Eukaryotic Gene Expression:
Introduction:
Structural organization of eukaryotic genes is more complex than prokaryotes. The regulation of gene expression is much more evolved and complex. Different types of genes are transcribed by different RNA-polymerases; there is a kind of division of labor among themselves. RNA-polymerase-I transcribes Ribosomal RNA genes, RNA-polymerase–II transcribes all structural genes and few sn-RNA, small interfering RNAs and sno-RNA genes, and RNA-polymerase III transcribes tRNA, 5s RNA, 7s L-RNA, 7Sk RNA, U6 RNA genes, and few others.
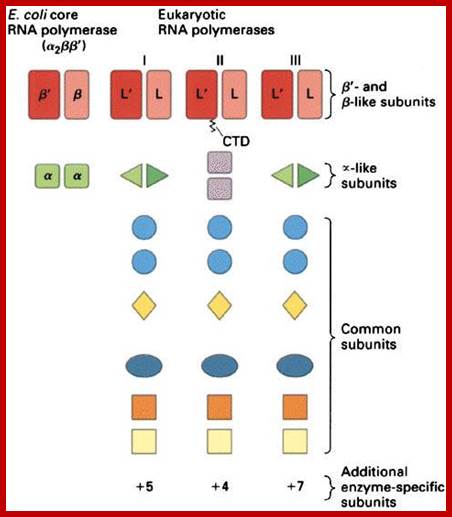
· Unlike prokaryotic, eukaryotic enzymes have an elaborate number of subunits and each of them differs in their characteristic features and functions.
These three types, in spite of many subunits as components, they can easily be distinguished by their sensitivity to α-Amanitin, a fungal poison. RNAP-I is insensitive to Amanitin, RNAP-II is very sensitive to low concentration of the drug. The RNAP’-III activity can be inhibited only at high concentrations of α-Amanitin.
· Millions of years of natural selection and evolution have fixed RNAP's subunits and their accessory proteins for specific function and the kind of genes they transcribe.
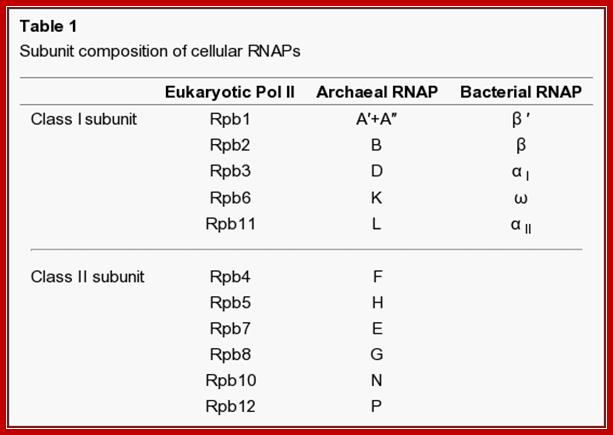
· The Archaea organisms too have similar RNAPs but not the same.
Parallel evolution can be observed in the structural organization of DNA segments at the promoter, activator and other upstream regions and also the number and kinds of transcription factors in the form of general, specific to tissues, specific to stimulus and specific to developmental stages.
· It is amazing as well as difficult to comprehend how this elaborate process has been designed and executed to perfection; and the process is still going on. Understanding the microcosm of the cell is as fascinating as the external macrocosm.
An
Introduction to the Control of
Gene Expression in Eukaryotes http://www.cbs.dtu.dk/
DNA-dependent RNA polymerases of archaea bacteria not only resemble the nuclear RNA polymerases of eukaryotes rather than the eubacterial enzymes in their complex component patterns but also show striking immunochemical, i.e., structural, homology with the eukaryotic polymerases at the level of single components. Thus, eukaryotic and archaebacterial RNA polymerases are indeed of the same type, but distinct from the eubacterial enzymes, which, however, are also derived from a common ancestral structure; J Huet, R Schnabel, A Sentenac, and W Zillig
It is very illuminating if one compares prokaryotic and archaeal with eukaryotic system in terms of structural features of genes and the promoter elements and upstream elements, the enzymes and its accessory proteins, which among them have perfected and the most evolved. Archaeal type of RNAPs is found to be in between prokaryotes and eukaryotes. Viral RNAPs, some are similar to bacteria, some with archaeal and some with eukaryotes. Unique among them are plus strand transcribing RNAPs and (-) strand transcribing RNAPs; they are RNA dependent RNA polymerases. Orthopoxviruses synthesize RNA using a virally encoded RNAP that is structurally and mechanistically related to bacterial RNAP In addition one finds RNA dependent DNA polymerases called ‘Reverse Transcriptase’. Most other viruses that synthesize RNA using a virally encoded RNAP use an RNAP that is not structurally and mechanistically related to bacterial RNAP, archaeal RNAP, and eukaryotic nuclear RNAP I-V. Many viruses use a single-subunit DNA-dependent RNAP that is structurally and mechanistically related to the single-subunit RNAP of eukaryotic chloroplasts and mitochondria and, more distantly, to DNA polymerases and Reverse transcriptases. Perhaps the most widely studied is single-subunit RNAP is bacteriophage T7 RNA polymerase. A DNA-dependent RNA polymerase is present in Vaccinia virus particles and the cytoplasm of infected cells. The purified enzyme is similar in overall size to eukaryotic RNA polymerase and also contains many subunits. Two large polypeptides of 130,000 to 140,000 daltons and at least seven smaller ones ranging in size from 13,000 to 36,000 daltons have been observed. An additional polypeptide of 77,000 daltons was associated with the RNA polymerase that was isolated from infected cells.
· From having one enzyme with few subunits plus specific sigma factors for simple systems, organisms have evolved to possess different enzymes for different set of genes, with a host of factors operating, is a biological accomplishment.
Evolution has, simplified some systems as perfectional, but in other systems, elaboration with multiplicity of structures as perfection; it is difficult to discern which one is advanced and which one is primitive.
· It is acceptable that any group of organisms originated nearly 3.8 to 3.2 billion years ago, yet survived that long and still prospering must have developed reasonable perfection in molecular and biochemical operative systems. Perhaps one can opine that the prokaryotes have achieved that perfection, yet they are lacking many things, where the eukaryotes are superior; but they are par excellent in organizing their genome and acquiring new means and modes for future development, but what future genomic development is perplexing and awful, ex. It is estimated and projected that the human Y chromosome is going vanish from human genome.
· Eukaryotes have developed division of labor among cells and cell types (living); but all interconnected in terms of adhesion and communication with each another.
Complexity in human system arises because human body consist of at least well designed and characterized ~210 cell types (acc. Wikipedia) or there can be more than 250 – 320 cell types (Sanger Institute- old information).
To understand gene expression and its regulation, first an understanding of cell types and their function is inevitable.
The cellular enzymes, their subunits, accessory transcriptional factors, and accessory proteins and structural organization of promoter elements of different genes, is essential before one looks into the mechanism and regulation transcription.
Estimates show that there are more than 200 different cell types, at this point of time.
· Genes are identified from cell types by their in-built structural features such as promoter, coding and terminal regions. Prokaryotes contain genes that remain active or silent because of the DNA bound specific proteins. The silent or repressed genes can be induced to be active and vice versa.
· Eukaryotes too contain genes in repressed or active state by the binding of pioneering proteins to their respective regulatory regions. The gene products themselves perform variety of functions on genes themselves. Such status of cell types is perpetuated till they are changed due signals or changed inputs. An identification gene with bound proteins in a chromatin is a herculean task.
· Most of the RNAPs are made up of a group of protein subunits, which are associated together, as tight complex.
· For example, an enzyme complex to be positioned at a particular gene and at a particular sequence at a pre-defined site; it also requires a group of accessory factors as complexes. They assemble on their respective site with specific sequences, and facilitate the positioning of the RNAP, to initiate transcription at a predefined site called START; whether this complex succeeds in initiation or not is a different matter.
Such a complex of proteins (products of genes), which associate at promoter, is called basal transcriptional apparatus (BTA), often called Preinitiation complex or PIC, which consists of RNAP core complex and accessory protein complexes named as TFs (Transcription Factors). Many proteins can act as regulatory factors. Their regulation beyond human imagination. Each class of genes requires, though the same RNA Pol II, but requires specific kind of accessory complex of proteins to perform transcription in cell specific mode or tissue specific mode or in response mode.
A list of cell types in Eukaryotes: (WIKIPEDIA).
Animal Cell Types:
Exocrine secretory epithelial cells- 28 Types; Hormone secreting cells-38 types; Epithelial cells lining closed internal body cavities-21 types,
Ciliated cells lining internal body cavities-21 types and Keratinizing epithelial cells-11 types and so on.
Wet stratified barrier epithelial cells-3 types; Nervous systemory transducer cells-20 types; Autonomic neuron cells-3 types; Sense organ and peripheral neuron supporting cells-12 types; Central nervous system neurons and glial cells-4 types.; Lens cells-2 types; Metabolism and storage cells-5 types; Barrier function cells (Lung, Gut, Exocrine Glands and Urogenital Tract); Kidney-18 types. Extracellular matrix secretion cells-21 types; Contractile cells-14 types.;Blood and immune system cells-21 types; Pigment cells-2 types.; Germ cells-5 types; Nurse cells-3 types.
Interstitial cells;. Interstitial kidney cells-1.
Plant Cell Types (Angiosperms):
Meristems: 3 types; apical, Intercalary, peridermal and vascular; Epidermis: 2 types: epidermis, hypodermis, epidermal outgrowths, Endodermis: Single layer of cells around V.B; Pericycle: Underlying endodermal layer, a potential meristem layer. Bundle sheath: Around monocot VB, Parenchyma: ~5 types- Cortical, Collenchyma, Spongy & Palisade, Pith, Seed; Sclerenchyma: 2 types- Sclerenchyma fibers and Stone cells; Xylem: 4 types: Xylem vessels, X. fibers, Sclereids, X. parenchyma; Phloem: 4 types: Sieve tubes, Companion cells, Parenchyma and Phloem fibers and many others.