Molecular Tools-II:
Restriction Enzymes.
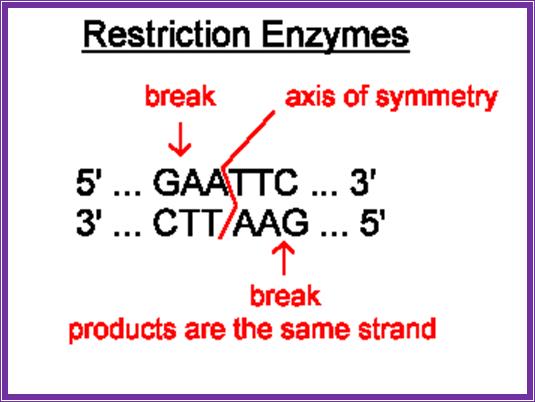
Restriction enzymes are characterized by -they are restricted to prokaryotic organisms (with certain exceptions); they act on foreign DNA, but not on their own; they act at a particular site or sequence specific; they cut the DNA into fragments in a specific way.
Genome mapping using Restriction enzymes;
http://utcinnovationlabs.blogspot.com/
Restriction enzymes: http://www.symposcium.com/
Most of the restriction enzymes now available are isolated from specific microorganisms.� Each of the microbes produces specific enzymes and the name of the RE�s is derived form the name of the organism.� Based on the structure and function they are classified into three types, Type I, II and III.
Type-I Enzymes:
Most of them are multimeric enzymes.� They undertake both methylation and restriction activities. Type-I enzymes have three groups of related genes. The genes concerned are �Host specificity of DNA� �Had� systems. Strain E.co-K shows three variants of �Had� system.�� E.co-B shows two variant of �Had�.
E.co K enzymes consist of three types of subunits called R, M, and S, exist as complex of 2 R subunits, 2 M subunits and 1 S subunit.� The genes for them are hsd-R, hsd-M and hsd-S.
The R subunit is responsible for restriction activity.� The M subunits are for methylation and S subunits for recognizing the site; on the contrary E.coli-B consists of only M and S subunits in 1: 1 ratio.� In this the S subunit recognizes the site.� This is followed either restriction or methylation, which events are mutually exclusive.
E.co-B and E.co-K genes are allelic, their target sequences are different, but in diploid state the S of one strain can direct the activities of R and M to the recognition site
Recognition site of both K and B are bipartite, each consists of specific 3 bp and 4 bp sequences separated by few bp.� So the recognition surface of both lie on the same side of the DNA.� The possess Adenine residue that can be methylated on the opposite strands.
5� TGA (N) 8 TGCT
3� ACT (N) 8 ACGA
Methylation of Adenine is performed by S-Adenosyl Methylase (dAM), Adenosyl carries methyl group.
Methylation of Cytosine is performed by Cytidyl Methylase (dCM).
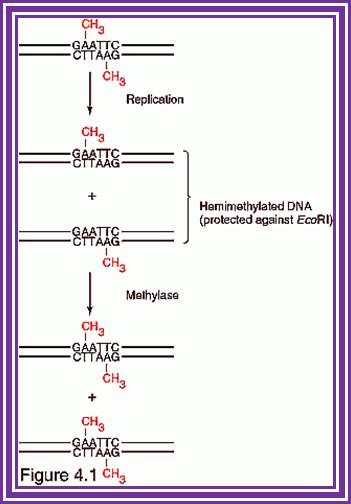
Whether the target to be cleaved or methylated depends upon the state of the site in the DNA.� If the target is fully methylated, the enzyme binds to and gets released without cutting.� If the target is hemi methylated the complex binds and methylates the site to fully methylated state and the enzyme is released from the site.� If the target is unmethylated the enzyme binds and gets triggered for cleavage activity.
� The cleavage is generally 400 to 1000 base pairs away from the binding site.� It is believed that when once the enzyme binds it pulls the DNA through second binding site (via loop formation) and recognizes the site for cutting.� Then after some time the enzyme gets released from the site.
����� DNA:� 5� -I-P-I---hsd-R-I-hsd-M-I-hsd-S-I-3�
If there are mutations in hsd system one obtains new phenotypes of bacteria.� Ex. Mutation in hsd-M prevents restriction and methylation as well and strains will be r^- and M^-.� If the mutation is in hsd-R then the strain will be r^-M^+, where they cannot restrict but can modify.� If the mutation in the gene hsd-S the strain is r^-m^-, they neither modify nor cut
|
Type |
Number of subunits, require Mg2^+; Mol.wt |
Genes/ Mol.wt |
Restriction site |
Restriction and methylation |
Function |
|
Type-I |
Bifunctional, 3 subunits, Mg2^+- yes |
R= 135 M= 62 S= 55 |
Bipartite, asymmetric sequences |
Mutually exclusive |
R=restriction, M= methylation, S= site recognition; cutting is 1000 bp away from the binding site |
|
Type-II |
Restriction and methylase functions are separate.� Mg2^+ yes |
|
4-6 and 8 bp, often palindromic |
Separate reactions |
Cut with in the site or adjacent to it |
|
Type-III |
Bifunctional, 2 subunits, Mg2^+ yes |
|
5-7bp, asymmetric sequence |
Simultaneous |
Recognize a region and cut 25-27 bp away from the site |
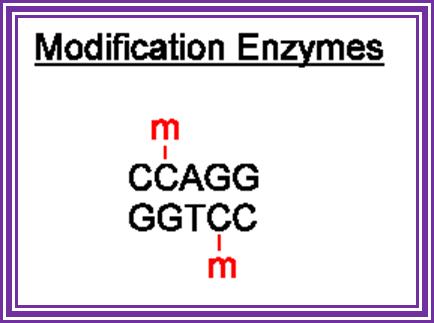
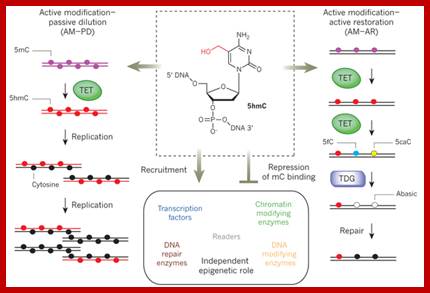
TET enzymes, TDG and the dynamics of DNA demethylation; DNA methylation has a profound impact on genome stability, transcription and development. Although enzymes that catalyse DNA methylation have been well characterized, those that are involved in methyl group removal have remained elusive, until recently. The transformative discovery that ten-eleven translocation (TET) family enzymes can oxidize 5-methylcytosine has greatly advanced our understanding of DNA demethylation. 5-Hydroxymethylcytosine is a key nexus in demethylation that can either be passively depleted through DNA replication or actively reverted to cytosine through iterative oxidation and thymine DNA glycosylase (TDG)-mediated base excision repair. Methylation, oxidation and repair now offer a model for a complete cycle of dynamic cytosine modification, with mounting evidence for its significance in the biological processes known to involve active demethylation. Rahul M. Kohli;� & Yi Zhang; http://www.nature.com
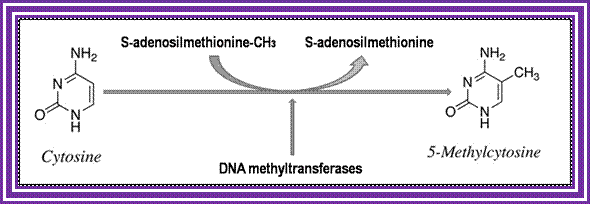
The reaction of DNA methylation; http://www.labome.com/
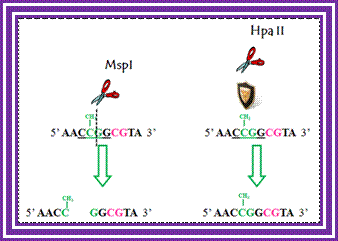
Determination of differential methylation with methylation-sensitive and methylation-insensitive restriction enzymes.; http://www.labome.com/
Type -III enzymes:
�These enzymes are rare.� Some strains of E.coli like P1 and P15 carry plasmids P1 and P15 and they carry genes for the said enzymes.� The Hinf plasmids found in Hemophilous influenzeae carries serotypes called R.
Each of theses enzymes carries two types of subunits. The R subunit for restriction (cutting), and MS subunits for site recognition and modification and they perform the function almost simultaneously.� Interestingly when the enzyme binds to its site (ATP dependent), methylation and restriction activities of the enzymes compete with one another.� Methylation takes place at Adenine at the binding site and cleavage takes place 24-27 bp away from the binding site and produces staggered or sticky tails.� However, unmethylated site favors binding and cutting.
The MS subunit has molecular mass of 75 KD and of R is about 108KD.
5�AGACC
����� 3�TCTGG for P1
����� 5�CAGCAG
����� 3�GTCGTC for P15,
����� AGA*CC-----GGTCT
TCTGG-------CCA*GA�� requires inverted repeats next to the recognition site.
Unmethylated site facilitate the binding and restriction;
����������� AGACC---GGTCT
����������� TCTGG---CCAGA
����������� ���� Restriction
����������� AGACC��� GGTCT
����������� TCTGG��� CCAGA
Type-IV Enzymes:
These are special kind of enzymes, which cut the sites only when they are methylated.� They use tetramer or hexamer sites but the A in the site should be methylated.�
Ex. Dpn-I from Streptococcus pneumoniae cuts > GATC, only if it is methylated, but the bacteria have no methylase activity.� Another strain produces a variety called Dpn-II, which cleaves GATC when it is not methylated, but the strain produces methylase enzyme.
Another enzyme from a phage PI cleaves TGATCA > only when the site is methylated at Adenine.� The enzyme is called Pac case.
Type-II Enzymes:
These enzymes are extensively used in recombinant DNA techniques.
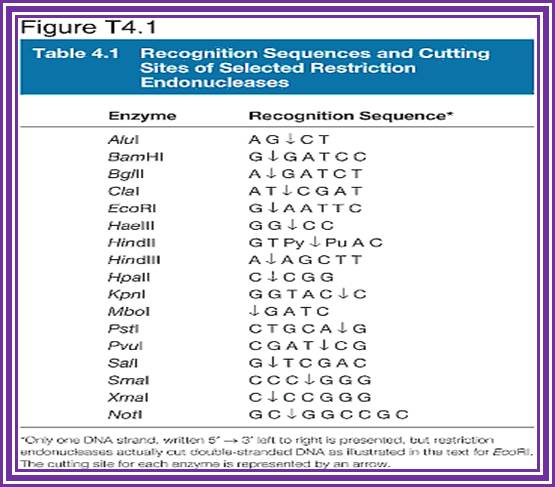
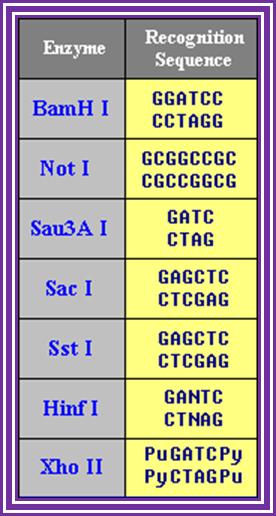
Restriction enzymes recognition sequences: http://www.vivo.colostate.edu/

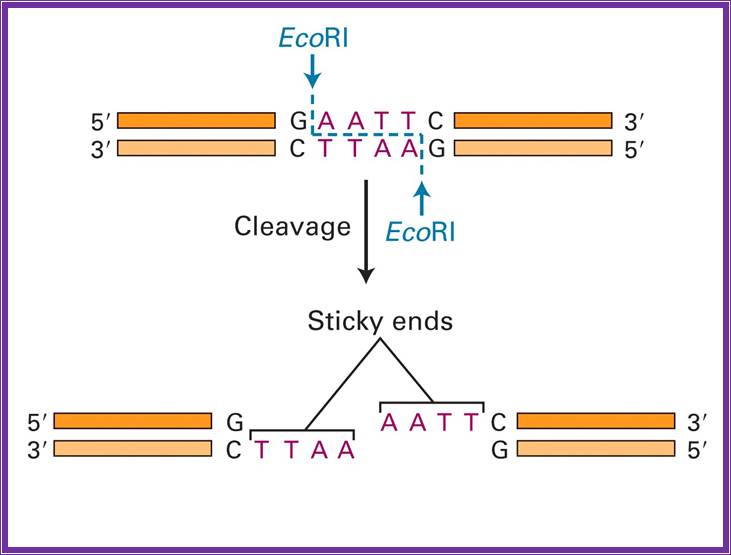
They are the most abundant group of enzymes.� Restriction and methylation are exclusively separate reactions.� They are made up of homo-dimmers. They recognize a set of sequences, which can be four base pairs or six base pairs; some times rarely the sequence can be 8 or more bases paired.� In most of the cases the sequence are palindromic in nature and actually they provide half sites for the enzyme binding and cutting. The enzymes use tetramer, hexamer or octamer sequences and can generate 5� overhangs, 3� overhangs or blunt ends, but they are strictly sequence specific.� Fully methylated sites are not cut with certain exceptions.� Hemi methylated sites are not cut but non-methylated sites are restricted.� However, methylase is a monomer, and methylates cytosine to 5-me-cytosine, or 4-me-cytosine or and methylates adenine to6-me-Adenoine
�
They are grouped as group -I, II, III and IV.� Each group contains subgroups. Subgroup A produces 5� overhangs, subgroup-B which generates 3� overhangs and sub group-C produces blunt ends.� Specific microbes or specific strains of a species synthesize all these enzymes.� Such organisms have been genetically manipulated for higher yield.� The organisms used for isolating restriction enzymes belong to aerobic, anaerobic, photosynthetic, diazotrophic, mesotropic, thermophilic or psychrophilic and they may be slow growing or fast-growing types.� Only few examples have been given.
Group-I:
Sub group-A generates 5� sticky tails:
(*= Position of cutting)
|
Name |
Sequence |
|
Sau 3A |
*GATC |
|
Nde I |
|
|
Nde II |
|
|
Taq I |
T*CGA |
|
Mbo I |
|
|
Nar I |
GG*CGCC |
|
Xma I |
C*CCCGGG |
|
EcoR I |
G*AATTC |
|
BamH I |
G*GATCC |
|
Bst I |
G*GATCC |
|
Bgl II |
A*GATCT |
|
Xho I |
C*TCGAG |
|
HinD III, Xba I |
A*AGCTT T*CTAGA |
|
Xba IT |
8CTAGA |
|
Nco I |
C*CATGG |
|
Not I |
GC*GGCCGC |
Sub group-B produce 3� sticky tails:
|
Name |
Sequence |
|
Hna I |
GCG*C |
|
Sac II |
CCGC*GG |
|
Sph I |
GCATG*C |
|
Kpn I |
GGTAC*C |
|
Sac I |
GAGCT*C |
|
Pst I |
CTGCA*G |
Sub group-C produce blunt ends:
|
Name |
Sequence |
|
Alu I |
AG*CT |
|
Hae III |
GG*CC |
|
Sma I |
CCC*GGG |
|
Eco RV |
GAT*ATC |
|
Dra I |
TTT*AAA |
|
Hapa I |
IGTT*AAC |
Group -II: Use interrupted palindromic sequences and generate 5�, or 3� or blunt ends (few examples).
Sub group-A:
Sub group-B:
Sub group-C:
Few Examples
|
Name |
Sequence |
|
Dcle I |
C*TNAG |
|
Sau I |
CC*TNAGG |
|
Sfi I |
GGCCNNNN*NGGCC |
|
Xmn I |
GAANN*NNTTC |
Group-III: They use slightly relaxed sequences; they also generate 5�or 3� or blunt ends (few examples).
Sub group-A:
Sub group-B:
Sub group-C:
|
Name |
Sequence |
|
EcoR II |
*CCA/TGG |
|
Xho II |
Pu*GATCpy |
|
Hae II |
PuGCC*py |
|
Hinc II |
Gtpy*puAC |
|
|
|
|
|
|
|
|
|
Group IV:� They use non-palindromic sequences.� They again generate 5� or 3� sticky tails or blunt ends (few examples)
Sub group-A:
Sub group-B:
Sub group-C:
|
Name |
Sequence |
|
Fok I |
GGGAGGATGN9* CTTCTN7* |
|
Hph I� |
GGTGAN8* CCACTN7* |
|
Mbo II |
GAAGAN8* CTTCTN7* |
|
Mnl I |
CCTCN7* GGAGN7* |
�
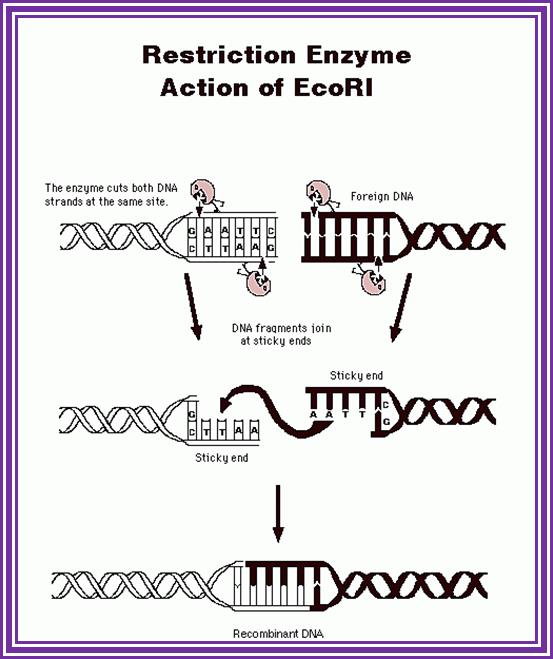
The most famous RE- is EcoR1; http://www.accessexcellence.org/
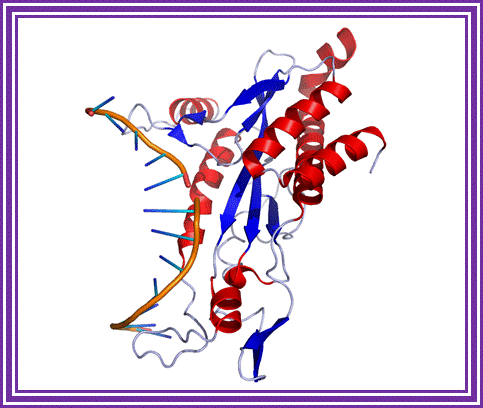
E.co R1 enzyme; http://www.austincc.edu/
�
Rare cutting Enzymes:
Such enzymes are very useful in generating genomic library.
Not I - one in 1000,000 bp
Sfi I - one in 1000 000 bp
Dma I- one in 60 000bp
Sac II - one in 60 000bp
Dra I- one in 20 000 bp
Xba I- one in 5000 bp
Isoschizemers:
They are different enzyme recognize the same sequence and cut differently.
Sma I/ Xma I / Cfrg I�� CCC*GGG
Msp I / Hapa II� C*CGG
SST I / Sac I GAGCT*C
Nde II / Mbo I / sau3A *GATC
�Nhe I / Sph I������ G*CTAGC.
Kpn I / Acc65 I
Mbo I / Sau3A I.
Sma I / Xma I.
Saph I / Bbu I.
Compatible ends:
Enzymes which produce similar sticky tails use the same sequence; they are very helpful in ligating the DNA fragments.
�
Sal I / Xho I cut with Taq I
Bgl II / BamH I cut with Mbo I
Xba I / Spo I cut with Bra I
Mbo I / BamH I cut with Mbo
Sau3AI / Bgl II cut with Mbo I
Bgl II/ Bcl I/Nde II
Sau3A I / Xho II�
Mbo I/Dpn I, Nde II, Sau3A I,
Dra I/ Aha II
Kpn I/ asp 718,
Pst I/ Nsci I
Sac I/Sst I
Sal I/Xho I,
Enzymes not inhibited by Dam methylation i.e. methylation of A in GATC sequence:
BamH I G/GAT*CC
Bgl II�������� A/GA*TCT
Sau3A I��� /GA*TC
Xho II�� �puG*TCpy
Enzymes not inhibited by DCM methylation i.e. methylation at C in a GG//CC or CGCG sequences:
BamH I G/GATCC*
Kpn I GGTAC/C*
Nar I GG/CGCC*
Hae III GG/CC*
Sfi I� GGCC*N5GGCC*
Dpn I GATC*
Restriction sites that contain ATG sequence:
Such sequences are helpful in cloning DNA in expression mode.� Such sequences can be introduced for initiating translation in generating proper reading frame.
Nco I C*CATGG
Nde I CA*TATG
Sph I GCATG*C
Fok I GGATG9*/13*/
Bbn I GCATG*C
Eco T121 ATGC*T
Microbes, their enzymes and their cutting sequence:
�
|
Microbe |
Enzyme |
Sequence |
|
|
Arthobacter luteus |
Alu I |
AG*CT |
|
|
Bacillus amyloliquefaciens.H |
BamH I |
G*GATCC |
|
|
Bacillus globigii |
Bgl I |
GCCNNNN*NBGC |
|
|
Bacillus globigii |
Bgl II |
A*GATCT |
|
|
Dienococcus radiophilous |
Dra I |
TTT*AAA |
|
|
Escherichia coli R1 |
E.coli RI |
G*AATTC |
|
|
E.co RV |
E.co RV |
GAT*ATC |
|
|
Hemophilous aegypticus |
Hae III |
GG*CC |
|
|
Hemophilous influenzae r. D 3 |
HinD3 |
A*AGCTT |
|
|
Hemophilous parainfluenzae |
Hapa I |
GTT*AAC |
|
|
Klebsiella pneumoniae |
Kpn I |
GGTAC*C |
|
|
Nocordia corallia |
Nco I |
C*CATGG |
|
|
Nocordia otidis cavarium |
Not I |
GC*GGCCGC |
|
|
Providencia stuartis |
Pst I |
CTGCA*G |
|
|
Streptomyces achronogenes |
Sac I |
GAGCT*C |
|
|
Streptococcus aureus 3A |
Sau3A |
*GATC |
|
|
Streptococcus caespitsus |
SCA I |
AGT*ACT |
|
|
Streptococcus fimbriatus |
Sfi I |
GGGCNNNN*NGGC |
|
|
Serratus marcescens |
Sma I |
CCC*GGG |
|
|
Xanthomonas badrii |
Xba I |
T*CTAGA |
|
|
Xanthomonas malvacearum |
Xma I |
C*CCGGG |
|
|
|
|
|
|
|
|
|
|
|
Application of Restriction Enzymes:
Enzymes have very wide use, especially in biochemistry, molecular biology, biotechnology, agriculture and medicine. They can be used in the following applications.
� Characterization of DNA.
� Size fractionation of genomic DNA.
� Cutting DNA at the required regions.
� Restriction site mapping.
� Can be used for RFLP studies.
� Restriction sites can be used for PCR amplifications using sequence specific primers.
� Can be used in recombinant DNA methods.
� Used in cloning and sub cloning and sequencing.
� Can be used in disease diagnosis.
� Can be used in creating in vitro mutagenesis.
� Many more.
�����������
����������� Restriction Site Mapping:
����������� It is a very useful technique, so one can analyze a DNA fragment where one uses it for marking a position; they act as flag post for molecular biologists.�
This technique along with end sequencing one can align fragments of DNA in a sequence, which is used for contigs alignment.
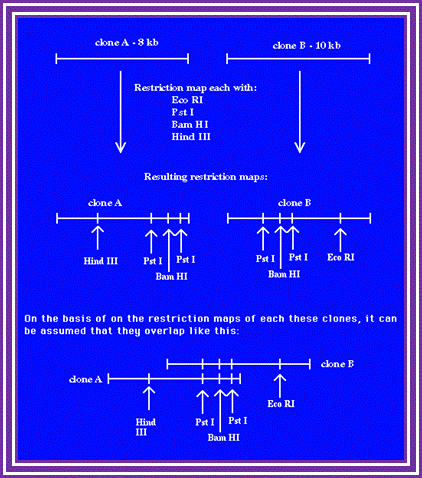
RE mapping; http://www.wormbook.org/
Take a piece of DNA from the clone, let us say it is 10kbp long or more label the 5�ends with radioactive 32P.� Clean the DNA free of the small 32P piece.
Assume the length of the DNA taken is 10KBp long and cut with E.coli I.� Similarly use another piece of the DNA cut it with Bam HI, Another with Pst I. Then separate each of the fragments cut by different enzymes on Agarose gel using different slots.� Note, in each of the cuttings only the fragment with label will show up in gel that is dried and auto radio graphed.� The other piece though found in the same slot does not show up as radioactive fragment.
For example, the DNA contains the label at one end.� When you cut with E. coli R1, it will cut at many site randomly (short period of time), wherever the sequence is found, but among these only one contains the tag.� Remember the digestion should be partial and should not be complete.� Partial means one enzyme cuts one of the several fragments at one site not more.� For this one has to adjust the concentration of the substrate and the enzymes.
����������� ����������������������� *I--1--2--3--4--5--6--7--8--9--I
Here the length is denoted by the number of nucleotide; 1=1000.2=2000 and so on.�� The astrich shows the position of the label.� Let us say E.coli has cut at position 2, 4 and 8. When you run the fragment on an Agarose gel and look for the radioactivity only 3 bands appear i.e. one with 0-2, 0-4 and 0-8, so three fragments are visible and we know where the sites for E.coR1 are present.� Similarly cut with Bam H I, let us say it has sites at 3, 5 and 9; you get 3 fragments.� Use another enzyme Pst I and let us say it cuts at 1, 6 and 7; this produces 3 fragments.
If such fragments cut by each enzymes are run on different slots, one can read the RE site of each and the size of each fragment and the sequence of each site.� One has to use the marker for the size of the size of the fragment.
�����������������������
|
Marker |
E.coR I |
Bam H I |
Pst I |
|
-------1 |
|
|
----- |
|
-------2 |
------ |
|
|
|
-------3 |
|
------ |
|
|
-------4 |
------- |
|
|
|
-------5 |
|
------ |
|
|
-------6 |
|
|
------ |
|
-------7 |
|
|
------ |
|
-------8 |
------- |
|
|
|
-------9 |
|
------ |
|
|
-------10 |
|
|
|
�
Now read the fragments and their site from the bottom.� The bottom most fragment is 9, it is Bam H 1, the next fragment is at 8, it is E.coR I, the next is at 7 i.e �at Pst1.� If you go on reading you get the sequence of RE sites like.�
I----Pst I-EcoRI-BamH-RI-BamH1-Pst1-Pst1-R 1-BamH1---I.�
Thus, one knows where the site for each of the REs.� This greatly help in cloning, sequencing, aligning each fragment in sequencing, creates mutations by deleting and filling up the cut ends, can be used for chromosomal waking.
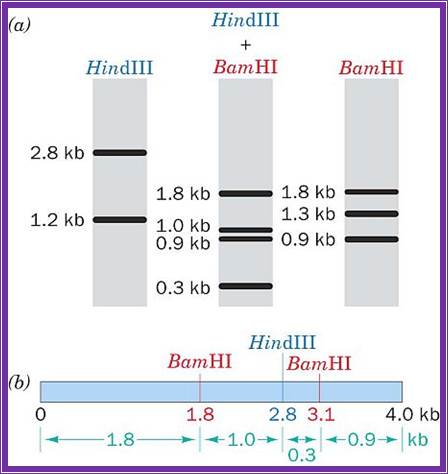
http://www.siumed.edu/
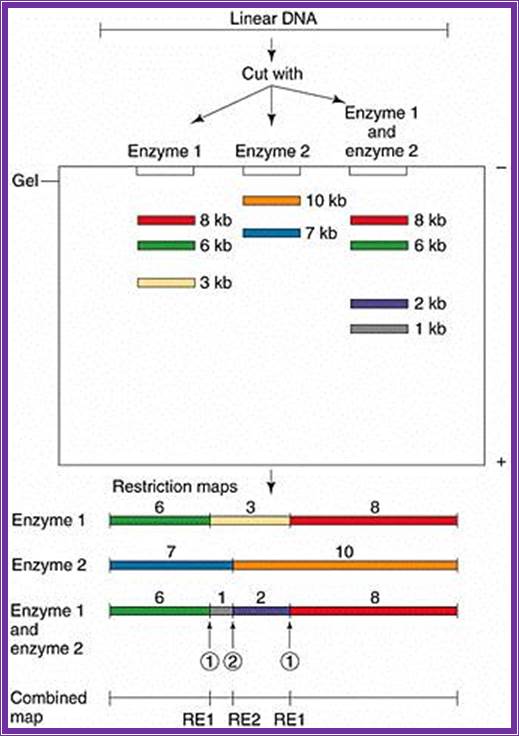
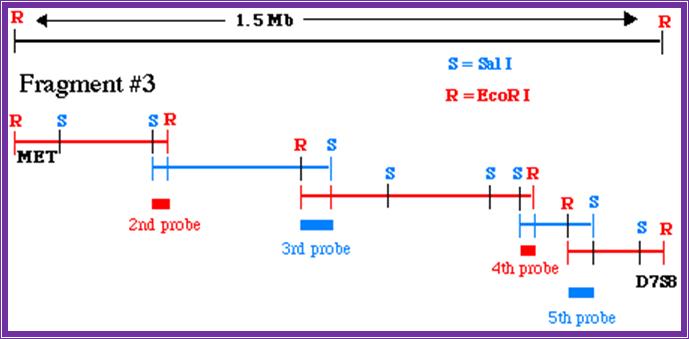
Chromosome walking; http://www.bio.davidson.edu/