Regulation of Hut and Histidine Operons:
And other related Operons.
Hut Operons:
Salmonella typhimurium causes food poisoning.� When nitrogen and carbohydrates are deprived of their sources, its hut operon gets activated.� The activated system produces enzymes, which uses freely available Histidine as a source of nitrogen.� It is a unique mechanism. Hut means Histidine Utilizing. Histidine operon means all the genes responsible for the synthesis of Histidine from a precursor.
����������������������� ����������� ����������������������������������������������� �����������
Histidine--H-->Uroconate---U--->Imadozole propionate---I--->
--->Form-immino glutamate--G->Glutamate
Gene: H, enzyme= histidinase,
Gene: U, enzyme= Urokinase,
Gene: I, enzyme= Imadozole propionate hydrolase,
Gene: G, enzyme= Form-immino glutamate hydrolase.
Glutamate, Formate and Ammonia are used as metabolites. The whole organization of the operon including the regulator is unusual.� The genes for G and I are under the control of one Promoter cum operator, and U and H are under another promoter cum operator.� Located in between these operons, Hut repressor gene is present with its own promoter.
When carbohydrate and nitrogen sources are available, the Hut repressor is constitutively expressed.� The hut repressor binds to both operators of I and G and U and H and blocks the transcription.
In the absence of glucose, adenine cyclase becomes active and by its action it generate adequate amounts of cAMP, which in turn binds to CRP; this complex binds to the activator region in the upstream of the promoter.�
Though RNAP complex is bound to their respective promoters, for its activation it requires an unusual component, non-adenylated glnA-synthase.�
In the absence of (CH2O) n and N2 sources and in the presence of Histidine, the amino acid Histidine binds to the repressor; the binding induces conformational change in the repressor and repressor falls off the operator of each of the operons.�
� By quirk of the nature�s design the hut repressor complex binds to its own promoter and blocks the production of hut repressor.
Once the repressors are removed from the operator, both gene clusters are expressed to produce the respective enzymes, which metabolize the Histidine and use the products.�
� This is an excellent example, where an enzyme is involved in activating an operon.� The non-adenylated form of Gln-synthase acts as an activator, but not the adenylated form of Gln-synthase.� But the activated form of Gln-synthase called Gln-synthase-A is responsible for the synthesis of glutamine from Glutamate.�
Active glutamate is adenylated or simply called Gln-A and non-adenylated as Gln.� And it is the non-adenylated form of Gln synthase is responsible for activation of Hut-operon.
��� Crp� P�� O��� I���� G������ Hu-R����� O ��P�� crp� P�� O��� H��� U�� L�� G�� M
---I�I------I---I--------I---------><-------I�--I-----I---I---I-----I---I----I---I----I-->
����� Gln���������������������������������������������������������� ������������ Gln
http://www.aist.go.jp/
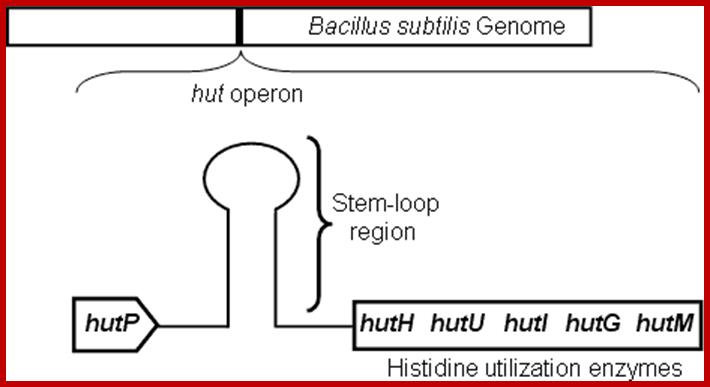
Schematic representation of the hut operon showing the arrangement of the hutP, terminator/antiterminator region (stem-loop), and structural genes; .http://www.aist.go.jp/
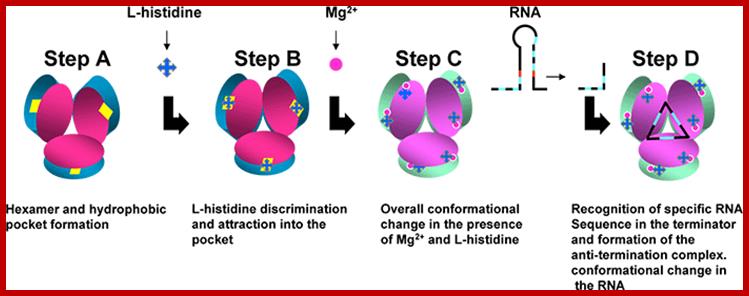
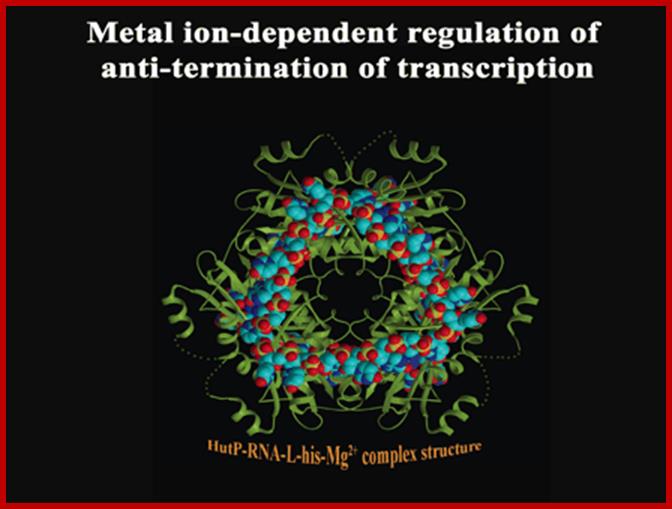
A schematic model proposed for Hut P antiterminator complex formation; http://www.aist.go.jp/
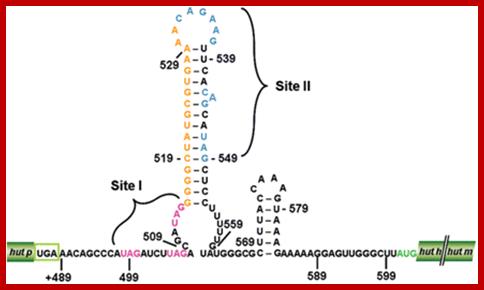
The proposed hut mRNA-terminator structure. Stem-Loop region of Hut operon; http://openi.nlm.nih.gov/
Histidine Operon:
Histidine is an important and an essential amino acid.� It is synthesized in cells using an elaborate biochemical step; starting from Phospho ribosyl pyrophosphate (PrPP) to L Histidine.� It requires nine to ten steps and nine to ten enzymes, and it means nine to ten genes. Interestingly most of the enzymes are monomers.�� All the genes are clustered into one operon under the control of one promoter-operator.� This type of organization is found, in both E. coli and S. typhimurium.
E.coli
��Organization genes in His operon in E.coli k12:
P1-O-> HisL-G(1)-D(10)-C(8)-p2-B(7/9)-H(5)-A(4)-F(6)-p3-I/E(3/3)
Escherichia coli K-12 substr. MG1655 Transcription Unit: hisLGDCBHAFI
�(-)35---(-10)p.box-(1)+1>(His-L) pppAUC----leader-----20ATG----uuuTer-160-170--> next ATG-at+1�for the first Gene (G) in the histidine operon----it has long translatable leader called hisL, where it contains attenuator sequences.
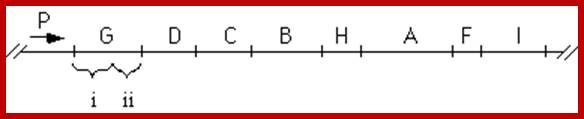
The order of genes (cistrons) in Histidine Operon. http://www.sci.sdsu.edu/
Numerical 1 to 8 indicates the steps in biochemical pathway in the synthesis of histidine.� But the order of genes in the operon not the same as the genes involved in synthesis. The number of citrons required for Histidine biosynthesis vary.
Transcription starts at +1 at and ATG in the leader starts at 20 and the leader terminates at +160-170� (leader sequence), and at 228 another ATG for histidine operon starts. This region acts as attenuator sequence.
E.coli Histidine Operon:
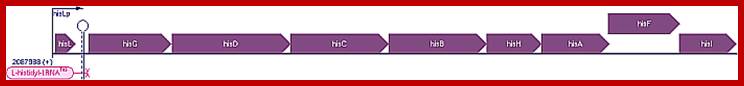
Gnetic map of E.coli histidine operon (8cistrons)
Organization of Histidine Operon in Salmonella typhimurium;
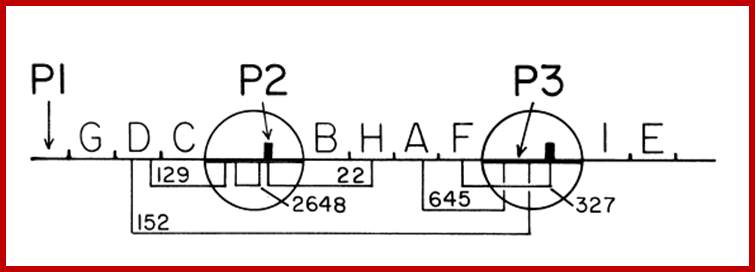
Genetic map of His operon in S. typhimurium with two internal promoter elements P2 and P3; P1 is the main promoter (9cistrons).
����������������������������
---(35)--(-10)TTTAGGTTA� +1ppp---ATGAAATG----GGCTTTTT-ter-//ATG-
Translated Leader: (His L) 5�ppp--M.T.R.V.Q.F.K.H.H.H.H.H.H.H.P.D--ter�M
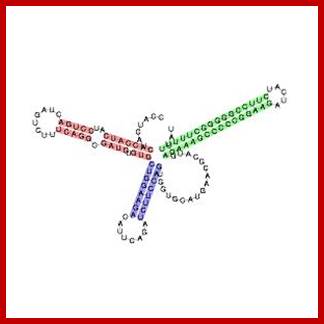
Histidine leader sequence conformation, in fact the last part of the leader i.e attenuator region assumes tRNA conformation. http://en.wikipedia.org/
Pathway- sequential reaction from pRpp to Histidine-E.coli and S. typhimurium:
�1. pRpp + ATP� phosphoribosyl ATP
(ATP phosphoribosyl transferase)- his (G)
2. N�-5-Phosphoribosyl ATP + H2O� Phosphoribosyl-AMP + ppi
(PPR�ATP pyrophotase)-his (I�)
3. N�5-Phosporibosyl-AMP +H2O� -> Phosphoribosyl-formiminoAICAR-P (AICARP= 5-amino imidazole-4-carboxamide ribonucleotide-p.
(Phosphoribosyl AMP cyclohydrolase) his (E��) (I?)
4.Phosphoribosyl-formiminoAICAR-P->Phosphoribulosylformimino-AICAR-P
Phosphoribosylformimino-5-amino-1-phosphoribosyl-4amidazole (Carboximide isomerase)-his-A
5.Phosphoribulosyl-formimino-AICAR+glutamate->D-erythroimidazole glycerol-P
(Imidazole glycerol phosphate synthase) his H and� his F
6. D-erythroimidazole glycerol-P -> Imidazole acetol -P
(Imidazoleglycerol phosphate dehydrtase) �his(Bc)
7. Imidazole acetol �P-> L histidinol-P
(Histidinol phosphate aminotransferase)-his (C)
8. Histidinol-P > Histidinol
(Histidinol phosphotase)-his B���
9. L histidinol--> Histidinal
( Histidinal isomerase) his D�
10. Histidinal-> Histidine
(Histidinal dehydrogenase) �his (D��)
P-L�G-------D------C-----B-------H------A------F-----I/I
����� 738 1065��� 1071� �1077� �1299����� 734 ��� 897 582/612���
�����
�36.8��������������� 36������� 38������� 39������� 39������� 27������� 41������� 28/30
These are the Genes in sequence of reactions.� In structural organization of individual gene in the operon, the first gene in the operon is gene-2 and the last is gene-1.
These are the sequence of reactions.� In structural organization of individual gene in the operon, the first gene in the operon is� gene-2 and the last is gene-1.
|
Gene |
Enzyme |
Name of the enzyme |
|
Gene-(G) |
Enzyme -1 |
PrPP pyro phosphorylase |
|
Gene-(I) |
Enzyme-2 |
Pr-AMP pyro phospho hydrolase |
|
Gene-(I) |
Enzyme-3 |
Hydrolase |
|
Gene-(A) |
Enzyme-4 |
Isomerase |
|
Gene-(F/H) |
Enzyme-5/6 |
Imidazole glycerol �P-synthase |
|
Gene-(B/c) |
Enzyme-7 |
IGP dehydratase |
|
Gene-(C) |
Enzyme-8 |
Histidinol-P aminotransferase |
|
Gene-(B/n) |
Enzyme-9 |
Histidinol phosphotase |
|
Gene-(Dn/Dc) |
Enzyme-10 |
Histidinol dehydrogenase |
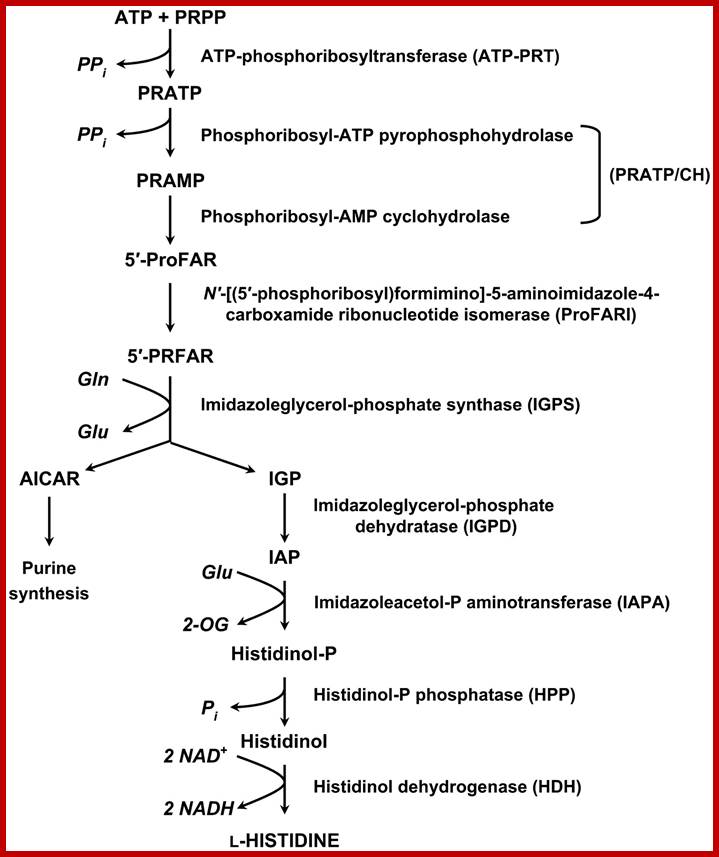 ����������������������� Histidine
Biosynthetic Pathway; Nine enzymes required; http://www.plantcell.org/
����������������������� Histidine
Biosynthetic Pathway; Nine enzymes required; http://www.plantcell.org/
�����������������������������������������������������������
Size of Genes and their products:
|
S.typhimurium pathway- Genes of His operon |
S.typhimurium Gene Size (bp)/ Protein Mol.wt (KD) |
|
E.coli operon gene sequence |
E.coli) Size(bp) |
|
|
G |
897/36.8 |
|
G |
738 |
|
|
I |
612/ |
|
D |
1065 (D1,D2) |
|
|
I |
582 |
|
C |
1071 |
|
|
A |
735/27 |
|
B |
1077 |
|
|
H / F |
10689/39 |
|
H |
1299 |
|
|
F |
774/41 |
|
F |
897 |
|
|
B |
1068/39 |
|
A |
774 |
|
|
C |
1071/68 |
|
F |
897 |
|
|
B |
1068/39 |
|
I |
582 |
|
|
D1 |
1065/75 |
|
|
609 |
|
|
D2 |
609 |
|
|
|
|
The leader sequence has 4 blocks with intra strand complementarity, thus they can form 4 stem loop structure, such as 1, 2, 3 and 4.
When His is low or absent translation stops at H codons, and 2 and 3 block base pair, thus allow transcription to continue.� If His is present translation proceeds beyond his codons and terminates in the second loop, this provides the formation of 3 and 4 form base pairing, which generates transcription terminator stem loop with uuuuu 3�ends
Some of Operons leader sequences used for attenuation:
Trp����������������� -14-���� MKAIFVLKGWWRTS
Phe-A������������ -16-����� MKHIPFFFAFFFTFP
His����������������� -16-����� MTRVQFKHHHHHHHPD
Thr���������������� -21-����� MKRISTTITTTITITTQNGAG
Leu���������������� -28-����� MSHIVRFTGLLLNAFIVRGRPVGGIQH
Ilv������������������ -32-����������������������� MTALLRVISLVVISVVVIIIPPCGALGRGKA
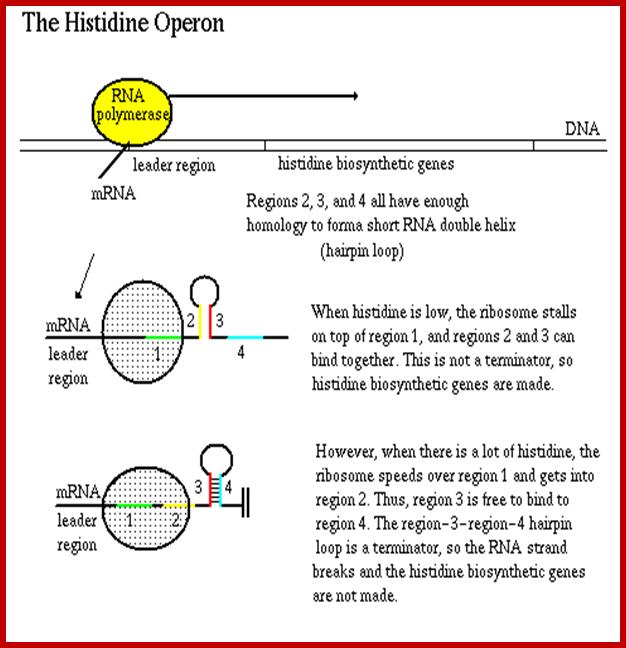
Attenuator sequences in histidine Operons
http://biocadmin.otago.ac.nz/
Enzymes in sequence:
G. PRPP �Phosphoribosyl transferase,
I.� Phosphoribosyl-ATP pyrophosphotase,
I.� Phosphoribosyl-AMP cyclohydrolase,
A.Phosphoribosylformimino-5-amino-1-phosphoribosyl-4-imidazolecarboxamide Isomerase,
F and H. Imidazole glycerol phosphate synthase
B. IGP dehydratase,
C. Histidinol phosphate aminotransferase,
B. Histidinol phophotase,
D. Histidinol dehydrogenase,
D. Histidinol dehydrogenase,
Expression of His operon enzymes are analyzed on� SDS- PAGE.
https://www.studyblue.com; www.cas.miamioh.ed
The Histidine operon is regulated similar to Tryptophan operon by attenuator mechanism.� Before the start of the first cistron the transcript has a long leader sequence, which can generate two stem loop structure, one of (4th) which contains stem loop with UUUU sequences.� This develops when Histidine present and the ribosome progresses through Histidine codons and stops at a terminator codon, this generates terminator or attenuator stem loop.� But when Histidine is absent the translating ribosome stops at �his� codons for the lack of his carrying tRNA, this generates hairpin loop between the 2nd and the 3rd part of the leader sequence, thus transcription is not terminated.� The attenuator mechanism is more or less similar to that of Tryptophan operon.
Arginine Operon:
P/O�A2-G7-C5-D6-E8-F1-I3-H4;
Gene products operate the pathway from enzyme 1-to 8
Arginine operon; http://microbiology.okstate.edu/
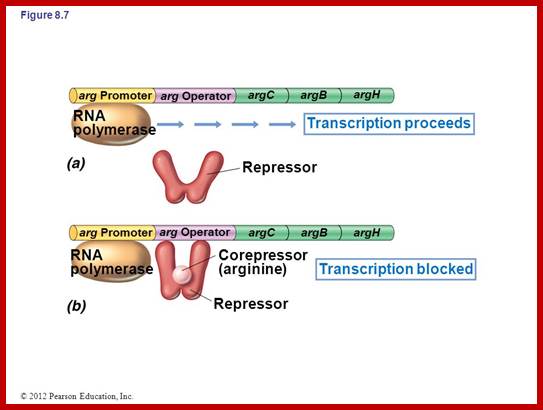
Regulation of Gene Expression:
(a) Transcription of the
operon occurs because the repressor is unable to bind to the operator.
(b) After a corepressor (small molecule) binds to the repressor, the repressor
binds to the operator and blocks transcription; mRNA and the proteins it ;
http://slideplayer.comencodes
are not made. In the case of the argCBH operon the repressor would be the
arginine repressor and the corepressor would be the amino acid arginine. http://202.204.115.67/
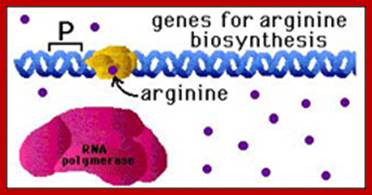
Transcription of Arginine operon; E. coli does this with a negative regulatory protein (a repressor protein) that binds to DNA near the promoter of the arginine genes. When there is no arginine present, the repressor is in its inactive conformation, and cannot bind to DNA. This means that the genes for arginine biosynthesis can be transcribed and translated. https://courses.cit.cornell.edu
![Schematic representation of the operon organization and regulation of the arginine metabolism and transport genes. Genes are represented by boxes. ARG boxes in the upstream region are shown by black arrows. The direction of the arrow indicates the direction of transcription. The linear pathway (in E. coli and V. cholerae) involves N-acetylglutamate synthase (argA) and N2-acetylornithine deacetylase (argE). The circular pathway (in other bacteria) involves N2-acetyl-L-ornithine: L-glutamate acetyltransferase (argJ). The common genes are acetylglutamate kinase (argB); acetylglutamate semialdehyde dehydrogenase (argC); acetylornitine delta-aminotransferase (argD); ornithine carbamoyltransferase (argF, argI); argininosuccinate synthase (argG); argininosuccinate lyase (argH); carbamoyl-phosphate synthase (carAB). The H. influenzae genome contains only argH, argG, argF and possibly argD orthologs. There are difficulties in identifying orthologs for argC, argJ and argB in D. radiodurans because there are several paralogous genes encoding proteins that can possibly perform these functions. The B. subtilisroc operons involved in arginine degradation are also regulated by AhrC, as well as anaerobic arginine catabolism genes arcABCD in B. licheniformis [14] (data not shown). The transporter genes are: periplasmic binding protein (white), permease transmembrane protein (light gray), ATPase component (dark gray).](Gene_Expression_I5E-Regulation_of_Hut_And_Histidine_Operons_files/image015.jpg)
Conservation of the binding site for the arginine repressor in all bacterial lineages: http://openi.nlm.nih.gov/
Biosynthetic Pathway
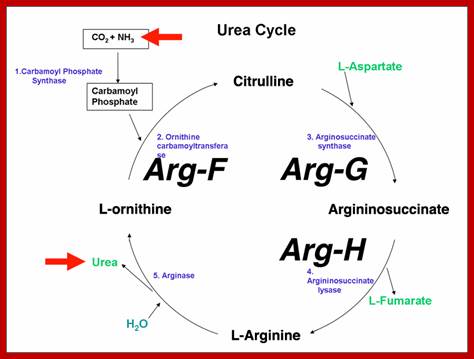
Inborn errors of the urea cycle are known in humans. We do not initially recognize these as arginine auxo trophy (arginine is a nonessential amino acid in normal humans), but rather as disorders in the disposal of waste nitrogen. We will use ornithine trans-carboxylase deficiency as an example. Ornithine trans-carbamylase (aka ornithine carbamoyl transferase) is encoded by the OTC gene, which is sex-linked. People with mutations in this gene lack the ability to convert ornithine to citrulline, the reaction that imports ammonia into the urea cycle. http://www.discoveryandinnovation.com
Arginine biosynthetic genes are scattered in different clusters, but they contain the same or similar Promoter operator box called ARG-Box.� This Arginine synthesis by different clusters by the same repressor activators, it is often termed as Arg Regulon.� Arginine acts as a co repressor and the repressor; it exists as a hexamer.� There are ~500 hexamer per cell.� Activated Arg receptor binding affinity to its operator is very strong.