Regulation of Gene Expression:
Almost all cells express, in any given living tissue, in any given organism. �Expression of housekeeping genes require for metabolic activity. Transcription, translation, transportation and others required for the up keeping the basal cellular activity.� This accounts for more than 50-60% of the total number of genes.� Among the rest of the genes 700-1000 (?) or more genes are expressed in tissue specific manner.� Among them few are expressed in high copy numbers in comparison to others.� Some genes are expressed during embryonic development, probably some of them are expressed only once in the lifetime of an organism, however substantial number of genes are moderately expressed in response to stimuli where their switch on and switch off is controlled. Tissue specific gene expression has been evaluated and the results with some reservation. Techniques like RT-PCR used is not good quantitatively, yet the results do show some interesting features. Retrotransposons comprise a large portion of mammalian genomes. They contribute to structural changes and more importantly to gene regulation. The expansion and diversification of gene families have been implicated as sources of evolutionary novelties. In human genome half or more of it is composed of interspersed repetitive DNA sequences. They contribute to structural changes and more importantly to gene regulation.
Eukaryotic Gene Density
So, it seems likely that the lack of an association between size of genome and number of functional genes, the C-value paradox is partially due to the number of retrotransposons accumulated in the genome.
Human genome:
- 3,200 Mb and 3000Mb (3.0-3.2x10^9bp).
- 90 Mb (3%) corresponds to coding sequences (it is too high).
- 810 Mb (27%) corresponds to associated sequences (introns, promoters, pseudo genes).
- 2,100 Mb (70%) junk DNA that is under little selective constraint.
- Unique sequences (1,680 Mb - 56%).
- Repetitive DNA (420 Mb - 14%).
Detecting and Profiling Tissue-Selective Genes:
Table: Comparison of several studies.
|
|||||||||||||||||||||||||||||||||||
Data coverage and results from several studies based on DNA microarray technology were compared. PCA, principal component analysis; HSD, honest significant difference. - Detecting and profiling tissue-selective genes; Shuang Liang , Yizheng Li , Xiaobing Be , Steve Howes and Wei Liu.
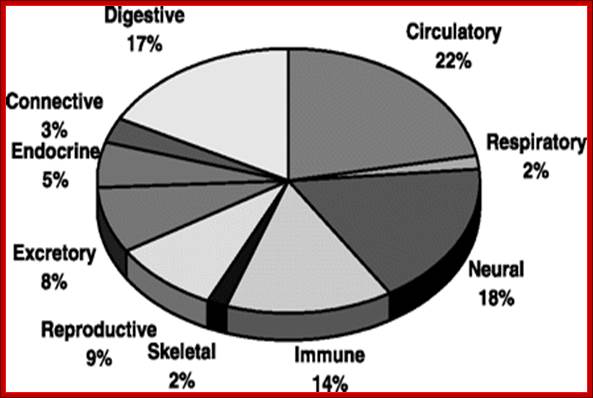
Overview of 10 tissue-selective genes: Gene-tissue counts are
organized by organ systems to illustrate the proportion of selective genes per
organ system; Shuang Liang , Yizheng Li , Xiaobing Be , Steve Howes and Wei Liu.
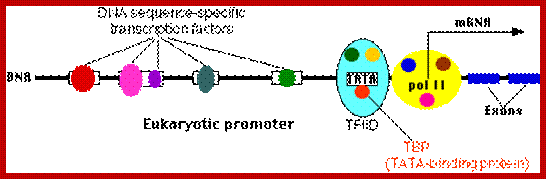
Regulation of gene expression in eukaryotes is more sophisticated and intricate, in comparison to prokaryotes.� Inexplicable structural functional cells/genes in Eukaryotic DNA is highly compacted and it is the greatest wonder and difficult to imagine, how it has developed. To function, these, have to be opened for transcriptional factors to access regulatory sequences of any gene to be expressed.� Organization of regulatory elements is elaborate and varied, complex and variable from one gene to the other.� Cells are also provided with a host of DNA binding proteins that can act as chromatin modifiers, activators, co activators, mediators, repressors, co repressors which have different but specific functions.� Cells receive a large number of signals and cells themselves generate quite a number of signals and all are integrated into regulatory system.� They also have inbuilt insulators or boundary elements to protect from spreading effects.� Otherwise, the regulatory mechanism is same from yeasts to mammals, with some differences, but overall theme is same.
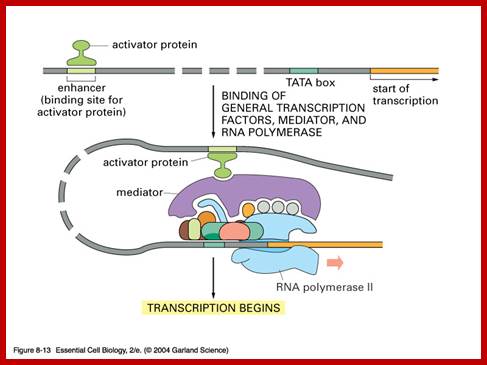
In general, once all the required transcription factors are assembled or facilitated to assemble as pre initiation complex with the RNA polymerase properly positioned, the PIC itself can initiate transcription by going through isomerization process.� In most of the cases, in general, some of the upstream factors like SP1 (GC binding), NFs, CTFs (CAAT binding factors), bind to the core promoter elements help in activating the transcriptional apparatus.� In fact, it is not inappropriate to call them core factors and core promoter elements.� But depending upon the tissue one finds many different factors, either singly or in the form of dimers or multimers operates either in activating specific genes or silencing specific genes but the whole set of house-keeping genes always on.� There is no simple unified mechanism that can be applied to all genes that can explain the process of regulation of gene expression or gene repression.
Repressors and Repression:
Making a gene to be switched off or disabling a gene or a group of genes from transcription is opposite to gene expression.� It is now easy to determine, how many genes are expressed and how many genes are repressed or not expressed in a given tissue at a given situation is discerned by DNA micro array and PCR methods.� There is so much of information on genome, yet information on how genes are regulated is yet to digged out.� All the time in a given tissue certain numbers of genes are expressed and certain numbers of genes are kept out of expression.� So also when a given condition changes a set of genes that are repressed hitherto are made to express and some are made to be silent.
In some case all the genes present in one specific chromosome are made silent.� In some cases, cluster of genes in certain regions of the chromosomes are switched off by certain changes in chromosomal structure, and surprisingly inactivation of genes can spread on either side of a locus.� This can lead to what is popularly called by classical genetists as �Epigenetics�.� Silencing is another form of repression, but act with slight variations.� An individual gene product that acts at specific promoters and bring about inactivation is generally referred to as Repression. Repressors can mediate with mediators and repress gene action. But certain chromosomal modifications locally or on a large scale make genes to switch off; called gene(s) called silencing.� This may due to deacetylation, methylation and dephosphorylation, which generally bring about condensation of chromosome so tightly; there won�t be any scope for any expression of any gene(s). The act of condensation, especially during mitotic cell divisions, is due to the act of a group of proteins called condensins. �While repression is at the level of individual gene(s), but silencing effect is often large scale and spreading.� Only insulator can prevent such spreading effect.
Repression through recruitment of HDA (Histone De Acetylase) is exemplified in regulation of GAL1 gene.� In the presence of glucose, the GAL1 gene is rendered silent by a protein called Mig1 that binds to upstream regulatory site (URS) that is found in between the UAS and the promoter elements.� Mig1 then recruits Tup1, which is a repressing complex.� Tup 1 recruits histone deacetylase and it may also interact with PIC of the gene.
GAL10}---/----[GAL7}------UAS---URS------P---+1>-----GAL1------
Chromosomal condensation and decondensation is a feature one can observe during cell cycle.� During interphase or G^o stage, chromosomes are relaxed and spread and bound to nuclear lamins via matrix proteins. Most of the genes specific to that cell type are actively expressed and those not required remain silent. Such state also allows gene expression in response to signals.� During transition from prophase to Metaphase, chromosomes undergo condensation, so tightly packed, all the genes are rendered inactive. Metaphase condensation and gene silencing is different from interphase localized chromatin condensation and gene silencing.
Such chromosomal changes, i.e., tight condensation and relaxation, express as heterochromatin and euchromatin at chromosomal level.� Such morphological states of chromosomes can be demonstrated by staining techniques such as G-band technique and R-band techniques.� Such morphological changes are due to changes in the composition of chromatin.� Phosphorylation of N-terminal region of histone 1 (H1) leads to condensation, which happens during metaphase.� Similarly, methylation of Lysine residues of histone 3 is a known cause for silencing.� Deacetylation can also cause repression.� Chromosomal dephosphorylation, methylation and deacetylation all cause gene inactivation either locally or enmass.� Localized demethylation, acetylation and phosphorylation of chromatin at histone N-tails at specific sites; all can lead to decondensation and perhaps gene activation but not enmass but at specific loci.
Repression through Chromatin Remodeling:
Methylation and demethylation is another method of activation and inactivation of certain set of genes.� Methylation of histone tails at lysine can bring about condensation of chromatin into heterochromatin and make it inactive.� Methylation of Histone 3 and Histone 4 can enforce silencing of gene expression.� Methylation of Histone 3 at lysine 9 recruits� specific proteins such as MeCP2, which on binding recruit�s histone deacetylase as well as methylases.� Methylation mediated silencing of genes can be propagated. Proteins, which have chromo-domains, such as Heterochromatin Protein (HP1), bind to methylated residues; HP is a component of heterochromatin. Even deacetylation of lysine 9 of Histone 3 (H3) attracts histone deacetylase enzymes to bind to the site that can lead to spreading of gene silencing effect. If this region of histone tails is methylated, they attract specific methyl binding proteins and make the chromatin heterochromatic or keep the chromatin in heterochromatin state; it can spread also.
Methylation at DNA level, i.e., methylation of Cytosine at 5th position in a sequence of CG, CG repeats induce heterochromatin in the region.� Any promoter region of a gene, if it contains such sequences; and if they are methylated, the downstream gene is rendered inactive; example β-globin genes. The enzyme is methyl transferase-A, by name Mecp1, binds to methylated C.CH3pG groups and prevents transcription.� The said protein directly interacts with BTA/PIC and represses the expression. Methylation of CG base pair mode of gene silencing can be propagated.� As soon as replication, one of the strands is methylated and the other is non-methylated (hemi-methylated state), this state of methylation is recognized by specific maintenance methylases and they methylate CpG base pairs on the newly formed DNA strand.� Such methylated residues recruit MeCP2 proteins, which in turn recruit histone deacetylases and more methylases for shutting down transcription.
Methylation can lead to the binding HP1 family of proteins that induces heterochromatization, where in some cases the whole chromosome becomes silent. Such process of silencing the whole chromosome is the basis of genetic imprinting. The whole chromosomal silencing in mammalian X chromosomes, involves Xist-RNA, which is a non-coding RNA transcribed from Xic-gene.� This leads to epigenetic expression and it is actually due to methylase and demethylase enzyme activity.
How methylation and demethylation works is exemplified by the incorporation of Azacytidne and an isomer kind of ddCytidine (dideoxy = dd) under in vitro cultures of muscle cells.� When cells incorporate Azacytidne in place of Cytosine, methylases cannot methylate Azacytidne.� So, some genes, not specific to muscle cells, start expressing.� The same drug can activate silent genes found on inactivated X-chromosomes.� So, hypo-methylation of sites provides scope gene activation.
In the case of globin genes, which are organized in clusters as a family of genes; the γ-globin genes are inactivated because of methylation in and around +90 from the START of the gene, but the removal all methyl groups from both upstream and downstream regions, relieve the gene from repression.� Though this is a general rule in higher organisms, methylation and demethylation mechanism does not operate in Drosophila; it is an exception to the rule.
The site or the locus at which methylation occurs contain CpG doublets.� Such doublets are found in blocks called islands.� In human genome ~45,000 such CpG islands are found, which accounts for ~20% of its GC content.� Such CpG islands are often found in stretches of 1 to 2kb, strangely some of them are also found in �Alu� elements, which are rich in GC content.� Excluding, Alu elements there are 15,500 groups of CpG in mouse genomes.� These regions show structural changes during gene expression, such regions have low levels of H1 contents., so the structure is less compact and sensitive to DNase-I; such regions are called DNase-I hypersensitive sites and histones are extensively acetylated.�
All housekeeping genes, which are constitutively expressed, contain
CpG islands, which accounts for 50% of the house keeping genes.� The other half of CpG islands is found in promoter regions that are regulated in tissue specific manner.� Methylation of Cytidine of CpG doublet causes the binding of certain proteins such as Mecp1, which generally binds to several, methylated group of CpGps.� But Mecp2 binds to single methylated groups of CpG pairs.� Binding of the said proteins prevent transcription, this happens in vitro conditions too.� Mecp2 interact directly with BTA/PIC and repress them from activating transcription.� Such Mecp2 groups of proteins also associate with Sin3 repressor complex, which has histone deacetylase enzymes.
Insulators:
There are sequences bound by specific proteins, block the passage of either repression or activation effects, they are deemed as Insulators.� In most of the cases they are positioned in between an enhancer and a promoter, where the action of an enhancer is limited to a specific promoter and prevent its effect to another promoter in the neighborhood.
����������� < +---Prom--<-------ENH--------->--Prom+>---
� < +-Prom-- < --ENH --Xgene <-P-I Insul I�P->gene Y- Prom>X
Prom = Promoter.
ENH = Enhancer.
Insul = Insulator.
X= blocks enhancer effect.
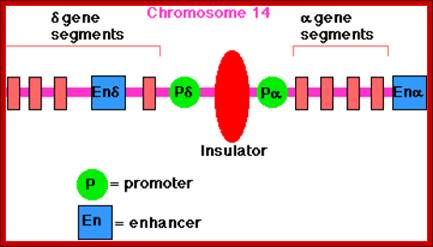
http://www.biology-pages.info
Similarly hetero chromatinization has spreading effect.� In such cases any gene(s) in the region get suppressed. If an insulator is present on one side or both sides of the active heterochromatin locus, it prevents the spreading effect of it.
X-XX-Gene < XXX -----<IIII [HChr] IIII>------XXX> Gene�XXX
�����������
< ---+ Gene < --- [Insul]-XX <IIII [HChr] IIIIII> ---->XX Gene XXX
HChr = Heterochromatin
Insul = Insulator
XXX =Inactive or inactivation,
XX = Insulator blocks HC spreading,
IIIIII> = HC can spread
http://dorim.mokpo.ac.kr
In Drosophila one finds such segments of DNA called SCs (24 base pair long blocks) and specific proteins called BEAF-32 bind them.� Such protein bound DNA sequences (repeats of CGAT) can be identified by fluorescence tagged IgGs, they look like inter-bands.� Nearly 50% of such inter-bands, found in polytene chromosomes, are found to act as insulators against the spreading effect of heterochromatization or enhancers that are located at adjacent regions.
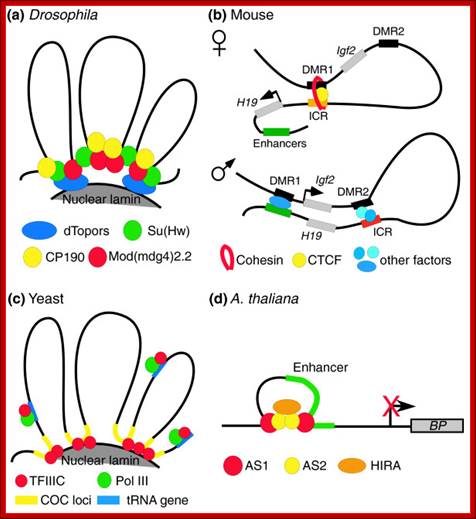
If �White� gene from Drosophila is placed in heterochromatin region, the gene becomes inactive, for that matter any gene if placed either at close vicinity of the heterochromatin or within heterochromatin region, becomes silent.� On the other hand, if the gene is placed in a heterochromatin region but with SCs blocks on either side, the gene remains active.� Insulators contain 26 bp long 12 copies in each segment. It demonstrates that SCs act as insulators against heterochromatin spreading effect and also multiple enhancers activating effect.� Insulator binding proteins like Su (Hw) and mod (mdg4) have been identified.�� Su�s (Hw) is zinc finger proteins bind to major groove of the DNA. The mdg (mdg4) protein controls the direction of insulator effect. The Su protein can have spreading insulator effect, but mod (mdg4) controls it. They are very important in ensuring insulator action and directionality of the insulator action.� Such proteins are also found in Drosophila and other systems.� The mode of working of the said proteins and insulator region is that the Su (Hw) binds to insulator regions that are found at several sites in a chromatin.� The mod proteins bind to the inner surface of the nuclear membrane in the form of a cluster and Su (Hw) proteins bind to mod clusters, so DNA segment in between Su bound regions loops out, thus the DNA is bound to nuclear matrix, such site is called matrix attachment regions (MARs), and such attachment sites may provide insulator effect.
Silencers:
Silencing can be deemed as position effect. Centromeric and Telomeric region consists of highly repetitive DNA and does not code for any functional products.� The chromosome in these regions exists in heterochromatic state.� The said regions have highly condensed state of chromosomes. �If any functional gene is introduced into such heterochromatin region, it becomes silent.� Histone modification such as deacetylation and methylation are few of the processes responsible for silencing. The region of DNA that is said to be silent has characteristic sequence module such as Growth Regulating Factors (GRF) sequence A/GA/CACCANNCAT/CT/C and ABF TATCATTNNNNACAG to which specific proteins such as GRF and ABF (ARS binding factor) proteins bind and make this region silent. The said proteins are also called Silencer Factors (SF-A), SBP1, SBP2 and SBP3, the latter bind to silencer sequences such as AGT/CCA? GC (it is called silencer DNA site). Such regions prevent the either spreading of chromatin activation or chromatin inactivation?� There is difference between insulator and silencer. Insulator region has sequence modules such as CCCTC to which proteins such as CTCF bind.� Insulator prevents the spreading action of Enhancers and/or spreading of heterochromatization.� But silencer makes the region totally inactive, e.g., silent mating factor sites in yeasts.
In yeast silent mating locus, CEN region and telomeric region are all silent.� Telomeric region contains 1-5 kbp long DNA with repetitive sequences.� This part is in highly folded state and it is very less acetylated than other regions. Mutation studies reveal that the genes responsible for silencing are RAP1 protein (Repressor/Activator factors), Silent information regulator (Sir) genes. SiR 2, SiR,3 and SiR 4 form complexes and they are found associated with silent chromatin.� SiR 2 is a histone deacetylase.� RAP protein binds first, then SiR proteins are recruited.� Deacetylated histones recognize SiR complexes, and this effect can spread, but the spreading effect is often stopped my other modification of histones.� Methylation of K9 of H3 tail and H4 are actually used for gene silencing.� However, recruitment of SiR complexes in Telomeric region is Rap1. SiR2 is a deacetylase; once it is bound it acts further and spreads its deacetylation activity. �SiR 3 and 4 bind to unacetylated histone tails; these events cause silence of gene expression in general. Telomeric heterochromatin is an excellent example.
Chromosomal state depends on the modification of chromosomal Histone proteins. If the Histone1 is phosphorylated at serine residues, chromosomes get tightly condensed, and there is no scope for expression of any genes.� If the same histones are dephosphorylated chromosomes relax, and there is scope for gene expression.� But if the chromosome is acetylated at histone3 and Histone 4 tails (at lysine) nucleosomal structure loosens, so some part of DNA is accessible for the assembly of transcriptional apparatus.� Deacetylation again makes chromatin condense, again leads to the gene switching off.� Similarly, methylation of Histone tails and methylation of Cytosine in a sequence of CGCGCG islands, make the genes at suc� locus becoming inactive, but demethylation makes the genes again to be active.
Decondensation of chromatin provides access to transcriptional machine to assemble through a set of identity factors, which provide the footprints which gene or genes to be expressed.� There is nothing like random; gene silencing and gene activation is highly specific, which genes, in which tissue should be active and which gene and in which tissue should be inactive it is regulated by specific factors, and by specific mechanism.
Enhancers:
Enhancers are elements similar to activator elements, but their location can be upstream of the promoter or downstream of it and it can be 100 base pairs or thousand base pairs away.� Many of the sequences found in enhancer region are similar to activator elements, for example GC rich, ATTAGTCAGC and CCCAAT sequences in promoter region.� Certain specific factors such as SP1 (specificity protein), AP1 activator factor and C/EBP, CCaAt binding acts as an enhancer factor respectively, they bind to specific elements, and interact with BTA and activate transcriptional apparatus.� Important feature of enhancers, is whichever position they are, whatever the distance of their location, they enhance the rate of transcriptional initiation by 50 to 100 to 200-fold.� If β-globin genes are inserted near any known enhancers, its transcriptional efficiency increases by 200 percent.� Again, their contact with BTA depends upon protein-protein interaction.� So, the proteins that bind to enhancers should have a DNA binding domain, a protein-protein interacting domain and an activating domain.
Enhancers are fond in some genes but not all genes.� They can act on any transcriptional apparatus provided the enhancer bound components interact with co activators which in turn interact with PIC components.� Some of the Enhancer elements and their binding factors:
AP1 =ATTAGTCAGC.
AP2 = GAAAGTCCCCAGC.
AP3 = GGTGTG.
AP4 = GCTGTGG.
OCTA = TATGCAAAG.
SP1 = GGGc GG
C/EBP = CCCAAT
In the enhancer region factors bind cooperatively to several proteins and form a complex and act on PIC, so the complex is called Enhanceosome.� Enhancers are modular structures for they too have promoter like elements to which similar factors bind.� The elements are found in clusters, so when proteins bind they form clusters. Enhancer found in one chromosome can activate another gene found in another chromosome by synaptic binding.� In the case of Gamma Interferon gene, the enhancer binding proteins form a complex of proteins such as HMG (Y), activators like KB, IRF and ATF-Jun.� These complex proteins interact with co- activators act with transcriptional apparatus.� The binding of HMG proteins to its site makes the DNA to bend into a structural form that facilitates binding of other factors like κβ, IRF and ATF-Jun.� Such complexes need not directly interact with RNAPs, but can interact through co activators complex, which may help in recruiting RNAP to its PIC complex.
Enhancers can change the overall structural feature of the DNA via remodeling of chromatin and may induce and increase the density of super coils. They may facilitate the location of DNA as matrix bound to nuclear membrane. They can also facilitate the entry site for RNAP or other proteins at the site in the chromatin.� Enhancer act as promoters from a distance, for they too contain sequence elements similar to activator elements.� Enhancer not only activates and enhances transcriptional efficiency of a gene found on the same chromosome, but they can enhance the activity of a gene that is found in another chromosome by what is called synaptic process.
Involvement of enhancers and enhancer binding factors in activation of human Interferon gene is very well documented. hu-INF is activated by the viral infection.� Viral entry triggers three activators NFκβ, IRF, and Jun/ATF.� These factors bind to their respective elements found in the Enhancer, which is actually located 1000bp upstream of the start site.� This complex is further assisted by the binding of High Mobility proteins called HMG-1 to the activator proteins that are bound to enhancer, thus they for a huge complex called Enhanceosome, where the activator components are brought to interferon promoter, for activating the gene.� The IRF binds to the central region of the enhancer, HMG1 binds on either side of IRF with space, into this space Jun/ATF binds at the distal end of the enhancer and NFkβ bind to the proximal side of the enhancer, thus the DNA is looped for the activators to contact BTA or assist the assembly of PIC components.
In the case of Drosophila enhancers are found far away (1Kbp to 100Kbp) from the start, located either downstream or upstream of the start.� Enhancer elements and their promoter elements are elaborate to accommodate the assembly of various factors either for inducing the gene expression or repressing the gene.� The sites are often called high affinity and low affinity sites, depending upon the number of particular sequence repeats found in it. A gene called �cut� is activated from a distance of 100 KBp away. In this case the protein called �chip� (this chip is different from immuno-technique called chip) facilitates enhancer factors with PIC.� The chip binds to multiple sites in between the enhancer and the promoter and facilitates the looping of the DNA.
Signal Transduction and Gene expression:
Cells and organisms are always impinged upon by a variety of signals such as environmental signals (heat, light, radiations etc) and chemicals in one or the other forms that one consumes or exposed to. Radiations that cause DNA damage induce the activation of p53 gene product, which in turn activate DNA repair enzymes and also induce few genes to prevent the cell to go into next stage of cell cycle.� Mitogens activate surface receptor kinase, which has a cascading effect that ultimately ends up in activation of several genes.� For example, specific Cdks-Cyclins that are activated due to mitogen signals phosphorylate retinoblastoma (RB) proteins.� RB proteins in unphosphorylated state bind to several E2F factors, which are actually Transcription factors for several genes that are required for cell cycle.� When RBs are phosphorylated, they undergo conformational change and release E2F factors, which no recruit few acetylases and with the association of dimer proteins activate several genes required for cell division cycle. So also, some kinases act on components that release transcriptional factors and also some phosphorylated factors enter the nucleus and activate certain genes.� Perhaps some of these are ultimate effects of signal transduction pathway.� In Drosophila Cactus binds to Dorsal a transcription factor in cytoplasm of its oocyte, but signal transduction from the cell surface receptor by activated kinase leads to phosphorylation of cactus; and cactus releases Dorsal, which enters the nucleus activates its gene by binding to its promoter elements.� Similarly in higher organisms, IκB binds to a transcriptional factor NFκB, but NFκB is released when IκB is phosphorylated by the kinases that are activated by signal transduction.� The NFκB enters into the nucleus of B-lymphoid cells to activate immunoglobulin-κ genes by binding to their respective promoter element.� NFκB is present in many cells and acts as a general transcriptional factor.� Activation of many genes by signal transduction pathway is executed through receptor kinase and other associated Kinases.
If an organism is subjected to either heat shock or high molecular weight metals such as mercury, certain genes respond and become active to overcome such harmful effects.� For example, if a person consumes a carcinogen like TPA inadvertently, specific genes respond and express to detoxify such chemicals.� Or if one consumes steroids or such hormones as treatment, they induce certain genes in response to them.� Even certain nutrients such as Galactose can induce Galactose utilizing genes in response to the specific carbohydrate.
In the case of heat shock, certain genes respond.� The genes called heat shock genes respond to such stimulus contain specific sequence elements in the upstream of heat shock genes.� They are called heat shock response elements, the sequence and the number and the position from the Start site vary from one heat shock gene to the other.�� Actually, in human beings there are 20 or so heat shock genes.� Each of them has specific response elements at different positions.� When an organism is subjected heat shock the, certain factors called heat shock transcriptional factors (HSTFs) get activated and bind to their respective elements and physically contact with transcriptional factors, probably including RNAP and activate the genes.� The products are essential for protecting cellular proteins from undergoing denaturation and destruction.
In the case of Galactose, in yeast cells, Gal utilizing genes are bound by GAL4 protein, which acts as repressor for it is bound by Gal80.� The GAL 80, when bound to GAL4 masks the gene activating domain of GAL4.� When galactose is provided, the carbohydrate binds to GAL3 which enters the nucleus and in turn binds to GAL80, thus GAL80 dissociates from GAL4 protein, unmasking the GAL4 activating domain, and thus the acidic domain interacts with transcriptional apparatus and activates Galactose utilizing genes.
When cells are activated by certain mitogens, the surface RTK proteins get activated with the binding of ligand.� The receptor protein becomes dimer and its cytoplasmic side of the protein becomes active as a kinase.� This kinase phosphorylates another kinase and thus a cascade of kinase-kinase leads to phoshorylation of certain proteins or TFs that enter into the nucleus and activate specific gene.�
When the cells take in heavy metals; genes to remove heavy metals gets activated.� The said genes are metallotheonein genes.� This gene responds to variety of stimuli, such as heavy metals, heat shock, and gluco-corticoids, and tumor inducing substances such as TPA.� The upstream elements of metallotheonein gene have elaborate elements placed in different positions and they are present in different numbers.� Even the upstream region contains Enhancer elements.� So, the gene gets activated to any of the chemicals or stimuli and the enzymes produced in response takes care of such components.
Similarly, body cells have genes, many of them, which respond to variety of chemicals, vitamins and hormones, such as glucocorticoids, sex hormones, growth hormones and others.� Specific genes to each of them respond to the hormones.� Each of these genes in their upstream, besides activator sequences, contains specific sequences called response elements similar to heat shock response elements.� Cells have already endowed with proteins to bind to such hormones; they are called receptor proteins (not a correct nomenclature).� The said receptor remains inactive because of the binding of an HSP protein.� When the lipid soluble hormone enters the cell, it specifically binds to its receptor protein, by displacing the HSP.� Thus, the receptor becomes activated and enters into the nucleus and seeks it respective promoter and activates respective gene, through interacting with TFs or through co activators.� Some of the hormone receptor protein when not bound to their hormones reside in the nucleus bound to their genes and repress the gene by interacting the TFs or co repressors.� They will be active only when their hormone binds to them.
Regulatory Transcription factors:
Positive regulation TFs: Constitutively produced:
SP1, CCAAT, NF1.
Regulatory Conditional: Developmental specific, Signal dependent and environmental signals.
Developmental specific:
GATA, HNFs, Pit1, MhoD, Bicoid, Hox, winged helix.
Signal dependent: Steroid super family, internal signals, cell surface receptor ligands.
Steroid family:
GR, ER, PR, TR, RAR, RXR, PPAR,
Internal signals:
SREBP, Steroid, Orphans( ?),
1. Cell surface receptor ligands, �
2. Resident Nuclear factors,
3. Latent cytoplasmic factors,
1. Cell surface receptor ligands: There are many- such as proteins and non-proteins which activate cell surface receptors.� The activated receptor-transducer signals through a cascade of kinases, some end up in activating specific genes through activating DNA binding proteins.
2. Resident Nuclear factors:
ETS, CREBp, ATMs, SRF, Fos-Jun, ME-2.
3. Latent Cytoplasmic factors:
STATs, sMads, NFkB/Rel, Ci/Cli (hh), NOTCH (NICD), Catenin (Wnt), TUBBy, NFAT
Some more:
AP1-Activating Protein, bind to enhancer elements.
ATFs 1 to 7: Activating TFs,
AR- Androgen receptor,
CR- Calcitrol receptor,
GR � Glucocorticoid receptor,
ARID1-4-X,
DAX-
DBP-
DMRT1-
Fos2-
Fox1-4-
GAPB-
GATA1-5
GCN4,
GAI- GA binding factor,
BCL-CBFA-
CCAT- enhancing factor,
CEBP-A, B, D, E and G-
CREB1-3-
GATA: A1-6-
GTFs-general transcription factors
GR- Glucocorticoid receptor,
AP1-L-zipper,
C.fos-L-zip,
NF-E2-Lzif
CRE Bp1-L.zip,
Activators� features:
Activators bind to DNA in sequence specific manner.
Activators can function directly with PIC,
Activators can function indirectly with co activators,
Activators recruit transcriptional factors,
Activators work though mediator complexes,
Activators work in concert with enhancer or upstream activator sequences.
Activators bound to enhancer work through co activators, mediator complexes and PIC directly.
Activators recruit chromosomal remodeling components, which can remodel the chromatin for accessing TF/RNAPs and other components.
Examples of Activator proteins:
GAC5, GcN4, HSP, NF, OCTA1, OCTA2, MADs, MAX, AP1, SP1, CREB, EGRF, SRF, NAP, and CTF; they bind to their respective sequence boxes and interact with others and PIC.
More examples:
NF-kB : Plays an important role in triggering immune responses.
IRF: Interferon regulatory, recruits histone acetylases to promoters.
ATF2-cJun: Recruits CBP/P300, SWI/SNF, make chromatin active.
HMG1: a family of proteins, bind to DNA and induce bending,
HNF1a and HNF4a: Hepatocyte nuclear factors induce chromatin remodeling n liver.
E2F1, E2F2, E2F3,: Bind to DNA in association with dimeric protein (DP) and activate transcription at G1/S transition stage, they recruit HATs,
Pho4: HLH proteins activate gene expression due to Phosphate starvation.
Swi 5: Zif protein complexes activate gene expression by chromatin remodeling.
SBF: Swi4 and Swi6 complex, activate gene at G1/S boundary,
GAL4: DNA binding protein, activates GAL response genes in yeast and other systems,
SP1, OCTA, and GATA: DNA binding interact with Pic and activate
Co activators� features:
Co activators are those proteins, which do not bind to DNA, but interact with protein that are already bound to DNA; they also facilitate the unwinding of DNA for the binding of proteins.� Some of the co activators are found to be acetylase; acetylation to Histone-3 and Histone-4 tails is known to loosen the chromatin and provide space for the binding of Transcriptional factors and others.� Co activators also have a function to bind to those proteins, which are already bound to specific regions of the DNA, and then interact with BTA and activate transcriptional apparatus.� Co activators are not just single monomeric proteins; they are dimers or multimers, example p300-CBP, PCAF, CAF and many others.
Co activator proteins (few):
RSC: remodeling of chromatin complexes.
PBP: a complex of TRAP, DRIP, and ARC, transcriptional activation by nuclear receptors.
SWI/SNF: ATP dependent chromatin remodeling complexes activate chromatin.
Swi2: SWI/SNF activating motor protein, an ATPase.
SAGA: histone acetyl transferase complex, acetylates specific histone lysines.
Gcn5; acetylating factor, catalytic subunit of SAGA,
PCAF: Histone acetyl transferase, transcriptional activation at cell cycle, and differentiation.
pRMT: Arginine methyl transferase of histone lysine and enhance transcriptional activation by nuclear hormone receptors.
CBP/P300: histone acetyl transferases, global transcriptional activators,
Snf1: a kinase, responds to cellular stress,
HATs: Histone acetylases
Mediator complex:
MC consists of 25 to 36 or more proteins interact with activators or co activators and PICs.
Their functions:
Simulate basal transcription; enable stimulation by activator and co activators
stimulate CTD domains and form stable complex with CD tail.
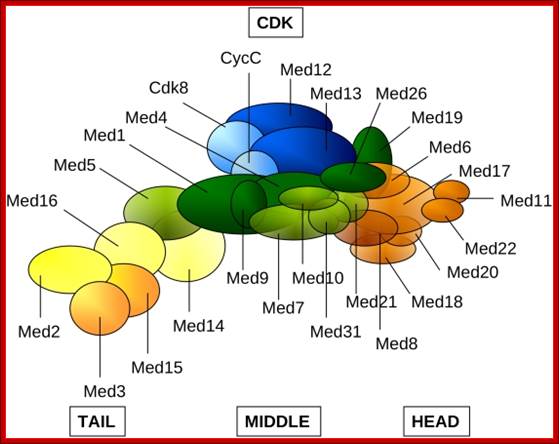
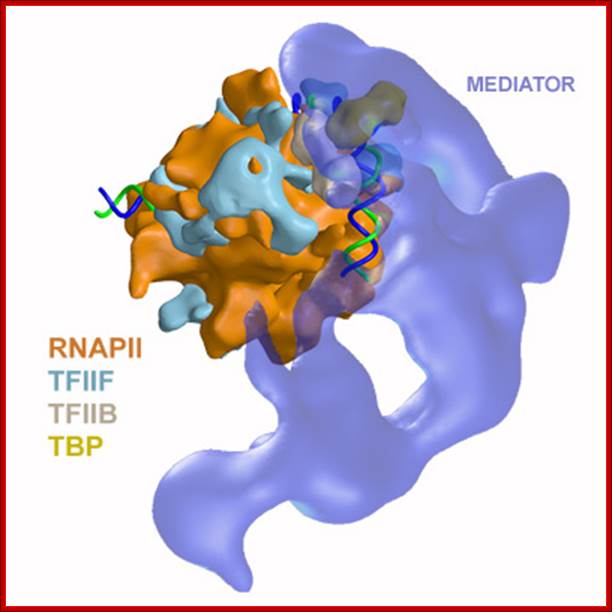
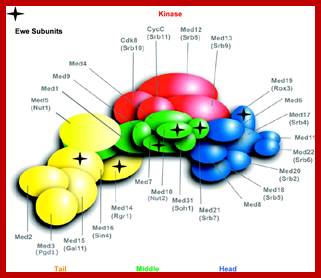
In the above figure of Mediator complex, there are 31 subunits grouped into Tail, Middle and Head components. http://www.antibodyreview.com/proteins
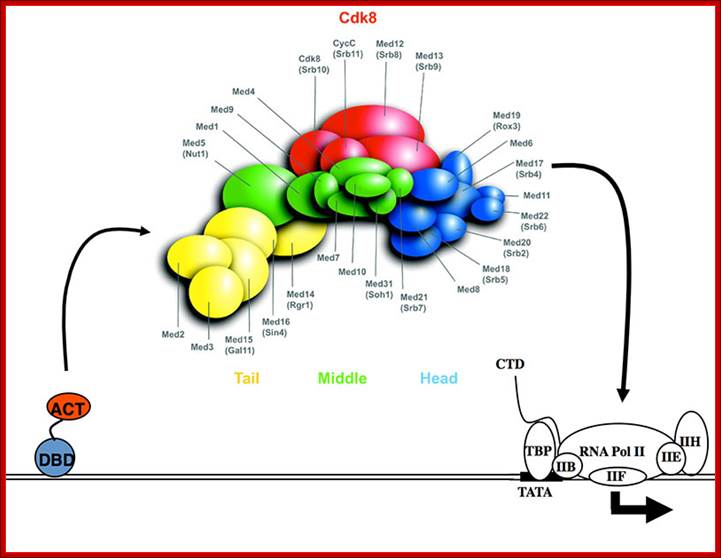
RNAP II complex for its activity requires mediator complex proteins; their composition varies from one tissue the other. Myers Laboratory; Geisel School of Medicine; http://geiselmed.dartmouth.edu/
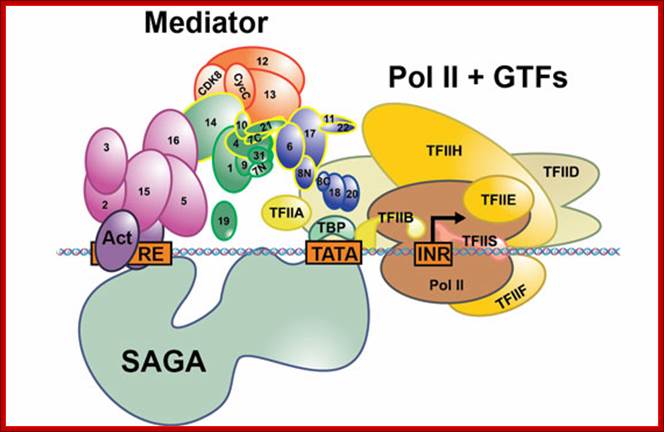
Schematic view of the Pol II preinitiation complex at the core promoter. Mediator bridges between activators (Act) bound to regulatory DNA elements (RE) and the basal transcription machinery (Pol II and the GTF). Pol II subsequently starts synthesizing RNA at the so-called Initiator DNA element (INR). Mediator modules are colored in blue (head), green (middle), magenta (tail) and orange (kinase). Individual Mediator subunits are shown. htts://www.elitenetzwerk.bayern.dep.
Cells, in most of the cases, have another set of proteins, whose composition differs from one tissue to the other; they are called mediators they interact with other proteins found on gene promoters� elements. They act in between activators on Basal Transcriptional Apparatus (BTA) as co-activators. For, the mediator complexes are unique for a given tissue. They mediate interaction between the proteins bound at distant sites and BTA; in fact, some of the components interact with specific components of PIC and trigger transcriptional initiation.� For example, yeast has certain mediator complex consisting of 20 or more protein subunits, such as Srb 2, 4, 5, 6 & 7, RGr1, Gal 11, Med 1, 2, 6, 7, pgd 1, Nut 1,2 and others.� In other systems different tissues produce mediators with different composition.� They mediate the action of activators, co activators, even TAFs with basal transcriptional apparatus.
In the case of metazoans, the mediator complexes consist of CRSP, NAT, ARC, DRIP, TRAP, SMCC, MED and PCL.
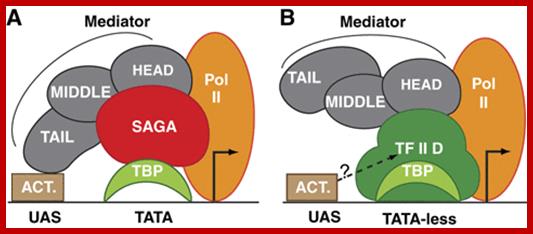
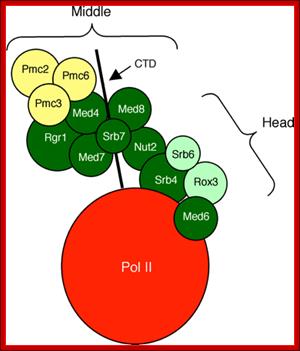
The diagram suggests the different regions of MC interact with RNAP tail and some with RNAP main complex. Distinct role of Mediator tail module in regulation of SAGA‐dependent, TATA‐containing genes in yeast; The evolutionarily conserved Mediator complex is required for transcription of nearly all RNA Pol II‐dependent promoters, with the tail module serving to recruit Mediators to active promoters in current models. However, transcriptional dependence on tail module subunits varies in a gene‐specific manner, and the generality of the tail module requirement for transcriptional activation has not been explored. Here, we show that tail module subunits function redundantly to recruit Mediator to promoters in yeast, and transcriptome analysis shows stronger effects on genome‐wide expression in a double‐tail subunit deletion mutant than in single‐subunit deletion mutants. Unexpectedly, TATA‐containing and SAGA‐dependent genes were much more affected by impairment of tail module function than were TFIID‐dependent genes. Consistent with this finding, Mediator and preinitiation complex association with SAGA‐dependent promoters is substantially reduced in gal11/med15Δ med3Δ yeast, whereas association of TBP, Pol II, and other Mediator modules with TFIID‐dependent genes is largely independent of the tail module. Thus, we have identified a connection between the Mediator tail module and the division of promoter dependence between TFIID and SAGA. Suraiya A Ansari et al; http://emboj.embopress.org
MC associated with RNAP II.
A Functional Module of Yeast Mediator That Governs the Dynamic Range of Heat-Shock Gene Expression; Harpreet Singh et al; http://www.genetics.org
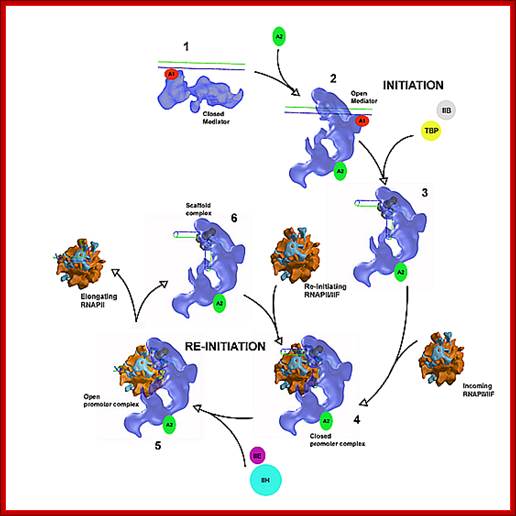
MC cycles through the activity of RNAP II
Function of MC: as mentioned above,
1. Mediator components stimulate basal transcription in association with GTF and RNAP II.
2. They enable stimulation through activators and co activators.
3. Stimulate CTD tail phoshorylation. It forms a stable complex with CTD tail.
Repressors� features:
Similar to activators there are proteins which bind to DNA and transcriptional complex and make it inactive.� Repressors can directly bind to DNA or many of them act as co repressors which are not bound to DNA, where they interact with repressors and possibly with RNAP and make transcription inactive.� There are many such proteins and they act in cell type and tissue specific manner. RAPs, Mig1, Tup1, HP1 family, MeCp, DNA- pC methylases, Histone methyl transferases, RiFs, SiR1, 2, 3 and 4, HDAC I and II families.
Repressors- work at activator sites and compete with activators,
Repressors- interact with Transcriptional factors,
Repressors- can mediate with mediator complexes.
Repressors- modify chromatin to inactivate genes at loci or genes enmass.
Repressors- have individual effects,
Repressors- have general effects,
Repressors- recruit histone deacetylases,
Repressors- can have enmass effect,
Repressors- gene silencing effect can spread.
Repressor proteins (few examples):
E2F4, E2F5: bind to DNA at GO� phase, repress gene transcription by reducing HAT activity.
pRB: Tumor suppressors, sequester E2Fs and prevent cell cycle gene transcription, recruit DHATs.
P130 and p107: RB related proteins; they form complexes with E2F repressors.
Ume6: DNA binding, recruits Sin3-Rpd3, targets histone deacetylation.
mSIN3B- HDAC: transcription repressor complex,
SiR1, 2, 3, 4:� Chromatin binding and repress.
SiR2 = Deacetylase
RAP: Repressor-activator proteins.
MeCp: methylated Cytosine binding proteins,
HP1 and family: bind to methylated histone tails; repressors.
GAL80: repressor of GAL4
Besides, the main transcriptional apparatus, including RNA polymerase II, when settles on the core promoter elements, it has to be activated.� Such activation requires interaction of the PIC components with other protein factors located in the upstream of the promoter.� In most of the cases PIC is activated by specific activators, which are located nearby at a distance.� Some of the activators such as yeast GAL gene UAS factors located upstream, far away from the PIC, stimulate and enhance the efficiency of expression by 150 to 200-fold. GAL4 facilitates the assembly of TFs on to the site but also responsible for activation.� They are specific-to-specific genes.
Some of the receptor proteins which bind to response elements also act as activators of genes.� They may act on their respective targets directly with bound ligands or act through specific co activators ligand bound or ligand activated.�
Some of the activators, when bound to DNA recruit chromatin remodulators such as Histidine acetyl transferases (HATs), SWI and SNFs.� They are responsible for acetylation of histone tails at specific amino acids such as Lysine or Arginine, thus cause loosening of nucleosome and make the DNA at promoter region accessible for the assembly of transcriptional and its related factors. Acetylases and remodulators uncover DNA from nucleosomes for the binding of specific proteins.� One of the important components that bind to such open binding sites is TF II-D, binds to acetylated nucleosomal region, for the proteins have what is called Bromo-domain. Activators also work with a complex of another set of proteins called Mediator complex, which contacts BTA elements including RNAP.�� Activators work with a variety of protein components, assist in the assembly of transcriptional apparatus, assist mediator complex of proteins, assist co activators and when every component required is assembled in the place, its activation domains induce the RNAP to get activated.
Activators not only work from near by activator elements in the promoter, but also work from far away distances, binding to enhancer elements ex. Human INF genes) and upstream activator sequences of GAL genes.
In such cases, the activator protein, can be a monomer or a dimer depending upon the kind of the factor, possesses a DNA binding domain by means of which it binds to specific sequence in the DNA.� The other domain has protein-protein interacting domain, by which it interact with certain components of Basal transcriptional apparatus (BTA).� The interacting domain brings proteins together, yet it requires another activator motif, which can prod and stimulates the PIC into activity or it can facilitate the assembly of additional factors and activate the BTA.� Again, the action of the activator depends upon its own nature.� Can the factor act on its own, or does it require binding of another factor or ligand to activate it or does it require phosphorylation by kinases for it to be active, it depends.� But what is important is that it has to physically contact the PIC.
If the activator binds upstream, then how it could contact the BTA?� It is known that the activators can contact with BTA by DNA looping.� Such looping is assisted, may be by another set of proteins such as IHS, HMG which on binding to DNA induces bending.� Even co activators work with activators and Basal transcriptional components, thus DNA loops over the transcriptional apparatus at Promoter/start site.
Activator domains, whether it is an activator or co activator, interacts with one of the many BTA complex of proteins.� Among the BTA proteins, TAFs are important for some of the TAFs are tissue specific, similarly TF II-B can also be involved and one or more subunits of RNAP II can participate in the interaction with the activator domain.� Some of the activator domains studied shows certain motifs rich in acidic amino acids (GAL4), or Proline rich regions (CTFs) or Glutamine rich motifs (SP1).� It is these motifs that when bind induce conformation changes in the whole set of protein factors, ultimately activating RNAP to initiate transcription.
To differentiate and delineate the difference between activator domain and DNA binding domain and protein-protein interacting domain, a recombinant DNA construct has come in handy in elucidating the process.� A simple but an elegant assay called Two Hybrid Assay.� In this experiment a reporter gene such as β-Galactosidase is used for expression; its expression is modulated by a combination of proteins.� A protein was constructed with the DNA binding domain and the other is for protein-protein interacting domain.� Another construct contains one domain for interacting with another protein and the other domain containing activating motif.� But this protein does not contain any DNA binding domain.� When such construct was expressed in the presence of a reporter gene containing the required promoter element, β-Galactosidase is expressed, which can be monitored by X-Gal.
Thus it is clear that the activators activate specific gene(s) expression directly, or through several other factors such as mediator complexes or through co activators, whether they are bound to upstream activator sites or bound to Enhancer regions, ultimately they act in concert with PIC especially TAFs and TF II-B and RNAP subunits by inducing allosteric transformation in their target proteins to become active.
Regulation of Gene Expression [few examples]:
Initiation of transcription, elongation and termination of it is described in other chapters.� Here few specific genes have been chosen to describe the mechanism of gene activation or gene repression.
It is important to remember those multicellular organisms are endowed with an elaborate but very complex network of gene regulatory system.� The common theme is every structural gene has its own built-in regulatory elements at the upstream region from the start.� The number and kind of elements are specific to specific gene.� These elements are called by different names such as activator, response elements, enhancer elements, insulator /boundary regions, LCR, repressor elements and so on.� These are found in complex of mix and match combinations, a combinatorial organization.� Similarly, each and every gene is taken care for its activation or for it repression in response to stimulus or tissue specifically or constitutive expression and for every eventuality, cells are provided with functional proteins which execute the process remarkably.� This network is so elaborate and complex, it may take few decades to unravel and elucidate the process.� Here few examples are given.
�
Cellular transcription factors, found either in cytoplasm or the nucleus are activated in several ways.� Few of them are briefly described here.
� Hmeo-domain genes produce Transcriptional factors de novo in response developmental signals, and then they are transferred into the nucleus, which in turn activate transcription of developmental genes E.g., Homeodomain proteins.
Certain transcriptional activator proteins are constitutively produced, but remain inactive in cytoplasm.� But when activated in the form of phosphorylation, they enter into the nucleus and activate its respective gene, e.g. heat shock proteins (HSTFs).
� In lymphocytes NFκB, a dimer of 60p and 65p is held in cytoplasm by an inhibitor protein called IκB, when the inhibitor is inactivated, NFκB is released and the same enters the nucleus and activates specific genes by binding to its upstream sequence elements.
Several transcriptional factors are sequestered and held in inactive state by specific proteins such as Retinoblastoma RB.� During mitogen induced cell division, specific Cdk-Cyclins get activated by mitogenic induced kinases; the activated Cdk-Cyclins phosphorylate RB proteins, thus the E2F are freed and they act as transcriptional factors, in association with dimeric protein called DP, activate genes required for cell cycle. Similarly, kinase activated Myc gene, by phosphorylation activates several genes required for cell cycle progression.
� In the case of steroid receptor factor remains in cytoplasm sequestered by a heat shock protein (HSP).� When a steroid (lipid soluble) moves into the cell through lipid layer, it binds to the receptor displacing HSP, thus the activated protein enters into the nucleus and binds to its response elements and activate transcription directly or through co activators.
A transcriptional factor remains inactive because of the partner protein, but when this partner protein is displaced with another factor, the dimer becomes active and activates a specific gene or genes.
Histidine Gene Expression:
Histidine gene 3 is expressed constitutively when amino acid level is normal.
� The promoter region has a two InR and start regions.�� One at normal position and the other at +12 position.�
In the upstream region it has 17 bp A/T rich sequences.� Such A/T rich blocks are found in many constitutively expressed genes.
� There is correlation between the length of the A/T rich region and the level of gene expression.� Increase in the length increases the level of gene expression.�
Such regions with A/T sequences fail to form nucleosomal structures.�
� In His 3 genes there are two such A/T blocks, and they work in either orientation, but act as activator sequences.
����������������������� ��������
-----A/T-------------A/T-----------------TATA--------+1>------+12>--->
���� �[TGACTC]�� [TGACTC]
�� ����������������������������������������������������������������
When conditions are normal, and amino acid levels are adequate, His-3 is expressed using +1 InR start point.
��
� Under starvation condition, a regulatory protein called GCN-4, General Control of Nitrogen genes, play an important role.
Under normal conditions, this protein, which is constitutively expressed, is in inactive state.� But under amino acid starvation the GCN4 gets activated.
GCN4 gets activated by GCN2 a kinase under nutrient deficiencies.
� The activated protein now binds to TGACTC sequence and activates the expression of the gene using the InR start from +12 regions.�
The protein has leucine zipper motifs, consisting of 281 amino acid (aa) long chain and they form dimers.�
� It has DNA binding domain at N-terminal region (1 to 60 aa) and it has an activator domain at 107 to 125 aa positions.
The activator domain contact RNA pol and activates the enzyme.
H2B Gene Expression in Sea Urchin:�
In the testis of sea urchin Psammechinus miliaris, the H2B1 gene is expressed, but its expression in somatic tissue is repressed.
����������� �
The promoter of this gene consists of a TATA box at �35 to �25 regions.� At �50, -70, -100 and �120 there are sequence boxes like OCTA, CCAAT, CCAAT and OCTA respectively (OCTA-ATTTGCAT)
--OCTA�CAAT---CAAT�OCTA---TATA-----InR+1>
Though the basal transcriptional apparatus binds to the promoter region, the gene expression fails, but the binding of CTFs and OCTA-1 factors to their respective sequences not only ����������� interact with one another but also interact with BTA or so called preinitiation complex (PIC) ���������� and initiate transcription with high efficiency.� In this interaction OCTA1 plays a very � important role.
� After fertilization, when the embryo starts developing, expression of H2B1 gene is repressed in cells.�
����������������������� This is because, in the embryonic cells, in the place of CTFs, another class of factors called CTF displacement factors (CDF) are produced.� This is an excellent example of differential expression of the same gene.� Different factors produced in different tissues are different, for the same gene.
� These CDFs bind to CTFs segments and displace CTFs. When CTFs are displaced though OCTA1 factors are present they won�t interact to form an active complex.� With CDFs in place OCTA1 frails to interact with transcriptional apparatus, and surprisingly even the BTA fails to assemble.�
����������������������������������� In this type of gene expression system, CTFs and OCTA-1 act as general TFs and CDP or CDFs act as gene repressors.� The expression of H2B1 gene in intestine is tissue specific, but � the factors are actually general TFs. But in embryonic cells CTF displacement factors act as general repressor factor for H2B1 gene.�
� While OCTA-1 works in most the general tissues, it does not work in lymphoid tissues.�
����������������������������������� For the expression of Immunoglobulins, OCTA-2 is required though the protein binding sequences are more or less is similar.
� The presence of sequences in the gene alone cannot initiate expression, it requires specific TFs.� Only lymphoid cells produce OCTA-2 TFs.
����������������������� What one can understand, from this simple example, is that each tissue produces specific �� factors. Some are for house keeping activities and others for tissue specific expression.�
� Genes may have upstream sequences, but if the factors required for the binding to express the gene are not present, then the genes won�t be expressed.
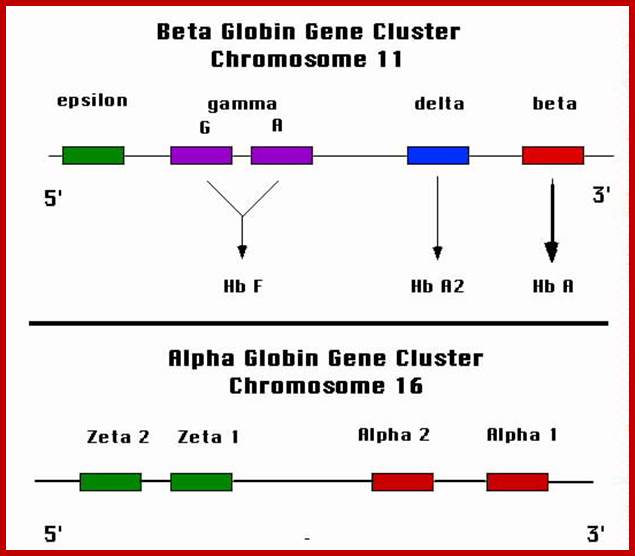
Globin Gene Expression:
Globin genes exist in many allelic forms.� They are expressed during development and finally some are expressed in tissue specific manner.��
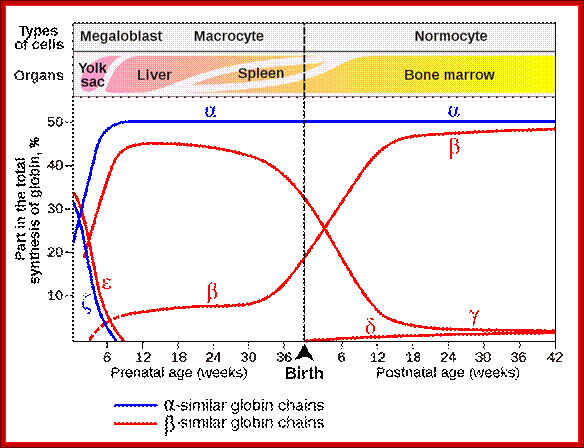
Different types of
hemoglobin, all derived from an original globin gene in early vertebrates;
�Postnatal_genetics.svg: original: Furfur,
File:Haemoglobin-Ketten.svg, derivation/translation: Leonid 2 derivative work:
Leonid 2 (Postnatal_genetics.svg) [CC-BY-SA-3.0 or GFDL], via Wikimedia Commons;
http://biobook.nerinxhs.org/
� All of them are organized as a cluster of genes in segment of 100,000 bp long, but their individual genes� expression is controlled by a region called locus control region (LCR).� The LCR contains lot of CG or GC rich regions and they are recognized by SP1 factors, which are ubiquitously produced.� These genes are expressed in temporal fashion in different tissues.� The regulatory proteins that bind to LCR region activate individual genes.� Individual genes have their specific promoter elements.� The LCR region shows 5 DNase-I hypersensitive sites on upstream of the globin clusters.
�
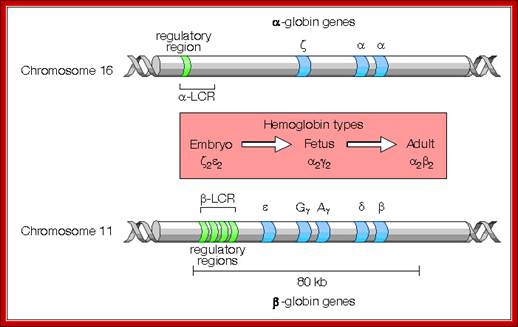
Differential and temporal expression of different globin genes; alpha globin are located on chromosome 16 and globin beta are located on chromosome 11.� Alpha globin cluster contains one control and the beta contains four control regions, Developmental Biology, Dr. Brain, E. Staveley. http://www.mun.ca/biology.
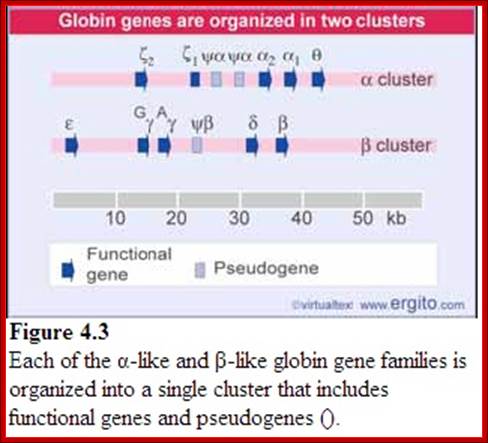
Globin alpha and beta clusters are formed by duplication and divergence; http://molecularstudy.blogspot.in/
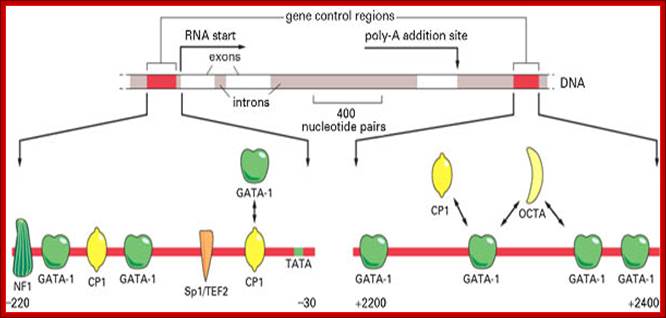
The diagram shows some of the gene regulatory proteins thought to control expression of this gene during red blood cell development. Some of the gene regulatory proteins shown, such as CP1, are found in many types of cells, while others, such as GATA-1, are present in only a few types of cells, including red blood cell precursors, and are therefore thought to contribute to the cell-type specificity of beta-globin gene expression. As indicated by the bidirectional arrows, several of the binding sites for GATA-1 overlap those of other gene regulatory proteins; it is thought that occupancy of these sites by GATA-1 excludes binding of other proteins. (Adapted from B. Emerson, In Gene Expression: General and Cell-Type Specific (M. Karin, ed.), pp. 116-161. Boston: Birkhauser, 1993.)
Globin gene regulatory elements; The gene control region consists of NF1, GATA, Cp1, GATA-1, Sp1/TEF2, Cp1 promoter elements to which specific factors bind and initiate transcription; globin gene consisting of 3exons and 2 introns. Role of GCN4 in histidine gene expression; Alberts et al, http:// garland Science.com
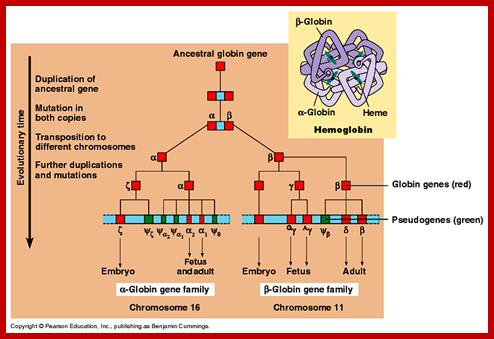
www.dianliwenmi.com/p; http://www.bio.utexas.edu
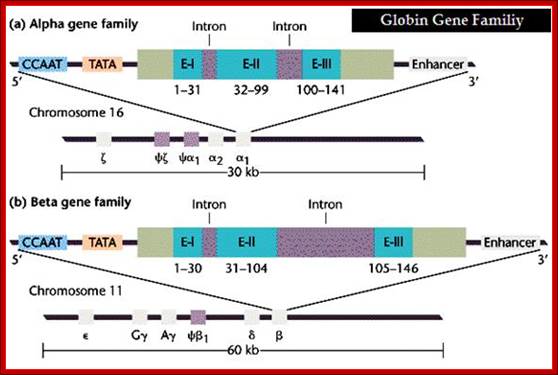
Alpha Globin genes: Globin zeta is expressed at very early embryonic stage; later alpha takes over and continues in the adult stage.
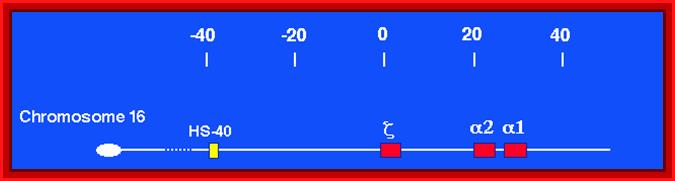
Alpha globin clusters are located in chromosome 16 (30kb region) consists of Chi and two alpha genes. http://sickle.bwh.harvard.edu
Alpha gene promoter elements:
5�--CCAAT�TATA�[-E1�In1�E2�In2�E3--]---Enhancer�3�
E = Exons: E1= 1-31, E2=32-99, E3= 100-141=~133
Globin alpha is expressed very early, after zeta.
https://sjhsrc.wikispaces.com
Beta Globins:
They are expressed even during embryonic development but at very low level, but after birth their level increases, and its level is maintained through out.� This gene is expressed in mature erythroid cells (in humans-RBAs).
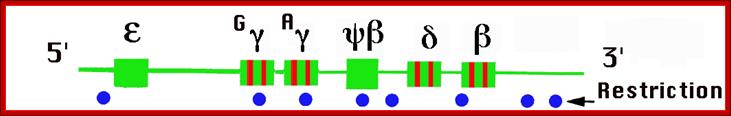
Beta globin clusters are located on chromosome 11 (60kb region). Errol L. Fields; http://sickle.bwh.harvard.edu/
Beta Globin Gene:
5�-GATA-OCTA-GATA-CCAAT--TATA+1---Ex1�In1�Ex2�In2�Ex3�enh-
E1 =1-30bp, E2=31-104bp, E3= 105-146 bp =144
Globin beta is expressed in adult cells for the GATA factors are produced, which are absent in younger stage. While in very early fetal stage the gene is not expressed by the lack of GATA factor and globin gamma somehow down regulates beta.
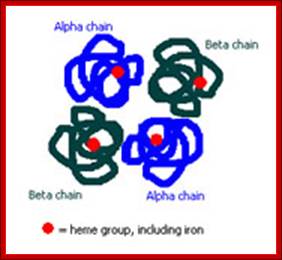
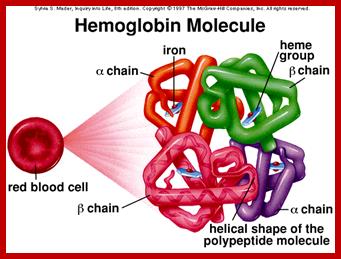
Tetramer-2alpha and 2beta subunits; http://carolguze.com
������������������ ��������� ������������� Two tertiary structures make up hemoglobin;
http://www.hinsdale86.org/
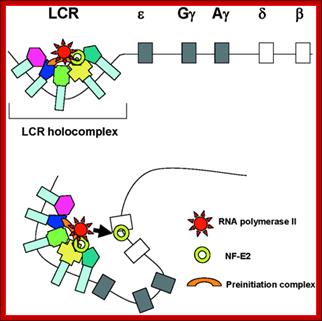
Globin Gene Expression �both LCR and Individual Promoters Involvement:
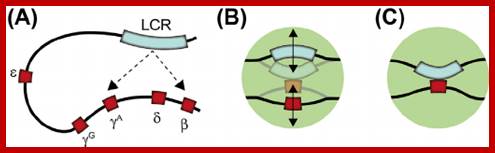
Model of transcription complex recruitment to the β-globin gene. According to a recent model, the LCR and other HS sites interact to form a chromatin hub. In the active chromatin hub, the expressed genes interact with the HS sites. We propose that transcription and other protein complexes are first recruited to a highly accessible LCR holocomplex in the context of the proposed chromatin hub. The genes come in close proximity to the LCR holocomplex by as yet unknown mechanisms that may involve local remodeling of chromatin structure at the active promoters. Transcription complexes are then transferred from the LCR to high affinity binding sites at the globin gene promoters. This transfer is facilitated by NF-E2 and/or related proteins.
Interactions within the beta-globin gene locus. (A) The stochastic looping mechanism of the interaction of the human locus control region (LCR) with different beta-globin genes in transgenic mouse erythroid cells. (B, C) The nature of the LCR-globin gene interactions. (B) The elements produce micro-oscillating movements within a small nuclear volume and can occasionally establish short-lived contacts. (C) The elements establish a stable long-lived contact. https://www.researchgate.net
Epsilon and Gamma gene expression-
�
They are expressed very early in the development.�
� Initially during the development globin epsilon is expressed.� Then gamma-A starts expressing and the level of Epsilon goes down.
Gamma-G and Gamma-A are synthesized during the early stages of development that is in the yolk sac and fetal liver.�
� This expression takes place from 6th week to 30 weeks, then onwards or later i.e. at the time of birth their synthesis goes down.
Globin gamma:
�����������
NF1�GATA�Cp�GATA�Sp1�Cp1------TTA-�+1
In fetal cells Cp1 is expressed and it is responsible for the expression of globin-gamma, but in adult cells NF-e is expressed and it blocks the expression globin-E
Delta globins:�
These genes are expressed at 36 th week after conception and its expression continues even after birth, but at very low level.
The Globin genes are found in a 100, 000 bp region and the locus control region (LCR) is about 50Kbp.
Beta Globin is expressed in adult Erythroid cells.� Its partner is alpha globins.� Expression of these genes is tissue type specific. Globin genes have several sequence modules in their promoter regions from the left i.e., from �220 upstream to down wards -30 TATA.� The modules are for the binding of factors such as in the same order.�
NF1, GATA, CP1, GATA, SP1/TEF2, CP1, TATA�nr+1>
General transcriptional factors available are CP1 and SP1, but the gene expression depends upon the GATA binding factor, and this factor is produced only in some specialized cells; among them are red blood cells (RBC).� It is the GATA factors that activate beta-Globins genes in Erythroid cells but not in other cell types; this is an excellent paradigm for cell type of tissue or cell type expression.
�Globin Gene Cluster:
����������������������������������� ���
|
��������� LCR��������������������������� |
ε .�� gγ.���� gγ.����� gα.���� δ����� β |
|
--I---I---I---I---I----I-----I---------I--- |
----I-----I--------I------I-----I------> |
|
-90 |
-75 |
-30 |
+1 |
|
GCCACACC------------- |
GGCCAATC------------- |
CATAA------------------- |
InR �>A->DPE |
��
Globin gene promoter regions:
I---NF1--GATA�Cp1-GATA-Sp1�Cp1-------TATA---- +1>-----
�
In fetal cells, expression of gamma globins, cell specific Cp1 factors are produced and they bind to their respective promoter elements and initiate transcription and produce gamma globin proteins.
In adult cells, as NF-E factors are produced, they displace Cp1 TFs and in turn NF-E binds to their respective sequences and block the transcription of Gamma Globins.
In adult erythroid cells GATA2 factors are produced, which bind to their respective sequences and induce the expression of Globin beta proteins.� Absence of GATA2 factors in fetal cells render them unexpressed.� While alpha globins are expressed after a transitory expression of Zeta genes.
Beta expression in fetal is blocked by NF factors, but in adult cells beta is expressed because GATA and CP1 factors act on the promoters.
�
hGCR complexes:
Grappling with the HOX network in hematopoiesis and leukemia: Glenda J. McGonigle1, Terence R.J. Lappin1, Alexander Thompson1; 1Haematology, Centre for Cancer Research and Cell Biology, Queen's University Belfast, 97 Lisburn Road, Belfast, BT9 7BL
Similar to LCR upstream regulators one finds such global regulators in homeaobox genes; they are called GCR
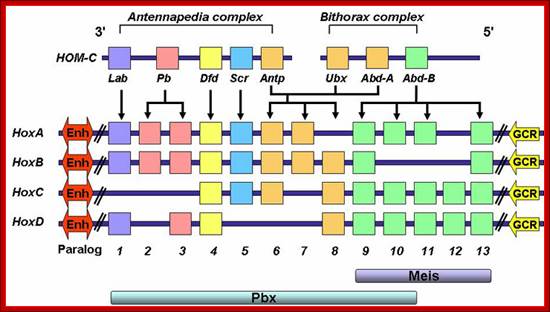
Figure. Schematic representation of conservation of the mammalian HOX gene network from DrosophilaHom-C depicting preferential binding of Pbx to paralog groups 1-10 and Meis to paralog groups 9-13. Downstream Enh (Enhancer) sequences and upstream GCR (global control region) elements thought to control global expression of individual clusters are represented.������������ � http://www.bioscience.org��������������������������������������������������� �����������������������������������������������������������������������
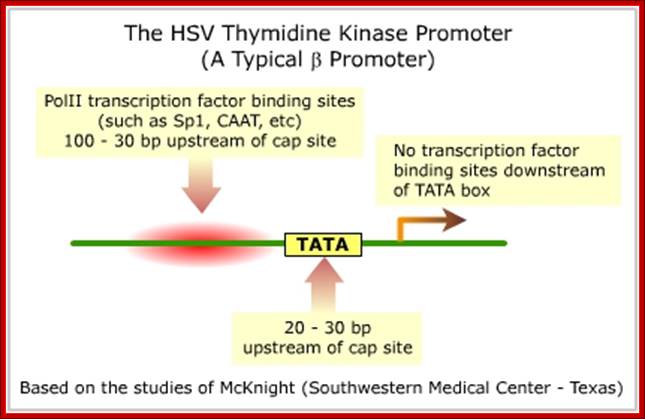
Expression of Thymidine Kinase:
This gene is found, not only in virus Herpes virus simplex, but also in most of the EK systems.�
� The promoter of this has many upstream elements -40(GC),
�-89(CAAT), -100(GC) and at �120 (OCTA).
---OCTA�GC---CAAT---GC-----TATA---InR>�
Transcriptional apparatus by itself can initiate transcription, but the binding of SP1 and CAAT factors increase the efficiency.
� CTF and Sp1 factors are general factors, which are ubiquitously found in most of the cells.� They increase the efficiency by interacting with the pre-initiation complex and activating the enzyme Interacting with what component of the RNAP-II complex.
|
-120 |
-100 |
-89 |
-40 |
-25/-30 |
+1 > |
|
OCTA----------- |
GC-------------- |
CAAT----------- |
GC-------------- |
TATA----------- |
�A>DPE |
Fig: Upstream sequence of the TK gene. Nucleotide I is the A of the initiation codon. mRNA starts around nucleotide -60. The box indicates the sequence highly conserved in mouse, hamster and human TK promoters. Underlined are the GGGCGG motifs (binding sites for transcription factor Spl, in direct and reverse orientation), broken lines indicate CCAAT-like sequences (in direct or reverse orientation), the wavy line denotes a possible binding site for transcription factor AP2, a sequence resembling the binding site for nuclear factor 1 (NF1, see ref. 23 for a review on trans-acting protein factors) is indicated by the dotted lines. The sequence has been deposited with the EMBL Data Library (accession number X12824).
Dr. Edward Wagner; http://darwin.bio.uci.edu/
Some more Factors:
In human species fetal gamma Globin expression is repressed in adults by the binding of NF-E factor.� Binding of this factor prevents the binding of CP1 to CAAT.� But in fetal cells CP1 is expressed but not NF-E.� The CP1 binds to its promoter elements, which results in the expression of gamma Globin gene.
Some of the GC box binding factors like SP1 compete among themselves.� SP1 of one tissue acting as a positive factor can acts as a negative factor for another gene in another tissue.
A component called FOS, as a hetero dimmer, like an inactive complex like C-jun-B-jun can replace jun-fos.� This competes with hetero dimer jun-fos.
The IkB an inhibitor can cause dissociation of TAFs from transcription complex.
A heat shock protein can interact with a Gluco-corticoid receptor, and makes it incapable of binding to DNA.
Certain trans-activating factors bind to DNA and interact with RNAP and other TFs and activate the expression.� During repression the activation domain of TAFs is made inactive. e.g.� binding of GAL-80 to GAL-4 makes GAL-4 inactive, even though it is bound to DNA.� The binding of Galactose to GAL 3 makes it active, which in turn binds to GAL-80; the activated Gal 80-gal3-galactose undergo conformational changes and they release from Gal4 thus Gal4 becomes active.
Silencer- Yeast Mating type:
Certain factors repress transcription from a distance, and they are called as silencers (opposite to enhancers).� They act in either orientation, e.g. �Mating type locus MAT is associated with two silent loci, one HMR located to the right side of the MAT and the other HML is located to the left of the MAT.� These silent loci can be alpha �α� and mating type �a� and they can replace MAT to bring about a change in mating type.� A silencer located at one Kbp upstream from mating loci represses both HMR and HML loci.�
------HML--------MAT-------HMR---
![]()
Map showing HMl alpha MAT alpha and HMRa loci and direction of transposition
Yeast mates signaling one another; fig.cox.miami.edu/
Fission yeast can switch between two mating types, P and M, by replacing the genetic information that specifies one mating type with that from another. This occurs through a recombination mechanism in which one of two silent donor loci, containing genetic information that specifies the two different mating types, replaces the information at the mating-type locus. In fission yeast, the mating-type locus is called mat1, whereas the two silent donor loci are referred to as mat2 (P) and mat3 (M). Importantly, the choice of donor locus is non-random in that recombination almost always results in a switch in mating type. H3-K9 methylation and Swi6, the S. pombe homologue of mammalian HP1, seem to function in mating type-switching by facilitating the spreading of a complex composed of Swi2 and Swi5 over the mat2 and mat3 region. In cells of the P-mating type, the Swi2�Swi5 complex associates with the boundary of the mat3 locus, but does not move inward towards mat2 (see figure). By contrast, this complex spreads over the entire mat2 and mat3 region in cells of the M-mating type. In the absence of Swi6 function, the Swi2�Swi5 complex associates with the mat3 boundary region, but does not move inward in M cells. In such cells, the mat3 locus is incorrectly used to replace the genetic information at the mating-type locus, therefore preventing a switch in mating type. This indicates that cell-type-specific spreading of the Swi2�Swi5 complex is important for mating-type switching and that Swi6, and therefore heterochromatin, is required for spreading. The mechanism by which Swi6 facilitates spreading is unknown. One possibility is that spreading is the result of direct physical interactions between these proteins that occur in a mating-type-dependent fashion.
These silencers are called as E and L or they are called as ER9 (HMR-E) and EL (HML-E).� Besides E and L silencers, there are several other trans activating factors involved in silencing activity, e.g. HMR-E has a binding site for TF-RAP-1 and another protein Abf-P is required at origin.� Both are required for repression.� Alpha-2 coded by one of the genes at HML loci binds to HML-E and represses mating type genes.� There are other SIR factors, like SIR1, SIR 2, SIR 3, and SIR 4, which are involved in silencing genes.
HPRT Gene:
Hypoxanthine phosphoribosyl transferase is a critical enzyme in nucleotide recycles.
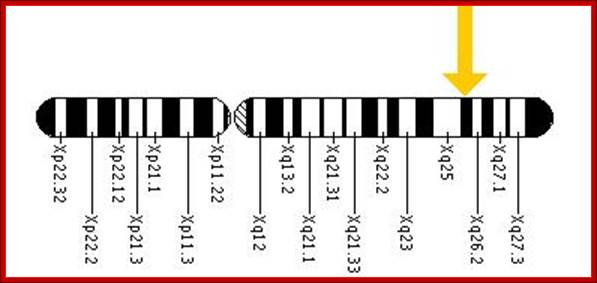
The HPRT1 gene is located on the long (q) arm of the X chromosome at position 26.1. More precisely, the HPRT1 gene is located from base pair 133,421,922 to base pair 133,462,361 on the X chromosome. >slideplayer.com/slide
Regulatory elements: (TATA less promoter):
---GC�GC�GC�GC�GC�GC� InR+1�(exon)8-(intron)7-
GC = CGCG
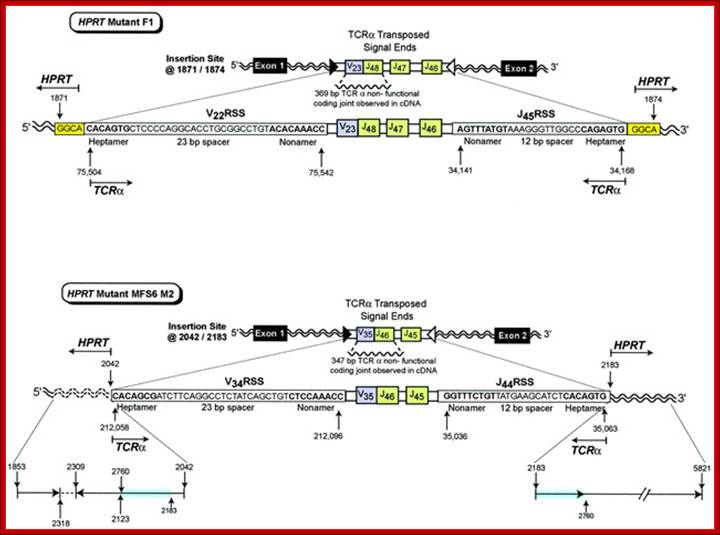
In vivo transposition of TCR![]() V-J signal ends from chromosome 14 into the HPRT gene on the X
chromosome. HPRT mutant F1 contains transposed TCR
V-J signal ends from chromosome 14 into the HPRT gene on the X
chromosome. HPRT mutant F1 contains transposed TCR![]() signal ends containing
signal ends containing ![]() 16.8 kbp of intervening sequence that includes the V22 RSS
(heptamer�23 bp spacer�nonamer), a V23 joined to J48 non-functional coding
joint, J47, J46, and the J45 RSS (nonamer�12 bp spacer�heptamer) inserted into HPRT
at bases 1871/1874. The insertion site is located in intron 1 of the HPRT gene.
A 4 bp target site duplication, GGCA, is shown at both sides of the insertion. HPRT
mutant MFS6 M2 contains transposed TCR
16.8 kbp of intervening sequence that includes the V22 RSS
(heptamer�23 bp spacer�nonamer), a V23 joined to J48 non-functional coding
joint, J47, J46, and the J45 RSS (nonamer�12 bp spacer�heptamer) inserted into HPRT
at bases 1871/1874. The insertion site is located in intron 1 of the HPRT gene.
A 4 bp target site duplication, GGCA, is shown at both sides of the insertion. HPRT
mutant MFS6 M2 contains transposed TCR![]() signal ends containing
signal ends containing ![]() 16.1 kbp of intervening sequence that includes the V34 RSS
(heptamer�23 bp spacer�nonamer), a V35 joined to J46 non-functional coding
joint, J45, and the J44 RSS (nonamer�12 bp spacer�heptamer) inserted into HPRT
at 2042/2183. The insertion site is located in intron 1 of the HPRT gene.
Reading 5' to 3', the 5' insertion site includes bases 1853�2318 followed by
the inverted sequence 2309�2123 and 2760�2042. There is normal sequence at the
3' HPRT insertion site from 2183 to 5821 that includes the same duplication of
bases from 2183 to 2760 observed at the 5' insertion site. HPRT bases that are
duplicated are highlighted in blue. The inverted segment is shown by an arrow
in the 3' to 5' direction. http://emboj.embopress.org
16.1 kbp of intervening sequence that includes the V34 RSS
(heptamer�23 bp spacer�nonamer), a V35 joined to J46 non-functional coding
joint, J45, and the J44 RSS (nonamer�12 bp spacer�heptamer) inserted into HPRT
at 2042/2183. The insertion site is located in intron 1 of the HPRT gene.
Reading 5' to 3', the 5' insertion site includes bases 1853�2318 followed by
the inverted sequence 2309�2123 and 2760�2042. There is normal sequence at the
3' HPRT insertion site from 2183 to 5821 that includes the same duplication of
bases from 2183 to 2760 observed at the 5' insertion site. HPRT bases that are
duplicated are highlighted in blue. The inverted segment is shown by an arrow
in the 3' to 5' direction. http://emboj.embopress.org
DHFR Gene regulation:
Dihydrofolate reductase is a critical enzyme involved in nucleotide bio-pathway.
IIn --1 �1�1�I�1- I- II- I -LE2F-e - - - - - - ATG 0o r.
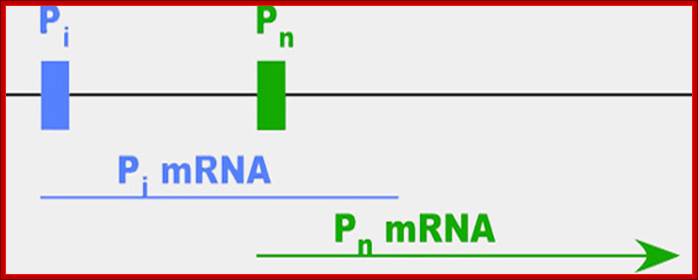
FIG: DHFR promoter transient expression vector. pDHF/CAT was constructed by cloning the hamster DHFR promoter fragment from nucleotide position -210 to -23 (relative to ATG = position 1) 5' to the bacterial CAT gene and simian virus 40 poly(A) signal in a pUC18 vector.
The major start site of DHFR transcription (bold arrow) is at nucleotide position -63, and nucleotide position -107 is the minor start site. GC boxes are indicated by stippled boxes and are numbered in the text I-IV, proximal to distal; open boxes indicate additional conserved sequence elements. Binding sites for the transcription factor E2F are indicated by the hatched box.
-GC-GC-GC-GC-GC-GC---+1 EXONS 6�Introns5�
Like many genes, DHFR has more than one promoter (diagrammed above)�the site at which proteins bind to kick off the process of making an RNA message (mRNA) out of the gene. When cells are dividing, the gene is active and 99 percent of the mRNAs start at the second promoter, which is called Pn for normal. When cells stop dividing, most of the mRNA starts at the first promoter, which we'll call Pi. The researchers were surprised to find, however, that these messages didn't actually contain the full sequence needed to make functional DHFR protein�instead, the stopped shortly after Pn.
Ribosome Protein Genes:
Ribosomal protein genes (RPG) in eukaryotes are found in multiple copies; the number of genes is about 73 (Hu) or 79 (drosophila and 78 for mitochondria). Average size of the genes is about 4.4kbp long and contains ~5.6 exons per mRNA. The largest gene is 25kb and the smallest is 0.9kb. The promoter elements contain several GC boxes.� There are no common motifs except the start site; the 3� end is 42-56 bp long. Ribosomal proteins account for 10% of the total cellular proteins. Similarly, the cytoplasmic rRNA accounts for 90% of the total RNA.� Regulation of ribosomal protein genes is still not clear, this is after fifty or more years.
5�-GC-GC-GC-GC-GC---TATA/�40 bp�pyC+1py-----------------//-T/t-3�
(1) transcription starts at a C residue within a characteristic oligopyrimidine tract; (2) the promoter region is GC rich, but often has a TATA box or similar sequence element; (3) the genes are small (4.4 kb), but have as many as 5.6 exons on average; (4) the initiator ATG is in the first or second exon and is within � 5 bp of the first intron boundaries in about half of cases; and (5) 5′- and 3′-UTRs are significantly smaller (42 bp and 56 bp, respectively) than the genome average. Comparison of RP genes from humans, Drosophila melanogaster, Caenorhabditis elegans, and Saccharomyces cerevisiae revealed the coding sequences to be highly conserved (63% homology on average), although gene size and the number of exons vary; Maki Yoshihama1, Tamayo Uechi1 et al.
Similarly, expression nucleolar gene for rRNA synthesis is often biased for the diploid genomes, derived from one species more prone for rRNA gene expression than the other.
Wilms� Tumor Gene Regulation (WT1):
In mammalian systems there are many factors that suppress the expression of certain genes. A good example is RB Retinoblastoma holding back genes involved in cell cycle regulation.
Wilms' tumour 1: Sitaram RT, Degerman S, Ljungberg B, Andersson E, Oji Y, Sugiyama H, Roos G, Li A - Br. J. Cancer (2010);
Wilms tumor (WT1) gene was discovered as a tumour suppressor gene. Later findings have suggested that WT1 also can be oncogenic. This complexity is partly explained by the fact that WT1 has a number of target genes.WT1 and its target gene human telomerase reverse transcriptase (hTERT) were analyzed in clear cell renal cell carcinoma (ccRCC). In vitro experiments were performed to examine the functional link between WT1 and hTERT by overexpression of WT1 isoforms in the ccRCC cell line, TK-10.WT1 demonstrated lower RNA expression in ccRCC compared with renal cortical tissue, whereas hTERT was increased, showing a negative correlation between WT1 and hTERT (P=0.005). These findings were experimentally confirmed in vitro. The WT1 generated effects on hTERT promoter activity seemed complex, as several negative regulators of hTERT transcription, such as SMAD3, JUN (AP-1) and ETS1, were activated by WT1 overexpression. Down regulation of potential positive hTERT regulators, such as cMyc, AP-2α, AP-2γ, IRF1, NFX1 and GM-CSF, were also observed. Chromatin Immunoprecipitation analysis verified WT1 binding to the hTERT, cMyc and SMAD3 promoters. The collected data strongly indicate multiple pathways for hTERT regulation by WT1 in ccRCC.
Acute myeloid leukemia in adults is a common and lethal malignant disease. Despite the tremendous efforts in the improvement of treatment in recent years, the survival of acute leukemia in adults remained poor. The Wilms� tumor (WT1) gene, located at chromosome 11p13, was identified as a gene responsible for Wilms� tumor, a kidney neoplasm of childhood. The WT1 gene is encoded by 10 exons with different transcripts that subjects to alternative splicing. WT1 gene encodes proteins isoforms with molecular masses ranging from 48 to 54 kDa with four zinc finger motifs. WT1 gene plays multiple and important roles in cell biology, such as cell and tissue development, cell proliferation, differentiation, and apoptosis It has been classified as a tumor suppressor gene�encoding a transcription factor. Expression of WT1 has been observed in different types of solid cancers, such as ovarian cancer, mesothelioma of the lung, melanoma, breast cancer, as well as in Wilms� tumor.� It has been reported that the Wilms� Tumor Gene (WT1) is expressed in leukemia blasts, irrespective of the subtypes of acute leukemia.� Early report showed that WT1 antisense oligonucleotides could induce apoptosis in myeloid cell lines.� In recent years, it has been found that WT1 could be used as a molecular marker to generate specific cytotoxic T cells (CTL) against leukemia cells.� The anti-leukemia activity of WT1-induced CTLs is reported to be HLA-A restricted and has been used as adoptive immunotherapy in some small scale clinical trials in patients with acute leukemia; Frederik Damm, Michael Heuser, Michael Morgan et al.
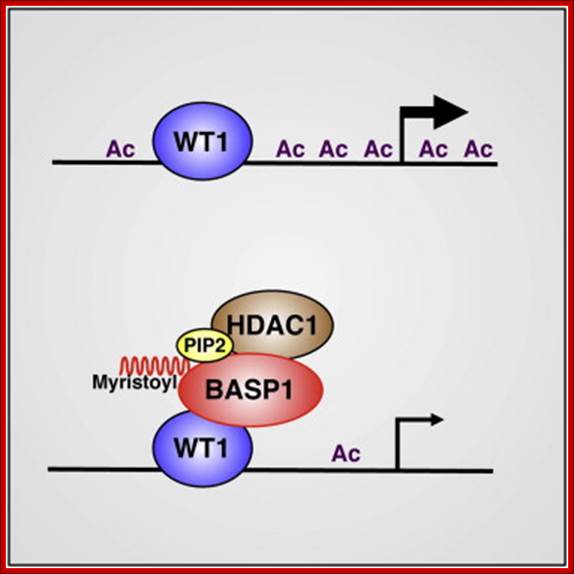
Frederik Damm, Michael Heuser, Michael Morgan et al. http://www.cell.com
The Wilms' tumor 1 protein WT1 is a transcriptional regulator that is involved in cell growth and differentiation. The transcriptional corepressor BASP1 interacts with WT1 and converts WT1 from a transcriptional activator to a repressor. Here, we demonstrate that the N-terminal myristoylation of BASP1 (Brain Acidic Soluble Protein) is required in order to elicit transcriptional repression at WT1 target genes. We show that myristoylated BASP1 binds to nuclear PIP2, which leads to the recruitment of PIP2 peroxisome induction pathway protein 2 to the promoter regions of WT1-dependent target genes. BASP1's myristoylation and association with PIP2 are required for the interaction of BASP1 with HDAC1, which mediates the recruitment of HDAC1 to the promoter and elicits transcriptional repression. Our findings uncover a role for myristoylation in transcription, as well as a critical function for PIP2 in gene-specific transcriptional repression through the recruitment of histone deacetylase. Eneda Toska1, Hayley A. Campbell2, Jayasha Shandilya1, Sarah J. Goodfellow2, 3, Paul Shore2, Kathryn F. Medler1 and Stefan G.E. Roberts
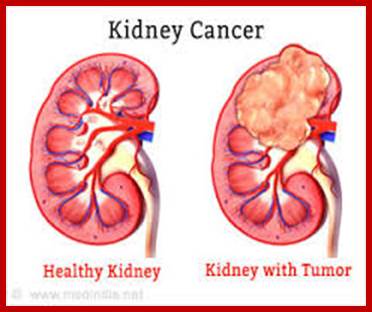
https://www.healthandholidays.com
Stages of development of kidney tumors; http://www.dranupramani.com
Wilm�s tumor gene WT1 (Nephroblastoma) is a master switch for the development of the genitourinary system and other organs. In mammalian kidneys, especially children, are susceptible for tumor development due to certain recessive mutations. Wilms� tumor affects 1:10,000 children and accounts for ~8% of all pediatric malignancies. �This disease has been identified by Max Wilms (1899) later called as Wilmer disease or Wilms tumor (WT) disease. The human WT1 gene spans ~50 kb and consists of 10 exons. The transcript undergoes alternative splicing and generates multiple forms of WT1 proteins. It is a TF contains C2H2 type zinc finger and binds to GC rich regions. �The WT1 protein (52-56kDa) is a repressor of Egr1 gene (Early Growth Response). EGR1 gene product acts as a transcriptional activator in inducing cell proliferation. If WT1 gene undergoes mutation, it fails to repress Egr1 gene.� WT1 gene is located in chromosome11 at p18 position. Continuous production of EGR1 can lead to cancer.
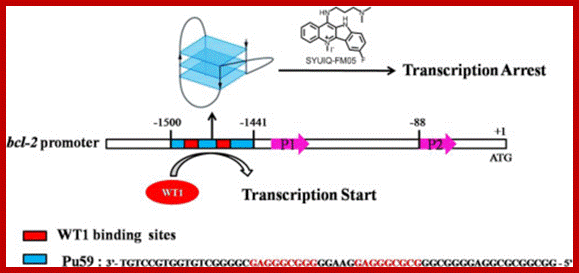
Fig. Sequence logo obtained after alignment of 28 potential WT1 binding sites from 12 human gene promoters regulated by WT1. For the purpose of this alignment, − 1500 segments of the coding strands of the promoters were searched for binding sites fitting the format 5′-N GNG NGG GNG NNN N-3′. The binding site for ZF1 (positions 11, 12, 13 and 14 from left) evidently has poor consensus, unlike the ones for ZF2�3�4 (positions 2 to 10). Note: The overall height of a stack indicates conservation at that position, while the height of symbols within the stack indicates the relative frequency of each nucleotide at that position. Lack of symbols indicates absence of conservation at a given position.
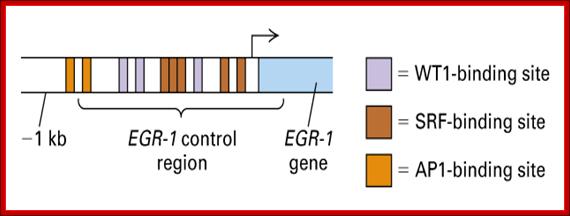
���������������������� 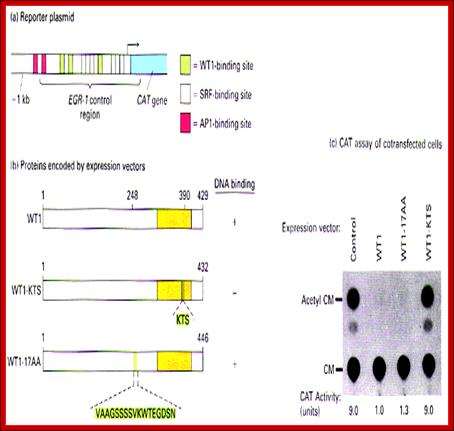
http://genatlas.medecine.univ-paris5.fr/
� The WT1 gene transcript consists of 10 exons.� The WT1 protein at C- terminal region has zinc finger domain with Cys2 and His2. The N- terminal region has repressor domain.�� The promoter of EGR1 has several sequence modules such as AP1 (x2, enhancer), WTR1 (x2), SRF1, and WTR1 (arranged in the same sequence) in the upstream region of the gene. ��The WT1 protein is constitutively expressed.� When the WT1 protein binds to WT1 element it represses the expression of EGR1 gene, thus tumor formation is suppressed.
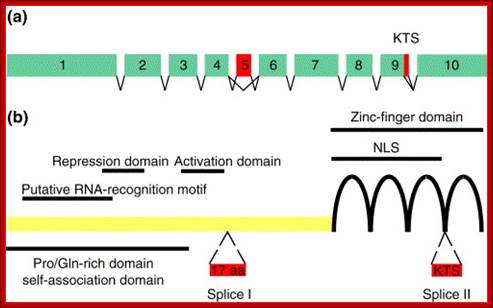
Fig. Structure of the WT1 mRNA (a) and WT1 protein (b). WT1 is encoded by ten exons. Alternative splicing of exon 5 and alternative usage of two different splice-donor sites at the 3′ end of exon 9 produce four different splice forms [28]. The inclusion of splice I (exon 5) leads to an insertion of 17 amino acid residues (aa) into the regulatory domain of WT1. The splice-II sequences at the 3′ end of exon 9 consists of the tripeptide KTS, which is inserted between zinc fingers III and IV. A Pro/Gln-rich region, which is almost identical to the self-association domain, and, as well as a repression and an activation domain, are indicated. WT1 seems to harbor multiple nuclear-localization signals (NLS), some of which are located within the zinc-finger region. A predicted RNA-binding motif resides in the N-terminus of the molecule. Christoph Englert �http://www.cell.com
The Wilms tumor-suppressor gene WT1was originally identified through its involvement in the development of a pediatric kidney tumor. Recent genetic data show that mutations in the WT1 gene cause a variety of other diseases, and new biochemical evidence suggests that the WT1protein is not only a transcription factor but might also act at the post-transcriptional level. But in patients with a mutation in the repressor domain of WT1 (homozygous recessive), the protein produced does not bind the promoter elements, so the EGR1 gene is allowed to express, so it causes tumors.� If the WT1 is produced in correct form, it represses EGR1 production and suppresses tumor formation.
Enhancer Modules, Sequences and Factors-SV40:
|
E-CBP |
EcbF; 60kd;LZ |
GGTGTGACG |
Enhancer Caat factor |
|
AP1 |
|
ATTAGTCAGCA ATTAGTCAGCA |
Sv40 enhancer binding, like sp1 |
|
AP2 |
|
TGGAAAGTCCC |
|
|
AP3 |
|
AGGGTG |
|
|
AP4 |
|
GCTGTGG |
|
|
NF Kb |
|
AGG (T/C) ATGCA |
|
|
SP1 binding |
|
GGGCGG |
|
|
C/EBP |
|
CCAAT |
Cat enhancer binding protein |
Gene expression at transcription level is controlled at initiation stage by the enzyme and other accessory factors. For efficient initiation, or increased rate of initiation the transcriptional apparatus requires few more additional factors; this in addition to the whole of RNAP-complex and TFII-complexes, which assemble as basal transcriptional apparatus or pre-initiation complex.
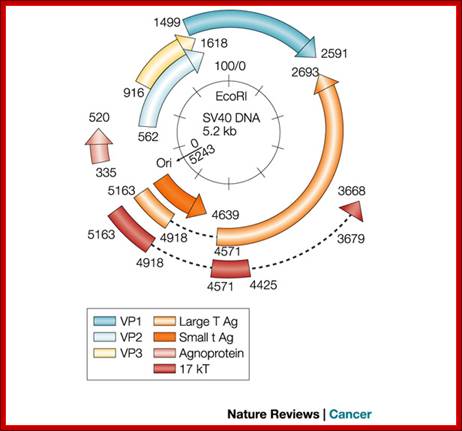
SV40 and human tumours: myth, association or causality? The simian virus 40 (SV40) genome is small (5.2 kb) and contains a limited coding capacity (see accompanying figure). It comprises three parts � a non-translated regulatory region of about 400 bp in size that contains the origin of replication (ori) and the promoters and enhancers that control replication; the early region that encodes the replication proteins (T Ag, t Ag and 17 kT protein) is expressed soon after the virus enters a cell; and the late region that encodes the capsid proteins (VP1, 2 and 3) and a maturation protein (agnoprotein), and is expressed efficiently only after viral DNA replication has begun.;SV40 regulator regions: Adi F. Gazdar, https://www.nature.com
Enhancer sequences are found in modules, one CCAAT, to which E/CBP binds and the second module is GGGCGG to which sp1 factors bind.� In SV40 enhancers facilitate the transcription of T antigen which leads to replication. The same enhancer module, found in the region of origin, also facilitates the binding of transcriptional factors to transcribe the late genes to produce viral coat proteins.� Enhancers act in either direction.� They can be found in the upstream region of the promoter or down stream of the promoter or in some it is found in an intron region.� Many a time�s activation of gene by the nearby enhancer elements is blocked by the insulator sequences to which insulator proteins are bound.� In SV40 the 72 bp enhancer contain 3 AP binding sites each with dyad sequence modules called A C B; A is bound by AP4-AP1, C is binds AP3-AP2 and B binds to OCTA1 and AP1
--72--&--72--- [GC]x 6 �AT-- rich-Pentamers-TATA-+1>�
Each Enhancer block contain 3 dyad sequences-
����������� ����������������������� [----]--[----]------[---]-[------]--------[---]-[---]-
[AP4-AP1]� [AP3-AP2]�[OCTA1-AP1]

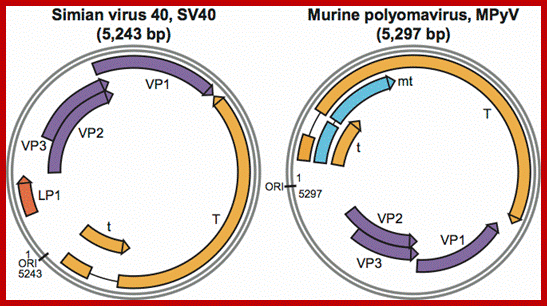
https://talk.ictvonline.org
Initiation in many genes requires some additional factors, which are ubiquitously produced in most of the cells. These are general transcriptional activators, which bind in the upstream of the start at different distances from the RNAP-TF complex.� All these put together form super complexes. Some of these modules are simple sequences, like GC box, CAAT box, OCTA sequences, including some nuclear factor (NFs) binding sequences.
�
There are many general factors such as sp1, which bind to GC boxes, CTF factors bind CAAT sequences, which are found as a family of factors, and OCTA binding factors bind OCTA eight-mers; all of them do contact with basal transcriptional apparatus, because most the binding sequences are found very near the RNAP-II-TFs. The position of each of them from the start and copy numbers of each module and the distribution and combination of them is specific for each gene and for each tissue.�
The module organization is a ubiquitous, but unique to each kind, whether they are expressed as house keeping genes or expressed in tissue specific manner or expressed stage specific or induced expression.� When compared to most of the house keeping genes with that of tissue specific, stage specific or induced in repose to stimuli, the promoter region in addition to the general sequence modules consists of another set of sequences located at different distances and with different combinations. Most of these modular elements are used for the binding of specific DNA binding proteins which bind in sequence specific manner, this apart, they interact among the other factors as well as with transcriptional apparatus. This prodding of the Transcriptional Apparatus, leads to activation of the enzyme to initiate transcription and increase the efficiency of initiation, or increase the rate of transcription from basal level to 200 or 1000 times the normal.�
The sequences, according to their functions, are named as activator elements, upstream activator elements, enhancer elements, or response elements.� The sequences come in different modules, positioned at different positions from the start site and all these are organized for that specific type of cell, or to specific situation.�� The number of each kind of elements and position in a given protein vary.�
It is not just the sequence modules present in the DNA of the gene alone works, it is the factors as small as metal ions, small organic molecules, proteins present or produced that makes the difference.� It is a masterly organized network of regulators that interact with one another with speed and precision.� Even the most superior super computer fails to do this kind of operation of network of gene regulation. After all computers and their programs came into existence only few decades ago, but the living system taken its origin 3.8 billion years ago and progressed and still progressing; in what direction; perhaps to perfection and variation to the good of its own species.
Enhanceosome in action in IFN beta gene:
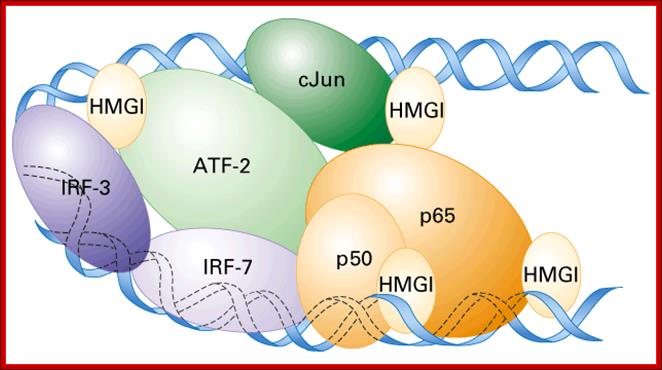
Enhanceosome- Interferon beta gene is activated by the binding of various factors where the factors such as HMG1 by binding to DNA bends the DNA so as other factors can contact with each other and activate RNAPII. Activation of IFN beta gene requires factors such as IRF (interferon response factors,) activating transcription factor (ATF), cJun and p50 and p65 are additional factor; all these come to interact with one another by bending of the DNA in the form of a loop by HMG1 protein. Note- this figute is repeated to show how factors that bind to enhancer regions enhance the transcription level. http://www.jbc.org
Mechanisms of X-chromosome inactivation: Samuel C. Chang, Tracy Tucker, Nancy P. Thorogood, and Carolyn J. Brown; Department of Medical Genetics, University of British Columbia, Vancouver, BC,
Epigenetic Regulation:
It has been over 30 years since DNA methylation was first postulated to be a heritable modification capable of influencing gene expression. The addition of a methyl-group to the cytosine base does not change the primary DNA sequence and is therefore considered to be an epigenetic modification, literally meaning to act �on top of� or �in addition� to genetics. The Human Epigenome Project was initiated as a mixed academic and industrial consortium in Europe, aiming to �identify, catalogue and interpret genome-wide DNA methylation patterns of all human genes in all major tissues." While DNA methylation is commonly agreed to be an epigenetic mark, other modifications of the chromatin structure remain more controversial. Part of this discussion may have been sparked by the recent support for epigenetics from the NIH Roadmap Initiative. The NIH project definition includes: �Epigenetics� is an emerging frontier of science that involves the study of changes in the regulation of gene activity and expression that are not dependent on gene sequence. For purposes of this program, epigenetics refers to both heritable changes in gene activity and expression (in the progeny of cells or of individuals) and also stable, long-term alterations in the transcriptional potential of a cell that are not necessarily heritable.� Looking at the controversy and concerns raised50 it becomes rapidly clear that arguments over the relevance of such a project are largely semantic.
Note: If you�d like to print out a hard copy of these reviews, you can find it at Zymo Research's website in their Publications.
Imprinting:
McGrath, Solter, Surani and their colleagues showed in the early 1980s that the maternal and paternal genomes are not equivalent and contained allele specific imprinting marks. Insulin Growth Factor 2 (Igf2) and its receptor (Igf2r) were the first imprinted genes to be discovered. A hallmark of imprinted genes is the monoallelic expression and parent of origin specific DNA methylation patterns. DNA methylation has been shown to be required to maintain monoallelic expression. Using conditional and reversible deletion of Dnmt1; certain genes have generated �imprint-free� ES cells and mice. The fact that most imprints are lost upon transient removal of Dnmt1, suggests that alternative mechanisms are either unstable or insufficient to propagate monoallelic expression; while chimeric mice from this system are viable, they are prone to develop tumors. Loss of imprinting (LOI) of Igf2 has been shown to increase the frequency of intestinal tumors and is frequently found in the normal mucosa of patients with colorectal cancer Jaensch and colleagues.
Mammalian X-Chromosome Inactivation
To achieve comparable X-linked gene expression levels in female (XX) cells similar to male (XY) cells (dosage compensation), they have to silence one of their two X-chromosomes. X-inactivation occurs shortly after implantation or after differentiation of female ES cells. The up-regulation of a long non-coding RNA, Xist, and the subsequent coating of the inactive X-chromosome is believed to be sufficient for the initiation of X-inactivation1. The spreading of Xist leads to chromosome wide transcriptional silencing and late replication of the inactive X chromosome (Xi). The silencing of Xi is further accompanied by histone modifications (H3K9me3 on the inactive and H3K4me3 on the active X) as well as DNA methylation. In a hierarchical model, the order of events would be as follows: Xist coating of paternal or maternal Xi, late replication timing, histone hypoacetylation, gain of DNA methylation1. Interestingly, the active X (Xa) displays more than two times as much allelic DNA methylation than the Xi. Most of this methylation is found within the gene bodies.
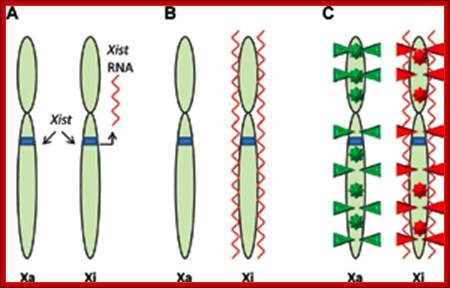
Epigenetic Regulation;http://epigenie.com
A) The non-coding RNA Xist is transcribed from the X inactivation center of the inactive X chromosome, Xi. B) Xist binds throughout the length of Xi. C) The silenced Xi displays suppressive histone modifications (red triangles) and DNA methylation at intragenic and promoter loci (red stars). The active X chromosome (Xa) displays activating histone modifications (green triangles) and gene body methylation (green stars).
The fact that all of these genes are hiallelically methylated prior to differentiation suggests a mechanism that leads to promoter hypomethylation and gene body hypermethylation. DNA methylation, histone hypoacetylation, and Xist act synergistically to maintain X-inactivation. While Xist seems largely dispensable for the maintenance of X-inactivation, DNA methylation and histone deacetylation are essential. Loss of DNA methylation leads to measurable reactivation of the Xi18. Additional evidence for the role of DNA methylation in regulation of X-inactivation comes from the human ICF (immunodeficiency, centromeric instability and facial anomaly) syndrome caused by a germ line mutation in the DNMT3B gene. In contrast to somatic cells, female ES cells maintain two active X-chromosomes. Interestingly, its has been shown that murine female ES cells show global DNA hypomethylation, which might be a result of two active X-chromosomes as well as lower level of Dnmt3a expression.
Imprinted X-Inactivation:
X-inactivation is imprinted during early development in placental mammals. The paternal X-chromosome is preferentially inactivated during the first lineage of differentiation that gives rise to the extra embryonic tissues, whereas X-inactivation in the embryo proper is random. Interestingly, in nuclear transfer experiments X-inactivation in extra-embryonic tissues specifically targets the silenced chromosome of the somatic donor, but remains random in the embryo proper. When female ES cells with two active X chromosomes were used as donors, the X-inactivation was random in all tissues. This work suggests that the imprinting marks set during gametogenesis are equivalent to those established during somatic X-inactivation.
Non-coding RNAs and the Mammalian Epigenome;
Several large non-coding RNAs that regulate epigenetic modifications in cis (Xist and AIR) or in trans (HOTAIR) have been previously identified and studied. A recent genome-wide analysis of K4-K36 domains (H3K4me3 marking the promoter and H3K36me3 marking the transcribed region) revealed a large number of conserved non-protein coding transcripts 29. Using published ChIP data, Guttmann et al. identified 118 non-coding RNA promoters that overlapped with Octa4 and Nano binding sites in ES cells. This new data suggests a possible role of large non-coding RNAs, in addition to small RNAs, in the regulation of the complex pluripotency network. Recently, two groups have shown an additional level of connectivity between non-coding RNAs and epigenetic modifications. The imprinted Air transcript is located in the second intron of Igf2r and regulates its expression in the embryo in cis. In the placenta, it has at least two additional targets Slc22a3 and Slc22a2. Recent work by Nagano et al. suggests that Air accumulates at the Slc22a3 promoter and directly recruits the H3K9 histone methyltransferase G9A. Loss or truncation of Air results in loss of imprinting and biallelic expression of the target genes. Air mediated repression of Igf2r in the placenta, however, appears independent of G9A. The second report has shown the involvement of a non-coding RNA (RepA) in initiation and spreading of X-chromosome inactivation. Loss of RepA results in the failure to induce full-length Xist and recruitment of PRC2 to induce H3K27 trimethylation on the inactive X-chromosome. Together these examples point to a general mechanism whereby RNAs can guide chromatin-modifying complexes to their specific sites of action.
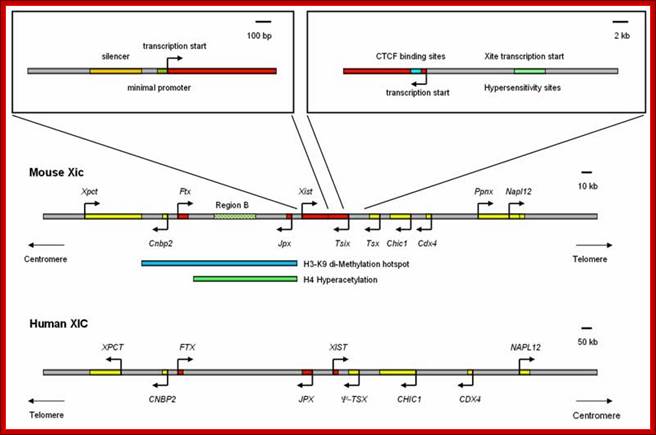
Figure. Transcription maps of the Xic/XIC regions in mouse and human. There are 11 genes in the mouse; the Transcription maps of the Xic region. http://www.bioscience.org
Xpct, Xist, Tsx, Tsix, Chic1 (formerly, Brx), Cdx4, NapIl2 (formerly, Bpx), Cnbp2, Ftx, Jpx, and Ppnx. Protein coding genes are represented by yellow boxes. Four of the 11 genes, Xist, Tsix, Ftx, and Jpx, are untranslated RNA genes and represented by red boxes. Region B, a non-coding expressed domain, is represented by a striped box. All the genes identified in mouse are conserved in human, except Ppnx and Tsix. Inhuman, however, Tsx has become a pseudo gene. The human region is approximately three times larger than the mouse. Despite this major change in size, the order and orientation of genes is conserved in human and mouse, except for Xpct, which is at the same location but in the inverse orientation. A histone H3 lysine 9 dimethylation hotspot and H4 hyperacetylation are represented by blue and green boxes below the transcription map of the Xicregion in mouse. Pillet et al. showed that the region -1157 to +917 has no in vitro sex-specific promoter activity. A minimal constitutional promoter was assigned to a region from -81 to +1. Deletion of the segment -441 to -231 is associated with an increase in CAT activity and may represent a silencer element. The choice/imprinting center contains tandem CTCF binding sites. Chao et al. proposed that Tsix and CTCF together establish a regulatable epigenetic switch for X-inactivation (49). Ogawa and Lee showed that Xite, located 10 kb from the Tsix transcription start, harbors two clusters of DNase hypersensitive sites.
Accumulation of chromatin changes during X inactivation. The timing of changes to the inactive X are ordered as observed in studies of early mouse development and ES cell differentiation. Silencing can result from Xist expression, but stabilization of the silencing requires additional changes. Reactivation occurs rarely as discussed in the text, and in general the inactivation status is very stably maintained once established. Figure: Accumulation of chromatin changes during X inactivation: http://www.bioscience.org.
The timing of changes to the inactive X are ordered as observed in studies of early mouse development and ES cell differentiation. Silencing can result from Xist expression, but stabilization of the silencing requires additional changes. Reactivation occurs rarely as discussed in the text, and in general the inactivation status is very stably maintained once established.
A). Active X chromatin is characterized by acetylation of H3 and H4 of the core nucleosome. There is another methylation of H3 lysine 4. B). Inactive X chromosome. Upon expression and localization of Xist there is macroH2A recruitment. It is unclear if these are bound together physically or are associated in some yet unidentified ribonuclear protein complex. The histone tails on the inactive X become hypoacetylated and methylated at H3 lysine 9 and 27, and H4 lysine 20. In addition, ubiquitination of H2A lysine 119 within the histone body is observed. DNA methylation is a late event in the inactivation process to lock in the inactive state. Note: it is not known to what extent the histone modifications are occurring on the same histone or within the same nucleosome, but at least some appear to be found in alternate domains.