RNA polymerases:
Bacteria and Archaea belong to prokaryotic group, both have common ancestry and both have similar structural and functional features, but also differ from one another.� Archaea, in some structural and functional features resembles that of bacteria and eukaryotes and has its own. Bacterial and archaeal RNA polymerases versatile enzymes for they transcribe all types of genes, i.e., tRNA, rRNA, mRNA and other genes. Though, in terms of promoter elements, each of them differs in contents, yet it is the same enzyme with some additional factors responsible for transcription of all the genes.
E. coli Bacterial RNAPs:
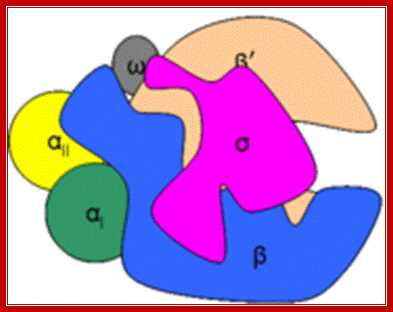
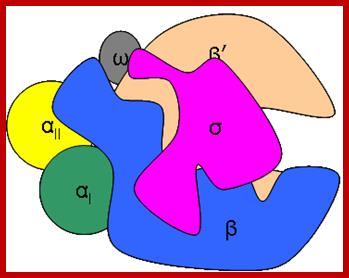
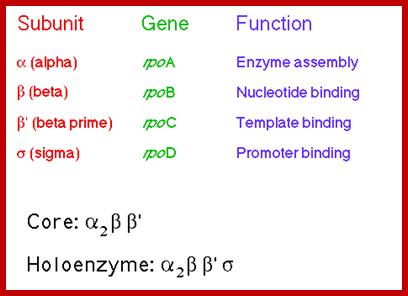
RNAP as Holozyme consists of core and accessory subunits having a molecular mass of 476 KD. �During transcription, the eubacterial RNAP holoenzyme, composed of a core enzyme containing the major subunits α2 (RpoA x 2), β (RpoB), and β� (RpoC), plays a central role, but requires a specific sigma-factor, selecting what gene to be transcribed. The core enzyme functions in RNA polymerization requires the σ subunit for specific transcriptional initiation at start site in the promoter. There are two alpha subunits, one beta, one beta� and one omega subunit, together they form the core structure.� Another factor called sigma binds and makes it as holozyme and guides the RNAP to its promoter elements.� Different kinds of sigma factors take the enzyme to different but specific gene promoters.� Without sigma factors the bacterial RNAP is an orphan and an aimless wanderer.� Though it can associate with DNA, it gets dissociated soon without its sigma factor.
Crystal structure of the Holozyme, up to 25� resolution has been obtained and it looks like a puckered palm, with fingers and the thumb on opposite sides can close or open. The groove or the channel found is about 25�, good enough for holding a ds DNA. There is also a groove running length wise, which is about 55� long, but shows branching in the region at which newly formed 5� end of the RNA threads through it?� The over all dimension is about (100 x 100 x160)-� (or 90x95x150 �).� The Holozyme exhibits three conformational forms called RP-O (open at higher TM.), RP-C1 (closed at low TM.) and RP-C2 state (closed at 20 0 C).
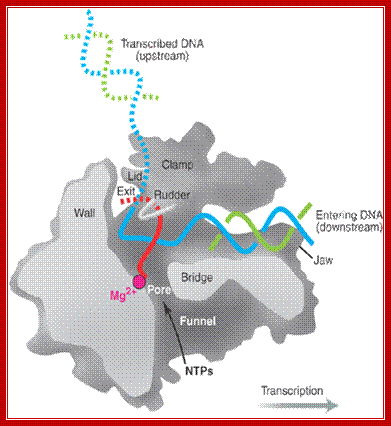
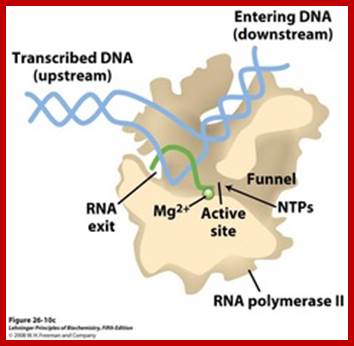
Positions of each subunits are labeled- the entry of DNA and the mRNA exit site; The structure of a bacterial RNAP: Two depictions of the three-dimensional structure of a bacterial RNA polymerase, with the DNA and RNA modeled in.
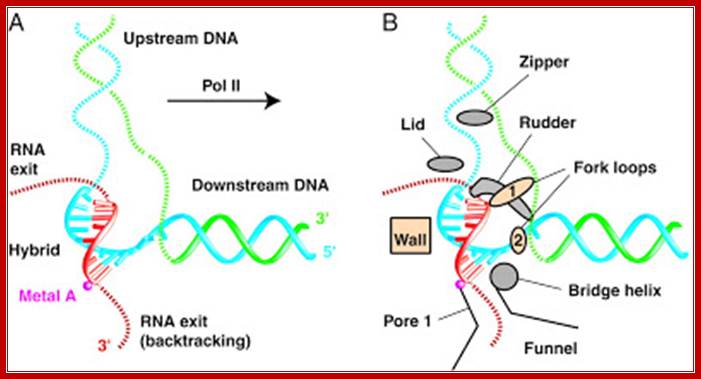
Top Fig: The "bridge" part of the enzyme is required for the translocation step. This is the step following addition of a ribonucleotide when the enzyme has to shift by one nucleotide (base pair) to the right. The new 3′ end of RNA has to be re-positioned at the active site during this shift. At the same time, one base pair of DNA is unwound by the "fork" region of the enzyme and one base pair is reformed at the back end of the bubble by the "zipper" region. For simplicity only the polypeptide backbone of the rudder is shown in the right-hand figure, and the DNA exiting from the polymerase has been omitted. Because the RNA polymerase is depicted in the elongation mode, the σ factor is absent. (Courtesy of Seth Darst); http://xray.bmc.uu.se/;http://sandwalk.blogspot.com; http://rationalwiki.org/wiki/
����������������������������
Second fig from the top; The enzyme has a complex subunit structure with two configurations designated as core group consisting σ2�ββ & ῲ subunits. The core group gets associated with sigma factor; this is the functional enzyme called Holozyme. The holoenzyme is involved in the synthesis of most of the cellular RNA. http://www.conservapedia.com/
Last bottom fig; This RNA polymerase is formed from four different subunits, indicated by different colors (right). The DNA strand used as a template is red, and the non-template strand is yellow. The rudder wedges apart the DNA-RNA hybrid as the polymerase moves. http://saypostna.host.sk/
For simplicity only the polypeptide backbone of the rudder is shown in the right-hand figure, and the DNA exiting from the polymerase has been omitted. Because the RNA polymerase is depicted in the elongation mode, the σ factor is absent. (Courtesy of Seth Darst); http://www.conservapedia.com/
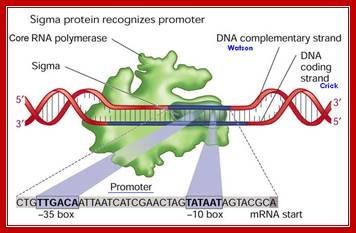
The Sigma Factor is involved in the Initiation step of Transcription in Prokaryotic life. It is a polypeptide subunit of the RNA polymerase. It serves to recognize specific binding sites on DNA molecules for initiation of RNA transcription.� The sigma factor is the subunit of the RNA polymerase complex that recognizes the specific promoter sequence of DNA that the RNA polymerase complex should bind to. This will be the initial site for DNA transcription, or where it initiates.
RNA Pol requires 2 copies of α subunits to work for synergistic transcription activation at bacterial promoters;
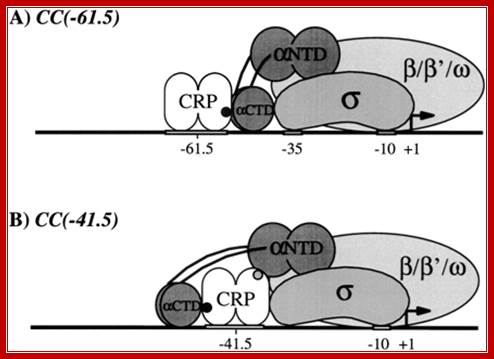
Transcription activation at class I and class II cyclic AMP receptor protein (CRP)-dependent promoters: published models.
(A) Ternary complex of CRP, Escherichia coliRNA polymerase holoenzyme (RNAP), and a class I CRP-dependent promoter [e.g., CC(-61.5); adapted from Blatter et al. 1994;Zhou et al. 1994a,b; Busby and Ebright 1999]. Transcription activation involves direct protein�protein interaction (black filled circle) between AR1 of the downstream subunit of CRP and the 287 determinants of α C-terminal domain (αCTD).
(B) Ternary complex of CRP, RNAP, and a class II CRP-dependent promoter [e.g.,CC(-41.5); adapted from Niu et al. 1996; Busby and Ebright 1999]. Transcription activation involves both (1) direct protein�protein interaction (black filled circle) between AR1 of the upstream subunit of CRP and the 287 determinants of αCTD, and (2) direct protein�protein interaction (grey filled circle) between AR2 in the downstream subunit of CRP and α subunit N-terminal domain (αNTD). http://genesdev.cshlp.org/
Interactions between RNA pol, CAP-CRP and CRP-dependent Promoters:
Interactions between RNA pol, CRP Cyclic AMP receptor protein (CRP) is an important transcriptional regulator in many Gram-negative bacteria. This protein controls the expression of many different genes in the cell in response to the cellular level of cAMP. Interaction of cAMP-CRP with RNA polymerase at the lac promoter and at the promoters of several other genes typically occurs at the C-terminal domain of the RNA polymerase alpha subunit (Text Fig. 10.9, class I promoters). With some other promoters affected by cAMP, CRP can also engage the N-terminal domain of the alpha subunit (Fig. 1, class II promoters). These protein-protein interactions stimulate RNA polymerase bound at the promoter to initiate transcription.
Three classes of promoters require CRP activation, with each class differing in where CRP binds. CRP binds class I, II, and III promoters at -60 bp, -41 bp, and -90 bp, respectively, from the start of transcription. The differences in binding location reflect the amount of �encouragement� a particular RNA polymerase�-promoter complex must receive from CRP to change from a closed promoter complex to the open promoter complex necessary for transcription. (Class I promoters (lacP) were discussed in Section 10. In Class II promoters, however, where CRP binding overlaps the �35 promoter region (Fig. 1), CRP also touches the N terminus of the alpha subunit, which, along with contact of CRP at the C terminus, is needed to stimulate transcription. CRP essentially strengthens a weak �35 site. Transcription of class III promoters requires activator proteins in addition to CRP; http://www.wwnorton.com/
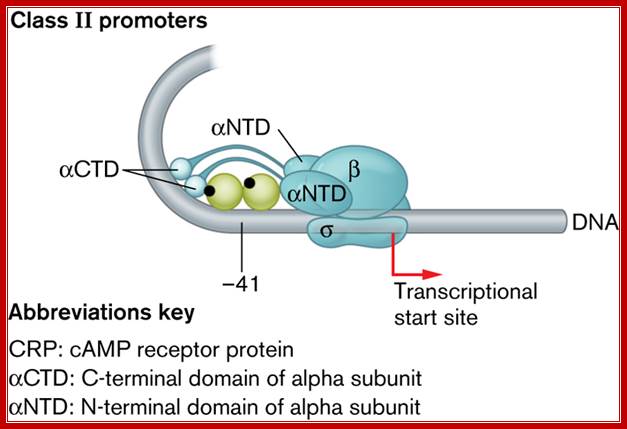
The CRP-binding site on class II promoters at -41 bp, overlaps the RNA polymerase�binding site, allowing CRP to interact with the C-terminal and N-terminal domains of the alpha subunit. CRP binds the third class of promoter (not shown) at -90 bp ;http://www.wwnorton.com/
�
Bacterial RNAP subunits:
The β� subunit of bacterial RNAP, which is largest subunit and it plays a role in catalysis, corresponds to the eukaryotic RNAP II subunit RPB1. The bacterial RNAP, β subunit, which is the second largest subunit and also takes part in catalysis and it corresponds to the eukaryotic RNAP II subunit RPB2. The αI and αII subunits of bacterial RNAP, which have identical sequences but different locations, with αI interacting with b and αII interacts with β�, correspond to the RNAP II subunits RPB3 and RPB11, respectively, and plays a role in RNAP assembly and transcriptional regulation (Minakhin 2000). Finally, the RNAP subunit �ω� corresponds to the RPB6 subunit of RNAP II. Table 1
|
Bacterial RNAP |
Eukaryotic RNAP II equivalents |
|
β� |
RPB1 |
|
β |
RPB2 |
|
αI |
RPB3 |
|
αII |
RPB11 |
|
ω |
RPB6 + |
Table: Conserved subunits of bacterial RNAP and eukaryotic RNAP II
Bacterial RNAP is approximately 150 � x 115 � size. The X-ray crystal structure of Thermus aquaticus bacterium core RNAP revealed that the enzyme has a shape similar to a crab claw, with the two arms of the claw defining a wide internal cleft between them (Zhang 1999). This cleft is about 27� in diameters, which can accommodate a double stranded nucleic acid molecule and contains the active center Mg2+. The b� subunit makes up one arm of the claw and a portion of the base of the cleft, while the b subunit makes up the other arm and a portion of the base of the cleft (Ebright 2000).
RNA polymerase resembles a �crab claw� with its active site at the base of the claw;
�����������������������������������������������������������
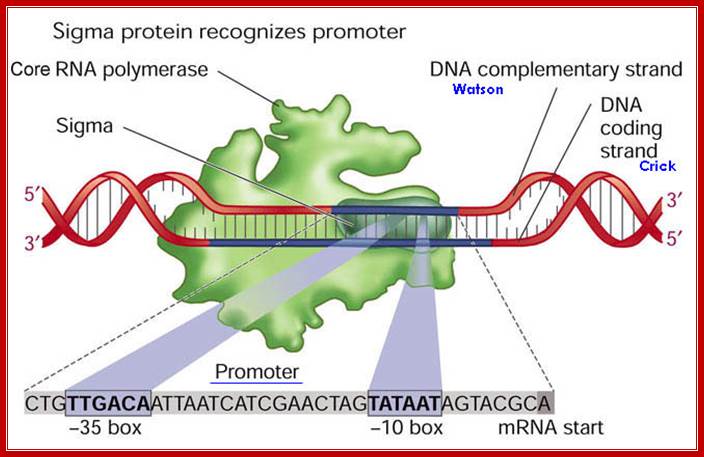
The sigma factor is a sequence-specific recognition protein that enables the enzyme complex to recognize and bind to the promoter, which lies just upstream of the start site.
�����������������������
A simplified transcription initiation pathway:
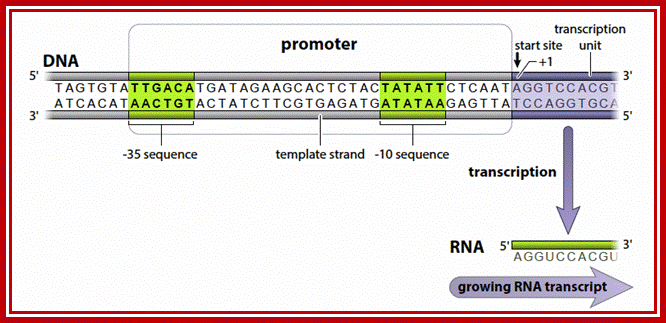
The Bacterial promoter contains -35 and -10 specific sequences, that recognize by the sigma factors.
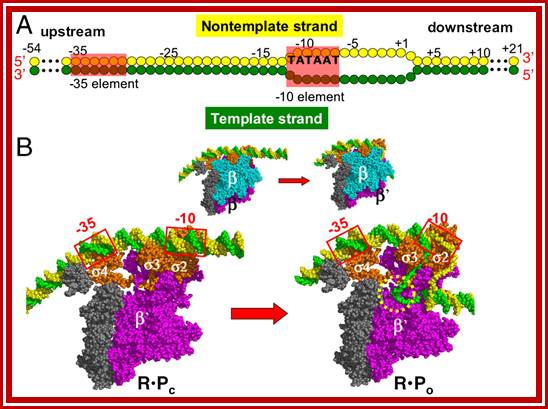
Structural models for the promoter DNA and RNAP. (A) Schematics of the base pairing between the template (green) and nontemplate (yellow) strands of the promoter. Nucleotide positions are numbered relative to the transcription start site, +1. DNA segments that interact with RNAP, -35 and -10 elements, are shaded red. (B) Structural models on top correspond to R�Pc (left) and R�Po (right) and are color-coded as β cyan, β′-magenta, σ-orange, αI, αII, and ω-gray, DNA nontemplate strand-green, and template strand-yellow. The β subunit is removed to show the interactions of the promoter with the σ subunits (bottom left) and the transcription bubble structure on the bottom right, while on the top row it is shown in full. https://www.pnas.org/
![]()
The R is RNAP, P is the promoter of DNA (Fig. 1A), and R�Pc and R�Po are the closed and open complexes. https://www.pnas.org/.
����������������������������������������������� ����������� 
Crystalline structure of the RNA Pol
The simulations of the kinetics of the R�Pc → R�Po transition show that the transcription bubble forms in three distinct steps. In step I,n step I, a region near the -10 element on the promoter DNA melts. In step II, the promoter DNA scrunches into the RNAP active channel, forming the transcription bubble and in step III, the downstream DNA bends. DNA bending is a result of downstream DNA relaxation, and occurs only after unwinding of the dsDNA. Widening of the channel needed to accommodate the dsDNA entry into the active channel requires transient expansion of key structural elements in the β subunit of RNAP, which implies that the internal enzyme dynamics plays an important role in the R�Pc → R�Po transition. https://www.pnas.org/
����������������������������������������������� ��������� 
D structural Model of RNA pol
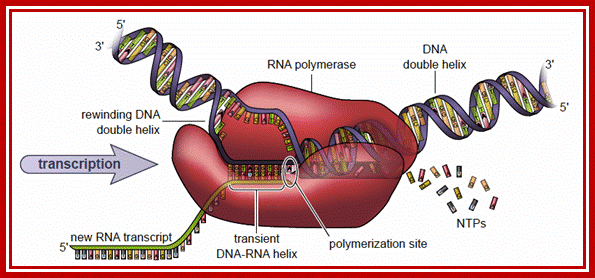
���������������������������� Transcription is catalyzed by RNA polymerase.
A model shown is RNA polymerase in the process of transcribing, that is, after the sigma factor (not shown) has directed the enzyme to the promoter. In the case of the −35 sequence, side chains of amino acids in the sigma subunit contact the edges of base pairs in the major groove. Interestingly, and in contrast to most DNA binding proteins, key contacts between the sigma factor and the −10 element are made as the two strands separate during the binding of RNA polymerase to the DNA to create the transcription bubble; the sigma factor and the −10 element are made as the two strands separate during the binding of RNA polymerase to the DNA to create the transcription bubble (Figure ).. Subsequently, a transition to the open R�Po complex occurs, characterized by separation of the promoter DNA strands in an approximately 12 base-pair regions to form the transcription bubble. https://www.labxchange.org/
The simulations of the kinetics of the R�Pc → R�Po transition show that the transcription bubble forms in three distinct steps. In step I,n step I, a region near the -10 element on the promoter DNA melts. In step II, the promoter DNA scrunches into the RNAP active channel, forming the transcription bubble and in step III, the downstream DNA bends. DNA bending is a result of downstream DNA relaxation, and occurs only after unwinding of the dsDNA. Widening of the channel needed to accommodate the dsDNA entry into the active channel requires transient expansion of key structural elements in the β subunit of RNAP, which implies that the internal enzyme dynamics plays an important role in the R�Pc → R�Po transition. https://www.pnas.org/
Initiation and fork with Repressed domain/ active domain.
Like bacterial RNAP, Euk-RNAP II has a shape similar to a crab�s claw, with the two arms of the claw forming a deep cleft between them (Cramer 2000). The fig above shows, the relative positions of the conserved subunits in bacterial RNAP and eukaryotic RNAP II are the same (Ebright 2000). Hence, RPB1 forms one arm of the claw and a portion of the base of the cleft, while RPB2 forms the other arm of the claw and a portion of the base of the cleft.
Activators and inhibitors: E. coli RNA polymerase has absolute requirements for divalent metal ions such as Mg++, Mn++, or Co++. Rifampicin (rifampicin), streptovaricins, streptolydigin, and sulfydryl reagents are among the inhibitors of E. coli RNA polymerase. Beta-subunit (rpoB) gene mutations were cross-screened against a number of other RNA polymerase inhibitors to correlate susceptibility with specific rpoB genotypes. The rpoB mutants were cross-resistant to streptolydigin and sorangicin A. In contrast, thiolutin, holomycin, corallopyronin A, and ripostatin A retained activity against the rpoB mutants. The second group of inhibitors may be of interest for drug development candidates.
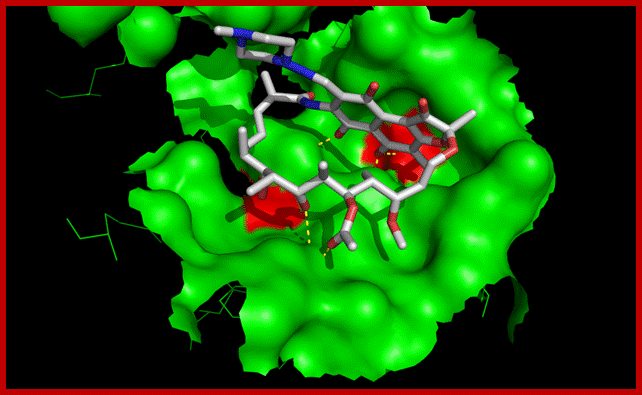
Binding of rifampicin in the active site of RNA polymerase. Mutation of amino acids shown in red are involved in resistance to the antibiotic. By binding next to RNA polymerase's active center (pink), the potent class of antibiotics called Rifamycin (red and yellow) prevents bacterial RNA from elongating. (Credit: Image courtesy of Rockefeller University; Rifamycin binding site Beta subunit is located near the center involving four of the subunits and the activating center. The inhibitor binds in a pocket of the RNAP β subunit deep within the DNA/RNA channel, but more than 12 � away from the active site. Resistance to Rifamycin arises due to mutations in Rif binding sites on RNApol, subunit Beta; http://en.wikipedia.org/
Concentration of RNAPs in the cell is approximately 7000 units per cell (1500 to 11,400) and that of sigma factors, is about 2300 and odd per cell.� At any given time more than 5000- 6000 enzymes are engaged in transcriptional activity. Some RNAPs are bound nonspecifically, some actively engaged in transcription and some in pausing state of transcription. The rate of synthesis under optimal conditions is about 25 to 75 ntds or more per second; the error rate is one in 10,00,000 ntds(?) incorporation.�
The enzyme perse, not any of its subunits, show explicit 3��>5� exonuclease activity, yet they have intrinsic ability to remove mismatched ntds from its 3� end by hydrolytic cleavage, which is a proof reading function.� They have high processivity and high fidelity. Another significant feature of PK is transcription and translation events are coupled.
RNAP components:
The Table below showing the subunits of RNAP
Assembly-2a�a2�>a2b�a2bb�->a2bb�sigma
|
Subunits |
Gene |
Mol.wt (kd) |
Numbers |
Function |
|
b� |
Rpo C 89.5 mpu |
155-160 1402 a.a |
1 |
DNA binding |
|
b |
Rpo B 89.5 mpu |
151-155, 1342 a.a |
1 |
Catalytic/nucleotidyl transferase |
|
a |
72 mpu |
36-40 329 a.a |
2 |
Assembly of the core and activation |
|
W (omega) |
Rpo Z 82 Mpu |
11 |
1 |
Regulation |
|
Sigma |
Rpo D 66.5 mpu |
70 |
1 |
Identity of TATAAT and� --35 region |
This is a model of bacterial RNAP shows specific subunit
positions and position of sigma factor. http://rationalwiki.org/wiki/Gene_expression
Bacterial_RNA_polymerase.png
(364 � 287 pixels, file size: 9 kilobytes, MIME type: image/png). https://www.studyblue.com
� Canada � York University
b Subunit:�
The protein, is coded by rpo-B gene, has a M.w. of 150-151 kDa.� It has two catalytic sites; one for the first nucleotide incorporation (at 5�end) and the second is for the addition of the next nucleotide for phosphodiester bond formation (called elongation site).�
The b subunit has Zn2^+ binding site.�� Rifamycin binds to chain initiation site.� Streptolydigin binds to translocation or elongation site.�
In the central region of the protein, a 200 a.a domain is involved in catalytic activity, but it is also known, that it also participates in transcriptional termination of both Rho dependent and Rho-independent modes.�
�
It�s COO^- terminal region is essential for the binding of sigma factor.�
�Its main function seems to be involved in chain initiation and catalysis of polynucleotide formation in template dependent manner.
b� Subunit:�
�
Its Mol.wt is155-160 KD, coded for by rpo-C gene. This is perhaps one of the largest proteins produced by the bacteria.� In general it has more basic amino acids, which mean it is positively charged.�
�It has two Zinc binding sites, and also contains DNA binding stretch and provides a 25 � groove for it.�� The strong affinity for DNA stems from its COO- terminal, which has a zinc finger motif, this region provides strong DNA binding forces.
�Beta and beta� together bind to both strands of DNA, but catalytic activity is located in b subunit; however, it requiresb� subunit for holding the non-transcriptional strand apart from the transcriptional template strand for its activity.� The subunits when assembled look like crabs� claws. The active center is located at the base of the cleft.� b subunit makes contact with non-template strand at �8, -9, -36 and �37 and binds to template strand at �32.� Together they also bind to �6 to +1, and from +2 to +11.
a Subunits:�
�Each RNAP consists of 2a subunits, whose Mol.wt is 34-36 KD each, they are coded for by rpo-A genes.� The C-terminal tail extends to -65 and can interact with activator proteins such as CAP/CRP
Omega subunit:
It is required for the complete assembly of all subunits into a complex.� Omega acts as chaperones.
In 3-D structure it exhibits little longish shape with globular region portion in the middle.� The proteins have more acidic amino acids. Alpha subunits are required for the assembly of the core complex in a sequence.�
These subunits are also required for the assembly of the enzyme as a complex on to the promoter, for these subunits recognize -35 sequences.� Activator proteins, that bind beyond upstream of the �35, interact with CTD domain (carboxyl end) of a units for the activation of the Holozyme.� Mutation or deletion of a subunits fails to activate the RNAP Holozyme.� The N-terminal and C-terminal regions are distinguished by their shape, where the N-terminal region called CDT tail contacts �35 region and beyond; and the NTD end, i.e. the N-terminal end is complexed with the other subunits of the RNAP.� The two ends ate connected with a hinge like connector.
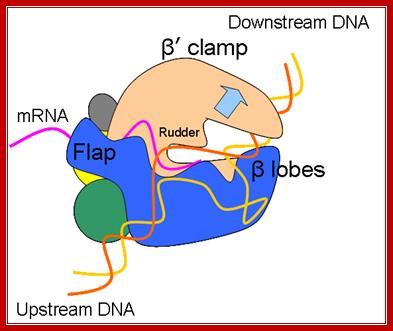
Bacterial RNAP with a flap of beta subunit and rudder of b� subunit; http://rationalwiki.org/wiki/Gene_expression
A simple view of RNAP bound to DNA and transcribing, showing the entry and exit of DNA and exit of RNA; http://xray.bmc.uu.se/
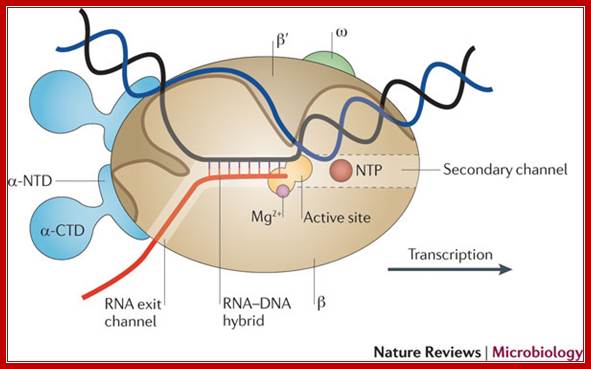
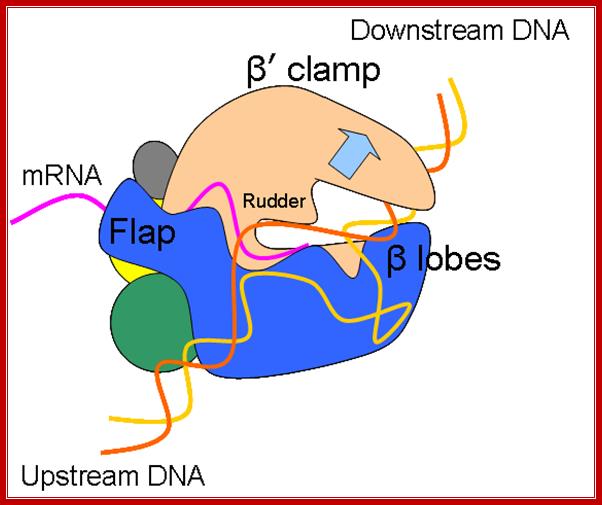
Core RNA polymerase (RNAP) (in bacteria, a complex composed of an α-dimer, a β-subunit, a β′-subunit and an ω-subunit) is bound to the DNA duplex composed of the template strand (black) and the non-template strand (blue), and the nascent RNA (red). The α-amino-terminal domains (α-NTDs) serve as a scaffold for complex assembly; the α-carboxy-terminal domains (α-CTDs) and ω-subunit play regulatory roles during initiation. The β- and β′-subunits jointly form the active site and make all the contacts to the nucleic acids. The substrate nucleoside 5′-triphosphate (NTP) (bound to a second Mg2+ ion) is thought to enter through the secondary channel. 12�14 bp of the DNA are melted in the transcription bubble. The non-template DNA strand is exposed on the surface, where it may interact with regulatory proteins. The nascent RNA is annealed to the template strand to form 8�9 bp of the RNA�DNA hybrid, which is the key determinant of elongation complex stability7, 98, 99. The upstream RNA is extruded through the RNA exit channel formed between the β-flap and β′-clamp; Thomas J. Santangelo & Irina Artsimovitch, Nature Reviews Microbiology.
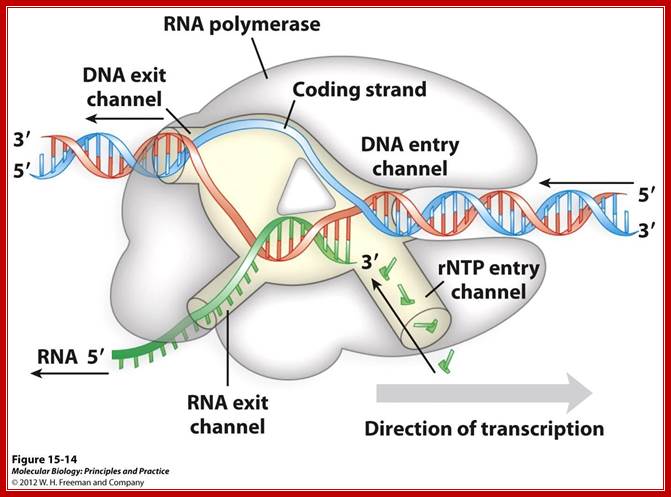
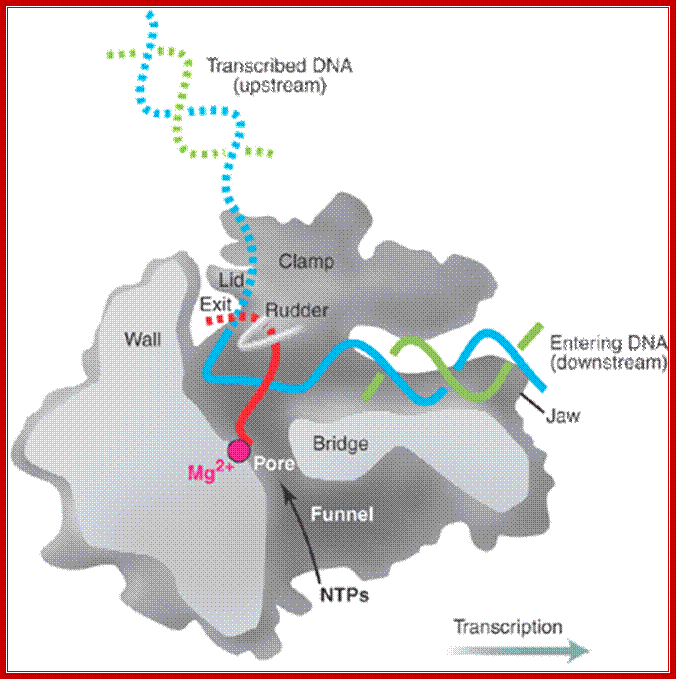
Distinct channels in RNA polymerase allow DNA to enter as dsDNA and peel apart within polymerase, so that 8bp form between template strand and growing RNA transcript -2 other channels provide entry for rNTPs and and an exit for the transcript; http://www.studyblue.com/notes/note
Sigma factors:
These proteins belong to a family of polypeptides and they have very interesting features and play important role in specific gene expression.
�They associate with RNAP subunits and they are also responsible for identifying the correct promoter and positioning the RNAP on DNA in proper context for initiating transcription. Otherwise, the RNAP without sig cannot function as an enzyme; it is like a driver without a rudder/conductor.�
Sigma factors in E. coli:
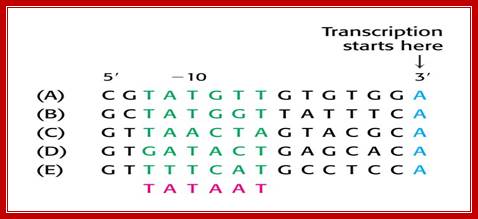
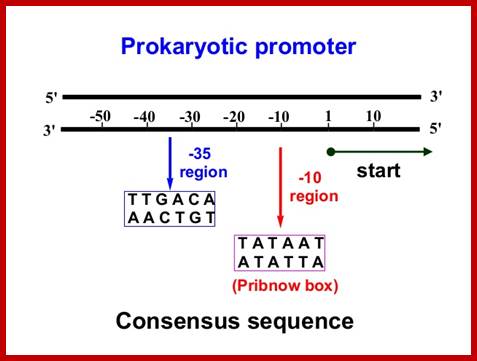
- σ70 (RpoD) - σA - the "housekeeping" sigma factor or also called as primary sigma factor, transcribes most genes in growing cells. Genes recognized by σ70 all contain similar promoter consensus sequences consisting of two parts; the consensus promoter sequences are characteristically centered at (�10 and �35) nucleotides before the start of transcription.
- σ19 (FecI) - the ferric citrate sigma factor, regulates the fec gene for iron transport
- σ24 (RpoE) - the extra cytoplasmic/extreme heat stress sigma factor
- σ28 (RpoF) - the flagellar sigma factor
- σ32 (RpoH) - the heat shock sigma factor, it is turned on when exposed to heat
- σ38 (RpoS) - the starvation/stationary phase sigma factor
- σ54 (RpoN) - the nitrogen-limitation sigma factor
RNAP by itself, can bind to DNA free from any proteins, it moves along the DNA by diffusion, and dissociate without performing transcription.� It can come across different genes and their promoters, but the enzyme fails to recognize which is which, even if it recognizes the promoter, it has no bias for a sequence, so it cannot initiate transcription.�
�In response to environment cells produce several kinds of sigma factors.� When conditions are favorable, for the growth, sig-70 is produced in plenty, it is this that drives most of the house keeping genes in active state and also few specialized genes.
�
When conditions change, ex. Nitrogen starvation, or increase in the temperature, changes the situation, where cell survival depends upon its ability to adapt to hostile conditions.� In such situations, signal specific sigma displaces sigma 70 and binds to specific promoters of genes and transcribe.� When conditions revert back to normalcy, sig-70 displaces them.�
�
Sigma is the key factor, that it not only recognizes promoter of the gene for transcriptional initiation, but also differentiate the genes to be transcribed.� Different bacterial systems produce their own specific sig factors. In many a situation, different types of sigma factors are produced in cascade manner, to the step wise or sequence wise developmental stages, ex. Sporulation in Bacillus subtilis.
Anti-Sigma factors:� Anti sigma factors bind to RNAP and inhibit its activity.� Such factors are found in E. coli, B. subtilis and T4 phages.� Anti-sigma factor 70 such as Rsd in E. coli is present in the stationary phase and blocks the activity of sigma factor 70 which in essence initiate gene transcription. This allows the sigma S factor to associate with RNA polymerase and direct the expression of the stationary genes.
Sigma 70:
The most common and ubiquitously produced sig-factor in E.coli is sig- 70; and in B. subtilis it is sig-43.�
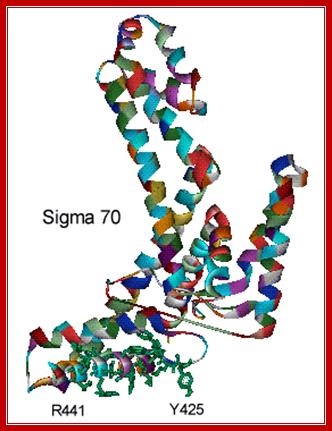
�There are at least 17 different types of sig-factors, which have been identified and their functions been studied.� In prokaryotes, binding of the polymerase's σ factor to promoter can catalyze unwinding of the DNA double helix. The most important σ factor is Sigma 70, whose structure has been determined by x-ray crystallography.
Sigma factor protein; http://www.web-books.com
/Sigma factor protein domains; http://genes.atspace.org/
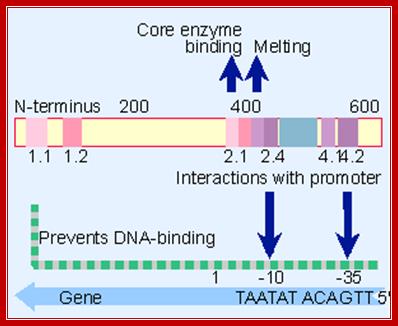
A map of E. coli sigma factor, conserved regions 2.1 and 2.2 contact core polymerase. 2.3 required for DNA melting and 2.4 and 4.2 contact -10 and -35 sequences.� But the N terminal region prevents 2.4 and 4.2 from binding to DNA in the absence of core enzyme.
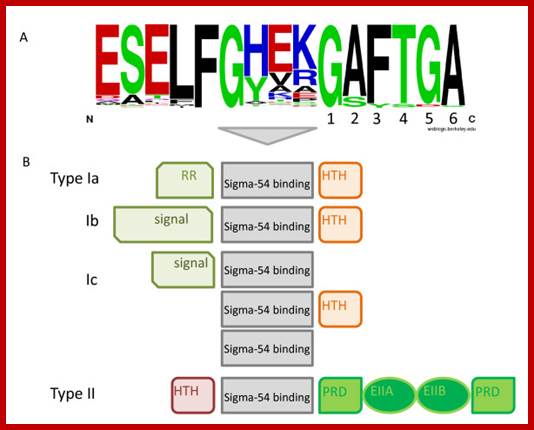
Sigma protein� specific amino acids contact specific DNA sequences; Previous comparative analyses of the Sigma-54 associated EBP54s made it clear that the Sigma-54 activators connect to a wide spectrum of input signals. Nevertheless, some generalizations can be made. All EBP54s possess a central activator domain, which is responsible for the interaction with Sigma-54 and provides the ATPase activity that is required to initiate transcription. In addition, most enhancer-binding proteins have one to several signal binding/recognition domains and a DNA-binding domain, although some EBP54s lack either the former or the latterTop fig. https://www.researchgate.net

Different Sigma factors� DNA binding sequences; www.slideplayer.com/slide; http://flylib.com/
Different kinds of sigma factors, in spite of structural and functional difference, do show some homology.�
The sig-70 is longish extended protein with four domains, called, from the N-terminal, as 1, 2, 3 and 4.� Among the sigma family members domains 2 and 4 are very well conserved and they are 75� apart.� It positions within the holozyme in such a way it recognizes different elements of promoter region.� The N terminal region is required for associating with beta subunit of RNAP.� The 2nd (2.4) domain has amino acid sequence, which is aromatic, does not generate any helix turn helix structures, yet it recognizes -10 TATAAT sequence and bind.� The C-terminal 4 th (4.2) domain recognizes �35 region in sequence specific manner.� This is also the site at which the N-end of the alpha subunits locates. In this process they intercalate into the major groove of the helix, destabilize it and open-up the helix, which means the DNA melts in that region.�
The C- terminal 4th domain has helix turn helix motifs and recognizes -35 sequences and bind.� The � 35 sequences vary from regulated promoters to those of housekeeping gene promoters.� The key components, in recognizing which gene to be identified, and the ability to distinguish one gene from the other, lie in the 4th domain of the sigma factor and the �35 promoter sequence.
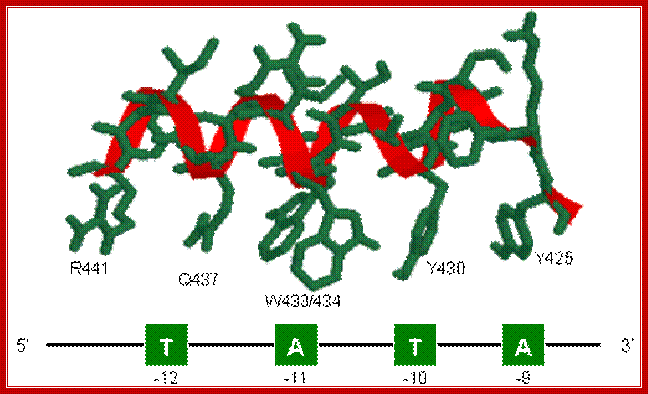
The structure of Sigma 70 and its DNA binding sites; (a) Structure of Sigma 70, residues 114 to 448. PDB ID = 1SIG. (b) A model for the binding between Sigma 70 and the promoter, based on biochemical studies. Residues Y425, Y430, W433 and W434 are directly involved in the unwinding (melting) of the double helix. http://www.web-books.com/
The recognition of sigma factor motif and the evaluation of relative promoter strength:
In the case of sigma 70 factors, the motifs are the −35 and −10 hexamers. Enclosing a spacer of length 15�19 bp.
Given a promoter sequence, the -10 and -35 hexamers are located by the total MSS of the two hexamers calculated by the position frequency matrices of the sigma factor binding sites, which are derived from Regulon DB. And the calculating process is subject to two constraints:
1. That the spacer length (the number of base pairs between the −35 hexamer and the −10 hexamer) should lie in the range (15�20);
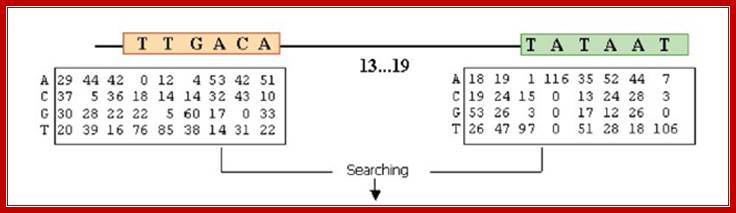
The consensus sequences of sigma 70 factor binding site;
The total MSS (our results are the sum of the scores for the −10 and −35 hexamers and therefore lie in the interval [0,2], with a score of 2 corresponding to the joint consensus TTGACA (−35) and TATAAT (−10).
Score(Promoter)=score(-10 box)+score(-35 box)+score(spacer between -10 & -35 boxes)
And the score of spacer length is calculated by algorithms proposed by Ryan K. Shultzaberger.el. in E. coli sigma70 promoters.8 But due to a lack of experimental data of promoter strength with both different motifs and spacer length, the weight of the total MSS and the spacer score is very roughly determined with few experimental data available. Currently our weight is determined with the promoter strength data in a literature16 to best fit the promoter score with promoter strength. Now the relative weight between the total MSS of the two motifs and the spacer score is 0.29:0.71.
In prokaryotes, the strength of sigma factor regulation for a particular gene often depends on how closely a particular site matches the consensus for the motif. The more mismatches to the consensus in a binding site, the less often the sigma factor will bind and therefore the less strength the promoter will have. Experiments have confirmed the hypothesis that the consensus promoter sequences is "best". We set the best promoter strength to 100% and calculate the relative strength of a given promoter by the Score (promoter). http://2013.igem.org/.
Sequential assembly of RNAP complex:
�
a + a-------> 2a,
2a + b -------> 2a 1 b,
2a b + b� -------> 2ab b� + ω.
Core enzyme + sig-70 -------> Holozyme.
The assembly is sequential and requires all the components and in equal concentrations.� Once transcription is initiated, it is believed the sigma factor dissociates form the RNAP.� But recent studies indicate that sigma factor remain bound but in dissociated form.
- Conserved in all E. coli genes transcribed by holoenzyme with sigma70
- Genes for more than 100 sigma proteins have been sequenced
- the proteins form a large family, the sigma 70 family of sigma factors
- one important exception is sigma54, which shows no homology to sigma70 proteins, and which interacts with core polymerase differently from members of the sigma70 family. �The C-terminal 2 sequence binds to -10 region of the promoter and the 4th segment binds to -35 region.
- Sigma 24 binds to -35 and -10
Members of the sigma70 family of sigma factors have 4 conserved regions (Figure adapted from Fig 7-3, Kornberg and Baker, DNA Replication,
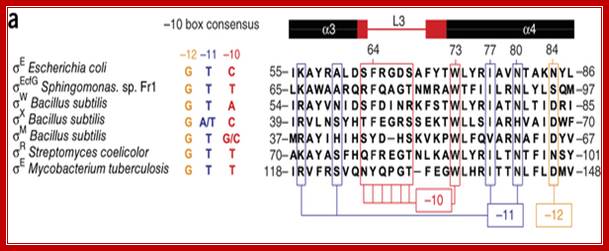
Structural basis for -10 promoter elements melting by environmentall induced sigma factors; �S�bastien Champagne, May E Marsh, Guido Capitaine,
http://www.nature.com
Region 4: a DNA binding activity that recognizes the -35-promoter motif; this is inferred from allele specific mutations that alter promoter recognition
Region 2: Sub region 2.3 and 2.4 form a DNA binding activity that recognizes the -10-promoter motif, region 2 also interacts with core enzyme components; this part of 70 is conserved with the eukaryotic factor RAP30
Region 1: masks the DNA binding activities that are present in regions 2 and 4.
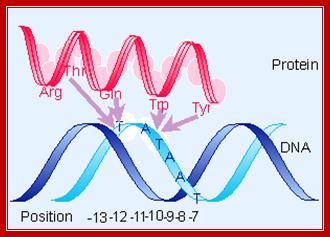
From X-ray structure of this portion of 70 (Malhotra et al., 1996, Cell 87:127-136) �determined-<> Acids (blue) - maybe contact backbone, <> Residues known to interact at -12 position (orange),<> conserved hydrophobic core (yellow) - structural integrity of this domain? ,<> clusters of aromatic amino acids that are solvent exposed (cyan) which may be used to intercalate between bases to stabilize single stranded DNA, thus contributing to melting the promoter region,<> red - promoter melting,<> green - mutations that perturb RNAP binding , <> yellow - exposed hydrophobic, highly conserved, for RNAP binding? <> blue - other exposed, highly conserved residues, <> all of these combined.
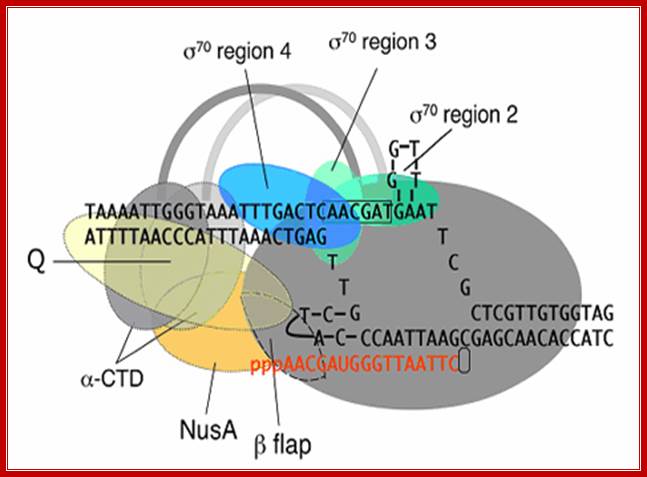
Positioning of different regions of sigma factor such as 2, 3 and 4, note that the 2nd segment binds to TATAAT box and 4th segment binds to TTGACA.� The "flexible flap" domain shown above of the RNA polymerase belongs to beta subunit. The flap interacts with conserved region 4 of sigma and triggers a conformational change that moves region 4 into the correct position for interaction with the -35 element. Because the flexible flap is evolutionarily conserved, this domain may facilitate promoter recognition by specificity factors in eukaryotes as well. The beta flap covers the exit of newly synthesized RNA. Nus factor facilitates elongation of transcription. jwr7@cornell.edu http://mbg.cornell.edu/
The sigma factor is the subunit of the RNA Polymerase complex that recognizes the specific promoter sequence of DNA that the RNA Polymerase complex should bind to. This will be the initial site for DNA transcription, or where it initiates. While the RNAP covers 60bp or more, sigma factor has restricted region to bind to the promoter and RNAP. Mathew-Hunter:� http://helicase.pbworks.com/ http://quotesfab.com
Schematic model of the elongation complex-Core RNA polymerase (RNAP) in bacteria, a complex composed of an α-dimer, a β-subunit, a β′-subunit and an ω-subunit) is bound to the DNA duplex composed of the template strand (black) and the non-template strand (blue), and the nascent RNA (red). The α-amino-terminal domains (α-NTDs) serve as a scaffold for complex assembly; the α-carboxy-terminal domains (α-CTDs) and ω-subunit play regulatory roles during initiation. The β- and β′-subunits jointly form the active site and make all the contacts to the nucleic acids. The substrate nucleoside 5′-triphosphate (NTP) (bound to a second Mg2+ ion) is thought to enter through the secondary channel. 12�14 bp of the DNA are melted in the transcription bubble. The non-template DNA strand is exposed on the surface, where it may interact with regulatory proteins. The nascent RNA is annealed to the template strand to form 8�9 bp of the RNA�DNA hybrid, which is the key determinant of elongation complex stability7, 98, 99. The upstream RNA is extruded through the RNA exit channel formed between the β-flap and β′-clamp; https://www.researchgate.net/
The RNAP by itself can bind to any promoter but it cannot position itself to a specific promoter and initiate transcription.�� Positioning of the enzyme on the promoter exactly in sequence context, in such a way, so as to initiate exactly and absolutely at the same nucleotide, which is predefined and called start site, requires sigma factor.
RNA pol at tarnation level; http://www.mun.ca
|
|
Gene |
Mol wt |
function |
Sequence binding -35 |
Sequence binding -10 |
|
E.coli |
|
|
|
|
|
|
Sig-70
|
rpoD |
70kd |
Most house keeping and few others |
TTGACA� <16 |
18> TATAAT |
|
Sig 54,(rpo N, Ntr A Glu F)/54 kd |
rpoN/ntrA |
54 |
N2 regulated |
-24TTGGNA
|
-12 TTGCA |
|
Sig 32
|
rpoH |
32 |
Heat shock regulated |
CCCTTGAA<13 |
15> CCCATNTA |
|
Sig 28 |
fli |
28 |
Flagillin & chemo taxis |
CTAAA�� <15> |
CCGATAT |
|
Sig 24 |
rpoE |
24 |
Extreme Heat shock/oxidative |
GAACTT |
TCTGA |
|
Sig S |
rpoS |
38 |
Stationary phase |
|
|
|
SigFecI, ecI |
fecI |
19 |
�irondicitrate transport |
|
|
|
B.subtilis |
|
|
|
|
|
|
Sig43 (sig A),rpoD,sigA |
sigA |
43 |
Most genes |
TTGACA |
TATAAT |
|
Sig 28,(Sig D) |
sigD |
28 |
Flagillin |
CTAAA |
CCGATAT |
|
Sig 29,(sig E1, spo II GB) |
sigE1,SPOii |
29 |
Sporulation |
TTNA |
CATTT |
|
Sig H,(sigH, spo H) |
sigH/spoH |
|
Sporulation |
-- |
-- |
|
Sig 32,(sig C ) |
SigC |
32 |
Het shock |
AAATC |
TANTGNTTNTA |
|
Sig 37,(sig B) |
sigB |
37 |
Unknown |
AGGNTT |
GGNATTGNT |
|
Sig G,(spo 3-G),sigG |
Spo3G |
|
Fore spore specific |
PyGNATpu |
CAN (T/A) NTA |
|
Sig F,(spo II AC),sigF |
sigF/spoiiAC |
|
Sporulation |
-35 PyGNATpu |
-10 CAN (T/A) NTA |
|
Sig K,(spo IV CB) sigK,
|
sigK/spoivCBIIIC |
27kd |
Sporulation mother cell specific |
� Not known |
� Not known |
Three of the E. coli s factors have regions of sequence similarity (s70, s32, and s28 ) whereas s54 is a distinctly different molecule that works rather differently.
|
Factor |
Gene |
Use |
‑35 |
Separation |
‑10 |
|
s70 |
rpoD |
General |
TTGACA |
16-19 bp |
TATAAT |
|
s32 |
rpoH |
Heat shock |
CCCTTGAA |
13-15 bp |
CCCGATNT |
|
s28 |
fliA |
Flagella |
CTAAA |
15 bp |
GCCGATAA |
|
s54 |
rpoN |
Nitrogen starvation |
CTGGNA |
6 bp |
TTGCA |
http://www.personal.psu.edu/
Sigma factor, binds to beta subunit of RNAP.� They place the RNAP on to a correct promoter and in correct position in the promoter.�� Once the correct promoter has been recognized, it induces conformational changes in the enzyme and activates it.� This leads to the tight binding of the enzyme to DNA and induce the melting of the DNA from �11 to +3 (about 12-14 base pair long) for initiation of transcription (amazing).�
Once the promoter is cleared by the RNAP the sigma factor dissociates from the RNAP, only to be associated with another core enzyme to initiate one more cycle of transcriptional initiation. The sigma 43 of B. subtilis also has similar sequences in promoter regions and similar domains in the sigma factor.� But, in the case of T4 phage sigma factor, it does not possess the C-terminal domain and it does not require �35 sequences for transcriptional initiation, by its T4 RNA polymerase.
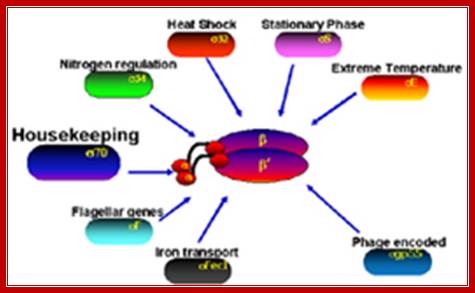
Different sigma factors in association with RNAP perform different functions in terms of transcription of different genes for different functions; Richard R.Burgess; http://mcardle.wisc.edu/
Omega protein:
It is coded for by the gene rpo Z; its Mol.wt is 11 KD. Even during purification of the enzyme, it is difficult to get rid of this omega protein. Absence of this protein from the Holozyme doesn�t affect the function of the enzyme.� But mutations in the gene affect the growth of the cells; growth rate reduces considerably.�
This phenotypic effect has been attributed to the disruption of a downstream gene called Spo-T gene. The spo-T gene is involved in the regulation of stringent response to amino acid starvation situations, where the rate of metabolism, rate of RNA synthesis and the rate of translation slow down and the whole cellular events are down regulated.��
A substance produced in such situations, called magic molecules, such as Guanosine tetra phosphate (ppGpp), requires omega protein for its activity.� Thus the omega protein has been implicated in transcription.
Rho protein:
This protein is coded for by rho gene; Mol.wt is 46.� The gene is mapped at 46 mpu position.� It assembles into a hexamer and binds to the 5� end of newly forming transcripts.� It acts like RNA dependent ATPase and slides along the RNA transcript, and interacts with RNAP at a particular region and terminates transcription.� In Lambda infected bacterial cell one finds such rho-dependent transcriptional termination.� This protein behaves like an RNA dependent helicase.
Nus A:
Molecular mass of this protein is 65kDa, coded for by nus-A gene. It is involved in termination and anti-termination of transcription.� Very strangely, as soon as the RNAP clears the promoter, sigma is released from the RNAP enzyme (how? and why?) And immediately Nus associates with RNAP.� In fact nus association with RNAP causes decrease in the affinity for NTPS.� But it also facilitates the binding of other proteins, which have anti terminator functions.� When the RNAP with Nus is released from the template DNA, sigma factor induces the release of the nus from the RNAP and never vice- versa.
RNAPs of Blue green algae: Cyanobacteria: They are gram negative bacteria (once called Blue Green Algae) and they perform oxygenic photosynthesis. It is the oxygenic photosynthesis that has provided oxygen for all respiratory organisms and food for their survival al organisms.� Blue green algae are made up of single cells to colonies and filaments, a fascinating group of bacteria; it is difficult to call them as bacteria. Fremyella diplosiphon RNAP is made up of five polypeptides (Synechocystis sp.)-RPoC1-161,000Da, RPoB-134,000Da, (91,000Da)?, RPoC1- 72,000 Da, and RPoA-41,000 daltons. Anacystid nudulins consists of four subunits- 147kD (b), 125kD (b�), p 86kD and 39kD (alpha).The presence of omega subunit (RpoZ) is not known.
There are three groups of sigma factors. Group 1 comprises a principal σ factor that is essential for cell viability. Group 2 (Hsp type) is similar to group 1 in molecular structure, but nonessential for cell viability. Group 3 σ factors are an alternative type, structurally different from the group 1 and group 2 σ factors, and are involved in the transcription of regulons for survival under stress. From a structural point of view, σ factors exhibit conserved domains known as regions 1 and/or 2, 3, and 4. Region 1 is subdivided into regions 1.1 and/or 1.2. Region 2 is subdivided into 2.1, 2.2, 2.3, 2,4, and 2.5. Region 2.4 (and 2.5) helps to recognize a −10 (and an extended −10 motif) element. Cyanobacteria Synechocystis contain nine sig factors.
Archaeal RNA Polymerases:
Three branches of life are shown and their inter relation is sketchy.� After life originated in hot springs on this planet, archaea came into existence around 3.9 x10^9 (billion) years ago.� Whether life molecules are still forming in such hot springs found all over the world is not known but possibilities are possible.� These places, ex. yellow stone in USA are like Urey� Millers lab. Archaea and Eubacteria are derived from Last Universal Common Ancestors (LUCA).� Eukaryotes developed from Last Eukaryotic Common Ancestors (LECA). Archaea cells are distinctly prokaryotic. Archaea and eubacteria have many resemblances and many differences.� Interestingly archaea in terms of certain enzymes they resemble eukaryotes. These organisms might be the precursor for eukaryotes.
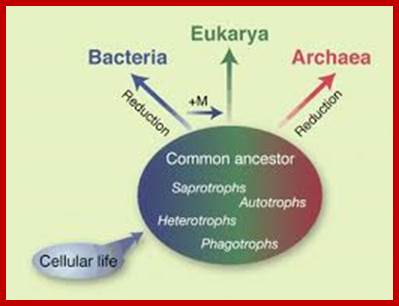
Origin of Archaea; Scientific American Blog Network

Archaea Asgard revealed unknown details of the origin of archaea and eukaryotes; http://earth-chronicles.com
Three branches of life are shown and their inter relation is sketched.� After life originated in hot springs on this planet archaea came in to existence around 3.9 x10^9 (billion) years ago.� Archaea and Eubacteria are derived from Last Universal Common Ancestors (LUCA).� Eukaryotes developed from Last Eukaryotic Common Ancestors (LECA).
Halobacterium, sp. strain NRC-http://en.wikipedia.org/
Methanococcus jannaschii, http://textbookofbacteriology.net/
Archaea were first found in extreme environments such as hot springs and volcanic eruptions. Archaea were first found in extreme environments, such as volcanic hot springs. Pictured here is Grand Prismatic Spring of Yellowstone National Park. There are thermophiles, Methanogens and halophile; Archaea bacteria; http://www.brighthub.com/
� Archaea once thought to be closer to bacteria, but now they are found to have characteristics of both bacteria and eukaryotes.
� The cell walls of archaea are chemically and structurally diverse and lack peptidoglycan.
� Archaea cell walls contain a pseudomurein that is different in structure from eubacterial murein, no peptidoglycan.
� Cells contain gas vesicles and glycogen granules.
� They don�t contain Nucleus,
� Archaeal cells contain Histone like (like H3+H4) tetramers that can compact DNA into nucleosomes (experimental demonstration) in methanobacterium sp.
� Archaeal nucleosomes protect ~60bp long DNA.
� They don�t have endoplasmic reticulum,
� They contain 70s ribosomes similar to that of bacteria; their proteins and translation factors are more like eukaryotes.
� The genomic DNA is circular, remains in supercoiled state.
� The genome size varies from 0.5Mb to 5.5mb (Methanosarcina barkeri), also contain significant number of plasmids.
� The genome in Methanococcus jannaschii is 1.7x 10^6bp, there are 1700 protein coding genes.
� Genes are organized into operons, mRNAs are polycistronic and no introns,
� rRNA operons with internal transcribed spacer, the number is 1-15.
� The archaeal RNAPs are now considered closely related to Eukaryotic RNAPII in terms of subunit composition and architecture.
� The transcriptional regulators activators and repressors encoded by archaeal genome are closely related to bacterial systems or bacterial-like regulatory mechanisms.
� Basal transcription factors for initiation and elongation are more or less same.
� RNAPs of this class are large and sophisticated, which interact with DNA/RNA scaffolds.
� Gene promoter structure in archaea is also more similar to that of eukaryotes than eubacteria in having �TATA� box, although, like the eubacteria, archaea have operons and transcribe these to polycistronic mRNA.
� Some polycistronic mRNA are cleaved to independent mRNAs and translated.
� Overall protein synthesis mechanisms of eukaryotes and archaea may be similar.
� The first amino acid incorporated is methionine not formyl methylated as in prokaryotes.
� Processing of tRNA and mRNA is same as PK and EK.
� 5� end of mRNA is processed by RNase P and 3� by RNaseZ.
� Introns in tRNA are spliced out and nucleotides are modified.
� In some cases mRNA contain short poly (A) tail.
� mRNAs contain 5� shine Delgarno sequence and binds to 30 ribosome subunit.
� In some cases translation initiation is like EK.
� Archaea have a single type of RNAP, responsible for the synthesis of all RNAs. Archaeal RNAP is structurally and mechanistically similar to eukaryotic RNAP II and nuclear RNAP IV and V, and it is especially structurally and mechanistically closely related to eukaryotic nuclear RNAP II.
� This 12-subunit complex play distinct role in RNAP assembly and catalysis (A�A��, B, D, K, L, f, H, E, G, N and P).� They are homologous to Eukaryotic RNAPII.
� The enzyme is not inhibited by Rifamycin, but can be inhibited by topoisomerase inhibitors.
� They don�t produce any endospores; they contain flagella and sex pili.
� Exhibit transduction and conjugation.
� Cells possess TCA cycle and electron transport and ATP is synthesized by chemiosmotic process.
� Ribosomes sensitive to Anisomycin (like EK) and insensitive to Chloramphenicol
|
Comparison between Eubacteria, Archaea and Eukaryotes |
|||
|
Eubacteria |
Archaea |
Eukaryotes |
|
|
Nucleus |
No |
No |
Yes: membrane-bound |
|
Nucleosomes/histones |
No |
Yes |
Yes |
|
Operons/polycistronic mRNAs |
Yes |
Yes |
No |
|
Introns |
No |
No |
Yes |
|
TATA Box binding protein |
No |
Yes |
Yes |
|
Organelles |
No |
No |
Yes: plastids, mitochondria, lysosomes, endoplasmic reticulum etc. |
|
Chromosomes |
One Circular |
One Circular |
More than one,linear |
|
RNA polymerase |
One (simple) |
More than one (complex) |
More than one (complex) |
|
Protein initiator amino acid |
N-formyl methionine |
Methionine |
Methionine |
|
Protein synthesis sensitivity to diphtheria toxin |
Insensitive |
Sensitive |
Sensitive |
|
Peptidoglycan |
Yes |
No |
No |
|
Protein synthesis |
|
||
� Archaea promoter elements have similarities to that of Eukaryotes.
� GATACACACA�AGATACAGAT�TATA-----+1>
� Compare to that of bacteria-
---TTGACA---TATAATT --GACACAACT----+1>
o Transcriptional factors required are TBP, TFB-B binding BRE factors.
|
Shared with Bacteria |
Shared with Eukarya |
Unique to Archaea |
|
No nucleus or membrane-bound organelles |
Cell wall structure (for example, some archaeal cell walls contain pseudomurein) |
|
|
Circular genome |
Cell membrane containing ether-linked lipids |
|
|
Genes grouped in operons |
Translation initiated with methionine |
|
|
No introns or RNA processing |
Similar RNA polymerase, promoters, other transcriptional machinery[39][40][41] |
Ribosomal structure (characteristics shared with both Bacteria and Eukarya) |
|
Polycistronic mRNA |
Similar DNA replication and repair[42] |
|
|
Cellular size (>100-fold smaller than eukaryotes) |
No fatty acid synthetase enzyme[39] |
� Transcriptional initiation is similar to that of Eukaryotes.
� Most of the mRNAs are polycistronic and contain smaller 5�UTR and 3�UTR.
� Some of the tRNA genes are transcribed as pieces and then they are trans- spliced.
� Leucine response factors are abundant.� It amounts to �regulons.
|
|
EK |
Archa (a) |
Bact |
|
classI subunit |
Rpb1 |
A�A�� |
B� |
|
|
Rpb2 |
B |
B |
|
|
Rpb3 |
D |
a1 |
|
|
Rpb6 |
K |
w |
|
|
Rpb11 |
L |
a2 |
|
classII |
Rpb4 |
f |
|
|
|
Rpb5 |
H |
|
|
|
Rpb |
E |
|
|
|
Rpb8 |
G |
|
|
|
Rpb10 |
N |
|
|
|
Rpb12 |
P |
|
|
classIII |
Rpb9^1 |
|
|
|
|
G down |
Rpo13 |
delta |
|
|
|
|
|
Just a comparison and equivalents of the subunits of Eukaryote RNAPII, archaea and Bacteria.
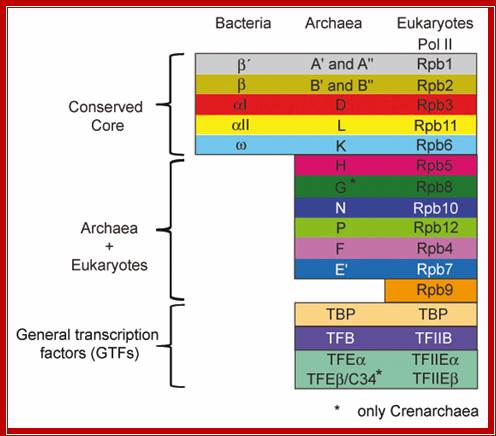
Werner F. University College London, Department of Biochemistry and Molecular Biology, Darwin Building, Gower Street, London WC1E 6BT, UK. werner@biochem.ucl.ac.uk.
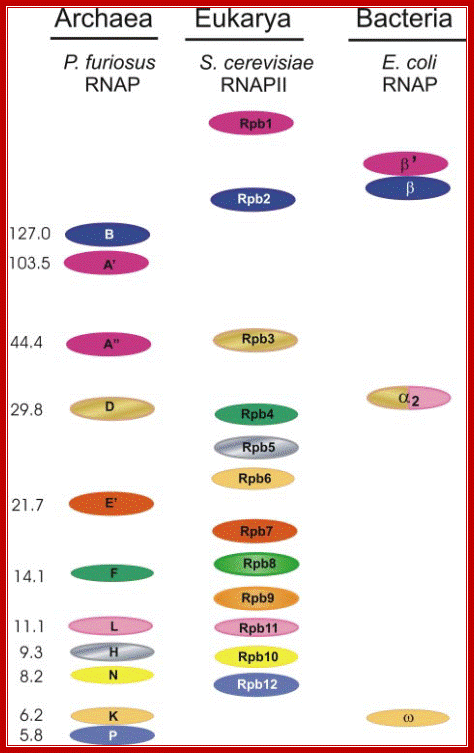
������������� Homologous subunits of RNAP from the three domains of life:
������������� The subunit pattern of the RNAP from P. furiosus, pol II of S. cerevisiae and of
������������� the E. coli enzyme are shown. Identical colors indicate homology. Note that
subunit Rpb1 of eukaryotes is split in Archaea into the two polypeptides, A_
and A_. Subunit _ of the E. coli RNAP is homologous to a part of subunit D of Archaea and of Rpb3 of yeast and to subunits L and Rpb11. The molecular
mass of the subunits of Pyrococcus RNAP is indicated to the left. http://www.biologie.uni-regensburg.de/
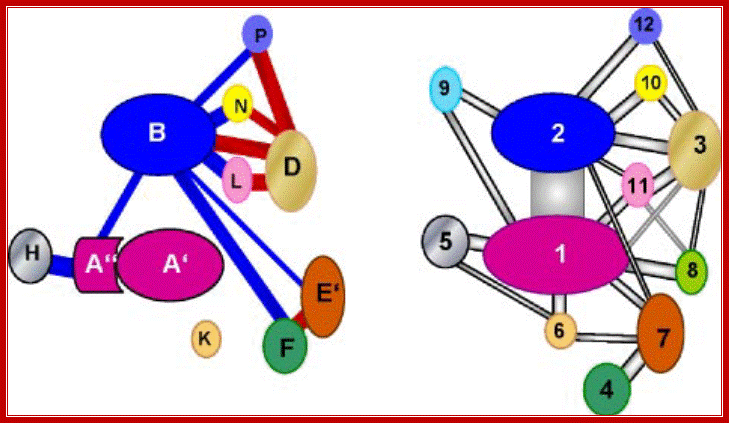
Left; RNAP subunuts of Archaeal� of eukaryotic RNAP II-http://www.biologie.uni-regensburg.de/;
Schematic representation of interaction of subunits in pol II and the archaeal enzyme: Right panel, modified interaction diagram of pol II subunits based on the crystal structure of pol II according to Ref. 1 and considering the interactions of Rpb3 and Rpb7 according to Ref. 2. The color code is the same as in Figs. 1 and 4. Left panel, interaction diagram of Pyrococcus subunits based on Far-Western analyses. The homology of subunits to pol II is indicated by the color code. The thickness of the lines connecting the subunits gives an estimation of the strength of interactions. Connecting lines in blue color indicate an interaction established by one labeled probe, connecting lines in red color indicate interactions established by both interacting partners as probe. Crit Rev Biochem Mol Biol. Author manuscript; available in PMC 2012 February 1.
Comparison of interaction networks in the
archaeal and eukaryotic RNAP.
Left, the diagram
illustrates the interactions between subunits of the Pyrococcus� RNAP based on
Far Western analysis. The thickness of the lines estimates the strength of
interaction. An interaction emanated by either one (blue line) or both (red
line) binding partners.
Right, a modified diagram based on the crystal structure of RNAP
II of S. cerevisiae (2,3) is shown. Homologous subunits are marked in the
same color
Comparison of Bacterial RNA polymerase and Eukaryotic RNA Polymerase II
RNA polymerase is an enzyme that is conserved in all living organisms.
Because bacterial, archaeal, and eukaryotic RNAPs are large enzymes comprising many
subunits; they are part of a conserved protein family called �multisubunit RNAP
family (Ebright 2000).� The bacterial RNAP core enzyme is made up of five
subunits (b�, b, aI, aII, and w), with a molecular mass of approximately 0.35
MDa (Minakhin 2001). Eukaryotic RNAP II consists of these five conserved
subunits and seven additional subunits, with a molecular mass of about 0.5 MDa.
It has been shown that the subunits of the bacterial RNAP core enzyme and the
subunits of eukaryotic RNAP II exhibit sequence, structural, and functional
homology (Minakhin 2001). Bacterial w subunit of RNAP is a homolog of
Eukaryotic RPB6.
Eukaryotes have multiple types of nuclear RNAPs, each responsible for synthesis of a distinct subset of RNAs. All are structurally and mechanistically related to each other and to bacterial RNAP (wiki).
- RNA polymerase I synthesizes a pre-rRNA 45S (35S in yeast), which matures into 28S, 18S and 5.8S rRNAs which will form the major RNA sections of the ribosome.
- RNA polymerase II synthesizes precursors of mRNAs and most snRNA and microRNAs. This is the most studied type, and due to the high level of control required over transcription a range of transcription factors are required for its binding to promoters.
- RNA polymerase III synthesizes tRNAs, rRNA 5S and other small RNAs found in the nucleus and cytosol.
- RNA polymerase IV synthesizes siRNA in plants.
- RNA polymerase V synthesizes RNAs involved in siRNA-directed heterochromatin formation in plants.
Chloroplasts in eukaryotic cells contain two types of RNAPs; one structurally and mechanistically similar to bacterial RNAP ("plastid-encoded polymerase with four subunits PEP "). Chloroplasts also contain a second type, structurally and mechanistically unrelated, RNAP ("nucleus-encoded polymerase"); member of the "single-subunit RNAP" protein family coded by nucleus (NEP).� Zea mays chloroplasts in green tissues contain nuclear coded sig1 and sig2 (50kDa in green and -60kDa in both etioplasts and green plastids) proteins. Sig 5 and sig6 are also found in Oryza sativa.
Mitochondrial RNAPs: Eukaryotic cells contain specialized symbiont organelle called mitochondria.� This has its genome with just 13 protein coding genes and 22tRNA genes plus two rRNA genes. During endosymbiontic process oxygen respiring bacteria has lost more than 99% of it genome to the eukaryotic cell, yet it has retain very crucial genes without which the parent cell is hapless and helpless. Mitochondria contain an nuclear coded single subunit RNAP which is structurally and mechanistically unrelated to other RNAPs, member of the "single-subunit RNAP" protein family, similar to that of T7, Sp6 �and T3 phage RNAPs.
RNA polymerases in Viruses:
Orthomyxovirus- It is structurally related to bacterial, archaea and eukaryotic RNAP I-IV. Some viruses use single subunit DNA dependent RNAP and related to eukaryotic mitochondria and chloroplasts.� There are single strand transcriptases or RNAPs. There are RNA dependent RNAPs, which convert (-) strand to positive strand and vice versa. There are DNA dependent Viral RNA polymerases.
Viral coded- RNA polymerases:
Picorna virus: ��The gene 3D codes for nonstructural protein that acts as viral RNAP. �The 3D codes for RNA dependent RNA polymerase.
T7 Phage:� codes for a RNAP of� 99kDa and it uses promoter sequences, even Sp6, T3 RNAPs have the same structural and functional similarities.
--I -------(-35) TTGACA----------(-10) TATAAT------+1>>>
Adeno virus:
There are different promoters such as early1A and 1B and late they are transcribed with time. The transcription is performed by ADV RNAP IIIc protein.
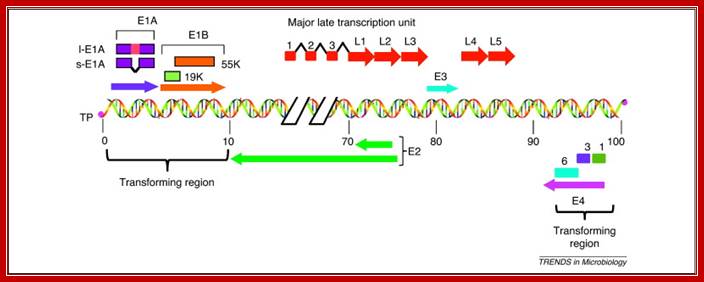
Figure 1. Schematic illustration of the HAdv5 genome, which consists of a 36 kb linear double-stranded DNA that is covalently linked to viral terminal proteins (TP) at the 3′ ends. The viral genome contains five early transcription units (E1A, E1B, E3, E4 and E2) and one major late transcription unit. The major late transcript is differentially processed into five groups of late mRNAs (L1�L5), to which three leader sequences are attached by RNA splicing. The late mRNAs code for various viral structural proteins. The left-hand 14% of the genome contains the E1A and E1B genes, which constitute the transforming genes. l-E1A indicates the large E1A protein and s-E1A indicates the small E1A protein. The early region E4 of different HAdv species encodes multiple proteins, of which at least three, Orf 1, Orf 3 and Orf 6 (indicated by 1, 3 and 6), exhibit transforming activities. http://www.sciencedirect.com/
Retro Viral coded RNA dependent DNAP:
There are two RNA genomes and they code for Reverse transcriptase, that use RNA to generate cDNA.� The enzyme uses promoter elements typical of eukaryotes.
The primary RNA transcribed from the provirus, which is identical to the viral genome, can be transported to the cytoplasm intact and serve as an mRNA for the gag and pol sequences or can be spliced (removing the gag and pol sequences) to a shorter mRNA from which the env sequences are translated. The gag proteins are translated as a polyprotein and proteolytically processed into the component capsid proteins. The pol ORF is in a different translational frame from the gag ORF, but is translated by "translational frame shifting" that occasionally occurs as the ribosome translates the gag ORF, resulting in a gag-pol fusion precursor. In this way, gene expression is controlled such that only 5% as much pol gene product is produced as are gag products. The env gene is also translated as a polyprotein and processed into its two component subunits after translation;
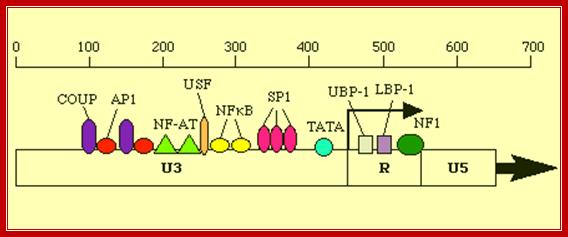
Retroviruses;Promoter elements� in LTR region of the retroviral genome. http://www2.gsu.edu/
5� �AP1-NFAT�NRF---NFkB-Sp1--CCAATTATA---TATA----R
Given that DNA and RNA polymerases both carry out template-dependent nucleotide polymerization, it might be expected that the two types of enzymes would be structurally related. However, x-ray crystallographic studies of both types of enzymes reveal that, other than containing a critical Mg2+ ion at the catalytic site, they are virtually unrelated to each other; indeed template-dependent nucleotide polymerizing enzymes seem to have arisen independently twice during the early evolution of cells. One lineage led to the modern DNA Polymerases and reverse transcriptase (RTs), as well as to a few single-subunit RNA polymerases from viruses. The other lineage formed all of the modern cellular RNA polymerases.� However there are Polymerases that adds CCA to tRNA 3�ends.� Some polymerases add Poly-A to the 3�ends of mRNAs.: M. F. Symmons, G. H. Jones & B. F. Luisi
In 1955, Marianne Grunberg-Manago and Severo Ochoa reported the isolation of an enzyme that catalyzed the synthesis of RNA. Their work built upon the earlier work of Jerard Hurwitz & J.J. Furth who performed experiments to see if isolated E. coli protein fractions could polymerize radioactively labeled nucleotides. Later Grunberg-Manago & Ochoa tested a protein fraction that could make RNA. For this work, Ochoa shared the 1959 Nobel Prize in Medicine with Arthur Kornberg (who received the Prize for his work on DNA polymerase- I). This enzyme works as polymerase without template.
This enzyme could convert ribonucleotide diphosphate into RNA:
(RNA)n + NDP --> (RNA)n+1 + Pi
However, the enzyme had a number of unsettling properties. It does not need a template; and, it could use as little as 1 NDP or as many as 4 NDPs as substrate. In fact, the sequence of the product RNA depended entirely on the number and concentration of substrate NDPs.
These are not the properties of an enzyme that must faithfully copy the genetic material for expression!
We now know that Grunberg-Manago and Ochoa had isolated the enzyme polynucleotide phosphorylase which usually catalyzes the breakdown of RNA - not its synthesis! i.e., it is a ribonuclease; it can polymerize nucleotides in vitro; Reference: Structure, 8:1215-1226, 2000
Description: PNPase is a disk-like trimeric exo ribonuclease. This side view of the trimer shows the interface between two subunits. The cores of subunits are constructed from two homologous domains shown in shades of red and blue. The lower and upper accessory domains are in green and grey respectively.
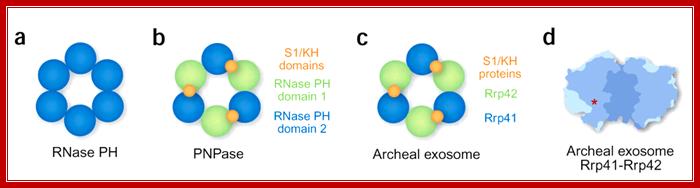
Structures of Phopholytic 3�exoRNase:
(a) Prokaryotic RNase PH is composed of six identical subunits forming a ring structure with a central hole. (b) In prokaryotic PNPase, the hexameric RNase PH�like ring is composed of three polypeptides, each contributing two PH domains. Only the second domain in each subunit is active. Every subunit also contributes two RNA-binding domains, which are all arranged on the top side of the hexameric PH ring. (c) The archaeal exosome resembles PNPase, except that it is composed of nine separate polypeptides. Three Rrp41-Rrp42 heterodimers form the hexameric PH ring. Only Rrp41 is active as a nuclease. Three additional subunits provide the RNA-binding domains arranged on the top side of the PH ring. (d) The hexameric ring of the archaeal exosome, cut open to reveal the central cavity. Red asterick indicates the location of one of three active sites, which is connected to the central cavity by an internal groove (not shown). RNA-binding subunits would be located at the top of the structure. Note the narrow constriction at this end of the central channel. Figure is adapted from ref. Elmar Wahle; http://www.nature.com/
PNPase: This view of the channel through the centre of the trimer channel shows side chains of the conserved 'FFRR loops' that may act as a mechanism to entrap RNA substrate and select against base-paired segments. This provides a possible structural explanation for both the high processivity and the regulation of the PNPase activity by RNA secondary structure.
As mentioned earlier, polynucleotide phosphorylase is a bifunctional enzyme. The mechanism of action of this enzyme can be represented by following reactions:
Polynucleotide phosphorylase is a bifunctional enzyme. The mechanism of action of this enzyme can be represented by following reactions:
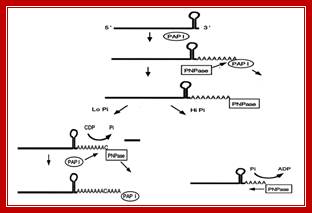
Schematic representation of the role of PNPase in poly(A) tail metabolism in E. coli . Mohanty B K, and Kushner S R PNAS 2000; http://nptel.ac.in/