Eukaryotic DNA Replication Mechanism:
Eukaryotic cells are endowed with a very complicated but sophisticated regulation of cell division.� Embryonic cells go through first seven to eight cell divisions very fast at an interval of 30- 40 minutes, but differentiated cells remain in resting stage i.e. G � stage.� Onion root tip cells divide once in every 12 hrs, but mammalian cultured cells take 24 hr for one cell cycle.� Resting cells, whatever may the cell type, initiate cell division when there is loss of cells in the tissue or by mitotic signals.� In order to replace the lost numbers, existing cells divide and redivide till the number is restored.
Control of Gene expression: http://sphweb.bumc.bu.edu/
Cells in cell division mode go through certain number of sequential molecular expressions and physical changes stepwise. We can call them as Interphase and Mitotic stages (M).� Interphase is further divided into G1, S and G2 stages in succession; each event is specific and interdependent. An event does not enter to the next cycle till the previous one is completed. The mitotic phase the M phase consists of Prophase, Metaphase, Anaphase and Telophase; all put together requires only 60 minutes or so. In 24hrs cycle the longest phase is G1 about 8-10 hrs., S phase requires about 6-8 hrs and G2 takes 4-6 hrs; �this is an approximation of 24 hrs cell cycle. But there are cell cycles such as early embryonic growth cell division takes just 4-6 hrs.
The DNA replication is activated at early G1 phase and replication is initiated at very early S-phase (DNA replication stage), and replication is completed at G2 (G stands for Gaps in interphase), till then cells don�t enter into M phase. �
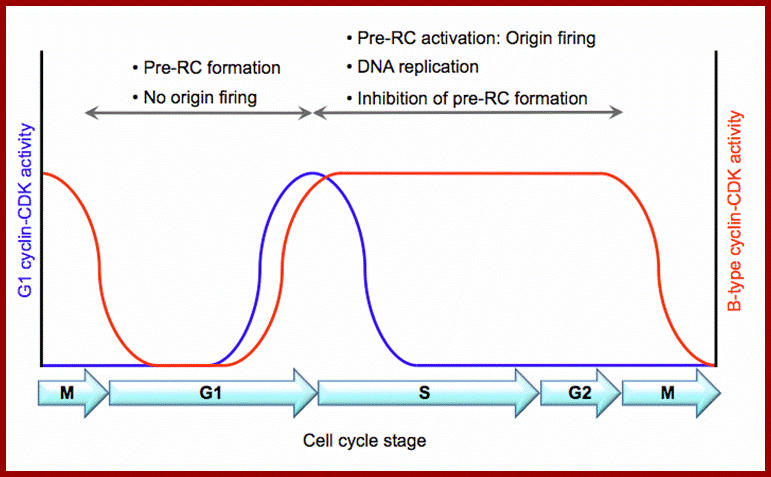
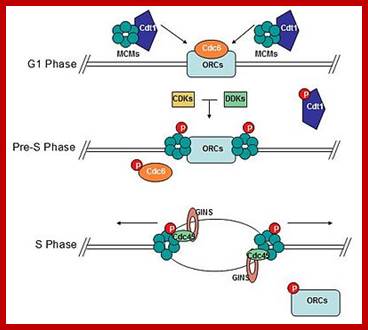
Licensing of origins of replication can only take place in G1 phase when Cdk1 is inactive, while origin activation only occurs during S phase when Cdk1 is active. See text for details. http://www.intechopen.com/
Replication initiation occurs only once in a cell cycle and second round of replication is prevented in the same cell cycle.�� In S.cerevisiae and S.pombe, gene products like Origin Replication Complexes ORC proteins, Cdc6 and Cdt1 respectively required for transition from G1 to S phase.� They are similar to Xenopus� p34 kinase.� There are different forms of P34 CdKs (Kinases).� One of them is cell cycle dependent CdK34, which is activated when specific S-cyclin binds to CDK.� The protein complex has now been purified; it consists of p34 kinase and Cyclin (or cyclin-B-CDK2), called Mitosis-Promoting Factor (MPF).� There are S-cyclins and M-cyclins, each have specific roles in S and M (Synthesis and Mitotic) phases. A START point or Restriction point is the stage at which commitment to initiation for S stage occurs at G1, when a whole battery of genes for DNA synthesis (DNA replication) is activated.� The regulatory pathway consists of a cascade of cyclin dependent phosphorylation and dephosphorylation events.� Until every bit of DNA sequence is replicated, this includes telomeric DNA synthesis, the cell doesn�t enter into M phase.
Initiation:
Replication initiation takes place at replication foci.� In yeast DNA in centromere initiate replication. The distance over which centromeres can influence origin activation time extends up to 19 kilobases. We further show that centromere-mediated early origin activation depends on the centromere's ability to recruit at least a subset of the proteins needed for chromosome segregation. This study thus provides the first direct functional link between kinetochore establishment and the mechanisms of DNA replication initiation (Thomas J.Pohl et al). This triggers replication initiation laterally.� The last part of the chromosomal DNA to be replicated is Telomeric DNA. Rest of the DNA Replication gets activated more or less simultaneously all along the length of the chromosomes. Eukaryotic DNA is linear and contains several Ori sites may be 50-200 or more distributed all along the length of each chromosomal DNA.� Taking SV40 DNA as a model contains only one Ori site.� It is a eukaryotic virus.� The viral replication is used as example of eukaryotic DNA replication. The viral circular DNA replication initiation starts with the binding of T-antigens as hexamers, to specific site called origin. There is only one Ori site in SV 40 DNA. �T-antigen also acts as a helicase (motor protein).� They bind to specific sequences called ORE (Origin Recognition Elements) and induce the opening of DNA in DUE (DNA unwinding Element). �The required replication factors coded for by the host cell, recruited and replication takes place.� Location of origin in different system differs, but initiation always takes place by the binding of specific factors such as origin recognition complexes (ORC) to specific sequences; this acts as replication initiation site.
Eukaryotic DNA is not like viral SV40 DNA, but compacted as chromatin/chromosome.� Chromatin DNA is associated with histones and non-histones.� Human chromosomes contain 50mb to 250 million base pairs long ntds� pairs.� Each of the chromosomes contains 50 to hundred or more origins some time they are clustered as Ori-Foci, Ori-Zones.� Each of Origins contain specific sites with specific sequences.� Look at diagrams below.� Yeast origins doesn�t hold good for higher systems.� As in bacterial systems there are no well-defined replication Ter sites.� There are many chromosomal zones where replication is not smooth perhaps contain barriers where replication slows down.� But replication of chromosomal terminal region is unique. Components of replication origins differ from one to the other. However, there are regions where there is no defined origin regions and replication initiates randomly, where ever the DNA sequence is A/T rich.
The ORC (Origin Replication Complex) is a protein binding complex, made up of 6 subunits (ORC p1, ORC p2, ORC p3, ORC p4, ORC p5, ORC p6) that has a binding specificity to DNA, more specifically to the origins of replication, in the presence of ATP, in the genomes of the eukaryotes. Theoretically this binding complex�s function is the initiation and/or regulation of the DNA replication in eukaryotes. More specifically, the ORC bound to the origins of replication act as the foundation for the assembly of the pre-replication complex (pre-RC), which is a protein complex essential for the initiation of the replication, in other words, the pre-RCs act as licensing factor of the chromosomes. That�s why ORCs do not work directly in DNA replication initiation but indirectly instead. In addition to this, the ORC is target of protein kinases which will phosphorylate some of it�s subunits in order to regulate DNA replication initiation and to block the reinitiation of the G2/M phases. (Blogspot). http://glossomics.blogspot.com
We the authors mentioned below report that a highly purified human origin recognition complex (HuORC) has intrinsic DNA-binding activity, and that this activity is modestly stimulated by ATP. ���HuORC binds preferentially to synthetic AT-rich polydeoxynucleotides, but does not effectively discriminate between natural DNA fragments that contain known human origins and control fragments. The complex fully restores DNA replication to ORC-depleted Xenopus egg extracts, providing strong evidence for its initiator function. Strikingly, HuORC stimulates initiation from any DNA sequence, and it does not preferentially replicate DNA containing human origin sequences. These data provide a biochemical explanation for the observation that in metazoans, initiation of DNA replication often occurs in a seemingly random pattern, and they have important implications for the nature of human origins of DNA replication, Sanjay Vashee1,4, Christin Cvetic2,4, Wenyan Lu3, Pamela Simancek3, Thomas J. Kelly3,6, and Johannes C. Walter2,5
Assembly of initiation factors at the S. cerevisiae origins; Frontiers in Bioscience; https://www.bioscience.org
Origin Recognition Complex; Stephen P. Bell; http://genesdev.cshlp.org/ and http://www.biochem.umd.edu/
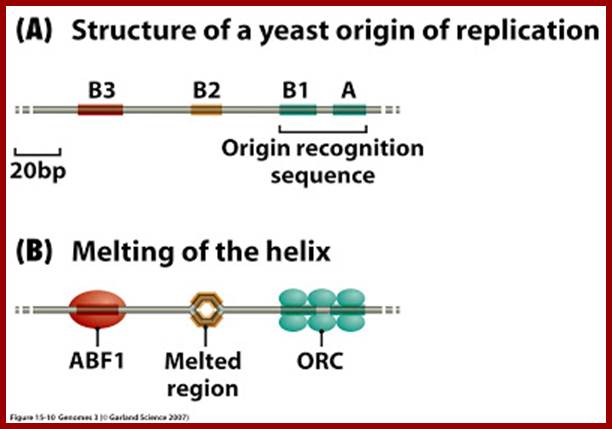
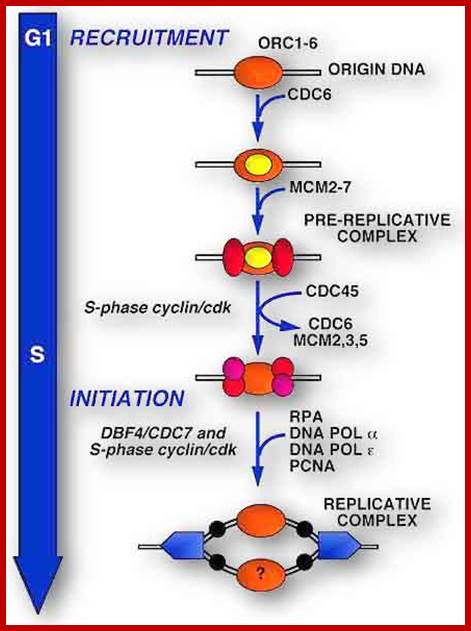
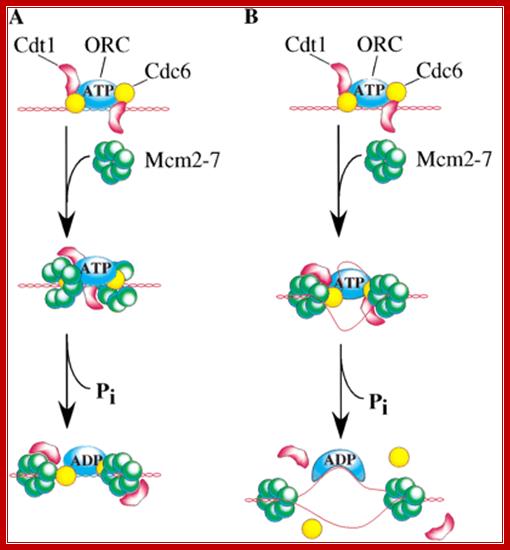
Origin recognition complex (ORC), a six member protein complex binds to Ori sequences; in yeast the ORS is 11bp long (�A� rich sequence), the binding to ARS is assisted by another protein, but Cdc6 may augment DNA binding; ORC and Cdc6p may act as clamp loader for hexameric MCM 2-7proteins. It is likely ORC/Cdc6p ATP binding coupled to ring opening of MCM and closing on to form Pre RC complex.
The MCM in open complex form is recruited by Cdc6/Cdt1 to ORC bound site and load the MCM; then MCM is activated to open the DNA into Replication bubble; For the function Cdc7/Dbf4 by phosphorylation of MCM leads to gate closing and activate opening; perhaps Cdc45 is required for to activate helicase activity. http://mmbr.asm.org/
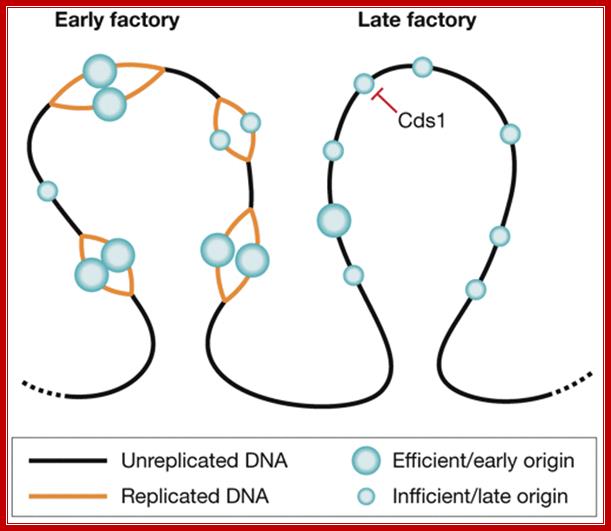
Organization of replication factories in fission yeast. Origins are found in clusters that replicate at different times in S phase and can be visualized as bright replication foci (factories). Origins can fire with different efficiencies. Efficient origins (large circles) fire early in S phase, whereas inefficient origins (small circles) can fire at different times throughout S phase, and so tend to be found in late factories. The S phase checkpoint kinase Cds1 might have a role in the replication timing programme by inhibiting the firing of defined clusters in early S phase. Silvia Costa, J Julian Blow; http://embor.embopress.org/content.
https://www.boundless.com; https://www.slideshare.net/
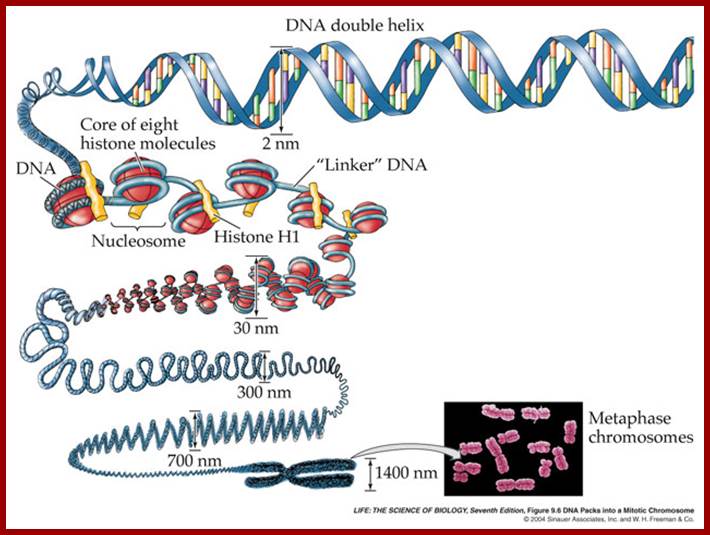
Chromatin during mitosis or meiosis undergoes structural changes.� During interphase the chromatin is relaxed and one can observe long nucleosomal thread bound to nuclear matrix at some sites.� Some regions of chromatin show 30nm structural features.� And one can observe certain regions of chromonemal threads show transcriptional factories and some regions are condensed and tightly packed, no transcriptional activity. While DNA replication is activated certain regions of chromatin remain transcriptionally active.� The chromatin becomes tightly condensed at metaphase, at which one does not find any transcriptional activity.
During DNA replication initiation ORC hexamer factors are bound to chromatin, probably at ORE regions.��� Binding of ORC complexes initiates the assembly of replication initiation and replication elongation factors.
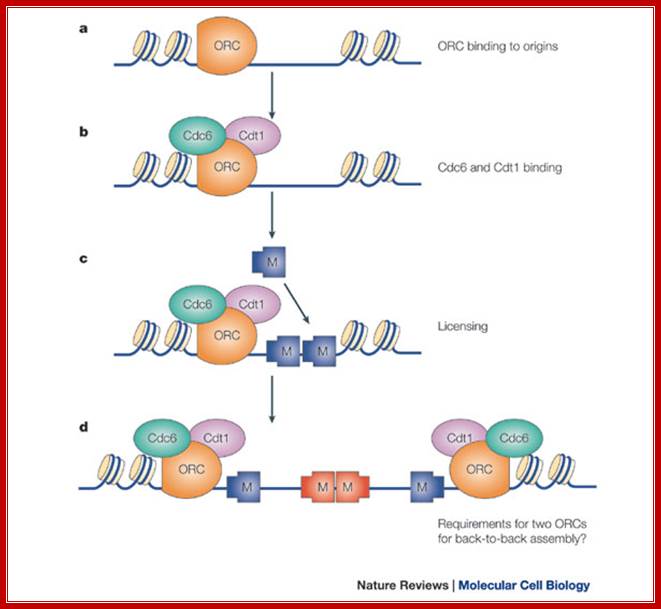
a | The origin recognition complex (ORC) is first recruited to the replication origin. b | ORC recruits Cdc6 and Cdt1. c | ORC, Cdc6 and Cdt1 act together to load multiple minichromosome maintenance (Mcm)2�7 protein hexamers onto the origin, which licenses the DNA for replication. d | Initiation-competent complexes are probably formed by the back-to-back assembly of two Mcm2�7 complexes. As the ORC is asymmetrical, this might require deposition of a second ORC molecule to load Mcm2�7 in the opposite orientation; J. Julian Blow & Anindya Dutta; Nature-2005.
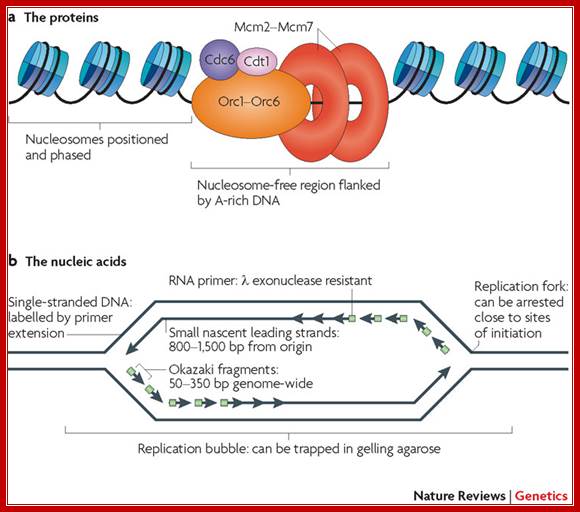
a | Pre-replication complexes consist of at least 14 different proteins conserved in all eukaryotes: cell division cycle protein 6 (Cdc6), DNA replication factor Cdt1, the heterohexameric origin recognition complex (composed of Orc1 to Orc6), and the heterohexameric mini-chromosome maintenance complex (composed of Mcm2 to Mcm7)1. The ORC competes with nucleosomes to bind to DNA and, once bound, is able to position nucleosomes in such a way as to leave sufficient space for MCM complex loading10. b | Summary of the unique nucleic acid features found near origins of replication. When cells that have been synchronized before the onset of S phase initiate replication in the presence of replication fork inhibitors, replication forks are arrested close to sites of initiation so that any DNA synthesized must be close to origins. The sites where forks are arrested consist of primed templates that can be labelled at the sites of arrest by extension. The leading strands of DNA synthesis quickly become larger than Okazaki fragments and can be isolated as small single-stranded molecules that can be verified to be nascent either by metabolic labelling or by virtue of the fact that nascent strands have small stretches of RNA at their 5′ ends that render them resistant to λ exonuclease55, 56. Finally, the physical structure of replication origins shortly after initiation is that of a bubble structure, which can be trapped in gelling agarose; Nature review David M. Gilbert
A very important event, of the most significance, happens in yeast. It is the role of licensing factors, which also control cell division via initiation of replication.����
In yeast, certain proteins like OBFs (Origin Binding Factors) or origin recognition complex (ORC), six subunits ORC1-PRC6, bind to ORE (Origin Recognition Elements) sequences, they act as positional factors, that facilitates the binding of other factors to induce melting of the DNA in that region, which takes place at A/T rich region (DUE). ORC1 and ORC5 interact with ATPs.� The binding of accessory factors or auxiliary factors to AUX-I AUX-II regions can further facilitate or augment melting of the DNA.
In yeast Cdc6 and Cdt1 activate MCM (Mini Chromosome Maintenance) factors to load on to the ORC (hexamer), which is already bound to origin site.� Yeast, autonomously replicating plasmid origin site, has been very well elucidated.
The binding of MCM2-7 to initiate opening of the DNA into Replication bubble; requires other factors such a Cdc7/DbF1. Phosphorylation of MCM2 by Cdc7/DbF1 activates its helicase activity to unwind the dsDNA.� MCM binds to ATP for it has ATPase domain.� Among the six MCMs, MCM 3, 5 and 6 are essential and others� role is not clear. For initiation of replication Cdc45 is required and the same is loaded on to fork joints. �At this point ORC, Cdc6 and Cdt1 are unloaded from the origin complex, the same are subjected ubiquitin- mediated degradation. Cdc45 is required for loading of DNAP alpha, Epsilon, RPA and PCNA on to chromatin. Cdc45 commits replication initiation. Note Cdc7-Dbf4 kinases are essential for loading Cdc45. Cdc7 is also called DDK. The unused MCMs are also degraded.�
Recruitment of MCM dimers, to Origin site, induces the MCM complex switch from ds DNA binding to ssDNA strands. Another complex essential is GINS (go, ichi, in, san-�5, 1, 2, 3� in Japanese language). GINs in association with Cdc45 and MCM act as coupling sites between helicase and polymerase, thus ensure unwinding and DNA synthesis go together in harmony.� Replication Factor Complex RFC is multi-subunit component, Rfc1-5 and they are ATPases.� RFC facilitates PCNA to load Pol delta at every end of OKAZAKI fragment.
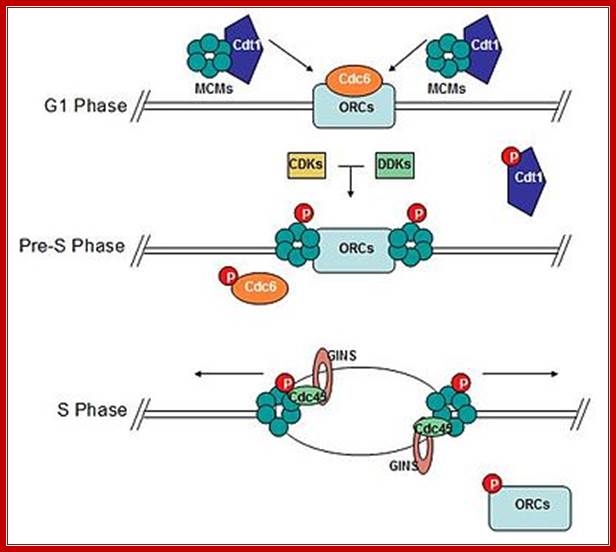
Diagram of the formation of the pre-replicative complex transforming into an active replisome. Mcm 2-7 complex loads onto DNA at replication origins during G1 and unwinds DNA ahead of replicative polymerases. Cdc6 and Cdt1 bring Mcm complexes to replication origins. CDK/DDK-dependent phosphorylation of pre-replicative proteins leads to replisome assembly and origin firing. Cdc6 and Cdt1 are no longer required and are removed from the nucleus or degraded. Mcms and associated proteins, GINS and Cdc45, unwind DNA to expose template DNA. At this point replisome assembly is completed and replication is initiated. "P" represents phosphorylation; MCM subunits organized in a sequence-Mcm3-7, Mcm2-6 ad Mcm4-5; WIKIPEDIA.
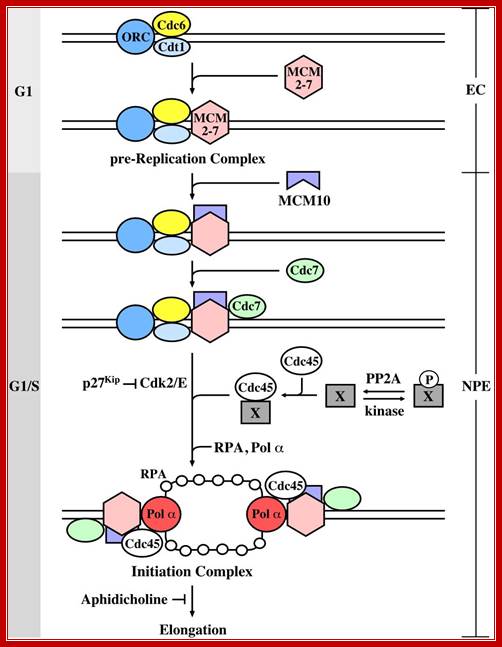
The initiation of chromosomal DNA replication in eukaryotes can be divided into two general steps.� The first occurs during the G1 phase of the cell cycle and involves the formation of a pre-replication complex (pre-RC) containing the origin recognition complex (ORC), Cdc6, Cdt1 and MCM. At the G1/S transition, cyclin-dependent kinase (Cdks) and the Cdc7-Dbf4 kinase convert the pre-RC into an active replication fork by a currently unknown mechanism. There is evidence that the substrate of Cdc7-Dbf4 may be the MCM complex (?), whereas the Cdk substrates are not known. To investigate whether PP2A is involved in the replication of chromosomal DNA, authors used Xenopus cell-free replication system and found that removal of PP2A from egg extract caused complete inhibition of DNA replication, and that PP2A is required for initiation but not elongation of DNA replication. (Stephen Kearsey, Regulation of eukaryotic DNA replication). Mcm10 binds to the complex depending on assembly of the CMG components. Loading of Mcm10 does not cause release of Sld3, Cut5 or Drc1. (iv) Mcm10 forms homo-multimers and promotes conversion of the Mcm2-7 complex from the dsDNA-bound double hexamer into two ssDNA-bound single hexamers through its interactions with multiple subunits of Mcm2-7. Once single stranded DNA templates are made available RPA bind to maintain the ssDNA strands till the new strand is synthesized. Gernot Walter; http://molpath.ucsd.edu/
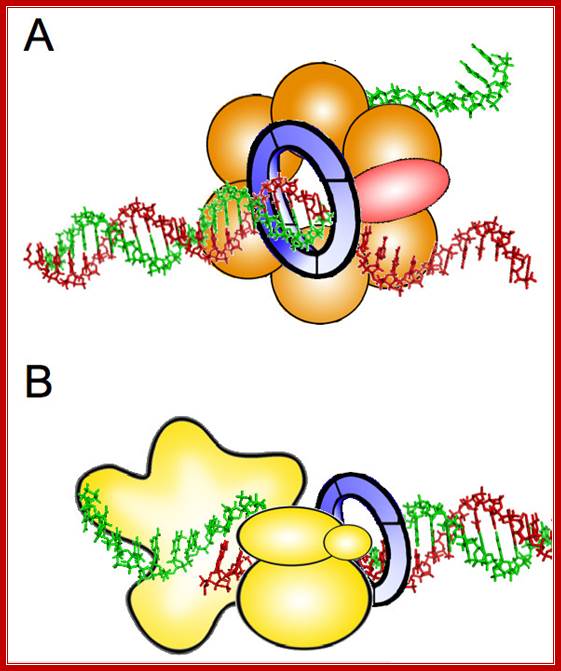
Possible roles of GINS during DNA replication. A. Structural component of the CMG complex that contains DNA helicase activity. Cdc45 protein is represented in red, Mcm2�7 proteins in orange, and GINS as a blue ring. B. DNA polymerase ε auxiliary factor. The different subunits of Pol ε are depicted in yellow. The relative position of the different components in each protein complex is speculative. See text for details. http://www.celldiv.com/
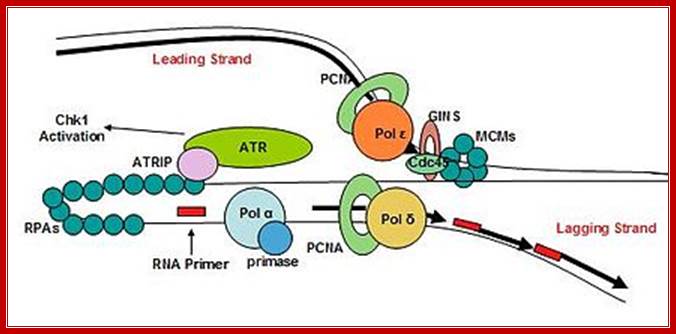
Replisome complex; Replication initiation steps and elongation- Pol alpha uses primers for elongation and pol Epsilon ahead of PCNA ring continues to replicate lagging strand. Pol delta replicates lagging strand producing multiple segments of newly synthesized short strands which are then ligated after removing primers; http://molpath.ucsd.edu/
Initiation requires a stepwise association of proteins with replication origins before DNA synthesis can begin. The origin recognition complex (ORC) binds to DNA and provides a site on the chromosome where additional replication factors can associate. An early step leading to initiation, called licensing or pre-replicative complex formation, involves the association of Mcm2-7 complex with DNA at ORC, in a process requiring Cdt1 and Cdc6. Mcm2-7 proteins provide helicase activity for DNA synthesis and loading of these proteins confers competence on the origin to fire in S phase. Onset of DNA synthesis requires the action of two protein kinases (cyclin dependent kinase (CDK) and Cdc7), which trigger the association of additional proteins with the origin, such as Cdc45 and GINS. During the process of initiation, DNA polymerases are also recruited and DNA synthesis starts. During replication, Mcm2-7 proteins move away from the origin and further assembly of pre-replicative complexes is blocked. This ensures that origins can only fire a single time per cell cycle. For further details see www.dnareplication.net. http://en.wikipedia.org/
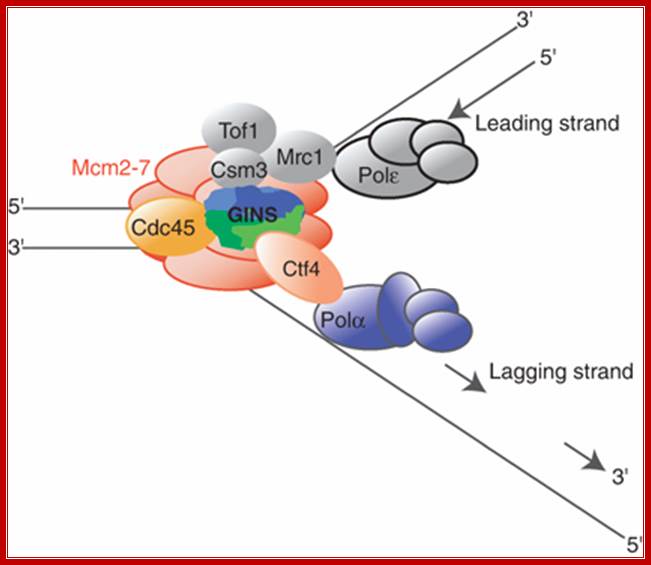
The role of Ctf4 in promoting replication fork stability
Ctf4 is the budding yeast And1 homolog and links polymerase alpha to the RF, specifically the helicase. Gambus et al. (2009) demonstrate in this paper that Ctf4 associates with helicase components (MCM proteins and also the GINS complex). They also show that cells lacking both Ctf4 and Mrc1 (delta-ctf4 and delta-mrc1 mutants) cannot survive, demonstrating how important the FPC is to cell health. If Mrc1 is not present (delta-mrc1), the Ctf4 component of the FPC becomes critical to holding the RF together. � 2009 Nature Publishing Group Gambus, A. et al. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase α within the eukaryotic replisome. The EMBO Journal 28, 2992�3004 (2009) doi:10.1038/emboj.2009.226. All rights reserved.
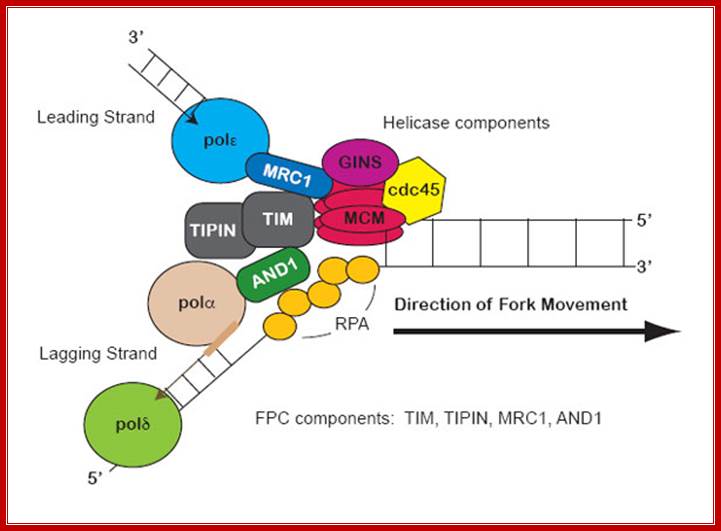
Replication fork components; The RF is a multiprotein complex with helicase and DNA synthesis activities. It is called a fork because the structure resembles a two-pronged fork. The helicase activities unwind DNA in front of the fork to create regions of singled-stranded DNA (ssDNA). The helicase components shown are the minichromosome maintenance (MCM) helicase 2-7 hexamer, CDC45, and associated GINS complex (simplified as a single entity here). The ssDNA is coated in RPA (yellow circles) to keep strands from reannealing. Fork protection complex (FPC) components shown are Timeless (TIM), Tipin (TIPIN), Claspin (CLASPIN), and And1 (AND1). Claspin (MRC1 in yeast) helps connect the leading-strand polymerase epsilon (light blue circle) to the helicase. And1 connects the lagging-strand polymerase alpha (tan circle) to the helicase. Pol-alpha is part of the primase complex, which synthesizes primers (thick tan line) on the lagging strand. These primers allow the polymerase of the main lagging-strand (polymerase delta, light green circle) to start synthesis. The direction of DNA synthesis and RF movement into DNA is indicated by arrows. � 2010 Nature Education All rights reserved.
In SV 40 the melting and stabilization of melted DNA is also facilitated by upstream transcriptional initiation factors like Sp1 (GC binding factors in SV 40) and other TFs.� The RNA Pol II Holozyme is involved in this process.� Probably enhancers too contribute to this event.
Chromosomal DNA is bound by histones and nonhistones.� The replication factors have to find their Ori sites to bind; it means the chromatin in the region should be remodeled to DNA free from chromatin proteins; Tomas Aparicio etal
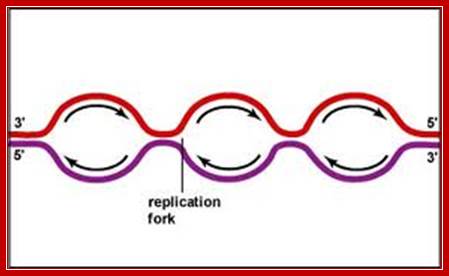
Replication proceeds in bi-directional manner, so each of the replication forks meet from opposite directions. Though replication is initiated in most of the origins simultaneously, replication of heterochromatin segments is delayed in comparison to euchromatin DNA. The last region that replicates is telomeric DNA.�
As in the case of SV 40, yeast and other Eukaryotes, replication initiation is controlled by what is called licensing factors RFM and RFL. The activity of these factors in regulating replication initiation events takes place only once per cell cycle.
At this stage, Cdc4-6 and G1 Cyclins are inhibited by Sic1p, but soon the inhibitor gets phosphorylated and degraded by SPF (S phase Promoting Factor) by ubiquitination process. In addition, few more Cdk-cyclin inhibitors are also inactivated.� Activated CDC2, 4/6-Cyclins phosphorylate Retino-Blastoma (RB) (tumor suppressor proteins) and release E2F transcription factors.� Similarly, p107 and p130, that have sequestered few other E2Fs, are also released.� The released E2Fs bind to their respective promoters and activate gene expression required factors for S-phase DNA replication events.
�
Cdc6 has a very short half-life say 2-5 minutes (in S.pombe, the fission yeast, it is called cdc18P). This is acquired only when the chromosomes are free from nuclear membrane i.e. between late M and G1.� The presence of phosphorylated Cdc6-P is essential for loading MCM 2-7 proteins on to the replication fork, one on each side of the fork.� The movement is like unwinding.
The Geminin by sequestering Cdc6/Cdt1 prevents reinitiation chromosomal DNA replication before the completion of Mitotic stage; later Geminin is degraded by APC1.� Once the replication bubble is formed and stabilized by ssDNA binding protein RPA, other replication components join.
The replication components are RPA 1, DNA Pol-α , RFC1, PCNA, DNA Pol �δ- and ε-proteins, they assemble at replication fork one at each fork as a complex.
At the end MCM proteins dissociate from DNA i.e. after replication but remain intact and stable, only Cdc6/Cdt1 proteins are destroyed in cell cycle dependent manner, that too by ubiquitination process.��
As long as these factors are not available as a pre initiation components, DNA replication is not initiated, hence Cdc6 and Cdt1 are called as licensing factor B and MCM are called licensing factor M.
The licensing factors are synthesized only once in a cell cycle and they are acquired by the nucleus when the nuclear membrane reassembles.� This happens only once in one cell cycle.�
Interestingly, if Cdc6 is made available by ectopic expression at G2 stage, MCM complex doesn�t bind to the ORC; it means that there is another control point with another licensing factor?
As the replication of DNA completes Cdh1 activates Anaphase Promoting Complex (APC) by Cdc20. They in turn mark Cyclin-A and Cyclin-B for ubiquitinated degradation.� They also mark the degradation of securins.� That leads the release of a protease which degrades cohesin components thus the daughter DNA molecules are released from one another at centromere, which is essential for the anaphasic movement of chromosomes.� Degradation of CyclinA and Cyclin-B prevent reinitiation of Mitosis before another round of DNA replication.
Opening of the DNA into ss DNA is stabilized by ss binding proteins RF-A (RPA).� They also prevent reannealing the DNA strands. The replication bubbles provide fork joints at which helicases bind.�
In SV 40, T-antigen, itself acts like a helicase (an hexamer) with 3�>>5� directional movement.�
In eukaryotic systems one MCM2-7 helicase complexes one each at fork found. As and when the replication bubble is formed, it is stabilized by SSB proteins, DNA pol-a, which has both 5�>>3� polymerase and primase activity, recognizes single stranded DNA and using specific sequences, binds to ssDNA and lays RNA primers on both leading and lagging strands in opposite orientation. This means, at central position one primer in each strand is laid, one for leading strand replication and another set of primers for lagging strands; however orientation of the primers on each of the strands depends upon the orientation the replicating strand. Leading and lagging strands are differentiated based on the� 5�-3� and 3� to 5� orientation of the strands.
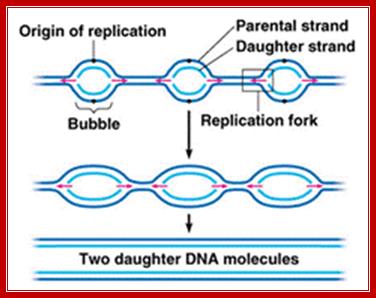
http://www.majordifferences.com/
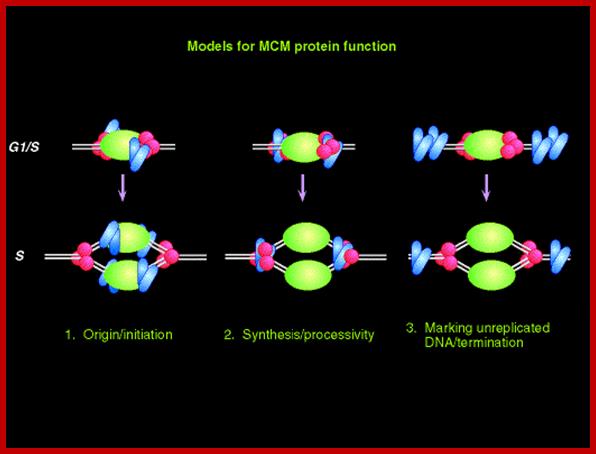
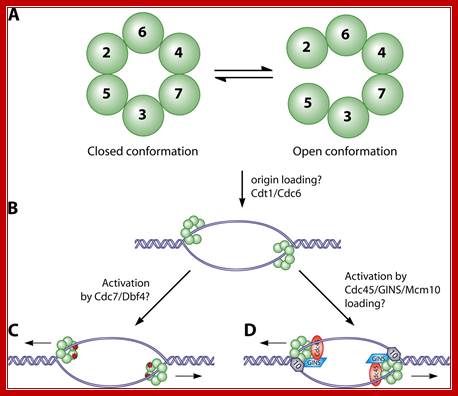
The required proteins are abundant, and exceed the stoichiometry of origins by 100-fold. In the cartoon above, a diagram is presented of these three models; the origin recognition proteins are shown in green; the replication machinery (polymerases etc) is shown in red, and the MCM proteins are shown in blue. Note that these models are not mutually exclusive. www-bcf.usc.edu
MCMs have homology to a wide class of putative DNA-dependent ATPases, and bind together as a hexamer. SV40 T antigen and bacterial DnaB protein are the helicases in their respective systems, and also have hexameric structure. However, in vitro helicase activity is only observed for a complex of Mcm4-6-7. Thus, there are some issues yet to be resolved. (1) All six MCMs are essential in eukaryotes, and evidence suggests that in the normal nucleus, they are present at 1:1:1:1:1:1 stoichiometry. Neighborhood of MCM is MCM3 withMCM7, MCM4 with MCM6 and Mcm2 with Mcm5. But the in vitro data doesn't account for Mcm2, Mcm3, or Mcm5. �
MCMs are amazingly abundant, far exceeding the stoichiometry of replication origins. Reducing the dose of MCMs has severe phenotypes, even though the cells can still synthesize DNA. Cytological data suggests that MCMs liberally decorate unreplicated chromatin, but do not co-localize with the "replication factories" that contain PCNA and other elongation factors.�
Model of the Mcm2-7 hetero-hexamer: http://www.jbc.org/
Pair wise interactions in vitro suggest this organization of the MCM is in the form of heterohexamer. The P-loop (Walker A motif) and SRF motif (arginine finger) are proposed to act together as ATPase domains. In isolation, this complex does not have helicase activity in vitro (see the text).
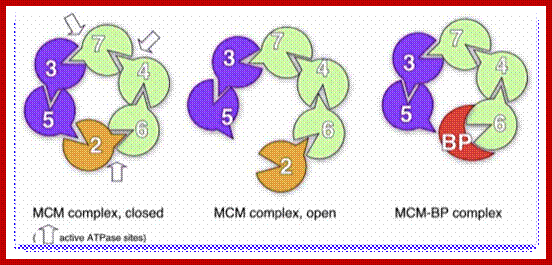
A cluster of MCM2-7 proteins bind and act as helicases in unwinding and progression of replication fork. http://www-bcf.usc.edu
Overview of LTag hexamer conformations and the β-hairpin structure in different nucleotide binding states; Three nucleotide binding states of LTag Hexameric helicase, seen from above (N-terminal face). Monomers are colored, with their central β-hairpins in red (equivalent to the PS1-hp of ssoMCM). The N-domain, D1, has been removed for clarity; https://www.ncbi.nlm.nih.gov
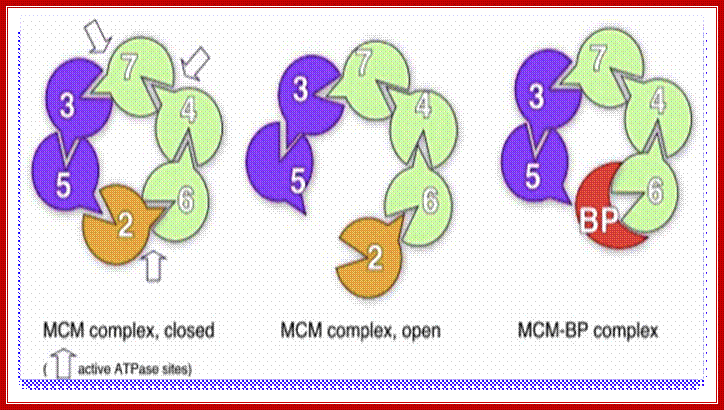
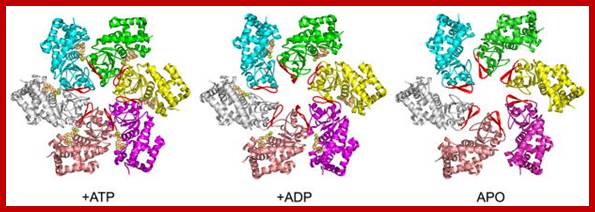
MCM complexes. Left, the MCM complex on chromatin hydrolyzes ATP at three of the six possible sites. middle, Mcm2 acts as a gating subunit. Right, in some cases, Mcm2 may be replaced by MCM-BP, which renders the complex inactive. This might be a way to remove the complex from the chromatin. http://www-bcf.usc.edu/
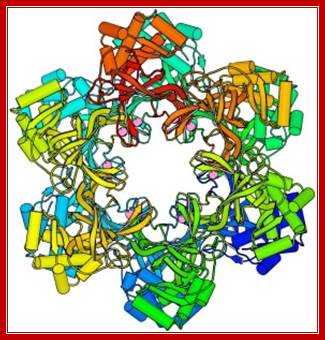
Reprinted from reference 55 with permission from the publisher: Aren't they pretty? This structure of the archaeal MCM protein hexamer comes from our colleague (Xiaojiang Chen) and Forsburg Lab Research: MCM Proteinshttp://www-bcf.usc.edu/
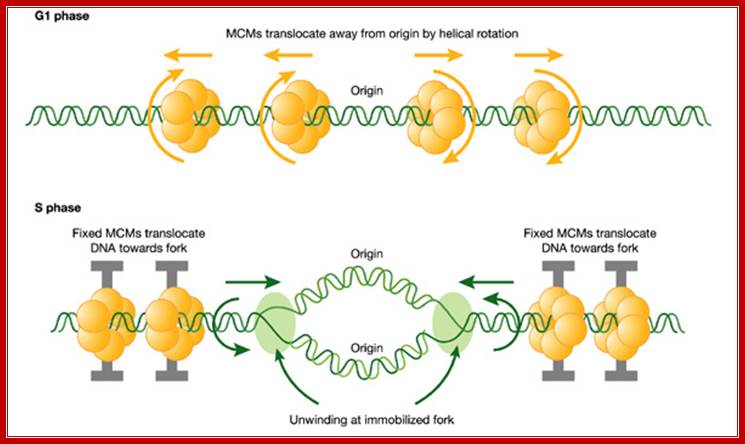
A hypothetical rotary pump model showing two stages in the distribution and function of MCM hexameric ATPase complexes. First, in G1 phase, MCM hexamers move spirally along the helical grooves of unreplicated DNA, away from ORC, which is required for their loading and orientation. Second, in S phase, MCMs become immobilized, so that exactly the same rotary mechanism moves the DNA instead of the MCM proteins. This would result in translocation of DNA towards the replication forks. As DNA is twisted by fixed MCMs in S phase, it would become unwound at the distant replication fork, which is itself immobilized in fixed clusters; A hypothetical rotary pump model showing two stages in the distribution and function of MCM; hexameric ATPase complexes. Ronald A Laskey, Mark A Madine;http://embor.embopress.org/
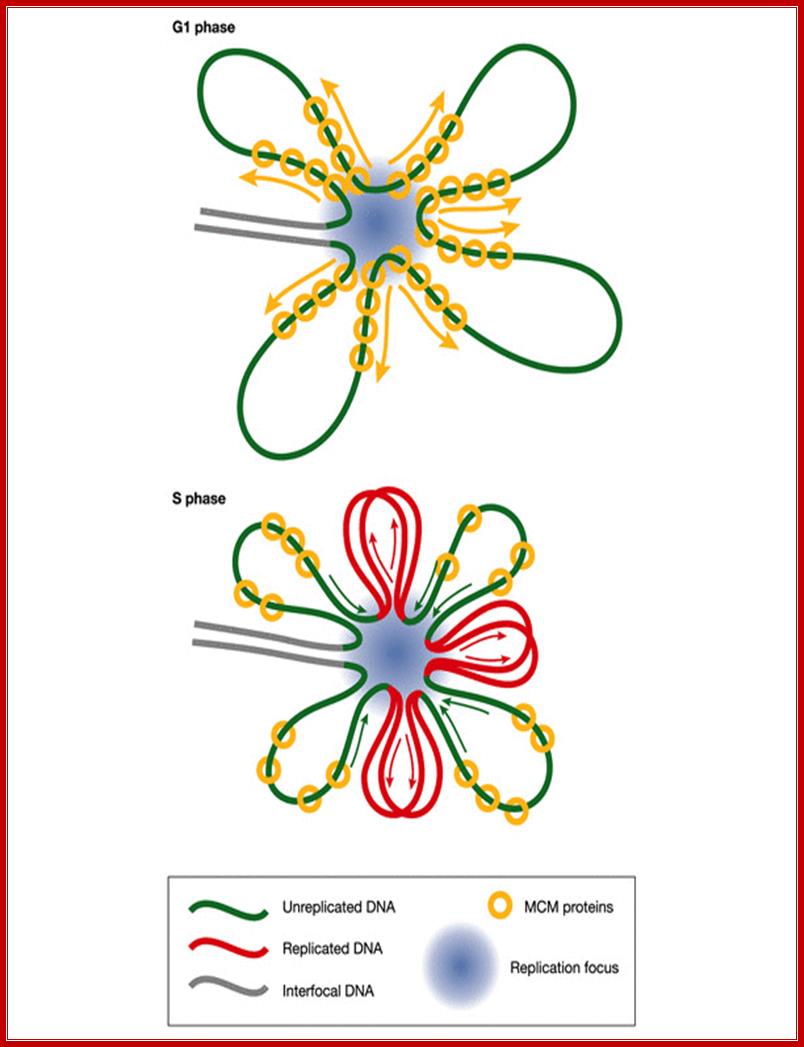
Implications of the rotary pump model for the clustering of several hundred replication forks into foci. Before DNA replication in G1 phase, MCM complexes (yellow) are distributed along radial loops of unreplicated DNA (green). During DNA replication in S phase, the forks remain clustered in foci (blue spot) and extrude replicated DNA loops (red). Arrows indicate growth and shrinkage of replicated and unreplicated loops, respectively, as replication proceeds. Ronald A Laskey, Mark A Madine; http://embor.embopress.org/
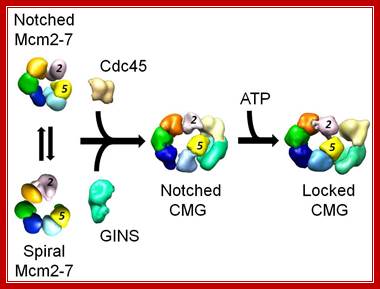
Figure: Activation of the eukaryotic replicative helicase. The isolated Mcm2-7 assembly exists in two forms, a planar, notched ring and a spiral state, with an opening between subunits 2 and 5. Binding of the Cdc45 and GINS activators to the side of the ring seal off the opening and stabilize the planar configuration. Nucleotide binding tightens the ring, giving rise to two topologically segregated conduits involved in tracking the leading and lagging strands at the replication fork, Alessandro Costa
������������������������������������������
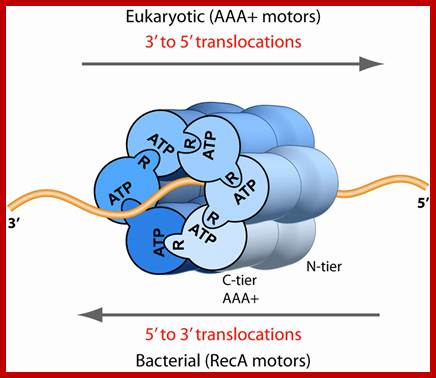
�DNA translocation polarity in the RecA-based and AAA+-based superfamilies of bacterial and eukaryotic hexameric helicases. ATP sites are at subunit interfaces in both the bacterial RecA-based motors and eukaryotic AAA+ motors. ATP signifies the Walker A site and R signifies the arginine finger. Crystal structure analysis of BPV E1 (12), E. coli DnaB (13), and E. coli Rho (3), members of the SF3, -4, and -5 superfamilies, respectively, show that DNA binds the motors in similar fashion, illustrated here by looking down the DNA axis from the 3′ terminus, and into the motors from the C-surface view; the P loops lie clockwise to the arginine finger.
�Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation; Eukaryoptivc (AAA+Motors);http://www.pnas.org/
First, in G1 phase, MCM hexamers move spirally along the helical grooves of unreplicated DNA, away from ORC, which is required for their loading and orientation. Second, in S phase, MCMs become immobilized, so that exactly the same rotary mechanism moves the DNA instead of the MCM proteins. This would result in translocation of DNA towards the replication forks. As DNA is twisted by fixed MCMs in S phase, it would become unwound at the distant replication fork, which is itself immobilized in fixed clusters.� The direction of MCM bound to single strand is 3�-5�.
�����������������������������������������������������������
Eukaryotic DNA replication:
Step
1: Primer synthesis by DNA polymerase-α
(Pol-α); step 2: replication factor C (RFC) displacement of DNA polymerase
alpha and recruitment of proliferating cell nuclear antigen (PCNA); step 3:
elongation by the newly recruited DNA polymerase-δ
holoenzyme (Pol-δ); step 4: strand
displacement.
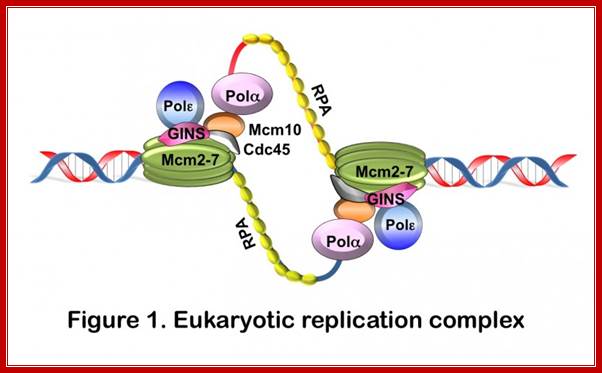
DNA replication is an integral part of the eukaryotic cell cycle, which coordinates the duplication of the entire genome during S phase. At the onset of S phase, origins of DNA replication are activated to "fire". Once DNA synthesis has been initiated, the cell is committed to undergo a complete round of division. Dysregulation of origin activation results, therefore, in uncontrolled cell proliferation, which is at the heart of diseases such as cancer. To initiate DNA replication, cells must assemble replication initiation complexes. Key components of these complexes are the minichromosomal maintenance (Mcm) proteins. Mcm2-7 comprise the core of the replicative helicase and Mcm10 is a scaffold protein that links polymerization to DNA unwinding (it connects Mcm2-7 with Polα; Anja-Katrin Bielinsky; http://www.cbs.umn.edu/;
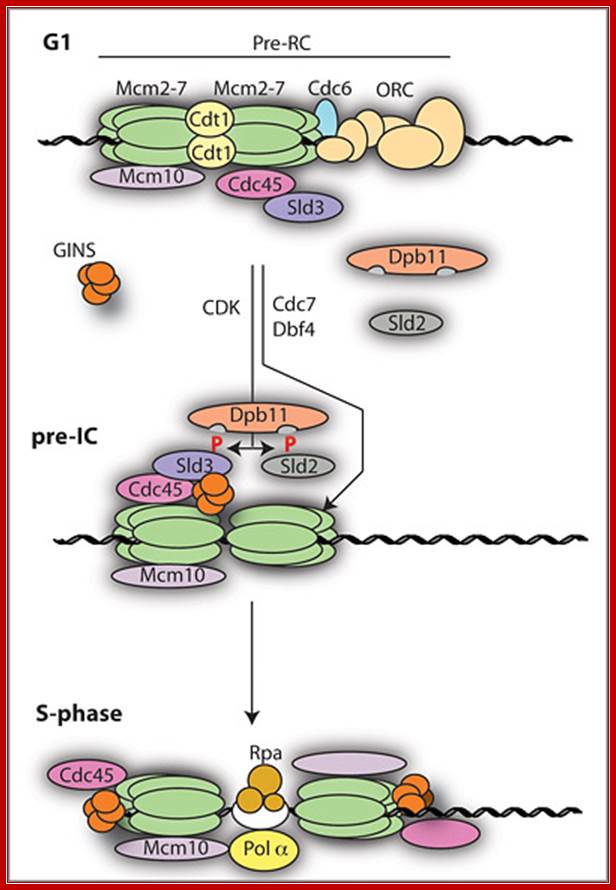
The sequence of eukaryotic replication initiation. Philip Zegerman
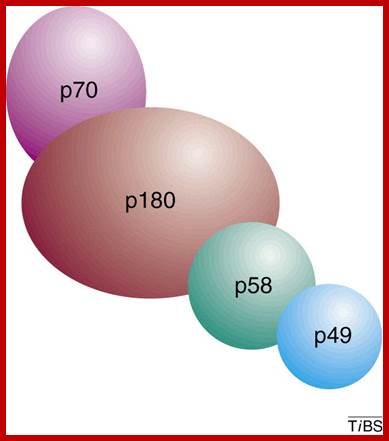
Eukaryotic DNA pol-primase: �Subunit 70 and 180 act as alpha-polymerase and subunits 58 and 49 act as primase; Eukaryotic DNA primase initiates the synthesis of all new DNA strands by synthesizing short RNA oligomers on single-stranded DNA. It also synthesizes DNA strands on lagging strands too. �It is also involved in DNA damage repair, telomere maintenance and epigenetic processes in high ordered chromatins. Additionally, primase helps couple replication and repair and is critical for telomere maintenance and, therefore, chromosome stability. In light of the many aspects of DNA metabolism in which primase is involved, understanding the unique features of the mechanism of this enzyme and how it interacts with other proteins will greatly advance our knowledge of DNA replication and repair. Bahram Arezia, ;http://www.sciencedirect.com
When Polymarase subunits with primase subunits bind where subunit 46 contacts MCM proteins for elongation of the strand
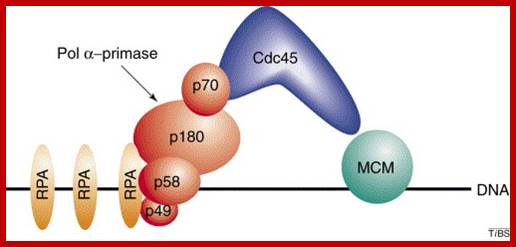
Eukaryotic DNA primase initiates the synthesis of all new DNA strands by synthesizing short RNA oligomers on single-stranded DNA. Additionally, primase helps couple replication and repair and is critical for telomere maintenance and, therefore, chromosome stability. In light of the many aspects of DNA metabolism in which primase is involved, understanding the unique features of the mechanism of this enzyme and how it interacts with other proteins will greatly advance our knowledge of DNA replication and repair. Eukaryotic DNA primase- Bahram Arezi
Robert D Kuchta; http://www.cell.com
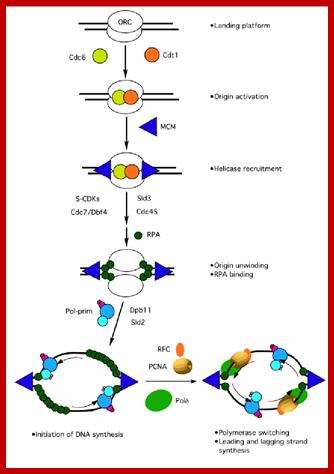
The assembly of the replication machinery at a yeast origin. The names of the indicated factors correspond to the components identified in S. cerevisiae, even though most of them are common in all eukaryotic cells; The maintenance of genome stability is of crucial importance for all living organisms. DNA must be replicated with high fidelity every cell cycle and different types of DNA damage must efficiently be repaired through the correct interplay among several DNA transactions (replication, repair, recombination, and transcription). ;https://www.researchgate.net
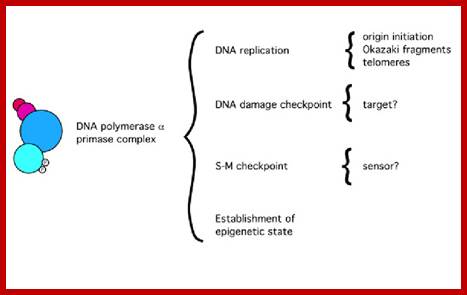
Functions of the Pol alpha-prim complex in DNA metabolism; https://www.researchgate.net
Elongation:
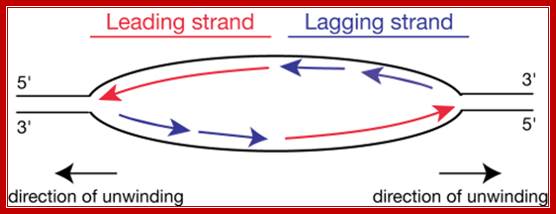
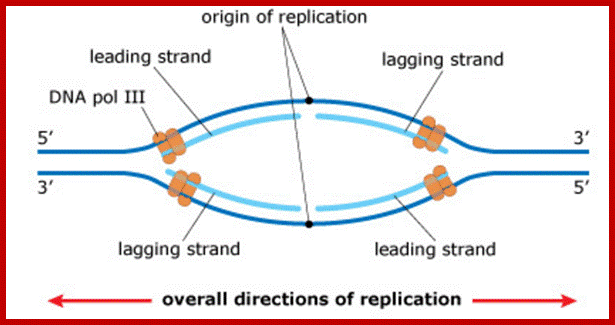
http://www.easynotecards.com
������������
by-Kistyn Dohn; www.bio.miami.edu; www.bio.miami.edu; https://www.slideshare.net
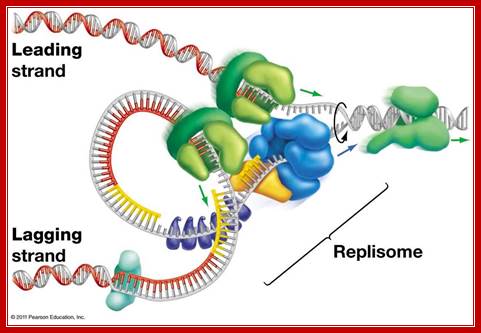
MCM proteins and checkpoint kinases get together at the fork;
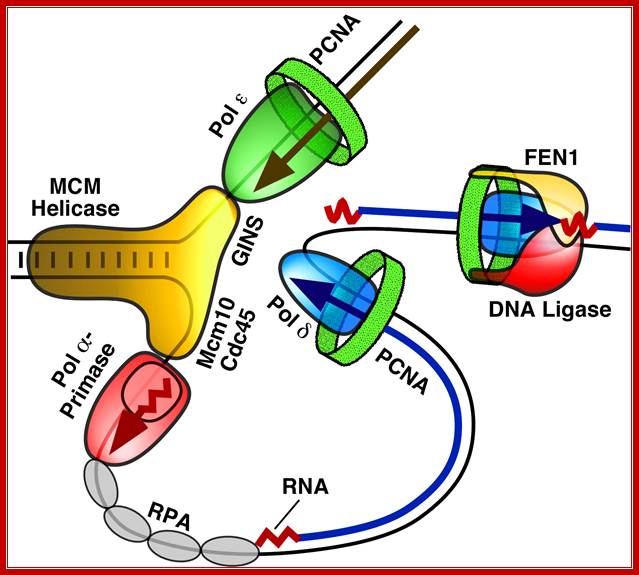
Figure. Diagram of the replication fork generated by MCM helicase, which separates the two strands of DNA, and single stranded binding proteins (RPA), which protect the strands. One of the strands is replicated in a continuous strand by DNA polymerase ε complexed with the protein PCNA (green). The second strand is replicated in shorter fragments, each beginning with the RNA/DNA primer made by pol-prim and extended by a DNA polymerase δ (blue) complex. Here you see pol-prim (Pol α-Primase - Red) bound to the DNA strand ready to displace RPA as it synthesizes a new primer. The fragments are linked together by the FEN1/DNA ligase complex. Image reproduced with the permission of Professor Peter Burgers, http://biochem.wustl.edu/~burgersw3/Replication.htm. Carol A. Rouzer, VICB Communications
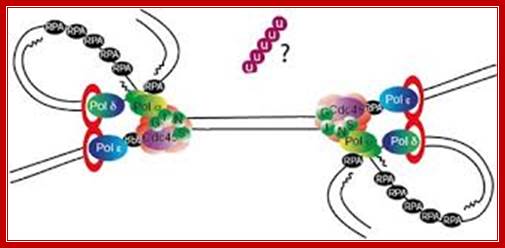
Chromosomal Replication; Dr. Aga Gambus; http://www.birmingham.ac.uk/
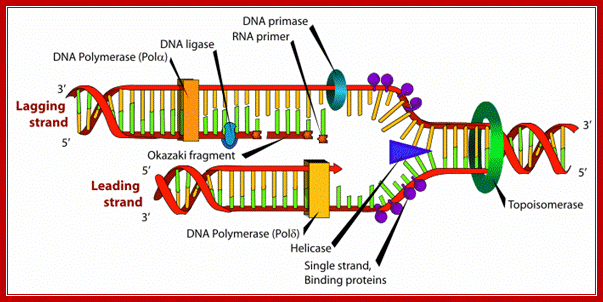
DNA Replication; www.wonderwhizkids
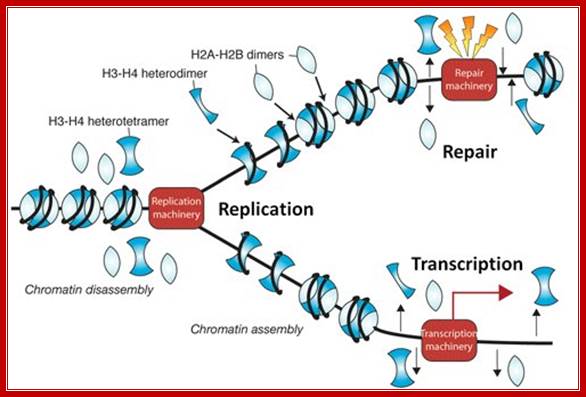
During chromosomal DNA replication chromatin associated proteins dissociate, but some ex. Some histone remain associated with DNA and as new DNA is produced the histone associate and nonhistones joint to form new chromatin. Prof Jessica Tyler; http://www.gla.ac.uk/
As the primers are laid, RF-C binds to POL-a; this leads to the extension of RNA primer into a short sequence of DNA called iDNA (initiator DNA).� The same enzyme uses dNTPs and assembles DNA segment of about 100 ntds long. Now RF-C clamp loading factor similar to E.coli beta clamp loader (i.e. li E.coli delta-gamma complex), loads the trimeric clamp called PCNA.
PCNA displaces the DNA Pol-a; and the RF-C now facilitates the loading of POL-E on to the iDNA. DNA Pol-ε loaded extends iDNA on leading strand.�
Similarly, primers laid on lagging strands are extended by PCNA-Pol-d complex.�� RF-C performs the loading of PCNA and Pol-delta every time when they dissociate on lagging strand.� The replication model is similar to that of prokaryote-�Trombone model�.
Whether the Pol-e -PCNA complexes are found as dimers at each fork or not is not clear, but it is logical that a pair of Pol-d-PCNA complexes to be present at each fork.� One of them synthesizes new strand on leading template and the other works on lagging strand in reverse orientation and produces DNA fragments in discontinuous manner.
DNA-pol a continues to assemble on lagging strand at frequent intervals and lays short primers, and RF-C activates the pol-a to extend it as I- DNA.�
Unlike the primase of prokaryotes, where the primase translocates with the helicase, the pol-a seems not to bind to any helicase or any other translocation protein on DNA.�
Similar to prokaryotes, primers in eukaryotes are removed by MF-1, FEN1 and RNase H proteins by their 3��5�exonuclease and endonuclease activities. The gaps are filled by pol-α and d polymerase.�
The gaps are sealed by DNA-ligases.� Mechanism of Ligases is similar to that of prokaryotes.�
The extended forks from opposite side�s meet they end up without any problem for the helicases dissociate.� Incomplete strands with 3�OH are extended and then ends are ligated. But the Telomeric ends replication requires Telomerase and its partner RNA for the extension of the ends.� This is delt separately.
Replication is not as smooth as we believe:
As we see the above drawing, line can observe what sort of obstacle come across, they have to be overcome by specific mechanism and then Replication progresses. Often DNA structural features act as hindrances and thus Replication gets slowdown. lifesci/departments/biotech;martin@post.tau.ac.il
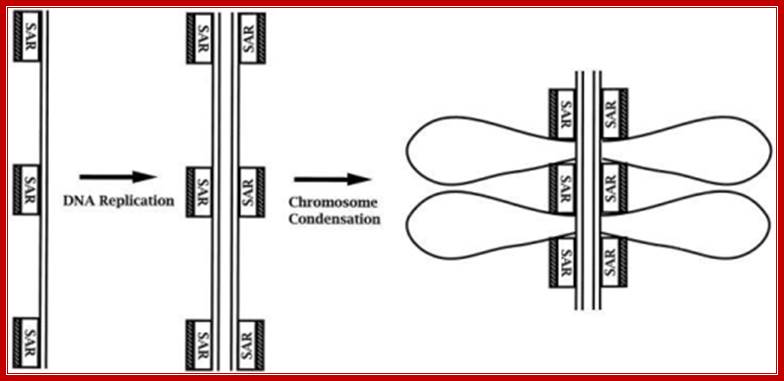
DNA Replication and Chromonemal Formation go Hand in Hand;
Figure; Model of the templating of the chromosome scaffold during DNA replication. In this model, we suggest that topo II (stippled bar) binds to new SAR sites as they are generated by DNA replication. At the termination of replication, this process yields sister DNA duplexes, each with independent scaffold elements that contain the information necessary to condense into sister chromatids independent of DNA decatenation.
Mentions: We present a model (Fig) in which topo II associates with the daughter DNA duplexes in a semiconservative manner during DNA replication. Topo II can bind DNA through SARs (Gasser and Laemmli, 1986; Adachi et al., 1989). We propose that topo II remains associated with SAR sites of the parental DNA strand and that topo II also associates with SAR sites that are generated on daughter DNA strands during DNA replication. In this manner, the process of DNA replication yields independent templates for condensation of the sister DNA duplexes, such as those we have observed in cells treated with ICRF-193 and 2-AP. This model suggests that autonomous organization of the sister duplexes is templated by their independent associations with topo II and are therefore preorganized for chromosome condensation. By this model, final condensation of the chromosome would involve compaction of the topo II�associated axial cores, bringing with them condensed DNA loops that are anchored to topo II. (Gasser and Laemmli, 1986; Adachi et al., 1989).
While chromosomal DNA goes through replication topoisomerases associate with DNA at SAR sites.� Action of TopII decatanation by TopII is not required to template the chromosome structure. We propose that the chromosome core structure is templated during interphase, before DNA decatanation, and that condensation of the two-armed chromosome scaffold can therefore occur independently of the formation of two intact and separate DNA helices; Andreessen PR, Lacroix FB, Margolis RL - J. Cell Biol. (1997).
Ligase mechanism
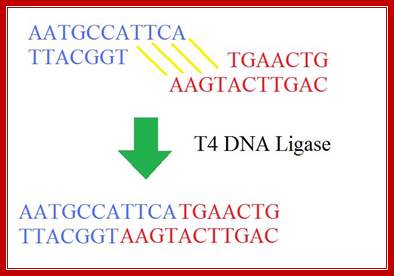
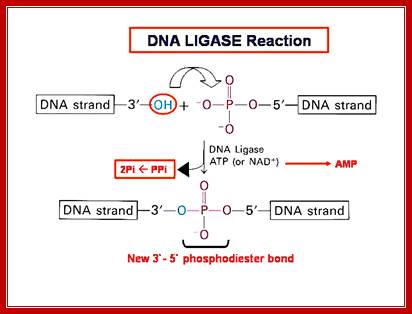
The mechanism of DNA ligase is to form two covalent phosphodiester bonds between 3' hydroxyl ends of one nucleotide, ("acceptor") with the 5' phosphate end of another ("donor"). ATP is required for the ligase reaction, which proceeds in three steps: (1) adenylation (addition of AMP) of a residue in the active center of the enzyme; pyrophosphate is released; (2) transfer of the AMP to the 5' phosphate of the so-called donor, formation of a pyrophosphate bond; (3) formation of a phosphodiester bond between the 5' phosphate of the donor and the 3' hydroxyl of the acceptor. [1] A pictorial example of how a ligase works (with sticky ends):
Ligase will also work with blunt ends, but requires higher concentrations �and different reaction conditions. http://en.wikipedia.org/
�����������������������
http://fhs-bio-wiki.pbworks.com/
Termination:
- Replication termination site or the process of termination in eukaryotic DNA is not clear.� However, when the replication forks from opposite directions meet one another, replication stops; and terminated.� By unknown means the entry of replication fork into newly formed DNA is prevented, perhaps through disassembly of MCM proteins and the action of Geminins.
If the replication machinery makes any error, DNA repair enzymes become active; even p53 induces gene expression for repair enzymes, and repair is done.� Till then the cell does not enter M-stage.� If the DNA damage is beyond repair, the p53 and other components induce the cell to Apoptosis.
Replication Ter sites in Eukaryotes; Molecular Cell Vol 39,Issue4,595-605; Dnielle Fachinetti
et al.
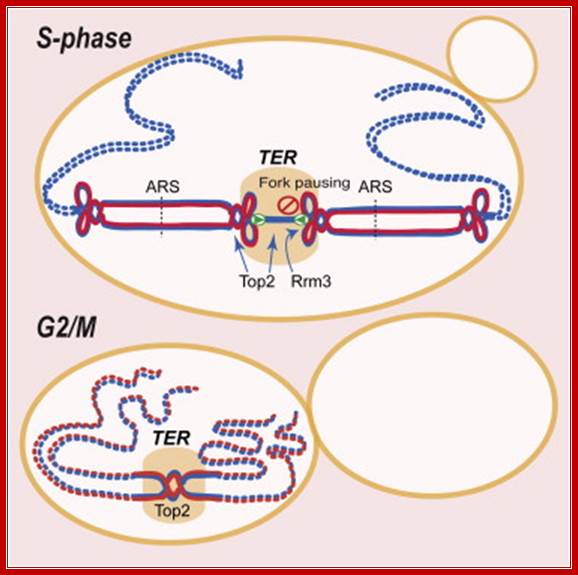
Replication termination occurs at fork pausing sites ► Rrm3 assists fork progression across termination regions ► Top2, but not Top3, facilitates fork fusion ► Top2 prevents genome instability at termination regions; Daniele Fachinetti, Rodrigo Bermejo
Chromosome replication initiates at multiple replicons and terminates when forks converge. In E. coli, the Tus-TER complex mediates polar fork converging at the terminator region, and aberrant termination events challenge chromosome integrity and segregation. Since in eukaryotes, termination is less characterized, we used budding yeast to identify the factors assisting fork fusion at replicating chromosomes. Using genomic and mechanistic studies, we have identified and characterized 71 chromosomal termination regions (TERs). TERs contain fork pausing elements that influence fork progression and merging. The Rrm3 DNA helicase assists fork progression across TERs, counteracting the accumulation of X-shaped structures. The Top2 DNA topoisomerase associates at TERs in S phase, and G2/M facilitates fork fusion and prevents DNA breaks and genome rearrangements at TERs. We propose that in eukaryotes, replication fork barriers, Rrm3, and Top2 coordinate replication fork progression and fusion at TERs, thus counteracting abnormal genomic transitions; Daniele Fanchonette, Rodrigo Bermejo, et al.
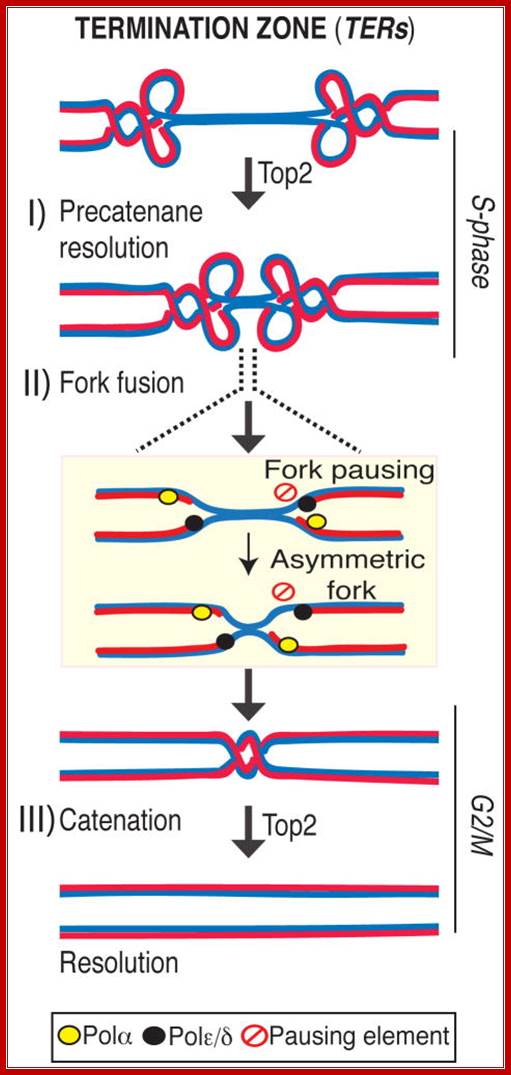
TERs contain fork pausing elements that influence fork progression and merging. The Rrm3 DNA helicase assists fork progression across TERs, counteracting the accumulation of X-shaped structures. The Top2 DNA topoisomerase associates at TERs in S phase, and G2/M facilitates fork fusion and prevents DNA breaks and genome rearrangements at TERs. We propose that in eukaryotes, replication fork barriers, Rrm3, and Top2 coordinate replication fork progression and fusion at TERs, thus counteracting abnormal genomic transitions.
Rrm3, Top1 and a fraction of Top2 travel with the fork (Azvolinsky et al., 2006; Bermejo et al., 2007). Rrm3 facilitates forks progression across pausing sites (Ivessa et al., 2003) while Top1 and Top2 are both needed to resolve the torsional stress ahead of the fork generated during fork progression: while Top1 resolves positive supercoiling ahead of the fork (Wang, 2002), also contributing to prevent interference between replication and transcription (Tuduri et al., 2009), Top2 likely acts behind the fork to resolve precatenanes (Lucas et al., 2001; Wang, 2002). When forks approach the termination zone, the topological constrains at converging forks can no longer be resolved by Top1 (Fields-Berry and DePamphilis, 1989) and therefore the only option for fork progression is to rely on Top2 activity. This is consistent with the observation that top2 mutants are selectively delayed in completing the last portion of replication but not the bulk of DNA synthesis. However, we cannot rule out that the topological architecture of the termination zone (e.g. chromosome loops) needs specifically Top2 activity for resolution. Indeed a subpopulation of Top2 is also bound to TERs in early S-phase, perhaps due to the affinity of Top2 for nucleosome-free regions (p= 2.10E-58). Moreover, other S-phase Top2 clusters have been recently suggested to correlate with the formation of chromosome loops (Bermejo et al., 2009). We found that the Top2 clusters at TERs are established already at the cdc7 dependent step and are not influenced by origin firing (data not shown), thus suggesting that TERs represent CIS chromosomal elements that undergo topological transitions requiring Top2 activity, Daniele Fachinetti, Rodrigo Bermejo et al
Replication Barriers-Replication Slowing Zones:
Eukaryotic DNA is so long, it is difficult to imagine how smoothly the entire lengths of 50 to 250 million base pair long nucleosomal DNA could be replicated and replicated DNAs are separated and organized into chromatin.� Replication perse in Eukaryotes is not smooth for it encounters many barriers, which slow down the process, yet they overcome.� Replication barriers are found in RDNA (rRNA), centromeric region, and DNA at telomeric region, LTRs and tRNA regions; understanding of this pausing or replication barriers is very confounding and interesting.
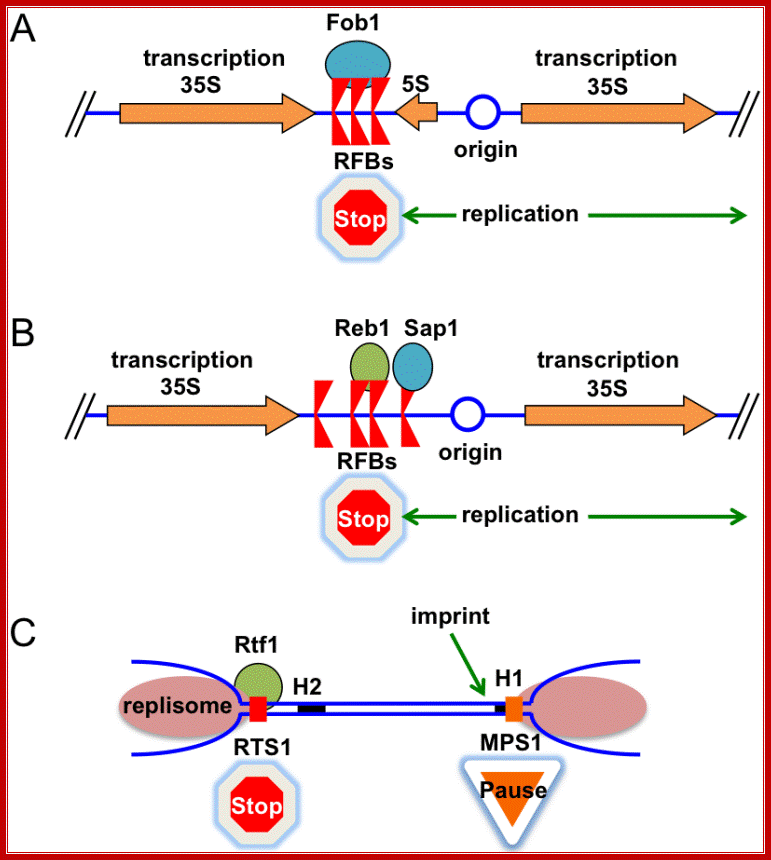
There are numerous programmed replication fork barriers (RFB).� Replication fork barriers are bound by proteins such as RFBs. In these circumstances, the Replisome and the RFB proteins must specifically interact to stop replication fork progression.� Once the barriers are overcome replication progresses to complete replication.
Replication of chromosomal terminal i.e Telomeric is dealt in the next section. Adam R. Leman �,* and Eishi Noguchi; http://www.mdpi.com/
Note: Refer to Cell Cycle and Its Regulation for DNA replication regulation;