Yeast Cells:
Transformation:
Yeast cells are unicellular eukaryotic systems, can be grown to high density and into large quantity. The duration of the cell cycle is hardly 60-90minutes. They can be mutated and the mutants can be maintained as biochemical and conditional mutants.� Genetic analysis is much simpler than multicellular organisms.� Mutations have yielded 50- 80 mutants involving cell cycle and other events.� Some of the biochemical mutants such as His, Leu are extensively used gene engineering processes. �Yeast is also used in understanding many gene regulations, example Yeast Galactoside metabolizing genes and their expression.
His3: Imidazole glycerophosphate dehydrase (Histidine synthesis).
His 1 and 4 are involved in the biosynthesis of Histidine.
Leu2:B-isopropylmalate dehydrogenase-leucinesynthesis.
Leu 3: is also involved in leucine biosynthesis
Lys2:� alpha amino adipate reductase- lysine biosynthesis.
Trp1: N-5� phosphoribosyl anthranalite isomerase- Tryptophan biosynthesis.
URA3: Orotidine 5�phosphate decarboxylase- involved in the synthesis of Uracil.
These genes can be used as markers.�� These cells can also be selected by using specific antibiotics like such as Kanamycin, Hygromycin, Spectinomycin, and herbicides like phosates.
Yeast Cells; http://www.inspirational-posters.com/
Advantages: Easy to handle, easy to transfect, selection, screening and propagation.� The proteins are secreted if there are any protein coding genes are used, which are super secretors.� Pep-4 mutants lack protease activity so the foreign proteins are more stable.� Protein folding is normal as in other system.
�
Disadvantages: If animal proteins are expressed, their glycosylation will be different.� Not all introns are spliced for they lack all splicing components as one finds in animal systems.
Cell Culture:
They are grown YPD medium overnight.� Wherever required they are to be supplemented with required substances.
YP medium:
10 gm Yeast extract,
20 gm Bacto peptone,
Make it to 1 liter.
YPD:
YP medium plus 0.1 volume of 20% glucose.
Minimal media:
10X YNB:
Yeast Nitrogen base without amino acids.� This should be filter sterilized.� Add 6.7 gm in 100 ml of DD water.� Use 1x of it with dilution with water.
Supplement this with required addition of Uracil, or Tryptophan or whatever required for maintaining mutant cell lines.� All these solutions should be filter sterilized.� They cannot be subjected autoclaving.
Density of culture solution can be measured by taking OD at 600nm.� Five to six OD at 600nm is equivalent to 10^8 cells per ml.
Yeast cell transformation:
There are methods such as-
1) Ca2+/ polyethylene glycol-sphaeroplast method,
2. Lithium chloride method for intact cells,
3) Electroporation method.
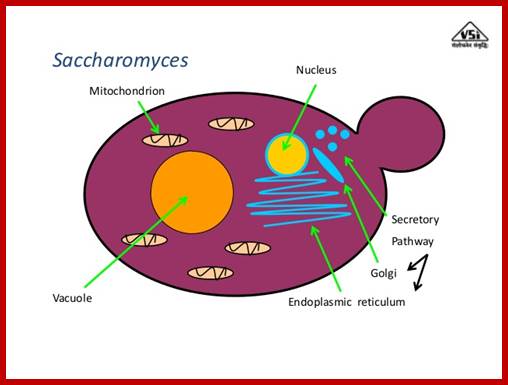
Yeast cell
Lithium chloride method:
~ Grow cells overnight with required medium and dilute it to 0.25 OD and grow further to 1OD.� This takes about 4-5 hrs.
Pellet the cells and suspend in TE buffer, keep the cells on ice.
Spin the cells and suspend in 2.5ml of 0.1M Lithium acetate buffer in TE �and incubate for 1hr at 30^oC.
These numbers of cells can be used for at least six to eight transformation experiments.
Take 300ul of cell suspensions and add 20ul of recombinant DNA i.e. ligated (recombinant) DNA (~ 10ug), and then add .7ml of 50% PEG 4000 (Polyethylene glycol-PEG is dissolved in TE buffer). Mix properly; incubate for an hour at 30�C at stationary condition. Then give heat shock for 5 minutes at 42�C. Pellet the cells and resuspend the same in 0.5ml of YPD. Grow them overnight to allow the expression of Neomycin marker gene. Pellet the cells, resuspend them in TE.
TE:
Tris-Cl 10M 7.4,
1mM EDTA
Then spread them on YPD+G418 agar plate containing 2% glucose and 50-100ug/ml Neomycin.
Only those cells, which have recombinant DNA, grow and other won�t.
Electroporation protocol:
Grow cells overnight on YPD.� YPD provides all the essential nutrients; one need not provide any supplements.
Dilute the culture to 0.25 OD and then grow the same to a density of 1.5OD at 600nm.
Pellet the cells.
Suspend them in 50ml of cold water.
Pellet and suspend in 4ml of ice-cold 1M Sorbitol.
Pellet again and suspend in .2ml of ice-cold 1M Sorbitol.
This can be used 4-5 transformation experiments.
These cells have to be prepared afresh and they cannot be stored at -70^C.
Pipette out 40ul of competent cells into cuvette kept in cold.
Add 100ng of recombinant DNA in a maximum volume of 5ul.� Keep them on ice for 5minutes.
Put the cuvette into electroporator and electroporate by giving a pulse at1.5Kv at 25uF and 200 ohms for 5milliseconds.
Then add 1ml of cold 1M Sorbitol. Mix and pipette out cells gently into 0.5ml of YPD containign1Msorbitol.
Incubate overnight at room temperature to allow the expression of antibiotic resistance gene.
Pellet the cells and suspend in sterile water and spread cells on an agar plate containing YPD+G418+2% glucose.
Transformation by Poly Ethylene Glycol (PEG) Method:
Prepare starter culture.
Use the starter culture and grow in fresh YPD at a concentration of 0.25 OD and grow the cells to a density of 1 OD.
Pellet cells and suspend the same in TE.
Keep them on ice for 5 minutes.
Pellet the cells and suspend in 0.1M lithium acetate in TE buffer.
Then culture the cells at 30^oC for 2hrs or little more.
Use about 300 to 400 ul of cells and add 10ug of recombinant DNA adjust the volume to 20 ul.
Then add 700ul of 50%PEG (from 4xPEG 4000), mix gently.
Incubate at 30^0C for 60 minutes.
Give heat shock to the above mix at 42˚C for 5 minutes.
Pellet cells and remove PEG,
Plate the cells on agar plate containing selection marker antibiotic.
Introduction of foreign DNA into eukaryotic cells is usually called "transfection". Intact �yeast cells treated with alkali cations took up plasmid DNA. Li+, Cs+, Rb+, K+, and Na+ were effective in inducing competence. Conditions for the transformation of Saccharomyces cerevisiae D13-1A with plasmid YRp7 were studied in detail with CsCl. The optimum incubation time was 1 h, and the optimum cell concentration was 5 x 10(7) cells per ml. The optimum concentration of Cs+ was 1.0 M. Transformation efficiency increased with increasing concentrations of plasmid DNA. Polyethylene glycol was absolutely required. Heat pulse and various polyamines or basic proteins stimulated the uptake of plasmid DNA. Besides circular DNA, linear plasmid DNA was also taken up by Cs+-treated yeast cells, although the uptake efficiency was considerably reduced. The transformation efficiency with Cs+ or Li+ was comparable with that of conventional protoplast methods for a plasmid containing ars1, although not for plasmids containing a 2 microns origin replication. Single- stranded vector DNA transforms 2-fold more efficiently than the dsDNA using the spheroplasting technique, whereas single-stranded vectors transform 1000-fold less efficiently with the LiAc/ssDNA/PEG method.
Some yeast cells used as host for transfection or transformation:
For a vector with 2u-ori-STB, the host cells used are Leu2-113, 112Ura3-52, trpi-289 and His3 induced by Galactose.� One can also use strains with PeP mutants, which lack protease gene.
Cells with vectors having GAL1 promoter should be grown in non-repressing carbon source like ethanol, lactate or Rafinose then induce by adding Galactose.
Vectors with ADH promoter; Glucose represses this promoter but it can be induced by ethanol, Glycerol or raffinose. Some of the strains such as Suc2 are secretors.
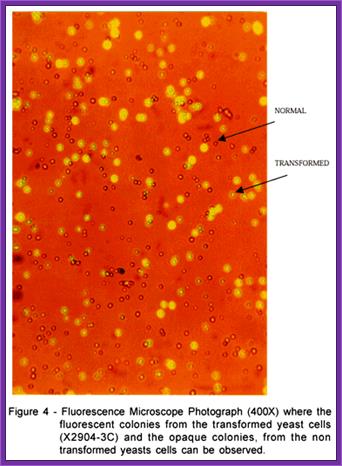
Transformed Yeast cell

Expression of Transformed Yeast cell