Eukaryotic Promoter Structure for RNA polymerase III:
RNA pol-III transcribes small molecular weight RNAs such as tRNAs, 5sRNAs, 7sKRNAs, 7sLRNAs, U6sn RNAs, few ncRNAs and it also transcribes some ADV, EBV and many eukaryotic viral genes.
� The 5s rRNA and tRNA genes have promoters within the coding region of the gene.
The promoter regions for 7S and U6sn RNAs, more or less, look like RNA pol-II promoters, with little differences.�
� Though the size of the genes is small ranging from 160 to 400 bp, their promoters are well defined for transcriptional initiation from their respective Start sites in the promoters.�
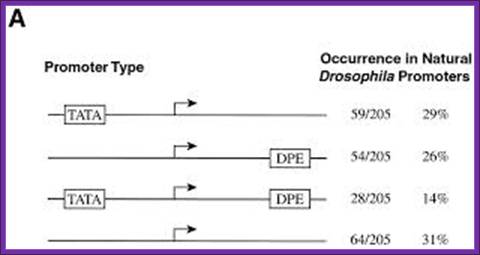
Statistics of use of TATA, and DPE and InR elements in Eukaryotic promoters in Drosophila; Molecular and Cell Biology; Alan K. Kutach and James T. Kadonaga: http://mcb.asm.org/
tRNA Genes:
In Eukaryotes tRNA genes are clustered into specific loci, where each locus consists of several tRNA genes organized into tandem repeats with spacers in between them.�
� In Drosophila, there are 50-60 loci, each locus is about 50kb long and contains 20-30 different tRNA genes.�
In X. laevis a 50kbp segment containing a cluster of tRNAs is found in more than 150 loci.�
� In rats, each of the 2kbp segments containing a cluster of tRNA segments found repeated at least 10 times.
In the above said tRNA gene clusters, each of the tRNA genes are separated by non-transcribing spacers?�
� All tRNA genes contain a specific start site, which is also a part of its promoter.�
A significant feature is that the promoter has no TATA box.
However, unlike other promoters, it has two-consensus sequence located deep in the coding region of the gene.�
� They are located at +10 to +20 and another at +50 to +60.�
They act as promoter elements for the RNA pol-III and their accessory factors to bind and to transcribe.
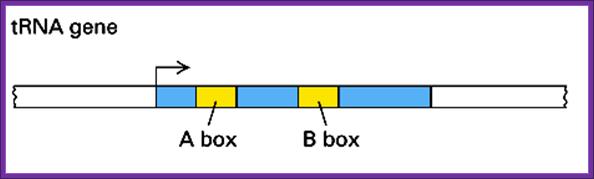

The promoters for tRNA genes-This simple line diagram shows internal ntds sequences
� +� >
--------------I----------I-----------------------I---------I----------�
�Start�������������������������� �� +10-----+20������������� ��������� +50��� 0
�A������������������������������� ����������������� B
�� �������������������� TGGCacAGCGG��� GGaTCGAaaCC
� ��������������������� TGCCCAG TGG���� GGTTCGAATCC
����������������������� TGGNNAG-TGG�� GGTTCGANACC
Each of the internal sequence represents certain tRNA domains, such as; A block representing D-arm and B block representing TUCG loop respectively.
�
� At the time of transcriptional initiation, a transcriptional factor TF-C made up of six subunits recognizes the sequence boxes and binds to them and positions the proteins in such a way one end of the protein is found at the start site.
![]()
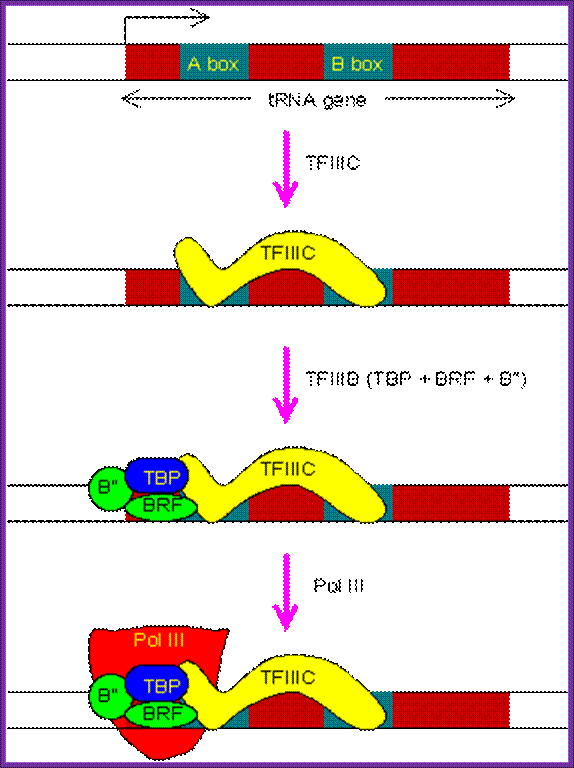
This illustrative diagram shows the assembly of RNA Pol-III Transcriptional complex on tRNA promoters. http://www.web-books.com/
Then this protein guides the TF-B, which is made up of several subunits, to be positioned at start site.�
� Then the RNA pol-III recognizes these proteins and binds to them and binds tightly and initiates transcription at the pre defined site.�
Here the role of a promoter is to provide recognition sequence modules for specific proteins to assemble in such a way; the polymerase is properly positioned to initiate transcription exactly at a pre-defined nucleotide, which is called start site.�
� If sequence motifs are not present, protein fails to bind and RNA pol fails to associate with accessory proteins and initiate transcription at specific site.
� In these promoters there is sequence such as TATA box for the binding of TBP, which acts as the positional factor.�
This is what the promoter is and what it is meant for; this is why promoter is required.
5sRNA genes:
Ribosomal RNAs, in eukaryotes consist of 28s, 18s, 5.8s and 5s RNAs.�
� The 28s, 18s and 5.8s rRNAs are synthesized as one block from nucleolar organizer region of the DNA, and the precursor 45S, larger than the final RNAs, is processed into 28s, 18s, and 5.8s RNAs, but no 5s RNA segment.�
Gene for 5s RNA are located elsewhere in the chromosomes, many times they are found just behind telomeres.�
� The number of 5s RNA genes in a haploid genome can vary from 200 to more than 1200, and all of them are tandemly repeated in the cluster and each of them are separated by non transcribing spacer.
5s rRNA genes have promoter elements located in the coding region, which suggests that promoter elements are the structural regions for the assembly of RNA polymerases and its associated factors for the initiation transcription at a defined position
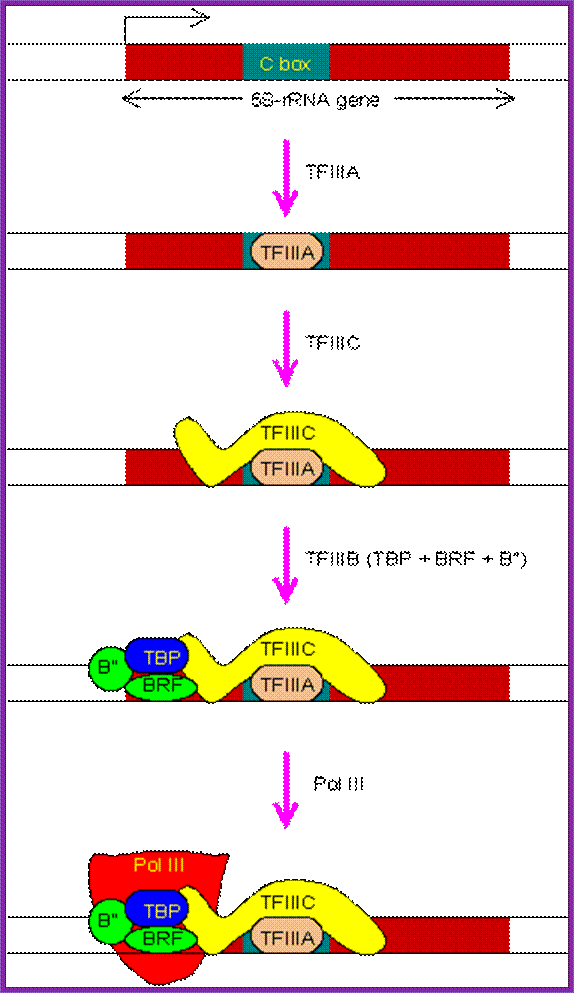
The PIC assembly for the transcription of 5S-rRNA gene. Note that TBP is involved in the transcription by all three types of RNA polymerases in eukaryotes.;This another diagram shows the assembly of Transcriptional apparatus for the transcription of 5s rRNA gene by RNA Pol-III; http://www.web-books.com/
In Xenopus, two types of 5SrRNAs are produced, one somatic RNAs and the other Oocyte specific RNAs.
� The size of the gene is about 250 to 300 bp long and it produces 120 ntd long rRNA, which gets associated with rRNAs at the time of ribosomal assembly.�
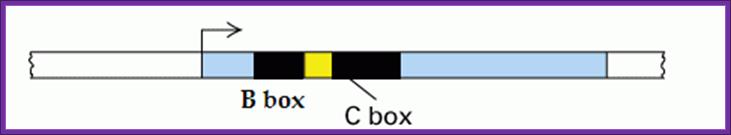
The promoter contains a start site, but no TATA box, which is an important feature of structural genes.�
� From +60 to +110 downstream of the start site, there are two conserved sequence elements.� These sequences are called A-block and C-block.�
The sequences in somatic 5s RNA and Oocyte 5s RNA have different set of sequences, but CTGGA appears to a common feature.
����������� ����������������������� �������� +50 ��+60���������� ������+ 81� �+99
-------I---------------I--------I---------------I---------I----------------
Start�������������� �� A ���������������������������������������� C
�����������
Somatic 5s RNA: A Block--� (+50�� aaGccaAGcaGG� +60)
�������� C-Block�( + 80 GGTTaGtactt �+91)
Oocyte 5s RNA: A-block= + 67 CCTGGt +71, +78CCTTGGA+83
C-block = +94 CCTGG +98, CTTGGA
The conserved sequences from +80 show variation among the species.
GCCTGG [TGGCCCAGTGG] AGGGCGTTAGACTGAAAA
CTGAAGGTCCCGT [GGTTCGAATCC] CGGCGGGGGCA
During transcriptional initiation, TF III A first recognizes the C box and binds, then TF-III-B containing TBP binds to the promoter using TF-III A and it positions at start site.
�
� Then the RNA-pol-III complex assembles at the start region and initiates transcription at the predefined site.
�
Again the role of internal promoters is to position the transcriptional factors and ultimately the RNA-pol so as to initiate at specified site.
� 5s RNA expression differs in Oocyte and somatic tissues.
�
Transcription factor TF III A, 40 KD proteins is produced in Oocyte specific manner.�
� This protein binding to internal site of the 5s gene activates the gene expression by facilitating the assembly of TF III-C and B and finally RNA pol-III.�
At a late stage of oogenesis, enormous quantities of 5sRNAs are produced, and the TF-III A binds to 5s RNA, thus all TF III-As get consumed and none of the factors are available for the activation of Oocyte specific 5sRNA gene.
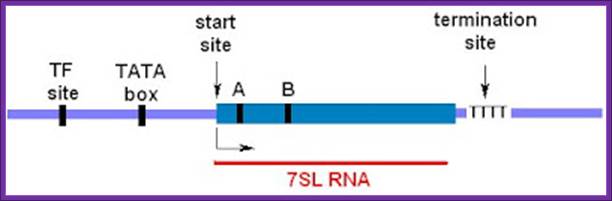
7sL RNA genes:
The size of the gene is about 350 to 400bp long.��
� The promoter contains a TATA box located at �35 to �30 upstream of the start.�
At �55 it has proximal sequence elements (PSE), increases the efficiency of transcription.�
� An octamer sequence is also found upstream to PSE.�
The TF III-B consisting of TBP and TAFs or TBP related factors (BRFs) bind directly to TATA box and facilitates the binding of RNA pol-III.�
� Binding of certain factors to PSE and DSE increases the efficiency transcriptional initiation.
���� +1>
---(DSE)-------OCTA----------------PSE----------------TATA-------
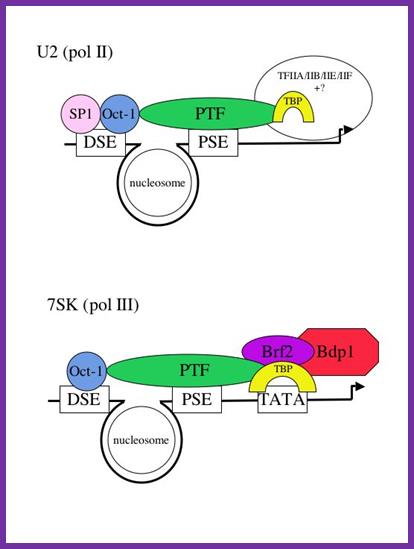
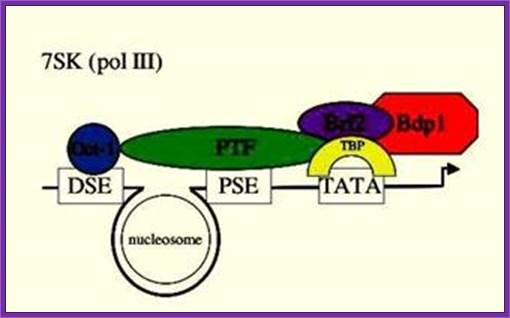
7SK gene is a representative of a group of genes that are transcribed by RNA pol-III. �The promoters more or less show some similarities to that of RNA Pol-II, but distinctly different even though it having TATA box for the transcription factors that bind TBP/TF-A/II-B/II-E/II-F. While U6, RNase P with 7sK though they have DSE and PSE and TATA elements the factors bind are STAF, OCT1, SNAP TFIIIB, BDP, BRF2 with RNA Pol-III; www.ploseone.org
Structure of snRNA genes and known transcription factors; The pol III-transcribed snRNA genes share components of the IIIB complex with tRNA genes that are also transcribed by pol III. Dr. S.Murphy; http://users.path.ox.ac.uk/
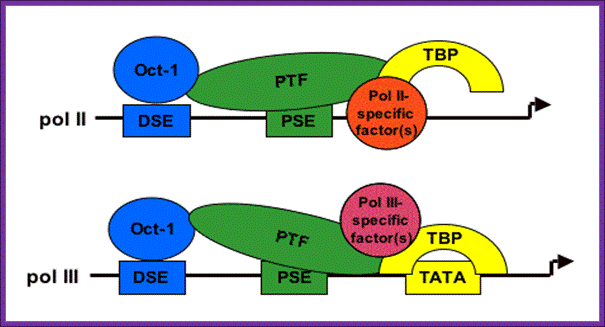
An "exclusion" model for polymerase specificity of snRNA genes. It is still unclear how the presence of a TATA box (often found in the promoters of protein-coding genes) switches the polymerase specificity to pol III. One possibility is that binding of TBP to the TATA box causes exclusion of a pol II-specific factor (eg TFIIB) from the promoter and allows a pol III-specific factor to bind (see Figure 2 and ref. 11). In support of this model we have shown that factor interactions occur with the region just downstream of the PSE of the pol II-transcribed U2 gene in vivo (ref. 11) and that TBP bound to the TATA box recruits Brf2 . Dr.S.Murphy; http://users.path.ox.ac.uk/