RNAP-III Promoter:
& Transcription.
RNA polymerase III: Transcription by RNA polymerase III: more complex than we thought; Robert J; http://www.nature.com.
It is also called Pol III, �transcribes specific DNA segments to synthesize ribosomal 5S rRNA, tRNA and few other small RNAs such as U6 spliceosomal RNA, RNase P and RNase mrp RNA, 7SL RNA (the RNA component of the signal recognition particles), Vault RNAs, Y RNA, SINEs (short interspersed repetitive elements), 7SK RNA, two micro-RNAs, several small nucleolar RNAs and many regulatory antisense RNAs.
The genes transcribed by RNA Pol III are in the category of "housekeeping" genes whose expression is required in all cell types and under most environmental conditions. Therefore the regulation of Pol III transcription is primarily tied to the regulation cell growth and the cell cycle, thus requiring fewer regulatory proteins than RNA polymerase II.
In the process of transcription (by any polymerases) there are three steps;
1. �Initiation; requiring construction of the RNA polymerase complex on the gene's promoter.
- Elongation; the writing of the RNA transcript.
- Termination; the finishing of RNA writing and disassembly of the RNA polymerase complex.
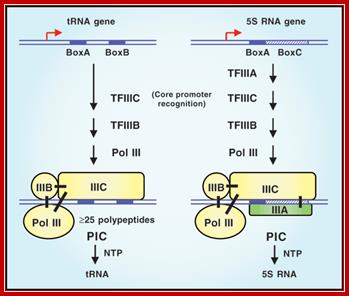
Initiation: The construction of the polymerase III complex on the promoter (compared to Pol II) requires no control sequences upstream of Start site of the gene, instead normally rely on internal control sequences i.e. sequences within the coding section of the gene although upstream sequences in some used in specific genes, e.g. U6 snRNA and 7sL RNA genes have TATA box �and PSE upstream of START site as seen in Pol II Promoters.
Promoters for RNAP-III have diverse structural features because the enzyme and its associated factors (different) transcribe at least four different kinds of genes, such as tRNA, 5s rRNA, U6 sn-RNA, 7SL-Sc-RNA, 7SK RNA and few others; thus, RNAPIII transcribes housekeeping genes required all the time.� The last three mentioned have TATA boxes in their promoters, at (-) 30 from the start, but the other two promoters lack in TATA boxes. The other two have promoter elements tucked within the coding region.� �The start is identified by (+) 1.
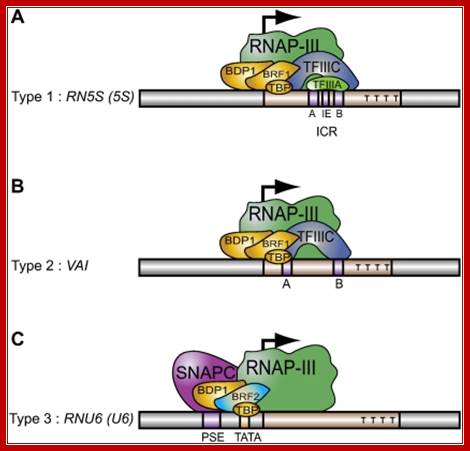
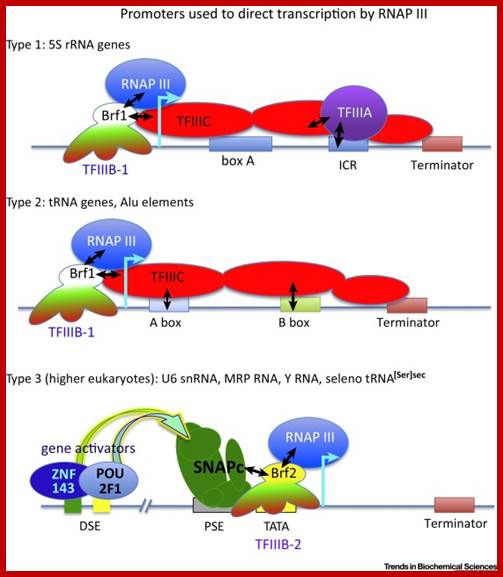
Three types of Pol III promoters and corresponding transcription factors. �(A�C) Structures of types 1, 2, and 3 promoters, respectively, as well as the factors they recruit. https://www.researchgate.net
Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors; Nature Structural & Molecular Biology-17
RNA polymerase (Pol) III transcribes many noncoding RNAs (for example, transfer RNAs) important for translational capacity and other functions. We localized Pol III, alternative TFIIIB complexes (BRF1 or BRF2) and TFIIIC in HeLa cells to determine the Pol III transcriptome, define gene classes and reveal 'TFIIIC-only' sites. Pol III localization in other transformed and primary cell lines reveals previously uncharacterized and cell type�specific Pol III loci as well as one microRNA. Notably, only a fraction of the in silico�predicted Pol III loci are occupied. Many occupied Pol III genes reside within an annotated Pol II promoter. Outside of Pol II promoters, occupied Pol III genes overlap with enhancer-like chromatin and enhancer-binding proteins such as ETS1 and STAT1. Moreover, Pol III occupancy scales with the levels of nearby Pol II, active chromatin and CpG content. These results suggest that active chromatin gates Pol III accessibility to the genome.
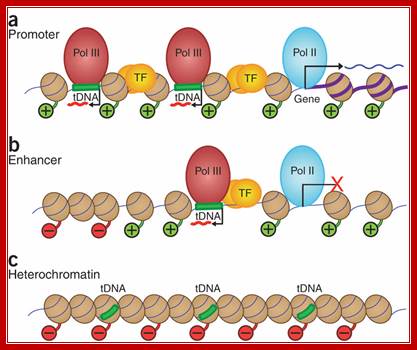
Nearby promoters are used for transcription in opposite direction by RNA pol-III and RNA pol-II, but they use different strands.
Andrew J Oler,� Ravi K Alla, Douglas N Roberts, Alexander Wong, Peter C Hollenhorst, �Katherine J Chandler, Patrick A Cassiday, Cassie A Nelson, Curt H Hagedorn, Barbara J Graves & Bradley R Cairns;; http://www.nature.com/
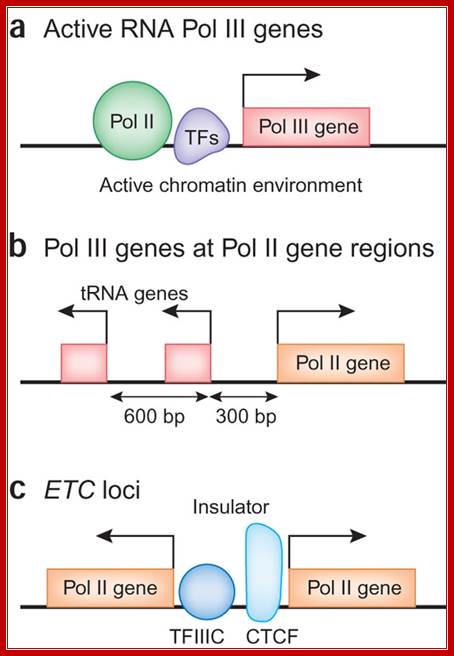
In this issue, three papers report the distribution of the RNA polymerase III (Pol III) machinery, including transcription factor IIIB, transcription factor IIIC and Pol III itself, across the human genome. These studies reveal cell type�specific expression of Pol III genes, functional interplay between the Pol II and Pol III transcriptional machineries and the potential involvement of Pol III genes in chromosome organization. The human Pol III transcriptome and gene information flow; Ken-ichi Noma & Rohinton T Kamakaka; http://www.nature.com/
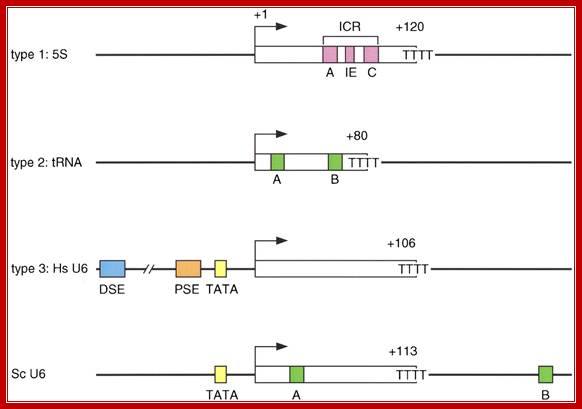
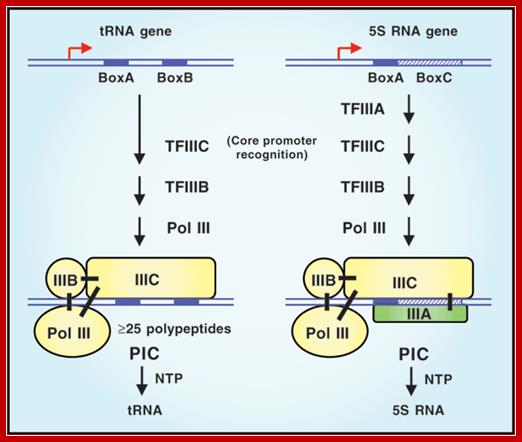
Recruitment of RNA polymerase III to its target promoters; Different types of Promoters for RNA polymerase III, Type1 is of 5s rRNA, Type 2 is tRNA ,Type 3 is 7sL RNA and U6 RNA; Laura Schramm and Nouria Hernandez1 http://genesdev.cshlp.org/ http://etd.lsu.edu/
Different types of RNA polymerase III promoters. The type 1 promoter of theXenopus laevis 5S RNA gene consists of an internal control region (ICR), which can be subdivided into A box (+50 to +60), intermediate element (IE, +67 to +72), and C box (+80 to +90). The type 2 promoter of the X. laevist RNALeugene consists of an A box (+8 to +19) and a B box (+52 to +62). The type 3 promoter of the Homo sapiens U6 snRNA gene consists of a distal sequence element (DSE, −215 to −240) that enhances transcription and a core promoter composed of a proximal sequence element (PSE, −65 to −48) and a TATA box (−32 to −25). ��
The Saccharomyces cerevisiae promoter is a hybrid promoter consisting of a TATA box (−30 to −23), an A box (+21 to +31), and a B box located downstream of the U6 coding region (from +234 to +244 relative to the start site of transcription,
The type 1 promoter of the Xenopus laevis 5S RNA gene consists of an internal control region (ICR), which can be subdivided into A box (+50 to +60), intermediate element (IE, +67 to +72), and C box (+80 to +90).
The type 2 promoter of the X. laevis example tRNA Leu gene consists of an A box (+8 to +19) and a B box (+52 to +62).
The type 3 promoter of the Homo sapiens U6 snRNA gene consists of a distal sequence element (DSE, −215 to −240) that enhances transcription and a core promoter is composed of a proximal sequence element (PSE, −65 to −48) and a TATA box (−32 to −25).
The type 4 Sc type: The Saccharomyces cerevisiae promoter is a hybrid promoter consisting of a TATA box (−30 to −23), an A box (+21 to +31), and a B box located downstream of the U6 coding region +113 TTT (from +234 to +244 relative to the start site of transcription.
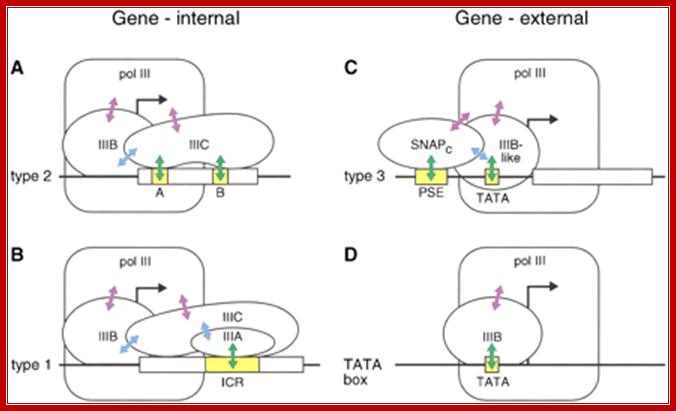
�� Different pathways for recruitment of TFIIIB and RNA polymerase III. The initiation complexes ������formed on type 2, 1, and 3 promoters, as well as on an artificial promoter consisting of just a TATA box, are shown. The green arrows symbolize interactions of DNA-binding proteins with promoter elements, the blue arrows protein�protein contacts among various transcription factors, and the purple arrows protein�protein contacts between RNA polymerase III and transcription factors; Genes and Development; Laura Schramm and Nouria Hernandez; http://genesdev.cshlp.org/.
Figure above:
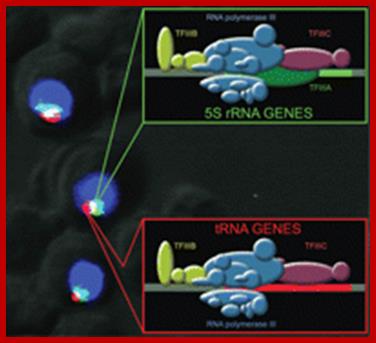
A: Type2 transcribes 5s rRNA genes; they contain ICR elements such as A and B.
B: �Type1 transcribes tRNA genes and they contain ICR.
C: Type3 transcription of NC RNAs, contain proximal sequence elements.
D: Type scU6-transcription U6 RNA, they use TATA box and also contain internal sequences in the coding region and 3� noncoding region.
Different pathways for recruitment of TFIIIB complex and RNA polymerase III is employed. The initiation complexes formed on type 2, 1, and 3 promoters, as well as on an artificial promoter consisting of just a TATA box, are shown above. The green arrows symbolize interactions of DNA-binding proteins with promoter elements, the blue arrows protein�protein contacts among various transcription factors, and the purple arrows protein�protein contacts between RNAPIII and transcription factors.
5s rRNA Gene Transcription:
I. Mechanism of Transcription of 5sRNA Genes:
Class I
The 5S ribosomal RNA (rRNA) genes in eukaryotes may occur either interspersed with the other rRNA genes in the ribosomal DNA (rDNA) repeat, or clustered in tandem arrays, or dispersed throughout the genome. In plants and animals, the 5S rRNA genes are generally organized into tandem arrays unlinked to the rDNA repeat units.� In some fungi, a 5S gene is embedded in the nontranscribed spacer (NTS) of the rDNA gene repeat unit.
In many members the 5srRNA gene is located outside the rRNA cluster. In Yeast the 5s rRNA gene is embedded within the 35s rRNA gene block and transcribed separately.� The 5S rRNA regions of most of the genes are of one Type 1; however, several other "isotypes" (Ii, y, 8, C, and ig) are also found. None of the 5S rRNA genes unlinked to rRNA tandem repeats studied maps close to the nucleolus organizer, the site of the genes that code for the three larger rRNAs. The 5S rDNA genes were not associated with the NORs or sex chromosomes. A highly conserved chromosomal location of these genes appears to characterize the Karyotype evolution of this �group.
Typical stages in 5S rRNA (also termed class I) gene initiation:
- TFIIIA (Transcription Factor for polymerase III A) binds to the intragenic (lying within the transcribed DNA sequence) 5S rRNA control sequence, C Block (also termed box C).
- TFIIIA serves as a platform (that replaces the A and B blocks of tRNA) for positioning TFIIIC in an orientation with respect to the start site of transcription that is equivalent to what is observed for tRNA genes.
- Once TFIIIC is bound to the TFIIIA-DNA complex the assembly of TFIIIB proceeds as described for tRNA transcription.
Though the main part of the rRNA gene is located in nucleolar organizer region and transcribed by RNAP-I, the 5S RNA genes are distributed elsewhere among the chromosomes.� But in yeast genome as shown below the 5srRNA genes located on either side of the main 35S rRNA segment.� They are transcribed by RNAPIII.
![[RNA_polymerase_III_complex.jpg]](Gene_Expression_II4-RNAP_III_Promoter_files/image006.jpg)
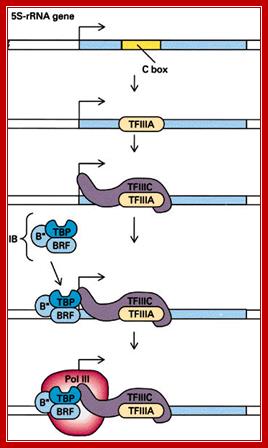
5s RNAgene promoter and Transcriptional complex bound; A cartoon drawing of the pol III transcription complex on a 5S RNA gene is shown above (Moran, Scrimgeour et al. 1994). The transcription factor TFIIIA binds to the internal control region (ICR). Another transcription factor, TFIIIC, binds TFIIIA and it, in turn, interacts with TFIIIB and RNA polymerase III. Transcription is initiated at a site (+1) upstream from the internal control region. Note that when the 5S RNA is produced it will contain the binding sites for TFIIIA. The significance of this fact will become clear in a few minutes: LAURENCE A. MORAN; http://sandwalk.blogspot.in/
This illustration depicts the assembly of RNAP-III and its related TFs for initiating transcription; http://www.igf.ib.unicamp.br
The temporal and spatial expression of specific genes is central to processes such as development, differentiation and homeostasis in eukaryotes, and is regulated primarily at the level of transcription. An understanding of the molecular basis for this regulation has presented, �a major challenge for the past 40 years.
This is yet another diagram showing the assembly of RNAP-III and its factors at their respective promoter elements of tRNA and 5s rRNA genes. Robert G Roeder http://www.nature.com/
� The size of the gene is about 300 bp and the final 5s RNA is 120 ntds.�
� The number of genes per haploid chromosome can be any where from 200 to 2000 and they are clustered.� Even so individual genes are transcribed as independent units.�
� Similar to tRNA genes, promoters are found internal to the start site, one at +60 called A and C boxes respectively.�
� These genes are also constitutively expressed for they are required for ribosomal assembly in the nucleolus.�
� First TF III-A recognizes A box, which facilitates the binding of IIIC which acts like a positional factor.� This leads to the assembly of TF III-B, which has TBP and Bs and BRFs.� They identify the start site for RNAP-III to initiate transcription, similar to that of tRNA genes.
Ribosome is a mega Dalton (2.6MD) mammoth RNA-protein structure.� 5S rRNA 120 ntd long is uniquely positioned in the central protuberance of the large subunit, so as to link together all of the functional centers of the ribosome. The availability of detailed structural information, biochemical assays, and molecular genetic systems make the ribosome a robust model for studying how the structures of complex molecules dictate function at the molecular level.
Analyses of static X-ray crystal structures have been useful in assigning the positions of the functional centers at the atomic level, revealing for example that the catalytic activity of the ribosome is mediated by RNA, and identifying the binding sites for antibiotics. Cryo-EM studies provide complementary information, showing dynamic views of intra-ribosome movements through many of the different phases of the translation cycle.� Perhaps it is the most dynamic cell organelle among the others.
Biochemical studies with E. coli ribosomes led to the hypothesis that 5S rRNA acts as a physical transducer of information, facilitating communication between the different functional centers and coordinating the multiple functional centers of the ribosome.
5S rRNA is a conserved component of the large ribosomal subunit that is thought to enhance protein synthesis by stabilizing ribosome structure.� In 70s ribosome it binds to L5, L18 and L25, but in Eukaryotes 5S rRNA molecule binds only ribosomal protein L5. Changes in the conformation of 5S rRNA cause alterations in principal functions of the ribosomal �Nano machine�.
II. Mechanism of Transcription of tRNA Genes:
Class II.
Unlike rRNA genes, tRNA genes are distributed as cluster among chromosomes.� Often, they are found at base of telomeres and some at the vicinity of centromeres.� Though dispersed in the linear genome, some colocalize with 5S ribosomal DNA and U14 small nucleolar RNA at the nucleolus. Nucleolar localization requires tRNA gene transcription-complex formation, because inactivation of the promoter at a single locus removes its nucleolar association. This organization of tRNA genes must profoundly affect the spatial packaging of the genome and raises the question of whether gene types might be coordinated in three dimensions to regulate transcription.
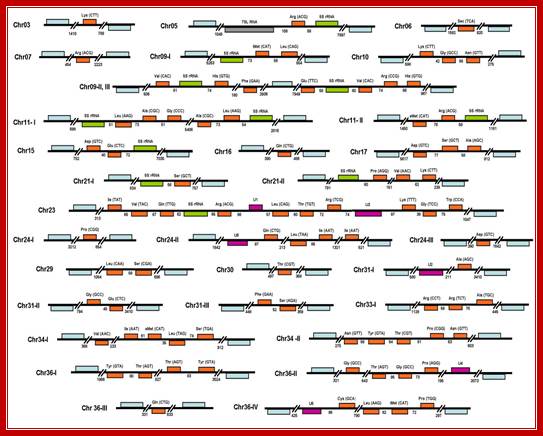
Organization of tRNA genes in L. major Padilla-Mej�a et al. BMC Genomics 2009 10:232
Organization of tRNA genes in L. major. The 83 tRNA genes found in the genome of the parasite are shown in orange. The predicted anticodons are indicated in parentheses. 5S rRNA, snRNA and 7SL genes are shown in green, purple and gray, respectively. Genes are drawn to scale, and the sizes of intergenic regions are indicated (in base pairs). Protein-coding genes that flank Pol III-transcribed genes are shown in blue (not to scale). The tRNA-Sec gene on chromosome 6 is located at positions 69,586 to 69,673, in the complement strand. Putative pseudogenes are not shown. For practical purposes, we regarded protein-coding genes as the limits of a particular Pol III locus. For that reason, we considered cluster chr09-II, III as two independent Pol III loci; Padilla-Mej�a NE, et al, http://openi.nlm.nih.gov/
In yeast there are 274 yeast tRNA genes which can be considered as active and grouped into 42 families of distinct codon specificity.
Most of the tRNA genes in Eukaryotic systems are clustered and distributed among the chromosomes.� Total number of tRNA genes range for 300 to 2000 per haploid genome.�
Archaeal/Eukaryal RNase P: subunits, functions and RNA diversification:
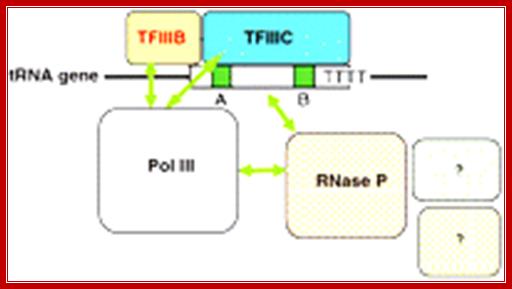
A model for association of human RNase P with Pol III and tRNA Genes, Nayef Jarrous1,* and 2 Venkat Gopalan2
Schematic of a tRNA gene with conserved internal boxes A and B regulatory elements bound by the transcription factors TFIIIC and TFIIIB. These two transcription factors recruit Pol III. Human nuclear RNase P associates with Pol III and with the tRNA gene. This may facilitate coordination of transcription and tRNA processing, and involve other tRNA processing and modifying activities.� �
Spatial organization of transcription by RNA polymerase III:
Rebecca A. Haeusler and 2David R. Engelke
In situ hybridization using short, oligonucleotide probes reveals that tRNA genes (red) overlap 5S genes (green) at the nucleolus. DAPI staining of chromatin is shown in blue; tRNA genes and 5S genes are both transcribed by RNA polymerase III and they also share the same transcription factors, although 5S genes require one additional factor, TFIIIA. Gene colors in the line drawings correspond to image pseudo coloring, while protein colors are arbitrary. Co-localization of tRNA and 5S genes may provide a basis for co regulation of their transcription with one another. Further, nucleolar co-localization of these genes might facilitate allows coordination of transcription with another heavily expressed component of the protein synthesis machinery, the large ribosomal RNA gene repeats that are by RNA polymerase I. With the current resolution it is not possible to conclusively interpret whether signals from tRNA and 5S genes are coincident or adjacent. Figure courtesy of Martin Thompson.
Typical stages in a tRNA (also termed class II) transcriptional initiation:
� Each of the tRNA genes is transcribed as individual units.
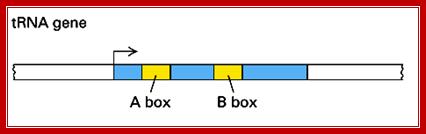
The promoter region is located with in the coding region at +10 to +20 and +50 to +60 from the start, called as A and B boxes.
� Actually, these regions code for tRNA D arm and TUCG loop.� All tRNA genes are constitutively expressed.
�
1. TFIIIC (Transcription Factor for polymerase III C) binds to two intragenic (lying within the transcribed DNA sequence) control sequences; the A and B Blocks are also termed as box A and box B.
2. Transcription factor TF III-C a multimer complex (six subunits), binds to both blocks in such a way one end of the protein is placed exactly at the start site. The TF III-C consists of 6 subunits, with a mol. wt of 600kd. �
3. The binding of III-C facilitates the binding of TF III-B, which contains one TBP, one B and one BRF. �For TBP to bind there is no TATA sequence, but acts as enzyme positional factor for RNAP III.
4. TFIIIC acts as an assembly factor that positions TFIIIB to bind to DNA at a site centered approximately 26 base pairs upstream of the start site of transcription. TFIIIB (Transcription Factor for polymerase III B), consists of three subunits: TBP (TATA Binding Protein), the Pol II transcription factor, TFIIB-related protein, Brf1 (or Brf2 for transcription of a subset of Pol III-transcribed genes in vertebrates) and Bdp1.
5. TFIIIB is the transcription factor that assembles Pol III at the start site of transcription. Once TFIIIB is bound to DNA, TFIIIC is no longer required. TFIIIB also plays an essential role in promoter opening.
TFIIIB remains bound to DNA following initiation of transcription by Pol III (unlike bacterial σ factors). This leads to a high rate of transcriptional reinitiation of Pol III-transcribed genes. TF III B�s and BRFs aid positioning of the TBP.
---I+1>>---- [+10 ---- +20] -------------[+50--- +60] -------TTTT--
A-Box=�� +10 to +20 = TGGCNNGTGG.
B-Box=� +50 to +60 = GGGTTCGAaacc
�
� Among these factors, TBP is a positioning factor for the RNAP-III.
When the polymerase binds, it can initiate transcription from the predefined start site.
� The tRNAs produced are precursors, which later are subjected processing and also base modifications.

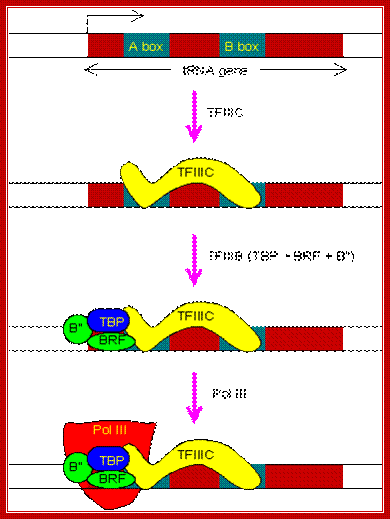
The regulatory region of tRNA gene contains A box and B box located inside the transcription unit. The PIC assembly begins with the binding of TFIIIC to both elements.; http;//www.web-books.com
Transcription of 5S-rRNA gene:
The regulatory region of 5S-rRNA gene contains a C box, also located inside the transcription unit. The PIC assembly begins with the binding of TFIIIA to the C box.
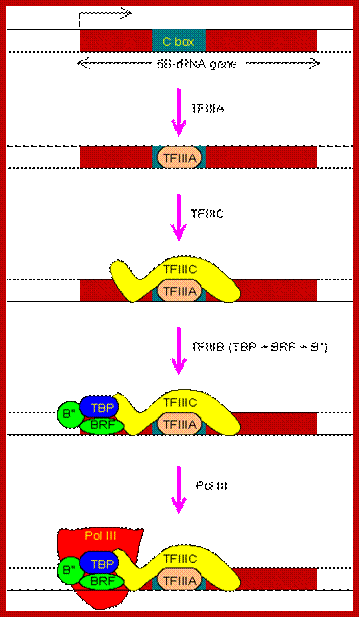
The PIC assembly for the transcription of 5S-rRNA gene. Note that TBP is involved in the transcription by all three types of RNA polymerases in eukaryotes. http://www.web-books.com
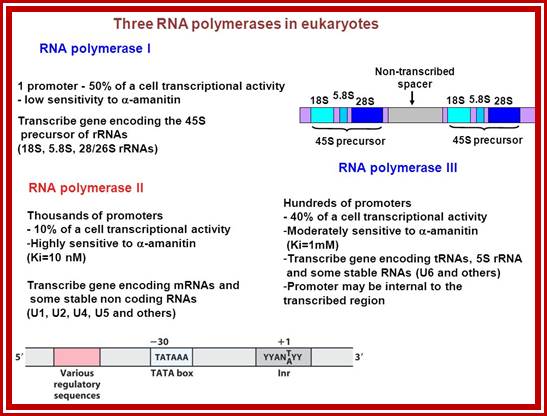
RNA synthesis in eukaryotes is divided among three RNA polymerases (RNAPs). RNAP III transcribes hundreds of tRNA genes and fewer additional short RNA genes. We survey recent work on transcription by RNAP III including an atomic structure, mechanisms of action, interactions with chromatin and Retrospons, and a conserved link between its activity and a tRNA modification that enhances mRNA decoding. Other new work suggests important mechanistic connections to oxidative stress, autoimmunity and cancer, embryonic stem cell pluripotency, and tissue-specific developmental effects. We consider that, for some of its complex functions, variation in RNAP III activity levels lead to nonuniform changes in tRNAs that can shift the translation profiles of key codon-biased mRNAs with resultant phenotypes or disease �states� RNA Polymerase III Advances: Structural and tRNA Functional Views; Aneesh Kumar et al. http://www.cell.com
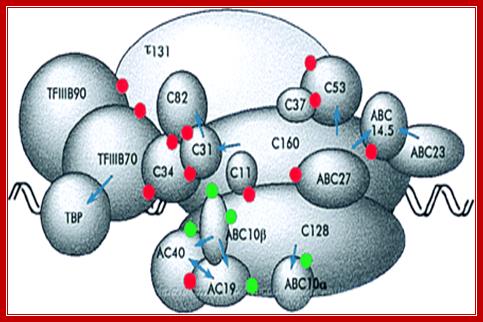
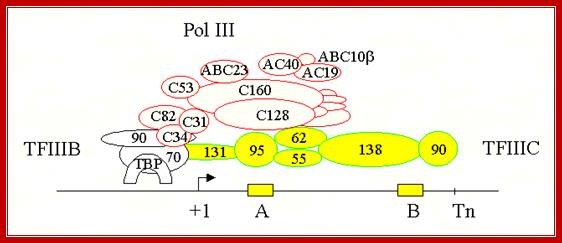
A model of the pol III transcription initiation complex of 18 subunits: Protein�protein contacts observed by using the two-hybrid system are indicated by red dots. Green dots indicate the interaction between AC40, AC19, ABC10α, and ABC10β with A190 and A135 pol I subunits homologous to C160 and C128. Genetic interactions observed by using multicopy suppression experiments of thermo sensitive mutations are indicated by arrows. The arrowhead points toward the subunit harboring the mutation that was suppressed. For the sake of simplicity, only the τ131 and τ138 subunits of TFIII C are represented. http://www.pnas.org/
.
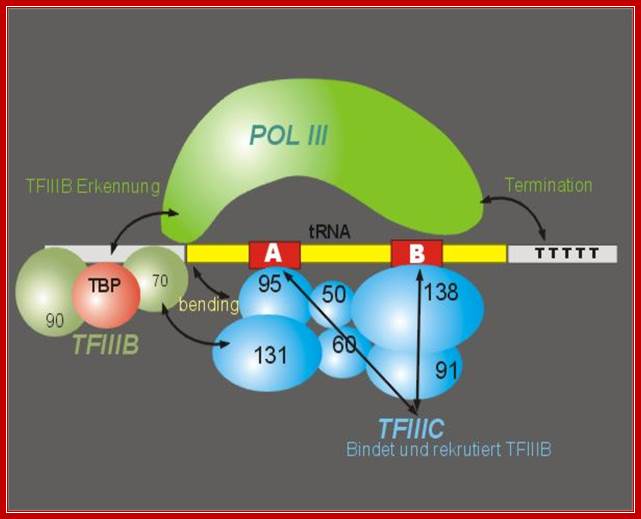
Yet another diagram showing the assembled components for transcription of eukaryotic tRNA genes.� TFIIIC directly assembles on to A and B boxes so as tyo facilitate the binding of TFIIIB. �TFIIIC acts as positional factor for the assembly of TBP and RNAP. http://www.image.3sir.net
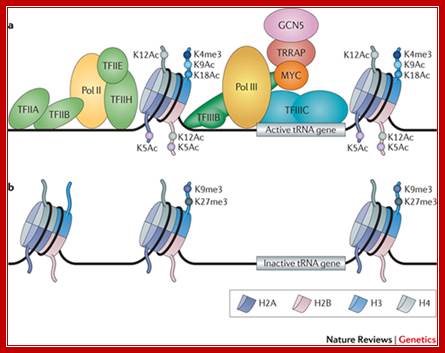
Transcription by RNA polymerase III: more complex than we thought; Robert J. White; http://www.nature.com/
�a | Active tRNA genes are occupied by transcription factor IIIC (TFIIIC), TFIIIB, RNA polymerase (Pol) III and often by MYC, which recruits the histone acetyltransferase GCN5 via the cofactor transformation/transcription domain-associated protein (TRRAP). Histones flanking active tRNA genes characteristically show euchromatic modifications, including trimethylation of histone H3 at lysine 4 (H3K4me3), and extensive histone acetylation � for example, acetylation of histone H2A at lysine 5 (H2AK5Ac), H2BK5Ac, H2BK12Ac, H3K9Ac, H3K18Ac and H4K12Ac. Pol II can often be detected ~200 bp upstream of active tRNA genes, along with basal Pol II transcription factors that include TFIIA, TFIIB, TFIIE and TFIIH. b | By contrast, Pol II and its basal factors are not usually associated with silent tRNA genes that are unoccupied by Pol III, TFIIIB and TFIIIC. Histones flanking such inactive tRNA genes show minimal acetylation and instead show heterochromatic modifications, such as H3K9me3 and H3K27me3. An example of these features is provided in HeLa cells by two tRNA-Leu (TAA) genes on chromosome 6, one active and the other inactive�.
Split tRNA precursors:
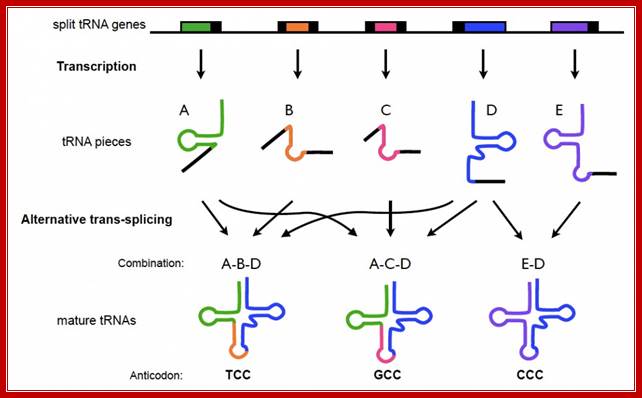
Representation of top/bottom half of tRNA in a cloverleaf and L-shaped structure; Individual DNA domains of tRNAs split and transcribed, the transcripts are brought together and processed to form active tRNAs by trnssplicing. Split tRNA genes are encoded at different loci of C. maquilingensis genome. tRNA fragments (A-E) are individually transcribed and assembled via trans-splicing based on the leader sequences (black) located at one or both ends. Sequences A and D are used multiple times in a different combination, as in a jigsaw puzzle. https://origins-of-life-fg.arc.nasa.gov
Split tRNA genes are encoded at different loci of C. maquilingensis genome. The tRNA fragments (A-E) are individually transcribed and assembled via trans-splicing based on the leader sequences (black) located at one or both ends. Sequences A and D are used multiple times in a different combination, as in a jigsaw puzzle.
In addition, a number of split tRNA genes have been recently found in the Desulfurococcales branch of archaea, expanding the population of split genes to diverse archaeal species. Examination of their gene arrangement combined with phylogenetic analysis has indicated that split tRNAs was a late acquisition, most likely created through local genome rearrangement. This means that split tRNAs in the archaeal genome might not be direct homologs but rather analogs of ancestral tRNAs
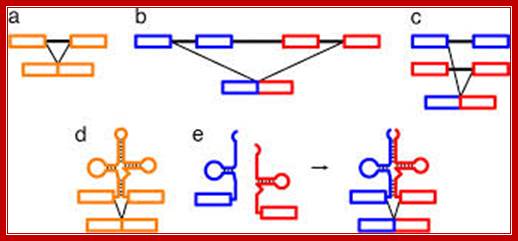
Individual segments of the tRNA gene segments are recombined and transcribed to generate a specific tRNA; RNA splicing pathways observed in eukaryotes (a�c) and engineered in yeast by Di Segni et al. (5) (dand e). (a) Canonical pre-mRNA cis-splicing yielding mRNA from pre-mRNA composed of two exons (orange boxes) and an intron (black line). (b) A transcription-induced chimera generated by read-through transcription from one gene (blue) into a downstream gene (red), followed by cis-splicing. (c)Trans-splicing of exons from two distinct pre-mRNAs (red and blue boxes). (d) Cis-splicing of a pre-mRNA embedded with a permuted tRNA. (e) Trans-splicing of two hybrid RNAs each composed of an mRNA fragment and a half tRNA. Note that in the tRNAs the aminoacyl acceptor stem is at the top and the anticodon loop is at the bottom (d and e). http;??www.pnas.org
A) Exon Theory of Genes� is a hypothesis introduced by Walter Gilbert, according to which exon shuffling in the primitive species gave rise to a variety of genes that produced different proteins. For example, recombination between gene A and gene B would create new mosaic genes C and gene D that partially possesses the previous exons. This process allows for creating various multi-domain proteins. (B) Two tRNA fragments are individually transcribed, but become connected due to complementarity in a long GC-rich leader sequence (orange). The exon-leader junction forms a Bulge-Helix-Bulge (BHB) motif, which is recognized and excised (arrow) by a tRNA splicing endonuclease and joined by RNA ligase.
Since tRNA is known as a universally conserved RNA molecule that decodes the genetic information for translation to proteins, the discovery of small genes that encode tRNA halves has brought forward an idea that it might be a ancestral trait of genes. This is in line with a hypothesis that ancient tRNA was once encoded as a simple hairpin RNA (minihelix) that served as an amino acid carrier in primordial translation or a donor of replication, and later became the top half of the present tRNA molecule. �Indeed, the top half sequence is highly conserved among different tRNA species in all three domains of life and recognized by various ribozymes and enzymes.
tRNA Processing:
1. tRNA is transcribed by RNA polymerase III. The transcript, the pre-tRNA, contains additional RNA sequences at both the 5� and 3�-ends. These additional sequences are removed during processing. Few nucleotides at the 5�-end are removed by an unusual RNA containing enzyme called eukaryote ribonuclease P (RNase P). Actually, PNase cuts the 5� site of the pretRNA (in prokaryotes also the similar homologous enzyme cuts at 5� end of the pretRNA located in the middle of the pre rRNA gene transcript).
2. Some tRNA precursors contain an intron located in the anticodon arm. These introns are spliced out during processing of the tRNA. Intervening sequences in eukaryotic tRNA genes were first described for yeast tRNATyr. With the availability of further sequences, rules for the location of introns within the coding sequences have been established, the mechanism of intron splicing in yeast has been elucidated and the components involved in this process have been characterized. Similarly, 5′- and 3′-end maturation and modification have been studied in great detail.
3. All mature tRNAs contain the trinucleotide CCA at their 3�-end. These three bases in some are encoded but in some they are not encoded in the tRNA gene. Instead, these nucleotides are added to tRNAs, that don�t contain CCA.� This is done during processing of the pre-tRNA transcript. The enzyme responsible for the addition of the CCA-end is tRNA nucleotidyl transferase and the reaction proceeds according to the following scheme:
tRNA +CTP --> tRNA-C + PPi (pyrophosphate)
tRNA-C +CTP --> tRNA-C-C + Ppi
tRNA-C-C +ATP --> tRNA-C-C-A + PPi
6. Mature tRNAs can contain modified bases up to 10% of the bases, other than the usual adenine (A), guanine (G), cytidine (C) and uracil (U). These base modifications are introduced into the tRNA at the final processing step. The biological function of most of the modified bases is uncertain and the translation process seems normal in mutants lacking the enzymes responsible for modifying the bases.
Holley, in 1968, awarded The Nobel Prize in Physiology/ Medicine 1968, for his pioneering work on tRNA �clover-leaf model�. The Nobel Prize was also shared in the same year by H.G Khorana (Indian) for his work on elucidation of genetic codon dictionary and M. Nierenberg for elucidating the first codon UUU for phenylalanine respectively.
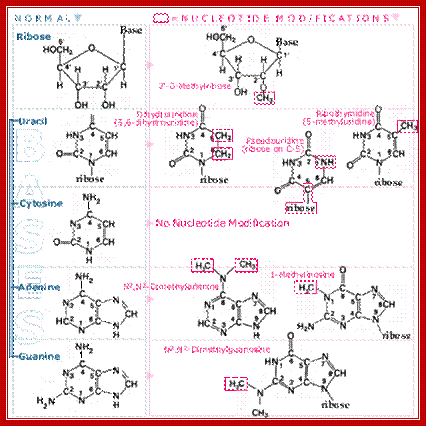
Base modification in tRNAs- few among the many. https://www.nobelprize.org/educational/medicine
III. Mechanism of Transcription of small molecular weight RNAs:
Class III:
Class III genes are Sn and Sc RNAs, such as U6, 7SK and 7SL RNA and some Sno RNA Genes. Many small molecular weights non coding RNAs are transcribed by RNAPII and some by RNAP III.
Typical stages in a U6 SnRNA (also termed class III) gene initiation (documented in vertebrates only):
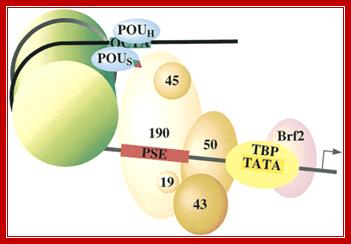
- SNAPc (SnRNA Activating Protein complex) (also termed PBP and PTF) binds to the PSE (Proximal Sequence Element) centered approximately 55 base pairs upstream of the start site of transcription. This assembly is greatly stimulated by the Pol II transcription factors Octa1 and STAF that bind to an enhancer-like DSE (Distal Sequence Element) at least 200 base pairs upstream of the start site of transcription. These factors and promoter elements are shared between Pol II and Pol III transcription of snRNA genes.
2. SNAPc acts to assemble TFIIIB at a TATA box centered 26 base pairs upstream of the start site of transcription. It is the presence of TBP at TATA box that specifies that the snRNA gene is to be transcribed by Pol III rather than Pol II. The TFIIIB for U6 snRNA transcription contains a smaller Brf1 paralogue; Brf2. TFIIIB is the transcription factor that assembles Pol III at the start site of transcription. Sequence conservation predicts that TFIIIB containing Brf2 also plays a role in promoter opening.
Fig. Shows a generalized assembly on snRNA genes with their specific RNAPs.� SNAP refers to snRNA gene activating factors. Snap factors 5 units, OCTA1, TBP and RNA polymerase form a transcriptional complex; William Henry, http://bmb.natsci.msu.edu/
I( -)100�OCTA/DSE (-)75---PSE(-)55--(-)30 TATA-->>+1�
Factors besides SNAPc required for pol III Transcription of snRNA Genes:
The key player for recruitment of pol II to a promoter is the factor TFIIB because it contacts the RNA polymerase directly. In the case of pol III, this role is played mainly by the multisubunit factor TFIIIB. TFIIIB was completely defined first in S. cerevisiae and consists of three subunits, TBP, a tightly associated subunit referred to as the TFIIB-related factor BRF1 (PCF4/TDS4) and a more loosely associated polypeptide called B" (TFIIIB90/TFC5/TFC7). This TFIIIB complex is involved in transcription from all types of yeast pol III promoters tested including the gene-internal tRNA-type promoters and the U6 promoter, which, as described above, contains a TATA box and A and B boxes .
In vertebrates the U6 RNA gene is Type3, contains an upstream promoter and no internal promoter. In this sense they resemble the typical class II genes (transcribed by RNA polymerase II), such as those that encode protein. The U6 snRNA gene, a component of the spliceosome [RNA Splicing: Introns and Exons] is the prototype gene of this category.
Promoters for the said genes have a TATA box at �30 from the start; some may not contain TATA box.
� At -55 regions the promoters have a sequence called PSE (proximal sequence elements) and in the upstream it may contain distal sequence elements DSE.
� At the upstream of PSE there is another box called OCTA, an octamer sequence.�
To initiate transcription, first TF III-B directly positions to the left of start on to TATA box directly.� The III-B has TBP and B and BRFs, which facilitate the binding of RNAP-III.
� There are factors such as PTF and DSEF that bind to ��their respective sequences. However the efficiency of transcription increases with binding of factors to PSE and OCTA regions.
In Saccharomyces, the U6 RNA gene from the yeast Saccharomyces cerevisiae (SNR6) has internal and downstream promoter elements that match the tRNA gene intragenic A- and B-block elements, respectively. Both vertebrate and yeast U6 RNA genes have an upstream TATA box element not normally found in tRNA genes. Substitution of the SNR6 TATA box altered the site of transcription initiation in vivo, while substitution of sequences further upstream had no effect on SNR6 transcription.
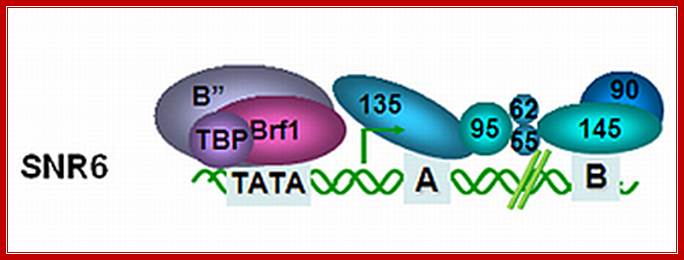
U6 RNA gene: U6 RNA with TATA sequence bound by TBP-BrF and B complexes; In vitro, TFIIIB can bind directly to a TATA box which is found upstream of some genes transcribed by RNA Pol III. http://www.webfiles.uci.edu
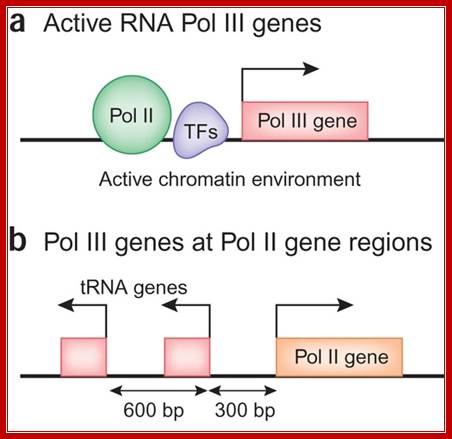
Some of the RNAP II and RNAP II transcriptional genes are placed side by side, but their transcriptional factors and positions vary and the direction of transcription each differ; Human Pol III transcriptome and gene information flow, Ken-ichi Noma, & Rohinton T Kamakaka ; http://www.nature.com/nsmb
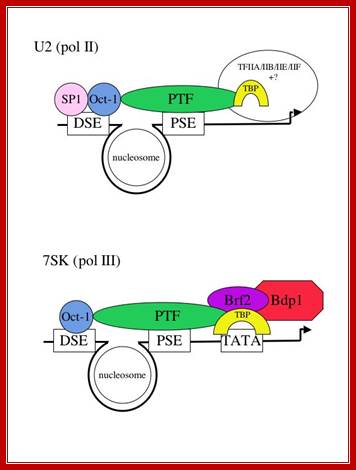
Structure of snRNA genes and known transcription factors;
The major difference between the pol II- and pol III-transcribed genes is the presence or absence of a TATA box-the rest of the elements are largely interchangeable (see refs. 2 and 3). Not all the factors required for transcription of these genes have been identified, as indicated by question marks. The pol II-transcribed snRNA genes share TBP/IIA/IIB/IIE and IIF with protein-coding genes that are also transcribed by pol II. The pol III-transcribed snRNA genes share components of the IIIB complex with tRNA genes that are also transcribed by pol III. In addition, these genes require an snRNA gene-specific factor-Brf2 (see refs. 5, 7 and 8). It is still unclear how the presence of a TATA box (often found in the promoters of protein-coding genes) switches the polymerase specificity to pol III. One possibility is that binding of TBP to the TATA box causes exclusion of a pol II-specific factor (eg TFIIB) from the promoter and allows a pol III-specific factor to bind (see Figure 2 and ref. 11). In support of this model, we have shown that factor interactions occur with the region just downstream of the PSE of the pol II-transcribed U2 gene in vivo (ref. 11) and that TBP bound to the TATA box recruits Brf2;
Sir.William Dun; Dr.S.Murphy: http://users.path.ox.ac.uk/
7SK RNA:
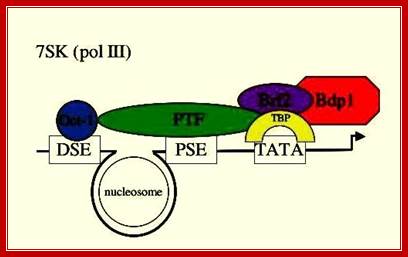
This is a simplistic diagram showing various components of 7s RNA, 7sK RNA, U6 RNA and other similar gene promoters and the components bind to initiate transcription.� Distal sequence elements (DSE) and Proximal sequence elements (PSE) binding proteins such as PTF are shown; 7SK gene is transcribed by RNAPIII which contains TATA box, which is bound by TBP; Dr.S.Murphy, Molecular Biology http://users.path.ox.ac.uk/
In all these promoters, the RNAP is positioned so as to initiate transcription at predefined nucleotide; this is aided by specific factors that bind to certain sequences, which may be present at the upstream of the start or in the down stream of the start.� Whatever may be the position, the promoter aids in the assembly of accessory proteins in stepwise manner and ultimately the RNAP binds to initiate transcription.� Thus, the function of the promoter, whatever the structure it has, wherever it is found, and its role is to assist the assembly of TFs factors which assist and guide the RNAP to position at a specific site to initiate transcription at pre defined site or nucleotide.
Type IV; Recall that 7SL RNA is the major component of Signal Recognition Particle (SRP). There are three genes for this RNA located on human chromosome 14 [Human Genes Involved in the Signal Hypothesis Pathway]. Since 7SL RNA is one of the small RNAs present in most cells it should not come as a surprise that its gene is transcribed by RNA polymerase III.
7sL RNA Genes
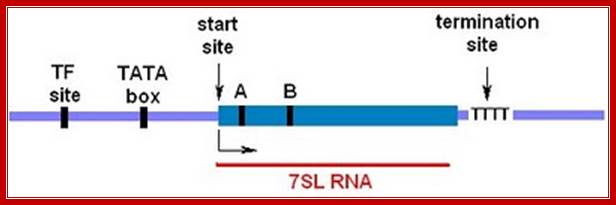
The RN7SL promoter has been characterized by Englert et al. (2004). The main features are shown in the diagram above. The solid blue box represents the gene�the DNA sequence corresponding to the 7SL RNA. There's a start site for transcription (+1) that's determined by the positioning of RNA polymerase III upstream. The transcription complex is assembled when TFIIIC binds to an internal control region specified by box A and box B. These are short DNA sequences (10 bp) that resemble the binding sites in tRNA genes. The upstream promoter region consists of a TATA box where RNA polymerase III binds and a regulatory site where unknown transcription factors (TF) bind. Transcription terminates at a short stretch of thymidylate residues (T) at the 3′ end of the gene. http://sandwalk.blogspot.in/
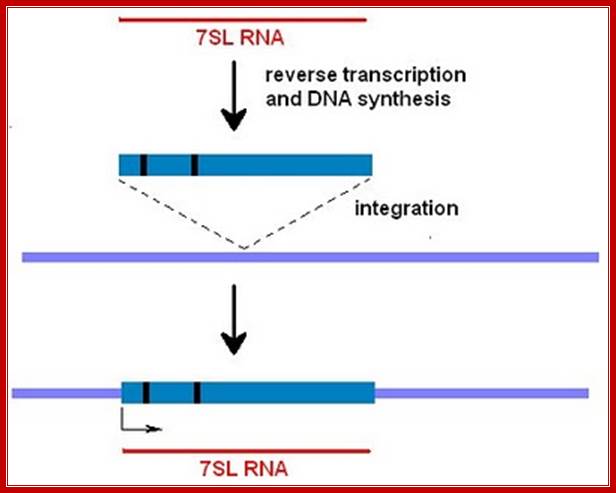
From time to time cellular RNAs are copied by an enzyme called reverse transcriptase to create a DNA: RNA double-stranded molecule. On occasion this hybrid molecule will become integrated into the genome through a nonhomologous recombination event. (Sometimes the RNA strand will be replaced by DNA synthesis to create double-stranded DNA corresponding to the 7SL sequence.) This creates something called a processed pseudogene. Most genomes have hundreds of these pseudogenes derived from hundreds of different genes. They have arisen from accidental events over the course of millions of years of evolution and since there's no pressure to eliminate them, they are retained in the genome as junk. http://sandwalk.blogspot.in/
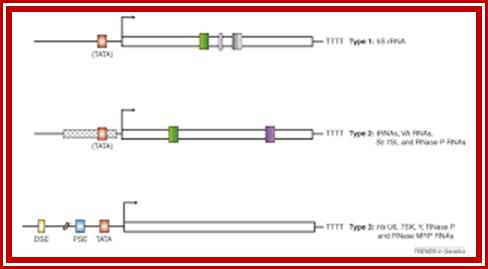
The class III genes can be subdivided into four types depending on the location of the promoter regions.
Finally, genes have both an internal region where the regular class III transcription factors bind and a promoter at the 5′ end of the gene where regulatory transcription factors bind to control transcription. The 7SL RNA gene is a type 4 gene.
����������������������� A recombinant 7sL RNA gene is placed in the vector to study its promoter and transcription; it contains two regulatory sequences immediately after the start site.
SnRNA genes:
�Preinitiation complex formation and polymerase selection in the transcription of human small nuclear snRNA genes��
These snRNA genes are found in large numbers spread over different chromosomes.� Many of them are transcribed by RNAP II and similarly others by RNAP III.
Small non-coding but functional nuclear RNAs perform a variety of essential roles in the control of gene expression in higher eukaryotic cells, including regulation of transcription and RNA processing. Authors (?) of this description were interested in how accurate and efficient expression of the genes encoding snRNAs is achieved. The mammalian genes encoding snRNAs have compact and simple promoter structures that can be divided into two groups since some are recognized by RNA polymerase II (pol II) (eg U2 genes) and some by RNA polymerase III (pol III) (eg the 7SK gene). Intriguingly, the promoter structures are very similar and all of the promoters� elements present in the RNA polymerase III-transcribed genes are also found in genes transcribed by pol II. �
In addition, the pol II-transcribed genes have a unique processing element (the 3' box) close to the end of the snRNA-encoding region. The 3' box is required for termination of transcription and only functions if transcription is initiated from an snRNA promoter and indicating that transcription by pol II is linked to processing of the transcripts. These genes are therefore ideally suited to study the mechanics of basic transcription factor interaction in both pol II- and pol III-transcribed genes and the link between transcription and RNA processing. Presently authors are working on dissecting the cis-acting sequences and the cognate trans-acting factors involved in these processes using a variety of molecular methods and a combination of in vitro and in vivo assay systems.
Several of the transcription factors involved in the expression of the human
snRNA genes have already been identified and characterized and therefore more
easily investigate their function. The promoter elements in a pol II- and a pol
III-transcribed snRNA gene are shown below with the cognate factors that have
been identified so far. In
vivo foot printing studies suggest that a nucleosome brings the DSE
and PSE together.
The major difference between the pol II- and pol III-transcribed genes is the
presence or absence of a TATA box-the rest of the elements are largely
interchangeable. �Not all the factors required for transcription of these genes
have been identified, as indicated by question marks. The pol II-transcribed
snRNA genes share TBP/IIA/IIB/IIE and IIF with protein-coding genes that are
also transcribed by pol II. The pol III-transcribed snRNA genes share
components of the IIIB complex with tRNA genes that are also transcribed by pol
III. In addition, these genes require a snRNA gene-specific factor-Brf2. �It is
still unclear how the presence of a TATA box (often found in the promoters of
protein-coding genes) switches the polymerase specificity to pol III. One possibility
is that binding of TBP to the TATA box causes exclusion of a pol II-specific
factor (eg TFIIB) from the promoter and allows a pol III-specific factor to
bind. In support of this model we have shown that factor interactions occur
with the region just downstream of the PSE of the pol II-transcribed U2 gene in vivo and that TBP
bound to the TATA box recruits Brf2.
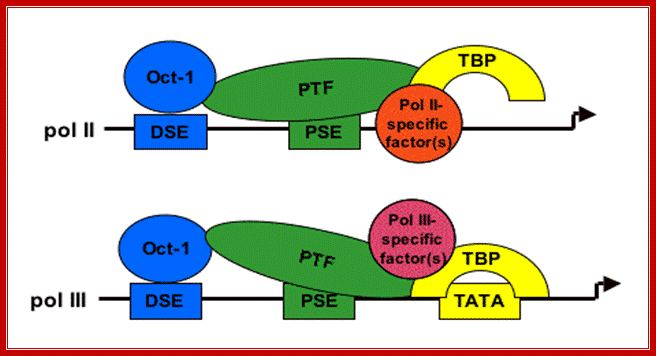
Figure:� Structure of snRNA gene�s
promoter elements and known transcription factors; Dr.
S. Murphy; http://users.path.ox.ac.uk/
��Characterization of factors for initiation of transcription of these genes in order is required to understand how initiation of transcription is regulated�.
Researchers are currently using a range of molecular techniques to identify the RNA processing factors and the factors linking processing of transcription. Recent results indicate that 3' box-dependent processing is activated by phosphorylation of serine 2 in the pol II CTD by the transcription elongation factor P-TEFb. �Since P-TEFb activates elongation of transcription of protein coding genes but not snRNA genes it is interesting to understand the molecular basis for the differences between snRNA genes and protein-coding RNA genes in the requirement for CTD phosphorylation for transcriptional elongation. Recent work indicates that elongation of transcription of the short introns-less replication-activated histone genes is also P-TEFb independent suggesting that short genes don't need the elongation function of P-TEFb unlike longer intron-containing genes.
Sn RNA genes by William Henry: Transcription of human small nuclear (sn) RNA genes as a model system is important to understand mechanisms of transcription and regulation of gene expression. Correctly regulated gene expression is important for healthy development and the maintenance of cellular homeostasis. Disruptions in transcriptional regulation are observed in many disease states including cancer. Moreover, viruses subvert the cellular transcriptional programs for their own benefit during viral infection.
Therefore, it is important to understand the transcriptional mechanisms that underlie control of gene expression. Human snRNA genes encode short stable RNAs that as part of specialized protein-snRNA complexes typically are involved in processing other RNA molecules. For example, snRNAs are essential for mRNA splicing and sno RNAs are responsible for rRNA and tRNA processing is different�
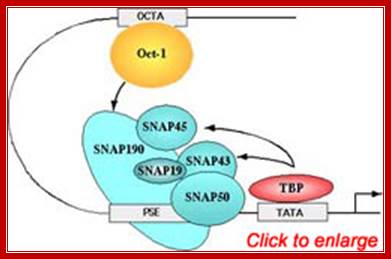
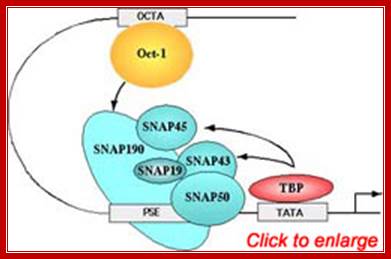
�The PSE is recognized by the basal transcription factor referred to as snRNA-activating protein complex (SNAPc). SNAPc is a multi-protein complex composed of at least five proteins SNAP19, SNAP43, SNAP45, SNAP50, and SNAP190. In addition, the TATA-box binding protein (TBP) can also associate with this complex. Each of these proteins is required for transcription of human snRNA genes by both RNA polymerase II and III. the diagram Shows a generalized assembly on snRNA genes with their specific RNAPs.� SNAP refers to snRNA gene activating factors. The diagram is repeated for understanding how snap components are involved in sn RNA transcription. William henry; http://bmb.natsci.msu.edu/
The diagram shows the cis-acting promoter elements and trans-acting factors involved in formation of a pre-initiation complex on a typical protein-coding gene. These include a TATA box, at �25 relative to the transcription start site, USEs (upstream sequence elements), generally located within approx. 200 bp of the start site and an Enhancer, which can be far upstream or downstream of the start site. USEs and Enhancers bind a range of sequence-specific DNA-binding factors and the TATA box is recognized by the general factor TFIID. The pre-initiation complex minimally comprises the general transcription factors TFIIA, TFIIB, TFIID, TFIIE, TFIIF and TFIIH. Initiation may also require interaction of the pol II CTD with the Mediator complex. The diagram shows the cis-acting promoter elements and trans-acting factors involved in formation of a pre-initiation complex on a typical snRNA gene. These include a DSE that is recognized by factors such as Oct-1 and Staf1 that are shared with protein-coding genes and pol III-transcribed snRNA genes and a PSE that is recognized by PTF, an snRNA gene-specific transcription factor. The pre-initiation complex minimally comprises TBP, TFIIA, TFIIB, TFIIE, TFIIF and possibly TFIIH. TAF100 is found associated with these genes. The involvement of other TAFs and TFIIH is yet to be demonstrated. Efficient initiation may also require interaction of the pol II CTD with the snRNA gene-specific Integrator complex.
Non-coding RNA (ncRNA) genes can be transcribed by either RNA polymerase I, II or III, depending on the individual ncRNA genes.
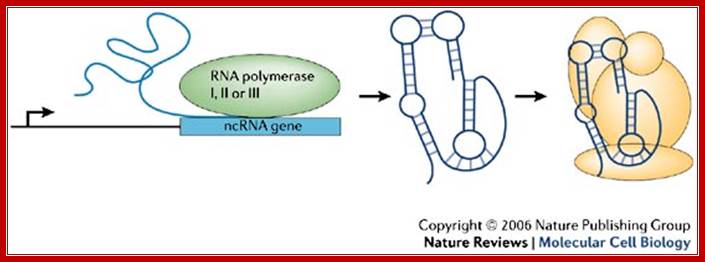
Non-coding RNA (ncRNA) genes can be transcribed by either RNA polymerase I, II or III, depending on the individual ncRNA. ncRNAs fold into specific structures that impart a function to the molecule. Often these RNAs are incorporated into large complexes (yellow) that contain proteins and sometimes other nucleic acids. These complexes then regulate biological reactions, and their function is strictly dependent on the presence of the ncRNA.Fig. 1: Non-coding RNA (ncRNA) genes can be transcribed by either RNA polymerase I, II or III, depending on the individual ncRNA (a generalized version). James A. Goodrich & Jennifer F. Kugel; �http://www.nature.com/
�ncRNAs� fold into specific structures that impart a function to the molecule. Often these RNAs are incorporated into large complexes (yellow) that contain proteins and sometimes other nucleic acids. These complexes then regulate biological reactions, and their function is strictly dependent on the presence of the ncRNA.
In eukaryotic cells, mRNA transcription is an intricate process that is tightly and temporally regulated in response to numerous signals, and involves the orchestrated interplay of DNA, many protein components and the RNA transcript itself. The transcription reaction is catalyzed by the Pol II enzyme, which synthesizes an mRNA copy of each protein-coding gene. Eukaryotic DNA and histones are packaged into a highly condensed structure that is known as chromatin, and the structure of chromatin dynamically participates in regulating mRNA transcription. The DNA elements that control transcription are contained in promoters. Promoters are the regions of genes that contain the sequences that bind to transcriptional activators and repressors, as well as to the Pol II general transcription factors that are thought to be required for the transcription of all genes. A diverse set of co-regulators mediates activation and repression, often bridging contacts between regulators and the general transcription machinery. All these components function together to set the level of transcription for each gene, which provides nearly limitless targets for regulation.
�The structure of human snRNA genes transcribed by pol II
The positive transcription elongation factor, P-TEFb, plays an essential role in the regulation of transcription by RNA polymerase II (Pol II) in eukaryotes.[1] Immediately following initiation Pol II becomes trapped in promoter proximal paused positions on the majority of human genes (Figure 1). P-TEFb is a cyclin dependent kinase that can phosphorylate the DRB sensitivity inducing factor (DSIF)[4]and negative elongation factor (NELF), as well as the carboxyl terminal domain of the large subunit of Pol II and this causes the transition into productive elongation leading to the synthesis of mRNAs. P-TEFb is regulated in part by a reversible association with the 7SK snRNP. Treatment of cells with the P-TEFb inhibitors DRB or flavopidirol leads to loss of mRNA production and ultimately cell death.
RNA polymerase II (Pol II) is responsible for synthesizing all protein-coding RNAs and most non-coding RNAs, including small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), microRNAs (miRNAs), cryptic unstable transcripts (CUTs) and stable unannotated transcripts (SUTs).�� RNA pol II assists RNA pol III in transcribing U6 RNAs, for both promoters are very nearby.
Spliceosomal snRNAs:
Mammalian RNAPII-transcribed spliceosomal snRNAs (U1, U2, U4, and U5) are intronless and nonpolyadenylated noncoding RNAs. Formation of their 3′-(3delta,epsilon^2) ends depends on specific snRNA promoters but does not require a poly(A) site or HDE, like histone mRNAs, but rather a 3′-box element located 9–19 nt downstream from the mature 3′-end of snRNAs. The 3′-box is important for proper transcription of snRNAs and was shown to be required for 3′-end processing of U1 and U2 snRNAs. After endonucleolytic cleavage downstream from the 3′-box, snRNAs carry an extended 3′-end that is trimmed after their export to the cytoplasm (for review, see Egloff et al. 2008).
Various examples of transcription activation by recruitment:
(A) A typical activator on a mRNA promoter contains a DNA-binding domain (DBD) and an activation domain (AD). The activation domain may perform a number of functions, among them recruitment of core promoter-binding factors, such as TFIID and TFIIA, and recruitment of the holoenzyme.
�
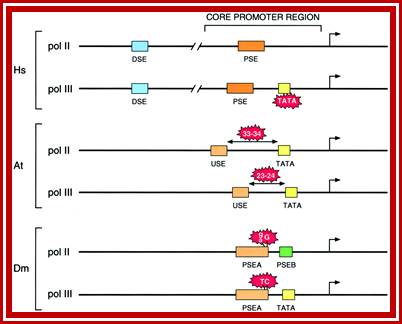
�(B) The Oct-1 activator contains a DNA-binding domain consisting of a POU domain and an activation domain. On snRNA promoters, the Oct-1 POU domain recruits the core promoter-binding factor SNAPc through a direct protein�protein interaction with a small region of SNAP190. How the Oct-1 AD activates snRNA gene transcription is not known. (C). The λ cI repressor activates transcription from the λ PRM promoter by binding as a dimer close to the binding site for RNA polymerase and recruiting, through a direct protein�protein contact, the core promoter�recognizing subunit of the RNAP and sigma factor shows the structures of snRNA promoters from Homo sapiens (Hs),Arabidopsis thaliana (At), and Drosophila melanogaster (Dm) and serves to illustrate the remarkable fact that although snRNA promoters have diverged during evolution, the close similarity between those recognized by pol II and those recognized by pol III has been conserved. In fact, in each of the examples in the Fig shown, RNA polymerase specificity can be changed by altering a single parameter, indicated in red on the figure.
In the human genes, the U1 and U2 snRNA promoters serve as the prototypic pol II snRNA promoters, and the U6 snRNA promoter serves as the prototypic pol III snRNA promoter. The human pol II snRNA core promoters contain only one essential element, the PSE, whereas the pol III snRNA core promoters consist of two elements, the PSE and a TATA box located at a fixed distance downstream. The DSE serves to enhance transcription from the core promoter. Both the DSE and the PSE can be interchanged between pol II and III snRNA promoters with no effect on RNA polymerase specificity, which is determined by the presence or absence of the TATA box. The A. thaliana pol II and III snRNA promoters contain an upstream sequence element (USE) and a TATA box, which are both interchangeable between the pol II and III snRNA promoters. RNA polymerase specificity is determined in this case by the exact spacing between the USE and the TATA box, which is 33�34 base pairs (bp) and 23�24 bp in the pol II and III snRNA promoters, respectively.
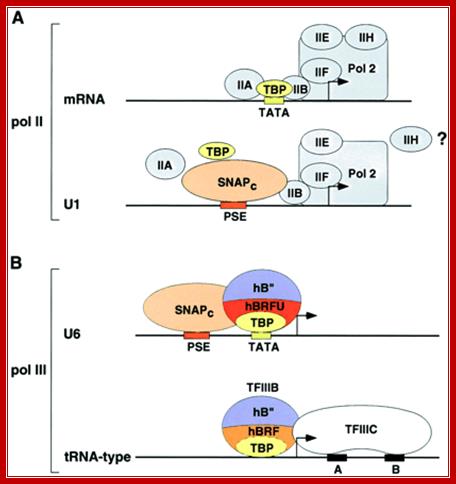
Composition of pol II and III transcription initiation complexes. A, pol II initiation complexes assembled on a TATA box containing mRNA core promoter and on the human U1 snRNA core promoter. B, pol III initiation complexes assembled on the human U6 snRNA promoter and a tRNA-type promoter. The placement of hBRF, hBRFU, and hB" is arbitrary. Adashed line separates TBP and hBRF to indicate that these factors are tightly associated with each other in solution. In contrast, there is no evidence that TBP and hBRFU are associated with each other in solution. http://www.jbc.org
Human U6 transcription initiation complex. TBP (yellow) and SNAPc (orange) bind cooperatively to the DNA, presumably through a direct protein�protein contact that involves a 50-amino-acid segment within the N-terminal region of SNAP190. TBP also binds cooperatively with Brf2. SNAPc and the Oct-1 POU domain (blue) bind cooperatively to DNA, through a direct protein�protein contact involving a glutamic acid at position 7 within the POUS domain (blue triangle) and a lysine at position 900 within SNAP190 (red triangle). This direct protein�protein interaction is mediated by a positioned nucleosome (green) that brings into close proximity the octamer sequence and the PSE. Adapted fromZhao et al. (2001).
Sno RNAs and their genes:
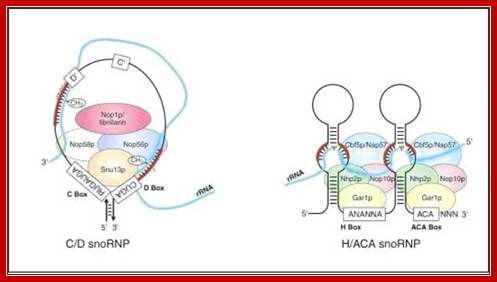
The above diagram shows the functional C/D snoRNA and H/ACA sno RNAs with their associated snoRNPs. http://www.web-books.com/
Small molecular nucleolar RNA-snoRNAs found in the nucleus play significant role in processing precursor rRNAs.� The sno RNAs are derived form many excised introns of pre mRNAs and many have their own small genes and they are transcribed by RNAPIII.
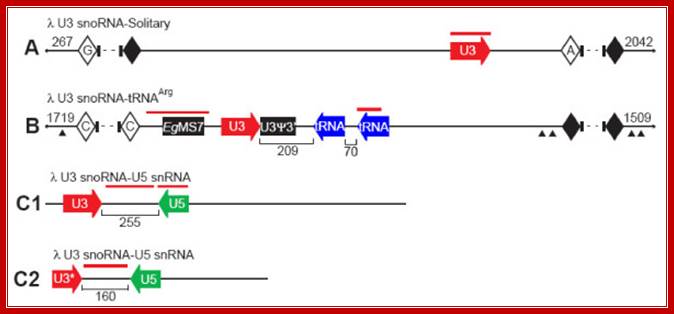
U3 snoRNA genes are multi-copy and frequently linked to U5 snRNA genes in Euglena gracilis; Charette JM, Grayhttp://openi.nlm.nih.gov/
Three different U3 snoRNA gene arrangements in λ clones of Euglena genomic DNA: Clone maps of (A) λ U3-Solitary (total size ~12 kbp; 4824 bp sequenced; U3-containing portion 2515 bp), (B) λ U3-tRNAArg (total size ~14 kbp; 6096 bp sequenced; U3-containing portion 2868 bp) and (C1 and C2) λ U3-U5 (total sizes 1772 and 516 bp, respectively) are shown. Terminal sequences of specified length (bp) do not contain any features of interest. Sizes of the intergenic spacers are indicated by the numbers between the various genes. Red lines demarcate the positions of probes used in Southern hybridization experiments. Repetitive elements, such as the Euglena EgMS7 microsatellite (filled rectangle), the U3 snoRNA pseudo-3'-end repeat (U3Ψ3' filled rectangle) and repetitive sequences also found within certain Euglena γ-tubulin introns (filled triangles) are indicated. Solid diamonds denote simple sequence repeats that precluded effective primer design for additional primer walking. Open diamonds with letters denote homopolymer runs (A, C or G) that prematurely terminated sequencing. Due to the technical challenges posed by the many repetitive sequence elements, the λ U3-Solitary and λ U3-tRNAArg clones were not sequenced in their entirety; dashed lines denote unsequenced portions. In the λ U3-U5-C2 clone, U3* refers to a 5' truncation of the U3 gene as a result of cloning. Clone maps not drawn to scale. Charette JM, Grayhttp://openi.nlm.nih.gov/
Yeast sno/snRNAs
The majority of snoRNAs fall into two structurally and functionally well-defined classes. SnoRNAs carrying the conserved box C and D elements function mainly as guides in the site-specific 2�-O-methylation of target RNAs, whereas box H/ACA snoRNAs direct pseudouridylation. While some snoRNAs are required for pre-rRNA processing, the majority of snoRNAs guide modifications of pre-rRNAs. �In mammals, these latter snoRNAs are encoded within introns of pre-mRNAs (while snoRNAs involved in pre-rRNA processing in humans, and most of the guide RNAs expressed in yeast, are transcribed by RNAPII from their own promoters.
Following cleavage by the endoribonuclease Rnt1 (yeast RNase III) or release from introns, the 3�-ends of snoRNA precursors are matured by exonucleolytic trimming via the exosome. In some cases, independently transcribed snoRNAs do not possess the RNA hairpin structures that are recognized by Rnt1 and may undergo endonucleolytic cleavage by the mRNA 3�-end formation machinery. After cleavage, co transcriptional association of specific factors and formation of sn/snoRNPs protect the mature 3′-end of sn/snoRNAs from further exonucleolytic trimming.
Involvement of mRNA 3� end processing factors in yeast snoRNA processing and termination
SnoRNAs and snRNAs are not polyadenylated. However, they require common factors from the cleavage/polyadenylation machinery; and thus, cleavage must be uncoupled from polyadenylation. Cleavage factors are, in fact, required for correct termination and processing of sno/snRNAs. Additionally, mutations in a sub complex of the cleavage/polyadenylation machinery, the APT complex (associated with Pta1 [homolog of symplekin]), lead to read-through transcription of certain sn/snoRNA genes. The APT factors implicated in sn/snoRNA termination include Pti1, a second yeast homolog of CstF-64 that also binds Rna14;, which interacts with the snoRNP-specific protein Nop1� the CTD phosphatase Ssu72; and Swd2, which is also a component of the Set1 complex Over expression of Pti1 inhibits polyadenylation without affecting cleavage, suggesting that Pti1 may function in uncoupling cleavage and polyadenylation during snoRNA 3′-end formation.
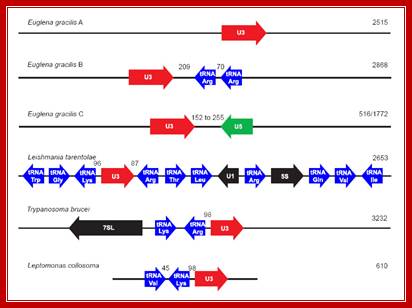
Organization of U3 snoRNA genes within Euglenozoa. The U3 snoRNA genes are shown in red, U5 snRNA gene in green, tRNA genes in blue and all other genes are shown in black. Euglena gracilis A, B and C correspond to the arrangements depicted in Fig. 2. Numbers shown between the various genes correspond to the sizes of the intergenic spacers separating the two genes. Numbers on the right hand of the figure indicate the size (bp) of the respective genomic region. Note that this figure is not drawn to scale. Organisms represented include Euglena gracilis, (Leishmania tarentolae ,Trypanosoma brucei, and Leptomonas collosoma [GenBank:L32919].
Transcriptional termination:
RNAP I- termination:
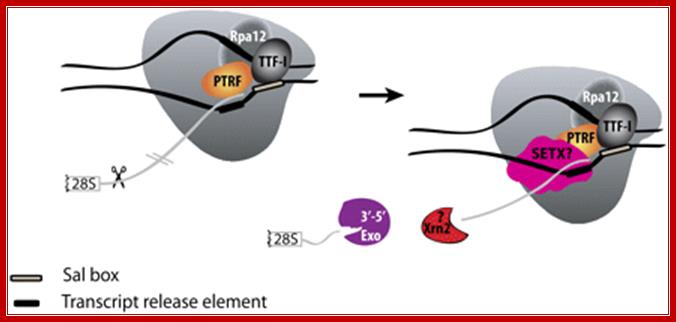
Mammalian RNAPI termination. Transcription terminates at the transcript release element composed of a stretch of Ts, upstream of the TTF-I-mediated pause site. The release factor PTRF, which interacts with RNAPI and TTF-I, recognizes the transcript release element and most likely binds the stretch of Us on the nascent RNA. The RNAPI-specific subunit Rpa12 also plays a role in yeast RNAPI termination that is possibly conserved in mammals. After cleavage of the precursor downstream from the 28S gene, Xrn2 degrades the downstream RNA, �catches up� with RNAPI, and participates in RNAPI release, possibly in conjunction with SETX. An unidentified 3′�5′ exonuclease might be involved in processing of the rRNA precursor. http://genesdev.cshlp.org/
Yeast RNAPII termination at snoRNAs genes.
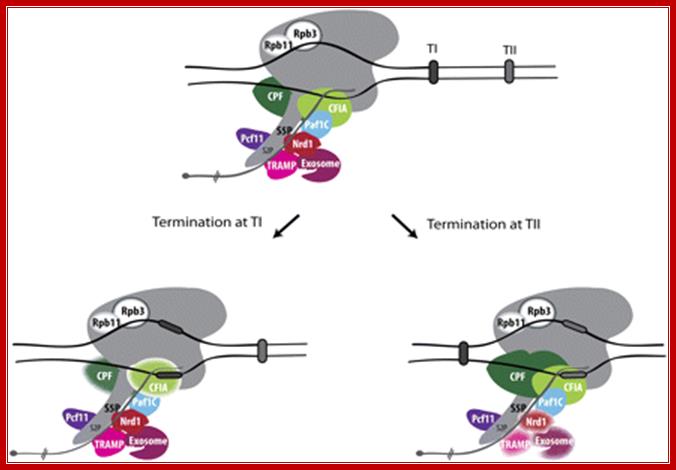
Termination at terminator I (TI), the major termination site, is Nrd1-binding-dependent. The Nrd1 complex interacts with the Ser5-phosphorylated CTD and with the TRAMP and exosome complexes. The cleavage/polyadenylation machinery (CPF and CFIA) is not required for termination at TI (drawn as blurry complexes). Termination at terminator II (TII) involves cleavage/polyadenylation factors, while the Nrd1�TRAMP �exosome complex appears inactive in that process (blurry complexes). SnoRNA transcription couples� termination with RNA surveillance.
Yeast RNAPII termination at snoRNA genes: Termination at terminator I (TI), the major termination site, is Nrd1-binding-dependent. The Nrd1 complex interacts with the Ser5-phosphorylated CTD and with the TRAMP and exosome complexes. The cleavage/polyadenylation machinery (CPF and CFIA) is not required for termination at TI (drawn as blurry complexes). Termination at terminator II (TII) involves cleavage/polyadenylation factors, while the Nrd1�TRAMP�exosome complex appears inactive in that process (blurry complexes). SnoRNA transcription couples� termination with RNA surveillance. After release of the precursor, Trf4 and Pap1 adenylates the pre-snoRNA that will be processed by the exosome. Other factors shown to have a role in snoRNA termination are indicated. These include Pcf11, Paf1C, and the RNAPII subunits Rpb3 and after release of the precursor, Trf4 and Pap1 adenylate the pre-snoRNA that will be processed by the exosome. Other factors shown to have a role in snoRNA termination are indicated. These include Pcf11, Paf1C, and the RNAPII subunits RPB3 and RPB1.http://www.genesdev.cship.org
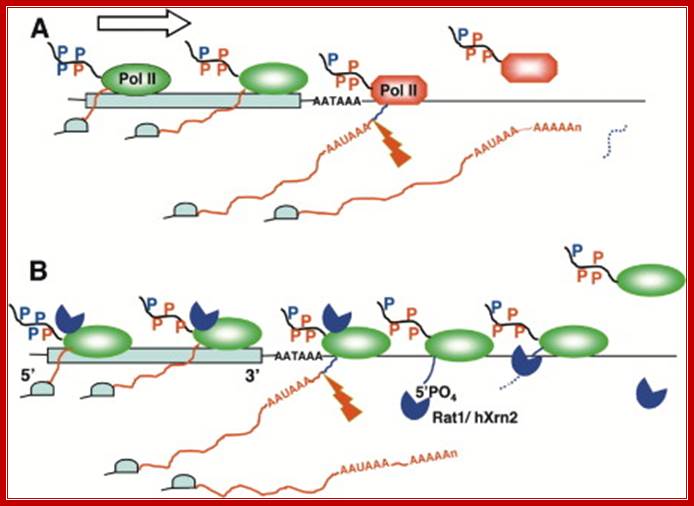
Models for Termination by RNA Polymerase II
(A) The allosteric model. During elongation within the gene (blue box), pol II is in a highly processive conformation (green oval) and is transformed by conformational change or covalent modification into a nonprocessive form (red octagon) after transcribing through the poly(A) site (AATAAA). Ser5 (blue P) and Ser2 (red P) phosphorylation of the pol II CTD (black line) are marked. (It is not known if CTD phosphorylation changes downstream of the poly[A] site.) The RNA transcript is red upstream of and blue downstream of the poly(A) cleavage site (lightning bolt). Dotted blue line denotes degraded RNA. The 5′ cap, added cotranscriptionally, is shown as a pale blue hat.
(B) The torpedo model. RNA downstream of the poly(A) cleavage site (blue line) is digested by a 5′-3′ exonuclease Rat1 in yeast and hXrn2 in humans (blue pacman), which tracks with pol II throughout the length of the gene. After poly(A) site cleavage, the exonuclease torpedo is guided along the RNA to its polymerase target and dissociates it from the DNA template. http://www.sciencedirect.com/
Polymerase III termination: �Polymerase III termination of transcription at small poly Ts stretch (5-6). In Eukaryotes, a hairpin loop is not required, as it is in prokaryotes.
� Though most of the RNAP-III transcribed genes are small, at the end of the gene, they have sequences, which generate GC rich complementary sequences with four Us, two of which are embedded in the stem structure and two Us at the 3� end.�
� The stem loop structure is similar to that of prokaryotic one.�
� The secondary structure interacts with RNAP and stalls the enzyme.
The weak bonds between Us in the transcripts and A s in the template, renders the enzyme fall off from the template.�
Polymerase III terminates transcription at small poly Ts stretch. In Eukaryotes, a hairpin loop is not required, as it is in prokaryote.
RNAPIII and RNAPI termination appear simpler than RNAPII termination. RNAPIII terminates transcription at T-rich sequences located a short distance from the mature RNA 3′-end and seems to involve at most a limited number of auxiliary factors
RNAPIII transcribes a variety of short, nuclear, and cytoplasmic noncoding genes (100�150 bp) including those encoding 5S rRNA, U6 spliceosomal snRNA, tRNAs, and the RNAse P, RNAse MRP, adenovirus-associated (VA), and 7SK RNAs (for review, see Deice et al. 2007). RNAPIII is the largest of the three RNAPs; yeast RNAPIII has an overall mass of 700 kDa and is composed of 17 subunits, all essential for cell viability. Nine subunits define the structural core, evolutionarily related to the other nuclear RNAPs, with only five subunits specific to RNAPIII (Jasiak et al. 2006). RNAPIII is a very efficient polymerase not only because of the short transcription units, but also because of rapid reloading of RNAPIII on the same transcription unit (Dieci and Sentenac 1996; Ferrari et al. 2004). RNAPIII can terminate efficiently with no apparent need of other factors.
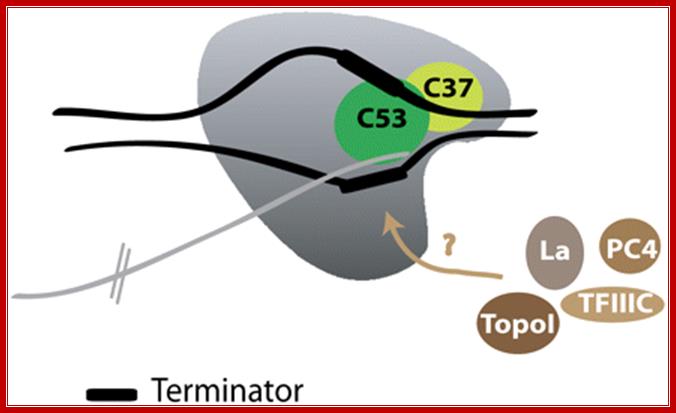
RNAPIII termination: Most of the termination activity is triggered by RNAPIII itself. The subunits C37 and C53 are essential for termination. The heterodimer C37�C53 might play a role in reducing the elongation rate of RNAPIII, allowing for an increased pausing time at the terminator, composed of a stretch of four to five Ts in the coding DNA strand. Several auxiliary factors, such as La, PC4, Topoisomerase I, or TFIIIC, are proposed to participate in RNAPIII termination in mammals. http://genesdev.cshlp.org
RNAP III and II Transcribed non-coding RNAs (list is not complete):
- Transfer RNAs,
- 5S ribosomal RNA,
- U6 spliceosomal RNA,
- RNase P and RNase MRP RNA,
- 7SL RNA (the RNA component of the signal recognition particle) ,
- Vault RNAs,
- Y RNA,
- SINEs (short interspersed repetitive elements) ,
- 7SK RNA,
- Several microRNAs,
- Several small nucleolar RNAs,
- Several gene regulatory antisense RNAs,
Genes and development: Different types of RNA polymerase III promoters. The type 1 promoter of the Xenopus laevis 5S RNA gene consists of an internal control region (ICR), which can be subdivided into A box (+50 to +60), intermediate element (IE, +67 to +72), and C box (+80 to +90). The type 2 promoter of the X. laevis tRNALeugene consists of an A box (+8 to +19) and a B box (+52 to +62). The type 3 promoter of the Homo sapiens U6 snRNA gene consists of a distal sequence element (DSE, −215 to −240) that enhances transcription and a core promoter composed of a proximal sequence element (PSE, −65 to −48) and a TATA box (−32 to −25). The Saccharomyces cerevisiae promoter is a hybrid promoter consisting of a TATA box (−30 to −23), an A box (+21 to +31), and a B box located downstream of the U6 coding region (from +234 to +244 relative to the start site of transcription).
www.sfsoo.com; https://fame-textile.ru
The expanding RNA polymerase III transcriptome:
Giorgio Dieci![]() Email the author Giorgio Dieci; Gloria Fiorino;�
Manuele Castelnuovo
Email the author Giorgio Dieci; Gloria Fiorino;�
Manuele Castelnuovo
Martin Teichmann; , ; Aldo Pagano
Cell.com� http://www.cell.com; The role of RNA polymerase (Pol) III in eukaryotic transcription is commonly thought of as being restricted to a small set of highly expressed, housekeeping non�protein-coding (nc)RNA genes. Recent studies, however, have remarkably expanded the set of known Pol III�synthesized ncRNAs, suggesting that gene-specific Pol III regulation is more common than previously appreciated. Newly identified Pol III transcripts include small nucleolar RNAs, microRNAs, short interspersed nuclear element�encoded or tRNA-derived RNAs and novel classes of ncRNA that can display significant sequence complementarity to protein-coding genes and might thus regulate their expression. The extent of the Pol III transcriptome, the complexity of its regulation and its influence on cell physiology, development and disease are emerging as new areas for future research.
Three RNA polymerases in eukaryotes. RNA polymerase III Hundreds of promoters - 40% of a cell transcriptional activity -Moderately sensitive to a -amanitin. http://slideplayer.com;
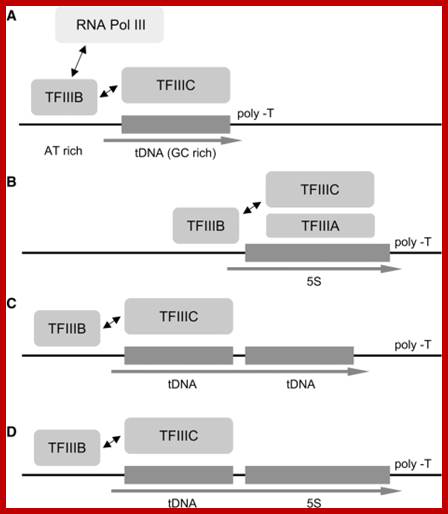
https://openi.nlm.nih.gov Dicistronic tRNA-5S rRNA genes in Yarrowia lipolytica: an alternative TFIIIA-independent way for expression of 5S rRNA genes.
Acker J, Ozanne C, Kachouri-Lafond R, Gaillardin C, Neuv�glise C, Marck C - Nucleic Acids Res.
Overview of the transcription mechanisms of tRNA, 5S rRNA and tRNA�5S rRNA dicistronic genes. (A) In S. cerevisiae, transcription of a tRNA gene by Pol III requires the assembly of transcription factor TFIIIC onto the tDNA (GC-rich) followed by that of TFIIIB (onto a AT-rich region) that finally recruits Pol III for multiple cycles of transcription. (B) A regular 5S rRNA gene is first recognized by its specific factor TFIIIA; then the 5S rRNA gene�TFIIIA complex is bound by TFIIIC and the next steps of transcription are identical to that of tRNA genes. In both cases, transcription stops and efficiently recycles when Pol III reaches the terminal T-track in the RNA-like strand. (C) Transcription of a dicistronic tRNA�tRNA gene by a unique TFIIIC molecule binding on the upstream gene. Assembly of TFIIIB onto the upstream gene (GC-rich) triggered by TFIIIC bound on the downstream gene to transcribe this only gene is penalized in vivo. (D) Hypothetical transcription of a dicistronic tRNA�5S rRNA gene may proceed similarly through the recognition of the promoter elements of the upstream gene by TFIIIC. In this case a single primary RNA (arrow) is produced and later matured into two functional products (tRNA and 5S rRNA) without the need of TFIIIA (see text for details).
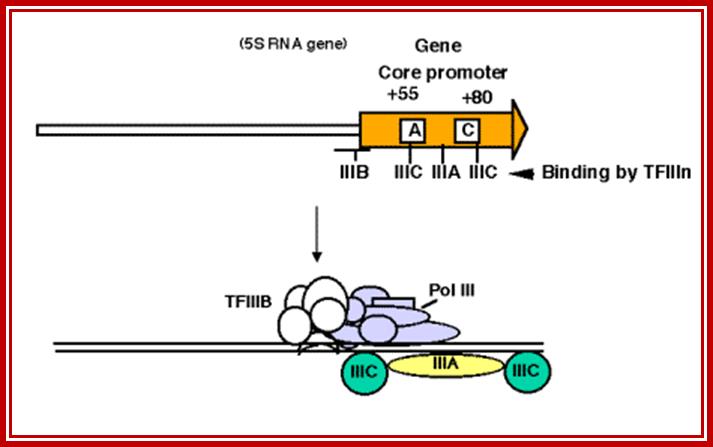
Binding of proteins for promoter for RNA polymearase III
This promoter has internal control sequences. Deletion of 5' flanking DNA still permits efficient transcription of (most) genes transcribed by RNA PolIII. Even the intial part of the gene is expendable, as is the 3' end. Sequences internal to the gene (e.g. +55 to +80 in 5S rRNA genes) are required for efficient initiation, in contrast to the familiar situation in bacteria, where most of the promoter sequences are 5' to the gene.
b. As discussed above, TFIIIA binds to the internal control region of genes that encode 5S RNA (type 1 internal promoter). TFIIIC binds to internal control regions of genes for 5S RNA (alongside TFIIIA) and for tRNAs (type 2 internal promoters). The binding of TFIIIC directs TFIIIB to bind to sequences (-40 to +11) that overlap the start site for transcription. One subunit of TFIIIB is TBP, even though no TATA box is required for transcription. TFIIIA and TFIIIC can now be removed without affecting the ability of RNA polymerase III to initiate transcription. Thus TFIIIA and TFIIIC are assembly factors, and TFIIIB is the initiation factor.
5S and
tRNA transcription
Section M Transcription in EukaryotesM3. RNA Pol III genes:5S
and tRNA transcriptionRNA polymerase IIItRNA genes5S rRNA genesAlternative RNA
Pol III promotersRNA Pol III termination
33 M3-1. RNA Pol III Contains at least 16
or more subunits
Section M Transcription in EukaryotesM3-1. RNA Pol III Contains
at least 16 or more subunits Is located in nucleoplasm Synthesizes the
precursors of 5S rRNA, the tRNAs and other small nuclear and cytosolic RNAs
34 Promoters for RNA polymerase III
May consist of bipartite sequences downstream of the start point,
with box A separated from either box C or box B. Or they may consist of
separated sequences upstream of the start point (Oct, PSE, TATA).
36 Section M Transcription in Eukaryotes
M3-2. tRNA genes. �The initial transcripts of tRNA genes need to
be processed to produce the mature tRNA.� The transcription control regions of
tRNA lies after the start site within the transcribed region. The two highly
conserved control sequences are called A box (5�-TGGCNNAGTGG) and B box
(5�-GGTTCGANNCC).
37 A box and B box also encode important
sequences in the tRNA itself, the D-loop and TyC-loop.
Therefore, the highly conserved sequence in tRNAs are also
highly conserved promoter DNA sequences.3. Two complex DNA-binding factors
required for tRNA transcription initiation: TFIIIC---binds to both the A and B
boxes, an assembly factor for positioning TFIIIB.
38 TFIIIB: (1) binds 50 bp upstream from the A box, but has no sequence specificity and the binding position is determined by the DNA bound TFIIIC. (2) consists of three subunits, one of which is TBP, the general initiation factor; the second is called BRF (TFIIB-related factor); and the third is called B�.
39 TFIIIC: A and B boxes binding and a
assembly factor to position TFIIIB
TFIIIB: DNA binding and RNA Pol III recruiting
40 Section M Transcription in Eukaryotes
M3-3. 5S rRNA ��of around 2000 genes.) Transcription control
regions (promoters) are organized similar to those of tRNA, except that C box
is in place of B box. C box: bp; A box:
41 3. Transcription factors: (1) The C box acts as the binding site for TFIIIA. (2) TFIIIA acts as an assembly factor which allows TFIIIC to interact with the 5S rRNA promoter. (3) The A box may also stabilize TFIIIC binding. (4) TFIIIC is then bound to DNA site near +1. (5) TFIIIB and TFIIIC interact to recruit RNA Pol III to initiate transcription.
42 TFIIIA,TFIIIC TFIIIB Pol III
43 M3-4. Alternative RNA Pol III
promoters.
Section M Transcription in EukaryotesM3-4. Alternative RNA Pol
III promoters; Many RNA Pol III genes also rely on upstream sequences for
regulation of their transcriptome. U6 snRNA and Epstein-Barr virus Use only
regulatory genes upstream from their transcription start sites.
44 U6 snRNA; The coding region contains a characteristic A box that is not required for transcription. The upstream sequence contains sequences typical of RNA Pol II promoters, including a TATA box at bases �30 to �23.Shares several other transcription factor binding sequences with many U RNA genes which are transcribed by RNA Pol II Suggestion: common transcription factors can regulate both RNA Pol II and Pol III genes
45 M3-5. RNA Pol III termination
Section M Transcription in EukaryotesM3-5. RNA Pol III
termination. The RNA polymerase can terminate transcription without accessory
factors. A cluster of A residue is often sufficient for termination. Xenopus
borealis terminator: 5�-GCAAAAGC-3�
46 M4. RNA Pol II genes: promoters and
enhancers
Section M Transcription in EukaryotesM4. RNA Pol II genes:
promoters and enhancers Pol IICis-acting elements -Promoters Upstream
regulatory elements -Enhancers
47 M4-1. RNA Pol II located in
nucleoplasm
Section M Transcription in EukaryotesM4-1. RNA Pol II located in
nucleoplasm catalysing the synthesis of the mRNA precursors for all
protein-coding genes.� RNA Pol Ⅱ-transcribed
pre-mRNAs are processed through cap addition, poly(A) tail addition and
splicing.
48 Secti
|
Reinitiation complex in eukaryotic transcription describes the assembly of transcription factors at the promoter before RNA polymerase binds. |
|
|
|
Key Concepts |
� RNA polymerase III has two types of promoters.� Internal promoters have short consensus sequences located within the transcription unit and cause initiation to occur a fixed distance upstream. � TFIIIA and TFIIIC bind to the consensus sequences and enable TFIIB to bind at the startpoint. � Upstream promoters contain three short consensus sequences upstream of the startpoint that are bound by transcription factors. � For both types of promoter, the factor bound at the startpoint has TBP as one subunit and enables RNA polymerase to bind. |
Recognition of promoters by RNA polymerase III strikingly illustrates the relative roles of transcription factors and the polymerase enzyme. The promoters fall into two general classes that are recognized in different ways by different groups of factors. The promoters for 5S and tRNA genes are internal; they lie downstream of the startpoint. The promoters for snRNA (small nuclear RNA) genes lie upstream of the startpoint in the more conventional manner of other promoters. In both cases, the individual elements that are necessary for promoter function consist exclusively of sequences recognized by transcription factors, which in turn direct the binding of RNA polymerase.
Before the promoter of 5S RNA genes was identified in X. laevis, all attempts to identify promoter sequences assumed that they would lie upstream of the startpoint. But deletion analysis showed that the 5S RNA product continues to be synthesized when the entire sequence upstream of the gene is removed!
When the deletions continue into the gene, a product very similar in size to the usual 5S RNA continues to be synthesized so long as the deletion ends before base +55. Figure 20.10 shows that the first part of the RNA product corresponds to plasmid DNA; the second part represents the segment remaining of the usual 5S RNA sequence. But when the deletion extends past +55, transcription does not occur. So the promoter lies downstream of position +55, but causes RNA polymerase III to initiate transcription a more or less fixed distance away.
When deletions extend into the gene from its distal end, transcription is unaffected so long as the first 80 bp remain intact. Once the deletion cuts into this region, transcription ceases. This places the downstream boundary position of the promoter at about position +80.
So the promoter for 5S RNA transcription lies between positions +55 and +80 within the gene. A fragment containing this region can sponsor initiation of any DNA in which it is placed, from a startpoint ~55 bp farther upstream. (The wild-type startpoint is unique; in deletions that lack it, transcription initiates at the purine base nearest to the position 55 bp upstream of the promoter. (641, 642))
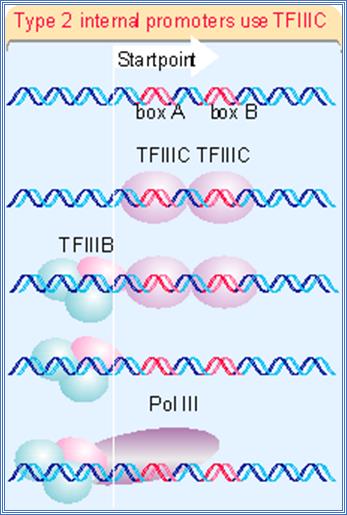
The structures of three types of promoters for RNA polymerase III are summarized in Figure 20.11. There are two types of internal promoter. Each contains a bipartite structure, in which two short sequence elements are separated by a variable sequence. Type 1 consists of a boxA sequence separated from a boxC sequence (1673), and type 2 consists of a boxA sequence separated from a boxB sequence (1674). The distance between boxA and boxB in a type 2 promoter can vary quite extensively, but the boxes usually cannot be brought too close together without abolishing function. Type 3 promoters have 3 sequences elements all located upstream of the startpoint (1675).
The detailed interactions are different at the two types of internal promoter, but the principle is the same. TFIIIC binds downstream of the startpoint, either independently (type 2 promoters) or in conjunction with TFIIIA (type 1 promoters). The presence of TFIIIC enables the positioning factor TFIIIB to bind at the startpoint. Then RNA polymerase is recruited.
Figure 20.12 summarizes the stages of reaction at type 2 internal promoters. TFIIIC binds to both boxA and boxB. This enables TFIIIB to bind at the startpoint. Then RNA polymerase III can bind.
The difference at type 1 internal promoters is that TFIIIA must bind at boxA to enable TFIIIC to bind at boxC. Figure 20.13 shows that, once TFIIIC has bound, events follow the same course as at type 2 promoters, with TFIIIB binding at the startpoint, and RNA polymerase III joining the complex. Type 1 promoters are found only in the genes for 5S rRNA.
TFIIIA and TFIIIC are assembly factors, whose sole role is to assist the binding of TFIIIB at the right location. Once TFIIIB has bound, TFIIIA and TFIIIC can be removed from the promoter (by high salt concentration in vitro) without affecting the initiation reaction. TFIIIB remains bound in the vicinity of the startpoint and its presence is sufficient to allow RNA polymerase III to bind at the startpoint. So TFIIIB is the only true initiation factor required by RNA polymerase III (643). This sequence of events explains how the promoter boxes downstream can cause RNA polymerase to bind at the startpoint, farther upstream. Although the ability to transcribe these genes is conferred by the internal promoter, changes in the region immediately upstream of the startpoint can alter the efficiency of transcription.
TFIIIC is a large protein complex (>500 kD), comparable in size to RNA polymerase itself, and containing 6 subunits. TFIIIA is a member of an interesting class of zinc finger proteins that we discuss later. The positioning factor, TFIIIIB, consists of three subunits. It includes the same protein, TBP, that is present in the core-binding factor for pol I promoters, and also in the corresponding transcription factor (TFIID) for RNA polymerase II (). It also contains Brf, which is related to the factor TFIIB that is used by RNA polymerase II. The third subunit is called B"; it is dispensable if the DNA duplex is partially melted, which suggests that its function is to initiate the transcription bubble the role of B" may be comparable to the role played by sigma factor in bacterial RNA polymerase (see 9.15 Substitution of sigma factors may control initiation).
The upstream region has a conventional role in the third class of polymerase III promoters. In the example shown in Figure 20.11, there are three upstream elements. These elements are also found in promoters for snRNA genes that are transcribed by RNA polymerase II. (Genes for some snRNAs are transcribed by RNA polymerase II, while others are transcribed by RNA polymerase III.) The upstream elements function in a similar manner in promoters for both polymerases II and III.
Initiation at an upstream promoter for RNA polymerase III can occur on a short region that immediately precedes the start point and contains only the TATA element. However, efficiency of transcription is much increased by the presence of the PSE and OCT elements. The factors that bind at these elements interact cooperatively. (The PSE element may be essential at promoters used by RNA polymerase II, whereas it is stimulatory in promoters used by RNA polymerase III; its name stands for proximal sequence element.)
The TATA element appears to confer specificity for the type of polymerase (II or III) that is recognized by an snRNA promoter. It is recognized by a factor that includes the TBP, which actually recognizes the sequence in DNA. The TBP is associated with other proteins, which are specific for the type of promoter. The function of TBP and its associated proteins is to position the RNA polymerase correctly at the start point. We discuss this in more detail for RNA polymerase II in the next section.
The factors work in the same way for both types of promoters for RNA polymerase III. The factors bind at the promoter before RNA polymerase itself can bind. They form a preinitiation complex that directs binding of the RNA polymerase. RNA polymerase III does not itself recognizes the promoter sequence, but binds adjacent to factors that are themselves bound just upstream of the start point. For the type 1 and type 2 internal promoters, the assembly factors ensure that TFIIIB (which includes TBP) is bound just upstream of the start point, to provide the positioning information. For the upstream promoters, transcription factors that directly recognize the upstream sites form a complex (including TBP) that is recognized by RNA polymerase III. So irrespective of the location of the promoter sequences, factor(s) are bound close to the start point in order to direct binding of RNA polymerase III (for review see 220).
This section updated 4-30-2001
http://www.biodados.icb.ufmg.br