����� Replication of Organelle DNA:
������ Chloroplast and Mitochondria:
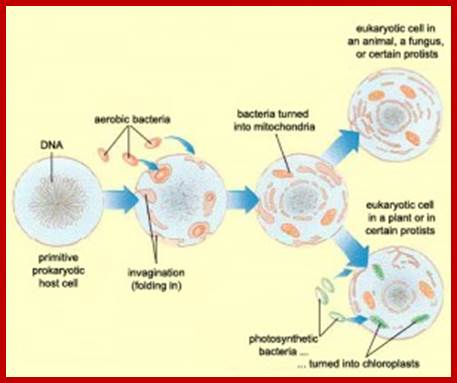
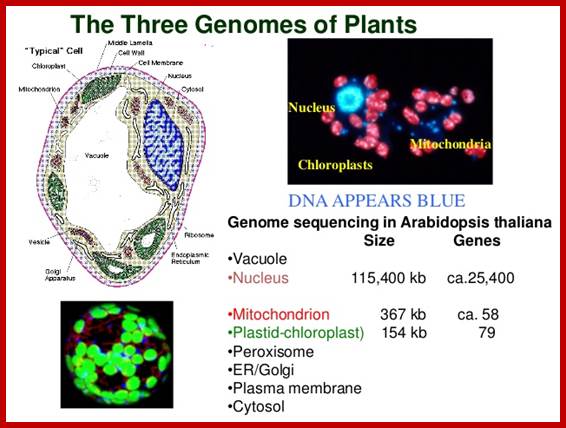
Only Eukaryotes have mitochondria as symbionts, and systems with only mitochondria turned out to be animal cells; and those that contain both mitochondria and plastids, are plants. �Both of them are involved in energy transducing functions.� Plastids, among them chloroplast are endowed with abilities to capture, the one and the only source of energy for all life systems i.e. Solar energy, and convert the same into chemical energy and conserve it in the form of chemical bonds.� Without photosynthesis, the biological world comes to halt for it is this process that provides biomass which is used an important source of energy for all other organisms, which are not autotrophic.� Mitochondria, on the other hand, uses the biomolecules containing chemical energy and oxidize them to capture the bond energy and the same is made available in the form of energy rich molecules like ATP, GTP NADH2 and NADPH2 for a variety of cellular functions without which cells cease to function.� Both these quintessential organelles are symbionts; long back; they were independent living prokaryotic organisms.� Chloroplast membrane is believed to be associated with ER similar to that of Mitochondria. Evidence for membrane contact sites MCSs between two membranes and we propose for the ER-chloroplast pair, that such tight associations are involved in bidirectional lipid trafficking between the two compartments, Mats X Anderson et al.� The chloroplast/ER nexus, play a role in protein flow within the ER?� �Studies of tobacco cells either constitutively or transiently expressing ER retained lumina.� Furthermore, both directional flow within the network and remodeling of the network are dependent on the actinomyosin cytoskeleton in non-dividing cells; Lawrence R. Griffing.� Transport of ER derived galactolipids (precursors) occurs at sites of physical contact between chloroplasts and a specialized plastid associated domain of the ER, called PLAM.
Chloroplasts:
The first plastid is thought to have originated through primary endosymbiosis, in which a photosynthetic cyanobacterium was captured by a heterotrophic protist and eventually transformed into an intracellular organelle. Molecular clock analysis suggests this key event in eukaryote evolution occurred approximately 1.7 billion years ago; early Devonian age.� Mitochondria originated from proteobacteria and chloroplasts from cyanobacteria.� Cyanobacteria originated around 3.5 by.ago.
Chemical Composition of chloroplasts (general):
|
S. No |
Chemical |
Percentage |
|
1. |
Proteins |
35 � 55% |
|
2. |
Lipids |
20 � 30% |
|
3. |
Carbohydrates |
Variable |
|
4. |
Chlorophyll |
9% |
|
5. |
Carotenoids |
4.5% |
|
6. |
RNA |
3 � 4% |
|
7. |
DNA |
0.5% |
|
8. |
Minerals |
0.2% |
Plant cell with 60-80 or more chloroplasts per cell; http://en.wikipedia.org/
Outer membrane, 2. Intermembrane space, 3. Inner membrane (1+2+3: envelope), 4. Stroma (aqueous fluid), 5.� thylakoid lumen (inside of thylakoid) �6.� Thylakoid membrane, 7. Granum (stack of thylakoids), 8. Thylakoid (lamella), 9. Starch, 10. Ribosome, 11. Plastidial DNA, 12.Plastoglobule (drop of lipids). http://www.answers.com/
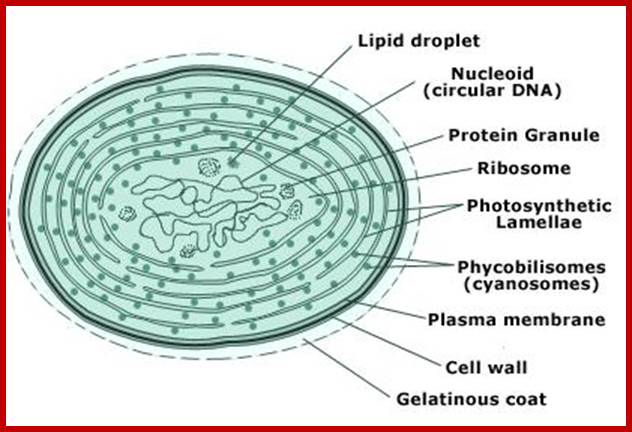
Structure of cyanobacterial cell.Membrnaea all-around the bacterial chromosome and they are studded with light capturing complexes and ATP synthesis machinery. �It is from these cyanobacterial cells plastids have been evolved. http://www.tutorvista.com/
Chloroplast DNA:
Plastids are considered as symbiont of past Cyanobacteria, where most of its DNA has been transferred into the nuclear genome and only 120-140 genes have been left out in chloroplast genome.� The symbiotic event happened somewhere 1.2 billion years (?) ago but mitochondrial symbiotic event is little older i.e., ~-1.7 billion years (?).
The number of plastids per cell varies, but in lower organisms the number, shape and sizes are constant and species specific.� Ex. Chlamydomonas contain only one chloroplast, which is cup shaped, in Spirogyra the number is 2 and they are spiral, in zygonema there are two chloroplasts per cell and they are star shaped.� In higher plants the number can range from 20 to 100, but in tropical plants like corn, though number range from 40 to 70, but coenocytic cells contain thousands and thousands. Plastids exhibit two forms one C3 kind in all and the other C4 kind exists with C3 in different cell types in monocots.�
Most not all plastids have double stranded, circular DNAs of 120-140kbps. But some show linear forms example Maize 10-14 days old plastid DNA is linear and exhibit.� The ends of linear genomic monomers and head-to-tail (h�t) concatemers within inverted repeat sequences (IRs) near probable origins of replication, Only 3-4% of cp DNAs are circular, �Delene J. Oldenburg,� Arnold J. Bendich. Chloroplast DNA exists in the form of protein associated loops called Nucleoids. The nucleoids are associated with 41kDa proteins (negative regulators). The nucleoids are also associated with many proteins of 12kDA to 30kDa. �Chloroplast DNA is compacted with histone like proteins. �Nucleoids of Chloroplast of Spinacia oleracea were analyzed for the protein content. They are looped plastid DNAs enriched with proteins involved in DNA replication, ����������� organization, and repair as well as transcription, mRNA processing, splicing and editing.
�Sequence of several plastids DNA has been obtained and number of genes they encode code range from 120-140. But chloroplast proteome is estimated to be in the range of 3500-4000.
For some inexplicable reasons, the size of plastid DNA varies from species to species, so also the number of DNA copies per plastid, though the function of all plastids is more or less same.� Plastid inheritance is uniparental- through female egg, like mitochondria from mother. �It is fascinating that during pollen grain germination pollen tube penetrates embryo sac with its cytoplasm, but only one of the two nuclei fuses with the egg; whatever that is contributed to the embryo sac and its cytoplasm contributes to its cytoplasmic inheritance; thus the plastid inheritance is maternal with certain exceptions. Though it is common Oenothera is different for it shows biparental inheritance.�
Size of plastid DNA and the copy numbers (only few).
|
Plant Name |
Size of the genome |
Copy nos./plastd |
|
Chlamydomonas |
110-130 (~180)kbp |
80 |
|
Liverwort |
121kbp (Marchantia) |
30-40 |
|
Tobacco |
155kbps |
40-50 |
|
Maize |
135 kbps (200kbps) |
40-60 |
|
Nicotiana tabaccum |
155.943 kbp |
60-80 |
|
E. gracillis |
120-170kbp |
58-600 |
|
Corynocarpus |
159,202bps |
|
|
|
|
|
|
|
|
|
Chloroplasts are the organelles of photosynthesis, an essential process for life on earth. It is this process that generated Oxygen in the atmosphere. It also fixed CO2 and provides food to the world microbes, animal and human population. Plastids are involved in fatty acid synthesis and amino acid synthesis. Today�s fossil fuel, a product of several billion years, is produced by cyanobacteria and plants in general. These organelles contain a genome that encodes several proteins for chloroplast function, but not enough so other thousands essential proteins encoded by the nuclear genome are imported into the chloroplast.
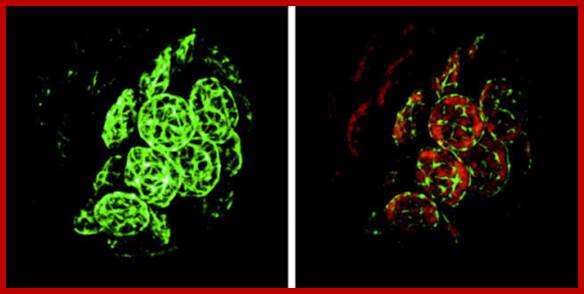
Ampicillin-treated cells of P. patens. Epifluorescence microphotographs of a DNA dye SYBR Green I-stained cells without antibiotic (left) and with ampicillin treatment (right) are shown. Chloroplast nuclei (nucleoids) are seen as yellow spots in the chloroplasts. Arrow indicates a septum. Bar = 20 �m.� Such structures are common in bacteria. http://openi.nlm.nih.gov/Mol Biol.
�����������
This genome varies widely in copy number, from a few copies in seeds and root cells, to very high copy number (up to 1,000 genome copies or more/per cell?) in young, rapidly growing leaf cells. As the leaves mature the genome copy number drops, suggesting a mechanism for control of cp (ct) DNA replication, but not linked to the nuclear genome replication or cell cycle. All components of the chloroplast DNA (ctDNA) replication machinery appear to be nuclear encoded, including the DNA polymerase(s) (mentioned above) and accessory proteins such as DNA primase, DNA helicase, SSBs, topoisomerases, and other factors. Research scholars have characterized ctDNA replication mechanisms in pea, spinach, soybean, Chlamydomonas and tobacco, and they have mapped specific sites of replication initiation.
The plastid DNA replication origins are located in between two inverted copies of rRNA genes. One D-loop has been mapped in the spacer region between the inverted 16S and 23S rRNA gene. The second D-loop has been located downstream of the 23S rRNA gene. Euglena gracilis rRNA genes exist in tandem copies, in addition it contains 16sRNA gene located upstream from the two copies. In Pea only one origin is present but in Tobacco plant chloroplasts there are two Origins
Some of the chloroplast DNA Pols mol. wt. in spinach is 105, wheat 110-180, pea 70-87, Chlamydomonas 110kDa.� The Pea DNA polymerase has 5� exonuclease activity.� But 3 > 5� exonuclease activities is restricted to its associated 20-43kDa accessory protein subunit.� There are two classes of DNAPs one PolIA responsible for replication, another Pol1B is for DNA repair. Plastid DNAPs are resistant to Amphidicolin. Chloroplast DNApols belong to gamma class polymerases are resistant to dideoxy ntds. Chloroplasts and mitochondria also contain gyrases. Inhibitors of bacterial DNA gyrases Novobiocin and Nalidixic acid also inhibit chloroplast DNA synthesis in higher plants.� Chloroplast helicases have been studied in Glycine max and Pea plants
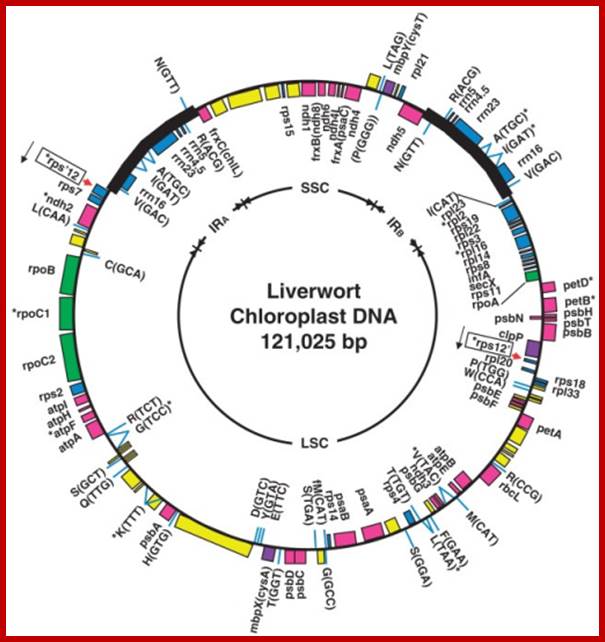
The complete nucleotide sequence of chloroplast DNA (121,025 base pairs, bp) from a liverwort, Marchantia polymorpha, has made clear the entire gene organization of the chloroplast genome. Quite a few genes encoding components of photosynthesis and protein synthesis machinery have been identified by comparative computer analysis. We also determined the complete nucleotide sequence of the liverwort mitochondrial DNA and deduced 96 possible genes in the sequence of 186,608 bp. The complete chloroplast genome encodes twenty introns (19 group II and 1 group I) in 18 different genes. One of the chloroplast group II introns separates a ribosomal protein gene in a trans-position. The mitochondrial genome contains thirty-two introns (25 group II and 7 group I) in the coding regions of 17 genes. From the evolutionary point of view, we describe the origin of organellar introns and give evidence for vertical and horizontal intron transfers and their intragenomic propagation. Furthermore, we describe the gene organization of the Y chromosome in the dioecious liverwort M. polymorpha, the first detailed view of a Y chromosome in a haploid organism. On the 10 megabase (Mb) Y chromosome, 64 genes are identified, 14 of which are detected only in the male genome. These 14 genes are expressed in reproductive organs but not in vegetative thalli, suggesting their participation in male reproductive functions. These findings indicate that the Y and X chromosomes share the same ancestral autosome and support the prediction that in a haploid organism essential gene on sex chromosomes are more likely to persist than in a diploid organism. http://openi.nlm.nih.gov/
The organization of the liverwort chloroplast genome; http://cc.scu.edu.cn/
The chloroplast genome organization is very similar in all higher plants, although some variations are found. In Chlamydomonas reinhardtii two distinct chloroplast genomes are found, one PS+ and the other PS-.
During plastid DNA replication, Pea plastid RNA primase 115-120kDa, (Nielsen BL,et al) produces RNA primers of 19-20 ntds long.� DNA polymerase extends 3�OH of the primers into complementary DNA strands. Then the primers are completely excised and replaced by DNA. In some, primers are not completely removed, thus plastid DNA is chimeric? �As a result, the plastid genome is a chimeric DNA-RNA molecule. Origins of plastid DNA replication have been identified.
- Origins as two D-loops on either side of the central space (the central space contain few tRNA segments) are formed in plastid DNA. They represent initiation of plastid DNA replication from opposite DNA strands.
- Plastid replication is not synchronous with nuclear replication. Young leaves have more ctDNA/cell than old leaves.
- The D loop regions contain few stem loops but rich in A+T sequences. In Pea plastids one loop is found in the spacer region between 16s and 23s segment of rRNA gene and the other is located, downstream of 23s rRNA gene
DNA of the Euglena gracllis, Pringsheim strain Z chloroplast genome. The arrows show replication bubble. https://www.slideshare.net
This circular DNA in E. gracillis is 143,170 bp. �It varies from 120, 000 to 170, 000bp depending upon the species. �Copy numbers per plastid varies from 58 to 600. Aging of plastids results in decreasing this number. There are genes for the 16S, 5S, and 23S rRNAs of the 70S chloroplast ribosomes, 27 different tRNA species, 21 ribosomal proteins plus the gene for elongation factor EF-Tu, three RNA polymerase subunits, and 27 known photosynthesis-related polypeptides. Several putative genes of unknown function have also been identified, including five within large introns.� This genome contains at least 149 introns. There are 72 individual group II introns, 46 individual group III introns; 10 group II introns and 18 group III introns that are components of twintrons (introns-within-introns), and three additional introns suspected to be twintrons composed of multiple groups II and/or group III introns, but not yet characterized. At least 54,804 bp, or 38.3% of the total DNA content is represented by introns, Richard B. Hallick et al.
Plastids are derived from cyanobacteria, but the number of genes of cyanobacterial genome has been estimated as ~2600 (from the analysis of A. thaliana genome), with time a large numbers have been transferred into the nucleus.� It has been estimated there are approximately ~3000 (Synechocystis data) proteins most coded for by the nuclear genome.� Interaction between chloroplast and the nucleus has been noticed. Note the above characters are not for all plastids.
Chloroplast DNA replication:
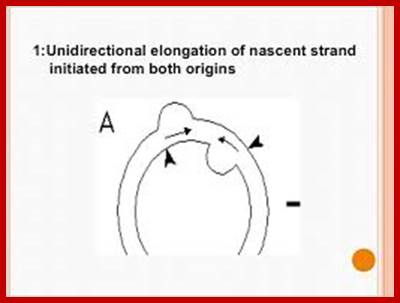
Most of the cp DNAs are circular and compacted as nucleoids. The dual D-loop model has been proposed for the initiation of chloroplast DNA replication. Unidirectional elongation of nascent strand initiated from both origins and the unidirectional fork move toward each other.� Fusion of D-loops forms Cairns-type intermediates; bidirectional, semi-discontinuous replication; resulting daughter molecules; the arrows denote the two D loop initiation sites (origins).
Complete gene map of the entire chloroplast genome. Inverted repeats of rRNA segments shown in orange red in opposite orientation; http://www.mun.ca/
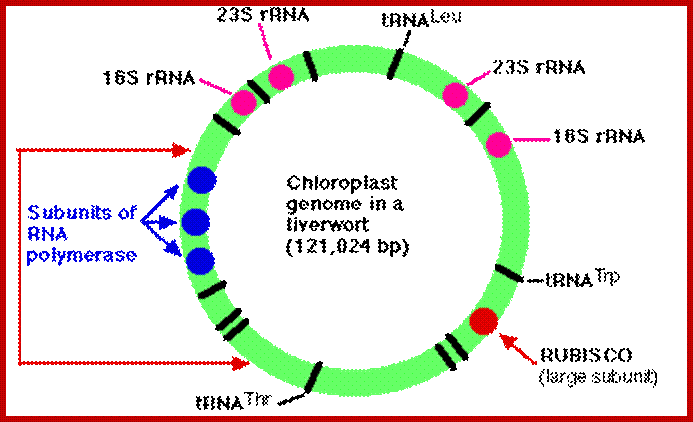
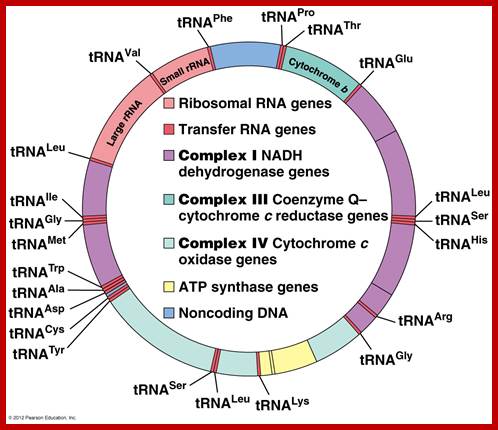
Marchantia polymorpha; encode 2 sets of rRNA genes, 37tRNA,4 RNAPs �one RUBISCO large subunit, 9 for Photosystem I and II, 6 for ATP synthase and ~19 for ribosome proteins; http://users.rcn.com/
����������������������������������������������������������������������������������������������������������
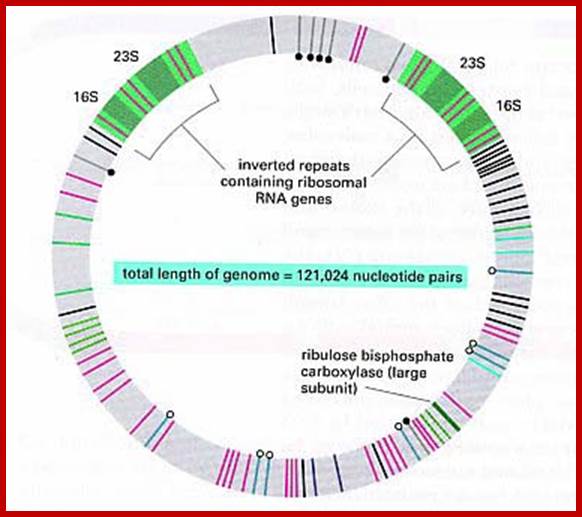
5.3Kbp--[--16s--23s >5s- rRNA]- -14.5Kbp- ---[5s-< -rRNA-23s---16s�]--5.3Kb]--
The spacer between two rRNA segments is of 14.5 kbp
Replication origins are located at the vicinity of each inverted repeats of 20-30kbp rRNA gene segments and 14.5kbp (~44kbp other system) apart. �Direction of rRNA genes orientation is shown as arrows.� The spacer is 14.5kbp and contains some tRNA segments.
In Tobacco Nicotiana tabacum- replication origins have been localized to minimal sequences of 82bp (pKN8, positions 137 683�137 764) and 243 bp (pKN3, positions 130 513�130 755) for oriA and oriB respectively.
Cp DNA polymerases: Spinach-105kDa, Wheat-110-180kDa, Pea-70-87kDa, subunit 20kDa 3�-5�exo, Chlamydomonas-110, CpDNA primase-115-120kDa. SSB 43kDa; Plastid DNAP are classified as DNAPol-1A- for replication and DNAPol-1B for repair.
cpDNAP ~90kDa, it is resistant to Amphidicolin, it is not inhibited by Tagetitoxin (inhibitor of cpRNAP).� Once replication is completed further replication takes place through rolling circle model. Rate of DNA replication -700ntds to 1000nts/second
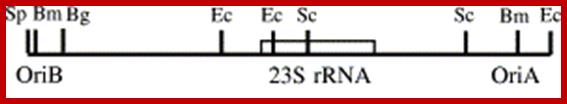
OriB and oriA on either sides of 23s rRNA gene; Plant science 166151.http://bricker.tcnj.edu/
Origins-
- OriB> Repeats ATTTTGTCC, Repeats ATTTCTTCCG-(Repeat2- Repeat3)
�spacer->130,152------23srRNA---137847<spacer>,
<OriA Repeats TTTCTTCCG�repeats TTTCTTCCG-,9Repeat1-Repeat2,3)
Ori binding proteins have been identified in Chlamydomonas as 28kDa (18kDA).� In some Ori binding proteins are found to be 18kDa-Fe-S proteins. In Chlamydomonas three DNA-binding polypeptides with apparent molecular weights of 18, 24 and 26 kDA, respectively are found bound to Origin and the proteins are associated with thylakoid membranes.� This suggests the plastid DNA is associated or bound to thylakoid membranes through such proteins.
Plastid and mitochondrial DNAP are homologous and share similarities with E.coli DNAP I. �Chloroplast primase protein is 115-120kDa and DNAP 90kDa.� Once the replication starting from their respective A and B origins is completed; the cp DNA can go through rolling circle mode.
The replication Origins contain stem loops rich in A and T sequences, they simulate ARS sequences.� The sites are recognized by 25 to 28kDa proteins.
The replication Origins contain stem loops rich in A and T sequences, they simulate ARS sequences.� The sites are recognized by 25 to 28kDa proteins.
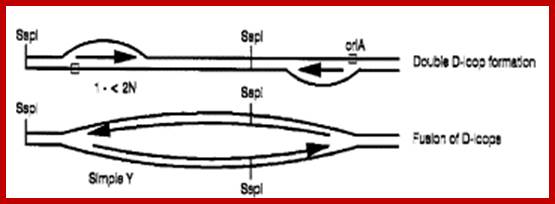
Double D-loop mechanism; https://www.slideshare.net
Double D-Loop; each origins are located some distance between the,� When replication starts and progresses the D-loops meet each other, but replication is opposite direction.
Ori A, spans about 0.7kbp located between 16s and 23s RNA segments.� Ori B located downstream of 23sRNA spans about 1.3kbp.� Once proteins bind to replication origins sites, the DNA opens into replication bubbles on either side spacer abutting rRNA repeat genes.� SSDNA binding proteins have not been identified, but found in mitochondria.� Plastid helicases 78kDa assist in opening from 3� to 5� direction.� The two loops open and the fork move in each other�s directions.� When they meet each other, they form Cairn�s like structures.� From this region the replication is bidirectional. Later these circular DNAs replicate in rolling circle mode.� The cpDNAP is 90-120kDA.� It is resistant Amphidicolin and dideoxynucleotides. It has 3� �5� exonuclease activity for it is associated with 25kDa component.� Okazaki fragment formation is absent and DNAs are synthesized as continuous strands.
--rRNA--23s-16s-4.5s-5s---spacer-tRNAs---5s-4.5s-23s rRNA-
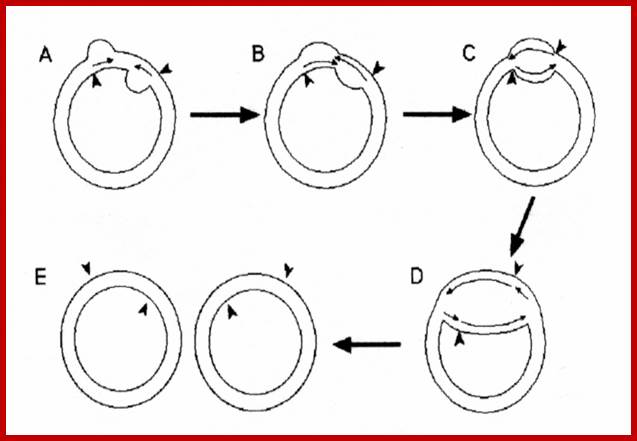
Fig: Schematic representation of the interaction of DNA helicase with chloroplast DNA during DNA replication following the double displacement loops (D‐loops) mechanism of chloroplast DNA replication. The chloroplast DNA (A) is a circular, supercoiled and double‐stranded molecule. B. In pea, chloroplast DNA replication proceeds by introducing two D‐loops. The synthesis from each origin is initially unidirectional (B), which expands towards each other, through unwinding by DNA helicase, and then they fuse and form a Cairns replicative fork structure (C). After fusion of the D‐loops (C), replication continues bidirectionally and forms two progeny molecules containing nick (D). After completion of daughter molecules (D), replication may continue by a rolling circle mechanism from either of the nicks remaining in the new molecules. The arrows indicate the direction of DNA synthesis and the polarity of DNA helicase movement. The involvement of a DNA helicase in chloroplast DNA replication can be inferred, but is as yet unproven. Explanation is different. �http://bricker.tcnj.edu/
Rolling Circle Mode of Replication:
Alternative mechanism has been suggested for the replication of circular DNA and this is called rolling circle mechanism, which explains multiplication of many viral DNAs and also explains DNA replication during bacterial mating. One of the evidence, in support of this model is the observation of long chains of polynucleotides which could measure several times the contour length of circular DNA. In this mode, due to nicking in one of the two strands, one strand becomes a polynucleotide with two ends, one end having 3'-OH group, the other having 5'p group. DNA synthesis starts by adding bases at 3'-OH end, displacing the 5' end, which rolls out as a free tail, and is then copied in 5'→3' direction in small segments using fresh enzymes system. These tails thus become double stranded and give rise to a circular DNA later on. A possible mechanism of replication of circular DNA using rolling circle model is shown in Figure.
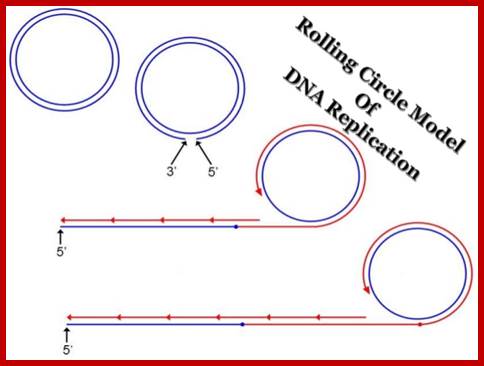
Rolling circle mode of replication; https://www.slideshare.net
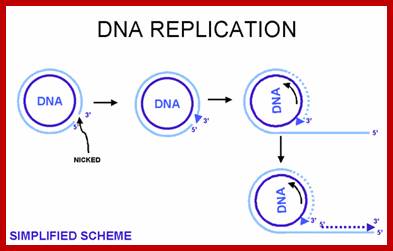
Rolling circle mode of replication of circular ds helical DNA, using a nick formation by an endonuclease; Fig; http://www.thailabonline.com/
Transcription:
Liverwort ctDNA contains two 25.08 kbp inverted repeats, which code for rRNAs.�� These are separated by one short single copy sequences and the other called large single copy sequence. Such inverted repeats are also found in most of the plastomes.� The genome contains more than 120 genes, which code for tRNAs and mRNAs.�
![]() �
�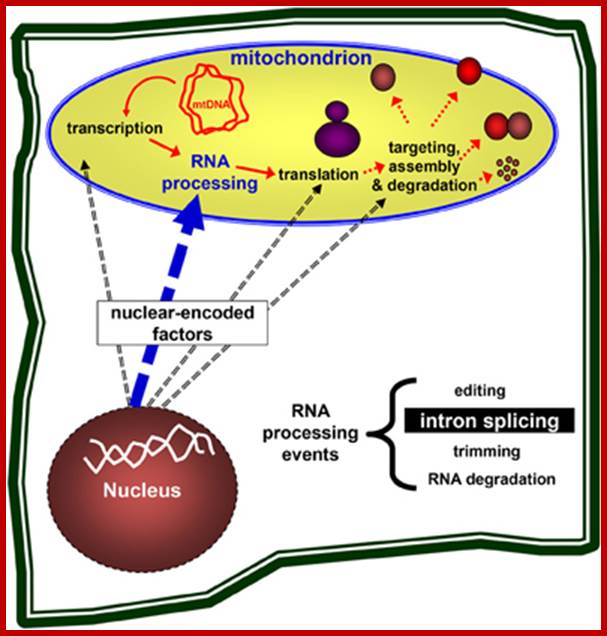
Nuclear genes control the mitochondria biogenesis. Nuclear-encoded genes are necessary for the expression of mitochondrial genes, including transcription, pre-mRNA processing, translation of the mRNAs into mitochondrial proteins, the assembly of ribosomes and respiratory complexes, and are also required for the targeting and degradation of organellar subunits. Plant organelles are excellent systems to study these processes in eukaryotes.;Gregory G. Brown et al;http://journal.frontiersin.org/
Plastid DNA of Arabidopsis thaliana contains 78 protein coding gene and 54 genes code for tRNA and 2 rRNA.� Chloroplast DNA encodes subunits for RNA polymerase (similar to bacteria) called PEP RNAPs.� Plastid also contains nuclear coded RNAPs called NEPs. Many of their genes are expressed as polycistronic RNA, and then they are processed. Most of the genes contain group II and group III introns (twintrons).� Recent analysis of cyanobacterial genome and plastid genome, as known today, it is estimated that nearly ~4500 gene have been transferred to nuclear DNA after symbiotic transfer.�
Plastid RNAPs:
Plastids contain two types of RNA polymerases; one multisubunit complex Plastid DNA encoded (PEP) similar to bacterial enzyme, and the other single subunit enzyme Nuclear coded (NEP) similar to bacteriophages.� The core subunits such as alpha, beta, beta prime and omega are encoded by chloroplast DNA.� There are sigma factors similar to sigma 70; six of them are encoded in nuclear DNA; after transcription and translation they transported into plastids.���
�����������
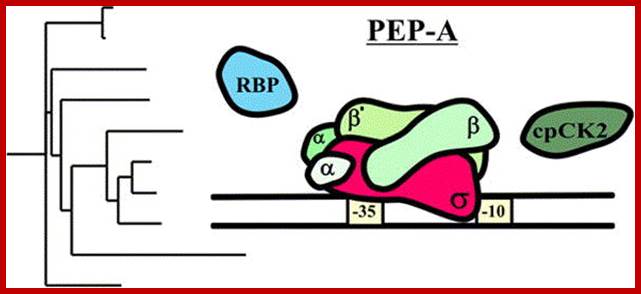
Proteomics-based Sequence Analysis of Plant Gene Expression � the Chloroplast Transcription Apparatus;
Heike Loschelder1, a, Anke Homann1, a, Karsten Ogrzewalla2, a and Gerhard Link:
The chloroplast transcription apparatus has turned out to be more complex than anticipated, with core polypeptides with multiple accessory proteins of diverse, and in part unexpected, functions. At least two different RNA-binding proteins and several redox-responsive proteins are components of the major chloroplast RNA polymerase termed PEP-A. One of the key-regulatory factors has been identified as a Ser/Thr-specific protein kinase that is sensitive to SH group modification by glutathione and this means it is able to modulate transcription. The cloned plastid transcription kinase from mustard (Sinapis alba L.) has been assigned as a member of the CK2 family and hence has been termed cpCK2 (mostly nucleo-cytosolic). Despite its apparent role in mustard chloroplast transcription, until recently no data have been available for other plant species. Using the web database resources, we find evidence for an evolutionarily conserved role of this redox-sensitive plastid transcription factor.
Database screening helped to identify regulatory proteins of chloroplast RNA polymerase, including a novel putative RNA-binding protein (RBP). The previously identified transcription kinase (cpCK2) is likely to play a common role in number of plant species.
Keywords: Plastid encoded RNA polymerase- PEP; It binds to promoter sequences similar to that of bacteria. The PEP RNAP consists of 2 alpha subunits one beta and one Beta prime subunits. Mol Biol
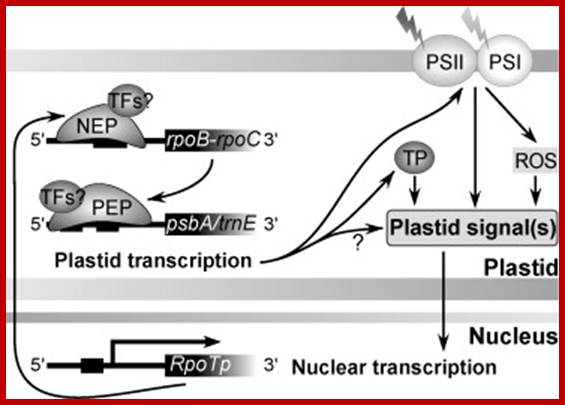
Exogenous and endogenous factors such as light, hormones, photosynthesis, plastid type and chloroplast development differentially modulate transcription of plastid genes. Cues with repressing action on plastid transcription are depicted with lines terminated with a bar while arrows represent promoting signals (see text for references; effects of respiration on chloroplast transcription: T. Potapova, Y. Zubo, Y. Konstantinov, T. B�rner, unpublished data). http://hos.ufl.edu/
Chloroplast RNA transcription; Yusuke Yagi1 and Takashi Shiina; http://www.frontiersin.org/
The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids; Peter T.J. Hajdukiewicz Lori A. Allison Pal Maliga
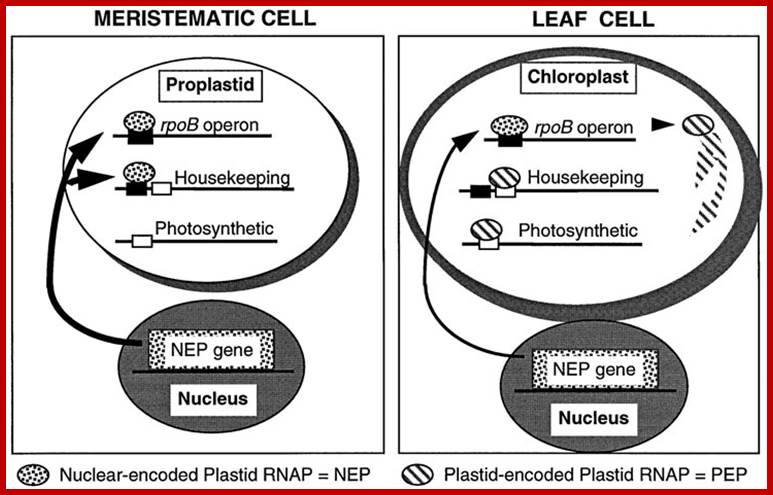
The general rule emerging from the data is that photosystem I and II genes are transcribed by the PEP polymerase, whereas most other genes have both PEP and NEP promoters. As to the role of NEP, we assume that it plays an important role in non‐green plastids, in tissues in which PEP is absent or is present only in limited amounts. A similar role for the two plastid RNA polymerases was proposed in the expression of plastid housekeeping genes from alternative promoters in photosynthetic and non‐photosynthetic tissues based on the study of tobacco tissue culture cells (Vera and Sugimura, 1995; Vera et al., 1996). Tissue‐ and cell type‐specific transcription from NEP promoters will have to be determined individually, by following accumulation of RNA and proteins for reporter genes in transgenic plastids. Selective transcription of the gene groups by the nucleus‐encoded and plastid‐encoded RNA polymerases was proposed as one possible mechanism of differential gene expression (Hess et al., 1993; Lerbs‐Mache, 1993;Mullet, 1993). Distinct NEP and PEP promoters reported here for a large number of transcription units provide a general mechanism for developmentally‐timed expression of groups of plastid genes by the two plastid RNA polymerases.
Proposed scheme for differential gene expression during chloroplast development based on recognition of distinct promoters by NEP and PEP (based on Mullet, 1993; Hess et al., 1993; this paper). Note that in proplastids, NEP transcribes housekeeping genes and the rpoB operon encoding the β, β′ and β′' subunits of PEP, whereas in chloroplasts both housekeeping and photosynthetic genes are transcribed by PEP. �Peter Hajdukiewicz Lori A. Allison Pal Maliga; �http://emboj.embopress.org/
Biogenesis and homeostasis of chloroplasts and other plastids; Paul Jarvis; & Enrique L�pez-Juez; Affiliations Corresponding author; Nature Reviews Molecular Biology 14,787-802 (2013)
Chloroplasts are the organelles that define plants, and they are responsible for photosynthesis as well as numerous other functions. They are the ancestral members of a family of organelles known as plastids. Plastids are remarkably dynamic, existing in strikingly different forms that interconvert in response to developmental or environmental cues. The genetic system of this organelle and its coordination with the nucleocytosolic system, the import and routing of nucleus-encoded proteins, as well as organellar division all contribute to the biogenesis and homeostasis of plastids. They are controlled by the ubiquitin�proteasome system, which is part of a network of regulatory mechanisms that integrate plastid development into broader programmes of cellular and organismal development.
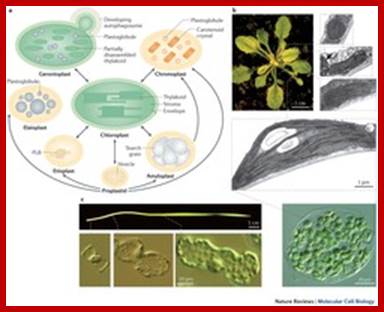
Diversity of plastid forms and their interconversions; http://www.nature.com/
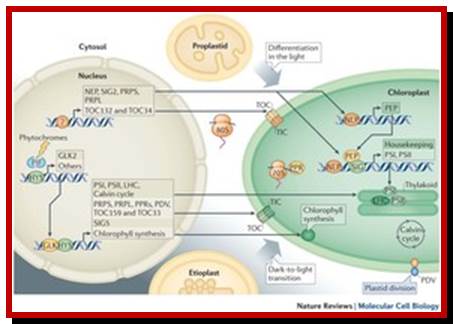
Light-mediated anterograde control of chloroplast development. http://www.nature.com/
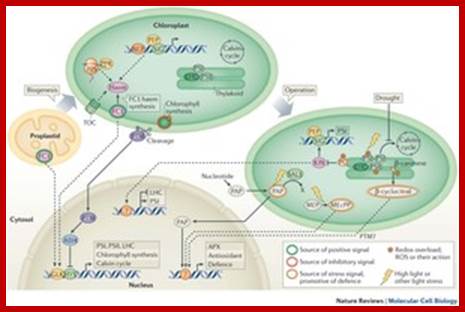
Plastid-to-nucleus or retrograde signaling pathways.; http://www.nature.com/
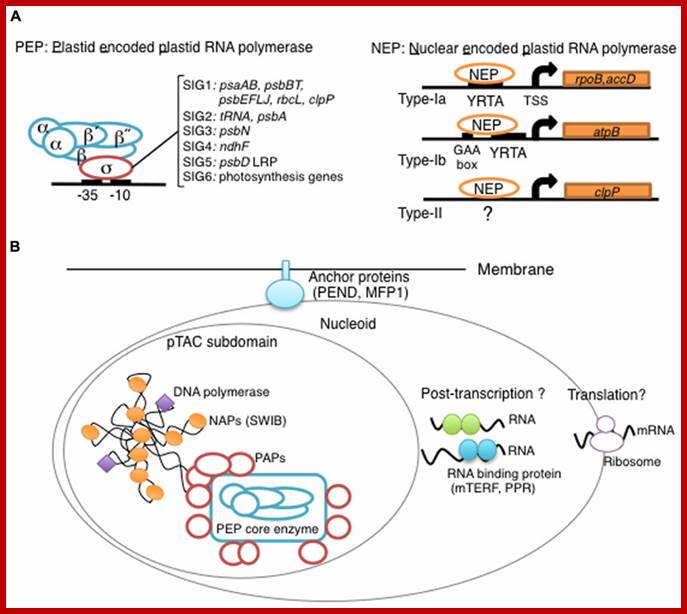
Overview of Chloroplast Transcription;
(A) Basic transcriptional machinery in higher plants. Higher plants have two distinct types of chloroplast RNA polymerase: plastid-encoded plastid RNA polymerase (PEP; left panel) and nucleus-encoded plastid RNA polymerase (NEP; right panel). PEP is a bacterial-type multi-subunit RNA polymerase composed of the core enzymatic subunits α, β, β′, β″ (blue) and a sigma subunit (red) that is responsible for promoter recognition. Plastid sigma factors are divided into six subgroups, SIG1�SIG6, and selectively recognize bacterial-type promoters in the plastid. NEP (right panel) is a monomeric enzyme that resembles mitochondrial T7-type RNA polymerases. NEP is involved in the transcription of housekeeping genes such as rpo genes for PEP core subunits, and ribosomal protein-coding genes. Positioned upstream of genes transcribed by NEP are three distinct types of promoter structures (Type-Ia, Type-Ib, and Type-II).
(B)The chloroplast nucleoid subdomain and its components. Chloroplast nucleoids are attached to the membrane (envelope or thylakoid) by anchor proteins (PEND and MFP1). The plastid transcription active chromosome (pTAC) is one of the nucleoid subdomains, which contains the transcription factory. Chloroplast genomic DNA is packed by chloroplast-specific nucleoid-associated proteins (NAPs; orange circle). The mature chloroplast contains a large PEP complex with several PEP associate proteins (PAPs; red circles). Recent proteome analysis suggested that chloroplast nucleoids contain additional subdomains, which regulate post-transcriptional RNA maturation and translation. http://www.frontiersin.org/
RNA Polymerase Subunits Encoded by the Plastid rpo Genes Are Not Shared with the Nuclear-Encoded Plastid Enzyme: Germ�n Serino and Pal Maliga
�Plastid genes in photosynthetic higher plants are transcribed by at
least two RNA polymerases. The plastid rpoA, rpoB,
rpoC1, and rpoC2 genes encode subunits of the plastid-encoded plastid RNA polymerase
(PEP), an Escherichia coli-like, core enzyme. The second
enzyme is referred to as the nucleus-encoded plastid RNA polymerase
(NEP), since its subunits are assumed to be encoded in the nucleus.
Promoters for NEP have been previously characterized in tobacco
plants lacking PEP due to targeted deletion of rpoB (encoding
the ![]() -subunit) from the plastid genome. To
determine if NEP and PEP share any essential subunits, the rpoA,
rpoC1, and rpoC2 genes encoding the PEP
-subunit) from the plastid genome. To
determine if NEP and PEP share any essential subunits, the rpoA,
rpoC1, and rpoC2 genes encoding the PEP ![]() -,
-, ![]()
![]() -, and
-, and ![]() "-subunits were removed by targeted gene deletion
from the plastid genome. We
report here that deletion of each of these genes yielded
photosynthetically defective plants that lack PEP activity while
maintaining transcription specificity from NEP promoters. Therefore,
rpoA, rpoB, rpoC1, and rpoC2 encode PEP
subunits that are not essential components of the NEP transcription
machinery. Furthermore, our data indicate that no functional copy of
rpoA, rpoB, rpoC1, or rpoC2 that could complement
the deleted plastid rpo
genes exists outside the plastids�.
"-subunits were removed by targeted gene deletion
from the plastid genome. We
report here that deletion of each of these genes yielded
photosynthetically defective plants that lack PEP activity while
maintaining transcription specificity from NEP promoters. Therefore,
rpoA, rpoB, rpoC1, and rpoC2 encode PEP
subunits that are not essential components of the NEP transcription
machinery. Furthermore, our data indicate that no functional copy of
rpoA, rpoB, rpoC1, or rpoC2 that could complement
the deleted plastid rpo
genes exists outside the plastids�.
At least two distinct RNA polymerases are involved in
the transcription of plastid genes in photosynthetic higher plants. One of
these contains homologs of the Escherichia coli enzyme, including
the ![]() -,
-, ![]() -,
-, ![]()
![]() -, and
-, and ![]() "-subunits encoded in the plastid rpoA, rpoB, rpoC1,
and rpoC2 genes, and is referred to as PEP. The promoters for
PEP are reminiscent of the E. coli
"-subunits encoded in the plastid rpoA, rpoB, rpoC1,
and rpoC2 genes, and is referred to as PEP. The promoters for
PEP are reminiscent of the E. coli ![]() 70-type promoters, and have two conserved hexameric
blocks of sequences (TTGACA or "-35" element; TATAAT or
"
70-type promoters, and have two conserved hexameric
blocks of sequences (TTGACA or "-35" element; TATAAT or
"![]() 10" element) 17 to 19 nucleotides apart.
Transcription from PEP promoters initiates 5 to 7 nucleotides downstream
of the "
10" element) 17 to 19 nucleotides apart.
Transcription from PEP promoters initiates 5 to 7 nucleotides downstream
of the "![]() 10" promoter element (Igloi and Kossel, 1992
10" promoter element (Igloi and Kossel, 1992![]() ;
Gruissem and Tonkyn, 1993
;
Gruissem and Tonkyn, 1993![]() ;
Link, 1996
;
Link, 1996![]() ).
Promoter specificity to PEP is conferred by nuclear-encoded
).
Promoter specificity to PEP is conferred by nuclear-encoded ![]() -like factors (Isonzo et al., 1997
-like factors (Isonzo et al., 1997![]() ;
Tanaka et al., 1997
;
Tanaka et al., 1997![]() ).
).
PEP promoter-�� ----35 TTGACA----17-19----- -10TATAAT�5-7--+1>
Several reports indicate the existence of a second, NEP
activity (Morden et al., 1991![]() ;
Hess et al., 1993
;
Hess et al., 1993![]() ;
Allison et al., 1996
;
Allison et al., 1996![]() ).
A candidate for NEP is an approximately 110-kD protein that has
properties similar to the mitochondrial and phage T3/T7 RNA polymerases that may be
part of a larger complex (Lerbs-Mache, 1993
).
A candidate for NEP is an approximately 110-kD protein that has
properties similar to the mitochondrial and phage T3/T7 RNA polymerases that may be
part of a larger complex (Lerbs-Mache, 1993![]() ;
Hedtke et al., 1997
;
Hedtke et al., 1997![]() ).
NEP promoters share a loose, 10-nucleotide consensus, ATAGAATA/GAA,
overlapping the transcription-initiation site, which is reminiscent
of promoters recognized by the mitochondrial and phage T3/T7 RNA polymerases (Hajdukiewicz
et al., 1997
).
NEP promoters share a loose, 10-nucleotide consensus, ATAGAATA/GAA,
overlapping the transcription-initiation site, which is reminiscent
of promoters recognized by the mitochondrial and phage T3/T7 RNA polymerases (Hajdukiewicz
et al., 1997![]() ;
H�bschmann and B�rner, 1998
;
H�bschmann and B�rner, 1998![]() ;
for review, see Maliga, 1998
;
for review, see Maliga, 1998![]() ).
).
NEP promoter elements-----ATAGAATA/GAA--------------+1>
Plastid RNA polymerase activities with
distinct sensitivities to inhibitors are present in higher plants in
multisubunit complexes (Pfannschmidt and Link, 1994![]() ). Sharing of essential subunits of
RNA polymerases
has been reported in yeast (Sentenac et al., 1992
). Sharing of essential subunits of
RNA polymerases
has been reported in yeast (Sentenac et al., 1992![]() ).
Therefore, plastid NEP and PEP could
be part of the same complex. To test if NEP and PEP share any
essential subunits, the rpo genes encoding PEP subunits were
removed by targeted gene deletion from the plastid
genome. Study of promoter activity in plastids lacking the rpoB
gene has shown that the PEP
).
Therefore, plastid NEP and PEP could
be part of the same complex. To test if NEP and PEP share any
essential subunits, the rpo genes encoding PEP subunits were
removed by targeted gene deletion from the plastid
genome. Study of promoter activity in plastids lacking the rpoB
gene has shown that the PEP ![]() -subunit is essential for PEP transcription activity,
but it is not required for transcription by NEP (Allison
et al., 1996
-subunit is essential for PEP transcription activity,
but it is not required for transcription by NEP (Allison
et al., 1996![]() ;
Hajdukiewicz et al., 1997
;
Hajdukiewicz et al., 1997![]() ).
).
These studies address the contribution of the rpoA, rpoC1, and rpoC2 genes to transcription from PEP and NEP promoters. We report here that deletion of each of these genes yields photosynthetically defective plants that lack PEP activity while maintaining transcription from NEP promoters. Therefore, rpoA, rpoB, rpoC1, and rpoC2 encode essential PEP subunits that are not components of the NEP transcription machinery. Furthermore, no functional copy of the rpo genes that could complement the deleted plastid rpo genes exists outside the plastid.
Chloroplast genome of N. tabacum contain more than sixty operons have been described and transcripts are polycistronic.� They are processed for translation.� Transcripts are stable because of their 5� and 3�UTRs.
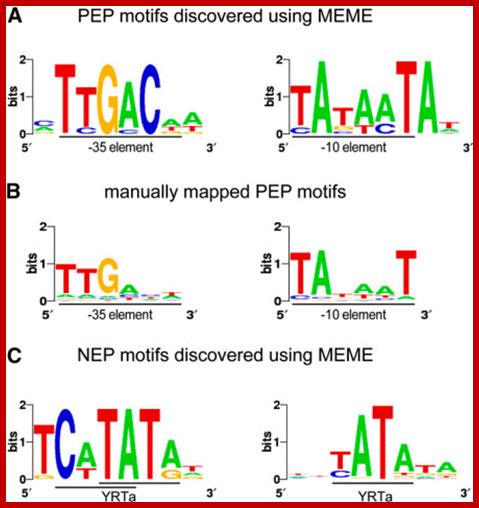
Promoter elements of Plastid and Nuclear
coded RNA polymerase genes. (Allison
et al., 1996![]() ;
Hajdukiewicz et al., 1997
;
Hajdukiewicz et al., 1997![]() ).
).
Transplastomic:
Plastids probably originated from formerly free-living bacterium-like organisms that were taken up by free living cells way back 2.7 billion years ago. The relationship is mutually beneficial: one example is the chloroplasts, a special group of plastids used by the plant to convert the sun�s energy. For their part, the plastids� forebears found a protected environment in the plant cells. They have retained some of their own genes, while others have over time become integrated into the cell nucleus of the plant cells. Interestingly none of the nuclear genes got transferred to Plastid genome.
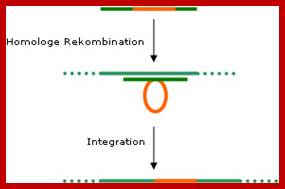
In principle, it is possible to integrate foreign (transgenic) DNA into the plastome. To do this, the target gene is �flanked� by sequences found in the plastome. Using the homologous recombination procedure (see diagram) the transgenic DNA is integrated into the plastome. Unlike in the plant�s nucleus genome, this process occurs naturally. The plants modified using this kind of plastid transformation is called �transplastomic�. In general, the pollen cells of flowering plants do not contain plastids. If a foreign gene is present only in the plastome it will not be passed on during propagation?
As well as the security aspect � the desire to confine the foreign genes to the transgenic plant � the increased activity of the foreign genes in the plastids is also interesting. The reason for this lies in their numbers: there are up to 100 plastids in each cell, each carrying up to 100 copies of their genetic material. The genetically modified plants carry the new gene on every copy in all plastids. This means that after successful transfer of a foreign gene, proteins produced by this gene accounts for 50% or more. This level of productivity cannot currently be achieved using traditional methods. However, implementing the procedure is proving to be a technological challenge.
Genetically engineering in plants; https://columbiasciencereview.com/
Step 1: The transgenic DNA (red) is flanked by plastomic sequences (gene constructs).
Step 2: The plastomic sequences of the gene construct are taken up by similar (homologous) DNA sequences in the plastid.
Step 3: The transgenic DNA is integrated into the plastome.
Introducing Foreign Genes into Plastids:
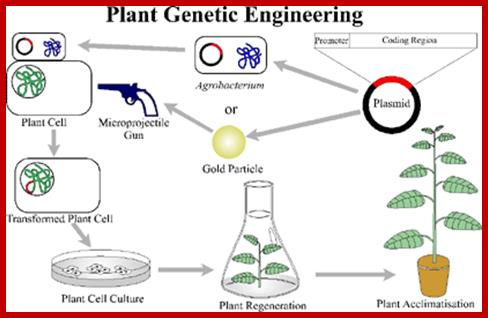
Established procedures are used to introduce the transgenic DNA into the cell/plastids � e.g. transformation using Agrobacterium, biolistic gun or microinjection. The important thing is that the tissue used should transform and regenerate easily and contain plastids, e.g. chloroplasts. Suitable tissues that have been used so far are leaves, microspores (unripe pollen with cell walls that are not yet fully formed), immature embryos and hypocotyls of young seedlings.
Plastid transformation is much more complex than cell nucleus transformation because there are up to 10,000 plastid genomes in each cell. Transplastomic lines are genetically stable only if all plastid copies are modified in the same way, i.e., uniformly. This is achieved through repeated regeneration under certain selection conditions. Plastid transformation has already been successfully performed in tobacco, potatoes, oilseed rape, rice, A. thaliana (Thale cress) and Lesquerella fendleri (bladder pod).
Plastid transformation for the production of genetically modified plants has advantages and disadvantages.
� Plastid transformation reduces the probability of the transgene spreading via pollen, since the plastids are usually inherited from the maternal plant, rather than via the pollen.
� Transformation and regeneration of transplastomic plants is technically more complex. The transformation efficiency rate is lower. Very high expression levels are limited to leaf tissue.
Results: A completed SiFo project succeeded in producing a transplastomic oilseed rape shoot (particle gun). If it proves productive, the shoot is to be investigated further, e.g. for stability and outcrossing behaviour of the transgene.
Construction of Marker-free Transplastomic Tobacco using the Cre-loxP Site-Specific Recombination System: Kerry Ann Lutz1, Zora Svab1 & Pal Maliga
Incorporation of a selectable marker gene in the plastid genome is essential to uniformly alter the thousands of genome copies in a tobacco cell. When transformation is accomplished, however, the marker gene becomes undesirable. Here we describe plastid transformation vectors, the method of plastid transformation using tobacco leaves and alternative protocols for marker gene excision with the P1 bacteriophage Cre-loxP site-specific recombination system. Plastid vectors carry a marker gene flanked with directly oriented loxP sites and a gene of interest, which are introduced into plastids by the biolistic process. The transforming DNA integrates into the plastid genome by homologous recombination via plastid targeting sequences. Marker gene excision is accomplished by a plastid-targeted Cre protein expressed from a nuclear gene. Expression may be from an integrated gene introduced by Agrobacterium transformation (Transformation Protocol), by pollination (Pollination Protocol) or from a transient, non-integrated T-DNA (Transient Protocol). Transplastomic plants are obtained in about 3 months, yielding seed after 2 months. The time required to remove the plastid marker and nuclear genes and to obtain seed takes 10�16 months, depending on which protocol is used.
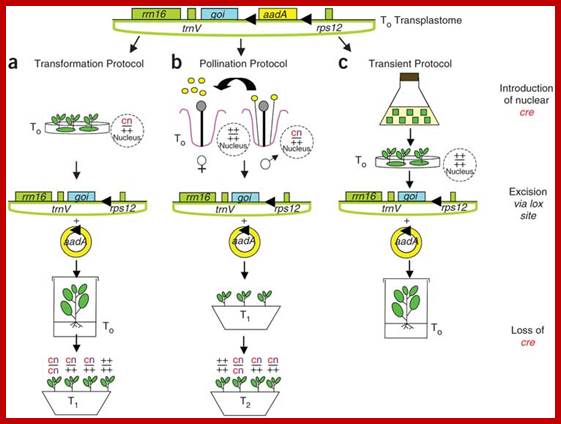
Removal of Marker gene: Incorporation of a selectable marker gene in the plastid genome is essential to uniformly alter the thousands of genome copies in a tobacco cell. When transformation is accomplished, however, the marker gene becomes undesirable. Here we describe plastid transformation vectors, the method of plastid transformation using tobacco leaves and alternative protocols for marker gene excision with the P1 bacteriophage Cre-loxP site-specific recombination system. Plastid vectors carry a marker gene flanked with directly oriented loxP sites and a gene of interest, which are introduced into plastids by the biolistic process. The transforming DNA integrates into the plastid genome by homologous recombination via plastid targeting sequences. Marker gene excision is accomplished by a plastid-targeted Cre protein expressed from a nuclear gene. Expression may be from an integrated gene introduced by Agrobacterium transformation (Transformation Protocol), by pollination (Pollination Protocol) or from a transient, non-integrated T-DNA (Transient Protocol). Transplastomic plants are obtained in about 3 months, yielding seed after 2 months. The time required to remove the plastid marker and nuclear genes and to obtain seed takes 10�16 months, depending on which protocol is used.; Nature �protocols �Kerry Ann Lutz et al
(a) The Transformation Protocol. (b) The Pollination Protocol. (c) The Transient Protocol. On top is the map of ptDNA containing the floxed aadA and the gene of interest, and the plastid rrn16, trnV and 3'rps12 genes. Triangles symbolize loxP excision sites through which circular aadA excision products form wild-type locus (+) or presence of cre (c) and kanamycin resistance (n, for neo) genes is indicated in the nucleus.
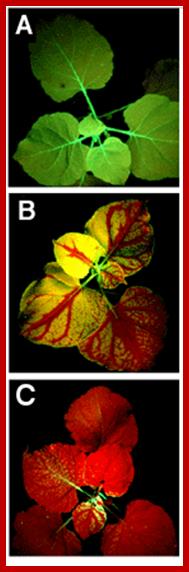
�Systemic silencing of GFP in transgenic tobacco plants. (A) Nonsilenced plants appear uniformly green under ultraviolet (UV) illumination. (B) Upon local activation in one single leaf, silencing of GFP spreads throughout the plant. The red color is from chlorophyll fluorescence under UV. (C) Extensive silencing caused by relay amplification of the systemic signal; www. scincemag.org���� �
.
Translastomic -N.excelsiana and N.benthamiana
Plastid Engineering; by Cody Cobb � November 3, 2009
Long ago � before you or anyone in your family photo albums were born � a small, unassuming cyanobacterium was busy being engulfed by another cell. The engulfing cell�s intentions were most likely along the lines of �Yum, food!�, but lucky for us the cyanobacterium was not consumed. Instead, it stayed there, establishing a new home inside the confines of its voracious captor. We now know this happy accident was a momentous first step towards a greener, more botanical planet, because our little cyanobacterium was the photosynthetic ancestor to that most remarkable organelle: the chloroplast.
(By law, any discussion of chloroplast origins compels me to mention the similar origin of the mitochondrion. With those requirements now met, let us now continue.)
The focus of this post will be more technological than biological, but there are a few basic facts we need to get out of the way before we can proceed.
Briefly:
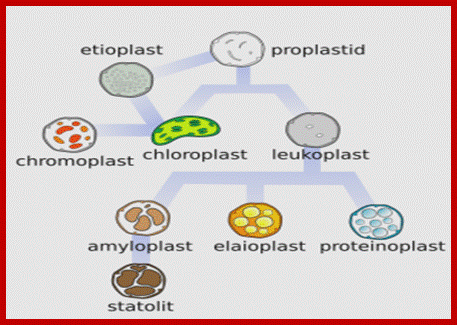
� Chloroplasts, along with leucoplasts, proteinoplasts, elaioplasts, amyloplasts, statoliths, and chromoplasts, belong to a class of organelles known as plastids. The names of these other plastids aren�t important so long as you realize the chloroplast isn�t the only game in town. That�s why the title of this post is �Plastid Engineering and not chloroplast.
Plastid Engineering, Cody Cobb; http://www.biofortified.org/
� Plastids replicate separately from their host cell, and in any given cell there can be 100 to 1,000 plastids. Moreover, plastids contain multiple copies of their genome (plastome) to the point where a single plant cell may contain 10,000 plastomes. By contrast, the nuclear genome has only one copy (this is manifestly untrue, but we�re talking orders of magnitude here).
� Plastids behave a lot like prokaryotes. Their genome is circular, their proteins aren�t glycosylated (i.e., have sugars attached to them), and they can process polycistronic mRNA (i.e., more than one protein produced from a single mRNA; most eukaryotic genes are monocistronic).
� Over history, most plastid genes have migrated into the nucleus, even though the protein produced might still accumulate in the plastid. Those proteins are instead brought back to the plastid by a specific targeting sequence. Quite a few genes have been lost from the original cyanobacterial ancestor, leaving only 50 to 200 of the original ~3,000 genes in most plastids today. In scientifically and agriculturally important species, these genes have all been sequenced and characterized.
� Plastids are inherited uniparentally, that is, from one parent and not the other. In most flowering plants, only maternal plastids are passed on. In some species, such as pine trees, paternal transmission in the pollen is the norm. There are exceptions.
Ideally as you pored over those facts your brain started piecing together the reasons why we would want to tinker with plastid � rather than nuclear � DNA. Uniparental inheritance is a big one: even people who know next to nothing about GM crops know there�s concern about, say, GM corn in one farmer�s field contaminating non-GM corn in their neighbor�s field. Crops with genetically engineered plastids (known by the awesomely retro-sounding name transplastomics don�t have this problem since plastids aren�t usually found in pollen. Of course, plant biology is, technically, a biological science, so there are exceptions that will to be need to be addressed.
Extreme polyploidy is another attractive feature: inserting a gene of interest (GOI) into the chloroplast genome means having up to 10,000 or more copies of that gene per cell. That translates (hah!) into very high levels of protein production indeed. And since most plastid genomes are already well characterized, we can know in advance where our inserted DNA will wind up.
Non-glycosylation differs in usefulness depending on the source of the foreign gene. Plants, mammals, fungi, and insects all have different patterns of glycosylation, with plastids and prokaryotes not participating in the ritual at all. So, proteins normally present in prokaryotes are produced identically in plastids, whereas proteins of eukaryotic origin might be missing structural elements crucial to their function (or the protein might find it does just fine without those extra sugars, you never know).
So what are some limitations and problems with plastid engineering? To answer that question, we must first learn how transplastomic plants are created.
Today, only a few species have had their plastids successfully transformed. The first transplastomic organism was created in 1988 using the unicellular alga Chlamydomonas reinhardtii, notable for having only one large chloroplast. Two years later, stable tobacco transplastomics were created. Since then, varying levels of success have been achieved with potato, tomato, rapeseed, cauliflower, poplar, rice, soybean, and a few others, but only in tobacco is plastid transformation routine.
The first step in plastid transformation is introducing the new genes to the old. Typically, this is done by particle bombardment (�biolistic� or the �gene gun�) or polyethylene glycol (PEG) treatment. In the latter, you remove the cell wall of a plant cell to create a protoplast and then subject it to a solution of DNA in PEG, whereas in the former you basically shoot the plant with DNA. Since particle bombardment is the more commonly used of the two, I�ll explain its mechanism.
First you need your gene of interest in a plasmid (a small circle of DNA that contains of a few genes and can be grown in and purified from bacteria). The plasmid will also contain a selectable marker (a gene that confers resistance to antibiotics like spectinomycin, streptomycin, or kanamycin) and a visual marker (green fluorescent protein or a derivative thereof). The GOI, selectable marker, and visual marker will be flanked by sequences taken from the plastid genome, carefully chosen so that the site of homologous recombination (see further reading) does not disrupt the function of normal plastid genes.
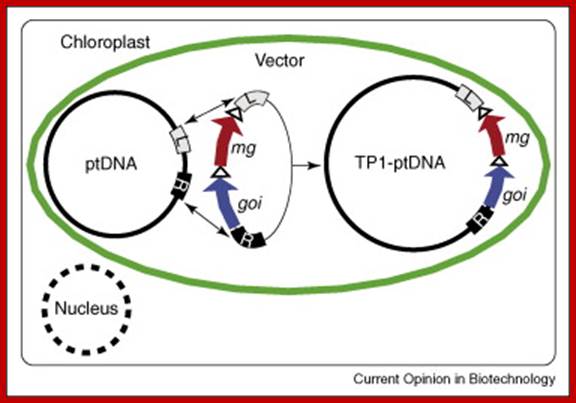
Plastid Engineering; Cody Cobb; ;http://www.biofortified.org/
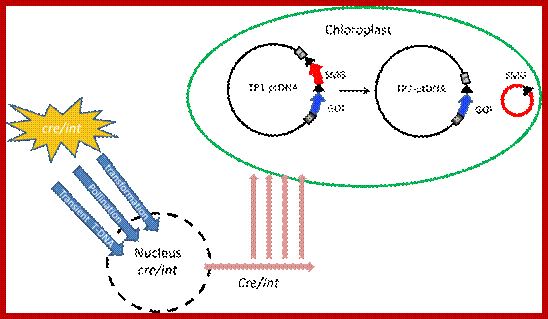
Marker gene excision from the plastid genome by Cre or Int site-specific recombinases. A site-specific recombinase gene (cre/int) introduced into the nucleus by transformation, pollination or transient Agroinfiltration, encodes a plastid-targeted recombinase that excises selectable marker gene (SMG) from TP1-ptDNA after import into plastids. Excision of the marker gene by phage recombinases via the target sites (black triangles) yields marker-free TP2-ptDNA carrying only the gene of interest (GOI) and one recombinase recognition sequence; http://www/.intechopen.com; Borys Chong-P�rez and Geert Angenon
Next, the plasmids are expressed to high quantities in bacteria and purified, then adhered to small particles of tungsten or gold, often to less than a millionth of a meter in diameter. A small section of leaf tissue is placed into a low-pressure vacuum chamber and bombarded with a volley of DNA-coated particles, obliterating most of it.
Regenerated Transplastomic shoot;
Plastid Engineering Cody Cobb;
A very small percentage of the remaining tissue will contain transformed plastids at this point. Worse yet, a surviving cell with a transformed plastid will still overwhelmingly contain untransformed plastids. The next steps are the lengthiest and most tedious part of the process, for now the bombarded tissue must be coaxed into regenerating into a wholly new plant while at the same time eliminating any untransformed plastids it may still harbor. Stringent antibiotic regimens are applied to emerging plantlets, and visual inspection of GFP expression reveals areas of transformed plastids. Those areas are then sliced away and grown on their own regenerative media. This process is repeated for about 20 cell divisions before a state of exclusively transformed plastids (homoplasmy) is achieved. Once reached, the plantlets are allowed to grow in the absence of antibiotic selection and set seed at maturity. If the progeny is shown to be homoplasmic, then the line is considered stably transformed.
So, you�ve created a transplastomic plant. Now what? Obviously that antibiotic resistance gene is no longer doing you any good, so you�ll have to find a way to get rid of it lest it sap precious metabolic resources and stunt your plant�s growth. And just how certain are we that plastid inheritance is uniparental? What if life, as renowned chaos theorist Ian Malcolm once gravely intoned, finds a way?� �Shouldn�t we run a few tests to determine the likelihood of plastid-transference via pollen? What about those cereals really important plants? Why are their plastids so difficult to transform?



Flouresacent, monkey by genetic engineering; Alba, left fluorescent rabbit; http://www.ekac.org/; The Nevada team has created environmental stress sensors by using rd29A and DREB1Cpromoters to express red fluorescent protein and other bio-fluorescent markers. When induced by environmental stress, plants carrying these genes can easily be detected by the farmer walking through his field or by a plane flying over acres of farmland. Promoter elements are short and easy to work with. They also allow for modification and specialization. DREB1C and rd29Aboth have multiple binding motifs in their promoter regions allowing for variation in expression levels and the particular stresses that induce them. 35S is a constitutive promoter that can be valuable for control groups in stress and other plant response research.An iconic image of plant genetic engineering 1986 of a glowing transgenic tobacco plant bearing Luciferace gene of fire flies striking; www. edinoformatics.com
Mitochondria:
Mitochondrial DNA:
�Mitochondrial inheritance in higher system is maternal as in the case of plastids.� Humans have inherited this genome from some of the African mothers called as Human-Mitochondrial Eves.� But in unicellular haploid yeast, often, the mitochondrial inheritance is biparental. �
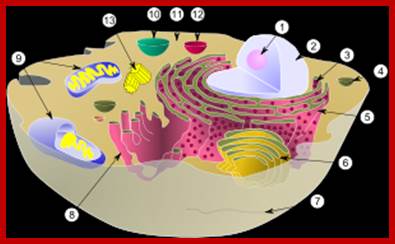
A typical animal cell diagram: Within the cytoplasm, the major organelles and cellular structures include: (1) nucleolus (2) nucleus (3) ribosome (4) vesicle (5) rough endoplasmic reticulum (6) Golgi apparatus (7) cytoskeleton (8) smooth endoplasmic reticulum (9) mitochondria (10) vacuole (11) cytosol (12) lysosome (13) centriole.; http://00kallus00581.blogspot.in/
The number of mitochondria per cell varies from tissue to tissue and the species. �Brain tissue contains ~10,000 mitochondria per cell, but liver cells contain ~1000-1200 per cell.� Flight muscles contain very high numbers.� Deletion of Mit genes in yeast exhibits petite character i.e. small colonies which grow slowly.
Mitochondrial ribosomes are unique for their size is 55s and made up of 39s and 28s ribosomes with 16s and 2s rRNAs respectively.� But the number of riboproteins can be ~80, the total mass of mitochondrions ribosomes is high.
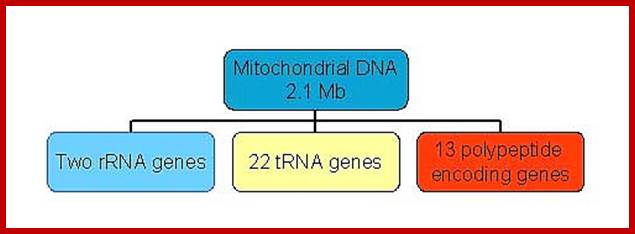
Size of the mitochondrial DNA surprisingly vary, even though they code for just 13 proteins and contain 7 �8 unrecognized reading frames (URFs); they code for two ribosomal RNAs and ~22 tRNAs.� The smallest mitochondrial genome is found in plasmodium (intracellular obligate parasite) is encoded with five proteins and ribosomal RNAs.� Looking at the genes encoding proteins among eukaryotes is not the same; there are some differences; ex. Cytochrome oxidase II gene subunits.� Moreover, many plant mitochondrial transcripts are edited like C to U and U to C.� Many of the ancestral mitochondrial genes after symbiosis are transferred to the nucleus through reverse transcriptase mechanism ?.

Figure of Mitochondria.; http://en.citizendium.org/
Mitochondria; http://www.proprofs.com/
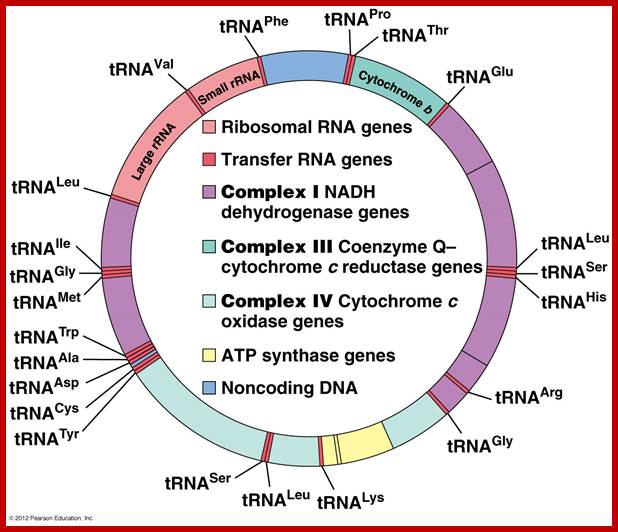
One strand is called H strand Guanine rich 28 genes and the other are called L strand with 9 genes. Department of Biology, Memorial University; Newfoundland; http://www.mun.ca/biology
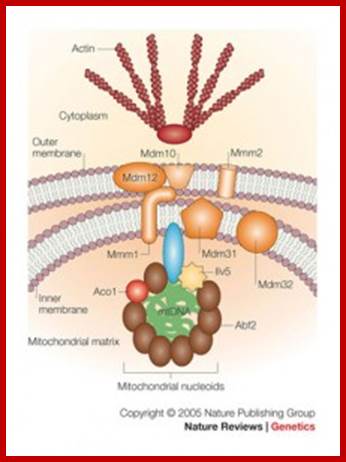
The DNA is packed into nucleoids and attached to inner membrane of mitochondria. Xin Jie Chen & Ronald A. Butow; �http://www.nature.com/
The core components of the segregation apparatus consist of the Mdm10�Mdm12�Mmm1 (mitochondrial membrane proteins) complex, which might also interact with Mmm2. This core complex forms the contact site between the inner and outer mitochondrial membranes. It is located primarily in the outer membrane, with the possible extension of Mmm1 into the inner membrane and its possible protrusion into the mitochondrial matrix. The inner membrane proteins, Mdm31 and Mdm32, are also proposed to be part of this double membrane-spanning complex, on the basis of genetic interactions with Mdm10, Mdm12, Mmm1 and Mmm2 (Ref. 100). This supramolecular complex mediates the attachment of mitochondria to actin cables in the cytosol and tethers mitochondrial nucleoids (mt-nucleoids) on the matrix side through as yet unknown proteins. This provides a mechanism for linking mt-nucleoids to the cytoskeleton so that mt-nucleoids move with mitochondria into the progeny cell during cell division7. How the Mdm10�Mdm12�Mmm1 complex, Mmm2, and the inner membrane proteins Mdm31 and Mdm32 interact has yet to be clarified;. Xin Jie Chen & Ronald A. Bu tow; /
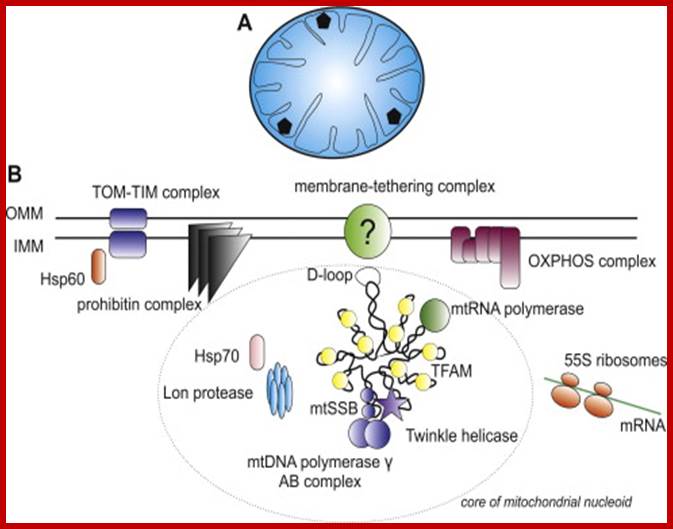
Mitochondrial Nucleoids and the structure of life; The eukaryotic nucleoid: (A) Schematic view of a transverse cross-section of the mammalian mitochondrion. Mammalian mitochondria contain multiple nucleoids (shown as black pentagons) and each individual nucleoid usually harbors more than one copy of the mtDNA. The total number of mitochondrial genomes per human cell ranges from hundreds to tens of thousands, depending on cell type. The human mitochondrial genome is considerably smaller than the E. coli one (16.5 kbp vs. 4.6 Mbp) and thus occupies a proportionally smaller volume in the mitochondrial matrix (rev. (Bogenhagen, 2011)). (B) A detailed view of a single human mitochondrial nucleoid. For simplicity, only one molecule of mtDNA is shown. MtDNA is usually packed with a single type of protein which often has several additional functions. In mammals, this is TFAM- transcription factor A of mitochondria, which also functions in mtDNA replication, transcription and maintenance. The D-loop is a regulation site for the replication and transcription of mtDNA and is thought to be bound to the inner mitochondrial membrane, probably through a multiprotein complex. Like the E. coli nucleoid, human mitochondrial nucleoids are believed to have a layered structure. MtDNA replication and transcription take place in the core part (circled) while the subsequent RNA processing and translation occurs in the outer part (rev. ( and )). Those proteins shown within the circle, TFAM, mtRNA polymerase, mtSSB, mitochondrial DNA polymerase γ complex, and Twinkle helicase, are thought to be components of the core of human mitochondrial nucleoids. Proteins outside the ellipse have been also identified in nucleoids. For a full list of proteins identified within human mitochondrial nucleoids to date, see (Bogenhagen et al., 2008). OMM � outer mitochondrial membrane, IMM � inner mitochondrial membrane, OXPHOS � oxidative phosphorylation. John Hewitt; http://phys.org/
Mitochondrial transcription factor A (TFAM) is the main factor packaging mtDNA into nucleoids and is also essential for mtDNA transcription initiation. The crystal structure of TFAM shows that it bends mtDNA in a sharp U-turn, which likely provides the structural basis for its dual functions. Super-resolution imaging studies have revealed that the nucleoid has an average diameter of ∼100 nm and frequently contains a single copy of mtDNA;Nucleoids are associated with inner membranes and highly packed; Mitochondrial Nucleoids are dynamic structures that are remodeled in response to metabolic cues. Christian Kuk at, Nils Goren Larsson; Trends in Biology.; Martin Kucej1,*, Blanka Kucejova1,�, Ramaiah Subramanian2, Xin Jie Chen1,� and Ronald A. Butow1; Journal of Cell Science.
Saccharomyces cerevisiae contain 100 to 200 mitochondria per cell and each mitochondrion contains about 30-50 copies of 84kbp size DNA molecules.� Plant mitochondrial DNA size vary from 1.5 X 10^5 bp to 2.5x^6 bp? Arabidopsis thaliana mit DNA is 367kbp long. Chara vulgaris 67,737bp, tomato 155,460, Potato 155,372bp. Human mitochondria has 16.59 kbp sized DNA and it is the most compact genome for there are no large regions that is said to be non-coding.� But Arabidopsis mitochondrial DNA is ~366,924 bp but in some cucurbit plants, largest mitochondrial Genome is ~2Mb.�
Most of the Mit genomes are circular, but some genomes found in Protists like Tetrahymena are linear and often concatemrs of 47,172 bp long, there are 44 genes; some protein coding genes and some are URFs. In trypanosomes the Mit DNA exists in the form of multiple maxi circles concatenated (interlocked DNA) by thousands minicircular DNA encoding guide RNAs which are used in editing transcripts.
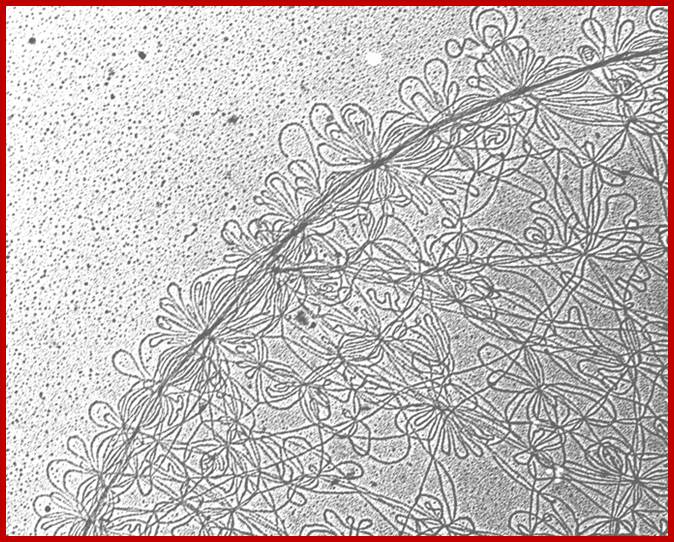
TEM kDNA network from C. fasciculata, small loops minicircle DNA; Kinetoplastid DNA; http://www.pnas.org/
|
Species |
Genome size in Mbps |
Copy no. |
|
Human |
16569 |
100-1000 |
|
S.cervisiae |
84 |
|
|
Neurospora |
108 |
|
|
Chlamydomonas |
16kbps linear |
|
|
Protist Reclenomonas |
69 |
|
|
Euglenozoan |
4kbp short, LSU and SSU |
Maxicircle and minicircles DNAs |
|
Apicomplexan protists Plasmodium sp. |
6kbp �five genes |
|
|
Oriza sativa |
490kbps |
|
|
Melon |
156013 |
|
|
Oenothera |
195kbps |
|
|
Turnip |
218 |
|
|
Musk melon |
2400kps |
|
|
Maize |
570kbps |
|
Mitochondrial DNA go through recombination, ex. A.thaliana
�
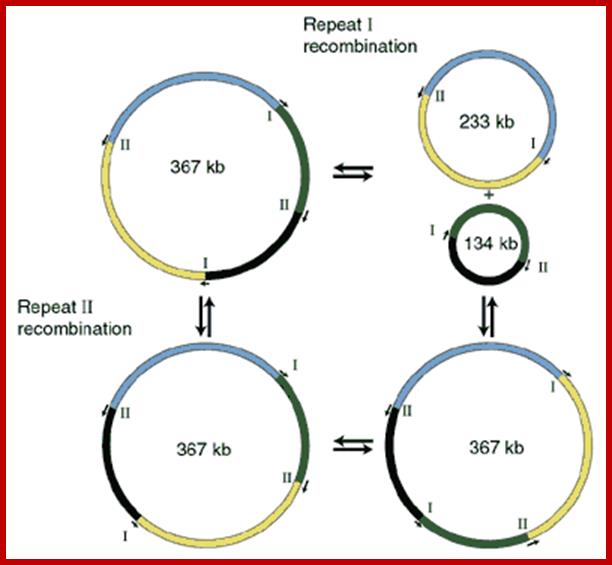
Web Figure; Recombination in mtDNA of Arabidopsis thaliana. The mtDNA contains a direct repeat (I) and an inverse repeat (II). Recombination at the two repeats will change the gene order and also split up the genome into two separate molecules. (After Unseld et al. 1997.) (Click image to enlarge.); Plant Physiology chapter 11.; Allan G. Rasmusson,et al;
Many of the tRNA genes or gene clusters are located between protein coding genes.� Genes are ordered in such a way, the end of one gene that is the last nucleotide of the gene can be the beginning of the next gene with one or two bases this side or that side.� In fact as many as five genes end with terminator codons with just two nucleotides such as U and UA, addition of poly-A tail produces ochre kind of TER codons.� In three codons the TER codon appears to have AGA, AGG sequence which normally code for Arginine.� It is only some mitochondrial genomes such as Neurospora and yeast; contain some unusual codons, which in character differ from universal codons.� Mitochondrial mRNAs have short poly-A tail of ~50 ntds long.�� In most of mRNAs, the initiator codons AUG, AUA or AUU lie just five to six bases down stream of the start nucleotide.� There are no 3� non-coding regions, but TER codon is the last.� Only ATPase 6 and Cytochrome oxidase-3 (CO-3) genes are contiguous and bi-cistronic, but the rest have tRNA clusters are found in between them.
Most transcripts, either from H-strand (heavily transcribed) or L-strand less transcribed), are single long strands and polycistronic. They are cleaved at tRNA sites to release tRNAs as well as monocistronic mRNAs.� Ribosomal RNAs are produced as precursor RNAs, but produced in large numbers. They are processed to functional rRNA segments.
Mammalian mtDNA:
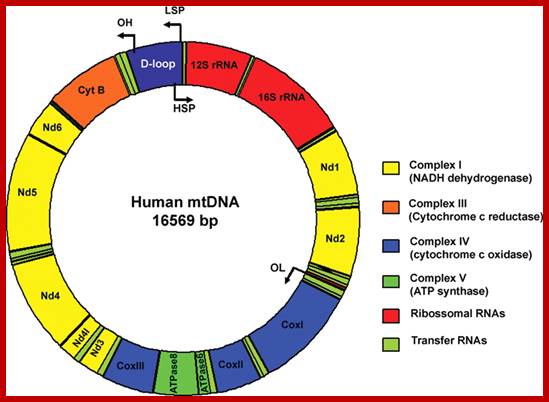
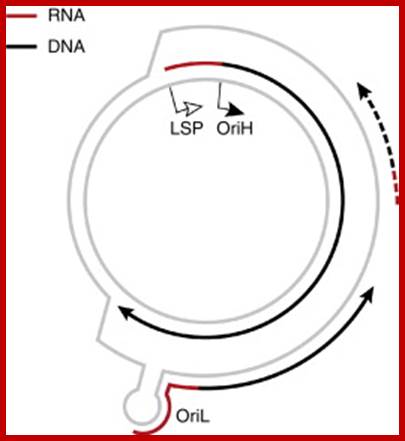
The human mtDNA genome encodes 13 polypeptides of the ETC;These include seven subunits of Complex I (ND 1�6 and ND4L), one subunit of Complex III (CytB), three subunits of Complex IV (COX I to III) and two subunits of Complex V (ATPase6 and ATPase8). It also encodes two rRNAs (12S and 16S) and 22 tRNAs (Anderson et al., 1981). The main control region is the D-loop which contains the H-strand promoter region (HSP), the LSP region and the origin of H-strand replication (OH). The second control region consists of only 30 bp and is located between ND2 and COXI and is the site of the origin of L-strand replication (OL). Copyright � 2014 European Society of Human Reproduction; tp://humupd.oxfordjournals.org/
Mammalian(human) mtDNA is a double stranded molecule, composed of a H (heavy) strand and a L (light) strand and is approximately 16.5 kb in size (e.g. human mtDNA is 16.569 kb; Anderson et al. 1981). The origin for H-strand replication (OH) and the HSP and LSP transcription promoters are located in the D-loop (displacement loop). The origin for L-strand replication (OL) is located two thirds away around the genome from the D-loop. MtDNA encodes thirteen proteins of the electron transport chain (ETC). Twelve genes are located on the H-strand and only one gene is located on the L-strand. mtDNA also has 22 tRNAs and 2 rRNAs involved in mtDNA transcript production and processing.
Mitochondrial DNA polymerase:
The enzymes for mitochondrial DNA replication are characterized from yeast system.� The yeast gene called MIP1 is equivalent to Eukaryotic pol-delta gene.
� Catalytic core �125kd,
� Subunits-two 35 and 47kd,
� It has 5�-3�polymerase activity,
� It has 3�-5� exonuclease activity,
� It has no primase activity,
� It has high fidelity and processivity,
� It has no repair functions,
� Primase-61-68Kda,
� Helicase-80x6,
� SSB-4mer-15kDa
As is shown in the human mtDNA map, several human disease-causing genes have been found to be localized in the mtDNA genome.
The strands of DNA are distinguished based on their coding ability.� The strand which codes for 13 proteins and 22 tRNAs and few URFs is called H-strand (heavily transcribed) and the other strand which codes for just 8 tRNAs and one URF is called L-strand (less transcribed).�� The origins for H-strand and L-strands are distinct and they are located at different positions, far away from one another.� The origin for H strand is in a region where transcriptions of H strand starts.� The origin for L strand is located at coding region containing 5 tRNA genes, nearly 2/3 distance from the origin-H.
Human mitochondrial genome inside:
Sci Tech Daily; http://www.biotechniques.com/
Mitochondrial DNA Replication:
|
||||
|
|
||||
|
|
||||
|
|
||||
|
|
||||
|
|
||||
|
|
||||
|
|
||||
|
|
||||
|
|
||||
|
|
||||
|
|
||||
|
|
||||
|
|
||||
|
|
||||
|
|
||||
|
|
||||
|
|
||||
|
|
Replication Origin� of Mitochondrial DNA:
- Mitochondrial DNA is ds, circular and exists in super coiled state.���
- �Though the size of the DNA varies from one system to the other, it codes for just 13 proteins.�
- It also contains seven or more unknown or unidentified reading frames (URFs).�
- Mitochondria contain genetic material in multiple copies, 20 or more per organelle.��
- Mitochondria multiply by elongation and fission of the existing mitochondria; mechanism is called fission, recalls bacterial mode of cell division.��
- The ORI site is located in a region called D-loop; this is where replication is initiated.
� In the D-Loop region the origin for H strand is located in a transcriptional start site between Cyt.B and 12s.rRNA genes.�
� The origin consists of three conserved sequences.�
- Similarly, origin sequence for L-strand is located at nearly 2/3 distances from the Ori-H.�
- Opening of the DNA in the origin-H is initiated by transcriptional process i.e. RNA synthesis, which initiates about 100 base pairs upstream of Ori-H.� The RNA transcript acts as a primer.
- As transcription proceeds into Ori-H region the L-strand is displaced as a loop called D-loop.
- RNase-H cleaves the transcript and provides a primer.
- Mitochondrial DNA polymerase (called gamma enzyme) starts extending the new strand i.e., it is the H-strand.�
- While the new strand grows it displaces the L-strand the D loop extends, so the loop is called displaced loop or D-loop.�
- The displacement and the replication of H strand continuous all along the length of the DNA till reach the end where it has begun.�
- When the displacement loop reaches 2/3rd distance from the origin, the displaced strand, which contains L-Ori sequence, between AN and CY tRNAs; it generates a hairpin structure because of complementary sequences, which acts as the landmark for the gamma polymerase to recognize and initiate replication of the L-strand.
Origin-Specific Initiation of Strand-Displacement mtDNA: Replication Leading-strand DNA synthesis is initiated from the heavy-strand origin (OriH) of DNA replication and passes OriL. The single-stranded OriL is exposed and adopts a stem-loop structure. OriL is the major initiation site for lagging-strand mtDNA synthesis, but other minor initiation sites also exist. In contrast, initiation of strand-coupled mtDNA synthesis occurs across a broad zone of several kilobases (Bowmaker et al., 2003).
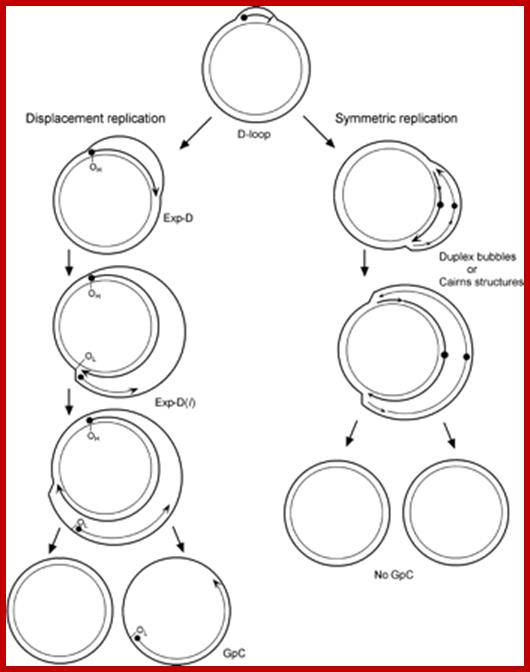
The asymmetric and strand-coupled models of mtDNA replication; Both models agree on the nature of the simple D-loop form of mtDNA. The displacement-model of replication is shown on the left and proceeds with single-stranded replication of the H-strand with further expansion and displacement of the D-loop. The intermediates are called expanded D-loops (Exp-D). This proceeds until the L-strand origin (OL) is exposed, with subsequent synthesis of the new L-strand in the opposite direction. Those intermediates are termed Exp-D(l). The asymmetry of strand synthesis leaves one segregated daughter molecule with an incompletely synthesized L-strand, called a gapped circle (GpC). The strand-coupled or synchronous model of replication is shown on the right. In this model, there is thought to be a zone of replication initiation within a broad area beyond the simple D-loop. Within this zone, both strands are synthesized bidirectionally as the double-stranded replication forks proceed through the length of the mtDNA. Exp-D, Exp-D(l), and GpC forms are excluded by this mode of replication; Timothy A. Brown1,2,6, Ciro Cecconi3, Ariana N. Tkachuk1,2, Carlos and Bustamante4, and� David A. Clayton1.
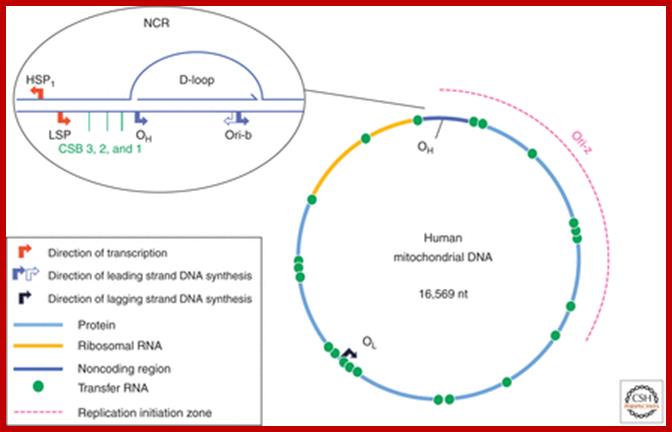
Schematic diagram of the human mitochondrial genome. Human mtDNA encodes 13 proteins that are essential for oxidative phosphorylation, and the ribosomal and transfer RNAs necessary for their translation. RITOLS replication initiates in the major noncoding region, at OH and Ori-b, and OL is a major initiation site of lagging strand DNA synthesis in mammals. Conversely, conventional bidirectional replication of mtDNA can initiate almost anywhere in the mitochondrial genome, with most such events occurring in a region of several kilobases adjacent to the major noncoding region (NCR), Ori-z (see main text for details). LSP, light strand promoter; HSP, heavy strand promoter; CSBs, conserved sequence blocks, Ian J. Holt and Aurelio Reyes
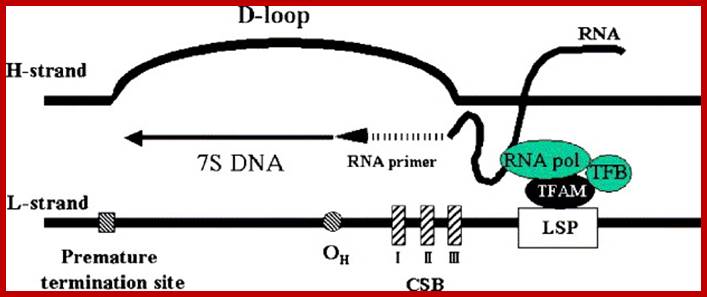
Schematic description of a D-loop region. A D-loop region is a major non-coding region in human mtDNA. TFAM, mitochondrial transcription factor B (TFB), and mitochondrial RNA polymerase (RNA pol) are assembled on a light-strand promoter (LSP), and then transcription is started. The transcript makes an RNA�DNA hybrid in the conserved sequence blocks (CSB) and is considered to serve as a primer for DNA synthesis (RNA to DNA transition). The transition occurs mainly in a region ranging np 190 to 110 (OH). Most of the DNA synthesis prematurely terminates about 700 bases downstream, which produces a characteristic triplex D-loop structure. In the strand-asymmetric model, the DNA synthesis proceeds to replication of mtDNA when the premature termination does not occur, Dongchon Kang,Naotaka Hamasaki
Replication of H strand in progress and the L strand replication just initiated. William C. Copeland, http://www.niehs.nih.gov/
- Does it require a primer? It uses its own RNAP, short 20-27nts
- Replication, whether it is H-strand or L-strand replication is unidirectional.
- H strand replication is initiated first; the L-strand replication is initiated later.
- Both strands are used for transcription, and the same are processed to generate tRNAs, rRNAs and mRNAs.
- Transcription produces more of rRNA than any other RNAs; It is required for the production of mitochondrial ribosomes?
Mechanism of Replication:
� The origin for H strand synthesis is located in a transcriptional start site located between Cyt.B and 12s.rRNA genes.�
� What signals actually trigger DNA replication though not clear, yet the number of mitochondria increases with the demand; Mitochondrial DNAs also multiply and produce more copies to the needs.��
� As the transcription initiates, the RNA is used as primer. The same is degraded by an enzyme called RNase-H. Mitochondrial DNA polymerase (called gamma unit), uses the RNA primer and extends new strand on H-DNA template.
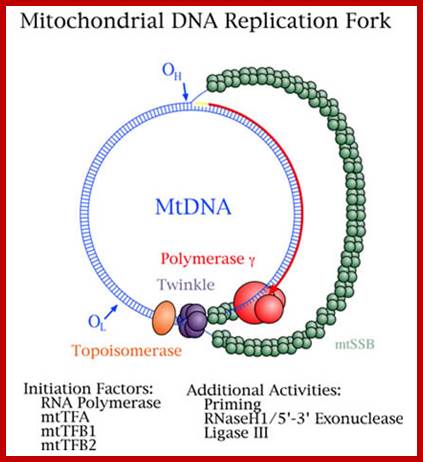
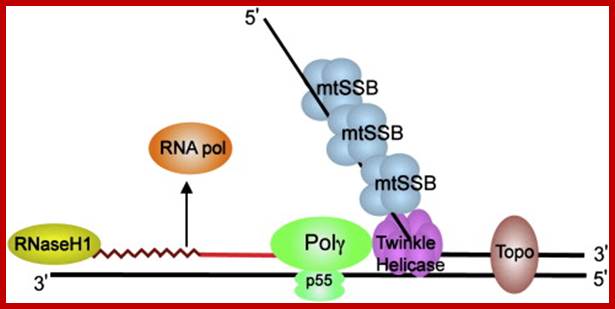
�Schematic diagram of a mitochondrial DNA replication fork showing the critical proteins required for DNA replication. The nascent DNA synthesized by pol γ (green) is shown as a solid red line, while the RNA primer (jagged red line) created by the mitochondrial RNA polymerase (orange) is being degraded by RNase H1 (yellow). The mitochondrial DNA helicase (purple) unwinds the downstream DNA forming a single-stranded loop which is coated with mtSSB (light blue). Topoisomerases (brown) work to relieve torsional tension in the DNA created by unwinding, Rajesh Kasiviswanathan, et al.
� While it is synthesizing, it progressively displaces the L-strand, which actually loops out, so the mechanism of replication is often termed as D-loop mechanism.��
� As replication passes through origin-L, the sequence in the L loop generates a secondary structure, which is what called origin for L strand.
� This region is recognized by the mitochondrial DNA-polymerase and starts L- strand replication.�
� Whether L-strand replication requires a primer or not, it is not clear.��
The region at which transcription initiates is also the site at which initiation of H strand synthesis begins.� So, this region is called D-loop region.
Mit DNA replication termination:
Not much is known about the replication termination for replication of H strand start from D loop and the fork from L strand meet each other from opposite side. Almost nothing is known about the termination of mtDNA replication beyond its location. Terminal replication intermediates contain two forks, one or both of which is advancing toward the other, creating so-called double Y arcs on 2D gels. When two replication forks meet, molecules mimic a four-way junction (or X structure), before separating into the two daughter molecules. Some mtTERFs have been identified, such as mtTERF, mtTERF1 and mtTERF3.
Mitochondrial transcription:
Mitochondrial RNA polymerase is encoded by the nuclear genome.� Translated RNA polymerase protein is transported into mitochondria as described in transcellular protein traffic.�� RNAPs from S. cerevisiae and Xenopus consist of a large subunit and a small subunit B (TFBM).� The large subunit has polymerase activity.� The large subunits show similarity with few bacteriophage RNAPs (T7 phage).
The promoter sites for RNAps consists of a start site and upstream A rich 15bp sequences.� There are two such sequences one for one H strand and the other for L strand.� Each strand is transcribed in entirety.� The long pre-RNAs are processed into specific rRNAs, tRNAs and mRNAs.� The mitochondrial mRNA transcripts are translated in organelle itself.
Most of the mitochondrial transcripts are polycistronic.� They are transcribed from H strand and L strand.� The H strand transcript gets processed into2mt-rRNAs, 14mttRNAs, and mt mRNA that encode 12 proteins.� The L strand generates polycistronic mRNAs after processing 8-mttRNAs and one mRNA for protein. Most of the mt mRNAs are polycistronic, but one is bicistronic for they have overlapping coding sequences.
Mitochondrial transcription starts at two positions one called H1 and the other H2.� H1 transcribes Phe tRNA terminate at the end of the tRNA transcript.� The H2 transcript produces 12mt-tRNAs. Most of the mRNAs lack 5� and 3�UTRs and no introns, no cap but contain 45-50 ntds long Poly-A tail fifty residues after the stop codon.� The above description holds good for human Mit transcripts.
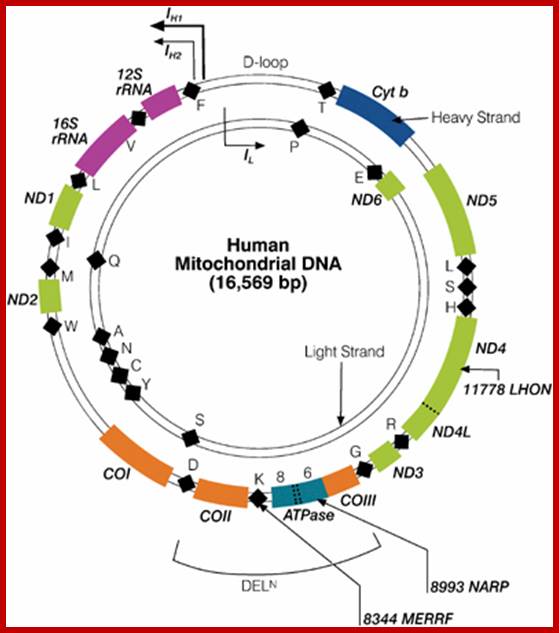
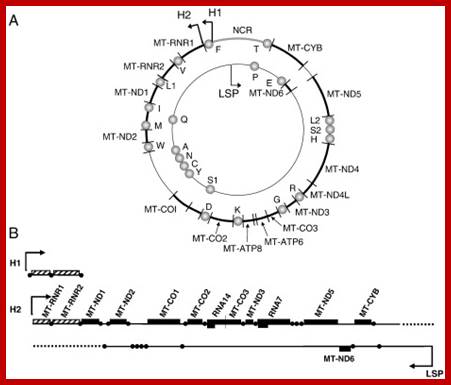
Top Figure: The human mitochondrial genome. This small (16 569 bp) genome is almost completely transcribed from both strands, initiating from one of two promoters (IH1, IH2) on the Heavy (H-) strand or the single promoter (IL) on the Light (L-) strand. All of these promoters and elements involved in replication initiation are found in the displacement (D-) loop, the only major non-coding region. The genome encodes 22 mt-tRNA (black diamonds), 2 mt-rRNA genes (fuscia) and 13 protein-coding genes (olive, ND1-6 encoding members of NADH:ubiquinone oxido-reductase; blue, Cytb encoding apocytochrome b of ubiquinol:cytochrome c oxido-reductase; orange, COI-III encoding members of cytochrome coxidase: aquamarine, ATPase 6, 8 encoding two members of Fo-F1 ATP synthase). Important mutations that are referred to in this article are indicated. Single letter code is given for each mt-tRNA-encoding gene. D S Kyriakouli, P Boesch, R W Taylor and R N Lightowlershttp://www.nature.com/
Bottom Figure: Map of human mitochondrial DNA and the RNA transcription units that are generated. (A) The human mitochondrial genome encodes 37 genes. Circles represent the 22 genes encoding mt-tRNAs depicted by their single letter code; the 13 genes encoding mt-mRNA and 2 mt-rRNA indicated using the nomenclature given on Mitomap (http://www.mitomap.org). The initiating/promoter positions of the polycistronic transcription units H1, H2 and LSP within the non-coding region (NCR) are indicated. (B) Transcription map of the human mitochondrial genome. The three polycistronic RNA units generated by transcription initiating at H1, H2 or produced from the Light Strand Promoter (LSP), are represented here in a linear fashion. The shortest of these commences from H1, with well-defined start and stop sites initiating upstream of MT-TF and terminating part way through MT-TL1[2]. The long transcription unit from H2 incorporates MTRNR1 and extends through the genome, terminating downstream of MT-TT. Transcription from LSP generates the second of the long unit that also extends throughout the genome, terminating downstream of MT-TP, however there is no consensus on the final termination site for either of the long polycistronic units [2], which are represented by dashed lines. RNA sequences are represented as follows; mt-rRNAs as hashed boxes, mt-mRNAs as black boxes and mt-tRNAs as circles. Nomenclature of each mt-mRNA or mt-rRNA corresponds to the gene nomenclature as described above, except for the two bicistronic RNA units RNA7 and RNA14 that encodes the proteins ND4L/ND4 and ATPase8/6, respectively; Richard J. Timperley, Mateusz Wydro, Robert N. Lightowlers, and Zofia M. Chrzanowska-Lightowlers; BBA. 2010.
Import of proteins into mitochondria directly with the binding of cytoplasmic polysomes on to mitochondrial outer membrane similar to ribosome binding to Endoplasmic Reticulum (ER);������� �(Kathleen S. Crowley and R. Mark Payne� (journal of Bilogical Chemistry 1998). �This transport is facilitated by GTP and �transit peptidise. Higher eukaryotic mitochondria contain ribosome receptors, possibly similar to the sec61p complex, which are active in initiating co-translational translocation. Mitochondria associated ER membrane MAM, which may comprise up to 20% of the mitochondrial outer membrane (BBA,2009,11, 13423-51).
The ABC of mitochondrial transcription:
Montreal Neurological Institute, McGill University, Montreal, Quebec H3A 2B4, Canada. eric@ericpc.mni.mcgill.ca
Two forms of a transcription factor, similar in sequence to an rRNA modifying enzyme, have been identified as the missing components of the transcription initiation machinery for human mitochondrial DNA (mtDNA). Transcription from mtDNA promoters can now be reconstituted in vitro with three recombinant proteins: mitochondrial transcription factors A and B and the core mitochondrial RNA polymerase.
Human mtDNA is a small circular genome encoding a handful of proteins that are essential components of the enzyme complexes of oxidative phosphorylation. Mutations in mtDNA cause a wide spectrum of multisystem human disorders with varying degrees of tissue specificity and severity. Transcription of mtDNA-encoded genes is initiated at two promoters on the so-called heavy and light strands of the molecule, and the resultant polycistronic transcripts are processed to produce mature rRNAs, tRNAs and mRNAs. A transcript generated from the light-strand promoter is also necessary to prime mtDNA replication, functionally coupling mitochondrial gene expression with genome maintenance.
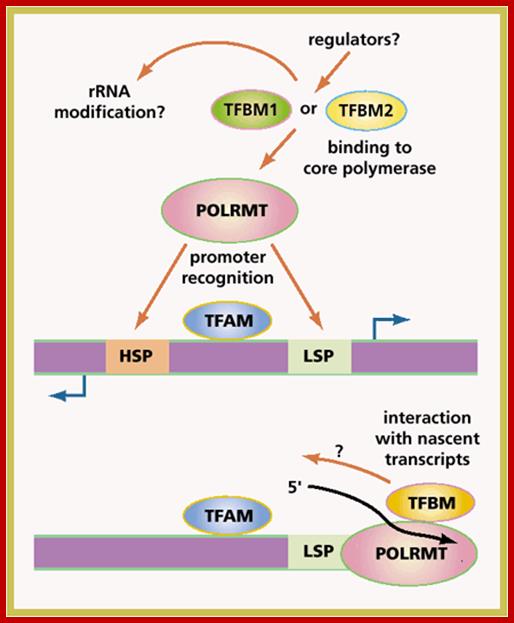
Time to transcribe: �Schematic drawing of
transcription initiation in human mtDNA showing potential roles of the TFBM
transcription factors. Transcription initiates from two promoters (LSP, HSP).
TFAM promotes bi-directional transcription by binding DNA upstream of the promoters
and unwinding the DNA template. TFB1M or 2 bind the core polymerase (POLRMT) in
a 1:1 stoichiometry. Either of the TFBM factors, in combination with TFAM and
POLRMT, is sufficient to initiate transcription from either promoter, although
TFB2M is an order of magnitude more effective. The factors that might regulate
this process are unknown. The TFBMs share strong sequence homology with
bacterial rRNA dimethylases; however, it is not known whether the TFBMs also
play a role in rRNA modification, or whether the RNA binding motif is important
in interacting with or subsequent processing of nascent transcripts. BOB CRIMI.
This comparatively simple transcription system requires an activator, mitochondrial transcription factor A (TFAM) and a core RNA polymerase; however, specific transcription initiation cannot be reconstituted from these components alone, suggesting that additional factors are necessary. Mitochondrial transcription factors like B1 and B2 activate transcription of human mtDNA. Maria Falkenberg and their colleagues reported on the identification of the missing factor and the development of an in vitro system that can be used to investigate the control of mitochondrial transcription and its coupling to mtDNA replication. Maria Falkenberg Martina Gaspari=, Anja Rantanen, Aleksandra Trifunovic2, Nils-G�ran Larsson1,2 & Claes M. Gustafsson.
Yeast/versus/man:
�In the yeast Saccharomyces
cerevisiae, specific mitochondrial transcription requires only two
components: a core mitochondrial RNA polymerase (Rpo41), which shares sequence
similarity with bacteriophage RNA polymerase, and a single transcription
factor, sc-mtTFB (Mtf1), which shares some sequence similarity with, but is
functionally distinct from, bacterial ![]() -factors.
The TFAM protein, which is essential for mammalian mtDNA transcription and
genome maintenance, is a member of the high mobility group (HMG)−box
family of DNA-binding proteins, and has no sequence similarity with the yeast
transcription factor. The yeast ortholog of TFAM, Abf2p, has an essential role
in mtDNA packaging, but does not seem to be important in mitochondrial
transcription. This difference in function between the yeast and human proteins
has been ascribed to a carboxy-terminal extension in mammalian proteins, which
is absent in Abf2p.
-factors.
The TFAM protein, which is essential for mammalian mtDNA transcription and
genome maintenance, is a member of the high mobility group (HMG)−box
family of DNA-binding proteins, and has no sequence similarity with the yeast
transcription factor. The yeast ortholog of TFAM, Abf2p, has an essential role
in mtDNA packaging, but does not seem to be important in mitochondrial
transcription. This difference in function between the yeast and human proteins
has been ascribed to a carboxy-terminal extension in mammalian proteins, which
is absent in Abf2p.
Attempting
to bridge the gap between the yeast and human studies, Bogenhagen and colleagues
carried out a series of studies on Xenopus laevis in which they
identified a biochemical activity distinct from TFAM that, in combination with
the core RNA polymerase, could support basal mitochondrial transcription.
Transcription in this system could be further activated by the ortholog of
human TFAM. These observations lent strong support to the notion that an
ortholog of Mtf1 might exist in vertebrates�.
The missing transcription factor:
�Using
PSI-BLAST and the Mtf1 ortholog from the fission yeast Schizosaccharomyces
pombe as the initial query sequence to search the database of the National
Center for Biotechnology Information, Falkenberg et al. identified two
human orthologs of Mtf1, which they called TFBM1 and TFBM2.
Using a similar strategy, Shadel
and colleagues also identified a transcription factor they called h-mtTFB,
which is identical to TFBM1.
To
show that these proteins are bona fide mitochondrial transcription
factors, Falkenberg et al. purified recombinant versions of both TFBM
proteins�TFAM and the mitochondrial RNA polymerase�and demonstrated specific
transcription from both heavy- and light-strand human mtDNA promoters in the
presence of either of the TFBM proteins, TFAM and the core polymerase.
Interestingly, TFBM2 was an order of magnitude more active than TFBM1 in the
transcription assay.
TFBMs: Bifunctional Proteins:
Remarkably, the TFBM
transcription factors share a striking sequence similarity with a family of RNA
modifying enzymes, the rRNA dimethyl transferases. While the studies of the
human TFBMs were in progress, the crystal structure of sc-mtTFB was solved, and
a search of the Protein Databank determined that it is part of a large family
of DNA/RNA methyltransferases, and not structurally related to bacterial ![]() -factors,
as had initially been thought. In particular, a C-terminal helix motif in
sc-mtTFB has so far only been observed in the rRNA methyltransferase, Erm.C'
from Bacillus subtilis.
-factors,
as had initially been thought. In particular, a C-terminal helix motif in
sc-mtTFB has so far only been observed in the rRNA methyltransferase, Erm.C'
from Bacillus subtilis.
These
observations raise the possibility that the TFBM family members are
bifunctional proteins that have roles both in mitochondrial transcription
initiation and RNA modification. The methyl donor of the rRNA dimethyl
transferases is S-adenosyl methionine (SAM). The crystal structure of
sc-mtTFB shows that many, but not all, of the amino-acid residues that bind SAM
have been conserved, and human TFBM1 has been shown to bind SAM in vitro11.
Specific methyl transferase activity has, however, not been demonstrated3,
and SAM is not required to initiate transcription in yeast. So, although the
jury is still out on this issue, it seems probable that a methyl transferase
protein has been co-opted for a different role.
This
may turn out to be a common theme in mitochondrial nucleic acid metabolism. The
small accessory subunit of the mitochondrial ![]() DNA
polymerase shares sequence similarity with another family of RNA-modifying
enzymes, the aminoacyl tRNA synthetases, and this is thought to relate to its
function in binding to a tRNA-like structure near the origin of heavy-strand
replication.
DNA
polymerase shares sequence similarity with another family of RNA-modifying
enzymes, the aminoacyl tRNA synthetases, and this is thought to relate to its
function in binding to a tRNA-like structure near the origin of heavy-strand
replication.
�
What,
then, is the function of TFBM, and why has an RNA modifying protein been
co-opted as a transcription factor? It seems probable that TFBM has a role not
in promoter recognition, but rather in activating the core RNA polymerase with
which it heterodimerizes. It is possible that the RNA binding function of the
methyl transferase protein domain is useful in this context, not in modifying
RNA per se, but in interacting with or stabilizing nascent transcripts.
The TFBM1 protein of mouse and human share significantly more sequence identity than do human TFBM1 and TFBM2, suggesting that the two proteins are the result of a gene duplication event early in mammalian evolution. Why do mammals have two factors that differ by about an order of magnitude in the strength of their transcriptional activation? One obvious possibility is that the two factors initiate transcription from specific promoters, but the available data do not support this suggestion. Another possibility is that regulation of mitochondrial gene expression in mammals requires the elaboration of different tissue-specific pathways, reflecting different metabolic demands. If, however, tissue-specific differences in mitochondrial transcription or replication are part of this equation, then this does not appear in the patterns of transcription of the two TFBM genes, which seem to be virtually identical by northern-blot analysis. It seems more likely that at least partially non-overlapping functions of both factors are important in all tissues in which they are expressed. This might involve the decision to process the nascent transcripts for DNA replication primers, or continue productive transcription if the light strand.
The�future�:
The basic components of the mitochondrial transcriptional machinery in mammals now seem to be in hands, but this only marks the end of the beginning. The really crucial questions concerning the control of mitochondrial transcription and DNA replication, and the regulation of mtDNA transcription copy number, remain to be answered. The mitochondrial genome has been pared down during evolution to the barest of essentials, and the investigation of its fundamental molecular biology continues to uncover surprises and evolutionary insights. It is to be hoped that some of the biological insights generated during the course of these studies will find application in the treatment of patients with mitochondrial-DNA-diseases.
Mitochondrial Diseases:
This list of few disorders associated with mitochondrial genes;
Cyclic vomiting syndrome,
Cytochrome c oxidase deficiency,
Lieber hereditary optic neuropathy,
Maternally inherited diabetes and deafness,
Mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes,
Myoclonic epilepsy with ragged-red fibers,
Neuropathy, ataxia, and retinitis pigmentosa,
Pearson marrow-pancreas syndrome,