T7 Phage DNA Replication:
T7 belongs to a group of odd phages such as T1, T3 and T5. The phages have an icosahedra head structures with a short tail, with a fiber. The genome size of T1, T3, and T7 is 40 kbp and that of T5 is 120 kbp. The genome of T7 phage is 39,936-7 bp, double stranded and linear. It has 160 bp long direct repeats at both ends. The genome is encoded with 56 genes with an ability to code for 55 proteins but only 44 have been identified so far. In this linear configuration, left end can be distinguished from the right end.

http://www.the scientist.com; http://www.uniroma2.it/
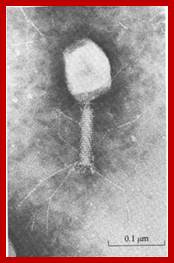
The electron micrograph of the virus (right side) shows the structural features of the T7 phage, where the elaborate neck and tail structures found in T4 Phages are missing (on the left side).
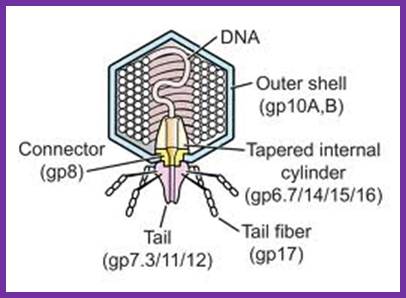
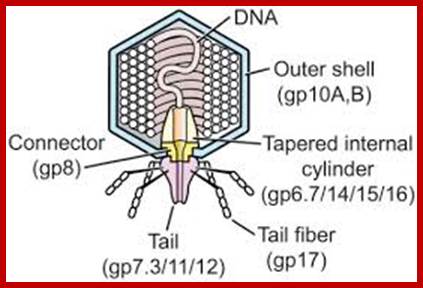
The structure of bacteriophage T7; The internal tapered cylinder and tail are shown attached to an icosahedral outer shell by a connector; http://www.biomedcentral.com/
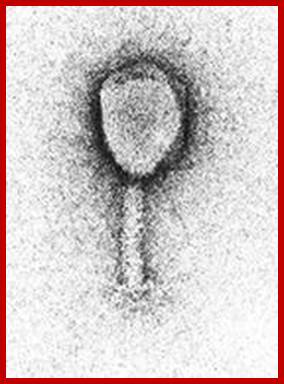
T1 phage https://www.mun.ca/
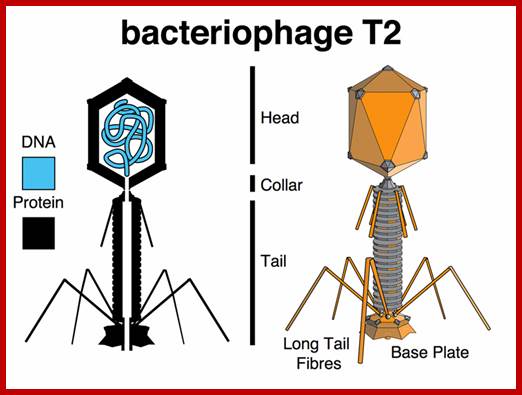
http://www.discoveryandinnovation.com/ttp://es-biotech.blogspot.in/
T2 phage; http://es-biotech.blogspot.in/
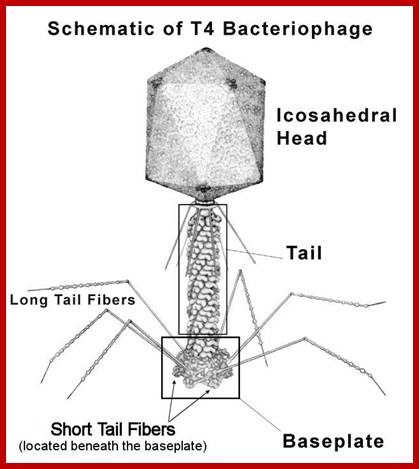
National Science foundation; https://www.nsf.gov;T4 phage
 https://www.nsf.gov
https://www.nsf.gov
T4 phage; http://www.phaget4.org/phage.html
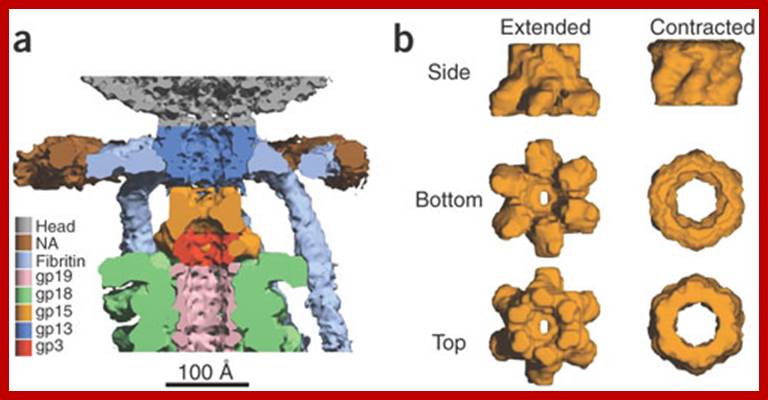
Serwer P, et al; https://openi.nlm.nih.gov
![]()
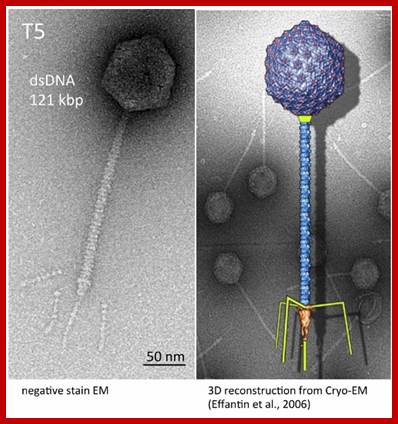
T5 phage;
http://www.i2bc.paris-saclay.fr/
The left region of the genome is injected into the bacterial cell in temporal fashion. The host RNA polymerase binds to the DNA for it has promoter elements like bacteria. There are three class of promoters- class I, II and III.
Transcription by host RNAP uses class-I type and ensue till it reaches rho dependent termination region at 1.3-1.4 mpu. The first phase of transcription accounts for 20% of the genome from the left end. This part of the genome doesn’t contain any restriction sites for the host REs (Restriction enzymes) to act.
The DNA end that enters first into the host cell during infection is the last part of that enters into the capsid at the time of phage assembly and maturation.
Class-1 Class-2 Class-3
5’AGG---II--------ori---I---------------I-----------------II---AGG3’
The genome is divided into 3 class of genes based on temporal expression of them. This is also based on the kind of promoters used and the kind of RNA polymerases employed for transcription.
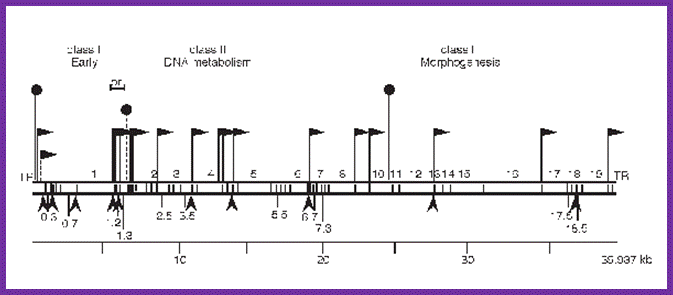
Bacteriophage genetic map: Mol.Biol
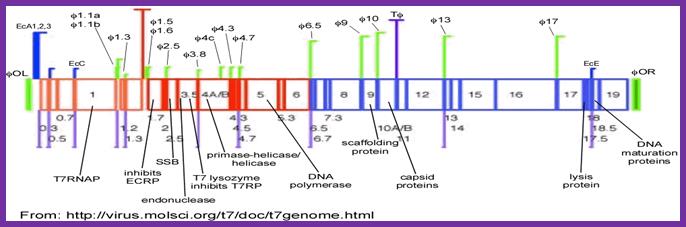
http:ww.lookfordiagnosis.com
This is the line diagram of the T7 genome showing class I promoter regions and Class II promoters. Host RNA pol transcribes using class I promoters (from 0.0 to 1.3mpu); from 1.3 to 7.3. Viral RNAps produced during earlier class I promoters transcribe the rest of the DNA.Mol.Biol
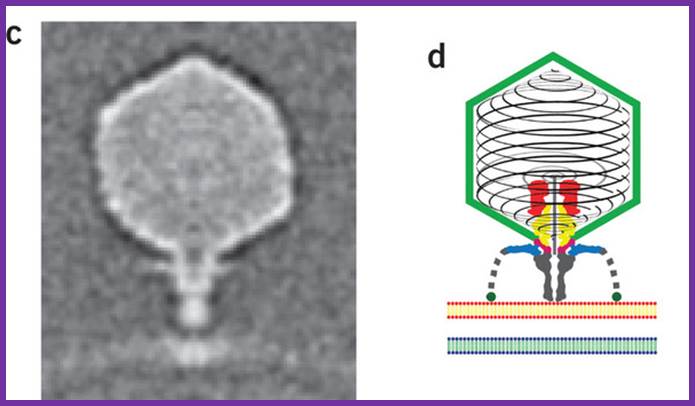
Podovirus P-SSP7 infects Prochlorococcus marinus, more or less similar to T7 viruses; Xiang liu et al; www.nature.com
First Phase:
In the first phase of infection, the phage binds to certain surface receptors facilitated by the tail fiber, then a channel is produced through which the left hand of the DNA containing early genes enters into the host cell. The DNA is injected slowly. This part contains few genes and using host RNA polymerase transcribes the same. The required promoter elements typical of E.coli are found at the extreme left end of the phage DNA.
Class I promoter:
--I -------(-35) TTGACA----------(-10) TATAAT------+1>>>
This part of the DNA that has entered (a part of the DNA) has no restriction sites for host restriction enzymes to act. The very early genes transcribed generate a transcript of 7000 ntds long and ends at Rho dependent termination at Ter region between 1.3 and 1.4 map positions. The transcript consists of cistrons separated by hairpin structures, which are cleaved by RNase III and release the individual cistrons for translation. In bacteria polycistronic RNA translated releasing the polypeptide chains individually. Here in the phage Individual cistrons are released.
http://www-personal.umich.edu/
The above diagram is simple representation of Transcription and translation process of a given gene.
The products are show in the fig. above are anti-restriction proteins (that inhibit host’s Eco-B and V restriction enzymes), a protein kinase (gp20) that inactivates host RNAPs by phosphorylating its B and B’ subunits. This segment also generates T7 RNAP and DNA ligase. Expression of these genes takes place with in first 6 minutes of infection. The proteins are called Class-I proteins and promoters as class-I promoters. Expression of T7 RNAP is important for the expression of rest of the genes, for the next part of the genome has promoters for T7 RNAP only.
0-----(-)-35—(-)10------------------------------ 1.0 -----1.3-Ter-1.4---I
III = Host RNAP-promoters.
1.0 mpu = T7 RNAP.
1.3-1.4 = Host RNAP terminator region.
The products of the genes transcribed at this (first) stage have very great impact on the production of phages. Among the five genes transcribed, the gene product RNAP is unique for the phage. It also produces a kinase (gp20) that inactivates host RNAP. Some products inhibit host restriction enzymes like E. coli-K and E. coli-B. As a result, they redirect the host’s cellular metabolism.
Second Phase:
As more DNA enters the cell, the DNA from mpu ~1.3 onwards contain promoters for T7 RNAP which are slightly different from host RNAP promoters and they are considered as week promoters. The consensus sequence of the class II promoter is as follows.
TAATCCG (-10) ACTCACTATA +1 GGGAG A>>>.
These promoters don’t have distinct regions like -35 and -10 sequences as found in host’s promoters. These promoters, nearly ten in numbers, are called class-II promoters and the products produced are also called class-II proteins. These are expressed in about 4-12 minutes of infection. These proteins are generally involved in DNA metabolism, i.e., DNA replication. One of the products acts as an inhibitor of host RNAP. The other proteins are DNA binding SSBs, Endonucleases, Lysozyme, Primase/Helicase, DNA polymerase and 5’Exonuclease. Phage endonucleases and 5’exonucleases degrade host’s DNA into individual nucleotides, which recycled into triphosopho ntds, which are used for the synthesis of viral DNA. SSBs provide stability for the separated DNA strands, primase provide the primers for viral DNA Pol (T& pol).
Third Phase:
At this phase, the genome is completely inside the host cell and the T7 RNAP uses class-III promoters and generates whole lot of transcripts for the production of structural proteins for head and tail. The sequences are compatible viral RNAPs. There are five such promoters and the consensus sequence is given below and transcripts produced are polycistronic
(-17) TAATACAACTCACTAT +1->AAGGAG (+6)
Transcripts generate proteins for head and tail and also for DNA maturation. Occasionally the T7 RNAP uses the class-II promoters and reads through till it reaches the end of the genome and produces a long polycistronic transcript of approximately 32000 ntds long, from 1.1 mpu onwards to the end of 20 mpu, which is processed by Ribonuclease III to release individual cistronic mRNAs for translation.
At this phase T7 DNA is replicated. As more and more number of dsDNA are produced, they are all in concatameric state. Such segments are used for transcription from class II and Class III promoters. Translation of them leads to the production of structural proteins for head and tail, but also proteins for capsid assembly and DNA maturation. Once the viral particles assemble T7 lysozyme causes cell lyses to release phage particles.
The genomic DNA is 36937 long and divided into 20 map units (mpu) like 1, 2 etc ending in 20 mpu. Class-I proteins are coded by the genes located from 0.1 to 1.3 mpu; Class-II proteins are coded by the genes located from 2.0 to 7.3 mpu and Class-III proteins are coded for rom genes located from 7.3 to the end of the DNA (20mpu).
Promoter for-I is located at the extreme left end of the genome and it is a tripartite structure and host RNA polymerase recognize it and its transcription is terminated at 1.3 in rho dependent manner. Viral T7-RNAPs start at phi 1.1A and extend up to phi 3.8
Replication Origin:
Replication origin is located in between 1.1b and ~1.3 mpu. It is located after two T7 RNAP promoters at AT-rich region.
First-promoter: +1>>-------5905; ---T7; Second promoter:
5905----TTAATACGACTCACTAT-CCTTAAGGTTTAACTTTAA’GACCC’TTAAGTGTTAATTAGATTTAAATTAAAGAATTACTAAG(5990)
-GAGAGGA—ATG—initiating site for 1.1 protein.
The GACCC is the primase binding site and the GAGAGGA is the Shine Delgarno sequence, ribosome binding sequence.
Origin:
The following segment forms the core of the Origin.
5’TTTAAGACCCTTAAGTGTTAATGAGATTTAAATTAAAGAATTACTAAGAGAGGACTTTAA3’
1.1b (A/T rich)1.1
Left 5’---------------I---I--------I-----I--------I=========I------II--
(61 bp)
The primary Origin of replication lies in between 1.1b to 1.1 mpu and it is AT rich.
On to the left of it there is a promoter for T7-RNAP at which transcription initiates and the same progresses into a noncoding region which happens to contain the origin sequences (AT rich); it opens into a bubble, and the same is used by replication components for initiating replication. The SSBs (viral products) bind to single strands and stabilize the ss DNA and the Primase (Gene 4 product) and Helicase (T7 product) binds to the lagging strand at each end of the fork. The primase lays primer for leading strands also. The Helicase moves at each end of the fork in 5—>3’ direction and unwinds the dsDNA ahead of the replication bubble. The replication bubble exhibits bidirectional movement in the beginning, but after the completion of the replication of left end of the genome, the replication will be unidirectional in the right part of the genome, one leading strand another lagging strand.
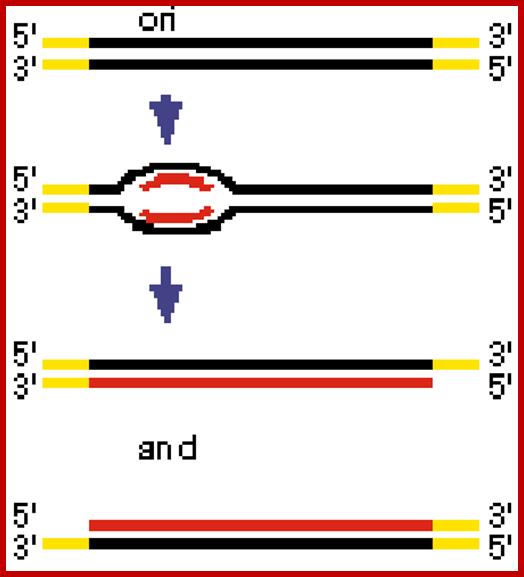
The replication origin is found at the left part of the T7 genome; initially the replication looks like bidirectional, but the left part of the genome is replicated early and the rest of the DNA replication appears as if it is unidirectional. mol.biol; http://www.kiveand.com/
http://iesbinef.educa.aragon.es/
T7 helicase is an Hexameric protein complex; DNAB Helicases: a family of Helicases; http://www.lookfordiagnosis.com/
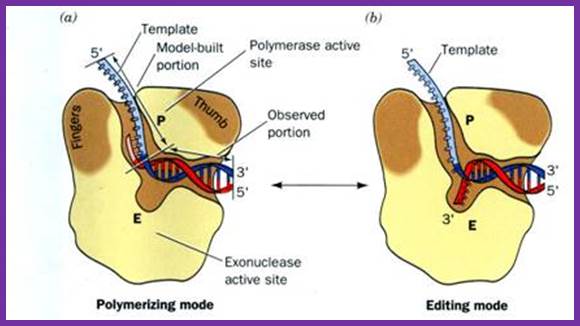
This provides structures for the assembly viral DNA polymerase whether host polymerase loading complex is required or not is unclear. The same enzyme as dimer at each fork operates, one on leading strand and the other on lagging strand is a question mark, but the replication is very efficient. The primers are removed and host DNA Pol-I fills the gaps and viral ligases join the nicks.
The Helicase-primase, bound to lagging strands, also uses specific sequences and generates four-nucleotide long (5’pppACCC3’ or 5’pppACCA3’) primers all along the length of the lagging strand as the replication complex progresses. These are used for synthesizing discontinuous strand.
The viral RNAP perse has cryptic RNAse-H activity, so it may nibble its own RNA transcript and provides a short piece of RNA, as a primer for continuous strand synthesis. Initially the replication is bi-directional. As the left part of the genome is hardly 20 %, its replication will be over in no time and the rest of genome is replicated as if it is unidirectional.
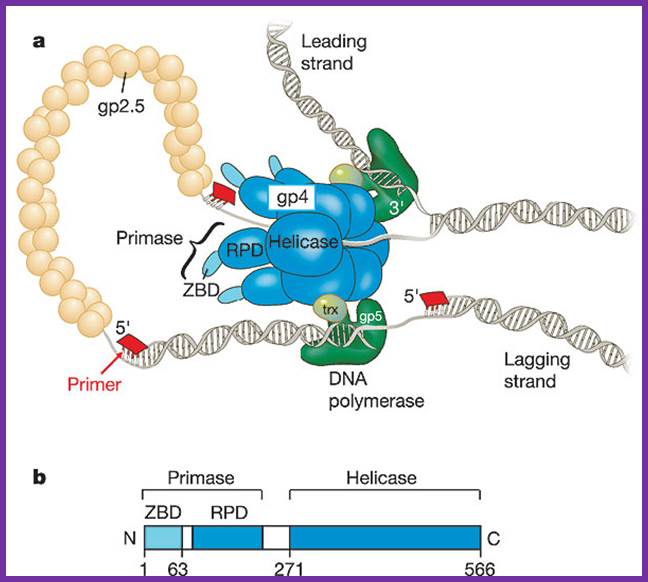
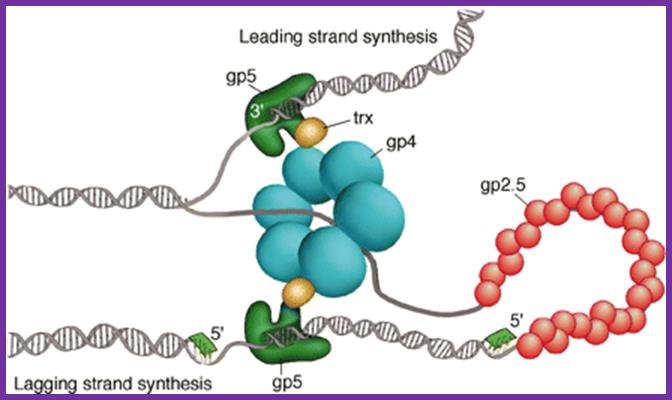
a, The bacteriophage T7 replisome consists of the hexameric T7 gene 4 protein (gp4) and two copies of the T7 DNA polymerase (T7 gene 5 protein (gp5) complexed with E. coli thioredoxin (trx)). T7 gene 2.5 protein (gp2.5), the ssDNA-binding protein, coats the transiently exposed ssDNA in the replication loop. b, Gp4 consists of a primase and a helicase domain, connected by a linker region. The primase domain consists of two subdomains: a zinc-binding domain (ZBD) and the RNA polymerase domain (RPD). DNA primase acts as a molecular brake in DNA replication; Jong-Bong Lee, Richard K. Hite, Samir M. Hamdan, X. Sunney Xie, Charles C. Richardson and Antoine M. van Oijen; https://bcmp.med.harvard.edu
T7DNA Polymerase (T7 Sequenase):
The Gp-5 with 98kd molecular mass is DNA polymerase and possesses 5’-3’ Pol activity, 3’-5’ exonuclease, but no 5’-3’ exonuclease activity. This protein is coupled to host 11.7 (13) KD theoredoxin in 1:1 ratio. This complexing makes it an efficient enzyme with high processivity and high fidelity. In combination with gp6 it exhibit properties similar to that of E.coli Pol-I, in having 5’-3’pol, 5’-3’exonuxlease, and 3’-5’ exonuclease activity. Purification of this enzyme without EDTA makes the enzyme to loose its 3’-5’ exonuclease activity. Selective oxidation of amino acids in the vicinity of iron binding sites also leads to the loss of 3’-5’ exonuclease activity. Iron is an essential component for T7-pol exonuclease activity. Mutants in gp5 lack 3’-5’ exonuclease property. Chemically modified T7 DNA polymerase does not discriminate between dideoxy-NTPs from dNTPs. Substituting Mn2+ for Mg2+ removes nucleotide bias.
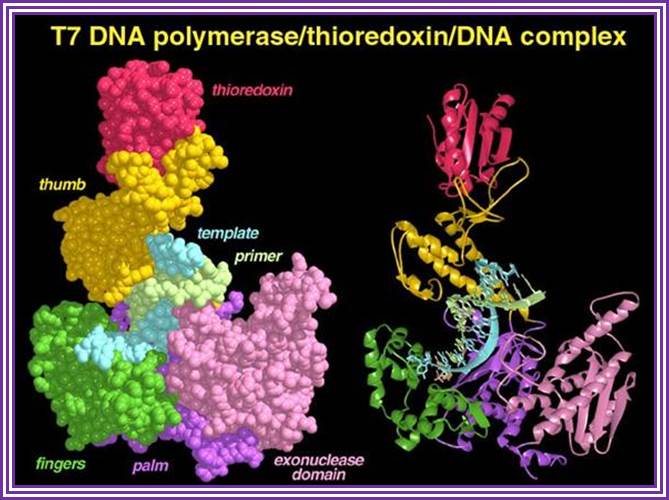
Omidconline.org; http://www.personal.psu.edu/
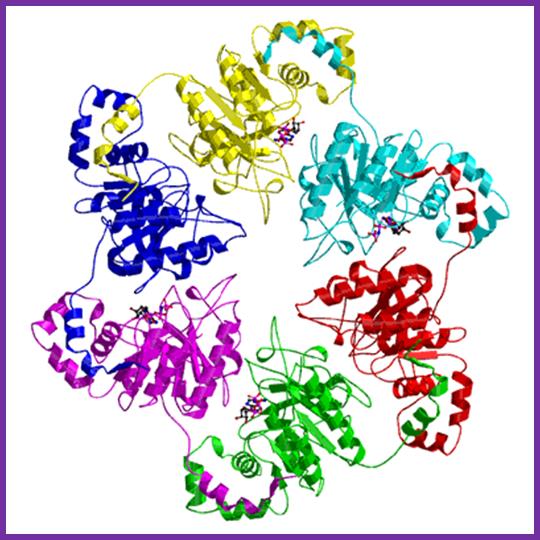
The above diagram is the 3-D model of T7 DNA polymerase; one ball stick model and the other is ribbon model showing the thioredoxin, bound DNA and the exonuclease domain property has made this enzyme so popular among molecular biologists; they love to use it in sequencing reactions. The modified enzyme is called Sequenase.
Host DNA-Pol-I removes primers and fills the gaps. Viral T7 DNA Ligases seals the ends. With the completion of the first round of replication, another cycle of replication is initiated and the process is repeated to generate substantial number of DNA molecules.
Fig.Model of the bacteriophage T7 replication fork. The T7 replisome consists of the DNA polymerase (gp5), the processivity factor E. coli thioredoxin (trx), the primase/helicase (gp4), and the ssDNA-binding protein (gp2.5). The gp5/trx complex synthesizes the leading strand continuously as the helicase unwinds the duplex. The lagging strand is synthesized as short Okazaki fragments. Synthesis of the lagging strand is initiated from RNA primers (green) catalyzed by the primase domain of gp4. A loop is formed on the lagging strand to align both gp5/trx complexes. Gp2.5 coats the ssDNA regions of the lagging strand generated as the helicase unwinds the DNA. https://bcmp.med.harvard.edu
However, the ends of each ds DNA gaps remain with single strands as tails (why?). The primers on 3’ of the parental template are removed by E. coli Pol-I, but there is no provision for filling up this gap so replication of Viral dsDNA leads to the generation sticky ends, but compatible; hence individual genomes with the compatible ends base pair and produce dimers or tetramers or concatemers.
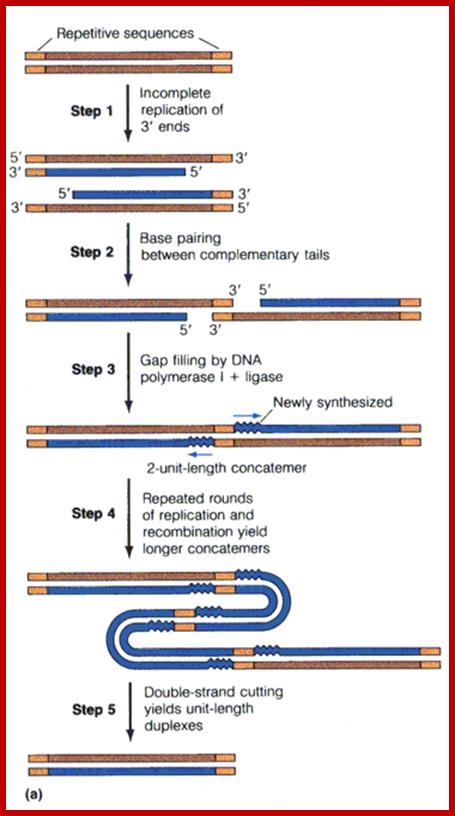
Replication produces Concatemeric DNA; http://www.keyword-suggestions.com/;
http://62.210.217.55/; http://deboj.club/topic/
As the replicated DNA ends have complementary over hanging tails, they get base paired and generate concatameric dimers or tetrameric or long concatameric fragments. These are used for packaging the DNA into preformed proviral capsids.
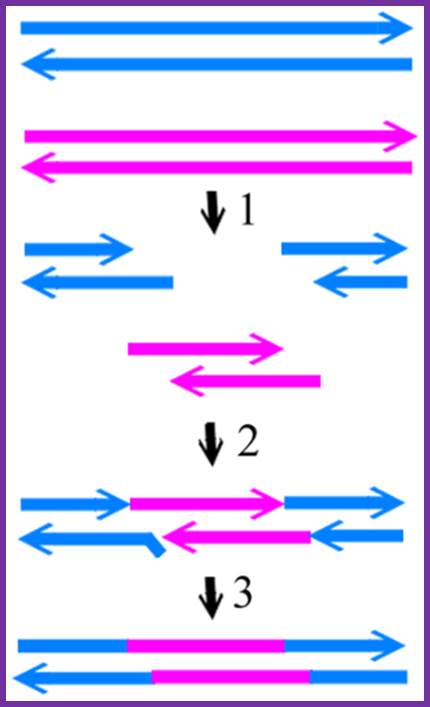
The mechanism of endonucleolytic cleavages at equivalent sites and strand annealing (CESA).
T7 and T3 DNA are shown in blue and pink, respectively. Arrows indicate the 3′ end. The steps are: 1, cleavage of T3 and T7 genomes by Endo I at equivalent sites; 2, annealing of the DSBs at both ends of T3 genome with the T7 DSB from the middle of the genome; 3, displacement and removal of the nonhomologous region, then filling the gap and ligation of the ends. http://www.plosone.org/
Out of 20 required proteins, seven are involved in replication and others have capsid building and one or two have regulatory roles; like gp2 has negative regulation over the expression of early genes.
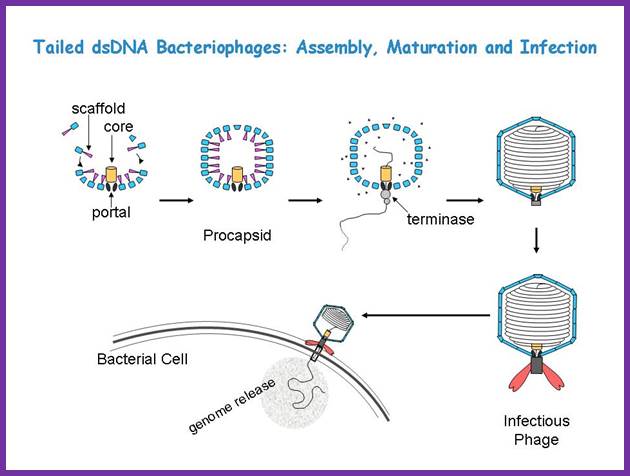
Though the above overall pathway is known for decades, the detailed mechanisms for the assembly, DNA packaing, and infection are still largely uncertain. We are interested in understanding these processes using a structural approach, primarily with cryo-EM imaging and 3-D reconstruction. Tailed ds Bacteriophages; Assembly, maturation and infection; http://jiang.bio.purdue.edu/
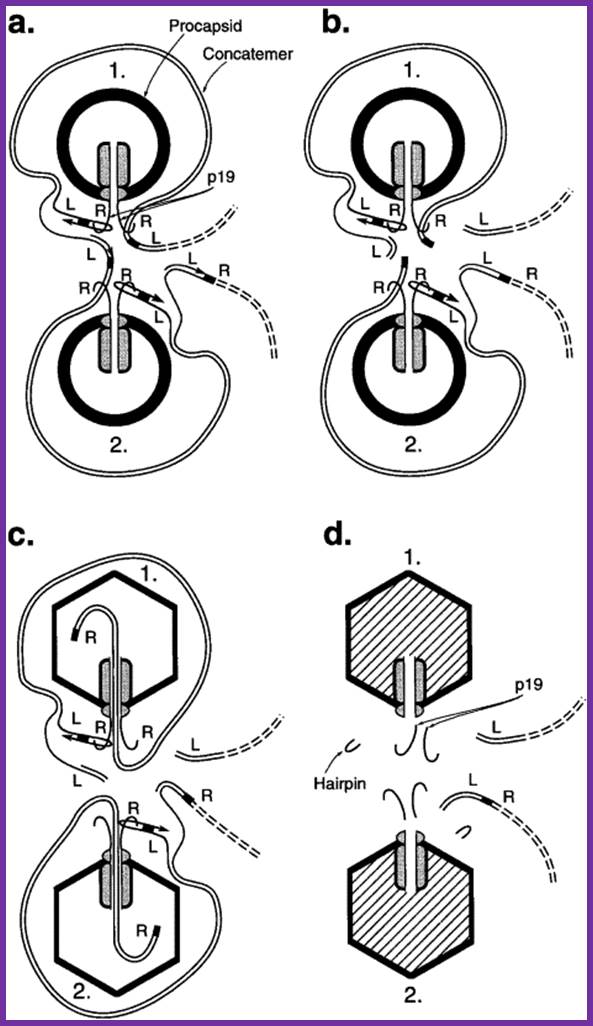
Packaging of Bacteriophage T7 DNA Concatemers in Vitro;
A hypothesized pathway that explains cooperativity during in vivo packaging of concatemer-associated T7 genomes. (a) A complex of a T7 concatemer and two procapsids (labeled 1 and 2) is shown. Each procapsid binds the concatemer at two identical sites. Each site is a previously described Pac B site ( Fujisawa and Hearing, 1994), one near the right end of each of two successive genomes in the concatemer. A hairpin-primed replicative branch duplicates the terminal repeat. The terminal repeat is indicated by filling the space between the two strands of a DNA double helix; an arrow within a DNA strand indicates replication of this strand. Each lobe observed by light microscopy corresponds to a capsid-associated loop of DNA drawn here. (b) After triggering that depends on formation of the complex in (a), the first of two cleavages occur to initiate DNA packaging. (c) A freshly cleaved right end enters while the procapsid, capsid I, converts to the larger, more angular capsid, capsid II. (d) DNA both finishes entry and undergoes a terminal cleavage near the hairpin formed by replication. L, left; R, right end of a T7 genome. The initiating cleavage of at least the first genome shown in (b) has previously been found to occur ( Sun et al., 1997) while the concatemer is part of a larger DNA network. In the figure, DNA network-associated initiating cleavages are not distinguished from initiating cleavages that occur after a concatemer is released from the DNA network. The events observed in Figure 3, Figure 4 and Figure 5 are both the entry of DNA and the terminating cleavage in (c) and (d). The complexes of Figure 3, Figure 4 and Figure 5 have additional capsids that, for simplicity, are not drawn here. In the pathway shown here, cleavage separates lobes before the separation is visible by light microscopy. In (c), end-first entry of DNA is drawn for simplicity. However, entry as a right end loop is equally compatible with the data. For visibility, the relative sizes of DNA, replicative branch, terminal repeat, and capsid have been altered. Mao Sun,Donna Louie, Philip Serwer; http://www.sciencedirect.com/
Note; Add on- some notes are taken from different authors and students update them.