DNA Profiling and Genomic DNA Editing;
Genetic polymorphism in human species and other living systems is the hallmark of variations. For example, hyper variable loci show extraordinarily individualistic for a given individual. Hyper variable loci are made up of variable number of identical sequences are arranged in tandem fashion. If such hyper variable (HPV) or variable number of tandem repeat (VNTR) regions are cut at their flanking region and analyzed on a n Agarose or acrylamide gel variation in individual lengths of such polymorphic DNA is visible. Such distinct number of bands differs from one individual to the other with the exception of monozygotic twins. The composition and variability of VNTR loci is an expression high degree of polymorphism.
The first HPV probe used for individual identification consisted of 33bp found in introns of Myoglobin gene. It actually had 10-15 bp long core sequence. When two such clones were used as probes 30-40 distinct bands could be discerned on a southern blot.

Physical finger printing, an age-old method, still being used

Imprinting machine; Retinal scanner

Retinal scanning
Iris scanning
Face imaging
Many of the countries use biometric methods for personal identifications. They can use retinal scanning (popularly done in USA when you enter any of the airports at Immigration desk). Many of the biometric techniques often use facial characteristics.
DNA profiling:
Alec Jeffrey and his colleagues (1985) developed this technique, which has far reaching impact in molecular biology. This technique is often referred to as “Digital Finger Printing” They found a sequence of nucleotides in human hemoglobin DNA was repeated several times. Furthermore such sequences are also found elsewhere in the genome. Such sequence elements, which are tandemly repeated in different loci, are called mini-satellites, which can be isolated by ultra centrifugation techniques. The sequences found were found to be made up of 2, 3, 4, 6, 7, 8 and 9 and ten or more nucleotide sequences. And each of the elements is tandemly repeated and the number of repeats for each species can vary from 10 to 100 or more and the number of repeats per locus varies from 5 to 50 or more. This provides variability and this variability of the said elements is species specific and fixed.
2bp: (GT / AC) n, (AT / TA) n, (CT / GA) n,
3bp: (CTG) n, (CAC) n, (GAA) n, (TCC) n,
4bp: (ATCC) n, (GATA) n, (GACA) n, (GGAT) n,
5bp: (AGAAG) n,
9bp: (GGG CAG GNG) n.
Perfect micro satellites (GT) n:
GT,
GT GT,
GT GT GT,
GT GT GT GT,
GT GT GT GT GT,
GT GT GT GT GT GT,
Interrupted MS:
GT GT GT GA GT GT,
Composite MS:
GT GT GT GT CT CT CT CT,
Cryptic MS:
GA GT GT CTT CTT GTC TG TGTTT G.
Y linked MS:
The denoted 'n' can be any number that can be from one to thousand tandem repeats. A given locus consists of a particular sequence and specific number of repeats, this is a constant feature of the locus and the number of repeats may vary in its homologous locus. Homologous chromosomes may contain many such loci having different characteristic features. Distinct loci with different sequences and with different number of repeats have been identified.
A 33bp probe was used for identification of introns in Myoglobin gene in human species. Using this probe people identified many clones from the human genomic cosmid library. Such sequences were found to be 10-15 bp long. One of the repeats such as CA / GT has been found to have 77 alleles in a population and the number of repeats has been found to range from 14 to 500. Such CA/GT has been found to occur in about 100,000 blocks in human genome. Even islands of GC/CG sequences are also found mostly in the regulator regions. Any such segments can be easily amplified by PCR.
For example, PYNH24 probe, developed from NIH (?), when used on 16 unrelated people detected 19 alleles and no two individuals contained the same pattern
Such repeats originate by unequal crossing over, which can spread this sequence allover, the chromosomal length. Some of the sequences may increase in their number and some sequences may shrink in their numbers. Nearly hundred of distinct min-satellite has been identified and they are distinctly different.
Let us look at the inheritance pattern of heterozygous alleles of these repeats. For example, a single locus contains two alleles, say male has 6bp and 9bp long and female consists of 5bp and 7bp long.
Multilocus fingerprints contain tandem repeat arrays and they are found in various region of the genome. When such Multilocus DNA the result appears as bar codes used in supermarket commodities used for identification of the product and its price.
Cross between two allelic genotypes:
|
Male/female |
6n |
7n |
|
5n |
5n/6n |
5n/7n
|
|
9n |
9n/6n |
9n/7n
|
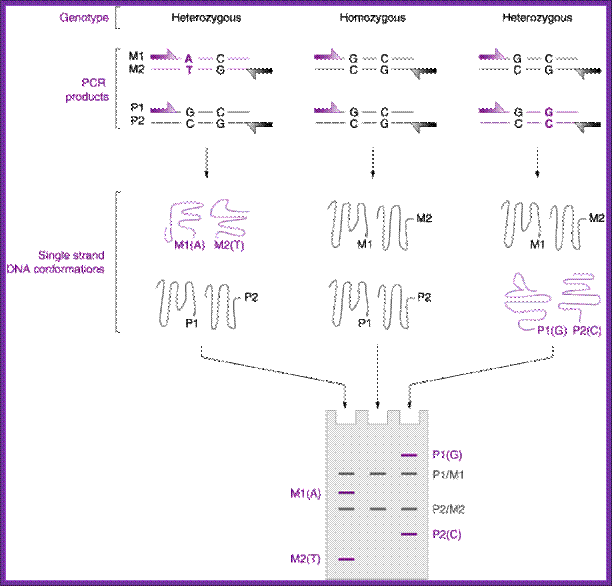
DNA profiling
If the number of alleles is more than two, which is actually the case, the identity of alleles of a single individual, becomes easier. For forensic analysis a large number of markers have been developed. They are short tandem repeats (STRs), which are derived from micro satellite DNA. They are made up of two to nine base pair long sequences and found in abundance in human genome, which is also true for other species. Using less than 1ng of DNA one can amplify it by PCR overnight.
Probes for Micro satellite and Minisatellite for single or multilocus have been developed.
The STR alleles consist of 100 to 350 ntds. Using multiplex PCR one can amplify many STR loci simultaneously. In USA 13 STR loci are employed for typing all felonies using National Data Bank called CODIS.
The 13 loci probes are- CSF1PO, D3S1358, D5S818, D7S820, FGA, THO1, TPOX, VWA, D8S1179, D135317, D16S539, D18551, D21S11. Perkin Elmer Company has developed three multiflour multiplex STR kits and Promega people have developed 8 loci STR kits. Each kit provides fluorescent dye-labels, a multiplex primer sets and reagents for PCR amplification.
Amplified DNA can be analyzed on PGA (poly acrylamide gel) or capillary electrophoresis; the later is good, faster and automated. Capillary tubes with the gel inside have high surface area to volume ratio, thus heat is dissipated. The bands are separated at high field strength and the results are computer analyzed and the data is stored. The capillary gel consists of Dimethyl polyacrylamide (POP4), in 100mM TAPs, 8M urea and 5% pyrolidine and chromogenic dyes such as 5-FAM-blue, JOE-green, ROX- red and NEO-Yellow. The enzyme used is Ampli-Taq-Gold.
Highest individualism has been obtained by using (CAC) 5 and its complement (GTC) 5.
Some of the sequences effectively used are-
(CA) n,
(CT) n,
(GAA) n,
(TCC) n,
(GGAT) n,
(GATA) n,
(GACA) n,
Different Individuals:
(CAC) 5
|
- |
|
|
|
|
= = |
- - |
= |
|
|
- = |
= - |
|
|
|
= - - |
- = = |
|
|
|
- - -
|
= = = |
|
|
|
= =
|
- -
|
|
|
|
= - = |
- = - = |
|
|
|
= - |
_ = |
|
|
Note- that none of the individuals in a group have copies of the same sequence pattern; the pattern distinctly varies from one individual to the other.
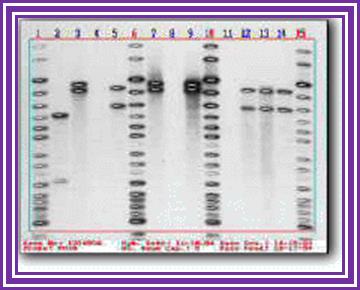
Twins -monozygotic twins have absolute similarities in terms of bands.
|
= |
= |
|
- - |
- -
|
|
= = =
|
= = =
|
|
= = |
= = |
|
- - - |
- - - |
|
= - = |
= - = |
|
= = = |
= = = |
|
= |
= |
|
= |
= |
|
= - |
= - |
Brother and sister:
|
= |
- |
|
= = = |
= = = |
|
= = - |
= - - |
|
= - |
- = |
|
= |
= |
In this case there are many common bands for they are inherited from their parents.
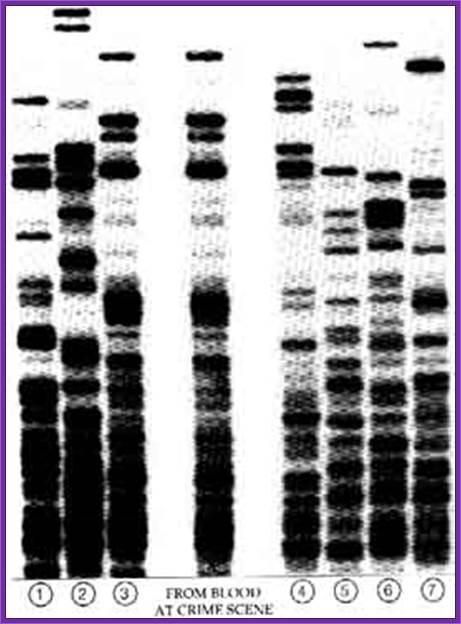
Analysis of DNA prints
Genetic finger printing-Gel analysis
Genotyping
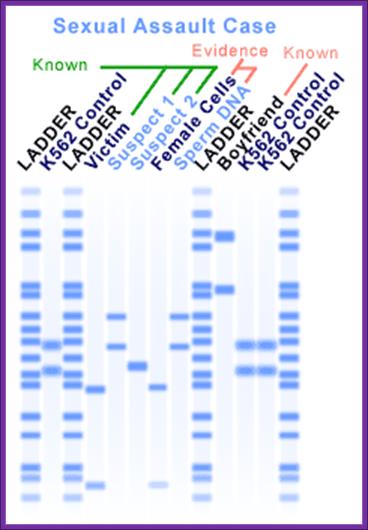
Sexually assaulted cases
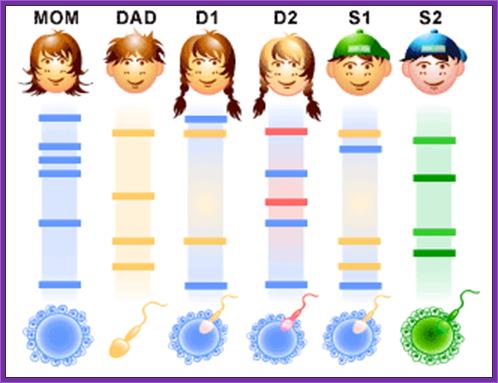
Family DNA finger printing
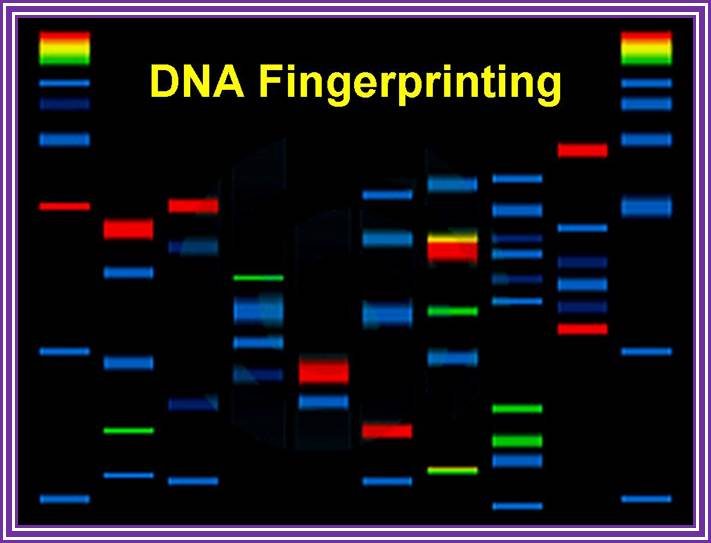
DNA finger prints
Polymorphic DNA refers to chromosomal regions that vary widely from individual to individual. By examining several of these regions within the genomic DNA obtained from an individual, one may determine a "DNA Fingerprint" for that individual. DNA polymorphisms are now widely used for determining paternity/maternity, kinship, identification of human remains, and the genetic basis of various diseases. The most far-reaching application, however, has been in the field of criminal forensics. DNA from crime victims and offenders can be matched to crime scenes, often affecting the outcome of criminal and civil trials.
Microsatellites, also known as Simple Sequence Repeats (SSRs) or short tandem repeats (STRs), are repeating sequences of 2-6 base pairs of DNA.
Microsatellites are typically co-dominant. They are used as molecular markers in genetics, for kinship, population and other studies. They can also be used to study gene duplication or deletion. Microsatellites are also known to be causative agents in human disease, especially neurodegenerative disorders and cancer.
For example, the following sample report from this commercial DNA paternity testing laboratory Universal Genetics signifies how relatedness between parents and child is identified on those special markers:
|
DNA Marker |
Mother |
Child |
Alleged father |
|
D21S11 |
28, 30 |
28, 31 |
29, 31 |
|
D7S820 |
9, 10 |
10, 11 |
11, 12 |
|
TH01 |
14, 15 |
14, 16 |
15, 16 |
|
D13S317 |
7, 8 |
7, 9 |
8, 9 |
|
D19S433 |
14, 16.2 |
14, 15 |
15, 17 |
The partial results indicate that the child and the alleged father’s DNA match among these five markers. The complete test results show this correlation on 16 markers between the child and the tested man to draw a conclusion of whether or not the man is the biological father.
Genome, epigenome and RNA sequences of monozygotic twins discordant for multiple sclerosis: Sergio Branzino et.al.
Monozygotic (MZ) or “identical” twins have been widely studied to dissect the relative contributions of genetics and environment in human diseases. In multiple sclerosis (MS), an autoimmune demyelinating disease and common cause of neurodegeneration and disability in young adults, disease discordance in MZ twins has been interpreted to indicate environmental importance in its pathogenesis1–8. However, genetic and epigenetic differences between MZ twins have been described, challenging the accepted experimental paradigm in disambiguating effects of nature and nurture. Here, we report the genome sequences of one MS-discordant MZ twin pair and messenger RNA (mRNA) transcriptome and epigenome sequences of CD4+ lymphocytes from three MS-discordant, MZ twin pairs. No reproducible differences were detected between co-twins among ~3.6 million single nucleotide polymorphisms (SNPs) or ~0.2 million insertion-deletion polymorphisms (indels). Nor were any reproducible differences observed between siblings of the three twin pairs in HLA haplotypes, confirmed MS-susceptibility SNPs, copy number variations, mRNA and genomic SNP and indel genotypes, or expression of ~19,000 genes in CD4+ T cells.
Figure.: Comparisons of methylation of genomic CpG sites in CD4+ lymphocytes and breast and lung tissue samples.
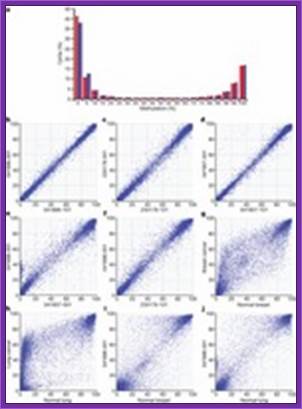
a, Frequency distribution of CpG site methylation in 041896-001 (blue) and -101 (red) using ELAND-extended. b–j, pairwise comparisons of CpG site methylation using ELAND-extended in CD4+ lymphocytes from monozygotic twin siblings 041896-001 and -101
Only two to 176 differences in methylation of ~2 million CpG dinucleotides were detected between siblings of the three twin pairs, in contrast to ~800 methylation differences between T cells of unrelated individuals and several thousand differences between tissues or normal and cancerous tissues. In the first systematic effort to estimate sequence variation among MZ co-twins, we did not find evidence for genetic, epigenetic or transcriptome differences that explained disease discordance. These are the first female, twin and autoimmune disease individual genome sequences reported.
DNA methylation profiles in monozygotic and dizygotic twins: Zachary A Kaminsky et.al,
Twin studies have provided the basis for genetic and epidemiological studies in human complex traits1,2. As epigenetic factors can contribute to phenotypic outcomes, we conducted a DNA methylation analysis in white blood cells (WBC), buccal epithelial cells and gut biopsies of 114 monozygotic (MZ) twins as well as WBC and buccal epithelial cells of 80 dizygotic (DZ) twins using 12K CpG island microarrays3,4. Here we provide the first annotation of epigenetic metastability of B6,000 unique genomic regions in MZ twins. An intraclass correlation (ICC)-based comparison of matched MZ and DZ twins showed significantly higher epigenetic difference in buccal cells of DZ co-twins (P ¼ 1.2 _ 10_294). Although such higher epigenetic discordance in DZ twins can result from DNA sequence differences, our in silico SNP analyses and animal studies favor the hypothesis that it is due to epigenomic differences in the zygotes, suggesting that molecular mechanisms of heritability may not be limited to DNA sequence differences. (Buccal cavity differences are local but not herited).
DNA Finger Prints: Technique of finger printing was perfected by British Genetist, Dr. Alec Jeffreys in 1984.
Fig. Identical Twins: DNA finger printing of identical and monozygotic twins did not show any variations in their DNA sequences.
Genomic/ DNA Finger Printing
DNA fingerprinting is a method used to identify an individual from a sample of DNA by looking at unique patterns in their DNA. DNA finger printing and Genomic finger printing is same. In humans 10 ^14 cells have the DNA, with some difference between Females and Males.
On average, about 99.9 per cent of the DNA between two humans is the same. But some amount varies between females and males.
However, minisatellites of humans show variations. DNA fingerprinting is a technique that simultaneously detects lots of minisatellites in the genome to produce a pattern unique to an individual. This is a DNA fingerprint. The first step of DNA fingerprinting was to extract DNA from a sample of human material, usually blood and cut with molecular scissors. These are electrophoresed, to assort pieces for comparison- All are labelled.
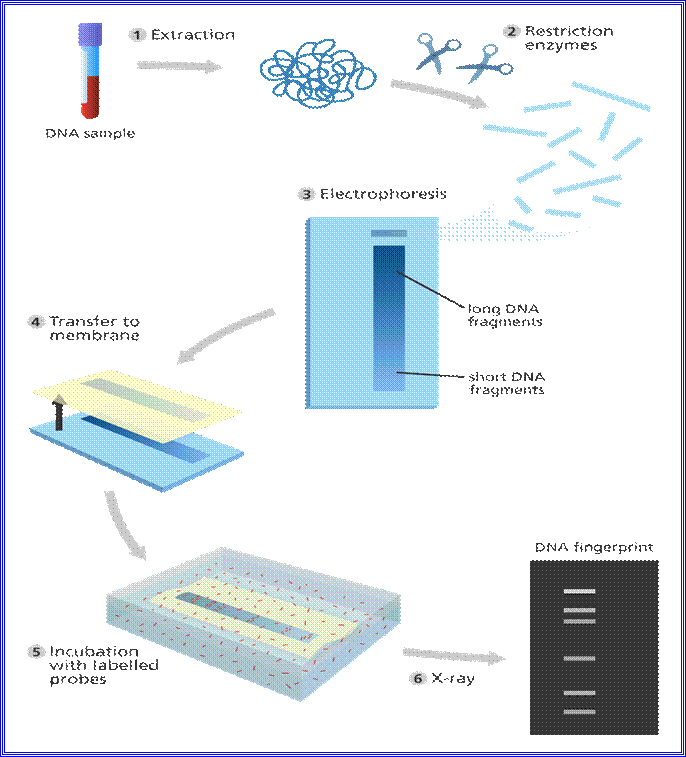
· Illustration showing the steps in DNA fingerprint
Microsatellites or minisatellites are used for DNA fingerprinting.
Microsatellites, or short tandem repeats (STRs), are the shorter relatives of minisatellites usually two to five base pairs long. Like minisatellites they are repeated many times throughout the human genome, for example ‘TATATATATATA’.Presently PCR is used and electrophoresed.
· 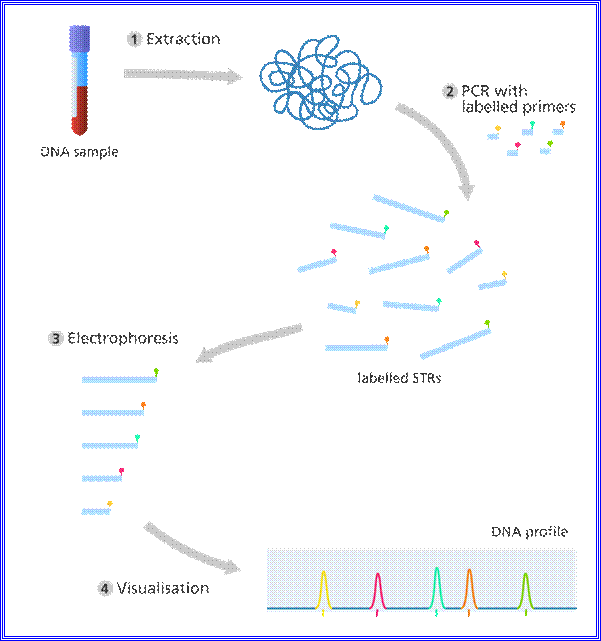
Illustration
showing the steps in DNA profiling.
Image credit: Genome Research Limited
For accuracy more STR ARE USED. Only one person in every 10 million (10,000,000,000,000) will have a particular STR profile. With the world human population estimated at only 7,100 million (7,100,000,000), it is therefore extremely unlikely you will share the same profile as someone else, unless you are an identical twin.
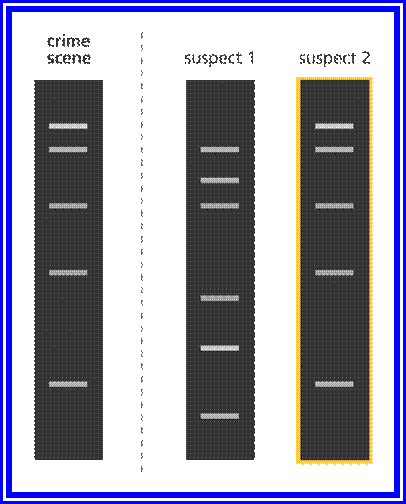
Illustration
showing a comparison of a DNA fingerprint from a crime scene and DNA
fingerprints from two suspects. The DNA fingerprint from suspect 2 matches that
taken from the crime scene.
Image credit: Genome Research Limited
The police may use this DNA evidence to support other evidence to help prosecute someone for a crime. Complete DNA profiles give very reliable matches and may provide strong evidence that a suspect is guilty or innocent of a crime.
DNA profiling can be used to help confirm whether two people are related to one another and is commonly used to provide evidence that someone is, or is not, the biological parent of a child. Illustration comparing the DNA profiles
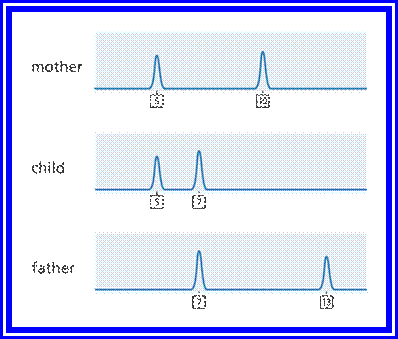
Image credit: Genome Research Limited
Illustration
comparing the DNA profiles of two
parents and their child. You can see which STRs in the child have been
inherited from which parent.
Is it ethical to have a national DNA database?
The National DNA Database has proved to be a valuable tool in the fight against crime. However, many people are concerned about how it has evolved from a database containing genetic information on convicted criminals to one that has information from a much wider group of people.

Electrophoresis is used for analyzing DNA samples
Genomic DNA Editing
In classical genetics, the gene-modifying activities were carried out selecting genetic sites related to the breeder’s goal. Subsequently, scientists used radiation and chemical mutagens to increase the probability of genetic mutations in experimental organisms. Although these methods were useful, they were time-consuming and expensive. Contrary to this, reverse genetics goes in the opposite direction of the so-called forward genetic screens of classical genetics. Reverse genetics is a method in molecular genetics that is used to help understanding the function of a gene by analyzing the phenotypic effects of specific engineered gene sequences. Robb et al. [68] defined and compared the three terms: “genome engineering”, “genome editing”, and “gene editing”. Genome engineering is the field in which the sequence of genomic DNA is designed and modified. Genome editing and gene editing are techniques for genome engineering that incorporate site-specific modifications into genomic DNA using DNA repair mechanisms. Gene editing differs from genome editing by dealing with only one gene.
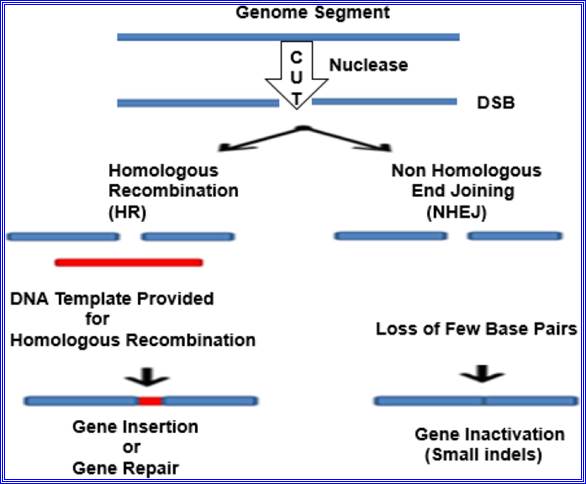
Genome editing.
Genome editing outcomes: Genome editing nucleases induce double-strand breaks (DSBs). The breaks are repaired through two ways: by non-homologous end joining (NHEJ) in the absence of a donor template or via homologous recombination (HR) in the presence of a donor template. The NHEJ creates few base insertions or deletion, resulting in an indel, or in frameshift that causes gene disruption. In the HR pathway, a donor DNA (a plasmid or single-stranded oligonucleotide) can be integrated to the target site to modify the gene, introducing the nucleotides and leading to insertion of cDNA or frameshifts induction.
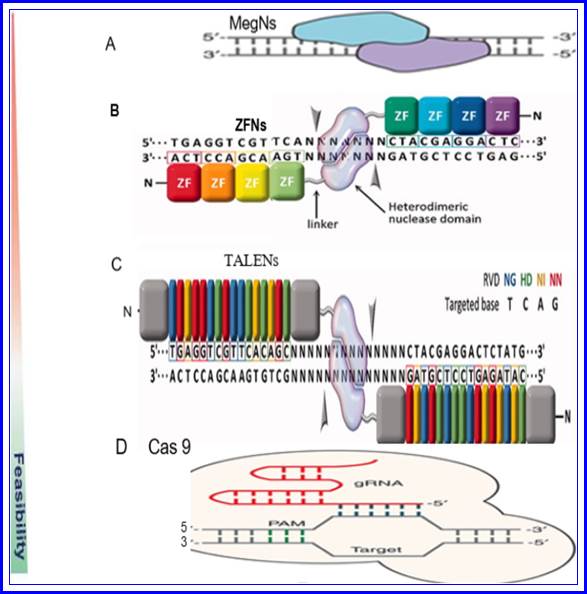
Schematic diagram of the four endonucleases used in gene editing technologies. a Meganuclease (MegN) that generally cleaves its DNA substrate as a homodimer. b Zinc finger nuclease (ZFN) recognizes its target sites which is composed of two zinc finger monomers that flank a short spacer sequence recognized by the FokI cleavage domain. c Transcription activator-like effector nuclease (TALEN) consists of two monomers; TALEN recognizes target sites which flank a fok1 nuclease domain to cut the DNA. d CRISPR/Cas9 system is made of a Cas9 protein with two nuclease domains: human umbilical vein endothelium cells (HuvC) split nuclease and the HNH, an endonuclease domain named for the characteristic histidine and asparagine residue, as well as a single guide RNA (sgRNA). Adapted from; Gaj et al..
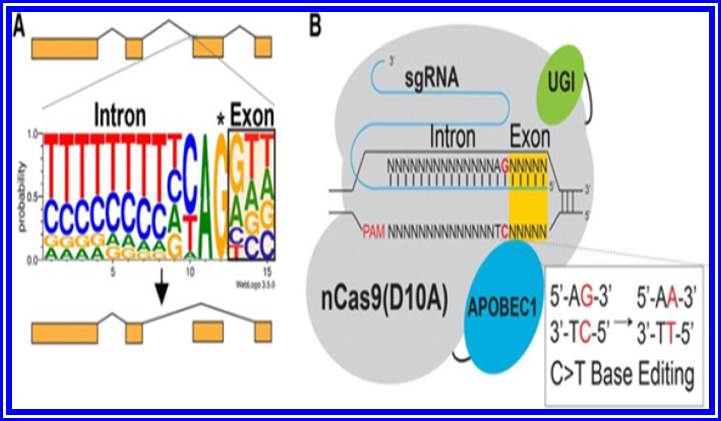
Base Editing
Essential steps in CRISPR-SKIP targeting approach: a. Nearly every intron ends with a guanosine (asterisked G). It is hypothesized that mutations that disrupt this highly conserved G within the splice acceptor of any given exon in genomic DNA would lead to exon skipping by preventing incorporation of the exon into mature transcripts base. B. In the presence of an appropriate PAM sequence, this G can be effectively mutated by converting the complementary cytidine to thymidine using CRISPR-Cas9 C>T single-base editors.
Editing the genome for a
specific purpose is not possible at this point of time.

