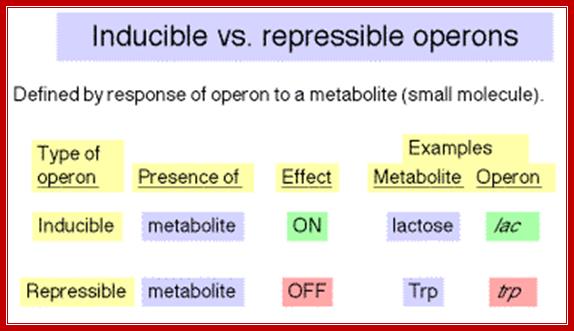
Lactose Operon:
E. coli K-12 is one of the prokaryotes which has been extensively studied.� It is often called �Cinderella� of prokaryotic Molecular Biology.� The genome size is 4.6x10^6 bp and it is organized into a circular genome. In the cell, its DNA in supercoiled state. It contains ~4288 (4466) genes for proteins, organized into ~ 258 operons. Based on the frequency and distance distributions, it is estimated a total of 630 to 700 operons in E. coli (Helaldia Salgado et al). And statistical analysis provides the information about 173 Regulons (Han Zang et al). There are seven rRNA genes, six ribosomal protein genes and 86 tRNA genes.� Coding density is very high (it has to be); intergenic distance is ~118 bp.� The genome also harbors some transposable elements. It also contains circular plasmids.
E. coli is a resident of human intestinal tracts.� There were reports in Europe, the bacteria have become a pathogenic bug, and this may be due to excessive use of antibiotics!?� Recombinant techniques were used to enable the bacteria to produce new genetic variants to treat intestinal diseases.
Engineered E.coli; http://newscenter.lbl.gov/
When, a cluster of independent genes coding for different proteins regulated concertedly with one promoter/regulator, then it is called an operon.� If more than one operon is controlled by one substance or regulator molecule it is called Regulon, ex. Leucine and NtrC.
�
In most of the situations, E. coli bacterial cells grow and grows well in glucose media.� In the presence of glucose all respiratory enzymes, carbohydrate metabolizing enzymes, intermediary metabolic enzymes, lipid metabolic enzymes, amino acid metabolizing enzymes, protein and nucleic acid metabolizing enzymes and many others involved in day-to-day functions are constitutively synthesized.�
In such cells one does not find any enzymes for utilizing non glucose sugars such as Lactose or Arabinose, even if one finds, their number is very few per cell. When the carbohydrate source from glucose is changed to, say lactose, a dramatic increase in lactose utilizing enzymes is observed.
Such increase is not due to increased rate of protein synthesis but it is due to the activation of a set of genes leading to the synthesis of specific mRNAs and proteins that utilize such carbohydrate sources.� Use of inhibitors of protein and RNA synthesis demonstrates the above changes. Another regulator, that shows dramatic increases and decrease, is cAMP.� When glucose is present its concentration is very low, and when glucose is absent its concentration increases substantially.�
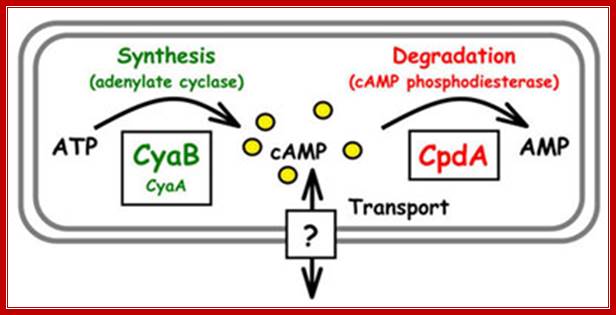
Mathew C. Wolfgang. http://www.med.unc.edu/
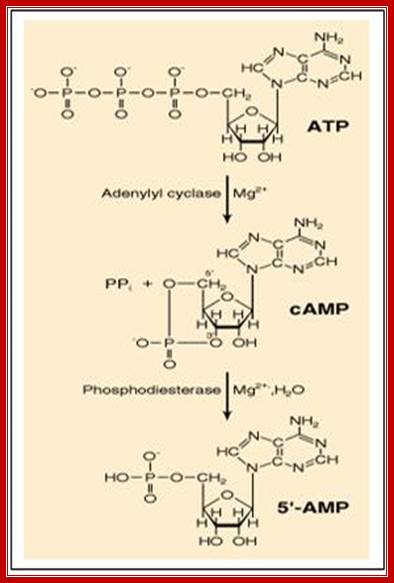
Figure:� The synthesis and degradation of cyclic AMP (cAMP).
A pyro phosphatase
makes the synthesis of cyclic AMP an irreversible reaction by hydrolyzing the
released pyrophosphate ![]() -
-![]() (not
shown. http://www.pharmainfo.net/
(not
shown. http://www.pharmainfo.net/
Two enzymes, which have opposing activities, are responsible for this.� When glucose is adequate in the cell, Adenyl phospho diesterases (specific) cleave cAMP phosphate bonds, but in absence of glucose, Adenyl cyclase acts on ATP and by cyclase activity, it generates cAMPs, which are considered to be very important second messengers in carbohydrate metabolic activities, perhaps they can be called as regulons.
Lactose is a disaccharide made up of a Galactose and a Glucose held by beta 1�4 glycosidic bond.� Lactose is a common sugar in milk; all newborn mammals depend upon this source of carbohydrate. Metabolism of lactose in bacteria is performed by two enzymes, one sugar-transporting enzyme called Permease and the other is beta Galactosidase which cleave the β 1-4 glycosidic bond.
�
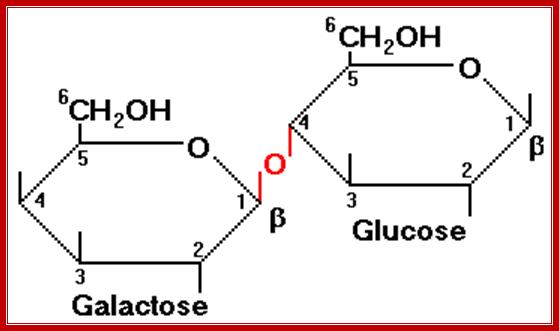
Permeases and β-Galactosidases are present in the cells even in the absence of lactose, but at a low concentration.� Permease is a plasma membrane transporter, which transports lactose into the cell. As soon as the lactose is transported into the cell, it is transformed into allolactose form by an Isomerase. Allolactose activates resident beta Galactosidase, which cuts Allolactose to Galactose and Glucose.
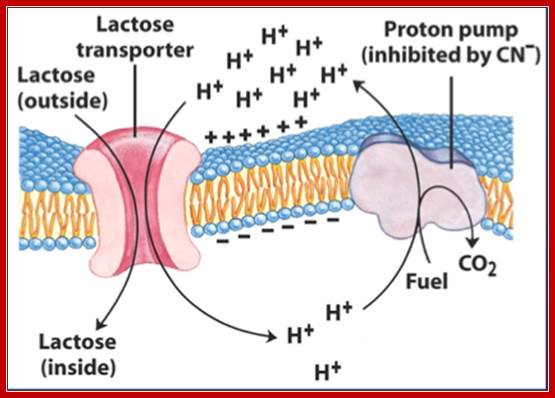
Lactose permease; mechanism of transport; http://www.studyblue.com/
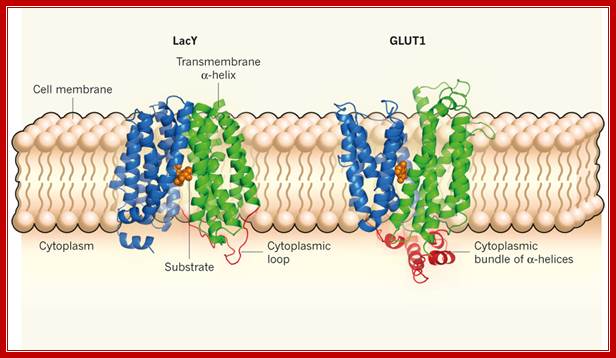
Lactose permease is transmembrane protein; http://www.nature.com/; http://www.rpi.edu/
����������� �����������
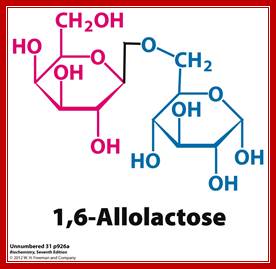
Allolactose β 1-6 linkage; http://www.pearsonhighered.com/
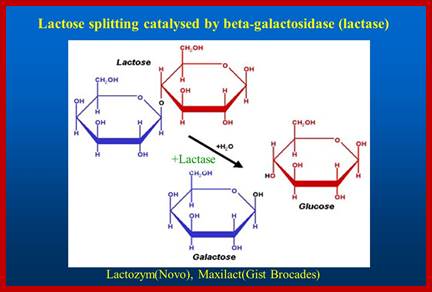
http://slideplayer.com
Lactose permease transports lactose from external media into the cell, where the lactose is converted to allolactose by the cellular Lactose isomerase. The allolactose induces the gene expression of lactose operon producing beta galactosidase, lactose permease and glucose transacetylase.
����������������������������
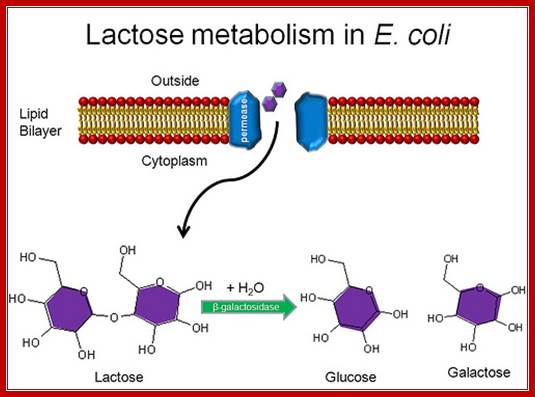
Transport of Lactose; Allen Gathmanhttps://www.flickr.com
Lactose hydrolytic products are metabolized as energy source.
Similar to Allolactose there are few other compounds which also acts as inducers, ex. Isopropyl-thio-Galactoside (IPTG) acts as a fortuitous inducer.� In bacterial cells IPTG is trans-acetylated by trans-acetylase (induced by lactose), it acetylates the 6� CH2OH group so as to form 6� acetyl, b 1� isopropyl thio-Galactoside.
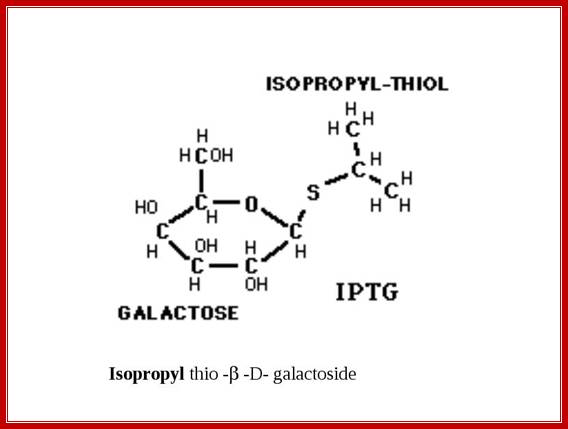
M/W Gerry Prody; http://shrdocs.com/
�����������������������������������
Another synthetic compound that can be used as a substrate is X-Gal, which is called as 5�-Bromo-4-Chloro-3-Indolyl b-D Galactoside. On hydrolysis of X-Gal by beta-Galactosidase, it gives intense blue color. This compound is used to find out whether the beta Galactosidase is expressed or not.
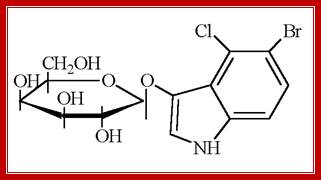
X-Gal: MagnetoWik; �ihttp://wiki.houptlab.org/

X-gal, when cleaved by β-galactosidase, yields galactose and 5-bromo-4-chloro-3-hydroxyindole. The latter then spontaneously dimerizes and is oxidized into 5,5'-dibromo-4,4'-dichloro-indigo, a deep blue colored product. http://parts.igem.org/;
When the cells are grown, in lactose or any other carbohydrate media without glucose, specific genes are expressed to utilize the given sources.� In absence of glucose, Adenyl cyclase becomes activated and it cyclizes ATP into cAMP, that in turn binds to inactive cAMP binding protein or CRP (cAMP Receptor Protein).� Binding of cAMP makes CRP active; which then binds to upstream activator region of the gene and bends the DNA so the CRP interacts with the CTD tail of alpha subunit of RNA Pol and makes the enzyme active, thus the operon is expressed.�
When such cells growing in non-glucose media is supplied with glucose, utilization of non-glucose substrates is suddenly stopped; this is called catabolitic repression.� This is because the glucose induces or activates cAMP phosphotases, which cleave cAMP.� So, no cAMP will be available and CRP remains inactive and so also the operons, this happens in spite of the RNA pol binding to their respective promoter sites.
Lactose operon:
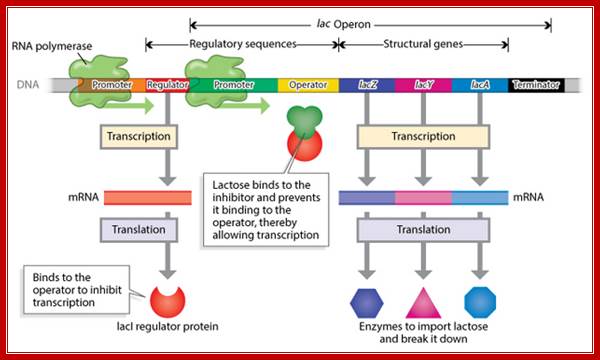
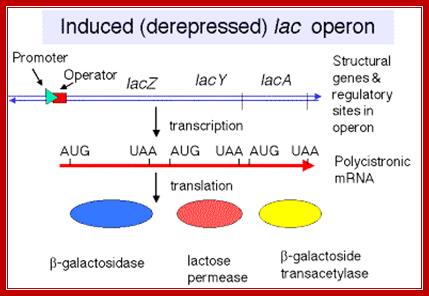
Utilization of lactose as an alternative carbohydrate source requires two enzymes, one Permease and the other beta Galactosidase. These two genes are linked to one another as two cistrons, but another gene Transacetylase is also linked to form polycistronic cluster of three.� This cluster is called Lac operon.
This operon consists of structural genes, βGalactosidase Z, Lactose permease Y and Transacetylase A, in the same order; each of these genes (cistrons) are separated by intercistronic spacers. The cell resident gene products such as permease transports Lactose into the cell. Similar resident enzyme converts beta Lactose into Allolactose.� The Allolactose binds to repressor and activates the expression of genes.
Overlapping on the left flank of the Z gene one finds promoter-operator segment.� The activator segment is found in the upstream of the -35 sequence.� Far away, from the lactose operon, in the upstream there is another gene called Lac-I, which produces a regulator protein called Lac-I a repressor.� The repressor as dimer (or tetramer) binds to operator sequences and prevents the RNA polymerase to be active in transcription.
Structural Features of Lac Operon:
http://biomoocnews.blogspot.com/
Remember, we don't want to expend energy for
things we don't need. Below is the size of the three gene products:
� β-Galactosidase 1,024 Amino Acids
� β-Galactoside Permease 418 Amino Acids
� β-Galactoside transacetylase 203 Amino Acids
Total Amino Acids, 1645. Using the assumption of 4 ATP per amino acid added to a protein, that makes 6580 ATPs needed just for protein synthesis. With this many amino acids, you are also looking at 4935 nucleotides (remember 3 nucleotides = codon = 1 amino acid), plus at least 3 stop codons. Each nucleotide added to a transcript takes the equivalent of 1 ATP, so you are looking at 4944 ATP minimum to make the transcript. Just to make one example of each protein (gene product), you are looking at 11,524 ATP. Do you just make one example of each protein? NO! Do you have though the idea that this is energy consumptive? Would you make it if there was no lactose around? http://biomoocnews.blogspot.com/

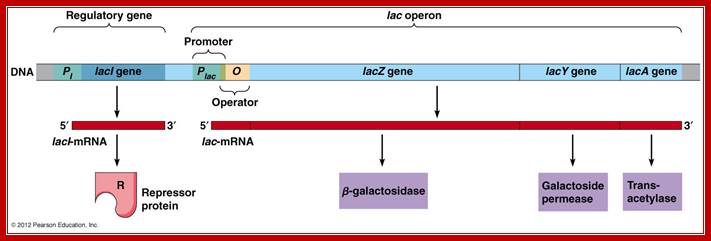
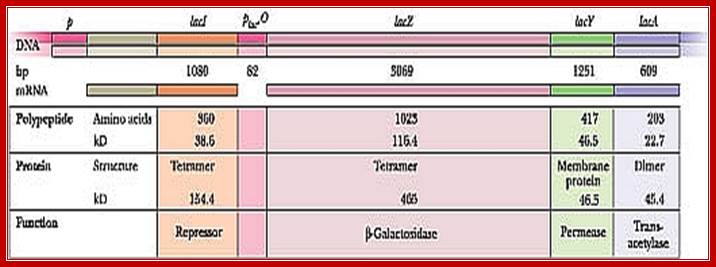
Diagram of a segment of an E. coli chromosome containing the lac operon, as well as the lac I coding region. The various genes and cis-elements are not drawn to scale; http://cubocube.com/; Principles of Cell biology; http://www.mun.ca/������
Lac I= repressor [32KD], tetramer,
Z= Beta-Galactosidase = [116kD], tetramer,
Y= Permease = [46kd]-, monomer,
A=Trans acetylase = [22kd], dimer,
|
Component |
Size (bp) |
Product |
Size (kDa) |
Function |
|
Lac-I |
1042 |
Repressor |
38 (360aa) |
Represses Lac-operon |
|
Promoter-operator |
82-112 |
|
|
Regulator regions |
|
Lac-Z |
3510 |
b-Galactosidase |
116 (1021aa) |
Cleaves lactose |
|
Lac-Y |
780 |
Lactose Permease |
30 (260aa) |
Transport of sugar into the cell |
|
Lac-A |
825 |
Trans-acetylase |
30 (275aa) |
Acetylates IPTG |
��� Note: a.a = amino acid
�
Bacterial consensus promoter elements are:
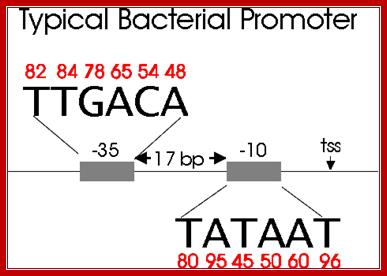
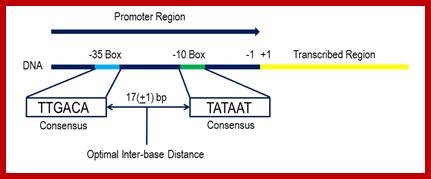
�Bacterial Promoter; https://www.oxfordgenetics.com
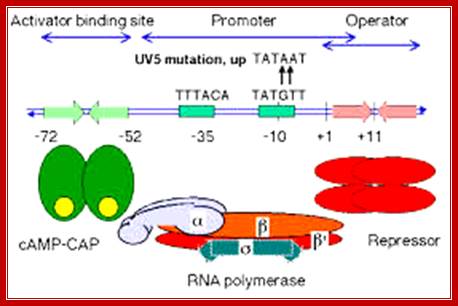
Lac Operon Promoter sequences: ��
(-)65�(-)55------ (-)35------ (-)10---------Start>
-65-72 = CAP-CRP binding site, GATGTG>�<CTCAC;
-35 RNAP alpha tail binds, sigma also binds, TTTACA,
-10 RNAP pribnow box, sigma also binds, TATGTT--+1>
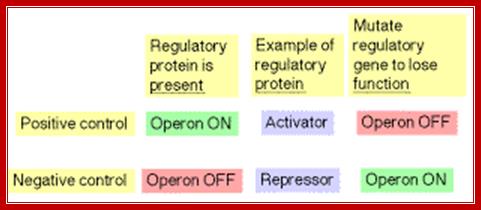
Lactose operon promoter sequence elements slightly deviate from the bacterial consensus sequence shown above.� Though RNAP with its sig70 binds to said sequences, it cannot be activated because of specific difference in Pribnow box and -35 sequences. In addition next to RNAP binding site another protein called Repressor is also bound, which hinders the RNAP movement, even if it is active.� There are two modes of regulation one positive and the other negative regulation. Negative regulation is operated by the repressor that binds to operator and blocks transcription, but positive regulation means gene expression is activated by a specific compound which removes the repressor from the operator.
http://www.personal.psu.edu/
-----35 GGCGCAA---17bp----10 CATGAT--- InR�
Lac Operon components; http://www.personal.psu.edu/
Lac operon Operator elements; http://www.personal.psu.edu/
�� 
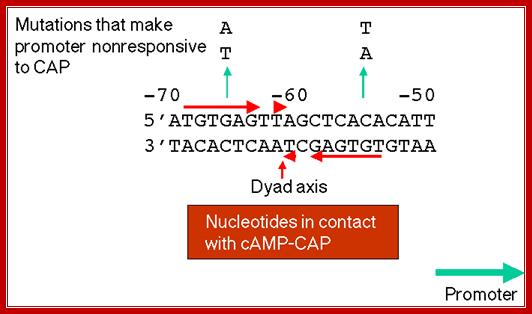
The upstream region at -65 to -55 region contains activator sequences.
When glucose is abundant, cells don�t produce any cAMPs and lactose utilizing enzymes are not made.� But the Lac-I gene is constitutively expressed, so the repressors interact with one another to produce tetramers, which are in active conformation and bind to operator region, as shown in the figure below.� This region is actually in the leader sequence of Lac-Z gene.
�
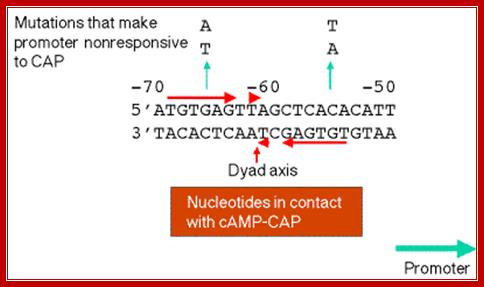
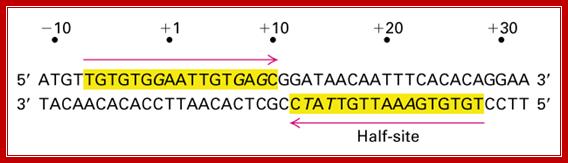
The operator region is made up of two half sites and they are palindromic; oriented in opposite direction and found in opposite strands.
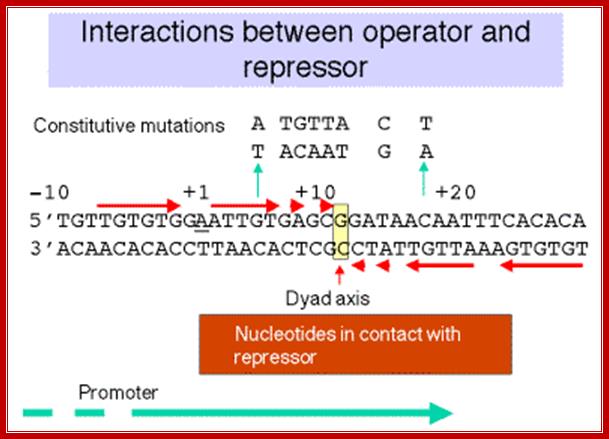
Lac Operon Operator sequences; http://www.personal.psu.edu/
On either side of the 17-18 bp operator one finds two more such palindromic sequences, one in the upstream of the first, another far downstream, -7 TGTGTG -3 ----//---+23 CACACA +28. When the repressor binds, it covers �6 to +30 region.� Such operator sequences are also found in the upstream of the main promoter and another downstream in the coding region.
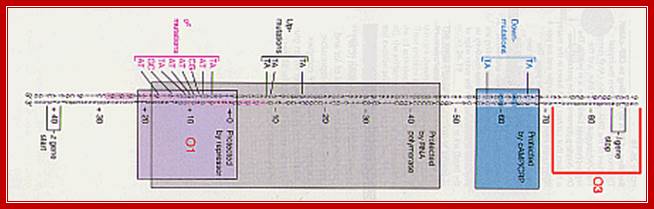
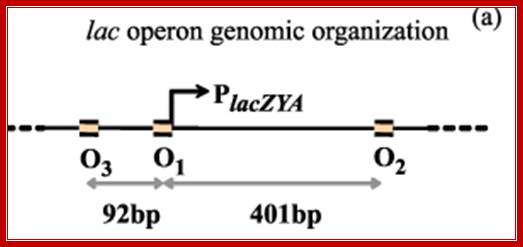
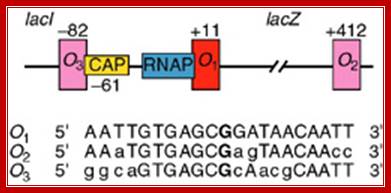
---- -82- O3--- (-) 7>---O1 InR+1---<(+)_10---+403- O2----
Operator half sites = O1--21bp>---(-10) to (+11)>- O2-<17bp (+410 to 430), and O3 -82 to -61
The Physics of Protein-DNA interaction networks in control gene expression, location operator sites; Leonor Saiz; http://iopscience.iop.org/
What you might not know is that the lac operator region is not as simple as shown in the above diagram. There are actually three lac operators. The one commonly shown in diagrams is called lac O1. The others are downstream (lac O2) and upstream (lac O3) from this one. O2 and O3 are called auxiliary operators. The true configuration is shown in the figure below: http://parts.igem.org/
The Physics of Protein-DNA interaction networks in control gene expression; Leonor Saiz; http://iopscience.iop.org/
Cartoon of the lac repressor (in gray with black contour line) bound simultaneously to the main (O1) and one auxiliary operator (Oid, with the sequence of the ideal operator), which are represented by white rectangles. Binding of the repressor to O1 represses the lac Z gene in the experiments. The simultaneous binding to O1 and Oid with free energies ΔGO 1 and ΔGOid, respectively, competes with the unfavorable process of forming a loop (ΔGl). By using this type of lac constructs, Muller et al measured the in vivo repression of the lac Z gene as a function of the distance between the centers of the O1 and Oid operators.
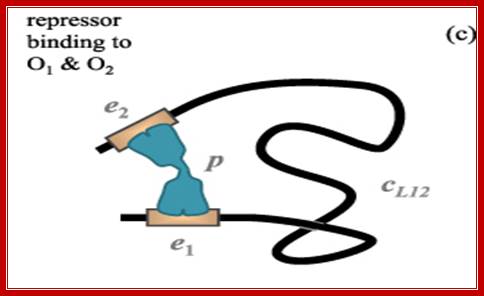
There are three repressor binding sites, you can call them as operators such as O1, O2, O3.� The O1 is at the start site, and the second O2 is located at 410 down stream of the start and the third O3 is at upstream - 83.� When repressors bind to O1 and O3 sites, where the O1 is a strong operator, proteins by protein-protein interaction they loop and produce strong repression.� The diagram below shows how the two-operator bound by repressors show looped configuration and tight repression.
The RNA polymerase binding requires a region from �55 to +15 or more (~10 bp).� Even in the presence of repressor and RNAP can bind the operator and TATAAT box including some part of the operator, it is squeezed; RNAP remains inactive, even if it initiates transcription, it cannot clear the promoter.� Thus, the repressor blocks transcription.� Such a control is called negative regulation.
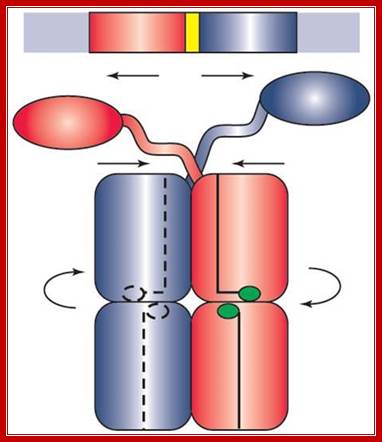
Getting a �Grip�; �Bringing Physics to Life�Lac I repressor when bound produces a loop and the repression is very strong; http://www.ks.uiuc.edu/
Repressor:
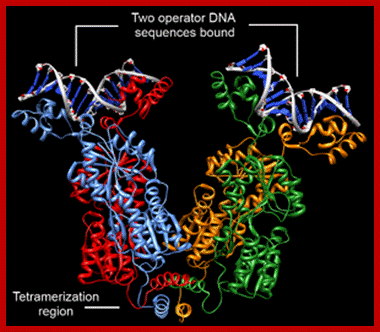
Lac Repressor forms a tetramer, with Allolactose; N-terminal Head like structure bids to specific sequences, in the hinge one van visualize the binding of Inducer; the C terminal domain is tetramerization domain; www.studyblue.com
It is 38kDa protein with 360 a.a It is a 38 KD protein with 360 amino acids.� It has two globular domains as core regions separated by a cleft.� The cleft is the site for the binding of inducer, which is Allo-lactose. But the N terminal region a globular structure binds to promoter elements.� The protein has DNA binding domain, protein-protein interacting domains and a ligand binding site; thus, it can become a tetramer.

������ Lac repressor domains; http://www.web-books.com
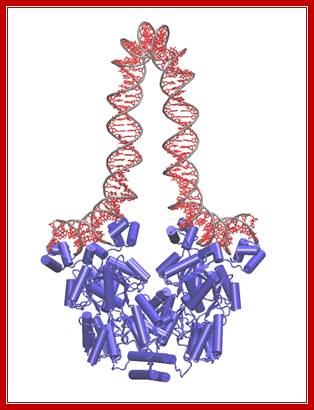
Tetrameric Repressor binding to their operator sequences produces DNA loop- Dimer-WIKIPEDIA;
� Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. �M. Lewis et al., 1996, Science 271 [5253]: 1247�1254. � 1996 American Association for the Advancement of Science.�
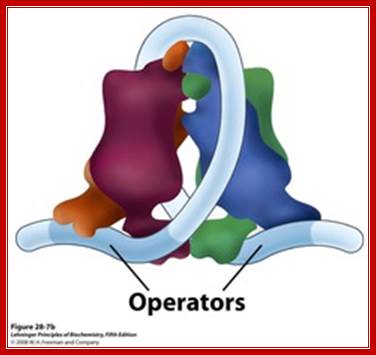
The Lac operon. The lac repressor binds to the main operator O1 and O2 and O3, apparently forming a loop, that might wrap around the repressor as shown in the figure; 11616580/chapter-28-slides-flash-cards; http://quizlet.com/
Lac repressor binds to its operator sequence in the form of a tetramer.� In the middle of the repressor protein, it has site for the binding of allolactose.
The tetrameric repressor in the absence of allolactose the dimeric protein binds strongly to O1 with their DNA binding motifs, but the other end of the dimer i.e the free end of the dimer interacts and become tetramer. The other end the tetramer has DNA binding domain, it too binds O2 but not so strong, thus the DNA loops around the tetrameric repressor. This type of repressor binding powers strong repressor activity.
At N- terminal region each subunits have helix-turn-helix motifs for the binding of protein to DNA, in sequence specific manner.� This is actually the region that makes contact with half sites of the operator, so two subunits bind to two half sites in the operator.�
The DNA binding domain of the repressor is connected to the core through a hinge, which is formed only when the protein is bound to DNA. Each of the core domains contains six parallel b sheets sandwiched between two helical motifs.�
The C-terminal also has two leucine rich heptad�s helices, which help in protein-protein interaction to form a tetramer
�LacI is a tetrameric protein, that can be thought of as a "dimer of dimers", connected by a bundle of 4 alpha helices. Each dimer consists of two 360 residue-long monomers tightly folded together and is morphologically divided into (1) a head group, which is the DNA binding domain, (2) a core which contains the lactose binding site between its N-terminal and C-terminal halves, and (3) the end helix that forms the 4-helix bundle when the protein is tetramerized. The helical bundle can act like a hinge for the dimeric "hands". LacI attaches itself to the genomic DNA so that each dimer binds a separate operator site. The DNA between the operators becomes folded into a loop. Several X-ray and NMR structures of LacI have been solved, but none of them contained all-atom information for the whole protein. We built an all-atom structure of LacI based on available PDB structures and molecular modeling and deposited the resulting structure to PDB�.
Among the four subunits in the tetramer, two actually bind to two half sites in the operator region, the other two are free on the other end of the repressor. This region can interact with another pair of repressors to form a tetramer, when bound to O1 and O2 sites produces a strong repression.
When the sugar binds, it binds with great affinity to the cleft region, so one molecule to each subunit at the cleft.�
Binding of Allo-lactose to repressor molecules induce conformational change in the protein, in such a way, the N-part of the protein easily dissociate from the DNA and the repressor falls off from the DNA, making it free from the blockade. And the RNA-Pol enzyme can now initiate transcription and progress onwards.
The mechanism of Lactose operon Regulation:
When bacteria are grown on glucose media, cells don�t produce any cAMP and lactose utilizing enzymes; yet cells produce cAMP binding proteins which remain inactive.
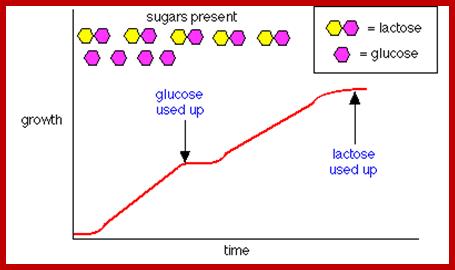
Cells grown on Glucose and lactose;
https://johnnydissidence.wordpress.com
Allolactose
IPTG
ONPG
X-Gal
A number of lactose derivatives or analogs have been described that are useful to work with the lac operon. These compounds are mainly substituted galactosides, where the glucose moiety of lactose is replaced by another chemical group.
- Isopropyl-β-D-thio-Galactoside (IPTG) is frequently used as an inducer of the lac operon for physiological work. IPTG binds to repressor and inactivates it, but is not a substrate for β-galactosidase. One advantage of IPTG for in vivo studies is that since it cannot be metabolized by E. coli its concentration remains constant and the rate of expression of lac p/o-controlled genes is not a variable in the experiment. IPTG intake is dependent on the action of lactose permease.
- Phenyl-β-D-galactose �is a substrate for β-galactosidase, but does not inactivate repressor and so is not an inducer. Since wild type cells carbon produce very little β-galactosidase, they cannot grow on phenyl-Gal as a energy source.
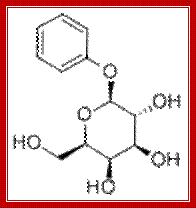
Phenyl b D Galactoside
- Mutants lacking repressor are able to grow on phenyl-Gal. Thus, minimal medium containing only phenyl-Gal as a source of carbon and energy is selective for repressor mutants and operator mutants. If 108 cells of a wild type strain are plated on agar plates containing phenyl-Gal, the rare colonies which grow are mainly spontaneous mutants affecting the repressor. The relative distribution of repressor and operator mutants is affected by the target size. Since the lacI gene encoding repressor is about 50 times larger than the operator, repressor mutants predominate in the selection.
- Other compounds serve as colorful indicators of β-galactosidase activity.
ONPG is cleaved to produce the intensely yellow compound, Orthonitrophenol, and is commonly used as a substrate for assay of β-galactosidase in vitro. Colonies that produce β-galactosidase are turned blue by X-gal (5-bromo-4-chloro-3-indolyl-β-D-galactoside).
- Allolactose is an isomer of lactose and is the inducer of the lac operon. Lactose is galactose-(β1->4)-glucose, whereas allolactose is galactose-(β1->6)-glucose. Lactose is converted to allolactose by β-galactosidase in an alternative reaction to the hydrolytic one. A physiological experiment which demonstrates the role of LacZ in production of the "true" inducer in E. coli cells is the observation that a null mutant of lacZ can still produce LacY permease when grown with IPTG but not when grown with lactose. The explanation is that processing of lactose to allolactose (catalyzed by β-galactosidase) is needed to produce the inducer inside the cell.
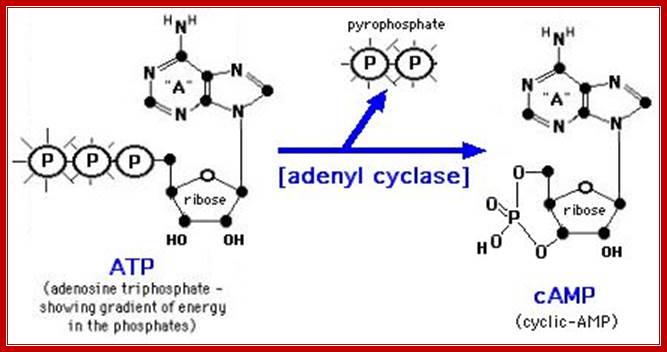
During activation Lac operon, rATP is converted to Cyclic AMP by adenyl cyclase which becomes active when glucose is absent. The binding of AMP to CRP protein makes it active and it binds to activator sequence of the upstream element of LaC operon. Mireille Capteaux; http://www.science-projects.com/
Cells produce Lac-I constitutively, which are repressors.� However repressor production is regulated by autonomous process.� The repressor is a tetramer and it has active conformation and binds to two half sites of the operator. �The RNA-pol holozyme, with its sigma 70, also binds to its promoter region, with some constraints; perhaps it is squeezed into the position.� The promoter elements at -10 and -35 in Lactose operon are not strong for tight binding of RNAP with its sigma 70 to initiate transcription by itself; this is in spite of the presence of correct sigma factor.� It is interesting to note sig-70 is an abundant factor involved in activating most of the house keeping genes. Perhaps the sigma does recognize the sequences, for the sequences are not consensus, so it cannot induce changes, thus fails to activate the RNAP.
Promoters are defined by sequences of base pairs upstream of the transcription start site. The RNA polymerase tends to recognize promoter sequences in which most of the base pairs match the promoter consensus sequence. The consensus sequence is a composite defined by the most common base to occur at each position. Base-substitution mutations can decrease or increase the efficiency of the promoter.
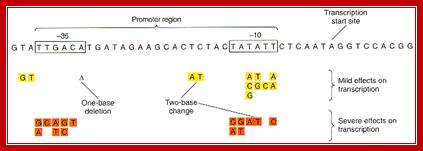
Bacterial consensus promoters include two regions of six base pairs each, at -10 and -35 bases upstream. However, no two promoters are exactly alike, and no promoter exactly matches the consensus sequence. Additional sites for environmental regulators can be found as far as -50 to -300 bases upstream. https://shwebook.com; http://www.gentarget.com; https://www.khanacademy.org/
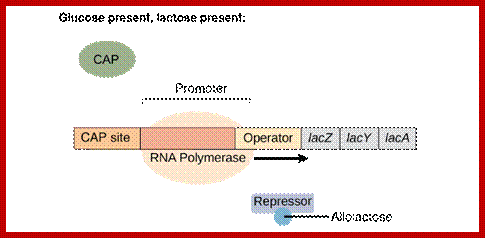
The diagram shows the sites for the binding of repressor, RNAP with sig7 and cAMP=CRP; https://bio.libretexts.org
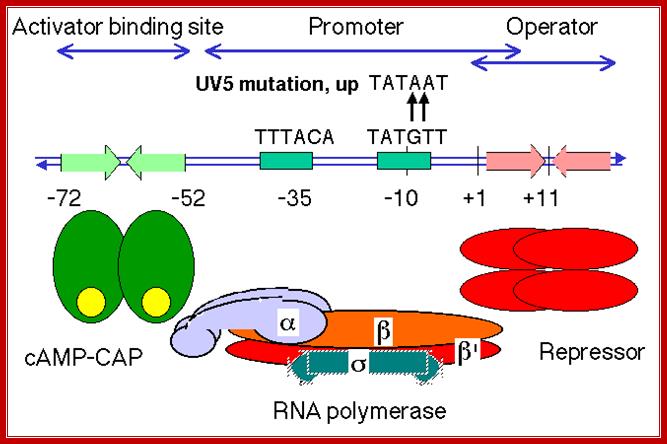
https://bio.libretexts.org
Presence of the repressor at operator region prevents the holozyme to initiate and translocate forward. When carbohydrate source is changed and provided with lactose, the residual permease as an exoplasmic enzyme present on the outer surface, transports�� lactose into the cell.
Few molecules of lactose transported into cell are converted by cellular enzymes into Allo-lactose.� The Allo-lactose is a correct structural form that can fit into its binding sites in the repressor.� The binding of one Allo-lactose to each of the monomers of the tetramer is cooperative and induces conformational change especially at N-terminal DNA binding domain and dissociates from the operator region.
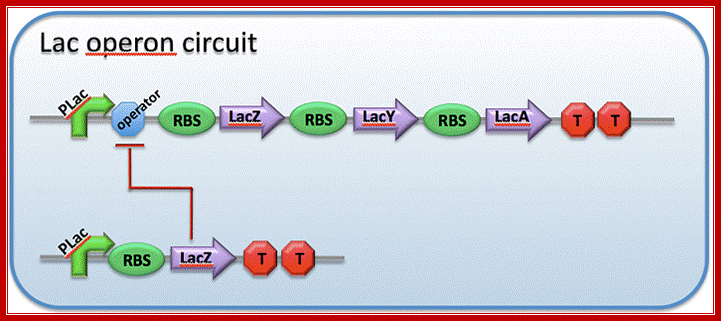
This diagram shows ribosome binding sites-during translation ribosomes bind to mRNA at the 5�end, once the translation of an a cistron over they get released then again bind to the intercistronic spacer and reinitiates translation; http://2009.igem.org/Team:Imperial_College_London
Each of the cistrons in mRNAs have an initiator codon and terminator codon at the beginning of the cistron and at the end of the cistron; http://www.personal.psu.edu/
Now the blockade is cleared off its obstacle, yet the RNAP-Holozyme cannot effectively initiate transcription.� The reason is that the enzyme binding to the promoter elements such as -10 and �35 sequences are not the strongest of the sequences for the enzyme to bind and undergo conformation changes like loose open binding complex to tight closed complex and then into tight open complexes for initiation. The -10 and -35 sequences are not consensus sequences; thus, the enzyme and sigma factors recognize the sequences, but poorly. So, the enzyme is still not active to transcribe at the expected rate.� That means it produces transcripts infrequently, not enough for the changed situations, where the enzymes are required in large numbers.
������������������������������������������������������������ �
Actually, from the experimental data, when cells are changed from glucose to lactose media, increase in the concentration of lactose utilizing enzymes takes place in few minutes, from just few molecules to several thousands.� Such a tremendous increase in rate and quantity is not possible from the RNA-Pol (Holozyme) that is sitting on the promoter, for the structural form is not in the best possible active state.
In the absence of glucose, cAMP is produced in large quantity.� cyclic AMP binding proteins, (CBPs or CAPs) that are already found in cells are activated by the binding of cAMP to them.� The cAMP binding protein is 209 a.a long with Mol.wt 22.5kDa. The cAMP-CAP complexes now join with each other to form dimers, which are in active state. These dimers now bind to ~ �72 to �55 sequences, called activator sequence.� The proteins have DNA binding domain, protein�protein interacting domain. The CTD ends of RNAP alfa subunits interact with CAP-cAMP. Binding of this activated CRP protein brings about a remarkable conformational change in DNA, it actually bends the DNA to 90˚ degrees or more, thus the CRP protein contacts the CTD tail alpha subunits of the RNAP enzyme.
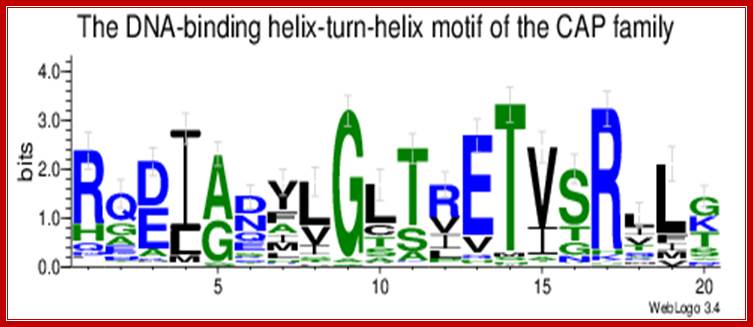

http://users.rcn.com/
CAP-CRP dimers bound to activator sequences;� Catabolite activator Protein as dimers with cAMP bind to the activator sequences and contact RNA pol through CTD domain and activate gene expression; http://weblogo.threeplusone.com/CAP consists of two identical polypeptides (hence it is a homodimer). Toward the C-terminal, each has two regions of alpha helix with a sharp bend between them. The longer of these is called the recognition helix because it is responsible for recognizing and binding to a particular sequence of bases in DNA.
The graphic shows a model of CAP. The two monomers are identical. Each monomer recognizes a sequence of nucleotides in DNA by means of the region of alpha helix labeled F. Note that the two recognition helices are spaced 34� apart, which is the distance that it takes the DNA molecule (on the left) to make precisely one complete turn.
This interaction between CRP and RNAP through a subunits of the enzyme induces the RNAP to an active state, from loose complex to tight closed complex and from tight closed complex to tight open complex, at which state, it can initiate transcription efficiently and clear the promoter efficiently, thus it can increase the efficiency of transcriptional initiation by several thousand fold. As the enzyme start elongation the sigma factor is released and another factor called NusA gets associated (an antitermination factor), thus transcription continues till the end of the gene.
![]()
The recognition helices of each polypeptide of CAP are, of course, identical. But their orientation in the dimer is such that the sequence of bases they recognize must run in the opposite direction for each recognition helix to bind properly. This arrangement of two identical sequences of base pairs running in opposite directions is called an inverted repeat. The operon; http://users.rcn.com/
CAP �CRP:
The recognition helices of each polypeptide of CAP are, of course, identical. But their orientation in the dimer is such that the sequence of bases they recognize must run in the opposite direction for each recognition helix to bind properly. This arrangement of two identical sequences of base pairs running in opposite directions is called an inverted repeat.
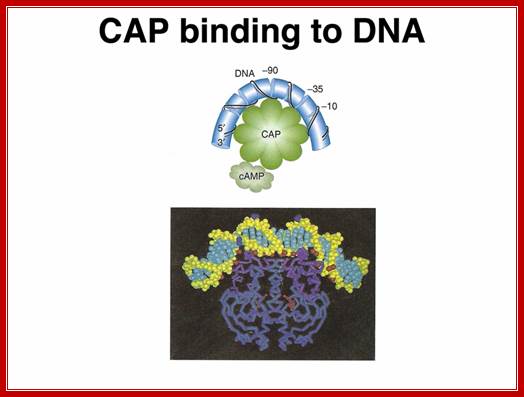
www.discoveryand innovation.com
The CAP dimer bends the DNA by about 90� in its complex. CAP
has a helix-turn-helix motif and a cAMP binding domain.
(Campbell p. 290); https://www.bio.cmu.edu
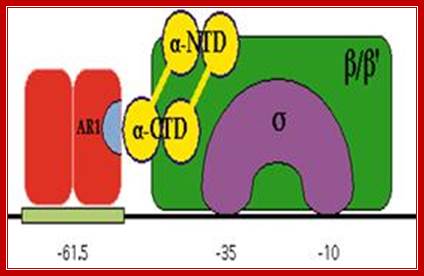
�����������
����������������������� RNAP when binds, it occupies almost -50 to +20
Activator bound to -61 to (-) 65 bp; region interacts with CTD tail of RNA pol and activates the enzyme. (mBio)
Molecular mechanism of negative auto regulation of Escherichia coli crp gene.
A Hana Mura and H Aiba
Institute of Biological Sciences, University of Tsukuba, Ibaraki, Japan.
* This article has been cited by another articles in PMC. Abstract*
Transcription of the Escherichia coli crp gene encoding cAMP receptor protein (CRP) is negatively regulated by CRP-cAMP complex that binds to a specific site located downstream from the transcription start site. The binding of CRP-cAMP to this site activates transcription from a second divergent overlapping promoter. The mechanism of this negative auto regulation of the crp gene has been investigated by in vitro transcription, gel shift, DNase I foot printing, and exonuclease III protection assays. Authors demonstrated that the crp and divergent promoters are reciprocally and coordinately regulated by CRP-cAMP. The abortive initiation assay revealed that the divergent RNA itself is not required for the inhibition of crp transcription. Detailed binding studies revealed that CRP-cAMP stimulates the binding of RNA polymerase to the divergent promoter and thus blocks the occupation of the crp promoter by RNA polymerase.
Elongation requires the assistance of nus A and other anti terminator proteins to overcome stalling and other obstacles.
Chain termination:
Chain termination takes place in rho-independent manner most of the time.
Here, at the end of the gene, the transcript has a sequence, which is a palindromic sequence with six Us in the 3� end. Palindromic sequence generates a stem-loop secondary structure, which interacts with the RNAP.� The enzyme stalls for a while.� But the strength of 4-6 Us base paired with As in the DNA is so week, the RNA falls off by itself. �This is further aided by the rewinding of the DNA from behind.� Thus, transcription by the enzyme stalls for a while Thus transcription is terminated.
Regulation of lactose operon employs positive regulation and negative regulation as well. Negative regulation implies that transcription by RNAP is blocked and it has to be removed.� The positive regulation means that a factor directly interacts with the enzyme or its associated proteins and activates the enzyme to its full efficiency.
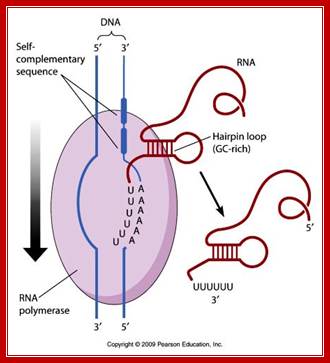
Terminal region of the mRNA generates stem loop structure with terminal Us, leads to the release of mRNA as well as RNAP; http://www.mun.ca/
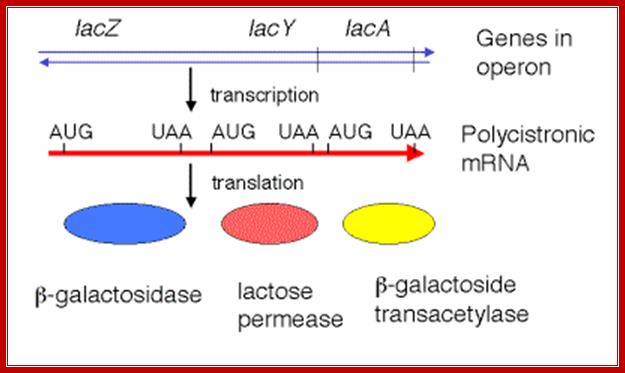
The transcript is poly cistronic and it is translated from the 5� end of the Z segment and falls off at the end of the Z transcript and reinitiates in the next segment Y, then it is reinitiated at A cistron.� So most of the times Z product i.e. beta Galactosidase is produced in comparison to other two products.� Between cistron one finds a spacer which facilitates the ribosome dissociate at TER codon and reassociate at Initiator codon of the next cistron.
The mRNA transcribed is polycistronic.� Translation always produced more of beta-galactosidase, because the first cistron is beta-galactosidase.� The least amount of protein produced is transacetylases for it is the last to be translated.� Between cistrons the mRNA contains spacer, which provides space for termination and also contain Shine Delgarno sequences just upstream of the start codon of each of the cistrons; thus each cistron is independently translated.
Regulation of LacI gene.
Lac repressor- autoregulation:
The Feedback Regulation of Lac Repressor Expression 1 in Escherichia coli
Stefan Oehler*June 2009
Negative feedback regulation, mediated through repressor binding site O3, which overlaps the lacI gene, could explain robustness of the weak expression of Lac repressor. Significant auto repression of Lac repressor has never been ruled out. In the present work, the degree of auto regulation of Lac repressor was determined. It is negligible.
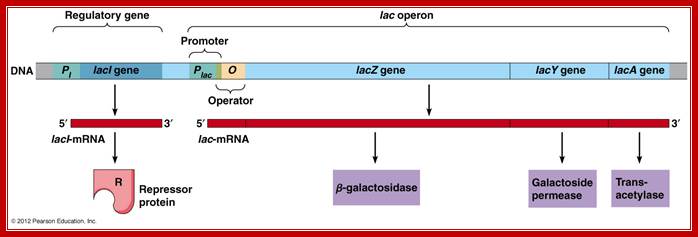
A general view of the Lac operon showing regulator gene LacI and other three genes and operator elements; http://www.mun.ca/
The lac operon is one of the classical model systems for transcriptional regulation. Expression of the tricistronic lac mRNA is negatively controlled (repressed) by the operon-specific Lac repressor (LacR) and positively controlled (activated) by the global regulator CAP. In recent years, the lacoperon has become a focus and testing ground for Systems Biology analyses.� Such, modeling can only deliver meaningful results when it incorporates all relevant features of the system The textbook picture of the lac operon has sustained substantial change over the years, and despite the wealth of information, many questions are still unanswered.
One of the traditional views that have become questionable holds that the lacI gene, encoding LacR, is constitutively expressed at low levels (in wild-type (wt) cells, LacR is present at about 10 tetramers per genome. It was established in the 1960s that Lac repressor expression is not coordinately controlled with the three genes of the lac operon. This did however not exclude auto regulation of LacR altogether. Indirect methods suggested that LacR auto regulation cannot be large. Gilbert and M�ller-Hill did not find an obvious difference between the yields of LacR purified from uninduced and induced cultures. Their recoveries were, however, not reproducible enough to detect a small several fold difference. An in vivo experiment with an E. coli strain temperature-sensitive for the production of LacR actually indicated a 2-fold inducibility of LacR expression, but the mutation used was not well characterized enough to allow conclusive interpretation of the data. Expression of the lacI gene is low but robust: there appears to be little stochastic fluctuation of LacR. Feedback regulation is a mechanism that would suppress such fluctuations. It was found that the third lac operator, O3, which lies upstream of the lac promoter and overlaps with the last nucleotides of the lacI gene, is not a pseudo-operator. It contributes through DNA loop formation to repression at the first lac operator, O1. It was later reported that transcription of lacI stalls at an occupied O3 and that the incompletely translated protein is tagged for degradation through the tmRNA pathway. LacR expression seemed to subject to negative feedback regulation, the extent of which could, however, not be determined from these data. This circumstantial evidence was so convincing that it has been Lac repressor auto regulation stated as a fact that 1 LacR is auto repressed.
LacI gene expression is auto regulated in the sense when there is excess of repressor it binds to its own promoter-operator elements and shuts off the expression of cI protein, but when the inducer concentration is low the repressor is produced.� But the expression of this and regulation of this is a kind of flip-flop.
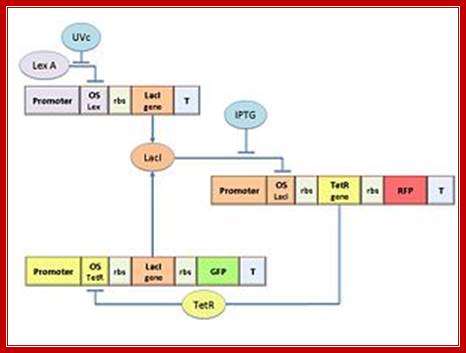
The molecular circuit in figure shows switch
between two different stable states (LacI-ON and TetR-ON), driven by two
external stimuli (UVc and IPTG). LacI-ON represents the stable state in which
LacI gene is active and LacI protein represses the TetR gene expression, with positive
feedback. Therefore, the LacI-ON state coincides with the TetR-OFF condition.
On the contrary, the TetR-ON represents the state with the TetR gene active and
the LacI gene silenced (LacI-OFF). Owing to the coexistence of two stable
states (bistability), this circuit is capable of serving as a binary memory
[Collins et al. 2000]. We denominated it a Flip-Flop since it works as a SR Latch:
LacI state is the ![]() output
and Tet-R state is the
output
and Tet-R state is the ![]() output.
output.
http://2008.igem.org/
LacI state is the ![]() output
and TetR state is the
output
and TetR state is the ![]() output.
Uvc is the set signal and IPTG is the reset signal. Indeed, IPTG stimulation
inhibits LacI repressor, thus can cause the transition from the LacI-ON state
to the TetR-ON. UVc radiation, inactivating LexA repressor through the SOS response [Friedberg et al. 1995] can
cause the opposite transition from LacI-ON to TetR-ON.
output.
Uvc is the set signal and IPTG is the reset signal. Indeed, IPTG stimulation
inhibits LacI repressor, thus can cause the transition from the LacI-ON state
to the TetR-ON. UVc radiation, inactivating LexA repressor through the SOS response [Friedberg et al. 1995] can
cause the opposite transition from LacI-ON to TetR-ON.
The core elements of the genetic program are two mutually regulated promoters. The promoter transcriptional strength and the repressor binding affinity to the promoter determinate such relevant circuit properties as the bistability and the response to inputs. To quote the promoters in terms of strength and sensitivity to repressor we resorted to the model-based analysis and numerical simulations.