RNAP-II Promoters:
And transcription complexes:
Almost all eukaryotic structural genes are monocistronic, with little exception like C. elegans and few protozoan parasites where certain percentages of genes expressed are polycistronic.
· Unlike the expression of rRNA genes, tRNA genes, 5sRNA genes, U6, 7SK RNA and 7SL RNA genes, Sc/Sn RNA genes, Sno RNAs and few others are expressed constitutively in all cells and in all tissue types; expression of protein coding genes is not all that constitutive.
Though a majority of protein coding genes are housekeeping genes, they are expressed constitutively in all cell types and at all times. There are other genes, where some are expressed only once in the life time of an organism and in stage specific manner ex. during development. Some are tissue specific. They are not expressed thereafter and remain silent. Is it a repression? If it so, How?
· There are genes, which are expressed in tissue specific manner. Each and every cell types in a tissue have one or more tissue specific genes, to be of special nature; they are expressed, which impart the structural and functional features of that tissue as special and specialized. A tissue means it contains several different cell types. One can identify them by their cell morphology and their cellular function.
· In cell types specific genes express, their expression is absolutely required for the structure and function of that cell type. At the same time new set of genes are expressed and some of the genes expressed before the cell type differentiation are switched off.
There are a number of other genes which express in response to a variety of stimuli, which can emanate with in the body or they can be from outside. The stimulus can be mechanical, radiations, chemical, light, temperature, nutritional sound or water deficiency or a change in the pH or ionic composition.
It is a good exercise to apportionate certain number of specific genes to each of the kinds of cell types. Nearly 96 million cells die per minute and in the same time 96 million cells divide and replace the dead cells. Half-life each of the cell types vary; for example, white blood cells live for about thirteen days, cells in the top layer of our skin live about 30 days, red blood cells live for about 120 days, and liver cells live about 18 months. It was believed that neuronal cells increase in number till the age of 5-6 yrs in humans, then those not used degenerate. No new cells develop. But it is now believed for hippocampus in adult rat brain produces new cell and they migrate to other parts of the brain. Now scientist are rethinking even human brain produces new brain cells and they spread, possibly they have a functions in learning and memory.
What does human genome project say about this? Looking at the number of tissue types, number of stages during development, number of cell cycle events, and number of stimuli whether inter-cellular, intracellular or extra cellular, gene or genes involved in all of them cannot be same and cannot be expressed by one general mechanism.
· Is there any correlation between the complexity of the cell types and complexity of cell type functions and the number of genes required for such cell types? Yes, it should be like that.
The modus operendi for each kind should be different, so also the structural organization of each kind of genes especially of their promoters should be different though not be unique. One need not be surprised, if there are any unique genes and unique mechanisms exist in the biological world.
· Almost all eukaryotic genes are split with noncoding Introns intervened in between coding Exon segments. The size of noncoding region of the DNA, some times, is many times more than the coding region of the DNA.
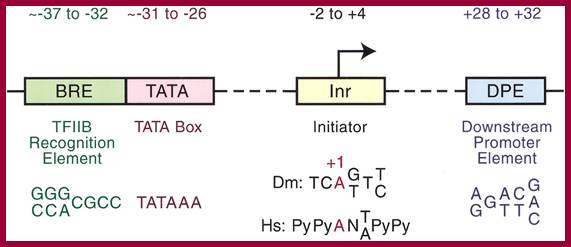
RNAP II core promoter components required for gene expression; Jennifer E.F. Butler1 and James T. Kadonaga; http://genesdev.cshlp.org/
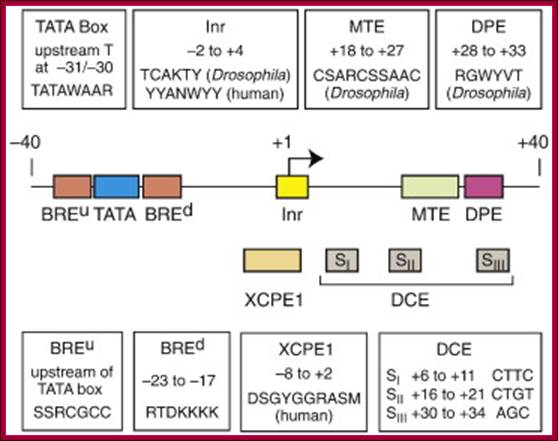
Core promoter elements. Some core promoter motifs that can participate in transcription by RNA polymerase II are depicted. Each of these elements is found in only a subset of core promoters. Any specific core promoter may contain some, all, or none of these motifs. The BRE is an upstream extension of a subset of TATA boxes. The DPE requires an Inr, and is located precisely at +28 to +32 relative to the A+1 nucleotide in the Inr. The DPE consensus was determined with Drosophila transcription factors and core promoters. The Inr consensus sequence is shown for bothDrosophila (Dm) and humans (Hs).
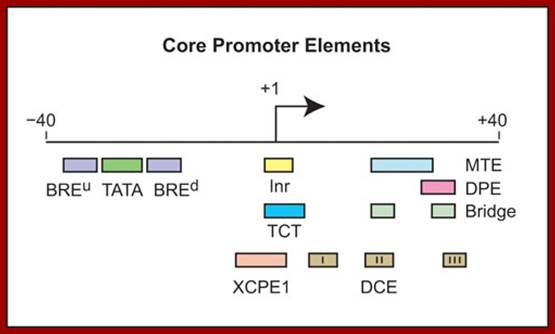
Core promoter elements; James T. Kadonaga ;http://www-biology.ucsd.edu/
“Depending on the presence or absence of specific core promoter elements, each core promoter has its own distinct properties. The best-known core promoter motif is the TATA box; however, the TATA box is present in only about 10 to 15% of human genes. In our studies of TATA-less promoters, we discovered two new core promoter motifs – the DPE and the MTE. Both the DPE and MTE are downstream of the transcription start site and are conserved from Drosophila to humans. It is interesting to note, for example, that the promoters of nearly all of the Drosophila homeotic (Hox) genes contain a DPE motif and lack a TATA box. [The promoters lacking a DPE motif are those associated with the evolutionarily most recent genes, Ubx and Abd-A. Hence, all of the more ancient Hox genes have TATA-less, DPE-containing core promoters.] Moreover, Caudal, a sequence-specific DNA-binding protein that is a master regulator of the Hox genes, is a DPE-specific activator. Thus, enhancer-core promoter specificity can be used in the regulation of gene networks”.
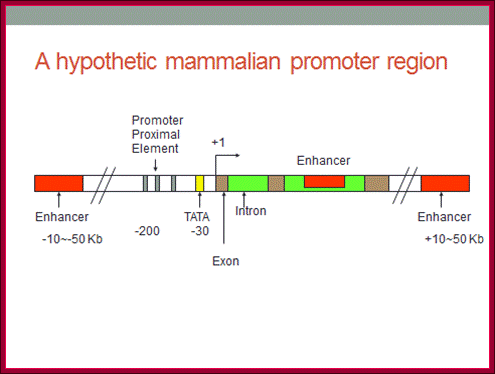
Whether the genes are split or not split, whether they are long genes or short ones, they all have one common feature that is all of them possess promoter and regulator elements. The promoters can be in the upstream of the start site or down stream of the start point (few).
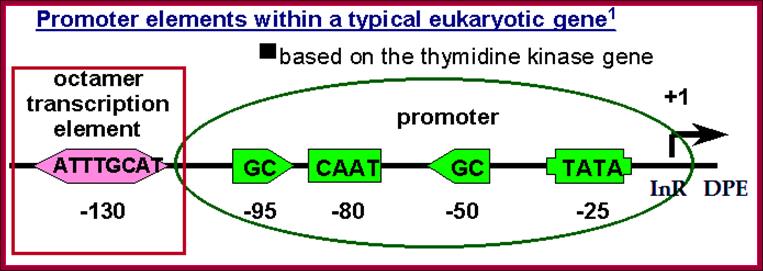
Substantial numbers of structural genes have promoters with TATA box, InR, MTE and DPE; they are the core components, but there are a large number genes >80% are TATA less promoters but contain only InR and DPE sequences. Not surprisingly there are few other genes without any TATA box and InR sequences but only MTE and DPE. MTE sequence elements are found in some and not in all. Besides the core region the upstream elements such as GC and CAAT sequences found close to core components can also be considered as core components; for example, globin gene have such arrangements. The following diagram shows different components of Eukaryotic Gene promoter elements involved gene expression.
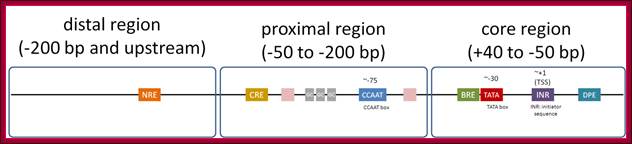
Eukaryotic promoter structure. Example human insulin gene promoter. The general structure is common to most eukaryotes, though the detailed sequence arrangements are highly variable.
;http://genocon.org/assignment-2012/assignment-a/basic-science-a-eukaryotic-promoter.
The core region is from -50 to +40; proximal -200 to -50 and Distal region- beyond -200. This is an approximate distances and positions. Distal regions (DRE) contain enhancer elements; they can be found in downstream, including some introns.
Distal Elements-DRE]—
Proximal elements-[-CRE--GC--CAAT-]
Core elements—[BRE-TATA---[-InR-]--MTE—DCE—-DPE]
The core promoter:
(-)45 BREu-TATA- BREd----InR--- DCE+DPE-+45
Note: there is no Universal Core Promoters (UCP);
Research scholars have identified two types of TATA sequences namely TATAAAAR as TATA1 canonical and the other is TATA (T/A)A(T/A)(A/G)C, as TATA2 regulatory. These sequences are called Goldberg- Hogness boxes. They are specific to specific genes not interchangeable. There are many TATA less promoters as in U1, U2, U4 and U5 sn RNA genes.

TATA 1; http://en.wikipedia.org/
TATA 2
Functional sequence elements in promoters are traditionally represented as consensus sequences that consist of the nucleotide(s) found most frequently at each position (see figure, top). The black print CG-[TATA-AT] A [AT-AG ] is weight matrix based sequence. Consensus sequences are useful for a high-level description of binding preferences; however, their use comes with certain potential pitfalls. First, they are typically reported in the initial identification of a new element, based on a handful of experimentally studied sequences, but often the number of sequences is too small to derive an unbiased (i.e. genome-wide, optimal) consensus. Second, a match to the consensus is either yes or no and does not reflect the actual binding affinity of a transcription factor to a functional element.
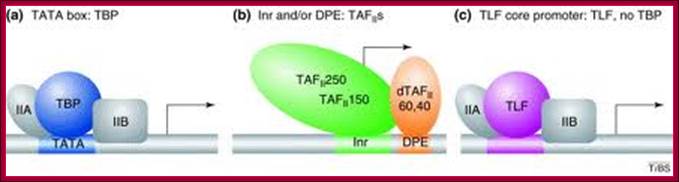
Gene-selective function of general transcription factors (GTFs) is mediated in part by core promoter elements. Factors binding to core promoter elements are shown [blue, TBP; purple, TBP-like factor (TLF); green, TBP-associated factors TAFII250 and TAFII150; orange, dTAFII60 and dTAFII40]. TFIIA and TFIIB, which increase the affinity of TBP for the TATA box are also shown (gray). (a) Core promoters featuring a TATA box require TBP for core promoter recognition and generally do not require TAFIIs for basal transcription in vitro. TATA-box-containing promoters can recruit TFIID (TBP plus TAFIIs) in vivo, but do not necessarily recruit TAFIIs – a subset of promoters has been shown to recruit TBP but no TAFIIs and . The in vivo requirements for TAFIIs most likely depend on both core promoter elements and interactions with transcriptional activators [37]. (b) Core promoters featuring an initiator (Inr) or downstream promoter element (DPE), or both, in many cases recruit and require TBP along with the TAFIIs that bind to these core promoter elements. Of the TAFIIs, only TAFII250, TAFII150, dTAFII60 and dTAFII40 are shown. (c) TLF-dependent core promoters do not require TBP in vivo. It is not known which core promoter elements are required for selective TLF recruitment and whether GTFs other than TFIIA and TFIIB are involved in TLF-dependent transcription and/or promoter recognition. It should be noted that core promoter elements can exist in different combinations and show sequence variation, and that additional core promoter elements could possibly exist, further diversifying core promoter architecture beyond the examples shown in this figure. Gert Jan C Veenstra and Alan P Wolffe,TBS
DSE or URE = Upstream or distal Regulatory Elements, they can be activators or enhancer sequences (-200 to -1000)
CRE, GC, CAAT = Activator sequences can be core components, TF IIB binding element (-50 to -200),
TATA = TBP binding (core component), TATA R=TATAAAA; TATA C=TATA(T/A)A(T/A)(A/G)TATA
BRE = TFIIB binding elements, BREu(G/CGCG/AGGCC an BREd(RTDKKKK); u= upstream of TATA, d=downstream of TATA
InR = TCANT/APyPy Initiator elements (core component) -2 to +4,
MTE = Motif Ten Elements (+18 - +28), CSARCSSAAC (where S is G/C; R is A/G).
DCE = Downstream Control Elements (+6 to +34); DCE I, II, III,
DPE =Downstream promoter elements (core component) found at +28 to + 32. Within DPE one finds MTE (motif ten elements) are found within DCE.
DPE and MTE sequences overlap DCE Downstream control elements.
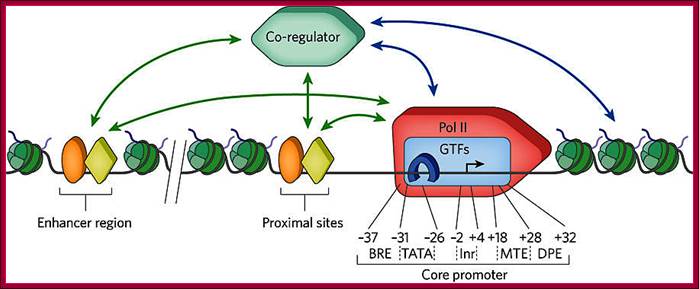
Transcripion regulatory interactionse: Defining mechanisms that regulate RNA polymerase II transcription in vivo; Nicholas J. Fuda, M. Behfar Ardehali & John T. Lis; Nature 461, 186-192(10 September 2009)
“General transcription factors (GTFs) bind to specific sequence elements in the promoter. These elements (the B recognition element (BRE), the TATA box (TATA), the initiator (InR), the motif ten elemental (MTE) and the downstream promoter element (DPE)) and their approximate locations relative to the transcription start site (TSS, black arrow) are shown. Recent investigations have revealed in some there another element called DCE (downstream control Elements) within the region of MTE and DPE. Transcriptional regulators (orange oval and yellow diamond), which are either activators or repressors, bind to specific DNA sequences located near the core promoter of the gene or various distant regions, called enhancers. The regulators can interact (green arrows) with GTFs, such as TFIID (blue rectangle) and TATA-binding protein (TBP, blue horseshoe), and the Pol II complex (red 'rocket') to enhance or repress transcription. They also interact (green arrows) with co-regulators (green hexagon) that can interact (blue arrows) with the general transcription machinery or chromatin-modifying factors, such as histone modifiers or nucleosome remodelers. The co-regulators can also bind to nucleosomes (green) with various histone modifications, stabilizing the co-regulator binding to the gene. Activators can recruit, stabilize or stimulate these factors, and repressors can disrupt or inhibit these factors”.
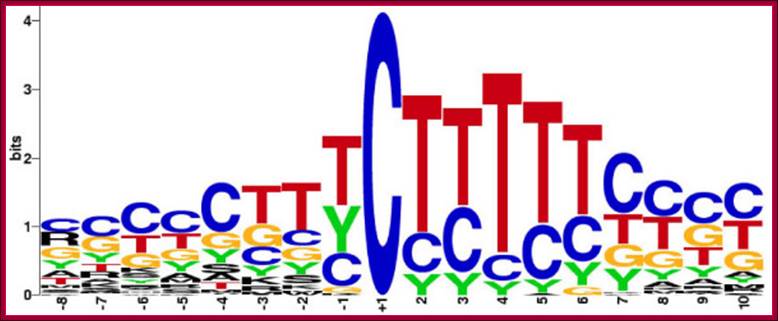
The start of transcription, initiation point, is mostly fixed in genes; not always. The start nucleotide in most of the genes is A (50% of the times), or G (25% of the times and C or U (25% of the times). The start nucleotides always bracketed with pyrimidines like, py py C A N T/A py py py. Organization of sequences as mentioned above at initiation region is called InR (Initiator Region). The InR is also called TSS (Transcription Start Site).
From the start, at –25 to – 30 upstream there is a sequence called TATAA/TA or it is also called Goldberg Hogness box (discoverer of this sequence) or simply called TATA box (A82%, A97%, T93%, A85%, A/T63%, A88% of the times). This box is similar to prokaryotic Pribnow box.
First TATA sequence by Goldberg-1979; [CG]TATA[AT]A[AT][AG].
Promoter elements are therefore now commonly represented in the form of weight matrices (see figure above and bottom), which describe the frequency of all four nucleotides at each position and which can be used to calculate scores that reflect the affinity of a transcription factor for a specific sequence. Weight matrices are often visualized as sequence logos, in which all four letters are shown at their relative frequencies.
Beyond TATA box, in the upstream regions, many genes contain some additional sequence motifs, which are used either for activating or repressing transcription or for increasing or reducing the rate of initiation; such sequences if they are very close to TATA are considered as a part of promoter element. Very often upstream nearby CAAT and GC sequence elements are considered as core elements.
TAFII150 and TAFII250 (i.e., TAF2 and TAF1; (Tora 2002) are the key subunits of TFIID that interact with the Inr
The MTE requires the InR, but functions independently of the TATA-box and DPE. Notably, the loss of transcriptional activity upon mutation of a TATA-box or DPE can be compensated by the addition of an MTE. DPE located at +28 to +32 with consensus sequence RGWYN. In addition, the MTE exhibits strong synergism with the TATA-box as well as the DPE. These findings indicate that the MTE is a novel downstream core promoter element that is important for transcription by RNA polymerase. DCE means downstream core elements.
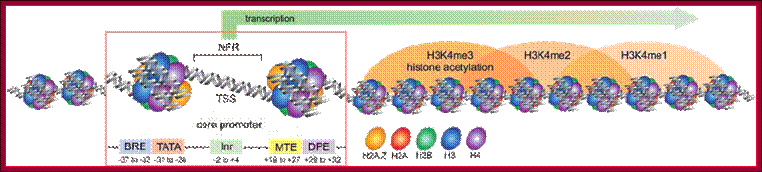
Schematic representation of an active promoter; Abbreviations: NFR, Nucleosome Free Region; TSS, Transcriptional Start Site; BRE, TFIIB Recognition Element; MTE, Motif Ten Element; DPE, Downstream Promoter Element. The nucleosome region comprises approximately 150-200bp. The MTE has been found only in Drosophila, where it can function synergistically with the TATA-box or DPE to initiate transcription. The MTE requires the Inr for its functionality. The ``Proximal Promoter Elements'' and the ``Distal Promoter Elements'' are not indicated here. Figure adapted from (139); http://nens.yellowcouch.org/
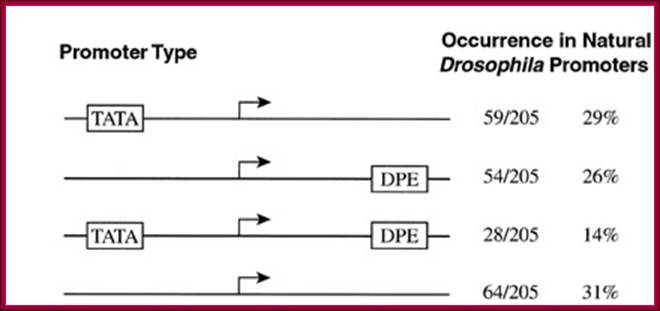
The Downstream Promoter Element DPE Appears To Be as Widely Used as the TATA Box in DrosophilaCore Promoters;;The DPE appears to be present in manyDrosophila promoters.,Drosophila, is not human speicies, but it shows variety of core promoter elements; http://mcb.asm.org/content
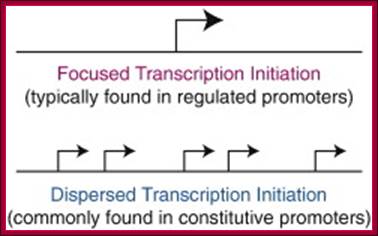
Fig: Focused versus dispersed transcription X initiation: In focused transcription, there is either a single major transcription start site or several start sites within a narrow region of several nucleotides. Focused transcription is the predominant mode of transcription in simpler organisms. In dispersed transcription, there are several weak transcriptions start sites over a broad region of about 50 to 100 nucleotides. Dispersed transcription is the most common mode of transcription in vertebrates. For instance, dispersed transcription is observed in about two-thirds of human genes. In vertebrates, focused transcription tends to be associated with regulated xpromoters, whereas dispersed transcription is typically observed in constitutive x-promoters in CpG islands.
Activators bound to enhancers can activate all the three genes. How? Activators contain acidic rich, glutamine rich or and proline rich domains. Such domains bring about physical contact between activators and the C-terminal domain of RNA polymerase II (Figure below by 9.27; Thompson et al., 1993; Kim et al., 1994), suggesting that rather than direct interaction between an activator and the pre-initiation complex, the signal is transduced by the mediator.
This hypothesis was strengthened when it was shown that the mediator possesses a protein kinase activity that enables it to phosphorylate the CTD of RNA polymerase II, stimulating promoter clearance.
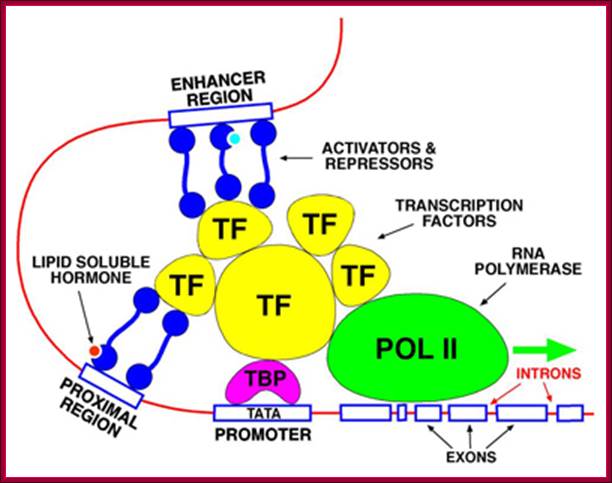
Upstream elements with specific sequences bind to activators or repressors which in turn interact with RNAP II and regulate the expression of the said gene; ;http://www.scilproj.org/
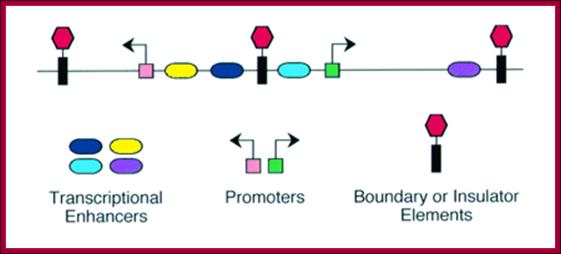
Enhancer - A type of regulatory sequence in eukaryotic DNA (rarely in prokaryotes, but there enhancer sequences even bacteria) that may be located at a great distance upstream or downstream from the promoter which it influences. Enhancers do not activate transcription but enhance the rate of transcription 10 fold to 50fold. Binding of specific proteins to an enhancer stimulates or decreases (silencer) the rate of transcription of the gene. Features thought to be unique to enhancers are that they function in either orientation and can function 5', 3' or within the gene. Enhancers work by binding transcription factors, of which we will see much more in future lectures. Transcription factors either have activating or repressive domains, or recruit activator or repressor proteins, which in turn influences gene expression. Blackwood and Kadonaga (1998). Science 281, 60-63; http://www.zoology.ubc.ca/~bio463
|
Element |
Consensus |
Location |
Factor |
Species |
References |
|
Inr* |
TCA[GT]T[CT] |
−2 |
TAF1, TAF2 |
All eukaryotes |
Smale and Baltimore, 1989 |
|
TATA box* |
[CG]TATA[AT]A[AT][AG] |
−31 |
TBP |
All eukaryotes |
M. L. Goldberg, PhD thesis, Stanford University, 1979 |
|
DPE* |
[GT]CGGTT[CG][GT] |
+26 |
TAF6, TAF9 |
Animals |
Burke and Kadonaga, 1996 |
|
MTE* |
C[CG]A[AG]C[CG][CG]A |
+18 |
TFIID (unknown TAF) |
Animals |
Lim et al., 2004 |
|
BREU |
[GC][GC][AG]CGCC |
−39 |
TFIIB |
Animals (adenovirus) |
Lagrange et al., 1998 |
|
BRED* |
[AG]T[AGT][GT][GT][GT][GT] |
−23 |
TFIIB |
Animals (adenovirus) |
Deng and Roberts, 2005 |
|
DRE† |
[AT]ATCGAT[AT] |
Upstream (variable) |
DREF-TRF2 complex |
Drosophila |
Hochheimer et al., 2002 |
|
Motif 1† |
[CT]GGTCACACT[AG] |
Upstream (variable) |
Unknown |
Drosophila |
Ohler et al., 2002 |
|
Motif 6† |
[CT][AG]GTAT[AT]TT[CT] |
||||
|
Motif 7† |
CA[GT]CNCT[AG] |
Eukaryotic core promoter elements:
Stats of promoter regions in human genome; Suzuki et ;al
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
The core promoter is usually defined as the −50 to +50
base pair (bp) region with respect to the start of transcription. Yet, despite
its short length, the core promoter harbors a number of conserved elements
including the TATA box, Initiator, downstream promoter element (DPE), TFIIB
recognition element (BRE), motif ten elements (MTE), and downstream core
element (DCE). The super core promoter” including four conserved core promoter
motifs (TATA box, MTE, DPE, and Initiator) exhibited significantly elevated
expression levels, both in vitro and in vivo.
Approximately
15-20-% of Human genes contain TATA box. Nearly ~46% don’t have both InR and
TATA. The presence of putative TATA and DPE motifs in Drosophila core
promoters was estimated to be as follows: ∼29% have a TATA box but
no DPE; 26% contain a DPE but no TATA box; 14% possess both TATA and DPE motifs;
31% do not appear to have either a TATA box or a
DPE. In TATA-less promoters, two new core promoter motifs – the DPE and the MTE.
Drosophila homeotic (Hox) genes contain a DPE motif and lack a TATA box. The analysis of TATA-less
promoters with unclustered, multiple start sites led to the identification of a
downstream motif termed MED-1 (multiple start site element downstream. Importantly,
we also identify genomic features called transcriptional initiation platforms (TIPs) that are
characterized by large areas of Pol II and GTF recruitment at promoters,
intergenic and intragenic regions.
Unidirectional
Vs Bidirectional promoters:
Majority of the genes exhibit
Unidirectional Promoters; i.e transcription starts at an Initiators region in
the promoter and transcribes in one direction. There are Bidirectional
Promoters too. The same promoter region also facilitates bidirectional
transcription. If one of the strands is used for the transcription in one
direction, the opposite strand in the same promoter region is used for the
transcription in the opposite direction. Authors mentioned below
downloaded 56,722 protein-coding gene annotations from UCSC genome browser hg 18
database. These collapsed into 25,147 unique and non-overlapping gene clusters.
Of these, 1,369 bidirectional gene pairs were present, defining bidirectional
promoters (for 2,738 genes). Each gene in a bidirectional gene pair formed a
head-to-head arrangement with its closest neighbour and the intergenic
distances between the TSS of a gene and its neighbour had to be within 1,000 bp. After excluding those pairs with too large an intergenic
distance and those with anti-sense overlap at the 5' ends of the transcripts,
we obtained 13,302 genes, which did not form head-to-head arrangement with the
closest neighbour. These were designated non-bidirectional promoters. We also
defined a negative control set. When a gene and its closest neighbour were
transcribed in convergent directions, ending within 1000 bp of each other, they
were designated as tail-to-tail regions Mary
Qu Yang and Laura L Elnitski.
 The Surf-1 and Surf-2 genes and their essential
bidirectional promoter elements are conserved between mouse and human; Lennard A, Gaston K, Fried M.
The Surf-1 and Surf-2 genes and their essential
bidirectional promoter elements are conserved between mouse and human; Lennard A, Gaston K, Fried M.
In the mouse,
the heterogeneous transcription start sites of the divergent Surf-1 and Surf-2
genes are separated by a maximum of only 73 bp (Williams and Fried, 1986). This
region contains a bidirectional promoter composed of three major factor binding
sites required for the efficient expression of both the Surf-1 and Surf-2 genes
(Lennard and Fried, 1991).
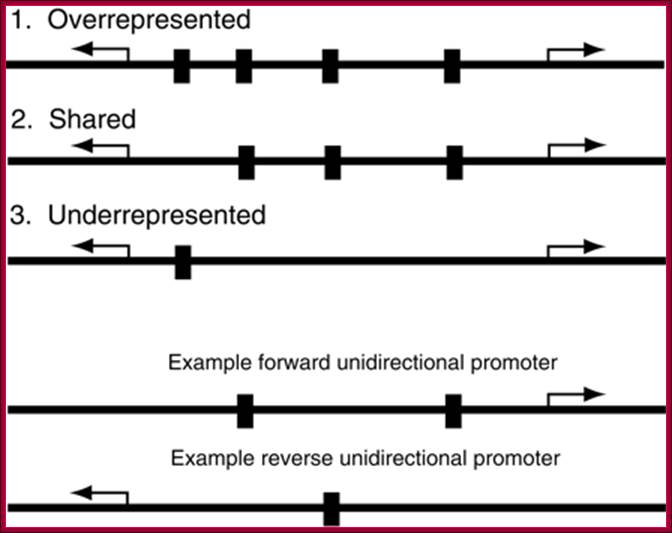
Three types of motif representation in bidirectional promoters.
Motifs in the overrepresented category occur more often (e.g., four times) than
the sum of occurrences in two unidirectional promoters (e.g., 2 + 1 = 3 times).
Motifs in the shared category occur as often (e.g., three times) as the sum of
occurrences in two unidirectional promoters. Motifs in the underrepresented
category occur less often (e.g., once) than the sum of occurrences in two
unidirectional promoters. http://genome.cshlp.org/
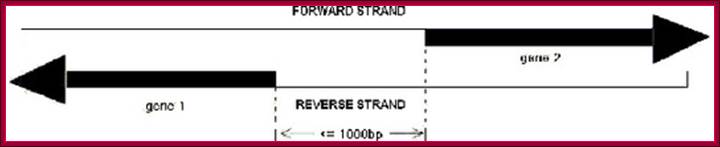
A
sketch map of bidirectional promoter. Gene 1 and 1 are almost on the same loci
on the chromosomes, but left diects transcription strand is opposite to the right-hand
side transcriptional strand. The distance between the two TSS is about 1000 bp.;http://openi.nlm.nih.gov/
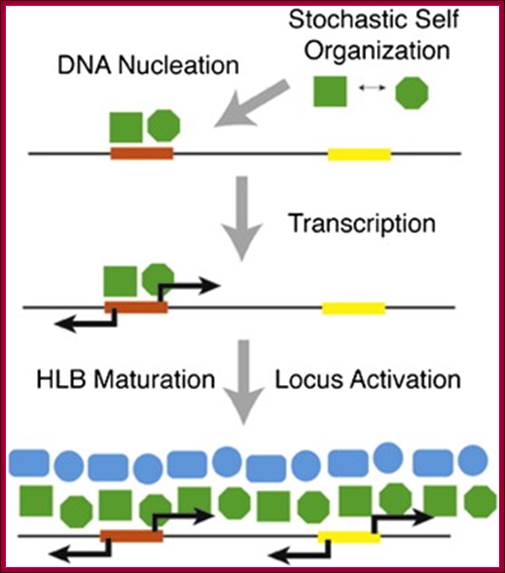
Drosophila H3-H4 promoters triggers Histone Locus Body assembly and Biosynthesis of replication coupled Histone mRNAs. Harmony R.Salzler et alhttp://www.cell.com/
Compartmentalization of RNA biosynthetic factors into nuclear bodies (NBs) is a ubiquitous feature of eukaryotic cells. How NBs initially assemble and ultimately affect gene expression remains unresolved. The histone locus body (HLB) contains factors necessary for replication-coupled histone messenger RNA transcription and processing and associates with histone gene clusters. Using a transgenic assay for ectopic Drosophila HLB assembly, we show that a sequence located between, and transcription from, the divergently transcribed H3-H4 genes nucleates HLB formation and activates other histone genes in the histone gene cluster. In the absence of transcription from the H3-H4 promoter, “proto-HLBs” (containing only a subset of HLB components) form, and the adjacent histone H2a-H2b genes are not expressed. Proto-HLBs also transiently form in mutant embryos with the histone locus deleted. We conclude that HLB assembly occurs through a stepwise process involving stochastic interactions of individual components that localize to a specific sequence in the H3-H4 promoter. Harmony R. Salzler; Deirdre C. Tatomer; , Pamela Y. Malek; Stephen L. McDaniel; Anna N. Orlando; William F. Marzluff; Robert J. Duronio.
Such an arrangement is previously defined as "bidirectional" and the divergent gene pairs are termed as "bidirectional genes", while the intergenic region between a "bidirectional gene pair" is often called a "bidirectional promoter" (Figure 1 – A sketch map of bidirectional promoter.).
A
basic analysis of the bidirectional promoter sequences allowed us to identify
sequence characteristics unique to this class. Many examples show GC-rich
promoters that function in both directions. Nearly 77% of the bidirectional
promoters are located within a CpG island, compared to 38% of non-bidirectional
promoters. Interestingly, only 8% of the bidirectional promoters contain a
strict TATA-box on either strand, which is not significantly different than
what one would expect by chance considering the nucleotide frequencies of the
bidirectional promoters, Nathan D. Trinklein,1 Shelley Force Aldred et al.
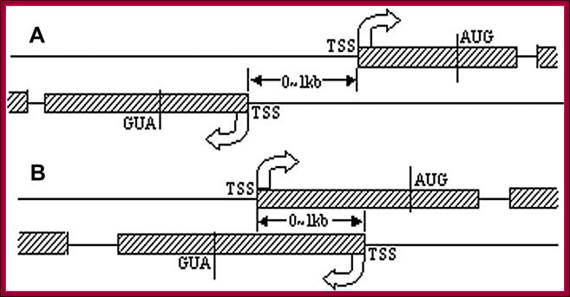
 A Schematic Illustration of Head-to-Head Gene Organization
A Schematic Illustration of Head-to-Head Gene Organization
Head-to-head (h2h) gene pairs with their Transcription Start
Sites (TSSs) less than 1kb apart were identified in five vertebrates: human,
mouse, rat, chicken and fugu. H2h Genes were identified with entrez Gene IDs
and official symbols, and were linked to genetic disorder information from OMIM
database. Basic analyses of their genome occurrences and structure properties
were performed.;Nonoverlapping (A) and overlapping (B) head-to-head gene pairs,
respectively DBH2H. http://lifecenter.sgst.cn/
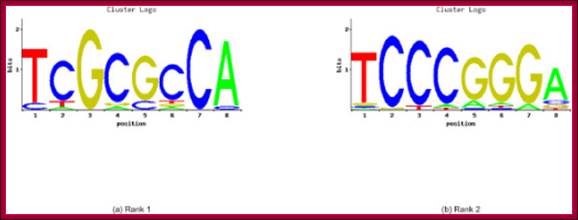
The genomic signatures include statistically
overrepresented words, word clusters, and co-occurring words.The robustness of
this method is confirmed by the ability to identify sequences that exist as
motifs in TRANSFAC and JASPAR databases, and in overlap with verified binding
sites in this set of promoter regions.The word-based signatures are shown to be
effective by finding occurrences of known regulatory sites.;http://openi.nlm.nih.gov/
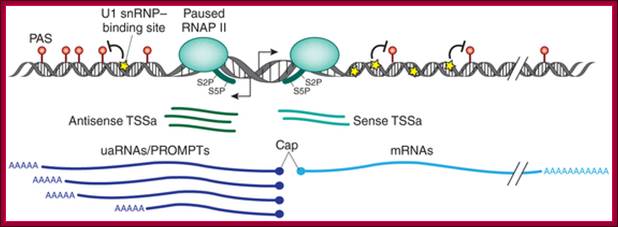
Polyadenylation signals and U1 snRNP–binding sites surrounding the TSS control transcriptional directionality. At the start of transcription, small uncapped TTS-associated RNAs (TSSa) emerge as a result of RNAP II pausing in both directions. If RNAP II continues transcription past these pause sites; Patricia Richard & James L. Manley; Nature Sructural and Molecular Biology.
Lambda gene Transcription of cI and Cro genes almost Bidirectional:
Regulation by cis-antisense RNA transcripts was first postulated in 1972 based on work in bacteriophage λ gene regulation. cI and cro are two essential genes coding for transcription inhibitors whose coding sequence lie adjacent to each other but in opposite directions. An alternative promoter for cI was discovered to be on the other side of cro, and initial studies confirmed the presence of cis-antisense cro RNA transcripts. This led the authors to hypothesize that this novel antisense RNA transcript might potentially serve a role in regulating cro gene activity (Spiegelman, 1972).
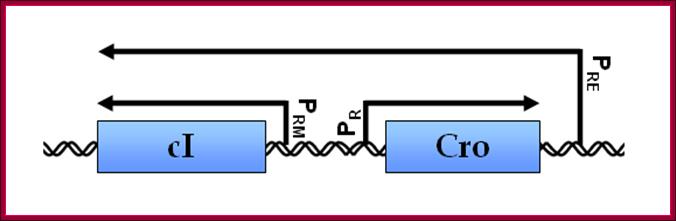
Lambda cI and Cro gene control region- Both sides are used for transcription in Bidirectional mode.
(1) SV40
(simian virus 40) infects monkey kidney cells, and it will also cause
transformation of rodent cells. It has a double stranded DNA genome of
about 5 kb. Because of its involvement in tumorigenesis, it has been a
favorite subject of molecular virologists. The early region encodes tumor
antigens (T-Ag and t-Ag) with many functions, including
stimulating DNA replication of SV40 and blocking the action of endogenous tumor
suppressors like p53 (the 1993 "Molecule of the Year"). The
late region encodes three capsid proteins called VP1, VP2 and VP3 (viral
protein n). A region between the early and late genes controls
both replication and transcription of both classes of genes.
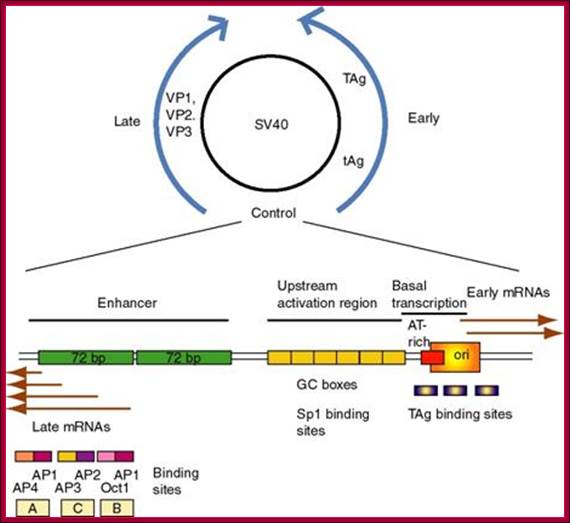
http://www.personal.psu.edu/
(2) The
control region has an origin of replication with binding sites for T-Ag.
In eukaryotes gene are embedded in chromatin, for their identification and transcription chromatin has to be loosened up and it is done various proteins such as histone acetylase, SAGA and SWI/SNF components.
Remodeling of Chromatin is essential for Transcriptional Initiation:
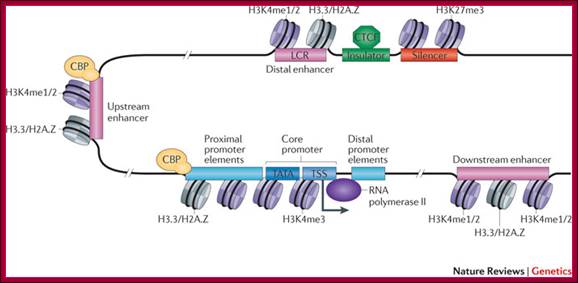
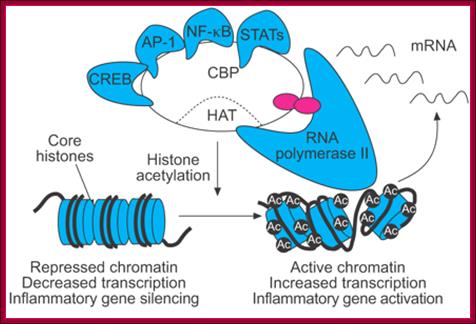
The promoter is typically comprised of proximal, core and downstream elements. Transcription of a gene can be regulated by multiple enhancers that are located distantly and interspersed with silencer and insulator elements, which are bound by regulatory proteins such as CCCTC-binding factor (CTCF). Recent genome-wide data have revealed that many enhancers can be defined by unique chromatin features and the binding of cyclic AMP-responsive element-binding (CREB) protein (CBP). H3K4me1/2, histone H3 mono- or dimethylation at lysine 4; H3K4me3, histone H3 trimethylation at lysine 4; H3K27me3, histone H3 trimethylation at lysine 27; H3.3/H2A.Z, histone variants H3.3 and H2A.Z; LCR, locus control region; TATA, 5′-TATAAAA-3′ core DNA sequences; TSS, transcription start site. Figure is modified, with permission, from Ref. 97 © (2003) Macmillan Publishers Ltd. All rights reserved., Chin-Tong Ong & Victor G. Corces
A) TATA, InR and DCE Containing Promoter Elements:
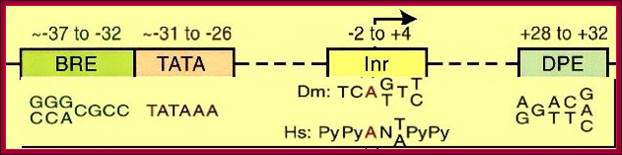
Start
Core promoter -Bre.TATA.Bre—InR (TsS)-DCE (MTE-DPE),
Note- There is no universal Core promoter.
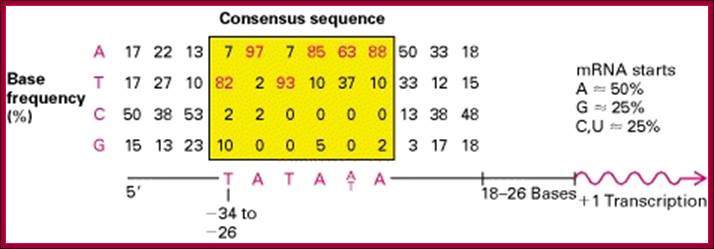
Top figure Comparison of nucleotide sequences upstream of the start site in 60 different vertebrate protein-coding genes; Each sequence was aligned to maximize homology in the region from −35 to −20. The tabulated numbers are the percentage frequency of each base at each position. Maximum homology occurs over a six-base region, referred to as the TATA box, whose consensus sequence is shown at the bottom. The initial base in mRNAs encoded by genes containing a TATA box most frequently is an A. [See R. Breathnach and P. Chambon, 1981, Ann. Rev. Biochem.50:349; P. Bucher, 1990,J. Mol. Biol.212:563.]
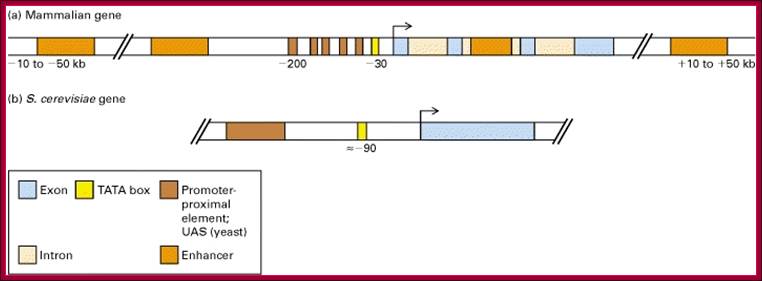
Figure. General pattern of cis-acting control elements that regulate gene expression in yeast and multicellular organisms (invertebrates, vertebrates, and plants); https://mcb.berkeley.edu
(a) Genes of multicellular organisms contain both promoter-proximal elements and enhancers as well as a TATA box or other promoter element. The latter positions RNA polymerase II to initiate transcription at the start site and influences the rate of transcription. Enhancers may be either upstream or downstream and as far away as 50 kb from the transcription start site. In some cases, promoter-proximal elements occur downstream from the start site as well. (b) Most yeast genes contain only one regulatory region, called an upstream activating sequence (UAS), and a TATA box, which is ≈90 base pairs upstream from the start site
Some of the upstream promoter elements can act as core elements.
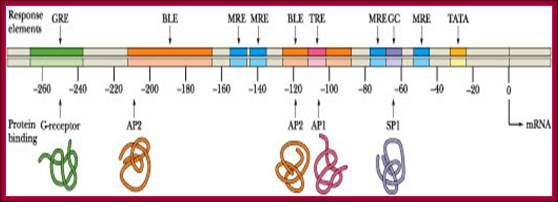
Image text transcribed for accessibility: The metallothionein promoter is illustrated below. How long is this promoter, in nm? How many turns of B-DNA are found in this length of DNA? How many nucleosomes (approximately) would be bound to this much DNA?.The metallothieonein gene possesses several constitutive elements in its promoter (the TATA and GC boxes) as well as specific response elements such as MREs and a GRE. The BLEs are elements involved in basal level expression (constitutive expression). TRE is a tumor response element activated in the presence of tumor-promoting phorbol esters such as TPA (tetradecanoyl phorbol acetate. There are ten upstream elements that regulate gene expression. ClammyHorse1501; http://www.chegg.com/homework-help.
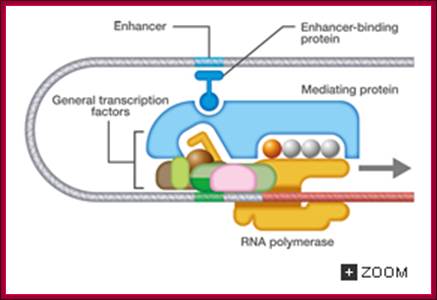
Eukaryotic transcriptional initiation from a promoter; http://csls-text3.c.u-tokyo.ac.jp/
Transcriptional promotion by an enhancer; http://csls-text3.c.u-tokyo.ac.jp/
B. TATA less InR Containing Promoters:
Computational analysis show 80% of human genes TATA less and 46% are both TATA and InR less
In promoters, where TATA box is absent but contain only InR sequence, which contain the start nucleotide and few surrounding pyrimidines.
· Nearly 50% of the genes have TATA-less promoters.
Majority of the house keeping genes have only InR elements, along with other upstream sequences but not TATA box.
· Some of the genes contain just InR sequences at initiator region.
TATA less promoters also contain down stream sequence elements called DPE (GACAC).
+>Start
--------GC--------CAATT----------I-InR-I-----DPE ->

DPE séquence-A/GGT/AC/T a/g
InR sequence- T/CTCAG/CTT/C ; DPE séquence-A/GGT/AC/T a/g
· The initiator sequences are enough for the assembly of basal transcriptional apparatus.
The binding of additional factors upstream facilitates transcription.
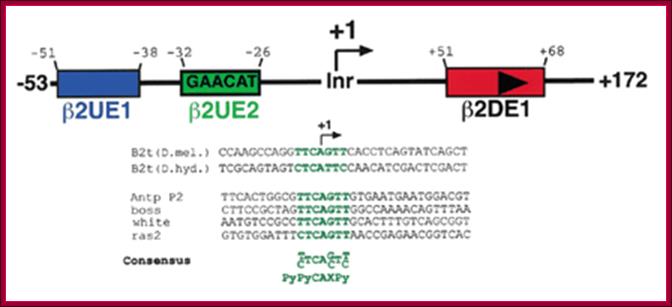
Schematic diagram of the structure of the Drosophila testis-specifc B2t promoter and showing a potential Inr element: It is TATAAA less promoter. The initiator element of the Drosophilaβ2 tubulin gene core promoter contributes to gene expression in vivo but is not required for male germ-cell specific expression -Ansgar Santel*, Jörg Kaufmann1, Ruth Hyland and Renate Renkawitz-Pohl.
The best-known core promoter motif is the TATA box. Yet, the TATA box is present in only about 10 to 15% of human genes. Authors therefore investigated transcription of TATA-less genes, and discovered two new core promoter motifs--the DPE and the MTE. Both the DPE and MTE are conserved from Drosophila to humans, and are located downstream of the transcription start site. Authors have found that there are sharp differences in the properties of TATA less, InR less TATA-dependent versus DPE-dependent core promoters. For example, Caudal, which is a sequence-specific DNA-binding protein that is a master regulator of the homeotic (Hox) genes, is a DPE-specific activator. Thus, enhancer-core promoter specificity can be used to create gene regulatory networks.
1. Present genome-scale computational analyses indicating that approximately 76% of human core promoters lack TATA-like elements, have a high GC content, and are enriched in Sp1-binding sites. We further identify two motifs - M3 (SCGGAAGY) and M22 (TGCGCANK) - that occur preferentially in human TATA-less core promoters. Evidence supporting numerous models of InR-mediated transcription complex formation exists, including the nucleation of a complex by InR-binding proteins, a component of the TFIID complex, or a specific upstream activator common to many TATA-less promoters, Sp1. A complex containing TBP, TFIIB, TFIIF, and RNAPII (DBPolF complex) is capable of forming on the promoter in an InR-dependent manner. A single point mutation within the InR that affects DBPolF complex formation diminishes beta-pol transcriptional activity; L Weis and D Reinberg.
And Takada S have characterized specific promoter elements where they identified new core promoter element called XCPE. This is a mammalian coding gene promoter element. The consensus sequence of this XCPE2 is A/C/G-C-C/T-C-G/A-T-T-G/A-C-C/A(+1)-C/T) that can direct specific transcription from the second TSS of hepatitis B viral gene. Scu genes are also found in human genes. Significance of this finding is that the XCPE is found in multiple TSS containing TATA less promoters.
Results from the authors show that XCPE2-driven transcription uses at least TFIIB, either TFIID or free TBP, RNA polymerase II (RNA pol II) and the MED26-containing mediator complex but not Gcn5. Therefore, XCPE2-driven transcription can be carried out by a mechanism which differs from previously described TAF-dependent mechanisms for initiator (Inr)- or downstream promoter element (DPE)-containing promoters, the TBP- and SAGA (Spt-Ada-Gcn5-acetyltransferase)-depended mechanism for yeast TATA-containing promoters, or the TFTC (TBP-free-TAF-containing complex)-dependent mechanism for certain Inr-containing TATA-less promoters. EMSA assays using XCPE2 promoter and purified factors further suggest that XCPE2 promoter recognition requires a set of factors different from those for TATA box, Inr, or DPE promoter recognition; Ramakrishnan Anish, et al. Nearly 72-76% of human genes are associated with CpG islands and have closely located TSS in an 50-100 bp. Transcription of the type of promoters require multisubunit TFIID complex. They lack TATA-like elements and contain high GC content and bound by SP1 factors.
C. TATA less, InR less, DPE (+) Promoters:
Many promoters have been identified which don’t contain either TATA box or InR sequences and both. But they contain DCE and DPE sequences, which help in recruiting the factors and the enzyme.
Example is the murine CD80 gene. In this preinitiation components assemble first on the upstream elements
· It means the initiator nucleotide in this region is not same. Initiator sites can be more than one. But the promoter region is rich in GC islands.
· It is this sequence that acts as a landmark for the assembly of RNAP-II and its accessory proteins.
Initiation take place in an extended region ranging from 20 to 200 nucleotides in the promoter region, but start is not fixed for it has been observed that mRNA generated from such promoters have different start nucleotide at its 5’ end.
· The genes transcribed show low rates of transcription. Very interesting is the fact that most of the genes transcribed are involved in intermediary metabolism.
The mode of
regulation of class II genes that lack the known core promoter elements is
presently unclear. Here, we studied
one such example, the murine CD80 gene. An unusual mechanism was
revealed wherein the pre-initiation complex (PIC) first assembled on an
upstream, NF-![]() B
enhancer element. Notably, this assembly independent of contributions from the
core promoter domain and resulted in a PIC that was competent for transcription
initiation. Positioning was subsequently achieved by exploiting the intrinsic
architecture of the promoter, by virtue of which the tethered PIC was spatially
juxtaposed with the transcription initiation site. Bridging interactions then
ensued, through protein–protein contacts, which then enabled the elongation
phase of CD80 transcription. Example is CD80 gene.
B
enhancer element. Notably, this assembly independent of contributions from the
core promoter domain and resulted in a PIC that was competent for transcription
initiation. Positioning was subsequently achieved by exploiting the intrinsic
architecture of the promoter, by virtue of which the tethered PIC was spatially
juxtaposed with the transcription initiation site. Bridging interactions then
ensued, through protein–protein contacts, which then enabled the elongation
phase of CD80 transcription. Example is CD80 gene.
---------------------------------------------I------I------I-----I
GC GC GC GC ------------>> +> +> +> DPE
Some InR Sequences:
+-------->
CCCT C A TTCT,
CAGGC A GGGA,-
TAGG C A ATCA,
GTTA C A TGGA,
GCCCC A AGGG,
ATGG C A ACCG,
TTGA C A GACT,
Consensus InR Sequence:
+>
Py Py C A N T/A py py,
-3-2 -1 +1 +2
TF-II D Binding Sequences:
CTTA C A ACCG, (-1 to+4)
CCTG C A TGGG,
CCGCC A AGCT,
BREu and BREd Sequences:
BREu consensus sequence: 5’ G/C-G/C-G/A-C-G-C-C 3’
BREd consensus sequence: 5’ G/A-T-T/G/A-T/G-G/T-T/G-T/G 3’
(TF-IIB binding sequences);
DCE
+6CTTC- CTGT- AGC+34
Motif Ten Elements (MTE) and Down Stream Promoter Elements (DPE) Sequences:
(+)18) C[CG]A[AG]C[CG][CG]A–(+) 26-32 A/GGA/TCGTG / [GT]CGGTT[CG][GT]
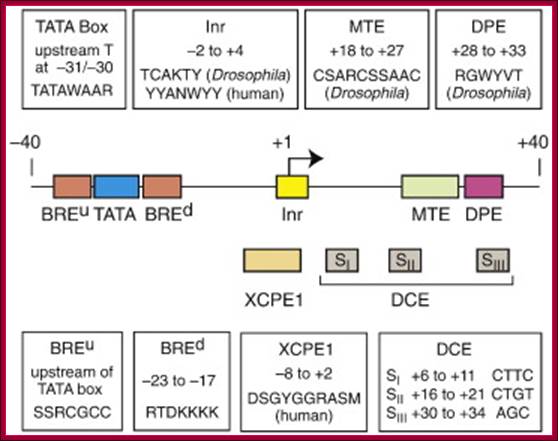
DCE- downstream core element (discovered in hu beta-globin prmoter, generally associated with core promoter containing TATA box in most of the time. TFIID binds to DCE and TAF1/TAFII.
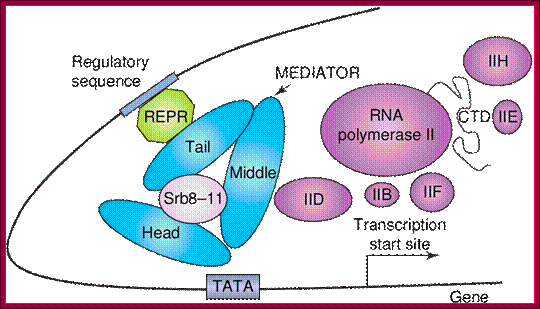
The role of Mediator in initiation of transcription. Panel A: Repressor interactions (green) and binding of the cdk8/CycC subunit (pink) to Mediator (blue) prevent it from binding to RNPII and the basal transcription machinery (purple). http://nens.yellowcouch.org/
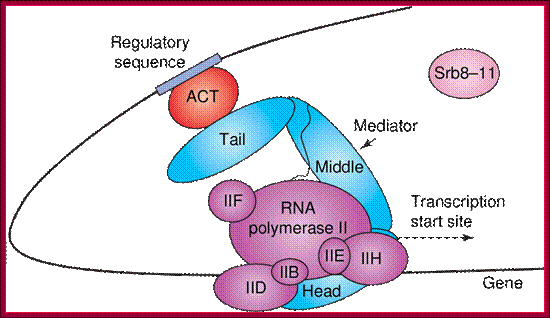
The role of Mediator in initiation of transcription. Panel A: Repressor interactions (green) and binding of the cdk8/CycC subunit (pink) to Mediator (blue) prevent it from binding to RNPII and the basal transcription machinery (purple). Panel B: Activator (red) interactions allow Mediator to adopt an open configuration, which results in binding of RNPII and the basal transcription machinery. This initiates gene transcription; http://nens.yellowcouch.org/
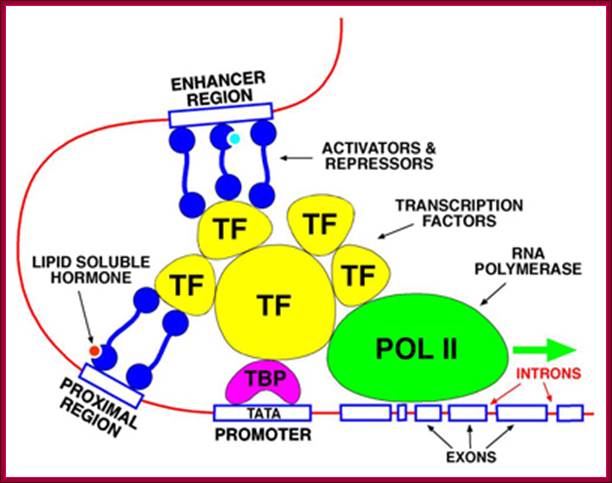
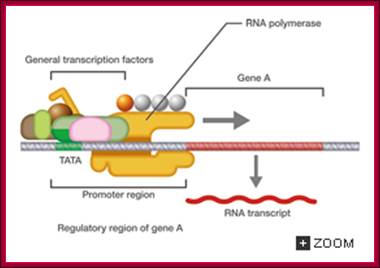
The promoter is the site for the transcription initiation complex, consisting of the transcription factors + POL II (POL II is RNA polymerase). Transcription occurs when POL II dissociates from the transcription initiation complex and moves downstream, transcribing the gene; (Repeat to emphasize).
In the promoter there is a highly conserved sequence of DNA called the TATA box. This is the site for binding TBP (TATA-binding protein). Once TBP is in place, more and more transcription factors assemble by binding to each other. Finally, when all the transcription factors have assembled, POL II is released and starts its journey along the DNA, transcribing the sequence as it does so
Some transcription factors which form part of the transcription initiation complex also bind to distant sites upstream (sometimes downstream) from the promoter site. These sites are called enhancer regions. The region immediately upstream from the gene is the promoter. There is a separate promoter for each eukaryote gene; http://www.scilproj.org/
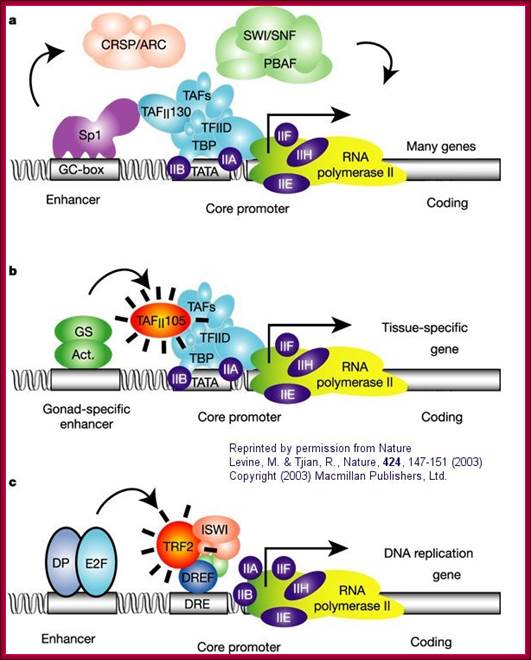
a. "The eukaryotic transcriptional apparatus can be subdivided into three broad classes of multi-subunit ensembles that include the RNA polymerase II core complex and associated general transcription factors (TFIIA, -B,-D,-E,-F and -H), multi-subunit cofactors (mediator, CRSP, TRAP, ARC/DRIP, and so on) and various chromatin modifying or remodeling complexes (SWI/SNF, PBAF, ACF, NURF and RSF)."
b, c. "Metazoan organisms have evolved multiple gene-selective and tissue-specific TFIID-like assemblies by using alternative TAFs (TBP-[TATA Binding Protein associated factors] such as the ovarian-specific TAF105) as well as TRFs (TBP-[TATA Binding Protein associated factors] related factors such as TRF2 in Drosophila and mice) to mediate the formation of specialized RNA polymerase initiation complexes that direct the transcription of tissue-specific and gene-selective programmes of expression." (Nature reference in figure above.)”; http://employees.csbsju.edu/
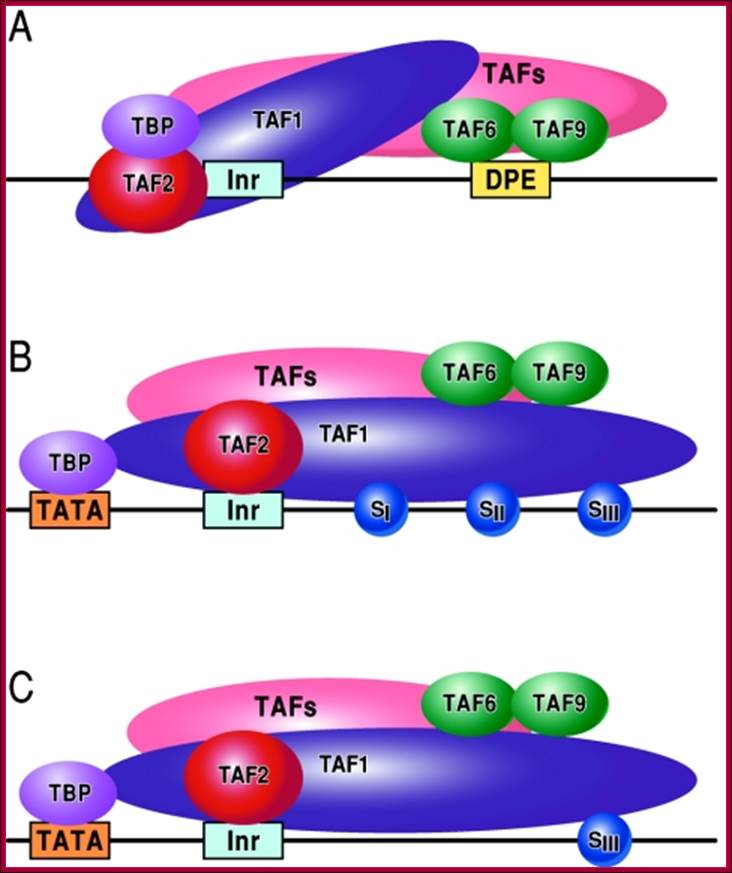
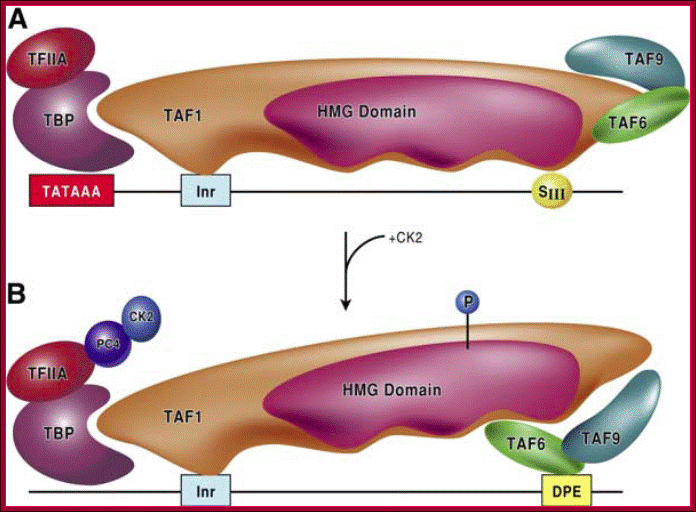
FIG. Models depicting the interaction of TFIID with the DPE and DCE: In the three models, TAF1 and TAF2 are jointly responsible for Inr element recognition (for a review, see reference 73). A. DPE sequence recognition is established by TAF6/TAF9 components of TFIID. This interaction results in a unique TFIID conformation that in turn results in the formation of a DPE-specific PIC (PICDPE). B. TFIID interacts with the DCE sub elements via TAF1. Again, this results in a TFIID conformation that is different than a TFIID/DPE interaction. This also leads to the formation of a DCE-specific PIC (PICDCE). C. A similar interaction of TFIID occurs with promoters containing only SIII of the DCE. In all three cases, these unique PICs consist of their own unique set of factors and cofactors that, in the end, manifest themselves as different regulatory phenomena. DCE downstream core elements or downstream control elements. http://mcb.asm.org/
Core promoter elements apart i.e. between –40 to +35, there are other regulatory elements located at different sites and positions; each of them are cognizable by their specific sequence elements, they are promoter proximal elements (PSE), upstream activator sequences (UAS), Enhancers elements, Response elements, Silencers sequences, Boundary elements and Insulators segments; all these contain specific sequence to which regulator proteins bind. These factors come in different forms, different sizes and with different functions; and certainly they are for housekeeping and tissue specific, stage specific and respond to stimuli. They can be upstream activators, enhancer proteins, mediator complexes, regulatory factors, they can be silencers or repressors or insulators or they can be chromosome-modulator or modifying proteins. These factors bind to their respective binding elements and regulate the expression of genes.
Assembly of General Transcription Factors (GTF) requires space at its promoter region, and the space is provided by the Acetylation of histones of nucleosomes in a specific region. Histone Acetylase (HATs) containing protein complexes facilitates this loosening or the removal histones from the DNA or loosening of nucleosomes for the assembly of transcriptional proteins. How does the HATs or HAT containing complexes know the site at which they acetylate the histone tails? Perhaps, it is sequence specific prebound factors provide the identity of the gene to be expressed or repressed.
The MTE, a new core promoter element for transcription by RNA Polymerase II:
Chin Yan Lim,1 Buyung Santoso,1 Thomas Boulay,1 Emily Dong,1 Uwe Ohler,2
and James T. Kadonaga1,3
The core promoter is the ultimate target of the vast network of regulatory factors that contribute to the initiation of transcription by RNA polymerase II. Here we describe the MTE (motif ten element), a new core promoter element that appears to be conserved from Drosophila to humans. The MTE promotes transcription by RNA polymerase II when it is located precisely at positions +18 to +27 relative to A+1 in the initiator (InR) element. MTE sequences from +18 to +22 relative to A+1 are important for basal transcription, and a region from +18 to +27 is sufficient to confer MTE activity to heterologous core promoters. The MTE requires the InR, but functions independently of the TATA-box and DPE. Notably, the loss of transcriptional activity upon mutation of a TATA-box or DPE can be compensated by the addition of an MTE. In addition, the MTE exhibits strong synergism with the TATA-box as well as the DPE. These findings indicate that the MTE is a novel downstream core promoter element that is important for transcription by RNA polymerase II. [Keywords: RNA polymerase II; core promoter; DPE; InR; TATA-box]
InR MTE (DCE) DPE
The MTE, a new core promoter element for transcription by RNA polymerase II; proposed hypothetical model on the mechanism establishing DPE specific transcription. http://www.med.nyu.edu/
Assembly of GTF as PIC can be assisted by various factors, it is only then the PIC is stimulated to express or it is repressed from expression. It is a universal fact that among 21000-22000 genes per haploid, found in each human cell; if the chromosome number is 2n, it means ~42000-44000 genes in any given cell. Only few thousand genes, say ~12000 genes in Brain tissues, ~10000 genes in Liver, ~7000 genes in muscle tissue, are expressed in tissue specific manner; but among them some are tissues specific and rest of them (>70%) are house keeping genes, which are expressed in all tissues. Therefore transcriptional factors, upstream factors required for house keeping genes might overlap for different tissues for tissue specific expression. The basal transcriptional apparatus may be same but tissue specificity is determined by tissue specific factors, for they are expressed in a given type of cell; this is preprogrammed.
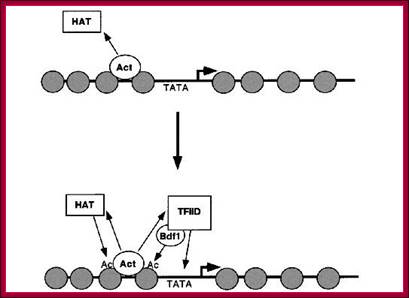
This diagram shows HATs acting at ACT site provide space for the assembly of TFs and RNAP components among them TFIID leads the way.
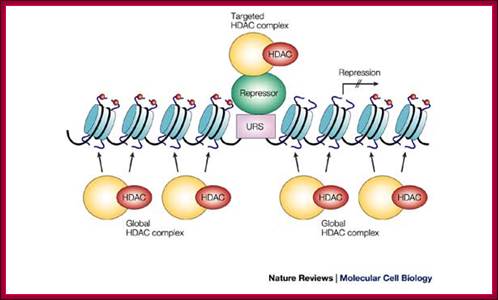
Gene repressed by DHATs and MeCps, prevent the assembly of TFs and RNAPs.
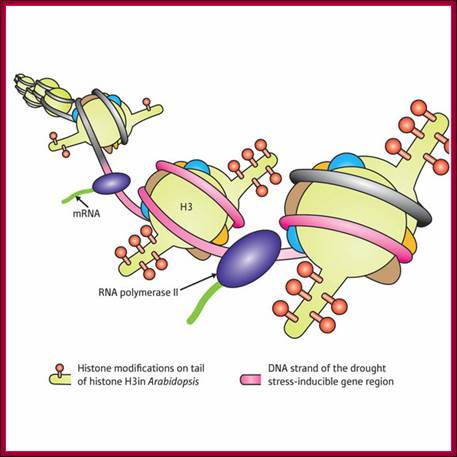
Nucleosome rearrangement for the binding of RNAP II-TF complexes
Genes are activated by HATs; HATS acetylate certain Histone tails, leading to the loosening of the chromatin and freed Nucleosomal thread provides space for the assembly of TFs and RNAPs. Acetylation of Histone tail specific amino acids, leads to increased negative charges on histone tails. As the DNA with Phosphate groups contain negative charges, these negative charges make the histones to loosen from the DNA.
Some of the upstream sequences are tissue specific. For example, GATA 1 binding factor is expressed in most of the tissue types, but GATA2 binding factor is expressed only in lymphoid cells. For the binding of these factors the promoter region should contain factor specific sequences. First let us try to understand the assembly of GTF into PIC. Later the expression and regulation of gene expression will be explained in brief.
Eukaryotic Ribosomal Proteins’ Gene Promoter:
Typically, mammalian cells contain ∼4 × 106 cytoplasmic ribosomes, which account for 80% of all cellular RNA and 5%–10% of cellular proteins. Nearly 73 RP genes found that their transcription starts at a consensus (1) (Y)2C+1TY(T)2(Y)3 residues within a characteristic oligopyrimidine tract; (2) the promoter region is GC rich, but often has a TATA box or similar A/T-rich motif, which should theoretically have TBP-binding . None of these RP genes contained a canonical TATA box in the -25 to -30 regions, although some had a "TATA-like" A/T-rich sequence elements; (3) the genes are small (4.4 kb), but have as many as 5.6 exons on average; (4) the translator initiator ATG is in the first or second exon and is within ± 5 bp of the first intron boundaries in about half of cases; and (5) 5′- and 3′-UTRs are significantly smaller (42 bp and 56 bp, respectively) than the genome average. Comparison of RP genes from humans, Drosophila melanogaster, Caenorhabditis elegans, and Saccharomyces cerevisiae revealed the coding sequences to be highly conserved (63% homology on average), although gene size and the number of exons vary.
GC-GC-GC--TATA--Py-Py +1C-py-py
The Human Ribosomal Protein Genes: Sequencing and Comparative Analysis of 73 Genes
|
Homo |
Fly |
Worm |
Yeast |
|
|
Gene length(bp) |
4316 |
922 |
742 |
764 |
|
CDS length (bp) |
541 |
524 |
520 |
498 |
|
Number of exons |
5.3 |
2.5 |
3.0 |
1.6 |
|
Exon length (bp) |
103 |
206 |
172 |
303 |
|
Intron length (bp) |
888 |
258 |
110 |
413 |
|
|
||||
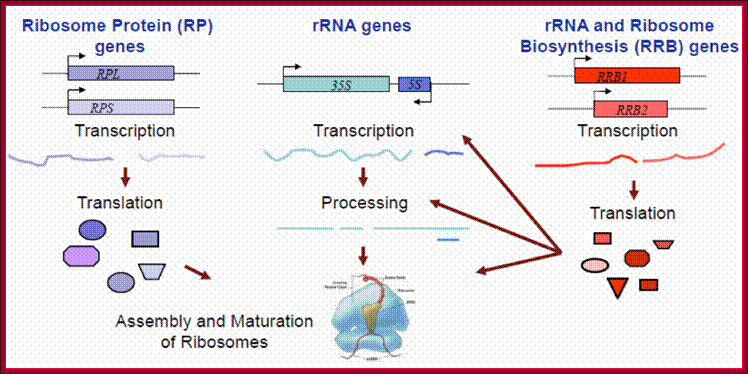
There are 73 to 79 RP genes, most of them are mono-genes, few are multiple genes and few of them are pseudo genes. Ribosomal protein gene expression and translation, and rRNA genes expressions and processing are coordinated to produce ribosomes.
Mammalian ribosomes contain 79 or more different proteins encoded by widely scattered single copy genes. Coordinate expression of these genes at transcriptional and post-transcriptional levels is required to ensure a roughly equimolar accumulation of ribosomal proteins
The consensus initiator sequence of mammalian rp genes: Seventy-nine pairs of orthologous human and mouse rp gene sequences were compared at positions -8 to +10 and the occurrence of each nucleotide or pair of nucleotides depicted by the height of the letters: A, G, C. T, Y = C/T, R = A/G, W = A/T, K = G/T, S = C/G, M = C/A. The tsp is the C at position +1; Robert P. Perry; http://www.biomedcentral.com/
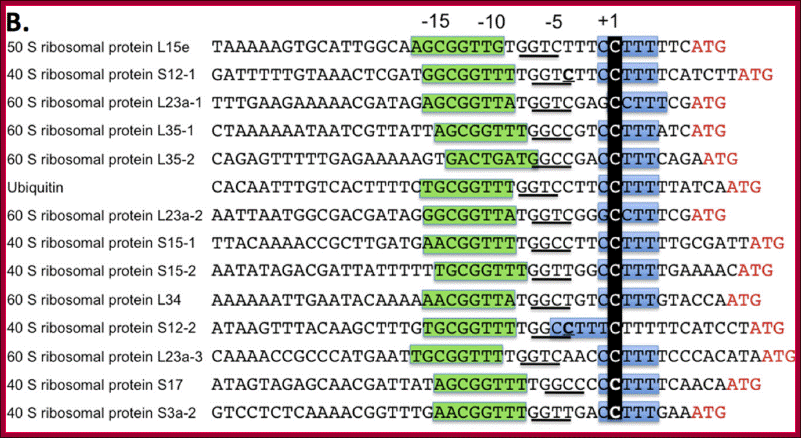
A view of the few promoter elements of some RP genes showing the Start site and the upstream elements.
The Human Ribosomal Protein Genes: Sequencing and Comparative Analysis of 73 Genes; Maki Yoshihama1, Tamayo Uechi1, Shuichi Asakawa2, Kazuhiko Kawasaki2, Seishi Kato3,4, Sayomi Higa1, Noriko Maeda1, Shinsei Minoshima2, Tatsuo Tanaka1, Nobuyoshi Shimizu2, and Naoya Kenmochi1,5,6.
The ribosome, as a mega Dalton catalyst for protein synthesis, is universal and essential for all organisms. Here we describe the structure of the genes encoding human ribosomal proteins (RPs) and compare this class of genes among several eukaryotes. Transcription starts at a C residue within a characteristic oligopyrimidine tract; (2) the promoter region is GC rich, but often has a TATA box or similar sequence element; (3) the genes are small (4.4 kb), but have as many as 5.6 exons on average; (4) the initiator ATG is in the first or second exon and is within ± 5 bp of the first intron boundaries in about half of cases; and (5) 5′- and 3′-UTRs are significantly smaller (42 bp and 56 bp, respectively) than the genome average. Comparison of RP genes from humans, Drosophila melanogaster, Caenorhabditis elegans, and Saccharomyces cerevisiae revealed the coding sequences to be highly conserved (63% homology on average), although gene size and the number of exons vary.
In mammalian cells, the biogenesis of cytoplasmic ribosomes requires assembly of 4 RNA molecules and 79 different proteins. With the exception of two proteins, all of these components are present as single copies within the ribosome. Typically, mammalian cells contain ∼4 × 106 cytoplasmic ribosomes, which account for 80% of all cellular RNA and 5%–10% of cellular proteins. Three different RNA polymerases are involved in production of these RNAs and proteins, RNA polymerase I (POL I) is involved in production of the 28S, 18S, and 5.8S rRNAs, POL II in production of ribosomal proteins (RPs), and POL III in production of the 5S rRNA.
The architecture of Mammalian Ribosomal Protein Promoters;
Mammalian ribosomes contain 79 different proteins encoded by widely scattered single copy genes. Coordinate expression of these genes at transcriptional and post-transcriptional levels is required to ensure a roughly equimolar accumulation of ribosomal proteins. To date, detailed studies of only a very few ribosomal protein (rp) promoters have been made. To elucidate the general features of rp promoter architecture, I made a detailed sequence comparison of the promoter regions of the entire set of orthologous human and mouse rp genes.
Those encoding the ribosomal proteins (rp genes) are single copy and scattered throughout the genome. In addition to the functional rp genes, all of which contain introns, mammalian genomes contain many nonfunctional intronless rp pseudogenes. The earliest determinations of mouse rp gene sequences and of transcriptional start points (tsp's) revealed a salient feature of rp genes, namely that transcription is initiated at a C residue within a polypyrimidine tract. A recent study by Kenmochi and coworkers has demonstrated that this is a general property of virtually all human rp genes. Because of this novel feature, the rp mRNAs contain a 5' terminal oligopyrimidine sequence (TOP), which is essential for their translational control.
RP- ribosomal proteins, RPG- ribosomal protein genes
The TCT motif, a key component of an RNA polymerase II transcription system for the translational machinery.
1. Trevor J. Parry1, Joshua W.M. Theisen1, Jer-Yuan Hsu1, Yuan-Liang Wang1, David L. Corcoran2, Moriah Eustice1, Uwe Ohler2,3,4 and James T. Kadonaga1,5
The TCT motif (polypyrimidine initiator) encompasses the transcription start site of nearly all ribosomal protein genes in Drosophila and mammals. The TCT motif is required for transcription of ribosomal protein gene promoters. The TCT element resembles the InR (initiator), but is not recognized by TFIID and cannot function in lieu of an InR. However, a single T-to-A substitution converts the TCT element into a functionally active InR. Thus, the TCT motif is a novel transcriptional element that is distinct from the InR. These findings reveal a specialized TCT-based transcription system that is directed toward the synthesis of ribosomal proteins.
--------BRE---------------TATA--------------TC/TT+1>--MTE—DPE
Many core promoters appear to lack the more extensively characterized core promoter motifs. We therefore sought to expand our understanding of core promoter elements. These studies led to the analysis of a core promoter motif at the transcription start sites of the Drosophila ribosomal protein (RP) gene family. This conserved sequence was originally found in mammalian RP gene promoters, and has been termed the polypyrimidine initiator (for example, see Perry 2005; Roepcke et al. 2006). Here we investigate the properties of this sequence, which we abbreviated as the TCT motif, based on the sequence of the pyrimidine nucleotides that encompass the C+1 start site. The TCT motif, which spans from −2 to +6 relative to the +1 transcription start site, overlaps with but is functionally distinct from the DNA encoding the 5′-terminal oligopyrimidine tract (5′-TOP), which is a polypyrimidine stretch in the 5′ end of RP mRNAs that is involved in the regulation of translation (for reviews, see Meyuhas 2000; Hamilton et al. 2006). Specifically, the TCT motif functions in a manner that is parallel to and distinct from the Inr element, and serves as a key component of an RNA polymerase II system that is directed toward the expression of RP genes as well as other genes encoding factors involved in protein synthesis.
RP gene Core Promoters and the TC/TT Motif: (Ohler et al. 2002; Ahsan et al. 2009)
To identify new sequences that contribute to core promoter activity, we analyzed promoters that appear to lack the BRE, TATA, InR, MTE, and DPE motifs. With this approach, we sought to find focused promoters that are driven by novel core promoter elements. To this end, we screened core promoter sequences (based on transcription start sites in Drosophila melanogaster) and identified a variety of potential new motif-containing core promoters. We then examined the promoter activity of 50 of these core promoters by in vitro transcription analysis of constructs containing sequences from −50 to +50 relative to the transcription start site. The transcriptionally active core promoters included two RP gene promoters: RpLP1 and RpS15. Transcription from these promoters is carried out by RNA polymerase II, as assessed by sensitivity to 4 μg/mL α-amanitin.
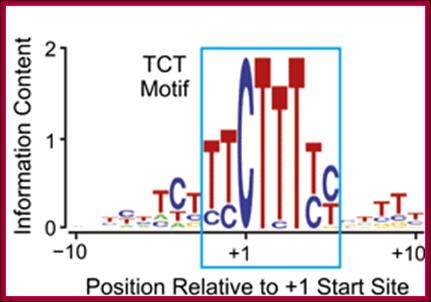
All of the mammalian ribosomal protein (rp) genes examined to date initiate transcription with high precision despite the fact that they do not contain a well-defined TATA box. The initiation sites are situated within polypyrimidine tracts that are flanked by both upstream and intragenic promoter elements. In the TATA-box region of each rp promoter, there is a functionally critical element with nuclear factor binding specificity that is distinct from that of a conventional TATA box.
Sequences -37 and -12 contain both an element that is essential for efficient expression and a specific binding site for a nuclear factor. Rap1 and more, recently Fhl1 were shown to bind upstream of many RP genes.
Cytoplasmic ribosomal protein genes of the fission yeast Schizosaccharomyces pombe display a unique promoter type: A suggestion for nomenclature of cytoplasmic ribosomal proteins in databases; Thomas Gross and Norbert F. Käufer*
Molecular dissection of ribosomal protein (rp) gene promoter regions of Schizosaccharomyces pombe revealed a promoter type which does not contain a canonical TATA-box. Instead, these promoters display a TATA-analogue named the Homol D-box. We showed that the Homol D-box, represented by the sequence CAGTCACA or its reverse complementary sequence TGTGACTG, is involved in determining transcriptional start sites and is the target of protein factor(s) binding. The binding of this factor cannot be competed with TATA-box containing oligonucleotides. The Homol D-box has been compared with the TATA-box with respect to its potential to form local sequence-specific structures which may contribute to the binding specificity of trans-acting proteins.
RNAP II Associated Transcriptional Factors:
Transcription Factors II (TFsII):
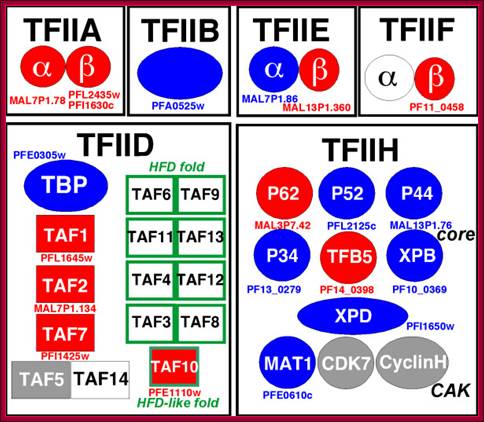
TF II-D. It consists of TBP and eight to twelve TBP associated factors called TAFs.
· TBP is a 30 KD protein, 180 amino acids long. It has a saddle shaped conformation which contains helix and beta sheet structures. It binds to TATA sequence through minor groove and sits like a saddle with stirrups.
• TF II-D: TBP-binding factor and TAF-II (10-12 subunits; tissue specific) 38-50, 27, 250, 150, 110, 80, 60, 40, 30(a), 30(b) ? ? Some of them have histone fold, TAF1, 2, 7, 5 and 14. HFD fold containing TAFs are TAF6, 9, 11, 13, 4, 12 , 3, 8 and 10. Some have acetylation characters, play arole in positioning of GTFs.
• TBP selects and binds to TATA box, disrupts the helix, bends the DNA, it further facilitates the binding of TAFs;
• TAFs in turn facilitate the binding of other TF. factors and RNAP binds to InR and DPE;
• The composition of TFs varies from species to species and tissue to tissue.
Schematic view of the predicted general transcription factors associated with RNA polymerase II in Plasmodium falciparum. Components which have been predicted in previous studies and in the present analysis, are displayed respectively in blue and in red. The components which have not been predicted from sequence analyses are shown in grey and white. Grey boxes indicate components for which potential candidates exist, but which cannot be discriminated from sequence analysis alone, due to the absence of specific domains. Green boxes indicate the HFD-containing TAF pairs which have not been identified in Plasmodium falciparum; TFIIA-2subunits, TFIIB-monomeric, TFIIE-2subunits, TFIID-1+14 subunits (includes TBP and TBP associated factors), IIFH->10; http://www.biomedcentral.com/

Structure of the human TBP core domain complexed with DNA as determined by x-ray crystallography. The DNA includes the TATA element. PDB ID = 1CDW.; TBP bound to the minor groove of TATAAA box and bends the DNA and little opens the helix; http://www.web-books.com/MoBio/
It contains 2 helical motifs and two parallel beta sheets organized into two domains.
The domain that binds TATA is C-terminal region of two direct repeats of 66-67 aa long, rich in basic amino acids (its N-terminal part is divergent).
· Its binding to GCTATAAAAGGGCA is through its amino acids Ile 52, leu163, asn 27 &117, phe 57, 74,148 and 165.
It distorts the DNA in the region of major groove and bends the DNA by 80^o and opens the DNA up to 6-8 base pairs long or 2/3 rds of the helix.
· TAFs are key components, they are tissue specific and they are involved in interacting with other TFs and also upstream factors in activation of transcriptional process.
![]()
Some of the components of TAFs interact not only with other TFs but also with RNAP elements. The complexes assemble at transcription initiation are shown above diagram.
· In the case of certain neuronal cells one finds what is called TRFs (TBP related factors), which assemble as nTAFs, and they work as alternatives to TF II-Ds specific to neuronal promoters.
Though TAFs are ubiquitous, they are unique, for example, neuronal cell have their own TAFs. In Drosophila there are eight TAFs and this number varies from 8 to 11 in other species. In Drosophila each of these proteins has been purified and the genes for them have been cloned.
· TAFII- 230 binds to TBP and mimics TBP in every respect, but it binds to N-terminal region of TBP.
TAF II-42 and TAF II-62 resemble histone H3 and H4 and they form hetero dimers, together with other TAFs, they form a structural complex similar to histone octamer.
· Some TAFs participate in interacting with upstream factors like Sp1 that stimulates transcription.
Some TAFs act as co activators. Sp1 factor interacts with TAF II-110.
· TAFs not only interact with promoter elements but also interact with gene specific TFs.
TAF 250 and TAF 150 facilitate TF II D complex bind to InR and to down stream elements in TATA less promoters, and enables the TBP to be positioned in right context to initiate transcription
at predefined start point.
· TAF II-250 and TAF II-110 help in binding to TATA less promoter that has GC boxes in the upstream region.
TAF 250 also has kinase and histone acetyl transferase activity.
· TBP is a location or site identifying factor and also a commitment factor for RNAP to position and initiate transcription at start site. But TAFs identify and place the TBP in right context. Binding of this ultimately leads to the positioning of the RNAP to initiate transcription at start site.
The N-terminal part of the protein interacts with other proteins.
· TAFs (TBP associated factors) are as many as eight or eleven in number with different Mol.wt.
The composition of TAFs varies from one tissue type to the other.
· TAFs have a role in interacting with other proteins, either to activate or to inhibit transcription.
Assemblies of TBP with TAFs occupy the promoter from-45 to +35.
The largest subunit of core RNAP consists of a variable length of CTD tail; the C-terminal tail, which consists of repeats of seven amino acid sequences YSPTSPS, this may little vary, but this seven a.a sequence is repeated from 22 (yeast) to 52 (humans). This tail associated complexes play a very important role in capping, splicing and poly-A addition and even regulation of transcription.
![]()
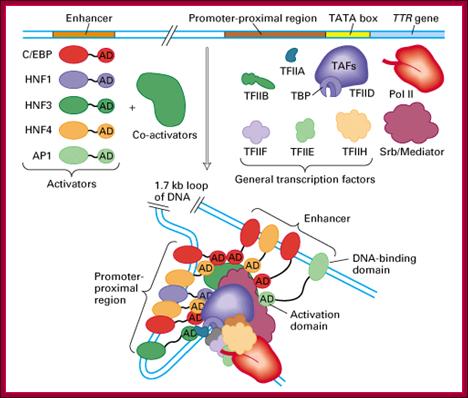
Transcriptional factors that are associated during transcriptional initiation; http://xray.bmc.uu.se/
Second complex:
The second complex is TF-II-A, it consists of two (alpha and beta) subunits and positions to the left of II-D.
· This may activate TBP by relieving it from repression by TAF II-230, which binds to the C-terminal region of the TBP. Some people consider TF II-A is a part of TAF –II factors. It covers further upstream of TF II-D.
TF II-A, 2 subunits, 56,14 in yeast; 3subunits-12,19,35 in (human), Binds to TBP upstream, may be required for TATA less promoters.
Third complex:
It is TF II-B, a small subunit of Mol.wt of 35kd and locates at the vicinity of start site and at the right side of II-D (-10 to +10).
· The protein has sequences similar to that of a sigma factor.
TF II-B-35 (H)(38 yeast) Acts like a bridge between TATA-TPB and adjacent DNA, ; this acts as a rate limiting factor; detects start and the orientation of promoter
It contacts DNA in sequence specific manner (how?) Binding of this is very important, for it facilitates the binding of RNAP, without which RNAP doesn’t bind. So this is a rate-limiting factor in the assembly process. It is also the target for specific upstream TFs.
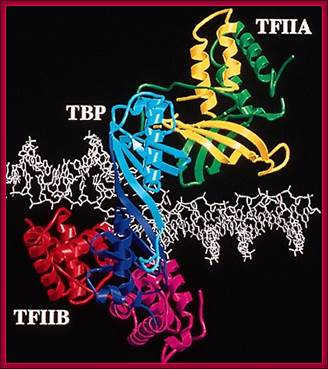
This ribbon model shows the assembled TBP on TATAA box, TFIIA and TFII-B on either side of the TBP. The DNA at the site of TBP bound region is slightly open.
Fourth complex:
It is TF II-F, consists of a sigma factor like subunits with Mol.wt of 30 (38) KD and binds to RNAP-II complex.
· At the same time another factor 74 KD, which is a helicase, also binds to the RNAP- II.
![]() Transcription
accessory factors
Transcription
accessory factors
These are referred to as RAP-I and RAP-II respectively.
· The RAP 30 has a weak homology to that of a sigma factor, yet it binds to RNAP-II tightly and plays a role similar to bacterial sigma factor in positioning the RNAP-II in proper context.
It interacts with TF II-D, TFs II- A and TF II-B.
· This complex settles on the already assembled factors. Here the II-F guides the multisubunit RNAP and positions on the TATA bound components.
In this assembly TBP and TAFs may interact with CTD domain of the large subunit of RNAP complex.
· The TBP and TAFs may also interact with TF II-E. The 74 KD proteins may help in the melting of DNA at transcriptional initiation point.
Now the assembled complex covers an area from-45 to +20 or more.
Fifth complex:
It is TF II-E consists of two subunits each of 56kd and another two subunits 34 KD each and they form homodimer of 56 and 34 KD. And the subunits assemble and position to the right side of the RNAP.
· The 56 KD proteins have zinc finger domains, which could bind to DNA. This protein can be stimulated by SP1. This assists the assembly of TF II-H.
![]()
Sixth Complex:
The sixth complex consists of many components 6 to 10 including TFII J and TFII H.
• TF II-J43 41 35 Kinase,
• TFII-H (9-12 subunits), Y=9=512H=9470
• XPG (XPD and XPB), XPF,Ercc1) XPC (C=RAD) binds to damaged DNA, involved DNA repair,
• Specific subunits:- such as Helicase and CTD Kinase required for unwinding and clearing the promoter, also involved in excision repair of DNA (if needed), during transcription.
![]()
![]()
It is TFs II-H, it has a helicase domain and kinase activities; it is associated with CDK 7 and perhaps it phosphorylates the CTD domain of the RNAP-II and activate the RNAP for promoter clearance or elongation. The kinase in the H factor is known as Mo15/CDK7.
· The RAD 25 has helicase activity, which is actually involved in promoter clearance. This TF is also involved in the repair of damaged DNA.
Seventh Complex: TF II-S:
The S proteins also contain kinase activity.
· Probably they may phosphorylate the CTD tails of the largest subunit of the RNAP-II. TF II-S is believed to stimulate transcriptional elongation and also limits RNAP pausing.
SII 38KDS III 15KD, 18KD, 110 KD’HSPT5-required for elongation, Stimulates elongation and limits RNAP pausing, involved in proof reading mismatch repair (28 subunits=1625kd)
Eighth Complex:
It is called K and it cannot be determined whether it is the last to associate, but it is found in the complex.
· This complex is believed to be essential for elongation. Few other proteins required are SRB proteins (belongs to mediator complexes), suppressors of RNA pol B (SRB 2, 4, 5, and 6) and perhaps involved in activation of transcription in vitro.
Even the mediator complex, specific-to-specific tissues, is also a co component of the General transcriptional factors, associated with especially RNAP.
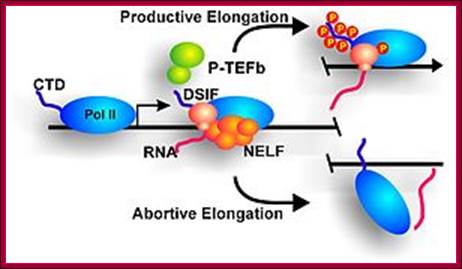
Productive or abortive Elongation complex
Mediator complex:
This complex consists of 15-36 subunits; some of them are tissue specific. They act between activators/co activators bound at upstream and BTA/RNAP complex. Components of MC are organized in different positions in the complex such as middle, head and tail regions.
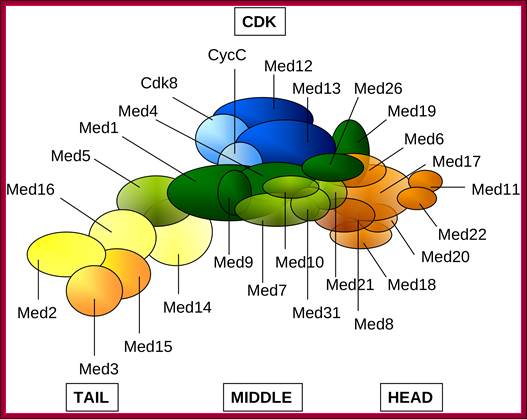
Schematic representation of the Mediator complex: Head (orange), Middle (green), Tail (yellow), CDK (blue). Subunits with higher than 50% average overall disorder (Med2, Med3 in Tail; Med9, Med19, Med26 in Middle and Med8 in Head) or subunits containing intrinsically disordered regions longer than 100 residues (Med12, Med13 of the CDK, Med1, Med9, Med26 of the Middle and Med15 of the Tail) in either Saccharomyces cerevisiae or inHomo sapiens are displayed by darker colors. Med19 and Med26 was assigned to the Middle module according to reference; Ágnes Tóth-Petróczy,et al, http://www.ploscompbiol.org/
The assembly of general factors and RNAP and other required components are triggered or facilitated by upstream factors such as activators (at UAS or enhancer regions), co activators and other factors. Some of the TAFs contain histone acetylation properties, thus loosen the chromatin in promoter region facilitating the assembly of transcriptional complex. All these components assembled at promoter have bound to one another in proper context and positioned on the promoter element in their specific position. The assembled complex at the promoter region is called Basal Transcriptional Apparatus (BTA) or preinitiation complex (PIC).
· In TATA box InR and DPE containing promoters, the guiding factor is the TBP and TAFs in TF II-D complex. TBP acts as the positional factor, aside it contributes to the melting and bending of DNA at TATA site.
In the case of TATA-less promoter, but InR and DPE containing promoter, it is again TF II-D, but here positioning of the TBP is facilitated by TAFs.
· In the absence of both TATA box and InR sequences in the promoters, the guiding elements are GC rich islands in the upstream that determine the assembly of the basal transcriptional apparatus.
All the general TFs organize on the promoter, which is promoted by activators or enhancer bound activators or both. Organization of BTA on a specific site itself is not enough for the initiation of transcription.
Activation requires mediator complex. The mediator complex composition varies from species to species, but some components are common. It acts between the co activators, enhancer complex and BTA
The whole assembled complex can undergo, if every thing is right, conformation changes and initiate isomerization and initiate conformational change from loose to closed complex to tight closed complex formation. Then it further undergoes conformational changes from tight closed complex to tight-open complex conformation; it is at this state, assembly of nucleotides on the template start.