Drosophila1-Developmental_Biology
Introduction:
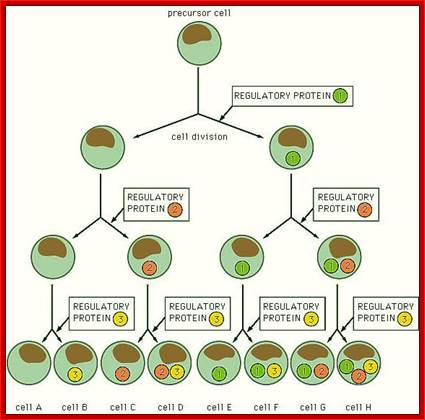
Development of multicellular organisms, whatever may be the organism, starts with single zygotic cell.� Unicellular cells as they are produced, they grow into their full size and reproduce to reborn as unicellular structures.� Whether an organism is unicellular or multicellular, cells go through a series of gene expressions to reach a particular stage of development; the reproduction and growth of an organism is all due to molecular expression in stepwise manner, from a young cell to an adult cell ready for reproduction.
During development, a single precursor cell with repeated cell divisions generates a series of progenitor cells, which in turn undergo programmed differentiation into specific fate.� Molecular basis of this differentiation and development is profound.� Initial stages of such events have been very well elucidated in Drosophila.
This illustrates the programming of the Zygote to divide and redivide and cell derivatives are programmed for differentiation into specific cell types.
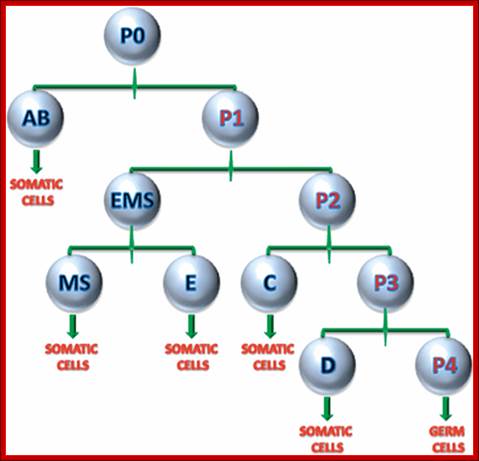
Repeated cell divisions in C. elegans as in diagram above (Drosophila); George J. Tserevelakis et al ; http://biomedicaloptics.spiedigitallibrary.org/
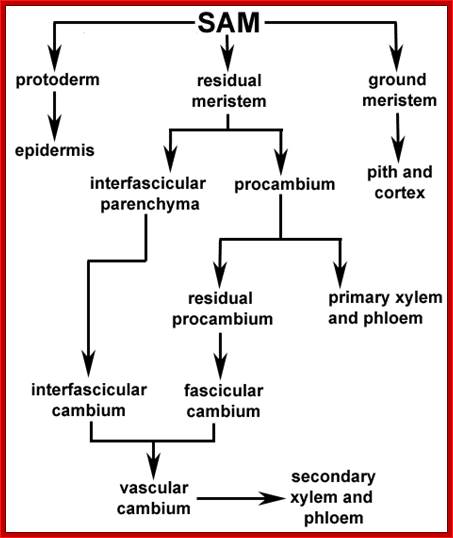
http://plantcellbiology.masters.grkraj.org/http://novszerv.elte.hu/
D. melanogaster has provided vital contributions to our understanding of biology, especially in the fields of genetics and developmental biology since Thomas Hunt Morgan discovered sex-linked inheritance studying Drosophila between 1910 and 1917 for which he was awarded the Nobel Prize in Physiology or Medicine in 1933 (Hunt Morgan 1919; Lewis 1998). Lewis (1998) suggests that Hunt Morgan initially �chose Drosophila because of its short life cycle [two weeks], ease of culturing and high fecundity,� characteristics which ensure its use as a model organism today. Manning (2008) suggest that more recently, it is used in developmental biology, looking to see how a complex organism arises from a relatively simple fertilized egg. Embryonic development is where most of the attention is concentrated, but there is also a great deal of interest in how various adult structures develop in course of time, In this case, mostly focused on the development of the compound eye, but also on the wings, legs and other organs.�
Life cycle of drosophila, vinegar fly; http://invitero.tumblr.com
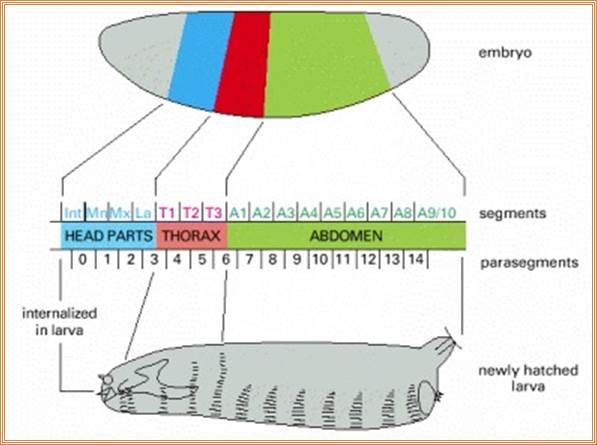
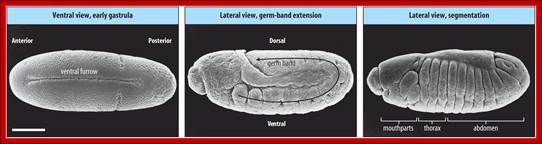
Figure 21-26The segments of the Drosophila larva and their correspondence with regions of the blastoderm
The parts of the embryo that become organized into segments are shown in color. The two ends of the embryo, shaded gray, are not segmented and become tucked into the interior of the body to form the internal structures of the head and gut. (The future external, segmental structures of the adult head are also transiently tucked into the interior in the larva.) Segmentation in Drosophila can be described in terms of either segments or para segments: the relationship is shown in the middle part of the figure. Para segments often correspond more simply to patterns of gene expression. The exact number of abdominal segments is debatable: eight are clearly defined, and a ninth is present vestigial in the larva, but absent in the adult insect.
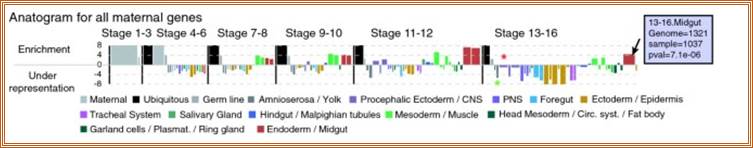
Normalized anatomical signature - the anatogram. A linear representation of the CV is used to show the enrichment of annotations within the set of all 3,334 maternally expressed genes versus the entire dataset of 4,759 genes expressed in the embryo. A vertical black line delimits stages, and each colored bar represents an individual CV term. The width of each bar is proportional to the number of times a term was used in our entire dataset, and the height represents the relative enrichment of the given term within the particular gene set (in this case, all maternally expressed genes). Enrichment is given in units of standard deviation above or below the expected sample count based on the background frequencies (z-score). Terms with bars below the zero line are under-represented in the sample. The green asterisk corresponds to the 'amnio serosa' term, while the red asterisk corresponds to the 'brain' term. On the web supplement [21], the user can place the mouse pointer over any bar in the anatomical signature (arrow on the midgut bar in stage range 13-16) and obtain the gene count for the term in the entire dataset, the gene count within the particular set of genes under study, and a statistical p value of statistical over- or under-representation within the set (shown in the black bordered lavender box). https://genomebiology.biomedcentral.com/
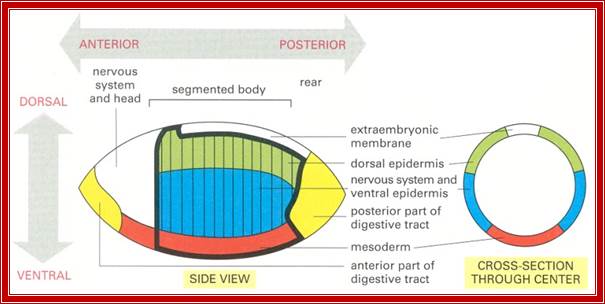
Figure-Fate map of a Drosophila embryo at the cellular blastoderm stage;
The embryo is shown in side view and in cross section, displaying the relationship between the dorsoventral subdivision into future major tissue types and the anteroposterior pattern of future segments. A heavy line encloses the region that will form segmental structures. During gastrulation the cells along the ventral midline invaginate to form mesoderm, while the cells fated to form the gut invaginate near each end of the embryo. Wilhelm Roux' Arch. Dev. Biol. 194:213�216, 1985.
All diploid multicellular organisms start with a single fertilized zygotic cell; which has all the encoded information in their chromosomes.� In the case of oogametic forms the maternally derived egg cell has more maternal genetic information, than the male sperm; for the egg contains, besides chromosomal complement, cytoplasmic informosomal elements in the form of proteins and mRNAs localized, some are spread over; (which are not contributed by sperm, the male complement and the most important sine qua non maternal �Mitochondria�.� Most of our cytoplasmic characters are inherited by our mothers.� Earlier embryologists, used histochemical methods to study development of organisms; which has provided anatomical details of development.� Though serial sectional views, either through optical microscopes with high resolution or Transmission Electron Microscopy or SEM of developmental stages, has provided a wealth of information about the cellular details. In this chapter a general view of cellular and molecular based understanding of development, is described.� Today the subject, molecular biology, has grown to such proportions, in terms of techniques and knowledge, it can provide many answers and explanations, hitherto remained unanswered.�� Molecular studies of growth and development, however, is in its embryonic stage, but it will grow with the development of more sophisticated methods and techniques.�� Genomic data of many organisms is now available; it by itself does not provide all the answers.�
Earlier Xenopus was used as the model for higher vertebrate, but soon they found Xenopus is too big and too complex an organism.� The fly, Drosophila melanogaster, a common fly in the kitchen, came handy to understand molecular basis of development. This fly has been extensively used by genetists over the century or more. �Then the nematode Caenorhabditis elegans became the darling for developmental biologists; for they can identify each and every cell derivative and their transformation into differentiated cell types through microscope, observe cellular structures under microscope easily.
The genomic content of each of the said organisms; Xenopus Leavis=3.1x10⁹ (n=10), D. melanogaster = 1.65 x 10⁸ (n=4) and C. elegans= 9.7 x 10⁷ (n=6, male=5A + X, female 5A +XX); consisting of (+/-) 215000 genes, 13601 genes and 18424 genes respectively.
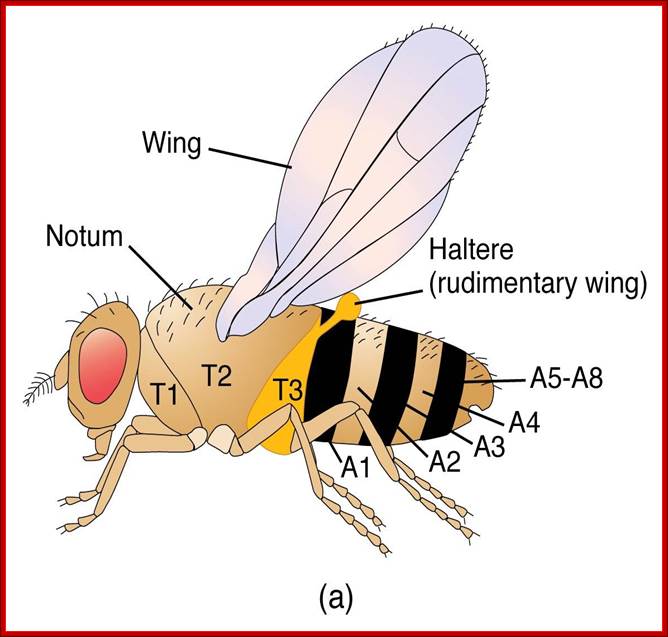
Flies (order Diptera), like other members of the class Insecta, have a number of serially homologous segments in a tri-partite body plan that comprises a Head or Cephalic region, a Thoracic region, and an Abdominal region. Each of the three pairs of legs is located on the ventral side of a different Thoracic segment. The wings are on the dorsal side of the second Thoracic segment (notum). In Diptera, a pair of rudimentary halters occurs on the next posterior segment: in other orders of Insecta, these are developed as a pair of fully functional wings. Abdominal segments 5-8 are compressed and modified for reproductive functions. The cephalic region includes segments that differentiate as paired antennae, eyes, and labial palps. All of the cuticular structures develop from imaginal discs present in the fly larva; Morphology of Drosophila; http://www.biopsychology.com/
The fruit fly belongs to:
Phylum: Arthropoda,
�Class: Insecta;
Division: Acalypterata;
Family: Drosophilidae,
Genus: Drosophila and
Species: melanogaster.�
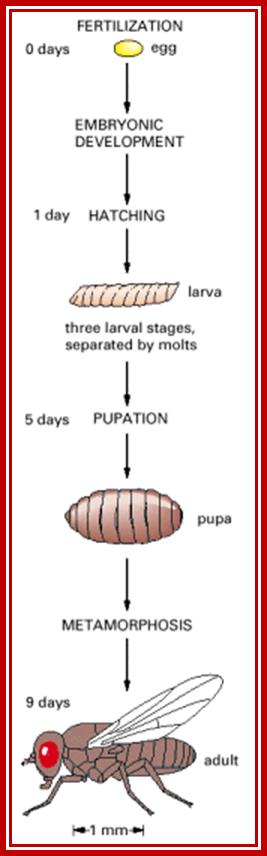
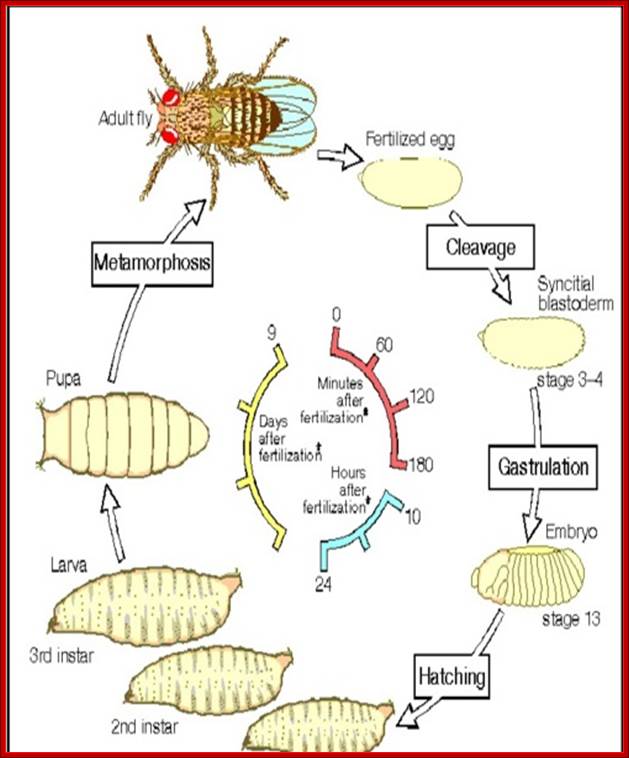
Synopsis of Drosophila development from egg to adult fly.
The timetable of Drosophila development, from egg to adult, is summarized in the above figure. The period of embryonic development begins at fertilization and takes about a day, at the end of which the embryo hatches out of the egg shell to become a larva. The larva then passes through three stages, or instars, separated by molts in which it sheds its old coat of cuticle and lays down a larger one. At the end of the third instar, it pupates. Inside the pupa, a radical remodeling of the body takes place�a process called metamorphosis. Eventually, about nine days after fertilization, an adult fly, or imago, emerges.
Development gets initiated soon after fertilization and first instar at 24hrs. Second instar at 48hrs, third at 72hrs, 5th at 118hrs, pupa at 6th day, and as it molts, insect emerges on 9th day.� Within six hours of emergence, it is ready for mating?� All the adult structures develop from mesoderm derived �fate cells or imaginal disc�, laid within the larval body.
The fly body pattern is simple with a head, thorax (3 segments) and an abdomen (8 segments).� Head consists of a pair of antennae, a proboscis and a mandible, a pair of compound eyes and bristles.� The thorax is made up of Prothorax (with first pair of legs and pair of humeri); a Mesothorax with a pair of wings and a pair of legs.� The Meta thorax consists of a pair of halters and a pair of hind legs.� Females have 7-8 segments in abdomen with vaginal plates and males have 7-8 segments with pigmented genital plates and also, they have chitinized black sex combs. �Flies show sexual dimorphism.
The fruit fly is considered as the �Cinderella� for molecular biologists and Genetists, similar to and more than E. coli for prokaryotes.�
The insect has been studied from 1890s onwards by Thomas Hunt Morgan (1910) and his students Sturtevant (1913); it has yielded important genetic information in the form of sex inheritance, linkage, gene lethal and many others of very importance and impact.� For any studies the fly offers great advantages; like very easy to handle, its life cycle is very short, develops population in good times; one can breed selectively, exhibits different stages of morphological changes which can be monitored using microscope and other methods and more importantly its salivary gland chromosomes offer vivid visual gene expression. Interestingly 50% of fly protein sequences have mammalian homologs? Genome size 1.5x10^8bp, containing~12,000 genes on haploid 4 chromosomes.
Its development starts in the space found within Germanium, where one of the progenitors of oocyte, called stem cell undergoes mitotic cell divisions to give rise to16 cells, where each of the cells are connected by one or two cytoplasmic bridges.� One of the cells which is connected to more than three nurse cell initiates meiotic division but remain arrested at meiosis I at metaphase. Even other cells, which are now called nurse cells, initiate meiotic division, but halt at some stage. How and why?� Not known!� Chromosomes in nurse cells become polytene which is a requirement for the development and maturation of the oocyte.� As the oocyte is developing, follicle cells surround the developing oocyte and show intimate contact and exchange of materials and information in the form of signals.
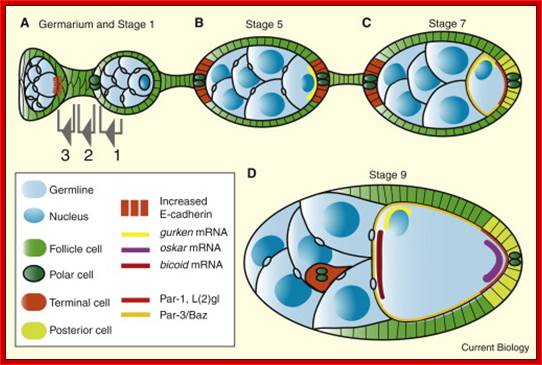
Current Biology; Rebecca Basock, Daniel St Johnston; http://www.cell.com/
An ovariole (A), expressing GFP tagged Staufen (green) and stained, with phalloidin, for actin (red), (Image courtesy Vitaly Zimyanin). The germarium to the left; egg chambers become more mature moving to the right. The Illustration of the germarium; shows the stem cells (yellow), which divide to produce a daughter cystoblast (blue). The daughter cells divide incompletely to produce a cyst of 16 cells (blue) joined by actin-rich ring canals (red), with a continuous fusome (grey). As the cyst matures it moves along the germarium and is surrounded by somatic follicle cells (green), which intercalate and pinch it off to form a discrete egg chamber. Columnar follicle cells (C), illustrating their polarized membrane domains, apical (orange), adherens junction (dark green) and basolateral (yellow), and cytoskeleton, with Actin (red) and microtubules (grey).
�����������������������
The illustration, below shows early stages of oogenesis, where the oocyte is in physical connection through cytoplasmic bridge with nurse cells.� During these process maternal cytoplasmic materials such as proteins and m RNAs are transferred and positioned with in the oocyte.� The process of egg formation starts at the base of ovary in germanium sites each of the individual body consists of nurse cells and one developing oocyte cell. �As the oocyte cell progressively mature, the oocyte gets surrounded by follicular cells as seen in the right part of the diagram.
The oocyte thus developed, is surrounded by a layer of follicle cells; enlarges and vitelline layer covers it.� Between the vitelline layer and the egg�s plasma membrane, the space found is called peri-vitelline space filled with a fluid.� The plasma membrane is studded with a variety of receptors like torso, toll and many others.
�
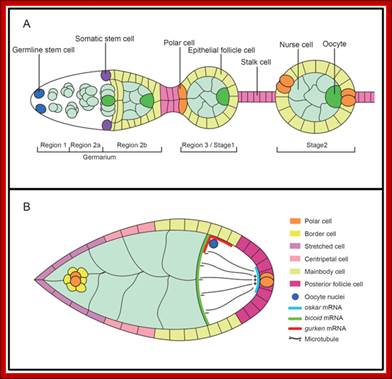
Schematic representation of early oogenesis (A) and stage 9 egg chamber (B). The germline and specified somatic cells are indicated individually. In both panels, anterior is to the left
Oocyte polarity during oogenesis; Qi Li, et al; http://www.nature.com/
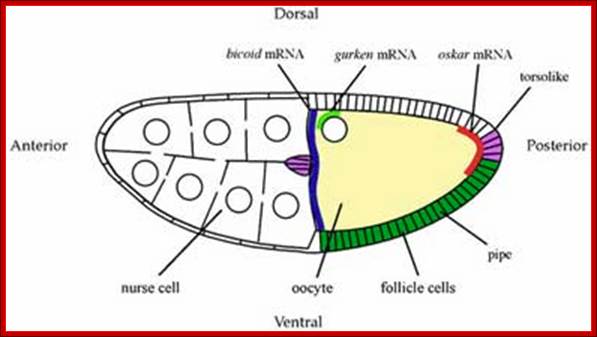
The oocyte in fact is physically connected to nurse cells at the anterior end by a huge cytoplasmic bridge called ring canal consisting microtubules and motor proteins, through which maternal components are transferred.� Cytoskeletons form a kind of network and connected to all cells; hence they are called fusome, which facilitate the movement of nutrients, proteins, mRNA other from nursing materials into oocytes.� The components thus transferred are distributed asymmetrically to form a gradient anterior to posterior (A ->P) and dorsal to ventral (D -- >V).
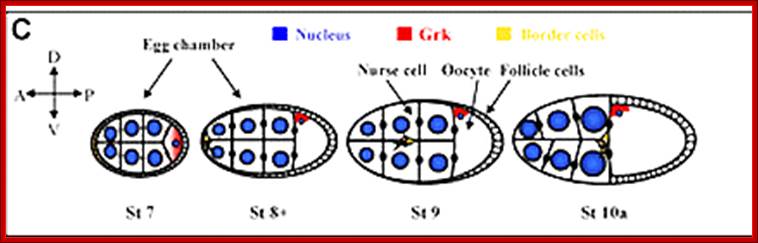
Antero-Dorsal; look at the Gurken mRNA. �Diagram above: A diagram of a stage 10a Drosophila egg chamber showing localized signals that polarize the AP and DV axes of the embryo. Bicoid mRNA blue, Oskar red gurken green pipe expression dark green and torso like expression magenta;�� http://webcache.googleusercontent.com/
����������������������������������� http://webcache.googleusercontent.com/
The oocyte grows in size and possesses all the maternally inherited information in the form of nutrients (yolk), mRNAs and a host of proteins.� Among the host of maternally derived mRNAs, to name few are, Acorn (Torso), Bicoid (bc), Caudal, Hunchback, Nanos, Gurken, Oskar and Telson, (Torso is a membrane receptor), for they have important roles to function.�
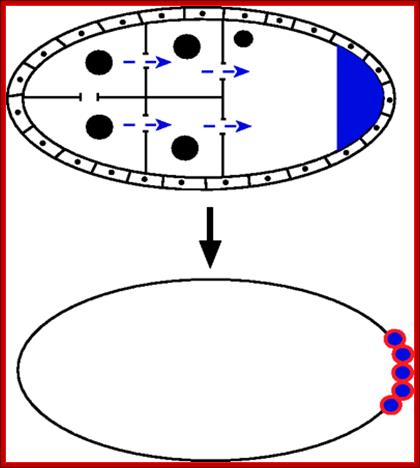
During oogenesis in Drosophila melanogaster, RNAs and proteins are synthesized by the nurse cells.� These products (blue) are transported through cytoplasmic bridges (blue arrows) to the oocyte. They become localized to the posterior of the ooplasm both by molecular anchoring at the posterior of the oocyte, and by posterior-specific translational and transcriptional regulation. This posterior ooplasm is the germplasm, or germ line determinant. During early embryogenesis, cells which inherit the germplasm become the primordial germ cells (PGCs; Red). http://www.devbio.biology.gatech.edu/
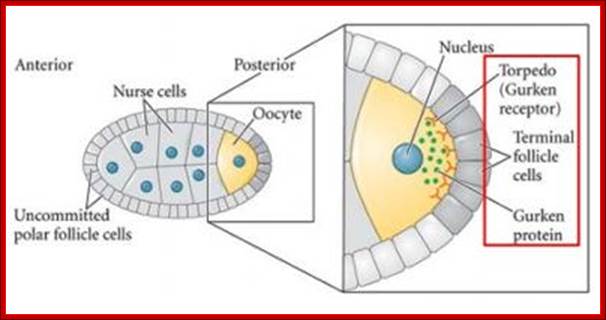
The oocyte moves to posterior region in the egg chamber and the nurse cells fill the anterior portion.� The terminal follicle cells express the torpedo receptor which interacts with Gurken expressed near the oocyte nucleus; Axis specification during development; http://mmg-233-2013-genetics-genomics
������������������������������������������������������������������������������������
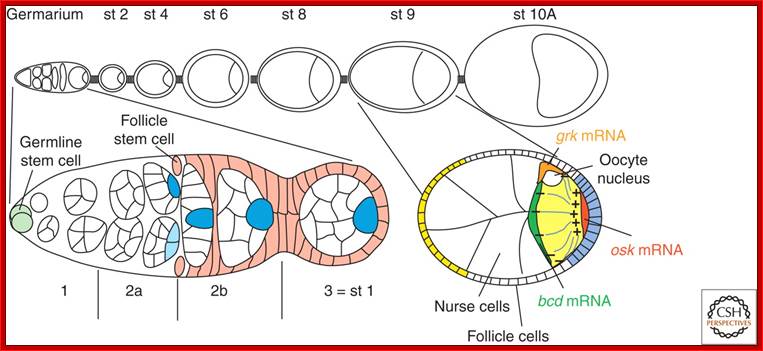
Symmetry Breaking During Drosophila-Oogenesis; �http://www.cshperspectives.net/
The ovariole. Top: schematic drawing of ovariole with the germarium at the anterior tip and egg chambers of increasing age. Bottom: Magnified view of germarium and stage 9 egg chamber Symmetry Breaking During Drosophila-Oogenesis; �
�http://www.cshperspectives.net/
Schematic representation of early oogenesis (A) and stage 9 egg chamber (B). The germline and specified somatic cells are indicated individually. In both panels, anterior is to the left. From-
Lethal (2)giant larvae is required in the follicle cells for formation of the initial AP asymmetry and the oocyte polarity during Drosophila oogenesis; Qi Li, Tanachi Xin, Wenlian Chen, Ming Wei Zhu and Mingfa Li
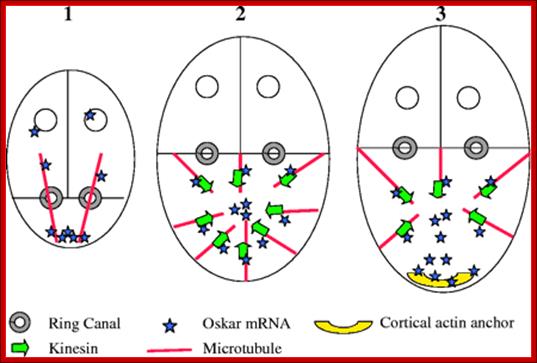
Transport and anchoring of oskar mRNA in Drosophila oogenesis. (1) In early oogenesis (stage 2-6), the MTs extend from the oocyte posterior to the nurse cells. Then, oskar mRNA is transported from the nurse cells, on the MTs, via ring canals, to the posterior cortex of the oocyte. (2) In mid oogenesis (stage 8), there is a re-polarization of the oocyte MTs, the MTOC at the oocyte posterior disassembles, and new MTs are nucleated from most of the oocyte cortex. The plus-end-directed motor kinesin transports oskar mRNA away from the cortex and towards the oocyte interior. (3) Subsequent destabilization of MTs at the oocyte posterior (late stage 8 and early stage 9) uncovers the posterior actin anchor, leading to the entrapment and concentration of oskar mRNA at the posterior pole [adapted from Cha et al. (Cha et al.2004); http://jcs.biologists.org/
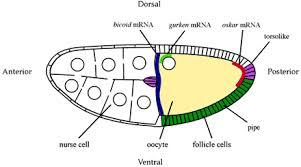
Fixation of Dorso-Ventral axis requires the action of gurken mRNA product, and the same is transported to antero-dorsal, where it is translated, whose product act as a surface protein which interacts with the follicle receptor called Torpedo. �In a series of reaction and interactions DPP is produced this form a gradient from Dorsal to ventral.� The Dpp prevents the dorsal (ds) function so, Dpp fixes the dorsal position determination, while the ds determine the ventral position of the embryo.
The intracellular localization of mRNAs is a general mechanism to target proteins to the regions of a cell where they are required, and plays an important role in the polarization of many cell types. A striking example of this phenomenon is provided by the Drosophila oocyte, where the localization of bicoid, oskar, and gurken mRNAs to three distinct positions within the cell determines the polarity of the anterior-posterior and dorsal-ventral axes of the embryo. Using the powerful genetics of Drosophila, we are using a combination of molecular, cell-biological and genetic techniques to investigate the mechanism of mRNA localization. In addition, we are studying how the two axes of the oocyte become polarized to define the destination of these transcripts, in order to understand the origin of polarity in Drosophila development; http://www2.gurdon.cam.ac.uk/
Transportation of mRNAs and protein particles:
Though most of the mRNAs, are transported from the anterior end through cytoplasmic bridge (ring canal), as ribonucleo-protein complexes. The Bicoids are placed in the anterior position, Gurken in antero-dorsal region, Nanos and Oskar at posterior region.� This specific directional transport of mRNA is aided by specific 3�UTR sequences bound proteins. They in turn bind to motor proteins found at the (-) ends of MTs. An array of microtubules and their associated motor proteins such as dyneins and kinesins act as cargo motors for transport in (-) to (+) direction of the MTs.� Thus, each of the mRNA populations is specified to its destination.��� Ultimately, they are transported to their specific destinations along MT tracks.� Thus, a gradient of mRNAs develops; this happens, even before fertilization.
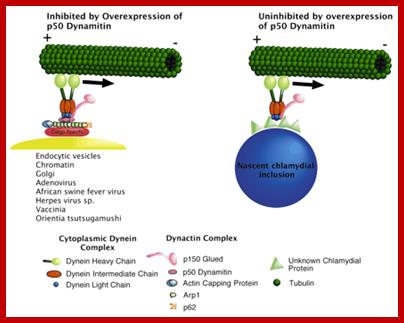
Model of molecular components in chlamydial migration. Model adapted from Hirokawa (Hirokawa, 1998) shows the differences between classical dynein-dynactin-dependent microtubule movement and that which we propose to be used by C. trachomatis. http://jcs.biologists.org/
Microtubules and motor proteins; Kinesins and cytoplasmic dyneins are microtubule motor proteins that generally move in opposite directions along a microtubule (A). These proteins (drawn here to scale) are complexes composed of two identical heavy chains.
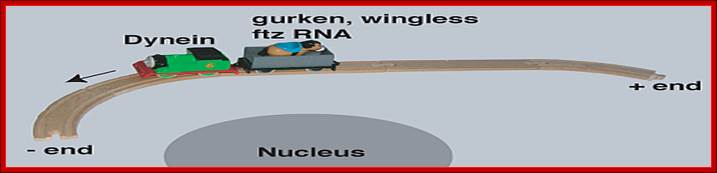
The cytoskeleton tracts are used for the transport of specific mRNAs, such as Gurken, wingless and ftz; dynein is the motor proteins. http://www.bioch.ox.ac.uk/

Intracellular mRNA transport illustrated with a train analogy. The train tracks represent the polarized microtubule cytoskeleton, engines represent molecular motors (dynein and kinesin) and the passengers represent different RNA cargo (gurken, wingless ftz and oskar mRNA); The microtubules act as tracts for the transport of Oscar mRNA, kinesin is the motor protein. http://www.bioch.ox.ac.uk/
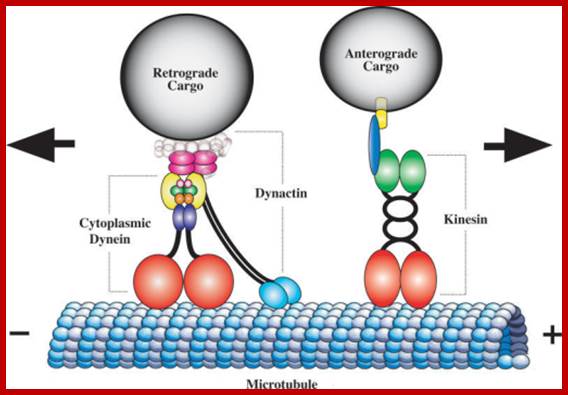
The genetics of axonal transport and axonal transport disorders.: Cytoplasmic Dynein and Kinesin Power Axonal TransportSchematic diagram of the microtubule motor proteins cytoplasmic dynein and kinesin. Cytoplasmic dynein transports cargo in the retrograde direction toward the minus ends of microtubules whereas kinesin transports cargo in the anterograde direction toward the plus ends. Cytoplasmic dynein is a large multimeric protein complex comprising two heavy chain subunits (red) that possess microtubule binding and ATPase activity, two intermediate chains (yellow), two light intermediate chains (indigo), and an assortment of light chains (light pink, green, orange) (reviewed in [7]). Dynactin, a large multisubunit protein complex of comparable size to cytoplasmic dynein, is proposed to link the dynein motor to cargo and/or increases its processivity. The largest dynactin subunit, p150Glued (turquoise), forms an elongated dimer that interacts with the dynein intermediate chain and binds to microtubules via a highly conserved CAP-Gly motif at the tip of globular heads. The dynactin subunit p50 (dark pink) occupies a central position linking p150Glued to cargo. The conventional kinesin holoenzyme, also known as kinesin-1, is a hetero-tetramer comprising two Khc subunits (red) with microtubule binding and ATPase domains, a central coiled stalk, and a tail domain that interacts with two Klc subunits (green). Klcs may mediate cargo-binding via an intermediate scaffold protein (blue) that binds a cargo transmembrane protein (yellow). Duncan JE, Goldstein LS - PLoS Genet. (2006);https://openi.nlm.nih.gov
Molecular Motors; Most forms of movement in the living world are powered by tiny protein machines known as molecular motors. Among the best known are motors that use sophisticated intramolecular amplification mechanisms to take nanometer steps along protein tracks in the cytoplasm. These motors transport a wide variety of cargo, power cell locomotion, drive cell division and, when combined in large ensembles, allow organisms to move. Motor defects can lead to severe diseases or may even be lethal. Basic principles of motor design and mechanism have now been derived, and an understanding of their complex cellular roles is emerging. Manfred Schliwa & G�nther Woehlkehttp://www.nature.com/
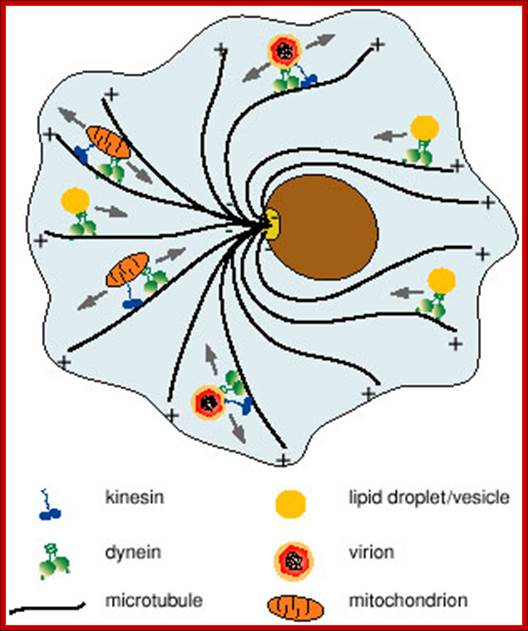
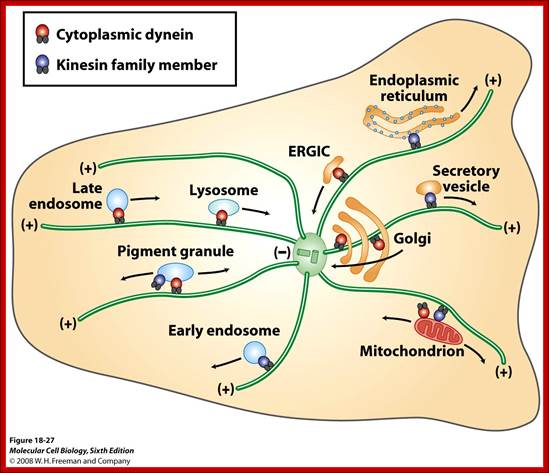
This is accomplished by a set of specialized motor proteins that haul the cargoes to where they are needed by stepping along a network of intracellular filaments (microtubules and actin filaments) (see figure 1). Examples of transported cargoes include mRNA particles, mitochondria, endosomes and phagosomes, various vesicles and lipid droplets. Cargoes are not always benign; pathogens like virus particles hijack the molecular motors of the cell that in turn deliver them to the nucleus where they can use the cellular machinery to replicate. Given the wide variety of cargoes that need to be transported, any failure of the transport machinery can affect many cellular processes and lead to disease. Yet, the molecular mechanisms relating failure of motor regulation to disease remain largely unknown. Microtubules are also used as rails for the transport of a variety of cellular structures including RNA, proteins, ER, mitochondria, lipid droplets and small vesicles. http://chaos.utexas.edu/
Oocyte is transcriptionally inactive at this stage.�
The egg cell membrane is also studded with a variety of receptor proteins, for example Toll, which is found all around the egg surface for signal transduction; the mechanism is not fully understood?� The oocytes have been known to be activated by calcium mediated signal transduction pathways. The positioning of these mRNAs provides information for anterior-posterior and dorsal-ventral polarity.
An overview of the developmental fates from the fertilized egg to segmentation, time scale is shown; Genetic Control of Segmentation; http://people.ucalgary.ca/
With male copulation and deposition of sperms in receptacles of females, perhaps immediately, triggers the deposition of the mature oocyte into uterus; it is at this point, the oocyte is activated through calcium mediated signal transduction mediated kinase activity.
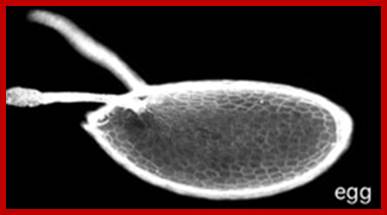
Mature egg; http://biology.mcgill.ca/
The oocyte 2n nucleus halted at first meiotic metaphase, now is triggered to go through meiosis to produce four nuclei and three of them are budded off into peri-vitteline space as polar bodies.� This event can happen even at the time of entry of sperm.�
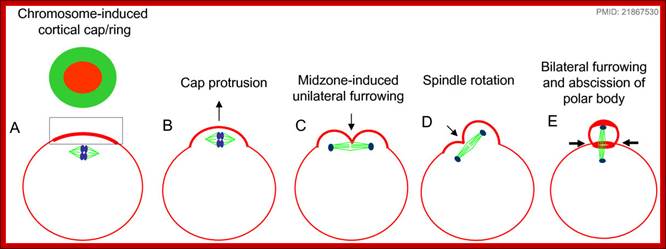
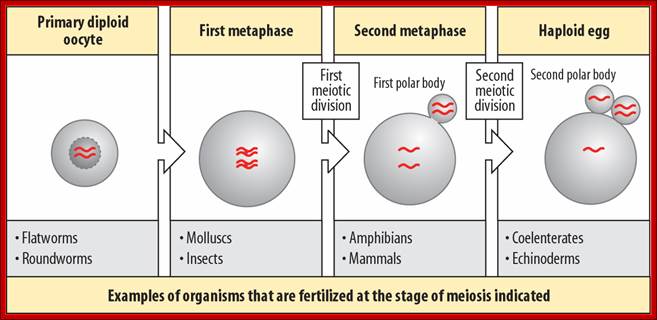
http://www.mun.ca/
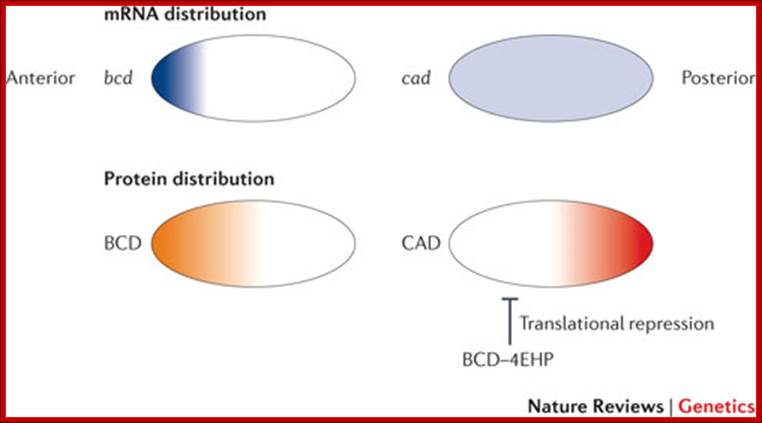
In the early Drosophila melanogaster embryo, translational regulation of maternal mRNAs establishes anterior-to-posterior pattern, and this is usually coupled with mRNA localization. After localization, mRNAs are translated in the region where they are concentrated and are repressed elsewhere. For example, bicoid (bcd) is transcribed maternally in the nurse cells and is transferred into the oocyte during late oogenesis, and its mRNA becomes distributed in a steep anterior-to-posterior gradient, leading to an anterior-to-posterior distribution of BCD protein, which is a transcription factor113. Somatic nuclei at the periphery of the embryo experience different concentrations of BCD and activate different sets of downstream genes, depending on the concentration of BCD that they receive, thus resulting in further patterning. Translation of caudal (cad) is repressed by the recruitment of 4EBP by BCD, resulting in a posterior localization of CAD protein. Translation of oskar (osk) and nanos (nos), which encode proteins that are essential for posterior patterning and germ cell development, is restricted to the posterior pole of the oocyte and syncytial embryo through coupled mRNA localization and translational control. 4EHP, eIF4E homologous protein; Nature Reviews, Jian Kong & Paul Lasko.
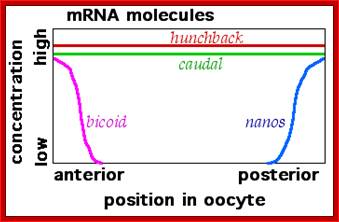
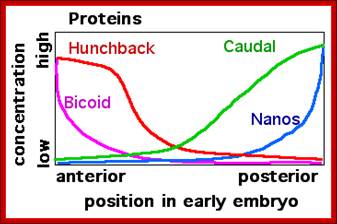
Left; mRNA distribution- right; distribution of proteins; http://en.wikipedia.org/
Sperms enter the egg through anterior pore.� Sperms of drosophila are really large and probably larger than human sperm.� They fuse with egg nucleus; an act of fertilization.
Fertilization triggers an avalanche of molecular activities.� All the informational molecules stored in the oocytes before fertilization get activated and initiate the process of development.� Such informational input at pre-fertilization stage in the egg cell is pervasive in all organisms, probably without any exception. The zygotic cell grows 300 or more times its original size in a very short period.� The 2n nucleus divides and redivides eight times without cytokinesis, leading to free nuclear condition (coenocytic condition), which is also called syncytial stage.�
Early embryogenesis: Drosophila eggs have a great deal of yolk at the center of the egg. The first 7 nuclear divisions occur in the center of the egg, and then most nuclei migrate out to the periphery. There is no cell division until cell cycle 14, so the early Drosophila embryo is a syncytium. This means that molecules can freely diffuse. The first cell divisions are very rapid, as the first occurs 20 minutes after fertilization and the subsequent divisions follow at 10-minute intervals. Each of the first 13 divisions is synchronous. http://www.zoology.ubc.ca/
At this stage polar granules, a complex of proteins, at the posterior end of the egg, generate polar cells; this requires Oskar and Telson activity. The follicle cells located at the posterior end provide signal information for the polar cells to develop into germ line cells. The germ line cells determine the sex of the animal, which depends upon inherent sex chromosomes. �Similarly, the anterior end is also defined by the formation of acorn mediated by the interaction between torso receptor and interactive follicle cells at the anterior position
Combination of 2A+XX determines the female sex and 2A+XY produce male animals.� In drosophila sex determination depends on the genic balance between the autosomes and sex chromosomes unlike higher organism where X-chromosomes determine female and Y-chromosomes determine male sex. Yet animals with 2A+X develop into males, but without the Y the insect remain sterile; which suggests that Y chromosomes have contributions to provide for the fertility of the insect.
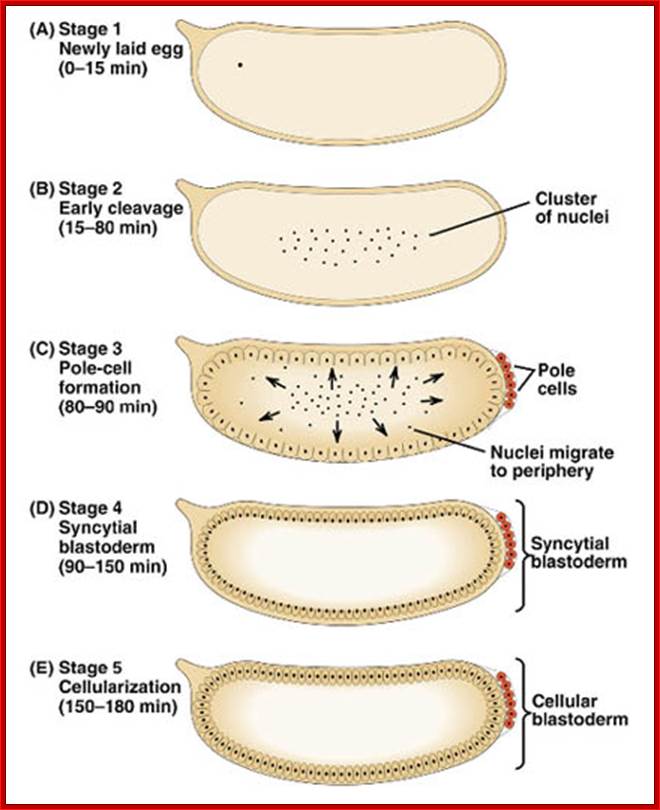
Time lapse from the time of fertilization to cellular blastoderm is 180-200 minutes.� At seventh nuclear division (about 90 minutes) nuclei are still in the central region, but diffused, and generate polar-plasm.� By about 150 minutes, after 9th nuclear division (512 nuclei), nuclei start migrating towards periphery; the stage is called syncytial blastoderm.� The directional transport of nuclei is perhaps aided by cytoskeletons with motor proteins. As they are reaching their target, they undergo two more divisions (10th division and 1024 nuclei).� All the moving nuclei are totipotent, but with reaching the peripheral cortex that is filled with cytoskeleton structures; the nuclei get signals from maternal components generated in anterior-posterior axis and dorsal-ventral axis and many more of them make them to acquire competence to transcribe specific genes genetically determined to certain cell-fate.
�
![]()
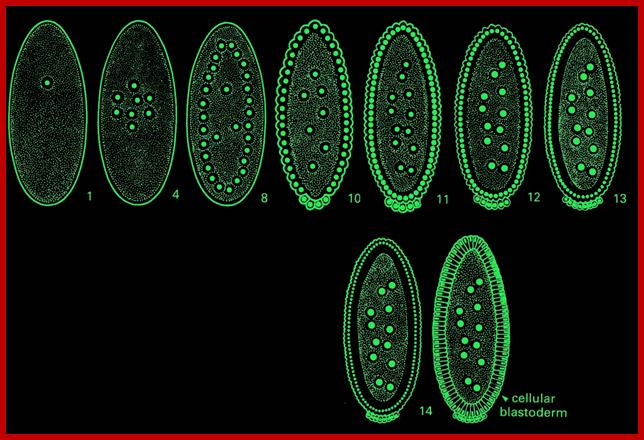
http://celldynamics.org/
![]()
Von Dassow and Shubiger 1994;A bud develops around each nucleus with bud protrusion at telophase,,.� This prevent MTs of other to fuse, this happens in syncytial stage of development. Psedo cleavage develop between each nuclei with plasmamebrane drawn down between each of the nucleus- called pseudo cleavage; Cell Dynamics, Univ. of Washington (Sullivan et al 1990; Postner et al 1992).Foe, Odell; http://celldynamics.org/
The syncytial Drosophila embryo contains nuclei in a single layer at the periphery incompletely surrounded by polarized plasma membrane domains. http://www.iiserpune.ac.in/
Nuclei migrate to cell periphery and cellular formation initiates and polar cells differentiate. http://www.discoveryandinnovation.com
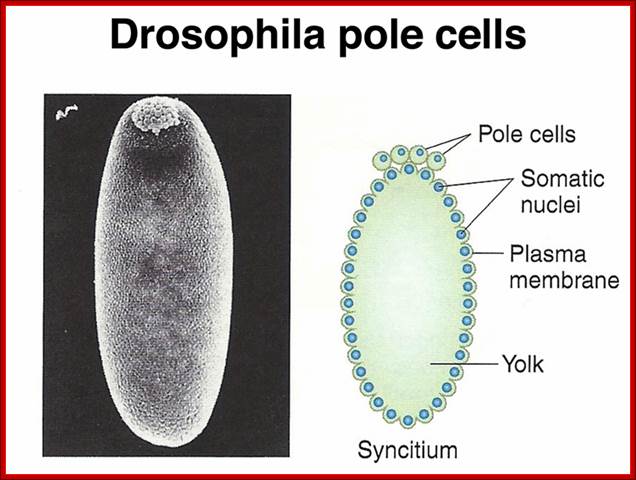
Pole cells, the future germ line, are set aside early in development. The image below shows a scanning electron micrograph of an early embryo with the posterior pole at the top of the image. The pole cells incorporate polar granules, particles of RNA and protein that are deposited in the egg by the mother during oogenesis.
����������������������������������������������������������
Antero-posterior Proteins and mRNAs located in antero-posterior position control positional information and dorsal to ventral gradient determine tissue differentiation like mesoderm, neural ectoderm and dorsal ectoderm.� At 13th division (8192 nuclei) and 14th division (16386 nuclei) the blastoderm is still in syncytial stage enmeshed in cytoskeleton elements. At least 6000 of the nuclei at the periphery, the cell membrane starts invaginating around each of the nuclei and encloses the nucleus and peripheral ooplasm containing informational molecules, into cells. �Thus, each of the cells acquires positional information and now they become competent to interact with one another through signal transduction. In this process the nuclei assume ellipsoid shape. �It is here, the totipotent nucleus acquires multi potent or monopotent characters depending on their position and interaction between the interacting cells.� By 195-200 minutes (after fertilization) the embryo becomes cellular blastoderm.
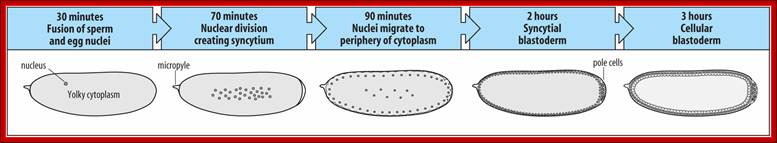
Drosophila melanogaster: early embryogenesis; The Drosophila egg is the shape of a sausage. It has a micropyle at the anterior end (site of sperm entry). With fertilization, the fusion of nuclei is followed by rapid mitotic divisions (90 minutes) and no cleavage. A syncytium is formed (many nuclei/common cytoplasm). After nine divisions, nuclei move to the periphery to form the syncytial blastoderm (2 hours). Drosophila melanogaster: embryogenesis-By 13 mitoses the membranes sprout to surround the nuclei to form cells (cellular blastoderm). ~15 cells at posterior (pole cells) are sequestered and become the germline. During first ~3 hrs. large molecules such as proteins can move between nuclei until the cellularization occurs. Single layer of cells give rise to all tissues. Gastrulation starts at ~3 hrs. 1) Mesoderm forms from ventral tissue. 2) Midgut from endoderm at the anterior and posterior ends. 3) Ectoderm remains on outside. http://www.mun.ca/
The embryo, now, is called blastula, at which the cells become programmed for certain development and the embryo launches a decisive and genetically programmed cascade of events that determines each and every structure of the fly.� This also leads to tissue development of different kinds.� At this stage of cellular blastoderm, a host of proteins in the form of transcriptional factors, activators, co activators or repressors, act in a combinatorial fashion.� Each of the genes whose promoters are exposed, contain elaborate regulatory elements in the form of high affinity, low affinity sites, in addition to their regular promoter elements; they may also contain repressor, insulator and other regulatory sites.
This intricate and complex process of gene activation and repression is so profound, it may take, for scientists, another century or so to decipher and delineate the complexity, created and fixed by three billion years of genetic engineering, natural selection and evolution at molecular level.
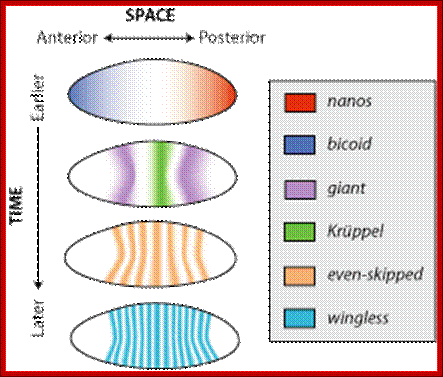
�The factors that form gradient in anterior to posterior axis and dorsal to ventral and those present at the extreme poles determine and differentiate cells into different cell types to tissues then into organs.� To cite few of the maternal factors localized are: Anterior end: Exuperantia, Swallow, Staufen, Bicoid, Hunchback and Caudel; Posterior end: Cappuccino, Staufen, Oskar, Nanos, Vasa, Tudor and few others; Ventral side: Pipe, Nudel, Dorsal, cactus and Spatzle; Dorsal side: Gurken and others in the dorsal side.
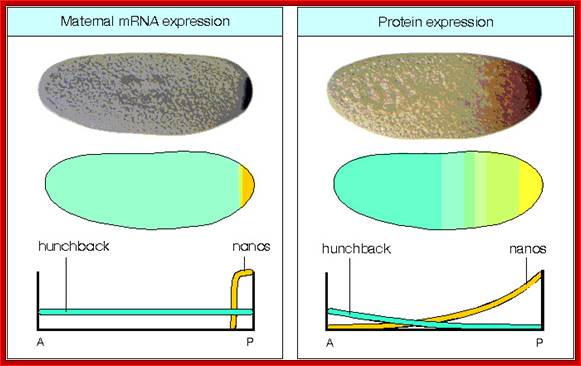
Dorsal gene mRNAs and hunchback maternal mRNAs are uniformly distributed. These are the core components of the oocyte for its developmental programme.� Among them Bicoid (bcd), nanos (nos) and hunchback (hb) act as morphogens.
Expression of different genes mostly as transcriptional factors, they are distributed locally, which ultimately control body pattern. http://www.zoology.ubc.ca
Bicoid evolution; http://scienceblogs.com/
Nanos on translation produce translational inhibitors at the posterior end. Important signal transducers are Toll at the ventral surface Torso at the terminal ends.� Bicoid and hunchback act as transcriptional factors. Among the 30 maternal factors involved, only few of them are cited here. There are many others, but this will suffice for our understanding of early developmental programmes. Effects of each of these genes have been identified by mutational phenotypes.� �This is the most fascinating and yet the most unknown territory; whatever that is not known becomes fascinating to scientist.
Anterior-Posterior Axis Determination:
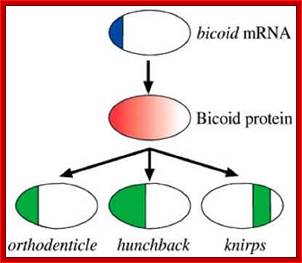
As the oocyte develops a very high concentration of maternal Bicoid (Bcd) mRNA is localized at the anterior end.� Hunchback (Hb) is another maternal mRNA, but it is distributed all along the length of the oocyte.� Nanos (Nos) mRNAs are localized at the posterior end and found in high concentration.� At anterior tip one finds Acron and at the posterior terminal region Telson.� All the said and other many more are maternal products.� Each of them on translation produces specific proteins and they have their own specific function.
Now the localized mRNAs start translation and generate their proteins.� As the Bicoid mRNA is found at high concentration, greater amount of Bicoid proteins are produced at the anterior end, but the proteins diffuse along the axis and generate a gradient.� Bicoid proteins are helix turn helix proteins and act as activators of transcription and bind to promoters of those genes that contain sequences such as TCTAATCC.� Difference in two bases can make high affinity binding site to bcd (or difference in four bases) as low affinity sites.
Bicoidal (bcd) proteins as monomers activate hb, genes in the anterior end, but they by binding to 3�UTR region of caudal mRNA inhibit translation of caudal protein. The bcd proteins, with helix turn helix motif act on other genes also involved in the activation of Pair rule genes. Note both caudal and hb mRNAs are uniformly disturbed as gradient all along the antero-posterior ends.
Maternal gene expression; http://www.mun.ca/
Nanos mRNA translation results in the accumulation of Nanos proteins at the posterior and they also diffuse towards the anterior end; they too form a gradient.� Nanos have greater affinity to the 3�UTR region of Hunchback mRNAs.
Maternal Hb mRNAs which are distributed uniformly from A-P axis, but their translation in the posterior region is inhibited, because of the binding of Nanos to their 3�UTR regions.� The 3�UTR region contain stem loop structure. This leads to deadenylation and degradation hunchback mRNAs in the posterior region.� Few more proteins assist these events.� But hunchback mRNAs found at the anterior end are translated to generate their proteins.� This differential translation creates a gradient from anterior to posterior and visa versa.
All the said protein products except Nanos are Transcriptional regulators, which bind to their specific promoter elements in sequence specific manner.� Their action on gene activation is concentration dependent.
Dorsal-Ventral Axis Fixation:
The ventral side is responsible for the development of mesoderm and including neurogenic ectoderm, so determining the ventral side of the oocyte is important.
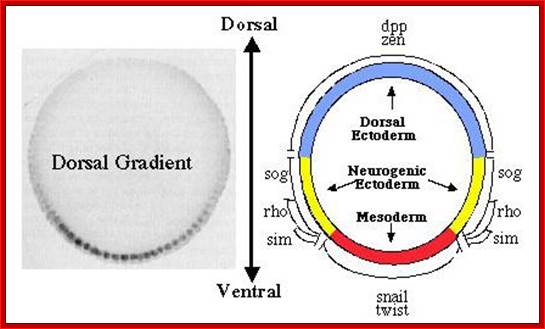
The Dorsal nuclear concentration gradient. The panel on the left shows a cross section through a blastoderm embryo stained with antibodies against Dorsal to reveal the concentration gradient. The panel on the right shows a fate map of the blastoderm embryo, illustrating the domains of expression of twi, sna, and rho, which are activated by Dorsal, and of dppand Zen, which are repressed by dorsal; http://www.biochemistry.ucla.edu/
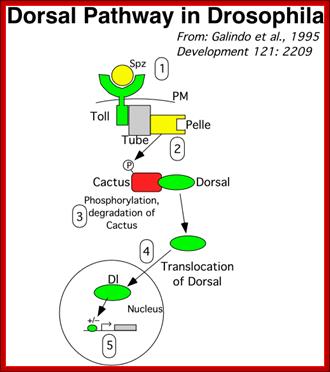
Lecture 21; http:// www.uvm.edu
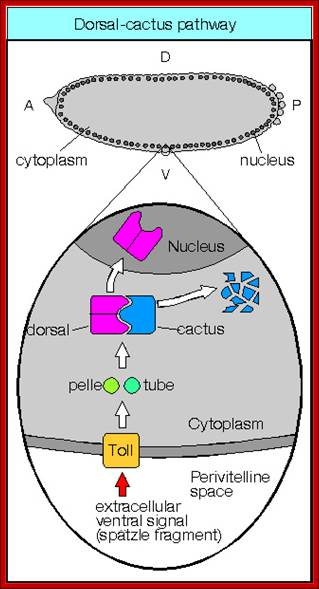
Activation of Toll receptors by sp�tzle at ventral side of the oocyte leads to a cascade effect leading to differentiation of ventral side of the embryo. Dorso-Ventral patterning; ;http://www.mindcreators.com/
Dorsal Group of Genes:
- Mutations in any of about 11 genes would produce a fly that produced defective embryos that exhibited dorsalized features, thus the naming of the pathway (Notes on the genes).
- A. Easter, Produces the D-V asymmetry in ligand with a higher concentration of Ea in ventral part of embryo,
- B. Sp�tzle, the asymmetrically accumulated ligand,
- C. Toll, a transmembrane receptor,
- D. Tube, a mysterious protein that is needed for coupling of the receptor to the effector kinase,
- E. Pelle, the effector protein kinase,
- F. Cactus, a cytoplasmic 'sequesterase' that binds Dl.
- G. Dorsal, a transcription factor,
- H. Various target genes give rise to the differences between dorsal and ventral cell type (target gene examples):
- Various target genes are expressed in the ventral portion of the embryo
- Disruption of this regulation can cause flies to develop 'upside-down,' in a sense, with dorsal-localized cells exhibiting ventral-cell-like features.
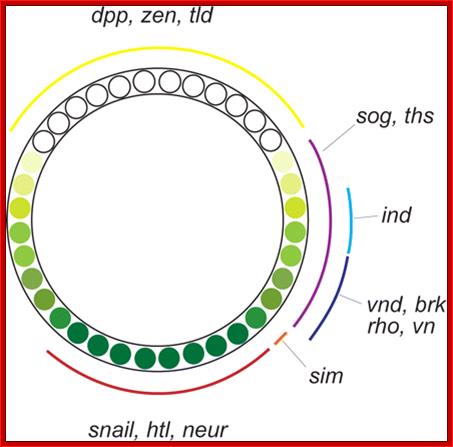
The Dorsal nuclear gradient generates diverse thresholds of gene expression. The Dorsal gradient regulates 60�70 target genes in a concentration-dependent manner across the DV axis of the early embryo. The expression domains of these genes are depicted on a diagram representing a cross-section through an early embryo. Filled green circles represent high levels of nuclear Dorsal protein, and shaded green and yellow circles represent intermediate and low levels, respectively. At least six different expression patterns have been identified. High levels of Dorsal activate Type 1 genes such assnail in the presumptive mesoderm. Type 2 genes such as rho and vnd are activated by intermediate levels of Dorsal gradient in ventral regions of the presumptive neurogenic ectoderm. In addition, type 1A and 2A expression profiles, represented by sim and ind, depend on Notch and EGF signaling and high and intermediate levels of the Dorsal gradient, respectively. Two type 3 expression patterns, those represented by sog and zen, are generated by the lowest levels of the gradient. The same low levels that are sufficient to activate sog repress the expression of dpp, zen, and tolloid. Thus, the Dorsal gradient generates three basic transcription responses, and these three thresholds produce a total of six different patterns of gene expression across the DV axis of the early embryo. This figure is adapted from the review by Stathopoulos and Levine; http://www.pnas.org/
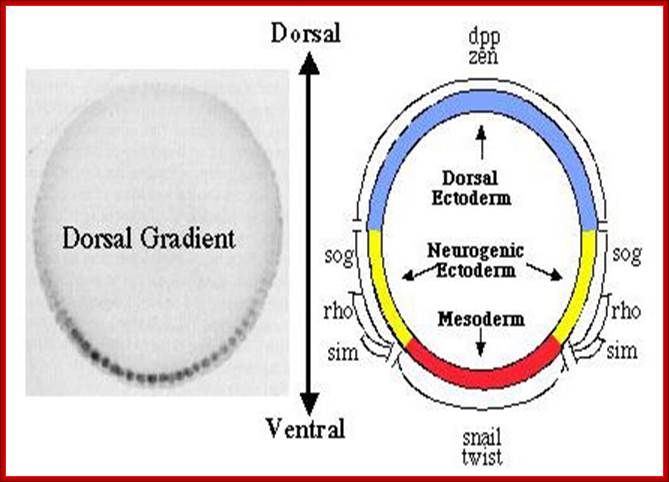
The Dorsal nuclear concentration gradient. The panel on the left shows a cross section through a blastoderm embryo stained with antibodies against Dorsal to reveal the concentration gradient. The panel on the right shows a fate map of the blastoderm embryo, illustrating the domains of expression of twi, sna, and rho, which are activated by Dorsal, and of dppand zen, which are repressed by dorsal; http://www.biochemistry.ucla.edu/
Determination of Ventral surface:
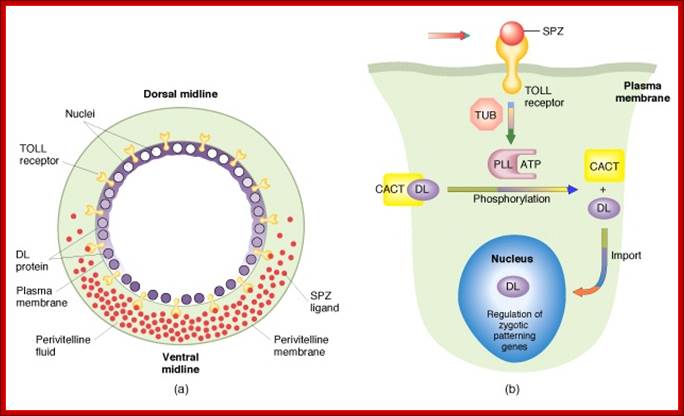
The Toll-Dorsal Pathway in Drosophila melanogaster; By James R: http://www.bio.miami.edu/
Dr. Brian E. Staveley; http://www.mun.ca/biology
Twist Gene expression:
Highest concentration Ds activates Twist genes in most of ventral layers of cells.� The twist gene promoter elements contain two low affinity binding sites for Ds. The ventral most layers of cells are precursors for mesodermal invagination; it is the beginning of gastrulation.
Ventral surface patterning:
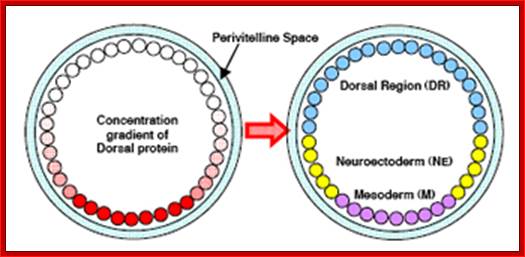
Early genetic assays suggested that the concentration gradient of activated Dorsal establishes between 4 and 7 regions of gene expression along the DV axis. More recently, whole genome assays relying on DNA microarrays suggest that Dorsal may have up to 57 target genes that exhibit localized patterns of gene expression along the DV axis. The function and interaction between at least 17 of the genes are known, however nearly 40 remain to be determined. While many of the Dorsal target genes� functions are not known, the establishment of regions of gene expression by Dorsal and its targets is a fairly well understood process. This section details the formation of regions of gene expression specified by a concentration gradient of activated Dorsal protein. The Dorsal gradient forms a bell-shaped curve with the highest concentration in the ventral-most region of the embryo. Near the maximum of the curve, the gradient of Dorsal is fairly shallow creating a broad region (12-14 nuclei) of maximum dorsal activation in the ventral region of the embryo which later specifies M. Immediately adjacent to the region of high Dorsal, the concentration of Dorsal decreases rapidly forming the region of steepest descent which becomes the boundary between M and NE. The concentration gradient of Dorsal along the lateral regions of the embryo specifies the NE and other boundaries within the NE. Following the descending region of Dorsal, the concentration is low and the gradient becomes shallow allowing the formation of the Dorsal. Review: Stathopoulos, et al. Dev. Biol. (2002). https://engineering.purdue.edu/
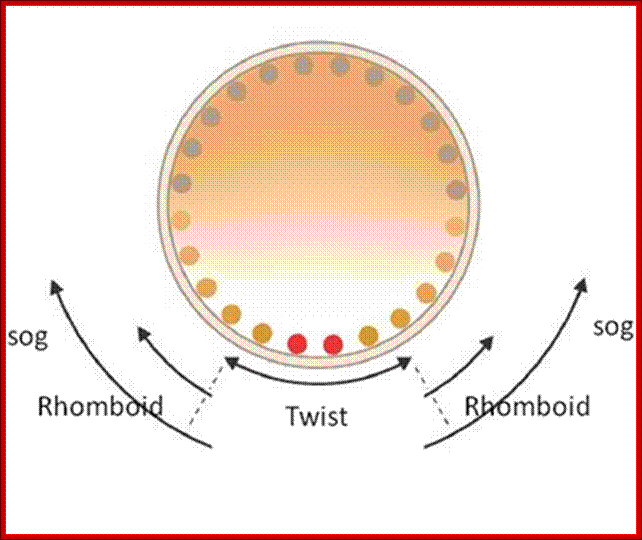
Drosophila embryogenesis; Drosophila embryo with gene expression pattern of Twist, Rhomboid and Sog; Anonymous2069; Drosophila Embryogenesis http://www.chegg.com/
Fertilization also leads to activation of some more protein complexes in the peri vitelline space, involving activation of certain proteases resulting in the release and activation of Spatzle.� Spatzle releasing components are found only at the ventral surface of the egg, and they are not found at the dorsal peri-vitteline space.� This happens only at the ventral surface of the egg.�
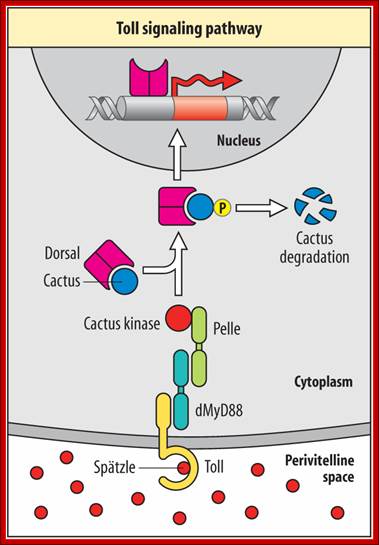
The Toll protein is transmembrane protein; it is another maternal product covers the entire surface of the egg plasma membrane.� This protein acts as a receptor for Spatzle.� When Spatzle, as a ligand binds to Toll receptor; its cytosolic domains become activates Pelle a kinase, and this results in a cascade effect of kinases leading to the phosphorylation of Cactus.� Phosphorylation of cactus releases dorsal Ds (ds) proteins and the cactus gets degraded by proteosome pathway.� This is similar to NFkB (Nuclear Factor kappa B), a transcriptional factor (TF) inhibited by an inhibitor NFIkB. �Phosphorylation of inhibitor protein, releases NFκB.�
As the Dorsal Ds proteins are released from the cactus only at the ventral side, they enter the nuclei found at the ventral side and to some extent they enter the nuclei found at the ventro-lateral side. The ds activates several genes that triggers ventral part of the embryo to differentiate.
Dorsal is transcription factor or an activator of transcription of specific genes.� Activation of such genes depends upon the concentration of factors and their promoters elements, which have binding sites such as high affinity sites and low affinity sites.
Almost simultaneously the Gurken mRNA first found at the posterior end, by its action and interaction with posterior side follicle components determines the posterior part of the embryo.� Then it is transported to dorsal side and positioned at antero-dorsal region near the nucleus.� The translated product of Gurken mRNA acts like TGF α protein.� This protein is secreted out of the zygotic embryo at dorsal side.� This protein binds to a set of receptors called Torpedo found located on the follicle cell membranes facing the dorsal side of the oocyte (Torpedo is somatic product).� This receptor gets activated and leads to a cascade of kinase activities similar to MAP kinase activity.� And the signals are transferred to dorsal region of the developing oocyte. This determines the dorsal side of the oocyte and the embryo.� In the dorsal side activated signal components inhibit the formation of Spatzle.
Dorsal side determination also requires factors called DPP (Deca Penta Plagic) and Zurkenn.� Expression of Dpp and Zn (Zurkenn) is unhindered because of the absence of free dorsal protein.� These are transcribed and translated; they are secreted into ooplasm, which forms a gradient from dorsal to ventral (high to low).� Dpp acts through receptor proteins to activate specific genes required for dorsal structures.� At the same time ds, which enters the nuclei at the ventral side, represses the expression of DPP and Zurkenn.� These two ultimately sets unmistakable dorsal and ventral sides.�
Absence of Toll, Dorsal and Spatzle at the ventral surface makes it as the ventral as dorsal side of the embryo.� Absence of Gurken and their associated products and DPP and Zw at the dorsal side makes the dorsal as the ventral side of the embryo.
Terminal Position Fixation:
Torso is membrane receptor protein found all over the surface of the oocyte membrane.� But the Torso receptor is receptor kinase activated only at Polar Regions.� When activated by external ligands, generated by polar follicle cells; it generates a cascade of kinase activity that leads to the activation of transcriptional factors such as Telson.� Oskar dependent Telson proteins determine the posterior terminal regions at their extremity. The polar granules produced at 7th nuclear division; by 9th division polar cells are produced, where the nuclei are extruded enclosed in a membrane. They act as germ line cells; and determine the sex of the animal depending upon its genetic constitution.� Similarly, Torso and Acron at the anterior end determine the anterior terminal structures.� Fertilization confirms and fixes the dorsal-ventral, anterior-posterior and terminal polarities in place.
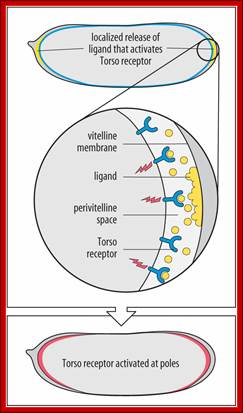
Determination of terminal regions-http://www.mun.ca/biology, Torso is a receptor tyrosine kinase (aka Torso RTK) uniformly located in the oocyte's plasma membrane. Torso Ligand is produced by follicle cells and present only at the termini. Huckebein and Tailless are also involved. Dr. Brian E. Staveley.
Development of Segments:
Proteins such as Bcd and Nanos act as morphogens, by virtue of their positions. They influence the development of positional tissues.� The Bcd is the maternal factor, hunch back is not only maternal factor but also the zygotic factor for its expression is activated by Bcd.� These i.e. Bcd and Hb in turn activate what is popularly called Gap-genes.
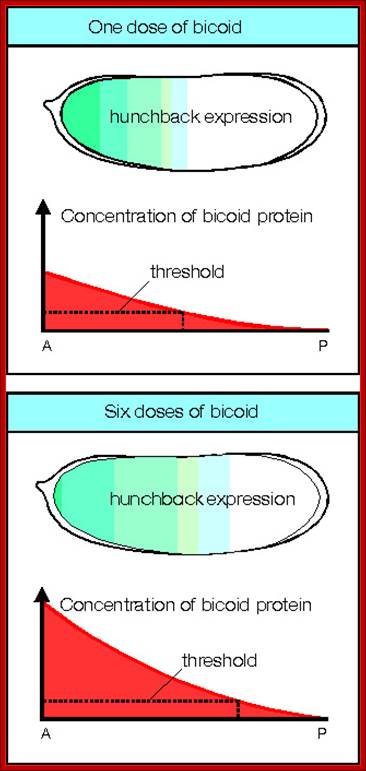
Cascade of Gene expression: maternal bcd protein induces the expression of hb which forms a gradient from anterior to posterior.� The hb together bcd induce the expression of Gap genes. By transgenic experiment it is possible increase bcd from n level to 6n level. The 2n level is normal.� If the bcd level increases more amount of bcd and hb is produced, that can be observed in successive figures. Molecular and Developmental Biology; http://www.mun.ca/
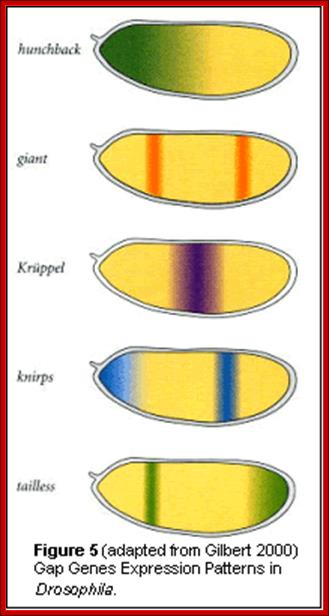
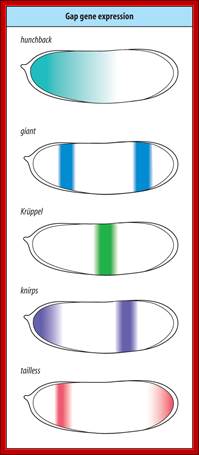
Molecular and developmental Biology; http://www.mun.ca/::� Gap gene and their significance in hunchback / Knirp double mutant; http://www.stolaf.edu/
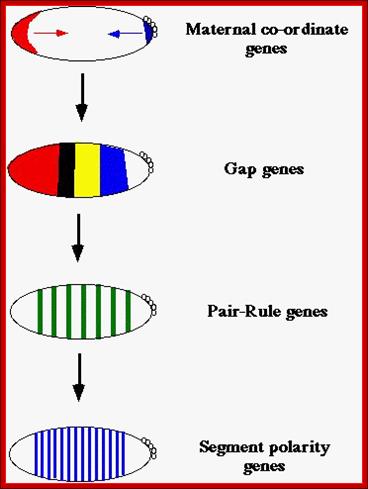
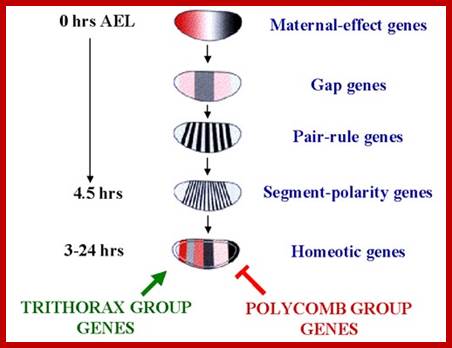
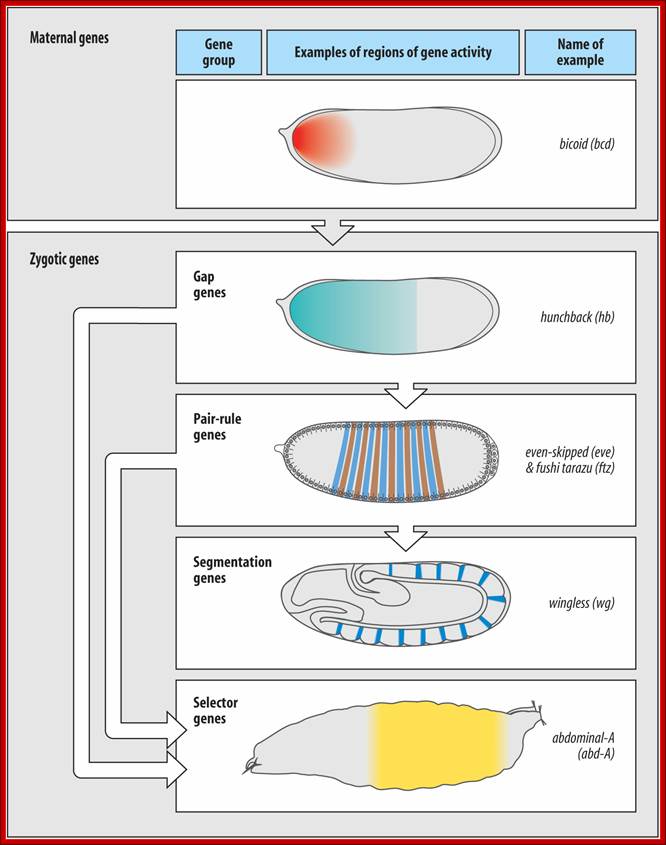
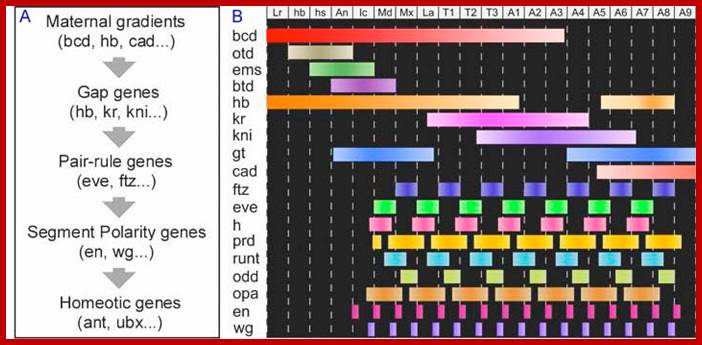
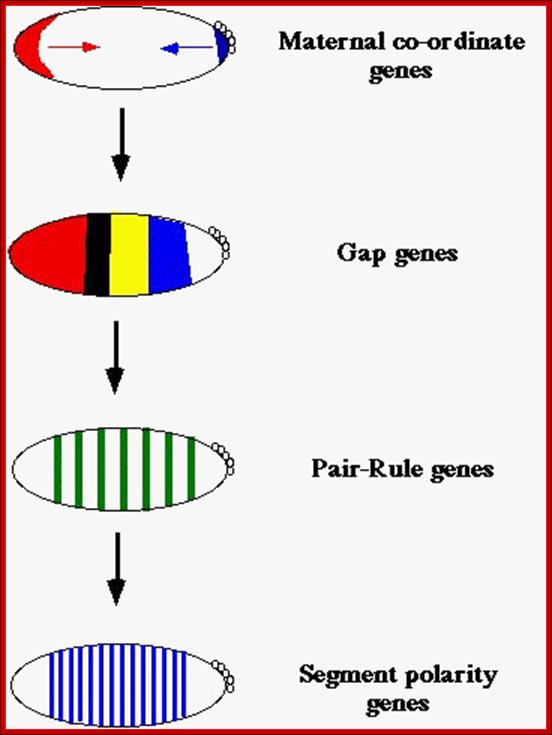
After the maternal genes have defined A-P and D-V axes, the drosophila embryo is organized into segments under the influence of three types of segmentation genes: gap genes, pair-rule genes and segment polarity genes. Gap gene products interact with pair-rule gene products to define Para segments (Carroll and Vavra 1989), areas of the embryo that include the future posterior section of the more anterior segment and the future anterior section of the more posterior segment. Segment polarity genes- define anterior and posterior compartments within each segment.
http://what-when-how.com/
Striped patterns of the pair-rule genes are differentially disrupted in embryos lacking the functions of individual maternal effect, gap, and other pair-rule genes (12-16). These experiments are difficult to interpret, however, because removing the function of a single gene causes pattern shifts and disruptions in many other genes. Several experimental approaches have been used to unravel the mechanisms involved in pair-rule patterning. Regulatory interactions can be inferred from transgenic experiments that ubiquitously mis-express segmentation genes under the control of a heat shock inducible promoter. Then short heat pulses are used and the timing of pattern disruptions carefully monitored, it is possible to determine if a gene directly affects the expression of a given target or if intermediary genes must be expressed to induce the response.� Ectopic expression is also possible using enhancers that activate position-specific expression during early development . Initially, promoter truncation and P-element transformation assays have identified two classes of cis-regulatory regions (enhancers) that are important for pair-rule patterning in the blastoderm. Enhancers of the first class specify the placement of individual stripes by responding primarily to gradients of maternal effect and gap proteins. Enhancers of the second class specify complete patterns of seven stripes but are active only after the first striped patterns are established. These enhancers respond to transcriptional cues provided by the pair-rule genes themselves and are thus important for mechanisms that maintain and refine initial patterns.
�������������
Gap genes and their significance on Hunchback.Knirp double mutants; http://www.stolaf.edu/
Gap gene products in turn activate pair rule genes and pair rule genes express segment polarity genes. Then selector genes take over the complete development of the larval body structure.� All the said gene products are transcriptional factors; so it is a cascade expression of TFs, one set activating another set, and the other activating another set, so on. Even in the larvae, adult insect structures are precisely and precociously formed in the form of �imaginal discs.� Once the larva turns into pupa and as the pupa transforms into insect; with all its adult structures.
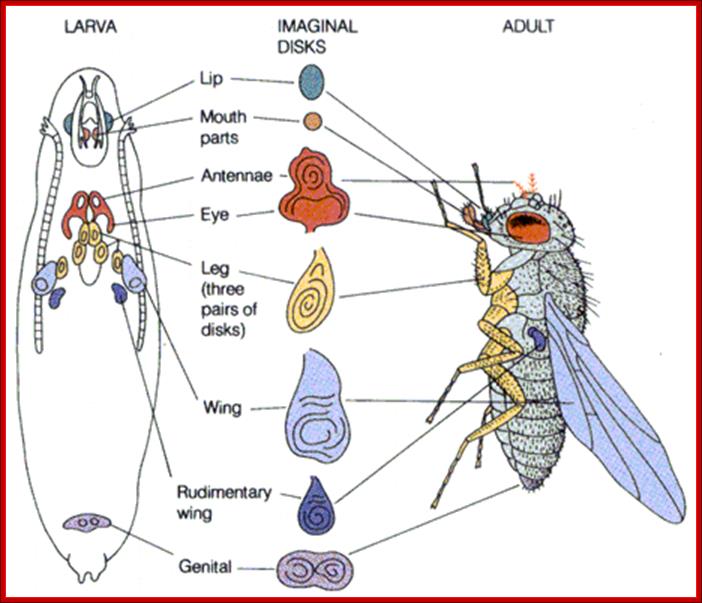
"Once a caterpillar has disintegrated all of its tissues except for the imaginal discs, those discs use the protein-rich soup all around them to fuel the rapid cell division required to form the wings, antennae, legs, eyes, genitals and all the other features of an adult butterfly or moth. The imaginal disc for a fruit fly�s wing, for example, might begin with only 50 cells and increase to more than 50,000 cells by the end of metamorphosis. �Scientific American�.
Imaginal discs are found in insect larvae at third instar stage; during pupal transformation they generate organs such as wings, legs antennae and other adult structures.� The fascinating part of imaginal disc function is, during pupal transformation many larval structure breakdown and each of the discs everts and develop, with central portion become distal part of the organs that develop.� In the larval stage the disc looks like undifferentiated mass of cells but programmed for development of specific structures.� Such discs can be taken out and they can be cultured for many generations and if such disc cells are implanted, they develop their predetermined structure.� It is a rare feature where preprogrammed cells can be grown and stimulate produces their programmed organ.
Maternal genes activate Gap genes.
(Anterior, Terminal, Ds and Posterior)
![]()
Gap genes activate Pair Rule genes.
(Gap genes-Knirps,Tailless,Giant Kruppel); Pair rule-
Genes-Even-skipped and
![]() Fushi
tarazu),
Fushi
tarazu),
Pair Rule genes activate Segment Polarity genes.
![]() Fushi Tarazu Ftz
and Even skipped Eve
Fushi Tarazu Ftz
and Even skipped Eve
Segment Polarity genes activate and facilitate Selector (Homeotic) gene. (engrailed, En and wingless wg, hedgehog, gooseberry, Armadillo arm),
Expression of Dorsal regulated Genes:
Receptor Toll activated by Spatzle, activate pelle/tube kinases which phosphorylate cactus bound to dorsal.� Once phosphorylated dorsal is freed and the cactus undergoes proteasome mediated proteolysis. �Though ds proteins sequestered, they are distributed all over the cytoplasm. Dorsals are released only in the ventral region, because Spatzle activate toll receptor at ventral surface; toll is a protein kinase.� The Ds is a regulatory protein; it enters the nuclei in Ventral region and regulates the expression of Ds specific genes.� Highest concentration of Ds is found at the ventral most layer of 18 cells; the concentration of Ds tapers of in the upper region of cells and very low in the lateral regions.
Twist gene enhancer elements:
- (ds)lARE---(ds)lARE--------------TATA--InR----DPE----
Snail gene enhancer elements:
---dslRE---dslRE-----ds.lRE---ds.lRE�tw---tw---TATA---InR---DPE----
Note- RE refers to response elements, L=low affinity, H=high affinity.
The proteins Ds with Twist activate the expression of Snail genes in the same layer of cells because its promoter elements contain binding sites for both Ds and Twist.� The role of Ds is to recruit acetylase and chromatin remodeling complexes.� The Twist stimulates transcription by interacting with mediator complexes and PIC. �
Here high affinity means the protein motifs bind to DNA sequences perfectly; low affinity means the DNA sequences are not perfect consensus elements; to bind to all of them Ds is required in high concentration. �Low affinity enhancers require high concentration of factors and High affinity binding elements requires lower number of factors. �Again the number of binding sites and the combination of them also contributes to the efficiency of transcription. Dorsal proteins bind to respective sites as monomer, but because of their protein-protein interactive nature they interact with other transcriptional complexes to activate transcription.
Combination of Ds and twist activate several genes synergistically in the ventral region.� And snail represses rhomboid and so go genes in ventral region.
Rhomboid gene expression:
Dorsal factors also activate rhomboid and sogo genes in the ventral region. But the snail proteins expressed at ventral most regions repress rhomboid genes and sogo genes for they have snail binding elements, but they are expressed in response to lower concentration of Ds but without snail proteins in ventro-lateral region; this is because Twist also binds to its promoter elements of them; thus, the expression of Rhomboid is facilitated. At moderate concentration of Ds at slightly dorsal to the 18 cells ventral stretch, the genes present are expressed for they contain the most high affinity binding sites for ds factors and 3 lARE and also contain binding sites for Twist.
Rhomboid enhancer elements:
--- lRE---snRE----hRE--tw--lRE--snRE--lRE�TATA�InR�DPE-
Sogo gene expression:
At low concentration of Ds factors at upper region of the ventro-lateral stretch, Ds activates Sogo genes in cells for their enhancer region consists of four high (response Elements (RE).� The sogo genes have enhancer elements with in the coding region. As enhancer region contains hRE elements for Ds, low concentration is enough for the activation of sogo genes.� Sogo product is a secretory protein; this creates a gradient opposed to Dpp gradient from dorsal to ventral.
Sogo enhancer elements:
----+1>-------hRE�hRE�hRE�hRE�TATA�InR---
Expression of Twist and Snail by ventral most layers of cells lead to the development of mesoderm layer.� Rhomboid expressed in ventro-lateral layer, leads to the development of ventral neurogenic ectoderm.�
Expression of Gap genes:
At the time of cellularization or little before, the Bicoid factors generated at the anterior position activate Hb genes in gradient fashion, highest at the anterior end and tapering at the mid region.� Bicoids are transcriptional activators of Hb genes, and they bind to sequence 5�TCTAATCC3�. as monomers with Helix turn helix motif.� Translation of Hb mRNAs at the posterior end are inhibited and destroyed by Nanos and its associated proteins.� So Hbs are absent from the middle to the posterior region of the embryo.�
Hbs have dual role in controlling gene expression.�
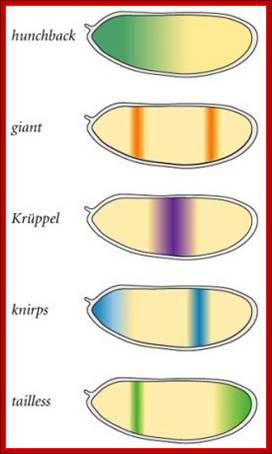
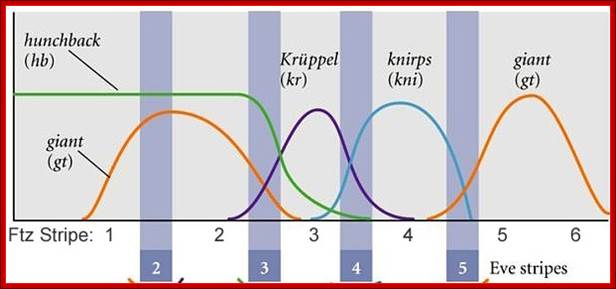
High concentration of Hb inhibits the expression of Kruppel genes at the anterior end, but medium concentration next to it or midway from the anterior end, activates Kruppel genes but the concentration of Hb is enough to inhibit Knirp and Giant genes in the anterior and mid-region, thus Hb acts as repressor of Kruppel, Knirp and Giant at the anterior end and acts as an activator of Kruppel in the middle region.� In the absence of Hb at posterior end Knirp and Giant genes are expressed.� Thus, four blocks of transcripts and their products are localized in an order from anterior end to the posterior end: they are Bicoid/Hunchback, Kruppel, Knirp and Giant in that order.
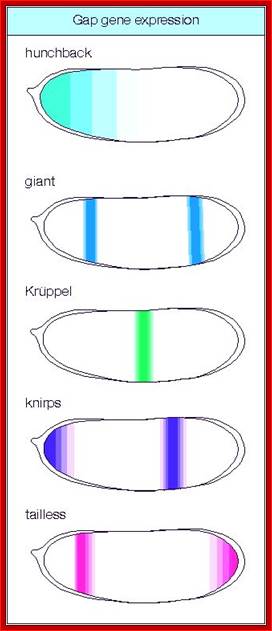
Maternal gene products present at the time of fertilization activate a set of genes, absence of whose products leads the loss of specific region of the larva, so they are called Gap genes.� The Gap gene products themselves are transcription factor, which by combination with other gene products activate another set of genes called pair rule genes. http://www.mun.ca/

Modeling the temporal evolution of the Drosophila gene expression from DNA microarray time series; Alexandre Haye et al; http://iopscience.iop.org/
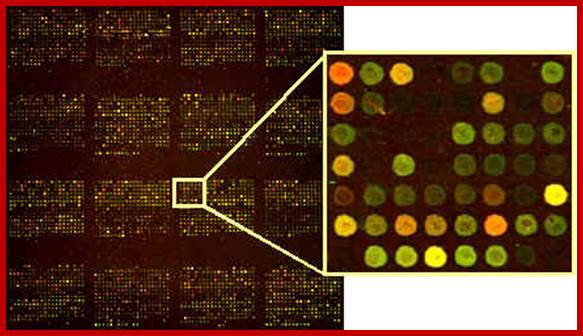
This diagram shows expression of different genes detected by DNA microarray or DNA chip methods. https://bspace.berkeley.edu
Interactive collection of cis-regulatory modules from Drosophila; https://bspace.berkeley.edu
In all the said genes, the promoters have elaborated regulatory elements, which respond to different concentration of regulatory factors for transcription and which gene(s) to be transcribed.� Regulatory elements have protein binding sequences and they may be present in one copy or in multiple copies.�
Modalities of gene expression:
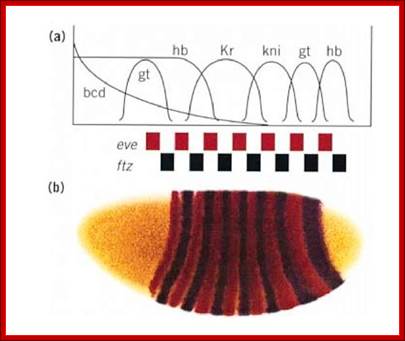
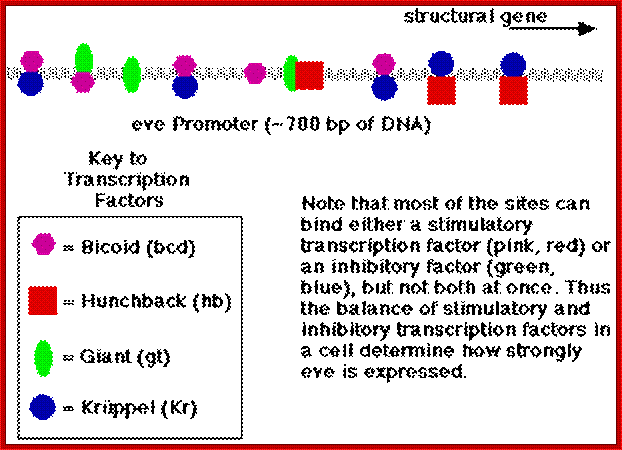
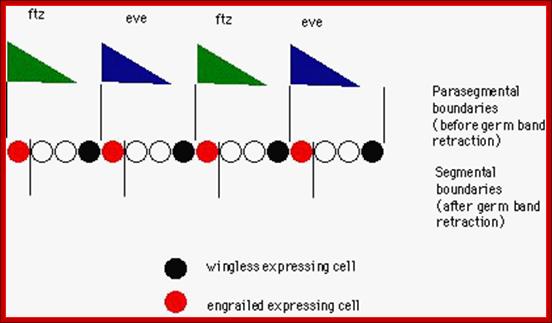
Most of the genes involved in developmental activities have elaborate regulatory elements upstream of their start site; some have regulatory elements within coding regions called intronic elements.� They have different sites including enhancer elements, for different transcriptional factors, all are sequence specific.� The same sequence may be found in one copy or more than one copy.� Based on them promoter elements are called as high affinity promoters, Low affinity promoters.� They also have repressor elements.� Many of the transcription or regulatory factors act on these elements in quantitative manner i.e. concentration dependent.� They in turn activate pair-rule genes in seven stripes each.� Even skipped (eve) are numbered from one to seven i.e. odd numbers and seven Fushi tarazu (ftz) stripes as even numbers.� This expression demarcates the body into segment forerunners.
A cascade of gene expression starting from maternal gene products to segment polarity genes; http://people.ucalgary.ca/
Expression of Pair Rule genes:
Gap gene products are organized in an order in the developing blastoderm embryo from anterior end to posterior end: Bicoid, hunchback, krupple, knirp, and giant.� These in their respective positions act on the nuclear genes such as pair rule genes.� The most important of all the pair rule genes are even skipped (eve) and Fushi tarazu (ftz) [but the primary pair rule genes are hairy and even skipped].
Eve and ftz are expressed in the form of stripes from dorsal to ventral region. �Seven stripes that are in odd numbers are called even skipped stripes and the even numbered ones are called ftz stripes.� Thus, they divide the body into segments.� Each stripe, to begin with is one cell thick and later it divides and redivide to 3-4 cells broad or thick.� Ftz genes are expressed very heavily at blastoderm stage.��� The expression of each of the genes in the form of stripes delineates the Para segmental boundary.� Para segments are distinguished by mesodermal thickenings and ectodermal grooves and they divide the embryo into 14 segments, determined by stripes expressed from pair rule genes.
Each of the stripe�s expression is regulated by the combinatorial action of kruppel, bicoid, hunchback, knirp, and giant factors in each of their regions.� Kruppel in combination with others expresses even skipped genes in its location.� Similarly other factors activate even skipped genes in similar fashion.� Even the expression of ftz genes follows the same pattern.��� The process of stripe formation takes place from anterior to posterior and it just takes 30 minutes to express all the stripes.
Gene expression patterns are regulated both spatially and temporally in embryos of Drosophila melanogaster; Wikipedia
Each of these genes has elaborate promoter regions with different binding sites for different gap and maternal proteins.� For example, the even skipped stripe 3 has binding sites for bicoid (bcd), Hunchback (hb) Kruppel (Kr) and gaint (gt) and the whole promoter region is 480 bp long.
Organizing the Embryo: Segmentation; http://users.rcn.com/
�
Expression of Segment Polarity genes:
Even skipped (eve) and Fushi tarazu (ftz) genes in each of the stripes activate another set of genes called segment polarity genes.�
II�Kru�Bcd�gaint�bcd�kru�bcd-hb-gaint-kru-bcd-II -+1
This happens after or during 13 division and cellularization. �At this stage cells have defined position and the cell fate is more or less determined.� Once the cells are formed by invagination of cell membranes around each of the nuclei, the cells start interacting with one another; communicate with each other through receptors, ligands, ECM (extra cellular matrix).�
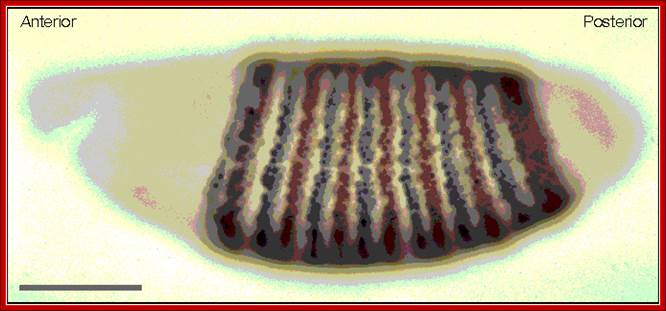
A lovely Segmentation diagram; fluorescent labeling (gpf) is used. http://www.mun.ca/
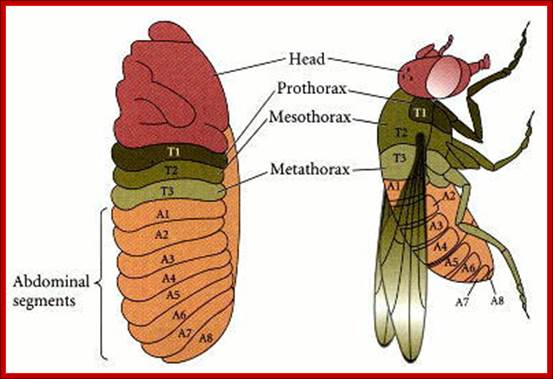
Segmental markings and the development of specific organs from them. courses.biology.utah.edu/
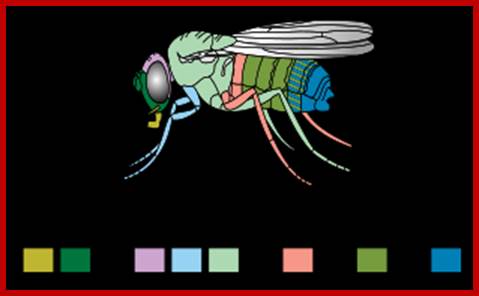
Matching colors to structure developed; http://en.wikipedia.org/
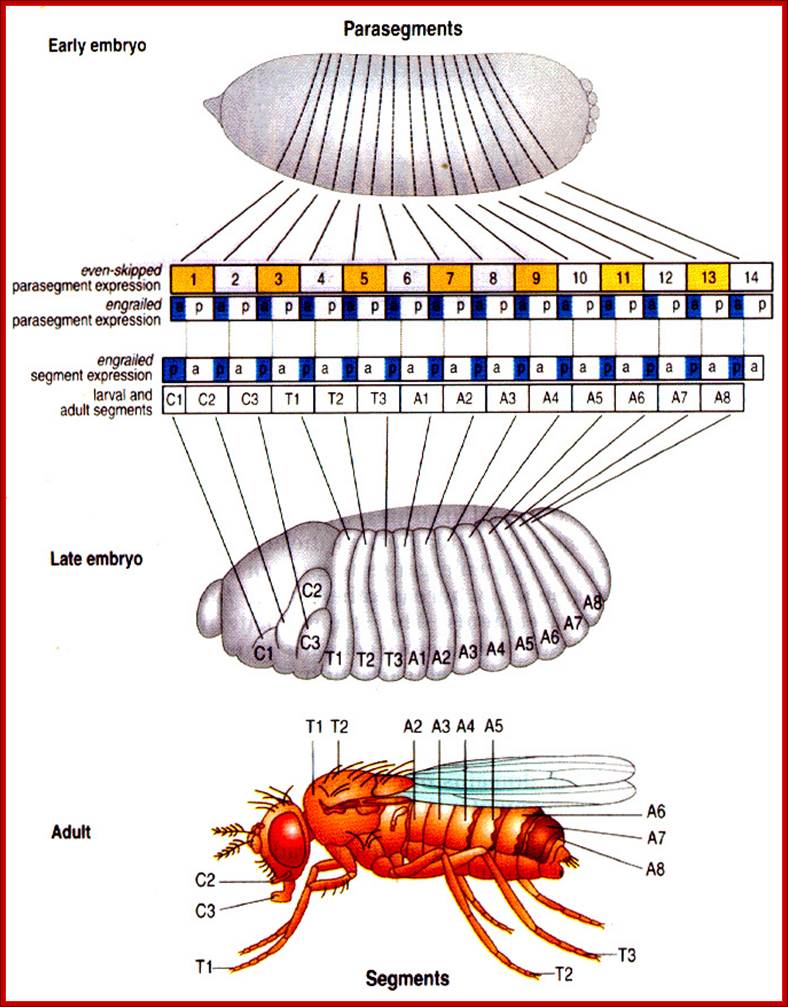
Molecular and Developmental; Biologyhttp://www.mun.ca/
Two of the seven segment polarity genes have significant effect.� The segment polarity genes establish cell�s fate in each of the arrangement, thus they establish anterior and posterior part within each segment.� They express genes for cellular communication in the form of transmembrane receptors, secreted proteins as ligands and inducers, kinases and several important transcription factors.
�
Jason N;http://www.studyblue.com/
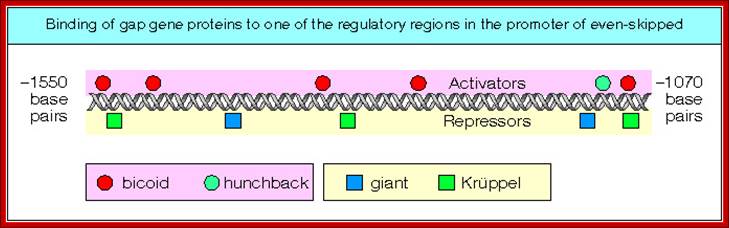
This is a diagrammatic representation of promoter elements for the binding of factors for the activation of gene expression; a protein binding element are shown in colors-spherical and squares; https://www.studyblue.com
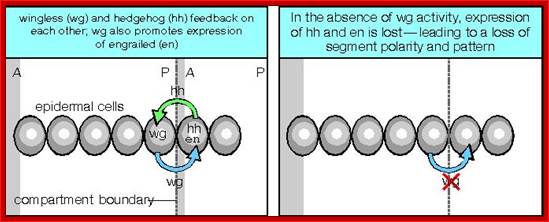
Among them Engrailed (en) and Wingless (Wg) genes have important role in determining the fate and polarity of cells in the region.� Engrailed expression is at the anterior part of each segment and the wing expression is at the posterior part of each segment, between them mesodermal thickenings and ectodermal groove are conspicuous.
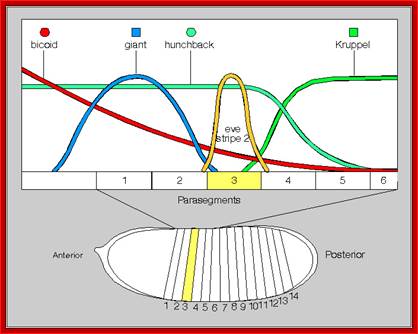
Expression of eve genes with the interaction of other genes; eve strip genes are expressed because of concentration gradient of different gene products due to repression and activation of certain genes. http://scienceblogs.com/
First engrailed is expressed and the expression of wingless requires engrail expression for induction.� These two genes are expressed in adjacent rows of cells and their expression is mutual to one another.� Engrailed proteins expressed in Para segments ensure anterior and posterior boundary of each segment.� Ftz and eve repress the expression of wing genes in each of the successive rows of cells.
Para segments:
---I-wg-I en-I--eve--I wg-I-en-I�ftz--I-wg I-en I�eve--I-wg-I�ftz--I---
In the parasegment delineation, wing and engrailed are expressed side by side in one cell thick adjacent rows and eve and ftz stripes are 3-4 cells thick.� In this combination wing row is always expressed anterior to the engrailed row.�
![]()
Molecular and Developmental Biology; http://www.mun.ca/
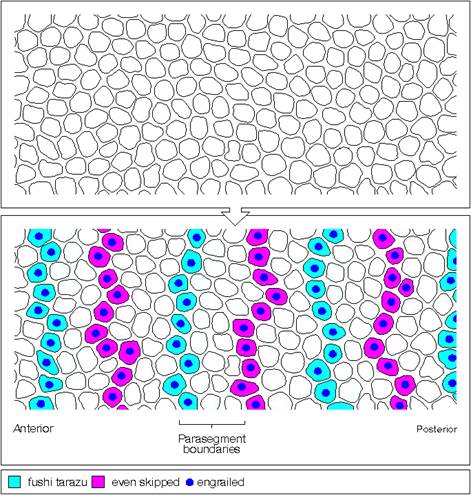
http://www.mun.ca/
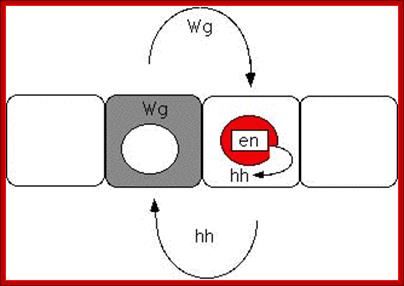
The diagrams above and below show Interaction between Hedgehog and Wing gene products in development of Para segmentation and segmentation; http://people.ucalgary.ca/
The pair-rule genes are already expressed in a periodic pattern, so it is easy to imagine how they establish the segment polarity gene expression in every Para segment. The expression patterns of the segment polarity genes engrailed (en)and wingless (wg) are established through positive and negative transcriptional regulation by the pair-rule genes. For example, the expression of en is activated by either ftz or eve in each Para segment, whereas wingless is repressed by ftz or eve in each Para segment. (How would these interactions be demonstrated, and how would it be shown that the interaction was direct or indirect?); Other pair-rule genes also control wg and en expression. For example, paired andodd-paired are responsible for the activation of engrailed AND wingless in alternating stripes. http://people.ucalgary.ca/
Wing is a secreted protein acts as ligand on engrailed cells.� Engrailed protein is a transcriptional factor that activates Hedgehog proteins, which move to next cell and activate wing genes.� The other polarity genes generate secreted proteins, transmembrane proteins, kinases, cytoskeletal proteins and many TFs.� These have great impact on further cell differentiation and development.
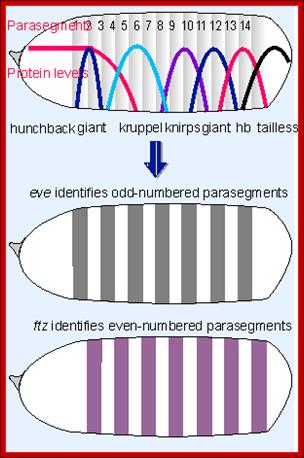
Figure: Expression of the gap genes defines adjacent regions of the embryo. The gap genes control the pair-rule genes, each of which is expressed in 7 stripes. http://genes.atspace.org/
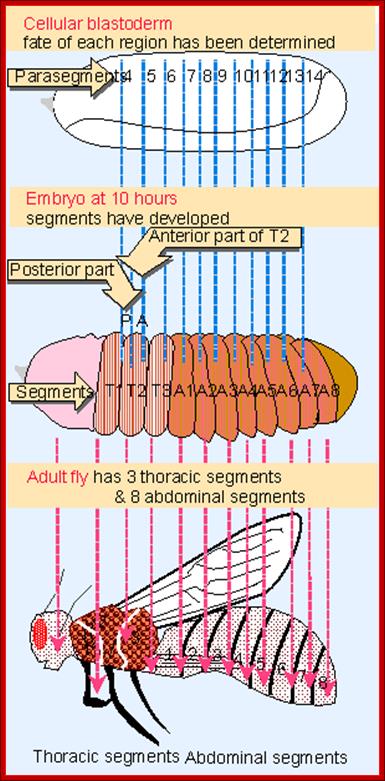
Figure: Drosophila development proceeds through formation of compartments that define Para segments and segments. http://genes.atspace.org/
Expression of Homeotic Genes and Selector genes:
Edward shared Nobel Prize, in 1995 with B Lewis, Christiane N�sslein Volhard and Eric Wieschaus for their work on homeotic and gene regulatory genes.� Expression of pair rule genes has great impact on segment polarity genes and homeotic genes in development, which happens at blastoderm stage.� Homeotic genes impose unique pattern of differentiation of each segment.� They are expressed temporally and spatially in terms of Para-segment.� Homeodomain genes do not create any pattern that are already determined by segment polarity genes, but modify the fates of them.� They act in cells where segment polarity genes and eve genes are still active.� Many of the Homeotic genes (HG) code for transcriptional factors.� One set of HG factors act upon another set of Homeotic genes, which in turn act upon one more set homeotic genes and other genes.� Their expression is temporal and time-lapse, that leads to the differentiation and development of structures from anterior to posterior direction of the body.� The Hg genes identify and elaborate in determining organs formation.
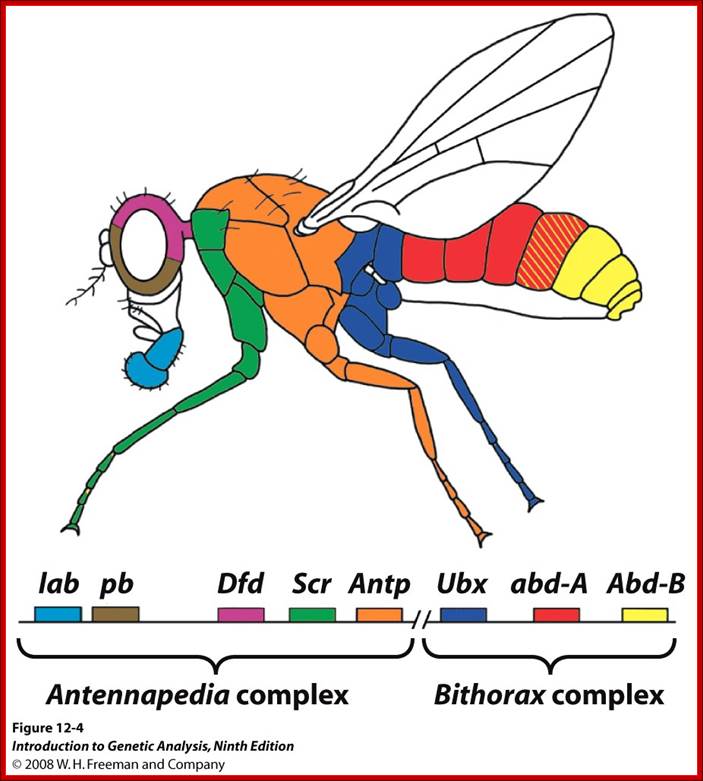
There are many homeotic genes and all of them are organized into clusters of Hg genes and occupy a locus or loci.� In the case of Drosophila they are organized into to two clusters called ANT-C and
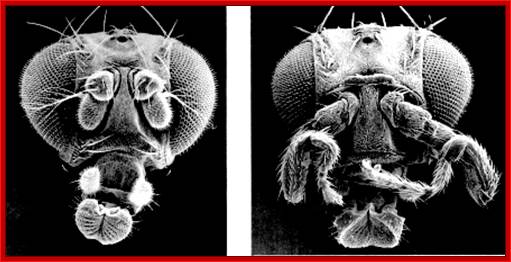
Antp-gene mutant and its effect on
morphological features;
https://neurophilosophy.wordpress.com
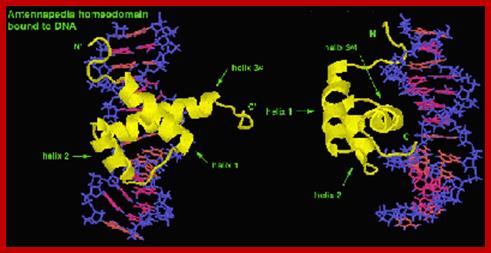
Antp proteins bound to DNA at its promoter; www.biology.kenyon.edu
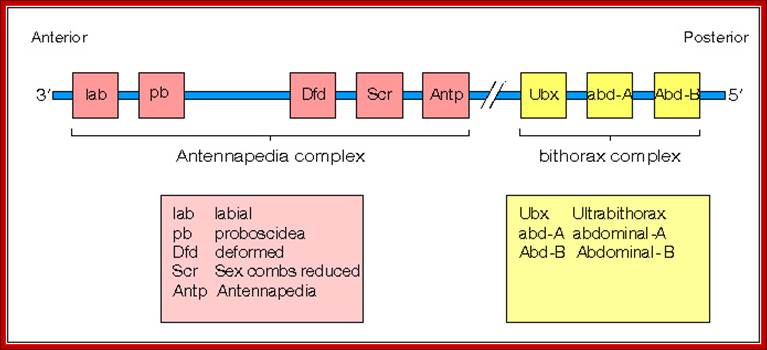
The homeobox gene complexes� map- view; Dr. Brian E. ��Staveley http://www.mun.ca/
BX-C.� Actually they are derived from single complex during evolution from Trifolium, a beetle.� There are other array of genes related to ANT-C and BX-C and they are located elsewhere.� ANT-C identifies the anterior most part of the insect (para-segment from1 to 4) containing structures such as labial (lab, proboscipedia (pb), deformed (dfd), sex combs reduced (scr) antennapedia (ant) and others, while BX complex contain genes for the development of thorax and abdominal segments from T2 to A8.
ANT-C:
ANT-C complex consists of genes for lab, pb, dfd, scr and Ant-P.� The genes are expressed successively more towards posterior sides.� The loci are nearly 350kbp long, but the Antennapedia gene is 103kbp long.� This gene transcribes from two promoters, p1 and P2.� It has 8 exons, but its open reading frame (ORF) starts only from 5th exon. And produces 48 KD protein (i.e. only 1% of the gene codes for the protein).
-I-lab-I--I-pb-I-zen�bcd-ama--I�I-dfd-I�I-ftz-I I�Antp�I350kbp
The two promoters are separated by 70kbp.� P1 is locate upstream of exon1 and the P2 is positioned upstream of exon3, both end at 8th exon terminal region.� Each promoter is used for tissue specific expression.
ANT-C:� controls the development of labial (lb) parts, antennapedia (Antp), sex combs reduced (Scr), proboscidia (pb) and thoracic segment two and three.� But mutations in ANT-C components affect labial parts, head parts, proboscidia, mandibular segments, sex combs and thoracic segments T2 to T3 and T3.� A dominant mutation leads to the development of a pair of legs in the place of antenna; the head behaves like a thoracic segment.� A recessive mutation expresses antenna in place of legs in second thoracic segment.
BX-C:
Bithorax locus controls thorax 2 and 3 segments and 1 to 8 segments of abdomen. It is 300 kbp long and only 1.4% codes for proteins.� It consists of ultra-bithorax containing Ubx transcriptional unit greater than 75kb.� Infra abdominal domain has Abo-A and Abd-B transcription units each 20kb.� The order of units in the BX-C locus from left to right coincides with body parts from left to right.
-cbx�abx�bx---bxd�pbx�iab2�iab3�iab4-iab5�iab6-iab8-iab9-
![]() -----------------------Ubx---------------���
I------abd-A----��������� ------- abd-B----
-----------------------Ubx---------------���
I------abd-A----��������� ------- abd-B----
Ubx produces 75 kb long transcript, abd-A produces 20kb and abd-B generates 20 kb long transcript.
It consists of three coding units.� Ubx:� contains cbx, abx, bx, bxd & pbx (ultrabithorax complex), which identifies and develop 3rd thoracic segment.� Abd-A and Abd-B control the eight abdominal segments.� Mutation makes it behave like T2 thoracic segment and develop a pair of wings in place of halters.�
The ubx, a homeodomain protein complex controls metathorax and represses the genes of mesothorax.� The ubx represses Antp expression in metathorax and restricts Antp expression in mesothorax.� Mutation in ubx, allows the expression of Antp in mesothorax (which is normal), but it also miss expression of Antp in metathorax, so metathorax becomes mesothorax, where halters are transformed into wings.
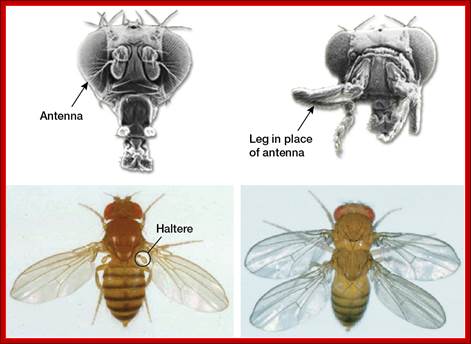
Homeotic Genes and Body Patterns; Top: (Left) Normal fruitfly; (Right) Fruitfly with mutation in antennapedia gene Bottom: (Left) Normal fruitfly; (Right) Fruitfly with a homeotic mutation that gives it two thoraxes. Bottom images courtesy of the Archives, California Institute of Technology. The diagram shows how mutations in specific hox genes affect the body parts.
�http://learn.genetics.utah.edu/
Genes c-bx and a-bx are the part of ubx cluster.� Mutation in cbx (counter bithorax) disrupts ubx without affecting the ubx�s protein, but causes misexpression in mesothorax; as a result, halters develop in place of wing.
Bxd causes development of halters, but mutation in bxd results in partial transformation of halters into wings at metathorax.
Pax-6 controls the development of eye in most of the animals, normally in the head and in bilateral position.� A mutation in this can change the pattern of expression. �A mutation can lead to misexpression producing an extra eye in wings or legs.� When pax-6 from squid is expressed in the fly, the eye develops either on the wing or on legs.� They share 30% of the sequences.
However mutation in abx, a part of ubx complex, causes repression of Antp and other genes for development of mesothorax, as a result mesothorax becomes metathorax.� So, wings are transformed into halters, the fly looks like an ant.�
Ubx acts as a suppressor of Antp and other mesothorax genes in the developing metathorax.� These Antp and Ubx genes are pattern-determining genes.� Some of their repressor activity is due to Alanine rich domains as found in even skipped proteins.
Antp determines mesothorax and it is expressed only in mesothorax, with a pair of legs and a pair of wings, but a dominant mutation in Antp causes (due to inversion it is brought under the control of head determining genes) expression of legs in place of antenna.� Antp produces a homeodomain protein.
Transgenic flies can be produced by placing whatever genes one likes under the regulatory elements (HSE) of heat shock protein genes. When transgenic larvae with Antp gene was subjected high temperature at 42 0C, the Antp genes are expressed in all tissues.� This causes the misexpression of Antp, resulting in head and thoracic segments becoming duplicated mesothoracic segments.� These embryos die but their morphology remains distinct.
Antps expression also shows denticles in all segments, which are normally present in mesothorax.
Ubx miss expression in transgenic fly results in the expression of denticles in all segments similar to the denticles normally found in metathorax.� But when ubx gene is modified to generate and function as an activator protein, when expressed in transgenic fly, results in conversion of all segments into mesothorax.
Mutational affects:
Abx- transforms T2/p/T3A to T2.
Bx- transforms T3A to T2.
Bxd- transforms A1A to T3A.
Homeobox containing genes and their sequences are also found in other organisms, which can be tested very easily by PCR or by DNA-DNA hybridization.� In other organism too homeobox genes are involved in identity of body parts from higher invertebrate to higher vertebrates; where all have segmentation as a theme for body construction including human body, which is more apparent in embryonic stages.� Homeotic genes of BX-C and ANT-C are often referred to as HOM-C genes.� In mammalian systems the cluster size ranges from 20-100kbp.� And they contain 9 to 10 gene.� One finds them as four duplicated clusters, but in drosophila it has two clusters.
Drosophila homeotic genes:
|
ANT-C |
Lab< |
Pb< |
|
Dfd> |
Scr< |
Antp< |
|
|
|
|
|
|
|
|
|
BX-C |
|
|
|
|
|
|
Ubx< |
Abd-A< |
Abd-B< |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Mammalian Homeotic genes:
|
|
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
|
Hox-A |
yes |
yes |
|
yes |
yes |
yes |
yes |
|
yes |
Yes |
Yes |
|
Yes |
Evx1 |
|
Yes |
Yes |
Yes |
|
Yes |
Yes |
Yes |
Yes |
Yes |
yes |
|
|
|
|
|
|
Hox-C |
|
|
|
Yes |
Yes |
Yes |
|
Yes |
Yes |
|
Yes |
Yes |
yes |
|
|
Hox-D |
Yes |
Yes |
Yes |
|
|
|
|
|
Yes |
Yes |
Yes |
Yes |
Yes |
Evx2 |
Fly�s ANT-C and BX-C correspond to group 1 to 9 of Hox-A, but group 10 to 13 in Hox-A have arisen due to tandem duplication and divergence of group 9 (abd-B).� Corresponding loci in each cluster are called paralogs, so Hox-A4 and Hox-B4 are paralogs.� These clusters of genes are expressed from one end of the cluster to other end of the cluster for the expression of one lead to the expression of the next and in the order to the last ending in the development of posterior part of the body.
Most of the Hox genes generate protein that contains a 180 base pair long region that codes for homeodomain at the carboxyl end of the protein.� Though there are several groups of homeodomain genes the proteins produced have 70-80% conserved sequences.� Homeodomain proteins do differ in their sequences and bind to different sequences of different promoters and thus responsible for the expression of different genes.� However, their specificity is further amplified and refined by their association with partner proteins.� Such Hox genes in humans are called HOM-C. gene clusters.
Developmental genes are expressed in hierarchical fashion, one after the other; one inducing the other, and the other inducing another and so on.� Maternal genes activate segmentation genes (which include gap genes which induce pair rule genes and they in turn activate polarity and selector genes in that order).� They all regulate one another by generating transcriptional factors, co activators, activators or repressors; all ultimately express structural genes for tissue specificity of an organism.� All of them act in combinatorial fashion.� The developmental genes have elaborate promoter regions with various elements in various positions.� The interplay of the factors with promoter element is the key in development, it is a sophisticated process.
Overall view of genes and gene products:
Four groups of genes control the development of drosophila starting from oocyte to early segmental larvae.� Maternal genes such as bicoid, hunch back, nanos, oskar and caudal, gurken and others, all are synthesized in nurse cells and transported into the egg cell and they are distinctly localized.� The proteins generated by them especially bicoid and hunchback are transcriptional factors; even dorsal is a transcriptional factor.� Similarly gap gene product activated by maternal products, hunchback, Kruppel, Knirp and giant, also turned out to be transcriptional factors; so also, pair rule gene products activated by gap genes and segmental polarity gene products, most of them, happen to be transcriptional factors.� The dominant and conspicuous homeotic gene products are also gene regulatory proteins, but characteristically, majority of them contain homeodomain motifs, hardly some are structural proteins.
Gap gene products are zinc finger proteins and they are transcriptional factors., they contact DNA in the promoter region in sequence specific manner.� Segmental and homeotic gene products have conserved motifs called Homeobox of 180 amino acids long.� The homeobox contains three helices, two run parallel to each other and the third one is placed across and this third one contacts specific DNA sequence, such motifs are called Homeodomain.� There are forty or more such genes contain such homeodomain motifs and most of them happen to be involved in transcription of genes in one way or the other.� Proteins containing homeodomain motifs have 80% conserved sequences and such domains are always found at the carboxyl end of the protein.� It is also interesting to note most of the homeobox containing genes are found to be in clusters like ANT-C, BX-C and other mammalian HOM-C genes.� Their effect on transcriptional activity is combinatorial and this could be quantitative or qualitative or both at different targets.� The most important feature of homeobox genes is that they identify body parts.
Sex Determination in Drosophila:
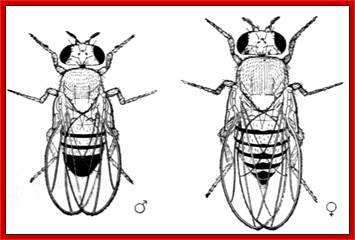
The Drosophila melanogaster sex determination hierarchy. The Drosophila sex determination hierarchy consists of a cascade of sex-specific alternatively spliced pre-mRNAs culminating in the production of sex-specific transcription factors encoded by doublesex (dsx) and fruitless P1 (fru P1). The primary determinate of sex is the X chromosome to autosomal chromosome (A) ratio. In females (X:A = 1), Sex Lethal (SXL) is produced and regulates the splicing of the pre-mRNA of transformer (tra), resulting in the production of TRA. TRA acts in conjunction with constitutively produced TRA-2, and regulates the alternative splicing of the pre-mRNAs of dsx and fru P1, leading to the production of the female-specific protein DSXF. In males (X:A = 0.5), SXL is not produced and dsx and fru P1 pre-mRNAs undergo default splicing, resulting in the production of the male sex-specific proteins DSXM and FRUM. In addition, in females SXL represses dosage compensation, the process by which transcription of genes on the single X chromosome in males is up-regulated to roughly equal that of the two X chromosomes in females. Lebo et al. BMC Genomics 2009 10:80 ; �http://www.biomedcentral.com/
RNA Binding Protein Sex-Lethal (Sxl) and Control of Drosophila Sex Determination and Dosage Compensation; he X:A signal and the control of sex determination, sexual behavior, and dosage compensation. The X:A signal targets the Sxl gene, controlling its expression. Autoregulation of Sxl is established only in embryos whose chromosomal constitution is 2X;2A (two X chromosomes; two sets of autosomes), but not in X(Y);2A (one X chromosome; two sets of autosomes) embryos. The autoregulatory feedback loop regulates Sxl expression throughout development and adult life. Sxl controls the expression of the traand msl-2 genes, whose products are required for control of somatic sex determination/sexual behavior and dosage compensation, respectively. The dotted lines indicate that the expression of the gene is �off,� and the solid lines indicate that the expression of the gene is �on�.http://mmbr.asm.org/
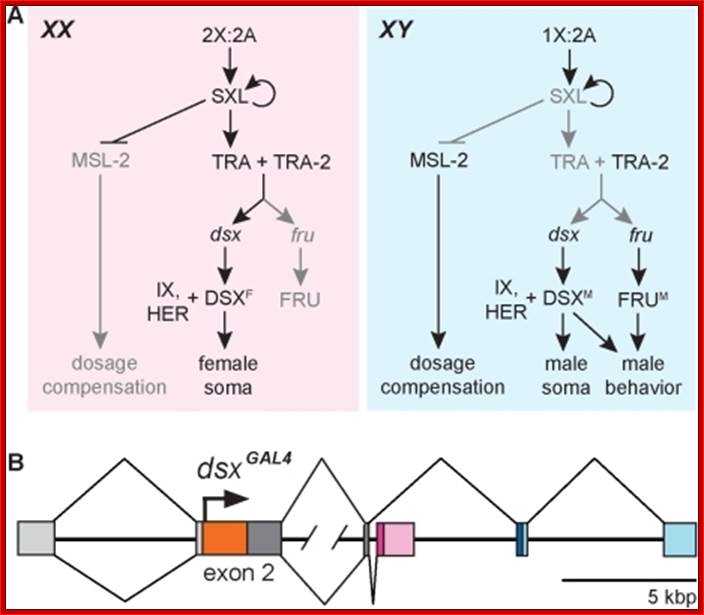
The Drosophila melanogaster sex hierarchy controls sexual differentiation of somatic cells via the activities of the terminal genes in the hierarchy, doublesex (dsx) and fruitless (fru). We have targeted an insertion of GAL4 into the dsx gene, allowing us to visualize dsx-expressing cells in both sexes. Developmentally and as adults, we find that both XX and XY individuals are fine mosaics of cells and tissues that express dsx and/or fruitless (fru(M)), and hence have the potential to sexually differentiate, and those that don't. Evolutionary considerations suggest such a mosaic expression of sexuality is likely to be a property of other animal species having two sexes. These results have also led to a major revision of our view of how sex-specific functions are regulated by the sex hierarchy in flies. Rather than there being a single regulatory event that governs the activities of all downstream sex determination regulatory genes-turning on Sex lethal (Sxl) RNA splicing activity in females while leaving it turned off in males-there are, in addition, elaborate temporal and spatial transcriptional controls on the expression of the terminal regulatory genes, dsx and fru. Thus tissue-specific aspects of sexual development are jointly specified by post-transcriptional control by Sxl and by the transcriptional controls of dsx and fru expression.
Evolutionary considerations suggest such a mosaic expression of sexuality is likely to be a property of other animal species having two sexes.These results have also led to a major revision of our view of how sex-specific functions are regulated by the sex hierarchy in flies.Thus tissue-specific aspects of sexual development are jointly specified by post-transcriptional control by Sxl and by the transcriptional controls of dsx and fru expression; http://openi.nlm.nih.gov/
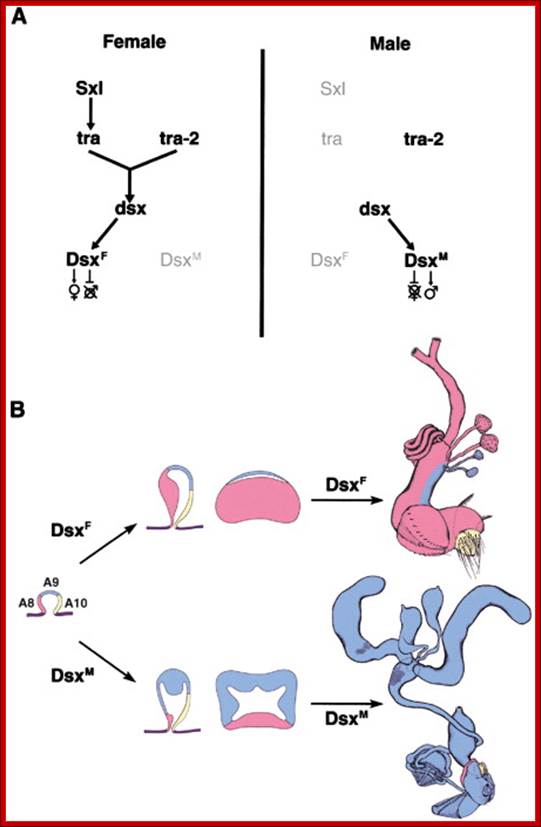
Male Drosophila hyper transcribes their single X, whereas female mammals silence one of their two X chromosomes: Expression of specific sex determining genes lead to the development of sex determining tissues.
Sex-specific expression of these genes is initiated at the time of gonad formation (stage 15), indicating that sexual identity in the germ line is established by this time. Experiments where the sex of the soma was altered relative to that of the germ line (by manipulating transformer) reveal a dominant role for the soma in regulating initial germ line sexual identity. Germ cells largely take on the sex of the surrounding soma, although the sex chromosome constitution of the germ cells still plays some role at this time. The male soma signals to the germ line cells through the JAK/STAT pathway, while the nature of the signal from the female soma remains unknown. The genes Ovo and ovarian tumor (otu) are expressed in a female-specific manner in embryonic germ cells, consistent with their role in promoting female germ line identity. However, removing the function of ovo and otu, or reducing germ line function of Sex lethal, had little effect on establishment of germ line sexual identity. This is consistent with findings that signals from the soma are dominant over germ line autonomous cues at the initial stage of germ line sex determination (Casper, 2009).
Stage 15; 12 hours after egg laying (AEL)- we correlate embryological and genetic evidence and propose that germ line could be allocated by the 4-cell stage and regulated by oocyte polarity, controlled cleavage planes and early transcription of sex-determining genes from pronuclear stages. �Subsequent steps including germ line segregation in early embryos and migration of primordial germ cells, and their possible role in the initiation of gonadal sex differentiation in the genital ridge are discussed.� Vital Genes That Flank Sex-Lethal, an X-Linked Sex-Determining Gene of DROSOPHILA MELANOGASTER-Janice A. Nicklas and Thomas W. Cline.
Global analysis of patterns of gene expression during Drosophilaembryogenesis
Network representation of tissue relatedness. Nodes represent collapsed annotation terms and edges represent the correlation between expression in each pair of terms. Only tissues that share a statistically significant number of genes are linked and the strength of the links is proportional to the number of genes the two tissues have in common. Tissues that share very few or no genes repel each other. The system is allowed to reach a low energy level in two-dimensional space under a physical spring model (force directed layout). Collapsed annotation terms are color-coded according to their organ system assignments as used throughout.
Global analysis of patterns of gene expression during Drosophilaembryogenesis

Network representation of tissue relatedness. Nodes represent collapsed annotation terms and edges represent the correlation between expression in each pair of terms. Only tissues that share a statistically significant number of genes are linked and the strength of the links is proportional to the number of genes the two tissues have in common. Tissues that share very few or no genes repel each other. The system is allowed to reach a low energy level in two-dimensional space under a physical spring model (force directed layout). Collapsed annotation terms are color-coded according to their organ system assignments as used throughout.
Global analysis of patterns of gene expression during Drosophilaembryogenesis

Network representation of tissue relatedness. Nodes represent collapsed annotation terms and edges represent the correlation between expression in each pair of terms. Only tissues that share a statistically significant number of genes are linked and the strength of the links is proportional to the number of genes the two tissues have in common. Tissues that share very few or no genes repel each other. The system is allowed to reach a low energy level in two-dimensional space under a physical spring model (force directed layout). Collapsed annotation terms are color-coded according to their organ system assignments as used throughout.
Global analysis of patterns of gene expression during Drosophilaembryogenesis

Network representation of tissue relatedness. Nodes represent collapsed annotation terms and edges represent the correlation between expression in each pair of terms. Only tissues that share a statistically significant number of genes are linked and the strength of the links is proportional to the number of genes the two tissues have in common. Tissues that share very few or no genes repel each other. The system is allowed to reach a low energy level in two-dimensional space under a physical spring model (force directed layout). Collapsed annotation terms are color-coded according to their organ system assignments as used throughout.
Global analysis of patterns of gene expression during Drosophilaembryogenesis

Network representation of tissue relatedness. Nodes represent collapsed annotation terms and edges represent the correlation between expression in each pair of terms. Only tissues that share a statistically significant number of genes are linked and the strength of the links is proportional to the number of genes the two tissues have in common. Tissues that share very few or no genes repel each other. The system is allowed to reach a low energy level in two-dimensional space under a physical spring model (force directed layout). Collapsed annotation terms are color-coded according to their organ system assignments as used throughout.
Global analysis of patterns of gene expression during Drosophilaembryogenesis
Network representation of tissue relatedness. Nodes represent collapsed annotation terms and edges represent the correlation between expression in each pair of terms. Only tissues that share a statistically significant number of genes are linked and the strength of the links is proportional to the number of genes the two tissues have in common. Tissues that share very few or no genes repel each other. The system is allowed to reach a low energy level in two-dimensional space under a physical spring model (force directed layout). Collapsed annotation terms are color-coded according to their organ system assignments as used throughout.
Global analysis of patterns of gene expression during Drosophilaembryogenesis
Network representation of tissue relatedness. Nodes represent collapsed annotation terms and edges represent the correlation between expression in each pair of terms. Only tissues that share a statistically significant number of genes are linked and the strength of the links is proportional to the number of genes the two tissues have in common. Tissues that share very few or no genes repel each other. The system is allowed to reach a low energy level in two-dimensional space under a physical spring model (force directed layout). Collapsed annotation terms are color-coded according to their organ system assignments as used throughout.