Molecular Tools-II
Vectors:
General:
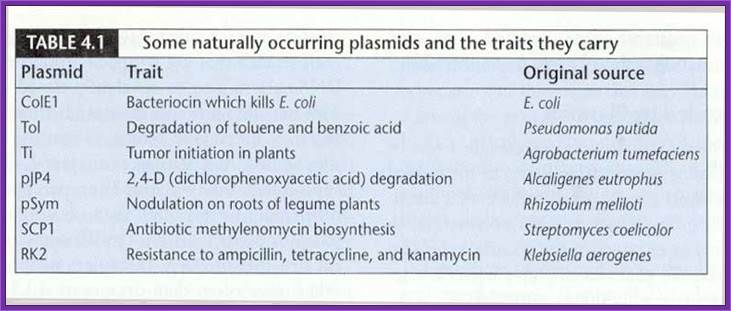
����� Vectors are carriers of a foreign DNA from one cell to the other or from one system to the other.� Once the foreign DNA is inserted, it can be propagated, it can be subjected in vitro mutation, it can be made to express, it can sequenced, and it can be subjected various modifications.
Most of the vectors are derived from different plasmids and they are then modified to needs of a scientist.
����� The kind of vectors used depends upon the molecular needs.� One can design a vector required.
�
In general, most of the vectors should have the following features:
� Vector DNA size should be as small as possible.
� It should contain an origin site, which is efficient and produce high copy numbers.
� The origin should be compatible to the kind of cell in which it replicates. Ori- Col E1 is good for bacterial systems.� Origin ARS is good for Yeast systems.� Some of eukaryotic viral origins such as SV40 Origin are good for animal systems.
� Such origins can be cloned and used.
� They should have a selection marker gene such as antibiotic resistant gene.
� They should contain unique cloning site(s) such as multiple cloning sites (MCS).� Unique in the sense such sites are not found any where in the vector DNA.
� There should be desirable promoter for expression of the gene at one end of the MCS and a transcriptional terminator sequence at the other end of the MCS.
� There can be additional DNA sequence for the regulation of gene expression such as response elements, operator elements, enhancer elements and possibly Lox sites for recombination by �Cre� enzymes.
� It is also important to have the knowledge of the other restriction sites and their position in the plasmid.
� The vector can also contain a reporter gene under the control of a regulator sequence.
� One can add few coding sequences before MCS in reading frame for generating fusion protein with specific sequences, so that they can either be secreted out or they can cut with specific enzyme to release the desired protein or sequence tags can used for the purification proteins such as his-tags.
��
�
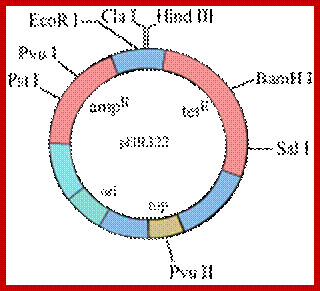
pBR322:
This vector was designed by Bolivar and F. Rodrigus in 1977.� Perhaps this vector was used for deriving various plasmid-based vectors.
pBR322- cloning vector-REs are marked; https://www.meritnation.com
Important features:
Size: 4.363 kbp,
ColE1 Origin,
Amp resistant gene,
Tetracycline resistant gene,
Rop- mediates RNase activity,
RE in Amp^+ = Pst-I, Puv II, Sca I,
RE in Tet^+ =HinD3 I, Bah I, Sal I,
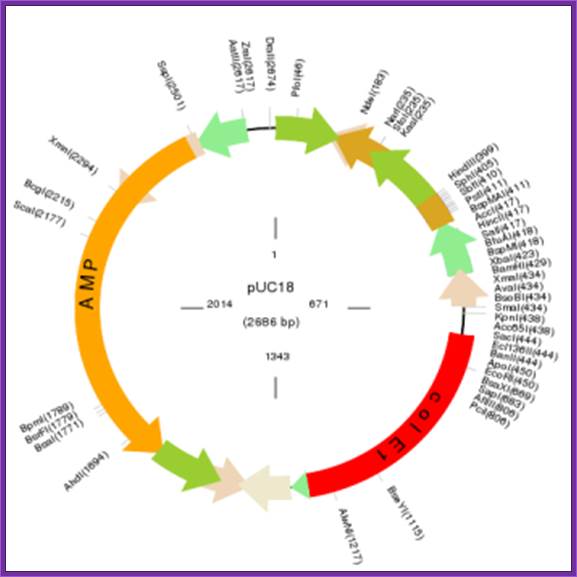
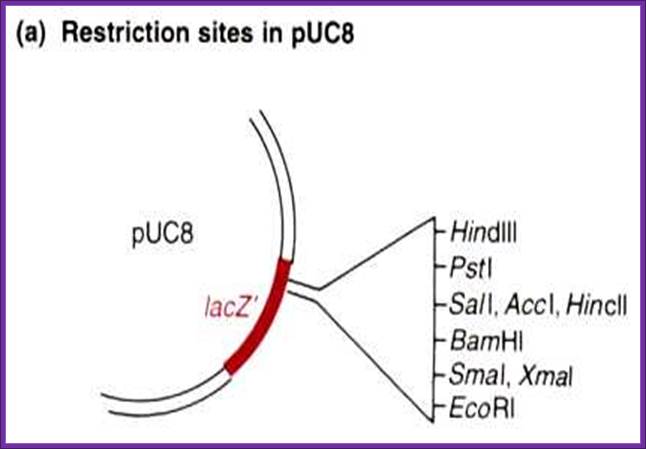
pUC 8/9 and their derivatives pUC18/19:
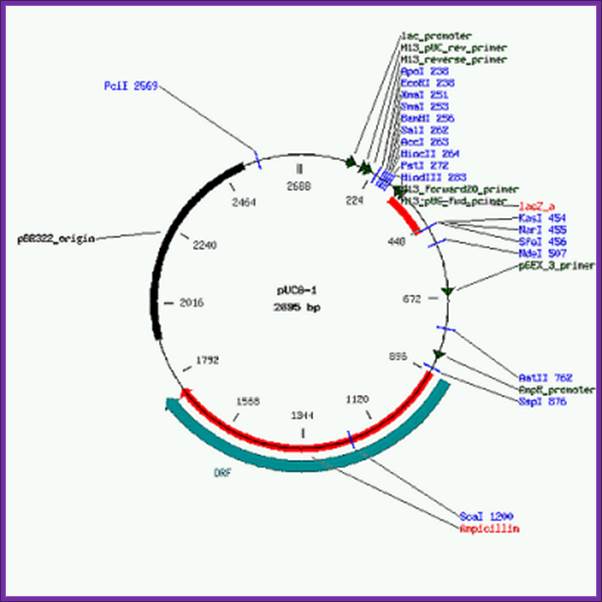
����������������������������������������� F. Pfeiffer, MPI, Martinized
pUC8 vector 2858bp
pUC8-1�2 2695 bp
pUC9/9-1 and 9-2
Ori-pBR322
Hosts: E. coli JM83, E. coli JM105, E. coli JM109, E. coli NM522
Size-2858,
LacZ laphasubunit,
Mcs
ColE1 ori pMB!
Ampicillin
|
Plasmid Name |
pUC8-1 |
|
Plasmid |
Size-2695 |
|
�� Note; |
These data and their annotation were supplied to GenBank by Will Gilbert under the auspices of the GenBank Curator Program. Assembled from pUC8 and pUV81a by F. Pfeiffer, MPI, Martinized The pUC vectors contain the beta-galactosidase gene under the control of the lac-promoter and lac-operator. Thus, they are usable as inducible expression vectors. To allow insertion of genes in any reading frame, the polylinker region was redesigned to contain the same restriction sites in all three reading frames: Trio '8' is pUC8, pUC8-1 and pUC8-2. Trio '9' is pUC9, pUC9-1 and pUC9-2. Hosts: E.coli JM83, E.coli JM105, E.coli JM109, E.coli NM522. Related vectors: pUC8, pUC91a, pUC8-2, pUC9-1, pUC9-2. (Information source: Vector DB.)
|
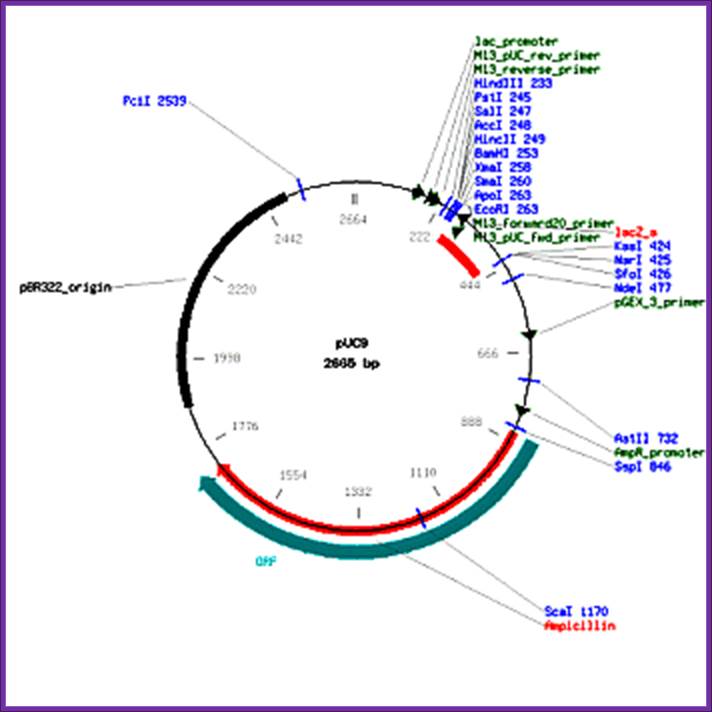
F. Pfeiffer, MPI, Martinized
pUC9 vector
2665 bp
Ori-pBR322
Hosts: E.coli JM83, E.coli JM105, E.coli JM109, E.coli NM522, E.coli.
|
Plasmid Name |
pUC9 |
|
Source/Vendor |
ATCC |
|
Plasmid Size |
2665 |
|
Notes |
These data and their annotation were supplied to GenBank by Will Gilbert under the auspices of the GenBank Curator Program. Assembled from pUC19 and M13MP9 by F. Pfeiffer, MPI, Martinized old Pharmacia vector. Expression vector with lacZ as an insertion detection marker. Medium is 1227 LB plus ampicillin. Hosts: E. coli JM83, E.coli JM105, E.coli JM109, E.coli NM522, E.coli. Related vectors: pUC7, M13mp9, pUC8. (Information source: VectorDB.) |
����� J. Messing and Vieira� �from University of California-1985 developed these vectors.� They are the most popular plasmid based vectors and contain most of the desirable features for day-to-day cloning experiments. U=University, C-California.
�����
Features:
����� Size: 2.686-2.725 kbp,
����� Contains ColE-I Origin,
����� Amp^+ gene,
����� MCS,
����� LacZ alfa
�����
����� Lac-Z operator promoter linked to Lac-Z gene only NH3 part of the gene called alpha subunit.� A DNA sequence has been introduced in this region, which has many restriction sites.� The sequences are in reading frame and when they are translated, they generate a part of LacZ protein.� Th host bacterial cell contains the omega subunit of the LacZ, �which is constitutively expressed. When the plasmid based LacZ expresses, it is complemented with the omega, so the fully functional LacZ protein is produced whose activity can be monitored by using X-Gal which on hydrolysis produces purple-blue color.� But if another DNA is introduced into any one of the sequences the protein produced is non functional.� Cloning sites are from left to the right. This vector has Lac I operator sequence.
�����
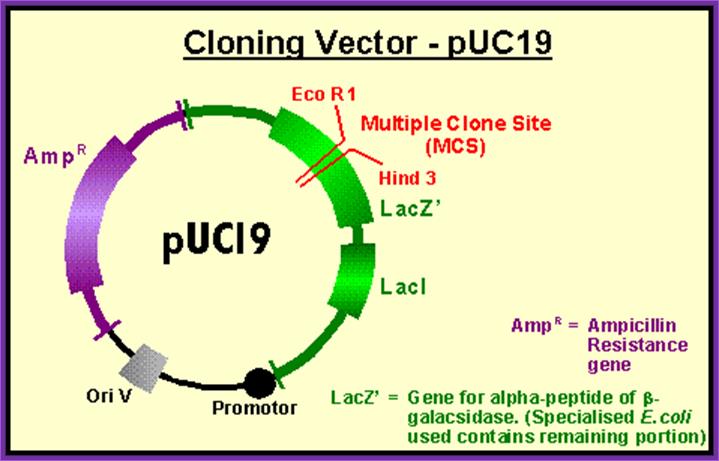
pUC 18/19 contain Lac I� adjacent to promoter. http://serc.carleton.edu
Thus plasmid can be used for cloning and sub cloning,
It can be used for cDNA bases cloning,
It can be used for Blue and white colony screening,
It can be used for in Vitro site directed mutagenesis,
It can be used for dsDNA sequencing.
Most of the shuttle vectors are created by using pUC vectors,
This is a kind of work horse in molecular techniques.
Host cells used for this vector-based transformation are Jm109, DH 5 alpha
pUC-18:
-ATG---ATG-----EcoR-I, Sma-I/Xma-I/Ava-I, BamH-I, Mlu-I/Hinc-II, Bfr-I, Not-I, Kpn-I, Apa-I, Sal-I, Xba-I, Sac-I, Sph-I, EcoRV-I, Nco-I, HinD3---------
pUC-19:
--ATG---ATG�HinD3-Nco I-EcoRV-I, Sph-I, SacI-Xba-I, Apa-I, Kpn-I, Not-I, Bfr-I, Hinc-II/Mlu-I, BamH-I, Ava-I/Xma-I/Sma-I, EcoR-I--------
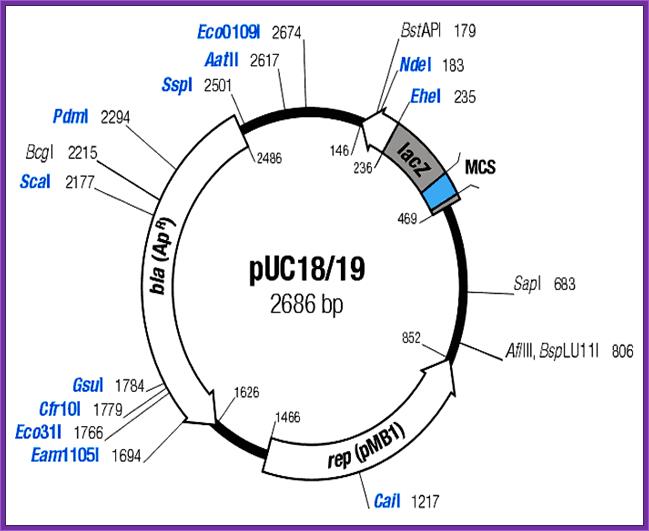


pU8/9 Mcs. pMB1 in E.coli Ori ; Institute of Bioinformatics and Biotechniques ; http://ibabmsc01.blogspot.com
The codons and amino acids they generate in mcs (multiple cloning sites) region:
pUC 18:
--ACA.GCT. ATG.ACC. ATG. ATT.ACG.AAT.TCG.AGC.TCG.GTA.CCC.
����������������������������������������� ��� M����� I������ T������ N���� S����� S������ S������ V����� P
GGG.GAT.CCT.CTA.GAG.TCG.ACC.TGC.AGG.CAT.GCA.AGC.TTG.GC
��� G������ D���� P����� L������ E����� S����� T������ C����� R����� H����� A����� S����� L����� A����
A.CTG.GCC.GTC.GTT.TTA.CAA.CGT.CGT.GAC.TGG.GAA.AAC.CCT
������ L������� A����� V���� V���� L����� Q����� R����� R������ D����� W���� E����� N����� P->LACZ
pUC19: MTMITTPSLHACRSTLEDPRVPSSDSLAVVLQRDWEDPG�Lac-Z
pACYC 184:
Developed by Chang A.C.Y and Cohen (1978):
Size: 4.244 kbp,
Kanamycin^+ gene,
Col E1 origin,
Tet^+,
Ori-p15A,
This plasmid is compatible with pBR322 and pUC plasmids.
Contain restriction sites in antibiotic genes.
����� M13/18:
����� This vector is derived from M13 filamentous phage.� The DNA is in replicative form.� The vector has all its 10 genes intact.� Next to the origin the entire Lac-Z operator�promoter and Lac-Z alpha subunit with MCS segment from pUC18 has been inserted.� In addition, it has Lac-I is also introduced to the left of the Lac-Z alpha complex or cassette. The sequence of MCS is ATG�ATG Eco RI to HinD3.
����� ----Ori (+)-ori (-)�Lac-I�Lac Z�I mcs I-------z alpha-�
����� McCS=�� ATG---ATG--- EcoRI--------Xba-I-------HinD3
M13 vectors are derived from the combination pUC and M13 phage ds DNA,
M13 plasmid for cloning
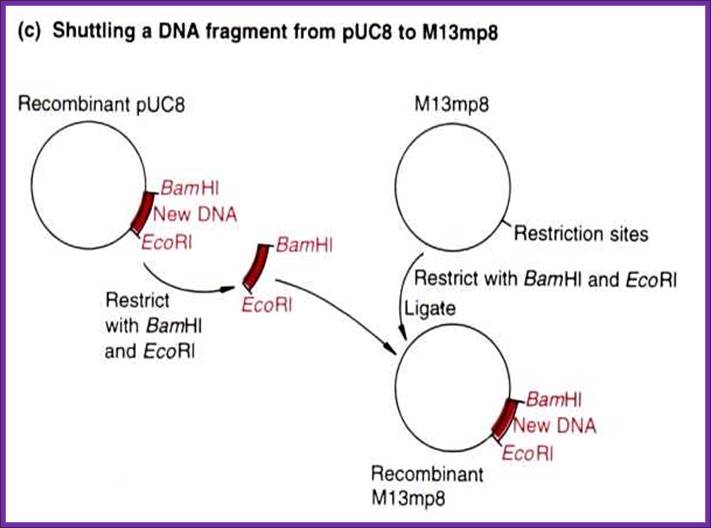
����������������������������������������� Deriving M13/18 vectors
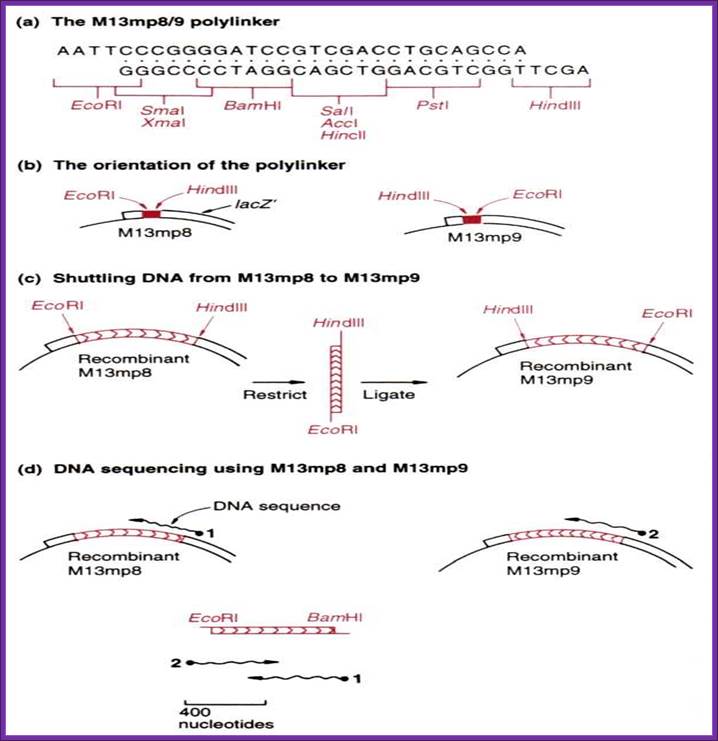
����� M13/19:
����� This is same as that of M13/18, but it differs from M13/18 in the orientation of MCS sequence, which is in ATG---ATG---HinD3 to EcoR-I.
----Ori (+)(-)---Lac I ----lacZ---ImcsI----lacz alpha--�
����� MCS = ATG---ATG---HnD3-------Xba-I--------EcoRI
����� These vectors can be used to generate single strand DNA either of plus sense or negative sense.�
����� This can be used preparing cDNA library.
����� Can be used for Blue and White colony screening.
����� One can use it for creating a library for antibodies, it the cloning is done into to the capsid proteins.
����� Host cells are again JM109, Dh5 alpha
M13/18 Phagmids:
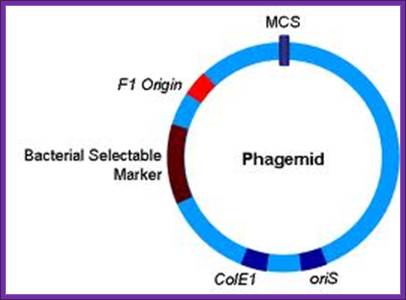
����������������������������� A general view of the phagmid
����� These Phagemids are derived from pUC vectors. They contain f1 or M13 phage derived Ori sequences.
---M13/18: ---ori (+)---lacz (P)>---II-mcs-II---lacZ alpha---,
The mcs sequence is from ATG�ATG�>EcoR I ---Xba I---HinD3->�----in both of M13/18(+) and M13/18(-) Phagmids.
Phagmid size = 2746-3200bp,
They contain Amp^+ gene,
They can used to generate ssDNA for sequencing,
They can be used for site directed in vitro mutations,
They can be screened as Blue and white colonies.
Host cells are JM109 and Dh alpha.
�M13/19 Phagmids:
![]() The mcs sequence is ATG�-ATG--Hind3-XaB1-- I----EcoR I-->, in both m13/19 (+) (-) phagmid.
The mcs sequence is ATG�-ATG--Hind3-XaB1-- I----EcoR I-->, in both m13/19 (+) (-) phagmid.
![]() Phagmid size = 2746-3200bp,
Phagmid size = 2746-3200bp,
They contain Amp^+ gene,
They can used to generate ssDNA for sequencing,
They can be used for site directed in vitro mutations
They contain AMP^+ gene,
They can be screened as Blue and white colonies.
Host cells used are DH5 alpha and Jm109.
pGEM-3Zf(-) and pGEM3Zf/+):
Size of the phagmid is 3199 bp,
Both are identical except for the presence of (+) or (-) origin sequences,
It has combination of a plasmid and f1 phage origin sequence either in + or - orientation,
It has an Amp^+ gene,
It has Lac-z P/O and Lac-z alpha subunit,
A mcs is located in the NH3 terminal part of the Lac-z subunit,
On either side of the mcs promoters like T7 and SP6 are present.
Host cells are JM109 and Dh5 alpha;
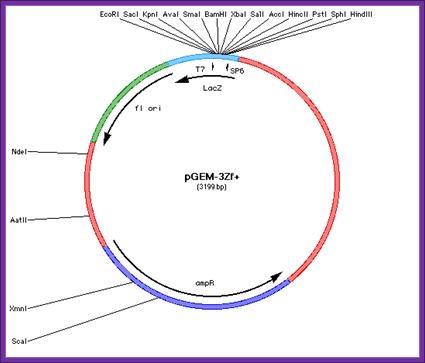
pGEM; Mcs
The mcs are:
�
SP6 promoter> HinD3-Sph I-Pst I-Hinc II-Acc I-Sal I-Xba I-BamH I-Sma I-Ava I-Kpn I-Sac I-EcoRI-T7 promoter,
pGEM-3Zf(-)/(+):� ----Lac-Z---I-SP6-- mcs-T7-I------------>
It can used for Blue and white colony screening,
It can be used for dsDNA sequencing,
Contains Amp^+ gene,
Cloned segment can be used for generating labeled transcripts, which can be used as probes.
They have promoters for SP6 and T7 RNA polymerase initiating sites.
They have Lac operon sequences including operator for regulation,
It can be used for production ssDNA for sequencing.
4-B.� For cDNA library
�����
Lambda DNA based Vectors:
���� Lambda DNA is 48.52kbp long.� The virus exhibits both lysogenic and lytic cycles.� Regulation of this is very fascinating and it has been extensively studied, for that matter it is one of the par excellent examples for understanding gene regulation.� It is a double stranded DNA with 12 ntds long sticky tails whose sequence is complementary, hence the term COS site.� The genome can be grossly divided into Left arm and Right arm and the central codong region.� The left has genes for head, tail, fiber, tail plate and other structural genes. Nearly 200 bp from the left end it has sequences for packaging the DNA into the head. The Right arm contains genes for replication.� The central region contains genes for recombination and regulation such as cI and others.� Some of the sequences in the central region are dispensable and one can replace this segment with foreign DNA.� Thus, one uses this DNA for cDNA or genomic library preparations.
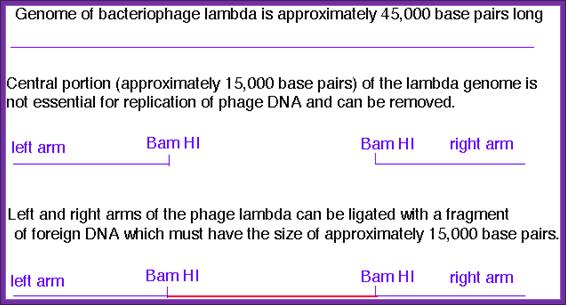
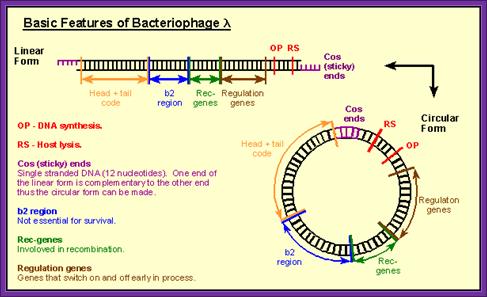
Lambda for cloning; Basic features.
Lambda gt 10:
Size if 43.34 kbp,
<I====================I====R1===I===============I>
<----==L==� >20kbp====I==C=14kbp=I===7-8kbp====R=---->
- R1 represents E.coR I site located within the cI region,
Insert size is 4 to 7.6kbp,
- Cloning site is E.coR-I located with in cI region (amp unit 32.71),
- Cloning is non-directional.
- The lambda DNA with an insert in the cI is disabled,
- This cannot be used for blue and white colony screening,
- It has no expression and regulatory sequences for the cloned DNA,
- The host strain used is LE 392, C600, and C600hfl, which lacks lysogeny function,
- This is very useful in screening with RNA or DNA probes.
- Only recombinant produce plaques for the cI is disabled,
- Non-recombinants don�t produce any plaques for the cI is still active.
- Cloning is non-directional.
Lambda gt11:
One can clone an insert of 4-to 7.6-kbp sizes,
It contains a segment of Lac-z gene with an E.coR I site at the carboxyl terminal region, so if the insert DNA expresses it will be in the form of a fused protein.
The Lac-z segment is introduced into the central stuffer region at 18.58 and 24.82 map unit position from the left end marked by Kpn I and Xba I sites.
There is one more restriction site called Sac I to the right of RI site.
The direction of the Lac-Z is from right to the left side.
L~~I===========I-�R1---lac-Z--I====cI====I========I~~~R
����������� 
Lac-Z has promoter and operator segments
Lac-Z has promoter and operator segments, and the expressed product is 116KD b-Galactosidase,
It carries an amber mutation, this leads to lyses deficient if the host cell is F^(-) an amber suppressor mutant.
In Y1090 host strains which is sup^(+) it forms clear plaques with DNA inserts,
In hosts such as Y1089, which is sup F^(-) and hfl-A mutant (high frequency lysogenic), with the insert DNA it becomes lysogenic.
This can be used for high-level expression of protein as fusion product.
The lambda DNA has a Tm sensitive cI 857 gene between 34.9 and 35.35.47 map unit position,
By increasing the Tm from 32^oC to 42^oC the cI repressor become inactive and lysogeny is prevented, even if the DNA is incorporated it will pop out of the chromosomal DNA,
The vector has Lac-Z gene with its promoter and operator, so it is regulated with host strain containing lac-Iq, which produces high amount of Lac-I repressor, which again is under the control of lac-z promoter-operator elements.
Recombinant produce clear plaques for the inserts make the lac-Z gene product nonfunctional.
The library can be screened with IgG or DNA probes,
Inserts can be released with E. CoR I enzymes.
Lambda DNA used for cloning and screening for the inserts
Lambda 11 Sfi-Not-1 Vector:
The vector is same as that of lambda gt 11, but it contains Not I and Sfi I restriction sites on either side of the E. CoR I site located with in the carboxyl terminal region of the Lac-Z gene.� It also contains Tm sensitive cI 857 mutant gene and also contain S100 amber mutation.
This can be used for Blue and white colony screening.
One can use IgG or DNA as probes for screening. One can do directional cloning for it has Sfi I and NotI in 5� to 3� direction of the genes.
The cloned genes if expressed one can obtain the protein as fused product.
Y1089 and Y1090 host strains can be used for cDNA library.
����������� ~~I============I<- ---ImcsI-lac-z-I---cI875---S100-----II----I~~
Lambda GEM 2 and GEM 4:
They are derived from gt10,
Multiple cloning sites with promoters have been cloned in the middle of cI gene,
����������������������� ~~~I===========I------I--cI--I----I===========I~~~~
GEM 2:
cI 434= ---Spe I-I P-Sp6�I-Xba I-Sac I-EcoR I IT7- PI Spe I---� �����������
GEM 4:
cI 434= ---Spe I-Amp^+I P-Sp6�I-Xba I-Sac I-EcoR I IT7- PI Spe I---
Difference between GEM and GEM4 is that in GEM 4 it has an Ampicillin gene.
Lambda DR2 expression vector:
This can be used as a shuttle vector and it can be expressed in human cells.
The construction of the vector is performed in the following way.
The central stuffer region of the lambda DNA is replaced with 10kbp long insert containing the following elements.
On either side of the insert are lox-P sites, which can be subjected to recombinase activity by Cre enzyme.
Lambda DNA:
~~~L~I========lox-P=claI====I-insertI===claI==lox-P======I~R~~
�
����� --lox-p-Cla------I-BamH I------I-Xba I------Cla I�lox-P--
--LoxP--ICla-I--EBNA1�EBV-Ori-->RSV-P-BamHI�insert�IXba I-SV40poly(A)->>-
<--poly(A) tK HSV � High�-P-HSV tK-�Amp^+-Ori--Cla I-Lox-P-cla I�
When this construct of lambda-based vector is infected into host strain AM-1 the enzyme Cre recognizes lox-p sites and brings about recombination and produces a circularized plasmid containing all the components present in between the lox sites.
If such a construct is transfected to permissive human cell lines the gene that is cloned can be expressed.
4-C.�� Vectors for Genomic Library:
1. Lambda based vectors:
Lambda Gem 11:
The insert can as long as 20-23kbp long.
In the place of central stuffer region it has cloning sites and promoters in reverse sequence.
����� ����������� ~~~I=====L=======I==C==I=====R====I~~
�I-SfiI-PT7-Sac I-Xho I-BamH I-Avr II-RI-Xba>< IXba I-R I-Avr II-BamH I-Xho I-Sac I P-Sp6-sfiI-
Lambda GEM 12:
In GEM 12, the central stuffer has different RE sites and promoters in opposite orientation.
>sfi-T7-Sac-Not-Bamh-R1-Xho-Xba>� <Xba-Xho-R1-BamH-Not-Sac-SP6-Sfi<
Lambda EMBL 3 and EMBL4:
They are similar to other Lambda used for genomic library, but they differ restriction site found on either side of the central stuffer region.� The insert size can any where between 20 to 23kbp long.
����� ����������������������� ~~~I======L=====I=C=I====R===I~~
GEM 3:
--I Sal I-BamH I-EcoR I>� <EcoRI-BamH I-Sal I---
GEM4:
--R I-BamH I-Sal I> <Sal I-BamH I-R I --
����� The size of eukaryotic genomes range from as small as 1000kbp to 1000, 000kbp long or more.� Cloning of such large size genome in vectors such as pUC plasmids and lambda gt -10, or gt-11 requires screening of millions of recombinant clones to represent at least 90% of the genome.
����� Example:
����� E. coli genome size is 4.7 X 10^6 bp.� If the size of the genome is large it is cut and the clones obtained contain 15000-20000 bp long, one has to obtain at least 230 clones multiplied by 10 times, that is about 2300 colonies or plaques.� This I achievable for the size of the bacterial genome for its size is small.
����� On the contrary if one wants make genomic library of human species, which is 3X10^9bp, if one uses the above sizes of the fragments for genomic library using lambda vectors requires at least screening of 1.5x10^6 multiplied by 10 times.� This is an enormous task.� Hence lambda based and pUC based vectors are not suitable for preparing genomic library of higher organisms.� So people have developed new vectors, which can take 40kbp, 90kbp or 100-300kbp long DNA fragments.� The vectors are so designed they can be manipulated not only in bacteria but also in higher organisms.
����� Host cells used for phage-based library are (in general) same as that of host cells used for lambda-based cDNA library, such as Y1089 and Y1090.
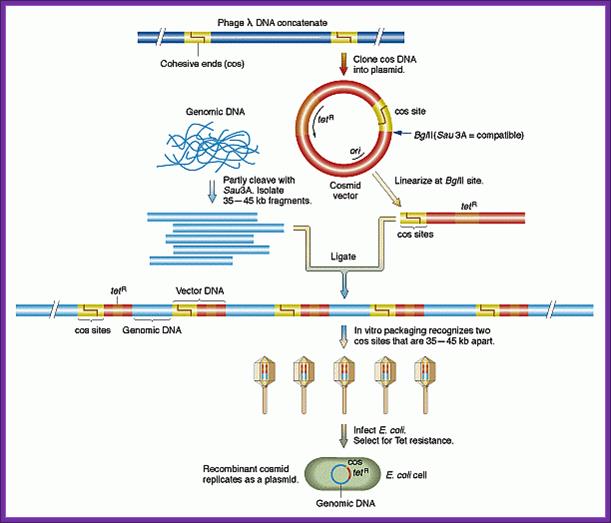
2. Cosmid Vectors:
����� pFLR vector:
����� Collins and Hahn were the first to develop cosmid vectors.� These vectors can carry an insert up to 35 to 45 kbp long.
� The size of the vector pFLR is 5.3 to 6kbp.
� It has two Cos sites, when cut with Terminase produces 12ntds long sticky ends, which are complementary to each other.
� On either side of the Cos site a stretch of 200 bp long sequence called packaging sequence which is required for recognizing the DNA and loading the DNA into lambda head till it recognizes the another such sequence.
� While it is packing concatameric lambda DNA when it contacts the Cos site, it cuts and recognizes the PKG sequence and loads the DNA into another head.
� It has origin site-Coli E1,
� Selection marker gene like Tet^+ or Amp^+ is also present.
� A cloning site BamH I is also present.
�����������������
---L�Cos---------BamH-I-������� -BamH-I�Ori�Tet�Cos---R--
---L�Cos------BamH-I-~~~~~~~~~~~~~-BamH-I�Ori�Tet�Cos---R�
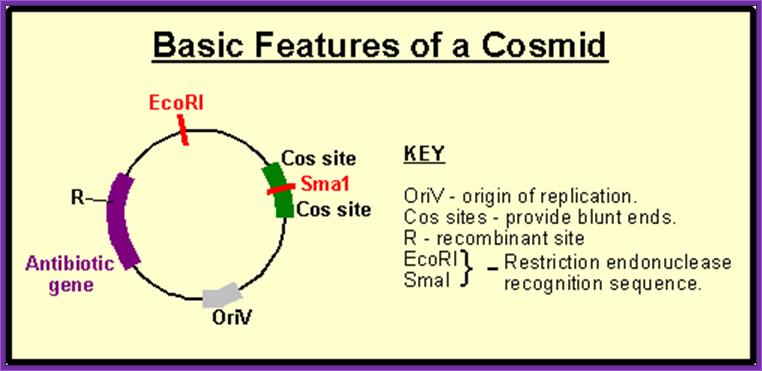
COSMID
� Cut the vector with BamHI and dephosphorylate the ends with Alkaline Phosphatase. Dephosphorylation prevents self ligation.
� Partially digest the genomic DNA with BamHI restriction enzyme and size fractionate.� Select the size of 45kbp long fragments.
� Ligate the fractionated genomic DNA with vector DNA.
� During packaging reaction, the Terminase enzyme present in the extract cut the Cos site and generate Cos ends and they anneal with one another and produce concatameric lambda DNA with inserts.
� These concatameric DNAs are recognized by the enzyme and using packaging sequences at he Cos sites load the DNA into viral prohead.
� While loading, as the enzymes come in contact with another Cos site at the other end of lambda fragment, the Terminase cuts at Cos site and holds the other packaging site and finds another prohead and loads the DNA till it reaches another Cos site.
� Fully packaged viral particles are then used for the infection specific host strains such as ED 8767.
� Then plaques are screened with DNA probe.
� One ug of such DNA yields about 10^9 plaques.
� Note there are few more cosmid vectors; they are Lawrist-4 and pWE15.� The Lawrist-4 vector has multiple cloning sites and has high copy number property.� It also contains Kan ^+ gene.
����������������������������������� Cloning into Lambda expression vector
� The pWe15 vector is a dual Cos vector and it has restriction cassette around the cloning site.� It is very helpful in releasing the insert and one can use this for restriction site mapping and chromosomal walking.
� If the size of the insert is 40 kbp the number of clones required to represent 90% of the total genome of human species is 82,500.� This is multiplied by ten equals to 825000 clones; one has to screen to get most of the genes.
Fosmids; �Fosmids are similar to Cosmids but are based on the bacterial F-plasmid. The cloning vector is limited, as a host (usually E. coli) can only contain one fosmid molecule. Fosmids are 40 kb of random genomic DNA. Fosmid library is prepared from a genome of the target organism and cloned into a fosmid vector. Low copy number offers higher stability than comparable high copy number Cosmids. Fosmid system may be useful for constructing stable libraries from complex genomes. Fosmids have high structural stability and have been found to maintain human DNA effectively even after 100 generations of growth.[2] Fosmid clones were used to help assess the accuracy of the Public Human Genome Sequence.
The fertility plasmid or F-plasmid was discovered by Esther Lederberg and codes for the formation of sex pilus to aid in bacterial conjugation. Conjugation involves using the sex pilus to form a bridge between two bacteria cells and to transfer a copy of the plasmid so that both cells contain identical DNA. It later was discovered that the F factor was the first episome and can exist as an independent plasmid making it a very stable vector for cloning. Conjugation aids in the formation of bacterial clone libraries by ensuring all cells contain the desired fosmid.
Fosmids-WiKipedia:
Fosmids are plasmids that use the F-plasmid origin of replication and partitioning mechanisms to allow cloning of large DNA fragments. A library that provides 20�70-fold redundant coverage of the genome can easily be prepared.
DNA Libraries
The first step in sequencing entire genomes is cloning the genome into manageable units of some 50-200 kilobases in length. It is ideal to use a Fosmids library because of it's stability and limitation of one plasmid per cell. By limiting the number of plasmids in the cells the potential for recombination is decreased, thus preserving the genome insert.
Fosmids contain several functional elements:
� OriT (Origin of Transfer): The sequence which marks the starting point of conjugative transfer.
� OriV (Origin of Replication): The sequence starting with which the plasmid-DNA will be replicated in the recipient cell.
� tra-region (transfer genes): Genes coding the F-Pilus and DNA transfer process.
� IS (Insertion Elements): so-called "selfish genes" (sequence fragments which can integrate copies of themselves at different locations).
An example of a mapped fosmid can be found here http://what-when-how.com/molecular-biology/f-plasmid-molecular-biology/
The methods of cutting and inserting DNA into fosmid vectors have been perfected. There are now many companies that can create a fosmid library from any sample of DNA in a very short period of time at a relatively low cost. This has been vital in allowing researchers to sequence numerous genomes for study. More than 180 organisms� genomes have been fully sequenced since 1995.[
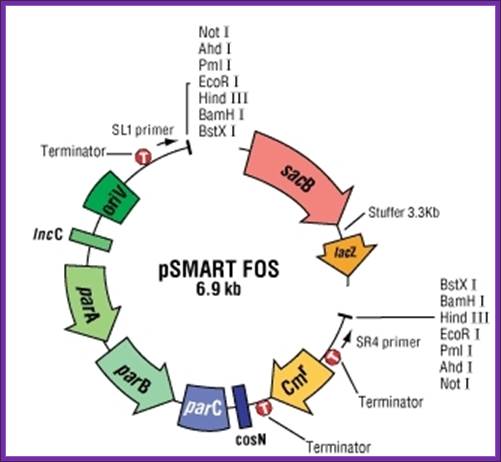
- More recombinants from less DNA. Conventional fosmid cloning systems have lower cloning efficiencies, requiring more target DNA, which can be difficult to obtain. The ultra-efficient CopyRight v2.0 Fosmid Kit gives high recombinant yields even when the amount of DNA is very limited (Figure 1).
- Zero background. A positive selection system.
- Dramatically reduced false positives/negatives. The pSMART FOS vector contains a lacZ/sacB stuffer region that is removed during processing
- No lost clones. FOS vector has the chloramphenicol promoter facing away from the cloning site.
- High DNA yields.� The Copyright v2.0 Fosmid vector and Replicator FOS cells feature inducible amplification of copy number**, increasing yields to as many as 50 copies per cell.
3. Pacmids:
pAD10:
Sac B11:
� Pacmid like pAD10 vectors are derived from p1 phage DNA elements and plasmid elements.
� This vector can take 90 - 110kbp long inserts.
� The size of the vector is about 30.3kbp and circular.
� It has two lox-P recombination sites. �
� They are oriented in opposite direction.
� There is one Pac I site outside lox-P site in counter clock orientation.� This site is required for packaging the DNA into viral head.
� On one side of the lox-P sites is pac I, Sca I, Xba I and a 11kbp segment of Adv 10.
� On the other side of the lox-P the vector contains multiple copies of ColE 1 Ori sites embedded in Amp^+ gene.
� Next it is Kan^+ gene derived from Transposon Tn 903.�
� Next to it is a pI plasmid lytic replicon copy.� Its activity is regulated by Lac operon-promoter.� However it is inactivated in the host that contains lac-IQ repressor.� But, by adding IPTG lytic activity can be activated.
� Between lox P site and PI replicon copy there is another segment, which has many important elements.
� Towards lox P there is Sac�B gene.� When cells are grown in 2% sucrose, the gene product converts sucrose into livens.� When livens accumulate in periplasmic space cells die.� But cells grow well in the absence of sucrose.
� On the other side of the Sac-B an E.coli promoter is located.
� The E.coli promoter is overlapped by PI phage repressor binding site.� This prevents the expression of Sac-B gene
� In between Sac-B and E.coli promoter there is cassette consisting of multiple cloning sites and two promoters.
�
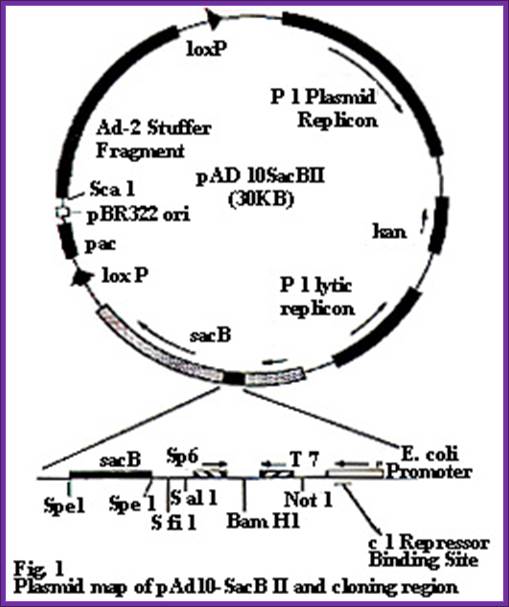
From left to right: -Sac-B----Sfi-I--P-SP6---Sal I�BamH I--- PT7, Not I�E.coli-P
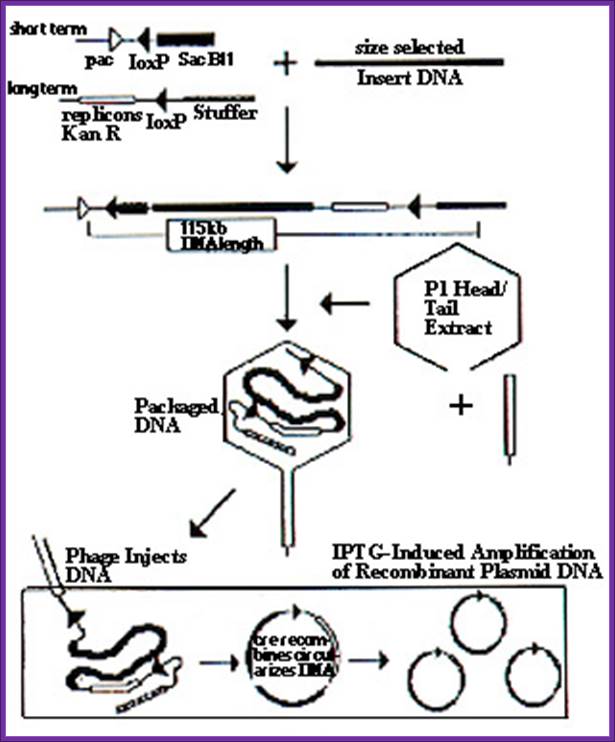
� Cut the vector with Sca I and BamH I: elements are arranged from left to right:
Sca I�pacI-lox-P-sac-B Sfi I Sal I P-SP6-IBamH I ~~ (4.3kbp)
~~~BamH I-P-T7-Not I-Ly-Rep-Kan^+ColE I-lox P-Ad10-Xba I- Sca I (circular)
� Cut the genomic DNA with BamH I and isolate genomic DNA fragments of 90-100kbp sizes.
� Cut the vector with BamH I and dephosphorylated.
� Ligate insert into vector at BamH I site.
� Use P I viral packaging extracts and pack the recombinant DNA into viral heads.
� Using the packaging sites called Pac-I sites (TGATCA* where A should be methylated for Paccase enzyme recognition), the DNA is cut and loaded into the head till the head is full, the rest is cut off.
����� ���� Then the tail components of the virus are added.
� Thus most of the recombinant DNAs are packed into viral structure.
� Specific host cells such as NS 3208 (Rec^-, hsdR^-, mcrA^ -, Pir^-m^-and Cre^+) and NS3210 (rec^-, hsdR^-, hsdm^-, mcrA^-, mcrB^-, Pi r^-m^- and Cre^+) can be used for the infection of viral particles.� The other host cells used for this kind of phages are NS 3529 (Cre^+, RecA ^ -, lacIQ^r, mcrABC^ -, mrr^ -) and NS 3622 which produces repressor that binds to E.coli promoter and inhibit transcription in the presence of sucrose less media.
� On infection host cell Cre recombinase bind to Lox-P sites and brings them together and recombines to generate lox-P containing circular plasmid containing the insert and other components.
� This, though of large size, replicate within the bacterial cells.
� The genomic insert DNAs can be used restriction site mapping and chromosomal walking and identification of genes using expression sequence tags.
� For generating full representative genome, one has to generate at least 33000 X 10, which is equivalent to 330000.
4. Bacmids:
pBeloBac-II:
� Bacmids are plasmids where very long fragments of DNA of 2x10^5 to 3x10^5 bp can be propagated, so they are called Bacterial Artificial Chromosomes and circular.� ^ = ten to the power of (10^6).
� Basically the vector contains F-plasmid Ori-S.
� It contains Rep-E, which controls plasmid replication unit.
� It also contains functional copies of DNA called par-A; par-B and par-C that controls copy numbers of vector DNA.
� A selection marker gene Chloramphenicol acetyl Transferase is present.
� A cloning cassette has been introduced into it.
� The cloning cassette contains Sp6 (P)-Not I lac-Z alpha Not I- and T7 (P).
� The cloning site HinD3 is introduced in the lac-Z alpha region.
� The cloned DNA will be stable and it can be propagated and used.
� Host cells can be Jm109 (STBL).
Clock wise>
~~-<-Cap�<-Ori-S�Rep-E�par-A->par-B�par-C�C.cassette-
Cloning cassette:���� --Sp6�Not I---Lac-Z--------I HinD3 I�Not I�T7�Circular
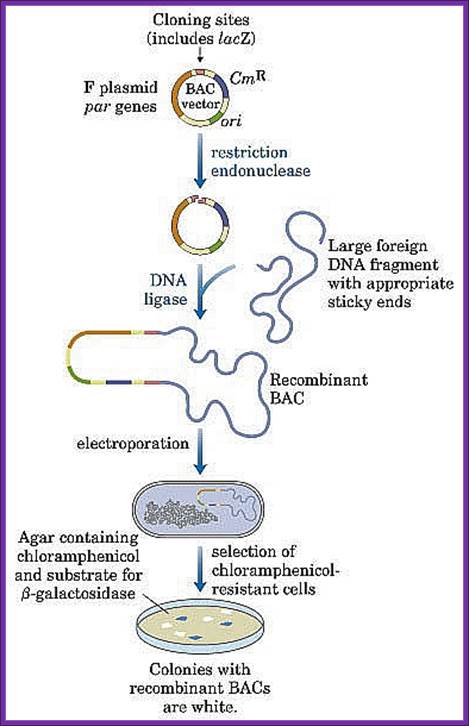
Bacmid expression vectors
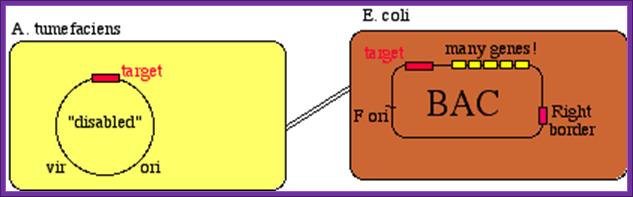
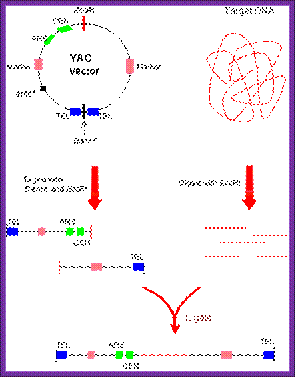
4-D.� Yeast Artificial Chromosome (YAC):
������������� Budding yeast; Budding yeast (Saccharomyces cerevisiae)
����������� Fission Yeast; Fission yeast (Schizosaccharomyces pombe)
� Yeast cells are unicellular, they can divide very fast and they can be grown in large quantities.� Yeast cells produce nutritional mutations and conditional mutants; and the same can be maintained.�� For genetic manipulation yeast system is very amenable.
� Some of the mutants that are extensively used in cloning are mutants such as Leu2, URA3, Trp1, Lys2, His 5 and few others.
� Basically, to create an artificial chromosome one has to have all the features of a chromosome, such as centromere for the movement of chromosomes during mitosis, Telomeres for the stability of chromosomes, and require efficient replication origin sequences.
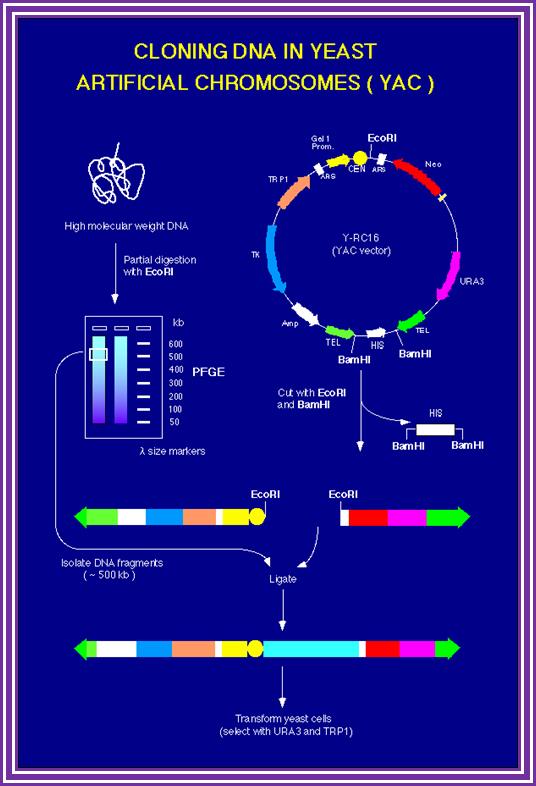
pYAC4 vector:
� The size of this vector is 11.2kbp and circular.
� It has left and right Tel sequences with bordered with Xho I and BamH I sites at their ends; in between His 3 gene is present.
� It has ColE I origin and Ampicillin resistant genes for manipulations in bacteria.
� It has a CEN sequence.
� Next to it is ARS sequence for efficient replication.
� It has selection marker gene URA3.
� The vector also contains a segment containing sup4 in which restriction site E. coli R1 and Sma I are found.� When cloned into this region Ochre suppression becomes non functional in hosts such as Ade-2 ochre cells.� A recombinant produces red colored colonies.
� When the vector is cut with BamH I and dephosphorylate the linear form is represented in the following diagram.
� It opens up as linear plasmid with telomere at their ends.� Then cut with EcoR1 and dephosphorylate for foreign DNA insertion.
YAC
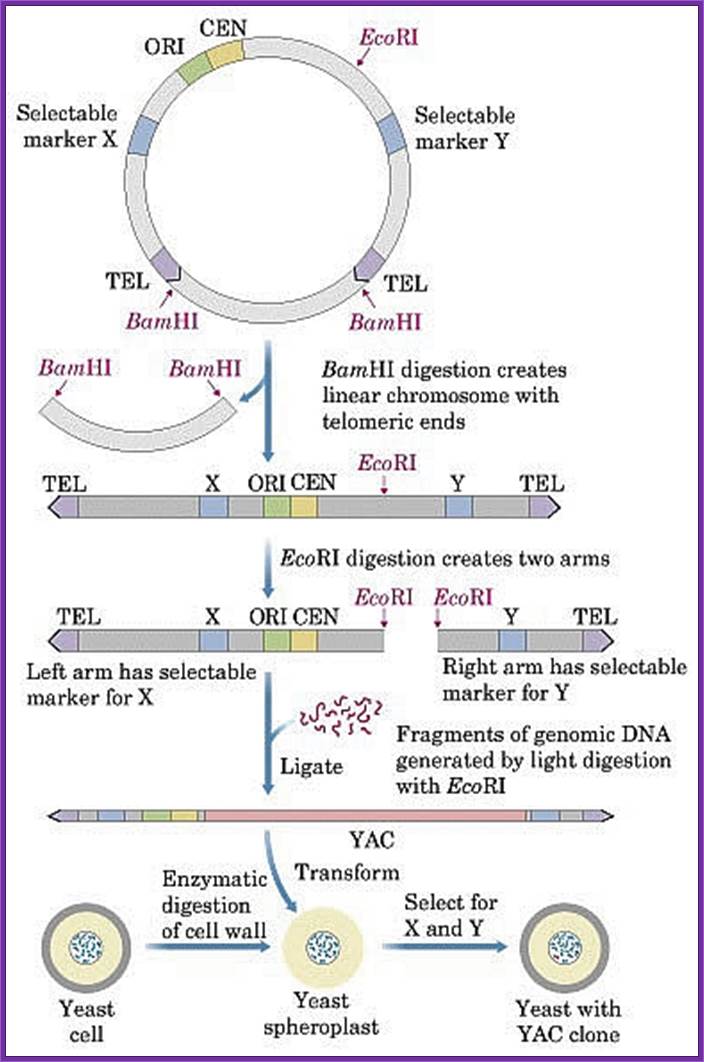
����� L/Tel�Amp�ARS/Trp�CEN----RI-Sma I�URA3�Tel/R
����� L/Tel�Amp�ARS/Trp�CEN---R1~~insert~~~R1-Sma-IURA3�Tel/R �(Circular)
Cut the YAC to linearize it and then cut it with for inserting the DNA and ligate
� Isolate pure nuclei, remove nuclear membrane Nonidet (non ionic) extract the chromosomal DNA.
� Prepare chromosomal DNA by pulse field gel electrophoresis and isolate DNA from the gel.
� Prepare genomic DNA by partial digestion with R1 and size fractionate.� Chose fragments of 10^6 to 10^7bp long and ligate them to R1 ends of the vector.
� The recombinant DNA is linear.
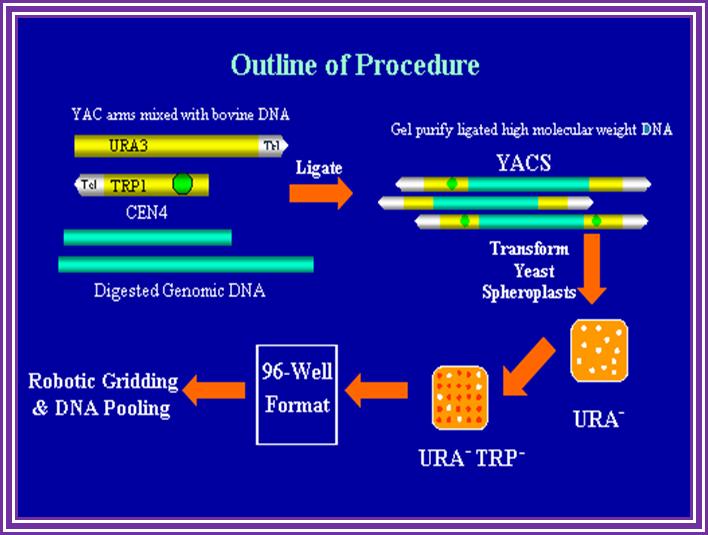
� This DNA is transfected or used for electroporation of yeast mutants URA3 cells.
� Transfected cells are grown overnight in YPD medium for recovery.
� Then the medium is removed and grown in minimal medium containing required antibiotic.
� Yeast cells are used for preparing human genome library and also Arabidopsis genomic library; this is the choicest system for genomic library.
� From such individual chromosomal library one can screen for the required genes.� This is achieved by restriction site mapping and sub cloning and sequencing and order them in overlapping fashion to develop what is called chromosomal walking.
� The same segments can be used for identifying the genes with known sequence tags.
Host yeast cells used are Endo-A and URA3^ - cells.
Many yeast DNAs - based vectors, YEP Yeast episomal vector), YRP (Yeast replicative vector) and YIP (Yeast integrative vector) vectors are also available.
Vectors for yeast
The 2 �m plasmid is a naturally occurring 6 kb episomal plasmid found in yeast.
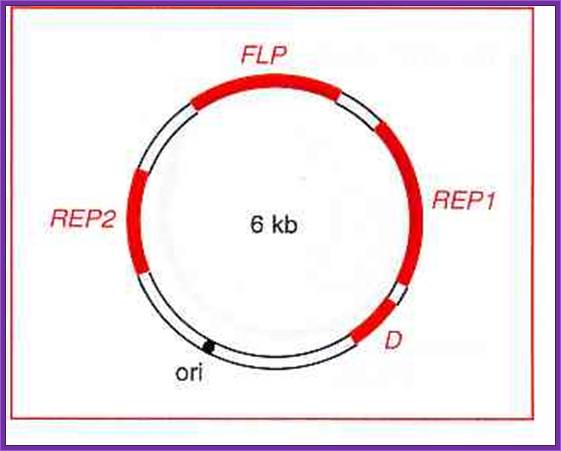
Yeast vectors
Selection is based on nutrition instead of drugs. Yeast cells that are auxotrophic (unable to synthesize an essential component, like an amino acid) can only grow when nutritionally supplemented, or when the missing nutritional function is provided for by genes contained on a plasmid or extra chromosome.
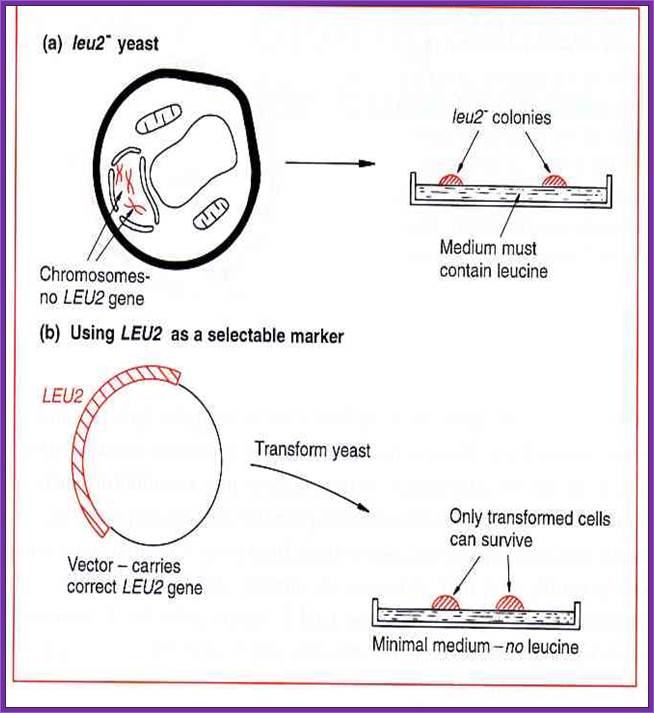
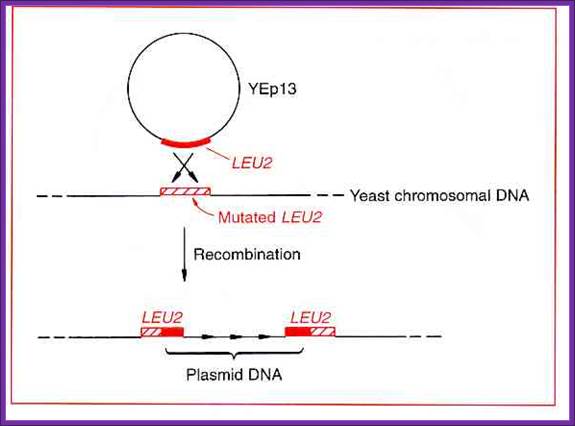
Yep vector- Episomal vector
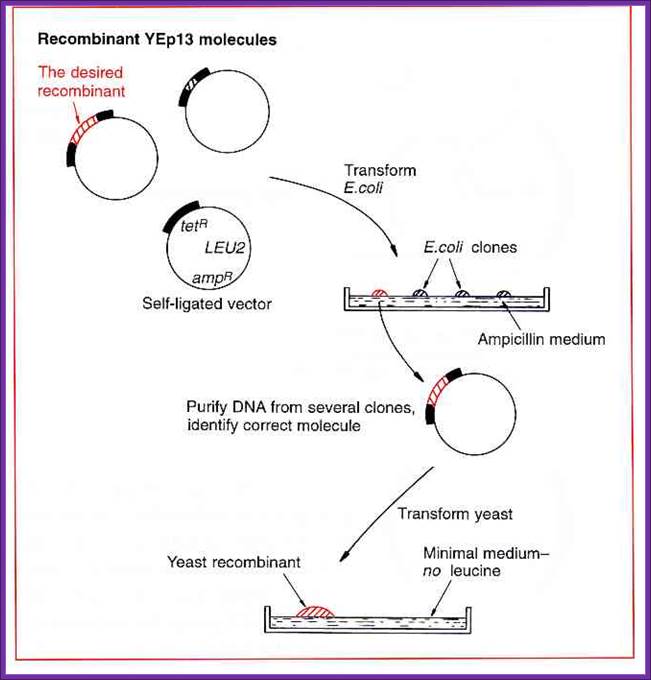
Derivation of Yep vector- Episomal vector

Yeast Integrative Vector (Yips):
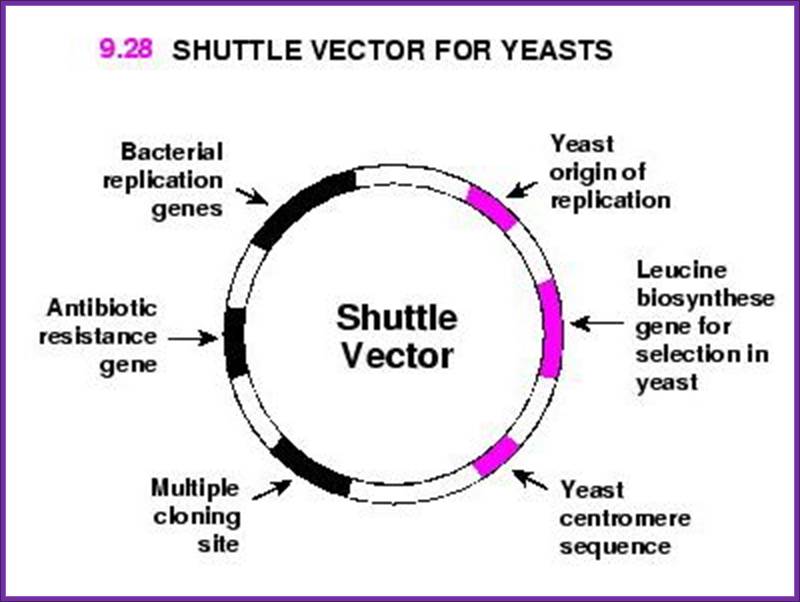
Shuttle Vector:
A shuttle vector is a vector (usually a plasmid) constructed so that it can propagate in two different host species. Therefore, DNA inserted into a shuttle vector can be tested or manipulated in two different cell types. The main advantage of these vectors is they can be manipulated in E. coli then used in a system which is more difficult or slower to use (e.g. yeast, other bacteria). Shuttle vectors include plasmids that can propagate in eukaryotes and prokaryotes (e.g. both Saccharomyces cerevisiae and Escherichia coli) or in different species of bacteria (e.g. both E. coli and Rhodococcus erythropolis). There are also adenovirus shuttle vectors, which can propagate in E. coli and mammals. One of the most common types of shuttle vectors is the yeast shuttle vector. Almost all commonly used S. cerevisiae vectors are shuttle vectors. Yeast shuttle vectors have components that allow for replication and selection in both E. coli cells and yeast cells. The E. coli component of a yeast shuttle vector includes an origin of replication and a selectable marker, e.g. antibiotic resistance, Beta lactamase. The yeast component of a yeast shuttle vector includes an autonomously replicating sequence (ARS), a yeast centromere (CEN), and a yeast selectable marker (e.g. URA3, a gene that encodes an enzyme for uracil synthesis, Lodish et al. 2007).
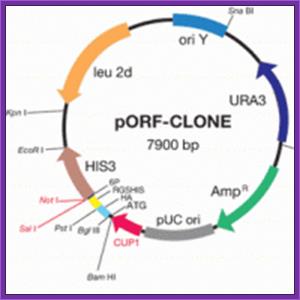
The yeast shuttle/expression vector pORF-CLONE has been specifically developed for an improved and facilitated selection of cloned cDNA inserts containing open reading frames (ORFs) thus allowing an enriched growth of clones expressing authentic polypeptides. This vector system is particularly useful for the development of a a high-throughput technique for the one-step generation of high-quality cDNA libraries in the yeast Saccharomyces cerevisiae and a direct, time-saving screening of random-primed cDNA libraries. In brief, the selection system is based on the HIS3 marker gene fused to the C terminus of the cDNA insert. The cDNAs cloned in-frame result in histidine prototrophic yeast cells growing on minimal medium, whereas clones bearing the vector without insert or out-of-frame inserts should not grow on this medium.
FEATURES:
Ideally suited for the construction of cDNA libraries enriched
for yeast clones possessing authentic ORFs.
Eukaryotic host enables highly efficient expression of soluble
proteins with correct modifications.
Easy for purification and detection of recombinant proteins
one-step screening procedure.
ORFEXPRESS Gateway Shuttle System
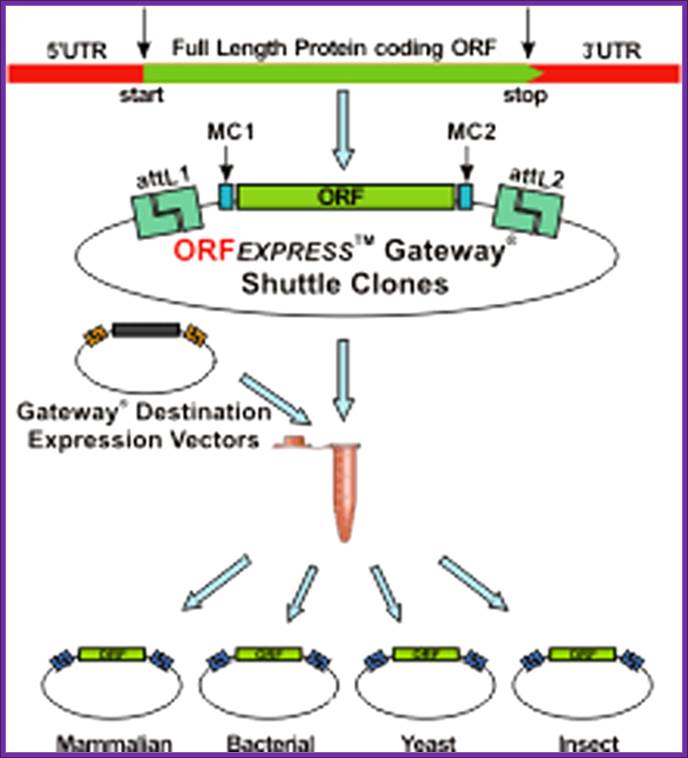
Description
The ORFEXPRESS� Gateway� Shuttle Clone product line currently contains over 20,000 unique clones derived from full length human genes that were chosen through a strict selection process. This process included the extraction, comparison, and validation of gene sequences and annotation information from multiple public and private sources; clustering and reduction of redundant gene sequences; filtering out error sequences or sequencing errors, plus several other manual curation steps. Once this stringent selection process was completed, the entire coding open reading frames (ORFs) of these genes were generated from sequence validated full length cDNA clones or high-quality human tissue cDNA libraries and then inserted into modified Gateway� Entry Vector using Gene-Copeia's proprietary high fidelity cloning technologies. These ORFs were manufactured to be compatible with Invitrogen Corporation's Gateway� Technology, an efficient universal cloning system developed by Invitrogen Corp .