Tobacco Mosaic Virus:
It is the first virus discovered to contain RNA as the genetic material. Dimitry Losifovich Ivanovsky (1892) a Russian scientist (Microbiologist), was the first to discover the virus as the causative agent in tobacco plants. Wendell Stanley (1948) was the first to crystallize the viruses, which showed infectivity properties. The crystallographer Rosalind Franklin worked for Stanley for about a month at Berkeley, and later designed and built a model of TMV for the 1958 World's Fair at Brussels. In 1958, she speculated that the virus was hollow, not solid, and hypothesized that the RNA of TMV is single-stranded Viruses can exist as living entities in living cells and also exist as crystal like particles outside living cells. It means they are in threshold of living and non-living objects.

Aaron Klug was born in 1926 in Zlvas, Lithuania, but he spent his early days in Durban, South Africa, where his family moved when he was two years old. While attending Durban High School he developed an interest in science, especially after reading Microbe Hunters, a popular book written by Paul de Kruif ;Nobel laureate; http://www.mediatheque.lindau-nobel.org
In 1971, David Baltimore of reverse transcriptase and future Nobel Prize fame published a review in which he described the “Expression of Animal Virus Genomes” as it was then understood.
Functionally grouping RNA viruses together.
He proposed 6 classes of viral expression. These were:
· Class I: all dsDNA viruses
· Class II: ssDNA viruses
· Class III: dsRNA viruses
· Class IV: ssRNA viruses whose mRNA sequences are subsets of the virion RNA sequence (ie: ss(+)RNA viruses)
· Class V: ssRNA viruses with genomes which are complementary in sequence to the mRNA sequences (ie: ss(-)RNA viruses)
· Class VI: ssRNA viruses which have a DNA intermediate in their lifecycle
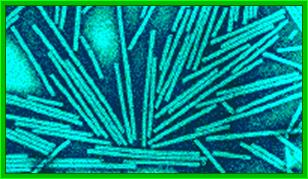
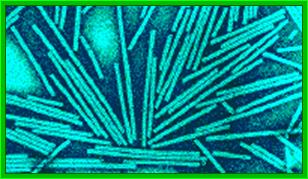
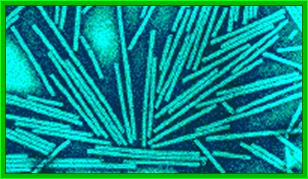
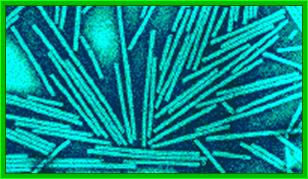
Crystal of TMV; http://rybicki.wordpress.com/

Diseased tobacco leaves; https://www.forestryimages.org

Diseased tomato leaves curling and wilting; http://www.shouragroup.com/images/galle http://www.helpfulgardener.com/
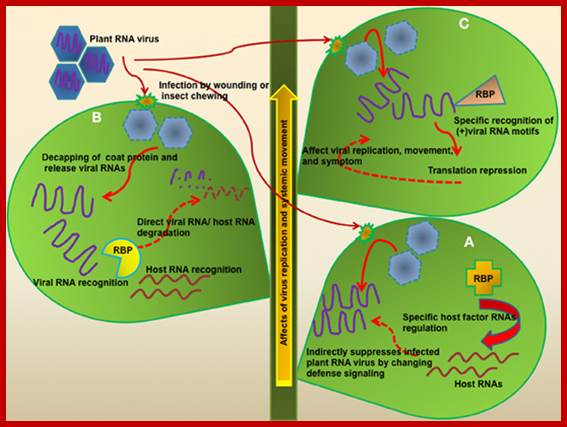
Plant RNA binding proteins for control of RNA virus infection; Sung Un Huh etal http://journal.frontiersin.org/
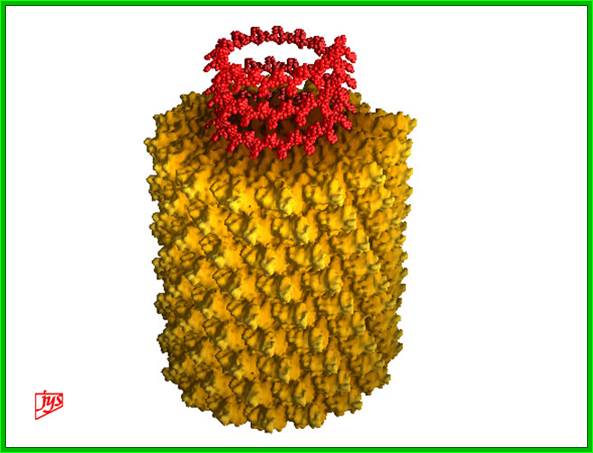
www.virology.wisc.edu
TMV rod shaped structure; http://www.virology.wisc.edu/

The figure shows one end view of the rod but helical structure of TMV; https://oak.ppws.vt.edu
Morphology:
The virus appears like a hallow cylinder of 300nm long and 18 nm thick. Mol.wt is 40 million KD mass. It is built up of 2130 similar subunits strung around axial but helically oriented RNA in circular fashion.
Each of the subunits is 158 amino acids long and 17.5 KD mass and consists of four large a helices, one small helix and 6 b sheets organized into a flat disc with N- and C- terminals found at the periphery. Nearly 16 and half subunits form one circular disc or organize into one turn, with 40 A ^o hallow and 140 A^o thick wall. On the inner surface of the disc RNA is bound to each subunit in each disc. The genomic RNA using its GAA sequences bind to Arginine residues found at 90 and 92 nd positions of the subunit protein. In all 129 discs are laid one above the other and they are aligned conformationally like a spiral string of subunits, where 6395 ntds long RNA is bound to each subunit from inside via 3 ntds (GAA). Each of the subunits is cross-linked to one another by salt bridges. At one end of the hollow rod is 3’ end of the RNA and at the other end of the rod the 5’ end of the RNA is found.
· Length of the rod is ~300nm,
· Width of the rod is 18nm,
· Thickness of the wall is 14 nm,
· Hollow of the cylinder is 4 nm,
· Capsids are organized into two layered cylindrical discs,
· Each disc is made up of sixteen and half subunits,
· Each subunit is made up of 158 aa, each subunit 17.5 kDa.
· Helix pitch is 23 A^o rise,
· Each subunit protein consists of 4 large and one small a helices and 6 b sheets, with N – and C- terminals towards periphery,
· The sequence GAA of the RNA binds to Arginine residues found at 90 and 92-nd position of the subunit.
· Total number of subunits found in each TMV rod like particle is 2130, bound from inside to 6395 nucleotide long RNA, with one 5’ end and the other 3’end.
· Uncoating of the protein subunits is from 3’end of the TMV RNA towards 5’end.
· But under alkaline condition the stripping of proteins is from 5’ end of RNA towards 3’ end.
· The rod is stable at low pH (5.2), and discs grow well at low pH. At higher pH the discs are unstable and fall apart.
· The genomic RNA is 6395 ntds long and it has positive sense, which means it can be translated as mRNA.
· Its 5’end has Methyl-GpppC and 3’ end has CpCpCpA-OH, this can be charged with Histidine in the presence of histidine tRNA synthetase.
· The nucleoprotein helix of TMV is right-handed and the RNA from 5’ to 3’ direction.
· The apex of this helix is composed of sloping subunits with a morphology of conves end of the viral particle and the RNA runs from the bottom to the top of the helix in 5’ to 3’ direction of the RNA in anti-clock direction.
· Under In vitro condition the assembly of the particles into a viral structure takes about 6 hrs, but under in vivo conditions it hardly takes 10 minutes.
· TMV can be crystallized.

The above picture shows the helical organization of capsomeres around the positive sense genomic RNA from inside; http://www.turbosquid.com/
Assembly:
Assembly in the cytosol takes place in sequence; first two layered discs forms. At cellular pH 7, nucleation of discs starts on an 18 base pair long helix organized at 1000 ntds from 3’ end of the RNA. The secondary structure, with base sequence of 5’ AGA AGA AGU UGU UGA UGA 3’ of 18 ntds long contains G at every three bases, surprisingly nowhere C ntds are found in that region. The RNA with the said secondary structure first binds to one of the discs from inside, then the second disc is laid from the top and the RNA is drawn into it for binding, thus the discs are laid one above the other till the 5’ end of the RNA is completely pulled upwards into the rod. The 3’ end of the RNA is still

http://www.wired.com/
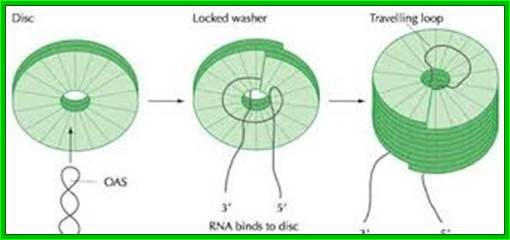
The diagram depicts how the capsomeres assemble into discs and how the discs get associated with the looped but coiled RNA; http://webappsr2.swau.edu/
hanging loose. During rod building process, the GAA sequence of the RNA binds to each of the discs to Arginine at 90 and 92nd position. Once the 5’ end assembly is completed, assembly of discs at lower end starts growing towards 3’ end of the RNA disc by disc. This type of assembly is highly favored at low pH, at which the rod like structure can be organized even without the presence of RNA. These discs as they organize or laid one above the other at the upper end (towards 5’ end) or one below the other at the lower end (towards the 3’ end), they conformationally change and slide to form one continuous strand in helical orientation.
Replication:
· The genomic TMV RNA is a plus RNA (coding) and 6395 ntds long consists of cap at 5’ end with 67 nucleotide leader sequence with AU rich sequences without any secondary structure, hence it is called ‘W’ sequence. The 5’End cap and structured leader sequence is a distinguishing feature of eukaryotic mRNAs, and TMV RNA has a tRNA like secondary structure at the 3’ end which accepts histidine in the presence of synthase. TMV RNA can be used as plant expression vector. Nobuhiko Takamatsu et al, https://www.ncbi.nlm.nih.gov.
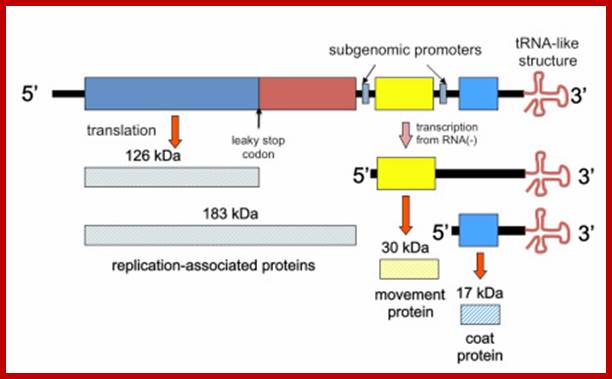
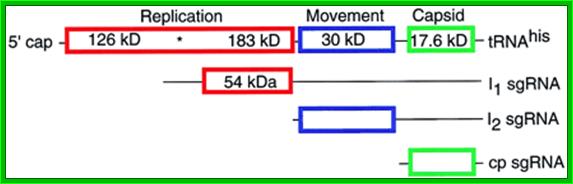
TMV genome, 6395 ntds with Transcription and translation modes; It has three major ORFs. They are found to be a unique hairpin loop-encoding sequence region for assembly initiation – nicely rounding out pioneering work by PJ Butler and colleagues and Genevieve Lebeurier and others – both published in January 1977, https://rybicki.wordpress.com; http://what-when-how.com/molecular-biology; https://rybicki.wordpress.com/

http://www.intechopen.com/
· The genomic RNA (30S) has positive sense; it is encoded with information to produce three polypeptides, one 126kDa and second 183kDa (183kDa), the third with 29.978/ 17.60 kDa proteins.
· The replicase protein is 126kDa, it produces negative strand or 183kDa produces positive strand on the negative strand.
· The first one, which is not fully characterized, is RNA dependent RNA polymerase, acts on positive strand. The second, a 29kd product acts as a transporter protein, which facilitates the movement of viral particles across the cells through plasmodesmota. The third 17.6 KD is the capsid protein, which is located at the 3’ end of the genome.
· Entry of the virus is through wounds, and once in the cell, its protein coat gets decoated.
Figure. Current Molecular Biological Conception of TMV, as Represented by a Schematic Diagram of its Genome Organization. https://hubpages.com/education
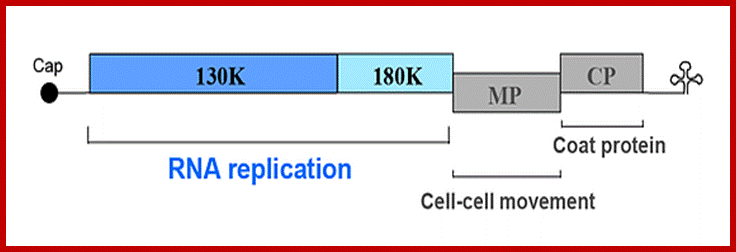
ToMV codes for two replication proteins, 130kDa and 180kDa movement proteins and coat proteins; 180kDa protein is produced through read though of 130kDa involved in RNA capping and RNA synthesis. http://www.komodak.com/

The genome is 6395 ntds) 6400ntds long, with 2130 protein molecules; around the RNA and 18nm diameter Genome with protein coding regions; http://users.aber.ac.uk/
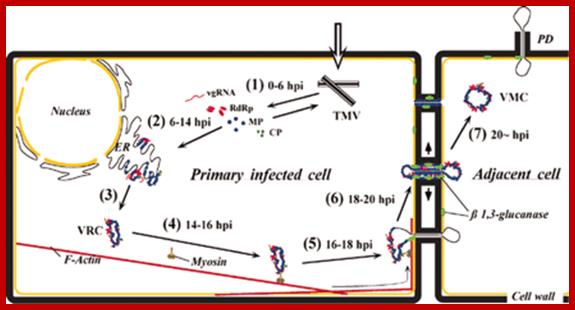
Richard S Nelson; http://www.pnas.org/content
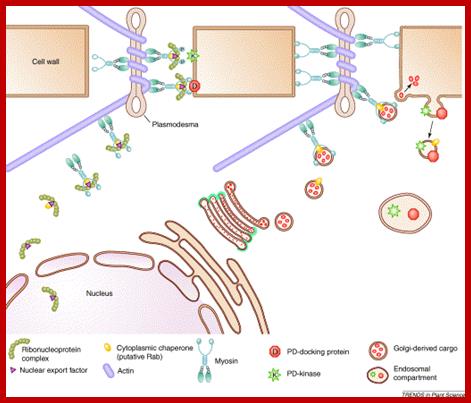
Successful systemic infection of a plant by Tobacco mosaic virus (TMV) requires three processes that repeat over time: initial establishment and accumulation in invaded cells, intercellular movement, and systemic transport. Accumulation and intercellular movement of TMV necessarily involves intracellular transport by complexes containing virus and host proteins and virus RNA during a dynamic process that can be visualized. Multiple membranes appear to assist TMV accumulation, while membranes, microfilaments and microtubules appear to assist TMV movement. Here we review cell biological studies that describe TMV-membrane, -cytoskeleton, and -other host protein interactions which influence virus accumulation and movement in leaves and callus tissue. The importance of understanding the developmental phase of the infection in relationship to the observed virus-membrane or -host protein interaction is emphasized. Utilizing the latest observations of TMV-membrane and -host protein interactions within our evolving understanding of the infection ontogeny, a model for TMV accumulation and intracellular spread in a cell biological context is provided. Chengke Liu; Richard S Nelson;
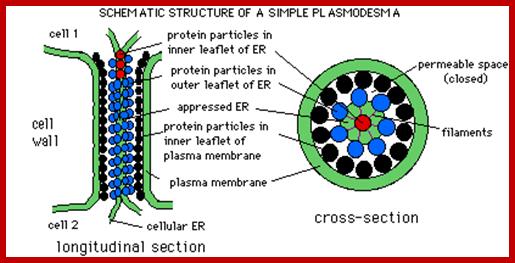
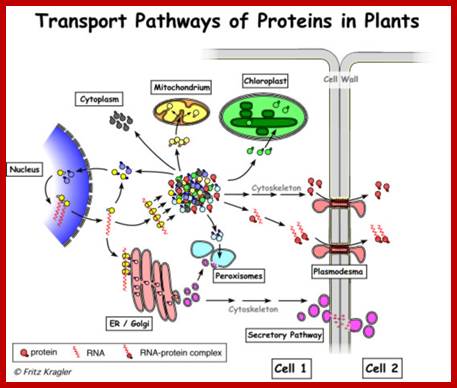
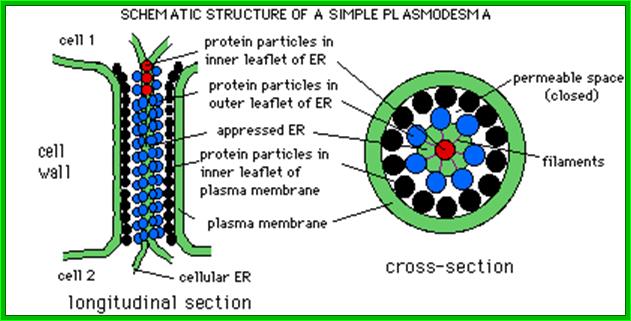
Plasmodesmata is a highly structured transport system which communicates from cell to cell; www.viralinfections.info; www.life.bio.sunysb.edu
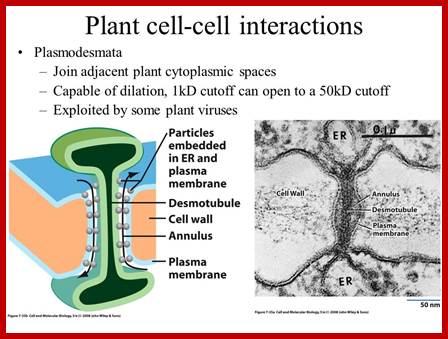
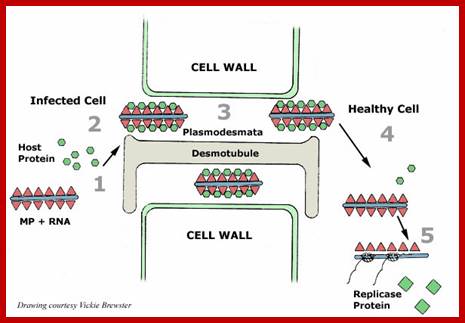
It is throgh plasmodesmata the viral particles/viran RNA tare transported from one cell to th other; http://slideplayer.com/
http://www.pinsdaddy.com
|
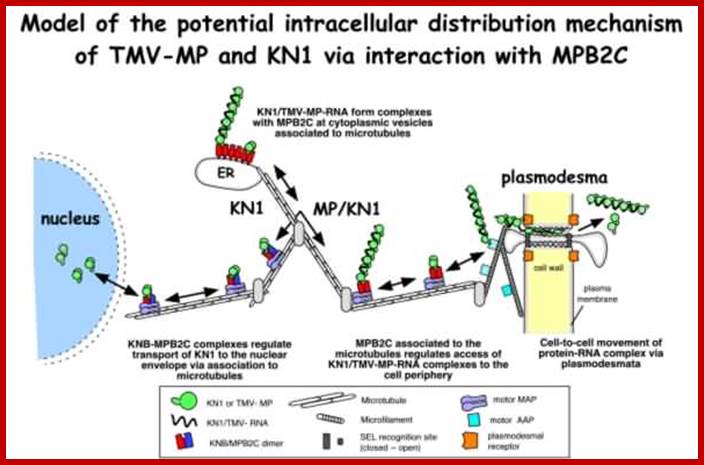
Many of the proteins transport across Plamodesmat is often regulated by microtubules associated proteins, MPB2C, (TMV-MP), KNOTTED1 (KN1) , KN1, (STM), even RNA and many others; Myosin plays an important role in transport KRAGLER LAB; www.univie.ac.a
Recent cell-biological studies have identified proteins involved in the directional trafficking of MCs to PD, and yeast two-hybrid studies have isolated novel host proteins that interact with viral movement proteins. Collectively, these studies are yielding important clues in the search for components that compose the plant intercellular MC trafficking pathway. Here, they are placed in the context of a functional model that links the cytoskeleton, chaperones and secretory pathway in the intercellular trafficking of MCs; Karl J Oparka;http://www.cell.com
|
|
|
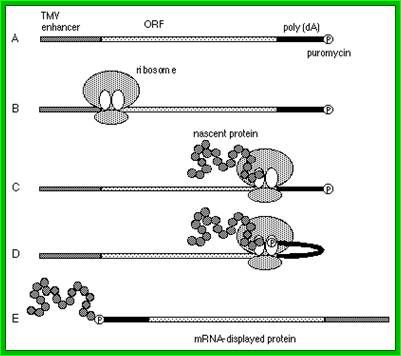
The 5′‐leader sequence (called Ω) of tobacco mosaic virus (TMV) functions as a translational enhancer in plants. A poly(CAA) region within Ω is responsible for the translation enhancement and serves as a binding site for the heat shock protein, HSP101, which is required for the translational enhancement. the functional interaction between Ω and other RNA elements known to participate in the recruitment of eIF4G, The use of a fractionated translation lysate revealed that of the two eIF4F proteins present in plants, eIF4F was specifically required for the activity of Ω. The data suggest that Ω is functionally similar to a 5′‐cap and a poly(A) tail in that it serves to recruit eIF4F in order to enhance translation from an mRNA. By Daniel R. Gallie*
The TMV RNA is 6395 nucleotides long and separate ribosome binding sites for the two genomic 3’ ORFs plus the already proven existence of sub genomic 3’-coterminal mRNAs, explained how the other proteins could be made.

Non-enveloped, rigid helical rods with a helical symmetry. Virion is about 18 nm in diameter, and 300-310 nm in length.
GENOME
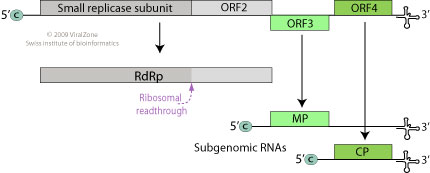
Monopartite, linear, ssRNA(+) genome of 6.3-6.5 kb. The 3’-terminus has a tRNA-like structure. The 5’ terminus has a methylated nucleotide cap (m7G5’pppG). http://viralzone.expasy.org/
The viroid RNA is infectious and serves as both as the genomic and viral messenger RNA. The 5’-proximal ORFs are directly translated to produce the viral constituents of the replicase complex. RdRP is translated through suppression of termination at the end of ORF1. The small replicase is involved in replication and acts as a suppressor of RNA silencing. The movement proteins and the capsid protein are expressed from separate sub genomic mRNAs.
· The genomic RNA has all the characteristic features of an mRNA. Its 5’end has 5’methylated cap structure and 3” end has tRNA like structure.
· The genomic RNA has open reading frame for RNA dependent RNA polymerase to act. The RNA polymerase is the product of TMV RNA.
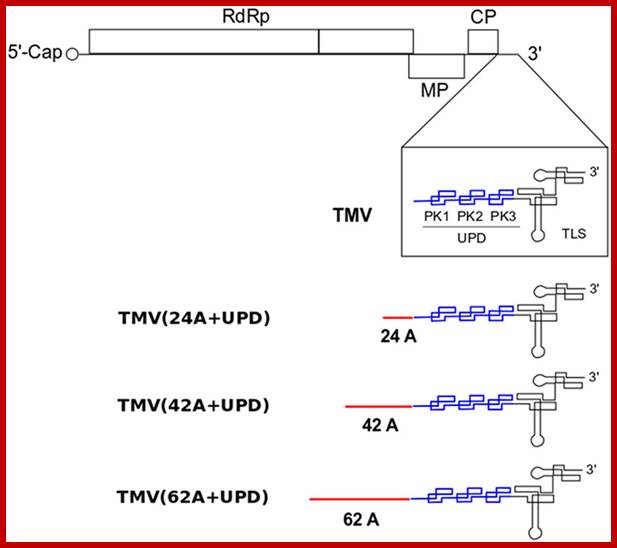
Schematic structure of TMV and its mutants with different lengths of internal poly(A) tract (24A, 42A and 62A) introduced before the upstream pseudo-knotted domain (UPD).
· Its 5’ end contains a cap structure-m7G5pppG, associates with cytosolic ribosomes and translates. The RNA has four reading frames first and second are often code for RNA dependent RNA polymerase (RdRP) for it has read through frame. Initially translation appears to terminate at the end of the first reading frame and the product is +-RNA dependent RNA polymerase, looks like a monomer.
· The read-through product 183kDa acts as negative strand replicase.

http://users.aber.ac.uk/
· The polymerase enzyme recognizes the tRNA like structural feature at the 3’ end of the genome and uses it as the primer for yet it is not clear how the replication initiation takes place on the plus strand to produce negative strand.
· Using tRNA like structures for replication is not an unique feature, for this feature is exploited by reverse transcriptases during retro viral genome replication.
· The RNA polymerase copies the positive strand into negative strand, the negative strand produced often complete or fragmented.
· Once the full length (-) RNA is produced and stabilized, it is again used by the different RNA polymerase or Replicase to produce the positive strand. In the beginning the products are sub-genomic (+) ve RNAs, in the sense only one reading frame length of the RNA is transcribed, no full length (+) RNA.
· Both the virus-encoded 126-kDa protein, which has amino-acid sequence motifs typical of methyltransferases and helicases, and the 183-kDa protein, which has additional motifs characteristic of RNA-dependent RNA polymerase, are required for efficient TMV RNA replication.
· Study of Arabidopsis mutants defective in RNA replication indicates that at least two host proteins are needed for TMV-RNA replication. TMV replicase complexes are located on the endoplasmic reticulum in close association with the cytoskeleton in cytoplasmic bodies called viroplasms, which mature to produce 'X bodies'. Studies have shown for the complete TMV RNA replication it requires two host proteins

- A dsRNA genome is synthesized from the genomic ssRNA (+).
- The dsRNA genome is transcribed/replicated thereby providing viral mRNAs/new ssRNA (+) genome.
- The RdRP recognizes internal sub genomic promoters on the negative-sense RNA to transcribe the 3’co-terminal sub genomic RNAs that will generate the capsid protein, movement protein and viral suppressor of RNA silencing.
· Most abundantly produced RNA is capsid mRNA for 17.6kd protein. The second most mRNA produced is of transport protein. The third mRNAs are for enzyme proteins.
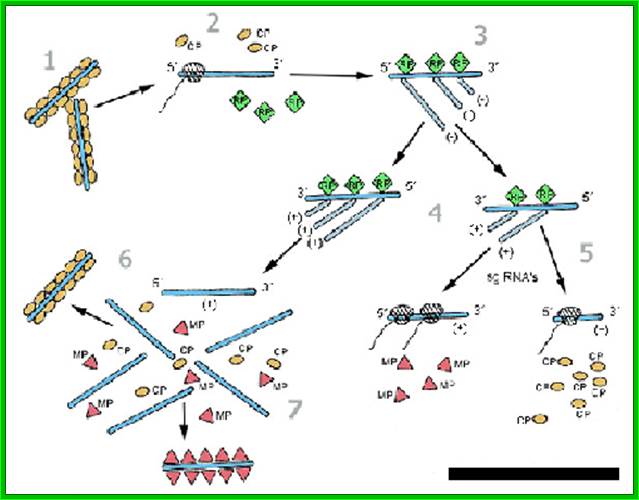
The RNA is first translated and the replication enzymes produced use the positive RNA strand for the synthesis of negative strand and then the negative strand is used for the production of positive strand, which associates with capsid discs for the assembly of TMV particles; https://www.apsnet.org.
· When capsid proteins are produced to an optimal level, the enzyme starts producing, full length (+) genomic RNA. How this switch from sub-genomic RNAs to full length genomic RNA synthesis takes place is not fully understood even after 80 years of its discovery i.e., in 1920s. Another way to explain the production full length (+) RNA synthesis and production of proteins, is explained, as the (-) RNA strand is used directly for the production of a full length (+) RNA, and it is then translated differentially, to synthesize large number of capsid proteins required for viral production, strangely this part of the RNA is found at 3’ end of the RNA.
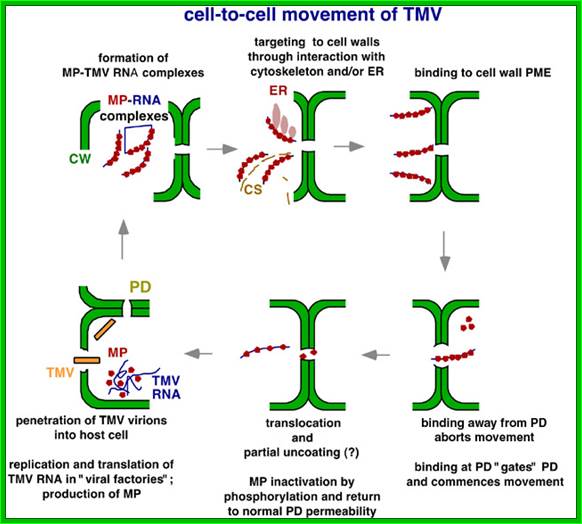
The above diagram shows how the TMV particles form complexes and bind to cell walls and enter through plasmodesma into cells; http://www.mindfully.org/
http://life.bio.sunysb.edu/
Cell to cell transmission is through Plasmodesmata; http://www.mcb.uct.ac.za/
Once TMV RNA and coat proteins are produced in sufficient Numbers and coat proteins bind to internal helical RNA strand which acts as nucleation center and capsid proteins associate one after another in bidirectional manner. Each layer of the disc consists of 17 capsid proteins and induce coiled RNA formation. Each capsid protein binds to 3 ntds of the RNA.
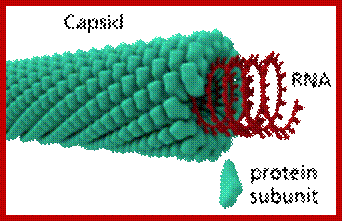
3-D model of a helical capsid structure of TMV virus; https://en.wikipedia.org
The TMV virus, though detected in Tobacco plants, first, causing yellow color of the leaves; the mosaic virus has been known to infect other plants especially Solanaceae members including tomato plants. As this disease can spread fast it can devastate a large area of crops in fields. The other important crop plant that is seriously affected by this virus is tomato, on which it causes Tomato Mosaic disease (ToMV disease); combined with tomato leaf curl virus affect 60 to 80 percentage of tomato crop and it is deadly in India.
Disease resistance to these viruses has been developed based on capsid mediated resistance and RNA mediated resistance. RNA mediated resistance is by ds RNA mediated mi/siRNA mode, another is ds mediated PKR mode and RNase L mode, and developed Transgenic Tomato plants for Tol CV and To MV, (GRK-Raj in IAHS, Bangalore).
Stem-cell-triggered immunity safeguards cytokinin enriched plant shoot apexes from pathogen infection; Muhamad Naseem et al.
MuhammadNaseem†, MugdhaSrivastava† and ThomasDandekar http://journal.frontiersin.org
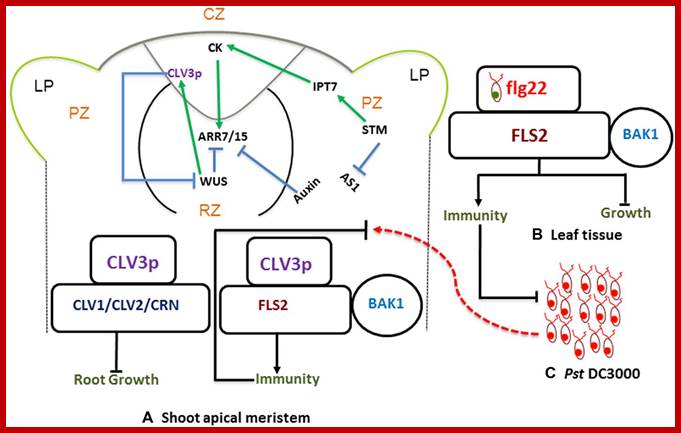
Shoot apical meristem has all the potential to ward of infections; the regulatory network is determined by Auxins and cytokinins; they develop innate immunity in the leaf tissue against; this is stem cell signaling immunity model and CK cross talk.
The SAM is a dynamic structure of a hemispherical collection of identical appearing cells with a stable organization that maintain a balance between the self-renewal of stem cell population and conversion of meristematic cells into aerial organs such as shoot, leaves, and flowers.
The Stem Cell Signaling Immunity Model and CKs Crosstalk; Plant organs deploy overlapping and distinct protection strategies. Despite vulnerability to a plethora of pathogens, the growing tips of plants grow bacteria free. The shoot apical meristem (SAM) is among three stem cells niches, a self-renewable reservoir for the future organogenesis of leaf, stem, and flowers. How plants safeguard this high value growth target from infections was not known until now. Recent reports find the stem cell secreted 12-amino acid peptide CLV3p (CLAVATA3 peptide) is perceived by FLS2 (FLAGELLIN SENSING 2) receptor and activates the transcription of immunity and defense marker genes. No infection in the SAM of wild type plants and bacterial infection in clv3 and fls2 mutants illustrate this natural protection against infections. Cytokinins (CKs) are enriched in the SAM and regulate meristem activities by their involvement in stem cell signaling networks. Auxin mediates plant susceptibility to pathogen infections while CKs boost plant immunity. Here, in addition to the stem-cell-triggered immunity we also highlight a potential link between CK signaling and CLV3p mediated immune response in the SAM.
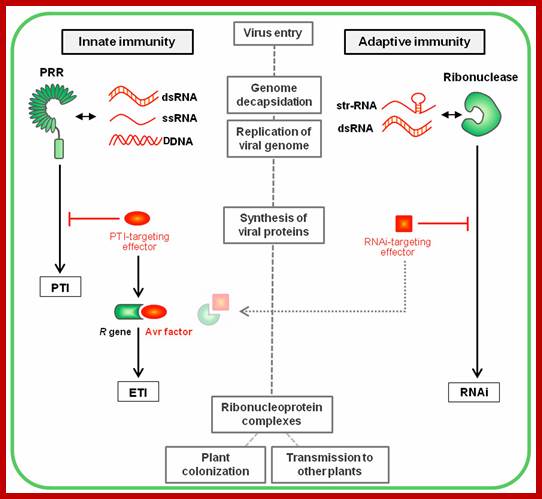
Plant immunity; http://journal.frontiersin.org/
Over the last decades, a concept revolutionizing in understanding of the immunity in plant pathology. This new concept stems from the ability for each organism to discriminate between self and non-self-molecules through the action of pattern recognition receptors (PRRs) perceiving specific microbial molecular signatures, named pathogen-associated molecular patterns (PAMPs). Perception of PAMPs by these immune receptors induces a downstream signaling cascade including PRR association with positive regulators, phosphorylation events, successive activation of cytoplasmic kinases (including the MAP kinases) and defense-related transcription factors, as well as specific defense genes expression. This range of fast, efficient and multi-layered defense responses is referred as PTI (Nicaise et al., 2009; Macho and Zipfel, 2014). To counteract this defense strategy, successful pathogens deploy effectors proteins, the primary function of which is to evade/interfere with PTI. In turn, some plant cultivars have evolved R proteins to block these effectors, leading to effector-triggered immunity (ETI; Jones and Dangl, 2006). Because both PTI and ETI defense layers rely on cellular actors already present primary to the infection, they are commonly classified into plant innate immunity, in opposition to the concept of adaptive immunity (e.g., gene silencing), where the defense responses are acquired following an infection and are adapted to the pathogen. Plant vaccination gambles on cross-protection, a phenomenon whereby the inoculation of a virus into a host plant prevents the multiplication of a subsequent challenge virus.
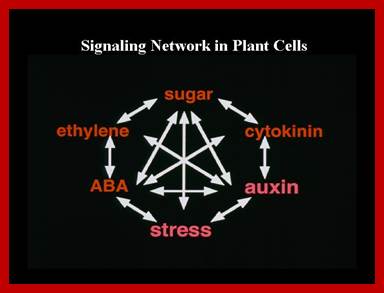
Genetic and molecular evidence from our studies suggest that plant signal transduction pathways connect to each other through many levels and by diverse mechanisms. Pant signaling; http://molbio.mgh.harvard.edu/
|
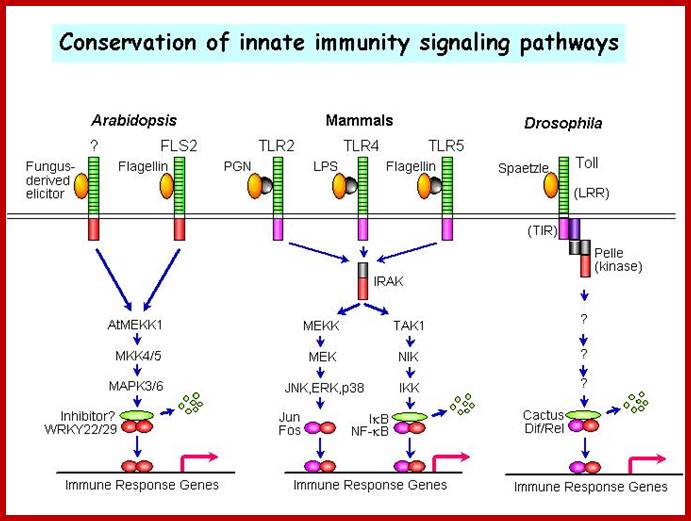
A MAPK cascade in Arabidopsis innate immunity; A model of innate immune signalling activated by LRR receptors in Arabidopsis, mammals and Drosophila. http://molbio.mgh.harvard.edu/ |