Replication of M13 phage DNA:
Bacterial viruses are many but there are few belonging to a class called filamentous forms, they are very interesting. They are single strand DNA (+ strand) phages. M13 phage is related to Fd and Fi class of phages.
M13 is 6407 ntds long.
Fi is 6408 ntds long.
FD is 6408 ntds long.
Pf1 form is a related to the above phages but pseudomonas is the host for the phage. The size of the phage is twice that of M13, but contains the same amount of DNA that is found in M13, it is because it is packaged into the viral capsid in extended form.
Structural Features:
M13 phage is 895-900 nm long and 6 nm thick, the central core is 2.5 nm wide.
The DNA 6407ntds long is packed in linear but in helical form and at one end it contains a 100 ntds long hairpin like structure, it is called intergenic region (IG). This region contains signals for viral packaging and also contains origin sequences for minus strand as well as plus strand synthesis.
All along the length of 6407 ntds, genomic DNA is enclosed by protein subunits called gp8, which are organized in shingle fashion. The gp8 protein subunits have basic and acidic ends with their charges and hydrophobic region is in the middle. The basic region of the protein is bound to phosphate groups of DNA backbone. The central hydrophobic core is responsible for protein � protein association. The acidic group is exposed outside. There are 2700 to 3000 subunits per a phage particle.
���������� Electron micrograph of the filamentous M13 phage; www.kennislink.nl
 �
�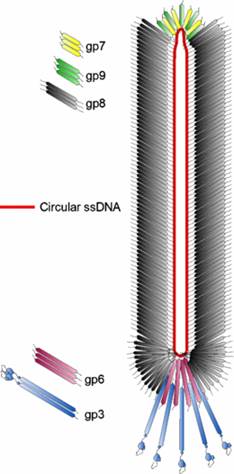
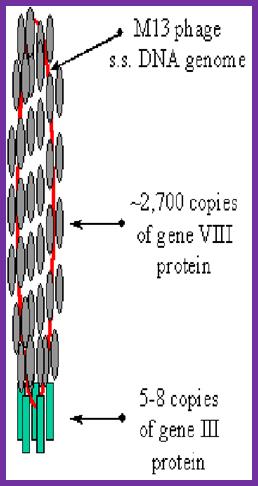
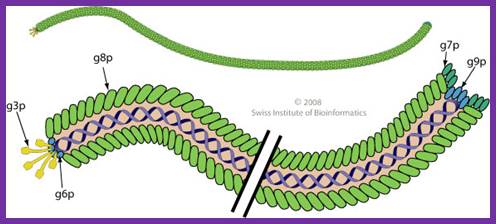
The diagram (the above and below) illustrates the organization of capsomeres on ssDNA and the gp3/ at one end of the particle. And gp7and Gp9 at the other end not shown. This another view of the M13 phage with its polar proteins; Martin Ploss and Andreas Kuhn
Fig- Right side; http://iopscience.iop.org/
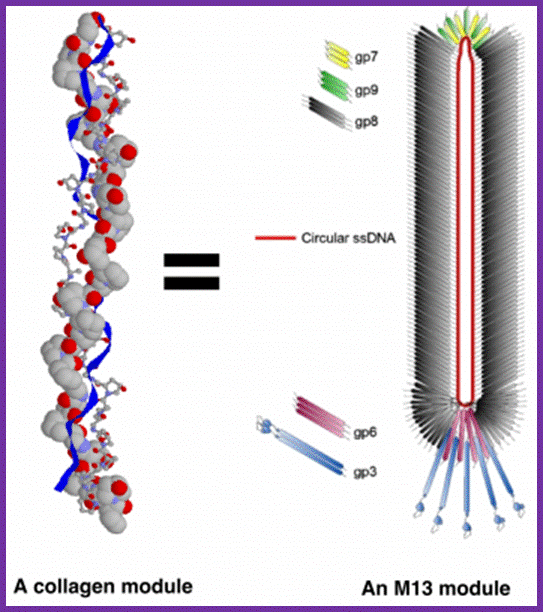
Each M13 virus acts as a synthetic module, equivalent to one collagen fibril. Source images: M13 - http://iopscience.iop.org and Collagen - http://www.cryst.bbk.ac.uk; http://urbantimes.co/
The diagram (the above and below) illustrates the organization of capsomeres on ssDNA and the gp3/gp6 proteins at one end of the particle. And gp7and Gp9 at the other end http://2011.igem.org; http://keywordteam.net; http://viralzone.expasy.org/
M13 phage particle, it is cute diagram; position of different capsid are illustrated. Non enveloped, rod of filaments 7nm in diameter and 700 to 2000nm long and , helical capsid with adsorption proteins on one end. http://viralzone.expasy.org/ ; This phage non-lytic, contains M13amp18 genome; pVIII is present at~27-00 copies per virion and 10% of them reliably fused to peptides or proteins; http://2011.igem.org/
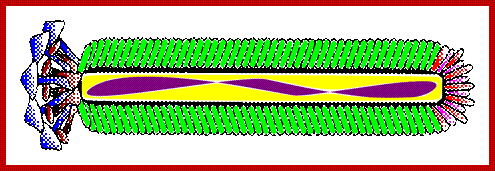
Blue: Coat Protein pIII; Brown: Coat Prote�n pVI; Red: Coat Protein pVII; Limegreen: Coat Protein pVIII; Fuchsia: Coat Prote�n pIX; Purple: Single Stranded DNA; https://en.wikipedia.org; www.mashpedia.com
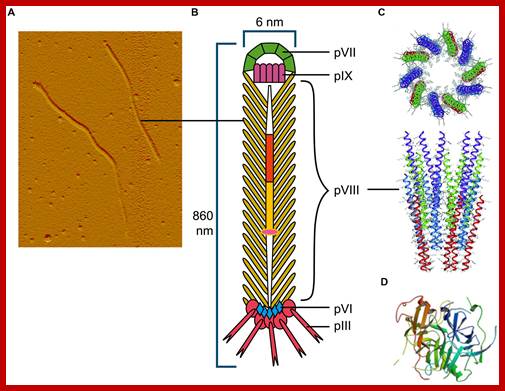
Dragana Gagic et al.;http://journal.frontiersin.org
Association of subunits is 4.5 subunits per one helical turn of the DNA. Its axial rise per turn is 1.5nm. But by increasing the size of phage�s genome, it is possible to extend the length 3 to 5 times the original size of the particle.
The phage shows certain morphological polarity, in the sense; it has two sets of distinct proteins localized at front end of the phage and another two sets at the opposite end. At an end at which one finds five gp-3 and five gp-6 proteins; (gp6 at the base of gp3), it is called attachment end (can we call it front end?). The other end has five gp-7 (at the periphery) and five gp-9 proteins (in the central region); the intergenic region with capsid packaging site viral coat assembly starts at the DNA packaging signals; this region is located in between IV and II genes called intergenic (IG) space. In this polarity, during infection, the phage end (gp3 and gp6) that binds first comes out last and the end that enters last (gp 7 and gp9) emerges out first during phage output.
A List of Genes and Gene Products of M13 Phages:
|
Gene |
Mol.wt |
Function
|
|
Gene 1 |
35 |
NS membrane protein, interacts with FiP-ferrodoxin,� Morphogenesis |
|
Gene 2 |
46 |
RF-replication, by nicking + strand |
|
Gene 3 |
42 |
5 copies, anterior Adsorption, and morphogenesis, binds to pilius, |
|
Gene 4 |
50 |
Required for morphogenesis |
|
Gene 5 |
9.8-10 |
Phage ssB, structural protein for replication, controls expression gp-2; controls switch from RF to ss form |
|
Gene 6 |
12 |
5 copies,, Minor coat protein, front end involved in adsorption and morphogenesis |
|
Gene 7 |
3.5 |
5 copies, Minor coat protein, binds to IG, required for packaging |
|
Gene 8 |
5.2 |
Major coat protein,2700-3000, which cover the entire length of DNA |
|
Gene 9 |
3.3 |
5 copies, Minor coat protein, binds to IG, required for packaging |
|
Gene X |
12 |
N-terminal psrt of gene2, 500/virus,Required for switching from RF to ss |
|
|
|
|
Infection:
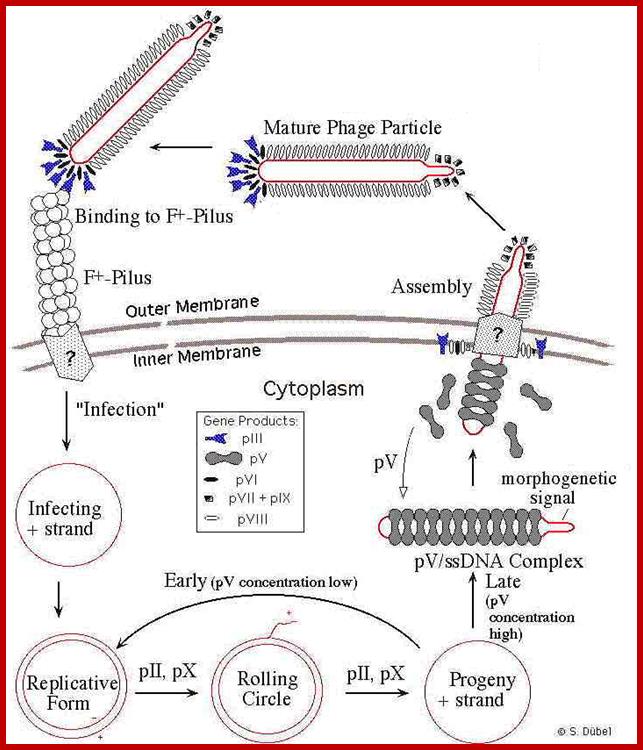
Phage infection takes place, first by binding of its minor coat P3 to the receptor at the tip of the F pilus sex pili through adsorption proteins located at attachment end that means they infect only male bacterial cells. Then it is through pili, phage DNA enters into the cell.
Infection of bacteria without sex pili also takes place, but with very low frequency. Many speculations are in air about the mechanism by which the DNA enters.
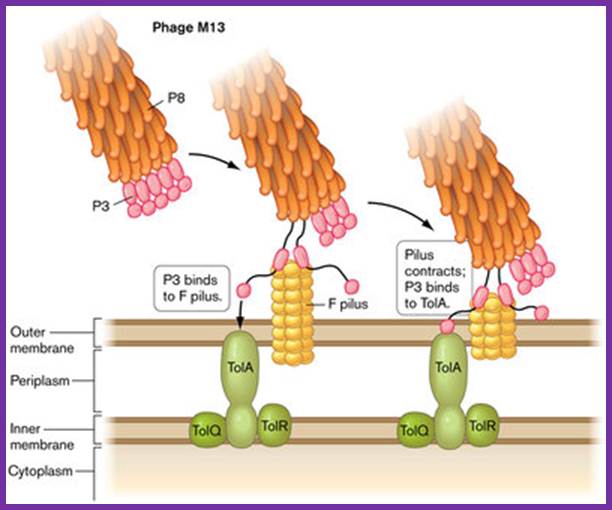
M13 adsorption to the pilus and TolA. Attachment to pilus of F+ cell. http://www.wwnorton.com/Slonczewski Foster
According to one hypothesis, DNA enters along the groove of pili. Second speculation is that the phage is guided by the pilus to an outer membrane receptor through which DNA enters the cell. The third group thinks, with adsorption to the tip of the pilus, the pilus undergoes depolymerisation at membrane side that is from its base, where it has anchored to the membrane,
With attachment of the gp6 and gp3 to the pilus the gp8 capsid proteins undergo conformational change in their helical structure. With more penetration they undergo more changes, by the time the DNA enters the proteins undergo full conformational changes, thus the DNA enters into the cytoplasm and gp8 capsid proteins enter into bacterial plasma membrane.
�
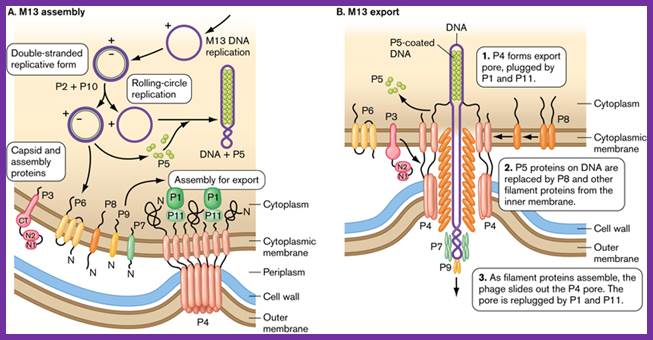
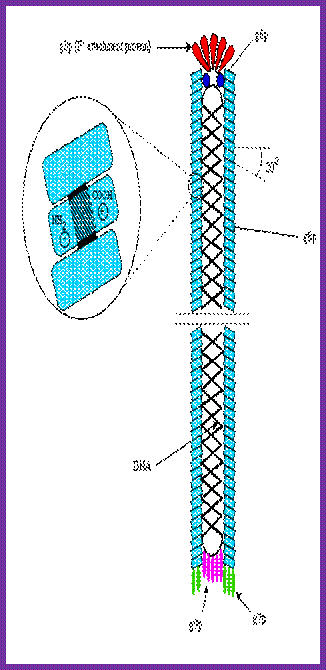
Assembly and export of M13 progeny phages. A. Replication of phage M13. Most assembly is conducted by phage proteins within the cell membrane. B. Assembly for export through the pore made of P4 monomers. P4 makes a channel used to export filamentous phages without lysing the bacterial host. The single-stranded DNA of the filamentous virus is coated by P8 subunits as it passes through the membrane, and then passed through the P4 channel to emerge as mature, infectious phages. http://www.wwnorton.com/
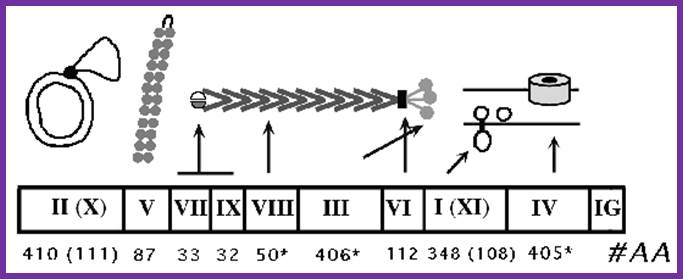
The above diagram shows the genes order. http://www.thebacteriophages.org
Filamentous phage f1/M13/fd: genes and gene products. pII binds to a sequence (the + strand origin) in the intergenic region (IG) of dsDNA and nicks the + strand; the original + strand is displaced by Rep helicase as a new + strand is elongated from the 3' end of the nick by host DNA polymerase III, using the - strand as template. pX, which is identical to the C-terminal third of pII, is required for the accumulation of ssDNA, as is pV. Dimers of pV bind cooperatively to single-stranded DNA, which collapses the circular genome into a flexible rod with the packaging signal (PS) exposed at one end of the filament. pVII and pIX are small coat proteins located at the tip of the virus that is first to emerge from the cell during assembly. pVIII is the major coat protein, several thousand copies of which form the cylinder that encases the ssDNA phage genome. pIII and pVI are located at the end of the virion where they mediate termination of assembly and release of the virion from the cell membrane. pIII is also necessary for phage infectivity. pI may hydrolyze ATP to promote assembly; pXI is identical to the C-terminal third of pI; it lacks the cytoplasmic domain and may play a structural role as part of an oligomeric pI/pXI complex. pIV is a multimeric outer membrane channel through which the phage exits the bacterium. - View larger tif in separate window
It is now clear that the place of viral attachment is the region where the outer and the inner membranes are adpressed to each other called adhesion region.
The �tol� gene product is believed to play an important role in infection. What is the role of gp3 and gp6 in infection apart from adsorption of phages to pili? Not clear! Once the particle is in contact with membrane at attachment site, �the DNA is pulled inwards, (how?), while coat proteins just enter in to the outer membrane and stay there till new phages emerge out. Note; that coat proteins have hydrophobic segments, thus they can easily move into the hydrophobic lipid membranes.
Phage is a real parasite, because, after gaining the entry it replicates and produces all the components required for viruses and finally the viruses are budded off. All these events happen with out killing the bacteria. Only bacterial cellular metabolism is affected, but the cell is yet not killed.
Metaphorically, virus is like a son-in law living in father-in-laws house, eats, recreate, reproduce, and do every thing he can do at his father-in-laws expense.
Replication cycle:
SsDNA (+)--> ds RF DNA (+/-),
ds RF---> gets super coiled in to RF1 form,
RF1 form----->Transcription and translation---> proteins,
RF1 ----> (+) ss DNA circles,
(+) SS DNA circles----> RF1 DNAs,
RF1DNAs------> (+) ss DNA -----> packaging at membrane bud off particles.
Structural Features of M13 Origins:
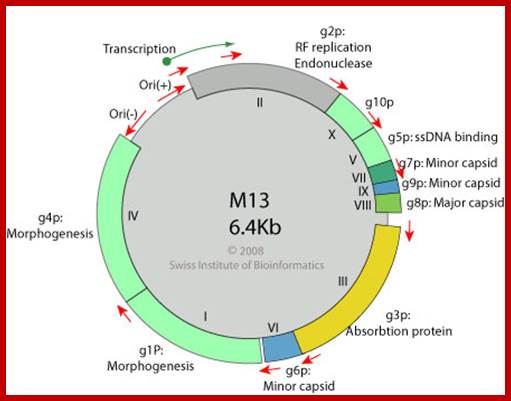
http://viralzone.expasy.org/
Both + and - origins are located in an intergenic region, located in between gene-2 (II)and gene-4 (IV). The size of the intergenic region is about ~450 nucleotides long.
It consists of 5 specific stem loop structure with elaborate sequence complementarity. They are called A, B, C, D and E, and each of them have their own specific secondary structural features.
The stem loop �A� is approximately 100 ntds long, B and C about 150 ntds long each, and D and E about 150 ntds long each. At one end of E certain sequences found act as enhancers.
The A stem-loop has viral capsid packaging sequence that is required for the formation of the phage. It is at this site viral coat proteins start assembling.
All viruses have such packaging sequences with which their genomic DNA or RNA recognizes specific capsid proteins and encapsulation takes place.
The B and C are little longer with some unpaired regions act as origins for (-) strand synthesis. The D and E are for (+) strand synthesis.
(-) Strand synthesis:
As the ss DNA enters into the cell, it gets coated with host ssB proteins and becomes super coiled. Super coiled DNA generates specific stem-loop structures at specific sites.
Host RNAP binds to the B and C stems and starts producing a primer from the basal region of the C stem towards B loop. The host DNA polymerase-III assembles on to the stem loop structure and uses RNA primer 3�OH end and extends it as complementary copy to the (+) strand, thus polymerase moves all the way round. DNA pol-I removes the primer and fills the gap. The DNA ligase seals the nick.
The replication is similar to that of continuous strand synthesis in E.coli.
The replication produces ds DNA called RF; it becomes super coiled which is called RF1 state.
Synthesis of (+) Strand:
The RF form is used to initiate transcription and it ensues translation. There are at least 5 or more promoters from which host RNAP can initiate transcription. The transcripts on translation produce their products. The genome continuously keeps on producing these components.
One of the proteins that is required for replication is gp-2. This protein has similar characteristic to that of gp-A of phi X-174 phage. Proteins Gp-2 recognizes D and E structural elements on the (+) strand and its activity is facilitated by host� histone like factors, which bind to AT rich IHF regions. This leads to proper binding of the gp-2 onto the D loop.
It cuts the (+) strand in the D loop to generate 3�OH and 5�p free ends. In this process the gp2 covalently binds to the 5�p end of the (+) strand through one of the tyrosine amino acids.
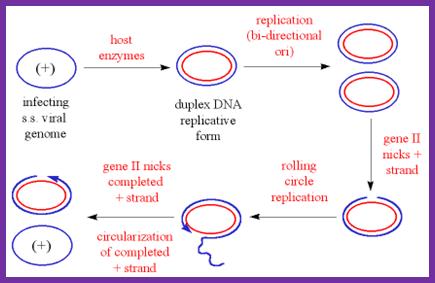
The diagram shows the outline of replication mechanism from plus strand to ds RF form, from that plus strand synthesis by rolling circle mode. The blue and red lines represent (+) and (-) strands respectively. http://www.mikeblaber.org/
It is at the nick region, primosomal complex assembles. The complex consists of Rep-A and DNA-pol-III. The Rep-A is bound to (-) strand and it acts as an helicase, but the movement is from 3�>>5� direction. Rep-A is bound to gp2. However, mutations in host DNA-A, Dna-B and Dna-G genes block phage growth; hence possible role of PriA, priB, PriC and DnaT and DnaB are implicated.
With the assembly of Rep-A bound to (-) strand and gp3 bound to the 5� of the (+) strand, the DNA pol-III (Holozyme) is loaded on to this complex.
The DNA pol-III uses the nicked end with 3�OH for extension. In this case the 3�OH group acts as the primer for the enzyme.
The gp-2 bound to 5� end also complexes with rep-A. The motor protein in this is Rep-A unwinds the RF DNA. In this Rep-A, gp-2 and DNA pol-III as complex move together. The DNAP-III extends the 3�OH end and Rep-A and gp-2 plows along the plus strand (+) loops out.
While DNA pol-III completing replication one round, as it passes through (+) origin site, it cuts the (+) strand while the gp-2 protein is still bound to the 5�end of the first nick. The cutting is exactly at the same site or position where it has first cut the + strand of RF DNA. The gp-2, as it nicks, it transfers the 5� phosphate bond to the 3�OH group of the same strand and releases a circular molecule.
While it transfers the 5� end to 3�OH group, the newly cut 5�P is transferred to one of its tyrosine amino acid residues, thus the protein binds to 5� end of the newly formed (+) strand.
This process can go on cycle after cycle. And the Circular (+) strands continue to produce more RF forms, so more transcription, more translation and component builds up. As the phage genome builds up with in bacterial cell, phage proteins are also produced.
Bluescript KS+ is an example of a phagemid cloning vector. Often times, phagemid vectors are used to prepare single strand DNA for in vitro mutagenesis protocols using oligonucleotide primers; http://www.biochem.arizona.edu/
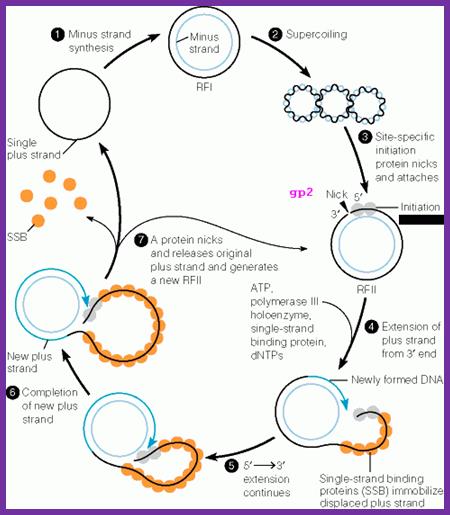
Rolling circle module of DNA replication; Dragana Gagic;www.keywordsking.com
One of the viral proteins gp8, on translation, is transported into plasma membrane. Concentration of gp1 builds up near the adhesion site at which the phage buds off. Gp-4 is also involved in catalyzing the assembly. The gp-5 binds to the cut 5� end of the (+) strand and as the (+) strand is peeled off in rolling circle mode, gp-5 covers the whole length and prevents the association of host SSB.
Once the circular (+) strand is generated, it is loaded with gp-5.� This region is moved to the adhesive region where both cell membranes are fused. This is where particles assemble.
Viral packaging site, which is sequence specific, is recognized by gp7 and gp-9. Then gp-8 found in the membrane, as if they are in the assembly line, are added on to DNA which is in linear, but super coiled helical state.
With continued assembly of gp-8, the DNA gets coated; whatever that is coated is extruded and the process of assembly and extrusion of the particles takes place simultaneously.
The proteins to be added last are gp-3 and gp-6. During infection gp-3 and gp-6 are the first to bind and disappear and the last to go are gp-7 and gp-9. During viral production the first to be added are gp-7 and gp-9 and the last be added are gp-3 and gp-6. The exact role of gp-4 and other minor proteins is not clear. The host cells can keep on producing viral particles for a prolonged period of time.
Mini defective phages:
Occasionally filaments of one-third size of M13 are generated. They contain large deletions and no intact gene is found. But they can be multiplied with helper phages, which provide the required components for DNA replication and phage particle assembly.
M13 phage DNA as cloning vectors:
M13 DNA or DNA from any other such filamentous phages can be used to clone foreign DNA into their IG region with out disturbing the sequence. By increase in the phage DNA size, the particle size can also be increased, but there is a limit to it. However the origins, either in plus or minus regions can be transferred to many popular vectors like pUC to generate, what popularly called �phagmids�. Thus one can generate ss DNA of either (+) or (�) stranded filamentous viruses. If a bacterial cell harboring a phagmid DNA, of any strand sense, is super infected with a helper phage M13Ko7, (carries a mutation Met40Ile in gene II, also contain ori from p15A, and contains Kanamycin resistance gene derived fromTn903 all localized in M13 origin), which is replication origin defective, but provides all the required structural protein components for the phagmid DNA, results in the production of packaged viruses containing either negative or positive single stranded DNA. They are then secreted. The secreted viruses can be harvested with PEG precipitation and its DNA can be extracted easily. Such phagmids can also be exploited for expression of cDNA clones in the form of coat proteins as a part of gp-8. Cloning at gp8 gene produces more number of recombinant proteins than cloning using gp3, gp6 or gp7 and gp9. Such proteins or clones can be easily identified and can be developed into monoclones. This has been achieved for generating a library of immunoglobulin clonal library. [Note: the M13Ko7 is capable of replicating in the absence of phagmid DNA, but in the presence of Phagmid DNA or f1 origin, single strand phagmid is packaged preferentially and secreted out of the cell].
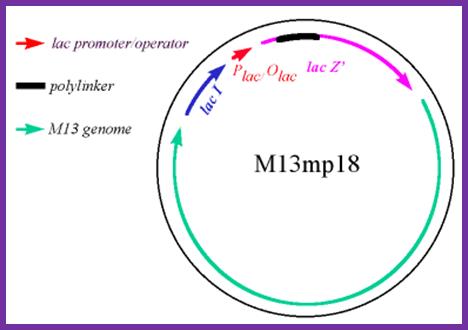
Development of M13 into a cloning vector.
M13 was developed into a useful cloning vector by inserting the following elements into the genome: a gene for the lac repressor (lacI) protein to allow regulation of the lac promoter: the operator-proximal region of the lacZ gene (to allow for α-complementation in a host with operator-proximal deletion of the lacZ gene): a lac promoter upstream of the lacZ gene; a polylinker (multiple cloning site) region inserted several codons into the lacZ gene.
*Paweł Kowalczyk et al ;http://www.czytelniamedyczna.pl/
The figure shows the M13 derived vector containing p18 multiple cloning sites; this kind vector can be used for creating point mutations, generating single stranded DNA for sequencing and general cloning purposes including cDNA library preps; it is as good as pUC plasmids. M13 phage can also be used to express cellular proteins by introducing gene at specific sites. These results in the expression of cloned genes and the same gene products can be purified and such proteins can be used as antigen for developing vaccines. In some rare cases a library can be developed, ex. Immunoglobulin library, and the same can be screened easily. Depending upon the position where the DNA is cloned, they express along pVIII and pIII.
���������������������������������������������������������� http://www.scielo.br/

Multiple cloning sites in both direction in mp 18 and mp19
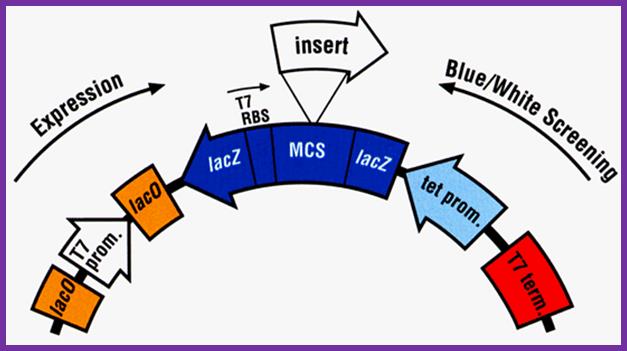
The expression cassette like the above can be inserted into M13 cloning vector for expression. https://www.researchgate.net
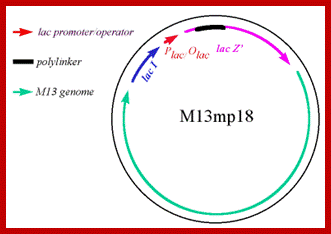
M13 phage DNA as a cloning vector; http://www.mikeblaber.org
M13 vector can be used for in vitro mutagenesis:
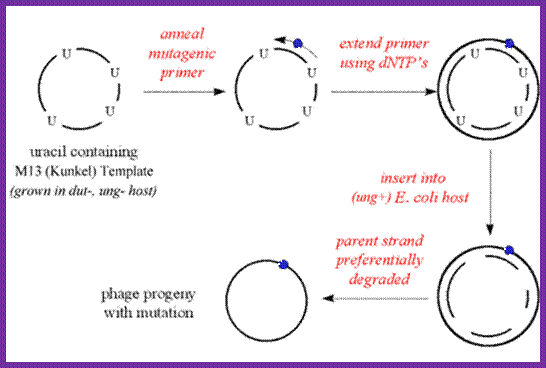
http://www.mikeblaber.org/
M13 phage can be grown in suitable bacteria and multiply phages in large numbers. The circular DNA (the vector M13 DNA can be annealed with primer with defective uracil at a particular site. Then extend the primer and ligate it. The said DNA can be used for bacterial cell transformation and the defective plasmid DNA can identified and isolated as mutant strain. Using this technique any gene can be subjected site-specific mutation, which can be used for its biochemical or phenotypic effects.
http://iopscience.iop.org
�����������������������
Recombinant proteins expressed along with phage proteins can be used for developing monoclonal antibodies.
Summary:
- Viral g3p protein mediates pilus-mediated adsorption of the virus onto host cell.
- Pilus retraction pulls the virion to the host internal membrane to allow and subsequent interaction of g3p with the integral membrane protein TolA (coreceptor).
- The proteins of the capsid perform the injection of the viral DNA through bacterial membranes into cell cytoplasm.
- Viral (+) strand DNA enters cytoplasm.
- Host polymerase convert the (+) ssDNA viral genome into a covalently closed dsDNA called replicative form DNA (RF).
- Complementary (-) strand is synthesized by bacterial enzymes
6. �DNA Gyrase, a type II topoisomerase, acts on double-stranded DNA and catalyzes formation of negative supercoils in double-stranded DNA
7. Final product is parental replicative form (RF) DNA
8. dsDNA transcription by host RNA polymerase gives rise to viral mRNAs.
9. Viral g2p protein nicks RF DNA strand at the origin of replication.
10. (+) strand replication occurs by rolling circle.
11. �New (+) ssDNA genomes are converted into new RF molecules, and further transcription occurs.
12. A phage protein, pII, nicks the (+) strand in the RF
13. 3'-hydroxyl acts as a primer in the creation of new viral strand
14. pII circularizes displaced viral (+) strand DNA
15. Pool of progeny double-stranded RF molecules produced
16. Negative strand of RF is template of transcription
17. mRNAs are translated into the phage proteins
18. Phage proteins in the cytoplasm are pII, pX, and pV, and they are part of the replication process of DNA. The other phage proteins are synthesized and inserted into the cytoplasmic or outer membranes.
19. pV dimers bind newly synthesized single-stranded DNA and prevent conversion to RF DNA
20. ����RF DNA synthesis continues and amount of pV reaches critical concentration
21. DNA replication switches to synthesis of single-stranded (+) viral DNA
22. pV-DNA structures from about 800 nm long and 8 nm in diameter
23. pV-DNA complex is substrate in phage assembly reaction
24. When enough g5p protein is synthesized, conversion into RF dsDNA is inhibited, as neo-synthesized genomic ssDNA is covered with g5p.
25. g5p are replaced by g8p proteins to trigger the assembly of the viral capsid.
26. New virions are secreted from host cell.
27. Infected cells continue to divide and produce virions indefinitely.