Cellular Protein Traffic:
Based on size of the cell and concentration studies of cellular proteins, it has been estimated that an eukaryotic calls contain about 8x10^9 molecules per cell and 2x10^6 molecules per prokaryotic cell.� Approximate number of a given protein in a given cell can be 10^5 per eukaryotic cell and 10^3 per prokaryotic cell. This is an approximate value estimated by F. Neidhard.
Most of the proteins destined to cell organelles and those to be cytosol resident proteins are synthesized free from ER.� In fact, cytosol is crowded with large number macromolecules including proteins and other organic and inorganic molecules.� A huge range of inclusions exist in different cell types, and they range from crystals of calcium oxalate or silicon dioxide in plants, to granules of energy-storage materials such as starch, glycogen, or polyhydroxy butyrate and lipid droplets. All these molecules small or large are bound by water-called �water of solvation�. Organelles fill the cytosolic space and make it like �too much� overcrowded space.� The mRNAs encoding cytosolic proteins can also be partitioned to the ER destined and non ER, suggesting that RNA partitioning in the eukaryotic cell is a complex process requiring the activity of multiple RNA-partitioning pathways; many of them RNAs are localized in the cell for specific functions ex. Drosophila developing oocyte oskar mRNA.
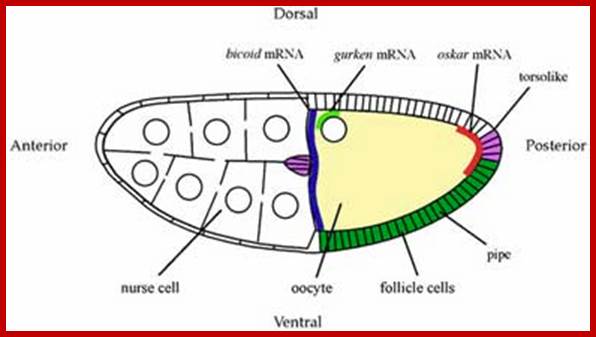
A diagram of a stage 10a Drosophila egg chamber showing localized signals that polarize the AP and DV axes of the embryo. bicoid mRNA (blue), oskar mRNA (red), gurken mRNA (green); pipe expression (dark green); and torsolike expression (magenta); http://cteappv.wordpress.com/
 ����
����
The cytosol, intracellular fluid is a crowded cytoplasmic matrix of many different types of structures such as microfilaments, microtubules, membranes and organelle that fills much of the volume of cells; http://en.wikipedia.org/
Transport of Proteins into Organelles:
Almost all cell organelle proteins are synthesized free from ER.� The mRNA-ribosomal machinery that is involved in translation may be held in cytoplasm by Actin filaments or microtrabacular elements!� They may not be localized as in the developing oocyte of Drosophila and other systems.
Mitochondrial genome has the ability to produce 13 polypeptide chains and also contain few URFs (7 unidentified reading frames).� In addition, the genome contains genes for all tRNAs and rRNAs. Chloroplasts, besides coding for 2 sets of ribosomal RNA genes and all its ~22 tRNAs, have the ability to synthesize at least 120-140 or more polypeptides. �Besides each organelle, Mts and Cps import at least ~1500 and 2800 proteins respectively from the nuclear coded transcripts.
- The rest of the proteins, few thousands required for the nucleus and Glyoxysomes or Peroxisomes are coded for by nuclear genes.� They are translated free from ER and translated products are transported to their destinations. Except nucleus the other organelle numbers double in mass at each cell generation.
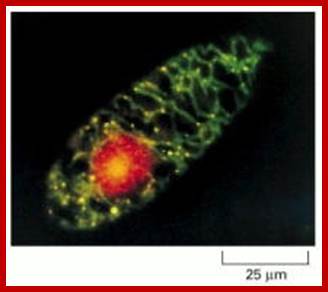
Mitochondrial and nuclear DNA stained with a fluorescent dye:� This micrograph shows the distribution of the nuclear genome (red) and the multiple small mitochondrial genomes (bright yellow spots) in a Euglena gracilis cell. The DNA is stained with ethidium bromide, a fluorescent dye that emits red light. In addition, the mitochondrial matrix space is stained with a green fluorescent dye that reveals the mitochondria as a branched network extending throughout the cytosol. The superposition of the green matrix and the red DNA gives the mitochondrial genomes their yellow color. (Courtesy of Y. Hayashi and K. Ueda, J. Cell Sci. 93:565�570, 1989. � The Company of Biologists.).
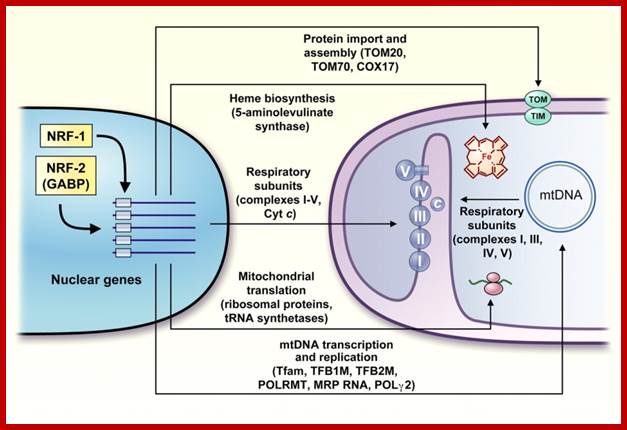
Richard C. Scarpulla; http://physrev.physiology.org/
Diagrammatic summary of the nuclear control of mitochondrial functions by NRF-1 and NRF-2 (GABP); NRFs contribute both directly and indirectly to the expression of many genes required for the maintenance and function of the mitochondrial respiratory apparatus. NRFs act on genes encoding cytochromec, the majority of nuclear subunits of respiratory complexes I�V, and the rate-limiting heme biosynthetic enzyme 5-aminolevulinate synthase. In addition, NRFs promote the expression of key components of the mitochondrial transcription and translation machinery that are necessary for the production of respiratory subunits encoded by mtDNA. These include Tfam, TFB1M, and TFB2M as well as a number of mitochondrial ribosomal proteins and tRNA synthetases. Recent findings suggest that NRFs are also involved in the expression of key components of the protein import and assembly machinery. Richard C. Scarpulla;
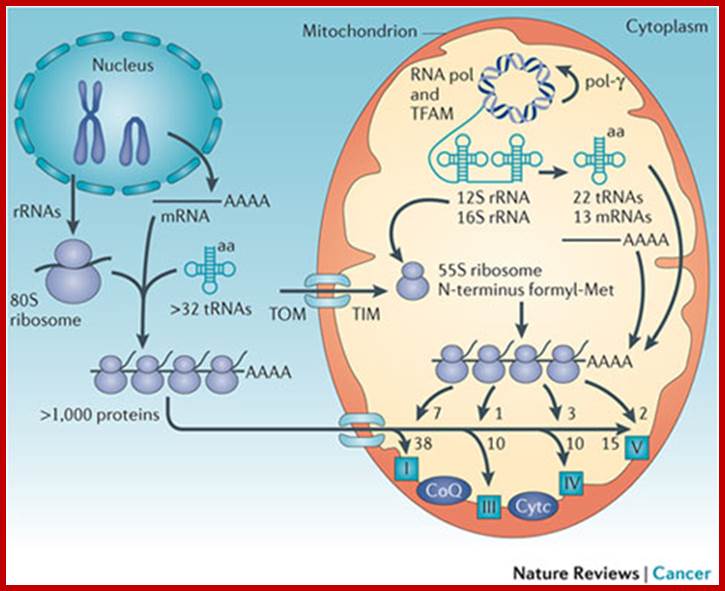
Mitochondrial genome and mitochondrial biogenesis.; Douglas C. Wallace; Nature Reviews-Cancer; http://www.nature.com/
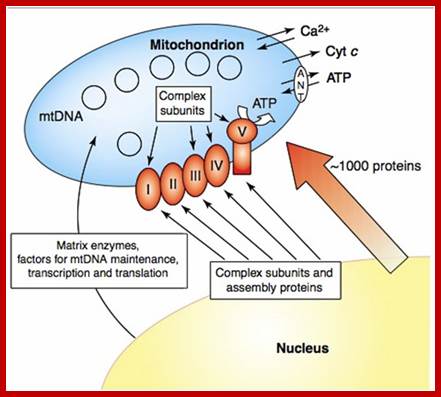
Transport of proteins (nuclear coded is specific transport system, dealt elsewhere;
Mitochondrial response to stress- mit. signaling; survival of death pathways.
James Watson and Vince Giuliano; http://www.anti-agingfirewalls.com/
The mitochondrial genome encompasses between one and two thousand nuclear-DNA-encoded mitochondrial genes and thousands of copies of the mitochondrial DNA (mtDNA). mtDNA has a high mutation rate, and de novo mtDNA mutations create a mixture of mutant and normal mtDNAs in cells, a state known as heteroplasmy. As the proportion of mutant mtDNAs increases, the energy output capacity of the cell declines until there is insufficient energy to sustain cellular function, termed the bioenergetic threshold. Mitochondria also constantly undergo fusion and fission, which permits complementation of mtDNAs in trans . The mtDNA encodes 13 proteins, 22 tRNAs, and 12S and 16S rRNAs. The mtDNA is packaged in the nucleoid and is replicated by DNA polymerase-γ (pol-γ). It is transcribed by mitochondrial RNA polymerase (RNA pol) symmetrically from both stands as large polycistron transcripts in which the larger transcripts are punctuated by the tRNAs. Cleavage of the tRNAs out of the polycistron transcripts creates the mature rRNAs and mRNAs, which are then translated on mitochondrial-specific chloramphenicol-sensitive ribosomes, in which the polypeptides are initiated by N-formyl methionine. The mtDNA encodes seven (ND1, ND2, ND3, ND4, ND4L, ND5 and ND6) of the 45 polypeptides of complex I; cytochrome b from the 11 polypeptides of complex III; three (cytochrome oxidase I (COI), COII and COIII) of the 13 polypeptides of complex IV; and two (ATP6 and ATP8) of the approximately 17 polypeptides of complex V. These proteins are central electron and proton carriers of the proton-transporting complexes and thus form the wiring diagram for oxidative phosphorylation (OXPHOS). All of the remaining mitochondrial proteins, including approximately 80 OXPHOS subunits and all four subunits of the non-proton-pumping complex II, are encoded by nuclear DNA (nDNA). The mRNAs from the nDNA-encoded subunits are translated on cytosolic ribosomes and the proteins are imported into the mitochondrion by transport through the outer (TOM) and inner (TIM) membrane complexes. TFAM, mitochondrial transcription factor A. Douglas C. Wallace
Mitochondria are associated with ER membranes called MAM. The MAM is recognized as a center for inter-membrane transport of phospholipids and for direct Ca (2+) transmission to mitochondria that activates the tricarboxylic acid cycle. However, MAM might be also involved in the inter-organelle transport of cholesterol, ceramides, ATP, and proteins as well as in proteasomal protein degradation and lipid droplet formation. Recent studies have begun to unveil the importance of inter-organelle communication in the innate immune response to virus infection. Thus, drug discovery aimed at regulating ER-to-mitochondria communication may open a new avenue in treatments of human diseases. This symbiotic relationship has probably developed 1.7 to 2 billion years ago?
- Each of these organelles is bound by outer and inner membranes except peroxisomes, Glyoxysomes and lysosomes which are enveloped by only one unit membrane. The said membranes are structurally and functionally distinct from one another. Majority of all cell organelle proteins contain 12 to 70 amino acid signal sequences one at their N-terminal and at the other at adjacent regions.� They can use two signal sequences one for the transport into the matrix and the other to transport into periplasmic or internal membranes of the organelles.
�
- The organelles membranes contain receptor cum protein channels where the outer and inner membranes are held to each other by transporter complexes.� These regions act as conduits or channels for the exchange of molecules of different sizes to and fro. Organelle proteins synthesized in cytosol are recognized by their sequences and organelle specific receptor protein and they bind to them.� Then they are transported to their respective destinations.
�The receptors located in membrane recognize the signal sequences of the protein, and contact to such sequences in a manner they are folded into a pattern that facilitates threading them into the channel.�� In this process chaperones help the proteins in threading the protein into organelles.
�
� While the protein is threaded through, it is in unfolded state and as and when it enters into the organelle, it again reforms into its native conformation.� In this process organelle HSPs subunits play important role in protein folding inside the mitochondria and other organelles.
�
� The signal sequence of organelle proteins is different from those of proteins destined to be secreted or destined to be the resident proteins of ER and other membranes.� There is little homology between them.� Once they are threaded through the channel, the signal sequences are cleaved by signal peptidases located at the inner side of organelle membranes.
� Chloroplast proteins have two sets of signal sequences, one for entry into chloroplast stroma and the other for thylakoid membranes.
� Moreover, the signal sequences between different organelles are different and there is little homology between them. However the leader sequences contain a stretch of uncharged amino acids interrupted by basic amino acids, but lack acidic amino acids.� The leader region may conformationally form a partial helical structure.
� If a cytosolic protein like di-Hydro folate Reductase (DHFR) gene is reconstructed with N-terminal leader sequences of mitochondria or chloroplast and expressed in eukaryotic cells like HeLa cells or any plant cells; the DHFR protein ends up either in chloroplast or mitochondria, depending upon the cell type.� This experiment clearly illustrates the importance of signal sequences in identifying the protein and delivering the protein to their respective targets.
Two ways that a sorting signal can be built into a protein; (A) A signal resides in a single discrete stretch of amino acid sequence, called a signal peptide, that is exposed in the folded protein. Signal peptides often occur at the end of the polypeptide chain, but they can also be located elsewhere. (B) A signal patch can be formed by the juxtaposition of amino acids from regions that are physically separated before the protein folds; alternatively, separate patches on the surface of the folded protein that are spaced a fixed distance apart could form the signal. In either case the transport signal depends on the three-dimensional conformation of the protein, which makes it difficult to locate the signal precisely.
The following table shows which part of the protein requires for identification of the target cell organelles.
|
Uptake target sequences that direct to specific location
|
�
Transport into Mitochondria:
Mitochondria are energy transducing organelles of eukaryotic cells.� They are bound by an outer and inner membrane.� The two membranes are adpressed to one another at regions of receptor cum channel proteins called attachment sites (old terminology).� It is through such protein complex transits mitochondrial proteins are recognized and transported into mitochondria.
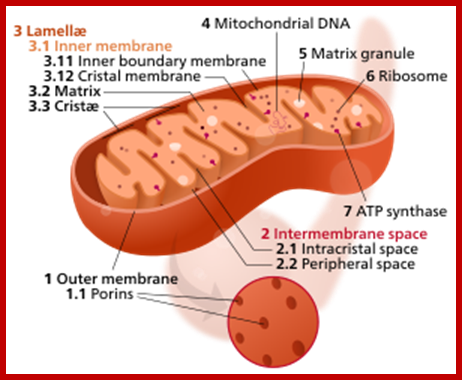
Power house of the Cell; WIKIPEDIA; http://clubhousenews.com/
Mitochondria by itself consist of four compartments, the central matrix, and inner membrane with cristae, outer membrane and peri-mitochondrial space.� Each of them contains different sets of proteins.
Mitochondria perse has its own genome.� The mitochondrial genome size varies from organism to another, but the number of genomic copies ranges from fifty to hundred or more.� But most of the mitochondrial DNA codes for ~13 proteins that gets integrated into inner membranes as protein complexes.� The genome also has few unidentified reading frames called URFs.� It also codes for mit-ribosomal RNAs and 22 or more tRNAs. Only in some, one or two tRNAs are imported. The mitochondria have its own prokaryotic translational machinery.� Yet for its structural and functional organization it requires more than another ~847 to ~1000 proteins (Human), which are coded for by nuclear genome, translated in the cytosol and transported or imported into the organelle. In some cases especially plants the some gene products, with certain modifications is transported into both mitochondria and plastids (dual targeting).
Mitochondrial proteins coded for by the nuclear genome possess signature sequences.� Some destined into matrix have one N-terminal sequence and those destined to inner membrane or peri mitochondrial space contain second signal sequences, one for the entry into matrix and the other directed to inner membrane or to periplasm.� Perhaps, mitochondria from all organisms from primitive to the most advanced ones may contain the same but a specific signal sequence for each and every mitochondrial protein.
How Mitochondria Replicate?
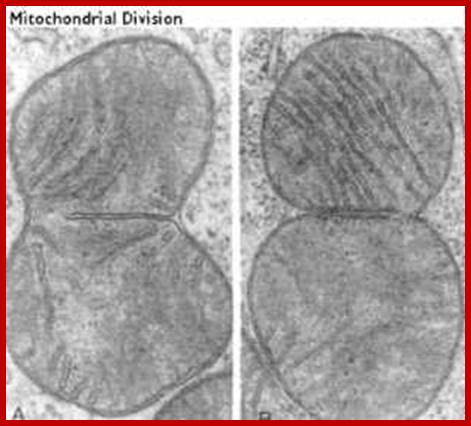
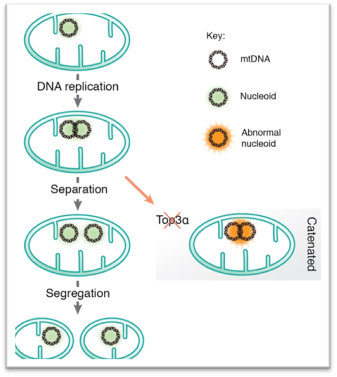
Separation and segregation of the human mitochondrial genome
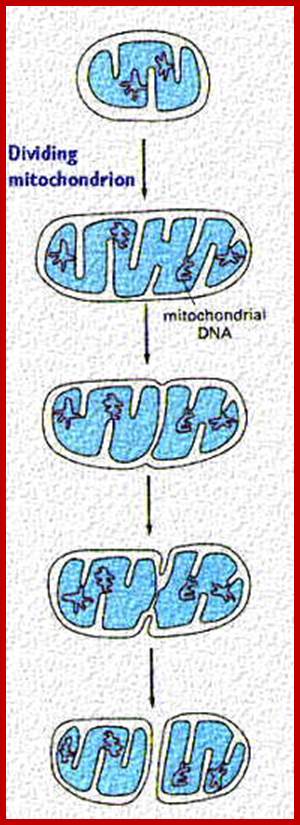
This process is diagrammed in the cartoon. Mitochondria replicate much like bacterial cells. When they get too large, they undergo fission. This involves a furrowing of the inner and then the outer membrane as if someone was pinching the mitochondrion. Then the two daughter mitochondria split. Of course, the mitochondria must first replicate their DNA. This will be discussed in more detail in the next section. An electron micrograph depicting the furrowing process is shown in these figures. The figure on the right was taken from Fawcett, A Textbook of Histology, Chapman and Hall, 12th edition, 1994; Mitochondria divide like a bacterial cell with fission process; http://www.cytochemistry.net/
- Mammalian mtDNA is replicated by proteins distinct from those used for nuclear DNA replication.
- According to the strand displacement model, replication is initiated from two distinct origins, OH and OL.
- Transcripts initiated at LSP provide the primer from which POLγ can initiate DNA synthesis at OH.
- OL forms a stem�loop structure and POLRMT initiates primer synthesis from the single-stranded loop region.
- The role of the mitochondrial D-loop is not understood.
- RNASEH1 and MGME1 play important roles in primer removal, but the details of this process are not fully understood.
- Top3α is required to resolve hemicatenane structures formed between new mtDNA molecules at the end of replication.
- mtDNA is not a naked molecule, but packaged into nucleoprotein complexes, nucleoids.
- Mitochondrial division is linked to active mtDNA synthesis. https://www.ncbi.nlm.nih.gov/pmc
Protein import into Mitochondria- TOM & TIM transporters:
��������������������������������� Principles of mitochondrial transport;
http://www.umanitoba.ca/
��������������������������������� Principles of mitochondrial transport;
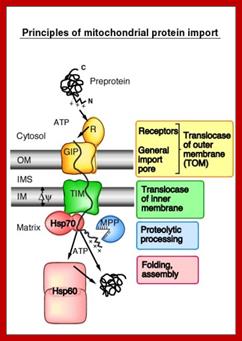
����������������������������������������������������������������������� http://www.bioch .freiburg.de/
We have identified a high molecular weight protein complex in
the outer mitochondrial membrane of the model organism, the yeast Saccharomyces cerevisiae. This
translocase of the outer membrane (TOM) contains three import receptors, Tom20,
Tom22 and Tom70, and a general import pore (GIP). Tom40 that is essential for
cell viability under all growth conditions forms a specific channel for
translocation of preproteins. Together with one receptor (Tom22) and three
small Tom proteins (Tom5, Tom6 and Tom7), Tom40 forms the stable core of the
TOM machinery, the GIP complex of 400 kDa. The interaction of the receptors and
GIP with the preproteins will be characterized in vivo, in organello and in
vitro. Since all Tom proteins are themselves nuclear-encoded, they are
synthesized as preproteins in the cytosol and must be imported into
mitochondria. First studies indicate that the assembly of Tom precursors into
the TOM machinery is a multi-step process, involving an ordered interaction
with nearly all preexisting Tom proteins. The aim of our study is to understand
the mechanism of preprotein recognition by receptors, the transport through the
GIP and the assembly of the translocase at a molecular level.
The transport of nuclear-encoded proteins into and across the
inner mitochondrial membrane is mechanistically independent of the
translocation across the outer membrane. The membrane potential is only
necessary for the translocation across the inner membrane. Two major
translocation pathways into the inner membrane can be distinguished;
(i) Preproteins with an amino-terminal signal sequence (presequence) are
translocated by the TIM23 complex. We have characterized three components of
this presequence translocase: Tim17, Tim23 and Tim44. Each protein is essential
for the viability of yeast cells and seems to be in direct contact with
preproteins during their translocation across the inner membrane. We could show
that Tim23 forms a specific, voltage-activated channel for preproteins.
(ii) Hydrophobic proteins with multiple internal targeting signals use the same
GIP of the outer membrane, but a different translocase of the inner membrane,
termed the TIM22 complex. Tiny Tim proteins in the intermembrane space guide
the hydrophobic proteins across this aqueous compartment to the
membrane-embedded TIM22 complex that consists of at least three integral
membrane proteins, Tim18, Tim22 and Tim54.
The aim is to understand the composition and mechanism of the import
machineries in the inner mitochondrial membrane at a molecular level.
A further major topic in our group is the characterization of the role of molecular chaperones in membrane translocation and folding of preproteins. The matrix heat shock protein Hsp70 associates with Tim44 of the inner membrane, binds the preprotein in transit and drives its translocation into the matrix. The co-chaperone Mge1 (mitochondrial GrpE) regulates the ATP-dependent reaction cycle of Hsp70. Mitochondrial Hsp70 and several partners, including Mdj1 (mitochondrial DnaJ), Mge1, Hsp60 and cyclophilin, are involved in the folding of imported proteins; http://www.biochemie.uni-freiburg.de/
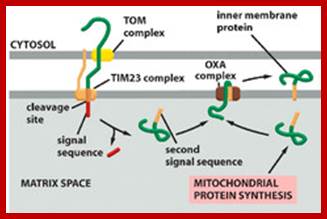
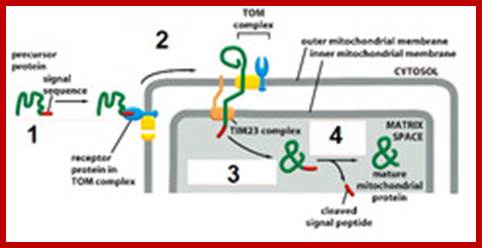
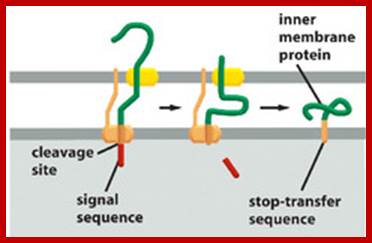
As mitochondrial protein is inserted across the transporter cum receptor, the protein, signal sequences are cleaved and the inner membrane protein gets integrated.� In some cases the inner membrane protein is transported into the matrix, which is then cleaves off the signal sequences and the protein gets integrated into the inner membrane. After cleaving N terminal signal sequences, many a times another inner membrane signal sequences found are used to incorporate the protein into inner membrane. In figure2 protein translocated into matrix is cleaved off its signal sequences and folds into its protein. 2.From Flash cards-Cell Biology lectures; Biogenesis of mitochondria; http://www.biochemie.uni-freiburg.de/ and http://www.studyblue.com/
Tom and Tim Transportes as shown in he diagram:
Summary of some of the overall steps of importation of a cytoplasmically-synthesized mitochondrial protein into a mitochondrion. The terms TOM and TIM stand for transporter outer membrane and transporter inner membrane, respectively. The red line is a newly-synthesized protein, and the green rectangle represents its signal peptide. As shown, the proteins are held in unfolded states by chaperone proteins, and the energy of ATP is required in order for them to release the protein to the mitochondrial receptor (not shown). At the entry site the inner and outer membranes are thought to be very near to one another, Cell Biol. Tutorials; http://classes.kumc.edu/
One can observe the mitochondrial protein on the top of the above figure with its exposed mitochondrial targeting sequence (the + signs near the N). The mitochondrial targeting sequence binds to the receptor (R) which passes the protein to the TOM (outer membrane) channel (shown as GIP in this picture). The protein is then threaded through both TOM and TIM (inner membrane), using the same.
������������������� 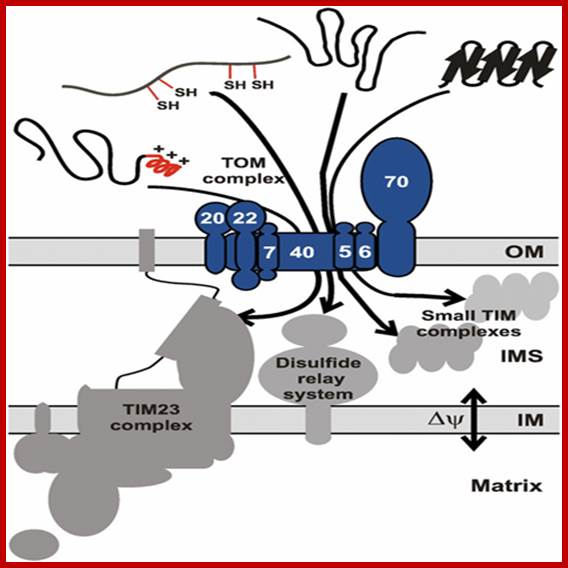
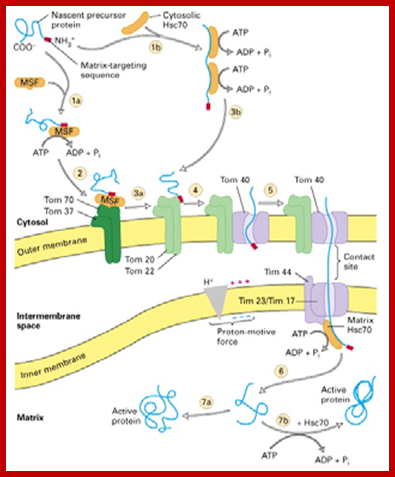
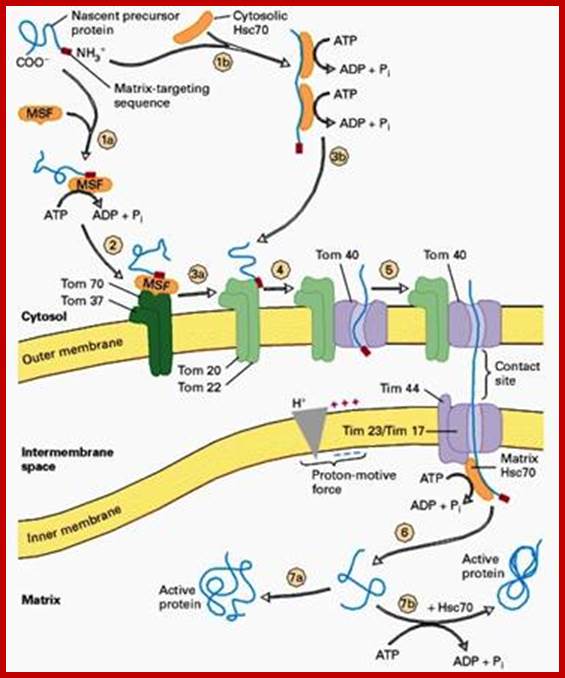
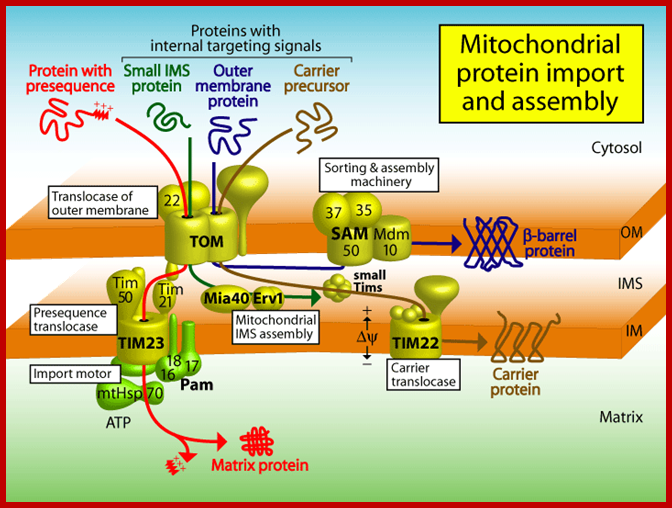
Proteins synthesized in free state-means not bound to ER; and guided to mit. Transporters using guides such as MSFs and Hsc70 proteins; Mitochondria are essential organelles of the eukaryotic cells that are made by expansion and division of pre-existing mitochondria. The majority of their protein constituents are synthesized in the cytosol. They are transported into and put together within the organelle. This complex process is facilitated by several protein translocases. Here we summarize current knowledge on these sophisticated molecular machines that mediate recognition, transport across membranes and intramitochondrial sorting of many hundreds of mitochondrial proteins.� �Dejana Mokranjac, Walter Neupert; http://www.cipsm.de/
In the cytosol, the soluble precursors of mitochondrial proteins (including hydrophobic integral membrane proteins) bind to one or more chaperones. These proteins use the energy released by ATP hydrolysis to keep nascent and newly made proteins in an unfolded state (see fig). Two chaperones, cytosolic Hsc70 and mitochondrial-import stimulation factor (MSF), have been shown to prevent the misfolding or aggregation of mitochondrial precursor proteins so that they can be taken up by mitochondria. MSF is also able to disperse aggregates of proteins.
Some precursor proteins, such as the inner-membrane ATP/ADP anti-porter, bind to MSF and the resulting complex then binds to a set of receptors called Tom37 and Tom70 on the outer membrane; Tom37 and Tom70 then transfer the precursor to a second set of receptors (Tom20 and Tom22) with release of MSF. Most precursor proteins bind to cytosolic Hsc70, which delivers the protein directly to the Tom20 and Tom22 receptors. These receptors are linked to Tom40, the actual channel in the outer membrane. When purified and incorporated into liposomes, Tom40 forms a transmembrane channel with a pore wide enough � about 1.5 to 2.5 nm in diameter � to accommodate an unfolded polypeptide chain.
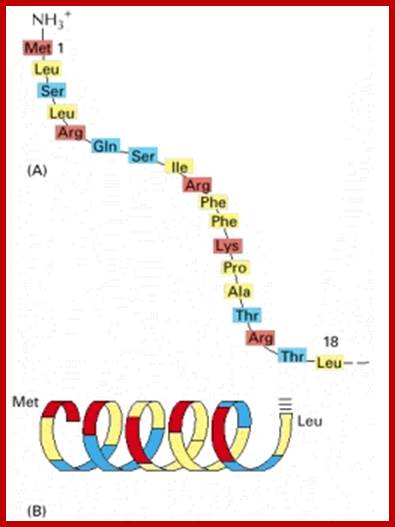
Signal sequence for mitochondrial protein import: http://www.woc.uc,pt
Cytochrome oxidase is a large multiprotein complex located in the inner mitochondrial membrane, where it functions as the terminal enzyme in the electron-transport chain (discussed in Chapter 14). (A) The first 18 amino acids of the precursor to subunit IV of this enzyme serve as a signal sequence for import of the subunit into the mitochondrion. (B) When the signal sequence is folded as an α helix, the positively charged residues (red) are seen to be clustered on one face of the helix, while the nonpolar residues (yellow) are clustered primarily on the opposite face. Mitochondrial matrix-targeting sequences always have the potential to form such an amphipathic α helix, which is recognized by specific receptor proteins on the mitochondrial surface.
Mitochondria membranes at attachment point consist of a channel protein complex; one complex is Transporter of outer membrane called TOM and another Transporter at inner membrane called TIM.� TOM consists of more than nine subunits of 500kDa mass.� The thickness of the channel is 188 � and the central channel is about 20 �.� Tim is a multi-protein complex consists of channel made up of 17kDa and 23kDa subunits.� The TOM 40 and TIM subunits are in direct line with one another but they don�t interact with one another.� Tom is associated with many protein subunits such as tom-5, 6, 7 (assembly factors) and also with Tom 37, 71 and 70 (one type of receptors) and Tom 22 and 22 and 22 (another type of receptor). Tim is also associated with several proteins such as Tim 17, 23, 44, 22-54, 9, 10 and 12), involved in transport of protein delivered by Tom 40.
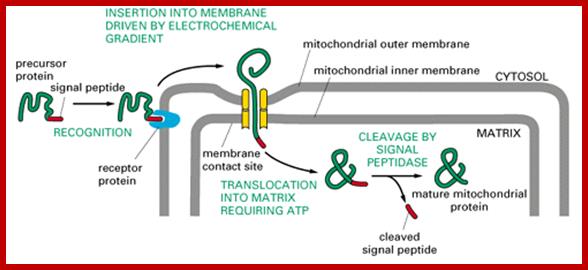
����������������������������������������������������������������������������� Protein import by mitochondria. The amino-terminal signal peptide of the precursor protein is recognized by receptors that reside in the outer membrane. The protein is thought to be translocated across both mitochondrial membranes at or near special contact sites, driven first by the electrochemical gradient across the inner membrane and then by ATP hydrolysis. The signal peptide is cleaved off by a signal peptidase in the matrix to form the mature protein; the free signal peptide is rapidly degraded.;http://www.unitus.it/scienze
To begin with, as polypeptide chain emerges out of the tunnel in large ribosomal subunit, the trigger factor and Srp complex monitor the emerging N-end of the protein.� When these proteins don�t contain such sequences for the binding of SRP they left free and they are delivered into cytosolic space and the proteins can assume to specific conformation guided by chaperone, but before the ends assume any structural form they are bound by HsP proteins to prevent such folding.�
At the same time there specific receptor proteins that bind to signal sequence and directs the protein to mitochondrial transport receptor located at the TOM outer region of the channel. During transfer receptors recognize specific mitochondrial signal sequences in the protein and binds to them, and then they are threaded into the channel by ATP dependent manner.� Then the N-terminal region of the protein is handed over to intermediate TIMs (Tim 9, 10 and 12 located in interspaces� or what is called peri-mitochondrial space), then the protein is transferred to TIMs 22-54 channel proteins and the protein is transferred to mitochondrial matrix.� But the entry is facilitated by the HsP70 and MGE (= Grp-E), facilitated by Tim 44, which not only binds to the N-terminal part of the protein but also pulls the protein inwards.� As the N-terminal segment of the protein enters the matrix, the signal sequences are cut.� If the protein destined to go into inner membrane the protein uses the second signal sequence.
���
The protein subunit IV of Cyt.C1 has 61 amino acids at the N-��� terminal end. This sequence is not recognized by SRP protein complex. The first 32 amino acids are used to transport into mitochondrial matrix and the next 19 amino acids are used for targeting protein into inner membrane.
�������� Cyt.C1: NH2-----32 aa-II-29aa-I�DHFR---------------------COOˉ
�
A recombinant DNA coding for DHFR containing two mitochondrial signal sequences, one for matrix and the second for peri-mitochondrial space in to a system, the DHFR ultimately lands up in peri-mitochondrial space.
Some mitochondrial proteins have two signal sequences, one at N-terminal end, which facilitates the entry into the mitochondrial matrix.� The matrix signal sequences are cleaved.� The second sequence facilitates the entry into peri-mitochondrial space, there water soluble protease cleaves the second sequence, but it requires Mg 2+. �Transport to each of the destinations requires ATP energy.� An electro potential gradient across the membrane is an important force required for the protein to be transported across the membranes.
�Role of energy in protein import into the mitochondrial matrix:
�
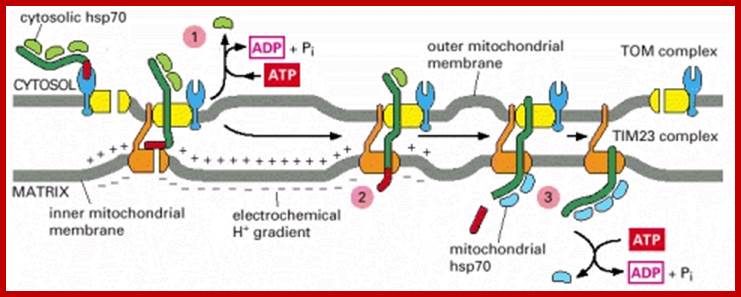
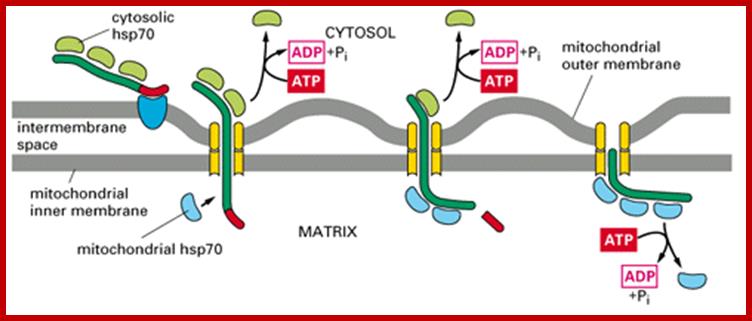
�(1) Bound cytosolic hsp70 is released from the protein in a step that depends on ATP hydrolysis. After initial insertion of the signal sequence and of adjacent portions of the polypeptide chain into the TOM complex, the signal sequence interacts with a TIM complex. (2) The signal sequence is then translocated into the matrix in a process that requires an electrochemical H+ gradient across the inner membrane, positioning the unfolded polypeptide chain so that it transiently spans both membranes. (3) Mitochondrial hsp70 binds to regions of the polypeptide chain as they become exposed in the matrix, thereby �pulling� the protein into the matrix. ATP hydrolysis then removes the mitochondrial hsp70, allowing the imported protein to fold; Requires two signal sequences; http://cc.scu.edu.cn/
Cyt.C oxidase subunit IV- leader sequence:
3HN-M - -+- + - +32 - - - - - - - -A-49-IS-T - - - - - -A-61-I- - - -
----- Charged---��� ----����� � ��uncharged---
Cyt.C oxidase:� NH2-MLSLRFFK PATRTLCSSRYLL//
Cyt. C1: MFSNLSKRWAQRTLSKSFYSTATGAASL-
�������� SGKLT//EKLVTAGVAAAGITASTLLYADSLTAEA// ( NCBI book)
Mitochondria perse lack SRP system for targeting proteins cotranslationally into mitochondrial membranes. Instead mit ribosomes directly bind to inner membranes to transfer proteins into membranes.� It requires Oxa1 insertase for ribosome binding to membrane and Mba1 ribosome receptor.� Transgenically FtsY, Srp and Ffth were introduced into yeast mitochondria, but there was no improvement in incorporation of motochondrially made proteins into their membranes (FEBS 2013, Funes S etl).
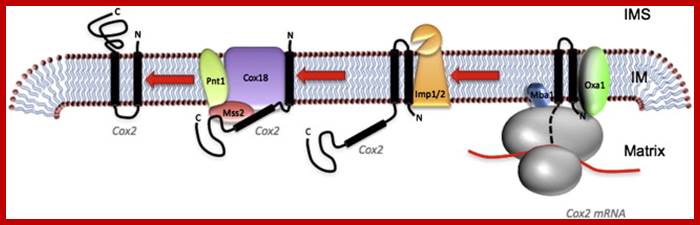
��������������
The role of Oxa1 and Oxa2 in the membrane biogenesis of the mitochondrial Cox2 protein. Cox2 is synthesized in a precursor form with an N-terminal signal peptide. The precursor protein is cotranslationally targeted to the membrane via the interaction of the ribosome�Cox2 nascent chain complex with Oxa1 and Mba1. After Oxa1 mediated the insertion of the N-terminal region of Cox2, Imp1 (of the Imp1/2 complex) cleaves off the N-terminal signal peptide. The large C-terminal domain of Cox2 is subsequently translocated across the membrane by Oxa2 (Cox18) together with Pnt1 and Mss2; � Peng Wang, � Ross E. Dalbey;BBA 2011
Transport into Plastids:
Plasmid perse develops from proplastids and requires many signals for the full development.� Among the signals light is very important.
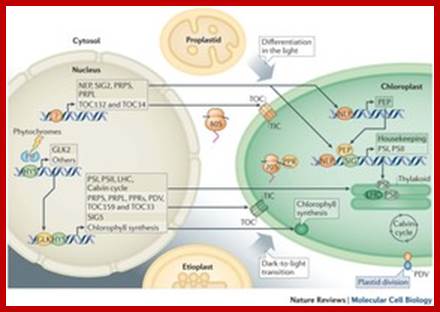
� Light-mediated anterograde control of chloroplast development. Paul Jarvis & Enrique L�pez-Juez
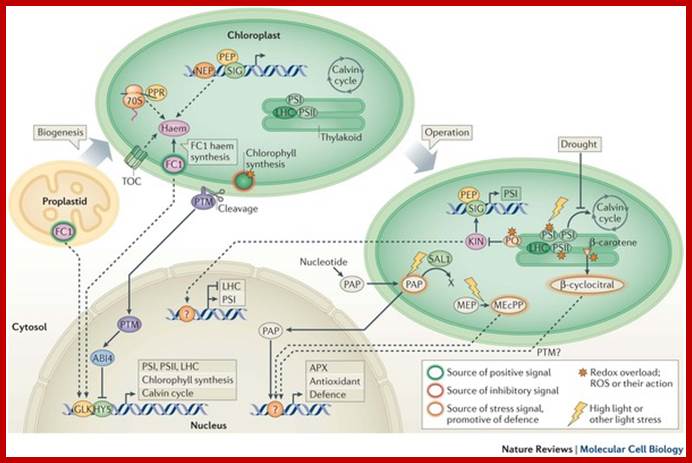
- Chloroplast and nucleus interaction; Chloroplasts are the organelles that define plants, and they are responsible for photosynthesis as well as numerous other functions. They are the ancestral members of a family of organelles known as plastids. Plastids are remarkably dynamic, existing in strikingly different forms that interconvert in response to developmental or environmental cues. The genetic system of this organelle and its coordination with the nucleocytosolic system, the import and routing of nucleus-encoded proteins, as well as organellar division all contribute to the biogenesis and homeostasis of plastids. They are controlled by the ubiquitin�proteasome system, which is part of a network of regulatory mechanisms that integrate plastid development into broader programmes of cellular and organismal development. �Paul Jarvis & Enrique L�pez-Juez; http://www.nature.com/
Plastids exist in different forms such as pro-plastids that develop into fully formed plastids, either as chloroplasts, chromoplasts, thioplasts, leucoplasts or proteinoplast.� A green plastid, the chloroplast is highly differentiated into stromal lamellae and granal lamellae.� These are embedded in a fluid called stromal fluid.� Chloroplast possesses its own genome, whose size varies from one system to the other, however, though the size, is limited, and they contain several copies of the genome.� The plastid genome encoded with 2 sets of rRNA genes (in inverted orientation), 27 or more tRNA genes and 120 or more chloroplast resident protein coding genes; many of the protein coding genes are organized in thylakoid membranes for light harvesting as photosystems PSI And PSII.� The rest of the proteins may be in many hundreds, located in matrix and membranes they have to come from cytoplasm as they are nuclear coded and translated in cytoplasm.
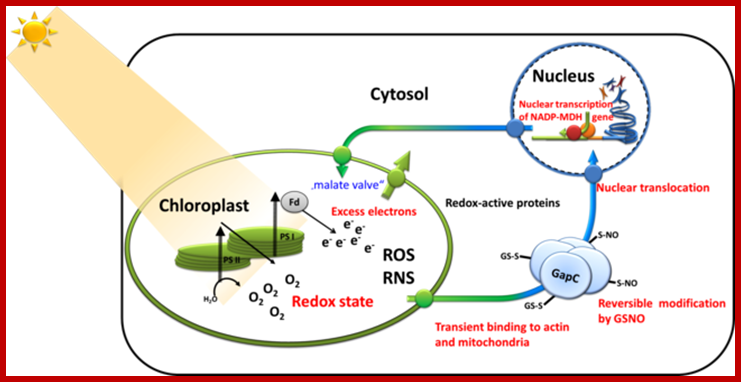
Chloroplast biogenesis and function depend on the concerted action of the nuclear and organellar genetic systems; While the majority of chloroplast proteins are nucleus-encoded, the chloroplast genome encodes a relatively small number of genes, the products of which are mainly required for photosynthesis and chloroplast gene expression. ;Jean-David Rochaix ;http://nano.nstl.gov.cn/ Retrograde signaling of chloroplast redox-state to nucleus. http://www.biologie.uni-osnabrueck.de/
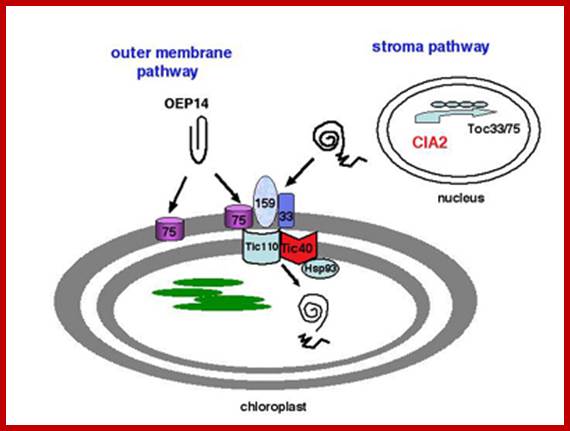
Transport into plastid stroma
�
These proteins as in mitochondria, as they emerge from the surface of large ribosomes, as they don�t have sequences for the binding of SRPs, they are threaded into cytoplasm. Many HSPs bind to such proteins to prevent unwanted folding.� At the same time chloroplast receptor proteins bind to specific signal sequences.� Then they are handed over to receptor cum translocator channels found at attachment site i.e. at junction of outer and inner membranes.� Such receptor-channel protein complexes are found in large numbers to facilitate the transport of proteins.� Matrix proteins require just one signal, but those proteins that are destined to thylakoid membranes, require two such sequences, one for the matrix and another for thylakoid membranes. �
According to studies, the thylakoid proteome consists of at least 335 (now it is known there are ~2800 proteins) different proteins. Out of these, 89 are in the lumen, 116 are integral membrane proteins, 62 are peripheral proteins on the stroma side, and 68 peripheral proteins on the lumenal side. Additional low-abundance lumenal proteins can be predicted through computational methods. �Of the thylakoid proteins with known functions, 42% are involved in photosynthesis. The next largest functional groups include proteins involved in protein targeting, processing and folding with 11%, oxidative stress response (9%) and translation (8% ) (Wikipedia). The electron transport protein plastocyanin is present in the lumen and shuttles electrons from the Cytochrome b6f protein complex to photo system I. While plastoquinones are lipid-soluble and therefore move within the thylakoid membrane, plastocyanin moves through the thylakoid lumen.
The lumen of the thylakoids is also the site of water oxidation by the oxygen evolving complex associated with the lumenal side of photo system II.
Lumenal proteins can be predicted computationally based on their targeting signals. In Arabidopsis, out of the predicted lumenal proteins possessing the Tat signal, the largest groups with known functions are 19% involved in protein processing (proteolysis and folding), 18% in photosynthesis, 11% in metabolism, and 7% redox carriers and defense.
Chloroplasts are derived from blue-green algae often called cyanobacteria; compare the thylakoid in the above diagram with bacterial cell membranes. Thylakoids (green) inside a cyanobacterium (Synechocystis); http://en.wikipedia.org/
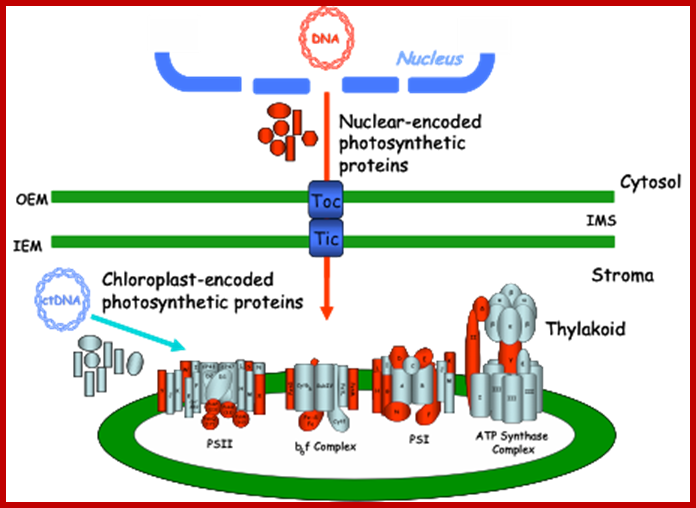
Biogenesis of the core photosynthetic complexes requires the coordinate assembly of both nuclear- and chloroplastic-encoded proteins. Nuclear-encoded photosynthetic complex proteins must first cross the envelope membrane using the Toc/Tic complex (in Blue) prior to their targeting and insertion into the thylakoid membrane. Nuclear-encoded photosynthetic proteins are shown in red and chloroplastic-encoded photosynthetic proteins are shown in aqua. Toc, translocon at the outer envelope membrane of chloroplasts;Tic, translocon at the inner envelope membrane of chloroplasts; OEM, outer envelope membrane; IMS, intermembrane space; IEM, inner envelope membrane proteins. Prof. John Froelich; http://www.prl.msu.edu/
Chloroplasts contain outer and inner membrane specific protein complexes which act as transporters. Once proteins are drawn into the stromatic fluid, many of them get integrated into granal membranes, which requires plastid SRP-RNA-protein complex for the movement into thylakoid membranes.
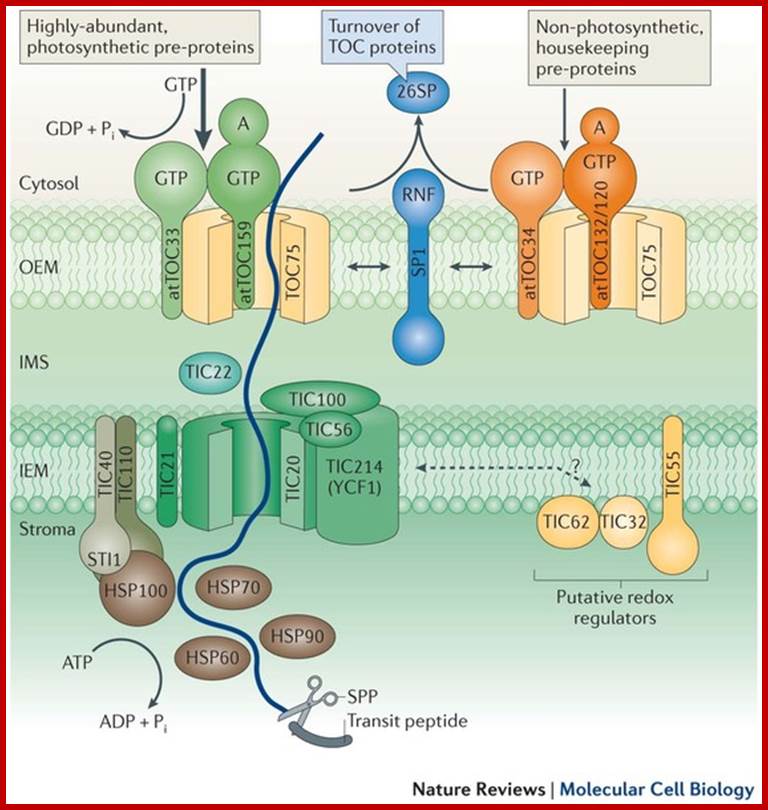
� Biogenesis and homeostasis of chloroplasts and other plastids; Paul Jarvis & Enrique L�pez-Juez; /http://www.nature.com/
In plants, nuclear coded proteins, for both mitochondrial and plastids are synthesized in the same cytoplasmic space, using specific signal sequences and specific receptors, they are differentially sorted out and targeted to their respective destinations.� Once they enter the inner space of the organelles many remain in the matrix and others are further targeted to their respective inner membranes, sometimes few specific proteins move into periplasmic space.
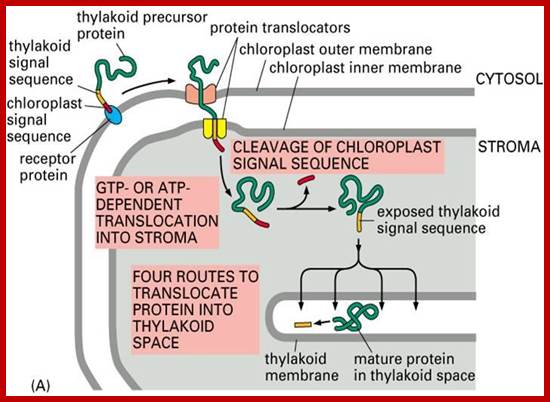
��������������������������������� Chloroplast proteins contain signal peptide for entry and another signal sequence ������������� �for the entry into Thylakoid membranes. http://webcache.googleusercontent.com/
Transport pathway into and within chloroplasts;
http://webcache.googleusercontent.com/
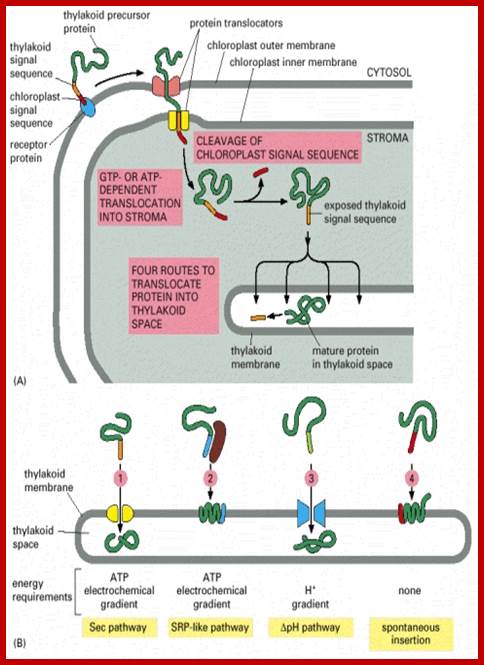
�Sorting all chloroplast proteins (fig above) and translocation of the same into the thylakoid membrane and space of chloroplasts: The precursor polypeptide contains an amino-terminal chloroplast signal peptide (red) followed immediately by a thylakoid signal peptide (orange). The chloroplast signal peptide initiates translocation into the stroma through a membrane contact site by a mechanism similar to that used for translocation into the mitochondrial matrix. The signal peptide is then cleaved off, unmasking the thylakoid signal peptide, which initiates translocation across the thylakoid membrane.
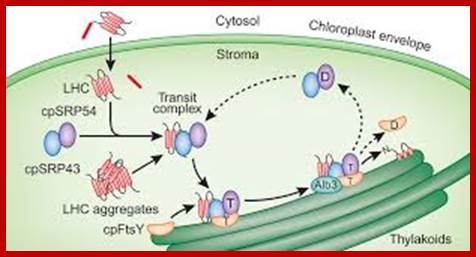
Cp SRP mediated Transport:
According one view chloroplasts contain SRPs similar to that of bacterial cytoplasmic SRPs, but the Cp.SRP RNA is 4.5s and associated with Ffh (FtsY). Cp.SRP protein (homologue of cytoplasmic 54kda protein).� The CpSRP located in stroma binds to the translocated protein and then it is guided by another protein FtSY (a homologue of bacterial system).� These complexes transfer the protein into thylakoid membranes.� Then they are released as 4.5S-(54)-48 unit and SRP denoted as CpFtsY (bacterial SRP receptor) also acts as SRP receptor in chloroplasts. These complexes transfer the protein into thylakoid membranes.
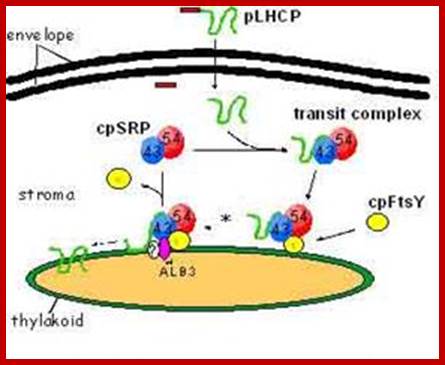
Cp-SRP mediated transport into thylakoid membranes; Hypothesised model for posttranslational LHCP localization buy SRP. GTP-bound and GDP-bound states (Tand D respectively) of cpSRP and cpFtsy are predicted by anlogy to ER targeting models (Keenaan et al 2001) and from adssays tat reconstitute� the soluble targeting complex (transit complex) in the absence of guanine nucleotides.� Membrane complexes containing cpSRP are described in Moore et al (Cell Biology.2003); mol. biol
Chloroplast SRP (cpSRP) contains a SRP54 homologue but differs strikingly from cytosolic SRP in various aspects of structure and function. In contrast to cytosolic SRP, it contains another novel protein subunit (cpSRP42-43) and lacks RNA. In higher plants plastid the Srp lacks RNA but has been detected the Srp RNA in some red algae.� cpSRP is also distinctive in its ability to interact with its substrate, ex. light-harvesting chlorophyll a/b-binding protein. �Furthermore, it is remarkable that the 54 kDa like subunit of cpSRP is also involved in the co-translational transport of chloroplast-encoded thylakoid proteins, and is therefore able to switch between the co- and post-translational means of interaction with its respective substrate proteins (S.Hohmann). Bacterial SRP54/Ffh homologs are cpSRP54 and cpSRP43.� Bacterial FtsY is bound to thylakoid membranes. The chloroplast SRP (cpSRP) is defined as a collection of four proteins that work together, including CpSRP54, CpSRP43, CpFTSY, and ALB3 (Henning Kirist). Chloroplast employs two distinct protein transport, one post translation and another cotraslational event.� Post translation process use srp54 and 43, but cotranslation process uses onlySrp 54.
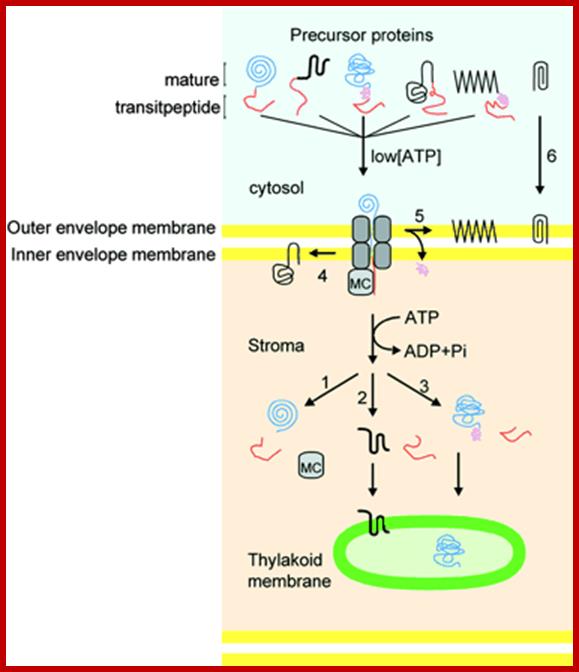
Schematic Representation of Pathways Responsible for Targeting Proteins to Their Proper Location within Chloroplasts. The large majority of precursor proteins have a cleavable transit peptide (shown in red). In most cases, the transit peptide includes a stromal-targeting domain that initiates transport of the precursor through the general import pathway (shown in gray). The driving force for protein translocation is thought to be provided by molecular chaperones (MC) that pull precursor proteins into plastids. Precursors lacking additional targeting information are thus deposited into the stromal space, where the stromal-processing protease removes the transit peptide (path 1). For precursors destined for insertion into membranes, the additional targeting information generally is contained within the mature region of the protein (paths 2 and 4), although some proteins may require a stop-transfer signal for localization to the outer membrane (path 5). Precursors for some outer membrane proteins lack a cleavable transit peptide and are inserted directly into the outer membrane without using the general import pathway (path 6). Some precursors destined for the thylakoid lumen require a second targeting signal (shown in purple), which is cleaved as the proteins enter the lumen (path 3). See text for additional details; Kenneth Keegstraa,1 and Kenneth Clineb
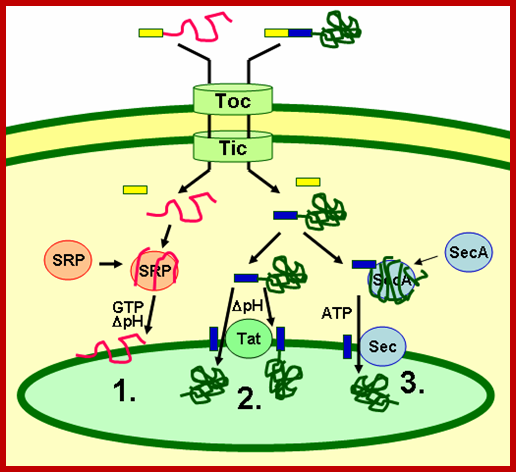
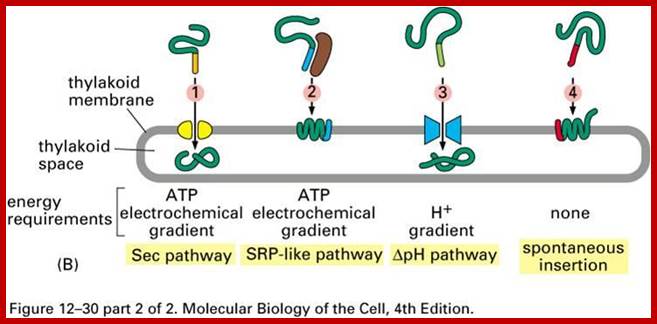
Three modes of transport into thylakoid membranes- Srp, Tat and Sec modes.
Proteins are inserted into the membrane via the SRP-dependent pathway (1), the Tat-dependent pathway (2), or spontaneously via their transmembrane domains (not shown in figure). Luminal proteins are exported across the thylakoid membrane into the lumen by either the Tat-dependent pathway (2) or the Sec-dependent pathway (3) and released by cleavage from the thylakoid targeting signal. The Sec (secretory) pathway requires ATP as energy source and consists of SecA, which binds to the imported protein and a Sec membrane complex to shuttle the protein across. Proteins with a twin-arginine motif in their thylakoid signal peptide are shuttled through the Tat (twin arginine translocation) pathway, which requires a membrane-bound Tat complex and the pH gradient as an energy source. Some other proteins are inserted into the membrane via the SRP (signal recognition particle) pathway. The chloroplast SRP can interact with its target proteins either post-translationally or co-translationally, thus transporting imported proteins as well as those that are translated inside the chloroplast. The SRP pathway requires GTP and the pH gradient as energy sources. Some transmembrane proteins may also spontaneously insert into the membrane from the stromal side without energy requirement. http://en.wikipedia.org/
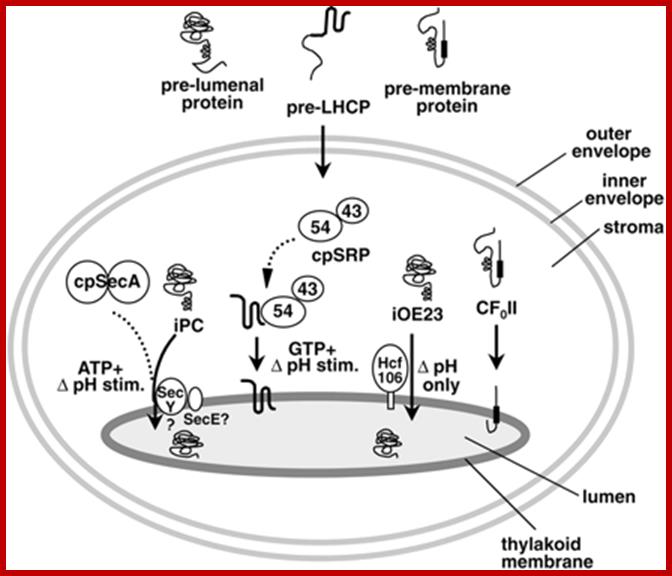
A Working Model for the Routing of Lumen-Resident and Integral Membrane Thylakoid Proteins via Four Precursor-Specific Pathways.
Proteins requiring cpSecA and ATP (e.g., PC [shown], OE33, PSI-F, and cytochrome f) are presumed to move across the membrane through a pore consisting of cpSecY and cpSecE. This pathway is referred to as the Sec pathway. Integration of the LHCP requires cpSRP, which consists of cpSRP54 and cpSRP43. Another stromal component and a membrane component (not shown) also are required. This pathway is referred to as the SRP-like pathway. OE23 (shown), OE17, PSII-T, and PSI-N require the membrane protein Hcf106p for transport. This pathway is called the ΔpH pathway because the transthylakoid ΔpH is the sole energy source for transport. A fourth pathway mediates the insertion of the membrane proteins CF0II (shown), PSII-X, PSII-W, and ELIP, presumably by a spontaneous mechanism. Evidence for pathway-specific groupings is described in the text. Question marks denote components that are presumed but not yet demonstrated to operate on a pathway. iOE23 and iPC are the stromal intermediate forms of OE23 and PC, respectively. stim., stimulated; Protein Import and Routing Systems of Chloroplasts ,Kenneth Keegstraa,et al
����������������������������� Chloroplast SRP are used to target proteins into thylakoids;
SRP: adapting to life in the Chloroplast; Once inside the chloroplast, transfer of thylakoid protein takes place in three modes- SEC mode, TAT mode and SRP mode. Ralph L Henry;http://www.nature.com/
Three modes of Transport- SRP, TAT and SEC mediated:
��Thylakoid proteins are targeted to their destination via signal peptides and prokaryotic-type secretory pathways inside the chloroplast. Most thylakoid proteins encoded by a plant's nuclear genome need two targeting signals for proper localization: An N-terminal chloroplast targeting peptide (shown in yellow in the figure), followed by a thylakoid targeting peptide (shown in blue). Proteins are imported through the translocon of outer and inner membrane (Toc and Tic) complexes. After entering the chloroplast, the first signal peptide is cleaved off by a protease processing imported proteins. This unmasks the second targeting signal and the protein is transported from the stroma into the thylakoid in a second targeting step. This second step requires the action of protein translocation components of the thylakoids and is energy-dependent. Proteins are inserted into the membrane via the SRP-dependent pathway (1), the Tat-dependent pathway (2), or spontaneously via their transmembrane domains (not shown in figure). Luminal proteins are transported across the thylakoid membrane into the lumen by either the Tat-dependent pathway or the Sec-dependent pathway (3) and released by cleavage from the thylakoid targeting signal. The different pathways utilize different signals and energy sources. The Sec (secretory) pathway requires ATP as energy source and consists of SecA, which binds to the imported protein and a Sec membrane complex to shuttle the protein across�.
����������������������������������������� ��������������������������� http://webcache.googleusercontent.com/� ���������������������������
Proteins with a twin arginine motif in their thylakoid signal peptide are shuttled through the Tat (Twin Arginine translocation) pathway, which requires a membrane-bound Tat complex and the pH gradient as an energy source. Few other proteins are inserted into the membrane via the SRP (signal recognition particle) pathway. The chloroplast SRP can interact with its target proteins either post-translationally or co-translationally. They are involved in transporting imported proteins as well as those that are translated inside the chloroplast. The SRP pathway requires GTP and the pH gradient as energy sources. Some transmembrane proteins may also spontaneously insert into the membrane from the stromal side without energy requirement.
Peroxisomes and Glyoxysomes:
Organelles that often coexist are Peroxisomes and Glyoxysome, twin organelles.� They are also called micro bodies.� Glyoxysome are more or less restricted to plants.� Peroxisomes are very small structure and they grow into large sized organelles. They are found both in plant and animal cells. �Average size is 0.3 - 1.2 � in diameter. They are distinguished into Peroxisomes and Glyoxysomes by specific staining protocols. The two types differ in their functions: peroxisomes are the sites of glyoxylate cycle and photorespiration, while Glyoxysomes are in charge of the mobilization of storage compounds (fats) into utilizable form of components. Both peroxisomes and Glyoxysomes contain crystalline inclusions. �These cell organelles are bound by one unit membrane.� The membrane consists of many integral and many channel cum receptor proteins.
Peroxisomes are vesicular structures containing proteins called Peroxins.� It has been estimated that more than 26 nuclear genes are involved in the development of peroxisomes.� Their main function is detoxifying peroxides or peroxide radicals into water by catalases.� But Peroxidase enzymes can also generate harmful hydrogen peroxides, but they are removed immediately by catalases.
RH2 + O2 � R + H2O
All cell organelles, which have membranes as their structural component, require specific pre-existing particles for their growth and multiplication.� Pre-existing peroxisomal membrane vesicular structures are required for the assembly and growth of peroxisomes. Peroxisomes or Glyoxysomes are bound by single unit membranes similar to that of Lysosomes.� Few studies that indicate that there is relationship between development of peroxisomes from the tips of ER for some of the enzyme components in the ER and peroxisomes are similar.
Microbodies are not endosymbiotic organlees. Peroxisomes associated with chloroplasts and mitochondria; http://plantphys.info/ http://www.pcsd.k12.ny.us/-http://plantphys.info/
Microbodies were the original name for small single-membrane bound organelles. In plants these have been renamed peroxisomes and glyoxysomes. The peroxisome is shown with its crystalline matrix of catalase below. This organelle mostly degrades glycolate, a 2-C acid produced in chloroplasts as the result of[KG1] [KG2] [KG3] RuBisCO combining with oxygen rather than carbon dioxide, in the process of photorespiration. The glycolate is transferred into the peroxisome. In degrading glycolate, oxygen is consumed and peroxide (H2O2) is produced. This toxic material is enzymatically degraded to water and oxygen by the enzyme catalase. This critical enzyme may comprise 40% of the protein in the peroxisome; little wonder catalase appears in crystalline form in peroxisomes. Mitochondria are a third-partner in photorespiration.;http://plantphys.info/
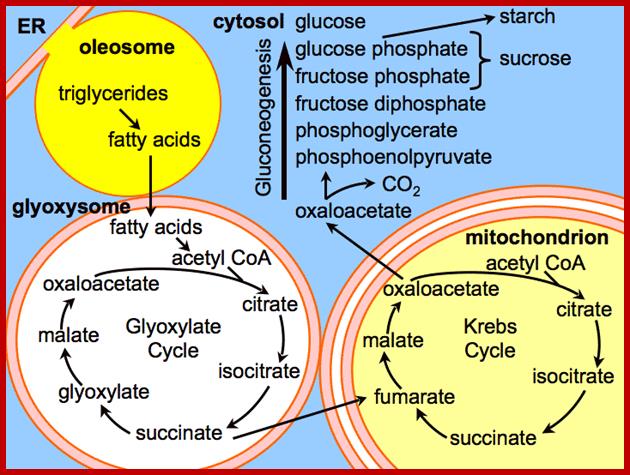
Glyoxysomes are abundant in oil storage tissues in plant.� These micobodies contain enzymes to degrade fatty acids into 4-C acids, which drives mitochondrial respiration and reverse glycolysis to make sugars for growth and development.
http://plantphys.info/plant_physiology/images/glyoxylatecycle.gif
Oleosomes: Note partners of Glyoxysomes are Oleosomes.� These organelles are spherical and bound by single layer of phospholipids.� Hydrophilic PL faces cytosol and hydrophobic end faces the interior. Hydrophilic surface possesses many proteins. These are spherical and they are bound by phosphor lipid layer.� Interior of oleosomes are loaded with hydrophobic oils from ER.� There are pheripheral proteins on the hydrophilic side of monolayer.� These proteins include oleocins which may help attach the enzyme lipase to this monolayer to initiate fat digestion for glyoxylate cycle.
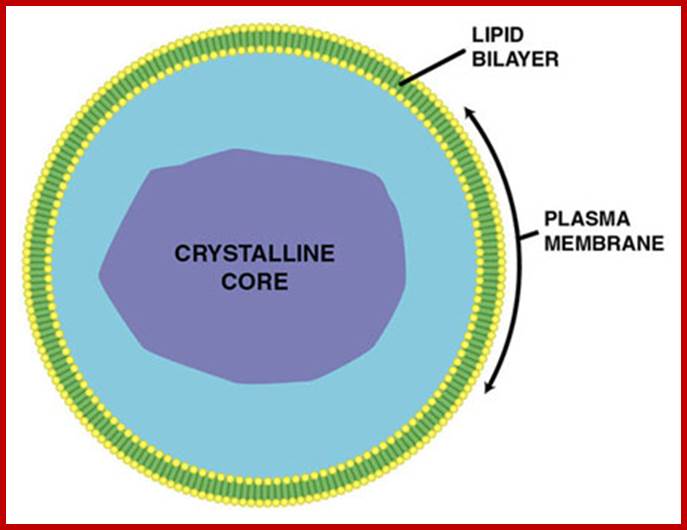
The crystalline core consists of protein aggregates; bound by a singl membrane; it breaks down metabolic H2O2; http://en.wikipedia.org/; http://de.academic.ru/
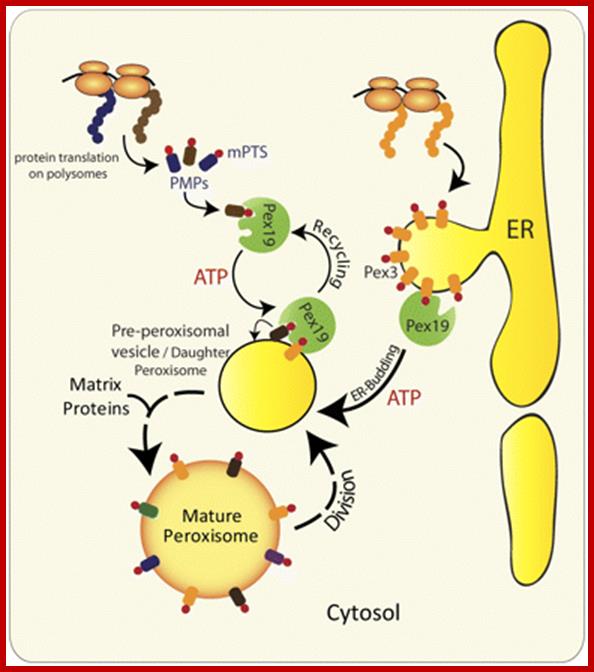
Alternative roles of Pex19 in the insertion of PMPs into the peroxisomal membrane. The role of Pex19 in peroxisome biogenesis and import of various PMPs has been clearly established in yeast and mammals, but its mechanism of action is still a matter of debate (Snyder et al., 1999; Sacksteder et al., 2000). Previous studies implicated Pex3 and Pex19 in the posttranslational insertion of PMPs. Pex19 serving as a chaperone binds and stabilizes newly synthesized mPTS-containing PMPs in the cytoplasm, and transports them to peroxisomes by docking to Pex3 present in the peroxisomal membrane (Muntau et al., 2003; Fang et al., 2004; Jones et al., 2004; Matsuzono and Fujiki, 2006; Matsuzono et al., 2006). However, subsequent studies in yeast show the requirement of Pex19 for the exit of most, if not all PMPs, including Pex3, from the ER (Fig. 2 B; Hoepfner et al., 2005; Lam et al., 2010; van der Zand et al., 2010; unpublished data). In the light of the Pex19-independent insertion of most PMPs into the ER and the role of Pex19 in mediating the budding of pre-peroxisomal vesicles, the role of Pex19 in the posttranslational import of PMPs is questionable for all PMPs that go to peroxisomes via the ER. http://jcb.rupress.org/

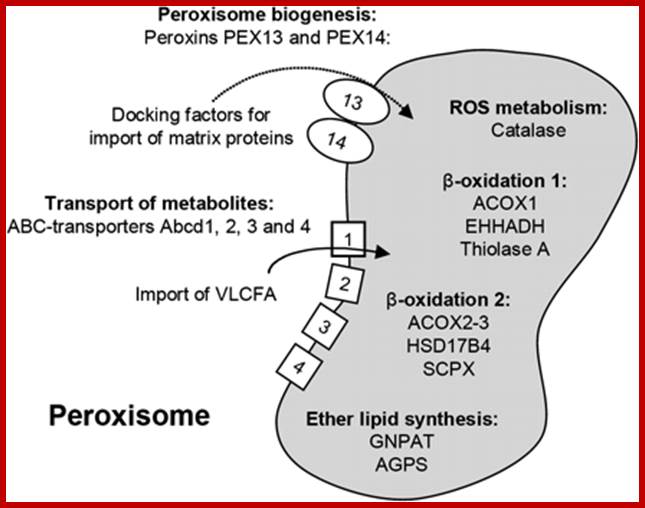
Scheme of Peroxisomes with the marker proteins used show peroxisomal proteins as groups representing main functions for biogenesis and metabolism of peroxisomes. http://imgarcade.com/
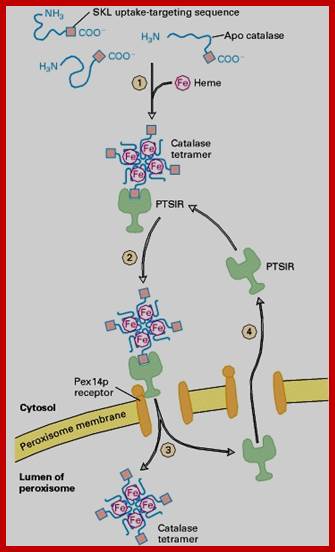
Newly made catalase subunits are released from polyribosomes into the cytosol asapocatalase, amonomer that contains a C-terminal SKL
uptake-targeting sequence (red) but lacks an iron-containing heme group. Step 1: Four monomers are assembled and heme is added,
forming the mature tetrameric catalase molecule. Steps 2 and 3: The cytosolicreceptor protein PTS1R binds an SKL signal and escorts
the catalase tetramer to the Pex14preceptor on the peroxisome membrane.
In the model depicted here, the catalase-PTS1R complex is transported across
the membrane and then dissociates in the lumen.
As-yet uncharacterized proteins on the peroxisomal membrane probably form part of thereceptor-transport
channel. Step 4:
PTS1R returns to the cytosol to pick up another peroxisome-destined protein.
In an alternative model, PTS1R delivers catalase to the Pex14p receptorbut
does not enter the lumen itself. ;Synthesis
of catalase and its incorporation into peroxisomes:
C-terminal signal sequences are SKL. http://board.212cafe.com/
Newly made catalase subunits are released from polyribosomes into the cytosol as apocatalase, a monomer that contains a C-terminal SKL uptake-targeting sequence (red) but lacks an iron-containing heme group. Step 1: Four monomers are assembled and heme is added, forming the mature tetrameric catalase molecule. Steps 2 and 3: The cytosolic receptor protein PTS1R binds an SKL signal and escorts the catalase tetramer to the Pex14p receptor on the peroxisome membrane. In the model depicted here, the catalase-PTS1R complex is transported across the membrane and then dissociates in the lumen. As-yet uncharacterized proteins on the peroxisomal membrane probably form part of the receptor-transport channel. Step 4: PTS1R returns to the cytosol to pick up another peroxisome-destined proteins. In an alternative model, PTS1R delivers catalase to the Pex14p receptor but does not enter the lumen itself. [See R. A. Ruchubinski and S. Subramani, 1995, Cell 83:525; J. A. McNew and J. M. Goodman, 1996, Trends Biochem. Sci. 21:54; and M. Albertini et al., 1997, Cell 89:83.]
Glyoxysomes:
Cells contain very small structures, often called micro bodies; one such structure is called Glyoxysome, mostly found in plants.� In plants they are interconvertible. They are involved in carbohydrate/fatty acid metabolism in association with mitochondria.
Glyoxysomes; http://tgesbiology.weebly.com/
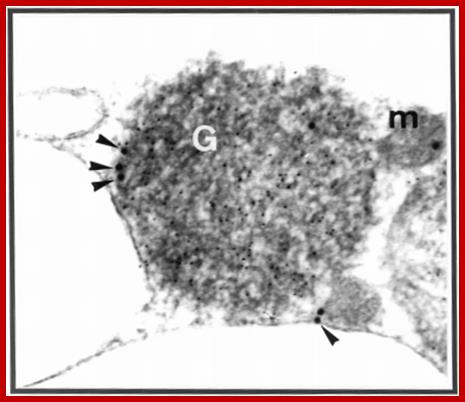
A representative electron micrograph of cucumber cotyledon thin sections illustrating results of indirect, double immunogold labeling with potato porin antiserum and anti-pumpkin seed catalase. The 15-nm gold particles show the peripheral localization of porin. The 5-nm gold particles show the matrix localization of catalase. G, glyoxysome; m, mitochondrion. The magnification is 90,000�. Glyoxysomes contain porin like-polypeptides in� the boundry membranes of oil seed Glyoxysomes; http://pcp.oxfordjournals.org/
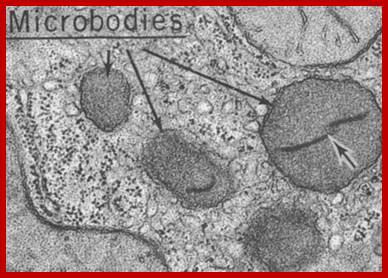
Microbodies
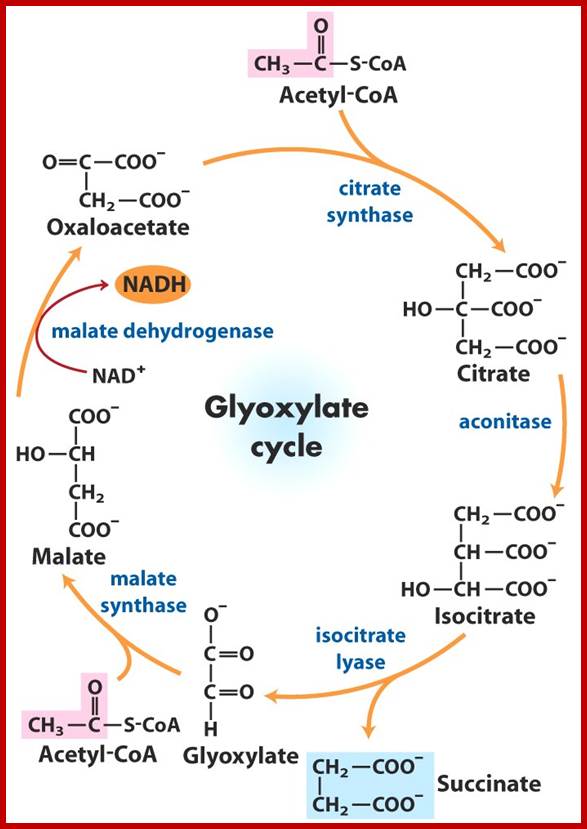
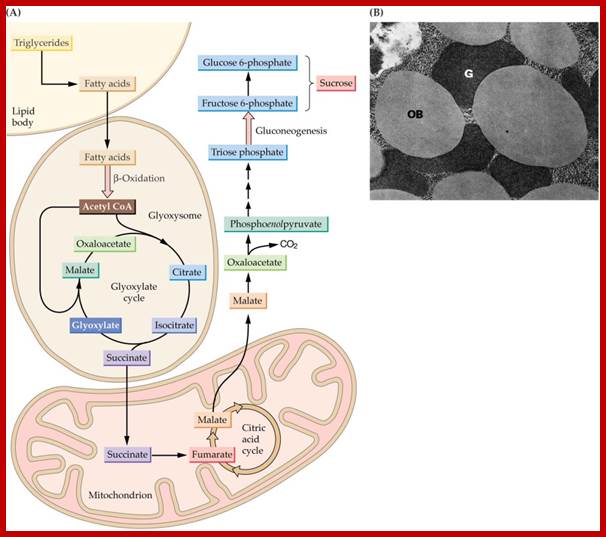
Glyoxysomes are specialized peroxisomes found in plants (particularly in the fat storage tissues of germinating seeds) and also in filamentous fungi. In glyoxysomes the fatty acids are hydrolyzed to acetyl-CoA by peroxisomal β-oxidation enzymes. Besides peroxisomal functions, glyoxysomes possess additional key enzymes of glyoxylate cycle (isocitrate lyase and malate synthase) which accomplish the glyoxylate cycle bypass.
Thus, Glyoxysomes (as all peroxisomes) contain enzymes that initiate the breakdown of fatty acids and additionally possess the enzymes to produce intermediate products for the synthesis of sugars by gluconeogenesis. Oil rich seeds such as castor, the seedlings of them convert oil/fat into glucose like compounds and use these sugars produced until it is mature enough to produce them by photosynthesis
Apart from the well-known compartments just discussed occur in a variety of so-called microbodies in plant cells. On average are they of 0.3 - 1.2 � diameter. They are distinguished into peroxisomes and glyoxysomes.
The two types differ in their functions: peroxisomes are the sites of photorespiration or oxidation of RH to H2O and R, while glyoxysomes are in charge of the mobilization of storage compounds (fats). Both peroxisomes and glyoxysomes contain crystalline inclusions.
The two compartment types are not distributed evenly. Peroxisomes are usually found in the vicinity of chloroplasts and mitochondria, glyoxysomes occur only near mitochondria.
A
glyoxysomes is a specialized peroxisome containing the enzymes of the glyoxalate
cycle. Glyoxysomes are particularly prevalent and important in germinating
seeds. Glyoxysomes are in charge of the mobilization of stored energy
compounds. Especially during growth, a large amount of carbohydrates are
required for the synthesis of new cell walls. The protein analysis of early
glyoxysomes and ER shows that glyoxysomes are derived from ER?
Energy Mobilization in Glyoxysomes:
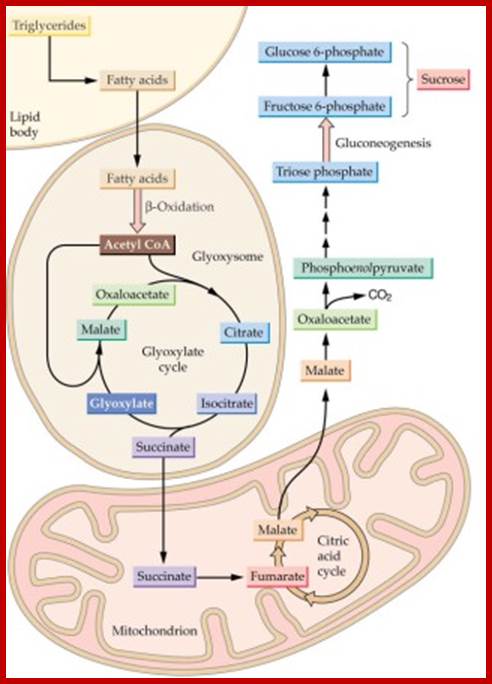
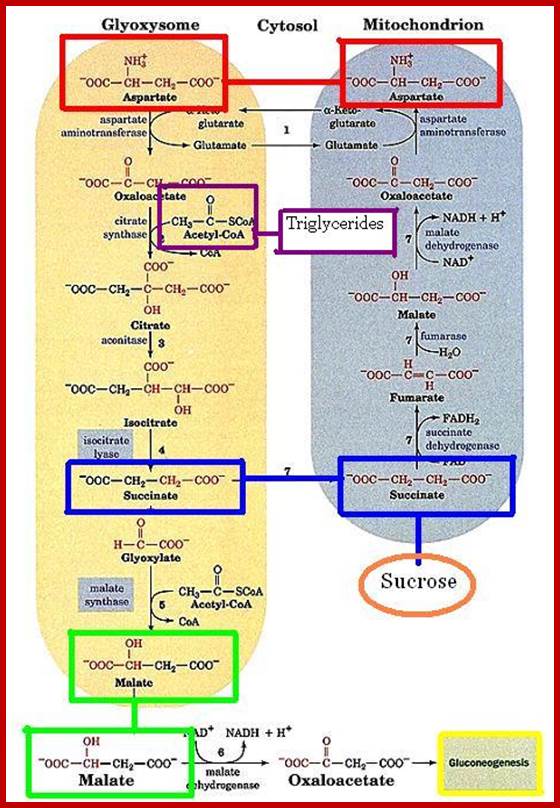
Triglycerides and aspartates enter the Glyoxysomes to be converted into succinate. The succinate is required in order for the energy-yielding process to continue through the mitochondria and finally to create sucrose in the cytosol. Malate can be shuttled directly into the cytosol to enter into a gluconeogenic pathway.
Glyoxysomes must borrow three reactions from Mitochondria:
Glyoxysomes do not contain all the enzymes needed to run the glyoxylate cycle; succinate dehydrogenase, fumerase, and malate dehydrogenase are absent. Consequently, glyoxysomes must cooperate with mitochondria to run their cycle. Succinate travels from the glyoxysomes to the mitochondria, where it is converted to oxaloacetate. Trans-amination to aspartate follows because oxaloacetate cannot be transported out of the mitochondria. Aspartate formed in this way then moves from the mitochondria back to the glyoxysomes, where a reverse transamination with alpha-ketoglutarate forms oxaloacetate, completing the shuttle. Finally, to balance the transamination, glutamate shuttles from glyoxysomes to mitochondria.
In plant cells glyoxysomes and mitochondria are in intimate contact with each other. http://www.biologyexams4u.com/; http://www.fns.uniba.sk/
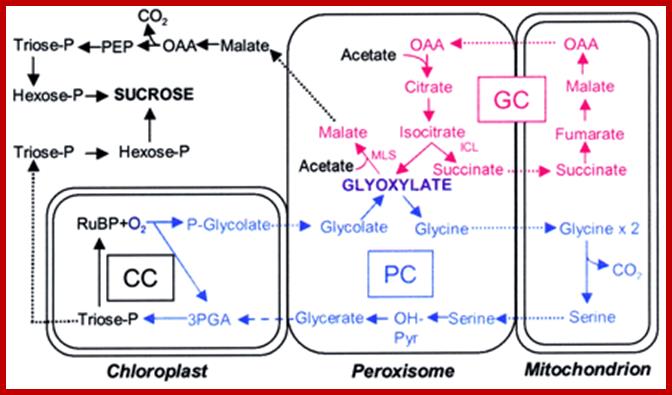
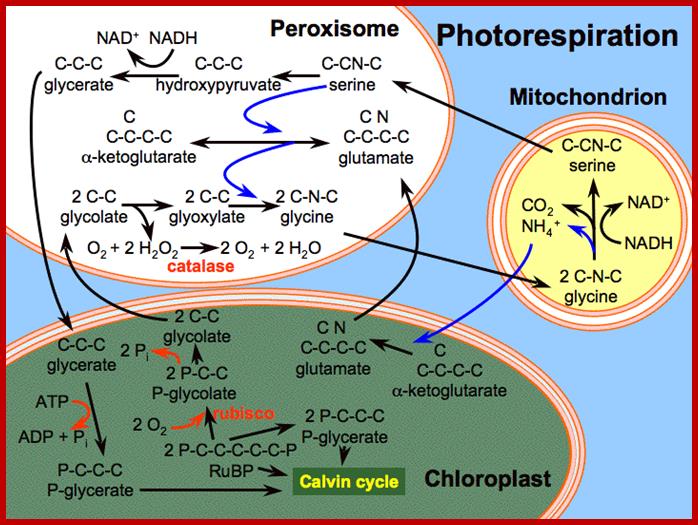
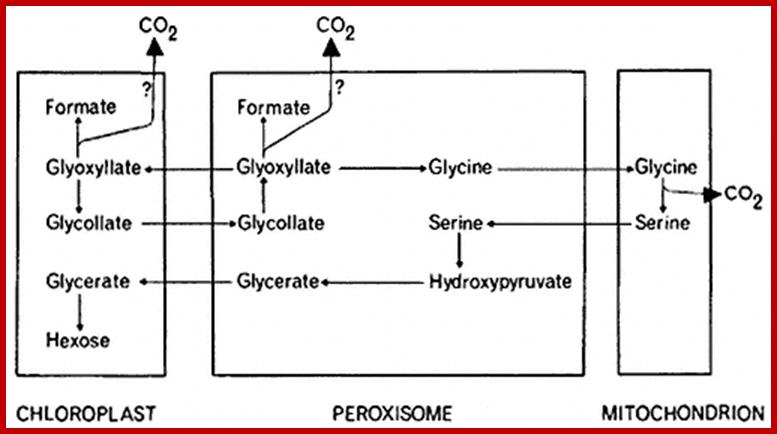
Alternative fates for glyoxylate in peroxisomes of Arabidopsis. Solid arrows indicate enzyme reactions, and dotted arrows indicate transport. The dashed arrow from glycerate to 3-phosphoglycerate (3PGA) is used because the location of glycerate kinase is unknown. CC, Calvin cycle; GC, glyoxylate cycle; OH-Pyr, hydroxypyruvate; OAA, oxaloacetate; PEP, phosphoenolpyruvate. http://www.jbc.org/
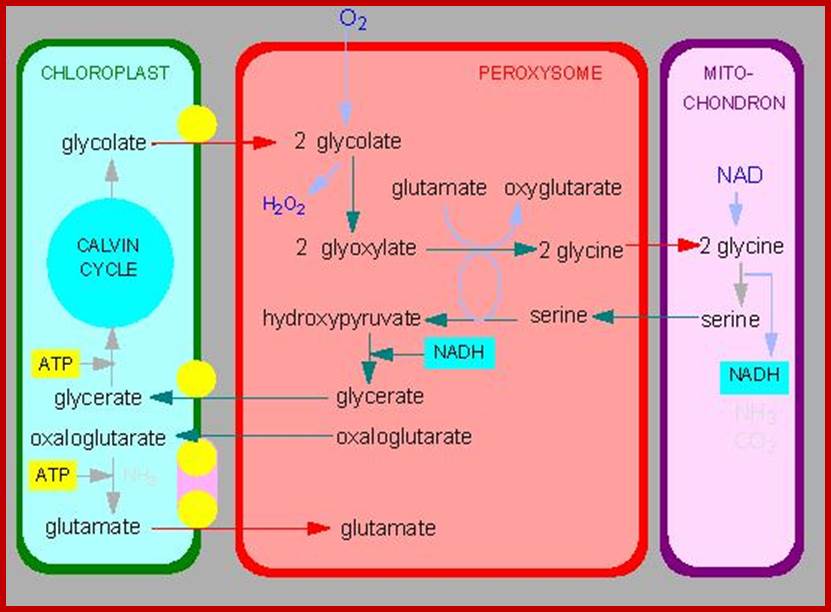
Photorespiration is a process at least partially opposing photosynthesis. In mitochondria develops carbon dioxide as a product of the serine synthesis. Serine is synthesized under light exposure and oxygen uptake from two glycerines. During the synthesis of glyoxalate from glycolate is the heavy cytotoxin hydrogen peroxide produced that is immediately afterwards broken down by the enzyme catalase. Catalase is an enzyme specific for peroxysomes. In addition is the fraction-1-protein involved, the enzyme that catalyzes carbon dioxide fixation during photosynthesis. It has two activities: Photorespiration involved cell organelles; http://www.biologie.uni-hamburg.de/
����������� In plants, during seed germination of oil rich seeds, mobilization of such reserve materials is very important.� This function is performed by Glyoxysomes.
The changes in activities of glyoxysomal and peroxisomal enzymes have been correlated with the fine structure of microbodies in cotyledons of the cucumber (Cucumis sativus L.) during the transition from fat degradation to photosynthesis in light-grown plants, and in plants grown in the dark and then exposed to light. During early periods of development in the light (days 2 through 4) the microbodies (Glyoxysomes) are interspersed among lipid structures and contain relatively high activities of glyoxalate cycle enzymes involved in lipid degradation. Thereafter, these activities decrease rapidly as the cotyledons expand and become photosynthetic, and the activity of glycolate oxidase rises to a peak (day 7); concomitantly the microbodies (peroxisomes) become preferentially associated with chloroplasts.
In seedlings grown in the dark for 10 days, the reserve lipid and the glyoxalate cycle enzyme activities persist for a longer time than in the light; correlated with this, there is a continued association of the microbodies with the lipid bodies. When these dark-grown seedlings are then exposed to 51 hours of the light-dark cycle, peroxisomal marker enzymes increase rapidly in activity, and the microbodies become oppressed to chloroplasts. We conclude that the characteristic association observed between glyoxysomes and lipid bodies reflects their mutual involvement in net gluconeogenesis through the conversion of fatty acids to carbohydrate, while the close spatial relationship observed between peroxisomes and chloroplasts at later stages of development reflects their mutual involvement in glycolate metabolism. Glyoxysomes are the site of the glyoxalate cycle that is tightly linked to the breakdown of fatty acids.
Although glyoxysomal enzyme activities are dropping rapidly while peroxisomal enzyme activities are increasing rapidly during the transition period in the light, the electron microscopic evidence does not indicate that glyoxysomes are being degraded or peroxisomes are being formed. Since in the dark-grown seedlings the activities of peroxisomal enzymes remain low and do not increase as they do in the light, an opportunity is afforded to compare quantitatively any changes in numbers of microbodies per cell with the changes in activities of glyoxysomal enzymes. It is found that the magnitude of the decrease in numbers of microbodies is considerably less than that of the decrease in glyoxysomal enzyme activities between days 4 and 10. When the cotyledons are exposed to light, peroxisomal enzyme activities increase greatly, but again there is no ultra-structural evidence for the synthesis of a new population of microbodies to accommodate this increase. These results allow us to conclude that the developmental transition from glyoxysomal to peroxisomal function almost certainly does not involve the actual replacement of one population of microbodies by another. Rather, the transition probably occurs within existing particles, either by a sequential functioning of two different kinds of microbodies or by a change in enzyme complement within a single population. Our findings with both light- and dark-grown cotyledons favor the latter possibility. The cytoplasmic invaginations into microbodies seen during greening of both light-grown cotyledons and etiolated cotyledons exposed to light may be morphological manifestations of the mechanism by which the microbodies lose or gain enzymes; Richard N. Trelease2, Wayne M. Becker, Peter J. Gruber3 and Eldon H. Newcomb
Photorespiration is a process at least partially opposing photosynthesis. In mitochondria carbon dioxide is generated as a product of the serine synthesis. Serine is synthesized under light exposure and oxygen uptake from two glycerines. During the synthesis of glyoxalate from glycolate is the heavy cytotoxic hydrogen peroxide produced that is immediately afterwards broken down by the enzyme catalase. Catalase is an enzyme specific for peroxisomes. In addition is the fraction-1-protein involved the enzyme that catalyzes carbon dioxide fixation during photosynthesis. It has two activities:
- Ribulose 1, 5 biphosphate carboxylase (RuBisCo) (photosynthesis).
- Ribulose biphosphate oxygenase (photorespiration).
Photorespiration is only indirectly dependent on light. The light is necessary to produce 3-P-glycerate via photosynthesis. 3-P-glycerate is the starting compound of photosynthesis. It has to be supplied constantly since it is quite short-lived.� The main product of photorespiration is the amino acid glycine and this again is the starting compound for the synthesis of the amino acid serine. �
http://imgarcade.com/
Figures above and below show metabolic interaction between Mitochondria and Glyoxysomes; glycollate reactions among organelles of the leaf cells. http://publishing.cdlib.org/
http://imgarcade.com/
Microbodies in the cotyledons of cucumber seedlings perform two successive metabolic functions during early post-germinative development. During the first 4 or 5 d, glyoxylate cycle enzymes accumulate in microbodies called glyoxysomes. Beginning at about day 3, light-induced activities of enzymes involved in photorespiratory glycolate metabolism accumulate rapidly in microbodies. As the cotyledonary microbodies undergo a functional transition from glyoxysomal to peroxisomal metabolism, both sets of enzymes are present at the same time, either within two distinct populations of microbodies with different functions or within a single population of microbodies with a dual function.
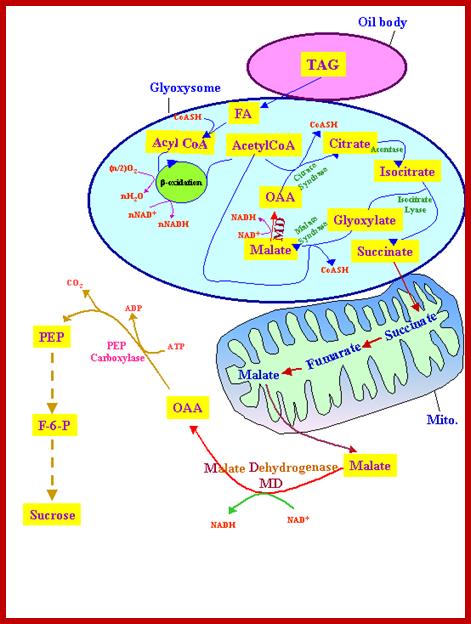
Gluconeogenesis (reverse glycolysis--);The operation of glycolysis in reverse - sort of! This metabolism is particularly prominent during germination of fat (oil) rich seeds (e.g. castor bean, sunflower). You will remember from the cell biology lectures that the glyoxylate cycle in the glyoxysomes takes fatty acids hydrolyzed from TAG�s in the lipid bodies and converts them to succinate. The mitochondria convert succinate to malate. In the cytosol the malate is converted to oxaloacetate and this to PEP. PEP is then converted to sucrose by gluconeogenesis. The sucrose is then used to support growth of the seedling until PS can take over:; Metabolism in germinating oil rich seeds; http://www.uky.edu
A Glyoxysome is a specialized peroxisome containing the enzymes of the glyoxalate cycle. Glyoxysomes are particularly prevalent and important in germinating seeds. Glyoxysomes are in charge of the mobilization of stored energy compounds. Especially during growth, a large amount of carbohydrates are required for the synthesis of new cell walls.
Triglycerides and aspartate
enter the glyoxysomes to be converted into succinate.
The succinate is required in order for the
energy-yielding process to continue through the mitochondria and finally to
create sucrose in the cytosol.
����������������������� Peroxisomes��������� ����������������������� ����������� Glyoxysomes
|
|
Glycosomes:
Trypanosomatids have invented their own �new cell organelle not present
in any other eukaryotic group! �The glycosome is a specialized microbody
that contains most of the enzymes for the glycolytic pathway. Unlike a
mammalian cell, the first nine reactions of glycolysis are
organelle-associated.
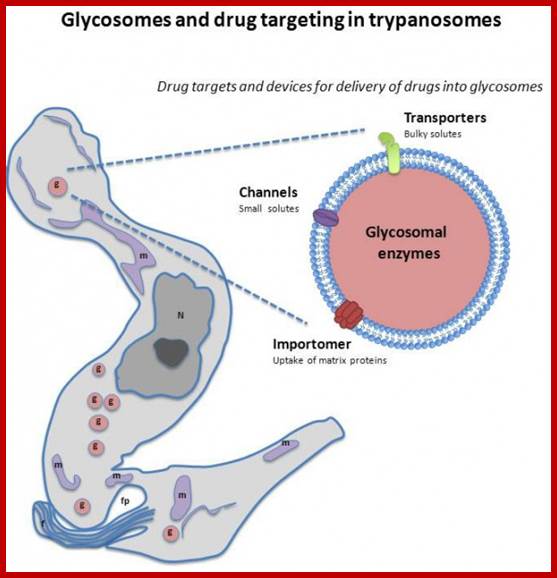
Glycosomes; The glycosome is a membrane-enclosed organelle that contains the glycolytic enzymes. The term was first used by Scott and Still in 1968 after they realized that the glycogen in the cell was not static but rather a dynamic organelle.[1] It is found in a few species of protozoa including the Kinetoplastida which included the suborders Trypanosomatina and Bodonina, most notably in the human pathogenic trypanosomes, which can cause sleeping sickness and Chagas's disease, and Leishmania. The organelle is bounded by a single membrane and contains a dense proteinaceous matrix. It is believed to have evolved from the peroxisome.[2] This has been verified by work done on Leishmania genetics.The glycosome is currently being researched as a possible target for drug therapies.Glycosomes are also found in hepatocytes responsible for storing sugar; http://en.wikipedia.org/ Channel-Forming-Activities-in-the-Glycosomal-Fraction-from-the-Bloodstream-Form-of-Trypanosoma-bruc�CC BY-SA 3.0 - http://globalmedicaldiscovery.com/key-scientifi.
Glycolytic
Steps Inside Glycosomes:
The purpose of Hannaert et al's work is to
attempt to find genetic links between trypanosome glycolytic enzymes and
plant or animal glycolytic enzymes. But the initial question they are trying to
answer is, "Why are the steps of glycolysis sequestered into an
organelle, the glycosome?" Below is a diagram of the nine steps that occur
within the glycosome and the shuttles that allow compounds in and out.
The glycosome is an undisputed member of the peroxisomes family (Martin and Borst), �as it has the same structure, bound by a single unit membrane, and also uses targeting signals. Enzymes normally found within a plastid, and at some point "abandoned", have found a home inside of the glycosome!
Lysosomes-Animals:
They are hydrolytic enzymes� containing membranous bags.� They are very small structures and their number increase or decrease depends upon the requirement. Their biogenesis and development depends upon two pathway; vesicles from phagocytosis and the other from enzyme loaded Golgi vesicles.� Lysosomes are contained in a single unit membrane similar to peroxisomes and glyoxysomes.� Lysosomes are loaded with a large number of hydrolytic enzymes and their activity is contained by maintaining acidic pH in the lysosomal sap.
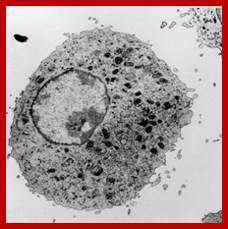
Artist Sketch of lysosome; TEM of a lysosome; http://dehistology.blogspot.in/
Based upon appearance alone, the lysosome looks simplest cellular organelle. Basically it is a bag of digestive enzymes. But looks can be deceiving. This group of lysosomal enzymes is capable of digesting essentially every type of biological molecule. For this reason, the lysosome was originally considered to be only involved in digesting organic components that the cell ingested through phagocytosis or pinocytosis. With more research it became clear the lysosome has many more cellular responsibilities. This was dramatically emphasized when it was shown that the absence of a single lysosomal enzyme in humans can lead to serious abnormalities, dementia and death.
The Lysosomes - Structure:
- A newly formed lysosome that has not yet been engaged in any cellular activity is called a primary lysosome; all others are generally classed as secondary lysosomes (e.g., digestive vacuole, residual vacuole, autophagic vacuole, etc.).
- A single biomembrane surrounds enzyme-rich matrix.
- Matrix varies in density: it is relatively homogeneous in primary lysosomes; in secondary lysosomes the matrix contains various inclusions (e.g., partially digested organelles or bacteria, etc.)
- Matrix consists of many different hydrolytic enzymes
- Enzymes can digest every cell component
- Acid Phosphatase = classic, marker enzyme; used to demonstrate the presence of lysosomes in animal tissues
Lysosome Membrane Proteins
Lysosomes are bound by a single unit membrane. The limiting membranes of lysosomes and late endosomes are enriched in a specific set of integral membrane proteins called lysosome associated membrane proteins (LAMPs) or lysosomal integral membrane proteins (LIMPs) (Eskelinen et al. 2003).
�LAMP-1 and LAMP-2 are type 1 membrane proteins. Both LAMPs are ubiquitously expressed and abundant. It is demonstrated that LAMP molecules are critical for the maturation of phagosomes and killing of the ingested bacteria (Huynh et al. 2007; Beertsen et al. 2008). LAMP-2 plays a role in intracellular cholesterol traffic, as unesterified cholesterol accumulates in late endosomes and lysosomes of cells lacking LAMP-2, or both LAMP-1 and LAMP-2 (Eskelinen et al. 2004; Schneede et al. 2009). Our results show that the accumulation is due to retarded export of cholesterol from late endosomes and lysosomes.
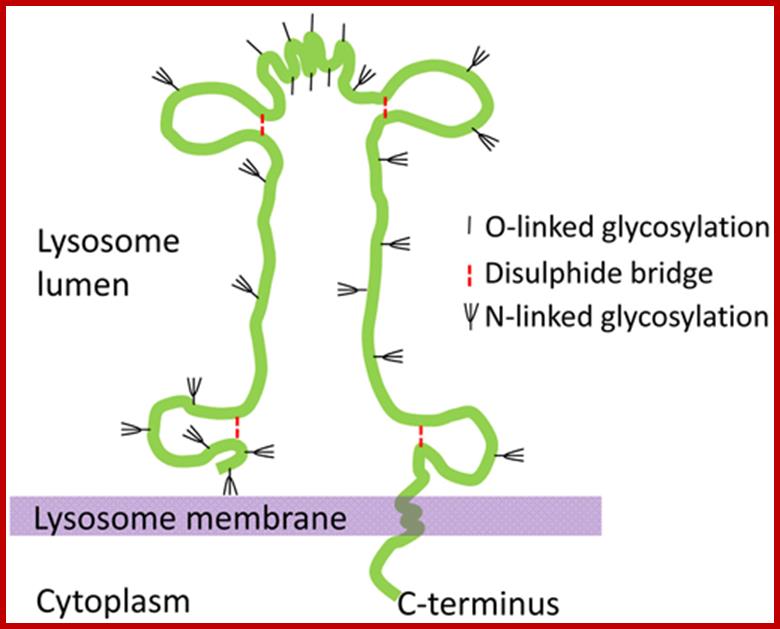
The schematic structure of the lysosome associated membrane protein LAMP-2. http://www.helsinki.fi/
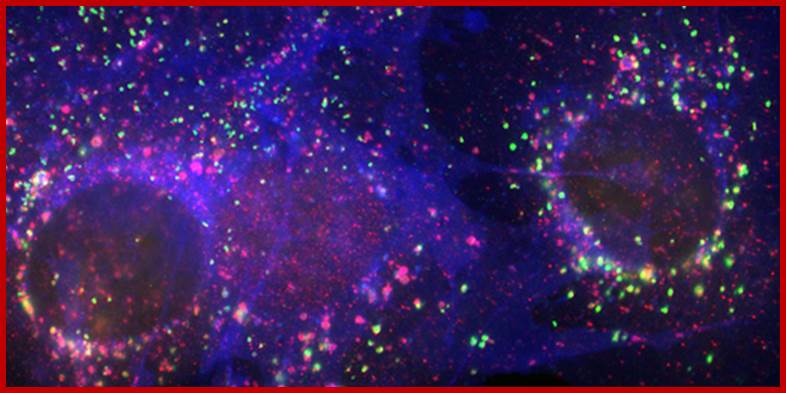
Figure: Immunofluorescence staining of LAMP-2 (green), the cholesterol transport protein NPC1 (red), and unesterified cholesterol (blue) in mouse fibroblasts. www.helsinki.fi
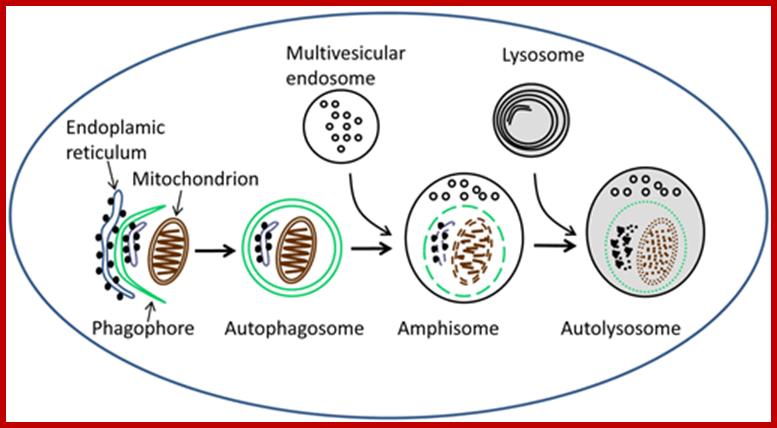
Autophagosomes undergo a stepwise maturation process including fusion events with endosomal and lysosomal vesicles (Figure 1). During the maturation process the segregated cytoplasm is delivered to the endo/lysosomal lumen, where it is degraded by lysosomal hydrolases. The degradation products are transported back to cytoplasm where they can be reused for energy production and biosynthetic reactions. Our studies showed that the small GTPase Rab7 and the lysosomal membrane protein LAMP-2 are essential for the maturation of autophagosomes (J�ger et al. 2004; Eskelinen and Saftig 2009). http://www.helsinki.fi/
Many of the enzymes are glycosylated with mannose in the rough endoplasmic reticulum and a mannose group is phosphorylated in the Golgi to target them to lysosomal vesicles. In the classic view of lysosomal biogenesis, prelysosomal vesicles that bud directly from the Golgi fuse to form mature, primary lysosomes. Recently it has been shown there are different routes to forming lysosomes. For example, the formation of late endosomes by the fusion of vesicles (early endosomes) from the cell membrane with vesicles from the trans-Golgi network can lead to lysosome formation. Our understanding of the details of endosomal events and lysosome biogenesis are still in their infancy (Mullins & Bonifacino, 2001. BioEssays 23: 333-343). The endosome formation and lysosomal digestion are due to receptor-mediated exocytosis.
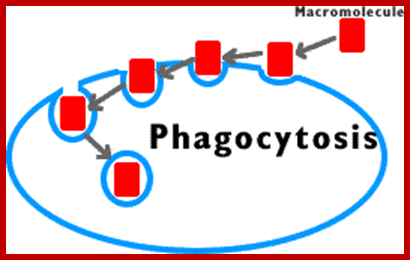 �
�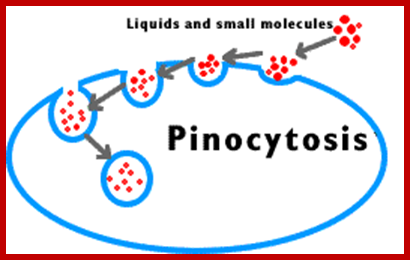
Phagocytosis � Cell eating large particles by forming vacuoles; and Pinocytosis- Cell drinking liquid material into cell� by vacuole formation; http://science.halleyhosting.com/
.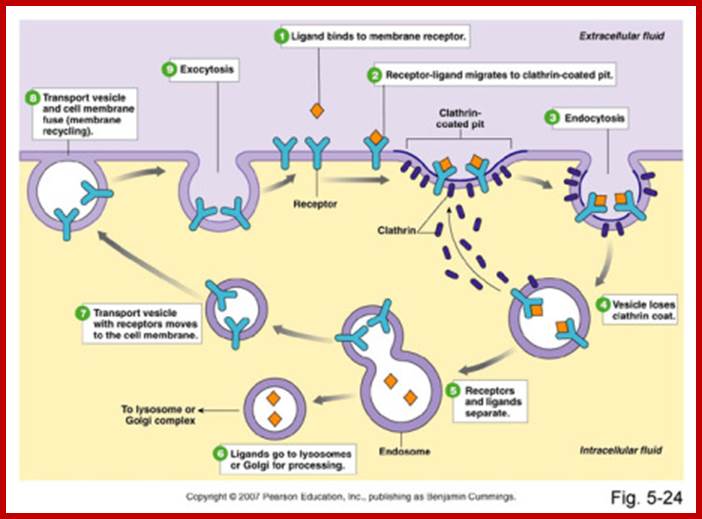
Receptor sorting is the major form of endosome sorting because the receptors engage in rapid recycling. The other two are clear enough without illustrations I guess. If not, then google them; http://animalcellbiology.wordpress.com/
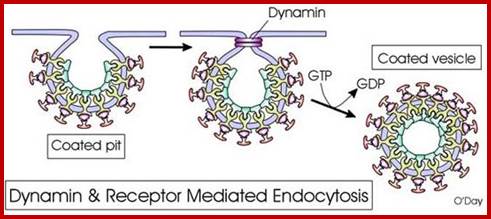
Endocytosis and exocytosis; http://www.utm.utoronto.ca/
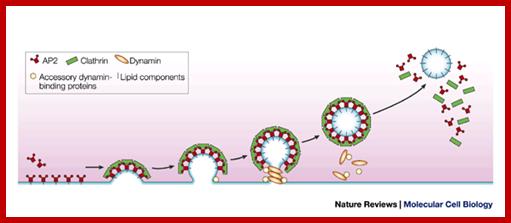
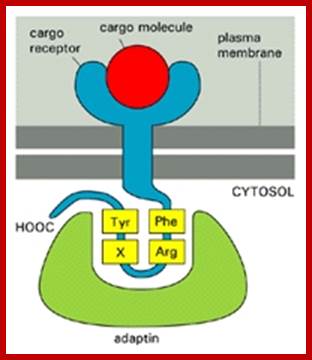
The molecular mechanisms of clathrin-mediated endocytosis; Eckart D. Gundelfinger, Michael M. Kessels and Britta Qualmannhttp://www.nature.com/
Generally, different forms of endocytosis, which can all occur through clathrin-mediated mechanisms, can be distinguished from each other. These processes are constitutive endocytosis, compensatory endocytosis and ligand-and its receptor induced endocytosis (for example, of the epidermal growth factor receptor). The general sets of proteins that are required for these types of endocytosis are largely identical. The formation of clathrin-coated vesicles at the plasma membrane is driven by the assembly of clathrin triskelia into polyhedral lattices or cages (for a review, see Ref. 135). Clathrin is recruited to the plasma membrane by the tetrameric adaptor protein (AP) complex AP2. The membrane association of AP2 is dependent on interactions with membrane phospholipids (phosphoinositides), the integral membrane protein synaptotagmin89 and specific sorting signals in the cytoplasmic tails of receptors and other cargo molecules (for a review, see Ref. 5). Furthermore, clathrin-coat formation at the plasma membrane involves AP180, which binds to both clathrin and AP2 and has been proposed to regulate vesicle size. The subsequent formation of a deeply invaginated constricted coated pit, and its sequestration and detachment from the plasma membrane requires various additional proteins (for a review, see Ref. 4). One is the large GTPase dynamin, which is targeted to the necks of endocytic coated pits where it participates in the fission reaction (for a review, see Ref. 58. Clathrin-coated vesicles liberated from the donor membrane are then uncoated rapidly by an uncoating ATPase and moved into the cytosol, where they undergo further cellular sorting. Eckart D. Gundelfinger, Michael M. Kessels and Britta Qualmannhttp://www.nature.com/
.
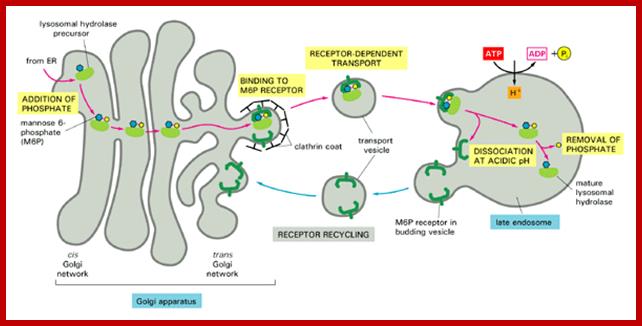
Biogenesis of Lysosome; Transfer of m6p marked lysosomal proteins to endosome; http://cc.scu.edu.cn/
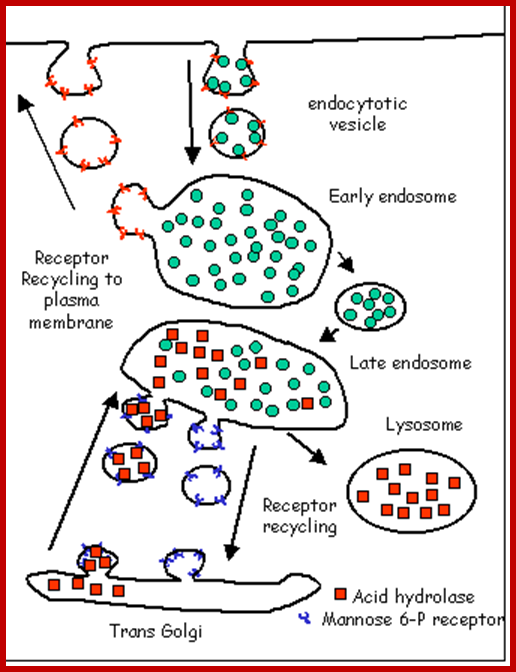
Recycling of cell surface receptors and mannose- 6 - phosphate receptors by endosomes. The green circles represent cargo molecules coming from formation of endocytotic vesicles at the cell surface. The red squares represent lysosomal enzymes carried from the trans Golgi network to the endosome by mannose-6 -phosphate receptors. Both types of receptors are recycled. Transport vesicles carry both lysosomal enzymes and macromolecules to be degraded to lysosomes.;http://www.zoology.ubc.ca/
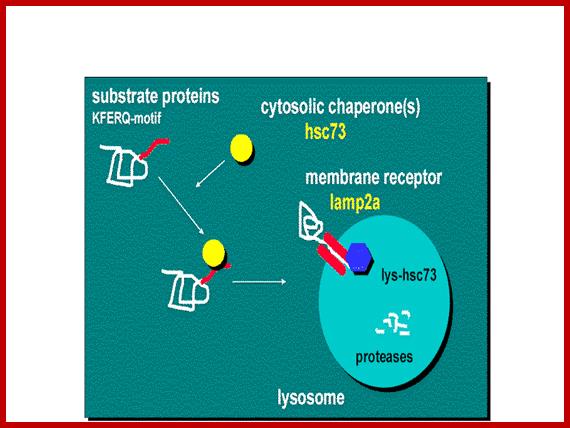
Chaperone-mediated autophagy (CMA) is responsible for the degradation of aproximately 30% of cytosolic proteins in tissues such as liver, kidney and in many types of cultured cells .
This lysosomal mechanism of protein degradation is mainly activated in conditions of stress such as nutrient deprivation or exposure to different toxin compounds. Under those conditions, the substrate proteins for this pathway, all those containing a motif biochemically related to the pentapetide KFERQ , are selectively recognized by acytosolic chaperone, the heat shock cognate protein of 70 kDa (hsc70) [1]. The interaction between the chaperone and the substrate in the cytosol targets the complex to the lysosomal membrane where it binds to the lysosome associated membrane protein type 2A (LAMP-2A), that acts as a receptor for this pathway ]. A second chaperone, the lysosomal hsc73 (lys-hsc70), is required for the complete translocation of the substrate protein into the lysosomal matrix [3] where it is completely degraded by the lysosomal proteases.Cytosolic proteins targeting lysosomal membrane bound receptors/translocons. Many lysosomal proteins are synthesized and released free in cytoplasm and they are immediately associate with chaperones and direct them to membrane bound receptor using signal sequences. Signal sequences for lysosome target proteins KFERQ. http://www.einstein.yu.edu/
Chaperone-mediated autophagy (CMA):
�Lysosomes are responsible for the degradation of approximately 30% of cytosolic proteins in tissues such as liver, kidney and in many types of cultured cells.
This lysosomal mechanism of protein degradation is mainly activated in conditions of stress such as nutrient deprivation or exposure to different toxin compounds. Under those conditions, the substrate proteins for this pathway, all those containing a motif biochemically related to the pentapetide KFERQ, are selectively recognized by a cytosolic chaperone, the heat shock cognate protein of 70 kDa (hsc70). The interaction between the chaperone and the substrate in the cytosol targets the complex to the lysosomal membrane where it binds to the lysosome associated membrane protein type 2A (LAMP-2A) that acts as a receptor for this pathway. A second chaperone, the lysosomal hsc73 (lys-hsc70), is required for the complete translocation of the substrate protein into the lysosomal matrix where it is completely degraded by the lysosomal proteases.
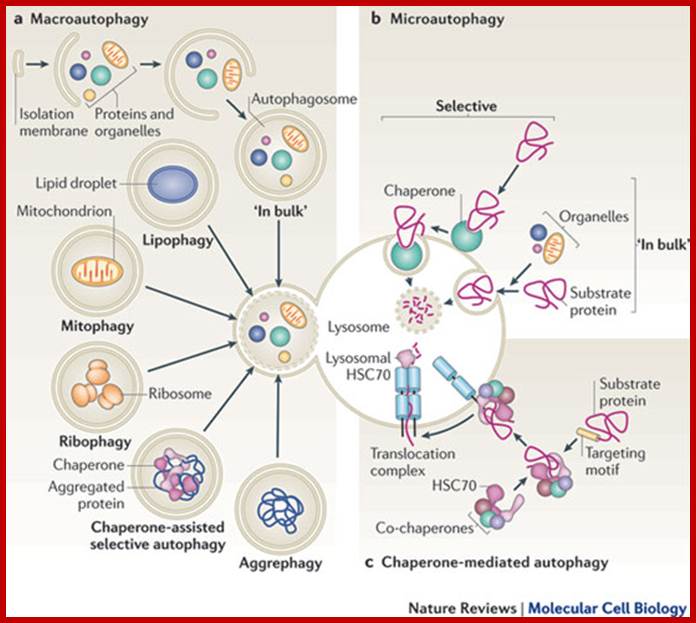
Distinct types of autophagy. Chaperone-mediated autophagy; http://www.nature.com/
Cytosolic proteins can enter the lysosome for degradation by at least three autophagic pathways. a | During macroautophagy, proteins are sequestered along with other cytosolic components and organelles by a de novo-formed isolation membrane that expands and seals to form a double-membrane-bound vesicle, the autophagosome. Degradation occurs when autophagosomes fuse with lysosomes. Selective variations of this process, in which distinct substrates (aggregate proteins or organelles) are targeted for degradation, and their names, are also depicted. b | During microautophagy, invaginations at the surface of the lysosome or late endosomes trap cytosolic material, including proteins, and are then internalized after membrane scission and degraded in the lumen of the organelle. Cytosolic material can be sequestered 'in bulk' or selectively with the help of a cytosolic chaperone that recognizes the substrates. c | During chaperone-mediated autophagy, soluble cytosolic proteins containing a targeting motif are recognized by the cytosolic heat shock cognate 70 (HSC70) chaperone and its co-chaperones, which deliver the substrate to the membrane of the lysosome. After docking onto the cytosolic tail of the lysosomal receptor, the substrate protein unfolds and crosses the lysosomal membrane through a multimeric complex. Substrate translocation requires a lumenal HSC70 chaperone and is followed by rapid degradation in the lysosomal lumen.; http://www.nature.com/
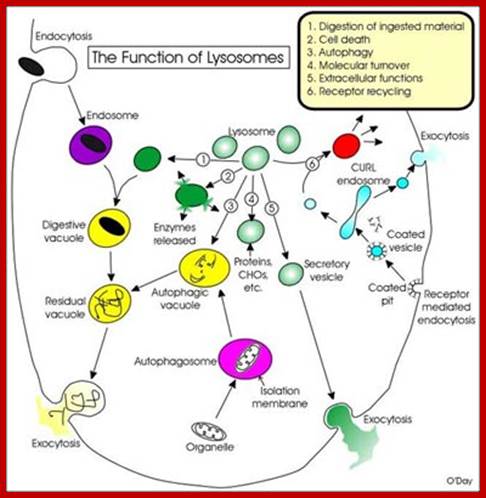
Intricate network of lysosomal structures and functions; http://www.nature.com/
- Digestion of Ingested Materials: Cells ingest materials by various endocytotic means including the classic phagocytosis ("cell eating") and pinocytosis ("cell drinking"). Inside the cell, the material that is taken up is enclosed in an endosome (phagosome or pinosome, respectively). Inside the cell the endosome fuses with a primary lysosome to form a digestive vacuole. In the digestive vacuole the hydrolases of the lysosome will act on the ingested material to break it down. After digestion is complete, the vacuole is called a residual vacuole because it is full of residual, indigestible components. The contents of the residual vacuole are released outside of the cell by exocytosis.
- Cell Death - Lysosomes mediate events in the controlled or programmed death of cells called apoptosis. They also come into play during necrosis, the pathologic death of cells and tissues. For example, meat becomes tenderized after the death of the animal because the lysosomes break down releasing their enzymes into the muscle causing the digestion of the contractile and other muscle proteins.
- Autophagy - The survival of cells requires that cellular constituents are constantly turning over. New molecules and structures are made while old unnecessary or worn out components are removed. During starvation, cells use autophagy to break down cellular components to provide energy for their survival. In the case of organelles, the mitochondrion, for example, is separated from other cellular constituents by an isolation membrane to become an autophagosome. The autophagosome fuses with primary lysosomes to form an autophagic vacuole within which the mitochondrion is digested. The resulting residual vacuole is exocytosed. The following picture shows the sequence of events in digesting mitochondria.
Lysosomes and Apoptosis: Controlled Cell Death:
Apoptosis is dealt with in more detail in several other courses including "Human Development, BIO380". Here's the URL for the lecture in which apoptosis is discussed: http://www.utm.utoronto.ca/~w3bio380/lecture3.htm
- Cell death is an important phenomenon
- Normal event in development of brain (neurons), ovaries (ova), hands
- Mediated by specific signalling pathways that lead to activation of "Death Genes"
- Specific characteristics are evident: blebbing of membrane, laddering of DNA, etc.
Apoptosis is characteristic by specific morphological features:
- Cell shrinkage: cells become smaller and lose cell-cell contacts
- Chromatin condensation: chromatin initially condenses to the periphery of the nucleus and ultimately nuclear fragmentation occurs. During these events DNA is digested in specific ways leading to what is called "laddering" in DNA gels.
- Cell membrane blebbing (small bulges at cell surface) occurs
- Cell fragmentation ("apoptotic bodies" are formed) and phagocytosis of these by macrophages.
Patterns of Cell Death. Methods Achiev: (Modified
from: Walker NI, Harmon B V, Gobe G C and Kerr J F, 1988. Exp. Pathol. 13:
18-54)
Apoptosis is regulated by a diversity of signalling pathways all of which involve caspases. There is a large family of caspases in humans that exist in an inactive form (pro-caspase) that becomes activated by limited proteolysis. Caspases are a family of cysteine proteases, protein-digesting enzymes that that cleave proteins after aspartic acid residues. The caspases work in cascades (a number of caspases working in sequence) digesting a diversity of proteins that underlie specific apoptotic events. Typically initiator caspases (e.g., caspase-2, 8, 9, 10) activate effector caspases (e.g., caspase-3, 6, 7) that digest specific proteins or activate other specific caspases (e.g., caspase-1, 4, 5, 11, 12, 13, 14) that have roles in inflammation. Effector caspases are sometimes called executioner caspases.
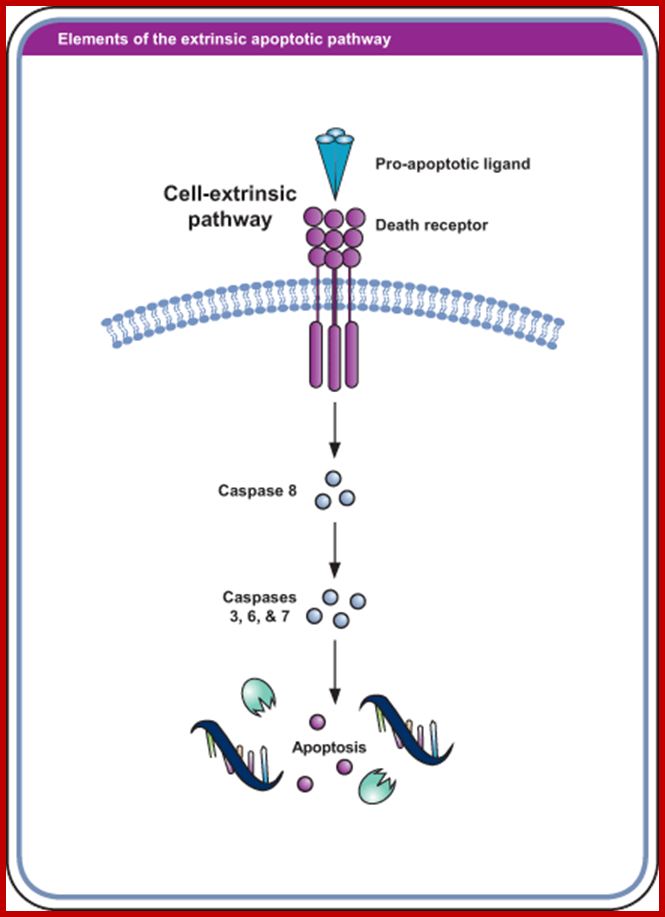
Elements of Extrinsic Apoptotic pathway; http://www.biooncology.com/
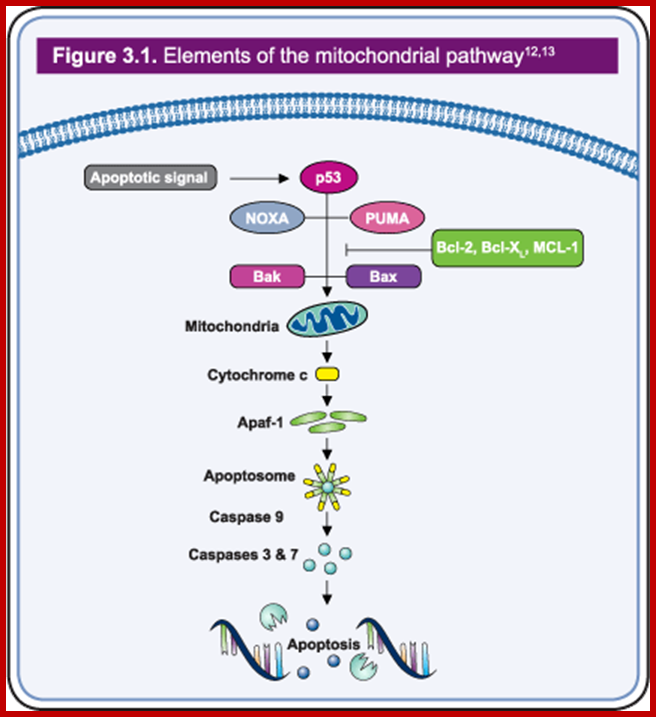
Intrinsic Pathway; The intrinsic pathway (or mitochondrial pathway) hinges on the balance of activites between pro-and antiapoptotic signals of Bcl-2 family; they regulate the permeability of mitochondrial membrane and determine whether a pro-or-anti apoptotic signal will be released inside the cell. Note the extrincic pathway begins outside the cell through activation of pro-apoptotic receptors and the cell surface. These are activated by molecules known as pro-apoptotic ligands; http://www.biooncology.com/
Extrinsic Pathway;

Elements of extrinsic pathway; http://www.biooncology.com/
Extrinsic pathway involves� transmembrane death receptors that are members of TNF factor receptor genes superfamily. They bind to extrinsic ligands and transduce intracellular signals that lead to to death of the cell.� The most well charactrised of these� receptors to date FasL, TNF-alpha, Apo3L and Apo2L, coreesponding eceptors are FasR, DR3, and DR4/DR5 respectively. This pathway involves several caspases which are protease with specific� cellular targets.� Once activated the Caspases affect seeral cellular functions s a process that results in the death of cells. http://www.biooncology.com/
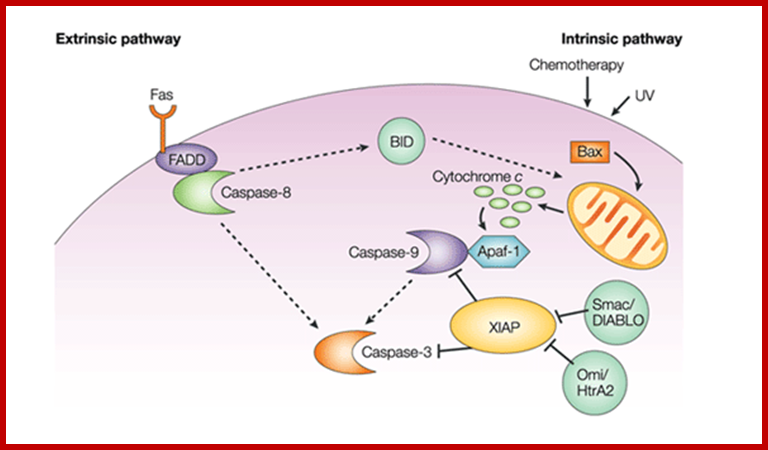
Extrinsic
and intrinsic apoptosis pathways, figure taken from� modified. Direct positive
feedback from caspase-3 on caspase-9 not shown.; June 2008, model of the month by Melanie
I. Stefan
Original model:Legeie
et al. http://www.ebi.ac.uk/
The extrinsic apoptotic pathway is triggered by ligand binding to cell-surface receptors, resulting in the recruitment of various proteins to form the death-inducing signalling complex (DISC). This complex promotes activation of caspase-8, which in turn activates caspase-3. Caspase-3 then induces the cellular changes that characterise apoptosis. The intrinsic pathway, in contrast, is triggered by cytotoxic stress, which induces the translocation of pro-apoptotic Bcl-2 family members, such as Bax, to the mitochondria. This leads to the release of mitochondrial cytochrome c into the cytosol, where it promotes the oligomerisation of the pro-apoptotic factor Apaf-1 into a complex called the "apoptosome". The aptoptosome recruits and activates caspase-9, which in turn promotes the activation of caspase-3. This process is further regulated by X-linked inhibitor of apoptosis (XIAP) protein, which inhibits the activity of both caspase-9 and caspase-3. Both extrinsic and intrinsic pathways have been reviewed in� and are shown in figure 1,
The model by Legewie et al. BIOMD0000000102) focuses on the intrinsic apoptotic pathway. This pathway displays some properties that make it an interesting target for closer examination with mathematical methods: Depending on cellular context, cytochrome c induces caspase activation either gradually or in an all-or-none fashion. In addition, caspase activation is reversible in some cellular contexts, but irreversible in others. This suggests that the caspase activation pathway is monostable (characterised by a gradual response and reversibility) in some cells, but bistable (characterised by an all-or-none-response and sometimes irreversibility) in others. In order for bistability to occur, the existence of some form of positive (or double negative) feedback loop is a necessary, but not sufficient condition (reviewed ). In the feedback loop 9additional pathway), the caspase-3 activates caspase-9 thus promotes its own activation.
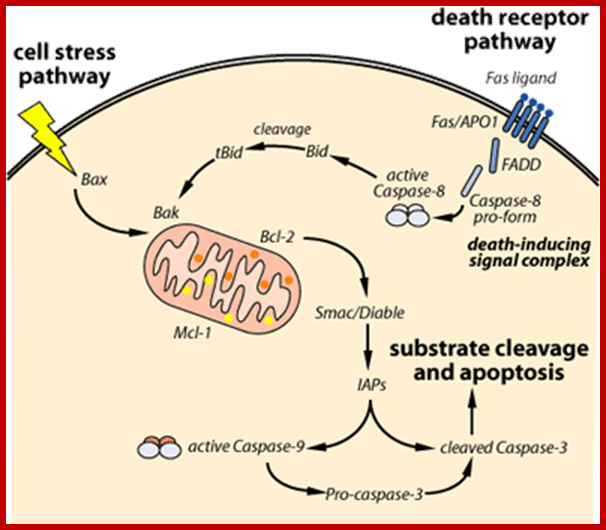
Apoptosis: he intrinsic and extrinsic pathways leading to apoptosis.Cancer
http://www.scq.ubc.ca/
�
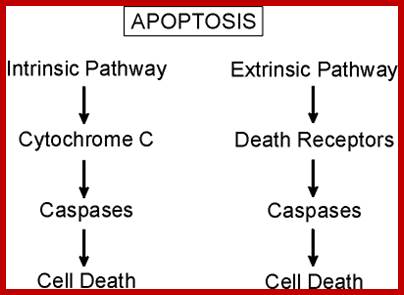
Apoptosis can be initiated by internal events (i.e., "Intrinsic Pathway") involving the disruption of mitochondria and the specific enzyme cytochrome C, in turn leading to the downstream activation of caspases. Alternatively, surface receptors can be activated by specific ligands that bind to "death receptors" (i.e., "Extrinsic Pathway"). Death receptors are members of the tumor necrosis factor (TNF)/nerve growth factor (NGF) receptor super family. They make up a subfamily characterized by the intracellular death domain (DD). The extrinsic pathway is typically mediated by immune cells, to initiate intracellular signaling and the downstream activation of relevant caspases. Some work suggests both Intrinsic and Extrinsic Pathways mediate the apoptosis during oogenesis and likely of aging eggs after fertilization (Gordo et al, 2002).
The following diagram shows some of the signaling events that are initiated when tumor necrosis factor alpha (TNF-α) leads to apoptosis. It should be noted that TNF-α also mediates other signaling pathways involved in normal cellular functions.
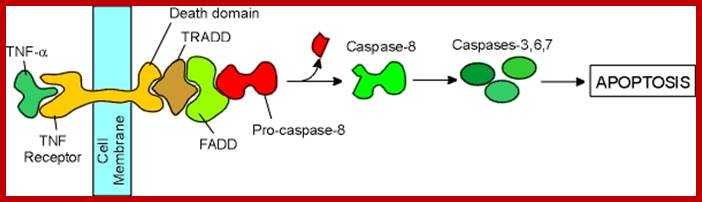
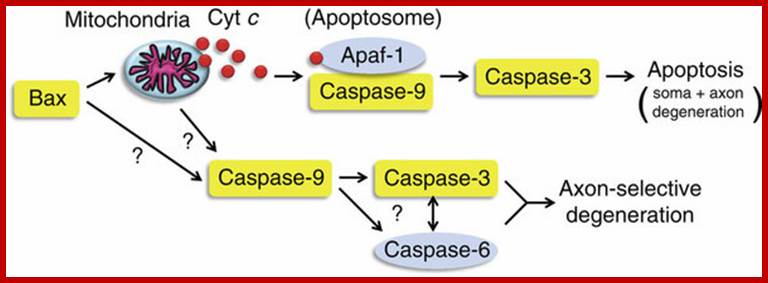
A simple representation of Apoptosis starting from signaling and its induced process to the activation of Caspases to cell death.
Distinct pathways mediate axon degeneration during apoptosis and axon-specific pruning; Both apoptosis and axon degeneration require Bax and caspases. Here we demonstrate that despite this overlap, the pathways mediating axon degeneration during apoptosis versus axon pruning are distinct; While Caspase-6 is activated in axons following nerve growth factor deprivation, microfluidic chamber experiments reveal that Caspase-6 deficiency only protects axons during axon-specific but not whole-cell (apoptotic) nerve growth factor deprivation. Strikingly, axon-selective degeneration requires the apoptotic proteins Caspase-9 and Caspase-3 but, in contrast to apoptosis, not apoptotic protease activating factor-1. Additionally, cell bodies of degenerating axons are protected from caspase activation by proteasome activity and X-linked inhibitor of apoptosis protein. Also, mature neurons restrict apoptosis but remain permissive for axon degeneration, further demonstrating the independent regulation of these two pathways. These results reveal insight into how neurons allow for precise control over apoptosis and axon-selective degeneration pathways, thereby permitting long-term plasticity without risking neurodegeneration. Corey L. Cusack, Vijay Swahari, W. Hampton Henley, J. Michael Ramsey ;& Mohanish Deshmukh; http://www.nature.com/
The binding of TNF-α to its receptor (TNF-receptor or TNFR) makes the receptors intracellular death domain available for binding to TRADD (TNFR-associated death domain). TRADD is an adaptor that in turn directs the binding of FADD (Fas-associated death domain) another adaptor that mediates the binding of pro-caspase-8 to this multiprotein complex. This leads to the proteolytic processing of the inactive pro-caspase-8 into the active caspase-8 enzyme. Caspase-8 is an initiator caspase that in turn proteolytically activates several other caspases. The activated caspases-3,6 and 7 are effector caspases that proteolytically digest a number of target proteins ultimately leading to apoptosis. There are a number of other apoptosis-specific pathways each of which involves unique sets of adaptor proteins and caspases and each of which is designed to direct apoptosis at a specific place or time in human development or other aspects of cell function.
Lysosomes & Cell Death in Normal Development:
The controlled killing of cells is an essential process that occurs during embryonic development as well as during our day to day lives. One classic example of programmed cell death is digit formation.
Programmed Cell death:
Top: http://www.smallerquestions.org; bottom: http://biokav.weebly.com/
- Lysosomes "kill" cells between digits
- Process of controlled cell death is called "Apoptosis"
- Ducks have no lysosomal activity between digits of toes = webbed feet; chickens have activity = no web
- Lack of apoptosis in humans can lead to webbed fingers called syndactyly as shown in the next figure
Lysosomal enzyme release from human monocytes in response to particulate stimuli: PJ Keeling and PM Henson
Lysosomal enzyme release from human monocytes was evaluated in response to opsonized zymosan, opsonized sheep erythrocytes, and latex beads. Monocytes were found to release lysosomal enzymes immediately upon challenge with all three phagocytosable particles. Cytochalasin B enhanced beta-glucosaminidase release from mononuclear cells challenged with opsonized zymosan or opsonized red blood cells, but inhibited the response to latex particles. Lysosomal enzyme release was found to be independent of protein synthesis, and in the absence of cytochalasin B required the stimulus to be presented either as a phagocytosable particle or immobilized on a surface. The kinetics of enzyme release and phagocytosis were also examined and found to be different, lending support to the hypothesis that lysosomal enzyme release may be a physiologic response to a biologic stimulus in vivo and not simply an "accidental" consequence of an ongoing phagocytic event.
Diseases & Mechanisms
|
PROTEOLYSIS: DISEASE & REGULATORY MECHANISMS |
|
|
Disease or regulatory mechanism |
Example |
|
� Proteolytic activation of enzyme |
|
|
� Proteolytic inactivation of enzyme |
|
|
� Protein expression 2� stabilized mRNA |
|
|
� Enzyme activity; Disulfide bond Δ |
|
|
� Enzyme activity; Loss of highly conserved arginine |
|
|
� Enzyme activity; Structural change near catalytic cysteine |
|
|
Gene promoter inactivation |
|
|
Docking site change on cysteine protease inhibitor |
|
|
Deletion of binding site for ubiquitination enzymes |
|
|
Degranulation from immune cells |
MMP-9: IL8 effect on neutrophils |
|
Glycosylation |
N-linked sugars � serine protease activity |
|
Response to inflammation |
� matrilysin & gelatinase B in GBS nerves |
Plant Central Vacuoles: Plant Lysosomes:
In plants one finds a large central vacuole covered by single unit membrane called Tonoplast as found in animal cells. The central vacuoles have many roles, one, it provides enzymes for hydrolysis of many compounds.� It also plays many important roles in plant cell turgidity, development and stores many components including proteins.� The central vacuole occupies more than 80-90% of the cell volume.� Its sap is acidic similar to that of animal cell lysosomes and contains raphide crystals (in many plants cells).
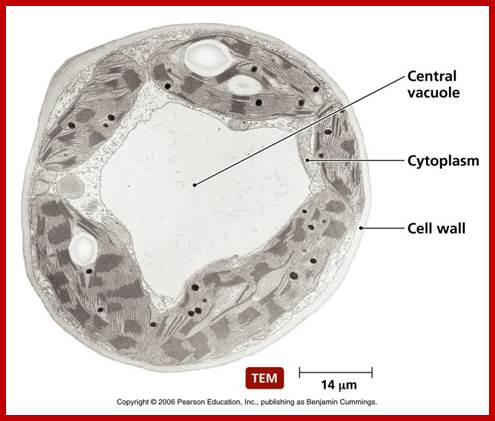
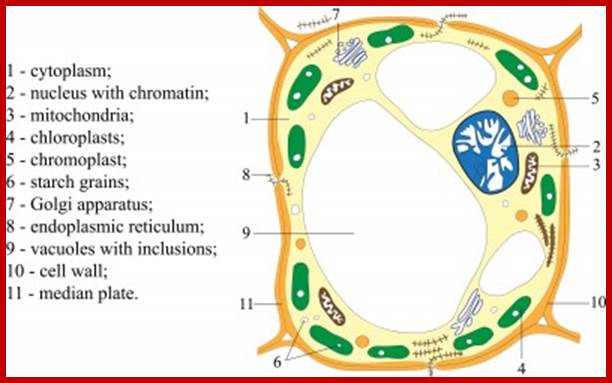
http://biology.tutorvista.com/
They are lytic compartments, function as reservoirs for ions and metabolites, including pigments, and are crucial to processes of detoxification and general cell homeostasis. They are involved in cellular responses to environmental and biotic factors that provoke stress. . In seeds and specialized storage tissues, they serve as sites for storing reserve proteins and soluble carbohydrates. In this way, vacuoles serve physical and metabolic functions that are essential to plant life.
The biogenetic pathways include (1) sorting of proteins destined for the vacuole away from those to be delivered to the cell surface after transit through the early stages of the secretory pathway; (2) endocytosis of materials from the plasma membrane; (3) autophagy pathways for vacuole formation; and (4) direct cytoplasm-to-vacuole delivery. Ultimately, sorting and targeting mechanisms ensure that specific proteins are faithfully assigned to conduct the vacuolar functions.
Seven basic pathways are used for the biogenesis, maintenance, and supplying of vacuoles. Pathway 1: entry and transport in the early secretory pathway (from ER to late Golgi compartments). Pathway2: sorting of vacuolar proteins in the trans-Golgi network (TGN) to a pre/provacuolar compartment (PVC) via an early biosynthetic vacuolar pathway. Pathway 3: transport from PVC to vacuole via the late biosynthetic vacuolar pathway. Pathway 4: transport from early secretory steps (ER to Golgi complex; pathway 1) to the vacuole via an alternative route with possible material accretion from Golgi (indicated by the asterisk). Pathway 5: endocytotic pathway from the cell surface to the vacuole via endosomes. Pathway 6: cytoplasm to vacuole through autophagy by degradative or biosynthetic pathways.� Pathway 7: transport of ions and solutes across the tonoplast. AV, autophagic vacuole; E, early endosome; ER, endoplasmic reticulum; PVC, pre/provacuolar compartment; TGN, trans-Golgi network.( Plant Vacuoles ,Francis Martya)
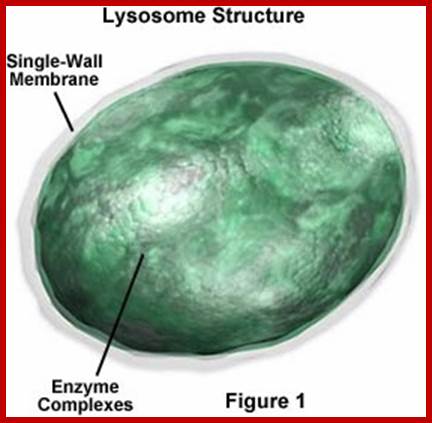
http://aclasen.weebly.com/
Electron micrographs show that developing vacuoles (provacuoles) in root tip meristem cells comprise complex membrane structures, including large numbers of vesicles and tubular membrane elements at various stages of fusion.� Clearly, vacuole biogenesis is a highly dynamic process that is difficult to interpret using static electron micrographs.
Although the vacuolar sap is very dilute, it contains the bulk of the cell's complement of K+, Ca2+, sugar, organic acids and other solutes, many of which must be actively transported against their electrochemical gradients. Hence, a considerable amount of energy must be expended to maintain the solute concentration inside the central vacuole. The two enzymes which directly transduce the energy for tonoplast transport are the vacuolar H+-ATPase (V-ATPase) and the vacuolar H+-pyrophosphatase (H+-PPase). Vacuole Biogenesis in Living Soybean Root Tip Cells; R. Howard Berg etal
Some enzymes found in plant vacuoles are called Cellulysin Cells, contain a-Mannosidase, 8-N-Acetylglucosaminidase, S-Fructosidase, a-Galactosidase, Phosphodiesterase, Nuclease, Proteinase; Total Protein2 (mg).� When the cell divides the daughter cells develop smaller vesicles from their membrane systems and gradually fuse with one another to develop into central larger vacuole.
Protein Transport Across the Nuclear Membrane:�
Presence and absence of the nucleus makes organisms distinguishable as prokaryotes and eukaryotes, one without the nucleus and the other with the nucleus respectively.� Nucleus is a vital cell organelle and contains genomes with all the genetic information for that cell and for the organism perse.�
Nuclear structure is highly organized and Nucleus executes all cellular functions through genetic regulation of host of genes by the way of transcriptional activation or repression and processing of transcripts and transport of the same.� A single mutation can render the nucleus into dysfunctional state.� Nucleus, with chromosomes, nucleolus and nuclear sap, consists of thousands of proteins, all have to be imported.� Many molecules of various sizes are transported into the nucleus; similarly various molecules are transported out into the cytoplasm.� Nucleus bound by a membrane encloses nuclear sap- a liquid.� Located in the nuclear sap are-Nucleolus, Cajal bodies, Nuclear speckles and PML bodies; none of them are bound by any membranes.
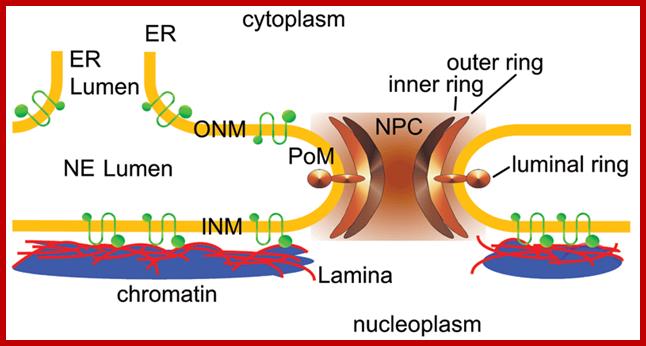
Figure . Organization of the nuclear and ER membranes and NPCs. The nuclear envelope is made of outer and inner nuclear membranes (ONM and INM), which are connected at the pore membrane (PoM) that encircles the nuclear pore complexes (NPCs). The ONM is continuous with the endoplasmic reticulum (ER) and studded with ribosomes. Underneath the INM is the nuclear lamin polymer. Together with associated integral membrane proteins of the INM this polymer is called the nuclear lamina and some unique integral single or multi‑spanning transmembrane proteins of the INM interact with chromatin and provide attachment points for the lamina. There are also integral proteins of the INM, PoM and ONM that are not as‑
sociated with the lamin polymer. Chromatin and soluble nucleoplasmic proteins may have specific interactions at the NE, but it is hard to distinguish these from contaminants because of the particular �stickiness� of many highly charged DNA and RNA‑binding proteins. Within the NPC most of the FG nucleoporins occur in the central ring so that they can extend into the central channel of the NPC, but some also occur in the outer ring facing the membrane. Due to the lack of structure in the FG regions they are thought to be mobile and sampling a wide space around the two channel rings and thus are presented as a less substantial haze. Several structural studies have indicated the presence of peripheral channels with a diameter
that may be as big as 100 �. NPCs are tethered to the PoM by three integral proteins that also form a ring in the lumen: gp210, POM121 and NDC1/NET3; http://www.landesbioscience.com/
.
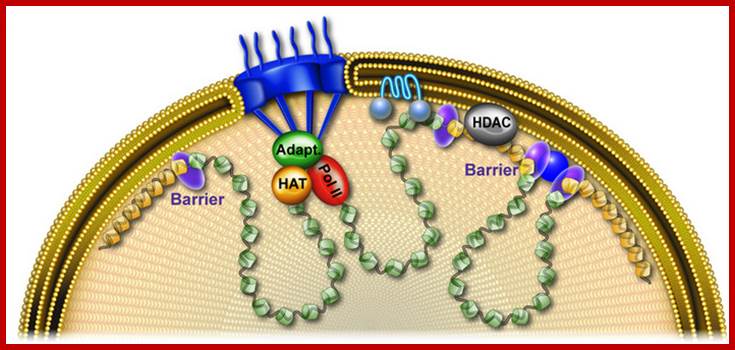
NPC and proteins of the inner nuclear membrane partition the genome into area of silent (yellow) and active chromatin (gren).� Gene-NPC interactions require various adapteorss including the SAGA histone acetyltransferase (HAT)-Kohler and Hurt, Mol.cell.http://www.mfpl.ac.at/uploads/tx_onmfpl/Figure_A.jpg
Structurally the vital contents of the nucleus are enveloped by a membrane system that is very complex and organized structure.� It is the membrane that acts as the gateway for the movement of proteins into the nucleus and out of the nucleus.� The structure in the membrane that acts as a portal for such movements is called Nuclear Pore Complex (NPC).
�
� �The nuclear pore complex (NPC) is the gate to the nucleus. Recent determination of the configuration of proteins in the yeast NPC at ~5 nm resolution permits us to study the NPC global dynamics using coarse-grained structural models. Although it is typically circular, the overall structure of the NPC has been observed to change in response to the presence of cargo. Timothy R. Lezon et al
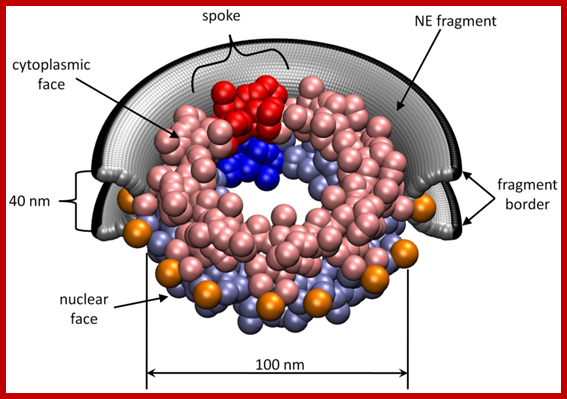
PLOS computational Biology; Timothy R. Lezon,et al; ;http://www.ploscompbiol.org/
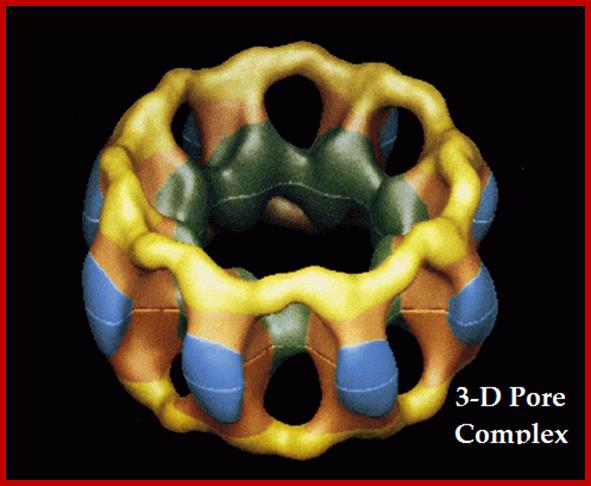
An artistic view of Nuclear Pore Complex-Molecular Basket.
Structural studies of the nuclear pore complex, conducted at Rockefeller, have revealed much about the inner workings of the huge 450-protein pore and have provided a glimpse into how the nucleus itself first evolved.;http://newswire.rockefeller.edu/
The nuclear pore complex is a huge structure (120nm in diameter) with hundreds of proteins; the molecular weight of the NPC, total mass is 125 million Daltons or more.� The pore complex is constructed like an annular concentric rings looks like a wine-barrel with an upper ring and lower ring of eight subunits each.� Supporting them there are outer spoke ring and inner spoke ring. The upper top ring faces the cytoplasm and lower or bottom ring faces the nuclear sap. Protein filaments emanate from the upper ring, so also from the bottom ring, but the filaments from the bottom ring are connected to each other in the form of a ring/basket.� NPC contains a central channel of 40 nm size runs all along the length of the barrel, perhaps held by outer coaxial ring of proteins by eight spokes.� The coaxial ring in turn is connected to nuclear membranes by radial arms.� The space between the radial spokes also provides free space for the movement of smaller molecules.� The central pore complex has cytoplasmic Iris and nucleoplasmic Iris connected to cytoplasmic ring and nucleoplasmic ring respectively.
A network of structural proteins called Lamins supports the nuclear membrane at its inner surface. The number of pore complexes can expand and contract according to the needs.� Average number of pore complexes per nucleus is ~3000. The number can increase or decrease when needed.
The pore complex is responsible for the import of several kinds of nuclear proteins/ RNAs-RNPs and export several species of RNAs/RNPs, ribosomal subunits and many snRNPs.� The number of proteins imported per pore complex per minute is remarkable.� One hundred Histones are imported per minute per pore complex.� Non-histones 200, and riboproteins 150 per minute imported.� The same pore complexes export ~5 ribosomes per minute per pore complex, mRNAs one per minute.� The database estimates 6000 to 10000 different proteins are imported from cytoplasm into the nucleus.
Molecules of 5 to 500 daltons size easily move across the pore complex, thus nucleotides, ions many other molecules have easy access into the nucleus.� But proteins of 10-50KD (size of ~5nm) cannot diffuse passively and they are transported in ATP dependent manner (Active process).� The central channel is exclusively used for the transport of larger cargo by expansion and contraction mode.� They can transport gold particles of size~ 20nm through central core.� The pore complex transports ribosome of 120 x 200� size.� The central channel has a cytoplasmic iris connecting cytoplasm and a nucleoplasmic iris connecting nucleoplasm.� Though the pore complex allows components passive diffusion, and facilitated. Active transport is always a facilitated transport. Cytoplasm contains 50-100 thousand different proteins, but only those proteins that are required are transported into and not others.� So also export is directed and facilitated.
Import of Nuclear proteins:
Transport of proteins into the nucleus is determined by specific signal sequences called Nuclear Localization Sequences (NLS).� It appears most of the nuclear proteins have a common signature sequences for the import. Similarly those proteins or RNA species destined for export should have similar signature sequences called Nuclear Export Sequences (NES).� Proteins involved in import are called Importins and those used in export are considered as Exportins.� They have signals sequences destined to the nucleus.� Export protein sequences have a conserved 10 amino acids with conserved Leucines.
Signal sequences of proteins for nuclear import and export;
SV40 T-antigen: PKKKRKV.
Polyoma T-antigen: PKKARED.
Nucleoplasmin: KR-10 aa-KKKK.
SV40-P1:� APTKRKGS.
Import of nuclear protein requires a host of proteins called Importins.� But the importin-α, β, and β3 are important.� The importins are located at the orifice of the pore complex. Importin-α binds to NLS sequences then transfers to importin-β, which in turn delivers to nucleoporins (Nuc-Ps) found at the nuclear membrane in the pore complex. They are involved in inward movement of cargo.� There are a many nucleoporins involved in translocation of proteins into the nucleus; it requires the input of ATP energy.� There is other group of importins called Importins-β and, another Transportins which not only binds to NLS sequences but also delivers the cargo to nucleoporins.� Transportin M9 is mainly responsible for the transport of hnRNPs.� Importinβ3 is believed to responsible for the import of riboproteins.� There are more importins, which carry signal containing molecules into the nucleus for activating transcription. Nucleoporins have repeated motifs such as GKFG, FG, and FxFG.� These motifs help in binding to importin-β and driving the cargo inside.�
Another cytosolic factor that supports translocation is Ran. It is a GTPase protein.� Ran is a monomeric G-protein.� The Ran-GTP complex is found associated with importins at cytosolic side and also found at the nucleoplasmic side.� The Ran-GTP is active, when hydrolyzed it provides energy for the movement. The Ran-GDP, is inactive, it will be activated only when GDP is exchanged with another GTP by another factor called GEF (GTP exchange factor).
Export of Nuclear Cargo:
Similar to importins there are Exportins molecules that are specific each type and they are responsible for the export of mRNAs, ScRNAs, SnRNAs, tRNAs, many proteins and ribosomes into cytoplasm.� Export of protein requires signal sequences such as NES, a ten amino acid residues stretch that is rich in Leucines.� Export of components is species specific and requires specific nucleoporins aided by Ran-GTP factors found at nucleoplasmic surface.�
Specific factors determine which RNA has to be exported, ex. HIV mRNA intact transcript (unspliced requires Rev Proteins which bind to sequences called Rev response elements (RREs).� Similarly most of the mRNA, which is spliced, requires cap proteins bound to the Guanine nucleotide found at 5�end for transport of processed mRNAs.� tRNA�s transport again requires specific factors for transport.� It is interesting to know some proteins such as poly-A binding proteins; PAB-II and RNAs (especially snRNAs) that are transported out of the nucleus are transported back.� Such transport requires specific importins and exportins, which act to and fro transport.
The nuclear pore complex becomes alive: new insights into its dynamics and involvement in different cellular processes; Joachim K�ser*, Bohumil Maco*, Ueli Aebi, and Birthe Fahrenkrog
Nucleo-cytoplasmic transport:
Whereas ions and small molecules move in and out of the nucleus by passive diffusion but transport of proteins, RNAs, ribonucleoprotein particles (RNPs), and ribosomal subunits is an energy-dependent and receptor-mediated process. Because many excellent reviews about nucleocytoplasmic transport have been published in the last few years (c.f. Corbett and Silver, 1997; Mattaj and Englmeier, 1998; G�rlich and Kutay, 1999; Kaffman and O'Shea, 1999; Conti and Izaurralde, 2001; Komeili and O'Shea, 2001; Macara, 2001; Weis, 2002; Quimby and Dasso, 2003; Fried and Kutay, 2003; Johnson et al., 2004; Xu and Massagu�, 2004), we (authors above) describe here only the best characterized classical receptor-mediated nucleocytoplasmic transport (see Fig below). Accordingly, cargo to be transported in or out of the nucleus harbors a nuclear localization signal (NLS) for nuclear import or a nuclear export signal (NES) for nuclear export. NLS and NES are recognized by soluble transport receptors known as importins and exportins, also called karyopherins. These receptors facilitate transport through the NPC by interaction with FG repeats that many nucleoporins harbor. The key regulator of the directionality of nucleocytoplasmic transport is Ran, a member of the Ras-related GTPase superfamily, by switching between its GDP- and GTP-bound forms. The nucleotide exchange factor, RanGEF (or RCCI) is bound to DNA and facilitates RanGTP formation in the nucleus, whereas the GTPase activating protein, RanGAP, �that is excluded from the nucleus, assures depletion of the cytoplasm of RanGTP in combination with Ran binding protein RanBP1 and RanBP2. Hence, interaction of Ran with its compartmentalized regulatory proteins RanGEF and RanGAP generates a gradient across the NE with a high RanGTP concentration in the nucleus and a low level in the cytoplasm, and vice versa for RanGDP.
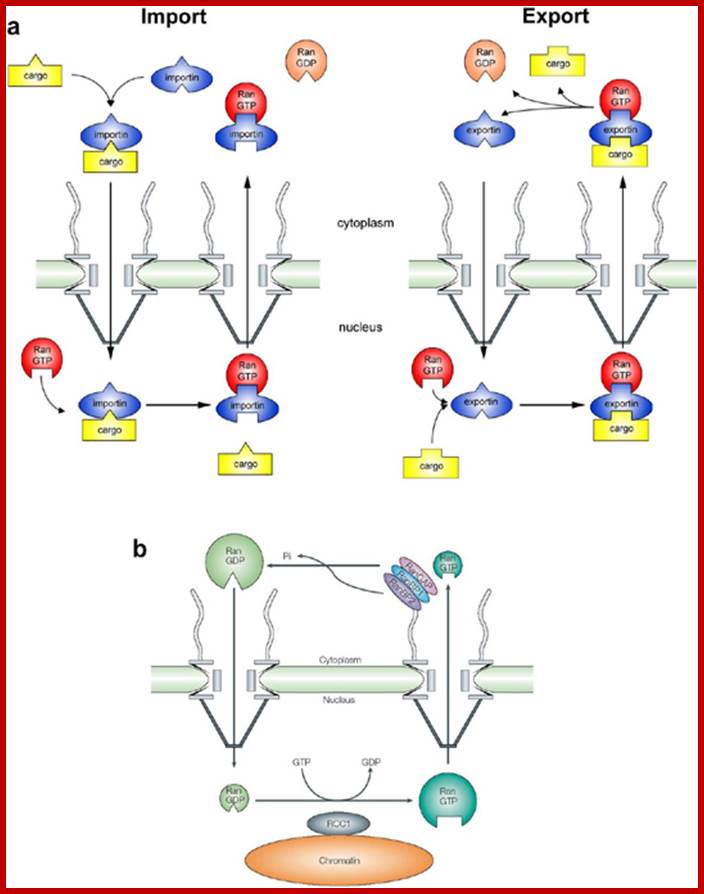
�Scheme of import (left) and export (right) of proteins (a) and control of the nucleocytoplasmic transport by Ran (b). Joachim K�ser et al ; http://atlasgeneticsoncology.org/
(a) In the cytoplasm an importin binds to cargo molecules containing a nuclear localization signal and mediate interactions with the NPC to translocate the complex into the nucleus. Nuclear RanGTP binds to the importin and induces cargo release from the complex. The importin�RanGTP complex is then recycled to the cytoplasm, where RanGTP is displaced from the importin by RanBP1 and/or RanBP2 and subsequently hydrolyzed by RanGAP. In the case of export, cargo with a nuclear export signal binds to an exportin induced by RanGTP in the nucleus and the trimeric complex is then translocated through the NPC to the cytoplasm, where RanGTP is removed from the complex by GTP hydrolysis and the exportin dissociates from the cargo. The exportin recycles back to the nucleus and is ready for the next round of the export cycle.
(b) Directionality of the nucleocytoplasmic transport by the RanGTPase system. Chromatin-bound RCC1 is the nucleotide exchange factor for Ran (RanGEF) and promotes exchange from RanGDP to RanGTP. This results in a high concentration of RanGTP in the nucleus. After translocation of importin-RanGTP complex back to the cytoplasm, RanGTP becomes hydrolyzed by cooperation of RanGTP-binding proteins (RanBP1 and RanBP2) and Ran GTPase-activating protein (RanGAP). This prevents the accumulation of RanGTP and results in its low concentration in the cytoplasm.
Intracellular Protein� Traffic: Via Membrane Bound Vesicles:
Whether proteins synthesized on ER or in free-state in cellular milieu, localized or not, each of the proteins have distinct destination and they have to be directed and targeted specifically and correctly.� A single cell can easily produce 50,000 to 70,000 or more proteins.� If alternative splicing of mRNAs, splicing of intein containing proteins and processing of poly-proteins all combined, the total number can be anywhere ~100,000 or more.� Cells have endowed with facility and innate ability to sort out each and every protein and transfer to their specific destination.� Though the process is complex and many of the events are yet to be understood; the overall or generalized mechanism is emerging from studies.
From PDB Database:
|
Experimental |
Protein/Nucleic Acid |
Other |
Total |
||
|
64155 |
1335 |
3147 |
2 |
68639 |
|
|
68639 |
967 |
185 |
7 |
9230 |
|
|
277 |
22 |
101 |
0 |
400 |
|
|
Hybrid |
42 |
3 |
2 |
1 |
48 |
|
Other |
138 |
4 |
5 |
13 |
160 |
|
Total: |
72683 |
2331 |
3440 |
23 |
78477 |
The PDB database is updated weekly (on Tuesday�s). Likewise, the PDB Holdings List is also updated weekly. As of 10 January 2012, the breakdown of current holdings is as follows:
58034 structures in the PDB have a structure factor file.
6,535 structures have an NMR restraint file.
304 structures in the PDB have a chemical shifts file
� Proteins, that are synthesized free and those threaded through ER, have several destinations like ER itself, Golgi, lysosomes, vacuoles, plasma membranes and secretion to extracellular periplasmic space.� The transport Path- from ER to Cis-Golgi to median-Golgi to Trans-Golgi, to transport vesicles and then to plasma membrane, forms the default pathway.
Proteins that are synthesized in free-state, localized or otherwise, are sorted and transferred to their specific destinations like the Nucleus, plastids, mitochondria, microbodies, lysosomes, central vacuoles and plastids.
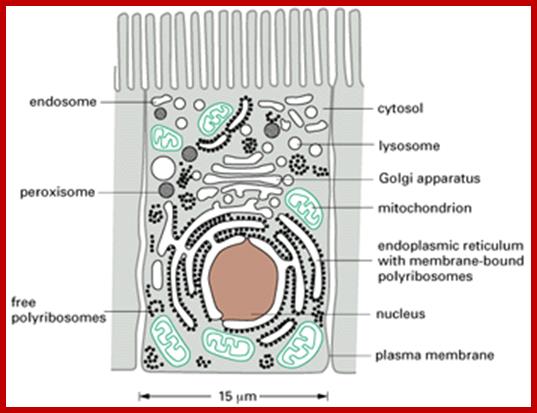
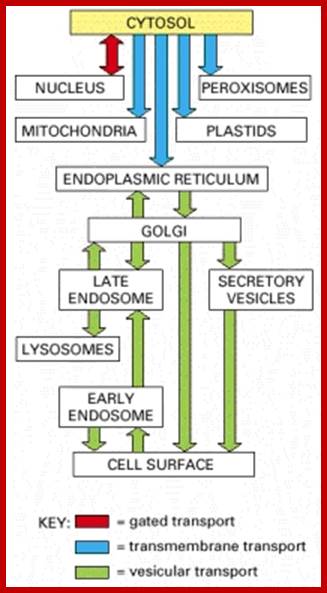
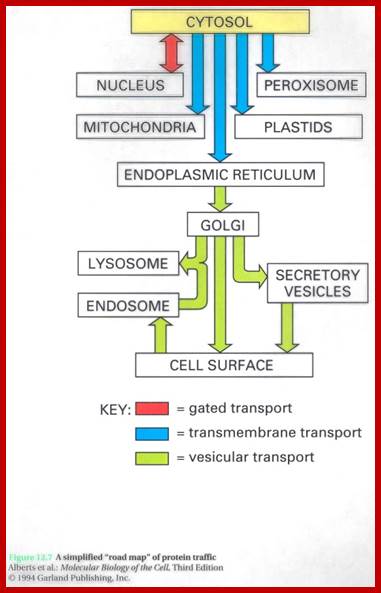
������� http://www.unitus.it/
A simplified "road map" of protein traffic: �Proteins can move from one compartment to another by gated transport ( red), trans-membrane transport ( blue), or vesicular transport ( green). The signals that direct a given protein's movement through the system, and thereby determine its eventual location in the cell, are contained in its amino acid sequence. The journey begins with the synthesis of a protein on a ribosome and terminates when the final destination is reached. At each intermediate station ( boxes) a decision is made as to whether the protein is to be retained or transported further. In principle, a signal could be required either for retention in or for exit from each of the compartments shown, with the alternative fate being the default pathway (one that requires no signal). The vesicular transport of proteins from the ER through the Golgi apparatus to the cell surface, for example, appears not to require any specific sorting signals; specific sorting signals therefore are required to retain in the ER and the Golgi apparatus those specialized proteins that are resident there.We shall use this figure repeatedly as a guide throughout this chapter and the next, highlighting the particular pathway being discussed. http://www.unitus.it/
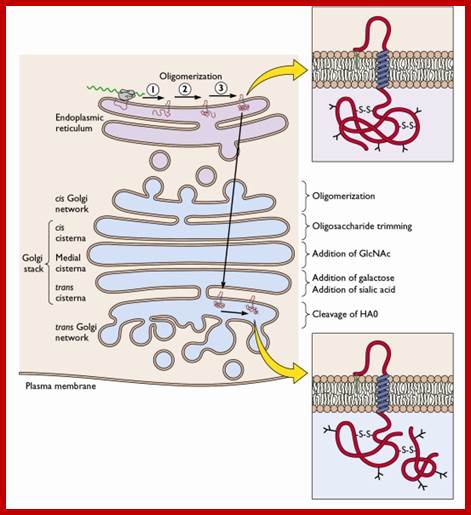
A protein that is entered ER reaches Trans-Golgi from where it loaded into
Vesicles and reach specific target; http://www.unitus.it/
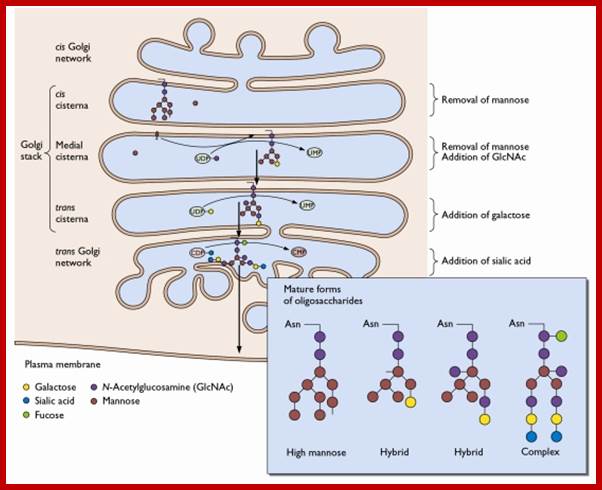
As the golgi membranes gradually mature, the resident enzymes perform various modifications progressively; removing certain glycans, adding sialic acid, transferring protein on to GPI and linking protein to serine (O-linked). Golgi membrane sacs contain several resident enzymes similar to that of ER. http://www.unitus.it/
A cell-free system demonstrating movement of protein from one Golgi cisternae to another:
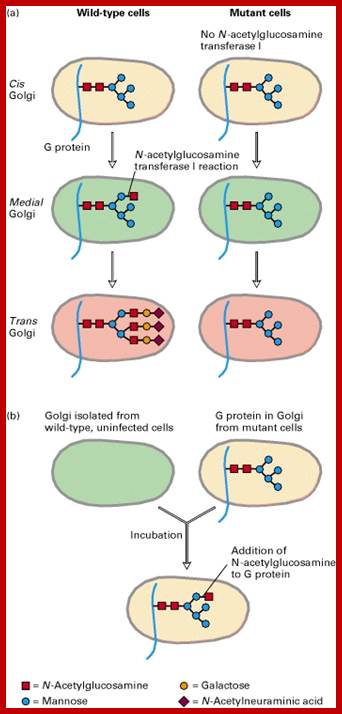
Vesicle Traffic;Protein sorting; Lodish; Molecular Cell Biology;
(a) A mutant line of cultured fibroblasts is essential in this type of assay. In this case, the cells lack the enzyme N-acetyl glucosamine transferase. In wild-type cells, this enzyme is localized to the medial-Golgi and modifies asparagine-linked oligosaccharides by the addition of one N-acetyl glucosamine. In VSV-infected wild-type cells, the oligosaccharide on the viral G protein is modified to a typical complex oligosaccharide, as shown in the trans-Golgi panel. In infected mutant cells, however, the G protein reaches the cell surface with a simpler high-mannose oligosaccharide containing only two N-acetyl glucosamine and five mannose residues.
�b) When Golgi cisternae isolated from infected mutant cells were incubated with Golgi cisternae from normal, uninfected cells, the G protein produced contained the additional N-acetyl glucosamine. Two interpretations of this finding are possible: (1) the G protein traveled from mutant cis-Golgi cisternae to wild-type medial-Golgi cisternae to receive the N-acetyl glucosamine residue, or (2) the transferase enzyme moved in the retrograde direction from the wild-type medial- to the mutant cis-Golgi cisternae where it modified the VSV G protein. The latter explanation is now believed to be correct, with transport mediated by COP I vesicles. [See W. E. Balch et al., 1984, Cell 39:405 and 525; W. A. Braell et al., 1984, Cell 39:511; and J. E. Rothman and T. S�llner, 1997, Science 276:1212.]
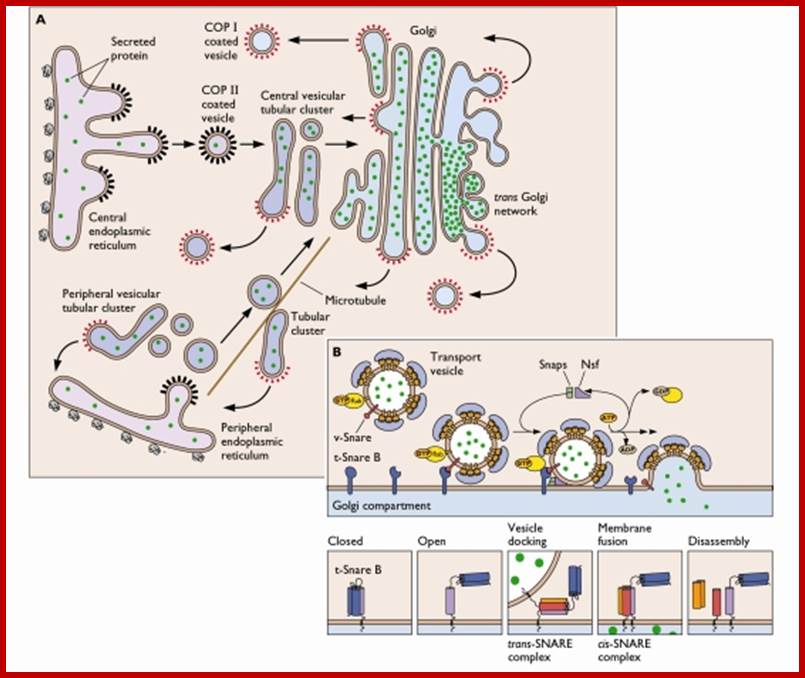
In this diagram movement of proteins from ER into coated vesicles (COPII) to cis golgi and the proteins are gradually modified, then from trans golgi the proteins are assorted and budded off by clathrin coated vesicles.� At the same time some of the ER resident proteins are retrieved by COPI coated vesicles.
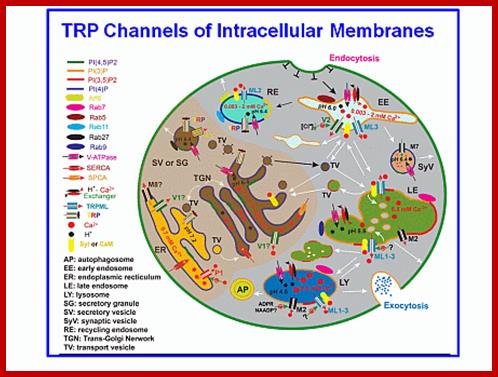
(TRP) channel superfamily have proved to be essential in maintaining adequate ion homeostasis, signaling, and membrane trafficking in the endosomal pathway. The unique properties of the TRP channels confer cells the ability to integrate cytosolic and intraluminal stimuli and allow maintained and regulated release of Ca2+ from endosomes and lysosomes.
Transient Receptor Potential (TRP) Channels; Dr. Hoaxing Xu; http://labs.mcdb.lsa.umich.edu/
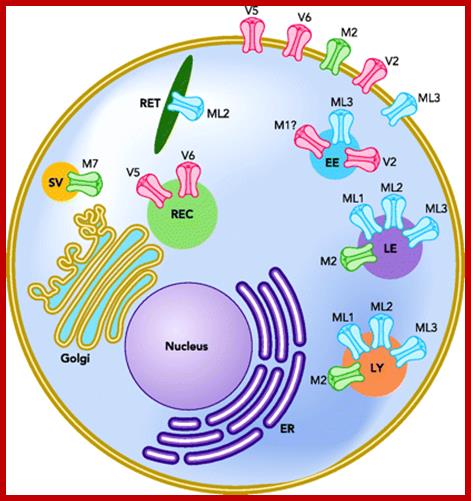
Members of some TRP subfamilies are widely distributed in various organelles along endolysosomal pathways. TRPML1 (ML1) is predominantly expressed in late endosomes (LE) and lysosomes (LY). Similarly, TRPML2 (ML2) is expressed in LE and LY but is also found on recycling endosomal tubules (RET). TRPML3 (ML3) is found not only in membranes of LE and LY but also in early endosomes (EE) and is distributed within the plasma membrane of the cell. TRPV5 (V5) and TRPV6 (V6) channels are found within the plasma membrane and recycling endosomal compartments (REC), but it is not yet known whether they are actively involved in this trafficking pathway. Recent reports indicate that TRPV2 (V2) is found to be expressed in both the plasma membrane and in early endosomes. TRPM2 (M2) is found in the plasma membrane, late endosomes, and lysosomes. TRPM1 (M1) is speculated to be present on endosomes, but conclusive evidence for its presence remains controversial. TRPM7 (M7) is thought to play a role in vesicular content release in neurons. Ken Abe , Rosa Puertollano http://physiologyonline.physiology.org/
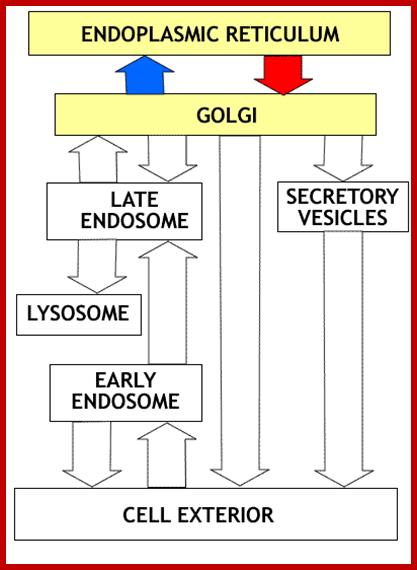
From ER to ECS and From ECS to intracellular (and ER?); http://www.unitus.it/
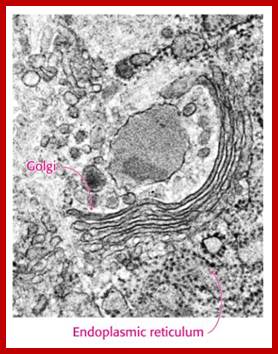
The diagram is an excellent EM of Golgi membranes associated with ER
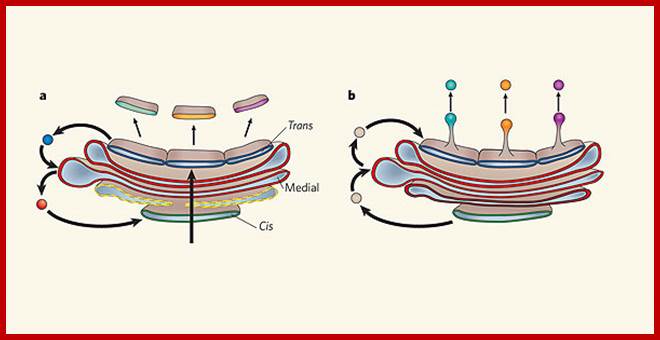
Figure: Two models of protein trafficking through the Golgi: (A) The cisternal maturation model of protein movement through the Golgi. As a new cis cisterna is formed it traverses the Golgi stack, changing as it matures by accumulating medial, then trans enzymes through vesicles that move from later to earlier cisternae (retrograde traffic). (B) The vesicular transport model, where each cisterna remains in one place with unchanging enzymes, and the proteins move forward through the stack via vesicles that move from earlier to later cisternae (anterograde traffic). Malhotra, V. & Mayor, S ;http://www.nature.com/
(A) The cisternal maturation model of protein movement through the Golgi. As a new cis cisterna is formed it traverses the Golgi stack, changing as it matures by accumulating medial, then Trans enzymes through vesicles that move from later to earlier cisternae (retrograde traffic). (B) The vesicular transport model, where each cisterna remains in one place with unchanging enzymes, and the proteins move forward through the stack via vesicles that move from earlier to later cisternae (anterograde traffic).
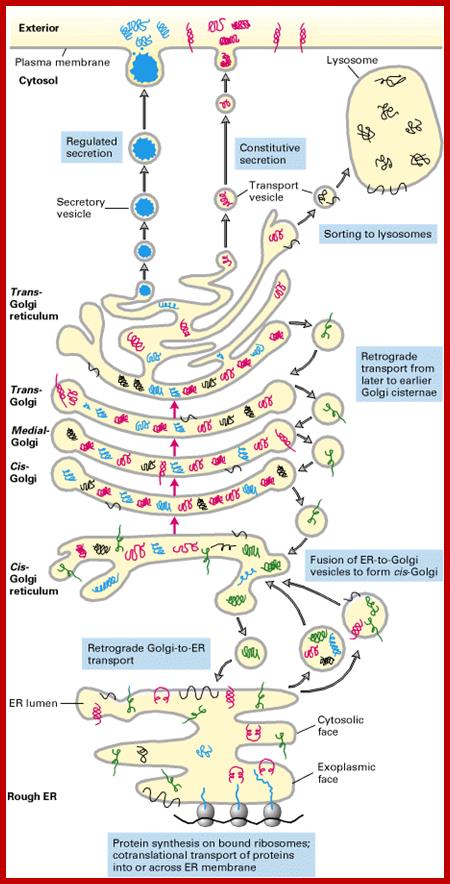
The secretory pathway of protein synthesis and sorting. Ribosomes synthesizing proteins bearing an ER signal sequence become bound to the rough ER. As translation is completed on the ER, the polypeptide chains are inserted into the ER membrane or cross it into the lumen. Some proteins (e.g., rough ER enzymes or structural proteins) remain resident in the ER. The remainder move into transport vesicles that fuse together to form new cis-Golgi vesicles. Each cis-Golgi cisterna, with its protein content, physically moves from the cis to the trans face of the Golgi stack (red arrows). As this cisternal progression occurs, many luminal and membrane proteins undergo modifications, primarily to attached oligosaccharide chains. Some proteins remain in the trans-Golgi cisternae, while others move via small vesicles to the cell surface or to lysosomes. In certain cell types (e.g., nerve cells and pancreatic acinar cells), some soluble proteins are stored in secretory vesicles and are released only after the cell receives an appropriate neural or hormonal signal (regulated secretion). In all cells, certain proteins move to the cell surface in transport vesicles and are secreted continuously (constitutive secretion). Like soluble proteins, integral membrane proteins move via transport vesicles from the rough ER to the cis-Golgi and then on to their final destinations. The orientation of a membrane protein, established when it is inserted into the ER membrane, is retained during all the sorting steps: Some segments always face the cytosol; others always face the exoplasmic space (i.e., the lumen of the ER, Golgi cisternae, and vesicles or the cell exterior). Retrograde movement via small transport vesicles retrieves ER proteins that migrate to the cis-Golgi and returns them to the ER. Similarly, cis- or medial-Golgi proteins that migrate to a later compartment are retrieved by small retrograde transport vesicles. [See B. Glick and V. Malhotra, 1988, Cell 95:883.]; http://www.unitus.it/
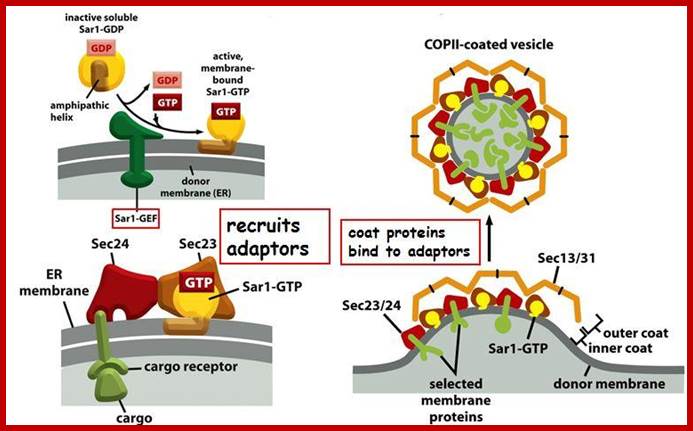
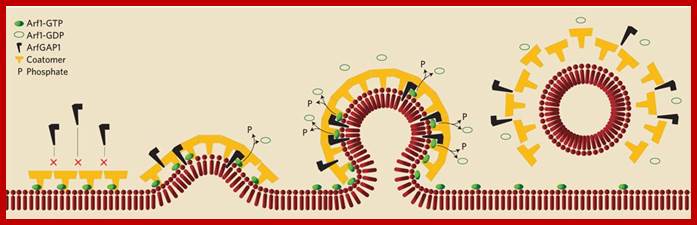
Top Figure: SAR1 regulates COPII coat formation by interactingwith GDP/GTP; GTP attaches to the membrane, adaptors are recruited to the membranes, Coatamers bind to adaptors, Coat is assembled and vesicles are pinched off, Coat is shed off soon, vesicles find their targets; COPII vesicles are associated with Plus Micotubules (MT) on which the vesicle move; http://www.studyblue.com/
Bottom Figure: Proposed model for the association and dissociation of COPI coatomer in vivo. Coat proteins, made up of seven proteins, they are recruited to the site of vesicle budding by membrane-bound Arf1 in its GTP form, and begin to deform the donor membrane. Sensing membrane curvature, ArfGAP1 is recruited to the budding site where it hydrolyzes GTP bound to Arf1, which then dissociates. As long at the budding site is attached to the donor membrane, the GTP form of Arf1 is replenished at the budding site. Once dissociated, the new vesicle lacks a fresh supply of Arf1-GTP. After all the Arf1-GTP has been hydrolyzed by ArfGAP1, the COPI coat dissociates from the newly formed transport vesicle or tubule.:http://www.the-scientist.com/
The Golgi apparatus is often found in close proximity to the ER in cells. Protein cargo moves from the ER to the Golgi, is modified within the Golgi, and is then sent to various destinations in the cell, including the lysosomes and the cell surface.� 2009 Nature Publishing Group Xu, D. & Esko, J. D. A Golgi-on-a-chip for glycan synthesis. Nature Chemical Biology 5, 612�613 (2009). doi:10.1038/nchembio0909-612. All rights reserved.
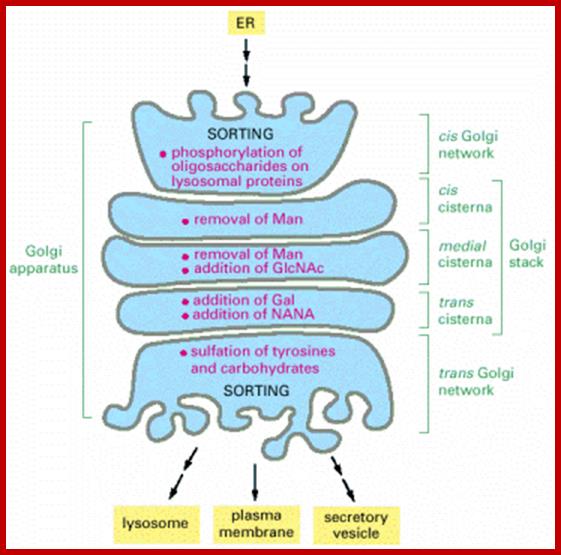
The picture depicts the relationship between the ER and Golgi membranes
and components. http://quizlet.com/
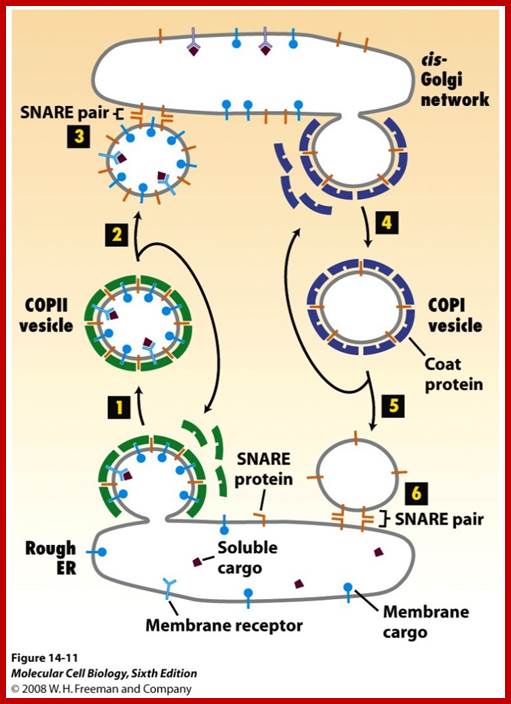
Used for transfers within the Golgi and for retrograde transfer to the ER as well as for transfer to the �cell surface via the constitutive secretory pathway; http://www.studyblue.com/
Cell organelles like chloroplasts and mitochondria themselves synthesize some of their own proteins, and they have to find their specific destination to organize into specific structures such as thylakoids in chloroplasts, cristae in mitochondria and soluble fluid within the organelle.� No other organelles have the ability to transcribe its own genomic DNA and translate and integrate like mitochondria and plastids.
If one visualizes the cellular microcosm and its components, which are produced and degraded every minute of its existence, it is as amazing and it is as bewildering as the macrocosm of the universe.
� Proteins that are synthesized in free form, i.e. not on ER, are targeted to different organelles and transported to their respective destinations. Nevertheless, some that are synthesized in free-state move into ER through special Sec protein channels.� �But proteins that are synthesized on ER have various targets.� First they have to be folded properly, if needed they are processed and modified in the sense they are added with carbohydrates, acetyl groups, or phosphates and localized in the membrane in specific orientation or they may be just placed in the lumen.�� Gradually as the more and more of proteins are transported into the ER lumen, with modification the membrane start extending into ER free from ribosomes, called SER.� It is during these transitions RER to SER the proteins are marked, modified or being in the process of modification and marking.� Endoplasmic membrane network is supported by microtubular skeletal network?
At a docking site of ER, the binding of one SRP-mRNA-ribosome complex, it is fairly clear at this site one kind of proteins is synthesized and threaded into the ER lumen and it accumulates at the site.� Once such cluster of proteins build up in a site they are modified and bunched together; then the ER membrane expands into a SER surface, and then it is pulled towards cytoplasm into a sac like structure called a vesicle. Many such vesicles from SER at various sites develop and pinched off.� Vesiculation from SER is performed by a set of proteins called COPs II (Coatamer proteins of specific kind).� Such vesicles are then fuse with one another or fuse to form proximal (cis) surface of Golgi complex, which is called cis surface.
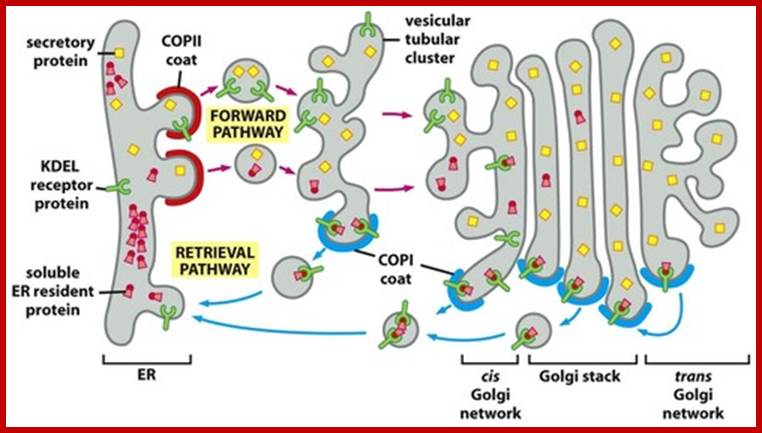
How do proteins made in RER get into the Golgi? How they resident
proteins are retrived; They use COPII and COPI coatamers for forward movement and retrival repectively:
�http://quizlet.com/
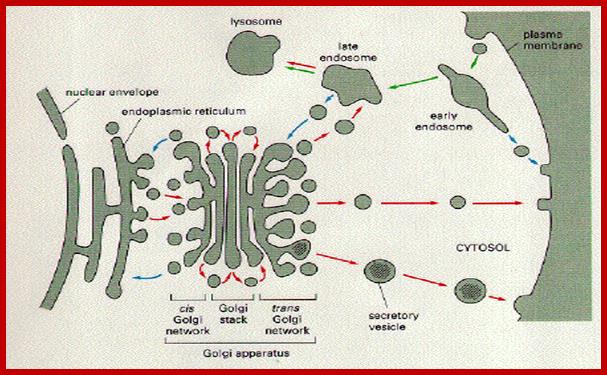
This is another default pathway i.e. from ER to Golgi, Golgi to ER and from Golgi to Plasma membrane, golgi to late endosome and back.� Pulse labeled proteins reach specific destination within 4-5 minutes.
� Golgi complex is a bunch of membranous sacs called cisternae, like saucers, stacked one above the other or one on the other.� The Golgi membranes near to SER are called cis Golgi and the membranes found at other extreme end are called Trans Golgi.� In between cis and trans-Golgi membranes many layers of sacs are found called median Golgi.� The Golgi membranes near the nucleus curved away from it, but membranes facing SER are curved towards them.� Each of the Golgi membranes can be identified by marker enzyme found in them.� The cis Golgi sacs contain many enzymes among them are glycosylamine acetyl transferase and mannosidase. Medial sacs also contain many such as N-acetyl glucosaminidase, mannosidase-II etc.� Trans and Trans Golgi network contain galactosyl transferase, mannose6-phosphate receptors, sialyl transferase and acid phosphatase.
As in the case of ER, which is supported by microtubules as shown below, for their stability and movement (?), the centrosome complex and its elements have a role in the stability of Golgi apparatus.� Golgi bodies are always in close proximity to centrioles. Use of colchicine and similar drugs to destabilize the microtubular assembly also disturbs the assembly and stability of Golgi membrane.� As in the case of ER, Golgi membranes also disperse into membrane vesicles during mitosis and meiosis.� Are these dispersed membrane vesicles do have some markers so as to reassociate to form the fully developed systems?!
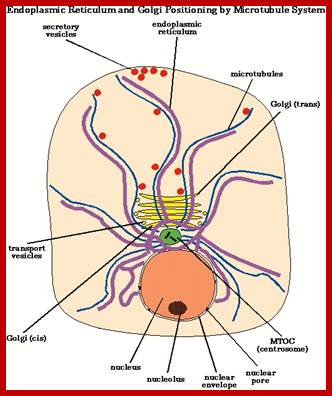
This diagram is a simplistic representation of reticular endothelium spread all over but supported by microtubules? Golgi apparatus is just sitting very adjacent to the nucleus; even the Golgi membranes are supported by cytoskeletal network?
How and why does the endoplasmic reticulum move?
(by Becky Bola and Viki AllaN)
Microtubule assisted ER movement
The ER (endoplasmic reticulum) is a fascinating organelle that is highly dynamic, undergoing constant movement and reorganization. It has many key roles, including protein synthesis, folding and trafficking, calcium homoeostasis and lipid synthesis. It can expand or contract in size when needed, and the balance between tubular and lamellar regions can be altered. The distribution and organization of the ER depends on both motile and static interactions with microtubules and the actin cytoskeleton. How the ER moves and consider why this movement may be important for ER and cellular function.
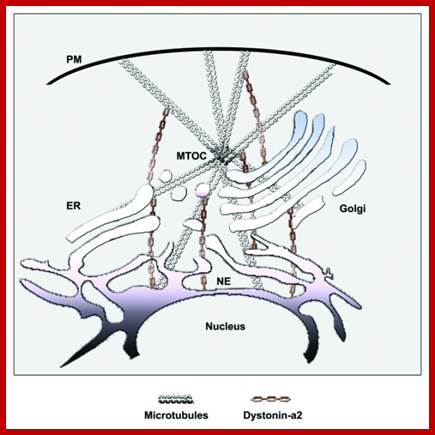
Steven P.Gross.http://iopscience.iop.org/;MT use Dynein and Kinesin for cargo movement even organelle transport; A novel role for Cyttoskeletal linker protein dystonin in maintenance of Microtubules stability and regulation of ER-Golgi transport. Scott D. Ryan et al; https://www.scienceopen.com.
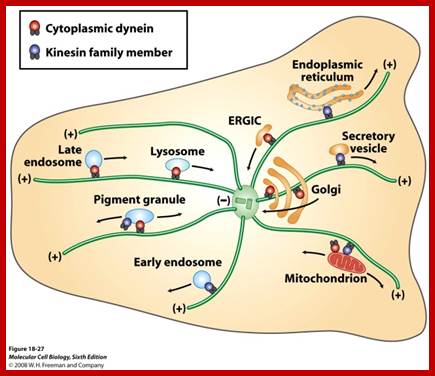
The network of microtubules connecting cellular organelles is a path for Kinesin and Dynein to transport materials within cells;http://www.bio.miami.edu/
Active transport is critical for cellular organization and function, and impaired transport has been linked to diseases such as neuronal degeneration. Much long distance transport in cells uses opposite polarity molecular motors of the kinesin and dynein families to move cargos along microtubules. It is increasingly clear that many cargos are moved by both sets of motors, and frequently reverse course. This review compares this bi-directional transport to the more well studied uni-directional transport. It discusses some bi-directionally moving cargos, and critically evaluates three different physical models for how such transport might occur. It then considers the evidence for the number of active motors per cargo, and how the net or average direction of transport might be controlled. The likelihood of a complex linking the activities of kinesin and dynein is also discussed. The paper concludes by reviewing elements of apparent universality between different bi-directionally moving cargos and by briefly considering possible reasons for the existence of bi-directional transport. Steven P.Gross.http://iopscience.iop.org/;
Crosslinking proteins maintain organelle structure and facilitate their function through the crosslinking of cytoskeletal elements. We recently found an interaction between the giant crosslinking protein dystonin-a2 and the microtubule-associated protein-1B (MAP1B), occurring in the centrosomal region of the cell. In addition, we showed that this interaction is necessary to maintain microtubule acetylation. Loss of dystonin-a2 disrupts MT stability, Golgi organization, and flux through the secretory pathway. This, coupled to our recent finding that dystonin-a2 is critical in maintaining endoplasmic reticulum (ER) structure and function, provides novel insight into the importance of dystonin in maintenance of organelle structure and in facilitating intracellular transport. These results highlight the importance of cytoskeletal dynamics in communicating signals between organelle membranes and the cytoskeleton. Importantly, they demonstrate how defects in cytoskeletal dynamics can translate into a failure of vesicular trafficking associated with neurodegenerative disease. Scott Rayan et al ;https://www.scienceopen.com
The ER (endoplasmic reticulum) is an extended network of lamellar membrane sheets and reticular tubules that spreads throughout the cell and is continuous with the nuclear envelope. In some cells, it is clearly divided into two types, rough ER and smooth ER, which are distinguished by the presence or absence of ribosomes on their surface. Proteins destined for secretion, or that function within the organelles of the secretory and endocytic pathway, are synthesized at the rough ER. The lumen of the ER is much more oxidizing than that of the cytoplasm, which allows the formation of disulfide bonds necessary for proper protein folding. Newly synthesized proteins can then be transported from the ER on towards the Golgi apparatus, or be retained in the ER. The ER is also a site for lipid and sterol synthesis, as well as being major intracellular calcium storage. Last, but not least, the ER is a highly dynamic organelle that is continually moving and changing shape.
The endoplasmic reticulum (ER) in Saccharomyces cerevisiae consists of a reticulum underlying the plasma membrane (cortical ER) and ER associated with the nuclear envelope (nuclear ER). This transcript was used a Sec63p-green fluorescent protein fusion protein to study motility events associated with inheritance of cortical ER and nuclear ER in living yeast cells. During M phase before nuclear migration, we observed thick, apparently rigid tubular extensions emanating from the nuclear ER that elongate, undergo sweeping motions along the cell cortex, and shorten. Two findings support a role for microtubules in this process. First, extension of tubular structures from the nuclear ER is inhibited by destabilizer of microtubules. Second, astral microtubules, structures that undergo similar patterns of extension, cortical surveillance and retraction, co-localize with nuclear ER extensions. During S and G-2 phases of the cell cycle, we observed anchorage of the cortical ER at the site of bud emergence and apical bud growth. Thin tubules of the ER that extend from the anchored cortical ER display undulating, apparently random movement and move into the bud as it grows. Finally, we found that cortical ER morphology is sensitive to a filamentous actin-destabilizing drug, latrunculin-A, and to mutations in the actin-encoding ACT1 gene. Our observations support 1) different mechanisms and cytoskeletal mediators for the inheritance of nuclear and cortical ER elements and 2) a mechanism for cortical ER inheritance that is cytoskeleton dependent but relies on anchorage, not directed movement.
At Least Three Types of Coated Vesicles Transport Proteins from Organelle to Organelle; COP1 coated, COPII coated and Clathrin coated:
The figure below depicts that all eukaryotic cells contain a plethora of small membrane-limited vesicles, many with a protein coat on their cytosolic surface. Three types of coated vesicles are known, each with a different type of protein coat and each formed by reversible polymerization of a distinct set of protein subunits under carefully regulated conditions. Each type of vesicle transports proteins from particular parent organelles to particular destination organelles. For instance, vesicles with a Clathrin coated form from the plasma membrane and the trans-Golgi network and move to late endosomes. Vesicles with a COP II coat transport proteins from the rough ER to the Golgi. Finally, vesicles with a COP I coat mainly transport proteins in the retrograde direction between Golgi cisternae and from the cis-Golgi back to the rough ER. Researchers have not yet identified the coat proteins surrounding many types of transport vesicles, such as those moving from late endosomes to lysosomes or the constitutive secretory vesicles that move proteins from the trans-Golgi to the plasma membrane.
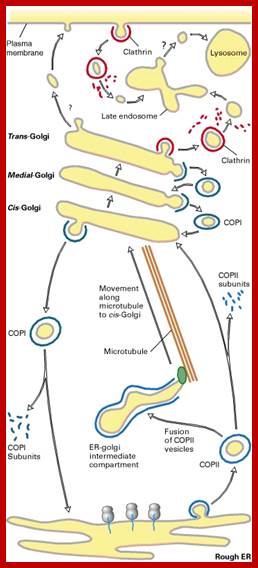
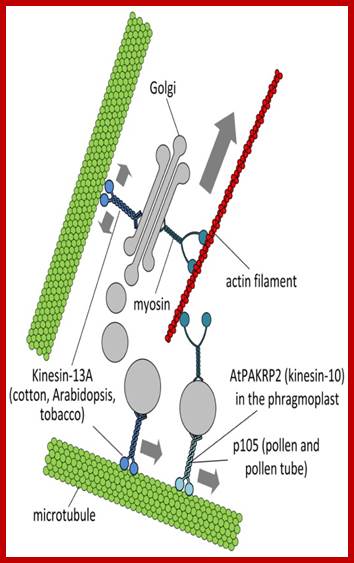
Traffic through Clathrin coated From PM and Trans-Golgi.� COP I coated and COP� II coated Golgi to Golgi and ER to Golgi respectively; the figure shows certain COPII coated vesicles are transported along microtubules mediated by dynein-ATP transporters. Lodish;� http://web.uconn.edu/
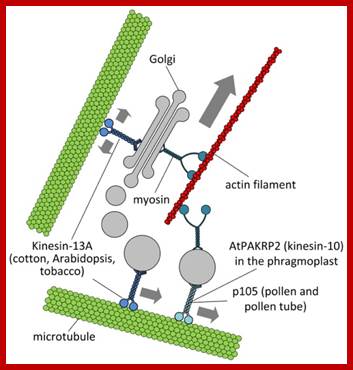
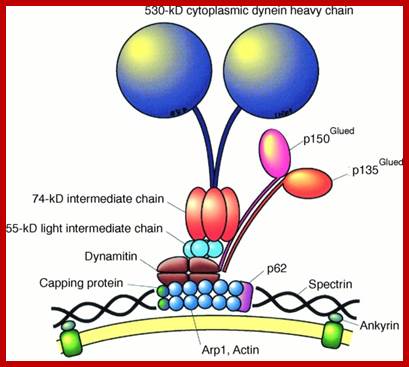
Giampiero Cai* and Mauro Cresti; http://journal.frontiersin.org/ Dynein bound to cytoskeletal tract; http://www.sciencemag.org/
Putative interactions between Golgi stacks/vesicles and motor proteins. Specific subclasses of myosin XI would be responsible for the long-range transport of Golgi bodies within plant cells. Members of the kinesin superfamily are likely involved in either the anchorage of Golgi stacks to microtubules (such as kinesin-13A) or the short-range transport of Golgi vesicles to defined cell positions (such as AtPAKRP2 and p105). The arrow length indicates the relative velocity as produced by motor proteins. Opposing arrows indicate an anchorage role for the motor.
![Cytoplasmic Dynein and Kinesin Power Axonal TransportSchematic diagram of the microtubule motor proteins cytoplasmic dynein and kinesin. Cytoplasmic dynein transports cargo in the retrograde direction toward the minus ends of microtubules whereas kinesin transports cargo in the anterograde direction toward the plus ends. Cytoplasmic dynein is a large multimeric protein complex comprising two heavy chain subunits (red) that possess microtubule binding and ATPase activity, two intermediate chains (yellow), two light intermediate chains (indigo), and an assortment of light chains (light pink, green, orange) (reviewed in [7]). Dynactin, a large multisubunit protein complex of comparable size to cytoplasmic dynein, is proposed to link the dynein motor to cargo and/or increases its processivity. The largest dynactin subunit, p150Glued (turquoise), forms an elongated dimer that interacts with the dynein intermediate chain and binds to microtubules via a highly conserved CAP-Gly motif at the tip of globular heads. The dynactin subunit p50 (dark pink) occupies a central position linking p150Glued to cargo. The conventional kinesin holoenzyme, also known as kinesin-1, is a heterotetramer comprising two Khc subunits (red) with microtubule binding and ATPase domains, a central coiled stalk, and a tail domain that interacts with two Klc subunits (green). Klcs may mediate cargo-binding via an intermediate scaffold protein (blue) that binds a cargo transmembrane protein (yellow).](Co_and_Post_Translational_Events5-Cellular_Protein_Traffic_files/image124.jpg)
Kinesins and Dyneins are super family of proteins involved in the intracellular traffic of vesicles
and organelle on cytoskeletal tracts; Duncan JE, Goldstein LS - PLoS Genet. (2006); http://openi.nlm.nih.gov/
Three modes of entry into the cell:
The general scheme of vesicle budding shown in figure below applies to all three known types of coated vesicles. During formation of these vesicles, the coat subunit proteins polymerize around the cytosolic face of a budding vesicle, thereby helping the vesicle to pinch off from the parent organelle. Some coat-protein subunits or associated adapter proteins select which membrane and soluble proteins will enter the transport vesicles as cargo proteins. These cargo proteins contain short signal sequences that direct them to a specific type of transport vesicle. A third feature common to formation of clathrin, COP I, and COP II vesicles is involvement of a GTP-binding protein that regulates the rate of vesicle formation. We begin our study of vesicular transport with a detailed look at the similarities and differences in the formation of the three types of coated vesicles, followed by a discussion of the mechanism of membrane fusion.
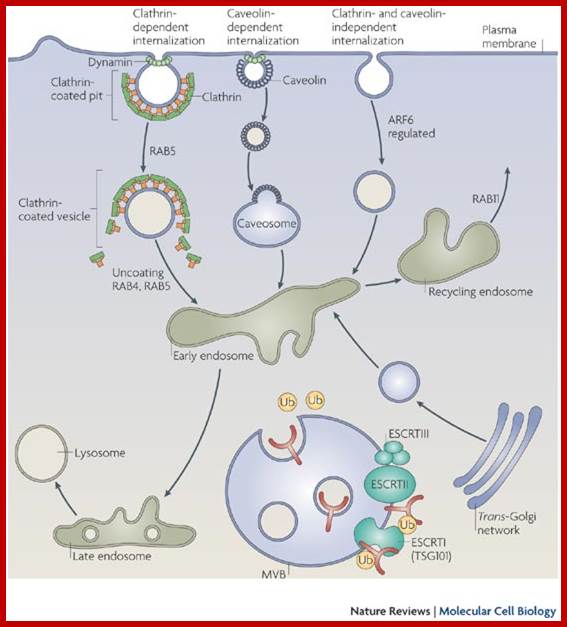
Three modes of transport from exterior into the cell; Clathrin dependent and Clathrin Independent pathways; http://www.nature.com/
Clathrin mediated:
Receptors, (in the above figure,) ligands, viruses and other endocytosed molecules are internalized using several routes. In the classic pathway Clathrin dependent endocytosis, receptors and their bound ligands are sorted into specialized domains of the plasma membrane through interactions with adaptor protein complexes. These areas invaginate and are coated with the scaffold protein clathrin. The interaction of the adaptors with the GTPase dynamin results in the formation of a ring of dynamin around the neck of the forming clathrin-coated pit, leading to scission and the formation of clathrin-coated vesicles. These vesicles then uncoated and fuse with early endosomes. This pathway is used by recycling receptors, such as transferrin and low-density lipoprotein receptors.
Caveolin Mediated:
Caveolin-dependent endocytosis is important for the uptake of toxins, viruses, bacteria and circulating proteins. This does not involve clathrin, but requires the scaffold protein �caveolin�. There are three kinds of caveolin proteins of which Caveolin 1 is important.� They produce cave like structures in plasma membrane and even in trans-golgi membranes.� Dynamin is also important for the scission of caveolin-containing vesicles, known as caveosomes, which can fuse with early endosomes.
Caveolin vesicles are clathrin independent vesicles carry some pathogens carry signal molecules and often associates with cholesterol, fatty acids and lipid droplets and is involved in its regulation.
GTP-ARF6 pathway:
A third, distinct mechanism of internalization does not use clathrin, caveolin or dynamin but requires GTPase ARF6 pathway. Cargo that is internalized by this pathway includes integrins, major histocompatibility complex molecules and glycosyl phosphatidyl inositol-anchored proteins. Once in early endosomes, receptors and other cargo can then be sorted either into late endosomes and lysosomes, or recycled back to the cell surface.
Mono-ubiquitylated membrane proteins destined for down regulation are recognized by a series of receptors containing mono-ubiquitin-binding domains. These receptors target mono-ubiquitylated membrane proteins through a series of trafficking steps that ultimately deliver them to be destroyed in the lysosome. This requires an invagination of the endosome membrane that forms multivesicular bodies (MVBs). Ubiquitin (Ub) functions as a key signal for sorting into MVBs. Endosomal membrane proteins that are destined for degradation in the lysosome are actively sorted into the intraluminal vesicles of a MVB, whereas proteins destined for the limiting membrane of the lysosome are retained in the limiting membrane of the MVB. ESCRT (endosomal sorting complex required for transport) complexes have a crucial role in this process. TSG101 is a subunit of the ESCRTI protein complex.
Rab proteins are members of the Ras superfamily of monomeric GTPases. Different Rabs regulate many facets of membrane trafficking, including vesicle formation and movement and membrane fusion, and are localized to distinct endosomal compartments. �Gwyn W. Gould & Jennifer Lippincott-Schwartz�
Protein Sorting and transport to their destination:
Sorting out of proteins, aggregate them into clusters and transport the same to targets via vesicular membranes, is a complex but defined process involving a variety of proteins. �There are specific proteins which recognize them individually through their receptors found in/on membranes ex. Plasma membranes and Endoplasmic reticular membranes. There are proteins that bind to variety of membranes on their cytoplasmic side, bind to receptor carrying cargo, pull the same into vesicles, ex. Endocytotic vesicles, vesicles at SER, vesicles at cis-Golgi, median Golgi and trans-Golgi membranes.� Such vesicles carry a group of enzymes or proteins, ex. Endosomes, synaptic vesicles, vesicles destined to plasma membranes, lysosomes and few others.� The proteins that bind at cytoplasmic surface for vesiculation differ from one type of vesicles to the other.� Such vesicles are marked by specific membrane proteins and their destination is determined by marker membrane proteins found at donor vesicles and target membranes.� Such vesicles are transported from the donor side to target by cytoskeleton mediated motor proteins (microtubules and microfilaments). Vesiculation and transport of the same involve variety of proteins.
Proteins Involved in Vesiculation:
Clathrins: Clathrins are made up of two subunits; large subunits 180KD and small subunits 35KD.� The large subunit, with small subunit, generates a network of triskelions linked to each other forming cage around the membrane with a hexagonal pattern.� Triskelions consist of three light chains and three heavy chains organized into tri-radiant structure that form network of polyhedral coat protein as a coat anchoring the vesicle.� Clathrin coated vesicles form at cytoplasmic surface of the PM containing coated pits.� Such clathrins are also involved in vesicle transport from trans-golgi to various destinations.� Composition of clathrin coated pits varies in their adapter proteins.
����������������������������������������������������������� ��������������������������������� 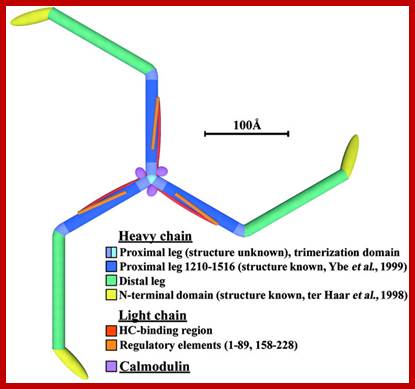 ���
���
Clathrin light and heavy chain interface; alpha helix binding superhelix loops via critical Tryptophans; Chih‐Ying Chen,et al; http://emboj.embopress.org/
Assembly unit of clathrin baskets. Three heavy-chains form a triskelion. Three light-chains (grey boxes) bind to the section of the heavy-chains proximal to the center. The N-terminus of a heavy-chain forms a globular domain separated from the leg by a flexible domain and projects inward.
P Schu, http://www.nature.com/
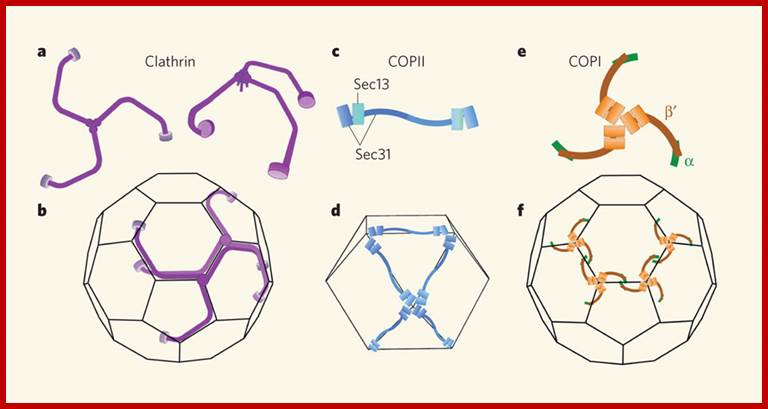
The basic structural elements of the assembly units of clathrin, COPII and COPI are α-solenoids (thick, curving lines) and β-propellers (cylinders). a, Two views of a clathrin triskelion. The N-terminal, β-propeller domains are at the tips of the legs. b, Packing of triskelions in an icosahedral clathrin coat, one of many lattices formed by clathrin5. There is a triskelion at each vertex, but only two are shown. c, The dimeric Sec31/Sec13 complex forms the coat of COPII vesicles6. The N-terminal, β-propeller domain of Sec31 is at the tip of the rod-like complex. A C-terminal segment of Sec31 is not present in the structure shown. d, Packing of Sec31/Sec13 in a COPII cuboctahedral lattice. The Sec31 β-propellers pack around the four-coordinated vertices6, 7. e, Lee and Goldberg1 report the crystal structure of the COPI assembly unit, a complex of the N-terminal half of the β′-chain and a central segment of the α-chain. f, The authors propose this model for the lattice of a COPI coat. Although the COPI triskelion is present in their crystal structure, the lattice symmetry and packing of triskelions within the lattice are speculative. The lattices are shown roughly to scale; the outer diameter of the clathrin coat in b is about 90 nm.;Schematic representation of coat proteins and their assemblies. Stephen C. Harrison
& Tomas Kirchhausen; http://www.nature.com/
Clathrin coat assembly
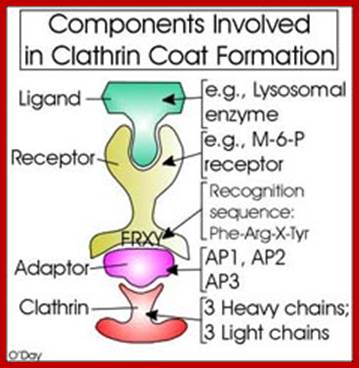
http://php.med.unsw.edu.au/
Coated vesicles in protein trafficking
|
Vesicle type |
Transport via |
Coat proteins |
GTPase |
|
Clathrin and its associated proteins |
Trans Golgi to endosome |
Clathrin+AP1 |
ARF |
|
��� |
TG to endosome |
Clathrin+GGA |
ARF |
|
�� |
PM to endosome |
Clathrin+AP2 |
ARF |
|
�� |
Golgi to lysosome, melanosome |
AP1 |
ARF |
|
|
Golgi to Golgi |
CopI |
|
|
|
|
|
|
From Mol Biol of the Cell (2006)
Clathrin triskelions assemble into a basketlike convex framework of hexagons and pentagons to form coated pits on the cytosolic surface of membranes
Under appropriate conditions, isolated triskelions spontaneously self-assemble into typical polyhedral cages in a test tube, even in the absence of the membrane vesicles that these baskets normally enclose
There are at least four types of adaptins, each specific for a different set of cargo receptors. Clathrin-coated vesicles budding from different membranes use different adaptins and thus package different receptors and cargo molecules. The formation of a clathrin-coated pit is driven by forces generated by the successive assembly of adaptins and the clathrin coat on the cytosolic surface of the membrane. The lateral interactions between adaptins and between clathrin molecules then aid in bud formation.
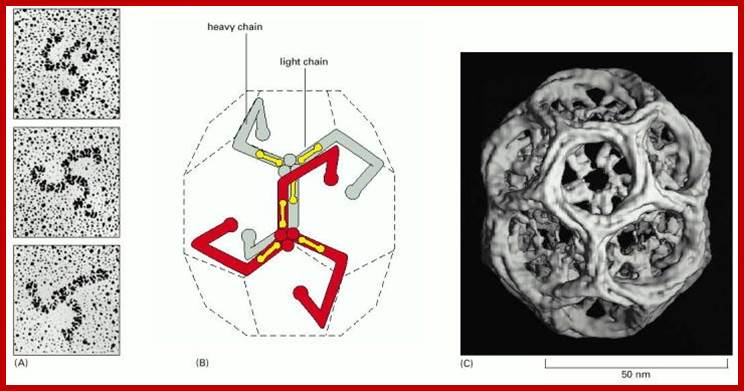
TEM of clathrin- Triskilions
Adaptor proteins:� These proteins are associated with clathrin and many of them act like clathrins.� There are at least four types of adapter proteins- AP-1, AP-2, AP-3 and AP-4.� Each adaptor protein complexes are hetero tetramers made up of adaptins.
Adaptins:
AP-1: μ1A, σ1 form the base; and γ1, β1 form arms; they are associated with mannose marked vesicles. In association with clathrins, AP1 is involved in transport from trans-golgi to endosomes; associated with ARFs.
AP-2: μ2, σ2 form the base and α, β2 form the arms; they are associated with Clathrin coated vesicles at Plasma membranes. �It is the most abundant class of coatomer.� In association with clathrins they transport vesicles from PM to endosomes; they are associated with ARFs.
AP-3: μ3, σ3; form the base and β3, δ, form the arms; they are associated with signal transduced secretory vesicles. In association with ARFs transport vesicles form from trans-golgi and move to lysosomes, melanosomes derived or platelet derived vesicles.
AP4: μ4, σ4 form the base and ε, β4 form arms; they are associated with clathrin coats at Trans-Golgi membranes involved in transcytosis in polarized cells.
Table: The subunit composition, localization and function of different coat complexes. AP, adaptor protein complex; ARF, ADP ribosylation factor; COP, coatomer protein; ER, endoplasmic reticulum; PM, plasma membrane; TGN, trans-Golgi network.
|
Coated vesicle |
Subunits of coat 1) |
GTPase 2) |
localization 3) |
Function4) |
|
AP-1 |
clathrin γ , β 1, �1, σ1 |
ARF1 |
TGN, early endosomes |
transport from TGN to endosomes/lysosomes |
|
AP-2 |
clathrin α, β 2, �2, σ2 |
ARF6? Rab5? |
PM, endosomes |
endocytosis |
|
AP-3 |
δ, β 3, �3, σ3 |
ARF1 |
TGN endosomes |
transport from TGN to lysosomes |
|
AP-4 |
ε, β 4, �4, σ4 |
ARF? |
TGN? |
? |
|
COPI |
α,β , β �, γ , δ, ε,x |
ARF1 |
ER-Golgi |
bidirectional Golgi-ER transport, intra-Golgi traffick |
|
COPII |
hSec23/24p hSec13/31p |
hSar1p |
ER-Golgi |
ER-Golgitransport |
|
1) (Serafini et al. 1991, Waters et al. 1991, Barlowe et al. 1994, Simpson et al. 1997, Dell�Angelica et al. 1999)2) (Donaldson et al. 1990, Robinson & Kreis 1992, Wong & Brodsky 1992, Stamnes & Rothman 1993, Rothman & Wieland 1996, Simpson et al. 1997, West et al. 1997 Dell�Angelica et al. 1999)3) (Serafini et al. 1991, Waters et al. 1991, Barlowe et al. 1994, Simpson et al. 1997, Kirchhausen 1999, Wieland & Harter 1999)4) (Letourneur et al. 1994, Schekman & Orci 1996, Le Borgne et al. 1998, Kirchhausen 1999, Hirst et al. 1999, Wieland & Harter 1999, Scales et al. 2000) |
||||
Among Adaptor proteins AP2 is the most abundant class of proteins, mostly found at plasma membranes.� AP4 is associated with Trans-Golgi membranes.� AP1 is generally associated with vesicles that transport lysosomal proteins with Mannose markers.� AP3 associates with signal response secretory vesicles such as synaptic vesicles that form at endosomes.� This type of APs is also found on secretory vesicles and those that transfer cargo from Golgi to lysosomes.
�����������
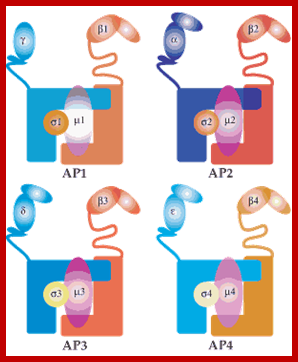
http://www.endocytosis.org/
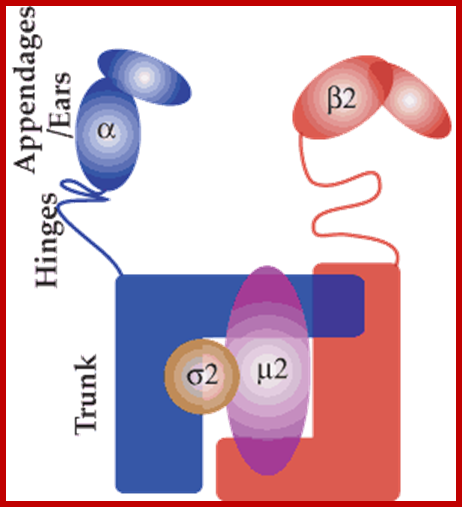
There are 4 different subunits (as illustrated in the model above based on the crystal structure) and thus they are sometimes called polymeric or hetrotetrameric adaptors. The trunk domain binds to membranes and to cargo via the mu and beta subunits. The appendage domains (sometimes called Ears) bind to accessory proteins. Structures of a number of appendage domains have been solved and binding motifs and partners characterised (seeappendages). The hinge domains bind to clathrin terminal domains. AP1, AP3 and AP4 adaptors have similar subunit compositions and bind to different cargo and accessory proteins (see Adaptor table 1).
The AP2 adaptor complex; cargo adaptors; http://www.endocytosis.org/
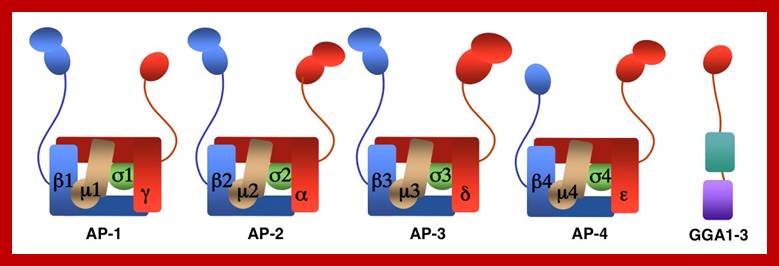
Structure of AP complex and GGAs; Eunice Kennedy Shriver; http://annualreport.nichd.nih.gov/
�����������������������������������
PM to endosomes, cells use clathrin and AP2 complex with ARFs. Trans-golgi to early endosomes�contain clathrin and AP1 (adapter proteins) complexes, associated with ARFs. Golgi to lysosomes, melanosomes and platelet derived vesicles contain AP3 complexes with clathrin proteins or with out them.
Cargo Adaptors are the proteins that link cargo into the
clathrin-coated pits. Adaptors can also be used to refer to the subset of
protein which we call 'accessory proteins' (the protein used in CCV formation
that do not bind to cargo.
There are many types of adaptor molecules:
Classical adaptors: AP1, AP2, AP3 and AP4 tetrameric
adaptor complexes (see below).
Alternative adaptors: A
single protein (in some cases homodimer) that links cargo into clathrin-coated
pits. Examples are GGAs, epsins and Hrs. Some of these proteins can also bind
to accessory proteins.
Accessory proteins: They are involved in clathrin
recruitment, membrane bending and scission of molecules etc. This can cover all
proteins involved in clathrin-mediated endocytosis except clathrin. But the
definition is more usefully applied when it neither covers clathrin nor cargo
adaptors.
There are 4 different subunits (as illustrated in the model above based on the crystal structure) and thus they are sometimes called polymeric or hetero-tetrameric adaptors. The trunk domain binds to membranes and to cargo via the mu and beta subunits. The appendage domains (sometimes called Ears) bind to accessory proteins. Structures of a number of appendage domains have been solved and binding motifs and partners characterized. The hinge domains bind to clathrin terminal domains. AP1, AP3 and AP4 adaptors have similar subunit compositions and bind to different cargo and accessory proteins.
There is a significant
homology between subunits of similar colors. Note that the gamma appendage
domain in the AP1 complex has only one sub domain like the GGA appendage domain
in the next panel.
AP1 complex: cargo selection from the
TGN and endosomes
AP2 complex:
cargo selection from plasma membrane
AP3 complex: cargo selection to
lysosomes
AP4 complex: cargo selection from trans-Golgi
in transcytosis in polarized cells.
COP proteins or Coatomers:
COP-I: Seven different subunits form a complex of ~700KD.� The Beta Cop protein has homology to that of adaptin beta proteins; they are associated with ARFs. They are involved in transport from Cis Golgi to ER � retrograde transport; and Cis Golgi to Trans-Golgi and vice versa. They are associated with ARFs.
COP-II: Sec 23p/ 24p (Tetramers 400KD), Sec13/ Sec 31(700KD), sec16, plus it is complexed with another protein called Sar 1p.� The Sar 1P is a monomeric G-protein. Sec 23-P is GTPase activating protein (GAP).� Sec 24 is recruiting protein; involved in transport from SER to cis golgi-anterograde transport. They are associated with Sar1.
The COP proteins exist in different forms having different functions, not all of them are well discerned.
The basic structural elements of the assembly units of clathrin, COPII and COPI are α-solenoids (thick, curving lines) and β-propellers (cylinders). a, Two views of a clathrin triskelions. The N-terminal, β-propeller domains are at the tips of the legs. b, Packing of triskelions in an icosahedral clathrin coat, one of many lattices formed by clathrin5. There is a triskelion at each vertex, but only two are shown. c, The dimeric Sec31/Sec13 complex forms the coat of COPII vesicles6. The N-terminal, β-propeller domain of Sec31 is at the tip of the rod-like complex. A C-terminal segment of Sec31 is not present in the structure shown. d, Packing of Sec31/Sec13 in a COPII cuboctahedral lattice. The Sec31 β-propellers pack around the four-coordinated vertices6, 7. e, Lee and Goldberg1 report the crystal structure of the COPI assembly unit, a complex of the N-terminal half of the β′-chain and a central segment of the α-chain. f, The authors propose this model for the lattice of a COPI coat. Although the COPI triskelion is present in their crystal structure, the lattice symmetry and packing of triskelions within the lattice are speculative. The lattices are shown roughly to scale; the outer diameter of the clathrin coat in b is about 90; http://www.nature.com/
Caveolin-mediated endocytosis of virus by host cells:
Caveolins are a family of integral membrane proteins which are the principal components of caveolae membranes and involved in receptor-independent endocytosis. Caveolins may act as scaffolding proteins within caveolar membranes by compartmentalizing and concentrating signaling molecules. Various classes of signaling molecules, including G-protein subunits, receptor and non-receptor tyrosine kinases, endothelial nitric oxide synthase (eNOS), and small GTPases, bind Cav-1 through its 'caveolin-scaffolding domain'. The Caveolin gene family has three members in vertebrates: CAV1, CAV2, and CAV3, coding for the proteins caveolin-1, caveolin-2 and caveolin-3, respectively.
Caveolins membrane proteins contain hairpin loops.� Their C and N terminals face cytoplasm side of the membrane. Caveolins are also expressed in endothelial, fibrous and adipose tissues.� They form small 50nm sized invaginations in the plasma membrane.� Caveolins also play a role integrin signaling (Wiki).
Internalization via caveolae is not a constitutive process but only occurs upon cell stimulation. Internalized viruses bound to their host cell receptor are delivered to the early endosome. It has been first thought that bound viruses were first taken to pH-neutral organelles in the cytoplasm called caveosomes and finally delivered to the ER, but this has been refuted in. Caveolae represent a low capacity but highly regulated pathway.
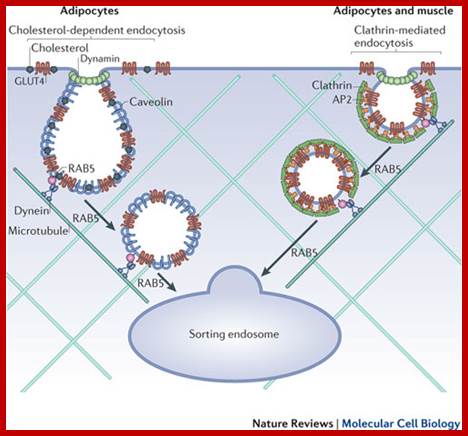
Dara Leto & Alan R. Saltiel; Molecular Mechanism of GLUT4 internalization
. 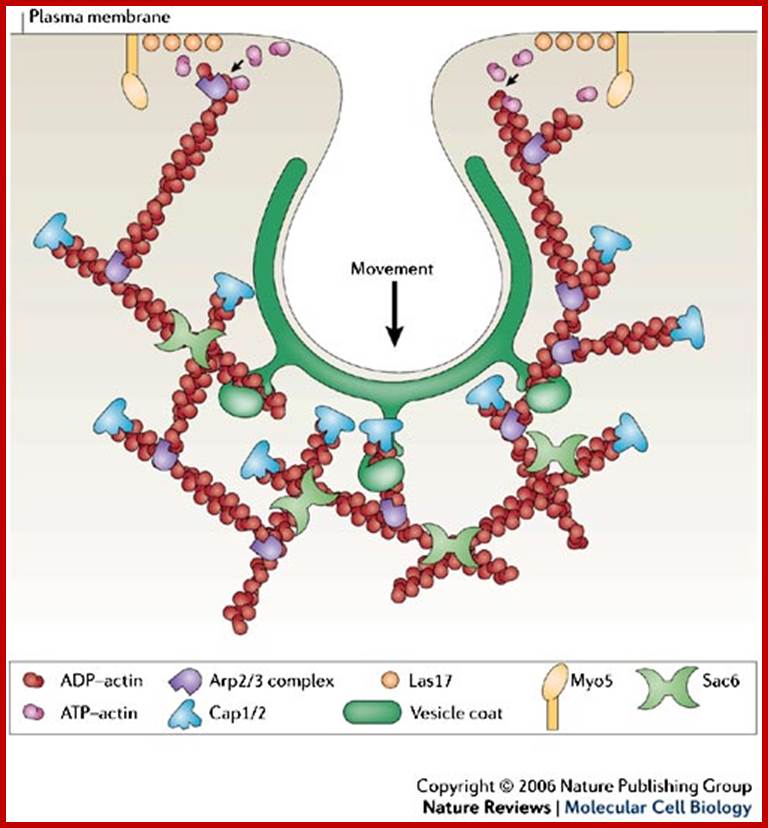
Actin polymerization often occurs at the plasma membrane to drive the protrusion of lamellipodia and filopodia at the leading edge of migrating cells. A role for actin polymerization in another cellular process that involves the reshaping of the plasma membrane � namely endocytosis � has recently been established. Live-cell imaging studies are shedding light on the order and timing of the molecular events and mechanisms of actin function during endocytosis. Harnessing actin dynamics for clathrn mediated endocytosis; Marko Kaksnenon et al; http://www.nature.com/
Glucose transporter type 4 (GLUT4) is internalized via clathrin-mediated and cholesterol-dependent endocytosis. Internalization of GLUT4 through clathrin-mediated endocytosis requires the adaptor protein AP2. AP2 coordinates packaging of this glucose transporter into endocytic vesicles by recruiting clathrin to the plasma membrane and by binding to an amino-terminal F5QQI sequence in GLUT4. Vesicle scission from the plasma membrane requires the GTPase dynamin, which assembles at the neck of the invaginating vesicle and, following GTP hydrolysis, constricts and breaks the membrane. Cholesterol-dependent endocytosis requires the membrane-deforming protein caveolin, which oligomerizes in lipid rafts and forms large cave-like structures, or caveolae. Cholesterol-dependent endocytosis is likely to also require dynamin to liberate caveolae from the cell surface. Vesicles carrying newly endocytosed cargo move towards the cell interior on dynein motors, which attach to the vesicles via RAB5. Endocytosed vesicles fuse with endosomes, where their cargo is sorted for degradation or recycling. Whereas clathrin-mediated endocytosis is the predominant mode of GLUT4 endocytosis in muscle cells, both clathrin-mediated and caveolin-dependent endocytosis have been implicated in GLUT4 endocytosis in adipocytes. In adipocytes, an insulin-stimulated switch from cholesterol-dependent to clathrin-mediated endocytosis may alter the rate of GLUT4 internalization. Dara Leto & Alan R. Saltiel
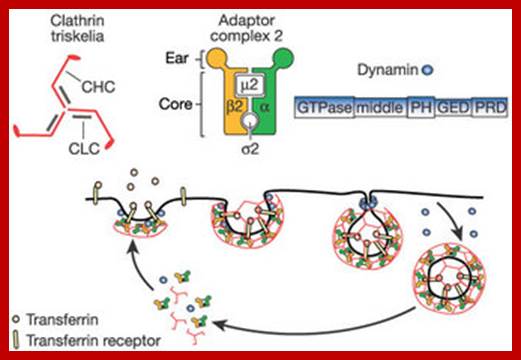
Core components of the machinery driving clathrin-mediated endocytosis
� 2003 Nature Publishing Group Tauber, A. I. Metchnikoff and the phagocytosis theory. Nature Reviews Molecular Cell Biology 4, 897�901 (2003) doi:10.1038/nrm1244. Susana Lopez, Ph.D. & Carlos F. Arias,http://www.nature.com/
Virus internalization by the host cell via caveolae, which are specialized lipid rafts that form 50-70 nm flask-shaped invaginations of the plasma membrane. Caveolins form the structural backbone of caveolae (CAV1/caveolin-1 and CAV2/caveolin-2 ; CAV3/caveolin-3 in muscle cells).
�
The following table shows few known sorting signal sequences that direct proteins to specific destinations:
|
Luminal signals: |
||||||||||||||||||||
|
|
Cytoplasmic sorting sequences |
||||||||||||||||||||||||||||||||||||||||
|
Note the amino acid symbols: K=lysine, D=aspartic acid, L=leucine, X+ any, N=Aspargine, E= Glutamic acid, P= proline, L=Leucine. Y= Tyrosine

This picture shows the mechanism of endocytosis of LDL receptors with cargo; once the cargo is delivered into cytoplasm the receptors are returned to the membrane surface. http://individual.utoronto.ca/
Formation Clathrin Coated Vesicles: Plasma membranes to endosomes:
Plasma membranes are studded with receptor proteins and many trans-membrane proteins; organized in the form of a patch.� Specific receptor proteins are associated with specific adaptor proteins; the adaptor proteins in turn are associated with clathrin skeletal proteins.� They are the fore runners of Clathrin coated vesicles.� Such membrane associated receptor regions are called coated pits.� Such coated pits with different adaptor proteins are found as patches all along the inner surface of the plasma membrane.� In clathrin coated pits β-adaptins bind to di-lysine (KK) sorting sequence of the receptor at the cytoplasmic face; at the same time they also bind to triskelions. Adaptins μ2 in AP2 complex recognize tyrosine (YY) based sorting signal sequences. In this case, phosphorylation of adaptins triggers internalization by clathrin-coated endocytosis.� Similarly α & γ including δ and ε involved in vesicle formation by clathrin mediated coats.
�
http://molar.crb.ucp.pt/
The peptide signal for endocytosis: The various cell-surface receptor proteins that are endocytosed in clathrin-coated vesicles are thought to share this signal, which is recognized by the adaptins that function in receptor-mediated endocytosis from

The receptors are different for different cargo proteins; http://molar.crb.ucp.pt/
There are 49 types of Clathrin-coated pits and vesicles:
This rapid-freeze, deep-etch electron micrograph shows numerous clathrin-coated pits and vesicles on the inner surface of the plasma membrane of cultured fibroblasts. The cells were rapidly frozen in liquid helium, fractured, and deep-etched to expose the cytoplasmic surface of the plasma membrane.
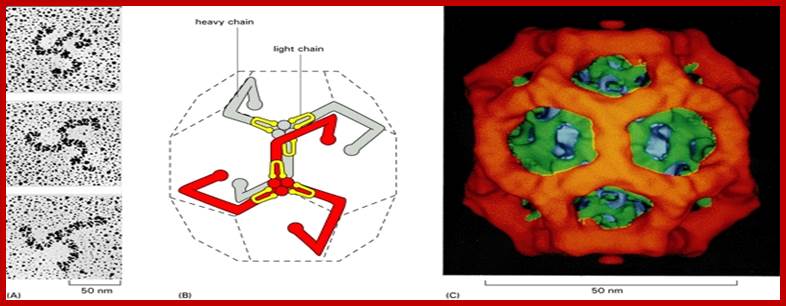
The structure of a clathrin coat
(A) Electron micrographs of clathrin triskelions shadowed with platinum. Although this feature cannot be seen in these micrographs, each triskelion is composed of 3 clathrin heavy chains and 3 clathrin light chains. (B) A schematic drawing of the probable arrangement of triskelions on the cytosolic surface of a clathrin-coated vesicle. Two triskelions are shown, with the heavy chains of one in red and of the other in gray; the light chains are shown in yellow. The overlapping arrangement of the flexible triskelion arms provides both mechanical strength and flexibility. Note that the end of each leg of the triskelion turns inward, so that its amino-terminal domain forms an intermediate shell. (C) A three-dimensional reconstruction of a clathrin coat composed of 36 triskelions organized in a network of 12 pentagons and 6 hexagons. The outer, red polygonal shell represents the overlapping legs of the clathrin triskelions; the intermediate, green shell, the amino-terminal domains of the triskelions; and the inner, blue shell, the adaptor proteins that we discuss later. Although the coat shown is too small to enclose a membrane vesicle, the clathrin coats on vesicles are constructed in a similar way from 12 pentagons plus a larger number of hexagons. (From J. Heuser, J. Cell Biol. 84:560-583, 1980, by copyright permission of the Rockefeller University Press.). �(A, from E. Ungewickell and D. Branton, Nature 289:420-422, 1981, � 1981 Macmillan Journals Ltd.; B, from I.S. Nathke et al., Cell 68:899-910, 1992. �Cell Press, from G.P.A. Vigers, R.A. Crowther, and B.M.F. Pearse, EMBO J. 5:2079-2085, 1986.)
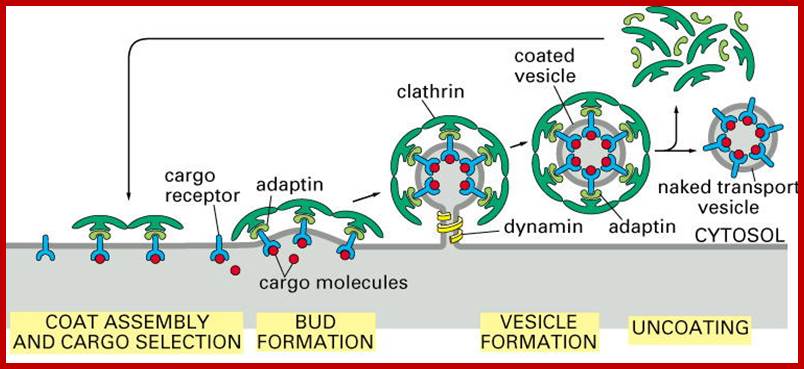
This diagram shows how a clathrin coated vesicle is formed and budded of into cytosol; http://www.zoology.ubc.ca/

Dynamin (Dark Blue) Assembled Around Neck of Budding Clathrin-Coated Vesicle; http://www.utm.utoronto.ca/
Dynamin forms a helical ring around the stalk of a budding clathrin-coated pit;
Dynamin binds and hydrolyzes GTP (GTPase activity), causing fission of the clathrin-coated vesicle from the membrane
- Several models have been proposed for dynamin-mediated vesicle fission:
- GTP hydrolysis results in a conformational change leading to dynamin constriction, generating the force necessary for scission of the vesicle (the �pinchase� model)
- the dynamin spiral around the stalk of the coated pit extends upon GTP hydrolysis, causing the vesicle to �pop off� the membrane (the �poppase� model)
- GTP binding activates dynamin to recruit downstream proteins which cause vesicle fission, although these proteins have not been identified (the �molecular switch� model)
Other proteins form an endocytic complex with dynamin:
- Amphiphysin assists in localizing dynamin to clathrin-coated pits by binding to the α adaptin subunit of AP2; it stabilizes the dynamin rings, stimulates dynamin�s GTPase activity, and regulates vesicle formation.
- Endophilin interacts with amphiphysin, facilitates membrane curvature, and recruits synaptojanin.
- Synaptojanin binds to endophilin and amphiphysin and regulates dynamin function; it dephosphorylates membrane lipids (phosphoinositide phosphatase activity) and is involved in vesicle uncoating.
- Many other proteins interact with dynamin, including syndapin and intersectin.

Phosphorylation and Ca2+ Regulate Dynamin-Mediated Synaptic Vesicle Endocytosis; http://www.utm.utoronto.ca/
- In neurons, the dynamin-dependent recycling of synaptic vesicles by endocytosis is regulated by phosphorylation and calcium (Ca2+):
- Dynamin 1 (a neuronal isoform of dynamin) is recruited to the membrane in its dephosphorylated form.
- Dephosphorylation of dynamin 1 is mediated by calcineurin �(Click here to learn more about calcineurin)
- Calcineurin activation is stimulated by Ca2+ influx during synapse depolarization.
- Therefore Ca2+ activates calcineurin, triggers dynamin 1 dephosphorylation, and allows synaptic vesicle recycling by endocytosis
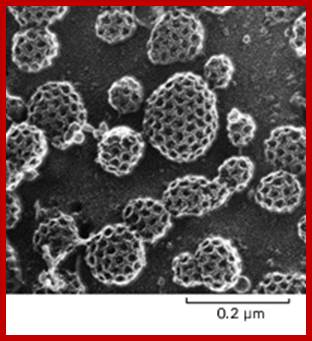
This figure is an SEM image looking at the inside of the plasma membrane of a skin cell. It shows many clathrin coated pits and vesicles forming on the inner surface of the plasma membrane.;Each of the polygonal rings on the surface of the pits is a complex of coat protein molecules. ;SEM pictures of clathrin coated vesicles;� Formation of Coated Vesicles; http://www.zoology.ubc.ca/
Cell surfaces contain receptors, which have external domains with sites for the binding of specific ligands.� Each cell, depending upon the cell type, bear thousands of different receptors.� Pit areas mark the cytoplasmic side of the receptors.� When the ligand binds, the membrane is pulled inwards carrying the ligand inwards as endosomes, by a process called endocytosis.� Sometimes cell surface bound components are engulfed as phagocytosomes or phagosomes. Another way of membrane invagination engulfing liquid from outside is Pinocytosis. The vesiculation and inward movement of vesicles is also controlled by a set of proteins.� From plasma membrane to lysosomes, from endothelial reticulum to Golgi complex, from Golgi complex to lysosomes, plasma membranes and secretion to extra cellular space is a designed traffic flow of membranes carrying proteins from one destination to the other delivering different proteins to different sites.� In this process membranes are budded off, membranes fuse with one another and in some cases membranes disperse into small vesicles only to be reformed at certain stages.� In this fascinating traffic cargos are delivered via membrane vesicles and emptied vesicles with any receptor return to their original site.
Current model for actin-driven endocytic internalization: Harnessing actin dynamics for clathrin-mediated endocytosis
Marko Kaksonen, Christopher P. Toret & David G. Drubin
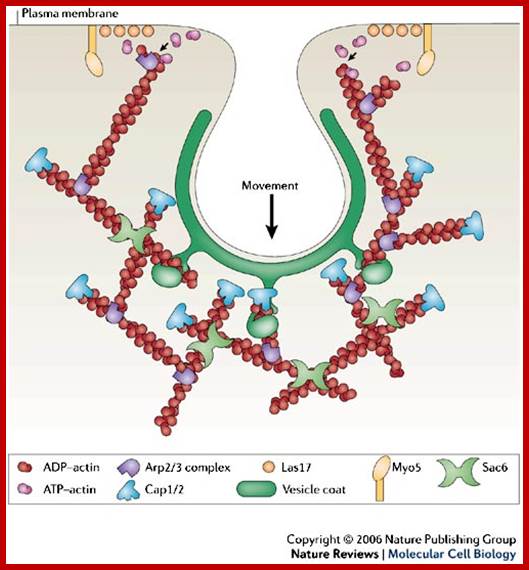
;Cureent model for actin driven endocytotic internalization; Marko Kaksonen, Christopher P. Toret & David G. Drubin; http://www.nature.com/
This schematic diagram illustrates putative functions of different actin-cytoskeleton proteins during endocytic internalization in Saccharomyces cerevisiae. Las17 ( Wiskott�Aldrich syndrome protein (WASP) in mammals) together with the myosins Myo3 (not shown) and Myo5 activate the actin-related protein-2/3 (Arp2/3) complex at the cell surface. Myosins might also generate force on the actin network or anchor the actin filaments to the plasma membrane through their motor domains. The activated Arp2/3 complexes form branched actin filaments that grow through the addition of ATP�actin monomers near the plasma membrane. Older filaments are capped at their barbed ends by capping proteins (Cap1/2). The branched filaments are further crosslinked by Sac6. The crosslinked actin network is linked to the underlying vesicle coat by actin-binding proteins such as Sla2 and Pan1, which are represented by green hand-like structures. The growth of the actin network leads to the invagination of the coated membrane. For further information on the proteins involved, Marko Kaksonen, Christopher P. Toret & David G. Drubin;
From RER/SER to Golgi:
RER is the region where ribosomes are bound and translation of proteins and insertion into ER takes place.� Also in this region proteins undergo glycosylation and folding.� The process of protein modification and some of the proteins bind to anchor proteins found in the SER and start assembling at the ends of SER.�
The protein cargo provides the surface for the assembly COPII protein complexes, which pulls the vesicle loaded with proteins.� Such vesicles formed may fuse with one another to form cis Golgi or the vesicles fuse with already existing cis Golgi membranes.
Once the cargo containing vesicles from SER fuse with cis Golgi, proteins go through further processing�s. Step by step proteins in median Golgi membranes are subjected designed modifications, they are also sorted out and collected at lateral regions of the membranes and they look like swollen structure. From the top Golgi membranes the proteins are processed and matured, again packed into vesicles and budded off from the trans-golgi surface.� As the processing is performed in stepwise manner the assorted proteins are packed into vesicle, mostly from the later side and budded off? Thus a series of vesicles are transferred from the Cis Golgi to the median Golgi one after another and finally to the trans-Golgi.
� In this process many of the proteins and enzymes transferred from SER, once their function is over, they are returned from Golgi in the form of vesicles back to ER.
� The stacked membrane sacs are interconnected (?) and fluid in them exchange. By the time protein reach trans surface they are already marked and they loaded into vesicles, which are released from the trans surface.� This vesiculation from SER to Cis Golgi to median Golgi and to trans Golgi is performed by a set of coat proteins. �These vesicles pinch off from the anterior membranes and handover them to the next membrane sac, then another set of vesicle develop from the cis Golgi membranes and they fuse with the next underlying Golgi membrane, thus the proteins are step wisely modified and processed and transported to the trans surface like a bucket brigade to be released ultimately to their specific destinations at the distal surface of the Golgi.� This type of movement from SER to Cis, from cis to median and from median to trans is called forward or anterograde movement.� But membranes from Golgi also move towards ER by a series vesicles.� This movement is called retrograde movement.� Again this process is performed by another set of coat proteins or cops.
There are two theories with regard to the maturation of Golgi vesicles from cis to trans.�
1. One view is that the golgi sacs are developed with the vesicles produced from SER.� Thus a series of Golgi membranes get stacked one above the other.
� While this process is going on the resident enzymes perform various processes in modifying proteins.� By the time these proteins fully formed they will be at trans-surface, from which sorted vesicles are budded off.
� In this process some of the enzymes and other proteins derived from SER, once their function is over they are returned by Vesiculation aided by COPI protein complexes.
2. Another view is that as vesicles produced from SER proteins in golgi membranes start undergoing modifications stepwise.� Then they are sorted and budded off to the next Golgi membranes; thus a series of vesicle produced join respective lower Golgi membranes.� The enzymes derived from SER are retrieved back to SER median golgi vesicles.
The time taken from loading to cis Golgi membranes, processing in the median Golgi and reach Tran Golgi and leave the same takes just less than 5 minutes.
The vesicles produced at trans-surface are loaded with proteins that are destined to specific location of the cell.� The proteins that pass through various stages of processing and various levels of Golgi are ultimately loaded into vesicles.� Some of the proteins that are transported from one level to the other are marked.� Example lysosomal proteins are phosphorylated at specific mannose residues, so phosphorylation of mannose is marker for lysosomal enzymes. ER and Golgi membranes contain transmembrane receptors for mannose phosphate residues, facing the lumen.� Thus the lysosomal proteins are held onto receptors.� Specific coatomer proteins called Cops bud off such clusters from cytosolic membrane surface.� Similarly other proteins are also sorted and loaded onto membranes, which are pulled away from the membranes in the form of vesicles, which are again coated with Cops.
� Vesicles carrying lysosomal proteins are transferred to early endosomes or late endosome to develop into lysosomes.� Vesicles loaded with lysosomal enzymes are marked with specific receptor proteins, so their destination is already determined.� Similarly the proteins for plasma membranes are carried by coated vesicles and transported to plasma membrane surface, where the vesicles fuse with plasma membrane.� Specific proteins again aid the fusion.� Similarly those proteins that are destined to secrete out of cell surface are transported as coated vesicles and they fuse with plasma membrane in such a way all the contents of vesicles are unloaded into exoplasmic region.� Some vesicles carrying certain cargo like acetylcholine or such compounds are transferred to synaptonemal membranes.� Some vesicles store certain components where they are stored as granules in storage vesicles.� Whatever vesicles generate trans-golgi membrane is coated with specific coat proteins, which have marked destination.� The most default path way is from ER to Golgi and from Golgi to plasma membrane.
Cell surfaces contain receptors, which have external domains with sites for the binding of specific ligands.� Each cell, depending upon the cell type, possesses thousands of different receptors.� Pit areas mark the cytoplasmic side of the receptors.� When the ligand binds, the membrane pulled inwards carrying the ligand inwards as endosomes, by a process called endocytosis.� Sometimes cell surface bound components are engulfed as phagocytosomes. Another way of membrane invagination engulfing liquid from outside is Pinocytosis. The vesiculation and inward movement of vesicles is also controlled by a set of proteins.� From plasma membrane to lysosomes, from endothelial reticulum to Golgi complex, from Golgi complex to lysosomes, plasma membranes and secretion to extra cellular space is a designed traffic flow of membranes carrying proteins from one destination to the other delivering different proteins to different sites.� In this process membranes are budded off, membranes fuse with one another and in some cases membranes disperse into small vesicles only to be reformed at certain stages.� In this fascinating traffic cargos are delivered via membrane vesicles and emptied vesicles with any receptor return to their original site.
� The flow of proteins from the site of synthesis to their destination follows two distinct pathways.� One that the proteins synthesized in free-state are directed to their respective destination such as chloroplasts, mitochondria, peroxisome and the nucleus, is more or less guided by HsPs and receptor proteins.� The other pathway is that though the protein synthesis starts in free-state, the N-terminal signal sequences of them determine, their further fate; they are placed on ER and the synthesis continues and all those proteins synthesized on ER are threaded into ER lumen.� The number of proteins synthesized on ER is substantial and their destination is also varied; their destination includes all the network of endothelial membranes, golgi bodies, microbodies, lysosomes, plasma membranes, exoplasmic space, probably all other.� Most of the proteins that are synthesized on ER and threaded into ER are transported through vesicles.� Not with standing, a large number of components from exoplasmic space find their way into cells, by inward flow in the form of membrane bound vesicles and the same are distributed to their destinations.
Not all thousands of proteins that enter from ribosomes into ER, from ER to Golgi are transported out of them.� Many, reasonably all the proteins required for structural organization of membranes, proteins required for protein modifications and marking other proteins for transport out of Golgi, remain in ER and Golgi bodies, they are called resident proteins.� These proteins have trans cellular domains, so their hydrophobic regions are anchored into the membranes.� One end of the protein, a major part of the protein, is towards the lumen and its other domain is towards cytoplasm.� Many of such proteins luminal domains act as receptors and they are responsible for binding to specific lumen proteins, they move along with the flow of membranes from one region to the other region, till they are released from their cargo.� Similarly anchored proteins with cytosolic domains facilitate the binding of coatamer- proteins.
The cytoplasmic domain has certain sequence that acts a signature for the movement of resident proteins from ER to Golgi and back to ER.� Specific carrier or transporter transfers all the inputs required for modifications of lumen proteins from cytoplasm into lumen.
� A set of related proteins are involved in binding to cytoplasmic surface of membranes and pull the membrane into a bud or vesicle carrying fluid and proteins.� This happens all the time at plasma membrane, at ER, at Golgi membranes and endosomes.� It is the vesicles carrying proteins are budded off from the said membrane surfaces, they are also transported to target sites and they fuse with their target membranes.� This kind of membrane flow, carrying protein as cargo, takes place in the cell all the time and each of these events take place at their own pace.
The vesicle budding off at plasma membrane surface and vesicles forming at trans-golgi are mediated by clathrin-coated structures.� Preformed clathrin coated pit areas determine their site at PM.� The RER transits into SER loaded with proteins; as cluster they are loaded at one side of the membrane and the same budded off carrying a group of proteins in the form of transitional vesicles.� Budding at SER surface is due to certain protein assembly called coatamers COP-II.� These vesicles fuse with cis-golgi membranes at which proteins are subjected to modification in terms of removal certain sugars and addition of certain sugars.� Addition of phosphate groups at certain sites of proteins.� Then the cis-golgi bud off vesicles, which fuse with median Golgi membranes, where more modification takes place.� Proteins with O-linked glycosylation take place only in Golgi cisternae not anywhere else.� From Cis-Golgi, proteins are clustered into similar kinds or same types and transferred to Trans-Golgi.� It is here at Trans-Golgi final sorting of proteins is done according to their structural or functional features.� Earlier the proteins are marked, as in the case of all lysosomal enzymes; they are phosphorylated at two mannose sites.� Mannose-P is a marker, the resident receptor proteins found in the membranes facing luminal side bind to mannose phosphate group, thus all lysosomal proteins are bound and localized at a given region.� Similarly other proteins are also localized and held onto to their respective receptor proteins.� From the Trans-Golgi membranes proteins are further grouped and loaded into membrane vesicles and budded off to generate a large number of vesicles at Trans-Golgi surface called Trans-Golgi Net Work (TGNW).�
The vesicles that develop from median and trans-Golgi towards cis-Golgi use another kind of coatamers called COP-I.� COP-I performs forward movement, but also backward movement i.e. from trans-Golgi to Cis -Golgi to ER.� But COP-II performs only forward movement is called Anterograde movement. Backward movement is called retrograde movement.
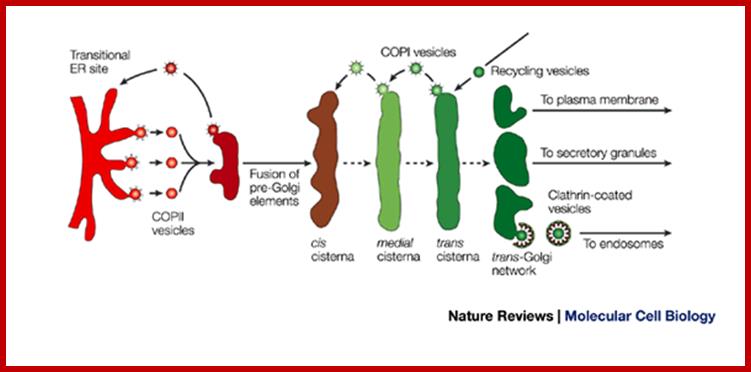
Golgi vesicle formation, maturation and vesicles leaving trans-golgi to various destinations.
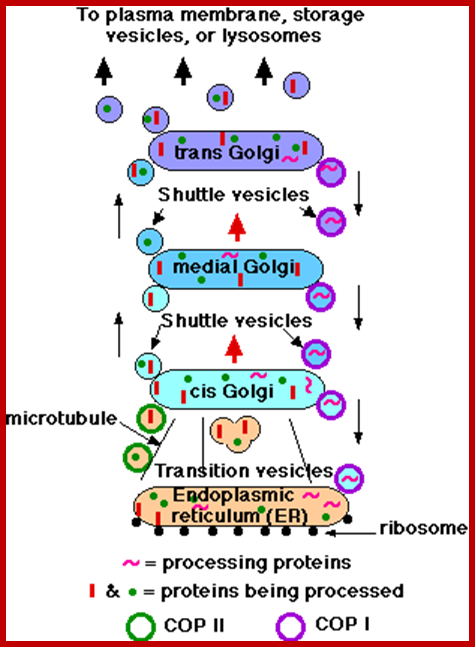
http://users.rcn.com/kimball
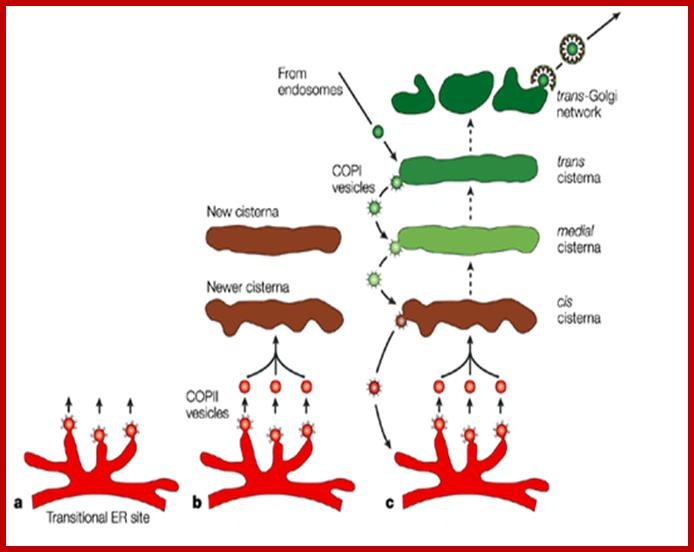
This picture shows how Golgi cisternae can be formed by budding vesicles from SER and fusion of the vesicles into cis surface of the Golgi complex.� Similarly traffic from one membrane of the Golgi to the other in forward and backward direction, then the matured and modified proteins are packed into vesicles at Trans surface of the Golgi complex and budded off.� Vesicle formation from SER requires COPII mediated protein structures.� But the reverse movement of vesicle is performed by COP1 mediated proteins.
Similarly AP1 complex is involved in producing a clathrin coated vesicles in carrying mannose-6 phosphorylated lysosomal proteins.� But a variant of AP1 complex containing μ1B instead of u1A is found in polarized epithelial cells, where the vesicle carrying a cargo is carried from apical to baso-lateral surface.� Again AP4 is associated with trans-Golgi membranes.� However all these contain an additional protein called AP180; this protein decides the size of the vesicle.� The above examples suggest that the adaptors are ultimate deciders, which cargo to be carried and to which destination.
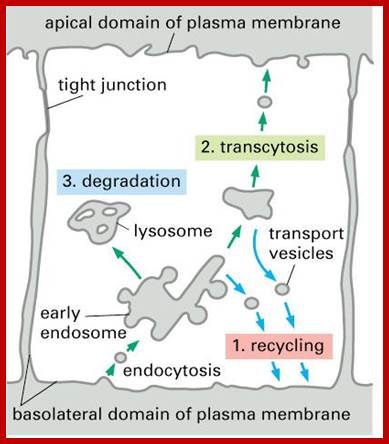
Transport within the cell;Transcytosis; http://www.bio.davidson.edu/
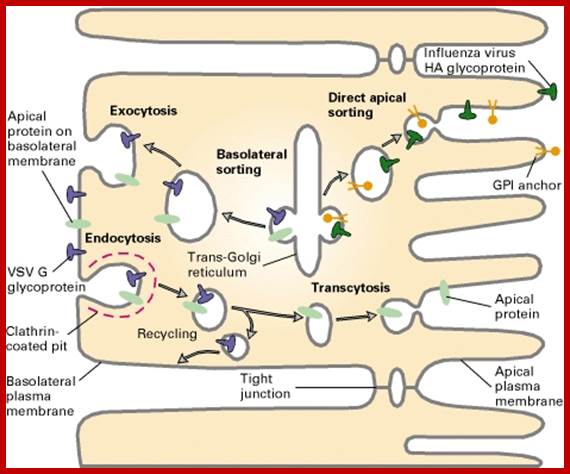
Proteins from Golgi sorted out and transported towards apical region or basal region�Transcytosis; http://www.bio.davidson.edu/
COP-1 Coated Vesicle Formation:
COP-1 coated vesicles contain a seven protein (coatomer) complex.� The coatomer forms two layers of proteins, the inner and the outer. One of the components of COP-1 is β-Cop. This protein has homology to clathrin coated adaptins β and β�-adaptins, which suggests that the β-cops play a similar role like clathrin by contacting the signal sequences of protein on one side and on the other side they contact outer cop-coat proteins; the outer coatamers are analogous to clathrin coatamers.� COP-1 coated vesicles perform both retrograde (mostly) and also anterograde (forward) movement from cis Golgi to ER and cis Golgi to trans-golgi and vice versa. They are associated with ARF GTPase.
����������������������������������������������������������������������������������������������������������������
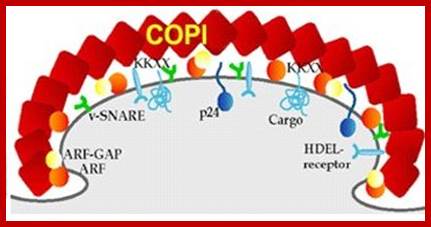
COP1 mediated Golgi-ER transport
Model for COP I�Coated Vesicle Formation from Golgi Membranes:
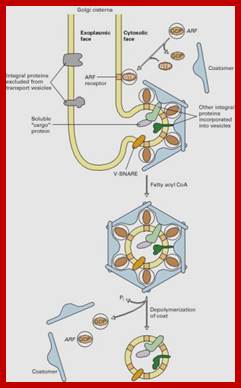
Budding is initiated when molecules of ARF protein exchange their bound GDP for GTP, a reaction catalyzed by an enzyme in the Golgi membrane. After ARF-GTP binds to ARF receptors on Golgi cisternae, coatamers bind to the cytosolic face of the Golgi cisterna and polymerize into a fibrous coat that induces vesicle budding. Because they can bind to coatomer, certain integral membrane proteins are incorporated into the vesicles. These include a V-SNARE, which functions in targeting vesicles to appropriate acceptor membranes. Soluble proteins in the lumen are selected for entry into these vesicles by binding to specific membrane receptor proteins. Fatty acyl CoA is essential for the final separation of the transport vesicle from the donor membrane, but how it functions is not known. Finally, hydrolysis of GTP bound to the ARF proteins causes depolymerization of the coat and release of coatomers and ARF-GDP. [Adapted from J. E. Rothman, 1994, Nature 372:55, and J. E. Rothman and F. Wieland, 1996, Science 272:227.]
COP-II Coated Vesicle Formation from ER:
COP-II coatamers consists of sec23p and Sec24p as one complex of 700KD and the other complex sec13p and sec31p of 400KD.� This also consists of Sar 1p.� Sar1p is a GTP binding protein, Sec1p is GTPase activating protein (GAP), and it acts on Sar1p.� But Sec1p is a protein responsible for recruiting the cargo proteins.� COPII complex are responsible for the vesicle movement from SER to Cis-Golgi.� Most of the COP coated vesicles have a size of 60-90nm.� They are associated with Sar1P GTPase.
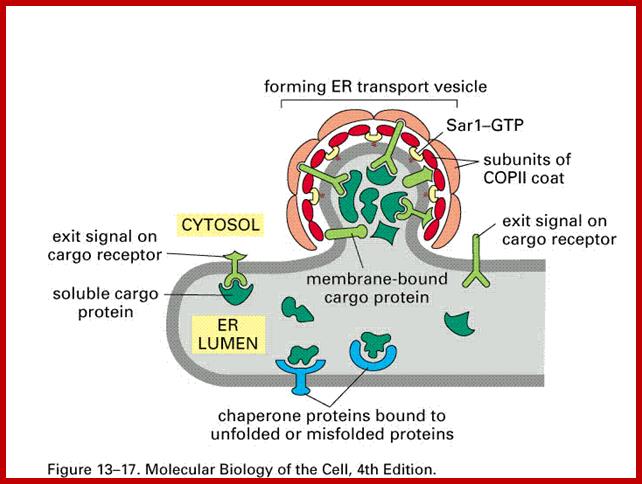
COPII mediated vesicles contain ER resident receptor attached to a cargo protein, budded with COPII subunits and Sar1-GTP
How Vesicles Budd off and Fuse:
Proteins at different membranes surfaces at different sites (patch) get loaded and they are pulled into coated vesicles of different kinds, enclosing the cargo.� Then they are transported (guided mode?) to their destinations, where they come in contact with another membrane surface either in the form of a specific vesicular membrane or a cell surface membrane; and fuse to deliver their cargo.� There are a number of proteins involved in budding and fusion.
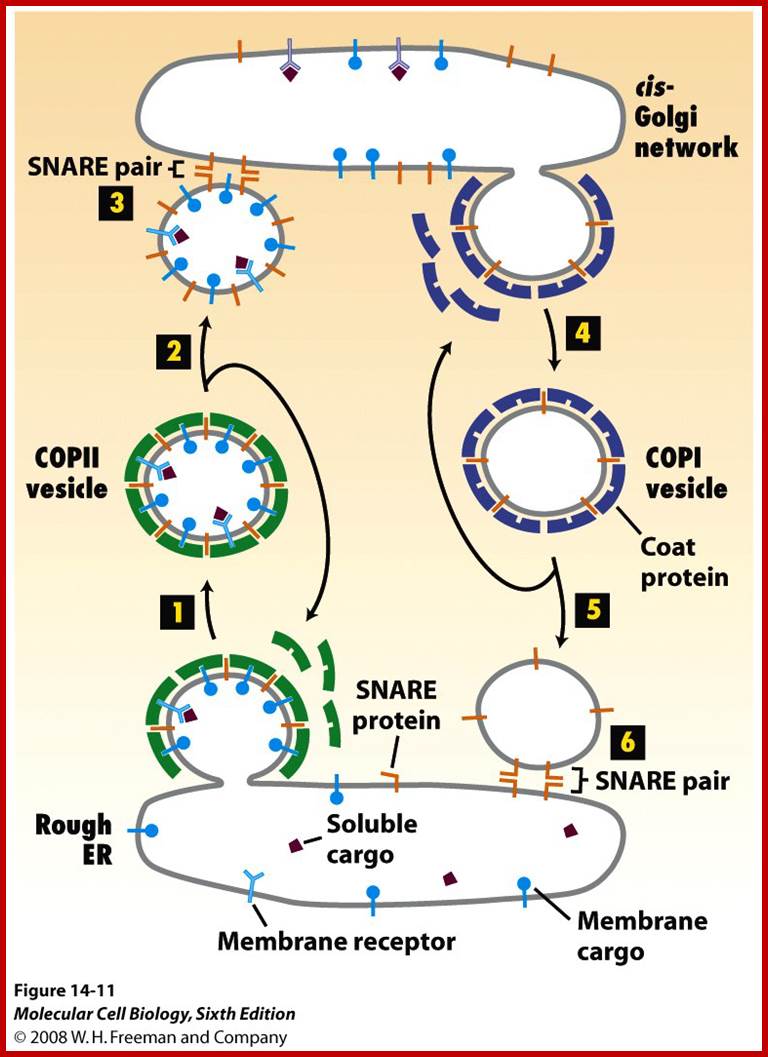
http://users.rcn.com/study; http://www.Blue.com
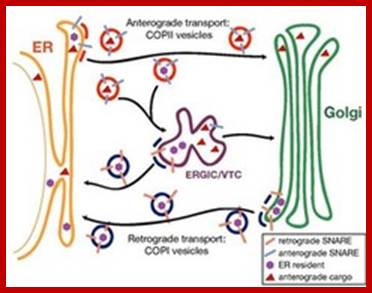
How does a vesicle join with a target membrane?
- vesicle
shed coat quickly after budding
- has v-snare on it that attaches to t-snare on target
membrane; http://quizlet.com/
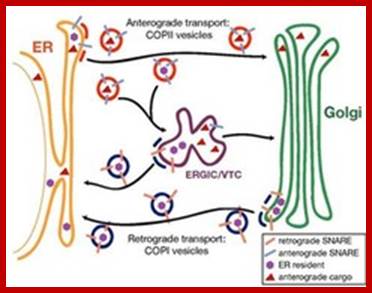
� - formed when vesicles from ER transport shed their COP II
proteins and fuse in the middle; - ERGIC fuses with cis-Golgi to deliver
contents
- formation of COP I coated vesicle is needed to
retrieve ER resident proteins; http://quizlet.com/
Budding Proteins and Budding and Vesicle transport and fusion:
ARF: �ADP-Ribosylation factor for COP I and Clathrin coated vesicles.
SaR1p: A related factor (Secretion associated Ras related protein) for COP II coated vesicles, which perform function similar to ARF.
ARF is a GTP binding protein. It has binding site at N- terminal region.� Its N-end can be myristoylated, provided the N-end opens out, so it can get inserted into a membrane.� But opening of N-end depend upon the binding of GTP. When it is bound by GTP the N-end of the ARF opens up; and it get myristoylated and it inserts into the membrane.� When the GTP hydrolyses to GDP, it becomes inactive and gets free from the membrane.� Binding of ARF to membranes provide surface for the binding of coat proteins. Whether the COP-I or Clathrin associated proteins bind to membranes in association with ARFs, the membrane is deformed and pulled into a sac like structure, but the pinching off of the vesicle is performed by another protein called Dynamin.� Dynamin is a contractile kind of protein.� In GDP form it binds to coat proteins, but it is activated by GTP binding.� Activation of Dynamin results in complete constriction and release of the cargo loaded vesicle.�
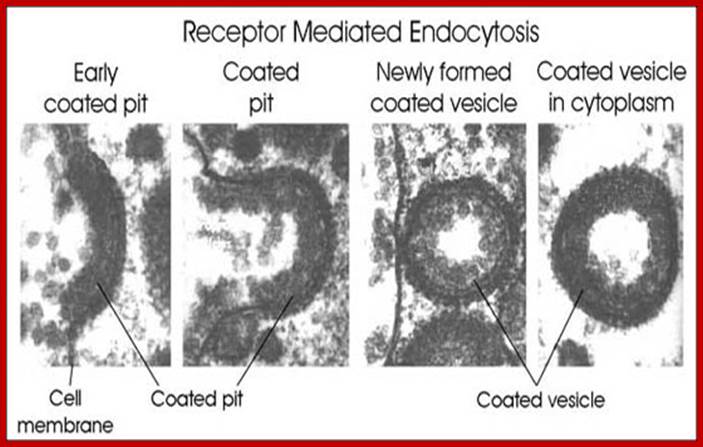
��������������������� � Initially a clathrin coated pit forms. The cargo receptors extend through the membrane and interact with cargo molecules in the cytoplasm. In this case the cells are chicken oocytes and the cargo receptors are interacting with particles containing protein and lipids that will be incorporated into the yolk. As the process continues, the vesicles rounds up and pinches off.������������ ; http://www.zoology.ubc.ca/
Various SNAP, Vamps and Syntaxins involved in intracellular compartments:
A figure below where an epithelial cell with its vesicle transport pathways and the localizations of selected Rab GTPase is shown.
RAB1, located at endoplasmic reticulum (ER) exit sites and the pre-Golgi intermediate compartment (IC), mediates ER�Golgi trafficking. RAB2, located at the IC, might also regulate Golgi�ER trafficking. The Golgi-localized RAB6, RAB33 and RAB40 mediate intra-Golgi trafficking. RAB33, together with RAB24, also regulates the formation of autophagosomes.
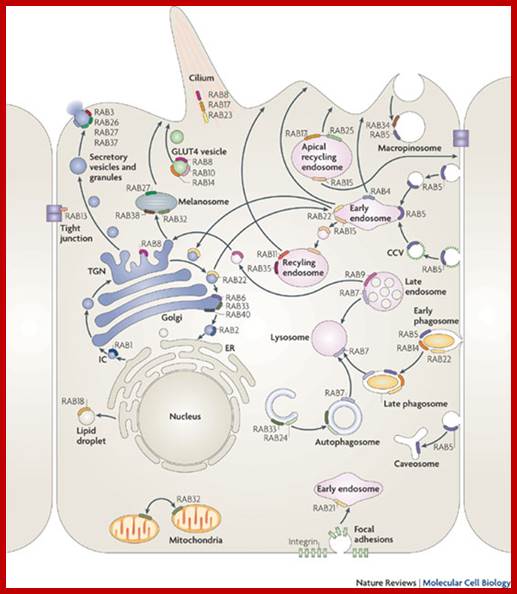
A general view vesicle movements in the cells: Nature Reviews; Molecular Biology
RAB8 mediates constitutive biosynthetic trafficking from the trans-Golgi network (TGN) to the plasma membrane and also participates in GLUT4 vesicle translocation (with RAB10 and RAB14) and ciliogenesis (with RAB17 and RAB23). RAB3, RAB26, RAB27 and RAB37 mediate various types of regulated exocytic events and RAB27 also mediates the translocation of melanosomes to the cell periphery. RAB32 and RAB38 are involved in the biogenesis of melanosomes and RAB32 also controls mitochondrial fission. RAB13 regulates the assembly of tight junctions between epithelial cells. RAB18 controls the formation of lipid droplets. RAB22 mediates trafficking between the TGN and early endosomes and vice versa. RAB5, which is localized to early endosomes, phagosomes, caveosomes and the plasma membrane, mediates endocytosis and endosome fusion of clathrin-coated vesicles (CCVs), macropinocytosis (with RAB34) and maturation of early phagosomes (with RAB14 and RAB22). RAB21 mediates integrin endocytosis. RAB11 and RAB35 mediate slow endocytic recycling through recycling endosomes, whereas RAB4 mediates fast endocytic recycling directly from early endosomes. RAB15 is involved in the trafficking from early endosomes to recycling endosomes and in the trafficking from apical recycling endosomes to the basolateral plasma membrane. RAB17 and RAB25 control trafficking through the apical recycling endosomes to the apical plasma membrane. The late endosome-associated RAB7 mediates maturation of late endosomes and phagosomes, and their fusion with lysosomes. Another late endosomal GTPase, RAB9, mediates trafficking from late endosomes to the TGN
The Role of Syntaxin 6 and Syntaxin 16 in Sub-cellular Membrane
Traffic and Endocytosis:
How is membrane protein sorting controlled? How does a cargo protein end up in its correct transport container? Different syntaxins populate distinct subcellular compartments (see figure), but how do they get there and how are they regulated? Such questions are fundamental to cell function. We are studying syntaxin 6 and 16 function as a paradigm for these questions. Our studies focus upon the role of Syntaxin 6 and 16 in Glut4 sorting in fat cells, and the role of syntaxin 6 in trafficking endocytosis machinery to the plasma membrane; ( Professor Dick Pagano , Mayo Clinic).
��������������������������������� �
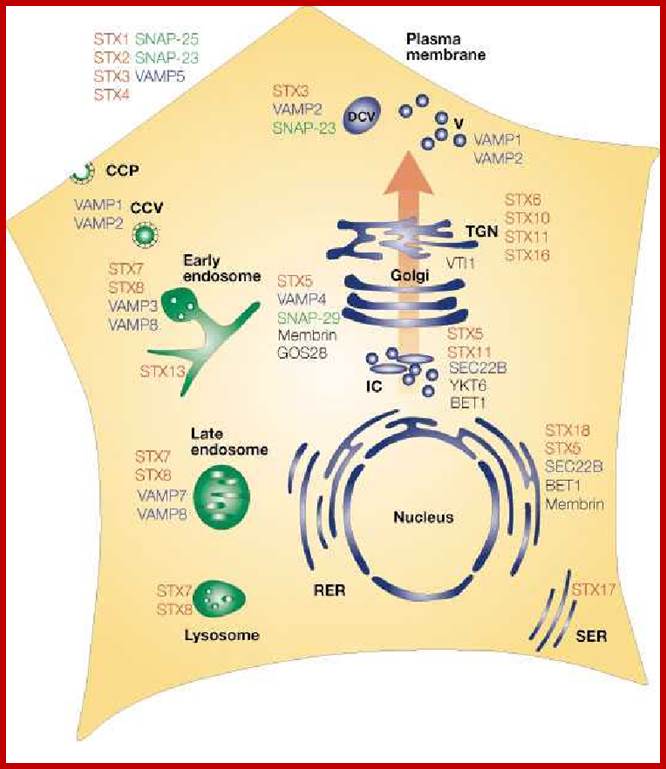
The figure shows various SNAP , VAMps and Syntaxins involved in membrane transport. Subcellular localization of mammalian SNAREs. The mammalian SNAREs that have been studied so far localize to distinct subcellular compartments in the secretory pathway. (Red, syntaxin family; blue, VAMP family; green, SNAP-25 family; black, others. CCP, clathrin-coated pit; CCV, clathrin-coated vesicles; DCV, dense core vesicles; IC, intermediate compartment; RER, rough endoplasmic reticulum; SER, smooth endoplasmic reticulum; SNAP-25, 25 kDa synaptosome-associated protein; TGN, trans-Golgi network; V, vesicles,� VAMP, vesicle-associated membrane protein. http://www.zoology.ubc.ca/
A similar process operates with Sar1p associated with COP-II proteins. It is also GTP binding protein, but it does not need GTP binding for insertion of its N-terminal end by myristoylation.� Activation of this protein by GTP leads to vesicle formation and hydrolysis of GTP by Dynamin pinches off the vesicle from the membrane.� ARFs in the released coated vesicles hydrolyze the GTP to GDP; this releases all the coat proteins from the vesicles.� Some more proteins like Amphiphysin and Endophilin can also cause vesiculation but requires phosphatidyl-inositol for interaction in the membrane.
Vesicle Fusion and Fusion Proteins:
Once vesicles are released they are transported to their targets.� Whether any actin microfilaments or microtubules are used for directed transport, though few years back it was uncertain, now it is known many types of vesicles are transported using cytoskeletal elements.� These microtubules and actin filaments act as tracts for vesicle transportation using specific motor proteins
Donor vesicles and the target membranes are recognized by specific proteins found on their surfaces.� One type of proteins is involved in recognition of specific membrane to membranes. Second type of proteins is involved bringing two membranes together.� The third type of proteins is actually involved in fusion.�
Rab proteins:�
Rab is monomeric G-protein.� Its C-end is prenylated and anchored in the membrane.� Rab proteins are related to Ras proteins (derived Ras like protein found in Brain); they are kinases.� There are as many as more than 70 Rab proteins.�
Rabs identify specific membranes of specific vesicles or the target membranes.� Rabs get activated when they are bound by GTP, but inactivated when GTP is hydrolyzed to GDP and Pi.� For activation it requires proteins like GAP and GEFs.� These Rabs are specific-to-specific types of membranes.� The following Rab proteins are involved in different transport vesicles (tentative).� Rab proteins have their respective Rab effector proteins or docking proteins on target membranes, for proper effector function.
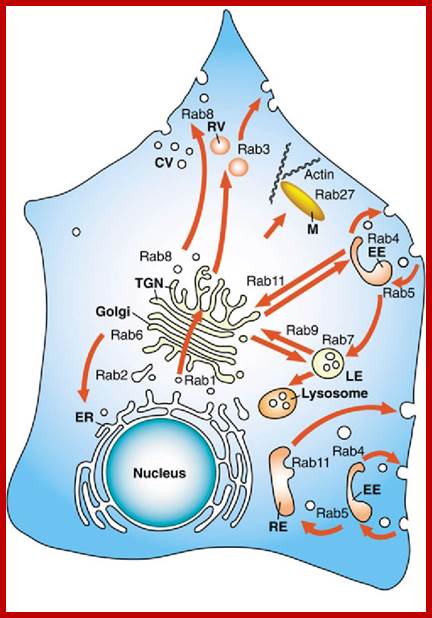
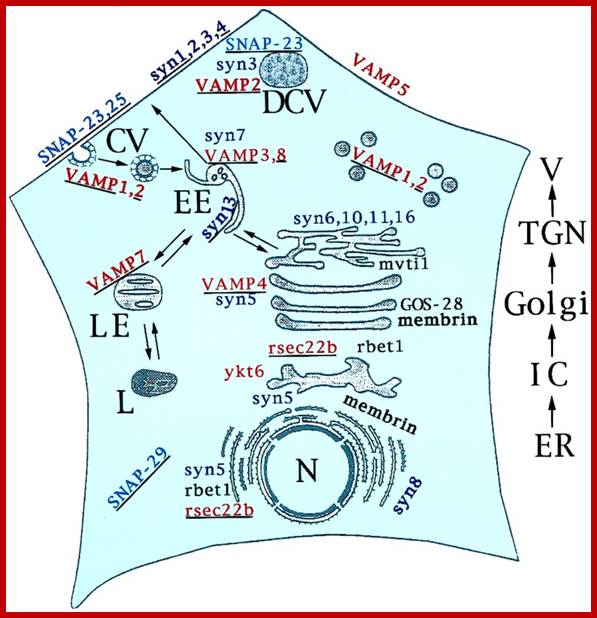
An overall view of intracellular transport involving Rab and VAMPs proteins; http://meddic.jp/
RABs:
Rab 1 and 2: From ER to cis Golgi.
Rab 6: From cis Golgi to ER.
Rab 30, 33b, and 36: From cis to median to trans Golgi.
Rab 10, 12, and 13: From trans-Golgi to trans-Golgi network.
Rab 9: From trans Golgi network to Lysosomes.
Rab 4 and 5: exocytosis and endocytosis respectively.
Rab 7:� From late endosome to lissome.
Rab 15, 18, 20 and 22: From early endosome to late endosome.
Rab 3 and 8: From TGNW to PM but regulated.
Rab 8 and 37: From TGNW to PM secretory (constitutive).
RB 11: From RE to exocytosis and from EE to TNG.
VAMPS: Vesicle Associated membrane proteins;
VAMP1, and VAMP2,called Sanaptobrevins a family of SNARE proteins,
VAMP3, called Cellubrevin, participate in exocytosis
VAMP4, transport from Golgi
VAMP5 and VAMP7-constitute exocytosis.
VAMP8, Particpates in endocytosis
VAMP
�Acronyms: SNARE, soluble N-ethylmaleimide-sensitive factor attachment protein receptor; v-SNARE, vesicle SNARE; t-SNARE, target SNARE; v-liposome, v-SNARE proteoliposome; NBD, 7-nitrobenz-2-oxa-1,3-diazole; POPC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine; Rh, lissamine rhodamine B; DPPE, 1,2-dipalmitoyl-sn-glycero-3-phosphatidylethanolamine; RT, room temperature; SUVs, small unilamellar vesicles; TIR-FM, total internal reflection fluorescence microscopy; SNAP-25, synaptosomal-associated protein of 25 kDa; VAMP, vesicle-associated membrane protein.
Tethering complex: It is a complex of proteins consisting of 5 to 8 subunits.� There are different kinds of tethering complexes.� Normally they are found very near to the membranes (i.e. target membranes).� These proteins actually recognize a specific Rab protein and bring the vesicle membrane and the target membrane together.� Rab protein provides the identity and facilitates tethering process.
20S complex:� This complex is located on the target membranes.� It consists of NSF protein that is sensitive to a Sulfhydryl agent called N-ethyl-maleimide.� SNF is a soluble ATPase. It is a homologue of yeast Sec18.� This complex is associated with another protein called SNAP; it is so called because it is a Soluble NSF Attachment Protein.� SNAP is anchored in the membrane.� The receptor for SNAP is called SNARE.
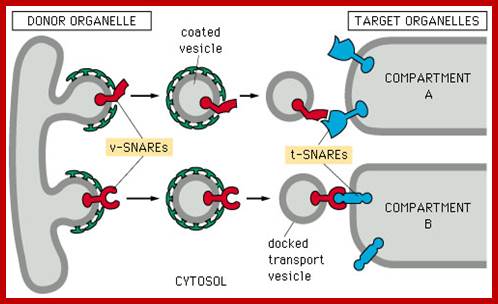
The figure simplistic representation of V-SNARES from the donor membranes with coated vesicles and t-SNARES with target membranes involved in transport and docking. http://www.zoology.ubc.ca/
SNAREs:� SNAREs are filamentous proteins associated with 20S complex of proteins.� SNAREs are localized (anchored) in the membrane.� There are two types of SNAREs, found at donor vesicle called v-SNARES and another is t-SNARES at target membranes, which are distinctly different from each of these membranes.� The t-SNARE consists of Syntaxin 1A and SNAP25B; note that this SNAP25B is distinctly different from the SNAP i.e. soluble NSF attachment protein).� Such SNAREs are also found in almost all systems.� The vSNAREs are made up of synaptobrevin and SNAP 25B and they lie parallel to their membrane surface.� v-SNAREs and tSNAREs are different from one type of membrane to the other and their sub components vary from one another.
Fusion Process:
Once the Rab mediated docking takes place with the assistance of tethering complex (each are specific to their transporter membranes),
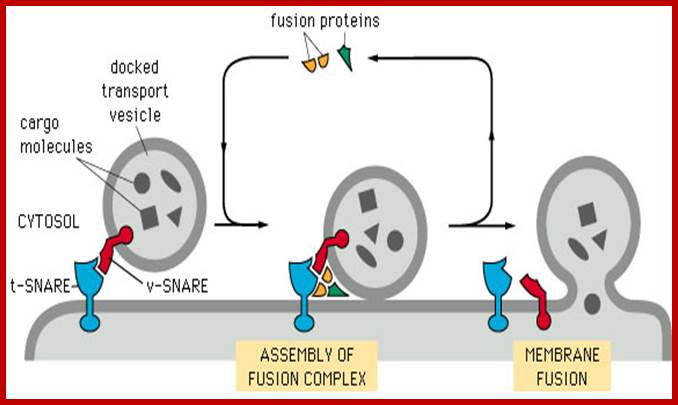
��������������������������������������������������������������
The diagram depicts the docking of vesicle with V-SNARE with the membrane containing t-SNARE; after docking fusion proteins join to perform membranes fusion, where fusion proteins and V-SNAREs and t-SNARES are released. http://www.zoology.ubc.ca/
The fusion of a transport vesicle with its target involves two types of events. First, the transport vesicle must specifically recognize the correct target membrane; for example, a vesicle carrying lysosomal enzymes has to deliver its cargo only to lysosomes. Second, the vesicle and target membranes must fuse thereby delivering the contents of the vesicle to the target organelle. Research over the last several years has led to development of a model of vesicle fusion in which specific recognition between a vesicle and its target is mediated by interactions between unique pairs of transmembrane proteins, followed by fusion between the phospholipid bilayers of the vesicle and target membranes.
Proteins involved in vesicle fusion were initially identified in James Rothman�s laboratory by biochemical analysis of reconstituted vesicular transport systems from mammalian cells.� Analysis of the proteins involved in vesicle fusion in these systems led Rothman and his colleagues to propose a general model, called the SNARE hypothesis, in which vesicle fusion is mediated by interactions between specific pairs of proteins, called SNAREs, on the vesicle and target membranes (v-SNAREs and t-SNAREs, respectively) (Figure 9.33). This hypothesis was supported by the identification of SNAREs that were present on synaptic vesicles and by the finding of yeast secretion mutants that appeared to encode SNAREs required for a variety of vesicle transport events. For example, transport from the ER to the Golgi in yeast requires specific SNAREs that are located on both the vesicle and target membranes. The formation of complexes between v-SNAREs on the vesicle and t-SNAREs on the target membranes then leads to membrane fusion, by mechanisms which remain to be fully understood.
In addition to SNAREs, vesicle fusion requires at least two other types of proteins. The Rab proteins are a family of small GTP-binding proteins that are related to the Ras proteins.� More than 70 different Rab proteins have been identified and shown to function in specific vesicle transport processes. They may function in several steps of vesicle trafficking, including interacting with SNAREs to regulate and facilitate the formation of v-SNARE/t-SNARE complexes.
SNARE proteins form a four-helical bundle complex that drives membrane
Fusion: a | VAMP (blue) on the vesicle interacts with syntaxin (red) and SNAP-25 (green) on the plasma membrane to form a four-helix bundle that zips up concomitant with bilayer fusion. b | The backbone of the SNARE complex is shown on the left52, with the central ionic layer (red) and 15 hydrophobic layers (black) that mediate the core interactions highlighted. Top-down views of side-chain interactions are shown on the right, with the four SNARE helices shown as
ribbons. The ball-and-stick structures represent the indicated amino acids; the dotted lines represent hydrogen bonds or salt bridges that stabilize interactions between SNAREs. QSNAREs and R-SNAREs are characterized by a glutamine (Q) or Arginine (R) residue, respectively, in the central layer of the SNARE complex (SNARE; soluble NSF attachment protein receptor, where NSF stands for N-ethyl-maleimide-sensitive fusion protein; SNAP-25).
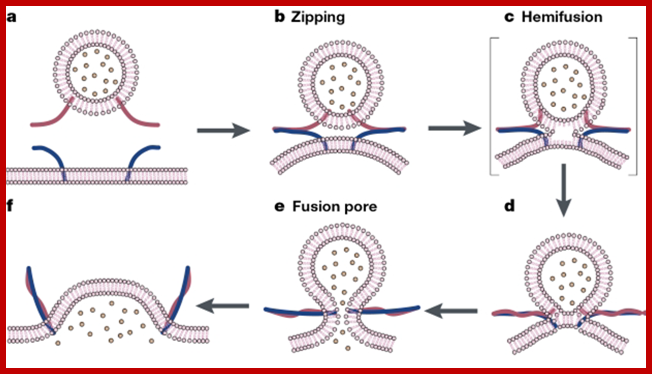
Model of SNARE-mediated lipid fusion. a | The two membranes are in the vicinity
of each other but the SNAREs are not yet in contact. b | SNARE complexes start zipping from the amino-terminal end, which draws the two membranes further towards each other. c | Zipping proceeds, causing increased curvature and lateral tension of the membranes, exposing the bilayer interior. Spontaneous hemifusion occurs as the separation is sufficiently reduced. d | The highly unfavourable void space at the membrane junction in (c) causes the establishment of contacts between the distal membrane leaflets. e | The lateral tension in the transbilayer contact area induces membrane breakdown, yielding a fusion pore. f | The fusion pore expands and the membrane relaxes. (SNARE, soluble NSF attachment protein receptor,
where NSF stands for N-ethyl-maleimide-sensitive fusion protein.);
Yu A. Chen & Richard H. Scheller ;http://www.nature.com/
The control of SNARE complex formation and function:
The ability
of insulin to stimulate glucose transport into muscle and adipose tissue is a
central facet of whole body glucose homeostasis. Insulin stimulates glucose
transport by inducing the translocation of the glucose transporter Glut4 from
an intracellular store to the cell surface. Individuals with insulin-resistance
and Type 2 diabetes exhibit a significant impairment in the ability of insulin
to stimulate glucose transport, largely due to a failure of the translocation
machinery. Hence understanding the molecular basis of this phenomenon is of
fundamental importance.
The insulin-dependent delivery of Glut4-containing vesicles to the plasma membrane is a specialized example of regulated membrane trafficking. The Glut4-containing vesicles are mobilized in response to a specific signal (insulin binding its receptor) and move to the cell periphery. Once at the cell surface, the vesicles dock and then fuse with the plasma membrane. This fusion step is mediated by the action of SNARE proteins.
�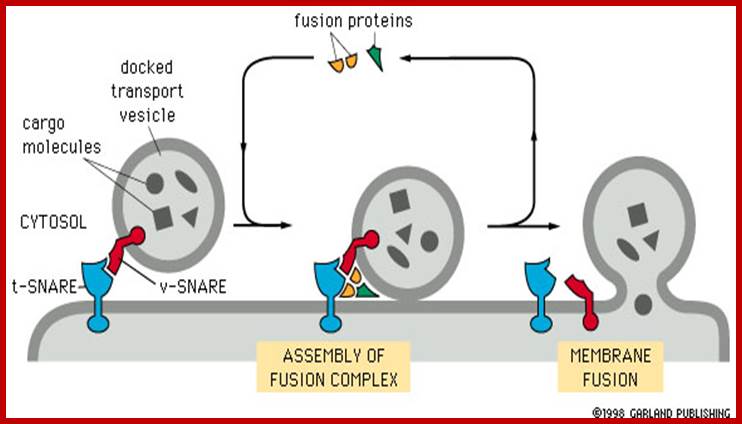
http://www.zoology.ubc.ca/
Hypothetical model for SNARE complex formation during regulated exocytosis:
Studies in both mammalian and yeast systems have led to the identification of a family of conserved proteins that function in membrane fusion using a mechanism highly conserved through evolution - this is known as the SNARE hypothesis. In its simplest form, the SNARE hypothesis states that the fusion of a donor membrane with its target is mediated by the specific pairing of target (t)-SNAREs with a cognate vesicle (v)-SNARE. Specific v- and t-SNARE combinations are involved in all membrane trafficking events in mammalian cells. Recently, reconstituted experiments in vitro and shown that the formation of the Syntaxin4/SNAP-23/VAMP2 ternary complex is sufficient to fuse membranes (see figure below). We are now studying how the formation of this complex is controlled, focusing upon phosphorylation as a potential regulatory mechanism, and also studying how SM processes control this process.
Following the formation of complexes between complementary SNAREs and membrane fusion, a complex of two additional proteins (the NSF/ SNAP complex) is needed to complete the process of vesicle transport. The NSF/SNAP proteins are recruited to membranes following the formation of v-SNARE/t-SNARE complexes, and they are not required directly for either vesicle/target pairing or for the fusion of paired membranes. Instead, the NSF/SNAP proteins act after membrane fusion to disassemble the SNARE complex, thereby allowing the SNAREs to be reutilized for subsequent rounds of vesicle transport.
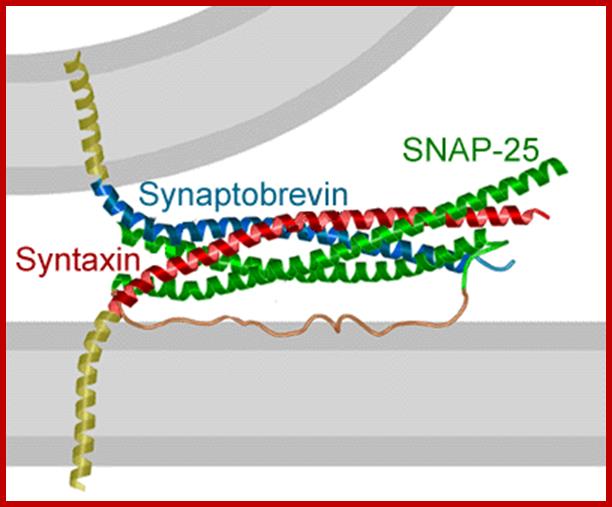
Neuronal SNARE proteins; Dirk Fasshauer, Christina Sch�tte, Olga Viteshttp://www3.mpibpc.mpg.de/
Proteins involved in membrane fusion are VAMP, Syntaxin, SNAP-25N and SNAP-25C
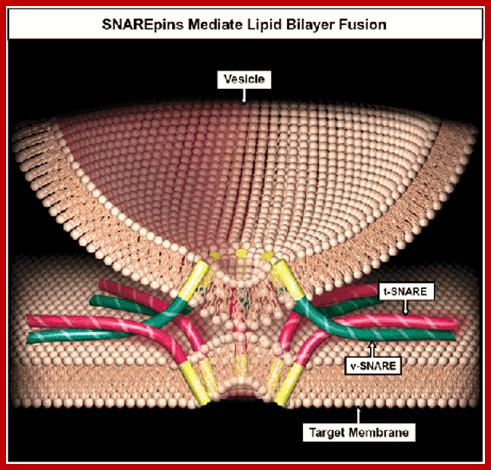
An artist diagram of t and V SNARES docked on to a membrane; Prof, Mayer Andreas http://www.unil.ch/ib
the SNARES from two membranes i.e. donor and target, coil to each other and pull the membranes close to each other in such a way the membranes bind to each other.� This leads to the fusion of lipid layers and exchange of lipid components they join to generate a joint membrane.� And release the cargo into the vesicle or to exterior space.� This leads to the release of the SNAREs.� Thus the proteins that are synthesized on ER are transferred into ER lumen and then they are modified at different levels and they do so while they are transported from SER to cis-Golgi.�
Proteins work together to mediate endosomal membrane fusion:
From the following article: Cell biology: A score for membrane fusion; Ruth N. Collins & Joshua Zimmerberg; Nature 459, 1065-1066(25 June 2009).
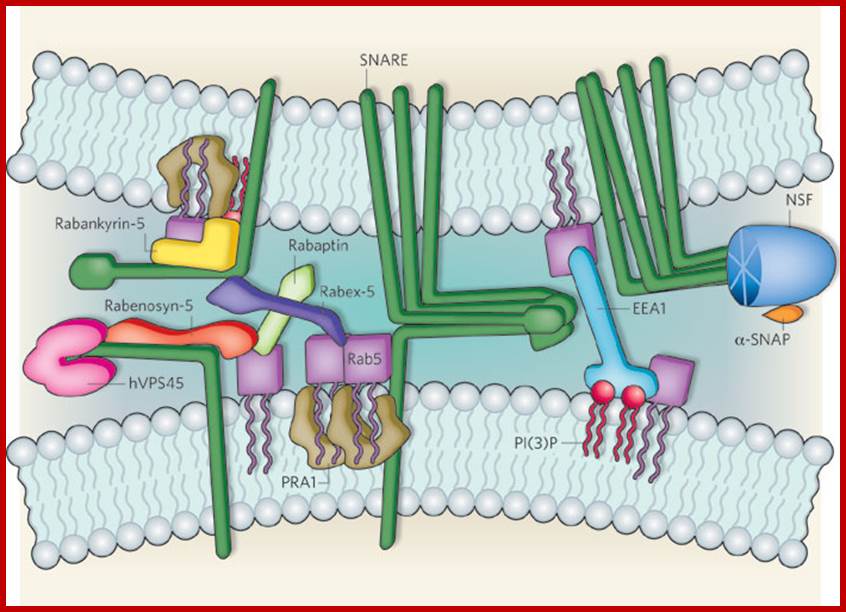
Ohya et al.
show that two protein systems, a Rab GTPase (Rab5) and the SNARE proteins,
combine synergistically to drive endosomal fusion. Rab5 and SNAREs are assisted
by multiple accessory factors. Rab5 effectors include rabankyrin-5,
rabenosyn-5, rabaptin, rabex-5 and EEA1. SNARE accessory factors include ![]() -SNAP, NSF and hVPS45. Not shown are the Rab�GDI complex (which
retains Rab5 in the cytoplasm by masking its prenyl group), as this component
is thought to function at an earlier stage, and another Rab5 effector, PI(3)K,
which is instead represented by its lipid product PI(3)P. The complexity of the
interactions observed in this supramolecular network is apparent; the
recruitment of Rab5 effectors is stabilized by SNARE proteins and SNARE-protein
accessory factors. For example, the Rab5 effector rabenosyn-5 also interacts
with the SNARE accessory factor hVPS45. Linker proteins such as PRA1 bind to
both Rabs and SNAREs. Ruth N. Collins & Joshua Zimmerberg; http://www.nature.com/
-SNAP, NSF and hVPS45. Not shown are the Rab�GDI complex (which
retains Rab5 in the cytoplasm by masking its prenyl group), as this component
is thought to function at an earlier stage, and another Rab5 effector, PI(3)K,
which is instead represented by its lipid product PI(3)P. The complexity of the
interactions observed in this supramolecular network is apparent; the
recruitment of Rab5 effectors is stabilized by SNARE proteins and SNARE-protein
accessory factors. For example, the Rab5 effector rabenosyn-5 also interacts
with the SNARE accessory factor hVPS45. Linker proteins such as PRA1 bind to
both Rabs and SNAREs. Ruth N. Collins & Joshua Zimmerberg; http://www.nature.com/
Overview of the Membrane Traffic:
This is an exotic 3-dimensional view of membrane transport to targets.
From the Cis-Golgi to median-Golgi to Trans-Golgi to Trans-Golgi network.� The proteins are marked by specific modification and specific cargo carriers called receptors hold them on to the membrane in the lumen; they are loaded and sorted to each kind or many of the similar kind as one lot.� This leads to the bundling of those sorted ones into a vesicle by specific coat proteins and they are budded off.� On the other end cargo is carried from the plasma membrane and budded of into the cytosol.� Such vesicles are transported to specific destinations where they are unloaded.� Some of the vesicles that unload their cargo return to their old destinations.� Example, lysosome receptors, secretory protein receptor that end up in plasma membranes return to their original locations.� So also SER vesicles after transferring to cis Golgi or median Golgi return to their SER loci.
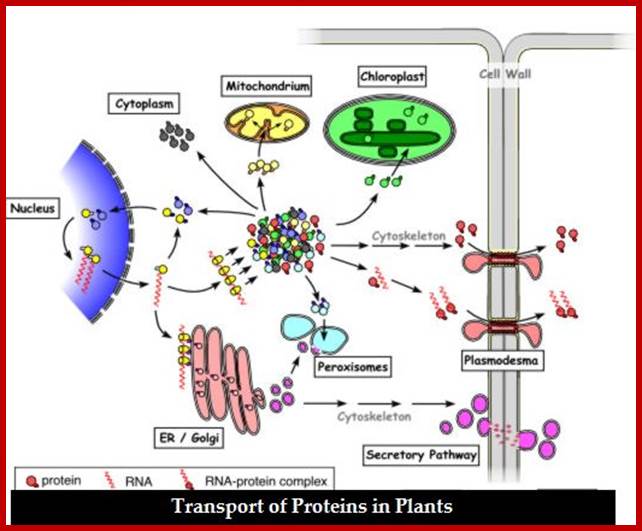
�Protein carrying membrane transport in Plants; a brief but a lovely self-explanatory diagram cum review of a plant cell: University of Vienna; Regulation of Cell-To-Cell Transport of RNA and Proteins via Plasmodesmata; Kragler Lab; http://www.univie.ac.at/
Synaptic Vesicles and Their Movement during Neuronal Synapses:
Basic Types: Bipolar, Unipolar, Multipolar and Pyrimidal cells http://www.solarnavigator.net
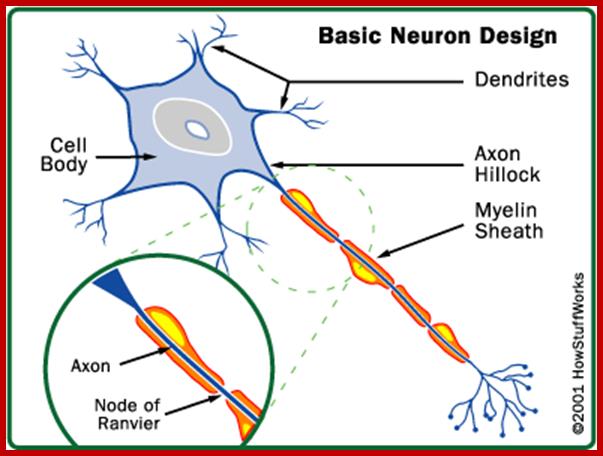
http://www. ocr.org.uk
�Our brain is made of approximately ~100 billion nerve cells, called neurons. Neurons have the amazing ability to gather and transmit electrochemical signals -- they are something like the gates and wires in a computer. Neurons share the same characteristics and have the same parts as other cells, but the electrochemical aspect lets them transmit signals over long distances (up to several feet or a few meters) in a fraction of second and pass messages to each other.
Neurons have three basic parts:
- Cell body - This main part has all of the necessary components of the cell, such as the nucleus (contains DNA), endoplasmic reticulum and ribosomes (for building proteins) and mitochondria (for making energy). If the cell body dies, the neuron dies.
- Axon - This long, cable-like projection of the cell carries the electrochemical message (nerve impulse or action potential) along the length of the cell.
- Depending upon the type of neuron, axons can be covered with a thin layer of myelin, like an insulated electrical wire. Myelin is made of fat, and it helps to speed transmission of a nerve impulse down a long axon. Myelinated neurons are typically found in the peripheral nerves (sensory and motor neurons), while non-myelinated neurons are found in the brain and spinal cord.
- Dendrites or nerve endings - These small, branch-like projections of the cell make connections to other cells and allow the neuron to talk with other cells or perceive the environment. Dendrites can be located on one or both ends of the cell.
Neurons also vary with respect to their functions:
- Sensory neurons carry signals from the outer parts of body (periphery) into the central nervous system.
- Motor neurons (motoneurons) carry signals from the central nervous system to the outer parts (muscles, skin, and glands) of body.
- Receptors sense the environment (chemicals, light, sound, touch) and encode this information into electrochemical messages that are transmitted by sensory neurons.
- Interneurons connect various neurons within the brain and spinal cord.
The simplest type of neural pathway is a monosynaptic (single connection) reflex pathway, like the knee-jerk reflex. When the doctor taps the right spot on your knee with a rubber hammer, receptors send a signal into the spinal cord through a sensory neuron. The sensory neuron passes the message to a motor neuron that controls your leg muscles. Nerve impulses travel down the motor neuron and stimulate the appropriate leg muscle to contract. The response is a muscular jerk that happens quickly and does not involve your brain. Humans have lots of hard-wired reflexes like this, but as tasks become more complex, the pathway "circuitry" gets more complicated and the brain gets involved.
The simplest possible creatures have incredibly simple nervous systems made up of nothing but reflex pathways. For example, flatworms and invertebrates do not have a centralized brain. They have loose associations of neurons arranged in simple reflex pathways. Flatworms have neural nets; individual neurons linked together that form a net around the entire animal.
Most invertebrates (such as the lobster) have simple "brains" that consist of localized collections of neuronal cell bodies called ganglia. Each ganglion controls sensory and motor functions in its segment through reflex pathways, and the ganglia are linked together to form a simple nervous system. As nervous systems evolved, chains of ganglia evolved into more centralized simple brains.
Major Divisions of the Brain:
- Spinal cord
- Brain stem
- Forebrain
- Diencephalon - thalamus, hypothalamus
Cerebral cortex:
Brains evolved from ganglia of invertebrates. Regardless of the animal, brains have the following parts:
- Brain stem - The brain stem consists of the medulla (an enlarged portion of the upper spinal cord), pons and midbrain (lower animals have only a medulla). The brain stem controls the reflexes and automatic functions (heart rate, blood pressure), limb movements and visceral functions (digestion, urination).
- Cerebellum - The cerebellum integrates information from the vestibular system that indicates position and movement and uses this information to coordinate limb movements.
- Hypothalamus and pituitary gland - These control visceral functions, body temperature and behavioral responses such as feeding, drinking, sexual response, aggression and pleasure.
- Cerebrum (also called the cerebral cortex or just the cortex) - The cerebrum consists of the cortex, large fiber tracts (corpus callosum) and some deeper structures (basal ganglia, amygdala, and hippocampus). It integrates information from all of the sense organs, initiates motor functions, controls emotions and holds memory and thought processes (emotional expression and thinking are more prevalent in higher mammals).
|
|
http://www.solarnavigator.net/
As you proceed from fish toward humans, you can see that the cortex gets bigger, takes up a larger portion of the total brain and becomes folded. The enlarged cortex takes on additional higher-order functions, such as information processing, speech, thought and memory. In addition, the part of the brain called the thalamus evolved to help relay information from the brain stem and spinal cord to the cerebral cortex.
In this diagram of the brain the different sections are shown. The Cerebrum are the two large hemispheres of the brain. Each hemisphere is further divided into lobes. Above is the break up of where each lobe is located and the structures under the cerebrum that make up the rest of the brain; ;http://www.brainhealthandpuzzles.com/
Human Brain: Royalty-free video and stock.
Map of Cortexes:
In this diagram of the brain the different sections are shown. The Cerebrum are the two large hemispheres of the brain. Each hemisphere is further divided into lobes. Above is the break up of where each lobe is located and the structures under the cerebrum that make up the rest of the brain.
;http://www.brainhealthandpuzzles.com/
Diagramed above are some of the major cortexes and their locations. The cortexes are the outermost layer of the brain. These areas of the brain surface control different functions of the body and the mind.
Internal Structures of the Brain
Basic components, architecture and Basic circuits in the Brain-being human-humannature; http://www.brainmuseum.org/
This diagram illustrates the principle that brain circuits travel up and down the neuroaxis, from one side of the brain to the other, as well as back and forth between many different brain components at several levels within the forebrain, thalamus, hypothalamus, midbrain, medulla, cerebellum and spinal cord. http://www.brainmuseum.org/
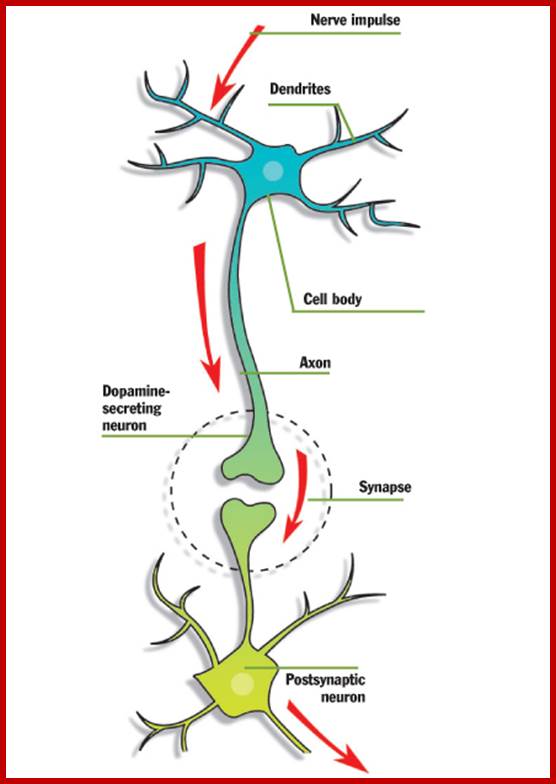
The science behind the Nobel Prize of Stanford's Thomas S�dhof; Synapses are junctions in the nervous system where information, in the form of chemical messengers called neurotransmitters, is passed from one neuron � or nerve cell � to another.; Nerve cell synapse; BRUCE GOLDMAN; http://news.stanford.edu/
����������������������������������������������������������������������� Nerve cells network-in trillions
Glial cells associated with neuronal cells; http://commons.wikimedia.org/
The Root of Thought, is all about the power of glial cells, which actually make up nearly 90 percent of cells in the brain. What do glial cells do? And why do we have so many inside our head?� Astrocyte is one of the glial cell type. Scientists have also discovered that astrocytes communicate to themselves in the cortex and are also capable of sending information to neurons. Finally, astrocytes are also the adult stem cell in the brain and control blood flow to regions of brain activity. Because of all these important properties, and since the cortex is believed responsible for higher thought, scientists have started to realize that astrocytes must contribute to thought. The glial cells surround neurons and provide support for them and insulation between them. Glial cells are capable of extensive signaling in response to a diversity of stimuli. Bidirectional communication exists between glial cells and neurons, and Human Brain.
� 
There are three main types of glial cells. Oligodendrocytes (1) send projections that wrap axons (2) � long, signal-carrying portions of neurons (3) � in sheathes of a fatty substance called myelin (4), speeding signal conduction. Microglia (5) are, essentially, the brain�s immune cells, but they also monitor neighboring brain cells for damage and gobble up debris, and they probably have other functions, too. Astrocytes (6) carry on a host of activities. Their long extensions can monitor levels of neuronal activity either along axons at synapses (7) � junctions that relay signals from one neuron to the next � and, when those activity levels are high, signal to local blood vessels (8) to dilate, increasing blood supply to hard-working neurons. Astrocytes also produce and secrete substances that have a major influence on the formation and elimination of synapses. http://med.stanford.edu/
Glial cells are the most abundant cell types in the central nervous system. There are three types of glial cells: Astrocytes, Oligodendrocytes, and Microglia. Glial cells outnumber the neurons by 1 to 10 ratio. The main function of the glia is to support the neurons in place as well as provide insulations. There are five different types of glia cells; however, some functions will overlap.
Astrocytes are concerned with neurotransmission and neuronal metabolism. Astrocyte - provides physical and nutritional support for neurons; digests part of dead neurons as well as regulates the extracellular fluid around the neuron.
�Oligodendrocytes are involved in the production of myelin, the insulating material around neurons.. And microglia are part of the immune system.
Oligodendroglia-Provides insulation for neurons (Myelin).
Macroglia- Digested part of dead neurons.
Satellite cells- Provides physical support for neurons (Myelin) in PNS.
Schwann cells- Provide physical support for neurons in the form of insulation (Myelin),
There
are differences between glia cells and neurons. Neurons can generate action
potentials whereas glial cells cannot.
Neurons have chemical synapses. Glial cells do not have synapses.� There are
more glial cells than neurons.
A scheme illustrating a
glutamatergic neuron (left) a glial cell (astrocyte) and a small blood vessel
(right) and the major components contributing to hypotheses 1 and 2 (H1 and
H2). Neural activity triggers release of the neurotransmitter glutamate that is
taken up into the astrocyte (via GLAST and GLT-1 transporters), and stimulates
the breakdown of glycogen, the uptake of glucose, and glycolysis, to produce
lactate. Rapid neuronal firing is sustained by the energy provided by the
astrocyte-neuron lactate shuttle. Energy demands are high during rapid (burst)
and maintained rates of neuronal firing. H1:
At times of increased neuronal demand, deficient lactate
results in decreased neuronal conversion of lactate to acetyl CoA, decreased
ATP formation, deficient ATPase function, delayed restoration of ion gradients,
elevated extracellular K+, deficient Na+-dependent
transport of glutamate into astrocytes that is required to drive glycolysis and
lactate release by the astrocytes. The result is that situationally appropriate
firing rates are achieved only episodically. Methylphenidate treatment results
in an increase of the extracellular levels of the catecholamine, NA (and DA)
that stimulate glycolysis and release of lactate from the astrocytes. This is
followed by glycogen replenishment, thereby correcting the energy deficiency,
and restoring appropriate firing rates. H2:
A deficient supply of lactate for oligodendrocytes in the
developing nervous system slows and reduces the synthesis of fatty acids
required for the synthesis of myelin. Poorly myelinated axons would transmit
action potentials more slowly, accounting for inefficient integration
(coherence) between brain regions and for slow reaction times. A number of
neurotransmitter receptors present on astrocytes are not illustrated (e.g.
muscarinic, α2,
DA D3, D4, D5 and receptors for several neuropeptides). Russell et al.
Behavioral and Brain
Functions 2006 2:30
doi:10.1186/1744-9081-2-30
Glial cells have been shown to directly participate to the genesis and maintenance of chronic pain in both the sensory ganglia and the central nervous system (CNS).
Attempting to explain the incredible complexity of the brain that is revealed, Smith said, �One synapse, by itself, is more like a microprocessor�with both memory-storage and information-processing elements�than a mere on/off switch. In fact, one synapse may contain on the order of 1,000 molecular-scale switches. A single human brain has more switches than all the computers and routers and Internet connections on Earth,� he said.
Glia functions can be divided into three categories. Astrocyte glia (so-called because they resembled stars to early anatomists) fill the spaces between neurons, providing energy sources and maintaining the chemical surroundings of neurons in which they can fire electrical impulses and communicate chemically through synapses.
Then, there are microglia that perform the role of an immune system within the brain. Since the brain is sealed off by a barrier from the rest of the body it needs its own immune system, killing bacteria and healing the brain after injury.
Thirdly there are glia that form the electrical insulation, called myelin, of the nerve fibers (axons) that in turn form the roots of neurons. Axons tightly bundled together and insulated with myelin form the �white matter� that makes up half of the brain.
White matter, for example, has been shown to change during experience, with myelin being formed by electrical impulses in the axons. As the speed of electrical transmission can increase 50 times in an axon insulated by myelin compared to one that is bare, this has a major impact on neural processing
Thomas Sudhof won a Nobel prize for his work in understanding how nerve cells communicate. http://news.stanford.edu/
Synapses computer artwork; http://fineartamerica.com/
Synapses: Computer artwork of synapses, the junctions between the ends (blue, swollen) of two nerve cells (neurons).
Nerve cells are responsible for passing information around the central nervous system (CNS) and from the CNS to the rest of the body. This information is transmitted from one nerve cell to the next as an electrical impulse (yellow flash). Each nerve cell comprises dendrites, nerve cell extensions which collect information from other nerve cells or from sensory cells, and an axon, through which information passes outwards to other cells, including other nerve cells and muscle fibers. Here, the axons are surrounded by a myelin sheath (yellow), which insulates the axon, allowing signals to be transmitted more quickly.
Development of Membrane Potential and Propagation of the same:
A diagram showing the progression in the development of a membrane potential from a concentration gradient (for potassium):
Green arrows indicate net movement of K+ down a concentration gradient. Red arrows indicate net movement of K+ due to the membrane potential. The diagram is misleading in that while the concentration of potassium ions outside of the cell increases, only a small amount of K+ needs to cross the membrane in order to produce a membrane potential with a magnitude large enough to counter the tendency the potassium ions to move down the concentration gradient
Membrane potential (or transmembrane potential or transmembrane potential difference or transmembrane potential gradient, is the electrical potential difference (voltage) across a cell's plasma membrane. The plasma membrane encloses the cell to provide a stable environment for biological processes. Membrane potential arises from the action of ion transporters embedded in the membrane which maintain viable ion concentrations inside the cell. The term "membrane potential" is sometimes used interchangeably with cell potential but it is applicable to any lipid bilayer membranes.
The membrane potential of most cells is kept relatively stable. Unlike most cells, neurons are specialized to use changes in membrane potential for fast communication, primarily with other neurons. When a neuron fires, the action potential travels down the axon to the synapses: the magnitude of the axonal membrane potential varies dynamically along its length. On reaching a (chemical) synapse, a neurotransmitter is released causing a localized change in potential in the membrane of the target neuron by opening ion channels in its membrane
Myelin Propagation of action potential;How neurons communicate; http://cnx.org/content
Communication at chemical synapses requires release of neurotransmitters. When the presynaptic membrane is depolarized, voltage-gated Ca2+ channels open and allow Ca2+ to enter the cell. The calcium entry causes synaptic vesicles to fuse with the membrane and release neurotransmitter molecules into the synaptic cleft. The neurotransmitter diffuses across the synaptic cleft and binds to ligand-gated ion channels in the postsynaptic membrane, resulting in a localized depolarization or hyperpolarization of the postsynaptic neuron. http://cnx.org/
A membrane at rest is more or less impermeable to positively charged sodium ions (Na+), but when stimulated it is transiently open to their passage. The Na+ ions thus flow in, attracted by the negative charge inside, and the membrane temporarily reverses its polarity, with a higher positive charge inside than out. This stage lasts less than a millisecond, and then the sodium channels close again. Potassium channels (K+) open, and K+ ions move out through the membrane, reversing the flow of positively charged ions. (Both these channels are known as voltage-gated, meaning that they open or close in response to changes in electrical charge occurring across the membrane.)
Over the next 3 milliseconds, the membrane becomes slightly hyperpolarized, with a charge of about �80 mV, and then returns to its resting potential. During this time the sodium channels remain closed; the membrane is in a refractory phase.
An action potential�the very brief pulse of positive membrane voltage�is transmitted forward along the axon; it is prevented from propagating backward as long as the sodium channels remain closed. After the membrane has returned to its resting potential, however, a new impulse may arrive to evoke an action potential, and the cycle can begin again.
Nerve system consists of cells that processes and transmit information; Direction of action potential conduction; http://bioserv.fiu.edu/
Gated channels, and the concomitant movement of ions in and out of the cell membrane, are widespread throughout the nervous system, with sodium, potassium, and chlorine being the most common ions involved. Calcium channels are also important, particularly at the presynaptic buttons of axons. When the membrane is at its resting potential, positively charged calcium ions (Ca2+) outside the cell far outnumber those inside. With the advent of an action potential, however, calcium ions rush into the cell. The influx of calcium ions leads to the release of neurotransmitter into the synaptic cleft; this passes the signal to a neighboring nerve cell.
Graded membrane potential:
A graded membrane potential is a gradient of transmembrane potential difference along a length of cell membrane. Graded potentials are particularly important in neurons that lack action potentials, such as some types of retinal neurons. Graded potentials that depolarize the membrane, increasing the membrane potential above the resting potential are important as "triggering potentials" that can spread along the surface of neuronal cell bodies to axon initial segments and trigger action potentials. Graded potentials that hyperpolarize the membrane potential to values more negative than the resting potential, this can inhibit the generation of action potentials. Graded potentials can arise at either portions of cells that function as sensory receptors or at synapses that are activated by neurotransmitters. These two types of graded potentials are called receptor potentials or synaptic potentials. Graded potentials are distinct from action potentials in that graded potentials spread electric potential changes along cell membranes without activating the kind of constant magnitude propagating signal that is characteristic of the action potential. Graded potentials are highest at a source and decay with increasing distance from the source.
Neurons that carry messages from the sense organs to the brain or spinal cord are called sensory (afferent) neurons. Neurons that carry messages from the brain or spinal cord to the muscles and glands are called motor (efferent) neurons. Interneurons (association neurons carry messages from one neuron to another. When the neuron is at rest, or at its resting potential, a slightly higher concentration of negative ions exists inside the membrane surrounding the cell body than outside, so there is a negative electrical charge inside relative to outside. At rest, a neuron is in a state of polarization. When an incoming message is strong enough, the electrical charge is changed, an action potential(neural impulse) is generated, and the neuron is depolarized. Incoming messages cause graded potentials, which, when combined, may exceed the minimum threshold of excitation and make the neuron fire. After firing, the neuron goes through the absolute refractory period, when it will not fire again, and then enters the relative refractory period, when firing will only occur if the incoming message is much stronger than usual. However, according to the all-or-none law, the impulse sent by a neuron does not vary in strength.
Electrical changes during the action potential.
Neurotransmitter molecules, released by synaptic vesicles, cross the tiny synaptic space (orcleft) between the axon terminal (or synaptic knob) of the sending neuron and the dendrite of the receiving neuron, where they latch on to a receptor site, much the way a key fits into a lock. This is how they pass on their excitatory or inhibitory messages.; http://cwx.prenhall.com/
What causes this change in potential to occur? The stimulus causes the sodium gates (or channels) to open and, because there's more sodium on the outside than the inside of the membrane, sodium then diffuses rapidly into the nerve cell. All these positively-charged sodium rushing in causes the membrane potential to become positive (the inside of the membrane is now positive relative to the outside). The sodium channels open only briefly, then close again.
Source:http://www.vet.purdue.edu/depts/bms/courses/bms511/coursework/nervous/n3.htm
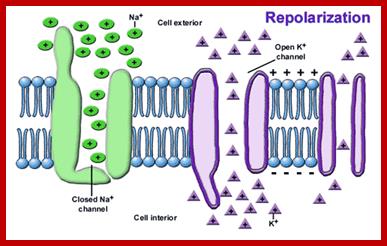
Source:http://www.vet.purdue.edu/depts/bms/courses/bms511/coursework/nervous/n3.htm
The potassium channels then open, and, because there is more potassium inside the membrane than outside, positively-charged potassium ions diffuse out. As these positive ions go out, the inside of the membrane once again becomes negative with respect to the outside.
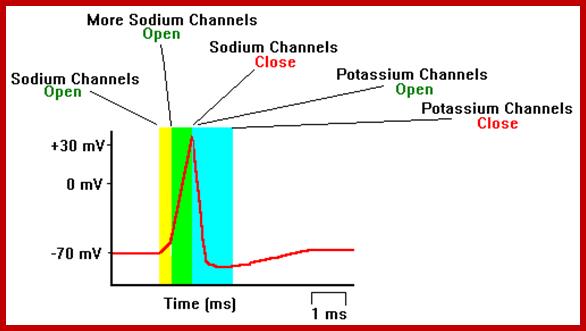
Source: http://faculty.washington.edu/chudler/ap.html
�
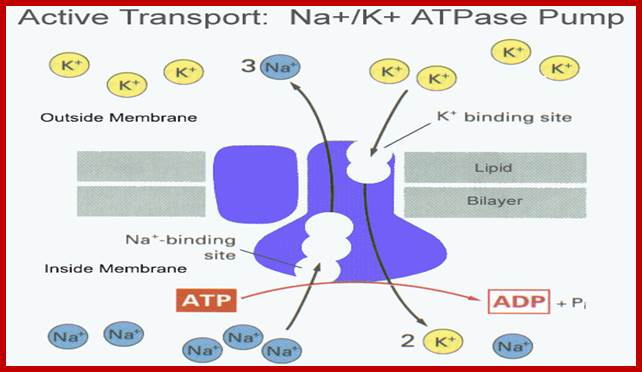
������������������������������������������������������������ Ion flow creates membrane potential
The net diffusion across a semi-permeable membrane occurs �down a concentration gradient. ��A. But this principle becomes more complex when you consider ions
������(=charged particles) because ions are influenced by both� 1) an ������electrical gradient and 2) a concentration gradient.� B. An abundance of anions (=large, impermeable proteins) on cytoplasmic side������ results in a charge asymmetry�
��1. K+ is much more permeable than Na+ so it is mostly found inside the cell������ a. Remember that like charges (+&+) repel; opposite charges (+&-) attract2. Both electrical & concentration gradients influence ion distribution (e.g. K+)
������� a. These forces often work in opposition (as in the case of K+)
� C. Charge across the membrane results.� Slightly positive outside and
�� negative inside--the result is a resting membrane potential IV. Electropotential Basis for Resting Membrane Potential: �A. Three ions are usually involved in establishing a membrane potential:1. Distribution of Na+, K+, Anions (=protein) across membrane
2. What determines that the ion distribution should be as such?�������������� a. Na+/K+ pump, but it is not that important�������������� b. Recall that the concentration gradient can oppose the electrical ������������������gradient for an ion, but an equilibrium is reached�������������� c. Equilibrium Potential (E) and the Nernst Equation
�������������������� 1) Limitations of Nernst Equation�������������� d. Goldman Equation considers differences in diffusion rate and ������������������������������������������� ����������better approximates resting potential�������������� e. Summary of resting potential (Animation)
������������������ Note: 3Na+/2K+ ATPase pump is not a major player hereB. What is the relationship of membrane potential to action potential? 
A diagram showing the progression in the development of a membrane potential from a concentration gradient (for potassium).Green arrows indicate net movement of K+ down a concentration gradient. Red arrows indicate net movement of K+ due to the membrane potential. The diagram is misleading in that while the concentration of potassium ions outside of the cell increases, only a small amount of K+ needs to cross the membrane in order to produce a membrane potential with a magnitude large enough to counter the tendency the potassium ions to move down the concentration gradient. http://en.wikipedia.org/
http://www.biologymad.com/
An action potential can be propagated along the length of an axon: (A) The voltages that would be recorded from a set of intracellular electrodes placed at intervals along the axon, whose width is greatly exaggerated in this schematic figure. Note that the action potential does not weaken as it travels. (B) The current changes in the Na+ channels and the current flows (orange arrows) that give rise to the traveling disturbance of the membrane potential. The region of the axon with a depolarized membrane is shaded in blue. Note that an action potential can travel only away from the site of depolarization because NA+-channel inactivation prevents the depolarization from spreading backward. In myelinated axons, clusters of NA+ channels can be millimeters apart from each other.
The impulse is received by the dendrites and passed through the cell body to the axon. An impulse is an electrical disturbance.; http://www.tutorvista.com/
Peptide transmission along neuronal axons;
Peptide transmitters and enzymes are synthesized in the cell body (1). Enzymes and propeptides are packaged into the Golgi apparatus. During fast axonal transport of these vesicles to the nerve terminal (2), the enzymes modify the propeptides to produce one or more neurotransmitter peptides (3). After vesicle fusion and exocytosis, the peptides diffuse away and are degraded by proteolytic enzymes.
The structure of the tubulin-kinesin complex, as revealed by image analysis of cryoelectron microscopy data. (a) The computed, three-dimensional map of a microtubule, (b) the kinesin globular head domain-microtubule complex, (c) a contour plot of a horizontal section of the kinesin-microtubule complex, and (d) a contour plot of a vertical section of the same complex. (Taken from Kikkawa et al., 1995. Nature 376:274-277. Photo courtesy of Nobutaka Hirokawa.)
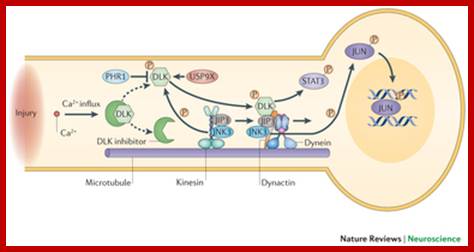 �
�
The extensive lengths of neuronal processes necessitate efficient mechanisms for communication with the cell body. Neuronal regeneration after nerve injury requires new transcription; thus, long-distance retrograde signalling from axonal lesion sites to the soma and nucleus is required. In recent years, considerable progress has been made in elucidating the mechanistic basis of this system. This has included the discovery of a priming role for early calcium waves; confirmation of central roles for mitogen-activated protein kinase signalling effectors, the importin family of nucleocytoplasmic transport factors and molecular motors such as dynein; and demonstration of the importance of local translation as a key regulatory mechanism. These recent findings provide a coherent mechanistic framework for axon�soma communication in the injured nerve and shed light on the integration of cytoplasmic and nuclear transport in all eukaryotic cells. Top fig; Early events in axon�soma communication after nerve injury. Bottom fig;Retrograde signalling via the JUN kinase pathway after an axonal lesion.
;Ida Rishal & Mike Fainzilber; http://www.nature .com
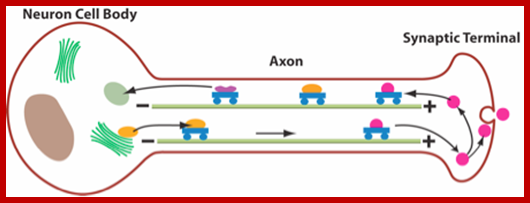
The microtubules serve as two-way highways for the transport of cargo. Most of the cargo is transported within �vesicles,� membrane wrapped containers. The vesicles carry essential cell nutrients and building materials (e.g., for neurotransmitters) to the far (distal) end of the axon, and return spent materials for recycling to the cell body. Keep in mind that in our bodies a single axon can extend more than a meter, with a diameter of less than 2 microns; thus, they are very long compared to their tiny width. In fact, if they were as wide as a single hair, they would be 30-50 meters long. So the molecular motors that move cargo along these very long narrow highways need to be pretty robust.
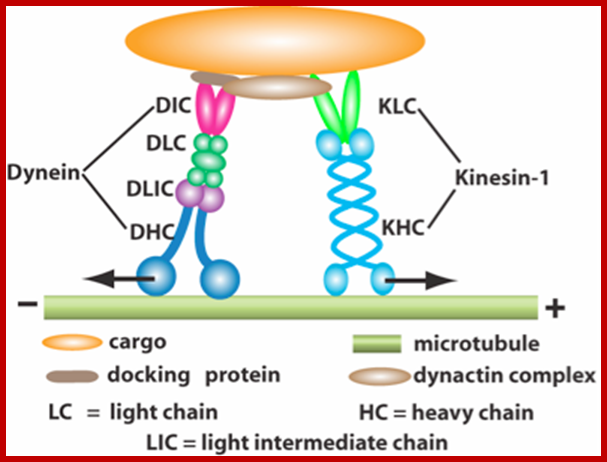
Sketch of motor proteins moving a single vesicle along a microtubule � A variety of different interlocking proteins provide the mechanism for transport of the vesicle along the microtubules. Kinesin protein complexes serve as motors pulling the vesicles toward the positively charged axon terminal. Dynein protein complexes drive the cargo assembly in the reverse, or negatively charged direction, toward the cell body. It is unclear how the movement of cargo is regulated, so that the right cargo is delivered to the right place at the right time. Dozens of different kinds of microtubule-associated proteins have been identified that may play regulatory roles.
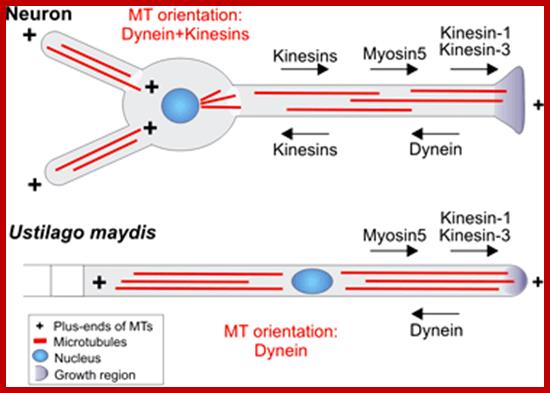
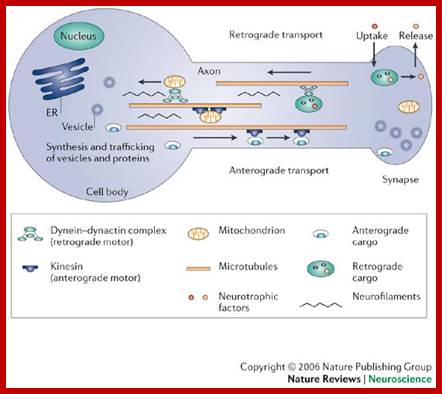
http://flipper.diff.org/
Vesicles for transport are sorted and loaded onto transport motors both in the cell body and the distal nerve terminal. The former are transported not only into the axon but also into dendrites. Those in the distal nerve terminals permit uptake and axosomatic movement of substances such as trophic proteins. Mutations in dynactin (humans), dynein (mice) and three different forms of kinesin all provoke motor neuron degeneration. Perturbation of neurofilaments through mutations and changes in phosphorylation and the physical structure of the axon could also adversely affect axonal transport. In transgenic mouse models, mutant superoxide dismutase 1 (SOD1) impairs anterograde axonal transport.
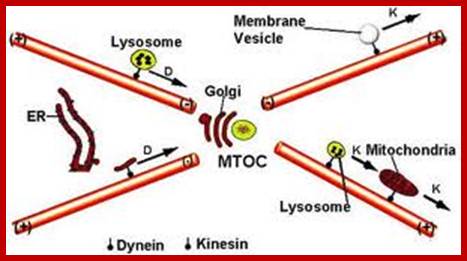
Motor proteins also move chromosomes along MTs during mitosis: kinesin pushes polar MTs and dynein pulls on astral MTs to help separate the chromosomes;http://www.angelfire.com/
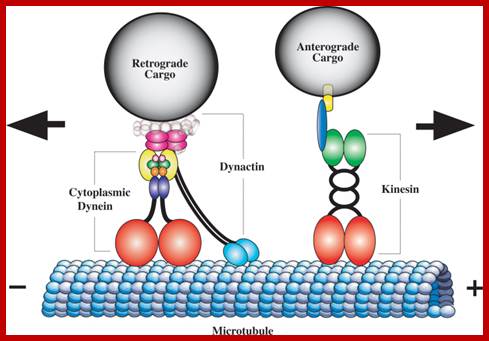
����������������������������������������������� http://www.plosgenetics.org/
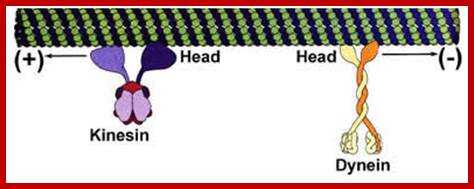
Both kinesin and dynein use the energy of ATP hydrolysis fror movement. The head domain binds MTs and hydrolyzes ATP while the tail domain binds cargo. There are two general types of dyenins: cytosolic and axonemal. Cytosolic dyenins are bound to cargo by a complex of MT binding proteins. http://www.angelfire.com/
Kinesin and Dynein Direct Organelle Movement along Microtubules in opposite direction.
Important advances in cell biology have often followed the introduction of a new experimental technique, and it was the improved ability to see small faint objects by video-enhanced light microscopy that led to the discovery of the microtubule motors responsible for organelle transport. Once it became possible to visualize single microtubules in an unfixed specimen, investigators could follow the movement of organelles and other particles along these microtubules in vitro. Alternatively, they could observe and measure the gliding movement of individual microtubules over glass surfaces coated with cell extracts.
Such in vitro motility assays were used to identify and isolate two classes of microtubule-dependent motor proteins - the kinesins and the cytoplasmic dyneins. Cytoplasmic dyneins are involved in organelle transport and mitosis and are closely related to ciliary dynein,the motor protein in cilia and flagella (discussed later). Kinesins are more diverse than the dyneins, and different family members are involved in organelle transport, in mitosis, in meiosis, and in the transport of synaptic vesicles along axons. Both the cytoplasmic dyneins and the kinesins are composed of two heavy chains plus several light chains. Each heavy chain contains a conserved, globular, ATP-binding head and a tail composed of a string of rod like domains. The two head domains are ATPase motors that bind to microtubules, while the tails generally bind to specific cell components and thereby specify the type of cargo that the protein transports
Bimolecular motors are proteins that convert chemical energy, Adenosine Tri-phosphate (ATP), into mechanical energy for transport inside cells. There are three types of motor proteins: Kinesin, Dynein, and Myosin. Research has discovered that Bimolecular Motor proteins are required for many cellular processes such as vesicle transport, mitotic spindle formation, and cellular motility. Due to the compactness of the cytoplasm in the cell, there is a need for the cell to maintain intracellular organization. Bimolecular motors are used to move large cellular components to certain destinations. Left figure shows an image of a Bimolecular motor that is carrying cargo.
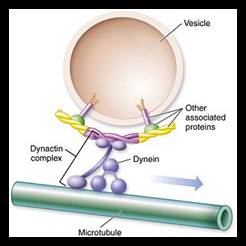
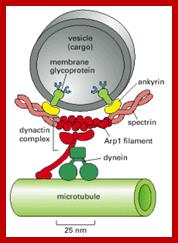
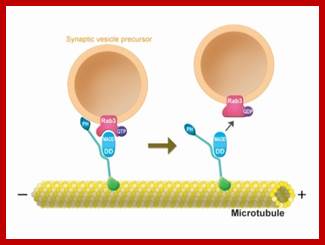
Cytoskeleton proteins with their motor proteins are involved in the transportation of vesicles at least in neuronal axons
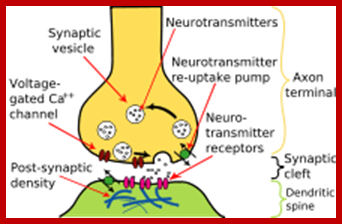
A diagram of a typical central nervous system synapse. The spheres located in the upper neuron contain neurotransmitters that fuse with the presynaptic membrane and release neurotransmitters into the synaptic cleft. These neurotransmitters bind to receptors located on the postsynaptic membrane of the lower neuron, and, in the case of an excitatory synapse, may lead to a depolarization of the postsynaptic cell. http://en.wikipedia.org/
A model of synaptic vesicle trafficking and release at the neurological synapse:
Molecular mechanisms of biogenesis and exocytosis of cytotoxic granules: Genevi�ve de Saint Basile, Ga�l M�nasch� & Alain Fischer
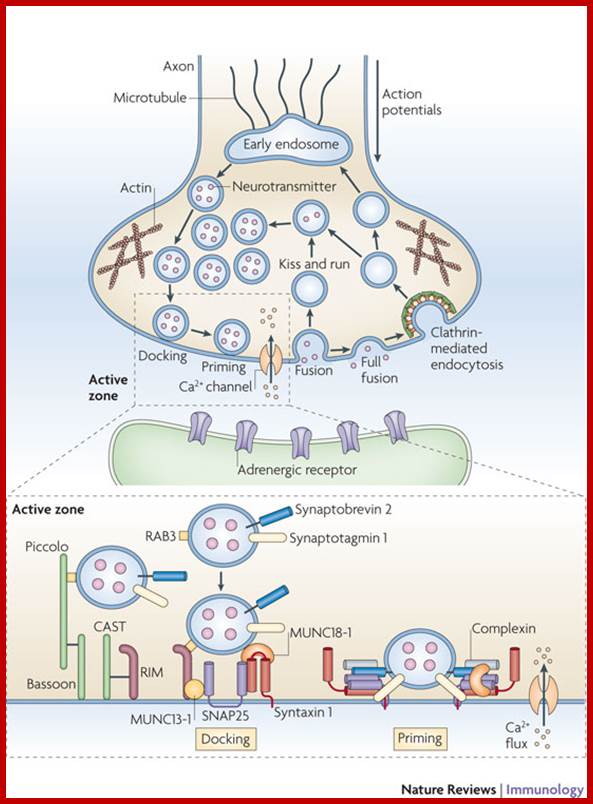
Synaptic vesicles undergo a trafficking cycle in the nerve cell terminal that involves a series of steps involving synaptic vesicle formation from endosomal compartments and storage of neurotransmitters by active transport into the vesicles, which is driven by a vacuolar proton pump. Synaptic vesicles accumulate in two different pools: a readily releasable pool that participates actively in exocytosis under conditions of physiological stimulation and a reserve pool that is activated only in response to strong synaptic stimulation and can rapidly replace vesicles that have released their contents. Synaptic vesicles from the readily releasable pool dock at the active zone, where they are primed to convert into a state of competence for Ca2+-triggered fusion-pore opening. After fusion, synaptic vesicles are endocytosed and recycled. Vesicles can be retrieved through two different pathways: in the first, vesicles undock and locally recycle, being refilled with neurotransmitters, a process known as 'kiss and run'; in the second, vesicles fully collapse with the plasma membrane, are then endocytosed through clathrin-coated pits and refilled with neurotransmitters either directly or after passing through an endosomal intermediate compartment. Different protein�protein interactions at the active zone mediate attachment of the vesicle to the target membrane. RAB3 in the vesicle binds to RAB3-interacting molecule (RIM), which also interacts with MUNC13-1, localized at the active zone. In addition, vesicles interact with a docking complex comprising MUNC18-1 and syntaxin 1 in the closed conformation. Then, MUNC13-1 induces the opening of syntaxin 1 to promote the formation of the synaptobrevin 2�syntaxin 1�MUNC18-1�SNAP25 SNARE complex. Synaptotagmin 1, the Ca2+ sensor for synchronous synaptic transmission, participates in this complex, as well as complexin, which maintains the complex in an activated but 'off' state, until Ca2+ enters and binds to synaptotagmin 1. MUNC18-1 also binds to syntaxin 1 in the open conformation, contributing to specificity and stimulating fusion of the trans-SNARE complex. Bassoon, Piccolo and CAZ-associated structural protein (CAST) are scaffold proteins for vesicle interactions that may link exocytosis and endocytosis of vesicles at the active zone. SNAP25, synaptosomal-associated protein of 25 kDa; �SNARE, N-ethylmaleimide-sensitive factor accessory protein receptor.
Synaptic Vesicles Form from Endosomes:
Nerve cells (and some endocrine cells) contain two types of secretory vesicles. As we have just discussed, these cells package proteins and peptides in dense-core secretory vesicles in the standard way for release by the regulated secretory pathway. In addition, however, they make use of another specialized class of tiny (~50-nm diameter) secretory vesicles that are generated in a different way. These synaptic vesicles store the small neurotransmitter molecules, such as acetylcholine, glutamate, and g-amino butyric acid (GABA) that serve for rapid signaling from cell to cell at chemical synapses (and for local signaling in some endocrine tissues). The vesicles are triggered to release their contents within a fraction of a millisecond when an action potential arrives at a nerve terminal, and some neurons fire more than 1000 times per second, releasing synaptic vesicles each time. This demands very rapid replenishment of the vesicles that are believed to be generated not from the Golgi membrane but by local recycling from the plasma membrane in the following way. It is thought that the membrane components of the synaptic vesicles are initially delivered to the plasma membrane by the constitutive secretory pathway and then retrieved by endocytosis and delivered to endosomes, from which they are reassembled and bud off to form synaptic vesicles. The membrane components of the vesicles include carrier proteins specialized for the uptake of neurotransmitter from the cytosol, where it is synthesized. Once filled with neurotransmitter, the vesicles return to the plasma membrane, where they wait until the cell is stimulated. After they release their contents, their membrane components are retrieved in the same way and used again (Figure 13-43). The whole cycle, from endocytosis to exocytosis, can be observed by adding a tracer molecule, such as peroxidase, to the external medium and following its fate as it is first taken up into endosomes and then returned to the cell surface in synaptic vesicles� NCBI
Synaptic Vesicles Can Form Directly from Endocytic Vesicles:
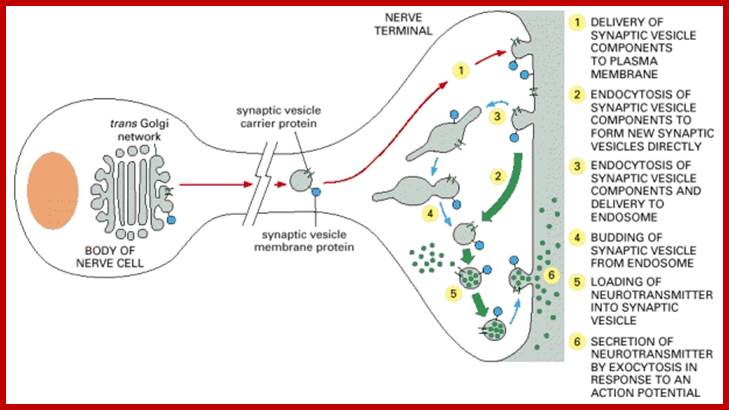
Presynaptic vesicles formation and discharge to inter-synaptic space
Nerve cells (and some endocrine cells) contain two types of secretory vesicles. As for all secretory cells, these cells package proteins and peptides in dense-cored secretory vesicles in the standard way for release by the regulated secretory pathway. In addition, however, they make use of another specialized class of tiny (~50-nm diameter) secretory vesicles, which are called synaptic vesicles and are generated in a different way. In nerve cells, these vesicles store small neurotransmitter molecules, such as acetylcholine, glutamate, glycine, and γ-aminobutyric acid (GABA), that mediate rapid signaling from cell to cell at chemical synapses. As discussed earlier, the vesicles are triggered to release their contents within a fraction of a millisecond when an action potential arrives at a nerve terminal. Some neurons fire more than 1000 times per second, releasing neurotransmitters, each time. This rapid release is possible because some of the vesicles are docked and primed for fusion, which will occur only when an action potential causes an influx of Ca2+ into the terminal.
Only a small proportion of the synaptic vesicles in the nerve terminal fuse with the plasma membrane in response to each action potential. But for the nerve terminal to respond rapidly and repeatedly, the vesicles need to be replenished very quickly after they discharge. Thus, most synaptic vesicles are generated not from the Golgi membrane in the nerve cell body but by local recycling from the plasma membrane in the nerve terminals. It is thought that the membrane components of the synaptic vesicles are initially delivered to the plasma membrane by the constitutive secretory pathway and then retrieved by endocytosis. But instead of fusing with endosomes, most of the endocytic vesicles immediately fill with transmitter to become synaptic vesicles.
The membrane components of a synaptic vesicle include carrier proteins specialized for the uptake of neurotransmitter from the cytosol, where the small-molecule neurotransmitters that mediate fast synaptic signaling are synthesized. Once filled with neurotransmitter, the vesicles return to the plasma membrane, where they wait until the cell is stimulated. After they have released their contents, their membrane components are retrieved in the same way and used again.
The formation of synaptic vesicles: These tiny uniform vesicles are found only in nerve cells and in some endocrine cells, where they store and secrete small-molecule neurotransmitters. The import of neurotransmitter directly into the small endocytic vesicles
These tiny uniform vesicles are found only in nerve cells and in some endocrine cells, where they store and secrete small-molecule neurotransmitters. The import of neurotransmitter directly into the small endocytic vesicles that form from the plasma membrane is mediated by membrane carrier proteins that function as antiports, being driven by a H+ gradient maintained by proton pumps in the vesicle membrane.
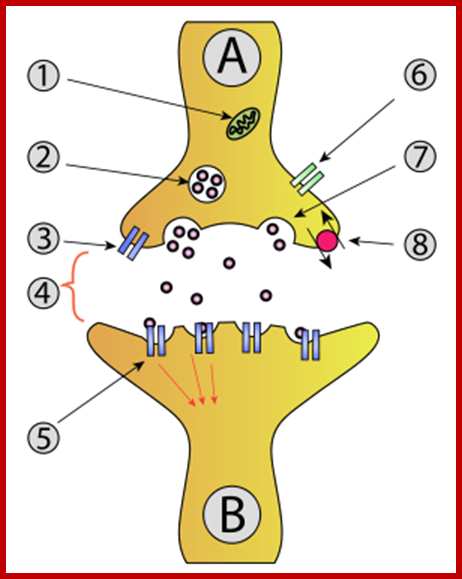
http://en.wikipedia.org/
In a neuron, synaptic vesicles (or neurotransmitter vesicles) store various neurotransmitters that are released at the synapse. The release is regulated by a voltage-dependent calcium channel. Vesicles are essential for propagating nerve impulses between neurons and are constantly recreated by the cell. The area in the axon which holds groups of vesicles is an axon terminal or "bouton". Up to 130 vesicles can be released per bouton over a ten minute period of stimulation at 0.2 Hz. In the human brain region V1 synaptic vesicles have an average diameter of 39.5 nanometers with a standard deviation of 5.1 nanometers.
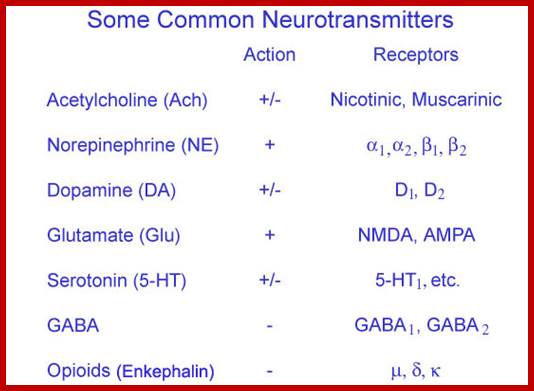
Neurotransmitters/Neuropeptides
There are various neurotransmitters and neuropeptides that are involved with the network of the brain. However, since many neurotransmitters and peptides co-relate in affecting one's body, it will be very difficult in defining one neurotransmitter/neuropeptide's function; henceforth, it will not be explained any further from the list.
Neuropeptides : Gamma-Aminobutyric Acid (GABA) , Glutamate , Aspartate , and glycine .
(There are more neuropeptides; visit �www.faculty.washington.edu/chudler/): Insulin ,Beta-endorphin , Neuropeptide Y and tonin
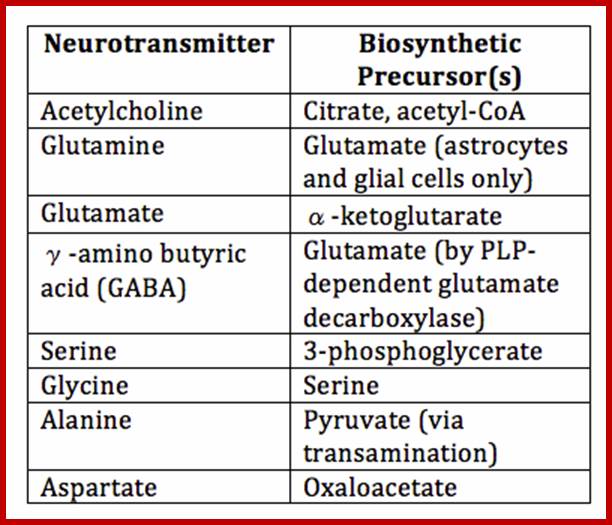
����������� ����������� ����������������������� Neurotransmitters
Neurotransmitter Release:
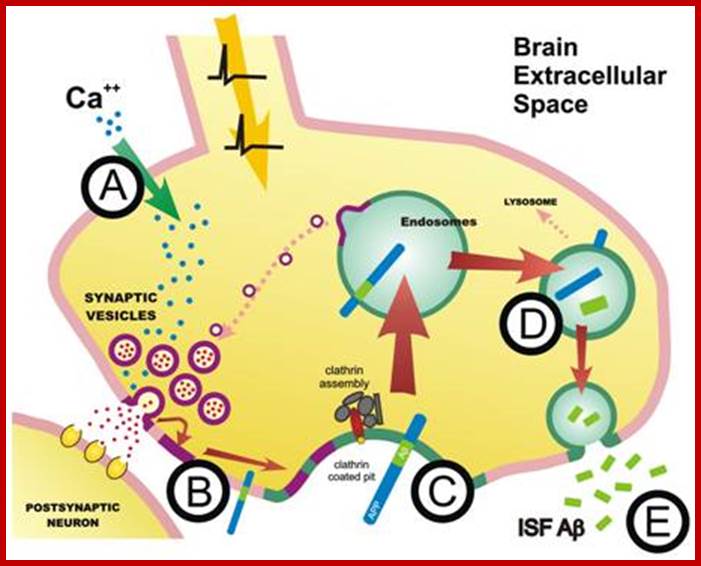
Entry of calcium activates the release of Neurotransmitters from presynaptic membranes
Model for the functions of SNARE proteins, complexins, and synaptotagmins 1 and 2 in synaptic vesicle exocytosis.
To identify key molecules involved in release, we initially set out to
characterize the proteins of synaptic vesicles and of the active zone. We then
used biochemical and biophysical methods to determine the properties and atomic
structures of these proteins, and targeted mouse mutants to examine their
functions. Although many questions remain, this combination of approaches has
elucidated fundamental mechanisms underlying key aspects of neurotransmitter
release (e.g., Ca2+ triggering of fast synaptic vesicle fusion).
Synaptic vesicle fusion is at least in part mediated by the assembly of three
synaptic SNARE proteins�the vesicle protein synaptobrevin/VAMP and the plasma
membrane proteins SNAP-25 and syntaxin/HPC-1�into a tight complex. Syntaxin
also interacts with another essential fusion protein, Munc18-1. SNARE proteins
and Munc18-1 perform multiple functions that are exquisitely regulated. For
example, during fusion, syntaxin changes from a closed into an open
conformation. We recently showed in mice that permanent "opening" of
syntaxin by mutagenesis destabilizes synapses, leading to massive epilepsy. As
another example, synaptobrevin is required for both spontaneous and evoked
synaptic vesicle fusion, but recent data reveal that the mechanisms by which
synaptobrevin acts in these two forms of fusion differ. In addition to this
dual function in exocytosis, synaptobrevin is also required for normal fast
endocytosis of vesicles. These findings suggest an unexpectedly economical
organization of the secretory machinery in which different steps are mediated
by the same molecules via different mechanisms.
At the synapse, Ca2+ triggers both a fast component and a slow component of
release. The fast component is induced by Ca2+ binding to the synaptic vesicle
proteins synaptotagmin 1 and 2. Recent data show that these two synaptotagmins
act similarly in stabilizing resting synapses and in triggering fast release
upon Ca2+ binding.
Based on the revealed physical properties, as well as genetic and
biochemical evidence, the investigators propose that sec1/syntaxin1 complex
communicates with Rab, a G protein on the vesicle membrane, and Rab-E, a
free-floating cousin, to recognize and guide the neurotransmitter-filled sac to
its appointed fusion site on the target membrane. They further propose that
syntaxin1a and Sec1 collaborate with VAMP, a velcro-like vesicle membrane
protein, and SNAP-25, a counterpart on the cell's membrane, to bring about
fusion itself.
In this model, nSec1 performs a "chaperone-like" role for syntaxin1a,
first shielding it from other proteins, and later facilitating its interactions
with them. When in its protective posture, nSec1 renders its partner physically
incapable of forming stable relationships with other proteins.
PRESYNAPTIC PROTEINS, SYNAPTIC VESICLE DOCKING & MEMBRANE FUSION1 ; http://neuromuscular.wustl.edu/
The steps to the dance are as follows (See diagram of model below):
B � The
vesicle's Rab and/or Rab-E proteins recognize the complex, signaling nSec1 to
change its shape and move outward, uncovering a key binding region of
syntaxin1a, and destabilizing it. Residues from the suddenly exposed binding
region signal SNAP-25 to begin making the core fusion complex, using nSec1 as a
platform.
C � Syntaxin1a's sharply bent end section moves away from its key binding
region, making way for SNAP-25 and VAMP to move in and bind.
D � VAMP/SNAP-25/syntaxin1a combine to form a long, straight, helical bundle,
the core complex. This pulls the vesicle and target membranes together, fusing
them, and releasing the neurotransmitter. After fusion, an enzyme breaks down the
complex and the SNAREs are recycled for a repeat performance.
Four years ago, Hamm and colleagues including Simon Alford, Ph.D.,
of the University of Illinois at Chicago, and Thomas F. J. Martin, Ph.D., of
the University of Wisconsin-Madison, reported that the beta-gamma subunit of an
inhibitory G protein blocked neurotransmitter release from large, easily
studied nerve cells of the lamprey eel. But they didn't know the mechanism �
until now.
In an elegant series of experiments, the researchers showed that the beta-gamma
subunit prevents vesicles from fusing with the membrane of the nerve cell. It
does this by preventing another protein from binding to the cell's �fusion
machinery,� a complex of proteins called SNARE that links the vesicle to the
membrane. In particular, the beta-gamma subunit prevents synaptotagmin, a
vesicle protein, from binding to the SNAP-25 protein in the SNARE complex.
The researchers identified the site of the subunit's action by injecting tiny
amounts of a form of the paralyzing botulinum toxin that chopped off the end of
the SNAP-25 protein. When that happened, the subunit could not block fusion,
indicating that the chopped-off portion was its binding site.
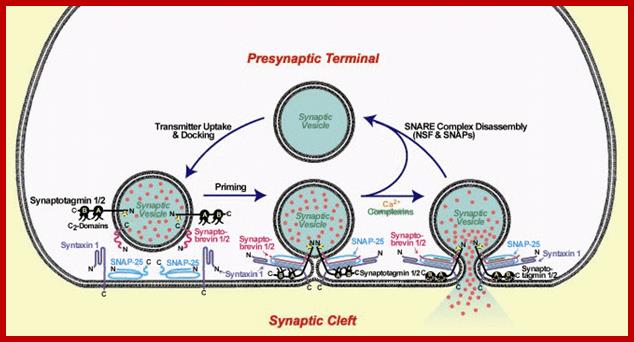
Neurotrophin-mediated maturation of neurotransmitter-release machinery at excitatory presynaptic terminals. Carlos Vicario-Abej�n et al; ;http://www.nature.com/
During maturation of the presynaptic terminal, neurotrophins might regulate synapse-related protein synthesis and/or modulate protein�protein interactions by regulating post-translational modifications. These actions could lead to an increase in protein incorporation into synaptic vesicles during vesicle biogenesis (1), vesicle accumulation into the reserve pool (2) and/or stabilization of the SNARE (soluble N-ethylmaleimide-sensitive fusion protein-attachment protein receptor) complex � SNAP25 (synaptosome-associated protein, 25 kDa), syntaxin and synaptobrevin. Brain-derived neurotrophic factor (BDNF) might also activate synaptic vesicle mobilization from a reserve pool to a docked pool, possibly through the BDNF-mediated regulation of synapsin 1 phosphorylation (3). This model is based on several studies that are discussed in the main text. Carlos Vicario et al
In docked vesicles (left), SNAREs and synaptotagmins are not engaged in direct interactions. During priming (center), SNARE complexes form, and synaptotagmins constitutively associate with the assembled SNARE complexes. The approximation of the synaptic vesicle and plasma membranes forced by SNARE complex assembly is proposed to create an unstable intermediate that is shown as a fusion stalk, but could have other structures. Subsequently, complexins (green) are bound to fully assembled complexes (right), and calcium influx (right) destabilizes the fusion intermediate by triggering the C2-domains of synaptotagmins to associate with, and partially insert into, the phospholipids. This is proposed to cause a mechanical perturbation that opens the fusion pore. Note that SNARE complex assembly in priming (center) is suggested to be reversible, whereas calcium triggering is not.
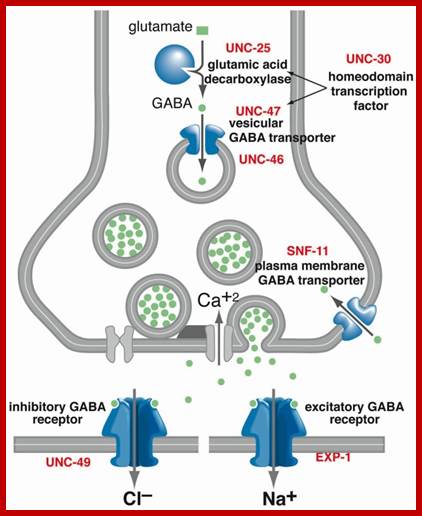
Entry of calcium ions in response to action potential leads activation of neurotransmitter loaded vesicle fuse with their own specific SNARE and release Glutamate.� The released neurotransmitters bind to post synaptic membrane resident receptors and allow ions to flow.
Neuronal presynaptic bulge consists of synaptic vesicles loaded with neurotransmitters. Some of the vesicles are fused and released neurotransmitter; http://www.cnsforum.com/
http://journal.frontiersin.org/
3. The filled vesicle is docked in the region of the synapse, where it awaits a nerve stimulation.
6. The vesicle again begins to accumulate hydrogen ions the cycle is repeated.
Synapsis vesicle circulation-formation and discharge and reformation; http://origins.swau.edu/