific enzymes.
PC2 and PC3 endo-proteases.
The intra-polypeptide sequences PC2 and PC3 act as endopeptidase that cleave at the said sites to generate two fragments, then the larger fragment is cleaved to produce two more fragments; then among them one of the large fragments is further cleaved to produce glucagons.
Two of the many gastrointestinal hormones have significant effects on insulin secretion and glucose regulation. These hormones are the glucagon-like peptides (principally glucagon-like peptide-1, GLP-1) and glucose-dependent insulinotropic peptide (GIP). Both of these gut hormones constitute the class of molecules referred to as the incretins. Incretins are molecules associated with stimulation of insulin secretion from the pancreas.
GLP-1 is derived from the product of the proglucagon gene. This gene encodes a preproprotein that is differentially cleaved dependent upon the tissue in which it is synthesized. For example, in pancreatic α-cells prohormone convertase 2 actions leads to the release of glucagon. In the gut prohormone convertase 1/3 action leads to release of several peptides including GLP-1. Upon nutrient ingestion GLP-1 is secreted from intestinal entero-endocrine L-cells that are found predominantly in the ileum and colon with some production from these cell types in the duodenum and jejunum. Bioactive GLP-1 consists of 2 forms; GLP-1(7-37) and GLP-1(7-36) amide, where the latter form constitutes the majority (80%) of the circulating hormone.

http://www.assignmentpoint.com
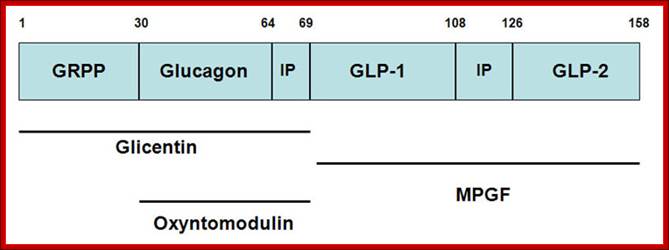
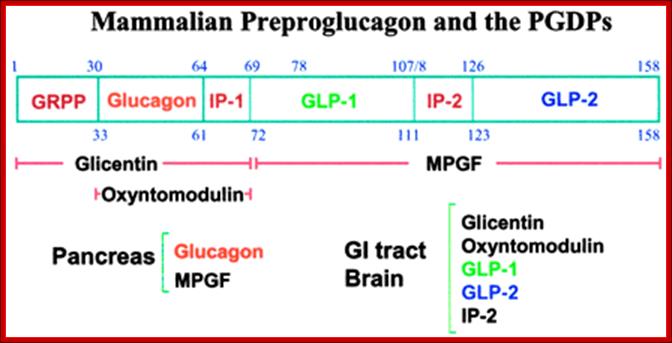
Structure of the mammalian pre-pro-glucagon product: GRPP=glicentin-related pancreatic peptide. IP=intervening peptide. GLP-2=glucagon-related peptide-2. Additional peptides are derived from the preproprotein including glicentin which is composed of amino acids 1�69, oxyntomodulin is composed of amino acids 30�69 and the major proglucagon fragment (MPGF) comprises amino acids 72�158.
�The primary physiological response to GLP-1 is glucose-dependent insulin secretion.� Inhibition of glucagon secretion and inhibition of gastric acid secretion and gastric emptying effect will lead to increased satiety with reduced food intake along with a reduced desire to ingest food. The action of GLP-1 at the level of insulin and glucagon secretion results in significant reduction in circulating levels of glucose following nutrient intake. This activity has obvious significance in the context of diabetes, in particular the hyperglycemia associated with poorly controlled type 2 diabetes. The glucose lowering activity of GLP-1 is highly transient as the half-life of this hormone in the circulation is less than 2 minutes. Removal of bioactive GLP-1 is a consequence of N-terminal proteolysis catalyzed by dipeptidyl peptidase IV (DPP IV or DPP4).
DPP4 was originally identified as the lymphocyte cell surface antigen CD26. In humans CD26 functions in many pathways that are not directly related to its peptidase activity nor to its role in incretin inactivation. DPP4 harbors adenosine deaminase-binding (ADA-binding) properties and is involved in extracellular matrix binding. Of importance to the immune system, CD26 expression and activity are enhanced upon T-cell activation. CD26 interacts with other lymphocyte cell surface antigens including ADA, CD45 and the chemokine receptor CXCR4 (notable is the fact that CXCR4 is a T-cell attachment site for HIV). As yet it has not been clearly delineated as to whether the enzymatic activity of DPP4 is essential for the T-cell activating and co-stimulatory functions assigned to CD26. Of significance, however, is that in gene knock-out mice lacking CD26 there is enhanced insulin secretion and improved glucose tolerance indicating that inhibition of DPP4 activity is a potential therapeutic target in the treatment of type 2 diabetes.
All of the effects of GLP-1 are mediated following activation of the GLP-1 receptor (GLP-1R). The GLP-1R is a typical seven-transmembrane spanning receptor coupled to G-protein activation, increased cAMP production and activation of PKA. However, there are also PKA-independent responses initiated through the GLP-1R. Other major responses to the actions of GLP-1 include pancreatic β-cell proliferation and expansion concomitant with a reduction of β-cell apoptosis (death). In addition, GLP-1 activity results in increased expression of the glucose transporter-2 (GLUT2) and glucokinase genes in pancreatic cells�.
http://www.medscape.org/
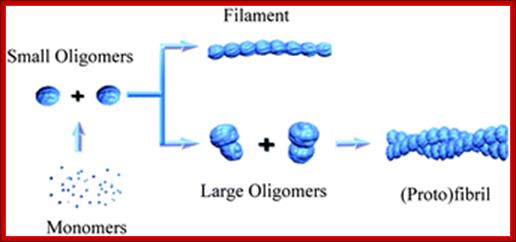
SAXS was used to provide a detailed structural description of the fibrillation of the 29-residue peptide hormone Glucagon at pH 2.5.
Recombinant Glucagon: 29 residue polypeptide hormone. �Glucagon was synthesized in a special non- pathogenic laboratory strain of Escherichia coli bacteria that has been genetically altered by the addition of the gene for Glucagon; A 29-(34) a.a residue Glucagon protein, a monomer; http://www.drugbank.ca/
Oligomerized Glucagon-semitransparent sphere-ab into model. http://pubs.rsc.org/
GRPP = glicentin-related pancreatic peptide. IP=intervening peptide. GLP-2 and 2 =glucagon-related peptide-1 and 2.
http://www.phoenixpeptide.com/
https://cancerdiagnostics.com
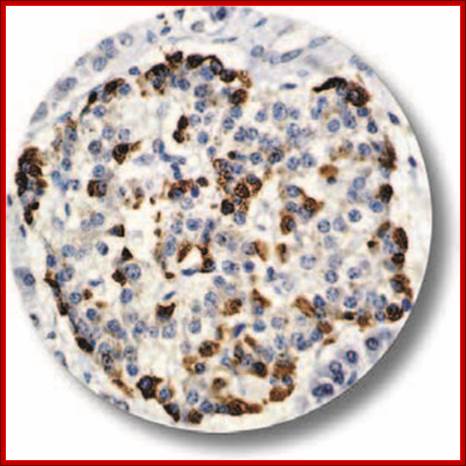
IHC of Glucagon on an FFPE Pancreas Tissue. Glucagon is a 29-amino acid polypeptide acting as an important hormone in carbohydrate metabolism. The hormone is synthesized and secreted from alpha cells of the islets of Langerhans, which are located in the endocrine portion of the pancreas
Where does protein digestion start?
In stomach, pepsin, the main digestive enzymes hydrolyze proteins. �All forms of substances; they are broken down into smaller molecules by a reaction with in which water molecules is split into OH- and H+ ions and each join with different product molecules.� This type of reaction is called hydrolysis.� This converts proteins in to shorter polypeptides then into amino acids.� Even the starch is converted to disaccharides, and then into monosaccharaides, which are transported into intestine via interstitial cells.
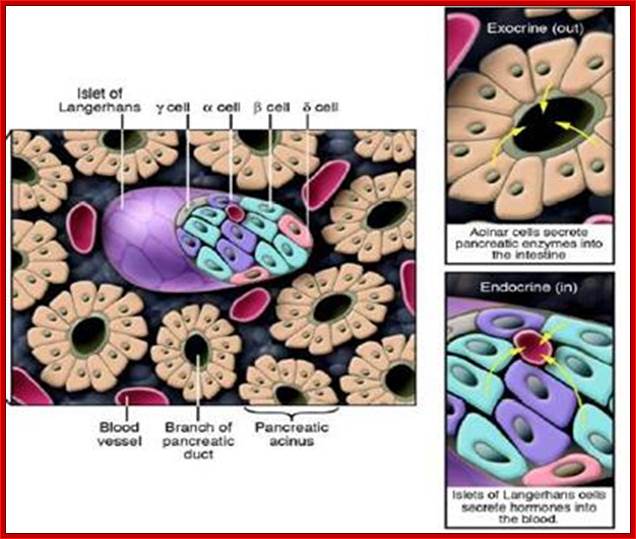
Cross-section of the pancreas.
The pancreas houses 2 distinctly different
tissues. Its bulk comprises exocrine tissue, which is made up of acinar cells
that secrete pancreatic enzymes delivered to the intestine to facilitate the
digestion of food. Scattered throughout the exocrine tissue are many thousands
of clusters of endocrine cells known as islets of Langerhans. Within the islet,
α cells produce glucagon; β cells, insulin; γ cells,
somatostatin; and δ cells, pancreatic polypeptide � all of which are delivered
to the blood. http://www.innovitaresearch.org/;
http://www.innovitaresearch.org/
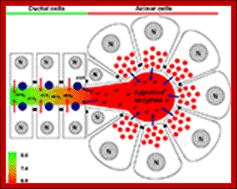
Changes of luminal pH in the exocrine pancreas during secretion. Under physiological conditions acinar cells secrete digestive enzymes and protons, the latter of which acidify the acinar lumen. In contrast, ductal cells secrete bicarbonate which will elevate the intraluminal pH. Our hypothesis is that protons may stimulate the ductal bicarbonate secretion via acid sensing receptors (ASR), which can elevate the pH in the ductal lumen setting the luminal pH to 8.0. (N, nucleus). http://journal.frontiersin.org/
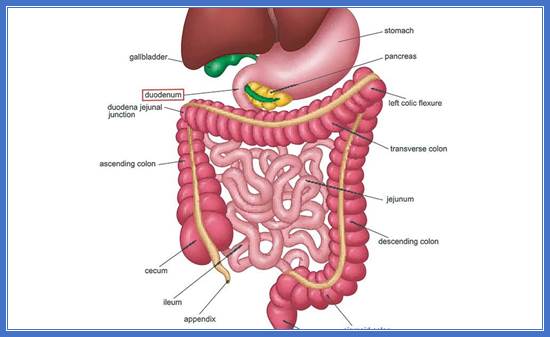
https://www.ncdhp.com/news/duodenum-an-overview
The major portion of �chemical digestion takes place when the chyme enters the first portion of the small intestine, the duodenum.� The chyme is semifluid which partially digested food in stomach.� It passes through sphincter on to dudenum.� The pancreas secretes digestive enzymes into the dudenum. Te pancreas is an elongated gland theat secretes both digestive enzymes and hormones as well, that helps to regulate the level of glucose in the blood.������������������
Four major groups of proteases:
� Serine proteases, which have a reactive serine residue, and act optimally at neutral pH
- The carboxyl or aspartyl proteases, which have catalytically important carboxyl�s and act at relatively low pH
� Thiol proteases have reactive cysteine residues
� Zinc proteases, which have a zinc in the active site and function at neutral pH
We will take a look at examples of each of these, beginning with the serine proteases: chymotrypsin, trypsin, and elastase, which catalyze the hydrolysis of peptides in the intestines.
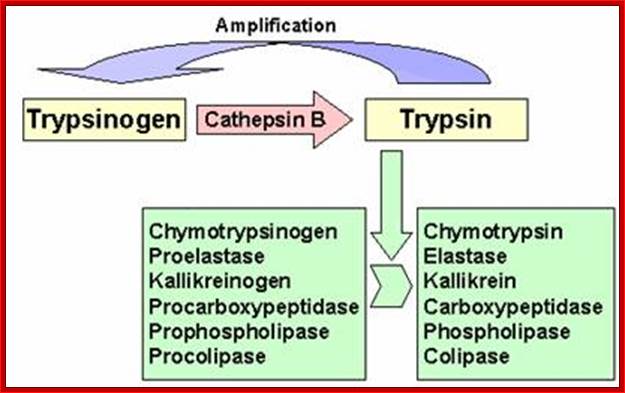
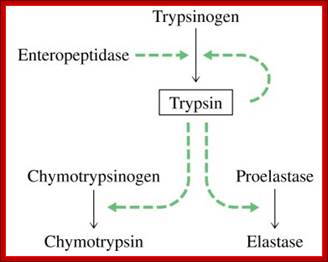
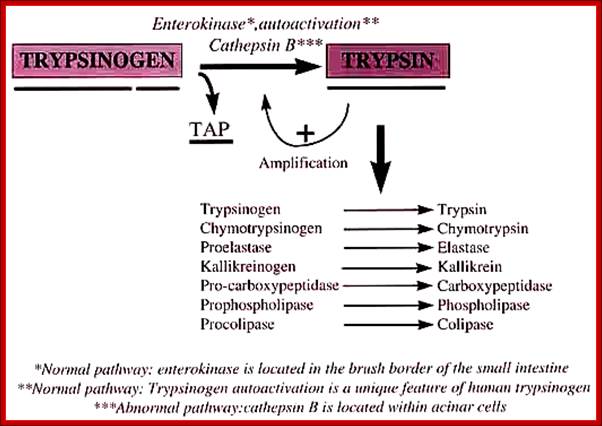
� Zinc proteases, An enter peptidase found in the duodenum activates trypsinogen by cleaving off the first six residues at the N-terminal end of the protein.
� The trypsin thus produced then activates chymotrypsinogen and pro-elastase by hydrolysis at several positions.
� It also can activate other trypsinogens in the same way. Here is an oversimplified picture of the activation cascade:
� An entero-peptidase found in the duodenum activates trypsinogen by cleaving off the first six residues at the N-terminal end of the protein.
� The trypsin thus produced then activates chymotrypsinogen and pro-elastase by hydrolysis at several points.
It also can activate other trypsinogens in the same way. Here is an oversimplified picture of the activation cascade:
http://chemistry.umeche.maine.edu/CHY431/Peptidase1.htm
Cathepsin B; Novel cysteine protease of papain family; http://www.pharmainfo.net/
The enteropeptidase itself a serine protease;
http://www.lookfordiagnosis.com
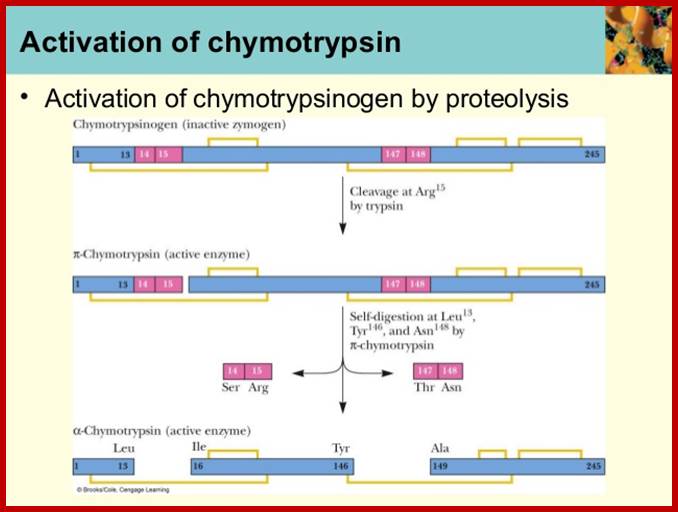
Chymotrypsinogen:
It is precursor protein stored as zymogen granules; when they are released they are processed by internal endopeptidase segments into active proteins.
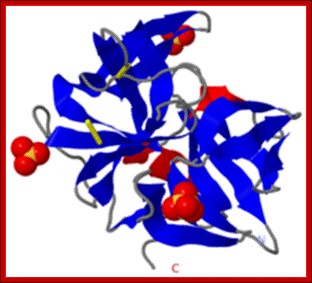
Crystal Structure of Chymotrypsin
A 245a.a long precursor protein is processed into pi-chymotrypsin and then into alpha-chymotrypsin (147-345a.a long). http://proteopedia.org/
1 campbell ch07-haddow-spring2013; �https://image.slidesharecdn.com/
Polyproteins:
There are several proteins that are synthesized as large precursor proteins, and then they are cleaved differently in different cell types to generate different but specific proteins, where each of them act as individual polypeptides and each have their own specific functions.�
Many viral proteins are produced as large precursor proteins and they are then cleaved into several independent proteins, such precursor proteins are called Polyproteins.
�There are several hormonal proteins, which are synthesized as precursors, but they are specifically in tissue specific manner, again such proteins are called Polyproteins.
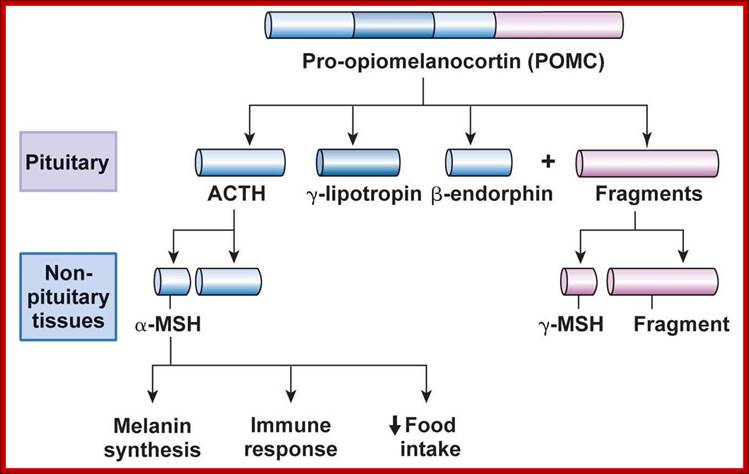
Pro-Opiomelanocortin (POMC):�
In rats this precursor protein is synthesized in both anterior and intermediate lobes of pituitary glands.� This single large protein contains seven important segments and each has specific destinations and functions as peptide hormones.
http://www.webmd.com/brain/pituitary-gland
www.studyblue.
�����������������������
The precursor poly-proteins are cleaved in both anterior and intermediary lobes.�
First it is cleaved into N-terminal fragment, ACTH fragment and B-LPH fragment.�
In intermediary lobes the N-terminal fragment is cleaved to produce g-MSH.
The ACTH is cleaved into α-MSH and C-LIP fragments.�
The β-LPH is further cleaved into γ-LPH and β-END in intermediate lobe.�
The cleaving is performed by specific proteases and cleaving generally occurs at the bonds between Lys-Arg, lys-lys and Arg Arg.
Following are the products of PoMC
ACTH = Adeno corticotropic hormone.
β-LPH = B-lipoprotein hormone.
γ-MSH = g-melanocyte stimulating hormone.
α-MSH = a-melanocyte stimulating hormone.
CLIP = Corticotrophin-like intermediate lobe protein
γ-LPH = g-lipoprotein hormone.
β-END = B-endotrophin.
Pre encephalin:
Pre encephalin is synthesized in adrenal medulla as precursor protein of 267 amino acids long.� The signal sequences contain 25 amino acid residues, which are cleaved while they transported across the membrane.� Then they are cleaved into segments such as B, E, and F.� The B is further cleaved into MRF and ME.� The E fragment is cleaved into LE and ME.� The F fragment is cut into 3 M. encephalins. L-stands for Leu encephalins and M-stands for met encephalins.
By genetic manipulation it is possible to construct a recombinant DNA containing different segments so as to generate a fusion protein.� Such constructs have been made using a segment from Maltose binding protein (MBP), and a segment from Pol Intein from Pyrococcus and Paramyosin from Pyrococcus.� The Pyrococcus is a thermophilous fungus. The DNA construct was introduced into E. coli. Under controlled in vitro conditions such as at elevated temperature, the fused protein under goes splicing through branch formation.� No splicing has been observed with in the cell.� Inteins resemble to that of intronic I-RNA, which belong to parasitic group of introns.
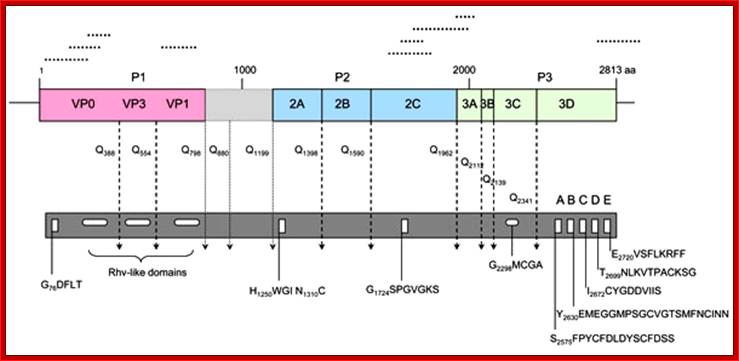
Picorna viral precursor proteins:
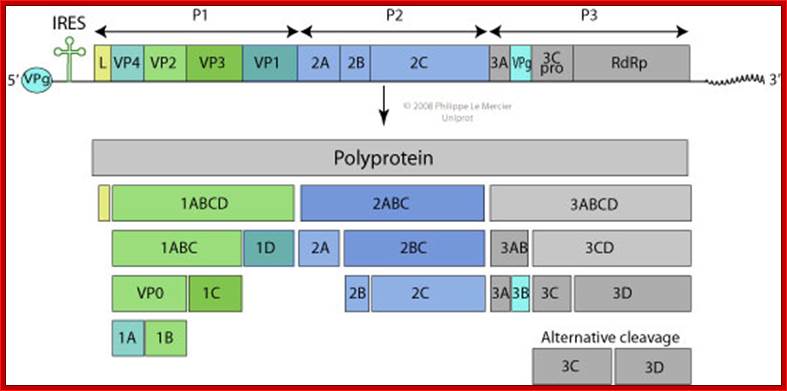
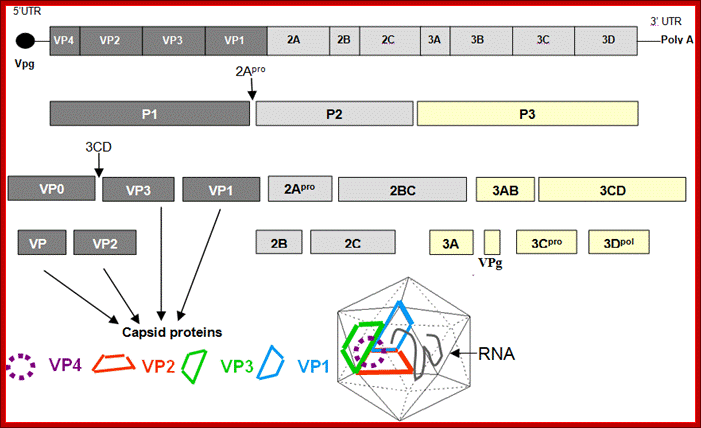
Picorna virus contains a positive sense RNA.� After infection the (+) sense translate to generate a protein with 2206-2207 amino acids.� The �protein undergoes self-cleavage to generate viral endoproteases-2A, 2B and 2C, coat proteins-Vp1, VP2, VP3 and VP4, cap protein- VPG and RNA replicase all from the same precursor proteins.
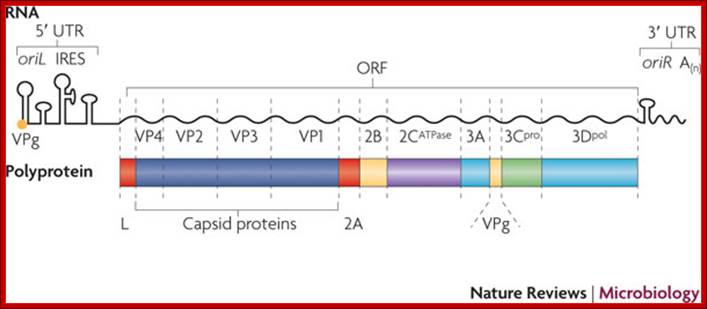
Picornaviruses possess a single-stranded positive-sense RNA genome (see the figure) comprising approximately 7,000�9,000 nucleotides. The RNA is packaged into an icosahedral capsid usually composed of four distinct proteins (VP1�VP4), but in some viruses VP4 and VP2 are fused (forming VP0). After productive contacts with specific cellular receptors, which differ between viruses, the genome is uncoated and enters the cytoplasm, where the main steps of viral reproduction occur. The viral RNA, which contains a single ORF (with one exception described in the main text), is translated in a cap-independent manner into a 2,200�2,500 amino acid polypeptide, which is eventually processed by limited proteolysis into a dozen 'mature' proteins. These proteins include: capsid proteins; an RNA-dependent RNA polymerase (3Dpol); a protein (VPg, or 3B) that serves as a primer for the initiation of RNA synthesis; an ATPase with a conserved superfamily 3 helicase motif (2CATPase) and an essential but poorly defined role in viral RNA replication; a chymotrypsin-like protease (3Cpro), which, as the mature protein or as a precursor, is a major factor in polyprotein processing; two hydrophobic membrane-binding proteins (2B and 3A) that participate in the generation of a virus-friendly environment; and, flanking the precursor of the capsid proteins, one or two highly variable proteins (L and 2A), the structure and functions of which are the subject of this Review. The entire coding region of the RNA is surrounded with untranslated regions (UTRs) harbouring regulatory elements that control viral translation (for example, the internal ribosome entry site (IRES)) and replication (the origins oriL and oriR), and this entire RNA sequence is flanked by the covalently linked viral protein VPg and a poly(A) tract at the 5′ and 3′ termini, respectively. There are four structurally and functionally distinct types of IRES131and many types of oriL and oriR. Picornavirus genes and proteins;
Vadim I. Agol & Anatoly P. Gmyl; http://www.nature.com/
Picorna viridae; �http://viralzone.expasy.org/
Organisation of the enterovirus genome, polyprotein processing cascade and architecture of enterovirus capsid the genome of enteroviruses contains one single open reading frame flanked by a 5′-and 3� untranslated regions (UTR). A small viral protein, VPg, is covalently linked to the 5′ UTR. The 3�UTR encoded poly(A) tail. The translation of the genome results in a polyprotein which is cleaved into four structural proteins (dark gray) and seven non-structural proteins (light gray and yellow). The sites of cleavage by viral proteinases are indicated by arrows. The four structural proteins adopte an icosahedral symmetry with VP1, VP2 and VP3 located at the outer surface of the capsid and VP4 at the inner surface. The single strand genomic RNA is located inside the capsid. �http://www.intechopen.com/
Novel
Novel Picorna viral genome:
Novel Picornavirus in Turkey Poults with Hepatitis, California, USA; Predicted turkey hepatitis virus (THV) genome organization based on sequence comparison to known picornaviruses. Dotted lines above the genome depict the location of the original sequences obtained by high-throughput sequence analysis. Conserved picornaviral motifs and predicted potential cleavage sites along the coding region are indicated in the bar below.
�Kirsi S. Honkavuori, H. L. Shivaprasad, Thomas Briese, Craig Street, David L. Hirschberg, Stephen K. Hutchison, and W. Ian Lipkin; http://wwwnc.cdc.gov/
VP1-acts as the binding protein to cell receptor.
VPo-cleaves into VP4 and VP2.
VP2ABC-cleaves into 2A, 2B, 2C; all are proteases.
VP3ABCD->cleaves into 3A-B and 3CD; then 3A-B cleaves into 3A, the 3B �no function assigned.
The 3-A is VPg protein.�
The 3CD are cleaved into 3C and 3D.�
The 3C is a protease, which cleaves most of the sites, and 3D is large protein and acts as RNA dependent RNA Replicase.
 �����������
�����������
FMD foot and mouth disease viral particle; Picornavirus � Apthovirus- www.virology.net
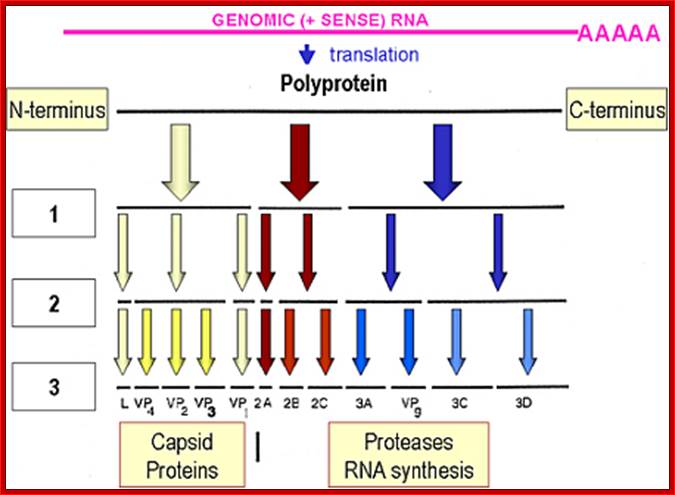
Cleavage positions of viral polyprotein;
A 245a.a long precursor protein is processed into pi-chymotrypsin and then into alpha-chymotrypsin (147-345a.a long). Microvirology.blogspot.com; http://proteopedia.org/
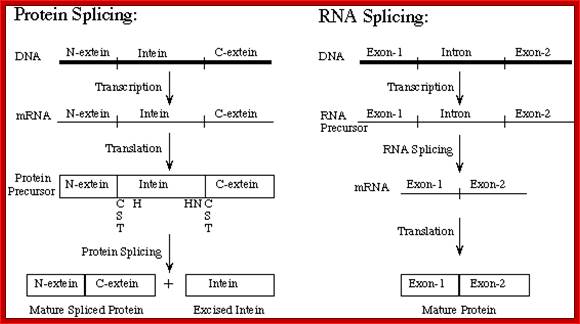
Protein Splicing: Inteins are protein introns:
In eukaryotes pre-mRNA undergoes various modes of processing.� One important process is alternative splicing, which generate multiple polypeptides from a single pre-mRNA. In certain systems, when mRNAs are translated, they generate a protein whose ultimate size is less than the coding capacity of mRNA.� The translated products, by itself, as it is cannot properly fold into functional protein, but by removing certain region or a segment from the protein, it can fold and function properly.� The region or a segment that is removed is called Intein.� The rest of the protein part involved in folding and function is called Extein.�
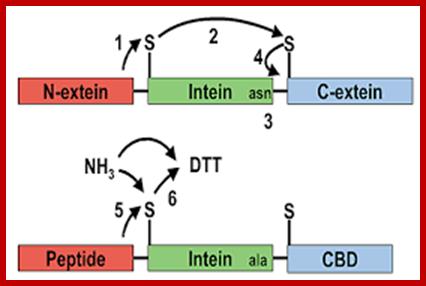
Schematic demonstrating the sequence of events leading to intein splicing and amide formation. A method for the amidation of recombinant peptides expressed as intein fusion proteins in Escherichia coli
Ian R. Cottingham, Alan Millar, Elisabeth Emslie, Alan Colman, Angelika E. Schnieke & Colin McKee; http://www.nature.com
Protein Splicing: Inteins and Exteins
Protein splicing with Inteins (protein introns), Extein (protein exons) was discovered some 20 years ago. Not to mention, this process is efficient and autocatalytic where the intein excises itself from the primary protein product (precursor protein) and then catalyzes the joining of the broken ends forming 2 protein products: 1) The mature protein 2) Intein itself. So, the protein fragments that are joined together to form the mature protein is called as Extein (Same as RNA exons), and Inteins are the protein introns.
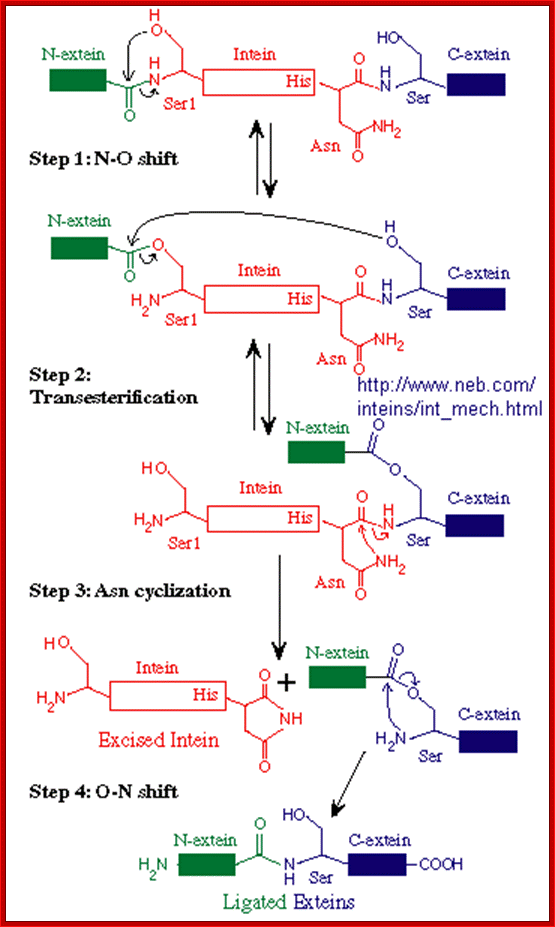
Mechanism of Intein splicing; In this scheme, the N-extein is shown in red, the intein in black, and the C-extein in blue. X represents either an oxygen or sulfur atom;
�http://biowiki.ucdavis.edu/
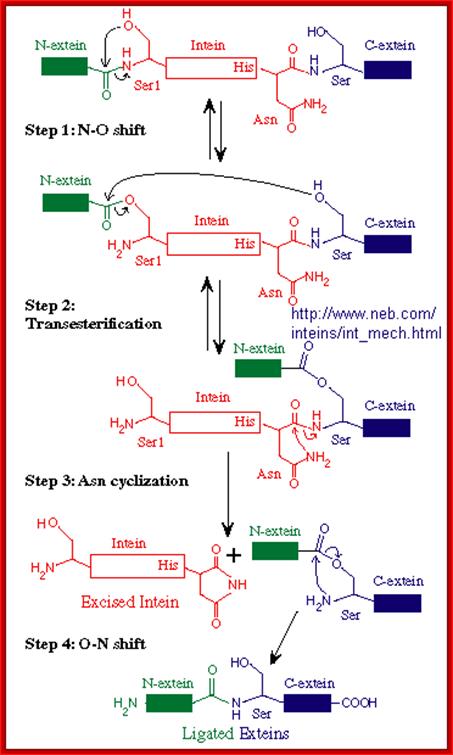
Inteins are often called selfish genetic elements.�� Actually, they are parasitic genetic elements. Data base search suggest there are 113 known inteins� in human system containing 138a.a to 844a.a.� The process begins with an N-O or N-S shift when the side chain of the first residue (a serine, threonine, or cysteine) of the intein portion of the precursor protein nucleophilic ally attacks the peptide bond of the residue immediately upstream (that is, the final residue of the N-extein) to form a linear ester (or thioester) intermediate. A transesterification occurs when the side chain of the first residue of the C-extein attacks the newly formed (thio)ester to free the N-terminal end of the intein. This forms a branched intermediate in which the N-Exstein and C-Exstein are attached, albeit not through a peptide bond. The last residue of the intein is always an asparagine, and the amide nitrogen atom of this side chain cleaves apart the peptide bond between the intein and the C-extein, resulting in a free intein segment with a terminal cyclic imide. Finally, the free amino group of the C-extein now attacks the (thio)ester linking the N- and C-extein together. An O-N or S-N shift produces a peptide bond and the functional, ligated protein; Noren CJ, Wang J, Perler FB (2000)
Fig reference: Elleuche S, P�ggeler S. Applied Microbial Biotechnology,2010.
All these inteins have sequence similarity with homing nucleases. Homing nucleases
are very interesting class of proteins that cleaves the DNA at a particular
recognition site. The recognition site is long enough (12 - 40 bp, as against
restriction enzymes that recognize 8 bp or less) to occur in a genome by chance.
Usually, the proteins encoding homing nucleases occur inside the DNA element
that is the recognition site for cleavage by themselves. �Thus, preventing the
cleavage of the DNA sequence that carries them, so they are a class of selfish
genes.� It is very interesting how these homing nucleases propagate
themselves to their non-allelic forms. The allele that has homing nuclease is
called as HEG+ and the one not having it is called as HEG-. Usually, the HEG+
alleles cleave the HEG- gene thus initiating homologous recombination DNA
repair. Once repair is initiated, HEG+ is copied at the HEG- locus thus
propagating it.
There are currently 4
structural domains of homing endonucleases:
1) LAGLIDADG: If present alone, needs a homodimer to act against a DNA sequence.
2) GIY-YIG: Acts as a monomer, occurs in the N terminus. 3)His-Cys box: 2 histidine�s
and 3 cysteines, acts as a monomer. 4)H-N-H: 2 pairs of histidine�s flanking
one asparagine.
Currently (June 2004) about 200 inteins are identified in more than100 different species and strains, in more than 50 various families of protein hosts (details here).
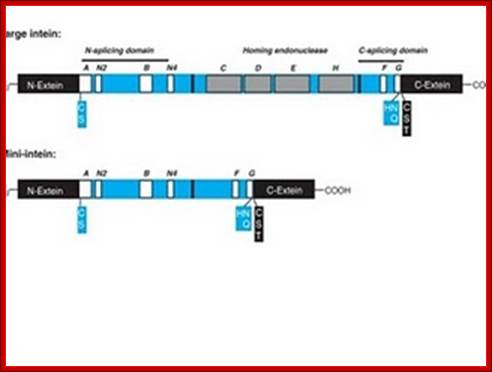
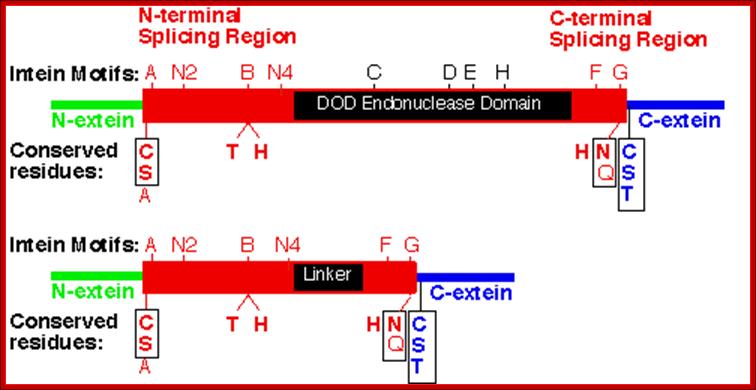
The size of the Intein (> 300 amino acids long) and the number of inteins vary from one species to the other.� Not all proteins have this Intein and Extein structural features and such feature is found only in some.� Inteins are similar but equivalent to introns in pre mRNAs, but the intein aminoacid sequences are found within the reading frame of the processed mRNA.� Some inteins have endonuclease activities, so they are termed as P1.� Most of the inteins have conserved motifs, but some are not well conserved.� The conserved motifs can be named as A, B, C, D, E, H, F and G or N1, N3, EN1, EN2, EN3, EN4, C2 and C1.� The CDEH motif in some proteins contain endonuclease (DOD) domain.� The endonuclease domains are also considered as homing sequences for the reason they cut the DNA and insert a segment of DNA which generate Inteins and splits the protein segment into exteins.�
Inteins structure and sequence vary from one protein to the other.�� Inteins are related to Hedgehog proteins for they contain auto-processing domains.� The splicing regions of inteins contain beta sheets as found in auto processing domain of Hedgehog proteins in Drosophila.� Hedgehog proteins are involved in embryonic pattern formation in many multicellular organisms.� Several genes in C. elegans have domains similar to Hedgehog & intein modules (HINT).� But the Hedgehog proteins and homing inteins have different N-terminal functional domains.�
However, both have some common motifs and common functions.� Both mediate auto-processing reactions initiated by acyl shift to form activated thio-ester bond, which is cleaved via transesteification by different thiol or hydroxyl containing molecules.� Inteins not only cut but also ligate flanking amino acids.� Modified inteins are used to ligate lipids to hedgehog signaling domains, which are used for compartmentalization at the cell surface
N-ext--I---------------intein-------------------------------I-extein-C
N-ext--IC, S, A-A-N2-B-IC D E HI- F-G-H, N, Q-I C S T-C-ext
�����������
C, S, A=N end intein splice site.
H, N, Q= C-end intein splice site.
C, S, T= N-end of extein-2 joint site.
NH2---extin1--I-S, C, A�intein--C, S, N�I-C, S, T---extein2�
�NH2---extin-1--I---extein 2------ COOˉ
At the borders Inteins, contain amino acid sequences such as cysteine, serine and alanine, at the N-terminal end histidine, glutamic acid and glutamine at the C-terminal end of the intein.� And also, extein at the right border of the intein contains some sequences such as C, S and T (cysteine, serine and threonine). The above-mentioned motifs differ from one protein to the other.� In some, in place of endonuclease domain, they contain Linker.� Serine, threonine and cysteine are generally found at both splicing junctions and they perform similar cyclising reactions.
Intein Structure:
by Kajoina, Florence
Inteins are intervening amino acid sequences in a DNA strand. Like introns within an RNA precursor, inteins are found within a mRNA strand. By the fact that these inteins are found within a protein and due to their similarity to introns, inteins are called Internal Proteins or protein introns. Just as introns are connected to RNA splicing, inteins are connected to protein splicing. Inteins are translated in frame with the host protein called the extein and then they splice themselves out of the translated proteins. Inteins can be divided into four basic classes and all have self-splicing activity. The classes are: maxi-intein, mini-intein, trans-splicing intein and alanine intein.
http://www.biocenter.helsinki.fi/
Intein proteins contain a number of conserved sequence motifs or blocks. The motifs can be grouped into three regions or domains according to their location and inferred function. The domains are an N-terminal splicing region, a central homing endonuclease region or a small central linker region and a C-terminal splicing region. The endonuclease domain is optional in inteins and thus not found in all inteins. The intein�s domain organization, deduced by sequence analysis exactly corresponds to the structural domains. Intein structures show that the inteins protein splicing and endonuclease active sites are formed from conserved motifs. Several conserved motifs have been observed by comparing intein amino acid sequence. There are two nomenclatures for these motifs: Blocks A, B, C, D, E, H, F, G or Blocks N1, N3, EN1, EN2, EN3, EN4, C2 and C1 respectively.
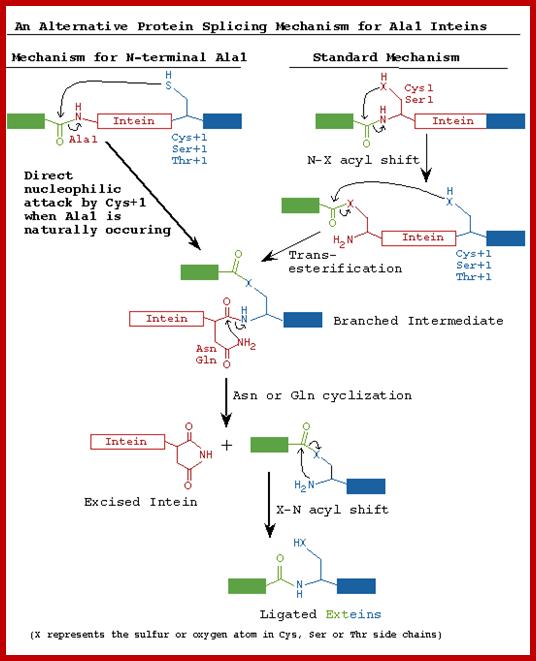
The maxi-inteins have an amino acid residue containing a nucleophilic side chain such as a cysteine or serine, at their N-terminus and the conserved intein motif blocks A and B. A mini-intein has the typical N- and C-terminus however the endonuclease domain is not present. Trans-splicing inteins lack a covalent linkage between their N- and C-terminal splicing domains. The alanine inteins contain a naturally occuring N-terminal alanine residue.
http://www.biocenter.helsinki.fi
The N-terminal splicing region is about 100 amino acids long and begins at the intein N-terminus and ends shortly after Block B. The intein C-terminal splicing region is usually less than 50 amino acids and includes Block F and G. The N-terminal splicing region and the C-terminal splicing region form a single structural domain, which is conserved in all inteins studied to date. There are four conserved splice junction residues: Ser, Thr or Cys at the N-terminus, the dipeptide His-Asn or His-Glu at the intein C-terminus and Ser, Thr or Cys following the downstream splice site. A few inteins have been identified with a C-terminal Gln (Q). There are four classes of homing endonuclease that are defined by their conserved signature sequence motifs: the DOD (dodecapeptide) or LAGLIDADG family the HNH (His-Asn-His) family the GIY-YIG family and the His-Cys family. Remarkably, inteins as small as 134 amino acids long and can splice out of precursor proteins. These �mini-inteins� do not have intein Blocks C, D, E, and H. Big inteins with as many as 608 amino acids have a larger linker region between inteins Blocks B and F that includes intein Blocks C, D, E, and H homing endonuclease motifs.
�����������������������������������������������������������������������������������������
Protein Splicing mechanism; Shmuel Pietrokovski;
FB Perler (New England Biolabs) and H Paulus (Boston, Biomedical Research Institute).� are the people contributed to the knowledge of Protein splicing; the diagrams below are just illustrations of the mechanism, that researches have understood. http://genome.weizmann.ac.il/
www.biocenter.helsinki.fi/bi/iwai/
http://www.biocenter.helsinki.fi
STEP 1: The N-terminal splice junction is activated by a N-O or N-S acyl rearrangement at the intein N-terminus that moves the N-extein to the side chain of the Ser/Cys at the intein N-terminus, forming the linear ester/thioester intermediate. A few Inteins have been identified with a N-terminal Ala (A) (see Splicing motifs), although splicing has not been demonstrated with these Inteins. Ala cannot undergo an acyl shift like Ser/Thr/Cys, since it doesn't have a hydroxyl/thiol side chain. However, these Inteins may be active if the residues facilitating the reaction are still making the splice junction peptide bond more labile and if the C-extein Ser/Thr/Cys is in the proper position to attack the splice site; in this case, the downstream splice junction Ser/Thr/Cys would directly cleave the N-terminal splice junction peptide bond (see splicing pathway A in Xu 1994) to form the branch intermediate.
STEP 2: The upstream ester/thioester bond is attacked during � transesterification reaction� by the hydroxyl/thiol group of the C-extein Ser/Thr/Cys, resulting in cleavage at the N-terminal splice junction and transfer of the N-extein to the side chain of the C-extein Ser/Thr/Cys, forming the branched protein intermediate.
STEP 3: The branch is resolved by cyclization of the conserved intein C-terminal Asn to form a succinimide ring, resulting in cleavage of the C-terminal splice junction. The succinimide can be hydrolyzed to form Asn or isoasparagine. A few Inteins have been identified with a C-terminal Gln (Q) (see Splicing motifs); although splicing has not been demonstrated with these, Gln is capable of undergoing a cyclization reaction just like Asn and should thus be able to substitute for Asn (Asn = Asparagine).
STEP 4: A spontaneous 0-N or S-N acyl rearrangement results in formation of a native peptide bond between the Exteins. http://www.biocenter.helsinki.fi