Eukaryotic Promoter Structural Elements for RNA Polymerase II
The RNA Polymerase II transcribes structural genes and also many non-structural genes such as U1, U2, U3, U4, U5, and U7 sn RNAs and few snoRNA, antisense RNA and Si and miRNA genes. The list of Nc RNAs is still growing.
The RNAP II promoter elements of protein coding genes in eukaryotes more or less, have the same structural features as those of prokaryotes, but their promoter sequence and nucleotides position vary.� However, the organization of regulatory upstream elements and context vary and complex.�
They have a InR start site, TATA like site, Down Stream Promoter elements (DPE), upstream activator elements/enhancer elements, repressor and even insulator and silencer sequences.� The promoter elements in EK are more elaborate and varietal than in PK.
General features;
The start nucleotide is always A, infrequently G. From the start site at �35 to �25 upstream there is a consensus sequence called TATA or Hogness box.
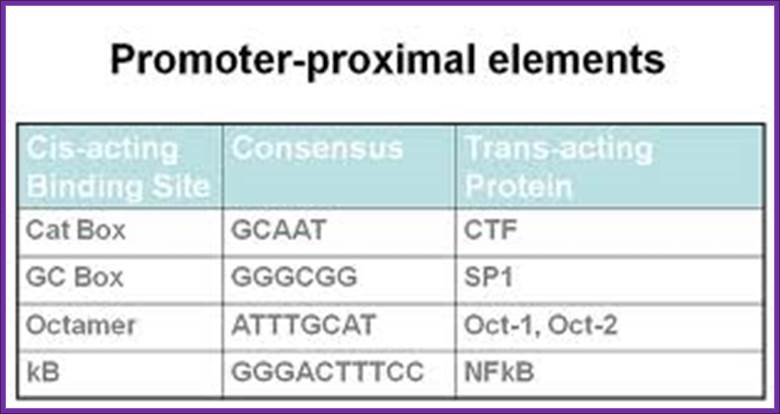
Next to TATA box, in the upstream region there are many sequence boxes positioned at different distances from the TATA box such as GC box and CAAT and other boxes. These boxes sequences to which transcriptional factors/regulators bind.
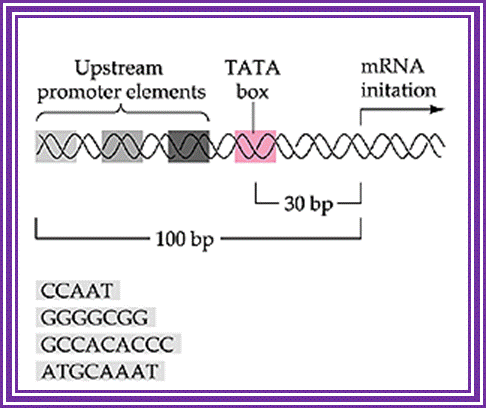
Typical promoter region for a protein-coding eukaryotic gene. The gene diagrammed here contains a TATA box and three upstream promoter elements. http://9e.devbio.com/
Sequences and positions are very important.� These sequences provide structural motifs for the binding of transcriptional factors.� Each gene has its own TF domains at specific positions.� Binding of the TF activates and enhances the efficiency of transcriptional initiation.
There are certain sequences found in the upstream, which could act as binding site for repressors, which block transcriptional initiation or activator for activation of transcription.
Nearly 200 or 1000 bp upstream from the start there can be an enhancer sequence. There are different sequence motifs specific-to-specific factors.� This sequence can be located in the upstream or down stream or in the middle of the gene or in introns of the gene or in between the genes.�� Binding of the factors to this region increases the efficiency of transcription by 100 to 200-fold.
In some genes, there are certain sequences located in the upstream region act as response elements, where a factor, activated in response certain environmental stimulus, or to a chemical stimulus, binds to the sequence and activates transcriptional initiation.� They act as sensors for environmental or nutritional or hormonal signals, which can be from outside or they can emanate from inside the cell.�� Some of these are tissue specific components. Binding of repressor to such response elements keep genes silent.� Activated factors either displace these repressors and bind to them and act as Coactivators.
Typical Eukaryotic Promoter of Structural Gene:
Sequence elements of a general eukaryotic promoter, the position of upstream elements and the kind of sequence elements vary among eukaryotic RNAP II coded genes.
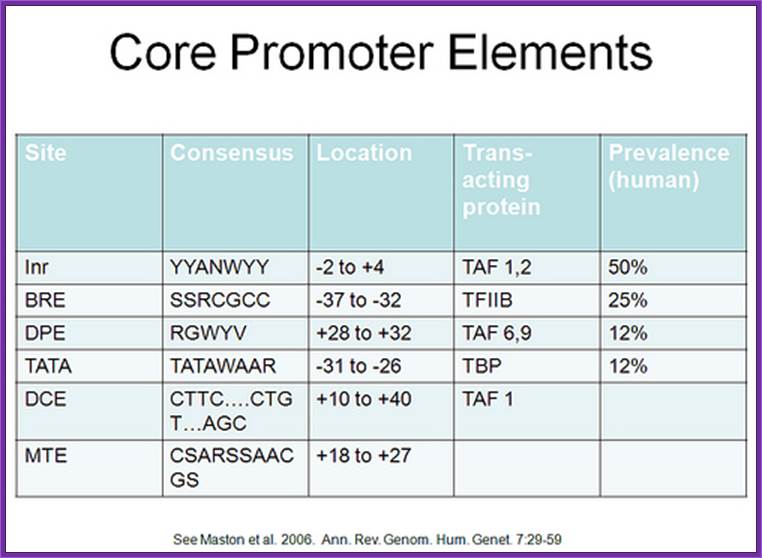
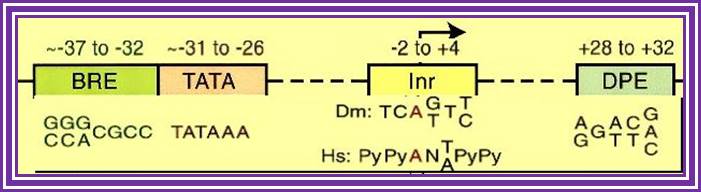
Core promoter elements. Some core promoter motifs that can participate in transcription by RNA polymerase II are depicted. Each of these elements is found in only a subset of core promoters. Any specific core promoter may contain some, all, or none of these motifs. The BRE is an upstream extension of a subset of TATA boxes. The DPE requires an Inr, and is located precisely at +28 to +32 relative to the A+1 nucleotide in the Inr. The DPE consensus was determined with Drosophila transcription factors and core promoters. The Inr consensus sequence is shown for both Drosophila (Dm) and humans (Hs); Jennifer E.F. Butler1 and James T. Kadonaga, http://genesdev.cshlp.org/
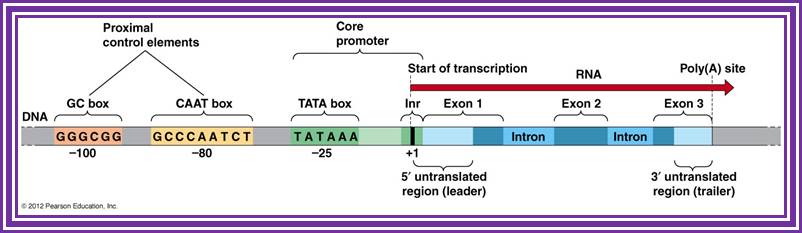
Generalized EK gene structural elements; http://www.mun.ca/biology/
�
TATA plus, InR plus and DPE Containing Promoters:
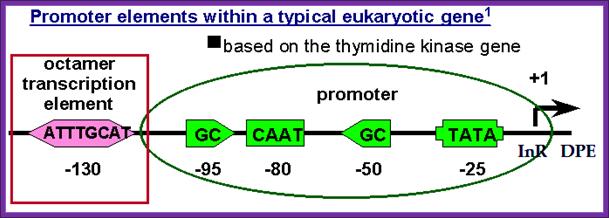
Thymidine kinase gene promoter elements; DPE is Downstream Promoter Element.; InR for chain Initiator Region.
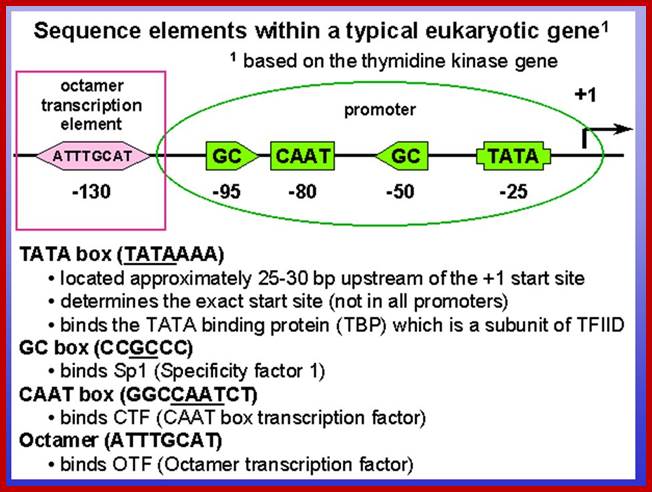
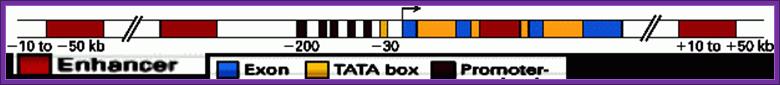
The color code tells you the components of the Gene including upstream elements and downstream gene terminal region-white. Coding region consists of exons and introns.

The regulator sequences are silencer, upstream activator elements and TATA box
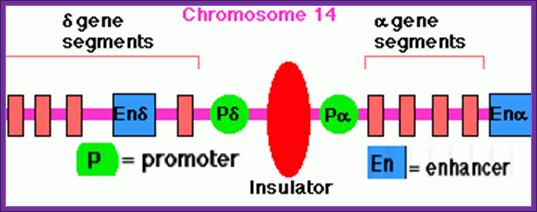
This diagram represents the promoter components of d and a Globin gene components with an insulator in between and enhancer on the other side of each of the promoters; Insulators block the continued transcription activated an enhancer to the next gene; http://www.uic.edu/
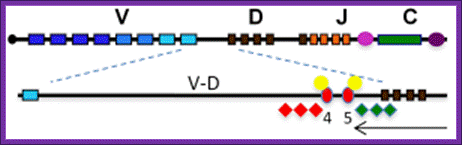
Position and role of insulators in the Igh V�D intergenic region. Key: V, D, J, Eu, C, 3′RR as in Fig. 1; V�D, 96kb V�D intergenic sequence; HS4 and HS5, red ovals; CTCF, yellow circles; active histone modifications, green diamonds; repressive histone modifications, red diamonds; antisense transcription, black arrow. http://www.babraham.ac.uk/
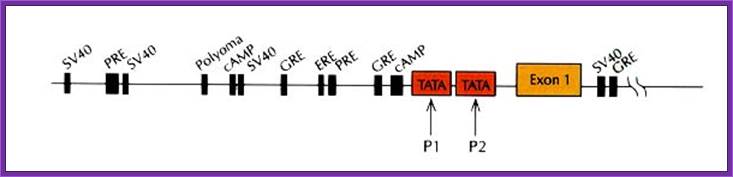
Control components of a representative eukaryotic Renin gene promoter contains several control elements including Response elements:
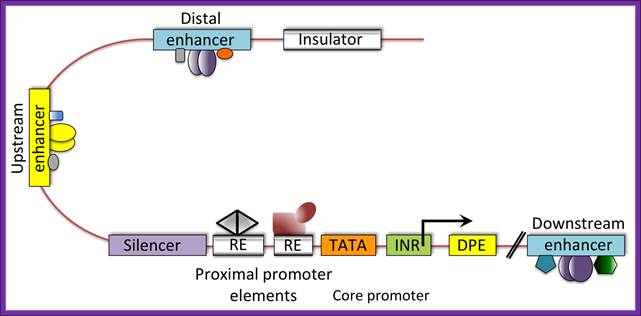
Metazoan regulatory modules controlling transcription. Shown is a diagram of a typical metazoan gene illustrating the complex interactions amongcis-acting modules and trans-acting factors regulating gene expression. Note that both positive and negative control regions are interspersed with promoter modules, all of which can be further influenced by distal regions regulating chromatin configuration, such as insulators. (Used with permission, Levine and Tjian, 2003); Valerie Reinke1�, Michael Krause2�, Peter Okkema
This diagram shows various elements operating upstream and down stream of the START site; They can be Enhancers, Insulators, Silencers nd Response elements. http://www.wormbook.org/
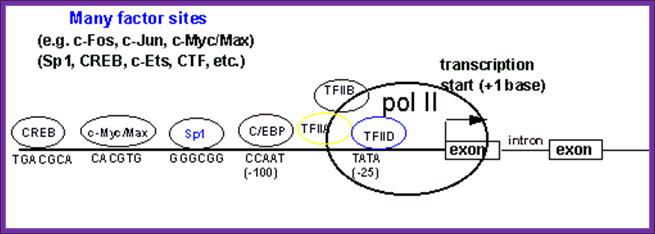
Structure of the upstream region of a typical eukaryotic mRNA gene that hypothetically contains 2 exons and a single intron. The diagram indicates the TATA-box and CCAAT-box basal elements at positions -25 and -100, respectively. The transcription factor TFIID has been shown to be the TATA-box binding protein, TBP. Several additional transcription factor binding sites have been included and shown to reside upstream of the 2 basal elements and of the transcriptional start site. The location and order of the variously indicated transcription factor-binding sites is only diagrammatic and not indicative as being typical of all eukaryotic mRNA genes. There exists a vast array of different transcription factors that regulate the transcription of all 3 classes of eukaryotic gene encoding the mRNAs, tRNAs and rRNAs. [CREB=cAMP response element binding protein] [C/EBP=CCAAT-box/enhancer binding protein]. The large green circle represents RNA polymerase II.; A promoter element containing several sequences boxes to which specific factor bind and interact with the Basal Transcriptional Apparatus. (BTA); http://www.1cro.com/mwking/gene-regulation.html
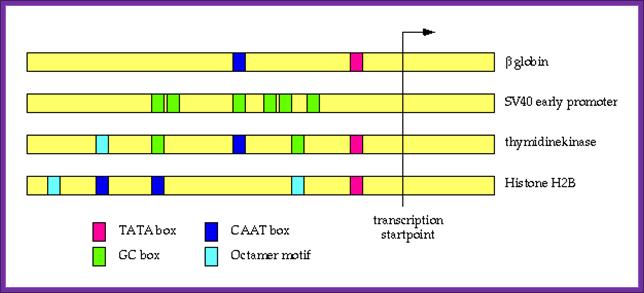
Position of several upstream regulatory elements in different Gene promoters with reference to start site; this suggests the upstream elements and their position from strat site differ.
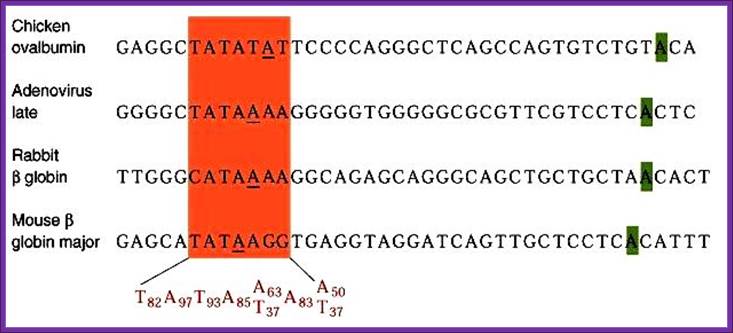
Position of TATA box and InR is not same, but vary slightly
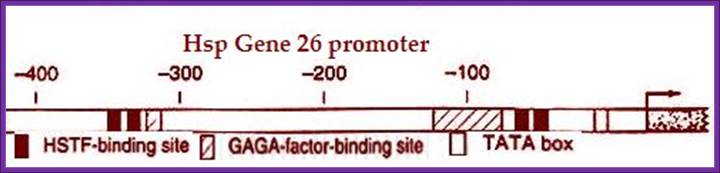
This is another typical representative of eukaryotic promoters containing Heat shock response elements; Heat shock TFs bind to these elements.
TATA less and InR plus�DPE Containing Promoters:
������������� OCTA���� ����GC������������ CAAT����� ���InR� +1
-------------BOX---------Box--------Box---------pyTCA G/TTT/C py 5-

Most of the house keeping genes, which constitutively express contains promoters with out TATA box; instead they have Initiator sequence (InR) sequences. The said sequences start with A, but they are always associated with a C preceding the A and on either side they are bracketed by 3pys and5pys; 3pys C A 5pys. The site or location for RNA polymerase binding is aided by other accessory factors which ultimately bring TBP preceding to InR region, there by RNAP-II can bind properly and initiate transcription at start site.���������������������������
TATA-less and InR less Promoters, DPE containing promoters:

This diagram shows lack of both TATAA box and InR elements, but contain DPE; the upstream elements contain many GC boxes
�����������������������������������������������
In the cases where both TATA box and InR sequences are missing, yet RNA pol-II binds with the assistance of accessory factors and initiates transcription at different positions.�� The upstream has many GC boxes.� The Sp1 factors that bind to GC sequences recruit TFII-D and RNAP II.
As there is no defined start site it is possible the mRNAs produced may have different 5� start nucleotide.
Ribosomal Protein Gene Promoter Elements- �rPLA peromoter;
-tgccctgttccg C >C >C >TTTTTACTCTACTACCAAGATGgtgagtag-
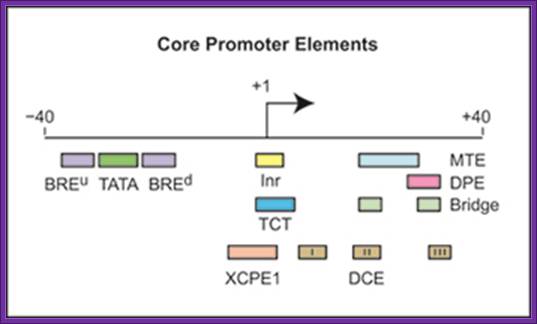
Core promoters contain DNA sequence motifs such as the TATA box, Inr, MTE, DPE, and TCT elements. Note, however, that there are no universal core promoter motifs. Specific protein elements specify and bind and interact with specific sequence elements. The best-known core promoter motif is the TATA box; however, the TATA box is present in only about 10 to 15% of human genes. In our studies of TATA-less promoters, we discovered two new core promoter motifs � the DPE and the MTE. Both the DPE and MTE are downstream of the transcription start site and are conserved from Drosophila to humans. It is interesting to note, for example, that the promoters of nearly all of the Drosophila homeotic (Hox) genes contain a DPE motif and lack a TATA box. [The promoters lacking a DPE motif are those associated with the evolutionarily most recent genes, Ubx and Abd-A. Hence, all of the more ancient Hox genes have TATA-less, DPE-containing core promoters.] Moreover, Caudal, a sequence-specific DNA-binding protein that is a master regulator of the Hox genes, is a DPE-specific activator. Thus, enhancer-core promoter specificity can be used in the regulation of gene networks, James T. Kadonaga
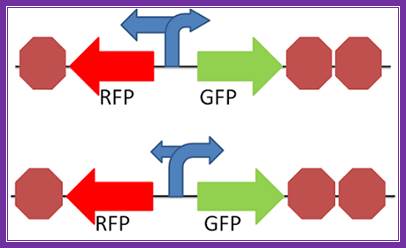
Not all promoters cause RNA polymerase to transcribe downstream in the expected "forward direction". Some promoters can cause RNA polymerase to go in the opposite direction from what is expected, or go in both directions. This is what we are trying to find out; do different promoters go in different directions, and what is the directional preference of different promoters. Bidirectional promoters are possible within the cell, and those designing promoters should be beware of palindromic sequences in their design and their potential effects on the initiation of protein within the cell. Further characterization of both biobrick promoters and palindromic sequences will yield more information of the prevalence of bidirectionality in promoters. To view the sequences, we plan to test in this construct, please visit our parts page. http://2012.igem.org/Team:Penn_State/Bidirectional_Promoters_Results
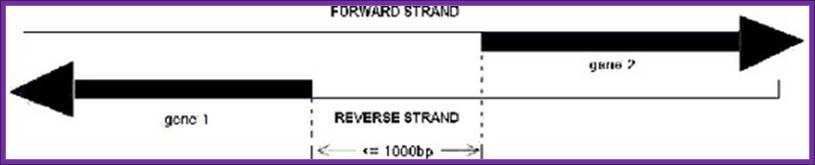 Such an arrangement is previously defined as
"bidirectional" and the divergent gene pairs are termed as
"bidirectional genes", while the intergenic region between a
"bidirectional gene pair" is often called a "bidirectional
promoter" (Figure 1 � A sketch map of bidirectional promoter.).; http://www.springerimages.com/
Such an arrangement is previously defined as
"bidirectional" and the divergent gene pairs are termed as
"bidirectional genes", while the intergenic region between a
"bidirectional gene pair" is often called a "bidirectional
promoter" (Figure 1 � A sketch map of bidirectional promoter.).; http://www.springerimages.com/
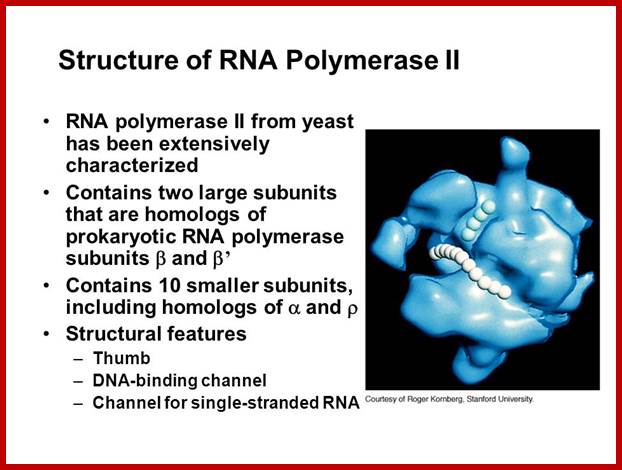
For RNAP II the RNAPII enzyme; Korenberd Stanford University; http://slideplayer.com/
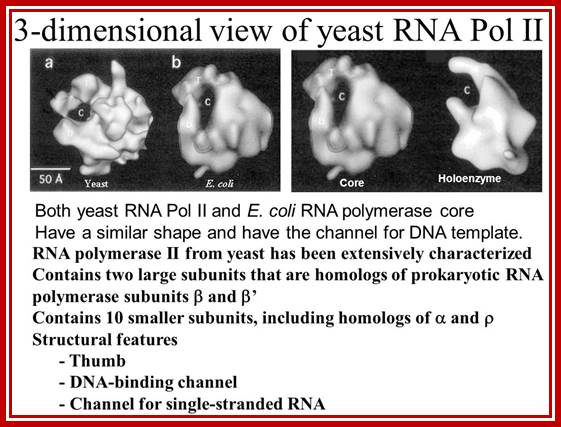
http://slideplayer.com

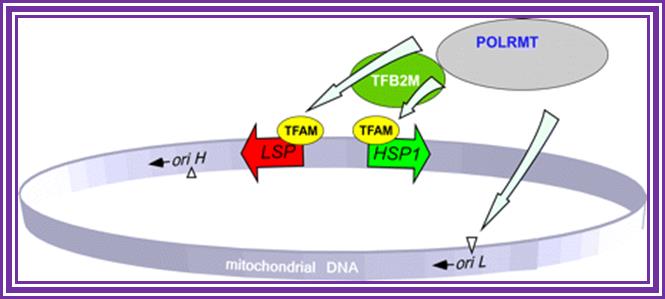
Schematics of the basal transcription initiation machinery in human mitochondria. Human mitochondrial genome contains two promoters located in the opposing DNA strands. The HSP1 promoter is responsible for synthesis of most mitochondrial genes and is activated by POLRMT-TFAM-TFB2M complex. During replication, when DNA region near oriL, becomes single-stranded and forms stem-loop structure, transcription by POLRMT generates short RNA primers. This initiation event is TFAM- and TFB2M-independent. Transcription from the LSP promoter generates primers for replication at oriH as well as the rest of the tRNAs and mRNA. This initiation event, similar to HSP1, requires cooperative action of POLRMT, TFAM, and TFB2M for efficient transcription and replication; Human mitochondrial transcription from the D loop goes through opposite direction complete one round; http://www.jbc.org/
End Region of the Genes:
When a gene starts at one position, it has to have an end also.� In eukaryotes there is no well-defined transcriptional terminator region, but transcription progresses well beyond 1000 or more nucleotides down stream from the last TER codon.�
How exactly and where exactly it is terminated is still an enigma to biologists.� But what they know is that a cleavage site, which is about 30-35 or even 100 nucleotides downstream of Terminator (UGA or UAA) sequence. This sequence in mRNA end is called poly-A+ signal (TTATTT in the DNA) that translates into AAUAAA in precursor mRNA.
\--UPE---TATA�-BRE--InR----DpE----//--//-----TTATTT---U(n)�G(n)---
The sequence present in the terminal region of the gene is TTATTT plus another sequence U is present 20 to 35 ntds down stream from the poly-A signal. �Down stream of U sequences, there can be another sequences G or GC rich sequences called GSR.� Present studies have revealed the presence of such sequences leads to the formation of quadruplex structures that are recognized by specific factors such as hnRNP-H.� Perhaps they help in recruiting poly adenylation protein complex. The said sequences are not used for transcriptional termination but they are used for mRNA processing at 3� ends.
The highly expressed mouse histone H2a-614 gene is located 800 ntd 5' of the histone H3-614 gene. There is a 140 ntd sequence located 500 ntd from the end of the H2-614 mRNA which has been defined as a transcription termination site for RNA polymerase II. Genes such as histones don�t contain Poly-A signal sequences and they are terminated beyond the TER codon sequence.� Rather, it is known that there is an extremely heterogeneous series of non-polyadenylated regions in Histone mRNAs. They are processed differently using UsnRNA/snRnp7.
A transcription termination site has been characterized between the mouse histone H2a-614 and H3-614 genes. There is a poly-RNA present in small amounts in the nucleus which ends 600 nucleotides 3' to the H2a-614 gene. Nuclear transcription studies demonstrate that transcription extends at least 600 nucleotides 3' to the gene but is greatly reduced 700 nucleotides 3' to the gene.� In contrast to polyadenylated transcripts, the 3′ processing of histone pre-mRNAs requires both conserved sequence and structural elements. The 3′ end of the mature mammalian histone mRNA possesses a highly conserved 26-nt sequence, encompassing a 16-ntd stem-loop, located 24-70 ntd downstream of the stop codon. The m RNA transcripts at 3� ends have a stem loop structure followed by a specific sequence.� These are used for 3� end processing of Histone precursor RNAs. (N Chodchoy, N.B.Pandey and W F Marzluff and (Marzluff 2005)
Cytoplasmic polyadenylation of maternal RNAs from the egg cell allows the cell to survive and grow even though transcription does not start until the middle of the 2-cell stage (4-cell stage in human). In the brain, cytoplasmic polyadenylation is active during learning and could play a role in long-term potentiation, which is the strengthening of the signal transmission from a nerve cell to another in response to nerve impulses and it is important for learning and memory formation. Cytoplasmic polyadenylation requires the RNA-binding proteins CPSF and CPEB, and can involve other RNA-binding proteins like Pumilo. Depending on the cell type, the polymerase can be the same called polyadenylate polymerase (PAP) that is used in the nuclear process, or the cytoplasmic polymerase GLD-2.