Gene Expression in Response To:
Biosystems live and perpetuate with in an ambient environment, whether it is inter -cellular or intracellular or it can be external.
· The kind of stimulus provided can be in the form of chemicals, change in the pH, change in the ionic concentration, change in the nutrients, change in the temperature, light, infectious attack from other organisms and many other situations like cell-to-cell contact and others; all of them act as stimulants which can have inducible effect and some have repressive effects.
A particular inducer for one gene can be a repressor for the other gene. Here inducer means a factor that induces the expression of a gene.
· All these effects are transduced through certain factors to the gene.
There is a time lapse between the time of stimulus application and the time at which response is manifested;” in space and time” a quote by the great Einstein.
· Genes are endowed with genetic facility to respond, by having a variety of sequence modules, located at specific distance from the start site- upwards or downwards.
Each module contains a specific sequence, located at specific position.
· There may be several modules, or combination of modules, positioned in combination.
The kind of modules, location of them from the start and combination of them are gene specific, cell/tissue specific and stimulus specific.
· Besides having response elements in their promoter regions for stimuli, cells do produce specific factors for specific genes and for specific response.
The cellular factors may be present in cytosol or in the nucleus or located in cell membranes or they may be synthesized in response stimuli.
· Whether the factors are in fixed position or free, they have many roles and many steps before they act on genes and express them specifically.
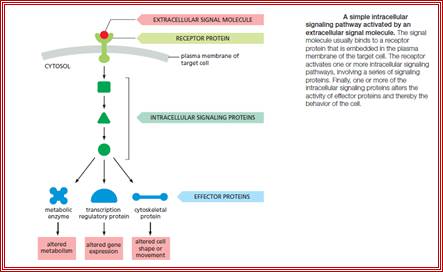
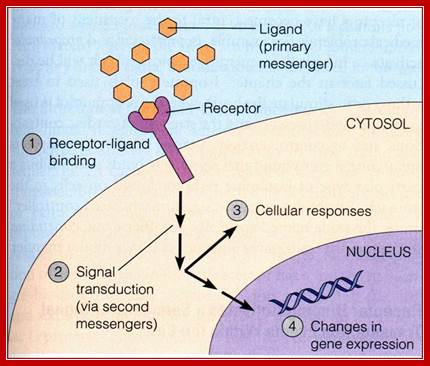
The stimulus, whatever kind it may be, first it should contact or impinge upon the cell or enter into the cell, by whatever means possible.
· Then they bind to certain factors (call them receptors), or bring about changes in the factors to activate or inactivate them, and perform certain reactions that ultimately lead to the entry of them into the nucleus.
In the nucleus they bind to respective DNA sequence elements or interact with proteins that are already bound to DNA. Then they recruit a variety of components such as co factors, mediator complexes and others. Once they bind, they contact or interact with the basal transcriptional apparatus elements (there are several of them, which are tissue specific or response specific and activate or modulate the RNAP enzyme to initiate transcription.
· However, the efficiency and the rate of transcription depend upon the kind of interaction they do.
Heat shock gene expression:
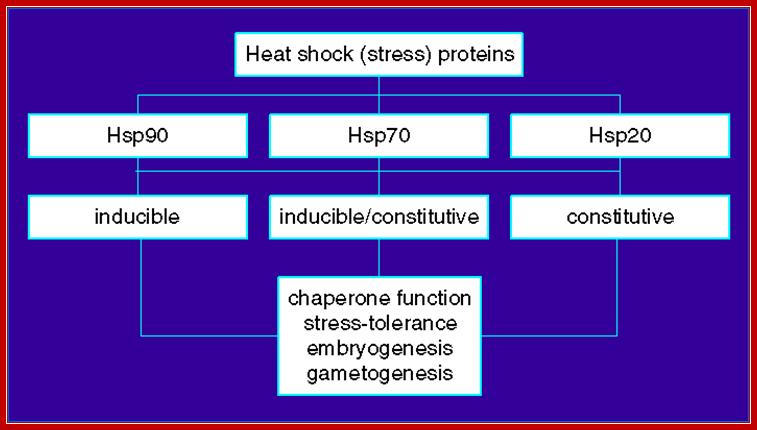
Both prokaryotes and eukaryotes contain specific kind of heat shock response genes, which produce heat shock proteins.
· When cells of PK or EK including humans, if subjected to elevated temperatures, cells respond by changing their pattern of gene expression.
Cells start producing few specific heat-shock proteins, called HSPs, and they are produced at higher level, while many other cellular gene expressions go down.
· In prokaryotes specific sigma factors become active and they in turn, switch on several heat-shock related genes.
Among eukaryotes, especially, Drosophila has been subjected to intensive studies to understand heat shock responses.
· When an 11th day larvae of this insect are subjected to elevated temperatures, with in minutes, specific changes in the form of gene expressions is observed.
At several loci, in the polytene chromosomes, chromosomal puffs develop. Some puffs already formed at some other loci start regressing. One finds gene regression and activation at different loci can be made out in salivary gland chromosomes.
· Chromosomal puffs are the sites at which chromatin has opened up for active transcription of specific genes in DNA loops, which is free from nucleosomes.
The system is ensured of proper and preferential transcription and translation of the heat shock mRNAs.
· The HSPs assure protection to several cellular proteins.
Such gene loci are called heat shock loci. Seven such heat shock loci have been identified in Drosophila.
· They are named as HSP 28, 23, 1, 26, 22, 4 and 5. All of them are clustered in a 15-kbp region called 67B locus in the insect chromosomes.
Heat Shock Gene Cluster:
<----Hs-28-I<----Hs23-I-1----->I-Hs26----->I-Hs22--->-<----Hs4-I-<---Hs5-I-----I--->
Promoter Elements of HSP 70 Gene:
Some of the heat shock genes are also expressed in stage specific manner during development. HSP 26 and 28 are expressed during oogenesis. HSP 22, 26 and 1, 4 & 5 are expressed at early and late pupa 3rd instar stage respectively.
· In higher systems there are ten different heat shock genes responsive to heat. All of them have similar sequence modules in the upstream of the start.
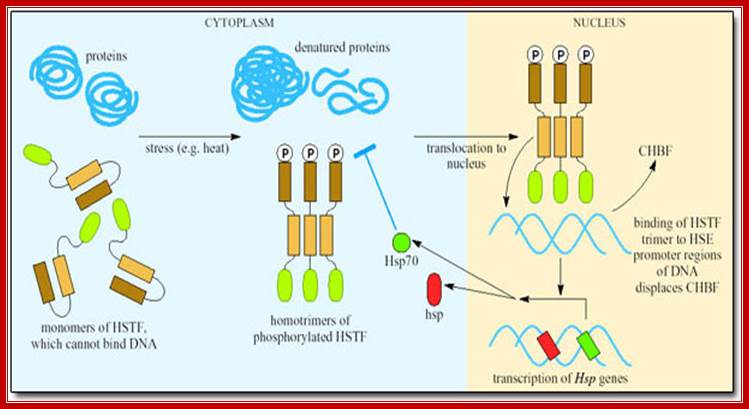
Heat shock transcription factors (HsTF) are constitutively synthesized at low level and they remain inactive, because a protein called ubiquitin binds them.
If the temperature of cytoplasm of in the cell is raised, the ubiquitin protein dissociates from the heat shock factors and the HsTFs become active and bind to specific upstream elements and activate transcriptional complex to initiate the synthesis of heat shock proteins.
Heat shock proteins under normal conditions are synthesized by the activity of HsTFs. However, many of the RNAPs when they are elongating the transcripts halt and paused at certain loci that are acetylated. But when the cell experiences heat shock HsTFs get activated and trimerize and enter the nucleus and activate transcription and also resume activation of the halted RNAP from transcription.
Heat shock proteins act as chaperones for a variety of proteins otherwise cellular proteins become inactive; they also help nascent polypeptide chains to under go proper folding.
HSE
----I -200I----------I -100I-------I -92I-------I -37I--------I -20I-----I+>
[C.GAA.TTC..G TATAAG]
www.open.edu
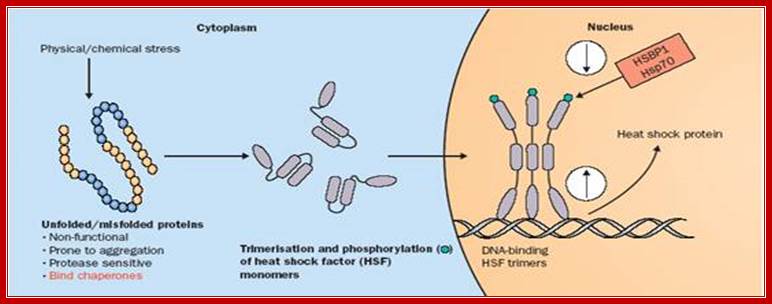
Heat activates disordered HsTFs with phosphorylation and they trimerize and enter into the nucleus, where they bind to their Heat shock response elements (HSRE). http://thelancet.com
HS HS HS TATA
------- -220---------- -150-------- -80----------- -40--------> +1
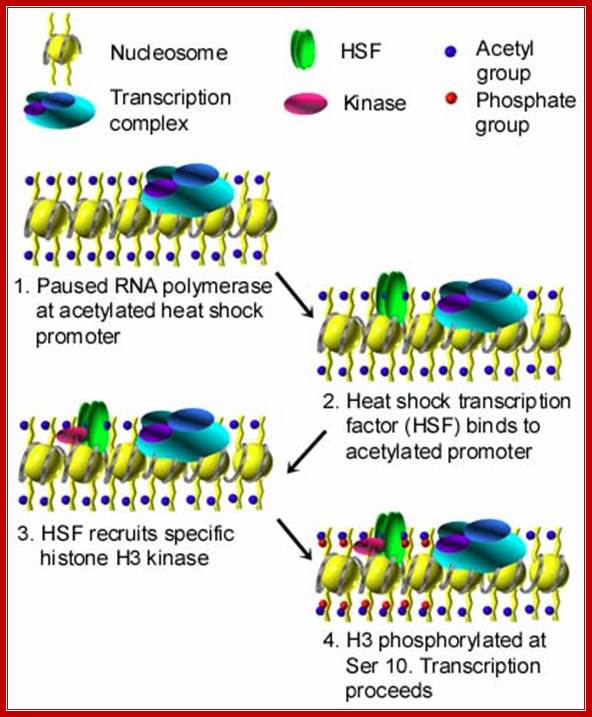
Transcription is paused at acetylated region, but when activated HSTFs bind to the acetylated region, it recruits H3 kinase; this leads to histone phosphorylation and the transcription resumes. www.biology.emory.edu
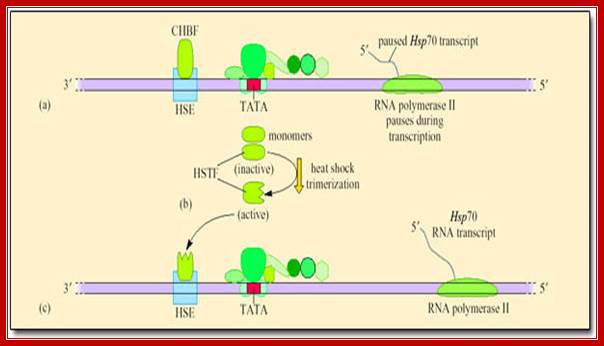
In many of the higher eukaryote’s HSTFs under normal conditions start transcriptional initiation but after certain length, transcription pauses, which will be activated only when the cells get heat shock and moves into the nucleus and binds the site and reactivates transcription that is stalled.
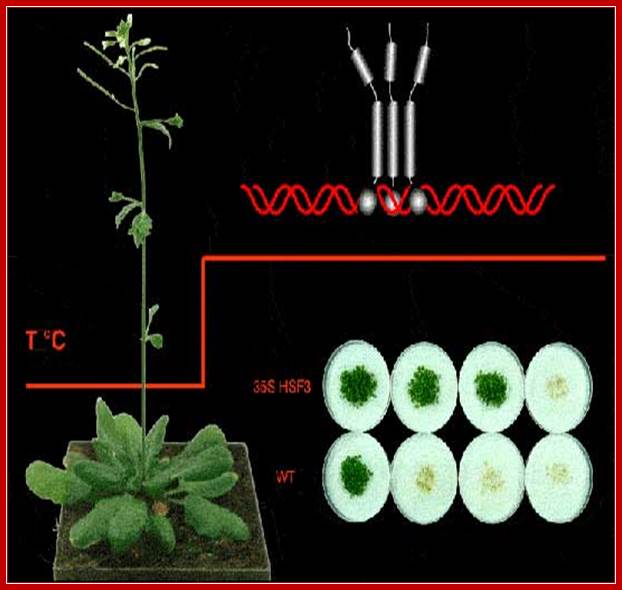
In plants (Arabidopsis) also, the Heat shock proteins operate under variety of stress conditions. Heat shock activates the HSTFs and they trimerize and move into the nucleus, where they bind to their response elements and activate transcription of heat shock proteins genes.
Plant Gene expression in response to Light:
Plants have genetically adopted to respond to light and dark, day and night, short days and long days, cold and hot temperatures, humid and dry conditions and many other environmental vagaries, for all of them grow under natural conditions.
· Plants and light are intertwined in their interactions, one as the influencing factor and the other as the responder. Plants are influenced by light in their growth, flowering, photosynthesis, transpiration, dormancy, germination, opening and closing of the of the stomata, opening of flowers, movement of leaves, shedding of leaves, pigment synthesis and many other metabolic processes.
Red light induces the expression of nuclear Ribulose Biphosphate Carboxylase (RuBisco -14KD) small subunit gene. There are at least 5-10 nuclear genes for RuBisCo’s. The RubisCos’ small subunit gene is a nuclear gene but the large subunit is coded for by plastid localized gene (55Kd belongs to chloroplast genome). The functional RuBisCo consists eight subunits.
· In the upstream of rbc-s gene promoter there are several sequence modules, which act as light response elements, called LREs. The TATA box is located at –30 and light response elements LRE at –166 and –149. There are three boxes in this region, called Box I, Box-II and Box-III.
Plant genes for RuBisco-S also have upstream elements at –410 and –160 called ARE (Age Response Elements) which respond to the age.
· In immature leaves RuBisco-s genes express at very high levels.
I-------- 410---- -166--II-------- -160----------140------------- -30--------I+>
ARE BOX-1 BOX-2 BOX-3 TATA
ARE : Age related response elements.
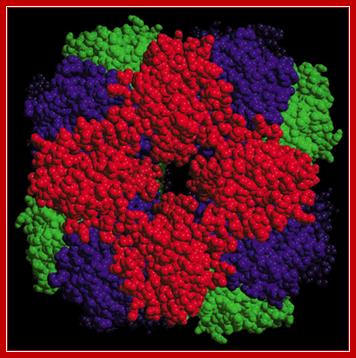
RuBisco is an octamer where the red colored protein is the small subunit coded for by the nuclear gene in response to light.
Flowering- in Response to Light:
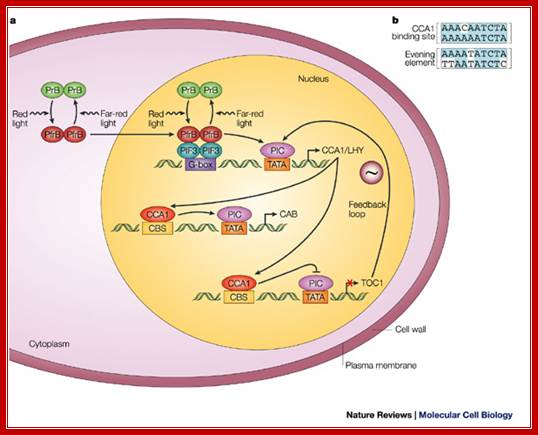
Light plays an important role in plants’ life, growth and ultimately flowering. One of the most important aspects of light on flowering is the requirement of cellular pigment called phytochrome. However there are other pigments such as cryptochrome which absorbs blue light. The flowering pigment phytochrome exists in association with a protein; on absorbing Red light at a particular wavelength for a particular period of time, undergoes conformational change from Pr to Pfr (cis-trans) P-Far-Red). This activated pigment protein complex enters the nucleus and binds to its response elements called G-box ‘GACGTG’ through dimeric PIFr factor and recruit transcriptional apparatus along with other required cofactors and initiate transcription and produces a transcription factor called MYB, which in turn activate another set of genes to induce flowering. This is a simplistic explanation of flowering. Phytochrome is also responsible for variety of plant growth and plant adaptation to day length.
---G box---TATA---InR+1----- Myb
---------MYB binding domain------TATA----InR+1----genes for flowering

https://www.pinterest.com; www.biochem-vivek.tripod.com
Expression of Genes- in Response to Cortisols/Steroids:
Pathway of biosynthesis of some sterols; http://circres.ahajournals.org
Different cell types in animals respond to different types of hormones.
· The hormones can be lipid soluble or some may be water-soluble. They act as signaling molecules. Steroid hormones are synthesized and released, which in turn act on the surrounding cells or work on their own or act on distant cells.
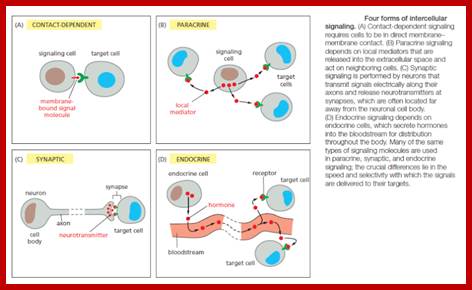
Autocrine: The released signals act on their own cells and stimulate their own cells to higher level.
Paracrine: Signal released act on nearby cells.
Endocrines: These are hormones; long distance targets.
Synaptic: Neurotransmitters released are transmitted via synaptic cleft or by synaptic mechanism.
Most signal-relay stations we know about were intelligently designed. Signal without recognition is meaningless. Communication implies a signaling convention (a “coming together” or agreement in advance) that a given signal means or represents something: e.g., that S-O-S means “Send Help!” The transmitter and receiver can be made of non-sentient materials, but the functional purpose of the system always comes from a mind. The mind uses the material substances to perform an algorithm that is not itself a product of the materials or the blind forces acting on them. Signal sequences may be composed of mindless matter, but they are marks of a mind behind the intelligent design.
The fundamental features of cell signaling have been conserved throughout the evolution of the eukaryotes. In budding yeast, for example, the response to mating factor depends on cell-surface receptor proteins, intracellular GTP-binding proteins, and protein kinases that are clearly related to functionally similar proteins in animal cells. Through gene duplication and divergence, however, the signaling systems in animals have become much more elaborate than those in yeasts; the human genome, for example, contains more than 1500 genes that encode receptor proteins, and the number of different receptor proteins is further increased by alternative RNA splicing and post-translational modifications.
Such contact dependent signaling is especially important during development and in immune responses. Contact-dependent signaling during development can sometimes operate over relatively large distances if the communicating cells extend long thin processes to make contact with one another. In most cases, however, signaling cells secrete signal molecules into the extracellular fluid. Often, the secreted molecules are local mediators, which act only on cells in the local environment of the signaling cell. This is called paracrine signaling (Figure B). Usually, the signaling and target cells in paracrine signaling are of different cell types, but cells may also produce signals that they themselves respond to: this is referred to as autocrine signaling. Cancer cells, for example, often produce extracellular signals that stimulate their own survival and proliferation.
Cell communication and signaling, evidences of design:
http://reasonandscience.heavenforum.org
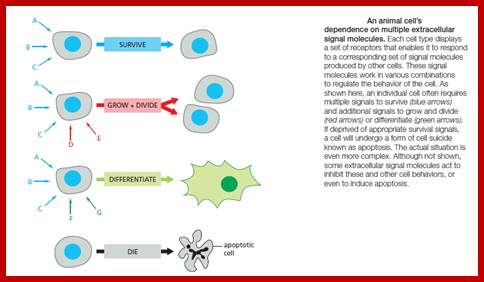
Signals can lead to variety of consequences such as cell survival or death, or induce cell division and differentiation, increase or decrease in metabolic rates.
Water soluble ligands; http://cc.scu.edu.cn
Cells are also endowed with specific receptors and they respond to specific signaling molecules such as hormone in a specific way.
· Some have general effects like growth hormone, cortisols, but some hormones are specific to certain tissues and their effect and response is hormone specific and tissue specific. For example, Glucocorticoids activate 100 or more genes, some of them respond very early and some respond late and long lasting.
Cortisols effect carbohydrate metabolism and increase blood sugar level and also have anti-inflammatory effects.
· Mineralocorticoids maintain water and salt balance.
Ecdysone acts on insect larvae and induces pupa formation, estrogens and androgens act as sex hormones in the development of sex organ. Estrogen activates ovalbumin genes in chicks. Vitamin-D helps in bone development and calcium metabolism.
Dennis A. Ausiello, MD, Editor er al; http://annals.org
· Retinoic acid (9-cis Retinoic acid) acts as a morphogen and it is also required for the function of vision in eyes. Thyroid hormones control basal metabolism and the rates of reactions.
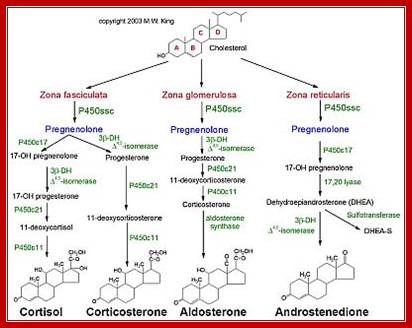
Steroid hormones are synthesized and released in response to a variety of neuro endocrine activities and effect major activities such as growth, tissue development and body homeostasis.
Adrenal glands synthesize nearly 30 or more steroids, majority of them are Glucocorticoid and Mineralocorticoids.
· Thyroid hormones, from thyroid tissue, in the form of iodinated Tyrosine such as T3 and T4 control the rate of basal metabolism.
Pituitary glands produce four hormones; by specific cell types within the same organ; islets of langerhans produce insulin, the others are somatostatin and two related hormones including glucagon.
· The release and the target tissue of action are classified into four forms.
· Peptide and non-peptide signaling molecules perform a variety of functions through, phosphorylation and dephosphorylation reactions. There are at least 1000 kinases and equal number of phosphotases. All of them are specific-to-specific biomolecules including proteins.
Phosphorylation of proteins can be via threonine (5-10%), serine (90-95%) or tyrosine (10%) and it is sequence specific. Besides, ligands’ activation and inactivation, proteins are regulated mainly due to kinase and phosphotases activities.
· The said enzymes are strongly influenced by the signal molecules. Most of the early reactions take place either at cell membrane level or in the cytoplasm before the factors move into the nucleus and activate specific genes.
Most of the lipid soluble hormones or signaling molecules have intracellular receptors, for ex. Cortisols receptors belong to a super family of receptors, which are intracellular, yet each is distinctly different. However, though different from one another, they show some characteristic features in their protein structures and functions. Intracellular receptors are classified into cytoplasmic (Type I) and Nuclear (Type II) and there are two more subtypes.
· All of them have sequence specific DNA binding domain that is to bind to response elements, dimerization domains, specific hormone binding domain, and an activator domain, which interacts with PIC and activates the RNAP.
The N-terminal region of all these receptors varies considerably in size and character; these regions contain activation domain.
· The dimer can be of the same kind to form homodimers or of different types to produce hetero dimers.
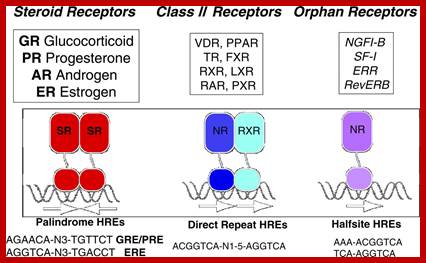
Each of these dimers (homodimer or heterodimers) bind to two half sites, one to one, found in response elements of the gene promoter regions. But some bind as monomers.
· The sequence elements in response genes are more or less show similarity, yet distinctly organized into dyads.
The receptors, many of them, just don’t bind to response elements as soon as they are bound by the ligand. After ligand binding, they may undergo further modifications or associate with other factors and co regulators, then they bind to DNA.
DNA Binding and other Domains of Few Receptor/ Inducers:
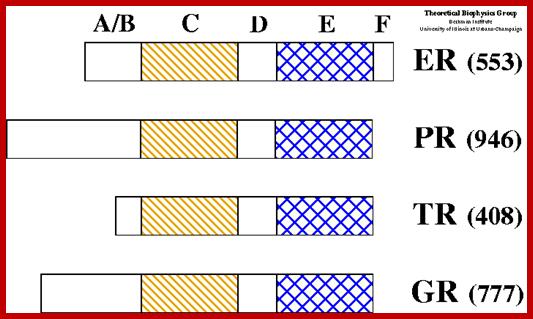
Nuclear Hormone Receptors;
Nuclear hormone receptors are ligand-activated transcription factors that regulate gene expression by interacting with specific DNA sequences upstream of their target genes. As early as 1968 a two-step mechanism of action was proposed for these receptors based upon the observation of an inactive and an active state of the receptors. The first step involves activation through binding of the hormone; the second step consists of receptor binding to DNA and regulation of transcription.
Receptor proteins contain N terminal activator domain. The middle contains DNA binding domain and the C terminal region contains ligand binding site and dimerization motifs. http://www.ks.uiuc.edu


Structure of nuclear receptors: nuclear receptors constitute a superfamily of dimeric C4 zinc-finger transcription factors. They are modular in structure and contain the following structural domains:
N-terminal regulatory domain (A-B): The A-B domain is highly variable in sequence between various nuclear receptors. It contains the activation function 1 (AF-1), whose action is independent of the presence of ligand. The transcriptional activation of AF-1 is normally very weak but it synergizes with AF-2 in the E-domain to produce an upregulation of gene expression.
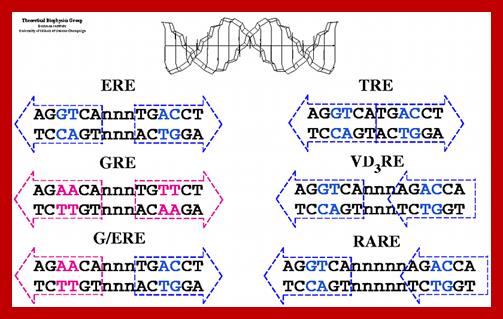
DNA-binding domain; DBD (C) : It is a highly conserved domain containing two zinc fingers that binds to specific sequences of the DNA called hormone response elements (HRE) as shown in Figure 3.
Hinge region (D) : It is the flexible domain that connects the DBD with the LBD. It influences subcellular distribution and intracellular trafficking.
Ligand binding domain LBD (E) : Its sequence is moderately conserved but it is highly conserved in structure between the various nuclear receptors. The structure of the LBD is referred to as an alpha helical sandwich fold in which three anti parallel alpha helices (the sandwich filling) are flanked by two alpha helices on one side and three on the other (the bread). The ligand binding cavity is within the interior of the LBD and just below is present three anti parallel alpha helical sandwich filling. Along with the DBD, the LBD contributes to the dimerization interface of the receptor. In addition, it binds coactivator and corepressor proteins. The LBD also contains the activation function 2 (AF-2) whose action is dependent on the presence of bound ligand.
C-terminal domain (F) : It is highly variable in sequence between various nuclear receptors. ;http://nptel.ac.in
Retinoic acid Induced gene expression:
“The development of many chordate- and vertebrate-specific characters is controlled, directly or indirectly, by retinoic acid (RA), a vitamin A-derived morphogen. In chordate embryos, too much or too little RA during early embryogenesis causes malformations, which are mainly due to a mispatterning of the embryo along the anteroposterior body axis. RA signaling is mediated by RA binding to retinoic acid receptors (RARs), which form heterodimers with retinoid X receptors (RXRs). During chordate development, important functions of RA signaling are mediated by Hox genes, at least some of which are direct targets of RA. In many animals, including most deuterostomes, Hox genes mediate anteroposterior positional patterning of the embryo. Moreover, in vertebrates, Hox genes have been suggested to play important roles in the patterning of all embryonic tissue layers.”
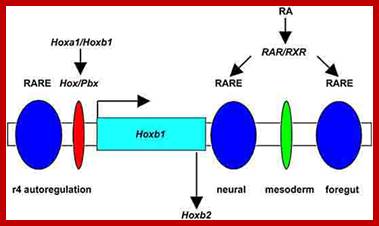
Regulation of Hoxb1 expression in the hindbrain.
The complexity in regulating Hoxb1 expression in r4 of the hindbrain is dependent upon Hox/Pbx (red oval) and retinoic acid response element (RARE) binding sites in the 5' autoregulatory region of the Hoxb1 locus that mediates auto-, para- and cross regulatory interactions. In addition, two other RAREs are locate in the 3' regulatory region of Hoxb1 which bind retinoic acid receptor heterodimers and are required to initiate the early broad expression of Hoxb1. Hoxb1 not only feeds back on itself in an autoregulatory loop but cross regulates the expression of Hoxb2 in r4.
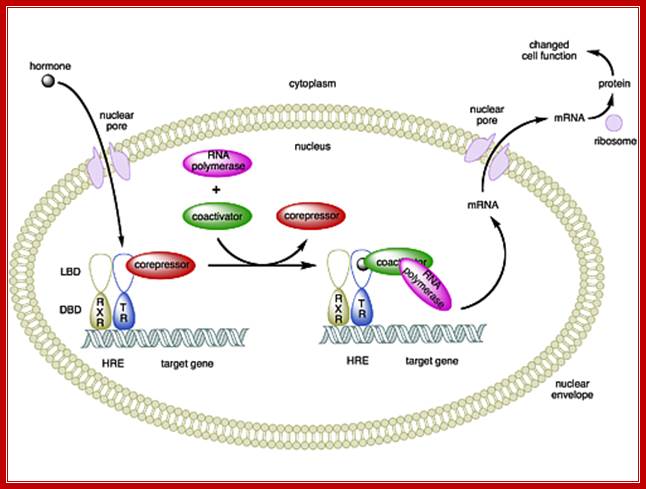
Retinoic acid induced gene expression, where retinoic acid enters into the nucleus and binds to receptors. The receptors till now by binding to DNA remain as repressors. With the binding of ligands the repressor become activators, which recruit co activators such as CBP/p300 and mediator complex and initiate gene transcription? https://www.bioscience.org/
1. Murine embryonic stem cells such as the AB1 cell line undergo differentiation in the presence of retinoic acid (RA) into an extra embryonic epithelial cell type. This results in the activation of genes such as Hoxa-1, Hoxb-1, laminin, collagen IV(α1), tissue plasminogen activator, RARβ, and CRABPII. The CRABPI gene is regulated in an unusual fashion; CRABPI message and protein levels are induced at low concentrations of RA, Anne C. Chen and Lorraine J. Gudas.
2. Retinoic Acid-Inducible Gene I Mediates Early Antiviral Response and Toll-Like Receptor 3 Expression in Respiratory Syncytial Virus-Infected Airway Epithelial Cells (ping Lu etal).
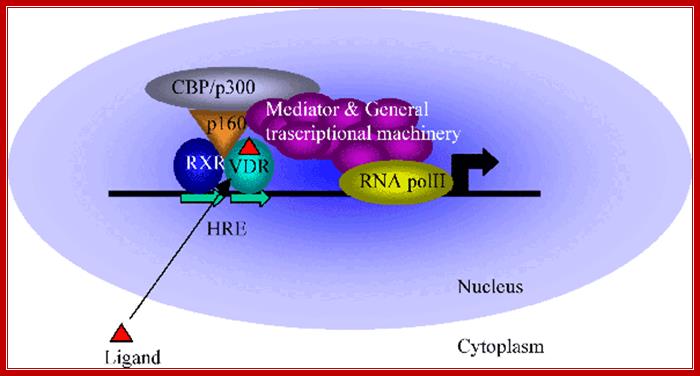
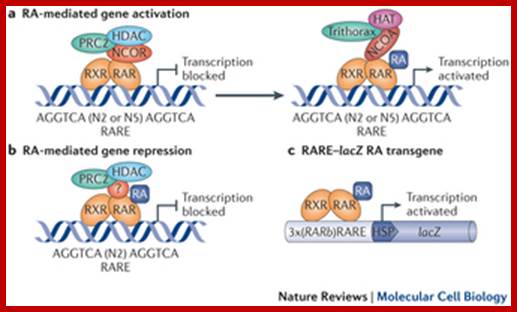
Retinoic acid (RA) signalling has a central role during vertebrate development. RA synthesized in specific locations regulates transcription by interacting with nuclear RA receptors (RARs) bound to RA response elements (RAREs) near target genes. RA was first implicated in signalling on the basis of its teratogenic effects on limb development. Genetic studies later revealed that endogenous RA promotes forelimb initiation by repressing fibroblast growth factor 8 (Fgf8). Insights into RA function in the limb serve as a paradigm for understanding how RA regulates other developmental processes. In vivo studies have identified RAREs that control repression of Fgf8 during body axis extension or activation of homeobox (Hox) genes and other key regulators during neuronal differentiation and organogenesis.
· Retinoic acid induced gene activation requires the association of heteromeric dimer receptor, with the binding of the ligand the activated receptors recruit CBP/p300 and involves mediator complex to activate the gene transcription. Mechanism of Retinoic acid signalling roles in organ development; Retinoic acid (RA) signalling has a central role during vertebrate development. RA synthesized in specific locations regulates transcription by interacting with nuclear RA receptors (RARs) bound to RA response elements (RAREs) near target genes. RA was first implicated in signalling on the basis of its teratogenic effects on limb development. Genetic studies later revealed that endogenous RA promotes forelimb initiation by repressing fibroblast growth factor 8 (Fgf8). Insights into RA function in the limb serve as a paradigm for understanding how RA regulates other developmental processes. In vivo studies have identified RAREs that control repression of Fgf8 during body axis extension or activation of homeobox (Hox) genes and other key regulators during neuronal differentiation and organogenesis. Thomas & Gregg Duester; ;http://www.nature.com
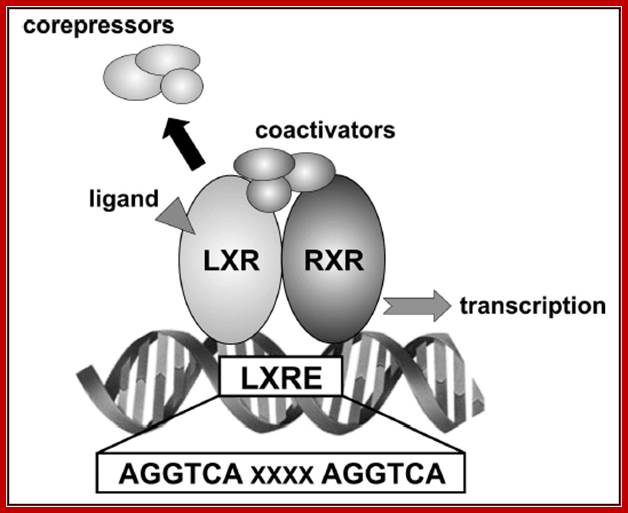
Retinoic acid induced genes contain response elements called RARE in dyads which are bound by dimeric receptors type II. In the absence of RA the RARE elements are bound by Repressor-co-repressor complex, which causes histone deacetylation which in turn causes gene repression. In the presence of RA, the hormones enter the nucleus binds to receptors and displace co repressors and recruits co activators and activate receptors and initiate transcription.
In chordates, which comprise urochordates, cephalochordates and vertebrates, the vitamin A-derived morphogen retinoic acid (RA) has a pivotal role during development. Altering levels of endogenous RA signaling during early embryology leads to severe malformations, mainly due to incorrect positional codes specifying the embryonic anteroposterior body axis. In this review, we present our current understanding of the RA signaling pathway and its roles during chordate development. In particular, we focus on the conserved roles of RA and its downstream mediators, the Hox genes, in conveying positional patterning information to different embryonic tissues, such as the endoderm and the central nervous system. We find that some of the control mechanisms governing RA-mediated patterning are well conserved between vertebrates and invertebrate chordates, such as the cephalochordate amphioxus. In contrast, outside the chordates, evidence for roles of RA signaling is scarce and the evolutionary origin of the RA pathway itself thus remains elusive. In sum, to fully understand the evolutionary history of the RA pathway, future research should focus on identification and study of components of the RA signaling cascade in non-chordate deuterostomes (such as hemichordates and echinoderms) and other invertebrates, such as insects, mollusks and cnidarians.; Ferdinand Marlétaz er al; http://www.ijbs.com; Int J Biol Sci 2006
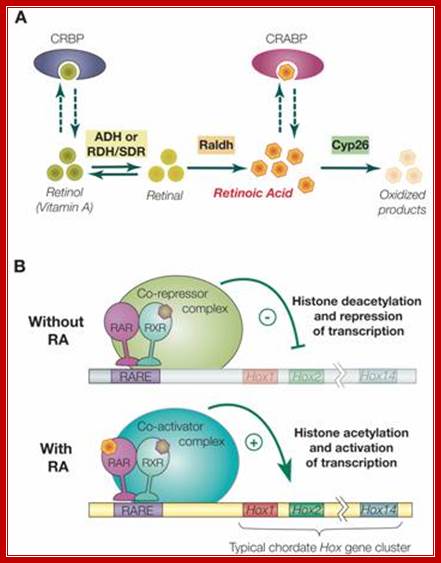
“Synthesis, degradation and mode of action of retinoic acid (RA):
A) The metabolic pathway for synthesis and degradation of endogenous RA is shown. RA is synthesized by oxidation of retinal by retinaldehyde dehydrogenases (RALDHs). In a reversible reaction, retinal is synthesized from retinol (vitamin A) by either aldehyde dehydrogenases (ADHs) or short-chain dehydrogenase/reductase (RDHs/SDRs). Cellular retinol binding proteins (CRBPs) can bind retinol, whereas cellular retinoic acid binding proteins (CRABPs) can bind RA. Finally, endogenous RA is degraded by CYP26 enzymes. B) The RAR/RXR heterodimer mediates the effects of RA. In the absence of ligand (RA), the RAR/RXR heterodimer is bound to DNA and co-repressors. This complex induces transcriptional repression through histone deacetylation. Binding of the ligand (RA) induces conformational changes and the binding of co-activators leading to histone acetylation and activation of transcription.
RA signaling is mediated by RA binding to retinoic acid receptors (RARs), which form heterodimers with retinoid X receptors (RXRs). This complex in turn binds to retinoic acid response elements (RAREs) in the regulatory regions of target genes. Only a few direct targets of RA signaling have been described. These include the RARs themselves, Hox genes, some other transcription factors, such as HNF-3α and Cdx1, plus genes involved in retinoic acid metabolism (e.g. CRABP1 and. In the mouse, there are 3 RARs and 3 RXRs (α, β and γ). Although knock-out of any one of these has only minor, tissue-specific effects on morphogenesis due most likely to functional redundancy among them, compound mutants with two or three of the genes inactivated are much more severely affected with defects in anteroposterior patterning of pharyngeal endoderm, hindbrain and neural crest cells.
Among the known targets of the RAR/RXR heterodimer are the Hox genes. Hox genes encode transcription factors that contain a highly conserved DNA binding domain of 60 amino acids, the homeodomain.”
Estradiol Induced gene expression:

Estradiol
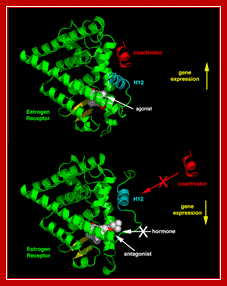
The 3D organization of estradiol receptor- active and inactive forms.

Endocrine therapies targeting oestrogen action (anti-oestrogens, such as tamoxifen, and aromatase inhibitors) decrease mortality from breast cancer, but their efficacy is limited by intrinsic and acquired therapeutic resistance. Candidate molecular biomarkers and gene expression signatures of tamoxifen response emphasize the importance of deregulation of proliferation and survival signalling in endocrine resistance. However, definition of the specific genetic lesions and molecular processes that determine clinical endocrine resistance is incomplete. The development of large-scale computational and genetic approaches offers the promise of identifying the mediators of endocrine resistance that may be exploited as potential therapeutic targets and biomarkers of response in the clinic
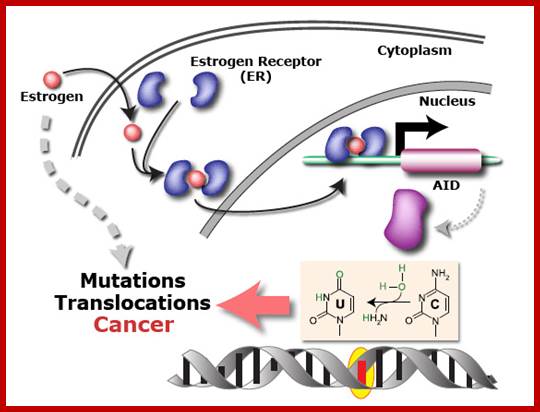
Oestrogen causes DNA mutation thus cancer; The evidence that the sex hormone oestrogen is involved in cancer is overwhelming.
For example, breast cancer is more common among women who take HRT and the Pill for long periods, both of which add oestrogen to the body’s natural levels. And this is the most common cancers; it is intimately linked to levels of the hormone over a woman’s life.
Oestrogen is also involved in cancers of the womb and ovaries, and, potentially, prostate cancer. ; http://scienceblog.cancerresearchuk.org
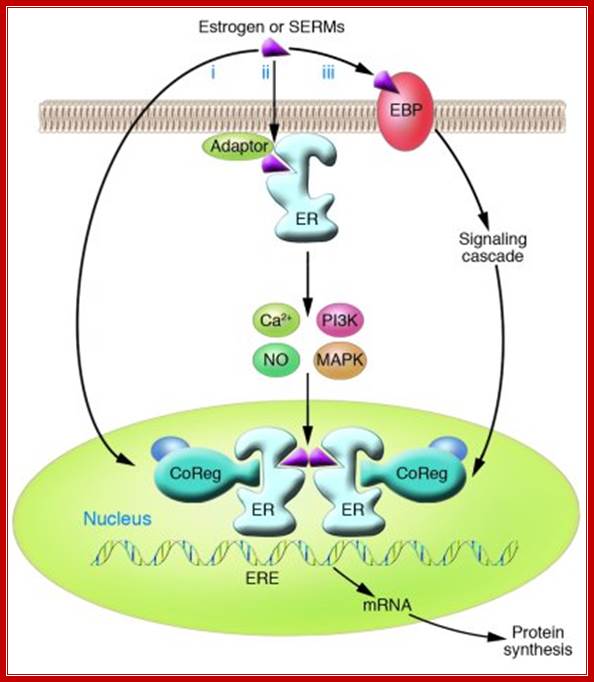
“Oestrogen action at the molecular level: Three distinct pathways of oestrogen regulation of gene expression. First, in classic oestrogens signalling, ligand-bound oestrogen receptor (ER) activates gene expression — either through direct binding of dimeric ER to specific DNA response elements, EREs, in complexes including co-activators (CoAs) and histone acetyl transferases (HATs), or through protein–protein interactions with other transcription factors, particularly members of the activation protein 1 (Ap1) and specificity protein 1 (Sp1) families — to facilitate binding to serum response elements (SREs) and activation of transcription (a). Second, ER can also be activated as a consequence of signalling events downstream of receptor tyrosine kinases (RTKs) such as the epidermal growth factor receptor (EGFR), ERBB2 (also known as HER2) and the insulin-like growth factor receptor (IGFR) (b). Phosphorylation (P) by the Erk or Akt serine/threonine kinases leads to ligand-independent activation of the ER. Third, signalling can be mediated through non-genomic mechanisms by ER that is localized at the cell membrane or in the cytoplasm (c). Ligand binding induces the assembly of functional protein complexes that involve other signalling molecules and that activate intracellular signalling cascades, resulting in transcription factor (TF) activation. Two recently characterized mechanisms that ultimately activate transcription independently of ER binding to DNA are illustrated: ligand-induced methylation (M) of ER and formation of an ER–PI3K–Src–focal adhesion kinase (FAK) complex that activates Akt (d), and activation of Erk by ER–Src–PELP1 complexes.
Sustained exposure to endogenous or exogenous oestrogen is a well-established cause of breast cancer, underpinning the use of anti-oestrogens and aromatase inhibitors in breast cancer prevention. At least 70% of breast cancers are classified as ER-positive breast cancers, and interfering with oestrogens action has been a mainstay of breast cancer treatment for more than a century. Early therapies included surgical removal of the ovaries, but the synthesis of competitive inhibitors of oestrogen–ER binding during the 1970s led to the first, and to date most successful, targeted cancer therapy: the selective oestrogen receptor modulator (SERM) tamoxifen. Adjuvant therapy with tamoxifen almost halves the rate of disease recurrence and reduces the annual breast cancer death rate by one-third, making a significant contribution to the 25–30% decrease in breast cancer mortality in the past two decades. Subsequently, other new, effective endocrine therapies have been developed that target oestrogen synthesis (such as aromatase inhibitors or ER signalling (such as other SERMs and 'pure' anti-oestrogens”.
http://www.ks.uiuc.edu
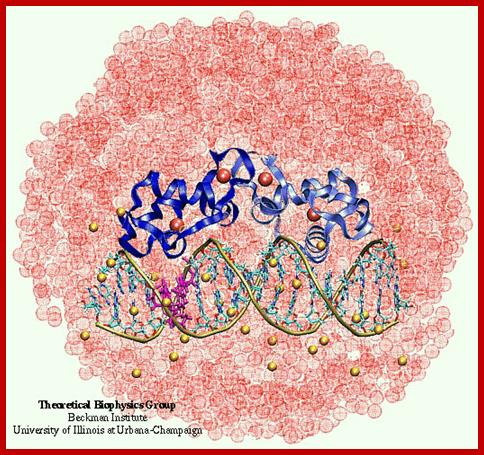
http://www.ks.uiuc.edu
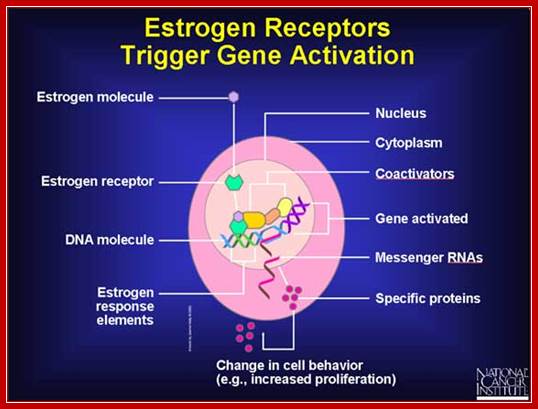
Estrogen Receptor Interacting with DNA; Nuclear hormone receptor proteins form a class of ligand activated proteins that, when bound to specific sequences of DNA serve as on-off switches for transcription within the cell nucleus. These switches control the development and differentiation of skin, bone and behavioral centers in the brain, as well as the continual regulation of reproductive tissues. The hormone receptor recognizes with markedly increased affinity a hormone response element (HRE). HRE is a specific DNA sequence that typically contains two consensus hexameric half-sites. The identity of a response element resides in three features: the sequence of the base pairs in the half-site, the number of base pairs between the half-sites and the relative orientation of the two half-sites. Thus each receptor protein dimer that binds the DNA has to recognize the sequence, spacing and orientation of the half-sites within their response element. ER-ERE system; ER-G/ERE system; Estrogen response elements and estrogen activation of transcription: Estrogen binds to the already DNA bound receptors and induce conformational changes, that leads to the recruitment of other co regulators whereby they modify the chromatin and activate transcription. In this picture one can see the methyl group bound to Arginine of the histone tail is removed. Tom Connor Bishop, Dorina Kosztin et al, http://advan.physiology.org
Autocrine: The released signals act on their own cells and stimulate their own cells to higher level.
Paracrine: Signal released act on nearby cells.
Endocrines: These are hormones; long distance targets.
Synaptic: Neurotransmitters released are transmitted via synaptic cleft or by synaptic mechanism.
Glucocorticoids’ induced gene expression:
Most of the steroid hormones have intracellular receptors, which can be located within the cytoplasm or in the nucleus.
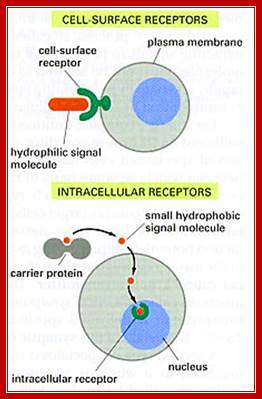
· Glucocorticoids receptors (Type I) are located in cytoplasm but sex hormone receptors and few others (Type II) are found in the nucleus.
In the case of cytoplasmic receptors, the steroids diffuse in to the cell through cell membrane because of its lipid solubility.
· The cytoplasmic receptors are anchored in the cytoplasm with an inhibitor; in the case of Glucocorticoid the receptor is bound by HSP.
The binding of the hormone to cytoplasmic receptor releases the inhibitor from the receptor and forms complex S-Rc (S=steroid, R=receptor, C=cytoplasm and n=nucleus)
· The receptors are also found in nucleus but maintain equilibrium status between cytoplasmic and nuclear.
The S-Rc in dimeric form enters into the nucleus, so it is now called S-Rn, which seeks response elements in the promoter region of the gene and binds and activates the RNAP in the PIC.
· However, activation by the S-Rn complex requires other additional proteins called co-activators.
The Co- activators are CBP protein or P300.
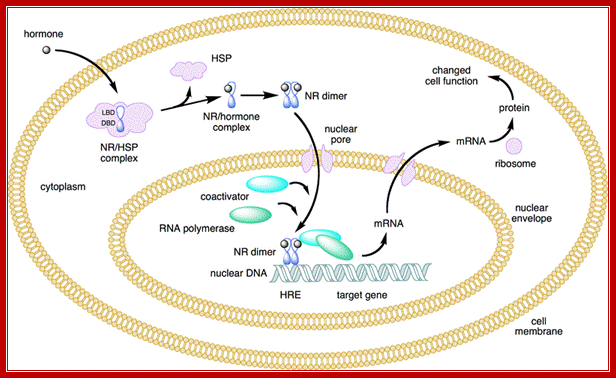
Corticoids in cytoplasm are bound by Hsp, but when the hormone enters into cytoplasm it binds to the receptor, where the Hsp is released and the receptor bound hormone enters the nucleus where they bind to their respective HREs and recruit several co-activators and initiate gene expression. Glucocorticoids induce 80-100 genes.
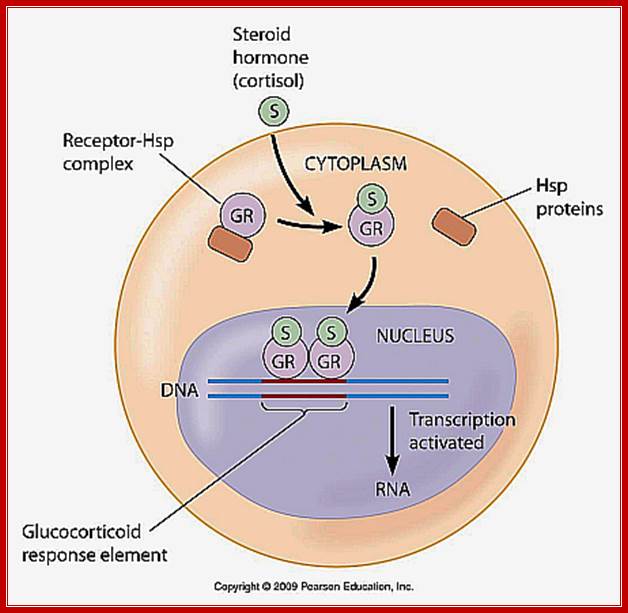

http://www.mun.ca
· The co-activator mediates between the ligand bound receptor, and another protein already anchored on to a DNA sequence and with one of the TFs of the transcriptional apparatus.
· The CBP has acetylating property, by which it can acetylate histone tails and make them to dissociate from the DNA, and facilitate the assembly of the transcriptional apparatus properly for initiation.
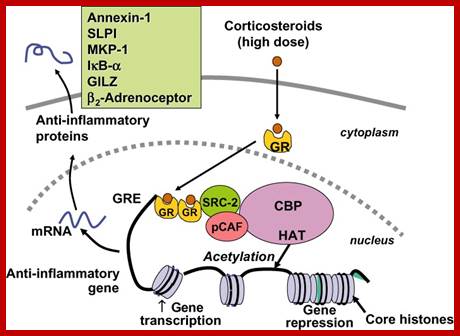
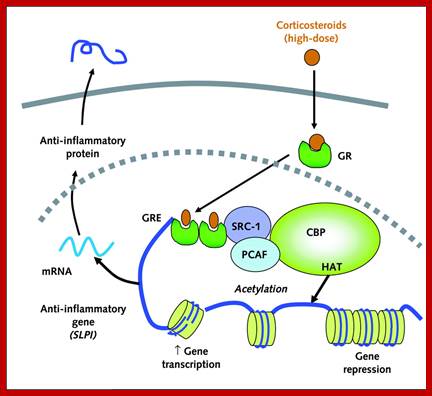
Top.fig; Glucocorticoid activation of anti-inflammatory gene expression. Glucocorticoids bind to cytoplasmic glucocorticoid receptors (GR) which translocate to the nucleus where they bind to glucocorticoid response elements (GRE) in the promoter region of steroid-sensitive genes and also directly or indirectly to co-activator molecules such as CREB-binding protein (CBP), p300/CBP activating factor (pCAF) or steroid receptor coactivator-2 (SRC-2), which have intrinsic histone acetyltransferase (HAT) activity, causing acetylation of lysines on histone H4, which leads to activation of genes encoding anti-inflammatory proteins, such as secretory leukoprotease inhibitor (SLPI), mitogen-activated kinase phosphatase-1 (MKP-1), inhibitor of nuclear factor κB (IκB-α) and glucocorticoid-induced leucine zipper protein (GILZ).
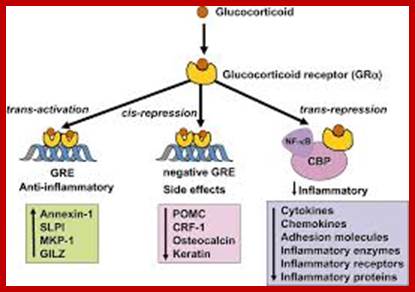
Bottom fig;.Glucocorticoids regulate gene expression in several ways. Glucocorticoids enter the cell to bind to glucocorticoid receptors (GR) in the cytoplasm that translocate to the nucleus. GR homodimers bind to glucocorticoid-response elements (GRE) in the promoter region of steroid-sensitive genes, which may encode anti-inflammatory proteins. Less commonly, GR homodimers interact with negative GREs to suppress genes. Nuclear GR also interact with co-activator molecules, such as CREB-binding protein (CBP), which is activated by pro-inflammatory transcription factors, such as nuclear factor-κB (NF-κB), thus switching off the inflammatory genes that are activated by these transcription factors. Other abbreviations: CRF, corticotrophin releasing factor; GILZ, glucocorticoid-induced leucine zipper protein; IκB-α, inhibitor of NF-κB; MKP-1, mitogen-activated kinase phosphatase-1; POMC, proopiomelanocortin; SLPI, secretory leukoprotease inhibitor. https://www.researchgate.net
Corticosteroid suppression of activated inflammatory genes: Inflammatory genes are activated by inflammatory stimuli, such as interleukin (IL)-1β or tumour necrosis factor (TNF)-α, resulting in activation of inhibitor of I-κB kinase (IKK)2, which activates the transcription factor nuclear factor (NF)-κB. A dimer of p50 and p65 NF-κB translocates to the nucleus and binds to specific κB recognition sites and also to coactivators, such as cAMP-response-element-binding-protein-binding protein (CBP) or p300/CBP-associated factor (pCAF), which have intrinsic histone acetyltransferase (HAT) activity. This results in acetylation of core histone H4, resulting in increased expression of genes encoding multiple inflammatory proteins. Glucocorticoid receptors (GRs), after activation by corticosteroids, translocate to the nucleus and bind to coactivators in order to inhibit HAT activity directly and recruiting histone deacetylase (HDAC)2, which reverses histone acetylation, leading to suppression of these activated inflammatory genes.
Inhibition of p38 mitogen-activated protein kinase (MAPK) by corticosteroids: p38 MAPK is activated by inflammatory stresses though activation of MAPK kinase (MKK)-3 and -6. p38 phosphorylates (P) MAPK-activated protein kinase (MAPKAPK)-2, which plays a role in stabilizing mRNA encoding several inflammatory proteins, such as tumour necrosis factor (TNF)-α, interleukin (IL)-1β, IL-6, IL-8, granulocyte-macrophage colony-stimulating factor (GM-CSF) and cyclooxygenase (COX)-2. This mRNA is characterized by adenine–uracil-rich elements (AREs) in the 3′-untranslated region, which make the mRNA unstable and rapidly degraded. ARE-binding proteins (AREBPs) stabilize these proteins and may be activated (probably indirectly) by MAPKAPK-2. Corticosteroids induce the expression of MAPK phosphatase (MKP)-1, which inhibits p38 and, thus, prevents the stabilization of multiple inflammatory proteins. GR: glucocorticoid receptor; GRE: glucocorticoid response element. ↑: increase; suppression.
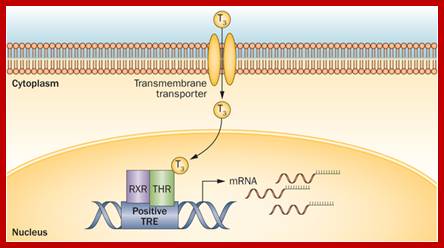
Thyroid hormone induced gene expression:
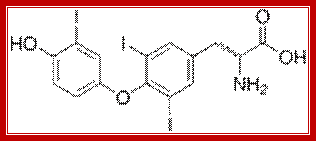
Thyroid is an important organ in human body. In response variety of factors, it releases thyroxine T3 and T4, in different states, which are finally activated and act on different tissues.
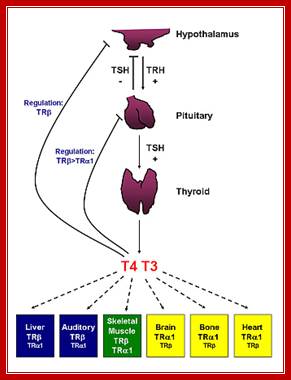
· Thyroid hormone receptors and resistance to thyroid hormone disorders; Thyroid hormone action is predominantly mediated by thyroid hormone receptors (THRs), which are encoded by the thyroid hormone receptor α (THRA) and thyroid hormone receptor β (THRB) genes. Patients with mutations in THRB present with resistance to thyroid hormone β (RTHβ), which is a disorder characterized by elevated levels of thyroid hormone, normal or elevated levels of TSH and goitre. Mechanistic insights about the contributions of THRβ to various processes, including colour vision, development of the cochlea and the cerebellum, and normal functioning of the adult liver and heart, have been obtained by either introducing human THRB mutations into mice or by deletion of the mouse Thrb gene. The introduction of the same mutations that mimic human THRβ alterations into the mouse Thra and Thrb genes resulted in distinct phenotypes, which suggests that THRA and THRB might have non-overlapping functions in human physiology. These studies also suggested that THRA mutations might not be lethal. Seven patients with mutations in THRα have since been described. These patients have RTHα and presented with major abnormalities in growth and gastrointestinal function. The hypothalamic–pituitary–thyroid axis in these individuals is minimally affected, which suggests that the central T3 feedback loop is not impaired in patients with RTHα, in stark contrast to patients with RTHβ. Tânia M. Ortiga-Carvalho,
Aniket R. Sidhaye ; & Fredric E. Wondisford; http://www.nature.com
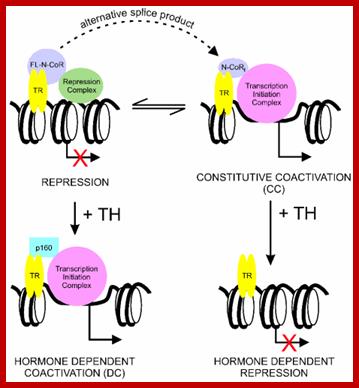
Thyroid hormone receptors, when not bound to thyroxine, remain bound to TRE or Thyroxin Response Element and repress gene activation or expression.
· But when thyroxine, lipid soluble compound, is present, it enters the nucleus and binds to thyroxin receptor. Binding of the thyroxin to the receptor, transforms it into an activator, which on recruiting cofactors activate its gene.
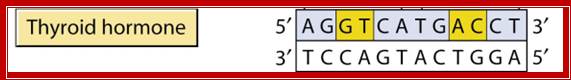
Binding receptor sequences.
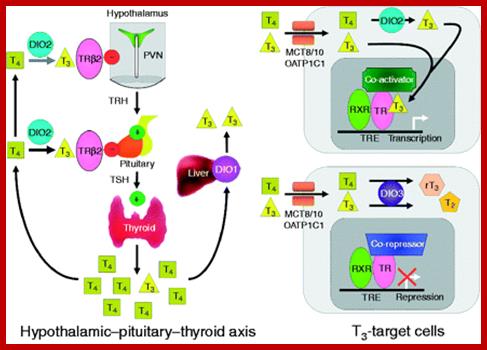
Systemic thyroid hormone concentrations are controlled by the negative feedback regulation of the hypothalamic–pituitary–thyroid (HPT) axis. TRH stimulates the release of TSH from the anterior pituitary, which then stimulates the synthesis and secretion of T4 and T3 by the thyroid gland. DIO2 converts the pro-hormone T4 to the active hormone T3, which binds and activates TRβ2 in the hypothalamus and pituitary, resulting in the feedback inhibition of TRH production and TSH secretion. DIO1 also converts T4 to T3 in the liver, contributing to the pool of circulating T3. Thyroid hormones enter target cells via specific cell membrane transporters and intracellular supplies of T3 to the nucleus of T3-target cells are regulated by the relative activities of DIO2 and DIO3. Expression of DIO2 results in the activation of T4 to T3, increased intracellular T3 concentrations and stimulation of T3-target gene transcription. Expression of DIO3 prevents the activation of T4 and inactivates T3, resulting in the repression of T3-target gene transcription. PVN, paraventricular nucleus; TRH, thyrotrophin-releasing hormone; TSH, thyroid-stimulating hormone; DIO1, DIO2 and DIO3, type 1, 2 and 3 deiodinases; MCT8 and MCT10, monocarboxylate transporters 8 and 10; OATP1C1, organic acid transporter protein-1C1; TR, thyroid hormone receptor; TRβ2, thyroid hormone receptor β2; RXR, retinoid X receptor; T4, thyroxine; T3, 3,5,3′-L-triiodothyronine; rT3, 3,3′,5′-triiodothyronine; T2, 3,3′-diiodothyronine.;Thyroxine can act as activator or repressor; http://joe.endocrinology-journals.org
When thyroxine binds the complex recruits co activators such as HAT and acetylate histone tails making the chromatin active for the binding of Basal transcriptional apparatus (BTA) and initiates transcription.
Gene expression containing CREs:
The CREBp, cAMP Response Element Binding protein, is another protein often binds to response elements CRE called cAMP Response Element, which is a common cytoplasmic protein, responds to cAMP mediated actions. Many neurotransmitters and few hormones bind to cell surface receptors, which are, not all, G protein linked receptors.
· When a ligand binds to such G linked receptors, its cytosolic side, activates Adenyl Cyclase. The enzyme acts on ATP and converts it to cAMP, which is an important second messenger.
cAMP in turn binds to cAMP dependent protein kinase (cAPK) and activates the catalytic subunit of cAPK.
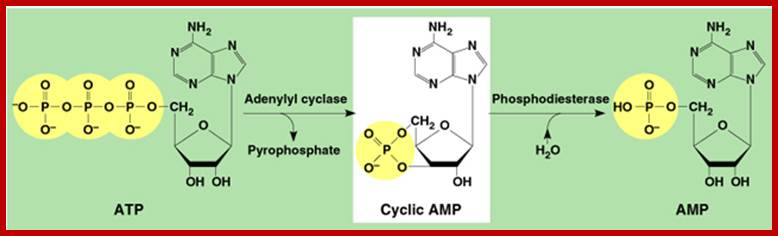
https://www.slideshare.net; fig.cox.miami.edu
· The catalytic subunit moves into the nucleus where it phosphorylates ser 133 of the cyclic AMP response element binding protein called CREBP.
The phosphorylated CREBP then binds cAMP response element (CRE) and contacts the PIC and activates the RNAP for inducting transcription; this may include recruitment of other chromosomal remodeling factors.
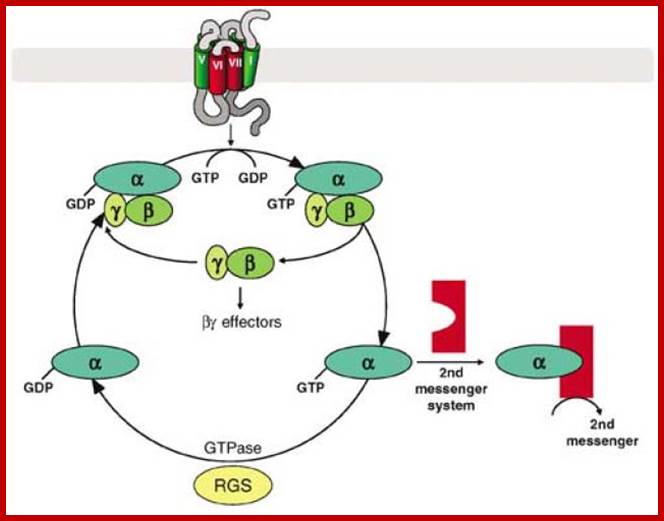
Signal transduction induces a signal cascade of kinases that results in different effects; some of the effects are given below in the form of diagrams.
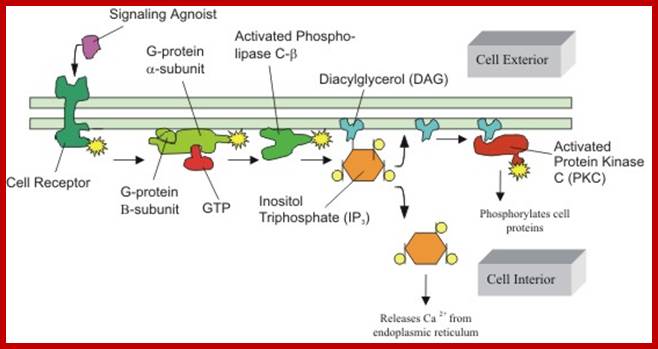
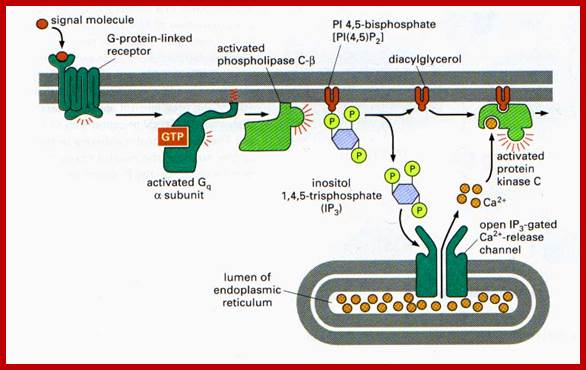
www.lifenews.com; https://www.slideshare.net
Higher levels of calcium ions also activate the CREBP. When there is an increase in Ca2+ ions, in response to cAMP, they bind to inactive Calmodulin. Binding of Ca2+ activates Calmodulin, which in turn activates CaM dependent CaM-kinase, which phosphorylates serine 133 of CREBP, which then binds to CRE elements and activates transcription. Synthesis of cAMP leads to a cascade effect on cellular metabolism and also ob gene expression.
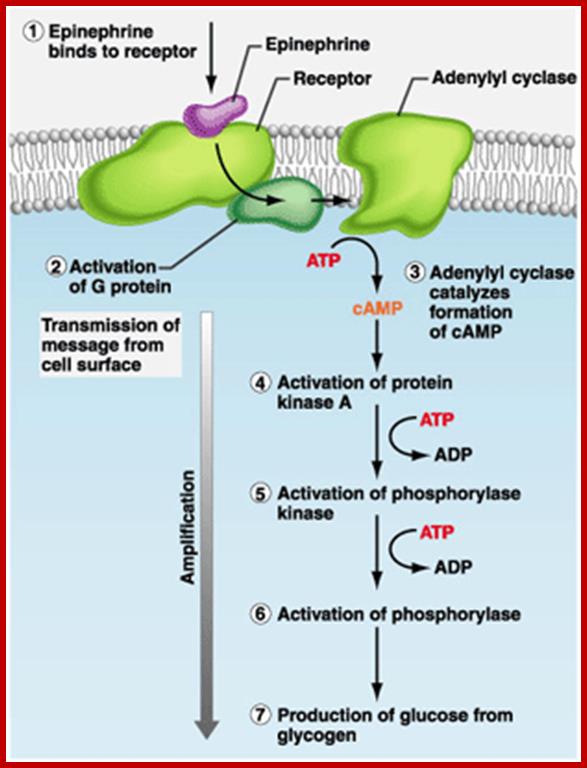
Signal of epinephrine transduction; https://www.studyblue.com
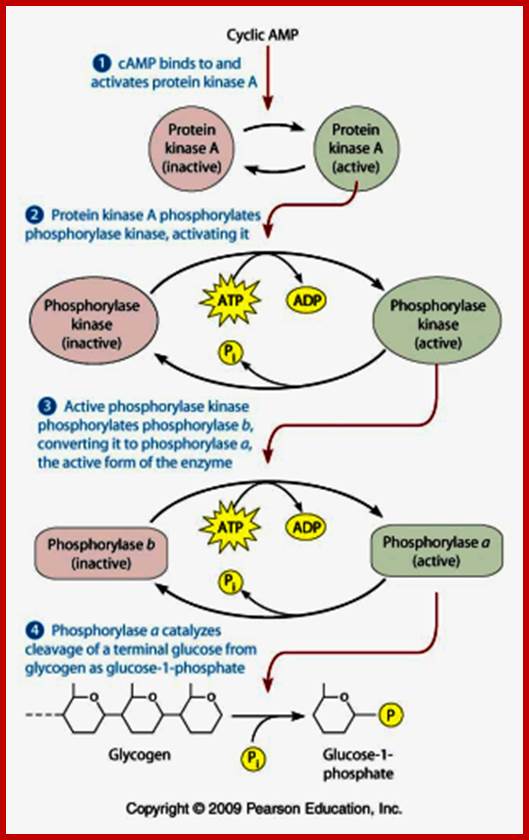
cAMP action on metabolic upregulation;
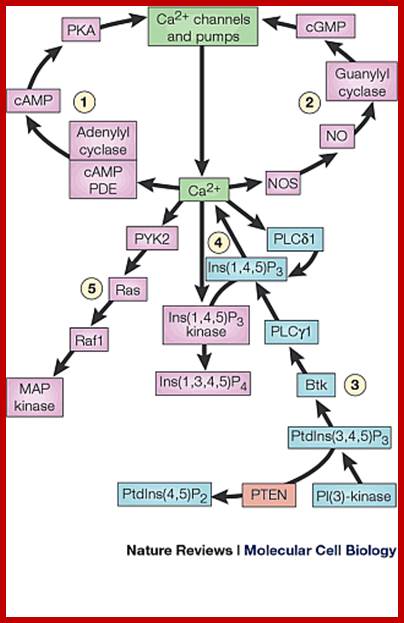
Effect of Ca+2 on diverse targets;
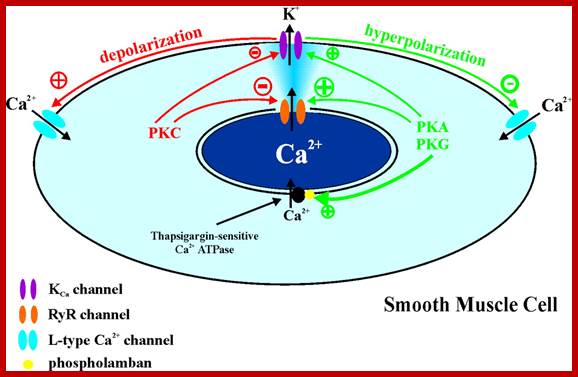
Possible mechanisms of action of PKA/PKG and PKC on Ca2+sparks, BK.Ca channels, and SR Ca2+-ATPase in arterial smooth muscle cells. Activation of PKA or PKG increases Ca2+ spark frequency and increases Ca2+ load of the SR (probably through activation of the SR Ca2+-ATPase via disinhibition of phospholamban). Increased Ca2+ spark frequency could occur due to a direct effect on the RyR channel and/or be a secondary effect from increased SR Ca2+ load. PKA/PKG activation also increases the activity of BKCa channels, which could manifest itself by an elevation in STOC amplitude and steady-state BKCa channel activity. Synergistic effect of increased Ca2+ spark frequency and a direct effect on BKCa channels will result in a significant increase of BKCa channel activity. Membrane potential hyperpolarization closes L-type Ca2+channels, which reduces Ca2+influx, lowering the cytoplasmic Ca2+ concentration, and leads to vasodilation (see Ref. 157). Activation of PKC reduces frequency of Ca2+ sparks and amplitude of STOCs, due to a direct inhibitory effect on RyR channels and BKCa channels, respectively. Additive effect results in decreased BKCa channel activity, a depolarization of the smooth muscle cell membrane potential, and activation of L-type Ca2+ channels. PKA, PKG, and PKC also modulate the voltage-dependent Ca2+channel, which would contribute to modulation of the Ca2+channel → RyR channel → BKCa channel pathways; Molecular Cell Biology; http://ajpcell.physiology.org
C-Fos gene expression:
Many growth factors such as EGF (epidermal growth factor) and platelet derived growth factor (PDGF); stimulate quiescent cells, under cell culture conditions, to enter into cell cycle from G-0 to G1 to S and G2 and back to G1.
· Stimulus, from growth factors, induces more than 100 genes.
One of the most important genes that respond very early to the stimulus is C-Fos gene, which in turn stimulates the expression of several other genes required for cell cycle.
· The regulatory region of C-fos gene has serum response elements (SRE). Also it has several other sequence modules, which respond differently for different signals.
Serum contains several growth factors; among them serum growth factor is one. The SGF activates some specific gene expression.
· When the ligand binds to cell surface receptors, the cytosolic region of the receptor gets activated by phosphorylation (auto-phoshorylation), which is called RTK pathway.
· RTK-Ras activates MAP- kinase (mitogen activating protein kinase).
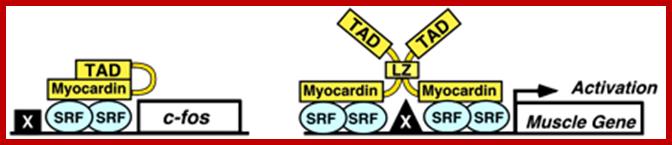
Fos-A model for the regulation of smooth muscle genes by SRF. Myocardia preferentially activates smooth muscle genes controlled by pairs of CArG boxes. The c-fos promoter contains a single CArG box and is not efficiently activated by myocardia. Dimerization of myocardia through the leucine zipper (LZ) may expose the TAD with resulting activation of muscle gene expression. Other promoter-specific factors (X) cooperate with SRF and myocardia. http://www.pnas.org
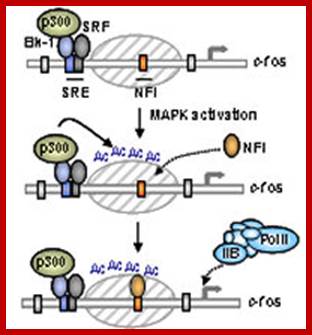
Model of proposed sequential activation of the c-fos promoter. MAP kinase pathway activation triggers Elk-1 activation and its associated p300. p300 subsequently acetylates the adjacent nucleosomes, causing a change in nucleosomal structure, NFI recruitment and the subsequent recruitment and activation of the basal machinery.
The activated MAP-kinase now enters into the nucleus and phosphorylates a protein called Ternary Complex Factor (TCF) at serine found at C- terminal part of the protein.
· MAP-kinase also activates another kinase called pp90. This protein in turn activates two SRF (serum response element binding factor) by phosphorylating ser at 103 position.
Unphosphorylated TCFs and SRFs can bind to the response element but cannot activate transcription.
· Phosphorylated SRFs and TCFs form a complex in the nucleus and bind to SRE and activate C-Fos gene.
The SRF-TCF factors are similar to yeast’s MCM1 and Ste-12 proteins.
Gene Expression in Response to Viral Infection:
Animal viruses as well as bacteria ad other pathogens infect through cell surface receptors or by endocytosis the pathogens are released into cell cytoplasm. The infected cells respond first by using the ds RNA produced during the replication or transcription of the viral genome. Cells infected with pathogens react to generate antiviral immunity by using different mechanisms. The ds RNAs activate cellular protein kinase (PKR), which phosphorylates translation initiation factor eIF-α at specific serine residue. The dsRNA can also be used as defense mechanism by producing si or mi RNA (RNA interference mechanism); depends on the kind of dsRNA produced. At the same time, they also induce Interferons, which on release activate cells to release different IFNs, which depends upon the type of cell that is infected. Several other genes are also activated, so the cell to be in antiviral state; this first defense mechanism called innate immunity, which leads to adaptive or cellular immunity.
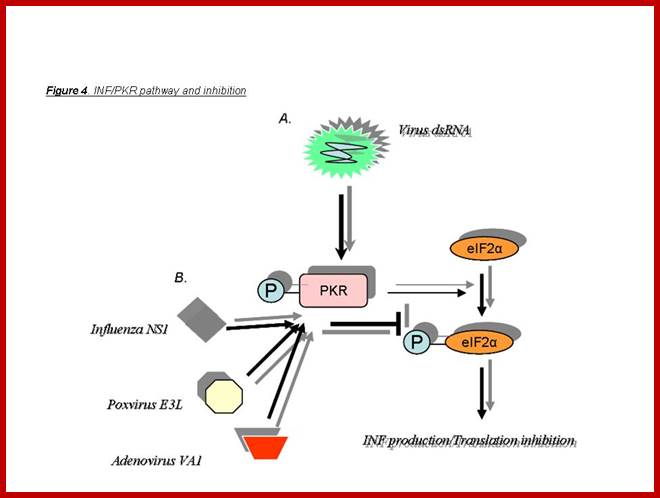
Aleksandar Masic; www.homepage.usask.ca
Immediately after infection, cells respond by innate immunity and make neighboring cells prepared for defense against the viral or any such pathogen attack. In the process the first cell lines infected with pathogens may die; self scarifies for the other sister cells.
Interferons:
Depending upon the infected cell types, they produce three different kinds of Interferons:
IFN-alpha: Produced by leukocytes, (165 a.a) prevent viral DNA or RNA replication, induces MHCI expression. The gene is located on hu-9p22 chromosome
IFN-beta: It is rapidly produced by fibroblasts, (166a.a), and acts on macrophages and activates natural killer cells (NKs). Induce MHC I expression.
IFN-gamma: Extension of IFNs activated lymphocytes produce IFN gamma, (146 a.a). They act on macrophages, activate NK cells. Activate MHCI and MHCII class of genes. They also activate T4 CD4 (cluster domain) helper cells and CD8T-cytotoxic cells.
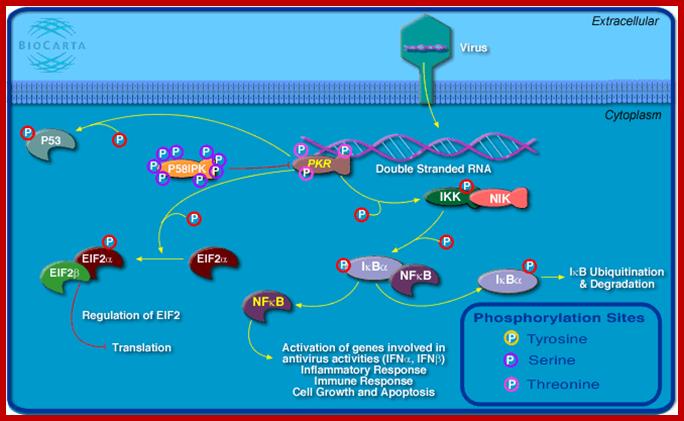
Viral infection leads to dsRNA synthesis during viral gene expression or during its viral genomic RNA replication. The dsRNA has multiple functions. It activates a cellular protein kinase. The activated PKR phosphorylates eIF α, which blocks cellular translation. The PKR phosphorylates Inhibitor (IkB) of NFkB, thus NFkB, a dimer is released, which enters the nucleus and activates several genes involved in antiviral activities; inflammatory responses and activation of several IFN genes (alpha and beta), activates immune response genes. PKR also phosphorylates p53, which acts as a transcription factor for several genes that can lead to the blocking of cell division and ultimately activates cellular apoptosis. Activation PKR can lead to activation interferon response factors that on binding to specific genes can lead to the transcription interferon proteins. When IFNs are released, they activate other cells and induce antiviral state is the said cells. www.bioon.com.cn
Viral infection leads to its transcription and replication leads to the production of RNA with double stranded structure that triggers the phosphorylation of PKR. The PKR has multiple phosphorylation functions. PKR by phosphorylating eIFa blocks cellular translation, phosphorylates P53, phosphorylated IkB thus it gets released form dimeric nuclear k Beta factor, which induces many inflammatory transcriptional activities. The p-lated IkB is subjected to ubiquitination and proteasome activity. NFkB also activates the expression of several Interferons genes, immune responses, cell growth and apoptosis.
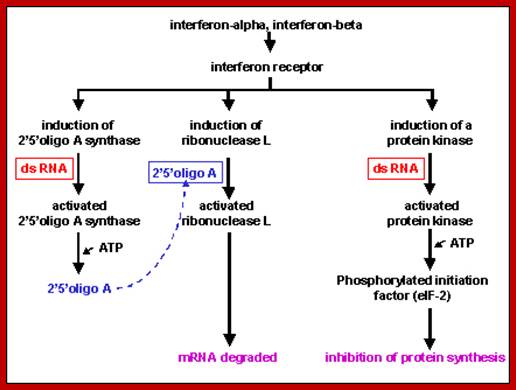
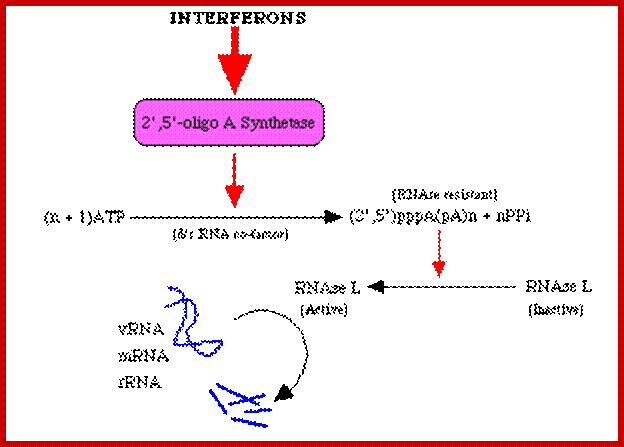
Viral infection also activates several interferon genes as a part of innate immunity. Interferons released from the infected cells, on binding to their specific cell surface receptors, activate genes for the synthesis of 2’5’oligo synthase, induce the synthesis of RNase L, and induce protein kinase; which all lead to blocking protein synthesis and initiates the degradation of all RNAs in the cell. The diagram below shows interferon induced 2’5’oligo synthase and its activity in degrading RNase through the activation RNaseL; their activity requires viral generated dsRNA; www.slideplayer.com
Based on the structure and functions, interferons are classified into several types. They are grouped into Type I , Type II and Type III. This classification is based on the structure and functions. They are very effective at very low concentration of 10x^-14M. They bind to wide variety of targets.
Type I: Consists of Interferon alpha (there are 13 (?) of them) and Interferon beta. Most of them are produced in Leucocytes and fibroblasts and such somatic cell types. They are effectors of innate and adaptive immunity. They activate NK cells, macrophages, MHC class-I protein producing cells which provide protection to CD8 cells from death
Type II: Consists of Interferon gamma, which is derived from lymphocytes; this leads to pervasive effects.
Type III: Consists of interferon lambda and few others.
· Animal cells have specific receptors for each of the mentioned interferons, which are glycoprotein types.
When IFNs bind to receptors; cells respond and develop antiviral defense. Activation of certain genes does this and their products act as defense systems against further viral attack. This acquisition of self defense is often called as “innate immunity”, which leads to Adaptive immunity.
ds RNA dependent PKR activity:
First, while the viral genome, it can be RNA or DNA, is replicated and as the genome is transcribed, certain sequences of RNA generated can fold into dsRNA configuration. This dsRNA activates protein kinase (PKR), which phosphorylates eIF-alpha subunit eIF a chain initiation factor of translation. The phosphorylated eIF-alpha binds to eIF-beta for GDP-GTP exchange, but as the eIF-α is phosphorylated and the eIF-β binds to alpha; it binds so tightly it prevents recycle function. Thus, all cellular eIF-α factors get sequestered; in this way protein synthesis of the infected cells is halted. This can lead to the death of the cell(ls).
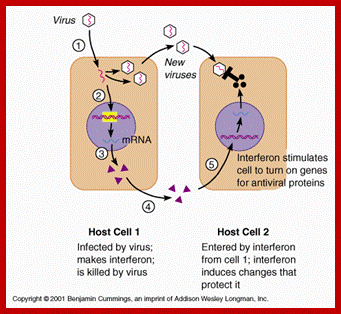
MICROBIAL INFECTIONS OF HUMANS leads to cell produce IFNs; www.slideplayer.com
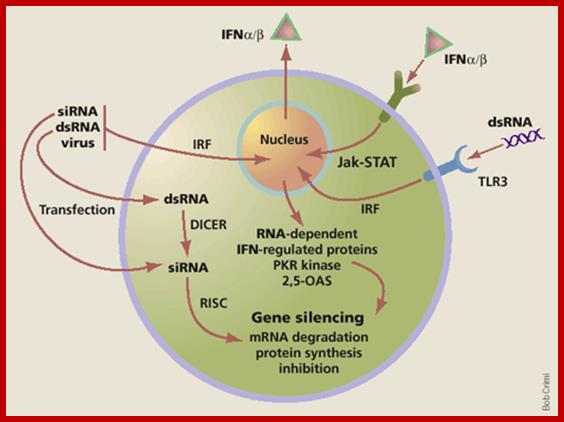
At the same time some dsRNA sequences can induce the synthesis of 2’-5’Adenine oligo synthase. These oligo’s produced activate RNase L, which simply degrades all RNAs; thus, viruses or for that matter most of the other pathogens cannot multiply or propagate and the infected cells die. Some of the dsRNA sequences can attract protein complexes such as Dicer; they in turn in association with RISC generate smaller si or mi RNAs which depends upon the kind of precursor dsRNA generated. The ssRNA strands bind to their own viral RNAs or mRNAs which are complementary to si/mi RNAs and degrade the same or inhibit translation; which is considered as the most potential RNAi mediated defense system.
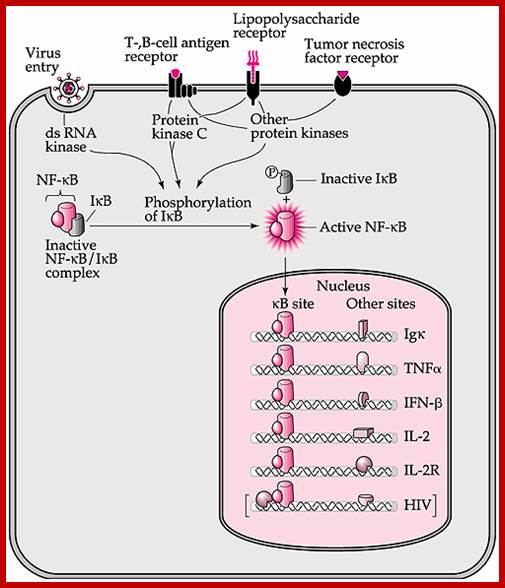
Viral infection can lead to dsRNA synthesis, which in turn activate ds RNA kinase which phosphorylates IkB bound to NFkB. Phosphorylated IkB (inhibitor of NFkB) dissociates form the NFkB. The released NFkB enters nucleus activates whole lot of genes such as IgK, TNF alpha, IFNbeta, IL2 IL2R and others.
Speaking of viral pathogenesis, it must describe the features and factors of viral pathogen, hosts and environment. Over its lifetime, an individual is exposed to many infectious agents, however, in most situations does not develop a disease thanks to factors such as physical and chemical host barriers. In other cases, pathogens circumvent these barriers and cause infection; however, a “biological war” will start between the determinants of pathogenicity and early host defenses. If the virus is able to overcome these first lines of defense, a type of highly specialized and specific protection will be activated. This defense will achieve, in most situations, the infection control and subsequent eradication of the disease. Furthermore, this process will initiate the generation of the immunological memory, enabling the individual with a more quickly and effectively response at the next contact with the same agent. On the contrary, if the foreign agent can overcome both defenses, the result is disease. In certain cases, the line of defense, when triggered, can also cooperate with the damage instead of healing, making the disease more severe. Thus, the immunopathology viral respiratory infection is a frequent consequence of the immune response against many of respiratory pathogens. Furthermore, if the infection is established, the factors or viral virulence determinants and physiological conditions of the host cell will determine; By Ma. Eugenia Manjarrez-Zavala et al-;https://www.intechopen.com
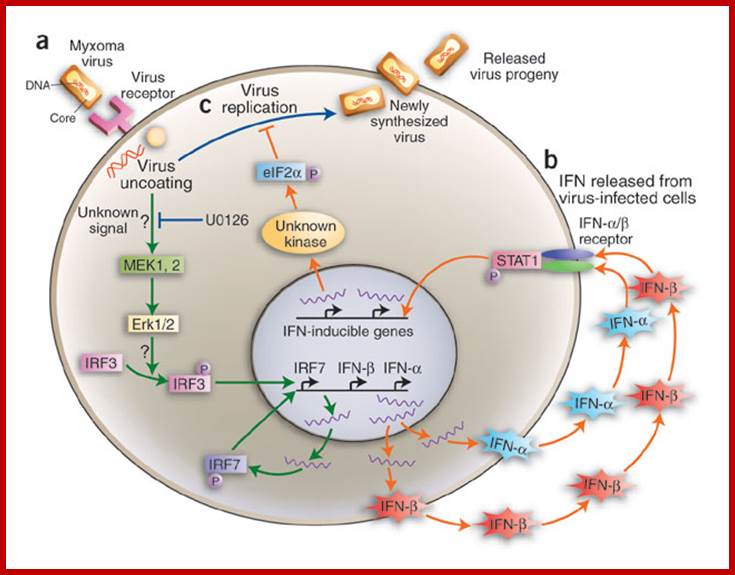
Infection like
Myxoma virus leads to signal transduction including MEK1,2 -> Erk1/2 bind to
IRF3 that activates IRF7 , IFN alpha and beta that binding to its specific
receptors in association with STATS activate more genes like ILs. Resistance
of nonpermissive MEFs to myxoma virus infection is mediated by Erk1/2
MAPK−triggered stimulation of IFN-![]() and IFN-
and IFN-![]() synthesis.
synthesis.
(a) Inoculation of MEF cultures with myxoma virus does not result in
efficient virus multiplication, as judged by the absence of late virus
products. However, inoculation with myxoma virus evokes Erk1/2 signaling,
activation of IRF3 by phosphorylation and de novo synthesis of IRF7.
Activated IRF3 and IRF7 translocate to the nucleus and trigger transcription of
IFN-![]() and IFN-
and IFN-![]() genes, followed by synthesis and secretion of IFN-
genes, followed by synthesis and secretion of IFN-![]() and IFN-
and IFN-![]() . (b) IFN-
. (b) IFN-![]() and IFN-
and IFN-![]() released from the cells that had encountered myxoma virus bind
to IFN-
released from the cells that had encountered myxoma virus bind
to IFN-![]() /
/![]() receptors on other cells in culture, which triggers STAT1
activation, resulting in synthesis of proteins that render cells resistant to
myxoma virus infection. Cellular blockade of myxoma virus replication
correlates with phosphorylation of translation factor eIF2
receptors on other cells in culture, which triggers STAT1
activation, resulting in synthesis of proteins that render cells resistant to
myxoma virus infection. Cellular blockade of myxoma virus replication
correlates with phosphorylation of translation factor eIF2![]() , which leads to inhibition of protein synthesis. The kinase
responsible for eIF2
, which leads to inhibition of protein synthesis. The kinase
responsible for eIF2![]() phosphorylation is apparently not the double-stranded
RNA−dependent kinase, PKR, usually implicated in interferon-mediated
antiviral actions, but another unknown serine-threonine kinase. (c) If
synthesis of IFN-
phosphorylation is apparently not the double-stranded
RNA−dependent kinase, PKR, usually implicated in interferon-mediated
antiviral actions, but another unknown serine-threonine kinase. (c) If
synthesis of IFN-![]() and IFN-
and IFN-![]() is disrupted in the myxoma virus−inoculated cells by the
MEK inhibitor U0126 or by other means, cells become permissive for myxoma virus
infection, resulting in full virus replication. Jan Vil
is disrupted in the myxoma virus−inoculated cells by the
MEK inhibitor U0126 or by other means, cells become permissive for myxoma virus
infection, resulting in full virus replication. Jan Vil![]() ek; www.nature.com
ek; www.nature.com
At the same viral infection via signal transduction also activates cytoplasmic interferon gene regulatory factors (IRFs) in the cell. The phosphorylated regulatory factors (IRFs) bind to their specific IREs’ elements of specific genes and recruit cofactors such as p300/CBP and activate IFN genes. The interferons thus synthesized are released into external space. The released IFNs bind to the receptors of neighboring cells and activate cells into antiviral state; this chain reaction results in cascade effect.
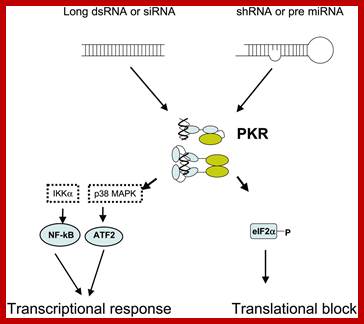
RNA interference in biology and disease; RNA interference (RNAi) is a conserved biologic response to double-stranded RNA that results in the sequence-specific silencing of target gene expression. Over the past 5 years, an intensive research effort has facilitated the rapid movement of RNAi from a relatively obscure biologic phenomenon to a valuable tool used to silence target gene expression and perform large-scale functional genomic screens. In fact, recent studies reported in this journal and others have demonstrated success using RNAi to address the role of oncogene expression in leukemia cell lines and to validate the therapeutic potential of RNAi for treating these blood disorders. In order to advance these applications and gain an appreciation for the future of RNAi both in basic research and in the treatment of diseases caused by aberrant gene expression, it is important to have an understanding of the process of RNAi and its limitations. Carol A. Sledz and Bryan R. G. Williams;http://www.bloodjournal.org
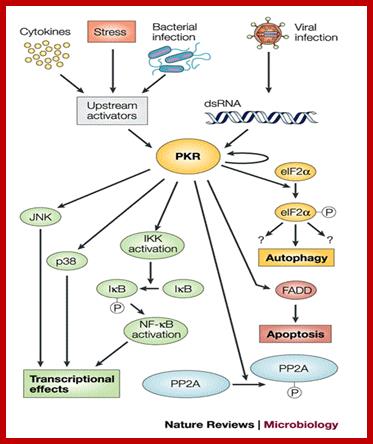
Infection causes dsRNA dependent activation of PKR which in turn has different effects such as activation NfkB, JNK, PP2A; all leads to inactivate the virus perse. www.blogqpot.com
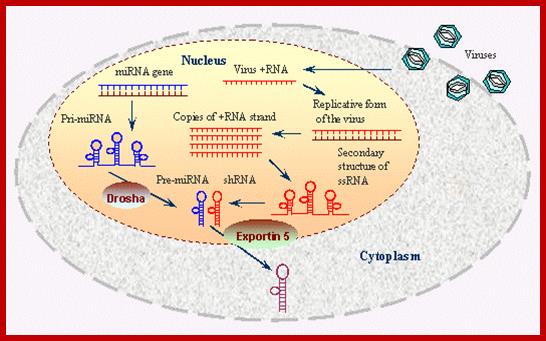
Viral generated dsRNA is used by Dicer and Risc protein complexes
to generate RNAi defense against invading viruses or any other pathogen.
www.bo.pianov.com
Expression of IFN beta gene:
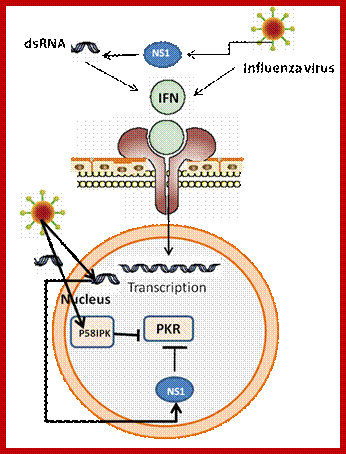
IFN genes contain certain regulator elements such as IREs. For example activation IFN beta requires a variety of factors such as NFkB, Jun/ATF and HMG for its activation. The most important feature of the IFN genes is IRE Interferon Response Elements to which IRF–Interferon Response Factors bind and activate IFN genes. Binding IRF to IRE leads to the binding of several other factors. There are several IRFs (at least 9 of them) of which IRF3 is important in activating IFN-beta. Viral infection induces dsRNA activation of proteins that leads to phosphorylation of IRF3. The PKR phosphorylates IkB (inhibitor of NF kB) which is bound to NFkB (a dimer protein). Phosphorylation of IkB, makes the IkB to be released and it is destroyed by ubiquitinated proteolysis. Then NFkB factors (65-50kd) move from cytoplasm into the nucleus. IRF3, NFkB, jun/ATF and HMG proteins bind to their specific sites in the promoter elements of IFN genes. High mobility proteins are a family of proteins; bind to the minor groove of specific DNA sequences and induce DNA bending. The binding of these factors recruit cofactors such as CBP/P300 and mediator complexes. The DNA then loops and generates a structure referred to as enhanceosome that activates the IFNβ gene. The mechanism for other IFN genes is more or less the same except for different IRFs and other cellular factors. IRF3 is involved in activating IFN alpha and beta. IRF7 is produced in lymphoid cells. Each of these genes has what is called positive regulatory domain (PRD) and negative regulatory domains (NRD). They are bound by NRDs before viral infection. Factors bound to NRD domains are released after viral activation and viral PRDs bind and activate the genes. Trans-retinoic acid and IFN-alpha inhibit cell proliferation and induce squamos carcinoma cell apoptosis.
IFN-alpha:
-HMG-Jun/ATF-(GAAAG/C)3 –CAAT-NFkB-HG—TATA--+1>
IFN-beta:
---Hmg---cJun/ATF—HMG-IRF3-IRF7—NFkB—HMG---TATA--+1>
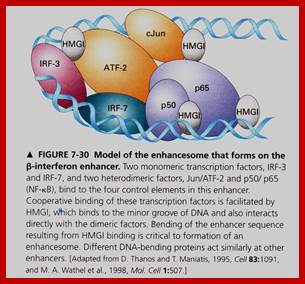
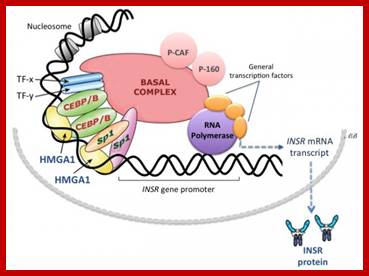
The IFN-β enhanceosome complex. Assembly of the IFN-β enhanceosome creates a stereospecific interaction surface for recruitment of CBP/p300 and the basal transcription machinery to allow multiple rounds of transcription. GTFsindicate general transcription factors. p160 refers to the SRC/TIF/pCIP family of coactivators.; Top Fig.CREB-binding Protein and p300 in Transcriptional Regulation ;http://www.jbc.org; Bottom Fig.https://globalmedicaldiscovery.com
With time, say 4-5 days after infection, the antigenic proteins processed by APC cells lead to activation of B and T cells. Synthesis of IFN alpha/beta also leads to activation T and B cells called adaptive immunity, where specific antibodies are produced against specific antigens.
First line of defence innate immunity leads to adaptive immunity where T and B- lymphocytes are activated to produce more antibodies against pathogens.
IFNs perse activate several cellular genes; can be 70-90 different genes, thus they have far-reaching effects. Each of the IFN regulated genes contain elaborates regulatory elements at its 5’ upstream.
Historically these IFNs were first observed by two Japanese workers in the University of Japan >54 years ago; they are Yasu-ichi Nagano and Yasuhiko Kojimma. They showed that untreated but viral infected skin cells inhibited second time viral infection. This discovery was published in a France journal ‘dela Societe de Biologie ‘(1954). In London Alick Isaacs and Jean Lindermann worked on heat killed flu viruses (1957). Injected heat killed viruses that induced resistance; they called the factors that are responsible for such resistance as ‘INTERFERON’, means interference with viral function.
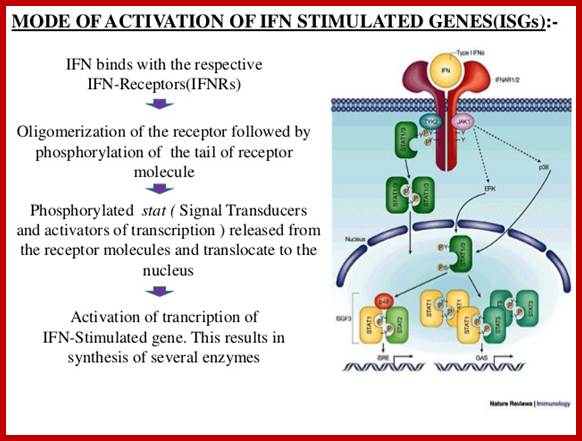
The IFNs are also called cytokines, most of them are glycoproteins. They will be circulating in blood stream and activate cells through their specific receptors and make these cells anti viral. Interferons do activate several cellular genes which have far reaching effects on cells. Cells respond to IFNs through specific receptors at cell surface. In order to activate IFN stimulated genes (ISGs) they require interferon regulatory factors (IRFs).
Interferons are called by that name, because they interfere with the viral multiplication and act as first and foremost defense systems against first and second viral attack. The IFNs are also called cytokines and they are glycoproteins. They are produced in response to pathogen infection within 2-3hrs of infection. Released IFNs act on the receptors found on several kinds of cells and make them immune for viral attack; this type response to viruses is called innate response. They are not viral species specific, but they specific to tissue types; murine tissue is specific to murine cells.
Interferons released from the infected cells circulate in the extra cellular fluid and act on variety of other cells. Different types of IFNs act on different types of cells, which is determined by the kind of IFN receptors they contain. The binding of IFNs to cell surface receptors is more or less specific. The binding leads to activation of receptor mediated kinase that leads to signal cascade in activating specific genes.
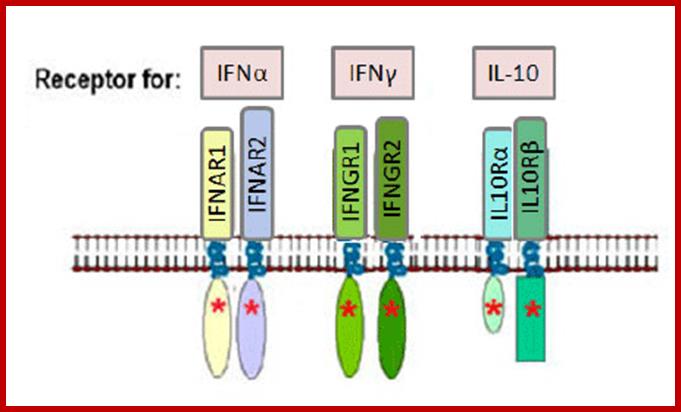
Type-I IFN-alpha and beta bind to IFNAR receptors and Type-II IFN-gamma bind to IFNGR receptors. Type III- lambda bind to different types of ILe receptors.
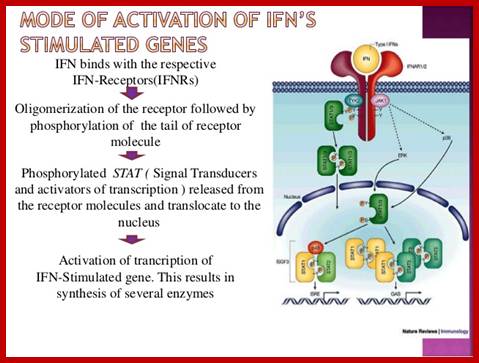
Investigations on IFNs receptors have revealed that there is a family of receptor associated with protein kinases. Inside one finds another family of transcriptional factors called STATS, meaning Signal Transducers and Activators of Transcription. Binding of IFNs to their cell specific receptors activate receptor endowed kinases; they may be tyrosine or serine or threonine kinases. They in turn activate STATs by phosphorylating specific STATs among the family of STATs.
The receptors for type-I IFNs are–IFNAR1/JYK2/-IFNAR2/JAK1; they phosphorylate STAT1-STAT2 which interact with IRF9. Receptors for type-II IFNs are IFNGR2/IFNGR1-JAK1-IFNGR1/IFNGR2-JAK1; they interact with STAT1 and STAT2. Type III IFN bind to receptors like IL10R2/Tyk2-IFNLR! /JAK1.
· They form a kind of link between cell surface receptors to gene expression.
IFN alpha, within 5-15 minutes of binding to cell surface receptor, induces the expression of some subset of genes by 20 fold.
The genes respond to IFN-alpha contain Interferon Stimulated Response Elements (ISRE) in their regulatory regions.
https://www.slideshare.net/ www.redcomovamos.org
Four different STAT proteins, (in combinations), have been found to bind to ISRE (Interferon Stimulated Response Elements) and their genes have been cloned.
· STAT-alpha is 91 KD proteins, STAT-beta is 84kd protein, and they are identical with exception that STAT1-alpha is 30 amino acids longer at C-terminal. The same gene codes both, but by alternate splicing they generate two different polypeptides.
The third is STAT-2 is 113kd protein. This protein shows 59% homology with STAT 1-alpha and STAT 1-beta.
· All these proteins have SH2 domains for protein-protein interaction to dimerize to produce homodimers or heterodimers among them.
The fourth protein, called P48 is actually the DNA binding protein and it is unrelated to the first three STATs and it has no SH2 domains.
· Only those Cells, which are stimulated by IFN-alpha, produce the above-mentioned STATs, which are found in cytoplasm.
On the contrary, whether the cell is stimulated or not, P48 is found both in cytosol and the nucleus. The P48 can bind to DNA to specific ISRE elements even without signals and the binding is independent of STATS.
· The receptors for all the three IFNs belong to a super family of receptors.
They don’t have intrinsic protein kinase activity, but the cytosolic domain of the receptor on binding to the ligand gets activated. In this state they associate with one or more cytosolic protein kinases and activate them. But receptors without ligand binding don’t do so.
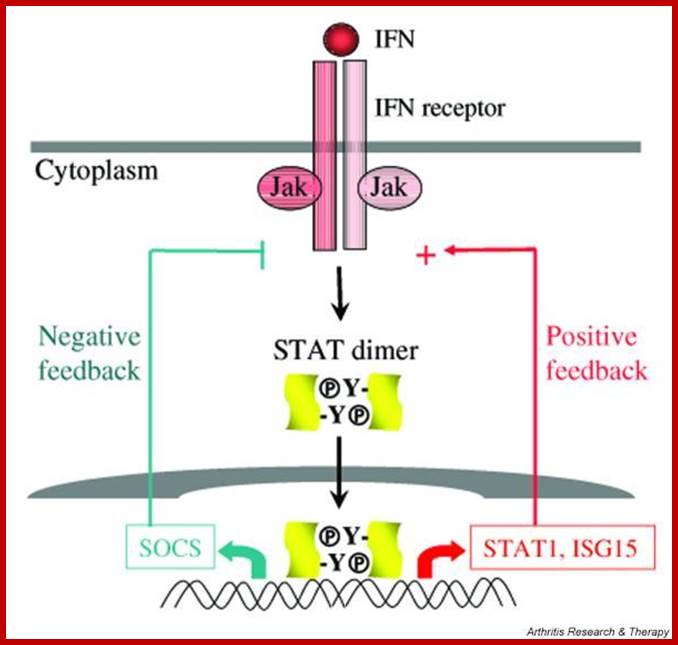
· The activated INF’s receptor is believed to act as TYK-2 and JAk-1 kinase (tyrosine kinase-2 and Janus kinase-1). Activated TYK-2 or JAK-1 phosphorylates STATS at Tyrosine residues.
Phosphorylated STATS interact with one another and generate heterodimers (one with phosphorylated protein dimerizes with another containing SH2 group). The dimerized STATs move into the nucleus and interact with P48, which is already bound to ISRE; this leads to activation of the transcriptional apparatus.
Each type of IFNs induces unique subset of genes by using IFNs-STATS signaling pathway. This specificity stems from the fact that each receptor is different and their associated proteins are different and the STATS are different and their ISRE elements may be different but may be located in different positions.
IMMUNOLOGY - CHAPTER THIRTEEN; CYTOKINES AND IMMUNOREGULATION; Dr Gene Mayer
Cytokines are a diverse group of non-antibody proteins that act as mediators between cells. They were initially identified as products of immune cells that act as mediators and regulators of immune processes but many cytokines are now known to be produced by cells other than immune cells and they can have effects on non-immune cells as well. Cytokines are currently being used clinically as biological response modifiers for the treatment of various disorders. The term cytokine is a general term used to describe a large group of proteins but there are other terms that are commonly used to describe particular kinds of cytokines. These include: NMonokines, Lymphokines, interleukins and chemokines and cytokines; http://www.microbiologybook.org/
· Specificity stems from cell lines, e.g. Mutants for TYK-2 are insensitive to IFN-alpha, but are sensitive to IFN-gamma, which suggests the TYK-2 is a component of IFN-alpha specific pathway.
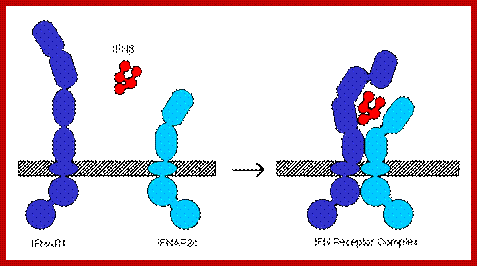
IFN alpha and beta receptors; http://www.invivogen.com
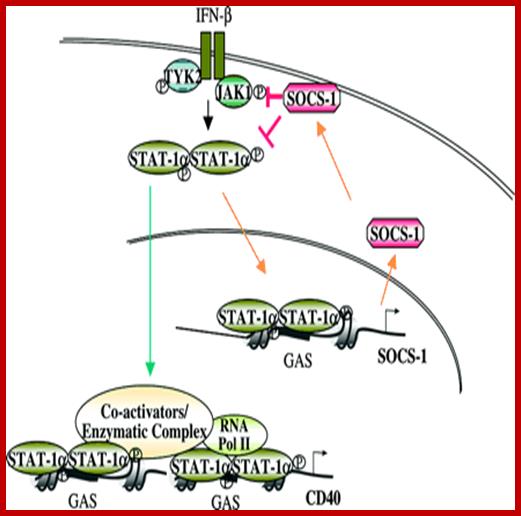
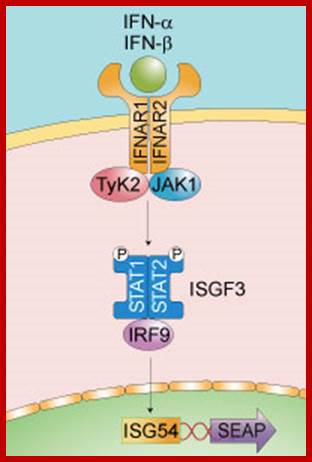
www.blogqpot.com/images
Activated IFN b self phosphorylates and then it phosphorylates STATs, the Stats then move into the nucleus and bind to response elements. This leads to the recruitment of coactivator and RNAP complex to the promoters for transcription.
Cell line mutants for JAK-1 are sensitive to both IFN-alpha and IFN-gamma, thus they play a role in both pathways.
· However, IFN-gamma receptors have shown to be
associated with second kinase called JAK-2.
Antigen stimulated T-helper cells, release or secrete IFN-gamma kind of Interferons or what is called interleukins.
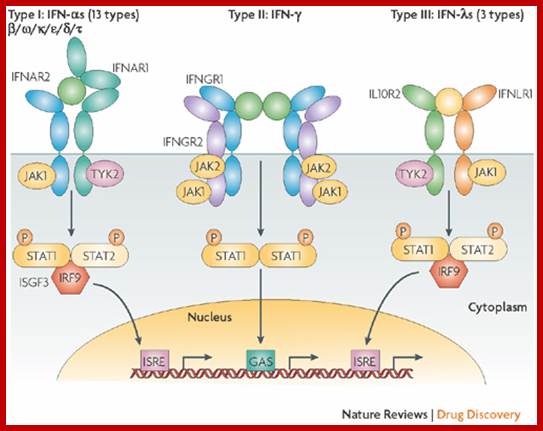
https://www.heighpubs.org
· Most of the other cells contain receptors for IFN-gamma.
When IFN-gamma binds to its cellular receptors; activated cells induce transcription of a set of genes rendering cells to be in antiviral state.
· Each of the genes that respond to IFN-gamma stimulation contains, in their regulator regions, the GAF Response Elements (GAF-RE). GAF means gamma-associated factors.
A transcriptional factor, required for IFN-gamma mediated activation, has been identified and cloned. Its Mol.wt is 91 KD and it is called STAT-1 (which is a signal transducer and activator of transcription).
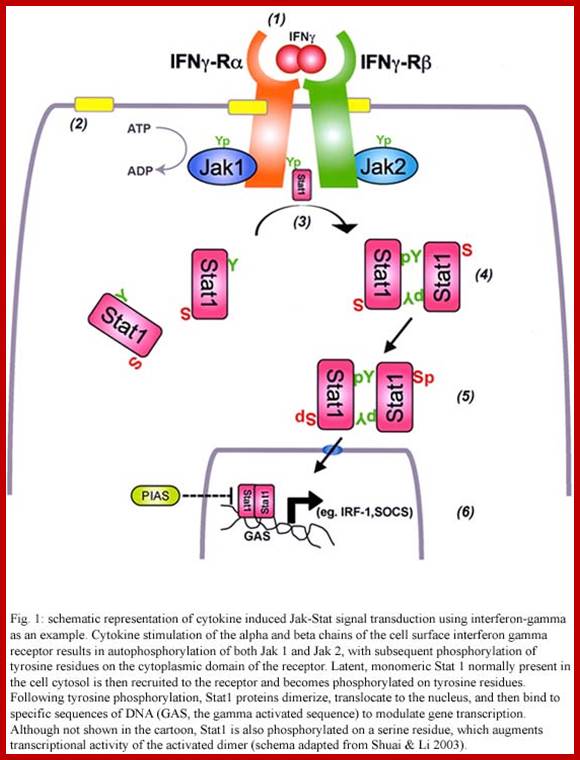
https://www.researchgate.net
· IFN-gamma transduces the signal by kinase activity, which results in the phosphorylation of STAT-1 at one of the tyrosine moieties. The phosphorylated STAT-1 dimerizes and moves into the nucleus and a bind to GAF, which is already bound to GAF-RE and the gene, is activated for transcription.
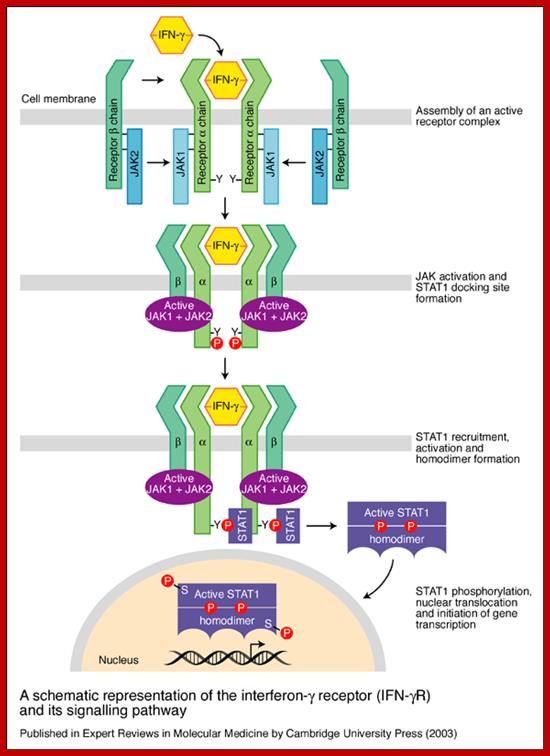
www.medscape.org
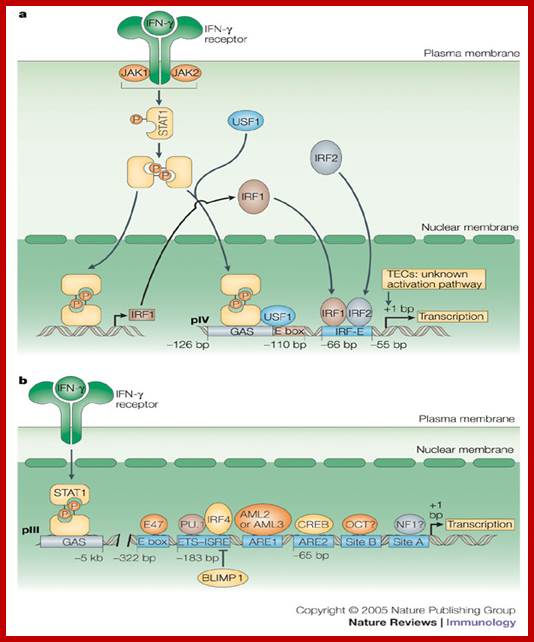
https://image.slidesharecdn.com
Cells, mutants for STAT-1 alpha, cannot be phosphorylated because of the absence of tyrosine residues and STAT-1 Alpha does not move into the nucleus, in spite of the stimulation by IFN-gamma.
· Interestingly, stimulation of cells by IFN-gamma, in the absence of IFN-alpha, leads to phosphorylation and dimrization of STAT-1 and they move into the nucleus and bind to GAF and stimulate only GAF bound to GAF response elements containing genes, it is an exclusive gene expression.
Signaling pathway of cytokine receptor families is more or less similar to interferon JAK-STAT or TYK-STAT pathways.
· In an generalized version, it know known, that binding of the ligand to a receptor causes dimrization of the receptor at cytosolic side, which interact with kinases of JAK family and activates them, which in turn specifically phosphorylate a set of transcriptional factors, which then move and bind directly to response elements or bind to a factor which is already bound the response element and activate the transcriptional apparatus.
This pathway appears to operate in other hormone stimulated gene expression, ex. EGF binding to the receptor gets activated to activate RTK, which then stimulate cytosolic STATs, which move into the nucleus and bind to c-Fos RE and activate genes, called serum inducible genes.