Molecular Tools-3:
Modifying Enzymes:
In recombinant DNA technology a substantial number of enzymes are required for modifying DNA for a variety purposes.� To modify the DNA, one should know what enzymes are required and their properties. It is sine quo non for any student of molecular Biotechnology.
Polymerases:
E.coli DNA pol-I:
Its Mol.wt is 109KD. This is extensively used for Nick translation and Random primer mode of labeling.� It is also used in the second strand synthesis during cDNA preparation.� However, it requires a primer or nick for its activity.
�
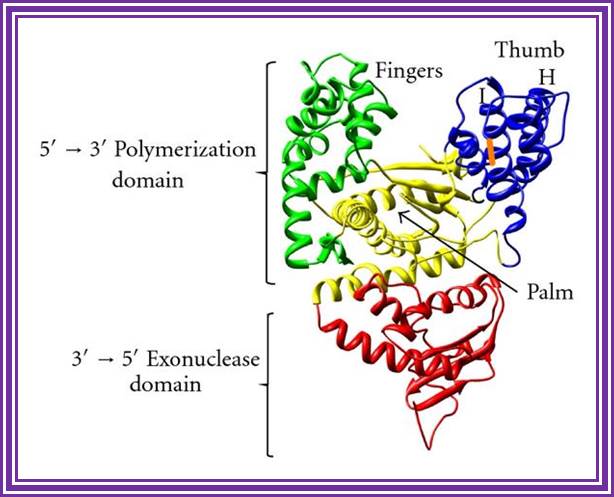
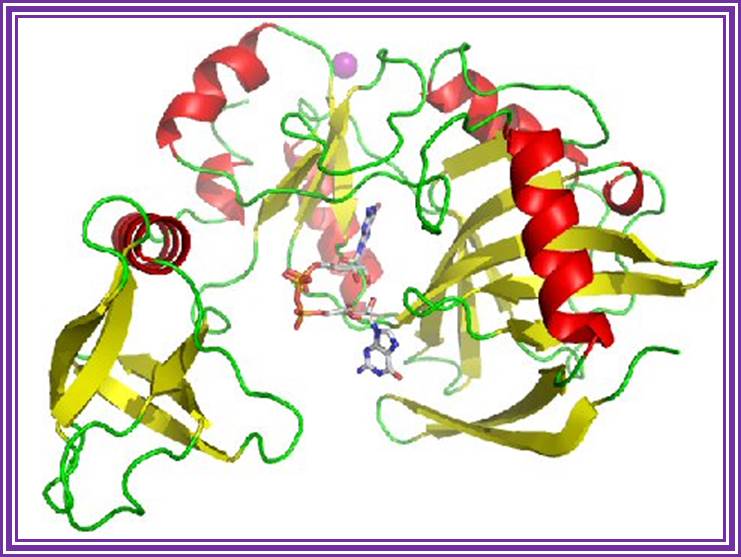
DNA Polymerase I; http://www.hindawi.com/
The enzyme from its NH3 end has 5�>3� exonucleases 3�>5� exonuclease and 5�> 3� Polymerase activity.� However the recombinant DNA pol-I is available as a truncated form called Klenow fragment (68KD), which has 3�>5� exonuclease and 5�>>3� Pol activity from its NH3 end.� One has to be careful about this enzyme for; the concentration of dNTPs provided should be optimal otherwise instead of polymerase it acts as an exonuclease.� If carefully used, one can apply this enzyme, for end filling or blunting dsDNA.� Using primers one can produce ds DNA by what is called primer extension method.� However, this enzyme has medium processivity and fidelity characters.
T7 DNA polymerase:
It mol wt is 84+12 KD (Pol + theoredoxin).� This polymerase is synthesized by the viral genome and the host bacterial cell provides the theoredoxin.
�
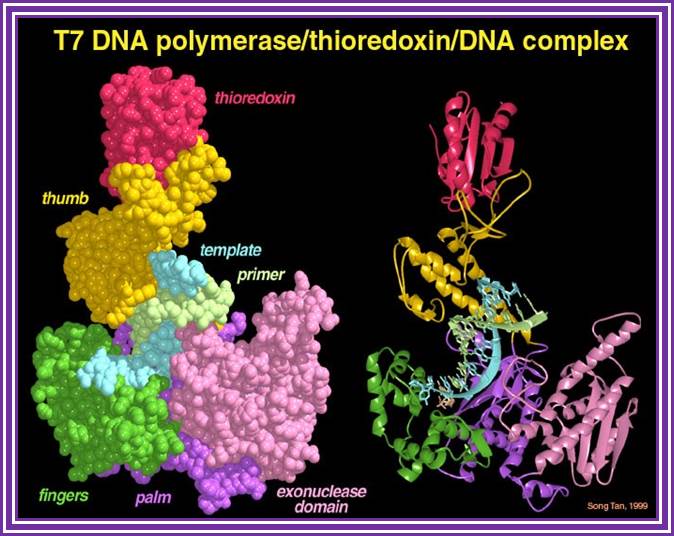
http://www.personal.psu.edu/
Together the enzyme is excellent in its polymerase activity.� It has 5�>>3� polymerase activity and 3�>>5� exonucleases activity.� Recombinant methods have produced T7 pol without 3�>>5� exonucleases property.� This enzyme is called �Sequenase� for this is an excellent enzyme for sequencing and primer extension protocols and even this can be used for labeling methods.� This has high processivity and high-fidelity properties.
T4 DNA polymerase:
It is produced by the T4 Phage viral genome.� Now it is available as recombinant product.� Its Mol.wt is 114KD and consists of single polypeptide chain. ��This is used for end filling and end labeling protocols.� It has 5�>>3� pol activity, 3�>>5� exonucleases activity.� The enzyme is quite stable and it can be substituted for DNA pol-I.� If the there are 3� over hangs it remove by 3� exonucleases activity and thus it produces blunt ends.� On the contrary if the ds have 3� recessive ends, the enzyme extends the ends by 5 > 3� polymerase activities.� However, it lacks 5�> 3� exonucleases activity.
Taq Polymerase:
This enzyme is isolated from thermophilic microbes, such as Thermophilous aquaticus, Pyrococcus species, and Thermococcus litoralis.� The enzyme Mol.wt is 85-95KD.� They have apparent 5�>>3� polymerase activity.� And they don�t exhibit 3�>>5� exonucleases activity.�� The enzyme is stable for 60 -70m minutes at 92^o temperature.� This enzyme is exclusively used for PCR amplification of DNA.� However, this enzyme has low processivity and low fidelity characters.� But recently pFu and ULTMA from Perkin Elmer/ Stratagene have been found to show high fidelity.� These enzymes are exclusively and extensively used in PCR based florescent-labeled sequencing reactions.
http://www.sharepoint.csiat,jmu.edu
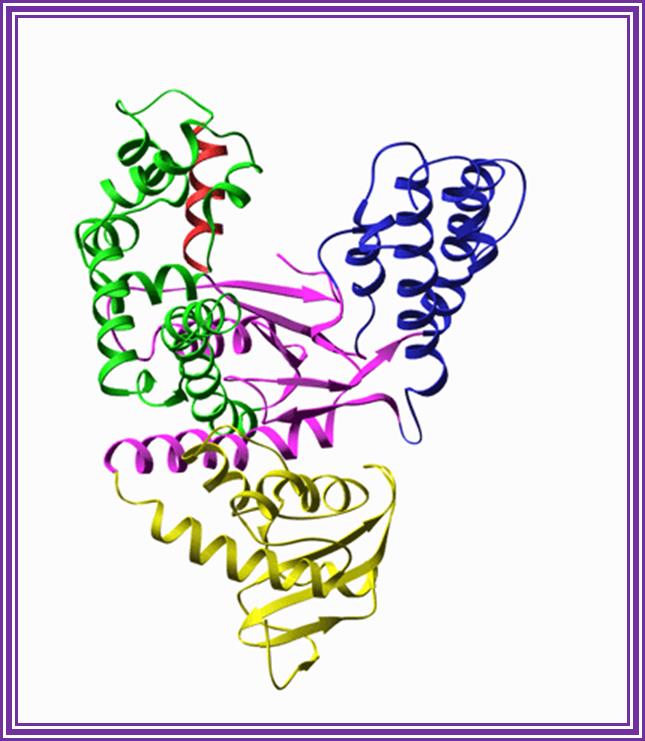
Taq polymerase: http://www.sharepoint.csiat,jmu.edu
These enzymes can also be used in Rt-PCR methods.� These are also used in pathogen detection protocols.� Also used in cDNA library preparations.� The amplified ds strands have one A base as extensions.� With suitable vectors the CDNA prepared can be directly ligated to dephosphorylated Vector DNAs.
RNA dependent DNA polymerase:
This is also called as Reverse Transcriptase.� It Mol.wt ranges from 85KD (from SDS-PAGE it shows 71KD) single polypeptide (murine melony leukemia virus) and 92 KD (two polypeptides called alpha 68KD and Beta 92KD) from Avian Myeloblastasis virus, all of them are retroviruses.�� These enzymes have RNA dependent DNA polymerase activity, but does not show any apparent 3�>>5� exonucleases activity.� It can also perform DNA dependent DNA polymerase activity. Another great feature of these enzymes is that they have RNase-H activity, where the enzyme removes by nicking or displacement of RNA when it is hybridized to DNA.� It can remove nucleotides from both ends i.e. from 5� and 3� ends.� The alpha subunit has contains both Polymerase and RNase -H activity.
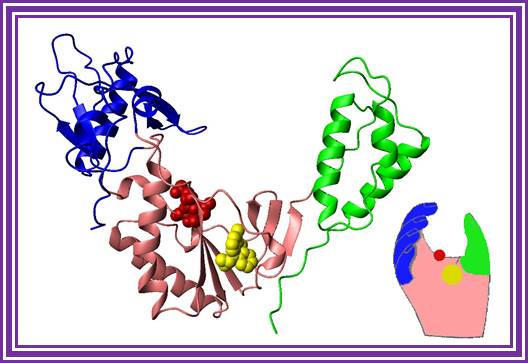
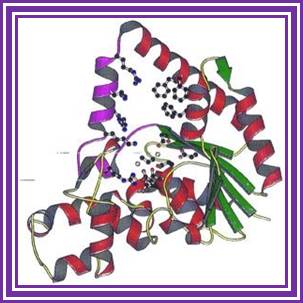
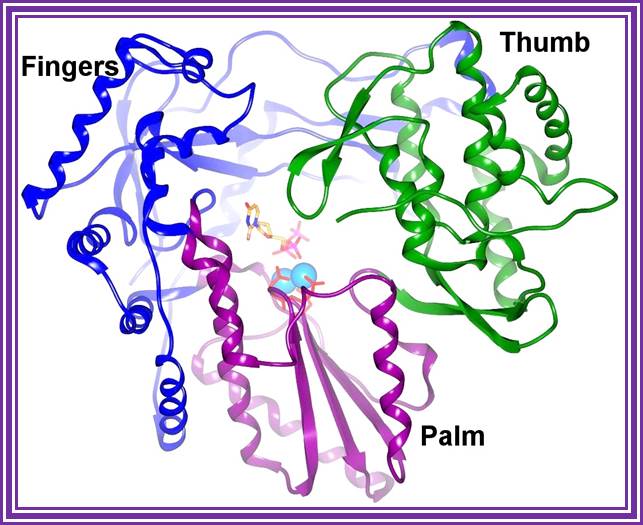
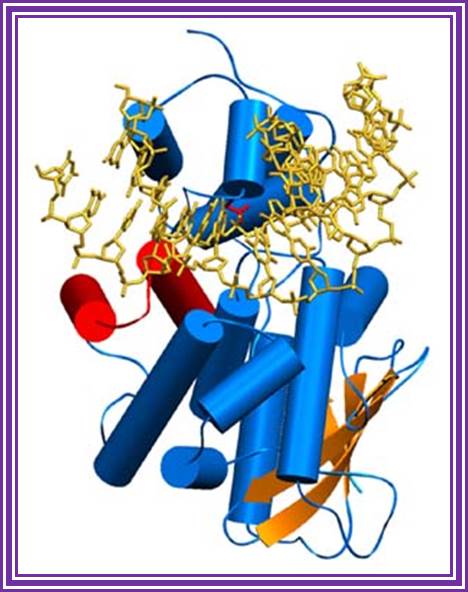
�A handy enzyme
This ribbon representation of the RT active
domain illustrates its hand-like structure, showing fingers (blue), palm (pink)
and thumb (green). The active site (red atoms), where DNA is elongated, is in
the palm region. Also shown is an RT-inhibitor drug (yellow) in the pocket
where it binds.
http://www.psc.edu/
This is extensively used during second strand synthesis of cDNA.� It requires RNA or DNA primers of 8 or more nucleotide long.� These enzymes show low processivity and low fidelity.� In most of the labs this enzyme is used for the preparation of first strand of cDNA.� Its high temperature dependence of 42^o is helpful in removing ant secondary structures of the template.� But Melony Murine leukemia enzyme is single stranded and it is mostly used in cDNA preparations.
Poly-(A) Polymerase.
This enzyme is a tetramer 33KD each.� It has unique property of adding adenine nucleotides to 3� end of any RNA.� Only few nucleotides are required at the 3� end of the RNA.�
Some of the viral RNAs don�t have poly (A) tail; in such situations one can add poly (A) tail, which is of great help in preparation of ds CDNA from them.
�
Polynucleotide Phosphorylase:
It is tetramer, 60-70KD each.� It can add rNDPs to 3� ends of RNA and require Mn2^+ or Mn2^+.� This can polymerize free nucleotides to generate polynucleotide chains. It can also act as nuclease.
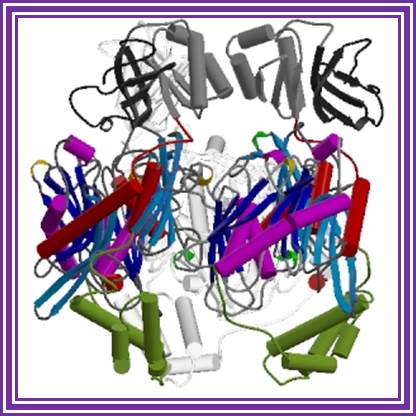
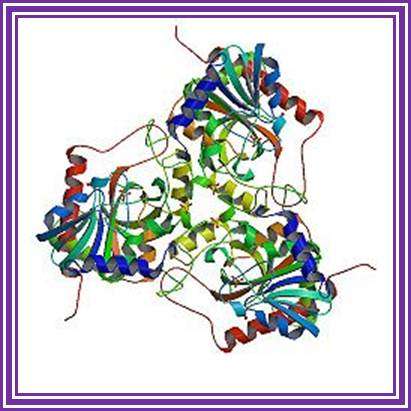
Polynucleotide Phosphorylase: http://www.creativebiomart.net/
Terminal Deoxynucleotidyl Transferase:
It is a dimmer consists of 26.5 and 8KD proteins.� It adds dNTPs to the 3� OH ends ss DNA or dsDNA.�� It is template independent.� This can be used for 3� end labeling.� This can also be used for generating 3�end tails of required nucleotides for cloning purpose.
T4 Polynucleotide Kinase:
It is obtained from T4 phages. Its molecular mass is 33KD, it is a tetramer.� It transfers gamma phosphate from the dATP to the 5� of ds DNA or dsRNA.� Even in the presence of 5� �P� group, it exchanges the gamma labeled phosphate, at the 5� end.� This is extensively used for 5�end labeling of DNA or RNA probes and the 5� labeled DNA can be used for sequencing reactions.
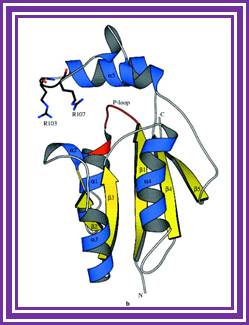
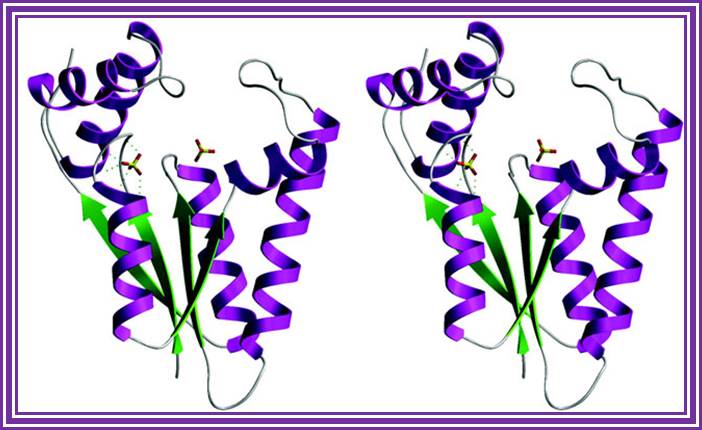
Structure an mechanism of T4 polynucleotide Kinase; an RNA repair emzymes; http://emboj.embopress.org/
RNA polymerase:
T7 RNA-pol (98KD), T3 RNA-Pol (100KD) and SP6 RNA-pol (96KD) can be used for synthesis of labeled RNA in large amounts provided if one uses their respective promoters in their constructs.
DNA Ligases:
There are different types of ligases of which E.coli DNA-ligase (74-77KD) and T4 DNA ligases (43KD) are extensively used.� The former is NAD dependent and the later is ATP dependent.� They are responsible for ligating the DNA ends.
http://www.biochem.umd.edu/
Alkaline Phosphotase:
Alkaline Phosphotase enzymes are available from bacteria and calf intestine.� They are used to remove phosphate groups from 5� ends of ssDNA or from dsDNA.
�
��������� 
Alkaline Phosphatase; �http://en.wikipedia.org/
�Removal phosphate groups in vectors prevent self-ligation.� However, one should be careful in using this enzyme for after treatment it has be removed completely by extracting the DNA 2 to 3 times in phenol-chloroform mode and then in chloroform once and precipitating in 95% alcohol in the presence of 0.15 molar sodium acetate.� Alkaline Phosphotase from calf intestine can be easily inactivated by heat.
Guanylyl Transferase:
�This enzyme used for capping of mRNA, which is with out it.� The enzyme transfers 7�CH3-GMP* from the 7�CH3-GTP to the 5 biphosphate or triphosphate by 5� to 5� phosphate bonding to generate 5�7�CH3 G-CH2-O-P*-O-P-O-P-O-CH2- ApXpXpX.
Guanylyl transferase: http://chemistry.umeche.maine.edu/
Nucleases:
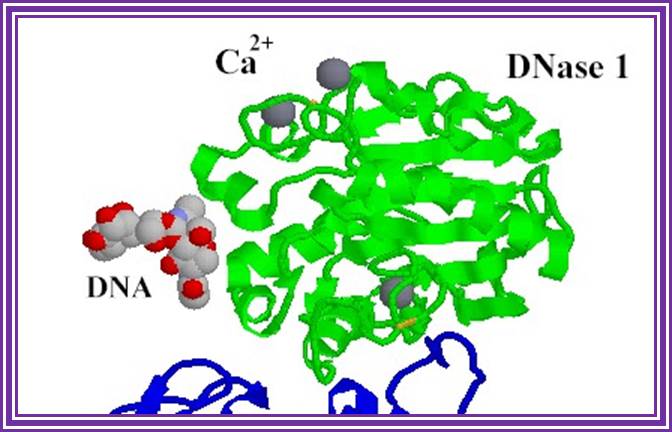
Dnase I: RNase free:
Dnase I from bovine pancreas, 37KD, free from RNase is used for preparation RNA free from DNA. It is glycoprotein.� It digests both ssDNA and dsDNA.
DNase 1;http://www.bms.ed.ac.uk/
This enzyme nicks double stranded RNA so it is used for labeling DNA by nick translation at very dilute concentration of enzyme.� It is also used for Dnase I foot printing and DNase I mapping.
Exonuclease III:
The enzyme is available as a recombinant product.� Its Mol.wt is 28KD.This enzyme has the ability remove nucleotides from 3� end of ds DNA either from blunt ends or from 3� recessive ends.�
T5 -5�exonuclease
Flap Endonucleases, 5'-3' Exonucleases and 5' Nucleases; http://www.sayers.staff.shef.ac.uk/
3�to 5� exonuclease; http://en.wikipedia.org/
It removes nucleotides after nucleotide to generate 5�P-mononucleotides.� This enzyme is extensively used for deletional analysis of promoters to find out which part of the DNA segment can act as the promoter.� If allowed for longer period it can generate single stranded DNA, but this enzyme does not act on single stranded DNA or RNA.� This enzyme can also remove Apurinic and Apyrimidinic sites by endonuclease activity.
Lambda 5� Exonuclease:
This has the ability remove nucleotides one after another from the 5� end of the ds DNA either from the blunt end or 5� over hangs.� This is also useful in generating single stranded DNA and also for deletional analysis of the promoter elements.
S1 nuclease:
It is obtained from Aspergillus oryzae.� It removes single stranded DNA or removes over hangs either from 3� or 5� end and produces a blunt ended DNA. Its Mol.wt is 38KD.� It is a glycoprotein.�
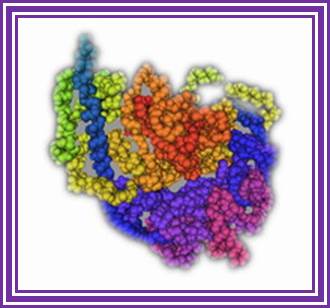
Aspergillus nuclease S (1); http://www.ebi.ac.uk/
�Double stranded RNA and RNA: DNA hybrids are resistant to the action of this enzyme.� This enzyme can be used S1 mapping of transcripts, one can remove 5� or 3� overhangs of DNA ends, and even they remove any hair pin structures.
Ribonuclease H:
The enzyme is obtained in large quantities from retroviral constructs.
�
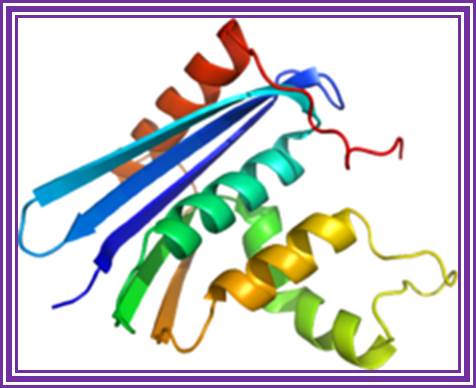
RNase H;� http://en.wikipedia.org/
The enzyme removes RNA by endonucleolytic cleavage of RNA from RNA: DNA hybrids. This enzyme used in the preparation of the second strand during ds cDNA synthesis.
Ribonuclease -A:
The main source is bovine pancreas.� Its Mol.wt is 13.7 KD.� In its pure form it is used for the preparation pure DNA free from RNA contamination in plasmid DNA preparation.
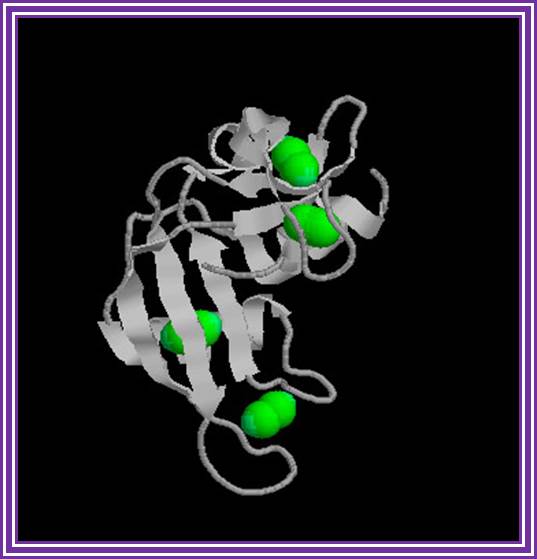
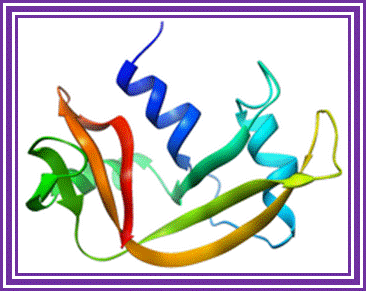
Ribonuclease A; http://en.wikipedia.org/
�It hydrolyses 3�-5� phophodiester bonds of RNA segments with 2�3�cyclic phosphates.
Protein kinase K:
During preparation of nucleic acids from whole cells containing lot of proteins; to free from other proteins this enzyme is used.� This gives reasonable nucleic acid preparations
Taq polymerase:
Reverse transcriptase:
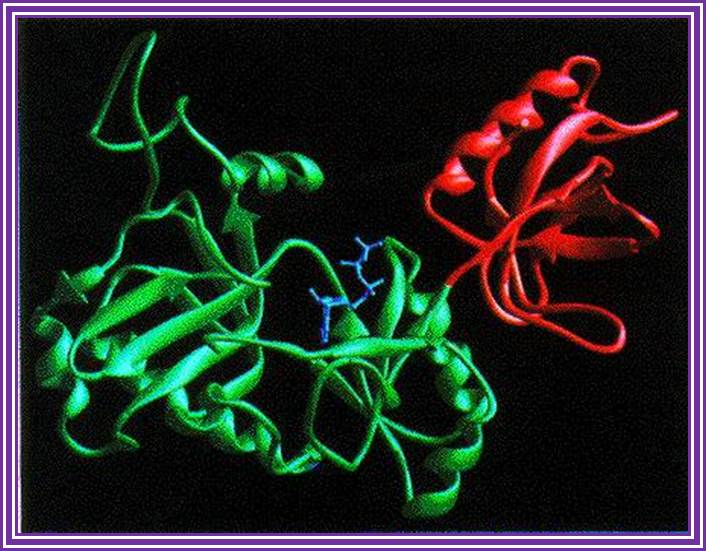
DNA ligase;� http://www.biochem.umd.edu/
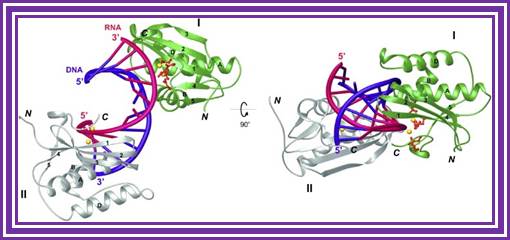
RNase H
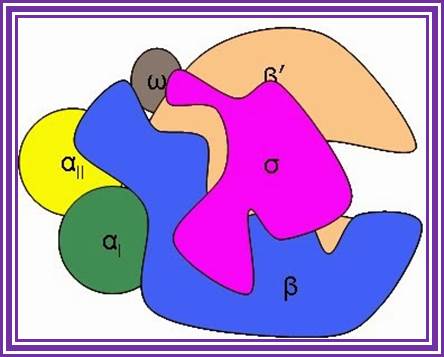
RNA polymerase?
Bacterial RNA Polymerase; http://small-collation.blogspot.com/
RNA Pol 1 (III); http://www.conservapedia.com/
RNA Pol II; http://www.conservapedia.com/
Proteinase K
http://scitechdaily.com/
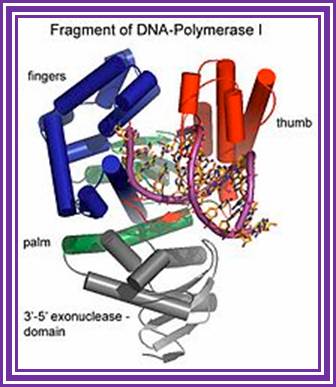
Fragment of DNA Pol I-3�-5� exonuclease; DNA Pol I also contain 5�-3� exonuclease activity http://en.wikipedia.org/