Prokaryotic Gene Expression:
Introduction:
Genes are �the essence� of life.� Life without genes is no life at all.� The size, the kind of genetic materials and the number of genes vary from one organism to the other; �variety is the spice of life�.� Structurally genes, both in PK and EK, have some basic pattern in common, but in details and context they differ.�� At molecular level, the kind of products they generate, has led them to be grouped into protein coding genes and RNA coding genes; Genes are the the basic components Life.� There are different types and kinds and size of Genes; tRNA genes, rRNA genes, sRNA genes, Sc/Sn RNA genes, Anti sense RNA genes, siRNA/miRNA genes, the latter three are non-coding eukaryotic ncRNA genes.
�
The EM shows normal prokaryotic E. coli cells. http://www.dounenberg.ui; Maryland
Genes are the basic structural components of life.� Genes are Molecular structures of life- called DNA, in some RNA. Expression of genes means the information in the gene is transferred into their respective products, nucleotide-by-nucleotide, exactly and absolutely free from error.� Organisms cannot afford to make any errors.� In the case of structural genes, the information is transferred in one step i.e., from DNA to RNA, but in the case of structural (protein coding) genes, first the information is transferred from DNA to mRNA, tRNA and other forms of RNA by a process called transcription, or coding; then the coded information is decoded called Translation to produce functional components.�
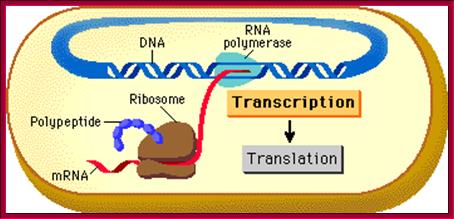
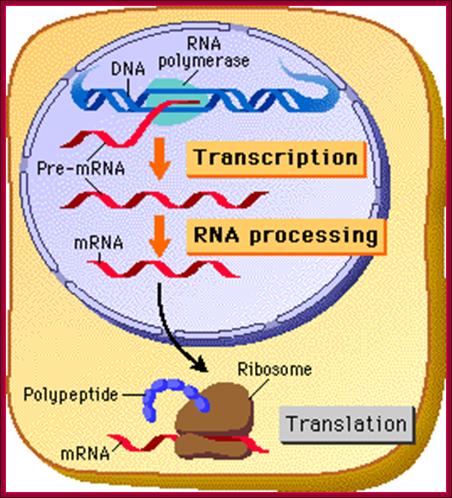
Fig-a; The upper fig. Cell not compartmentalized-simple representation of a prokaryotic cellular DNA going through replication, transcription and Translation. http://www.phschool.com/ Bottom Fig; Cell is compartmentalized where replication takes place..
In the case of genomic RNAs, if the RNA has positive sense, then it is directly translated or if it is of negative sense, then it generates positive strands, which is then translated and replicated into negative sense RNA.� Replication of TMV is possible only when it infects the Tobacco eukaryotic cell. �Exception to this rule is retroviral genomic RNA, after infection, which is first converted to ds cDNA and then inserted into the host genome (mostly immuno-cells).� Later if the infected cells are activated, the insert genetic DNA transcribes to produce a full-length RNA; that in turn generates proteins and encapsulation of RNA results in viral production.
The replication of tobacco mosaic virus (TMV) RNA involves synthesis of a negative-strand RNA using the genomic positive-strand RNA as a template, followed by the synthesis of positive-strand RNA on the negative-strand RNA templates using host cytoplasm. �Intermediates of replication isolated from infected cells include completely double-stranded RNA (replicative form) and partly double-stranded and partly single-stranded RNA (replicative intermediate), but it is not known whether these structures are double-stranded or largely single-stranded in vivo. This is possible only in the infected cells. The synthesis of negative strands ceases before that of positive strands, and positive and negative strands may be synthesized by two different polymerases. The genomic-length negative strand also serves as a template for the synthesis of sub genomic mRNAs for the virus replication. �Both the virus-encoded 126-kDa protein, which has amino-acid sequence motifs typical of methyltransferases and helicases and the 183-kDa protein, which has additional motifs characteristic of RNA-dependent RNA polymerases, that are required for efficient TMV RNA replication. Purified TMV-RNA polymerase also contains a host protein, serologically related to the RNA-binding subunit of translational initiation factor, eIF3. Study of Arabidopsis mutants defective in RNA replication indicates that at least two host proteins are needed for TMV RNA replication. The tomato resistance gene Tm-1 may also encode a mutant form of a host protein component of the TMV replicase. TMV replicase complexes are located on the endoplasmic reticulum in close association with the cytoskeleton in cytoplasmic bodies called Viroplasms, which mature to produce 'X bodies'. Viroplasms are sites of both RNA replication and protein synthesis, and may provide compartments in which the various stages of the virus multiplication cycle (protein synthesis, RNA replication, virus movement, encapsulation) are localized and coordinated. Membranes may also be important for the configuration of the replicase with respect to initiation of RNA synthesis, and synthesis and release of progeny single-stranded RNA.
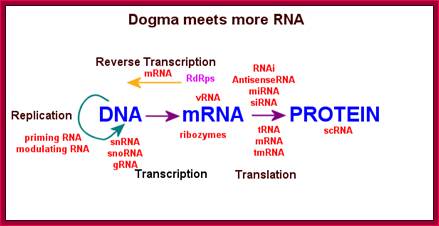
This diagram shows the reverse of Cricks� Central Dogma for the RNA can produce DNA by reverse transcription, even though it deviates from Cricks Central Dogma, it is an accepted as a fact-�The reverse of Central Dogma�. http://people.musc.edu/
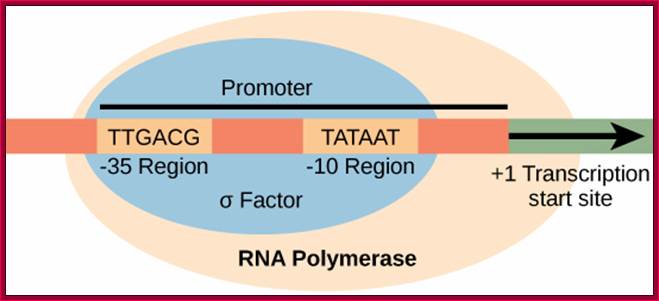
Prokaryotic Promoter Elements; https://www.boundless.com
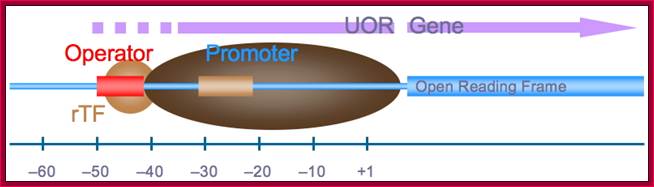
Archaeal Gene structure Pyrococcus furiosus; http://rscott.myweb.uga.edu/
��� ��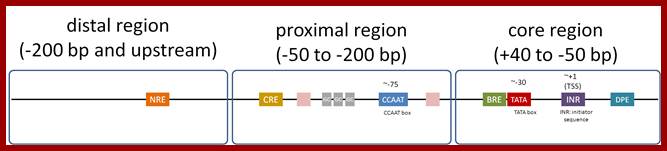
Eukaryotic Pol-II promoter elements; http://genocon.org/
Prokaryotic Genes:
Genes, in prokaryote genomes, are tightly packed; the genomes are generally small ranging from 10⁵ to 10^7, which means there is not much of scope for the spacers DNA or non-coding DNA in between the genes or within genes.� In most of the cases some of the functionally related genes or and structurally related genes are clustered together and they are under the control of one regulator region.� Each of these composite structures is called operons. Many of the operons are regulated by a common protein/RNA, called Regulon. The RNA products are either RNA or protein coding i.e., polycistronic; cistron means a segment that code for a single polypeptide. �That doesn�t mean there are no monocistronic genes, which, in fact, are in plenty.
�All genes, structurally consist of a 5� non coding region called 5�UTR with regulator and promoter sequences, a central coding region, which can be polycistronic or monocistronic and terminal non coding region 3�UTR, at which transcription terminates.�
The start site means it is the nucleotide at which the first rNTP is incorporated into RNA during transcription, which is normally an A and less frequently G, it is referred to as InR.
Any nucleotide found to the left of start site is termed as upstream and referred as �5, --10 so on.� Any nucleotide found to the right of the start is referred to as downstream and named as + 10, +20 and so on.��
Invariably, all housekeeping and other regulated genes, contain a sequence box at -10 regions, called Pribnow box named after the discoverer. It has a consensus sequence of TATAAT. At �35 to �30 there is another important sequence TTGAcat.� In the upstream of the TTGACAT, there can be other regulatory sequence components in the upstream, which depends on the kind of the gene; they can be activator/repressor or enhancer sequences (enhancer increases the efficiency of transcription).� Another sequence called Operator region exists around InR region.
5�--Regulatory�TTGACAT�TATAAT---InR-(codons)�UGA/UAA-3�
The -10, -35 and any other upstream regions, which are involved in initiation of transcription can be called promoter.� These sites attract the binding of Transcription factors and RNA pol; once they bind transcription is initiated.
The coding region, in general has bracketed on either side by a sequence called 5� untranslated region (UTR) and 3� untranslated region (UTR). �The Translated region of the polycistronic gene is partitioned into several cistrons, which are separated by intercistronic spacers.
The region from the start ntd to the first initiator codon is called leader sequence, which plays an important role in translation of mRNA.� The region from the last terminator codon to the end of the transcript is non-coding terminal region, often involved in regulation.
The coding region has sequences for specific amino acids, where the codons are arranged continuously with out any gap and this message ends in a nonsense codon, which is the end of coding region. �
The size of the coding region depends upon the kind of protein it generates. �In prokaryotic the coding region is not split into coding and non-coding segments; thus, the genes are not split. But the coding region may be made up of one or more individual coding segments, called monocistronic or polycistronic respectively. However, each of the cistrons are separated by spacers, which are used for reinitiation of translation.
�The first codon used in the reading frame is mostly AUG and infrequently GUG.� The last codon where the message is terminated or ends in what is called nonsense codons or terminator codons; they are UAG (Ochre), UAA (Amber) and UGA (Opal).
�
In the promoter region the TATAAT (-10) and TTGACA (-35) sequence play a significant role in recognizing and binding of RNA polymerase and its associated factors for initiation of transcription. The sigma factor guides RNA polymerase to the site-specific sequence and bind promoters to initiate transcription, but also there are specific proteins, which can prevent RNAP from initiating transcription. It is the sigma factor decides which gene to be transcribed and which is not.� There are different sigma factors depending upon situations they bind to RNAP and bind to the promoter elements and initiate transcription.
In many cases though RNAP binds to the promoter in correct sequence, but by itself cannot initiate transcription; in such cases it requires the activation by another factors, called activators.�
In some situations, the transcription is just at basal level, but certain proteins, in response to signals, bind to upper promoter regions and increase the efficiency of transcriptional rate by 100 to 200-fold.� They can be called enhancers.�
Activation in response to the stimuli may result in activating and increasing the rate of transcription of a gene.� In most of the prokaryotes, though one finds different kinds of genes, such as rRNA and tRNA genes and mRNA genes of different kinds, transcription process is executed by only one kind of multimeric RNA-polymerase enzyme.� It is the sigma factors that decide which gene should be expressed and which genes no to be expressed.
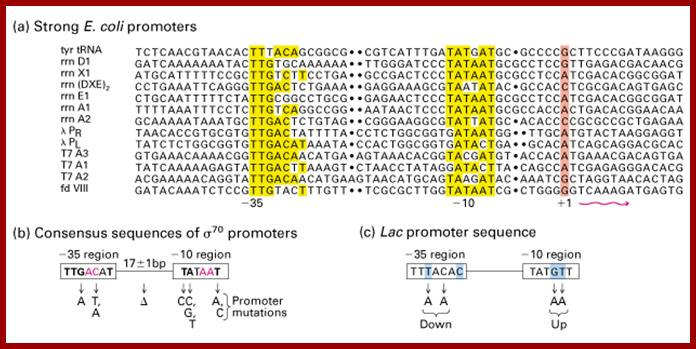
Prokaryotic upstream promoter elements- consensus sequences; http://archive.cnx.org
����� 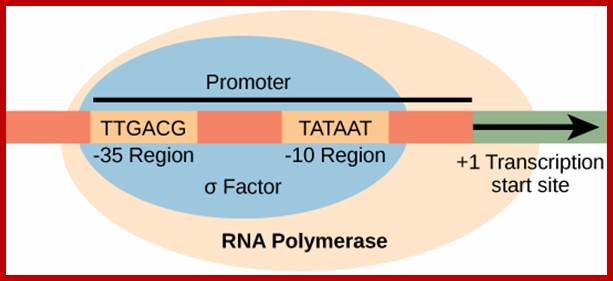
The σ subunit of prokaryotic RNA polymerase recognizes consensus sequences found in the promoter region upstream of the transcription start sight. The σ subunit dissociates from the polymerase after transcription has been initiated.The consensus sequence of promoter region (not universal); https://www.boundless.com
It is fascinating to know how the same enzyme identifies which gene to be transcribed and which gene not to be transcribed. �Under normal and favorable conditions, a large number of genes are expressed, but not all genes. �Constitutively expressed genes are called housekeeping genes, which run all metabolic activities, others have different functions. �Regulation of gene expression also depends upon the structural features of the Gene.
Under certain changed conditions or in response to stimulus or in developmental stages, certain genes are expressed. Whether it is a constitutively expressed or induced gene, the enzyme that transcribes is the same. �So, the enzyme must be regulated so as to recognize which gene is what.
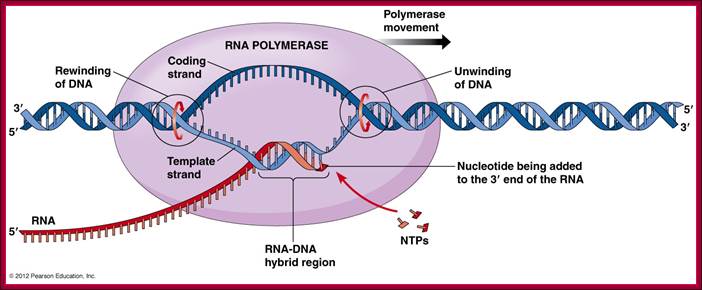
RNA pol recognizes specific promoter elements because of specific sigma factors bind to RNA pol at different situations. http://www.mun.ca/
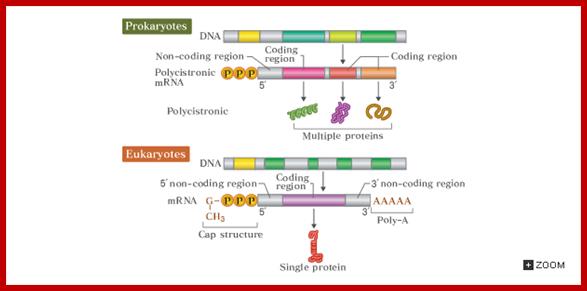
Comparison of promoter and mRNAs of prokaryote and eukaryotes; http://csls-text.c.u-tokyo.ac.jp
������������������������������������������ Incomplete- go to the next.