Prokaryotic DNA Replication:
Introduction
Duplication or copying of the DNA-to-DNA, nucleotide by nucleotide, is one of the fundamental properties of �molecule of life�. From one cell to one cell. It cannot afford to make mistakes in producing 10^14 in the development of Human body from one cell to one cell.� The process is called replication and replication takes place once in one cell cycle.� The initiation point and duration differs from one system to other.�� When conditions are favorable cell cytoplasmic mass increases and when cytoplasmic mass reaches a ratio to that of the cell size to 2:L, bacterial cell division is triggered.� To complete its cell cycle it requires hardly 30-40 minutes.� Several genes involved in different steps of cell division have been characterized(?).� Par-excellent system to understand molecular process of prokaryotic cell division is E.coli.� The size of the E.coli chromosome is 4.6x10⁶ base pairs, but it is circular and compacted by the binding of histone like proteins.
�
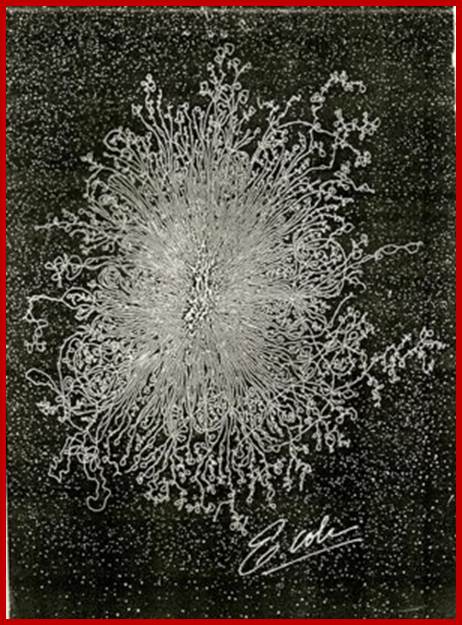
E.coli genome- free from the cell wall; http://utminers.utep.edu/
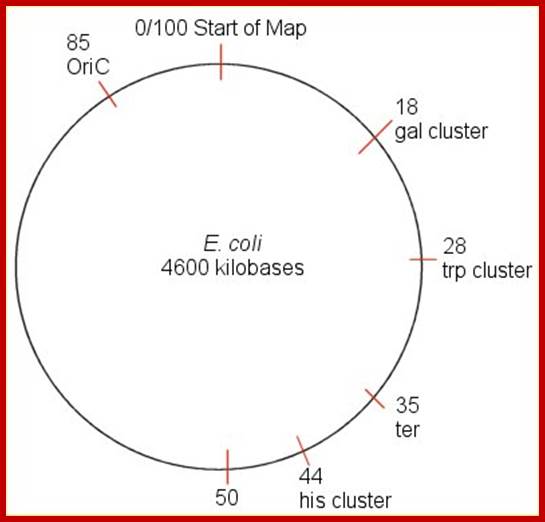
The �100-minute map 10x10^6 bp long is a time-based map of the E. coli genome. Based on the assumption/observation that it takes 100 minutes to replicate the genome, but replication takes place in about 30 minutes.� The map is a listing of at what points in time a particular gene is copied; in this case, it is looking at clusters of genes (it is important to note that most genes are not clustered). From the map, we can say,
�At time X, genes for traits A, B, and C have been copied�; conversely, we can say, I need to wait X minutes for my desired trait to copy.
The figure shows the basic circular model of the chromosomes with map positions such as Ori C and TER regions.
�http://utminers.utep.edu/
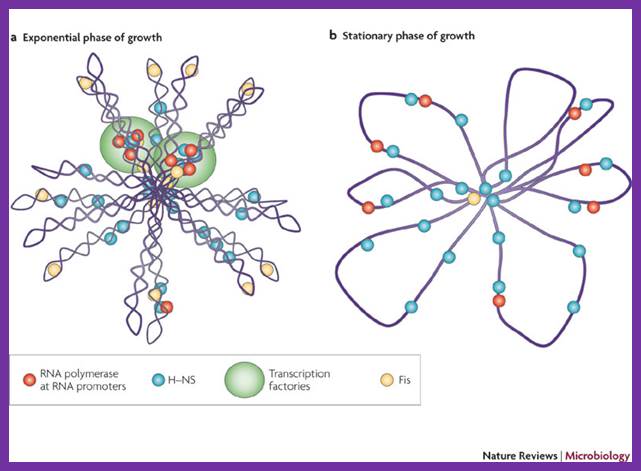
The folded chromosome is organized into looped domains that are negatively supercoiled during the exponential phase of growth. In this phase, the abundant nucleoid-associated proteins histone-like nucleoid-structuring protein (H-NS) and factor for inversion stimulation (Fis) bind throughout the nucleoid and are associated with the seven ribosomal RNA operons. As shown here in two cases, these are organized into superstructures called transcription factories. b | In stationary phase the rRNA operons are quiescent and Fis is almost undetectable. The chromosome has fewer looped domains, and those that are visible consist of relaxed DNA; Shane C. Dillon & Charles J. Dorman.
First, the nucleoid i.e. the chromosome frees from the membrane and unwinds.� Such DNAs initiate replication and completes in a matter of 15-20 minutes.� Then the cytoplasm starts dividing almost in the center of the cell by central septal ring formation.� It is at this time the daughter DNA molecules are partitioned and segregated to the two compartments.� The central peri-septal plasma membrane ring that starts 1-2 minutes after DNA replication now furrows inwards and divides the cytoplasm in the middle region and cell wall material is deposited in between the membranes.� Then mid-wall splits in the middle and daughter cells are set free, hence the name Schizomycetes to E. coli bacteria.
The picture shows bacterial cells � some divided and some yet to be divided; http://www.manucornet.net/
Eukaryotic cell division is more complex and highly regulated.� The paradigm for it is yeast, for it is a unicellular system, grows fast and requires hardly 60 minutes for one cell cycle.� One can obtain different mutations like conditional and null or auxotrophic kinds.� In culture cells, the duration of one cell cycle is about 12 hrs.�� Onion root tip cells divide once in 12 (24hrs) hrs. ��Whereas embryonic cells in its early stages require just 30 minutes for one cell cycle and they go through 6 to 8 such cell divisions very fast.
Replication of DNA in cell division is an important phase.� The replication is very accurate and it is nucleotide by nucleotide and there is no provision for making any mistakes, even if mistakes are made they are immediately corrected or repaired, otherwise consequences are serious and deleterious to the cell.� In general DNA replication is precise, exact and regulated and involves initiation, elongation and termination steps, but rate of replication vary from one system to the other.
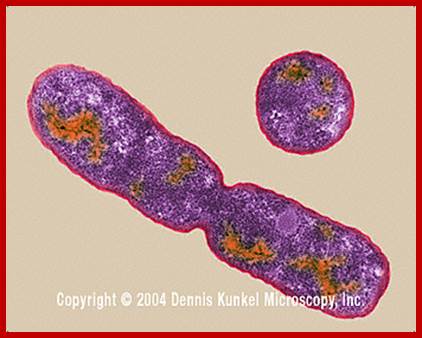
TEM structure of E.coli cell which shows constriction
�during cell division; http://www.cs.sandia.gov/; http://biology.kenyon.edu/
Replication is Semi Conservative:
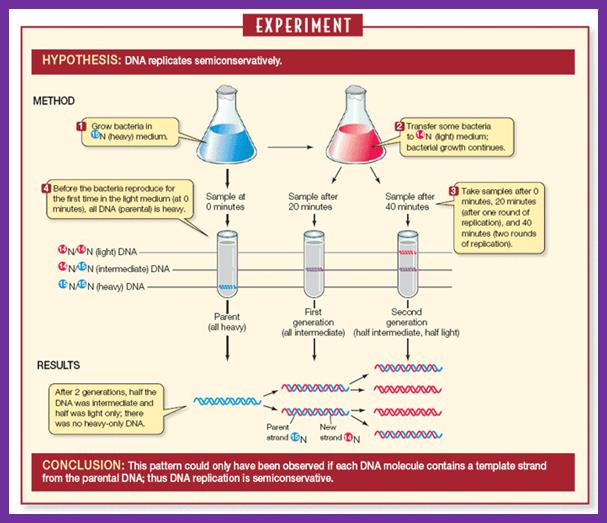
Meselson and Stahl, showed that DNA replicates in semi conservative mode.� There were some doubts about the mode whether it is conservative, dispersive or semi conservative.� But Meselson and Stahl used nonradioactive heavy Nitrogen isotope (15N), as the source of Nitrogen.� When cells were grown on such source, 15^N nitrogen bases get incorporated into DNA, thus the DNA becomes heavier than the DNA that is grown in normal (14N) nitrogen media.� When two such DNAs containing bacteria, one grown in the presence of 15*N source and the other in 14N (normal), are extracted separately and stained with ethidium bromide, then subjected to equilibrium density centrifugation at high-speed of ~42000rpm for about 24�36 hrs. (?), the DNAs, separates as two distinct bands, the one that is heavier (15N) is the lower band than the normal (14N) upper band DNA.� If cells grown in heavy isotopes are shifted to normal Nitrogen source and grown for one or two cell generation and if such DNA is isolated and if one combine this with the first two DNA sources and subject them to equilibrium density ultra centrifugation (using cesium chloride or cesium sulphate), one can observe three bands, the top DNA band is from cells grown in 14N without isotope, the second band is the hybrid band with one strand 15^N and the other with normal 14N-Nitrogen and the third band is with heavy nitrogen where both the strands are labeled with 15N.��� This experiment unambiguously proved that DNA replicates in semi conservative mode.�
Robert Taylor using root tips of Vicia faba, labeled with 32^P radioactive isotopes or 14N labeled, demonstrated semi conservative mode of replication at chromosomal level.� Semi conservative means that the two daughter molecules produced at the end of replication, each of the daughter chromosome retains one parental strand and the other one is the newly made one. Subsequent chromosomal replication leads one with radioisotope labelled and another without label.� So it is called semi conservative.�� The mode of replication is absolutely semi conservative irrespective the kind of DNA, whether it is ds DNA or ssDNA, even RNA also replicates semi conservatively, whether it is linear or circular, and the basic mechanism is more or less similar; each of them uses their own specialized mechanisms.
http://www.nature.com/http://www.uic.edu/


Meselson and Stahl/J. Kruper; http://www.apbiowiki.com/
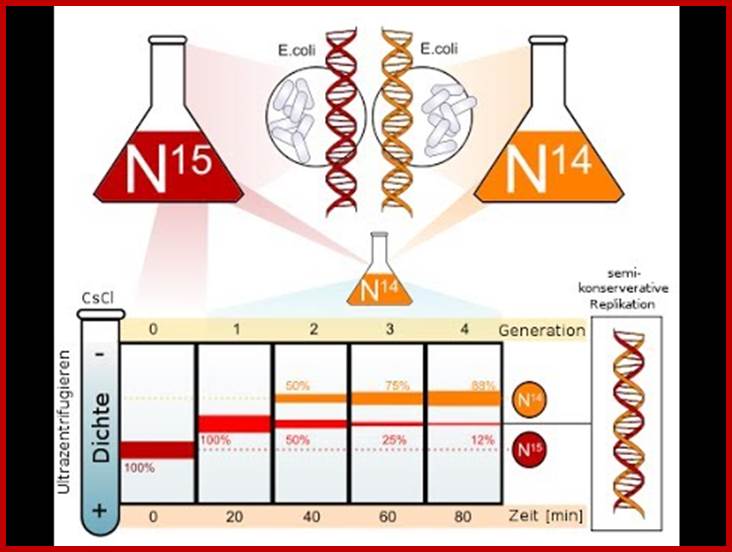
Replikationsmodelle und der Meselson-Stahl-Versuch;https://www.youtube.com
Experimentally semi conservative mode of replication has been shown by density gradient centrifugation using non-radioactive isotopes such as 15N and 14N sources.
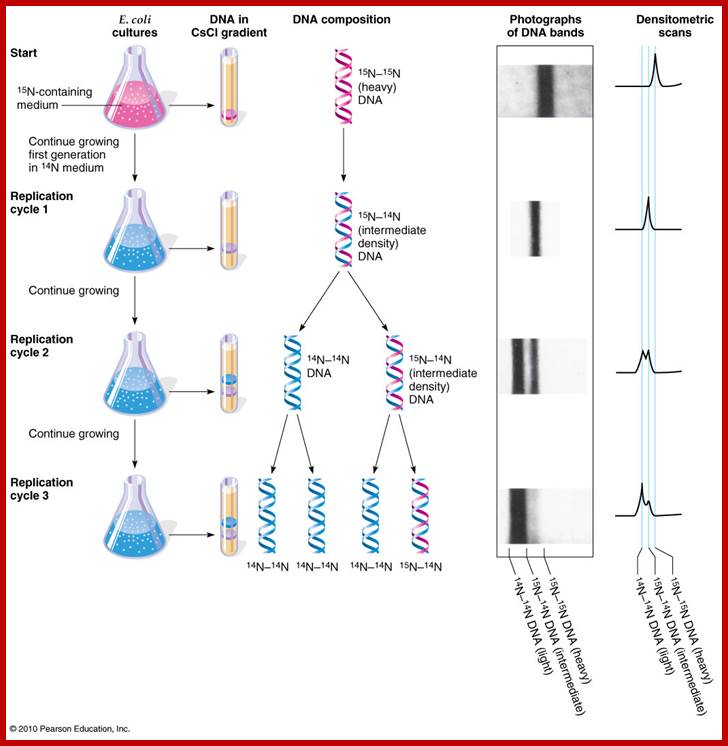
The Meselson - Stahl experiment: Proof of semi-conservative replication; https://www.mun.ca/biology
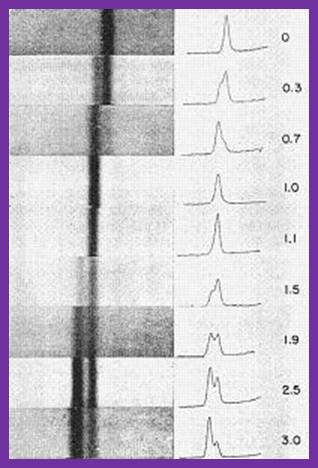
Overview of the Meselson-Stahl experiment supporting semi-conservative replication; http://www. saburchill.com
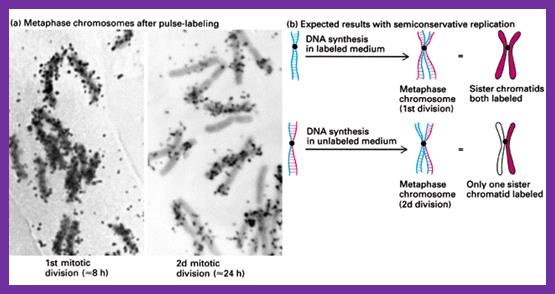
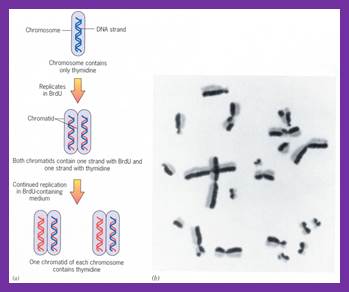
In vivo radioactive 14N isotope labeled plant chromosomes replicating in semi conservative fashion. Right diagram; http://yxsj.baiduyy.com/