More Promoter Elements:
Regulation of Gene Expression in Plants:
Refer to my website �Plant Cell Biology� for details�:
Plants are enigmatic in the sense, unraveling molecular events appears to be bogged down, one for the lack interest in plants, second lack of funds for research, third it is difficult and frustrating for the simple reason as soon as you put the knife plant respond with huge amount of polyphenol oxidases.� Unlike animal systems the cellular protective components are hard and rigid, difficult to work with.� In spite of it, one can do some good work, but people in the field are more interested only talking but not interested in working.� Yet there are labs which have done great work in understanding molecular aspects of plants.� Little is known but what one can perceive is that plant molecular aspects do not have different sets of gene structures and enzymes and mechanism; they do have more less the same components and mechanisms, perhaps with little difference.
Plant hormones are produced in different parts and structures of the plant and they are transported to their respective destination and bring about the effects.� All plant hormones to begin with function through signal transduction events and bring about changes in metabolism and furthermore they do effect differential gene expression to achieve the function.
They also interact with one another bring about the desired phenotypic expression.� Each has their own pathways. Most of the phytohormones use signal transduction pathway for eliciting their effect; however, elucidation of plant signal transduction pathway is not fully unraveled; it reflects on the quality and the caliber of plant scientists; I think they are the same all over the world with some exceptions and they are exceptional.
https://mm737.wikispaces.com
http://gardeningunlimited.com/
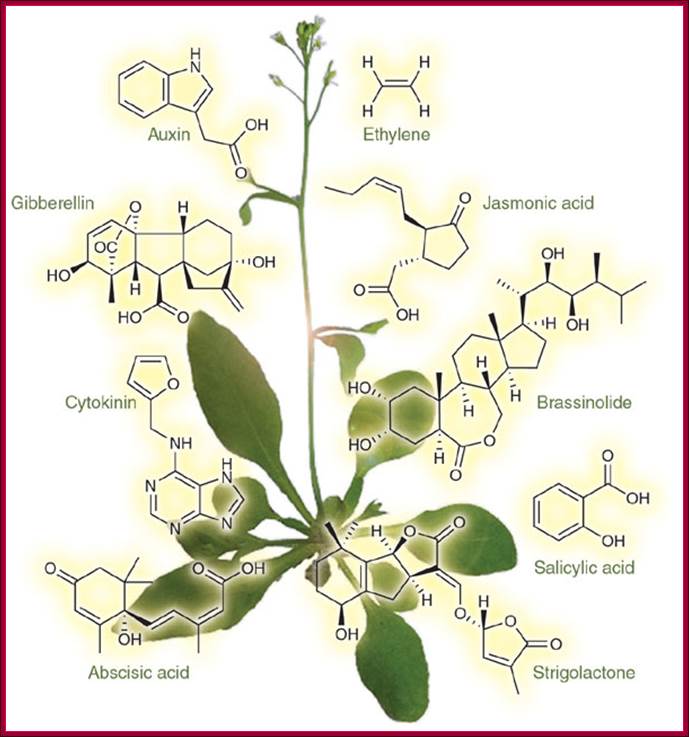
Phytohormones namely Auxin, Abscisic acid, Bassinosteroids, Cytokinin, Ethylene, Gibberellin, Jasmonates and few others, appear to regulate similar and opposite processes through signal transduction and activation of different sets of genes. This corresponds to observations that certain phytohormone responses being independent of other phytohormones, and the fact that a total deficiency in one phytohormone cannot be fully rescued by the application of any other phytohormone.
One should remember, while one conducts research; one should routinely follow the following points.� Discoveries are made by some people with great intuitions like �Avant Garde� or in regular course by what is called �Serendipitous�.
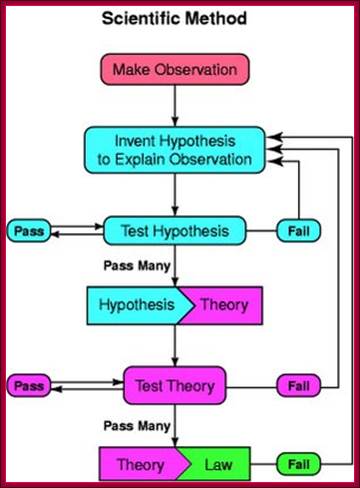
There are two reasons why the Biology textbook does not show a graph like this: a) it is not applicable to biology, and b) it is wrong.; Observation to > hypothesis to > Theory and finally to > Law; https://blogs.scientificamerican.com

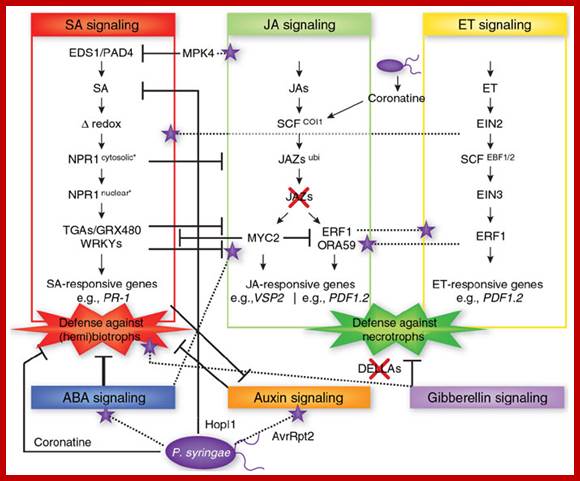
Network (web like) of phytohormones; all interact with one another. Networking by phytohormones in the plant immune response. Cross-communication between hormone signaling pathways provides the plant with a large regulatory capacity that may tailor its defense response to different types of attackers. On the other hand, pathogens such as P. syringae produce effector proteins (for example, coronatine, HopI1 and AvrRpt2) that manipulate the signaling network to suppress host immune responses and promote virulence. The SA, JA and ET signaling pathways represent the backbone of the defense signaling network, with other hormonal signaling pathways feeding into it. Only those signal transduction components that are relevant to this review are shown. , negative effect; purple stars, positive effect. https://www.researchgate.net
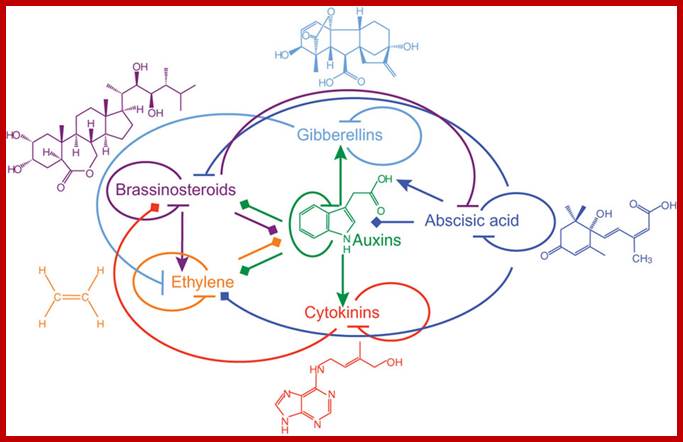
Interaction between phytohormones; Phytohormone structures and functional interactions. Lines with arrowheads, upregulation of hormone biosynthetic genes or downregulation of genes involved in hormone inactivation; blocked arrows, downregulation of genes involved in hormone biosynthesis or upregulation of genes involved in inactivation of a hormone; diamond arrowheads, changes in gene expression with ambiguous outcome. https://www.researchgate.net/
�����������
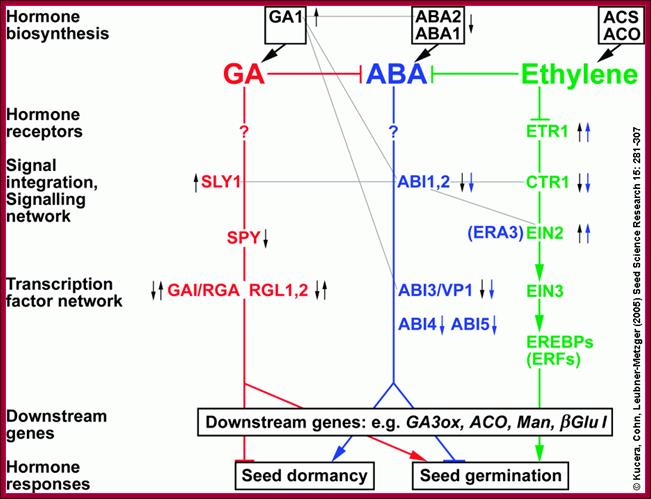
Hormonal interactions at different stages of plant development especially seed dormancy and seed germination; http://www.seedbiology.de/
�����������������������
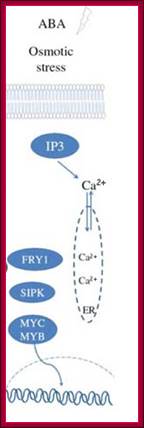
Osmotic stress leads to ABA synthesis and its effect.


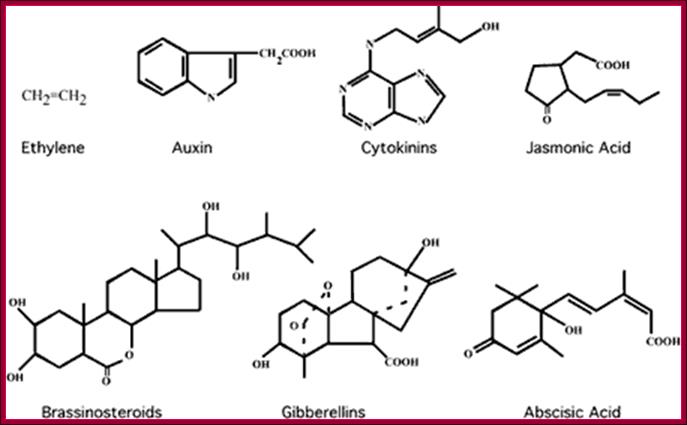
Phytohormone�s chemical structural features; http://www.biologyexams4u.com
In this chapter gene regulation in plant is restricted to phytohormone induced gene expression.� Hormones in Greek mean �stimulate�.� Basically land plants are built on structural features that provide two important sites as structural domains for growth and development; they are stem apex and root apex, which have stem apical meristem and root apical meristem called SAM and RAM respectively.� Stem apical meristem is covered with an epidermis below which one finds cell with potential to develop into leaves, axillary branches or flower buds.� Root apical meristem is generally covered with a root cap.��
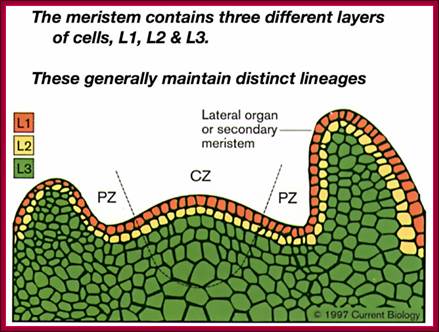
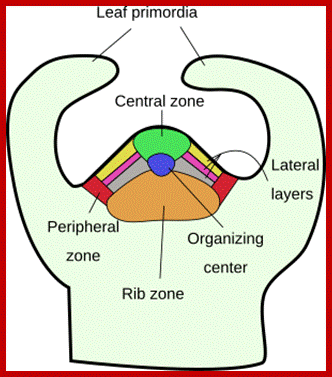
Plant�s Stem Apical Meristem. Common spatial territories in the shoot apical meristem (modified from Risopatron et al., 2010; �https://mmegias.webs.uvigo.es
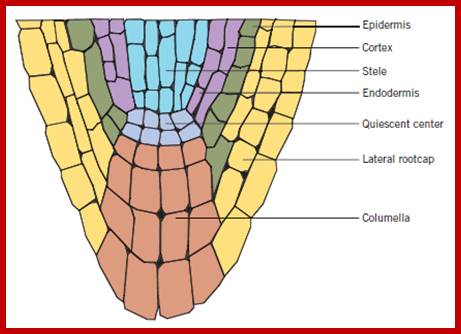
The CLE40-ACR4-WOX5 signaling in root apical meristem. Both CLE40 (light blue) and ACR4 (light green) are expressed in differentiating columella cells. CLE40 peptide acts through the ACR4 receptor in the columella root cap and restricts WOX5 expression in the quiescent center (QC) (purple). WOX5 promotes columella stem cell fates (indicated by the black arrow). https://www.researchgate.net
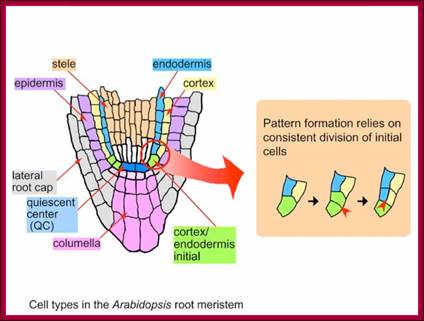
Structural regions of Root apex; www.en.ppt-online.org
Many plants have longer life than the most long living animals; ex. Sequoia sempervirens, lived nearly ~3500 years and diameter 25m and grown to the height, nearly ~379 feet and it has weathered, perhaps all kinds of environmental turmoil and many civilization have come and gone, yet the plant survived and still growing. Longest living plant is Pinus longaeva 4844-4846 years, germinated in 2832 BC in Inyo County in California. Old Tjikko, a 9,550-year-old Norway Spruce, is the oldest? The plant that is found in the National Park of California is Sequoiadendron giganteum (3226yrs). Tallest plant is redwood tree Giant sequoia 364ft. Plants� growth and reproductive cycles are intertwined with environment and plant hormones they produce within them.� The main phytohormones, about which most of the text books such as Plant physiology and Plant biochemistry are based, are Auxins, Gibberellins, Cytokinins, Abscisic acid and ethylene.� There are others like Bassinosteroids, Jasmonics etc.� Each of the hormones has specific effects, but they interact with each other to produce wholesome plant either in favorable or unfavorable conditions.� The fascination of plants is they live in a given environment and adapt to the changes, grow and reproduce.� It is these living systems that can undergo genetic changes and evolve.� One can observe such changes in one�s life time or two.� In this chapter we will succinctly provide basic information; which hormones act how.
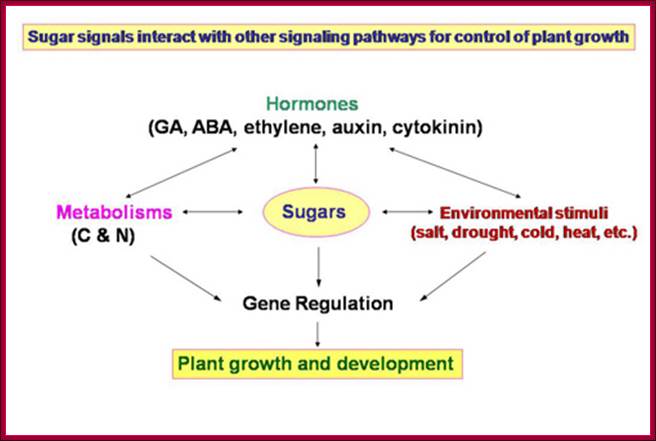
Plant growth and development depends on the availability of carbohydrate source in the form of sugars; which control various plant development pathways including the most sophisticated flowering. www.dreamhome.id/g/gene_expressioni
Auxins:
Auxins are made up of Indole compounds common component of asafetida used for flavoring cooked food- ex. Indole Acetic Acid (IAA), a natural plant hormone. There are few synthetic Auxins, which have been in use for a long time.
The master phytohormone, Auxin is involved in controlling virtually all aspects of growth and development in plants, including, directional growth, responses to external cues (phototropism and gravitropism), de novo organogenesis of leaves, flowers, floral organs, and lateral roots (Benkov� et al., 2003), the formation of vascular tissue in stems and roots and leaves (Mattsson et al., 2003; Scarpella et al., 2006), and the maintenance of meristem identity in shoot apical meristem (SAM) and root apical meristem (RAM). It also prepares cells for cell division in response to cytokinins. Apart from the above functions, it also responsible for cell elongation, apical dominance, parthenocarpy, newroot formation and impose apical dormancy.
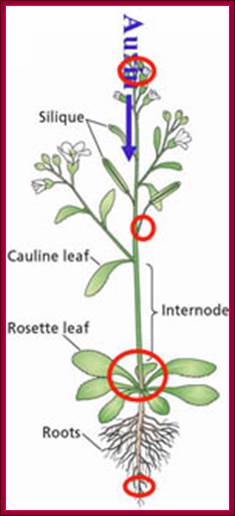
The figure shows plant�s important structural features.� Stem apex is the prime site of synthesis of IAA that is transported from stem apex to stem-root joint and then from there Auxin is transported basipetally to root apex. http://www.plantcell.org
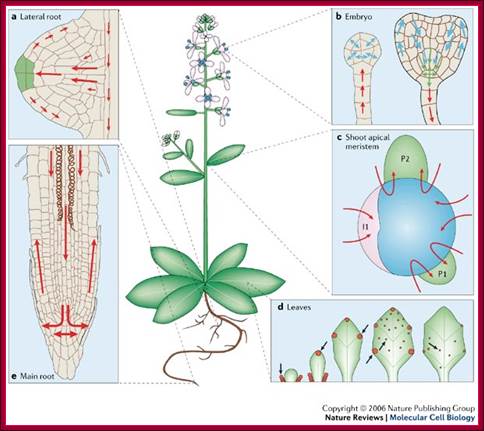
a | Lateral root. PINs conduct auxin from the centre of the root (stele) to the new root tip (auxin is indicated in green and auxin transport is indicated by red arrows), and then away again through the epidermis. This forms the basis of the 'fountain' model of lateral root formation62. b | Embryo. Auxin is taken to the very young embryo by PIN7 (left). At a later stage (right), the auxin flux is reversed as PIN1, PIN4 and PIN7 conduct auxin out of the embryo. Transport by PIN1, PIN4 and PIN7 is indicated by blue, green and red arrows, in corresponding order. c | Shoot apical meristem. Auxin is redirected towards the site of new leaf formation (primordial P1 and P2 and the incipient primordium I1) in the epidermal layer. The shoot apex is indicated in blue. d | Leaves. Auxin mediates vascular tissue development (indicated as uninterrupted green lines) and patterning in the developing leaf through non-polar PIN1. The arrows indicate sites of auxin production and the red circles indicate auxin accumulation. e | Main root. PINs determine the flux of auxin towards the root tip in the centre of the root, and back again in the epidermis. This movement forms the basis of the root's ability to respond quickly to gravity. Parts a�e adapted, with permission, from Refs 62,125,153�155 � (2003) Cell Press, (2005) Company of Biologists Ltd, (2005) Current Biology Ltd, (2004) Kluwer Academic Publishers, and (2005) Scandinavian Society for Plant Physiology, in corresponding order.; Auxin in action: signalling, transport and the control of plant growth and development; William D. Teale, Ivan A. Paponov and Klaus Palme; http://www.nature.com/
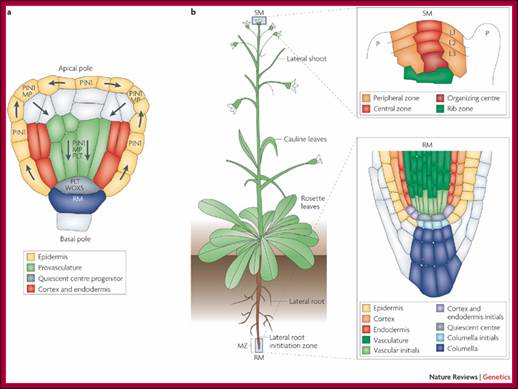
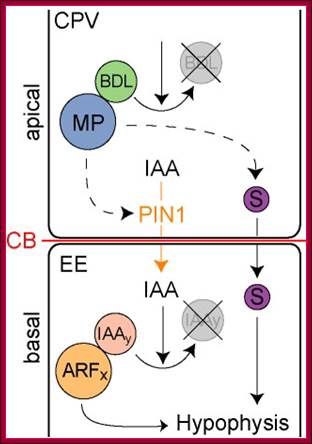
Embryo and meristem organization. a | The plant embryo at late-globular stage. Directional auxin transport requires MONOPTEROS (MP)-promoted expression of the auxin efflux protein PIN-FORMED 1 (PIN1) in provascular cells. The direction of transport is indicated by arrows. This mechanism leads to auxin accumulation at the basal pole, where the root meristem (RM) is established. PLETHORA (PLT), and WUSCHEL-related homeobox 5 (WOX5) are MP-dependent transcription factors. b | The location and structure of the root meristem and the shoot meristem (SM). The locations of the meristems in an adult plant are shown, with the different types of cells in the meristems illustrated in the expanded boxes. L1, L2 and L3, meristem layers; MZ, meristematic zone; P, primordium or primordium initiation site.
Survival of the flexible: hormonal growth control and adaptation in plant development; Hanno Wolters & Gerd Jurgens; http://www.nature.com/
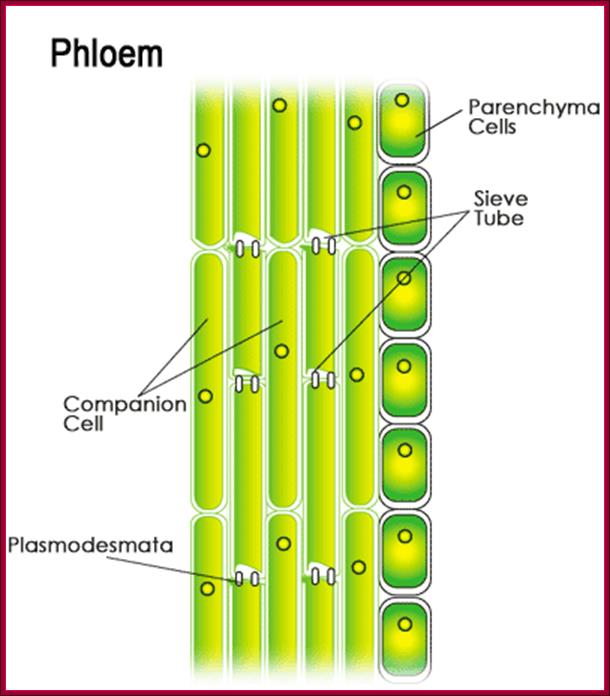
They are synthesized in very young stem tips that include young buds and budding leaves, and then they are translocated through sieve tubes to different parts of the plant that includes root tips.� Transport of auxin in plants is a facilitated process and active too.
www.slideplayer.com
http://plantphys.info/
They elicit effects on plants, such as cell elongation, apical dominance, phototropism, gravitropism, induce parthenocarpic fruits, involved in wound healing, delay senescence, induce new or adventitious roots and induce ethylene synthesis. To initiate such changes first they have to be transported to their specific destination via Auxin carrier proteins across the membranes; i.e. they have to be transported from the site of synthesis to the site of action; once they are transported they have to bind to their specific receptor proteins through them they will have to find specific genes in the nucleus and activate them for specific effects on plants.
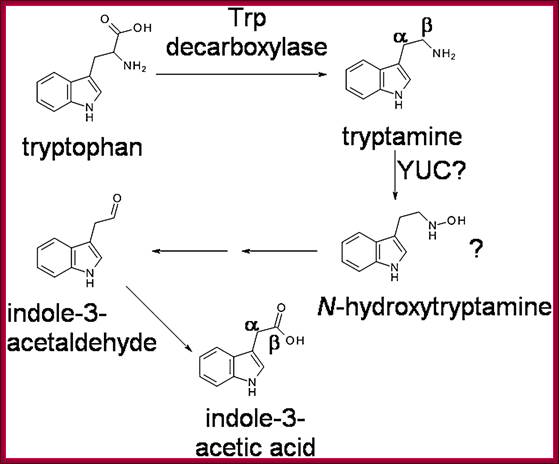
Biosynthesis-pathway; The previously proposed pathway for tryptamine-dependent IAA biosynthesis. The involvement of N-hydroxy tryptamine was first suggested by Zhao et al. (2001). The positions of some carbon atoms in Tryptamine and IAA are indicated; http://www.plantphysiol.org/
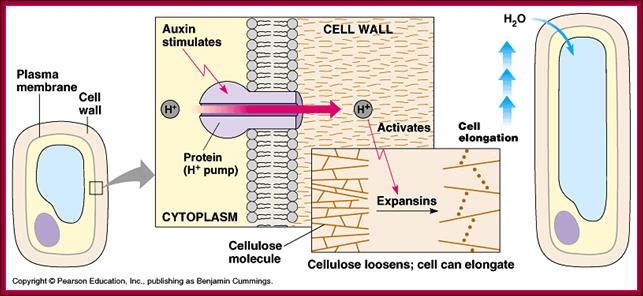
Cellular Elongation: This requires positive turgor pressure and increased elasticity of the cell wall. Turgor pressure in cells results from the presence of dissolved solutes. Elasticity of the cell wall is increased by the IAA.This is further explained by the acid-growth hypothesis:IAA stimulates H+ pumps in the cell membrane.H+ pumps secrete H+ into the cell wall, decreasing its pH.This acidifies the cell wall which activates pH-dependent enzymes and breaks bonds between cellulose microfibrils. The wall "loosens" because of the broken bonds and the turgor pressure expands the cell. http://www2.mcdaniel.edu/
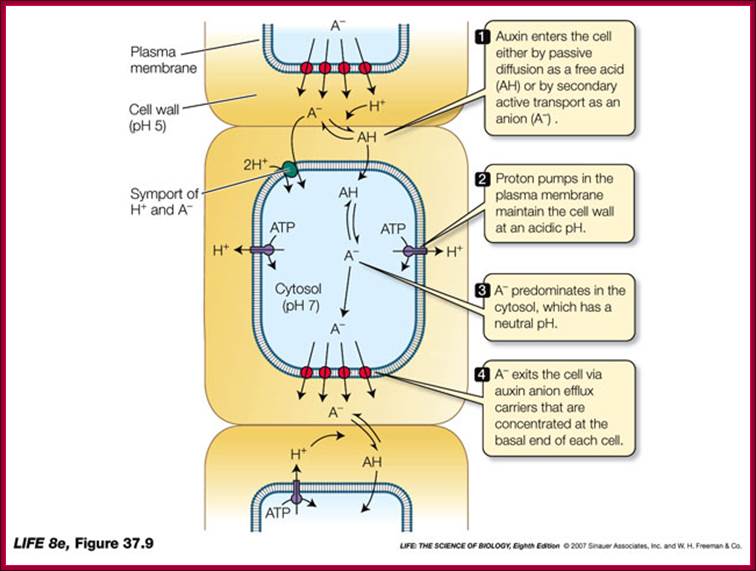
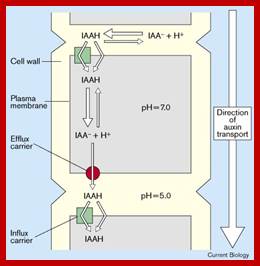
Auxin transport across the cells; http://www.vi.cl/
IAA enters (influx) parenchyma cells in a protonated form via passive transport, or as a anion through an influx carrier protein. Once inside the cell, IAAH dissociates because of the higher pH within the cytoplasm to become an anion. Ions are the only form of auxin that can leave the cell (efflux) through a efflux carrier protein. http://quizlet.com/
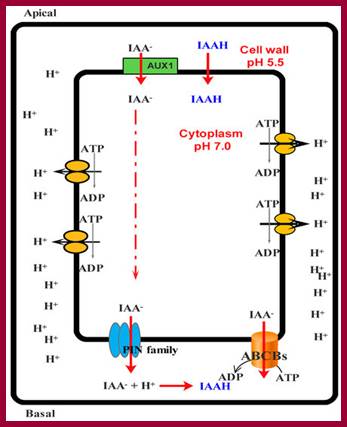
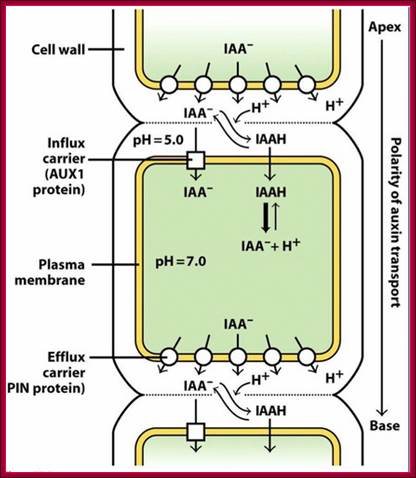
Simplified model for the entrance auxin into cell and response of cell to it. ABCB: ATP-binding cassette subfamily B; PIN: PIN-formed protein; AUX1: Auxin transporter protein 1. http://www.livescience.com
 �
�
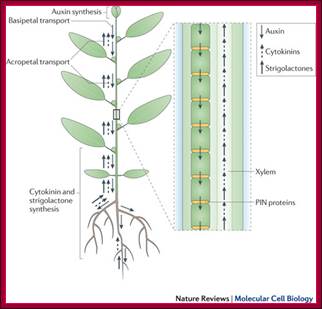
Auxins, cytokinins and strigolactones (or strigolactone derivatives) are three classes of hormones that are involved in the control of bud activation. These hormones are transported throughout the plant, forming a systemic network that allows integration of information between different plant organs. Auxin is mostly produced in the young expanding leaves of growing shoot apices and is actively transported basipetally in the polar auxin transport stream, involving basally localized PIN-FORMED (PIN)-type auxin efflux carriers, in particular PIN1. Strigolactones and cytokinin are mainly produced in the root, but also locally in the shoot, and are transported acropetally in the xylem. Malgorzata A. Domagalska & Ottoline Leyser; ;http://www.nature.com/
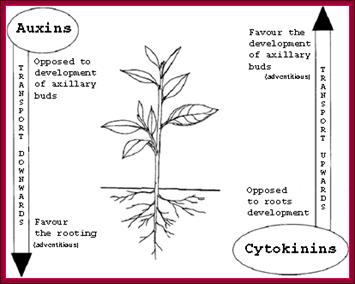
http://www.seattlecentral.edu
Auxin binding protein: http://www.rcsb.org
Plant hormones: Ins and outs of auxin transport; Ottoline Leyser; Regulated transport has long been known to play a key part in action of the plant hormone auxin. Now, at last, a family of auxin efflux carriers has been identified, and the characterisation of one family member has provided strong evidence in support of models that have been proposed to explain gravitropic curvature in roots.
PIN is Auxin efflux network controller. PIN genes collectively control Auxin distribution to regulate cell division and cell expansion in the primary root.� The joint action of these genes has an important role in pattern formation by focusing the Auxin maximum and restricting the expression domain of PLETHORA (PLT) genes, major determinants for root stem cell specification. In turn, PLT genes are required for PIN gene transcription to stabilize the auxin maximum at the distal root tip. Our data reveal an interaction network of auxin transport facilitators and root fate determinants that control patterning and growth of the root primordium. Different endogenous and environmental signals can modulate PIN activity and thus modulate auxin-distribution-dependent development. Arabidopsis has eight annotated PIN genes, of which six have been functionally characterized up to now: PIN1 [6], PIN2 [7-10], PIN3 [11], PIN4 [12], PIN5 [13], and PIN7 [14]. PIN6 and PIN8 are still awaiting characterization. http://www.cell.com
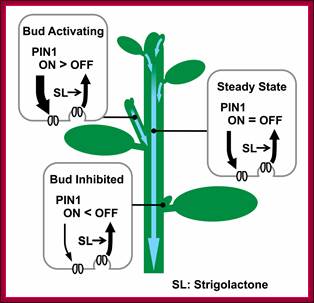
Schematic representation of PIN1 protein regulation by strigolactone and its effect on bud activity.
Strigolactones, signalling via MAX2, depletes PIN1 from the PM of cells in the shoot, for example by promoting clathrin-mediated endocytosis. Strigolactones acts systemically, influencing PM PIN1 levels throughout the shoot. In the main stem, PIN1 on the PM is at steady state. In an activating bud, canalization is underway, with rapid PIN1 insertion, outstripping PIN1 removal. In an inhibited bud, PIN1 insertion is slower than PIN1 depletion, such that PIN1 does not accumulate on the PM [20],[23]. Systemically higher Strigolactones levels will reduce the number of active buds and the steady-state levels of PM PIN1. Systemically lower Strigolactones will have the opposite effect. http://www.plosbiology.org/
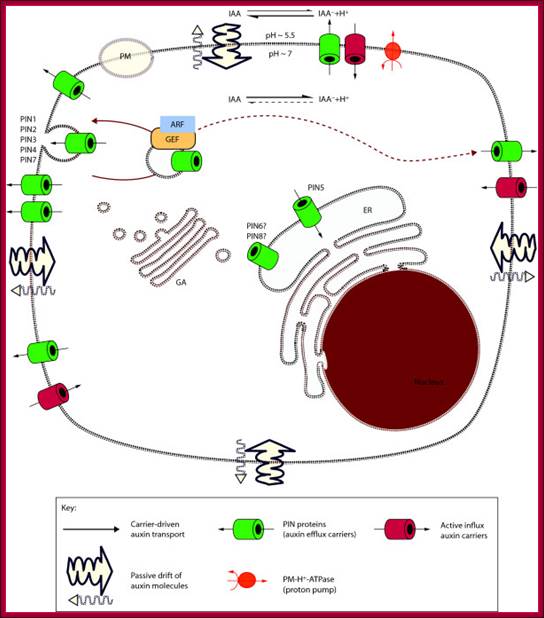
Schematic diagram of an idealized plant cell and the role of specific PIN proteins in auxin management at the cellular level. The low pH in the apoplast (the region outside the cell membrane comprising the plant cell wall) is maintained by the activity of the plasma membrane H+-ATPase. In the acidic environment of the apoplast, a relatively high proportion of auxin molecules stay protonated (un-ionized; indole-acetic acid (IAA)) and these can enter the cell directly via passive diffusion. In its ionized (dissociated) form (IAA- + H+), auxin cannot cross membranes by passive diffusion; it needs to be actively transported by carriers. Ionized auxin molecules can enter cells via active transport by auxin-influx carriers. In the relatively higher pH of the cytoplasm, auxin molecules undergo almost complete dissociation. The asymmetric positioning of the auxin-efflux carriers from the 'long' PIN subfamily at the plasma membrane then determines the direction of auxin efflux from the cell. Localization of AtPIN5 (from the 'short' PIN subfamily) at the membranes of the endoplasmic reticulum leads to compartmentalization of auxin into the lumen of the endoplasmic reticulum, where it undergoes metabolic conversion. PM, plasma membrane; ER, endoplasmic reticulum; GA, Golgi apparatus.;http://genomebiology.com/
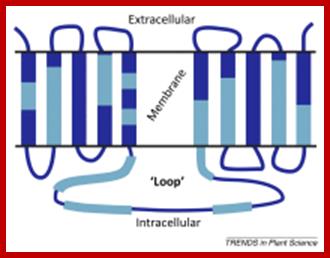
PIN is ten transmembrane Protein involved in efflux transport.
�PIN transporters and ABCB are efflux carriers that transport Auxins from cell to cell.� The PINs are involved in efflux from cell base to the next cell i.e as Auxin is pumped asymmetrically to the side of root of stem, it results in asymmetric growth (one side growths faster) and bending of the root or stem in response to the stimuli and also involved in intracellular transport. The PIN-formed (PIN) protein family of auxin transporters; The PIN-FORMED (PIN) proteins are secondary transporters acting in the efflux of the plant signal molecule auxin from cells. They are asymmetrically localized within cells and their polarity determines the directionality of intercellular auxin flow. PIN genes are found exclusively in the genomes of multicellular plants and play an important role in regulating asymmetric auxin distribution in multiple developmental processes, including embryogenesis, organogenesis, tissue differentiation and tropic responses. All PIN proteins have a similar structure with amino- and carboxy-terminal hydrophobic, membrane-spanning domains separated by a central hydrophilic domain. The structure of the hydrophobic domains is well conserved. The hydrophilic domain is more divergent and it determines eight groups within the protein family. The activity of PIN proteins is regulated at multiple levels, including transcription, protein stability, subcellular localization and transport activity. Different endogenous and environmental signals can modulate PIN activity and thus modulate auxin-distribution-dependent development. A large group of PIN proteins, including the most ancient members known from mosses, localize to the endoplasmic reticulum and they regulate the subcellular compartmentalization of auxin and thus auxin metabolism. Further work is needed to establish the physiological importance of this unexpected mode of auxin homeostasis regulation. Furthermore, the evolution of PIN-based transport, PIN protein structure and more detailed biochemical characterization of the transport function are important topics for further studie . The PIN-formed (PIN) protein family of auxin transporters (PDF Download Available). Available from: https://www.researchgate.net/publication/40869265_The_PIN- formed_PIN_protein_family_of_auxin_transporters; Tom Bennet http://www.cell.com
, ; http://dev.biologists.org
�
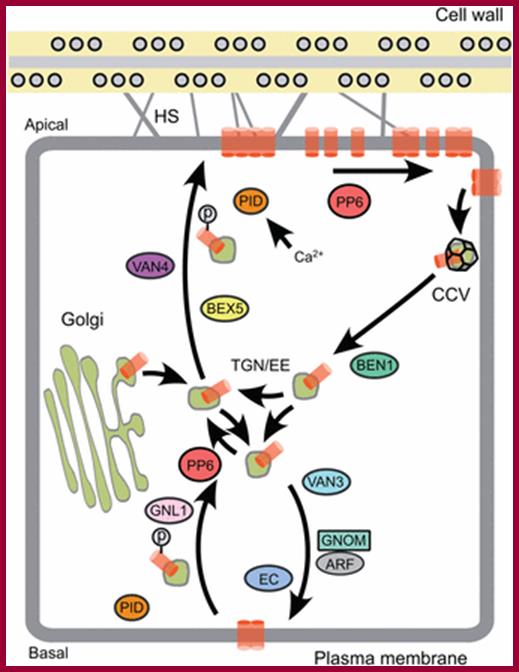
The regulation of plasma membrane protein recycling and polar localization. The ADP-ribosylation factor (ARF) GTPase guanine-nucleotide exchange factor (GEF) GNOM plays a key role in the recycling of PIN proteins (orange cylinders) between TGN/EE compartments and basal plasma membrane domains. In addition, regulators such as the Rab-type GTPase BEX5 and the ARF-GEF proteins BEN1 and GNL1 are implicated in PIN trafficking. This process also depends on the activity of the exocyst complex (EC), which is suggested to modulate the exocytic sorting of PINs. PIN targeting to the apical plasma membrane domain has been linked to PIN phosphorylation (p), presumably involving the activity of the AGC3-type protein kinase PINOID (PID), which in turn appears to be under the control of Ca2+ and phosphoinositide signaling. The activity of the PP6 phosphatase antagonizes PID, promoting dephosphorylation and PIN sorting to basal domains. PIN targeting to the plasma membrane is suggested to involve �super-polar� exocytosis, as reflected in the accumulation of PIN protein clusters at polar plasma membrane domains. Lateral diffusion of such protein clusters appears to be slow and depends on as-yet-unidentified crosstalk with cell wall components. For example, Hechtian, strands (HS) that bridge the space between the cell and plasma membrane have been suggested to act in this process. Adjacent to polar plasma membrane domains, clathrin-mediated endocytosis enforces internalization of PINs in clathrin-coated vesicles (CCVs), preventing their further diffusion within the plasma membrane. , ; http://dev.biologists.org.
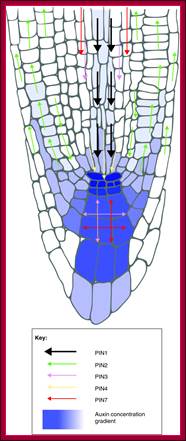
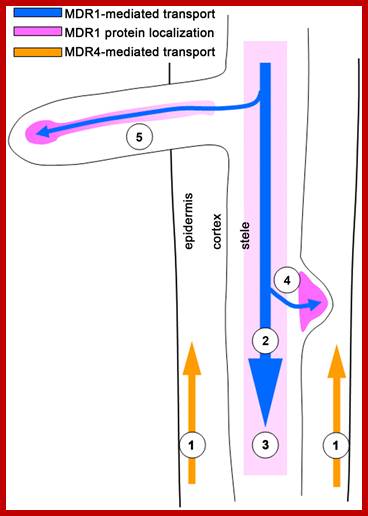
PIN-dependent Auxin-transport routes in the Arabidopsis thaliana root tip (black arrows). Auxin distribution is depicted as a blue gradient. The localization of Auxin transporters is based on proteins tagged with green fluorescent protein. Green arrows indicate Auxin flow mediated by a particular PIN transporter.
The low pH in the apoplast (the region outside the cell membrane comprising the plant cell wall) is maintained by the activity of the plasma membrane H+-ATPase. In the acidic environment of the apoplast, a relatively high proportion of Auxin molecules stay protonated (un-ionized; indole-acetic acid (IAA)) and these can enter the cell directly via passive diffusion. In its ionized (dissociated) form (IAA- + H+), Auxin cannot cross membranes by passive diffusion; it needs to be actively transported by carriers. Ionized Auxin molecules can enter cells via active transport by Auxin-influx carriers. In the relatively higher pH of the cytoplasm, Auxin molecules undergo almost complete dissociation. The asymmetric positioning of the auxin-efflux carriers from the 'long' PIN subfamily at the plasma membrane then determines the direction of auxin efflux from the cell. Localization of AtPIN5 (from the 'short' PIN subfamily) at the membranes of the endoplasmic reticulum leads to compartmentalization of auxin into the lumen of the endoplasmic reticulum, where it undergoes metabolic conversion. PM, plasma membrane; ER, endoplasmic reticulum; GA, Golgi apparatus.
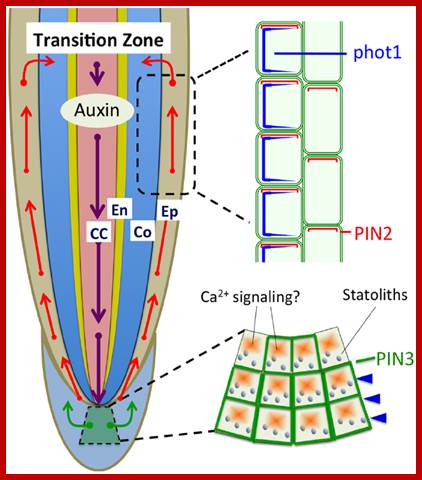
Polar auxin transport based on PIN1, PIN2, and PIN3 is light sensitive and involved in the light-induced negative phototropism of roots (Friml et al., 2002; Wan et al., 2012; Zhang et al., 2014). PIN1 is involved in the acropetal (rootward) auxin transport, PIN3 in the lateral auxin transport in statocytes, and PIN2 in the basipetal (shootward) auxin transport in epidermis and cortex cells. CC, central cylinder; En-endodermis; Co-cortex; Ep-epidermis. http://journal.frontiersin.org
ABP1 as Auxin Receptor Transducer- Hypothetical AUXIN Receptor:
Auxin binding protein 1 (ABP1) is a 22-kDa glycoprotein that is present in all green plants. A Hypothetic Model for the ABP-1 cell�surface Auxin Receptor is two subunit receptor; A GPI-anchored protein (CBP1) is proposed to bind the C-terminus masking the C-terminal KDEL ER-retention signal and facilitating ABP1 secretion to the PM. Auxin binds to C-terminal of ABP.� CBP1 alone is unlikely to transmit Auxin signaling across the PM.
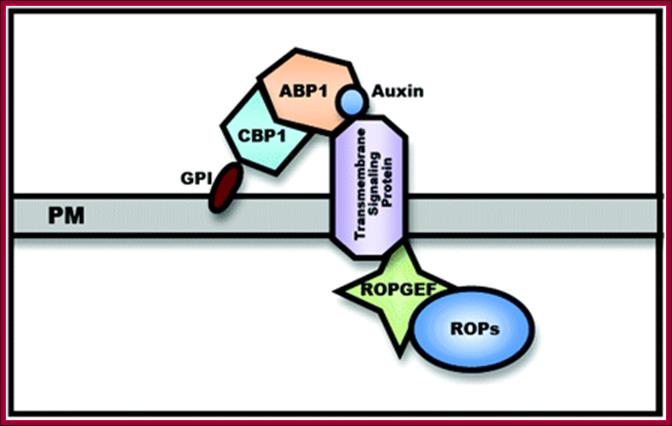 A Hypothetic
Model for the ABP1-Based Cell�Surface Auxin Receptor Complex. An GPI-anchored
protein (CBP1) is proposed to bind the C-terminus masking the C-terminal KDEL
ER-retention signal and facilitating ABP1 secretion to the PM. CBP1 alone is
unlikely to transmit auxin signal across the PM. A transmembrane protein is
speculated to act as an auxin co-receptor with ABP1 to transmit the auxin
signal to PM-localized RopGEFs that activate ROPs at the PM. ;A
general view of ABP1 mediated signal transduction; http://mplant.oxfordjournals.org/
A Hypothetic
Model for the ABP1-Based Cell�Surface Auxin Receptor Complex. An GPI-anchored
protein (CBP1) is proposed to bind the C-terminus masking the C-terminal KDEL
ER-retention signal and facilitating ABP1 secretion to the PM. CBP1 alone is
unlikely to transmit auxin signal across the PM. A transmembrane protein is
speculated to act as an auxin co-receptor with ABP1 to transmit the auxin
signal to PM-localized RopGEFs that activate ROPs at the PM. ;A
general view of ABP1 mediated signal transduction; http://mplant.oxfordjournals.org/
Auxin is perceived by distinct classes of receptors: transport inhibitor response 1 (TIR1, or auxin-related F-box (AFB)) and auxin/indole-3-acetic acid (AUX/IAA) coreceptors, that control transcriptional responses to auxin. The auxin-binding protein 1 (ABP1), that controls a wide variety of growth and developmental processes. To date, the mode of action of ABP1 is still poorly understood and its functional interaction with TIR1/AFB�AUX/IAA coreceptors remains elusive. Alexandre Tromas, S�bastien Paque et al. We find that ABP1 is genetically upstream of TIR1/AFBs; ABP1 knockdown leads to an enhanced degradation of AUX/IAA repressors, independently of its effects on endocytosis, through the SCFTIR1/AFB E3 ubiquitin ligase pathway. Combining positive and negative regulation of SCF ubiquitin-dependent pathways might be a common mechanism conferring tight control of hormone-mediated responses; but its main activity apparently lies in influencing events at the plasma membrane. ����
A trans- membrane protein is speculated to act as an Auxin co-receptor of ABP1 to transmit the Auxin signal to PM-localized RopGEFs that activate ROPs at the inner face of the PM.� ABP1 it is an important Auxin receptor for embryonic development.� ABP1 is required for activating two antagonizing ROP GTPase signaling pathways involved in cytoskeletal reorganization and cell shape formation, and participates in the regulation of Clathrin-mediated endocytosis to subsequently affect PIN protein distribution. It might be coupled with G-protein and lead to an activation of phospholipase and subsequent elevation of free Ca2+ and receptor operated IP3-regulated channels. These exciting discoveries provide indisputable evidence for the Auxin-induced signaling pathways that are downstream of ABP1 functions, and suggest intriguing mechanisms for ABP1-mediated polar cell expansion and spatial coordination in response to Auxin.� This aspect is very important in Auxin induced adventitious root initiation.
Another Intracellular Receptor- TIR1/AFB:
�Transport inhibitor response1 protein (complexed with Skp1/Cullin/F-box (SCF)-AFB.� There are more number of TIR1 and AFBs (AFB1-AFB5).� TIR1 and AFB consist of a family of proteins.� Recent genetic studies have indicated that TIR1, AFB1, AFB2 and AFB3 act as positive regulators, and that AFB4 is a negative regulator, of Auxin signaling.
Auxin binds to TIR1.� Auxin bound TIR1 recruits-SKp/Cullin/F box SCF-AFB and moves in to the nucleus and interacts with Auxin/IAA repressor that is already bound to Auxin response factor that is bound to the promoter of Auxin response genes.� Note there are several AFBs such as AFB2, AFB3, AFB4 and AFB 5.� This is a generalized view, as if all the Auxin inducible genes are bound by repressor such as Auxin/IAA proteins.
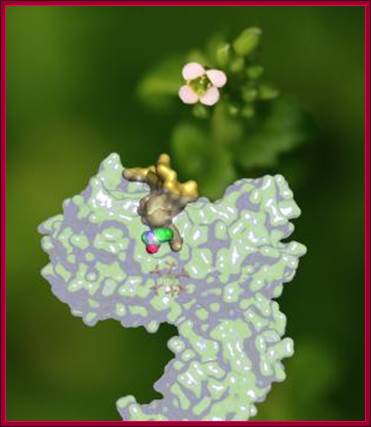
For the first time, scientists from the University of Washington School of Medicine, Indiana University Bloomington and the University of Cambridge have determined how a plant hormone -- auxin -- interacts with its hormone receptor, called TIR1. Their report, on the cover of Nature in April, also may have important implications for the treatment of human disease, because TIR1 is similar to human enzymes that are known to be involved in cancer. Superimposed on a photo of Arabidopsis, the plant hormone receptor TIR1 and its hormone, auxin. Until recently, how TIR1 and auxin interacted was unknown. (Credit: Ning Zheng); Superimposed on a photo of Arabidopsis, the plant hormone receptor TIR1 and its hormone, auxin. Until recently, how TIR1 and auxin interacted was unknown.� http://newsinfo.iu.edu/
Auxin Response genes:�
Auxin elicits fasted growth response in terms gene expression within 5-10 minutes of its application. Changes in protein level detected around 15-30min, Daniel Schenk et al. It is known that the AFB (Auxin binding F-box proteins, AFB 1 to AFB5) proteins activate the expression of auxin-induced, primary-response genes by affecting the steady-state level of a large family of transcription factors that directly or indirectly mediate hundreds of auxin-regulated genes, A general mechanism of Auxin induced gene expression has been conceptualized by Thomas J. Guilfoyle.� This was proposed long back, yet this proposition is believed to operate and good.� Why? The raison de et re is the lack of competent research work in the field of plant molecular biology.
Auxin per se induces many genes; it is to be noted that surprisingly small number of genes that appear to be coordinately regulated; in some cases by more than one phytohormone. Auxin induction of gene expression is very fast for the expression is within 15-20 minutes of application.� In our work (GRKRaj) on Auxin induced adventitious root initiation we found increase in protein synthesis without any increase in transcription in hypocotyls of P.vulgaris at 30 minutes of treatment.� Auxins may regulate some cellular responses such as cell expansion or polarity through direct effects on membranes or cytoskeletal functions but auxins also activate several genes at transcription level (Qing Tial et al). We predicted that increased protein synthesis without mRNA synthesis� due to activation preexisting activatable mRNAs. Auxins also induce short lived nuclear proteins within 6-8 minutes. Such proteins can act as either activators or repressors, (Abel S, et al)
Auxin R�ponse Promotor El�ments :
-----------RE�RE�RE---TATA�InR+1>---DPE
Response elements-
-----TGTCTC--------TATA---InR+1>----DPE�
Variants of REs- variants TGTCCC, TGTCAC),
Some promoter and upstream sequences- general view-
--TGACGcag� (TGA3)--TGACGTGG (TGA2)---TGACGTaa (TGA1)-----+1---ATG,
--GGTCCAT-84-GGACC-CTCAfrATATA-32 -54 TATATA�� --10 TATAAATA---+1
SAUR small auxin Up RNA- genes-
--TCTCTC and CCTCCCAT, TGTCTC and GGTCCCAT--TATAAAT--+1>,
GH3 (Gretchen Hagely) gene- promoter elements of Soybean-
--TGACGCAGAP1/CREBP-TGACGTGG--TGACGTAA-TATAAATA-34 to �27----+1>
�� More Promoter elements:
-TGA-element (AACGAC--AuxRR-core�GGTCCAT-AuxRE, TGTCTC)-TATAAAT--+1>
-271----TGACGAATGCGATGACC--84[GGTCCATI- -54[TATATA] --TATAAATA]--+1>-
MYB77� with ARF7activates- lateral root initiation-
---MYB77+ARF7 TGTCTC AuxREs�activates +1>,
Eight types of motifs and their variants exist for Auxin induced gene promoters:
They include, the ACGT-box, OCS-element, W-box, GT1-element, GATA-box, CAAT-box, PU-element, and YY1-element that are commonly present in highly expressed plant genes.
Models of Auxin induced gene expression:
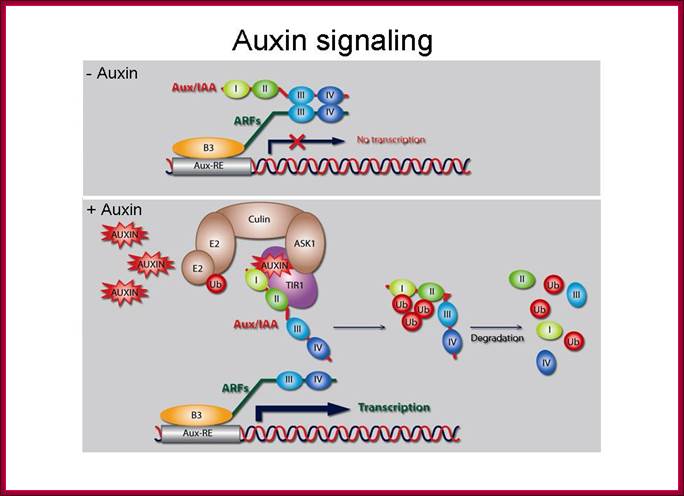
Plant
hormones control most aspects of the plant life cycle by regulating genome
expression. Expression of auxin-responsive genes involves interactions among auxin-responsive
DNA sequence elements, transcription factors and trans-acting transcriptional
repressors. Transcriptional output from these auxin signaling complexes is
regulated by proteasome-mediated degradation that is triggered by interaction
with auxin receptor-E3 ubiquitin ligases such SCF(TIR1). Auxin signaling
components are conserved throughout land plant evolution and have proliferated
and specialized to control specific developmental processes.
Mechanism of Auxin-Regulated Gene Expression in Plants (PDF Download
Available). Available from: https://www.researchgate.net/publication/26747190_Mechanism_of_Auxin-Regulated_Gene_Expression_in_Plants;Mechanism
of Auxin-Regulated Gene Expression in Plants; https://www.researchgate.net; www.dreamhome.id/g

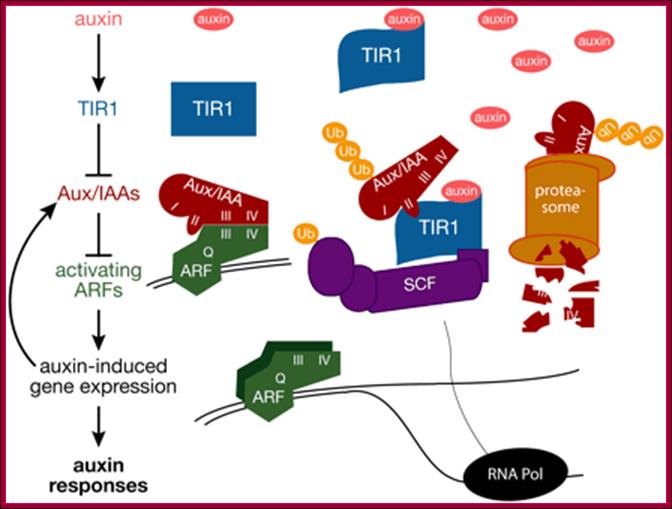
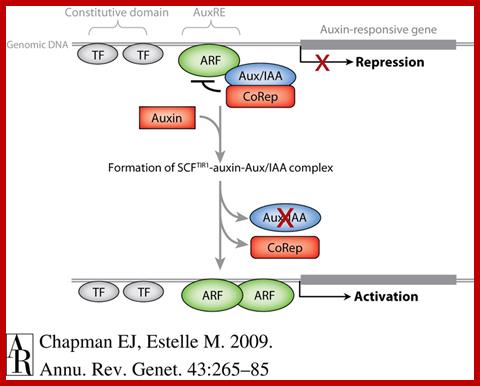
A Model for Auxin Response through the TIR1 Auxin Receptor Pathway: Transcriptionally activating ARFs (green) with Gln-rich (Q) middle regions are bound to auxin-responsive promoter elements but are counteracted (blunt arrows) by heterodimerization with Aux/IAA transcriptional repressors (dark red) via two domains (III and IV) conserved between ARF and Aux/IAA proteins.
A simple model for Auxin induced gene expression; Auxin is transported across via Transport inhibitor response1 protein TIR1; this protein has F-box protein mediated ubiquitin ligase motif (E3 ligase). On the entry with auxin TIR1-AFB interacts with a repressor called IAA/auxin protein; this repressor protein is ubquitinated and degraded by proteasome. This degradation of IAA/auxin repressor make the ARF (auxin receptor factor) active, which is already bound to Auxin response elements in a said gene.� This recruits all the required cofactors and MCs and BTA.� This assembly leads to activation of that specific gene. Critical factor that determines which Aux/IAA protein; there are several of them and they might be specific to specific to auxin responsive gene to be activated.� Here is an example where a repressor bound to regulatory sequence of the gene and it is perpetuated till it is removed by another factor for the activation of a gene.
The key components in auxin perception and signaling in Arabidopsis. ARF proteins contain a non-conserved AD or RD flanked by an N-terminal DBD (composed of a B3 domain, a dimerization domain: DD, and a Tudor-like ancillary domain within the C-terminal region of the flanking domain: FD) and a C-terminal PB1 domain (previously referred to as domain III/IV). Parts of the DD and FD are found both N-terminal and C-terminal to the B3 domain. In this pathway, the TRANSPORT INHIBITOR RESPONSE 1/AUXIN SIGNALING F-BOX proteins (TIR1/AFBs) are F-box proteins that, together with other proteins (ASK1, CUL1, RBX), form the ubiquitin protein ligase complex, SCFTIR1. At low auxin levels (A), the Aux/IAA proteins form multimers with ARFs and recruit TPL to the chromatin. High levels of auxin (B) promote ubiquitination and degradation of Aux/IAAs through SCFTIR1∕AFB and the 26S proteasome (Kim et al., 1997; Guilfoyle and Hagen, 2007, 2012; Szemenyei et al., 2008; Boer et al., 2014; Guilfoyle, 2015; Korasick et al., 2015; Salehin et al., 2015). https://www.ncbi.nlm.nih.gov;Plant cell Biol ?
Auxin induces global gene expression within 5-15 minutes of application; this is considered to be the fastest regulation of gene induced transcription known (Abel et al 1994). There are 31, ~47 and 25 genes that respond to auxins in Maize, Arabidopsis and Rice respectively.� Induction and global expression is approximately twenty fold in hypocotyls. Genes like IAA3 and IAA6 were induced within minutes and returned base line after 2hrs.� Others such as AXR2/IAA7 and IAA8 are induced slowly (Quing Tian et al). �Auxin responding genes are bound by ARFs (Auxin response factors) which are transcription factors specific to specific Auxin responding genes.� Some data set shows that there are 224 ARF-relatd protein sequences in most of the plant systems, (Cedric Finer etal).� They have a range of amino acid sequences and bind to different promoter elements activated by Auxins. �Ryoung Shin et al observed Auxin induced gene expression responses require another factor called MYB77, whose activation domain interacts with C terminus of ARF III and IV domains.� This has synergistic interaction between these two factors in Auxin induced gene expression.
ARFs contain N terminal DNA binding domain (DBD) and middle Q- rich x activator domain / other repressor domains (MR) and 3rd and 4th contain protein-protein interacting domains at their C-terminal where Auxin/IAA proteins 3rd and 4th domain bind covering the ARFs activator domain thus represses the activity of ARFs.� ARF1 is 665a.a long (~73kDa protein) contain four domains as described above.
Some of the Auxin induced genes are ARFs. Auxins in general induce the expression of some short lived nuclear proteins involved in elongating hypocotyls, induce ACS genes, induce cytokinin degradase enzymes, induce GA2oX and GA3ox genes, increase beta-1,4-glucanase tenfold, and up regulates the expression of IAA1, IAA19, PIN1, GH3.2, GH3.3, SAUR-AC1, HAT2, and MYB77 within 0.5 to 24hrs after application of the hormone.� This increase is knocked out by MYB 77 knock out. There are more ARFs- ARF5, ARF6, ARF7 and ARF8. Auxin responsive genes contain ARE promoter elements which differ from gene to gene.
Auxin/Indole-3-AceticAcid (AUX/IAA) proteins, considered to be auxin regulated ARF repressors, are 50-55 kDa proteins; there are more than 25 of them, they contain domains which bind to ARF III and IV domains and inhibit the activator domains of ARFs.
The most quoted Auxin regulation Model.
In addition to modulating auxin responsiveness, plants regulate auxin biosynthesis, transport, and storage forms. The auxin precursor indole-3-butyric acid (IBA) is converted into active auxin (IAA) by a peroxisomal process similar to fatty acid β-oxidation. Several peroxisomal enzymes appear to be necessary for IBA-to-IAA conversion and mutants in these enzymes have revealed that IBA-derived auxin has diverse roles for in plant development and a greater input into the pool of auxin than anticipated. Understanding the role of IBA-derived auxin and will allow us to deepen our knowledge of how the plant uses auxin and auxin precursors to regulate growth and development.
Similar to the active auxin IAA, the auxin precursor IBA is transported long-distance through the plant. However, examined IAA transporters do not transport IBA. In addition, transporters that move IBA out of the plant are unable to transport IAA. Thus, transporters necessary to move IBA long-distance through the plant are unidentified. We are using forward genetics in Arabidopsis to identify additional factors required for the transport of IBA. Identifying these factors and understanding their roles in IBA transport and metabolism will allow us to gain new insights into the complex mechanisms used by plants to control auxin homeostasis.
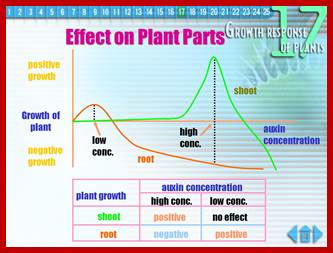
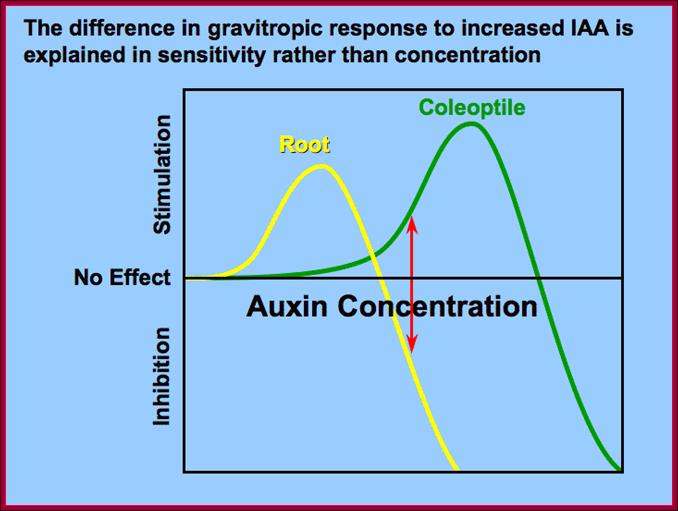
Differential response to Auxin concentration; however he GRN Gene regulatory network is not established. www.slideplayer.nl; /https://pages.wustl.edu
The basic premise of Auxin action is depicted in the following pathway.
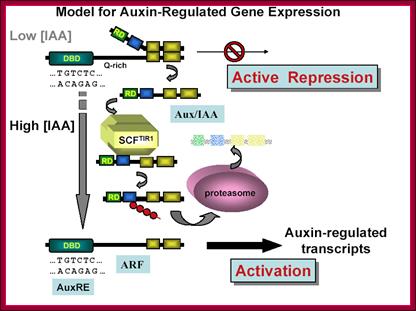
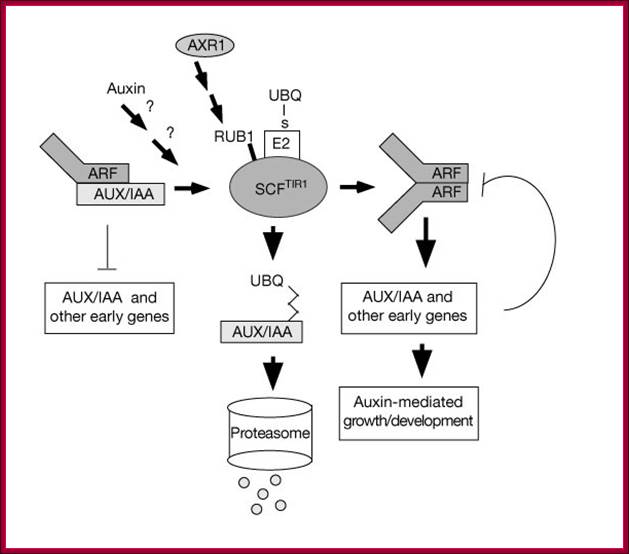
This model is also similar to the above except it was proposed by Guilfoyle; here he has proposed that the repressor cum activator ARF contains four domains, The N-terminal domain binds to AUXIN REs, the next domain has Gl activation domain, and the next two domains are involved in dimerization with Auxin/IAA binding repressor protein.� The IAA/Auxin repressor too contains four domains; it has 2 c-terminal domain binds to C-terminal domain of ARF and covers the activator domain of ARF. The N terminal domain has the motifs for interacting with TIR1 protein that carries Auxin.� When the TIR1 protein binds to Auxin it is transported into the nucleus, where it interacts with Auxin/IAA protein and subjects to ubquitinated proteolysis, thus the ARF becomes active and activates the transcription of the said gene.� This is the most quoted paper. Auxin/IAA proteins are found in different forms and context such as Auxin/IAA 1,3,7,9 etc.� They determine which gene to be transcribed.� If it is so the response element or other upstream activator sequences determine which of the auxin responsive gene to be activated. www.mfacourses476.web.fc2.com plant cell biol/?
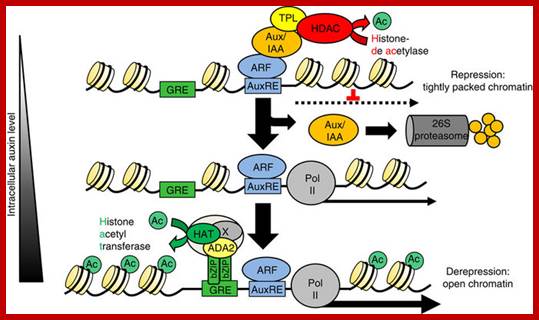
The Arabidopsis transcription factor bZIP11 activates auxin-mediated transcription by recruiting the histone acetylation machinery, In higher plants, the hormone auxin orchestrates a diverse array of developmental and environmental responses mainly exerted via transcriptional control. In its absence, auxin-mediated transcription is postulated to be repressed by histone deacetylases, which convert chromatin into a highly packed inactive state. Here we present a converse mechanism where Arabidopsis bZIP11-related basic leucine zipper (bZIP) transcription factors interact via an amino-terminal activation domain with ADA2b adapter proteins to recruit the histone acetylation machinery to specific auxin-responsive genes. Gain, loss-of-function and pharmacological approaches as well as chromatin immunoprecipitation experiments addressing various components of the recruitment and acetylation machinery substantiate the proposed mechanism. Importantly, G-box-related cis-elements, frequently found in auxin-induced promoters, are shown to bind bZIP11-related bZIPs and to function as quantitative modulators of auxin-induced transcription. In conclusion, we describe a regulatory activation mechanism that serves as a rheostat to modulate auxin-mediated responses Christoph Weiste, & Wolfgang Dr�ge-Lase; https://www.nature.com
Nothing is known how G-protein mediated signaling pathway activates auxin induced gene expression. But the other pathway that is TIR1-FBP has been described.� Auxin first binds to TIR1 at membrane level. Auxin responsive genes have Auxin response elements upstream of the InR.� In the absence of Auxin the ARF factor is bound by IAA/AUX repressor protein, thus the response factor remains inactive. When the Auxin bound TIR1-complexed with AF-box complex moves into the nucleus and interacts with IAA/Auxin protein a repressor.� This leads to the release of IAA/Auxin protein which is degraded by SCF mediated proteolysis. The so called response factor now becomes activator.� It recruits cofactors and other required factors (not characterized) and activates specific genes.
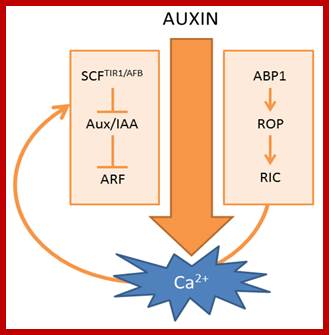
Scheme of auxin-induced Ca2+ signals. (Left) Canonical SCFTIR1/AFB-mediated auxin signaling; (Right) ABP1-mediated auxin signaling. The curved arrow represents a hypothetical model in which Ca2+acts as a connecting signal between ABP1 and SCFTIR1/AFB signaling cascades.; http://www.mdpi.com
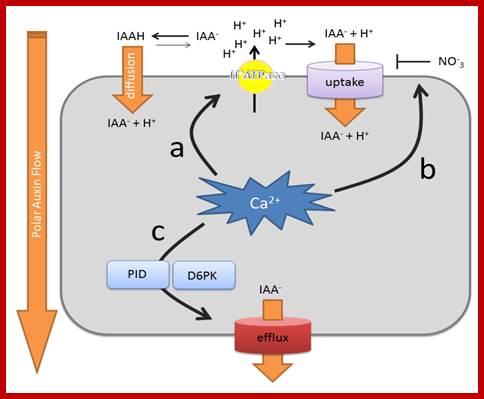
Summary of the effects of Ca2+ on polar auxin transport rates. (a) Cellular Ca2+ signaling impacts on auxin uptake mechanisms via effects on the abundance and activity of the plasma membrane H+ ATPase. The amount of protons in the apoplast determine the auxin uptake rate via diffusion of protonated indole-3-acetic acid (IAAH), as well as H+/IAA-(indole-3-acetic acid) symport; (b) Ca2+ can change the affinity of NRT1.1 for nitrate and auxin uptake; (c) Ca2+ controls the activity of the auxin efflux machinery by modulating the kinase activity of PINOID (PID) and, possibly, also D6PKs. PID also impacts on PIN-formed (PIN) polarity (not depicted). Uptake refers to active uptake mechanisms. Efflux refers to active auxin efflux mechanisms. http://www.mdpi.com
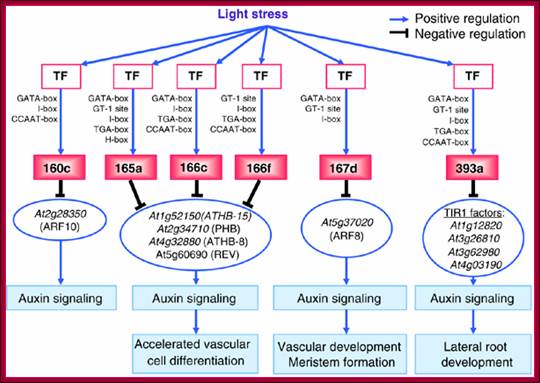
Diagram shows how light stress acts through auxin signaling; www.cs.wustl.edu/~zhang
Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins; The plant hormone auxin is central in many aspects of plant development. Previous studies have implicated the ubiquitin-ligase SCFTIR1 and the AUX/IAA proteins in auxin response. Dominant mutations in several AUX/IAA genes confer pleiotropic auxin-related phenotypes, whereas recessive mutations affecting the function of SCFTIR1 decrease auxin response. Here we show that SCFTIR1 is required for AUX/IAA degradation. We demonstrate that SCFTIR1 interacts with AXR2/IAA7 and AXR3/IAA17, and that domain II of these proteins is necessary and sufficient for this interaction. Further, auxin stimulates binding of SCFTIR1 to the AUX/IAA proteins, and their degradation. Because domain II is conserved in nearly all AUX/IAA proteins in Arabidopsis, we propose that auxin promotes the degradation of this large family of transcriptional regulators, leading to diverse downstream effects. William M. Gray et al; http://www.nature.com/
Auxin Induced Adventitious Root Initiation:
Hypocotyls, or stem cuttings like Rose, croton or any other stem cuttings, when dipped in asafetida, an Indole compound, for some period of time and plant it in the soil.� Then place a small lump of cow dung on the top of the cut stem. In about eight to ten days the stem cuttings produced roots.� This type of vegetative propagation has been done since time immemorial in rural area, even now we do it. Young terminal stems of the explants with one or two very young leaves (other are removed), found to root very well.
Auxins such as IBA and NAA are found to be very good in inducing new roots in stem cuttings. Phaseolus vulgaris hypocots, when treated with IBA, the hypocots show adventitious protuberances with in 36hrs and by 48 hours root projections can be observed very well.� Anatomical studies show, within 12 hrs. of treatment, pericycle cells found in between two split exarch xylem bundles exhibit cell divisions with islands of cells as the precursors of root primordial.� It is known that some of the cells in pericycle act as founder cells and they are induced by Auxins.� The cell division probably induced by Cdk-A and Cyclin-A and CdkB and Cyclin-B at G1 and S and G2 and M stages. These Cdk-Cyclins have an activation effect on E2Fs, which are required for the activation cell cycle, but it is not possible how the plane of cell division is controlled.� Any role played by microtubule associated proteins (MAPS) is discerned for the orientation of mitotic apparatus during cell division requires microtubules and actin filaments
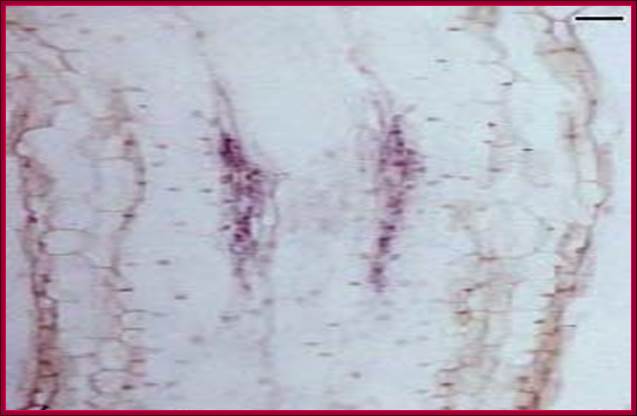
The in vivo stained diagram shows the stages of activation of cells in the pericycle into root initial-primordial (research done by by grkraj.blr, the author of this websites). www.grkraj.org
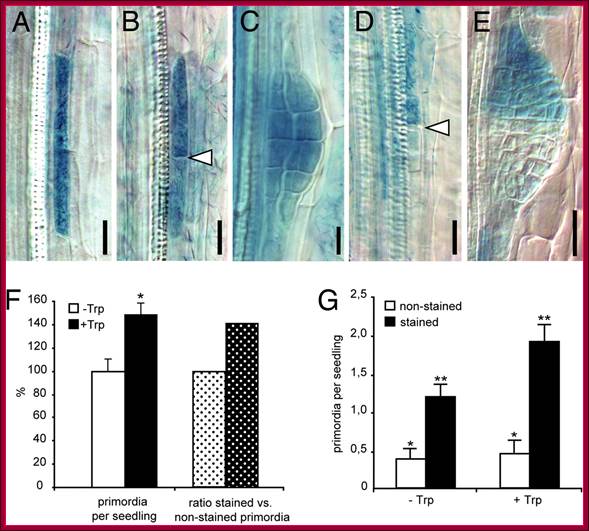
Auxin acts as a local morphogenetic trigger to specify lateral root founder cells; Local auxin production in pericycle cells triggers primordium formation. (A�E) Examples of sectors of GUS-marked iaaM-expressing cells induced by heat shock. (A) Sector of a single xylem-adjacent pericycle cell is shown. (B) After cell division two longitudinally abutted cells are formed; arrowhead indicates the new cell wall formed. (C) Such activation leads to formation of a fully stained primordium. (D and E) Activation of iaaM expression in only one of two longitudinally abutted cells can recruit the neighboring cell as the second founder cell and leads to staining of the progeny of only one founder cell (D) and development of a half-stained primordium (E). Arrowhead in D shows the position of end walls of a heat shock-activated and an independently specified pericycle founder cells. (F) Seedlings with random sectors of iaaM expression initiate 48% more primordia when auxin synthesis is induced by Trp treatment (black bar) as compared with untreated controls (white bar) (Student's t test; *, P < 0.0008). Accordingly, the proportion of GUS-positive primordia (indicating auxin-producing sectors) is 40% higher in Trp-treated seedlings (dotted black bar) as compared with untreated controls (dotted white bar). (G) Scoring of stained (iaaM expressing) and unstained primordia reveals that Trp treatment does not affect the iaaM-independent initiation of primordia, as the number of unstained primordia is not changed compared with the untreated controls (white bars) (Student's t test; *, P = 0.6,). The significant increase in the number of stained primordia (black bars) upon Trp treatment (Student's t test; **, P < 0.005) suggests that the local increase in the auxin level as a consequence of iaaM activity in pericycle sectors stimulates primordia initiation. The number of primordia was scored in 35�40 seedlings from two independent experiments (mean � SE). http://www.pnas.org/
Hypocotyls treated with IBA produce 50-70 roots (concentration dependent) per hypocotyl, but if they are treated with Cytokinin alone roots are not produced.� On the other hand, if the hypocotyls are provided with both Auxin and Cytokinin such as IBA, formation of adventitious root formation is inhibited.��� These are documented with anatomical evidences. In Auxin combined with Cytokinin treated hypocots, one can observe multilayer of pericyclic cells but no root initials. Auxin induced cell elongation is also inhibited by Cytokinins. IBA treated cells show cellular changes such as increase in the size of Nuclei and the nucleolus is so large it occupies more than 70% of the nuclear volume, an indication of ribosomal RNA and Ribosomal protein transcriptional activity.
Interestingly molecular studies show IBA induces activation of protein synthesis in the presence of ActinomycinD that inhibits transcription; which means IBA activates certain stored mRNAs; the mechanism is not known. However, if the hypocotyls are treated with Cytokinin alone or Cytokinin plus IBA, the increase in protein synthesis is only marginal.� But after 36-48 hrs, one finds significant increase in poly-A RNA in all the treated hypocotyls. But the poly-A RNA isolated from IBA treated hypocotyls at 36hrs, when translated we found some transitory protein of high Mol.wt, which is not found in Cytokinin and Cytokinin-IBA treated segments.� It clearly indicates a transient protein that is produced in response to Auxin may be responsible for activation of pericyclic cells into root initials but not confirmed. And Cytokinin blocks some Auxin mediated gene expression for the said transitory regulatory protein.
The role of Auxin in inducing adventitious roots or new roots requires many factors including cell cycle proteins; which has been depicted in the underlying diagram.
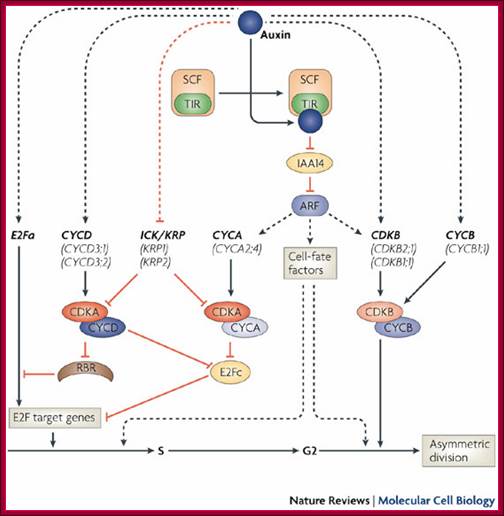
Lateral root formation starts when pericycle founder cells of the root are induced by auxin to re-enter the cell cycle at the G1�S transition. Transcript profiling studies reveal that this transition is realized by several co-acting mechanisms at the level of cell-cycle regulation. The transcription factor E2Fa and D-type cyclins (CYCD) are upregulated early, which is indicative for the activation of the canonical retinoblastoma-related (RBR) protein pathway and activation of E2F target genes. By contrast, ICK/KRP genes are transcriptionally repressed. This repression might release the activity of A-type cyclin-dependent kinase (CDKA)�CYCD and/or CDKA�A-type cyclin (CYCA) complexes, which, in turn, might mediate the degradation of E2Fc, an inhibitor of cell-cycle progression. Although recent data suggest that the expression of certain CYCA and B-type CDK (CDKB) family members might be controlled directly by auxin, further research is required to validate this observation. The transcriptional mechanism controlled by auxin is based on the binding of transcriptional activators, auxin response factors (ARFs), to auxin response elements in the promoter of downstream genes. In the absence or at low levels of auxin, ARFs dimerize with Aux/indole-3-acetic acid (IAA) proteins that function as transcriptional repressors. When auxin levels in pericycle founder cells increase, the degradation of Aux/IAAs is stimulated by the auxin-mediated recognition of Aux/IAAs by the SCFTIR complex, an E3 ligase complex that tags these proteins for degradation. Whereas CYCA is required for G1�S transition, CDKB�B-type cyclin (CYCB) might be involved in the G2�M transition. Interestingly, cell-cycle progression per se is not sufficient to drive lateral root initiation. Besides the cell-cycle regulators, auxin probably induces as-yet-unidentified cell-fate-specification factors. Such factors might be essential to install asymmetric divisions of the pericycle founder cells, a prerequisite for organogenesis to occur.; Lieven De Veylder, Tom Beckmann & Dirk Inez; http://www.nature.com (www.grkraj.org).
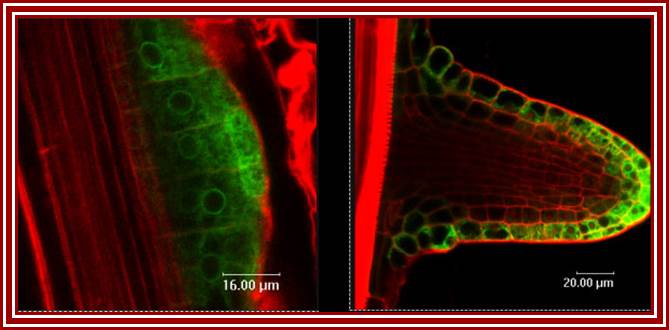
Phytohormone (IAA or IBA) induced new root formation (Taken from different source); wikivisually.com
Auxin-induced cell wall loosening and expansion:
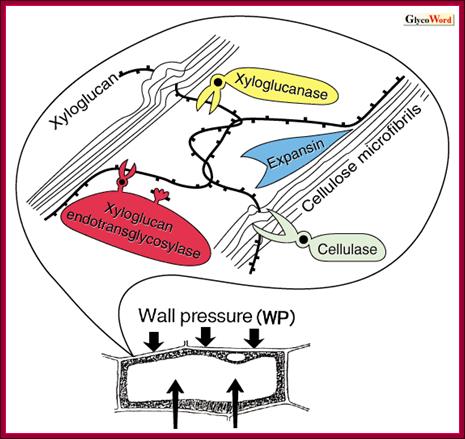
Looseniong of cell induced by auxin; http://www.glycoforum.gr.jp
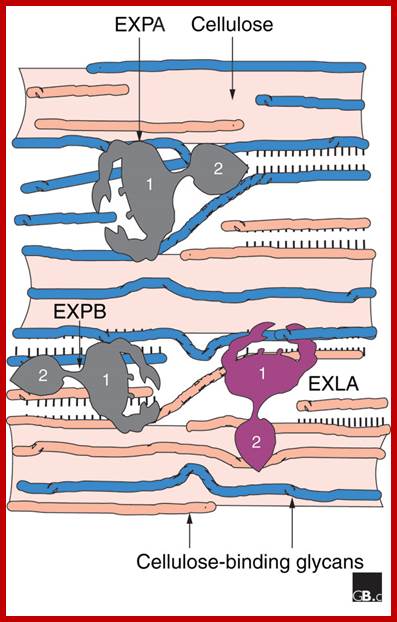
The expansin superfamily
of plant proteins is made up of four families, designated alpha-expansin,
beta-expansin, expansin-like A and expansin-like B. alpha-Expansin and
beta-expansin proteins are known to have cell-wall loosening activity and to be
involved in cell expansion and other developmental events during which
cell-wall modification occurs. Proteins in these two families bind tightly to
the cell wall and their activity is typically assayed by their stimulation of
cell-wall extension and stress relaxation; no bona fide enzymatic activity has been
detected for these proteins. Alpha-expansin proteins and some, but not all,
beta-expansin proteins are implicated as catalysts of 'acid growth', the
enlargement of plant cells stimulated by low extracellular pH. A divergent
group of beta-expansin genes are expressed at high levels in the pollen of
grasses but not of other plant groups. They probably function to loosen
maternal cell walls during growth of the pollen tube towards the ovary. All
expansins consist of two domains; domain 1 is homologous to the catalytic
domain of proteins in the glycoside hydrolase family 45 (GH45); expansin domain
2 is homologous to group-2 grass pollen allergens, which are of unknown
biological function. Experimental evidence suggests that expansin loosen cell
walls via a nonenzymatic mechanism that induces slippage of cellulose
microfibrils in the plant cell wall.
The expansin superfamily (PDF Download Available). Available from: https://www.researchgate.net/publication/7414466_The_expansin_superfamily
[accessed Jun 1, 2017].;http://www.glycoforum.gr.jp http://www.researchgate.net
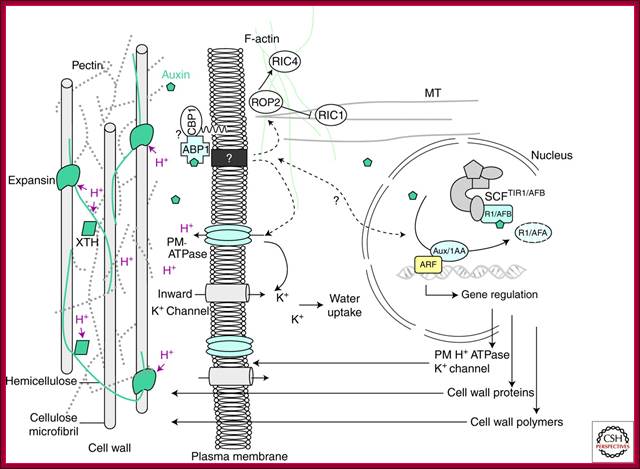
Cellular Responses to Auxin: Division versus Expansion; Auxin-induced cell wall loosening and expansion. The scheme represents the cell wall/plasma membrane/cytoskeleton continuum and the consequences of auxin action. Auxin is perceived by the auxin receptor ABP1, which interacts with unknown membrane-associated proteins at the plasma membrane (such as the putative candidate GPI-anchored protein CBP1) (Shimomura 2006). This activates the proton pump ATPase, provoking the acidification (H+) of the extracellular space, the activation of cell wall proteins such as expansins and xyloglucan endotransglycosylase/hydrolases (XTH), which mediate cell wall loosening by acting on the cell wall polysaccharide network. Polysaccharides forming the cell wall are cellulose microfibrils, cross-linked hemicelluloses, and pectins. Activation of the H+ ATPase also induces hyperpolarization of the plasma membrane and activation of K+ inward rectifying channels, essential for the uptake of water sustaining cell expansion. Auxin also enhances these effects by inducing the expression of genes encoding plasma membrane ATPase, K+ channels, expansin, and cell wall remodeling enzymes and promotes exportation of new cell wall material. Auxin is likely to act on actin microfilaments and microtubules via the modulation of ROP GTPases. http://cshperspectives.com/
The above illustrated scheme represents the cell wall/plasma membrane/cytoskeleton continuum and the consequences of Auxin action. Auxin is perceived by the Auxin receptor ABP1, which interacts with unknown membrane-associated proteins at the plasma membrane (such as the putative candidate GPI-anchored protein CBP1) (Shimomura 2006). This activates the proton pump ATPase, provoking the acidification (H+) of the extracellular space, the activation of cell wall proteins such as expansins and xyloglucan endotransglycosylase/hydrolases (XTH), which mediate cell wall loosening by acting on the cell wall polysaccharide network. Polysaccharides forming the cell wall are cellulose micro fibrils, cross-linked hemicelluloses, and pectins. Activation of the H+ ATPase also induces hyperpolarization of the plasma membrane and activation of K+ inward rectifying channels, essential for the uptake of water sustaining cell expansion. Auxin also enhances these effects by inducing the expression of genes encoding plasma membrane ATPase, K+ channels, expansins, and cell wall remodeling enzymes and promotes exportation of new cell wall material. Auxin is likely to act on actin microfilaments and microtubules via the modulation of ROP GTPase.
�����������
Gibberellic Acid:
Gibberellins were first discovered in Japan; first they found in rice plants in fields infected with a fungus called Gibberella fujikoroi.� The infected plants were found to be tall and thin and non-productive; called foolish seedlings.� Later American scientists identified the causative agent and called the substance as Gibberellins or Gibberellic Acid. They are ubiquitous both in flowering and nonflowering plants. �GA's are widespread and so far ubiquitous in flowering (angiosperms) and non-flowering (gymnosperms) plants as well as ferns.
Guang-Long Wang et al. ;https://www.nature.com
Some functions:
- Stimulate stem elongation by stimulating cell division and elongation.
- Stimulates bolting/flowering in response to long days.
- Breaks seed dormancy in some plants which require stratification or light to induce germination.
- Stimulates enzyme production (a-amylase) in germinating cereal grains for mobilization of seed reserves.
- Induces maleness in dioecious flowers (sex expression).
- Can delay senescence in leaves and citrus fruits.
- Promote seed germination,
- �Stimulate stem and root growth.
- �Regulate the transition from juvenile to adult Phases.
- �Influence sex determination.
- �Promote pollen development and tube growth.
- �Promote fruit set and parthenocarpy.
- �Promote early seed development.
Biosynthesis:
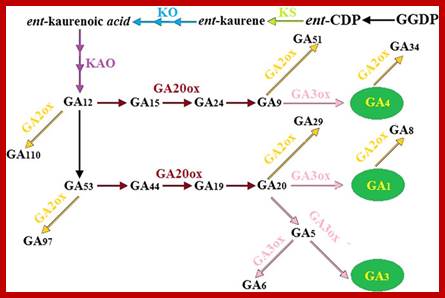
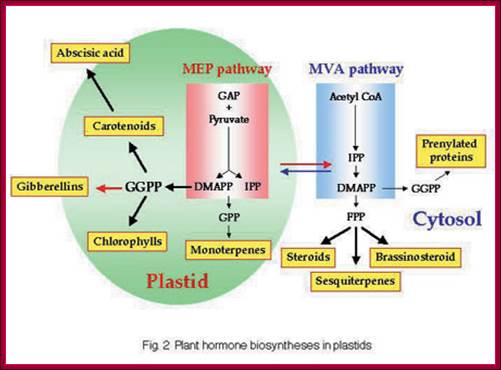
GA is derivative of isoprene compounds.� It has many isoforms, perhaps more than 152 or so.� The most common found among the tissues is GA3.� GAs are synthesized via terpenoids in chloroplasts and then released to cytoplasm and then they are transported along sieve tubes.
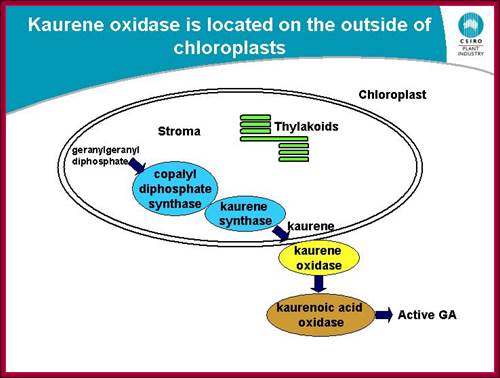
Plastids synthesize many components including Kaurine; https://www.slideshare.net/
GA synthesis pathway; www.jegol.ir/article/
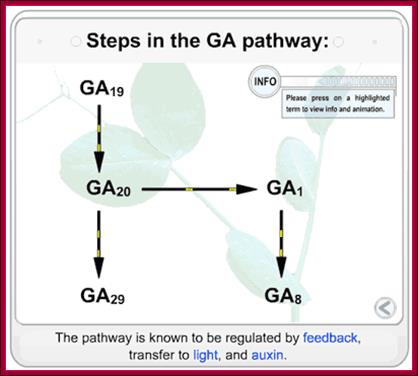
http://5e.plantphys.net/
����������� Basically all plant hormones function through membrane bound receptors or cytosolic receptors or combination of both.
Effects of GA in plants are many, some mentioned above; among them the important ones are: 1. Overcomes genetic dwarfism, 2. Induces Alpha Amylase gene expression on Aleurone cell layers of germinating barley seeds, 3. Induces bolting and flowering in long day plants that require cold treatment (substitute photoperiodism and vernalization).
4. Promotes fruit setting, 5. Increase sugar content in sugarcanes and 6. It also augments concentration of auxin. The most dramatic effect of GA has been found to be in germinating cereal grains.
Gibberellin (GA) and Alpha amylase Gene Expression:
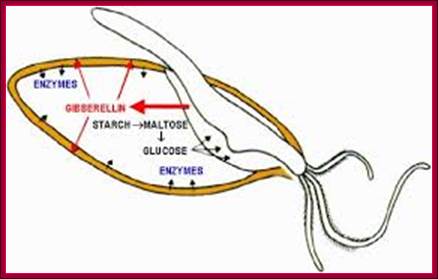
Most of the cereals� (monocotyledons)-embryo is enclosed in an envelope that is separated by scutellum from sugar laden endosperm.� Many cereals endosperm cells contain amylase stuck to the starch grains, but in some system grains of endospermal cells are free from a-amylase. They are synthesized in Aleurone cells in response to GA released from the embryo.� Activation of GA dependent amylase genes in aleurone cells and the release of the enzyme into endosperm, activate the hydrolysis of endospermal starch.� Starch consists of a-1,4 glycosidic linked straight chains, but also contains branched starch (amylopectins) that consists of 1-6 linkages to generate branched starch.� These starch complexes are degraded or hydrolyzed by a-amylases and b-amylases; where a-amylases hydrolyze the linear chains. The b-amylases generate alpha-maltose from the non-reducing ends.� In barley the beta-amylases are already present associated with starch grains, but in rice the b-amylase is synthesized in aleurone cells in response to GA and the same is released into endosperm.� Debranching enzymes hydrolyze a-1,6 bonds and the alpha-glycosidase catalyze the hydrolysis of a-1,4 glucan bonds of irreducible ends of starch and maltose to release glucoses.� Note the enzymes involved in mobilizing starch resources from the endosperm are a-amylase, b-amylase, debranching enzymes and a-glycosidase.
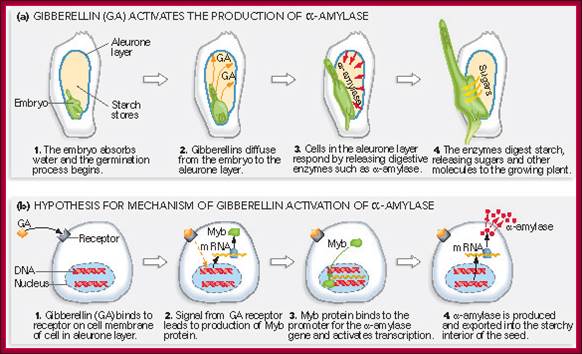
The GA induced pathway in activating alpha amylase gene and releasing the amylase into endosperm.� This operates in rice grains, but in barley alpha amylase is already bound to starch grains in dry grains. http://vinafer.vn/
Hydrolase action and modification in barley; The key physiological events of malting, which determine the quality of the final malt, include rapid and uniform germination, the synthesis of hydrolytic enzymes in the scutellum and aleurone tissues surrounding the endosperm and finally the degradation of endosperm cell walls, described as modification. Gibberellin, which regulates these physiological events, is a plant hormone produced by the germinating embryo. http://www.crc.dk/
�����������
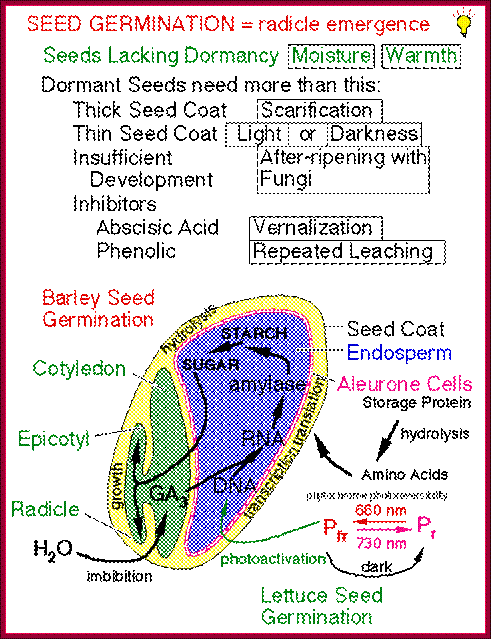
The role of GA in germination of dormant seeds in response to photoactivation, where Pr is converted to PfR, which in turn acts together with GA to generate the expression of amylases in converting starch into utilizable sugars. http://xphd.net/
During seed development and seed germination, two plant hormones act in apposite ways; antagonistic.� During maturity seeds loose water and become more or less dry retaining only small amount of moisture.� In this dry state the seed remains viable. In 6-8% moisture conditions how cells retain their viability is astounding? �In this process of grain maturation ABA plays an important role in dehydration of the seed cells.� At the time of germination of grains, absorption water triggers a process that leads to the awakening of the embryo and it grows out of the seed coat (germination).� This requires GA induced amylase synthesis which in turn hydrolyzes starch to provide free sugars for the developing embryo.
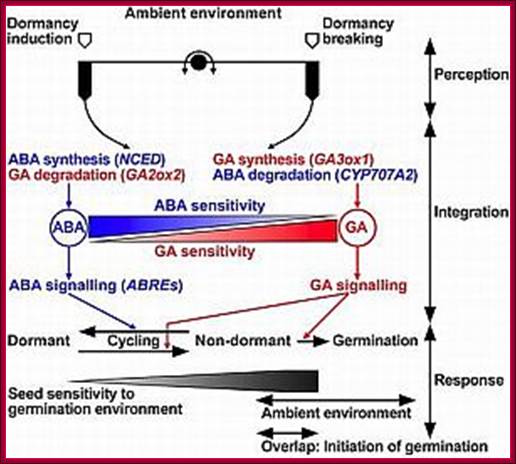
Counter acting hormones in seed maturation and seed germination, while ABA induces dormancy, GA induces germination; http://link.springer.com/
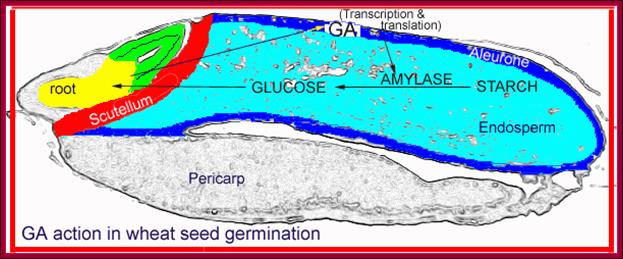
In the case of cereal grains the embryo is separated from the endosperm by a layer of cells called scutellum an epithelial layer of cells through which GA is transported across and sugars are transported in.� And the entire grain is covered by dead cell wall, but underneath of it one finds vibrant aleurone layer of cells where protein grains are stored (nutritious).�
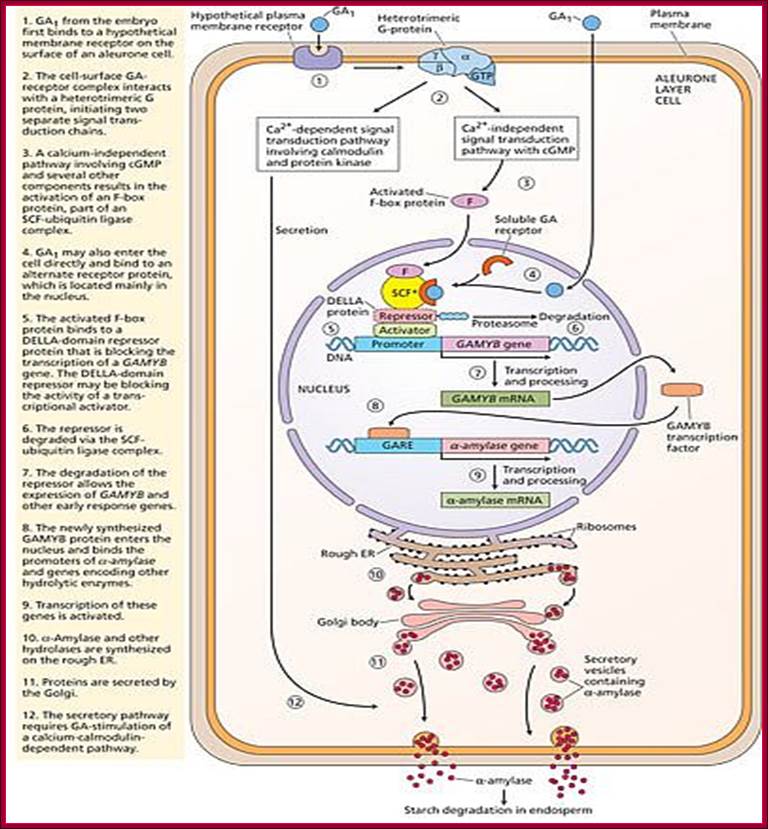
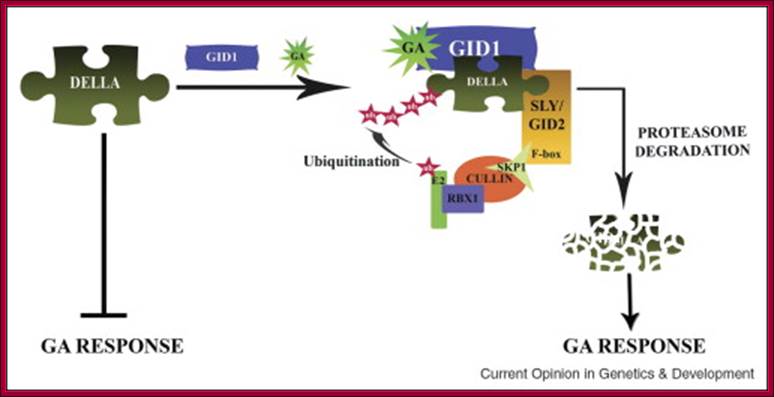
With absorption of water the embryo wakes up and GA1 is released.� It enters the aleurone layer of cells, where it first binds to membrane receptor which is bound or associated with trimeric G-protein complex. The G-protein complex consists of three subunits alpha, beta and gamma subunits. The activated alpha subunit activates Cyclases that produces cGMP similar to that of cAMP.� The c GMP can go through CAM pathway or the GA activated proteins can go through Ca2+ independent pathway that acts in combination with cGMP; it this that activates F-box proteins.� The F-box protein (skp1, Cullin and F-box protein) enters the nucleus where it interacts with the DELLA domain of the repressor (it is also called DELLA) that is bound to the activator (TF) called GID1 (Gibberellin Insensitive Dwarf 1 or 2) bound to GA-MYB promoter.� It is a proteasome mediated degradation of DELLA releases the Transcriptional factor from repression.� This mechanism does not involve GA binding GID proteins.
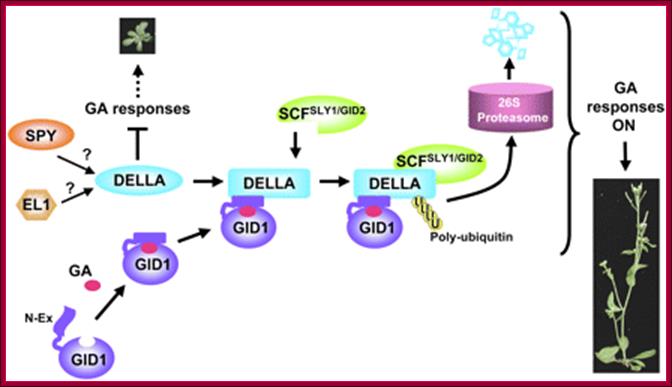
Model of GA signaling in plants. Bioactive GA binding induces a conformational switch in the N-Ex of GID1 for DELLA binding, which in turn promotes a conformational transition in the GRAS domain of the DELLA protein for SCFSLY1/GID2 recognition. DELLA protein will then be polyubiquitinated and degraded via the ubiquitin-proteasome pathway. SPY (an OGT) may activate DELLA by GlcNAc-modification, whereas EL1 (a casein kinase in rice) may phosphorylate and activate DELLA, Tai-ping Sun
Another view is that GA enters the cell by transporter or just diffuses into the cell where it binds to its receptor.� This complex moves into the nucleus and binds to a repressor containing DELLA motif.� The receptor can be GID1 (GA insensitive Dwarf1).� The GID1- GA binds to Transcriptional repressor.� This leads to the recruitments of F-box proteins (SCF-SLY1/GID) that lead to the degradation of DELLA proteins.� Thus, GID1 frees from repressor that is bound to GA response elements called GARE of the MYB gene.� The transcript of the MYB goes through processing and transported into cytoplasm where it gets translated.� The MYB proteins move back into the nucleus using NLS motif.� Now the MYB protein binds to alpha amylase gene promoter and recruits the required cofactors and RNAP complex and activates the transcription of alpha amylase gene.� The translated alpha amylase is released into endosperm.� MYB in animals is procancer protein, but in plants like Arabidopsis there are more than 100 MYBs involved in regulating plant gene expression.
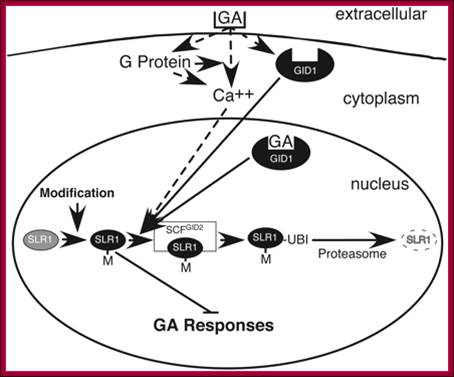
A Model of Rice GA Signaling from the Receptor to SLR1. A previously proposed GA signaling pathway involved GA binding to an extracellular receptor with signal transduction possibly via a heterotrimeric G protein and/or Ca2+ (dashed arrows) triggering interaction of active SLR1 with the SCFGID2 complex and subsequent destruction by the proteasome. Posttranslational modification (M) of SLR1 by phosphorylation or addition of O-GlcNAc activates the repressive activity. The model in which binding of GA to GID1 triggers association of active SLR1 with the SCFGID2 complex (solid arrows) leading to destruction of SLR1 is also shown. The possibility that the extracellular receptor acts through GID1 is indicated by the dotted arrow from this receptor to GID1. Note that GID1 could bind GA in either the cytoplasm or nucleus, Lynn M. Hartwick
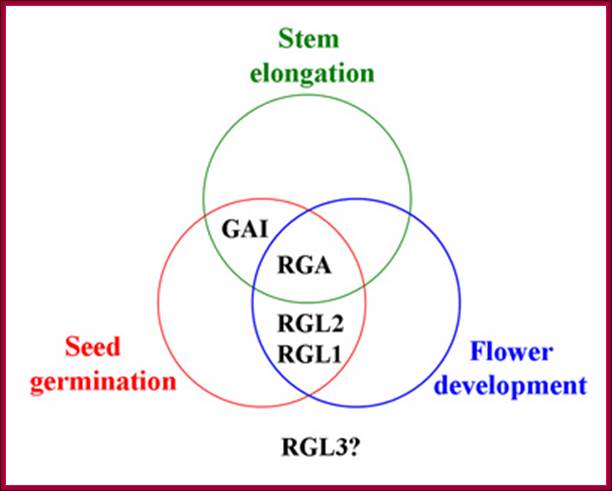
The overlapping functions of Arabidopsis DELLA proteins in control of GA-stimulated stages in plant development. RGA negatively regulates stem elongation, seed germination, and flower development. GAI is involved in stem elongation and seed development. RGL1 and RGL2 are involved in seed germination and flower development. The role of RGL3 is unknown. http://5e.plantphys.net/
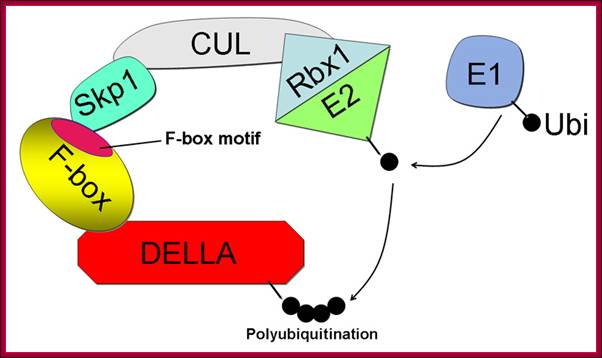
The structure of the SCF E3 ubiquitin ligase complex based on the SCFSKP2 crystal structure (Zheng et al. 2002). The DELLA protein is the target for destruction. The F-box protein SLY1 binds DELLA via its C-terminal domain and interacts with ASK1 through the N-terminal F-box motif. The Cullin (CUL1) backbone binds the ASK1 and RING-finger protein, RBX1. RBX1 binds the E2 ubiquitin conjugating enzyme, the source of ubiquitin. Ubiquitin (Ub) binds E1 (or ubiquitin activating enzyme) via a thioester bond to the E1. Next, the ubiquitin moiety is transferred to the E2, which binds to the SCF E3, which catalyzes transfer of Ub from E2 to the DELLA substrate; GA induces DELLA protein degradation. Components of F-box protein components; http://5e.plantphys.net/
http://vinafer.vn/
According to this model GA1 binds to its cell surface receptors and activates, this leads to the activation G-protein where alpha subunit binding to GTP separates from the complex and activates Cyclase to generate cGMP.� This leads to the activation of F-box proteins that move into the nucleus.� This complex binds to DELLA repressor that is already bound to the activator bound to GA response elements.� The activated F-box proteins ubiquitnate the DELLA proteins and the same is degraded leaving the activator free for the activation of the said MYB (GAMYB) gene.� The MYB transcript is processed and moves into cytoplasm and translated.� The translated MYB proteins move back into the nucleus and bind to GARE elements of alpha �amylase gene and activate the transcription.� The alpha Amylase protein is secreted into endosperm.
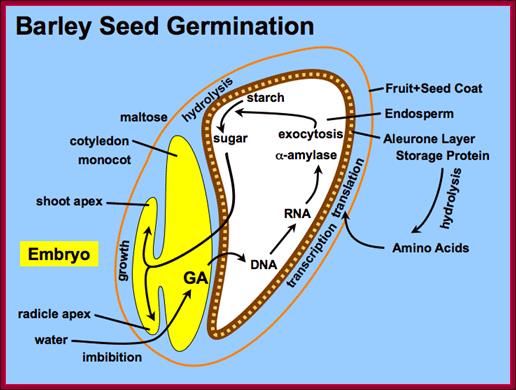
Barley seed endosperm region goes through metabolism to provide needed energy for the germination of the seed. http://vinafer.vn/
Note, in the nucleus there are two GA responsive genes, one is called GA-MYB (similar to mammalian Myeloblastoma protein) and another is alpha amylase gene.� The MYB gene promoter is bound by an activator cum repressor, thus the activator remain repressed and so the gene.� The alpha amylase genes also contain GA responsive elements called GARE (GC box and G box elements). They are also bound by repressor proteins.
Once the activated F-box protein enters the nucleus, the soluble form of GA binds to the GA receptor and binds to SCF complex that is already bound to Repressor-activator complex.� Now the activated F-box protein ubiquitinates the Della domain protein and the same is subjected proteasome degradation.� As the GA-MYB promoter is freed from the repressor, the hitherto remained inactive protein becomes an activator, which is bound to GC box and G-box (GACGTG) elements of amylase gene which recruits co activators and the MYB gene is transcribed.� The transcripts are transported out and the MYB protein produced acts as an TF.� It enters the nucleus and binds to GARE elements found in alpha amylase gene and activates the gene expression.� Now the transcripts enter the cytoplasm and produces alpha amylase. �Translation of the transcripts leads to secretion of the same into endosperm cells.
MYB was first identified in Myeloblastosis virus as TF. Human MYB mRNA is 3414ntd long code for 71kDa protein. There are a large number of such MYBs, they are found in plants and animal systems. In legume, soybean 244R2R3-MYB genes belong to 48 families. They are very important TFs.� One of the MYB mRNA is of ~2120 ntd long and goes through alternative splicing; one of the MYB TF is 72.34kDa. The term derived from its domains, such as 4 imperfect repeats called R0, R1, R2 and R3.� Each of them consists of a 3a helix with second and third contains HtH motifs. �Each repeat may contain 52-53 amino acid residues.� The second and third helices are made up of HtH with three regularly spaced Tryptophan residues, this forms a hydrophobic core.�� The third helix of each core the recognition helix� contacts DNA and intercalate.
MYB induces several hundred genes-some examples, induces CD4 gene, Maize P genes, and has role in GA induced alpha amylase and flowering.
Promoter elements of alpha Amylase gene promoters:
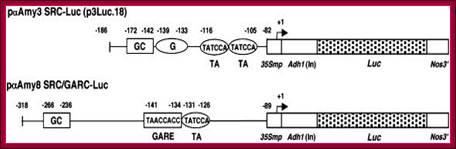
Schemes of Constructs for αAmy3 and αAmy8 Promoter Analysis;
The αAmy3 SRC (�186 to �82 relative to the transcription start site) and αAmy8 SRC/GARC (�318 to �89) were fused upstream of a cauliflower mosaic virus (CaMV) 35S minimal promoter (35Smp)�alcohol dehydrogenase1 (Adh1) intron (In)�Luc�Nos 3′ chimeric gene. Relative positions of cis-acting elements, including GC, G, the GARE, and TA boxes, in promoters are indicated.
http://www.plantcell.org/
In cereals alpha amylase synthesis plays an important role in germination of seedlings.� For alpha amylase genes, such as amylase-3 (a-amy3) and amylase-8 (a-amyl-8) are regulated differentially to sugars by GAs.� The sugar responsive and GA non-responsive amy-3 gene in embryo contains GC box; the G box TATTCCA and TATCCA (TA) sequence are found in the upstream of TATAA and the Start site.� In the case of GA responsive genes, amy-8 in endosperm consists of GC box and GARE and TA box upstream of TATA and start site.� These are controlled by SRC and GARC complex respectively.
�Amylases are processed through endoplasmic reticulum and Golgi bodies and finally secreted into endosperm. The secretion of alpha amylase executed via GA activated Ca-Calmodulin dependent pathway.� The endosperm contains stored starch and the same is degraded so the sugars released enter the embryo and the embryo uses for its growth.
Interactions of Two Transcriptional Repressors and Two Transcriptional Activators in Modulating Gibberellin Signaling in Aleurone Cells:
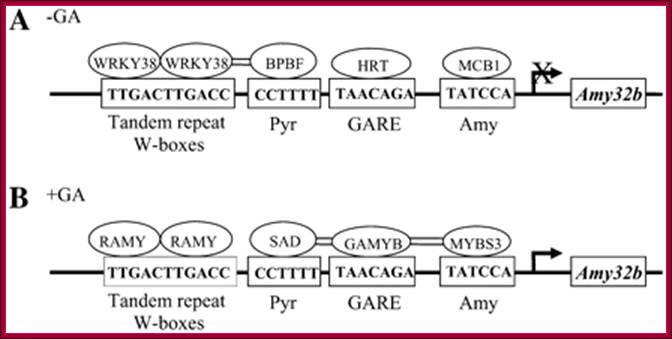
Figure; Hypothetical model for the
control of Amy32b
α-amylase gene expression in aleurone cells: A, Negative
regulators (HvWRKY38, BPBF, HRT, and HvMCB1) bind to their corresponding
cis-acting elements in the absence of GA. B, Positive regulators (RAMY, SAD,
HvGAMYB, and HvMYBS3) bind to corresponding cis-acting elements in the presence
of GA. Double lines between proteins indicate that their physical interactions
have been detected by BiFC. The arrow denotes the transcription start site; X
over the arrow means the transcription of Amy32b
is off or at a very low level in the absence of GA.
http://www.plantphysiol.org
GA on Oriza sativa OsEXP4 Expansin Gene Promoter Elements;
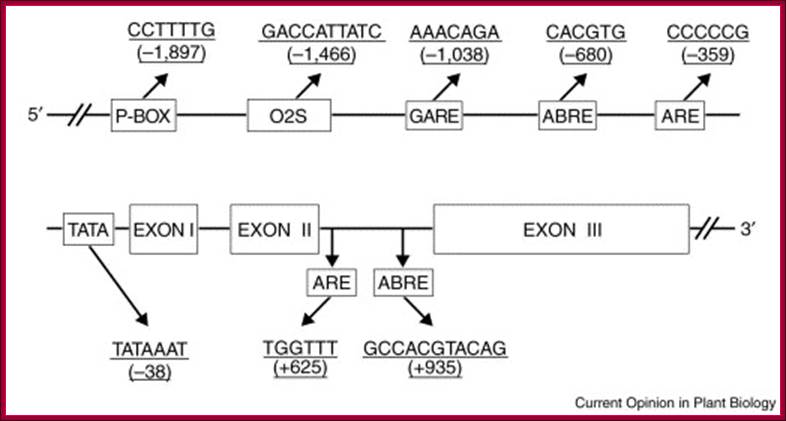
The P-box, O2S, and GARE are cis-acting elements that are involved in GA regulation of gene expression. ABRE and ARE represent abscisic-acid- and anoxia/hypoxia-responsive elements, respectively. These hormone-responsive elements were identified using the PLACE and Plant CARE databases. The numbers in parentheses indicate the distance to the putative transcription initiation site. ABA counteracts the effects of GA.
GAs induce expansin genes. More recently, it has been realized that expansins belong to two protein families, the α-and β-, and that they appear to be involved in regulating, besides cell expansion, a variety of plant processes, including morphogenesis, softening of fruits, and growth of the pollen tube of grasses through the stigma and the style. The Arabidopsis genome contains 26 α-expansin genes and the rice genome at least 26. There are more β-expansin genes in monocots than in dicots, at least 14 in rice and five in Arabidopsis. Expansin genes are differentially regulated by environmental and hormonal signals, and hormonal regulatory elements have been found in their promoter regions.
Cereal amylase Gene Promoter elements;
�O2S�CCTTTT---- TATCCATGCAGTG. 5�-(CCGATAACAAACTCCGG-) 6 time repeat---+1>-
GA induces Flowering:
GA induces many genes, alpha amylase in cereal grains, Expansin mediated hypocotyl elongation and many, among them it also activates floral initiation, especially in short day plants. A potential target for GAMYB in transcriptional regulation of flowering is the LEAFY gene. The LFY promoter of at least two dicots contains a potential MYB-binding motif that is required for normal LFY promoter activity. �In addition, consistent with the role for GAMYB in regulating LFY,� LtLFY is induced after LtGAMYB during the floral transition at the shoot apex of L. temulentum. In most of the plant genes the upstream region extends 300-1000ntds.
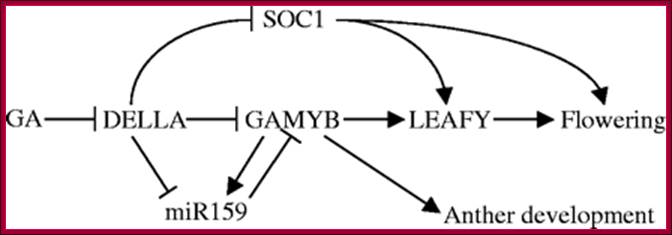
The GA-DELLA signalling system regulates floral development by modulation of miR159/GAMYB and SOC1. . Schematic representation of the regulation of the floral transition in SD photoperiods via the GA-DELLA signalling pathway. GA relieves the DELLA repression of GAMYB (e.g. MYB33) and SOC1 and enhances activity of the downstream floral meristem identity gene LEAFY. This activation is moderated by the GA activation of miR159, a post-transcriptional regulator of GAMYB transcript levels. This GA-dependent homeostatic mechanism provides sensitive regulation of the floral transition in SDs and anther development via the regulation of GAMYB levels.;http://dev.biologists.org
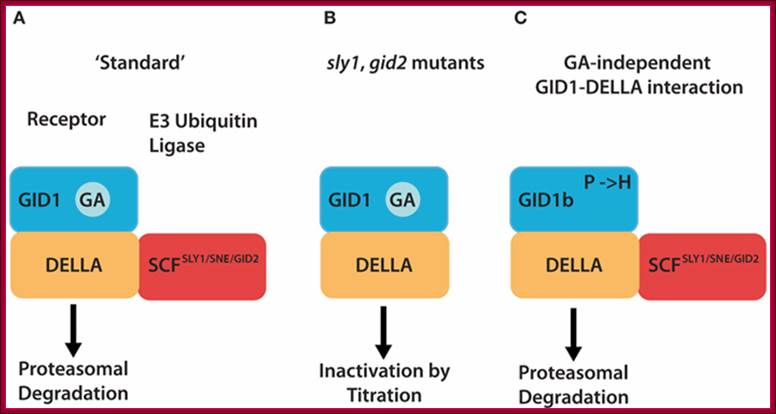
Different mechanism serve to inactivate DELLA repressors of the GA signaling pathway. (A) In the �standard� situation, GA-bound GID1 proteins interact with DELLA repressors and induce their ubiquitylation and degradation via E3 ubiquitin ligases such as Arabidopsis SCFSLY1/SNZ or rice SCFGID2. (B) DELLA ubiquitylation and degradation are defective in E3 ubiquitin ligase mutants such as sly1 or gid2. There, the GA-promoted GID1�DELLA interaction is sufficient to inactivate DELLAs and relieve DELLA-imposed growth restraints. (C) GID1 variants with a substitution of a conserved proline (P) residue can interact with DELLAs in a GA-independent manner and promote GA signaling independent from the hormone. Arabidopsis GID1b is a naturally occurring GID1 protein that has a histidine instead of the proline (P → H). GID1 mutant analyses additionally revealed that P → A or P → S substitutions render GID1 GA-independent. http://journal.frontiersin.org/
PIFs directly activate expression of genes with a role in cell elongation by binding a G-box element in the promoters of these genes.
In the light, active forms of PhyB migrate into the nucleus and target PIFs degradation by the 26S proteasome pathway.
DELLAs repress PIFs transcriptional activity by binding the bHLH DNA-recognition domain of these factors, hence sequestering them into inactive complexes unable to bind DNA. Bioactive GAs induce elongation growth by destabilizing these repressors, which allows accumulation of free PIF4 in the nucleus and activation of PIF4-regulated genes.� It is now reported that light activated otherwise increased temperature causes bHLH TF PIF4 directly binds to promoter elements of Flowering locus T ( FT) gene, which is involved in triggering floral initiation in Arabidopsis thaliana.
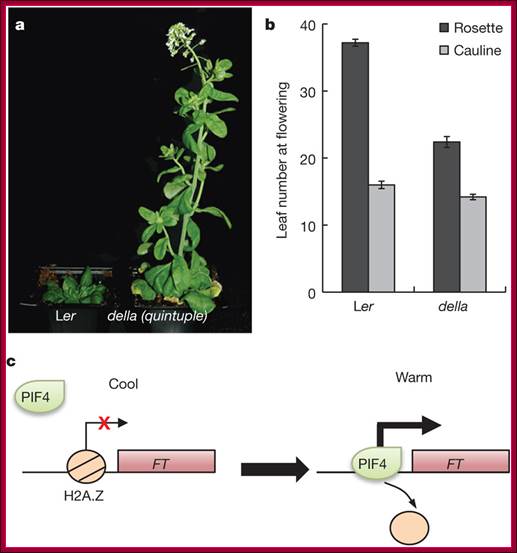
Thermosensory mediated PIF4 activates FT gene; Transcriptional regulation of PIF4 by temperature. Ten-day-old Col-0 seedlings grown at 12, 17, 22 and 27 �C under short photoperiods were analyzed for PIF4 expression by qPCR. Data shown are from three biological replicates. Plant growth and development are strongly affected by small differences in temperature1. Current climate change has already altered global plant phenology and distribution2, 3, and projected increases in temperature pose a significant challenge to agriculture4. Despite the important role of temperature on plant development, the underlying pathways are unknown. It has previously been shown that thermal acceleration of flowering is dependent on the florigen, FLOWERING LOCUS T (FT)5, 6. How this occurs is, however, not understood, because the major pathway known to upregulate FT, the photoperiod pathway, is not required for thermal acceleration of flowering6. Here we demonstrate a direct mechanism by which increasing temperature causes the bHLH transcription factor PHYTOCHROME INTERACTING FACTOR4 (PIF4) to activate FT. Our findings provide a new understanding of how plants control their timing of reproduction in response to temperature. Flowering time is an important trait in crops as well as affecting the life cycles of pollinator species. A molecular understanding of how temperature affects flowering will be important for mitigating the effects of climate change. S. Vinod Kumar, Doris Lucyshyn, Katja E. Jaeger, et al Nicholas P. Harberd ;& Philip A. Wigge;Elizabeth Alvey, http://www.nature.com/
The inductive role of GA on wheat flowering is further supported by the upregulation of GA-biosynthetic genes in the apices of plants transferred from SD to LD, and in photoperiod insensitive and transgenic wheat plants with increased FT transcription under SD. The upregulation of GA-biosynthetic genes was not observed in the leaves of the same genetic stocks. Based on these observations we propose a
model in which FT is up-regulated in the leaves under LD and is then transported to the shoot apical meristem where it simultaneously induces the expression of VRN1 and GA both are required for upregulation of early floral meristem genes SOC1 and LFY for timely development of wheat spike, Stephen Pearce1,, Leonardo S Vanzetti3, 4 and Jorge Dubcovsky. In VRN1 producing wheat plants application of GA (exogenously) accelerates spike development in SD plants.� The presence of both VRN1 and GA induce expression of floral meristem identity genes such as SOC1 and LFY.
Cytokinins:
Cytokinins are essential plant hormones that control cell division and cell differentiation.� Together with Auxin, Cytokinins can reprogram terminally differentiated leaf cells into pluripotent stem cells and support shoot regeneration indefinitely in plant tissue culture.
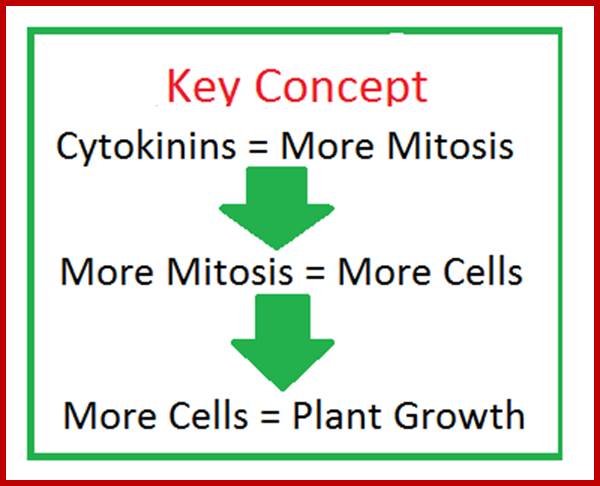
Cytokinins in Plants: Function & Concept; http://study.com
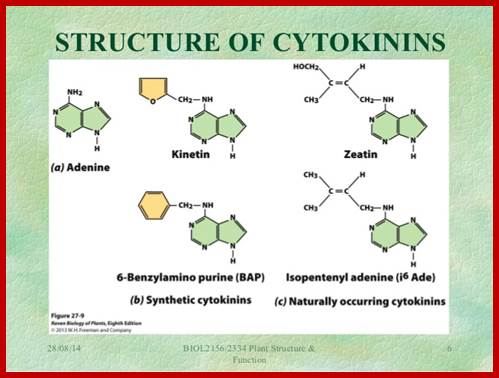
Nitin Mantri,IPA; https://www.slideshare.net
Isopentinyl-adenine ribonucleotide (IPA), Zeatin and Benzyl adenine are nucleotide derivatives.� Cytokinins have a role in activation of cell division, where as Auxin prepare the cell for cell division. Cytokinins execute cell division and they are involved in breaking bud dormancy, delays leaf senescence; in tissue culture it induces shoot regeneration in combination with auxin. Most of the Cytokinins are synthesized in root apical meristems (RAM), and then translocated to other parts via xylem stream. Note xylem and phloem elements are the main conductors and distribution of water, minerals, food resources, organic molecules, hormones and perhaps even signal transducing molecules.
Biosynthesis and Metabolism of Cytokinins:
Cytokinins generally found in higher concentrations in meristematic regions and growing tissues. They are believed to be synthesized in the roots and translocated via the xylem to shoots. Cytokinin biosynthesis happens through the biochemical modification of adenine. The process by which they are synthesized is as follows.
A product of the mevalonate pathway called Isopentenyl pyrophosphate is isomerized. This isomer can then react with adenosine monophosphate with the aid of an enzyme called Isopentenyl AMP synthase. The result is Isopentenyl adenosine-5�-phosphate.� There are 36 forms of synthetic cytokinins.
Some Functions:
- Stimulates cell division.
- Stimulates morphogenesis (shoot initiation/bud formation) in tissue culture.
- Stimulates the growth of lateral buds-release of apical dominance.
- Stimulates leaf expansion resulting from cell enlargement.
- May enhance stomatal opening in some species.
- Promotes the conversion of etioplasts into chloroplasts via stimulation of chlorophyll synthesis.
- Induce parthenocarpy.
- Prevent premature senescence and sex expression.
- Break dormancy,
- High Cytokinin to Auxin ratio causes differentiation of shoots.
� A low ratio of Cytokinin to Auxin causes root formation.
� Intermediate Cytokinin to Auxin ratio causes formation of roots as well as shoots.
� Intermediate Auxin to low Cytokinin causes growth of large amount of callus.
� Cytokinins are plant growth promoting hormones involved in the specification of embryonic cells, maintenance of meristematic cells, shoot formation and development of vasculature.
� Cytokinins have also emerged as a major factor in plant�microbe interactions during nodule organogenesis and pathogenesis.
� Plant-borne cytokinins systemically induce resistance against pathogen infection.
Spatial distribution of cytokinin-related gene expression in Arabidopsis. Data are based on studies of promoter:GUS (Miyawaki et al., 2004) and promoter:GFP (Takei et al., 2004a) fusions of AtIPTs (red), Type A ARRs (violet; D'Agostino et al., 2000; Kiba et al., 2002, 2003; Ferreira and Kieber, 2005; Yokoyama et al., 2007), Type B ARRs (blue; Mason et al., 2004; Tajima et al., 2004), and AtCKXs (green; Werner et al., 2003, 2006). http://jxb.oxfordjournals.org/
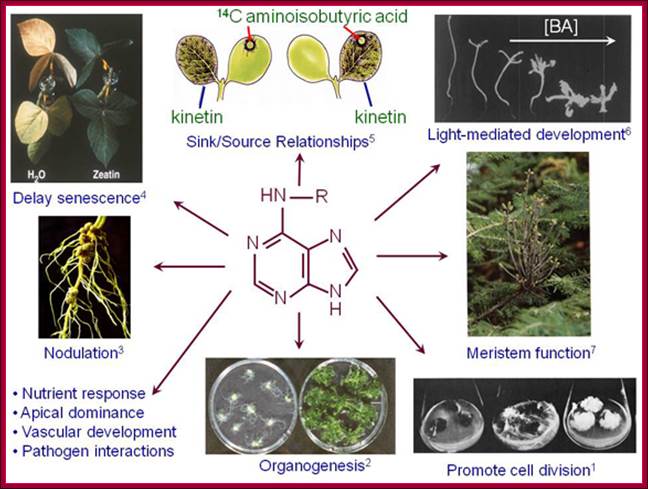
Illustration of some the functions of cytokinins in plant growth and development.
References: 1) Skoog, F. and Miller, C.O. (1957) Chemical regulation of growth and organ formation in plant tissue cultured in vitro. Symp Soc Exp Biol XI: 118�131; 2) Kakimoto T. (1996) CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science 274:982-5; 3) Tirichine, L., Sandal, N., Madsen, L.H., Radutoiu, S., Albrektsen, A.S., Sato, S., Asamizu, E., Tabata, S. and Stougaard, J. (2007) A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315 : 104�107 & Murray, J.D., Karas, B.J., Sato, S., Tabata, S., Amyot, L. and Szczyglowski, K. (2007) A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315: 101�104. 4) 5) Plant Physiology, 4th edition, Taiz and Zeiger, eds.; 6) Chory, J., Reinecke, D, Sim, S, Washburn, T., Brenner, M. (1994) A role for cytokinins in de-etiolation in Arabidopsis. Plant Physiol. 104:339-347. 7) Plant Physiology, 4th edition, Taiz and Zeiger, eds.
Perception and Signal Transduction:
Cytokinins are mostly synthesized in root meristems, and then translocated in Xylem in water-mineral stream to apical meristems and lateral organs. Cytokinins are perceived by membrane resident / anchored receptors.� The receptors are two components system (similar to Bacteria and receptor PHYs) Histidine kinases. They are called by their acronyms such as AHKs (A stands for Arabidopsis).�� There are many of them such as AHK2, AHK3, and AHK4/CRE1/WOL.
These receptors on binding to cytokinins on the outer surface of PM get activated.� This leads to auto phosphorylation on their cytosolic side of their proteins.� This leads to phosphorylation of cytosolic intermediary transferors called AHPs.� They move into the nucleus and activate Cytokinin response regulators (ARR) or transcription activators by phosphorylation.� Activated ARR join TGA3 and bind to their respective gene promoter elements activate specific genes.
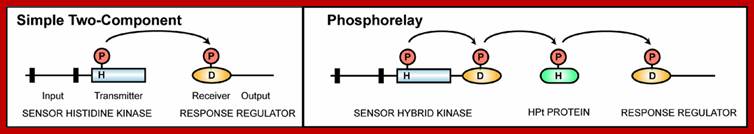
Plant� two component system of signal transduction;
Schematic representation of the two-component and the multistep phosphorelay signalling systems. (A) The basic phosphorelay system uses a single phosphoryl transfer event between a His protein kinase and its cognate response regulator. (B) The multistep His-to-Asp phosphorelay system, in which a His-containing phosphotransfer protein (HPt) serves as a phosphoryl acceptor and donor between the hybrid protein kinase (HK) and the response regulators (RR). The activation of the type-B RRs results in transcriptional activation. Type-A RRs attenuate signalling activity. Transcriptional activation of repressive type-A RRs establishes a negative feedback loop to the pathway.Top fig; Diagram of a simple two-component system compared to a phosphorelay. Histidine kinase domains are indicated by rectangles, receiver domains by ovals, HPt proteins by rounded rectangles, and transmembrane domains by black bars. Sites of phosphorylation upon histidine (H) and aspartic acid (D) residues are indicated.; http://labs.bio.unc.edu.
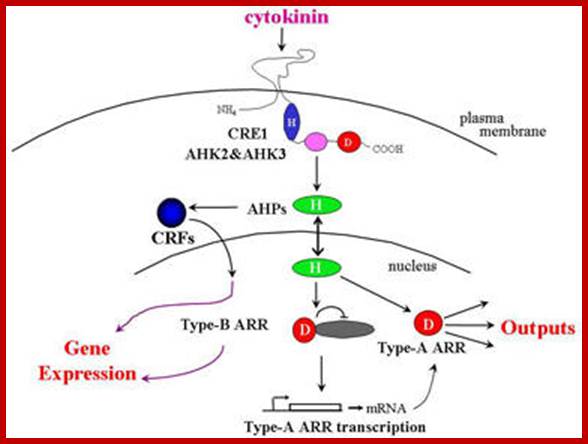
������������������������������������������������������������������������������������� Model for cytokinin signaling. Cytokinin binding to the AHK cytokinin receptors results in autophosphorylation on a His residue. The phosphoryl group is then transferred to an Asp on the fused receiver domain (red) and then is shuttled to the receiver domain of the type-A and type-B ARRs (red) via an AHP intermediate (green). The phosphorylated, and thus activated, type-B ARRs in turn regulate the transcription of numerous genes, including the type-A ARRs. The type-A ARRs negatively regulate the pathway via an unknown mechanism. The CRFs act in parallel with the type-B ARRs to modulate cytokinin-regulated transcription ������ ;http://labs.bio.unc.edu
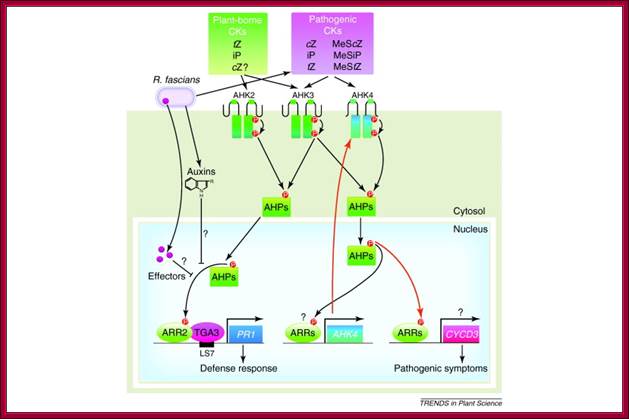
Modulation of plant cytokinin signaling by R. fascians-originated cytokinins. Rhodococcus fascians produces various cytokinin species, including non-degradable 2-methylthio (2-MeS) derivatives of cZ, iP and tZ (MeScZ, MeSiP and MeStZ). Perception of this cytokinin mixture from R. fascians by a cytokinin receptor AHK3 results in abnormal, constitutive activation of cytokinin signaling, including transcriptional induction of AHK4. Accumulation of AHK4 enhances sensitivity to pathogen-derived cytokinins. This results in the de-differentiation of plant cells and leafy gall formation. Although the direct targets of pathogen-derived cytokinins are still elusive, cytokinin-inducible cyclins such as CYCD3 and expansins might trigger de-differentiation as indicated by ectopic induction ofKNAT1. It is also possible that R. fascians secretes effectors or auxin to specifically suppress AHK2 and AHK3 � and ARR2-mediated defense responses. Red arrows indicate hyper-activated signaling cascades by R. fascians-derived cytokinins. Abbreviations: AHP, ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEIN; cZ, cis-zeatin; iP, isopentenyladenine; tZ, trans-zeatin.; http://www.cell.com/trends/plant-science
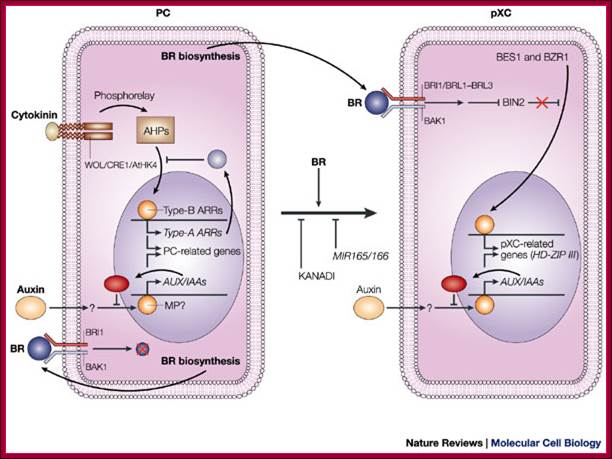
Signals that control plant vascular cell differentiation; In procambial cells (PCs), the coordinated signalling by cytokinin and auxin induces the expression of genes that are involved in the maintenance of procambial activities. The auxin-signalling pathway might involve gene expression of auxin-response factors, such as MONOPTEROS (MP), that also function as transcriptional activators, and their repressors, the AUX/IAA proteins. Cytokinin might be perceived by the WOL/CRE1/AtHK4 cytokinin receptor, which, in turn, transmits an intracellular signal that is mediated by a His�Asp phosphorelay mechanism to PC-related histidine-containing phosphotransfer factors (AHPs) and then to PC-related type-B response regulators (ARRs). The type-B ARRs might function as transcriptional activators of PC-related genes including the genes of their repressors, the type-A ARRs. The presence of repressors in auxin- and cytokinin-signalling pathways might allow cytokinin and auxin signalling to be temporal. Brassinosteroids (BRs in the figure) are biosynthesized actively in PCs and secreted, but brassinosteroids do not work as a signal for the maintenance of procambial activities. Instead, brassinosteroids, in the presence of auxin, might initiate differentiation of procambial cells to precursors of xylem cells (pXCs) after recognition by a receptor, which might be a heterodimer composed of either brassinosteroid-insensitive-1 (BRI1) or one of the BRI1-like proteins (BRL1�BRL3), plus BRI1-associated receptor kinase-1 (BAK1). The Brassino-steroid signal inactivates the negative regulator BIN2 (brassinosteroid-insensitive-2), which allows the unphosphorylated form of bri1-EMS-suppressor-1 (BES1) and brassinazole-resistant-1 (BZR1) to translocate to the nucleus and to promote pXC-related gene expression. Among the most important pXC-related genes that are induced by Brussino-steroids might be the HD-ZIP-III-homeobox gene family, which might function in further xylem cell differentiation. KANADI and the microRNAs MIR165 and MIR166 might suppress differentiation of PCs to pXCs. The suppression by the microRNAs might be caused by the rapid degradation of the HD-ZIP-III gene mRNA through RNAi machinery. Hiroo Fukuda; http://www.nature.com/
Arabidopsis contain several response regulators (ARRs), at least ten, such as ARR4, ARR5, ARR6 and ARR7.� All these and more are Cytokinin inducible.� They contain receiver domain and C-terminal extension.� They act as transcriptional factors.� The C-terminal domain binds to Cytokinin response elements (CRE) of DNA in sequence specific manner- such as 5�-AGATT-3�, 5�GAT(T/C)3�.� Some of the Cytokinin response regulator factors have been identified as AP2/ERF family.
Cytokinins and Riboswitches: In plants, some Riboswitches act as hormone receptors.� Cytokinin binding Riboswitches have been detected in Arabidopsis.
Cytokinins induce several genes, some of them are-
Cytokinin induce genes such as cig1 and cig2.
Cig1 is also induced by ABA,
Cig2 induction independent of ABA,
Induce ribosomal proteins,
CKs activate ARR6 genes,
Activate Sinapis alba MADs genes in SAMs.
Affymetrix- analyses- show at least 30 genes are upregulated and 40 are down regulated; like response regulators and many of them are TFs.
Abscisic Acid:
History of Abscisic Acid; In 1963, abscisic acid was first identified and characterized by Frederick Addicott and his associates. They were studying compounds responsible for the abscission of fruits (cotton). Two compounds were isolated and called Abscisin I and Abscisin II. Abscisin II is presently called abscisic acid (ABA) (Addicott, 1963).
It is a sesqui-terpenoid (15-carbon) which is partially produced via the Mevalonic pathway in chloroplasts and other plastids that is in leaves.
The production of ABA is accentuated by stresses such as water loss and freezing temperatures. It is believed that biosynthesis occurs indirectly through the production of carotenoids. Carotenoids are pigments produced by the chloroplast which have 40 carbons. Breakdown of these carotenoids occurs by the following mechanism:
�Some of the physiological responses known to be associated with Abscisic acid :(Davies, 1995; Mauseth, 1991; Raven, 1992; Salisbury and Ross, 1992).
- Stimulates the closure of stomata (water stress brings about an increase in ABA synthesis).
- Inhibits shoot growth but will not have as much effect on roots or may even promote growth of roots.
- Induces seeds to synthesize storage proteins.
- Inhibits the effect of gibberellins on stimulating de novo synthesis of a-amylase.
- Has some effect on induction and maintenance of dormancy.�
- Induces gene transcription especially for proteinase inhibitors in response to wounding which may explain an apparent role in pathogen defense.
- Developmental processes including seed dormancy, germination, growth, and stomatal movements.
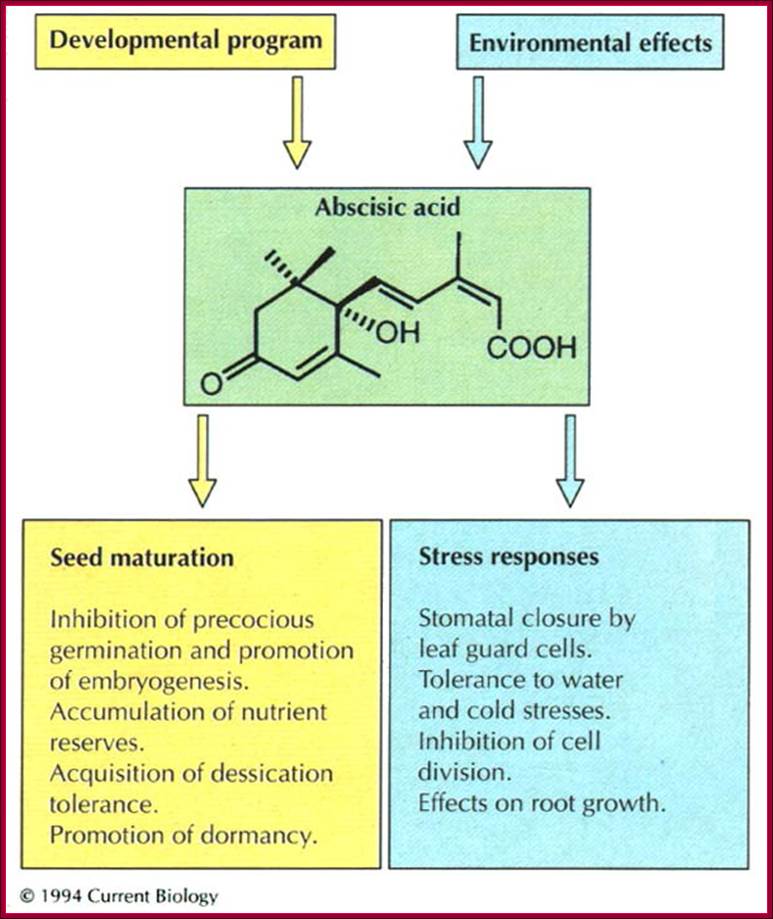
Abscisins are also derived from isoprene units.� It is also called as stress hormone. The site of synthesis is again chloroplasts, where GA synthesis is initiated.� GA and ABA biosynthesis, branch out in the middle of the pathway.� ABAs induces bud dormancy, seed dormancy prevents early germination of seeds, they are also involved in stomatal closing, promotes senescence, accelerates abscission. Phasiec acid is another abscission inducing substance. www.current Biology
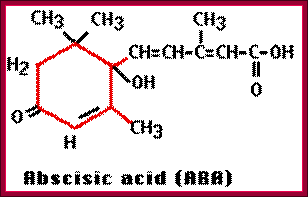 �
�
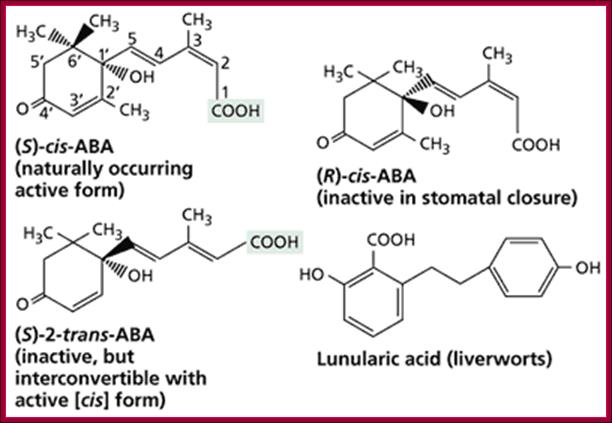
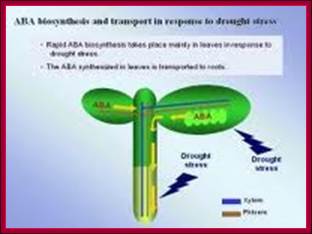
http://www.biol.se.tmu.ac.jp/
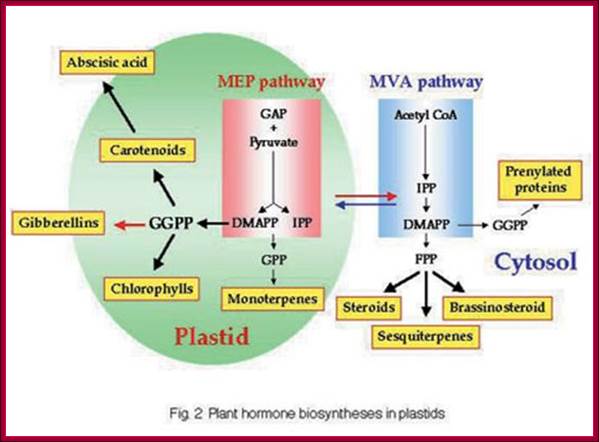
Pathways for the synthesis of GA and some steroids; ;http://vinafer.vn/
Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1mutant :
The phytohormone abscisic acid (ABA) plays important regulatory roles in many plant developmental processes including seed dormancy, germination, growth, and stomatal movements. These physiological responses to ABA are in large part brought about by changes in gene expression. To study genome-wide ABA-responsive gene expression we applied massively parallel signature sequencing (MPSS) to samples from Arabidopsis thaliana wild type (WT) and abi1- 1 mutant seedling. We identified 1354 genes that are either up- or down regulated following ABA treatment of WT seedlings. Among these ABA-responsive genes, many encode signal transduction components. In addition, we identified novel ABA-responsive gene families including those encoding ribosomal proteins and proteins involved in regulated proteolysis. In the ABA-insensitive mutant abi1-1, ABA regulation of about 84.5% and 6.9% of the identified genes was impaired or strongly diminished, respectively; however, 8.6% of the genes remained appropriately regulated. Compared to other methods of gene expression analysis, the high sensitivity and specificity of MPSS allowed us to identify a large number of ABA-responsive genes in WT Arabidopsis thaliana. The database given in our supplementary material (http://jcs.biologists.org/supplemental) provides researchers with the opportunity to rapidly assess whether genes of interest may be regulated by ABA. Regulation of the majority of the genes by ABA was impaired in the ABA-insensitive mutant abi1-1. However, a subset of genes continued to be appropriately regulated by ABA, which suggests the presence of at least two ABA signaling pathways, only one of which is blocked in abi1-1. Key words: ABA, Gene expression, abi1, MPSS, Stress response, Growth; ; Stefan Hoth1, Michele Morgante2, Juan-Pablo Sanchez1, Michael K. Hanafey2, Scott V. Tingey2 and Nam-Hai Chua1,*
ABA Perception and Signal Translocation:
Minimal abscisic acid (ABA) signalling pathway.
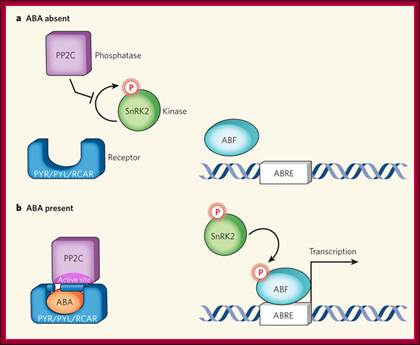
a, In the absence of the plant hormone ABA, the phosphatase PP2C is free to inhibit autophosphorylation of a family of SnRK kinases. b, ABA enables the PYR/PYL/RCAR family of proteins to bind to and sequester PP2C (see Figure 2, overleaf, for mechanistic details). This relieves inhibition on the kinase, which becomes auto-activated and can subsequently phosphorylate and activate downstream transcription factors (ABF) to initiate transcription at ABA-responsive promoter elements (ABREs);
Laura B. Sheard & Ning Zheng;http://www.nature.com/
Mammals contain ∼23 distinct Gα, six different Gβ, and at least 12 Gγ subunits, in addition to numerous GPCRs, making G protein signaling a highly versatile and prominent signaling mechanism.
By contrast, the Arabidopsis genome contains a single Gα gene, GPA1, a single Gβ gene, AGB1, two genes for putative Gγ subunits, AGG1 and AGG2, and, based on sequence similarity as the criterion, a single putative GPCR, GCR1.
ABA has two routes, one is the ABAs synthesized within cells, and the other is it binds to GPCR like protein complex at plasma membrane.� The GPCR like membrane complex consists of 8-10 trans membrane proteins linked to cytosolic G-proteins.� G proteins consist of alpha bound to GDP and other two are beta and gamma subunits.� The binding of ABA at surface receptor activates G-protein complex and the alpha unit dissociates with exchange of GTP to GDP.� The activated alpha subunit� by activating Cyclases produce cAMP, which in turn activates Ca+ release and also the release of Phophatidinyl inositol which also acts on Ca2+ transport.� These events lead to activation ABA response factors which bind to their ABA response element in their respective genes and activate specific genes.� The other ABA that has direct entry across the membrane bind to specific receptor and block ABA mediated gene expression.
ABA inhibits /represses GA inducible Amy32b alpha amylase promoter.� ABA decrease 245 genes and increase gene expression of 213 genes.� The cDNA microarray analysis showed that many ABA-inducible genes were induced after drought- and high-salinity-stress treatments, and that there is more crosstalk between drought and ABA responses than between ABA and cold responses.
Few examples of ABA induced genes:
- ABA induces ABA glucosyl transferase.
- Rice WRKY 51 genes induced by ABA and repressed by GA,
- ABA induces MAP kinase (mitogen activated kinase).
- Plant HVA22 is a unique abscisic acid (ABA)/stress-induced protein first isolated from barley (Hordeum vulgare) aleurone cells. This potein inhibits the GA activated gene expression.
- Barley Hordeum vulgare a dozen proteins are induced by ABA-LEA one-Late embryogenesis abundant; this provides tolerance to desiccation.
- ABA induces Ca2+ increase in the guard cells, and cause closing via cyclic adenosine 5*-diphosphoribose (cADPR). K+ ions exported.
- In aleurone cells at least 12 genes are expressed in response to ABA in about 4-8hrs.
- ABA induced-LEA proteins are late embryonic proteins abundant in higher plant embryos. It has been that LEA genes are a family of genes related to stress.
- ABA also inhibits DNA replication, RNA pol, and rRNA synthesis, tRNA in many plants such as barley, maize, beans and radish.� It also represses the synthesis of GA induced alpha-Amylase.
ABA Gene Response Elements;
They vary from gene to gene and species to species.
Cereals-
HVA1- GCGTG (CE3)�ACGTGGC A2,
HVA22-GCCACGT (A3)-CCACCG CE1
ABA response complex consisting of a G-box, namely, ABRE3 (GCCACGTACA), and a novel coupling element, CE1 (TGCCACCGG), is sufficient for high-level ABA induction.� Replacement of either of these sequences abolishes ABA responsiveness.
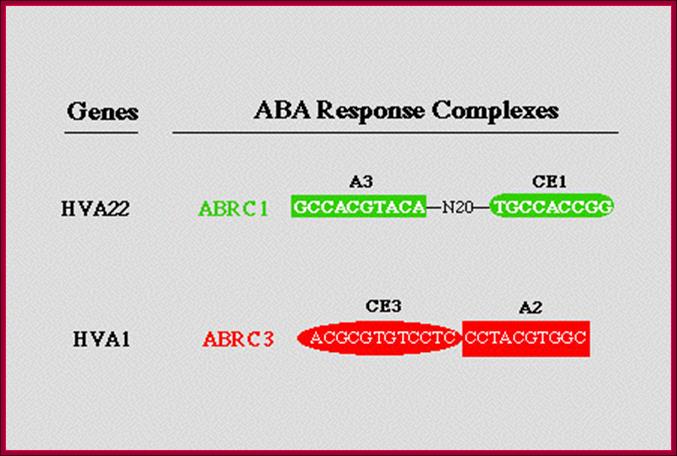
ABA response complex consisting of a G-box, namely, ABRE3 (GCCACGTACA), and a novel coupling element, CE1 (TGCCACCGG), is sufficient for high-level ABA induction, and replacement of either of these sequences abolishes ABA responsiveness. http://www.biology.wustl.edu/
ABA response- In Hordeum vulgare:
--- ABRE3 (GCCACGTACA),-- CE1 (TGCCACCGG) CE v coupling agent-,
----G-box (ABRE2; CGCACGTGTC----CE?
�A3-GCCACGTACA�N20-CE-TGCCACCGG, -CE3-ACGCGTCTCCTC-A2-CCTACGTGGC,
Coumeric acid response-
G box-TGCACGTATA-N7�H box-CCTACCN&CT
UV light �G.box-
TCCACGTGGC�N11 Box 1-GTCCCTCCAACCTAACC,
�Box II- -CTTCACTTGATGTATC�N 14- G box-TGTACGTGGGA,
White light- I box- AACGATAAGATT N16�G box-TCCACGTGGC,
Prediction and Validation of Promoters Involved in the Abscisic Acid Response in Physcomitrella patens:
Sequence Logos of Putative ABA-Responsive TFBS: Gerrit Timmerhausa,b,Sebastian T. Hankea,c, Karl Buchtaa and Stefan A. Rensing
(A) was identified within 10 A. thaliana promoters that contain ABREs. (B) (putative P. patens ABRE) and (C) (putative P. patens CE1) were predicted from the promoters of eight genes that are strongly induced by ABA (P. patens-positive promoters, (D, E) Sequence logos of ABRE and CE1 as published in the Transfac database. Reference numbers are shown on the left side. Logos in (D) refer to ABRE logos and logo (E) to a CE1 logo. The ACGT core motif of the ABREs is highlighted with a gray block.
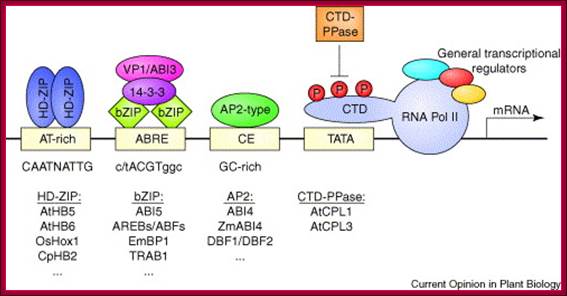
Major cis-regulatory elements and transcription factors of ABA-regulated gene expression. The transcriptional complex consists of RNA-polymerase II (RNA Pol II) in association with general transcriptional regulators including the TATA-box binding protein. The transcriptional machinery is controlled by the binding of regulatory TFs to promoter-specific cis elements. The ABA-regulatory element (ABRE) is contacted by dimeric bZIP TFs, such as ABI5 of Arabidopsis, which associate with the transcriptional regulators ABI3 (maize VIVIPAROUS1 [VP1]) and 14-3-3 protein. The coupling element (CE) is targeted by AP2-type TFs, such as ABI4, which enhance ABA-regulated transcription. The HD-ZIP proteins AtHB5 and AtHB6 of Arabidopsis bind to AT-rich pseudopalindromic sequences. RNA Pol II is regulated by the phosphorylation of its carboxy-terminal domain (CTD). Dephosphorylation by a CTD-specific protein phosphatase (CTD-PPase), such as AtCLP3 of Arabidopsis, downregulates the expression of ABA-inducible genes. Axel Himmelbach, Yi Yang, Erwin Grill
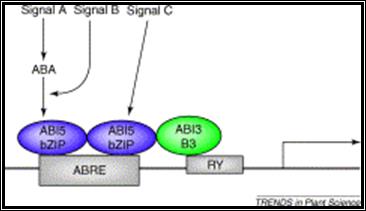

A hypothetical model for the signals that trigger abscisic acid-responsive element (ABRE)-mediated transcription. The black bar represents a promoter of an abscisic acid (ABA)-inducible gene. ABRE and RY are the cis-acting elements involved in ABA-mediated transcription. The corresponding transcription factors ABI3 and ABI5 bind to the cis elements. B3 and bZIP are the DNA-binding domain of ABI3 and ABI5, respectively. Signals A, B and C are the hypothetical signals that can trigger ABRE-mediated transcription. Signals B and C can trigger ABRE-mediated transcription without altering endogenous ABA content.
Minimal abscisic acid (ABA) signalling pathway. a, In the absence of the plant hormone ABA, the phosphatase PP2C is free to inhibit autophosphorylation of a family of SnRK kinases. b, ABA enables the PYR/PYL/RCAR family of proteins to bind to and sequester PP2C (see Figure 2, overleaf, for mechanistic details). This relieves inhibition on the kinase, which becomes auto-activated and can subsequently phosphorylate and activate downstream transcription factors (ABF) to initiate transcription at ABA-responsive promoter elements (ABREs). Laura B. Sheard & Ning Zheng; http://www.nature.com/
ABA and Stomatal Opening and Closure:
Plants are endowed with ventilation in leaves.� Leaves are not only the structure to harvest CO2 and harvest sun light energy to synthesize carbohydrates (global food), which are then transported and distributed and converted to various form of metabolic products.� Leaves contain stomata through which air; H2O, O2 and CO2 diffuse in and out.� Stomata responding to environmental conditions, they open or close.
Stomata structurally consist of two guard cells with thick inner wall and thin outer walls.� The guard cells are connected accessary cells.� Transport between them is aided by Ca2+ and K+ transporters. When conditions are conducive to stomatal opening (e.g., high light intensity and high humidity), a proton pump drives protons (H+) from the guard cells causing negative electrical potential. As a cause of this, K+ ion channels open.� Increase in solute concentration leads to diffusion of water that causes increased turgor pressure and stomata open.
Drought or dry atmosphere induces ABA production which induces stomatal closure. Cells treated with ABA induce closing by H2O2 production.� H2O2 activates Ca2+ channels in ABA induced stomatal closure.
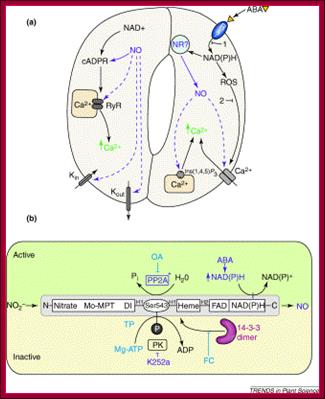
Model of a link between the regulation of nitrate reductase activity and guard cell abscisic acid (ABA)-signalling cascade. Regulation models: (a) Model of ABA-induced stomatal closure. Purple broken arrows indicate nitric oxide (NO) effects previously reported in guard cells; purple unbroken arrows indicate effects of NO reported in different animal signaling pathways, but not yet described in plants. (b) Nitrate reductase (NR) and its cofactors. The gray box represents a NR protein with its eight sequence domains: (i) N-terminal; (ii) molybdene-molybdopterin (Mo-MPT); (iii) dimer interface (DI); (iv) Hinge 1 (H1) where the phosphorylated Ser543 is located; (v) Heme; (vi) Hinge 2 (H2); (vii) flavin adenine dinucleotide (FAD) binding, and (viii) NAD(P)H binding at the C-terminus [47]. The upper green half of the figure contains those components that raise NR activity and okadaic acid (OA), a protein phosphatase type-2 A inhibitor (PP2 A). The lower cream half contains those effectors that inactivate NR and K252a, a protein kinase (PK) inhibitor. Purple components denote elements that positively regulate the ABA-dependent stomatal closure (an asterisk indicates conditional). Blue components negatively regulate ABA-induced stomatal closure. Abbreviations: 1, abi1�1; 2, abi2�1; cADPR, cyclic ADP ribose; FC, fusicoccin; ⊤, inhibition; Ins(1,4,5)P3, inositol (1,4,5)-trisphosphate; K252a, protein kinase inhibitor; NR, nitrate reductase; R, ABA receptor; ROS, reactive oxygen specie; RyR, ryanodine sensitive channels.
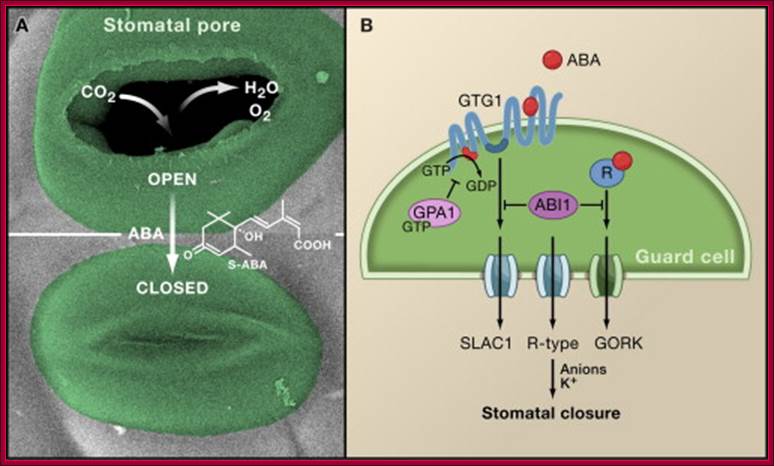
Arabs GTG1 and GTG2 are membrane receptors of ABA; it is GPCR type; IN vitro they bind to ABA; Julian I. Schroeder, June M. Kwak and Gethyn J. Allen
Components and pathways of guard cell ABA signaling:
Guard cell abscisic acid signaling and engineering drought hardiness in plants;
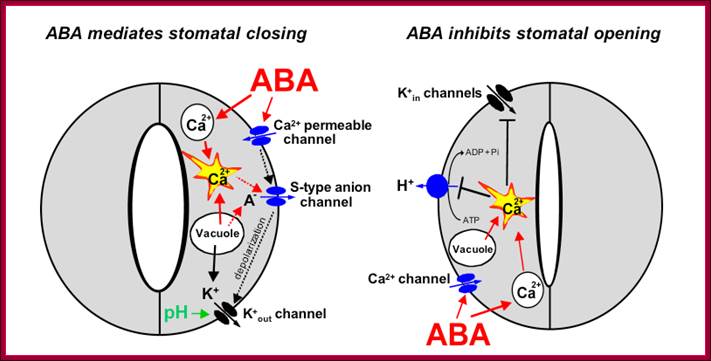
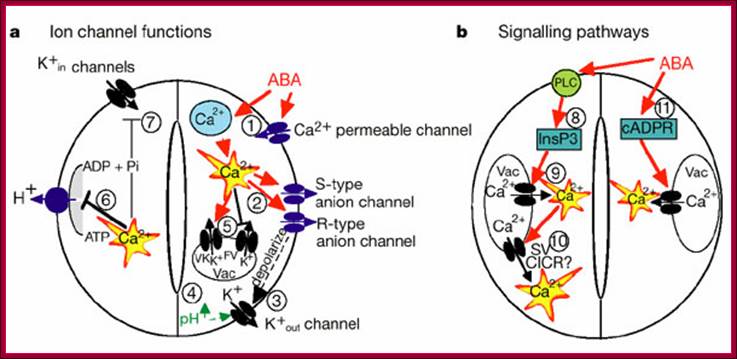
a, ABA is detected by as yet unidentified receptors (right guard cell) and induces cytosolic Ca2+ elevations (1) through extracellular Ca2+ influx and release from intracellular stores (for reviews see refs 2, 8, 9). (2) [Ca2+]cyt elevations activate two types of anion channel that mediate anion release from guard cells, slow-activating sustained (S-type) or rapid transient (R-type) anion channels3, 8. (3) Anion efflux causes depolarization, which activates outward-rectifying K+ (K+out) channels and results in K+ efflux from guard cells2, 3. (4) ABA causes an alkalization of the guard cell cytosol, which enhances K+out channel activity44. Overall, the long-term efflux of both anions and K+ from guard cells contributes to the loss of guard cell turgor, leading to stomatal closing3. Over 90% of the ions released from the cell during stomatal closing must be first released from vacuoles into the cytosol. (5) At the vacuole, [Ca2+]cyt elevation activates vacuolar K+ (VK) channels, which are thought to mediate Ca2+-induced K+ release from the vacuole8. In addition, fast vacuolar (FV) channels can mediate K+ efflux from guard cell vacuoles at resting [Ca2+]cyt45. ABA also inhibits ion uptake, which is required for stomatal opening (left guard cell). (6) [Ca2+]cyt elevations inhibit the electrogenic plasma membrane proton-extruding H+-ATPases46 and K+ uptake (K+in) channels2, 3, 44 (7). Initiation of ion efflux (1�5, right guard cell) and inhibition of stomatal opening processes (6, 7, left guard cell) provide a mechanistic basis for ABA-induced stomatal closing8.
b, Ca2+-releasing second messengers that mediate ABA-induced stomatal closing. Second messengers have been implicated in ABA signalling in guard cells, including InsP3 and cADPR. (8) Biochemical evidence indicates that ABA stimulates InsP3 production in guard cells2. (9) InsP3 can release Ca2+ from intracellular stores, inhibit K+in channels and cause stomatal closure (10) [Ca2+] cyt elevations may be amplified by a Ca2+-induced Ca2+ release (CICR) mechanism from the vacuole through activation of the Ca2+-dependent slow vacuolar (SV) channel8, 45. Data questioning this SV model for CICR47 and recent other data supporting it48 indicate the potential for molecular genetic analyses. (11) ABA stimulates the production of cADPR in plant cells49, which also increases [Ca2+]cyt in guard cells20, 45 and promotes stomatal closing. http://www.nature.com/
LONG DISTANCE SIGNALLING BETWEEN THE GUARD CELLS AND THE PHLOEM OF THE MODEL PLANT � ARABIDOPSIS THALIANA;
�The cell that is doing the signaling is the guard cells of Arabidopsis which are specialized cells found in a pair located on the epidermis of the leaves of plants. These cells aid in regulation of transpiration. They perceive stimuli from light, carbon dioxide, drought, temperature and plant hormones to trigger a cellular response. The cell that will be receiving the signal from some initiation created by the guard cells is the phloem. It is tissues in plants that conduct food made in the leaves to the roots and other parts of the plant. It comprises of various specialized cells called the sieve tubes, companion cells, phloem fibers and phloem parenchyma. http://biochemunrated.wordpress.com/ cells.
The long distance communication between the guard cells and the phloem against pathogen attack and herbivore consumption is initiated due to the accumulation of thioglucoside glucohydrolase 1 (TGG1). This is one of two known functional myrosinase enzymes in Arabidopsis. The enzyme catalyzes the hydrolysis of glucosinolates into compounds such as isothiocyanates, thiocyanates, and nitriles that are toxic to various microbes, insects and herbivores. The glucosinolates are organic compounds that contain sulfur and nitrogen and occur as a secondary metabolite in the model plant Arabidopsis. http://biochemunrated.wordpress.com/
Ethylene:
This is the only plant hormone that exists in gaseous state. Its synthesis is enhanced by auxin; it is involved in epinasty and often induces apical dominance.� The most dramatic effects such as Triple response characterized by inhibition of hypocotyls and root elongation, thickening of hypocotyl and exaggerated apical hook.� Another effect called �Climacteric Effect� during fruit ripening.
Ethylene H2C=CH2
Ethylene Synthesis:
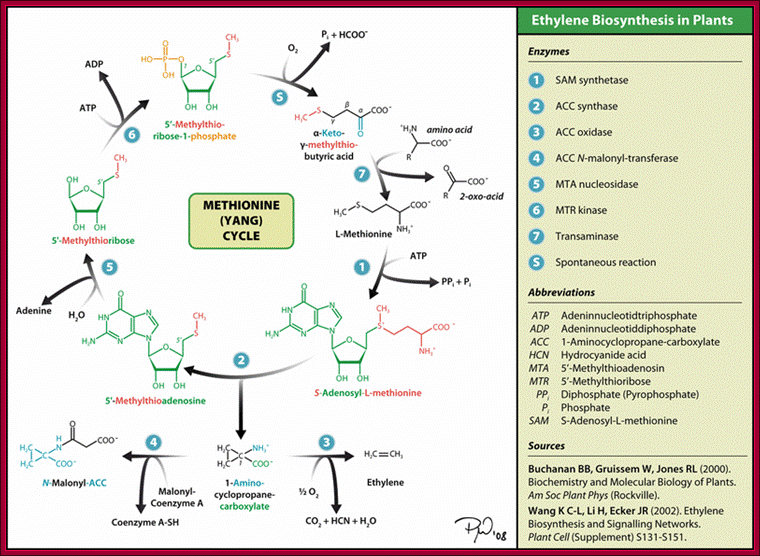 The diagram shows steps involved and
the enzymes involved; http://en.wikipedia.org/
The diagram shows steps involved and
the enzymes involved; http://en.wikipedia.org/
Plant Responses to Ethylene:
- Seedling triple response, thickening and shortening of hypocotyl with pronounced apical hook.
- In pollination, when the pollen reaches the stigma, the precursor of the ethylene, ACC, is secreted to the petal, the ACC releases ethylene with ACC oxidase.
- Stimulates leaf and flower senescence
- Stimulates senescence of mature xylem cells in preparation for plant use
- Induces leaf abscission
- Induces seed germination
- Induces root hair growth � increasing the efficiency of water and mineral absorption
- Induces the growth of adventitious roots during flooding
- Stimulates epinasty � leaf petiole grows out, leaf hangs down and curls into itself
- Stimulates fruit ripening
- Induces a climacteric rise in respiration in some fruit which causes a release of additional ethylene.
- Affects gravitropism
- Stimulates nutational bending
- Inhibits stem growth and stimulates stem and cell broadening and lateral branch growth outside of seedling stage (see Hyponastic response)
- Interference with Auxin transport (with high Auxin concentrations)
- Inhibits shoot growth and stomatal closing except in some water plants or habitually flooded ones such as some rice varieties, where the opposite occurs (conserving CO2 and O2),
- Induces flowering in pineapples,
- It slows stem elongation, Thickens stem (stronger),
- Induces stem curvature- stem starts growing horizontal to soil,
- Fruit ripening,
- Stimulates the release of dormancy.
- Stimulates shoot and root growth and differentiation (triple response)
- May have a role in adventitious root formation.
- Stimulates leaf and fruit abscission.
- Stimulates Bromeliad flower induction.
- Induction of femaleness in dioecious flowers.
- Stimulates flower opening.
- Stimulates flower and leaf senescence.
- Stimulates fruit ripening.
- Triple response-( Stunting of growth, Twisting of plants, Abnormal thickening of stems (the triple response).
- Stimulates the release of dormancy.
- Stimulates shoot and root growth and differentiation (triple response)
- May have a role in adventitious root formation.
- Stimulates leaf and fruit abscission.
- Stimulates Bromeliad flower induction.
- Induction of femaleness in dioecious flowers.
- Stimulates flower opening.
- Stimulates flower and leaf senescence.
- Stimulates fruit ripening.
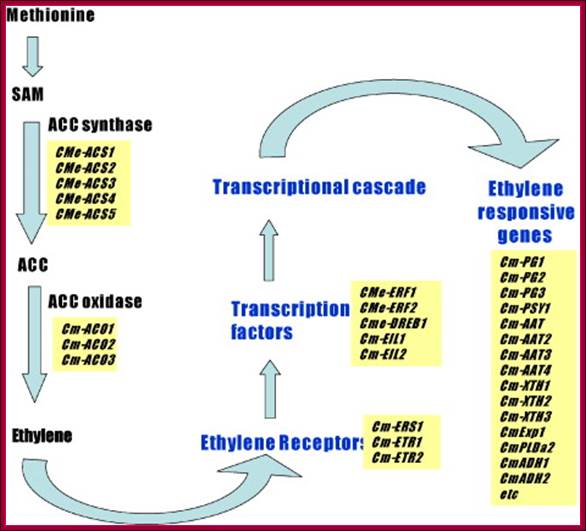
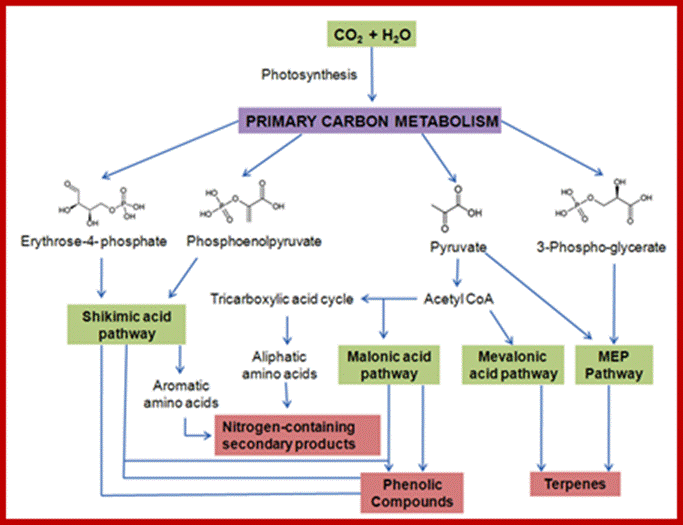
Plants using CO2 and sunlight and water produce first Carbohydrates and a variety of metabolites required for the plant;
Schematic representation of key events during ethylene biosynthesis, perception and signal transduction, finally leading to the regulation of ethylene-responsive genes in melon fruit. Ethylene is produced from methionine, which is converted to S-adenosyl-methionine (SAM). SAM is then converted to 1-aminocylopropane-1-carboxylic acid (ACC) by either of the five ACC synthases. ACC is then converted to ethylene by either of the three ACC oxidases. The ethylene is then perceived by a family of ethylene binding receptors. Upon ethylene binding, these receptors are most likely inactivated and in turn represses the yet to be identified downstream components thus activating the ethylene signaling pathway. The downstream components regulate among others the transcription of several members of the family of transcription factors which triggers a transcriptional cascade and as a result, the transcription of numerous ethylene-responsive genes are regulated. www. nptel.ac.in
Ethylene Signaling and Transduction:
Ethylene is produced in nature as well as in plants itself. Ethylene is perceived by plants by their cell surface membrane bound receptors.� It is also perceived within its cytoplasm. Ethylene is produced in gaseous form, but the gaseous form cannot bind to plant receptors, so the gas complexes with copper ions.� This metal-ethylene complex binds to the receptor ETR1.� Copper ions are bound to the receptors at active site via the Cys65 and His69 residues. 1-methylcyclopropene (1-MCP) has emerged as a viable inhibitor and has been implemented commercially. Solutions of 1-MCP will extend the shelf life of bananas for an additional 12 days
Despite its simple two-carbon structure, the olefin ethylene is a potent modulator of plant growth and development (Ecker, 1995). The plant hormone ethylene is involved in many aspects of the plant life cycle, including seed germination, root hair development, root nodulation, flower senescence, abscission, and fruit ripening.
The production of ethylene is tightly regulated by internal signals during development and in response to environmental stimuli from biotic (e.g., pathogen attack) and abiotic stresses, such as wounding, hypoxia, ozone, chilling, or freezing.
In Arabidopsis there ~five ethylene receptors, ETR1, ETR2, EIN4, ERS1, and ERS2. �ETR1 and ERS1 contain three transmembrane domains and a conserved histidine kinase domain, and have been shown to function as homodimer. Only ETR1, ETR2, and EIN4 have receiver domains at their C termini.� ETR2, EIN4, and ERS2 have four membrane-spanning regions and a degenerate Histidine Kinase domain. E2H2 binds at N terminal end with Cu2+ bound to Cys sites.
ETR receptor 79 kDa exist in dimeric transmembrane proteins.
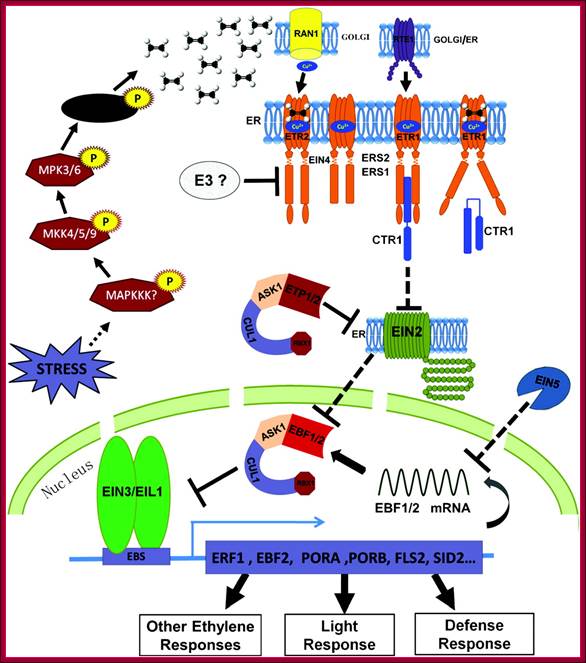
A Schematic Model for Ethylene Signal Transduction and the MAPK Pathway in Ethylene Biosynthesis.
Ethylene gas is perceived by the ER-integrated receptor proteins including ETR1, ETR2, ERS1, ERS2, and EIN4 (Bleecker et al., 1988; Hua and Meyerowitz, 1998; Sakai et al., 1998; Voet-van-Vormizeele and Groth, 2008). A Golgi-localized protein RAN1 (RESPONSIVE-TO-ANTAGONIST 1) is a P-type ATPase copper transporter that delivers the copper ion to the receptors to facilitate ethylene binding (Woeste and Kieber, 2000). RTE1 (REVERSION-TO-ETHYLENE SENSITIVITY 1), another membrane-located protein, promotes the transition of ETR1 from active to inactive state likely through modulating the action of ETR1 N-terminus (Dong et al., 2008; Resnick et al., 2008). In normal growth conditions in which the ethylene level is low, the unoccupied receptors remain in the active state and associate with CTR1, which, in turn, represses the downstream signaling pathway. When plants encounter stress conditions, the MAPK cascade composed of MKK4/5/9 and MPK3/6 can be activated, which then phosphorylates ACS2/6. The phosphorylated ACS2/6 become stabilized and consequently enhance the production of ethylene (Liu and Zhang, 2004). Upon binding by ethylene, the receptor complexes disassociate, and CTR1 released from ER membrane is somehow inactivated (Kieber et al., 1993; Clark et al., 1998; Huang et al., 2003). Therefore, the downstream ethylene signaling pathway including EIN2 is de-repressed (Alonso et al., 1999; Bisson et al., 2009). EIN2 is a short half-life protein targeted by SCFETP1/2 for degradation. Ethylene promotes the accumulation of EIN2 probably by down-regulating the level of ETP1/2 protein through an unknown mechanism (Qiao et al., 2009). In the nucleus, two transcription factors (EIN3 and EIL1) are both necessary and sufficient for the activation of ethylene-regulated gene expression and diverse responses (Chao et al., 1997; Solano et al., 1998; Alonso et al., 2003). EIN3 and EIL1 are also short-lived proteins that are targeted by SCFEBF1/2 for degradation (Guo and Ecker, 2003; Potuschak et al., 2003; Gagne et al., 2004). The ethylene signal is transmitted via the action of EIN2 to stabilize EIN3/EIL1, probably by promoting the proteasomal degradation of EBF1/2 proteins (An et al., 2010). EBF2 is a direct target gene of EIN3, which activates EBF2 transcription to form a negative feedback loop (Konishi and Yanagisawa, 2008). EBF1/2 mRNAs are subjected to negative regulation mediated by a 5'→3' exoribonuclease EIN5 (XRN4) (Olmedo et al., 2006; Potuschak et al., 2006). EIN3/EIL1 also directly regulate the expression of a diverse array of genes including ERF1 (ETHYLENE RESPONSE FACTOR 1), PORA, PORB, FLS2, and SID2, which initiate various interplays between ethylene and other signals, such as light and innate immunity (Chen et al., 2009; Zhong et al., 2009; Boutrot et al., 2010).
;http://mplant.oxfordjournals.org/
There are five ethylene receptors in Arabidopsis, ETR1, ETR2, EIN4, ERS1, and ERS2. ETR1 and ERS1 contain three transmembrane domains and a conserved Histidine kinase domain, and have been shown to function as homodimer. ETR2, EIN4, and ERS2 have four membrane-spanning regions and a degenerate Histidine kinase domain. Only ETR1, ETR2, and EIN4 have receiver domains at their C termini. Ethylene binding occurs at the N-terminal transmembrane domain of the receptors, and a copper co-factor is required for the binding. RAN1, a copper transporter, is involved in delivery of copper to the ethylene receptor. In the absence of an ethylene signal, ethylene receptors activate a Raf-like kinase, CTR1, and CTR1 in turn negatively regulates the downstream ethylene response pathway, possibly through a MAP-kinase cascade. Binding of ethylene inactivates the receptors, resulting in deactivation of CTR1, which allows EIN2 to function as a positive regulator of the ethylene pathway. EIN2 contains the N-terminal hydrophobic domain similar to the N-ramp metal transporter proteins and the novel hydrophilic C terminus. EIN2 positively signals downstream to the EIN3 family of transcription factors located in the nucleus. EIN3 binds to the promoter of ERF1 gene and activates its transcription in an ethylene-dependent manner. Transcription factors ERF1 and other EREBPs can interact with the GCC box in the promoter of target genes and activate downstream ethylene responses.
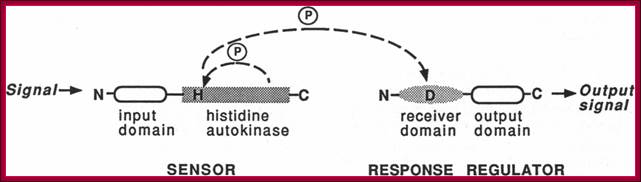
Ethylene binding occurs at the N-terminal transmembrane domain of the receptors, and a copper co-factor is required for the binding. RAN1, a copper transporter, is involved in delivery of copper to the ethylene receptor
When Ethylene binds to copper bound dimeric transmembrane receptor, the receptor gets activated and by auto phosphorylation Histidine is phosphorylated by Histidine kinase.� The receptors are like bacterial two component systems.� Binding of ethylene inactivates CTR1 (constitutive-triple-response1), which allows EIN2 to function as a positive regulator. The CTR1 product acts downstream of ETR1, ETR2, ERS1, ERS2 and EIN4 in the ethylene signal transduction pathway and corresponds to a negative regulator of other cascade components identified in Arabidopsis, including the ethylene-insensitive genes (EIN2-transporter like regulator and EIN3- F-box proteins) and the ethylene-response-factor (ERF1) (Giovannoni, 2004; Stepanova and Alonso, 2005).
All ethylene receptors have a sensor domain subdivided into transmembrane domain and a GAF domain (cGMP-Phosphodiesterase, and Adenylate Cyclase and TF) and Histidine kinase domain and a response domain.
In the process of Ethylene response first EIN3 enters the ncleus and binds to PERE elements and activate transcription of ERFs.�� There are at least 60 ERFs in Arabidopsis. The downstream Ethylene Response Regulator factors called ERFs, once activated by phosphorylation, bind to specific ERE domains and activate the genes required for certain ethylene functions such as fruit ripening and others.
Ethylene induced genes �some examples- chitinases, beta-1,3-glucanase,
Ethylene Response elements- identified as GCC box (AGCCGCC),
ERF1 to ERF4 bind to these response elements and initiate transcription.
ACG Box-GCCbox-
--AGCCCGCC (ACG box) ----------CATAGT----
GCC box-AGCCGCC, ------TATATAAA-----+1
TERF1 binds to these elements
Core sequence -35 to +35 A prototype;
A prototype 13-bp TATA-box sequence, TCACTATATATAG,
ABA ERE- TCCATGCATGCAC
Light-ETR-GGATTCAAGGGGCATGTATCTTGAATCC
CATGGGCGCGG
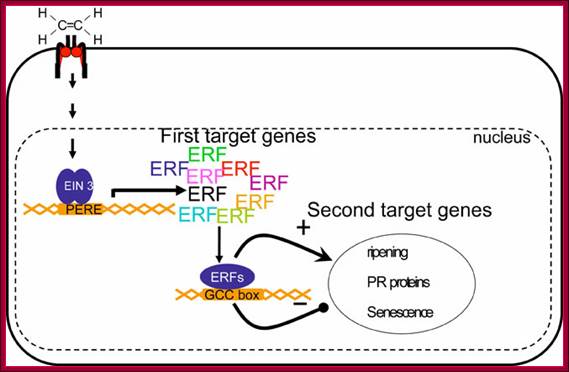
ERFs are the last actors of ethylene transduction pathway. ERFs diversity (more than 60 on Arabidopsis plant model) may explain the diversity of response to ethylene.
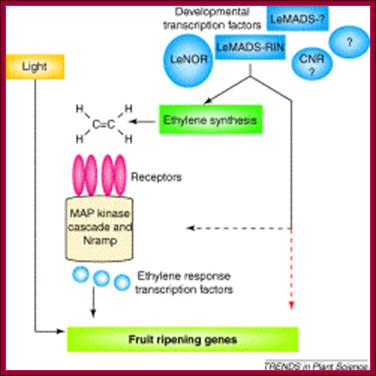
Model for regulation of climacteric ripening via coordinated signaling pathways: Transcription factors including LeMADS-RIN, LeNOR, likely additional MADS-box proteins, CNR and factors remaining to be discovered (?) represent the developmental signaling system that initiates ripening in climacteric fruit. Some components, such as those homologous to LeMADS-RIN can be used in non-climacteric species as well. The developmental signaling system regulates ethylene synthesis that is itself autocatalytic, in addition to non-ethylene-mediated ripening responses (represented by the red broken arrow). Light influences ripening, at least in tomato, only in relation to carotenoid accumulation and through activity of the DET1 (hp2) and DDB1 (hp1) gene products.
Fruit Ripening Process:
Fruit ripening process and execution depends upon the fruits classified into Climacteric fruits such as (e.g tomato, Apple, Melon, Banana, Mango etc) and non-Climacteric fruits (orange, mosambi, grape fruit, litchi, watermelon etc).
Process starts with small changes in the color of the fruit skin and hardness of the fruit gradually change.� During climacteric stage i.e. at the peak of respiration, there is a burst of Ethylene production.� Changes in fruit green to red (chloroplasts become chromoplasts with the synthesis of carotenoids), fruits gradually become soft with cellulose degradation, and then fruit becomes soft with conversion starch into sugar with flavor. Poly galcturonase enzyme degrades middle walls.� As the fruit ripens abscission at stalk takes place leading to the fall of fruits.
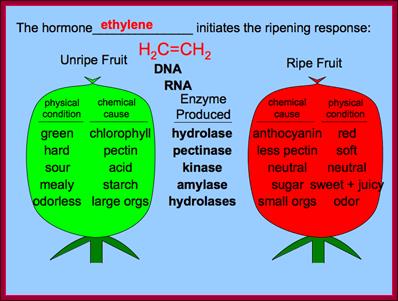
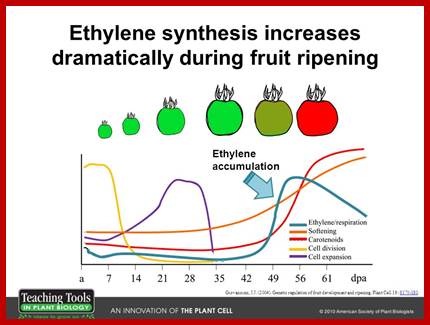
http://slideplayer.com
Climacteric fruits are:
|
*Mango |
*Banana |
*Papaya |
|
*Guava |
*Sapota |
*Kiwi |
|
*Fig |
*Apple |
*Passion fruit |
|
*Apricot |
*Plum |
*Pear |
Non-climacteric fruits are:
|
*Orange |
*Mousambi |
*Kinnow |
|
*Grapefruit |
*Grapes |
*Pomegranate |
|
*Lichi |
*Watermelon |
*Cherry |
|
*Raspberry |
*Blackberry |
*Strawberry |
Flowering Hormones:
The elusive compound or compounds, that are responsible for initiating flowers, are often called Florigen, the term coined long long ago.� It is only now scientists were able to understand certain proteins as transcription factors are involved in transforming SAM apical meristem into floral meristem.� All floral structures are transformed apical meristem, nodes and leaves.�� In flowers leaves assume the structure of sepals, petals, stamens and ovary. Many plants when they grow to a particular stage, they induce flowering irrespective of the environ conditions but some plants have to go through certain period of exposure to light, or to cold to flower.� In this process sugar, light, cold, GA play important roles in transforming SAM (stem apical meristem) into Floral meristem (FM).
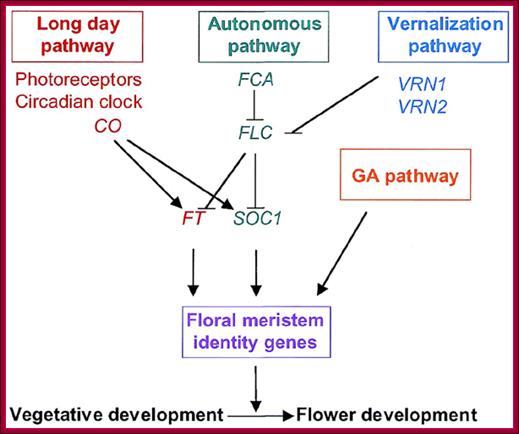
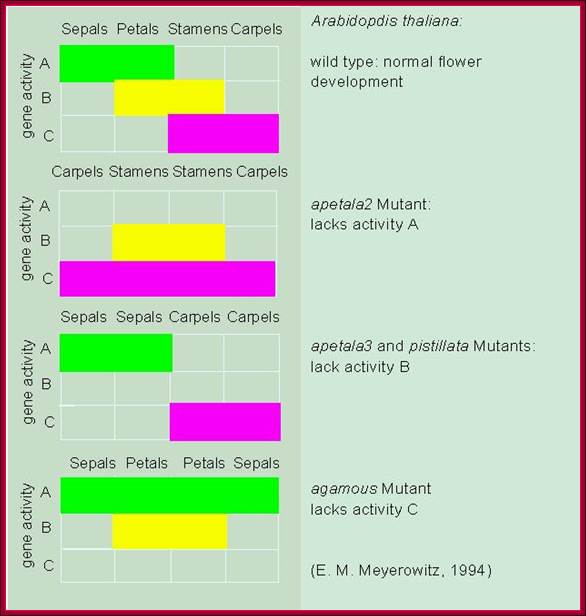
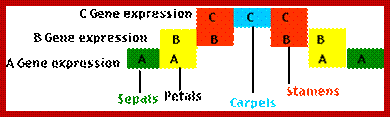
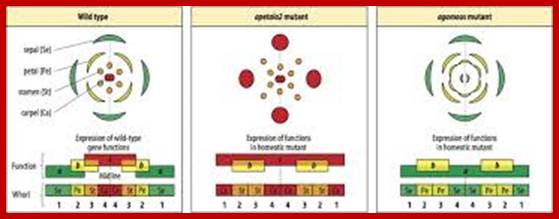
�������������� http://www.mun.ca ; Dr. Brian E. Staveley Combinatorial action of A, B and C modular genes
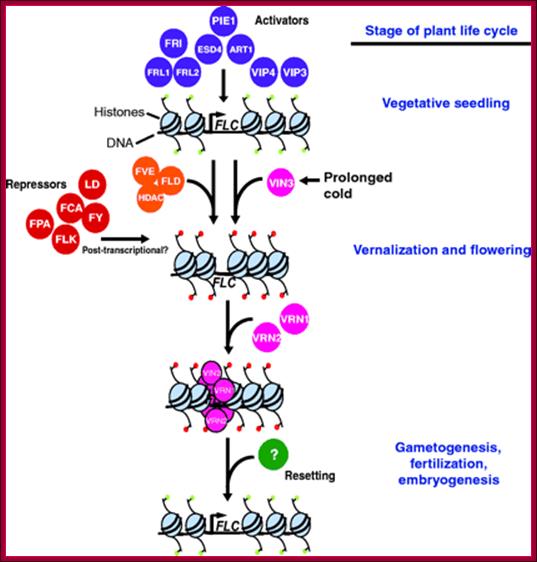
Model for the regulation of the floral repressor FLC throughout theArabidopsis life cycle. During seedling growth, a group of genes encode proteins that function as activators ofFLC expression (shown in purple); these genes include FRI, FRL1, FRL2, ESD4, ART1, PIE1, VIP3 and VIP4. These proteins may maintain FLC chromatin in an active state (indicated by an open structure and the presence of active histone tail modifications shown in green). The autonomous pathway functions antagonistically to the activators to repress FLC expression. The RNA-binding proteins FCA, FPA and FLK, and the polyadenylation factor FY, may function post-transcriptionally to achieve this and are shown in red. The FVE/FLD proteins act with a putative histone deacetylase (HDAC; all shown in orange) to promote an inactive FLC chromatin state, represented by a closed structure with inactive histone tail modifications (red). FLC is also repressed by exposure to long periods of cold (vernalization). The proteins acting in the vernalization pathway are shown in pink. Prolonged cold induces VIN3 expression, which promotes an inactive FLC chromatin state. Subsequently, the VRN1 and VRN2 proteins are recruited to FLC, and are required for the methylation of FLC histones and the maintenance of silencing. These marks may promote the association of silencing factors with FLC chromatin that reinforce its repression. During meiosis, gametogenesis or early embryogenesis, FLCrepression is overcome, thus resetting its expression in the next generation.;Vernalization causes the depletion of florigen repressors and activates floral inducing genes. http://dev.biologists.org/
Phytochrome mediated Flowering:
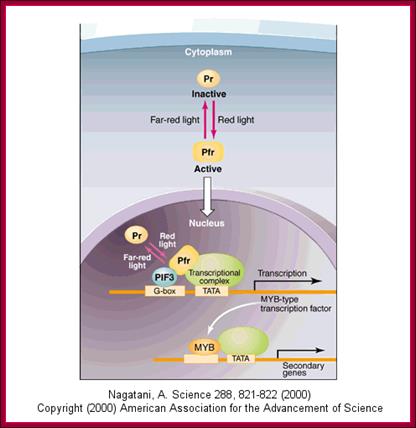
������������� http://biowiki.ucdavis.edu/
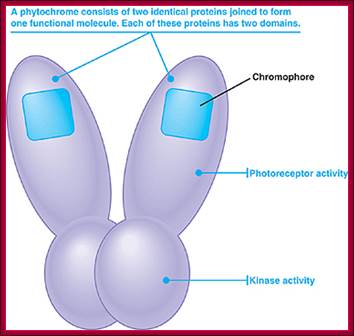 �
�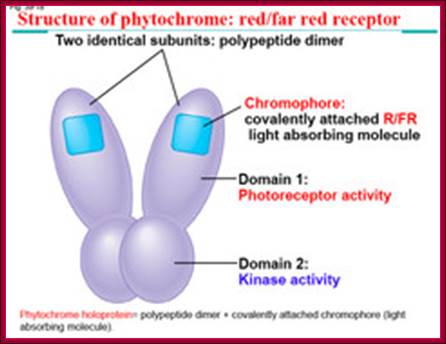
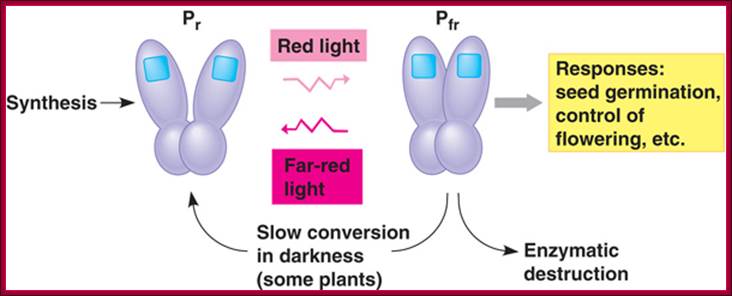
www.bio.miami.edu; slideplayer.com
A general mode Phytochrome mediated flowering is depicted in the above diagram.� The PhyA protein in Phy-Pr form absorbing red light for a period of time undergoes conformational change because the pigment is bound by the protein.� The pigment absorbs light at specific wave length and the energy (quantum of given wave length) is transferred to protein, so the protein undergoes conformational changes. This pigment protein is called Phy-Pfr form.� The activated pigment-protein complex performs auto phosphorylation to generate Phy-Pfr-p; this enters the nucleus; there complexes with Pfr receptor protein called Phytochrome Interacting Factor PIF3, which is already bound to�� MYB-like gene promoter.� This complex activates the MYB-like genes by recruiting activators, co activators and other proteins and the gene is transcribed.� The transcript is processed and transported into cytoplasm where it is translated into a MYB type transcriptional factors; that again enters the nucleus and binds other genes to activate light induced genes.� MYB is family gene activators.
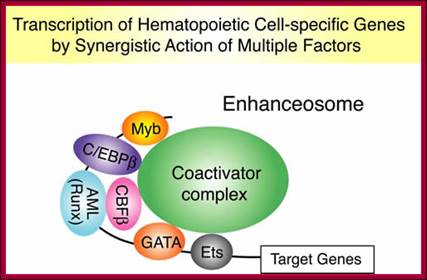
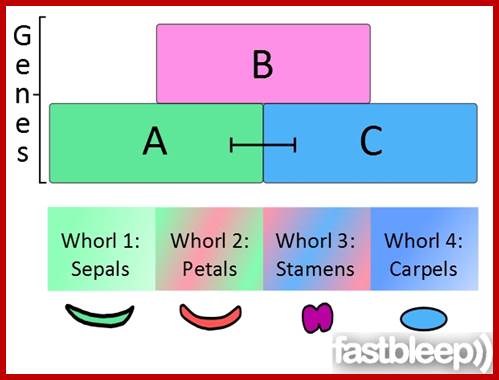
�Development of Flowering; Ruqya Patel; https://www.fastbleep.com
Generally, it is the PHYB is involved in inducing flowering rather than PHY-A, but now PHYA plays an important role in flowering. �The light induced Phy-Pfr form activate the trimeric (Heteromeric G-protein) that executes three separate pathways, both have confluencing effect on flower induction.� One pathway is G-protein induced CaM pathway that leads to the activation of Y� and the other is cyclic GMP activated pathway called X.� The Phy-Pfr-p now binds to LRE (light response elements} through PIF3 (Phytochrome integration factor) and activates the gene.� The Phy-Pfr-p acts as dimer and binds to dimeric PiF3 activator-repressor protein.
Another description of light induced gene expression describes that activated Phy-A-Pfr auto phosphorylates and binds to FHY1-FHL dimeric protein.� This complex now enters the nucleus and binds to their respective LRE and activates gene expression related to photo-responsive genes including flowering genes.� As and when PhyA accumulates, it blocks the expression of FHY3-FAR1.� However, the said dimers already existed bind to their promoters and activate the transcription of FHY1 and FHL; the protein products, both remain in cytoplasm till the activation of Phy-Pfr. The activated Phy-Pfr induces the gene expression and cycle is repeated. It is an established fact that PhyA triggers plant/light responses such as germination and flowering.
Two
proteins known as FHY3 and FAR1 bind to DNA in a plant cell and cause
production of two other proteins known as FHY1 and FHL. When hit with far red
light, a light-sensitive protein called phytochrome A (phyA) changes its shape.
http://phys.org/news115053032.html#jCp
http://phys.org/
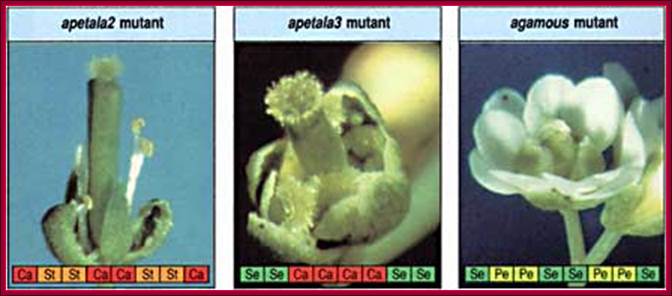
Floral gene mutants and their effects on their morphology.
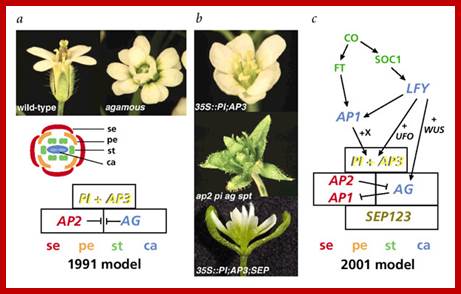
Progress
in our molecular genetic understanding of flower development:
a, The ABC model was proposed on the basis of the phenotypes of
homeotic floral mutants such as agamous (ag) and genetic interactions
among such mutants25, 26, 27. In this model, the combinatorial
activity of three classes of homeotic gene products specifies floral organ
identity: A (AP2) = sepals (se); A + B (AP3 + PI) = petals (pe); B +
C (AG) = stamens (st); and C = carpels (ca). Loss-of-function alleles of these
genes result in homeotic alterations; for example, in ag mutants,
petals develop in the positions normally occupied by stamens, and another
flower meristem replaces the carpels. b, The cloning of these genes
allowed gain-of-function mutants to be constructed. For example, when both AP3 and PI are
constitutively expressed (35S:PI;AP3), flowers form that have petals where
sepals normally develop, and stamens where carpels normally develop43. But
leaves are not converted into floral organs unless a SEP gene is also
expressed ectopically59, 60. Shown here is 35S:PI;AP3;SEP, in which
all leaves (except the cotyledons) are converted into petals. Loss of all
floral homeotic gene activities plus the gene SPT, which promotes carpel
development, results in flowers consisting totally of leaves (ap2 pi ag spt)212.
Thus, leaves can be converted into floral organs and floral organs into leaves. c,
Current understanding of the activation of floral organ identity genes. LEAFY (LFY)
and APETALA1 (AP1) act in conjunction with other spatially restricted
factors (X, UFO and WUS) to activate the B and C class genes
directly47, 48, 49, 213, 214, 215. The meristem
identity genes LFY and AP1 are themselves activated
directly or indirectly by factors (such as CO, FT and SOC1)
that mediate floral induction, only some of which are shown for simplicity216, 217.;Robert E. Pruitt, John L. Bowman & Ueli Grossniklaus;
Nature Genetics 33, 294
- 304 (2003); www.nature .com
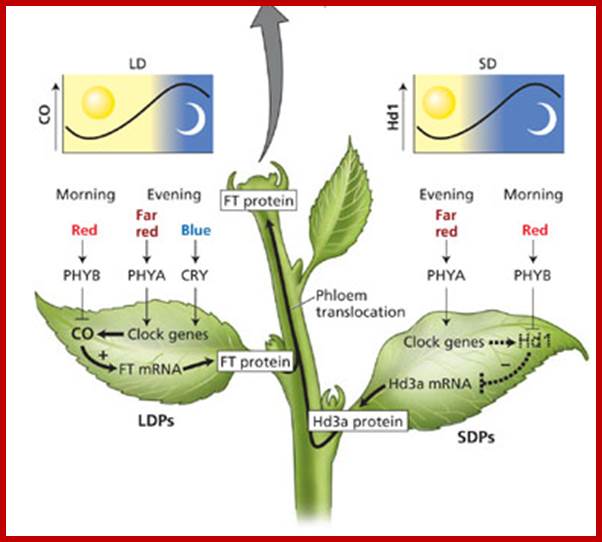
http://www.howplantswork.com/
The diagram explains what LD and SD�s induce specific proteins which get translocated along the sieve tubes to the base floral meristem where they bring about a whole series of gene expression for flowering. Once they reach the base of SAM they trigger membrane bound receptor and activate the flowering process.� How?
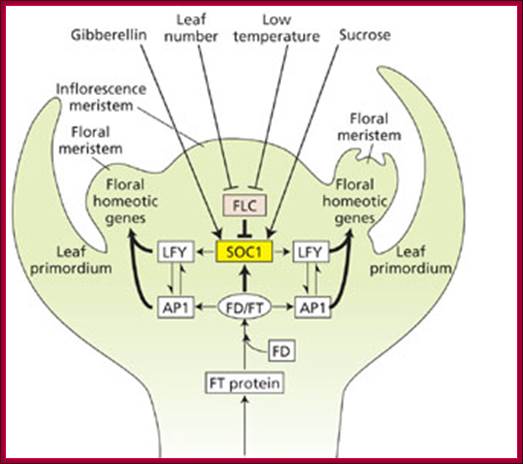
Multiple developmental pathways for flowering in Arabidopsis:photoperiodism, the autonomous (leaf number) and vernalization (low temperature) pathways, the energy (sucrose) pathway, and the gibberellin pathway. The photoperiodic pathway is located in the leaves and involves the production of a transmissible floral stimulus, FT protein. In LDPs such asArabidopsis, FT protein is produced in the phloem in response to CO protein accumulation under long days. It is then translocated via sieve tubes to the apical meristem. In SDPs such as rice, the transmissible floral stimulus Hd3a protein accumulates when the repressor protein, Hd1, is not produced under short days, and the Hd3a protein is translocated via the phloem to the apical meristem. In Arabidopsis, FT binds to FD, and the FT/FD protein complex activates the AP1 and SOC1genes, which trigger LFY gene expression. LFY and AP1 then trigger the expression of the floral homeotic genes. The autonomous (leaf number) and vernalization (low temperature) pathways act in the apical meristem to negatively regulate FLC, a negative regulator of SOC1. The sucrose and gibberellin pathways, also localized to the meristem, promote SOC1 expression. (After Bl�zquez 2005. It is simplified diagram that shows that the FT protein (once called elusive Florigin), in association with FD performs a cascading gene regulatory activity that leads to flowering. http://5e.plantphys.net/
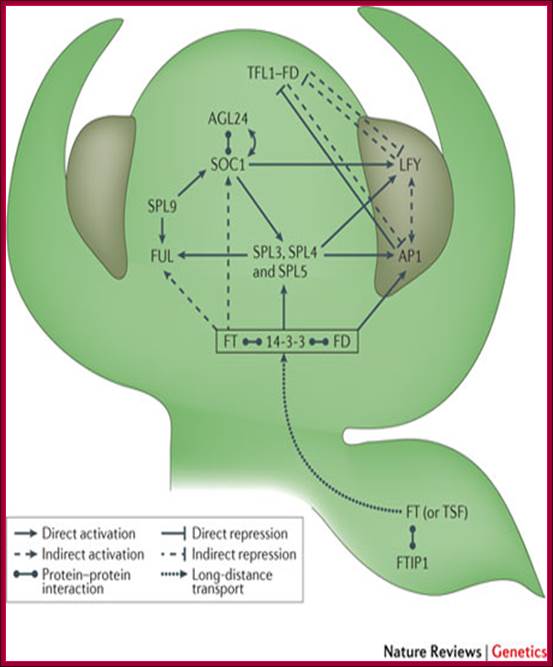
FLOWERING LOCUS T (FT) is made in the companion cells of the leaves and is transported from the leaves to the meristem through the phloem sieve elements. Recently, movement of FT from the companion cells to the sieve elements was shown to require the interaction between FT and a novel endoplasmic reticulum membrane protein called FT-INTERACTING PROTEIN 1 (FTIP1)40. TWIN SISTER OF FT (TSF) is a closely related protein and probably acts in a similar way to FT. At the shoot meristem, genetic data indicate that the FT�FLOWERING LOCUS D (FD) complex activates expression of flowering genes shown as a network in the meristem. In ft tsf double mutants and fd mutants, transcription of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) in response to long days is delayed in comparison to wild-type plants34, 72. SOC1, which encodes a MADS box transcription factor, is the earliest gene shown to be upregulated in response to long days at the meristem, and mutations in the gene cause late flowering115, 116. Strikingly, inactivation of both SOC1 and the related MADS box gene FRUITFULL (FUL; also known as AGL8) almost entirely suppresses the extreme early flowering caused by overexpression of FT from different heterologous promoters, suggesting that SOC1 and FUL are essential for the promotion of flowering by FT117, 118. When SOC1 is expressed in the meristem, it interacts with AGL24, another MADS box transcription factor, and promotes the activation of transcription of LEAFY (LFY), which is a meristem identity gene that is involved in the initiation of flower development119. SOC, FUL, LFYand APETALA1 � another floral meristem identity gene � are also activated by SQUAMOSA BINDING PROTEIN LIKE (SPL) transcription factors, which are expressed in the meristem in response to FT and FD and were recently proposed to be direct targets of FD on the basis of chromatin immunoprecipitation experiments. Interestingly, SOC1 also binds to the SPL genes, suggesting that FD might act at different layers of the hierarchy to upregulate both SOC1 and SPL gene transcription. Similarly, the FT�FD complex has been proposed directly to activate AP1, which is expressed in the cells that will give rise to the flower and confer floral identity on this primordium45. However, recent detailed analysis of the AP1promoter questioned whether FD binds directly123, Nevertheless, in rice, FD was also proposed to bind directly to the promoter of the AP1 homologue MADS15 via a similar element46. The expression of both meristem identity genes AP1 and LFY is antagonized by TERMINAL FLOWER1 (TFL1), which is a protein that is related to FT, preventing their ectopic expression in the centre of the shoot meristem. In young floral primordia, AP1 and LFY repressTFL1 transcription. Recently, TFL1 was also shown to depend on FD to trigger the transcriptional repression of its targets, suggesting a pivotal role of FD, depending on whether it interacts with FT or TFL1; Fernando Andr�s & George Coupland-Nature2012. http://www.nature.com/
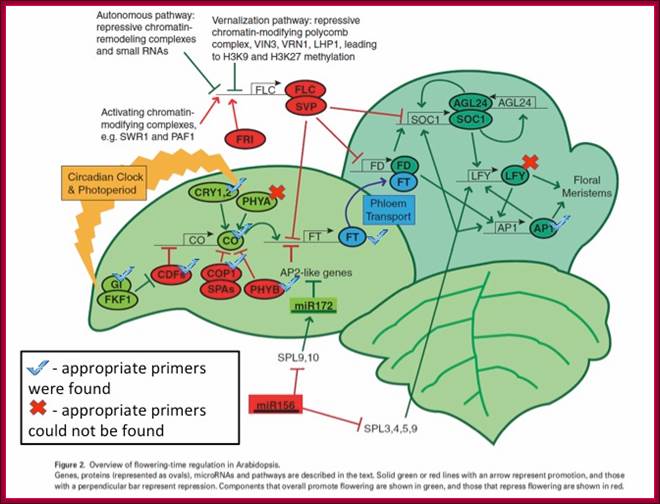
http://gcat.davidson.edu/
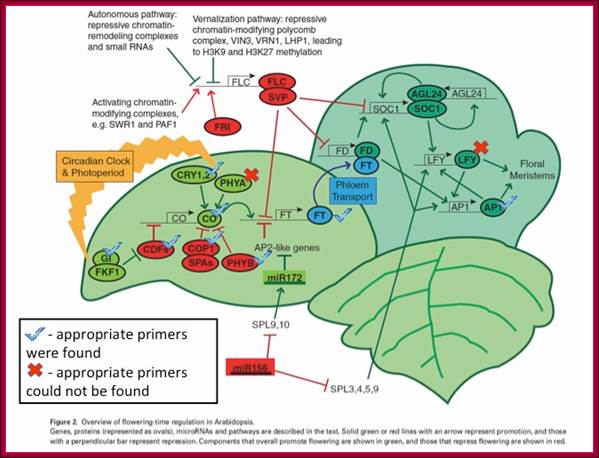
http://gcat.davidson.edu/
Flowering genes and pathway:
- FT from leaves; transported to the base of SAM,
- FTmRNA associated with some proteins and FT proteins are transported across the companion cell to sieve tubes and transported along protoplasmic stream to the base of SAM.
- Entry of these into cells of the SAM, either transported across the cell using specific signal induced transporters or bind to receptors and act.
- FT is like RAF kinase inhibitor.
- FT combines with FD,
- FT-FD acts on AP1 and SOC1 genes,
- SOC1 activates LFY genes,
- LFY-Leafy genes induce floral homeotic genes,
- SOC1 is inhibited by another protein called FLC (cold response protein),,
- Factors like GA and sucrose activate SOC1 and overcome FLC,
- Activated SOC1 activates LFY genes, which in combination with AP1 activate floral meristems to induce different floral structure in gene specific manner.
- FT Gene-The FT product is 19-20kDa protein.
FT Gene Structue:
�5� UTR-----------A/G-------- [3 exons] -------TER-----3�UTR.
Morphactins:
They are synthetic growth regulators and act in variety of means and ways.� Phenoxyalkanecarboxilic acid, substituted benzoic acid, maleic hydrazide, flurene (?), chloroflurenol, chlorofluron, methyl benzylate, and diflurenol are few of the morphactins.� They are involved in seed germination and inhibition, involved in dwarfing effect, induced apical dominance , increase tillering, prolong bud dormancy, prevent flowering in short day plants.� Each kind has its own effect.
Wound hormone:
Traumatic acid is called wound hormone, when there is wound it heals the wound.� Others hormones are triacontanol, Brassins, (steroid), Xanthoxin, Batasins, some vitamins.