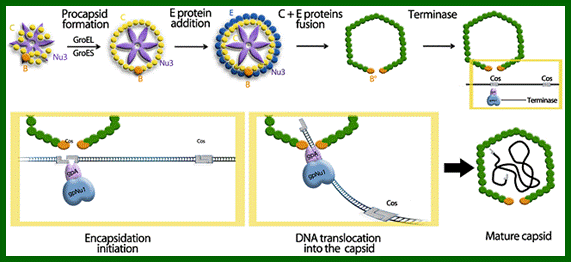
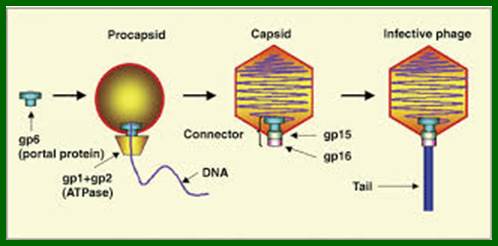
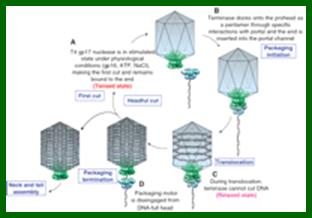
Lambda Phage DNA Replication:
Enterobacteria phage λ (lambda phage, coliphage λ) is a temperate bacteriophage that infects Escherichia coli.
It is a medium sized phage, double stranded and linear inside the viral capsid, but circularizes when it enters the bacterial cells. It belongs to a group of temperate E. coli phages; Lambda (48.5 kbp), P22 (43kbp), P2 (33kbp), P4 (11.5kbp), P1 (88kbp) and MU (38kbp). Some of the above said phage pictures have been shown below. It belongs to a Group I, dsDNA phages, Order-Caudovirales, Family-Siphoviridae, Genus- lambda like viruses and Species lambda phage.
Easter Lederberg discovered lambda phage in E. coli K12 strain in 1951. If the phage genome gets integrated into bacterial chromosome, it is called prophage. The phages that can go lytic mode or lysogenic mode are called temperate phages. The cell that possesses phage is called lysogen. Vegetative phase of the virus that lyses cells are called virulent. Prof. Delbruck (PhD in Physics) has done an extensive work on lambda phages for which he was awarded a Nobel Prize. Robert Edgar and William Wood have done invaluable studies in understanding the assembly of individual protein subunits into fully formed lambda phage.
Usually, a "lytic cycle" ensues, where the lambda DNA is replicated many times and the genes for head, tail and lysis proteins are expressed and assembled to generate fully formed phages and phages are released by host cell lyses.
Under certain conditions the phage DNA may integrate itself into the host cell chromosome in the lysogenic pathway. In this state, the λ DNA is called a prophage and stays resident within the host's genome without apparent harm to the host. Except for one gene all other phage (~48 genes?) genes remain repressed. The host can be termed a lysogen when a prophage is present. Only under Stress conditions like starvation, poisons (like antibiotics), or other factors that can damage or destroy the host; the prophage gets activated and it is excised from the DNA of the host cell by one of the newly expressed gene products and enters its lytic pathway.
A List of Temperate Phages:
|
Phage name |
Size of the genome and form |
Features |
|
Lambda |
48.5kbps, linear |
Showsspecialized transduction |
|
P22 |
43kbps,linear |
Salmonella-lysogenic |
|
P2 |
33kbp |
Tran activates with P4 |
|
P4 |
11.5 |
Satellite with P2 |
|
P1 |
88kbps |
Transduction can be maintained-plasmid state at low copy numbers. |
|
Mu |
38kbps |
Multiple integration sites, replication via transposition, invertible sequence |
|
Phi80, 21,434 and lambda7 |
|
They are called lambda -a phages |
|
|
|
|
|
|
|
|
https://en.wikibooks.org
://bioinfo2010.wordpress.com/

Podo virus
http://en.wikibooks.org/
https://commons.wikimedia.org
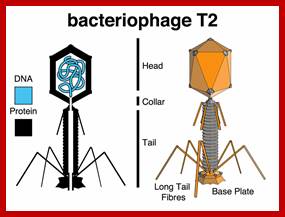
T2 phage EM picture; Jeff smith; http://www.discoveryandinnovation.com/
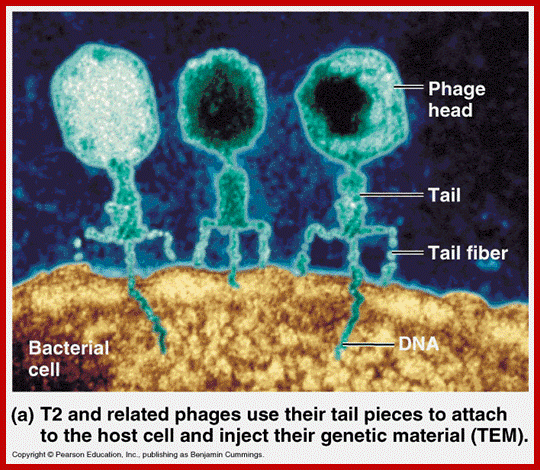
T4 phage EM picture; http://www.abovetopsecret.com/
P22 (T7) phage EM; https://www.asm.org
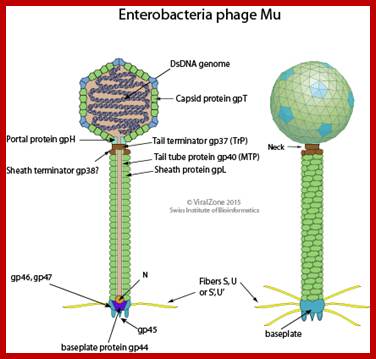
mu phage morphology; http://viralzone.expasy.org/
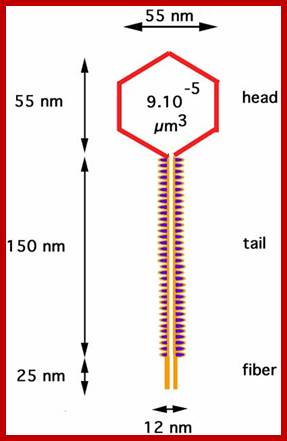
http://skiencestuff.blogspot.in/
http://genemol.org/genemol/lambda.html
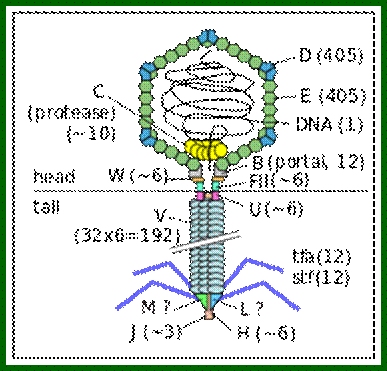
Bacteriophage lambda virion (schematic). Protein names and their copy numbers in the virion particle are shown. The presence of the L and M proteins in the virion is still unclear. http://en.wikipedia.org/https://commons.wikimedia.org
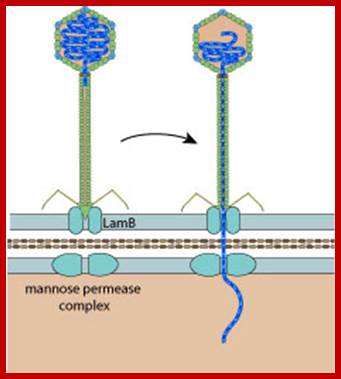
http://viralzone.expasy.org/
Virology: The gene weavers; Lambda DNA; http://www.nature.com/
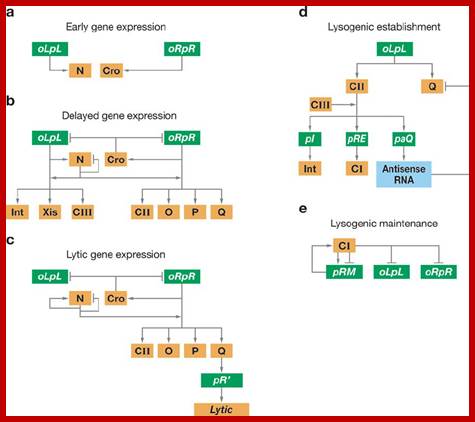
Description; Genetic network of the λ lytic and lysogenic; https://www.researchgate.net

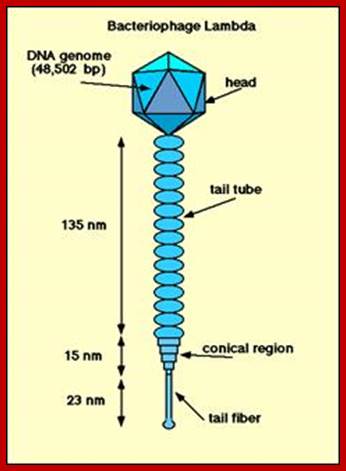
http://genemol.org/ and http://www.bioscripts.net/
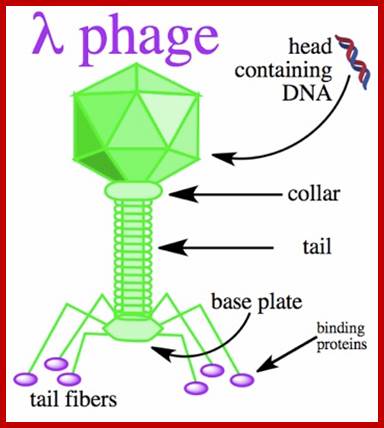
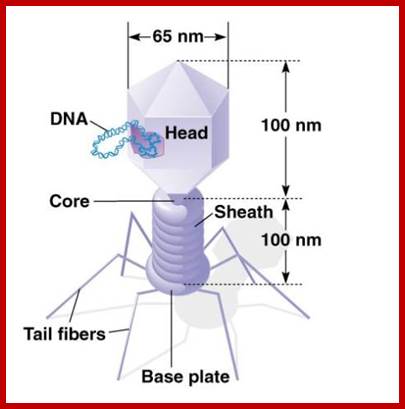
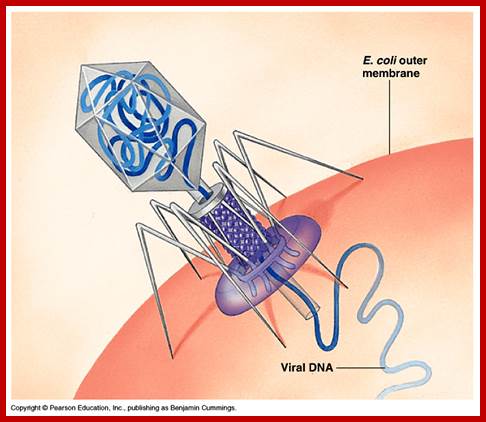
There are hundreds of strains of lambda virus. They have an isometric head (55nm, made up of gpE and gpD) and a tail tube 135nm long (gpV), it is attached to 23nm long tail fiber via a joint of 15nm long. It has a neck or connector made up of B, B*, C, F-II, and W proteins. Two proteins gpU and gpZ act as coupling the connector to the tail tube. The tail fiber is connected to tail tube by a group of proteins called gp G, H, L and M. A short fiber made up of ‘J’ is attached to the base of connector. Tail tube perse is made up of 32 rings of hexagonal gp V proteins. Detailed description has been given below.
The genome is ds linear DNA. Its genome size is 48502 bp to 48512 bp; there are some variations in genome size of different strains. It is linear, double stranded and its ends have 12 ntds long single stranded sticky tails, which are complementary to each other.
Infection: Infection:
The virus infects E. coli K12 host. The tip of the tail fiber J-protein contacts an outer membrane receptor/porin. This happens to be a host gene Lam-B product at the outer membrane, which is the maltose transporter called malto-porin and, it has regions to bind to terminal domain of the fiber, which is the product of gp-J. Binding of the fiber to the receptor activates its phospho-transferase domain (host p-I gene product), which is responsible for maltose transfer and the same transporter is also involved in the transfer of viral DNA which is in linear form, but packed inside phage capsid. Transport across the inner membrane is facilitated by sugar transport protein (ptsG).
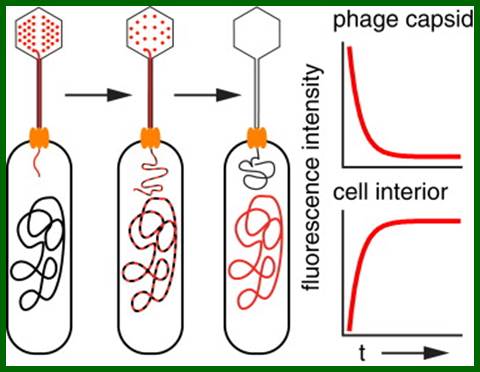
A Single-Molecule Hershey-Chase Experiment; Infection through Malto porin, David Van Valen5 , David Wu5 , Yi-Ju Chen,, Hannah Tuson , Paul Wiggins and , Rob Phillips; http://www.cell.com/
http://www.bio.utexas.edu/
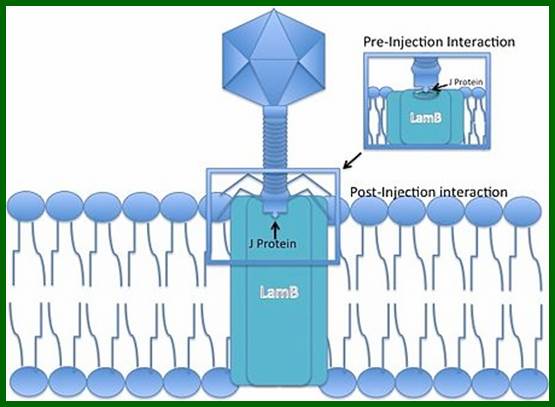
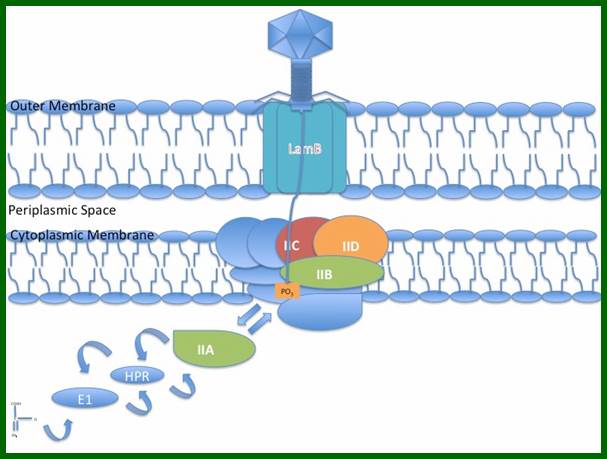
Lambda phage J protein interaction with the LamB porin; Lambda phage DNA injection into the cell membrane using Mannose PTS permease a sugar transporting system in the inner membrane, as a mechanism of entry into the cytoplasm; http://en.wikipedia.org/
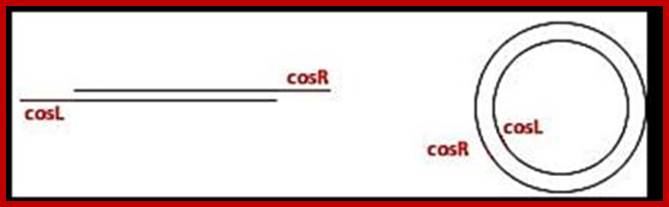
The viral DNA that enters the cell is the cos right (R2/Rz) end of the cosR and it is this end that enters the head last, similarly the end that enters the prohead first is cos left (nu2); it is the last to come out of the head at the time of infection. This is true for many viruses.
The gp-J facilitates the process, so it is called pilot protein. The gp-H located at the connector to tail fiber is speculated to enter into the cell along with the DNA? As soon as the linear DNA enters, its sticky tails base pair and then host NAD-dependent host DNA ligase ligates them. The circular DNA soon becomes supercoiled with the association of specific host proteins; super coiled state is an essential feature for most of its DNA activity. It is the ‘super structural model’ of any DNA that is endowed with regulatory activities.
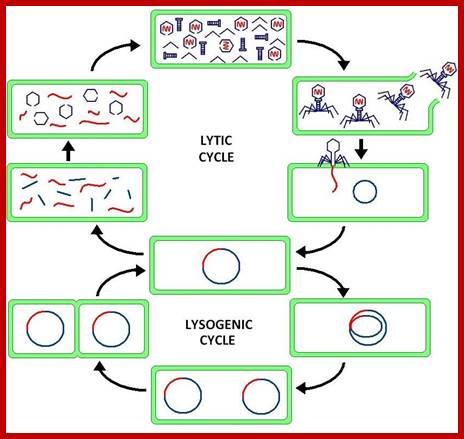
This line diagram depicts the events of lytic and lysogenic phases. http://en.wikipedia.org/

Lambda plaques; cleared areas are the regions where infected bacterial cells are killed-plaques; http://bio.classes.ucsc.edu/
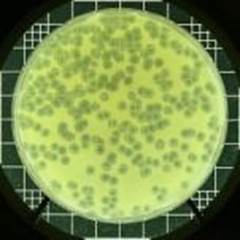
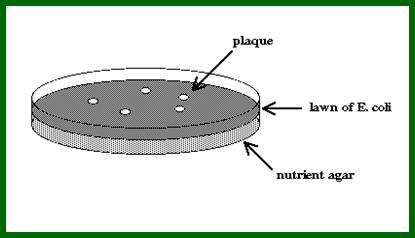
The clear areas in the plate are Plaques; http://faculty.plattsburgh.edu/
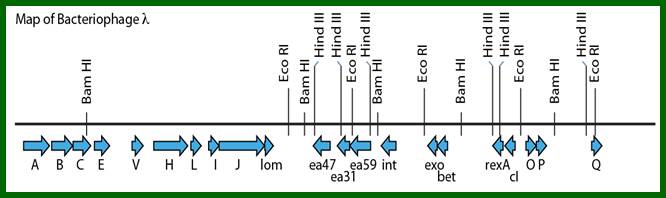
Linearized lambda DNA:
Cos (left)--I-Nu1-head-tail-I-A-W-B-C-E-FI-FII-//-S-R-R1z—cos(right)
![]()
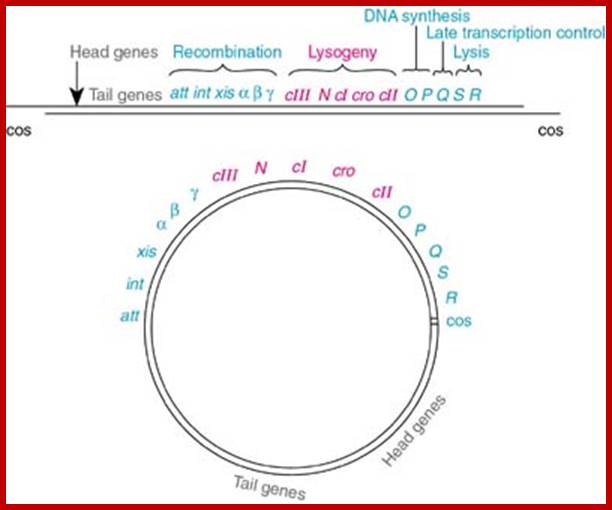
A map showing a group of genes involved in different functions, such as head and Tail, recombination, regulation, replication, and regulation II and Lysis.
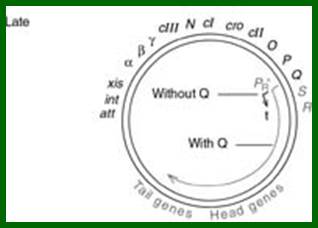
L.cos-Head-Tail-b-att-int-xis-aby-cIII-N-cI-cro-cII-OPQSR-R.cos
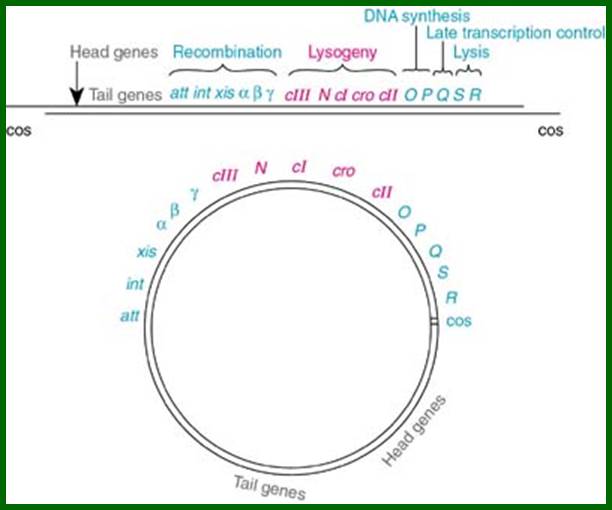
Diagram shows genome in circular mode, but cI has been positioned in the center for convenience to explain the events for it plays an important role in lysogeny or lytic functions. http://www1.nttu.edu.tw/
Restriction site mapping of lambda DNA; how the lambda DNA is protected against bacterial endo nucleases- (restriction enzymes?). http://courses.bio.indiana.edu/
Cos site:
It is the region where the lambda genome consists of single stranded sticky tails that get base paired to generate circular form of DNA inside the bacterial cell. When lambda infects the linear DNA that enters into the cell gets circularized by the base pairing of the cos sequences. Similarly during the packaging of the DNA into prohead the concatameric DNA is cut at this site to generate sticky tails.
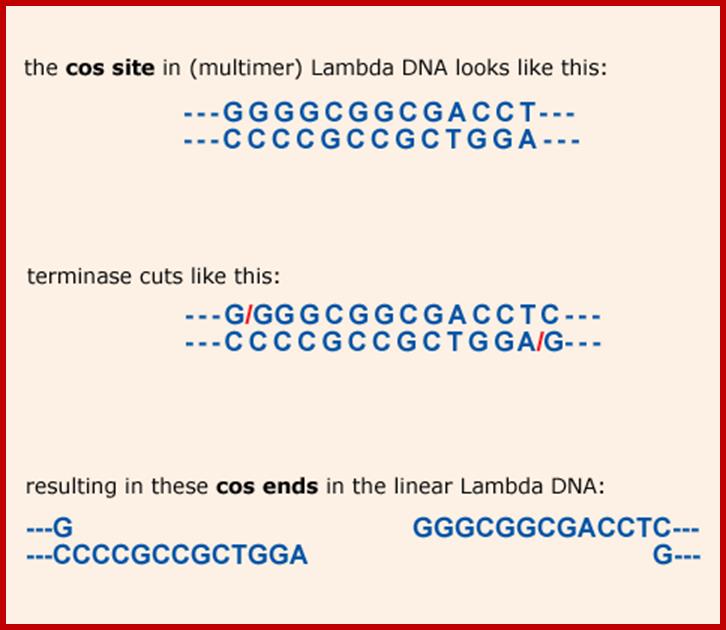
Cos site sequence: http://www.bioinformatics.nl/
L------- XXXXXX GGGCGGCGACCTXXXXXX---- R
R--------XXXXXXCCCGCCGCTGGA XXXXXX------l
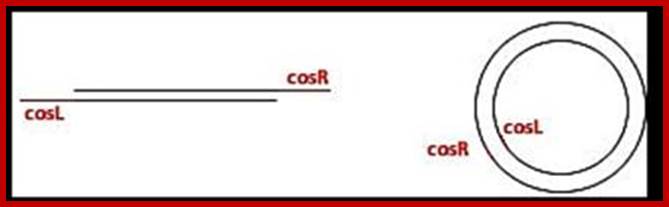
Sticky tails of cos in cos-L and cos-R region:
In circular module, the cos region, found in between Nu1 on the left side and Rz on the right side, consists of nearly 200bp, but it is subdivided into three regions based on their sequence and functions. From left end of the cos to the right end, it consists of sequences which are divided into- Left---cos-B---cos-N---cos-Q-cos-Right. Cos N has the palindromic sequences can be cleaved or joined.
left cosN --------><-----cosN right-
3’GGGCGGCGACCT------
--------CCCGCCGCTGGA3’
Cos N
Cos-B consists of R1-R2-I1-R3 sequences, gpU1 binds to R3, terminase binds to R1 and IHF (integration host factor) binds strongly to I1 found in between R2 and R3. The N-region is the site for terminase, nicking it generates 12 ntds long hangers. The cos-B region enters the head first and cos-Q on the other end enters the head last. The binding of IHF to DNA makes DNA to bend by 90° or more.
The ends can base pair complementarily to circularize, so the ends are called cohesive ends, so the name ‘cos’. The left end of the ‘cos’ of the genome has packaging sequences. Packaging of DNA into head capsid is initiated with the binding of Nu1 (Terminase-A) and IHF host factor. They are associated with a group of ATPase pumps which pump DNA into the prohead. Such energy dependent DNA pumping complexes are called ‘PACKASOMES perhaps consisting of multimeric motors of helicase-translocase family of proteins. Lambda genome contains about 50 cistrons; most of them have been more or less characterized but all of them are organized into polycistronic clusters. Nearly 48500 bp long DNA is packed into 55nm x 55nm sized head at the rate of 600bp s^-1 as compressed negatively coiled structure.
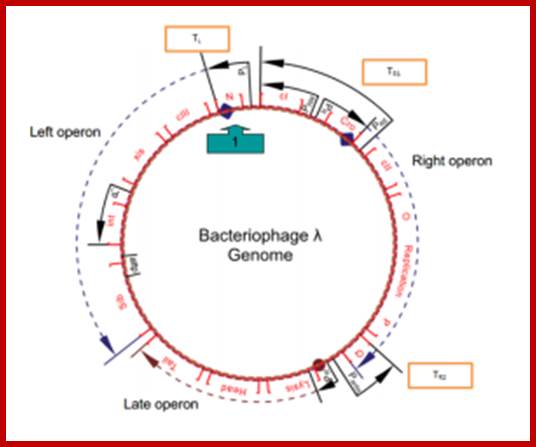
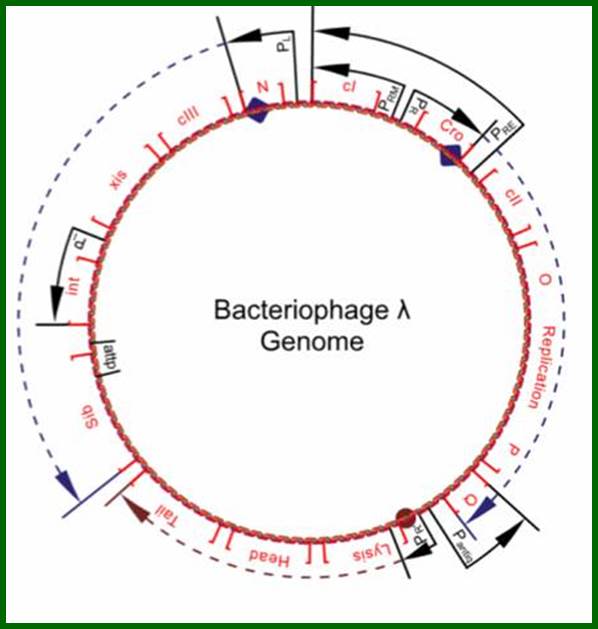
All the important promoters, terminators and genes are shown on this diagram. https://sciknowledge.wordpress.com
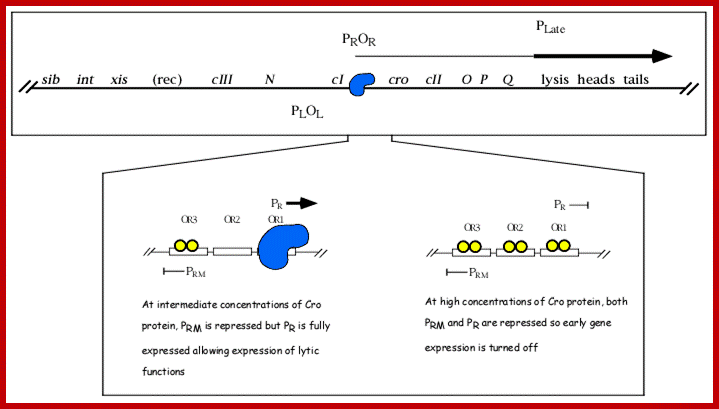
The above diagram drawn to scale (?) showing the positions of regulatory sites and genes and the arrows show the direction of transcription from their respective promoters. The O and P are the regions where the lambda DNA contain replication initiation site. Look at blue broken arrow from cro ends in PO; another broken blue arrow runs from N to the Blb. The third broken red arrow runs the start of Lysis ends in at the end of Tail. These arrows mentioned are transcription start and ends. Starts sites are in the form of respective blobs; one at N, second at Cro the third at lysis.
In circular module, the zero position in terms of map unit can be used for aligning the genes in an order. Most of the genes are organized into clusters, for temporal expression during its replicative lytic phase or to non replicative lysogenic phase. Regulation of the lytic and lysogenic phases provides a fascinating molecular expression, a par excellent paradigm to understand regulation of genetic modulation at molecular level.
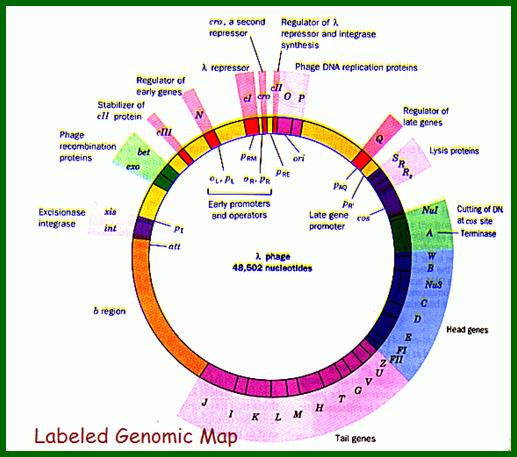
From this map one can identify 35 genes; labeled outside the circular map are genes and labeled inside are sites for promoters and operators. http://www.tulane.edu/
Phage Genome:
Left of cos- Mpu 0.0 -Nu1-A-W-B-NU3-C-D-E-F1-F2-Z-U-V-G-T-H-M-L-K-I-J----b---att-int-Xis<p---exo-beta-gamma-CIII-N<p-cI-p>--pRM-Cro--pRE-CII-O-P-Q>-<qap--pR’-S-R-Rz- mpu 100. Right of cos.
From gene Nu1 to gene F2 code for head proteins, from Z to J code for tail proteins, from Att to Xis code for recombination events, from N to Q regulation and immunity, from cro to Q regulation (O and P are involved DNA replication). Genes S, R and Rz involved in cell lyses. The Cos site found between Nu1 and R2/Rz is cut by the Terminase to generate linear molecule with sticky tails. The genes or the genomic segment found in between J and att-‘int’ is called ‘b’, is not important and this region is dispensable.
Cos = ~ 200 bp consists of-cosB (R1R2R3)—cosN—cosQ-
Cos B- initiates packing of DNA into head,
Cos N- nicking site to generate 12ntd sticky ends,
Cos Q- packaging ends.
Sticky tails of cos in cos-L and cos-R region: www.grosirbajusurabaya.top
Lambda DNA; this picture is little puzzling for there more three replication bubbles; https://www.biochem.wisc.edu/
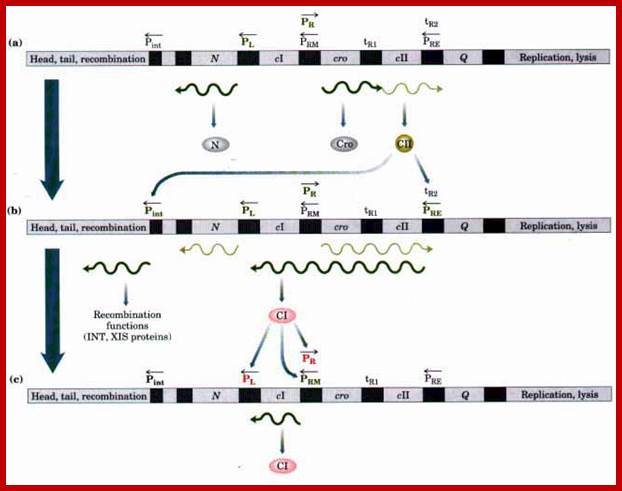
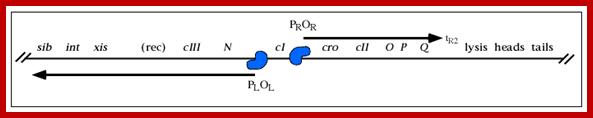
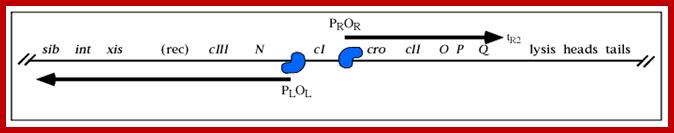
The line diagram below shows some important regulatory sites for early transcription, such as N, and Cro (on either side of ‘I’ or their promoter elements in specific direction; the diagram also shows the expression of gene for lysogenic or lytic pathways. Let us use cI (clear I) as the central point for our reference.
As soon as the DNA enters into the cell, the linear DNA circularized and get negatively supercoiled, hich opens up AT rich region; then a set of genes are transcribed, some very early, some delayed early and some late and some at the time of cell lyses. Expression of genes is all temporal and highly regulated. Lambda genes have promoters similar to bacteria. The host RNAP with sigma 70 binds to such promoters provided with the promoter regulatory elements are free from repressors. At certain promoters the RNAP requires activators for initiation of transcription.
The virus does not produce any polymerases or replicases or topoisomerases. It is an absolute parasite and completely depends on host components.
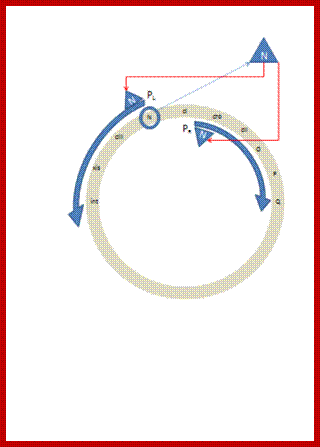
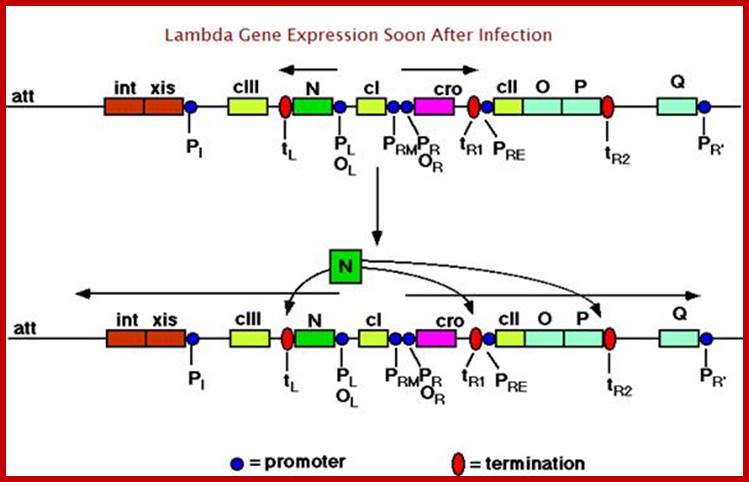
Early activation events involving N gene;Vhyr Qhan; https://commons.wikimedia.org
![]()
A map showing a group of genes involved in different functions, such as head and Tail, recombination, regulation, replication, regulation II and Lysis. Cos left- head and tail genes; cos right- lysis genes; www.slideplayer.com
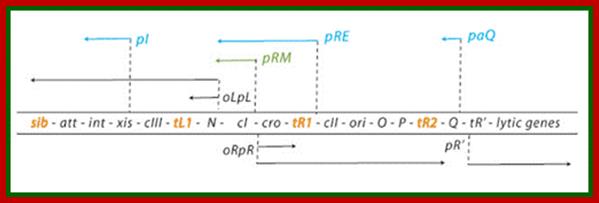
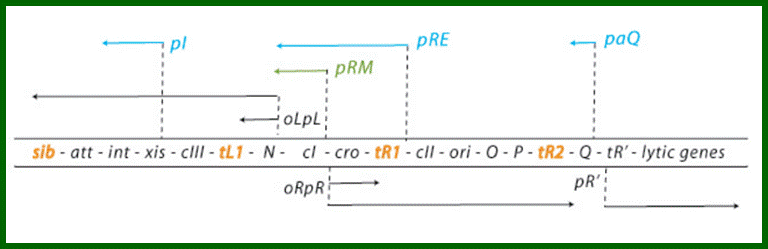
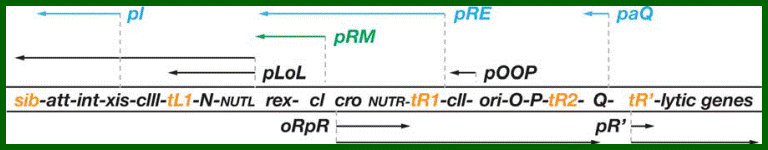
Regulatory genes of lambda phage;Gene and transcription map of λ. Genes are shown in the shaded rectangle. The early transcripts for pL and pR promoters are shown as red arrows. The late transcript from pR′ is indicated with black arrows. The CII-activated pI, pRE, and pAQ transcripts are indicated with blue arrows. The pRM transcript activated by CI is a green arrow. Transcription terminators (t) are shown as red letters among the genes. The tI terminator is indicated in parenthesis because it is contained within the larger sib processing site. The operators OL and OR where CI and Cro bind are shown next to the pL and pR promoters. http://2008.igem.org/
Left of cos- Mpu 0.0 -Nu1-A-W-B-NU3-C-D-E-F1-F2-Z-U-V-G-T-H-M-L-K-I-J----b---att-int-Xis<p---exo-beta-gamma-CIII-N-CI-Cro-CII-O-P-Q<p-S-R-R2- mpu 100. Right of cos.
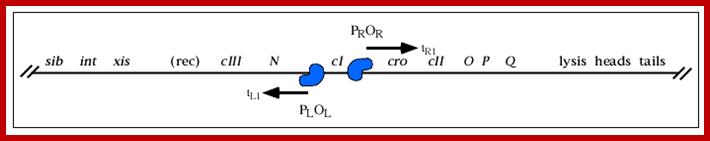
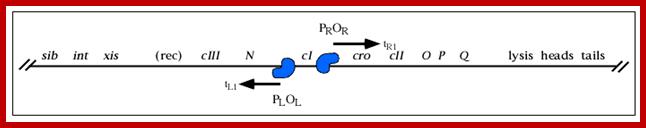
A part of the linear genome showing cI, promoters / operators on either side of it; cI is taken as the central gene for understanding the temporal gene expression of lambda. pLoL Promoter L for N gene and oL operator for N gene respectively. Similarly one finds pR and oR to the right of cI for cro gene.
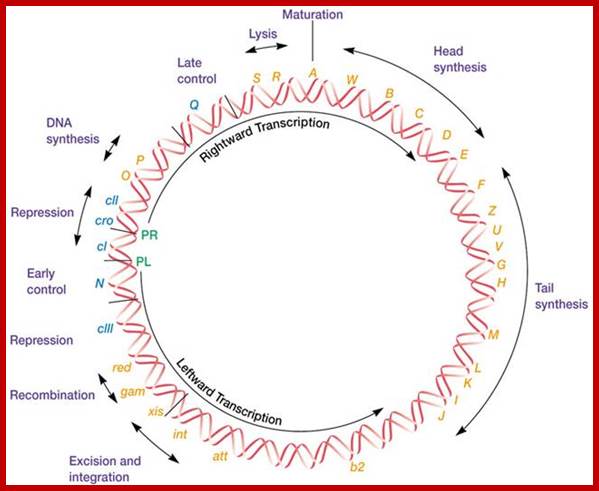
The Lambda Phage Genome. The direction of transcription and location of leftward and rightward promoters (PL and PR) are indicated on the inside of the map. The positions of major regulatory sites are shown by lines on the map and regulatory genes are in blue. Lambda DNA is double stranded, and transcription proceeds in opposite directions on opposite strands. http://210.44.48.210/
http://www.taq-dna.com/

Genomic map showing promoters and transcriptional start sites and the direction of transcription; PL and Pi left of cI; PRM and PR to the right of cI; PRE and Panti transcribe in left handed direction; PL, P’I, PRM, PR, PRE,Panti, P’R
Top fig; This is a lovely diagram showing all the segments of the genome coding for different proteins required for lytic and lysogenic evcIIents. It also shows sites for promoters and operators which are involved in regulation. Lower Fig; All the important promoters, terminators and genes are shown on this diagram. I highly recommend referring back to this when reading the text. sciknowledge.wordpress.; https://sciknowledge.wordpress.com/; http://www.taq-dna.com/
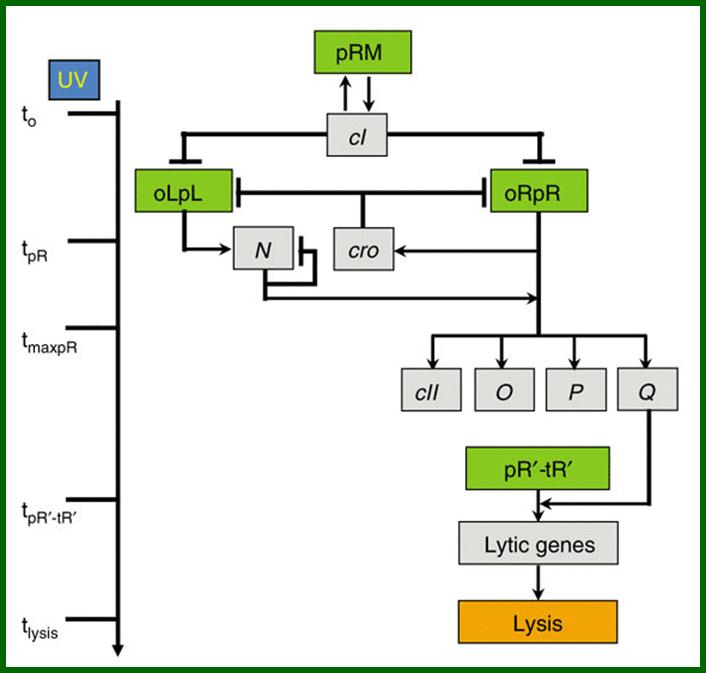
Noise in timing and precision of gene activities in a genetic cascade; Schematic model of lambda induction. Lambda promoters are colored in green and genes in gray. The lambda induction cascade is carried out in three stages. In the early stage, UV irradiation results in a decrease of CI levels, activating the pR and pL promoters at time tpR , leading to the expression of the cro and Ngenes. During the delayed early stage, RNAP is modified so as to override transcriptional terminators, allowing the continuation of both transcripts and the expression of CII, O, P and Q. During the late stage, the Q protein modifies RNAP, initiating transcription from the pR′ late promoter at time tpR′‐tR′, to become resistant to transcription terminators present downstream, and allowing the expression of late genes that encode proteins for phage morphogenesis and host cell lysis. During the late stage of the cascade, the late gene products assemble phage virions and lyse the host at time tlysis; Amnon Amir Oren Kobiler
Expression of Very Early Genes:
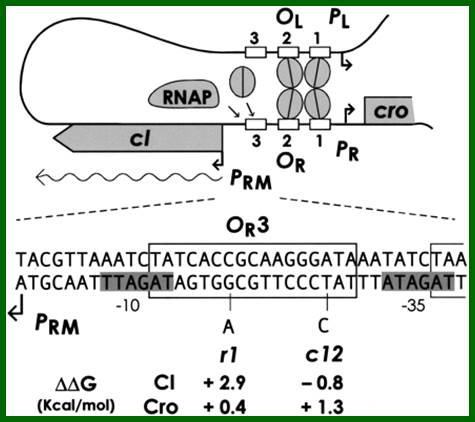
They are those expressed as soon as the DNA gets circularized and supercoiled. Host RNAP is used for initiating transcription. The genes transcribed are N from pL (promoter left of the cI ), i.e. leftwards of cI and cro (control of repressors and operators) from pR rightward of cI. N is an antiterminator and cro is a negative regulator. Promoter and operator elements are found on either side of cI gene. PL and OL (PL-OL1, OL2 and OL3) are found on the left side of cI gene and they transcribe N gene and beyond cIII; they are promoter-operator elements; similarly, one finds PR, OR (OR3, OR2 and OR1-PR) on the right side of the cI. They are operator/promoter elements for ‘cro’ gene. The successful expression of the genes on either side of the cI genes decides whether the lambda DNA goes into lytic or lysogenic cycle. But it depends upon cellular environ factors.
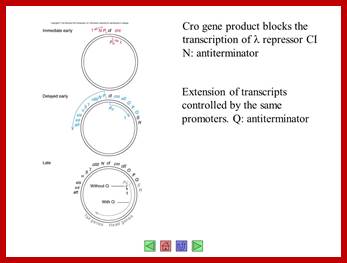

Very Early genes (within 5 minutes of infection) are transcribed from pL and pR coding for N and Cro respectively; N is an antiterminator and cro is a regulator gene. (O’ stands for operator to which repressor binds and P stands for promoter of a gene. The nut’L and nut’ R are anti- terminator sites to which antiterminator proteins bind. There is another promoter element pRM, it has different role, i.e. maintaining lysogeny, which will be explained later. Nut L and Nut R, are N protein (antiterminators) utilizing sites. While RNAP transcribing this region it recruits several host proteins (Nus) form a complex and skips termination sequences. The tR1 and tL1 are transcription termination sequences. http://slideplayer.com/
Expression of Delayed Early Genes:
This expression takes place at about 5-10 minutes after infection. Delayed early genes that can be expressed are cII right, replication genes (O and P) and another antiterminator called Q on the right of cI. The cIII and recombinase genes are on the left of N.
http://www1.nttu.edu.tw/

Genes left of cI
P/L for N and cIII, nut/L antiterminators for N. tLl transcription terminator for N; P1 promoter for Xis and int next to it is t1 (transcription terminator).

Genes-right to cI
PR promoter for cro, then it has nut/R anti terminator, then it has tR1. Next to it is P/RE then cII, O and P genes then t/R2. Next is another promoter P/aQ after Q there is another terminator tR3; at the end of Q gene there is another promoter that transcribes the opposite strand of the Q strand. PR promoter for right ward direction; tR1, tR2 transcription termination sites; t’R3- antitermination site.
PRE-Promoter for Repression Establishment.

PRE is the promoter for repression establishment with the expression of cI gene transcription. The DNA strand used for transcription is the opposite strand of cro gene template. Transcription succeeds to the end of cI gene and it is translated and cI protein i.e. repressor is produced. Once the repression is established, the PRM another promoter used for maintenance of repression by generating cI repressor protein. P/RE is the promoter for establishment of Lysogeny; P/RM is the promoter for cI expression and maintenance of Lysogeny.
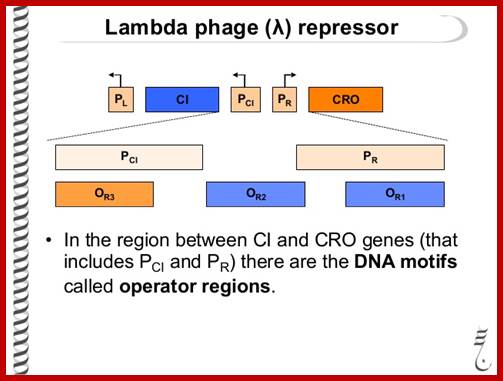
https://www.slideshare.net
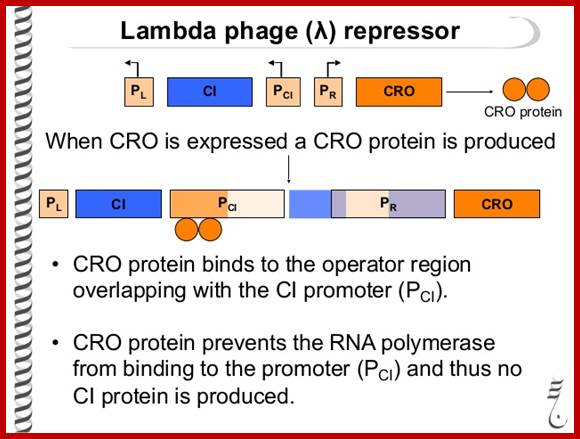
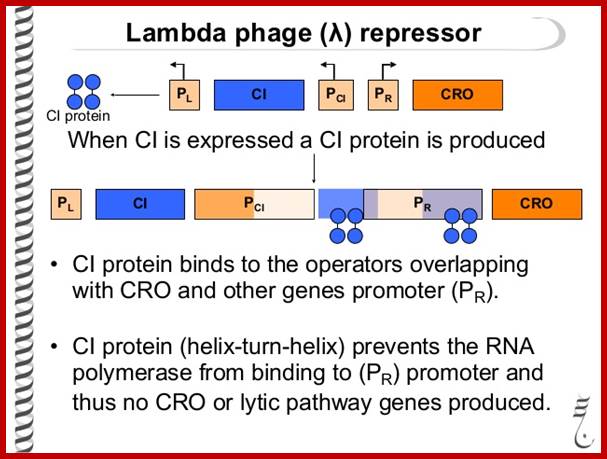
Phage Repressor; https://www.slideshare.net
Expression of Late genes:
From another promoter late promoter pR’ transcription leads to the production of 10 viral head constituting genes, 11 viral tail forming genes and 2 lyses genes. Anti terminator Q acts at t’R3 and transcription extends leading to synthesis of head and tail proteins. Lyses ends with the expression of gpS, gpR and gpRz.

Promoter PR’ is for late lytic phase. The ‘qut’ site is anti-terminator site for gpQ, this facilitates by passing tR’ and continues transcription up to the end of the genome.
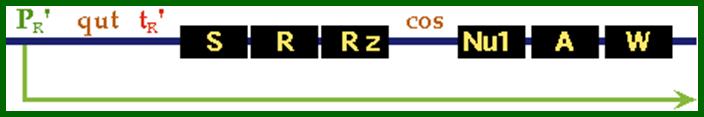
PR’ is the late promoter site used for the lytic phase and for the production of head and tail proteins. The qut is antitermination site for Q protein.
It is good to know about regulatory genes and structural genes, and their location in linear module than circular module of the genome. Regulation of gene expression during lytic cycle or lysogeny; use circular module of the genome. Let us be familiar with some important regulatory genes and their promoter-operator elements and transcription terminator regions. The understanding of this gives us the transcriptional start sites and termination sites and also gives us an understanding of transcript size and cistrons and their products.
Promoter/Operator elements:
Promoters are sequences to which bacterial RNAPs with their associated factors bind for initiating transcription. Promoter associated sequences are also involved in regulating the expression of genes. They can be operator elements or activator elements. The following is description of genes and the order of gene expression. The cI and Cro genes are used as central components for our understanding of lysogenic and lytic phases. They are regulatory genes and regulatory elements for the said gees are on either side of cI gene. The arrows show the direction of transcription.
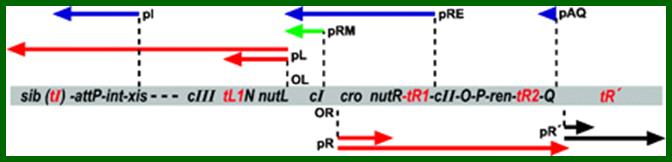
[-<-sib-att-t’iL’-int-Xist<p1-//-t’L’-N--- nutl.--<-pL-cI-]
[cI -pR>-cro>nutR-t’R1- -cII-O-P-t’R2--Q -t’R3-pR’->qut. tR’SRRz-cos->Nu1->A ->W.]
Rightward Transcription:
Rightward Transcription:
pR- cro—cII, O, P –Q,
pR’- S,R,Rz-nu1 A-W,
Leftward transcription using opposite strand of cro template;
cIII-N—<-pL
att-int xis <-p1,
cI-< -pRM,
cI ßpRE,
Q--<-paq,
In the linear order of genes shown above, the sites of all promoters are shown. Symbol ‘p’ represents promoter elements, such as pL for N gene (antiterminator) and promoter pI for Integrase to the left. The transcription of pRM is leftwards, pR for the right side, pRE leftwards, paQ-leftwards and p’R2 to the right. In our description, the cI gene is used as reference point.
The promoter elements have -10 TATAAT and -35 TTGACA sequences similar to that of bacterial promoters. Some variations are found in pRM and pRE. The enzyme is bacterial RNAP and the activator factor is the same sig70.
PRM—Repressor maintenance:
--<-(-10) TTAAATC-A_T--(-)35)-TATAGATT for leftward transcription.
|
PROMOTER Promoter PRM |
SEQUENCE
<-10-TTAAATC-A_T--(-)35)-TATAGATT for leftward transcription.
< TATAGATT---13-- TTAAATC |
|
PRE |
< TTGC GTTTGT TTGC <-- 13 --> AAGTAT |
|
PI |
< TTGC GTGTAA TTGC <-- 13 --> TGTACT |
|
PaQ |
< TTGC GAGCAC TTGC <-- 13 --> TAGTAT |
|
Conserved CII binding sites are shown in red; -35 sequences are in green; -10 sequences are in blue. The transcripts generated from pRM are used for the transcription of cI (not stable), Transcript produced form pRE that is anti-strand to cro generates cI protein. The transcript from paQ produced is an anti-strand for Q and inhibit Q production and stops production of head, tail and other genes. Transcript from p1 generates xis and int required for phage DNA integration. |
|
The pR is the promoter for cro (rightward transcription), pRM pRE is the promoter for cI (leftward transcription) and pRE is the promoter for cI leftwards assisted by cII gene product. The pRE- lacks consensus sequence at -35 and -10 region, so it is a poor promoter, but binding of cII makes it efficient in initiating transcription. Interestingly the same sequence in the opposite strand is used by cII for initiating transcription in opposite direction from pRE transcribing ‘cro’ opposite strand and full length cI gene.
Among the seven promoter elements pL and pR play very important roles. The two promoter elements are also associated with regulator-operator elements such as operator oL and oR located on either sides of cI.
Operator elements:
Operator elements represent the sequences for the binding of repressors which prevent the binding of RNAP to induce transcription. There are only two such operator site one at poL and the other at poR; located on either ends of cI gene. RNAP binding sites in these promoters often overlap.
----pL-oL1,2,3---------cI-------oR-pR-oR-1,2,3
-N--<-pL-o1L-o2L-o3L--cI--o3R-<pRM-o2R-o1R-pR>-cro>pRE-cII-
Operator element oL is subdivided into <oL1, oL2and oL3 of the N-gene. Similarly, oR is subdivided into oR3, oR2 and oR1> cro, in the same sequence towards right side of cI for cro. Each of these consists of two ~16-17bp half sites, to which repressors or activators bind.
Most of the operator elements found on either side of cI have split sequences or called half sites-TACCTCTG-G-CGGTGATA. These sequences are recognized by cI repressor (a dimeric protein) or activators/RNAPs.
Transcription termination sites:
Transcription termination sites are represented by ‘t’, i.e ‘tL’ to the left and ‘tR’ to the right of cI
-<tL1-N-NutL<-pL-oL--cI—oR-pR>—nutR-tR1>-cro->cII—tR2>—Q->-qut->t’R>-S-R>; note nutL and nutR are N utilizing sites (N is early antiterminator), similarly qut is Q utilizing sites (Q is late antiterminator). Transcription terminator sites have specific sequences for Rho dependent and Rho independent termination. Each of the sequences generates a specific stem loop structure. These are used by N and Q antiterminator proteins.
Transcription starts sites and their termination sites and the direction of the transcript and the products:
L stands for leftward transcription from the reference point cI and R stands for the rightward direction from the reference point cI. The ‘t’ denotes possible transcription termination region. pL for antiterminator gene N. pI for expression of xis and Int genes. pR for expression of cro gene. pRM means promoter for repression maintenance. pRE means promoter for repression establishment. pQ refers to promoter for Q an antiterminator. Then pR’ for lysis genes.
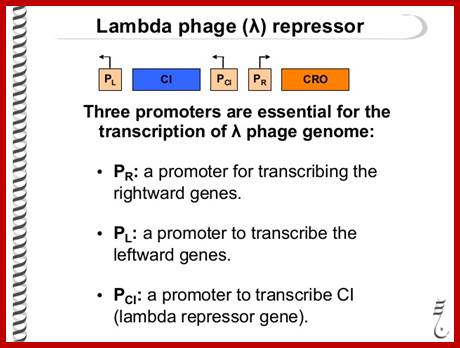
There are 7 promoters that are active at different stages of the bacteriophage lambda life cycles and which govern expression of bacteriophage lambda.
- PR expresses the replication genes as well as the anti-repressor, Cro; the transcriptional activator CII, and the anti-terminator Q protein.
- PL expresses anti-terminator, N, and the CIII protein.
- PR' expresses the lysis proteins, and the head and tail proteins.
- PRE expresses the repressor gene, cI, to establish lysogeny.
- PRM expresses the repressor gene, cI, to maintain lysogeny.
- PI expresses the int gene to synthesize the Integrase protein.
- PQ antiterminator leads to the expression of Head and tail genes
- PaQ drives synthesis of a short anti-sense RNA which blocks translation of Q gene mRNA.
pL>N>-NutL->—cIII-Xis- -tl1; the transcript is called L1.
p1 >>Int->att-sib; The transcript is called L2.
pR >cro—NutR-->tr1 , the transcript is called R1.
pR> cro> -cII-O-P-tr2; and Q the transcript is called R2.
p’R >>> Q >>t’R3, is called R3
p’R >> qut>> tR’ the transcript is called R4.
P’R >> - S-R late genes-the transcript is called R5
-cI--<pRM, the transcript is cI; late expression, it produces a repressor.
-pR > cro > cII >-O-P—tR2; generates cII
-cI-<--< pRE; the transcript antisense to cro > cI repressor
Depending upon the start site and termination sites, transcripts are named as L1, L2, R1, R2, R3, R4 and R5
L1: from pL to Lt1 (early transcripts)
-tL1<-------N-----<pL—cI— left of cI,
L2: from pL to b (delayed early transcripts)
-b<—<-att--int--xis--cIII—<-----N-----<pL—cI—left of cI
R1: from pR to tR1 (early transcripts),
--cI---pR>--cro---> >tR1- right of cI
cI: transcript- from pRM to the end of cI -leftward transcription from pRM to the end of cI.
---<--------cI-------<pRM
R2: from pR to tR2 (delayed early transcripts),
--cI---pR>---cro---cII>---O-P>-> right of cI, produces cro and cII * very important,
---pRE----cII—>O-P –tR2 right ward generates cII.
oR-<-----cI---<-cro<-nutR-<---pRE, leftward transcription from pRE, generates transcripts anti to cro and pro cI.
R3: from pR to tR3 (little more delayed transcripts),
--cI—pR->--cro---cII--->->-O—P—Q-->- transcription to the right of cI.
R4: from p’R to t’R,
--p’R->---qut t’R
R5: from p’R to head and tail genes.
--p’R-->>---qut--->>>-->> >S—R--> late genes.
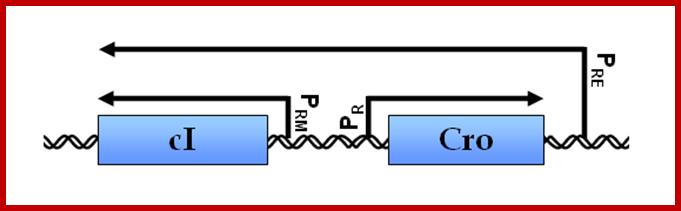
There are two major promoter elements, which are located on either side of c-I gene (cI = is a repressor protein Gene); they are called p-L and p-R (p= promoter, pL promoter left and pR=promoter Right). These Lambda promoters are prominent and host RNAPs recognizes the same provided they are free.

Discovery:
Regulation by cis-antisense RNA transcripts was first postulated in 1972 based on work in bacteriophage λ gene regulation. cI and cro are two essential genes coding for transcription inhibitors whose coding sequence lie adjacent to each other but in opposite directions. An alternative promoter for cI was discovered to be on the other side of cro, and initial studies confirmed the presence of cis-antisense cro RNA transcripts. This led the authors to hypothesize that this novel antisense RNA transcript might potentially serve a role in regulating cro gene activity (Spiegelman, 1972). The functional significance of cis-NATs was not validated until a decade later when an antisense RNA, RNA I, was found to regulate the copy number of plasmid ColE1 by the inhibiting the maturation of a primer essential for DNA replication.
http://mcmanuslab.ucsf.edu/Lambda gene transcription at cI


Genes left of cI;
P/L for N and cIII, nut/L antiterminators for N. tLl transcription terminator for N; P1 promoter for Xis and int next to it is t1 (transcription terminator). And
Genes-right to cI
PR promoter for cro, then it has nut/R anti terminator, then it has tR1. Next to it is P/RE then cII, O and P genes then t/R2. Next is another promoter P/aQ after Q there is another terminator tR3; at the end of Q gene there is another promoter that transcribes the opposite strand of the Q strand. PR promoter for right ward direction; tR1, tR2 transcription termination sites; t’R3- antitermination site. http://mcmanuslab.ucsf.edu/Lambda gene transcription at cI
In most of the situations host RNAPs with sigma70 initiate transcription at both promoters. The one that starts at p-L transcribes the N gene, if it is not terminated at terminator sites nut-L; all the genes beyond N are transcribed and possibly end in cIII and beyond.
The other transcription initiates at p-R, if allowed by passing respective terminator sites, transcribes all the way up to Q, promoter for Q and the transcription leads to all the genes up to the end of J gene.
However the transcription of these genes mentioned at the earlier stages is not continuous, but in pieces and regulated by antiterminator products at their respective termination sites. If transcription succeeds in the very early stages and delayed early genes, the transcripts produced, towards left and right of cI, unhindered decides to go for lytic. If the early transcription leads to the production cI repressor by the activity of cII, it leads to lysogenic state. Most of the transcripts are polycistronic. On translation generate individual protein subunits.
Fate of the viral DNA depends upon the host conditions and the success of early transcripts and its translated products. The viral DNA can go towards Lytic phase or Lysogenic phase; depends on its environmental conditions. Each of the phases has equal chances but under favorable conditions most of the time lytic pathway predominates (cyclic AMP absent). It is also possible if the conditions are unfavorable (cAMP present), the phage can enter into lysogenic pathway. Once lysogeny is executed, it will be maintained by auto regulation.
Successful expression cI results in lysogenic phase and the sustained expression of Cro leads to lytic phase.
In lytic phase the DNA successfully replicates, produces all the required proteins and generates 100 or more viral particles with in about 22-30 minutes of infection, and then viral particles are released by cell lyses. This happens when conditions are favorable and cells are happy to be killed.
The most critical components, that initiate either lytic pathway or lysogenic pathway, are cro, cI, cII, cIII and antiterminator products like N and Q and nutritional conditions. The critical events that can go either way is simplified in the following diagram.
Lytic phase:
When the phage DNA enters the cell, it gets circularized by ligating the 12nts long complementary sequences. Then it has two options either go into lytic (vegetative growth) or lysogenic state. When conditions are favorable, promoters and operator regions of pL/oL and oR/pR on either side of the cI gene are free. This facilitates the binding of host RNAP with its sig 70 to their respective promoters and initiates transcription from pL (to the left of cI) towards N gene and from pR (to the right of cI gene) towards Cro gene.
<pI-tL1--N--<pL-oL1-oL2-oL3--cI--oR3 pRM < -oR2-oR1-pR>-cro>tR1 >>
The N gene and Cro genes have transcription terminator sites at their ends. But the N gene product can act as an antiterminator at transcription termination sites such as tL1 and tR1 and tR2.
Promoter cro -35 TTGACT -----10 GATAAT- >—cro
-N--- 10 gATAAT ---35 ACAGTT-- Promoter for N gene
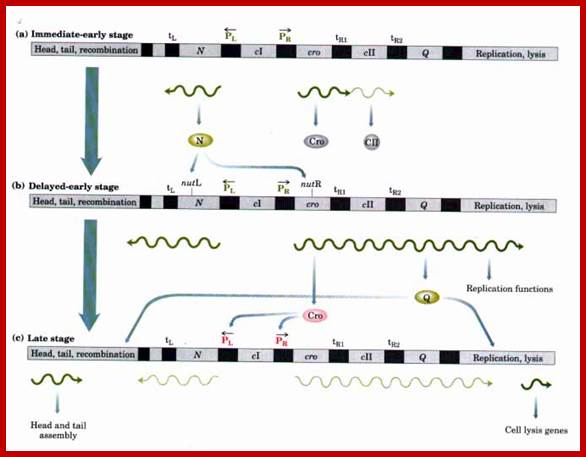
Regulation of gene expression in the lytic pathway of bacteriophage λ: An abbreviated map of bacteriophage λ is shown; key genes and regulatory sites involved in the lytic pathway are highlighted. Note that this map is not drawn to scale; the regulatory region is disproportionately large. Temporal regulation of gene expression is accomplished in a cascade, in which one gene product produced at each stage is required to stimulate gene expression in the next stage. The key gene products are the N protein, produced from an immediate-early gene (a), and the Q protein, produced from a delayed-early gene (b). The N protein stimulates (b) delayed-early and the Q protein stimulates (c) late gene expression. The promoters PL and PR are critical for immediate-early and delayed-early transcription. Other regulatory sites, including the transcription terminators (tRl, tR2, tL), are described in the text. (c) The stimulation of late gene expression by Q protein is complemented by repression of PR and PL transcripts by the Cro protein. The action of CII protein is described in Fig. 27-27. Components involved in activation, as well as new mRNAs synthesized at each stage, are shown in green; components involved in repression are in red. http://www.bioinfo.org.cn/
Regulation of gene expression in the lysogenic pathway of bacteriophage λ. (a) RNA transcripts initiated at PR continue through tRiabout 50% of the time, resulting in synthesis of CII protein as an early gene product. (b) The CII protein activates transcription at PRE and Pint, resulting in production of the CI protein (a repressor) and proteins required for recombination. (c) The CI repressor is an antagonist of the Cro repressor (see Fig. 27-28), and it effectively shuts down bacteriophage gene expression except from PRM. Early production of CII and then CI in sufficient amounts tips the balance toward lysogeny rather than lysis. Continued cI expression maintains the λ genome in the dormant state. (Red and green are used as in Fig. ;http://www.bioinfo.org.cn/
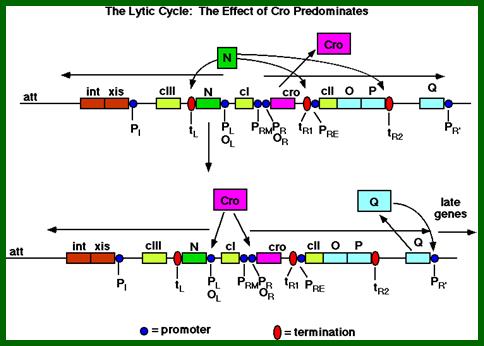
The above diagram illustrates the activity of several genes resulting in lytic pathway. The diagram shows expanded structural features of lytic pathway. In this cro gene expression and degradation of cII facilitate the expression of O and P genes and progress at Q and transcription from PR’ and beyond leads to lytic phase. http://www.escience.ws/
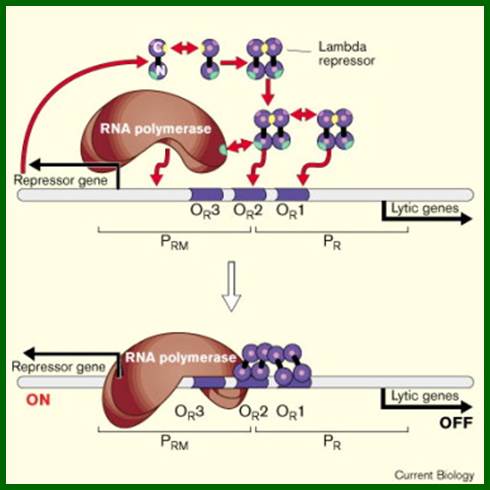
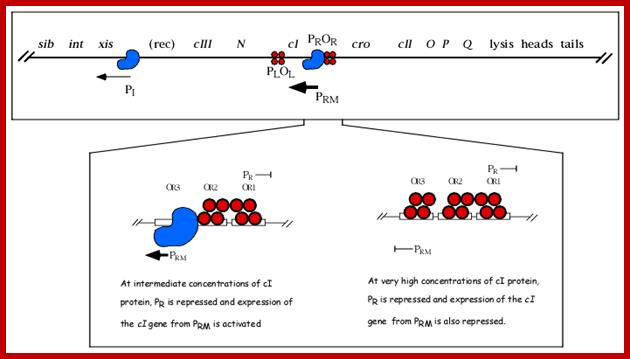
As the cro product is produced, it binds to O.R3 operator elements, recruits more RNAPs and activates more transcription of Cro and beyond. When cro is produced in sufficient numbers it binds oR3 and shuts off transcription of cI from pRM. But it can also act as an activator of its own transcription so more cro is produced. This transcription assisted by Q product leads to the transcription of head, tail and lytic genes.
Lambda's 'genetic switch' responds in a dramatic all-or-none fashion to an environmental signal, activating transcription of certain genes as it turns off others. The switch illustrates principal features of many biological regulatory processes.; http://www.nature.com
If the transcription of N gene continues, the N product bypasses the ‘ter’ sites at tL1, tR1 and tR2 leading to the transcription of cII, O, P and Q genes. The Q gene product is also another antiterminator. The Q product acts as an antiterminator at t’R, this leads to the transcription onwards. Genes O and P products are required to initiate replication of lambda DNA. Replication of the genome is very essential for Lytic phase.
However, the transcription of cII from pR, the cII acts as a regulator for the transcription of cI and the genes from promoter p’I left of cIII gene. In this situation cI is also transcribed, from pRM which is located at the left end of oR2, but the transcript is not translated efficiently for the lack of proper leader sequence, so hardly any molecules of cI are produced.
The cII to act as a regulator for it, it has to be rendered stable. The cII is a very unstable protein. Host gene products such as hlF1 and hlf2 act on cII and degrade, thus incapacitate cII activating the transcription of cI gene.
Note that pRM and pR on the right side of cI, have bidirectional promoter elements and they use opposite strands.
Anti-termination sites of N and Q are shown. http://www.escience.ws/
Transcription beyond N and transcribing cIII is not very effective in inducing lysogeny, for cII is more or less degraded as fast as it is synthesized. The action of cII is required at p’I to transcribe xis and int genes and onwards for they are required for integration and lysogeny. The product of cIII gene is required for cII to be stabilized at PRE for transcription of cI to establish lysogeny.
Delayed early gene transcription leading to the expression of gpO and gpP is essential to initiate DNA replication. The replication of DNA and transcription of the above said genes provide all the inputs for the production phage proteins for assembly of phage particles. When viral particles are produced, transcripts from p’R lead to the production of enzymes for cell lyses, thus lytic phase are achieved. As and when the phage particles are released they bind to neighboring host cells and infect and cause lyses. The colonies produced are called plaques and they are very clear. Within 50 to 60 minutes of infection 100 to 150 phages are released (very high titer), in our experiments we got the titer of 50 to 60.
Lysogeny: Establishment and Maintenance:
The diagram below is a simple illustration of establishment of lysogeny. Lysogeny normally initiates when conditions are unfavorable i.e. nutritional deficiency which induces the synthesis of more and more cAMP which is known to be global regulator of gene expression.
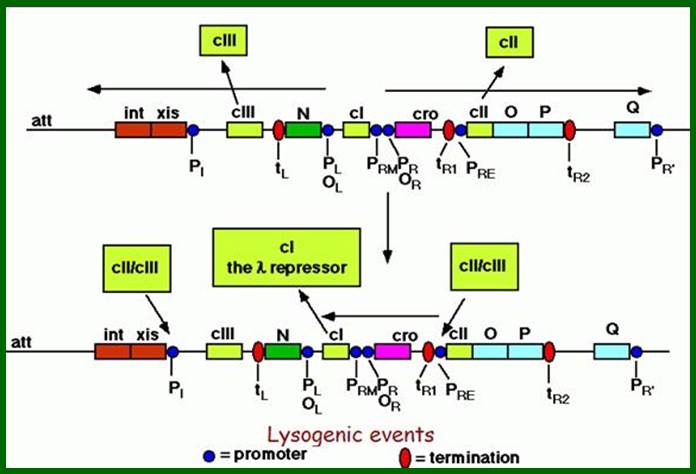
On either side of cI genes there are promoter operator segments called PL,oL3,oL2,oL3 same order, on to the left and on to the right are PR, oR1, oR2, o3 in the same order. Rest of the promoters and nuts and transcriptional terminators are the same as explained earlier. http://www.escience.ws/
Promoters to the left (pL) of cI- are for N gene and P1 promoter after cIII for xis and int genes. Promoters to the right of cI are PRM and PR . pRM overlaps O3and O2. The PR is for cro gene and onwards. After tR1 there is a promoter called pRE to the left of cII for cI gene; transcription from pRE leftwards and uses opposite strand of cro, generates an mRNA (with long leader sequence) for cI. The PR’ to the right of Q is for head and tail and lyses genes. Successful expression of cII and cIII aided by cAMP facilitates the expression of cI from PRE.
|
PRE |
< TTGC GTTTGT TTGC <-- 13 --> AAGTAT |
The same cIII and cII also uses paQ from the middle of the Q gene and transcribes p1 of Xis and int and other left6 of it. The transcript produced from paQ is anti-strand to pQ.
|
PaQ |
< TTGC GAGCAC TTGC <-- 13 --> TAGTAT |
The cI product dimers bind to operators’ elements oR and operator elements oL and block cro and N genes respectively. Blocking oR3 prevents cro expression and oL prevents the expression of N. Later once the lysogeny is initiated cI is expressed from PRM, leads to the maintenance of lysogeny.
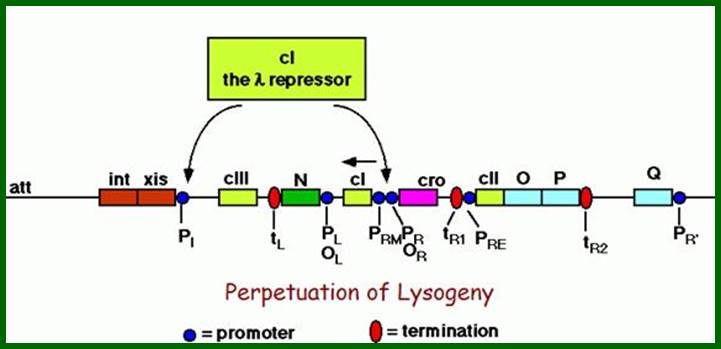
Genetic map and transcriptional units of the phage regulatory region: Key genes and signals discussed in the text are shown in their map order between the parallel lines. The early transcripts, the extended delayed early transcripts, and the late transcripts are shown in black arrows. The transcripts initiated from the pI, pRE, and paQ that are required for lysogeny are shown in blue. The pRM promoter, which is activated by the CI regulator, is required for maintenance of the lysogenic state, which is shown in green.
Critical transcription terminators are marked in orange, including the sib region containing the tI terminator. Leftward promoters are indicated above and the rightward ones below the map. pL and pR
are the early promoters and pR is the late lytic promoter. The role of the pOOP promoter is not fully understood. The operators oL and oR, cognate to pL and pR respectively, are also shown. The immunity module of the λ chromosome encompasses pLoL, rex, cI, oRpR, and cro. ori is the origin of O- and P-mediated phage DNA replication (Table 1). Int carries the site-specific recombination reaction, and Int and Xis support the excision reaction. http://2008.igem.org/
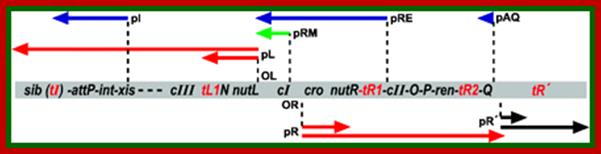
Gene and transcription map of λ. Genes are shown in the shaded rectangle. The early transcripts for pL and pR promoters are shown as red arrows. The late transcript from pR′ is indicated with black arrows. The CII-activated pI, pRE, and pAQ transcripts are indicated with blue arrows. The pRM transcript activated by CI is a green arrow. Transcription terminators (t) are shown as red letters among the genes. The tI terminator is indicated in parenthesis because it is contained within the larger sib processing site. The operators OL and OR where CI and Cro bind are shown next to the pL and pR promoters. . http://2008.igem.org; http://mcmanuslab.ucsf.edu/
Gene expression from pL and pR takes place early. pL produces antiterminator protein N that acts at tL1 and also at tR1 at the end of cro and tR2 at end of O and P (replication genes); and the the N gene continues to transcribe into cIII. And xis and ‘int’ are also transcribed de novo from the p1 promoter (they are responsible for integrating the lambda DNA into host cell DNA at specific site by site specific recombination process). At tR1, the N acts and the transcription continue beyond cro gene, cII and enter O and P and terminate at tR2. Translation of O and P looks very week.
The N protein by-passing the tR1 and transcribing cII gene is critical for lysogeny. The cII is a very unstable protein; its half-life is less than one minute. In normal conditions, it is degraded by cellular hflA and hflB gene products. Mutants in hfl genes lead to high lysogeny.
Though cI gene is transcribed from (if the operators are free) pRM, the mRNA for cI does not contain proper leader sequence for efficient translation. This renders transcription of cI is of no consequence.
Under nutritional unfavorable conditions cAMP is produced which destabilizes hflA and haflB gene products. Thus, the cII is not degraded. More to it, as cIII (left of N) is also produced because antiterminator factor promotes transcription beyond N and into the gene cIII. The c III stabilizes cII. Now the cII/cIII binds to pRE promoter region recruits RNAP and transcribes in leftward direction and transcribes opposite strand of cro and cI in opposite direction.
|
PRE |
< --TTGC GTTTGT TTGC <-- 13 --> AAGTAT |
(Note how the same dsDNA sequences are used to transcribe both the strands in opposite directions and produce different proteins which have different functions). This is an excellent example for bidirectional promoter cum transcription. As it uses the opposite strand, the early part of the transcript is anti sense to cro. If any cro mRNA is produced, it is blocked by antisense RNA. But the cI transcript produced is efficiently translated for it has a good leader sequence and good ORF. As cI, 27KD protein accumulates; it binds to operator’s oR1 as dimer with great affinity and then binds to oR2 and oR3 with decreasing affinity by protein-protein cooperative interaction.
This cI repressor protein (27kDa) also binds to oL1, oL2, and oL3, thus block the transcription of N and other genes beyond N. The protein cI repressor’s N-domain contains helix turn helix motifs but its 2 and 3 helixes bind to DNA major grooves. The N domain and C-domain are linked by a narrow groove like linker. The repressor binds to operator as dimers to half sites of operator elements. As cI binds strongly to oR1 and co- operatively to o2 and o3 operators repress transcription of cro.
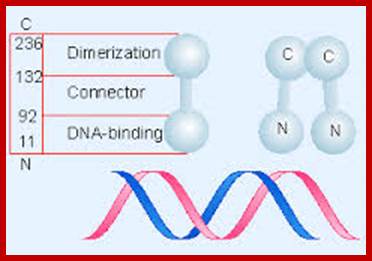
http://flylib.com/: The DNA binding repressor domain for dimerization and DNA binding domains; repressor 27kDa.
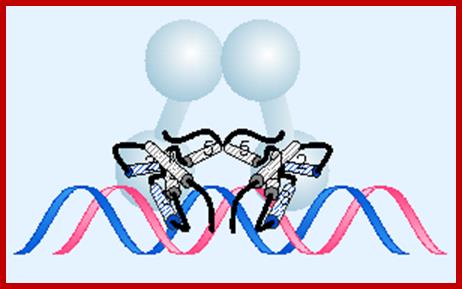
DNA binding dimer helix model for the protein bind to DNA; http://flylib.com/books
The cI protein has dimerization and tetramrization domains. It also contains cleavage site in the middle and has an activator motif in the DNA binding domain.
When oR3 site is free, it can be occupied by RNAP and it can be activated by the cI repressor bound at oR2 to produce more cI proteins, which keeps the lysogeny on. The production of cI is synergistically activated by the interaction of cAMP-CAP-with CTD tail of RNAP makes the RNAP to be active transcription of cI gene from pRM promoter element.
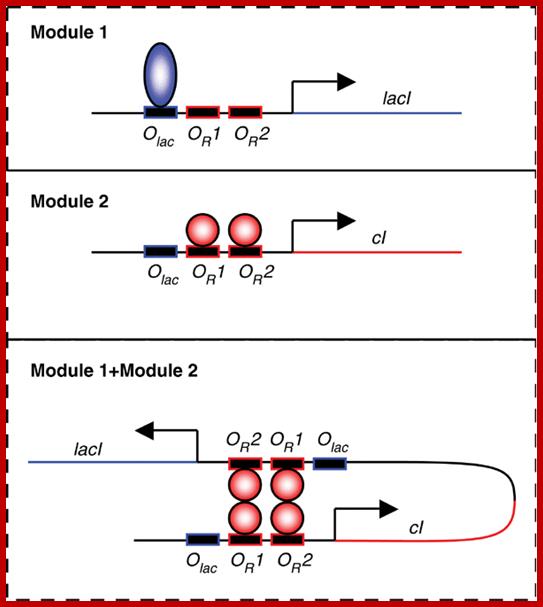
Modularizing gene regulation;modular cross linking in synthetic gene networks Jose MG Vilar; http://msb.embopress.org/
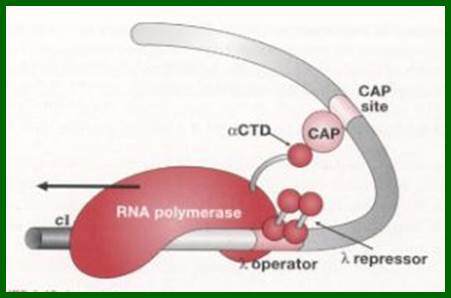
Synergistic activation of RNAP by CAP-CPR maintains lysogenic state. The CAP bound to to CAP binding site of lambda DNA interacts with alpha CTD tail of RNAP keeps it activate and cI are produced constantly.
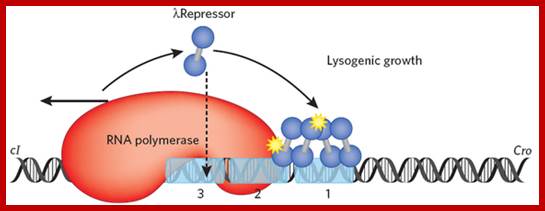
The binding of cI repressors activate interacts with RNAP and activate RNAP to transcribe cI gene which maintains lysogenic state.Mol.Biol4students
Though it is strong repressor, it activates its own gene-self regulatory for the repressor have activator domain which interact with RNAP and activates transcription of cI gene and maintains lysogeny. Ref@?
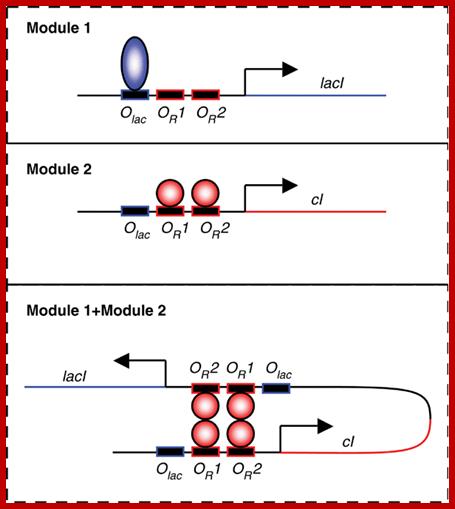
Modular cross talking in synthetic gene network, an example; A promoter with binding sites for Lambda cI dimers OR1 and OR2 and for Lac repressor; http://msb.embopress.org/
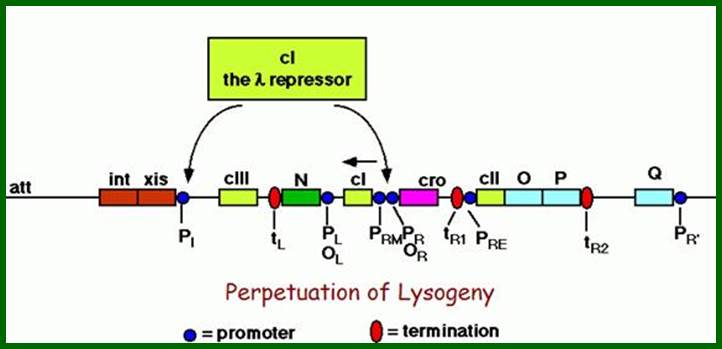
The cI not only blocks transcription of cro and the genes on to the right, but also blocks p1 for xis and int onwards. The cI is a strong repressor and shuts off all required for lysis. http://www.escience.ws/
The binding of dimeric repressors cooperatively to each other recuit RNAP to its promoter site and initiate transcription leftwards, which is translated to produce more and more of cI repressor protein. The repressor proteins also contain active sites for the activation of RNAP ( shown in the above dumbel sahped repressor protein in the above figure). Thus all other genes are repressed except cI gene, thus the repression is maintained. The dimeric cI repressor proteins have another domain for interacting to form tetrameric complexes, thus the repressor proteins bound at L operators and R operators force interaction and bind to each other by the looping of DNA. Thus the produce very tight complex.
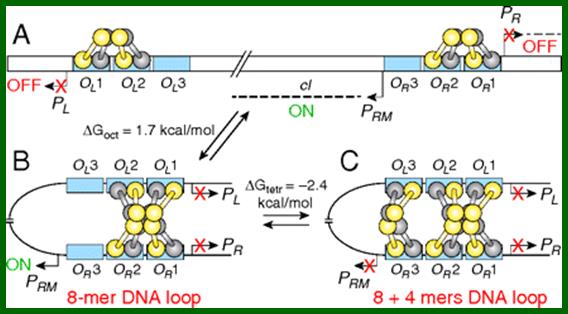
Multilevel autoregulation of λ repressor protein CI by DNA looping in vitro;
Models of cI regulation by DNA looping: Detailed conformations of the structures are not known and maps are not drawn to scale. (A) Promoters (PR, PL, and PRM); operators OL (OL1, OL2, OL3) and OR (OR1, OR2, OR3) are in blue rectangles; CI dimers (one monomer is shown in yellow, the other is in gray). The bent arrows show the transcription start points of promoters. The dashed line indicates transcripts from PR, PL, and PRM. The cI gene is transcribed from PRM. (B) DNA looping and octamer formation (8-mer) by CI tetramer binding to OL1 ∼ OL2 interacts with that at OR1 ∼ OR2. (C) Octamer and tetramer (12-mer) of CI binding to OL and OR. Red X means promoter is turned off. ΔGoct and ΔGtetr values for octamer and tetramer loops stability are from Zurla et al. (25), measured on linear DNA under different conditions. http://www.pnas.org/content/
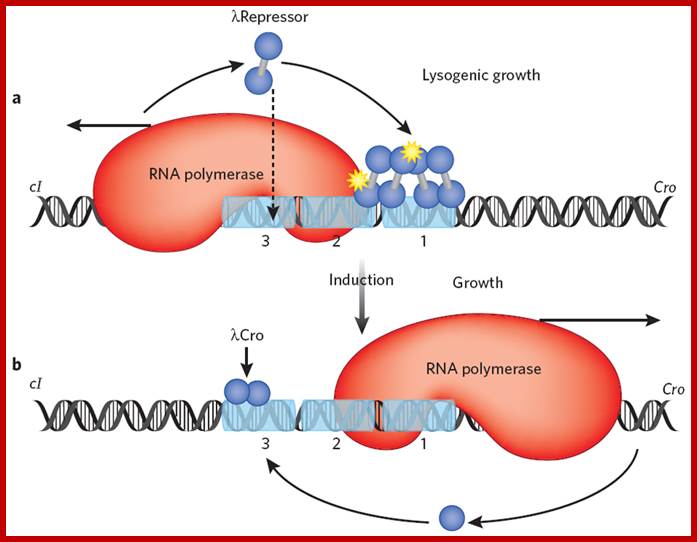
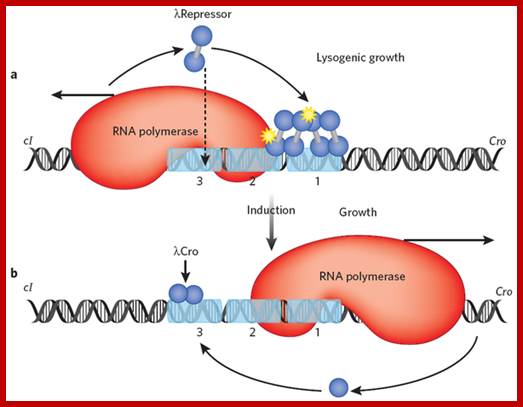
The Lambda switch; principles of a switch; (a) In a lysogen, lambda repressor (blue dumbbell) preferentially occupies two adjacent operator sites, labeled 1 and 2. This preferential occupancy is determined by two factors: site 1 has the highest affinity for repressor, and a repressor dimer binds there cooperatively with another repressor dimer binding site 2. In this state, repressor activates transcription of its own gene (which proceeds leftward in the figure) as it represses transcription of the cro gene. With lower efficiency, repressor also binds the weak site 3 and thereby turns off transcription of its own gene. Binding of the third site is facilitated by interaction with another repressor dimer bound some 2,500 base pairs away, in an example of cooperative binding accommodated by DNA looping13 (not shown). The yellow stars indicate protein-protein contacts of about equal strengths, one mediating cooperative binding of repressor dimers, the other mediating recruitment of RNA polymerase by repressor. (b) UV irradiation results in cleavage of repressor and the onset of transcription of cro and other lytic genes. Cro binds the same three operator sites, but in an order opposite that of repressor: it first binds site 3 and turns off expression of the repressor gene. Later in the lytic cycle, Cro decreases or stops transcription of its own gene by binding sites 1 and 2. Cooperativity has no role in Cro binding, but this is an exceptional case; Mark Ptashne; http://www.nature.com/ Repeat for emphasis
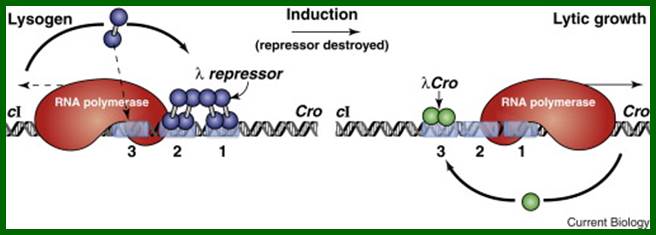
The lambda epigenetic switch. Two states of the switch are shown: on the left the repressor gene (cI) is transcribed but the Cro gene is not, and vice versa on the right. The scenario on the left is found in lambda lysogens, bacteria that carry an otherwise dormant phage lambda. Inactivation of repressor (induction) results in lytic growth of the phage, an early stage of which is shown on the right. Repressor and cro turn each other's genes off by blocking binding of RNA polymerase to the other's promoter: repressor covers the Cro gene promoter when bound at sites 1 and 2 as shown on the left, and cro covers the repressor-gene promoter when bound at site 3, as shown on the right. Repressor bound at sites 1 and 2 activates transcription of its own gene (cI), as it represses transcription of Cro. Repressor maintains its concentration below a specified level by binding, at higher concentrations, to site 3 (as indicated by the downwards arrow), and turning itself off. All of these effects — auto-activation and repression by repressor, and the opposing effects of repressor and cro — are effected by simple binding reactions with suitably adjusted binding constants. The figure indicates that the switch can be flipped by a dose of UV light which results indirectly in cleavage of repressor. An additional set of interactions involving repressors bound here and at a site some 2000 base pairs away has been omitted.
This is an epigenetic switch- where one set of genes i.e., lysogenic genes are on Lytic genes are rendered off; and vice versa. Here the pattern of gene expression is self-perpetuated. The switch can be flipped by environmental signals. The signals such as UV can trigger the breakdown of repressor proteins and the repression is rendered off and the lytic phase is expressed.
As the repression is initiated, pII/pIII with host RNAP initiates transcription from p1, this leads to the production of Xis and Int products.
|
PI |
< TTGC GTGTAA TTGC <-- 13 --> TGTACT |
Production of cII and cIII is very important in lysogenic pathway. The N-domain also contains an activator domain for cro-protein. With the binding of cI to oR1, oR2 and oL1 and oL2, strong protein-protein interaction leads to the looping of the DNA generates a strong repression and transcription of gene towards N and Cro is totally blocked. Thus it generates tight repression.
Look at the promoter sequences for pRM <-10 TTAGATA < -35 TTAGATA for the binding of RNAPs with sig70. Transcription is initiated at +1A; this sequences act as promoter of pRM. The pRM is located in oR3.
pRM-- ßTAGATT (-10) ---13—TTAGATA (-35)
The cI not only blocks transcription of cro and the genes on to the right, but also blocks p1 for xis and int onwards. http://www.escience.ws/
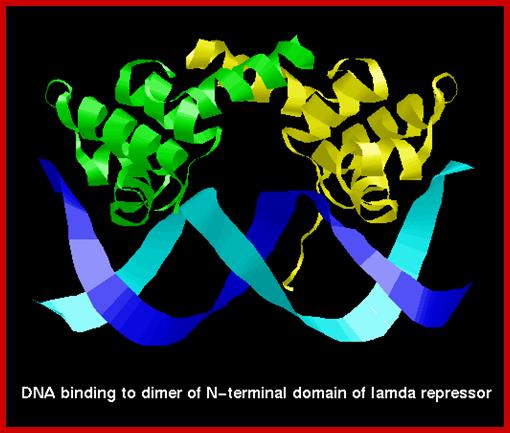
cI repressor bound to DNA is sequence context, the N-terminal helix turn helix domain intercalates into major groove of the operator sequence and binds to repress the gene activity. http://www.cryst.bbk.ac.uk/
The 3-D model of Lambda repressor bound to DNA elements. The cI repressor binds to oR1 very tightly; but its binding to o2 and o3 is cooperative, not that strong.
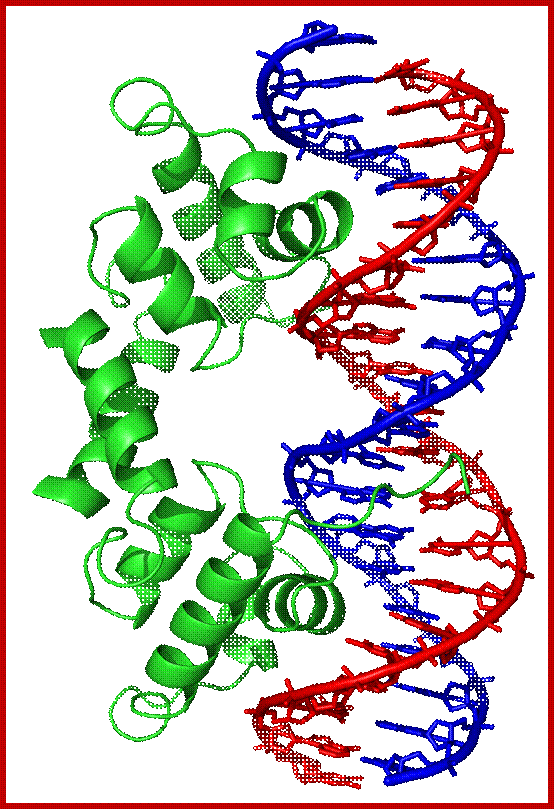
Repressor bound to the DNA grooves;
https://commons.wikimedia.org
Multilevel autoregulation of λ repressor protein cI by DNA looping in vitro: Dale Lewis et al
The prophage state of bacteriophage λ is extremely stable and is maintained by a highly regulated level of λ repressor protein, CI, which represses lytic functions. CI regulates its own synthesis in a lysogen by activating and repressing its promoter, PRM. CI participates in long-range interactions involving two regions of widely separated operator sites by generating a loop in the intervening DNA. We investigated the roles of each individual site under conditions that permitted DNA loop formation by using in vitro transcription assays for the first time on supercoiled DNA that mimics in vivo situation. We confirmed that DNA loops generated by oligomerization of cI bound to its operators influence the auto activation and auto repression of PRM regulation.
Protein interactions that lead to either Lytic or Lysogenic cycles for Lambda phage gene expression. The lambda repressor forms a ‘binary switch’ with two genes mutually exclusive expression. The repressors are self-assembling proteins as dimers called cI. The dimer binds to DNA in helix turn helix (HLH) motif. This protein regulates the transcription of cI and Cro proteins. Lambda phage remain in lysogenic state if cI predominant but it changed to Lytic state if cro protein predominates. The cI proteins bind to any of the three operator sites in the order oR1>OR2. They bind in cooperative manner, but binding to O3 is when the level of repressor is high.
cI when its concentration is high it also binds to OL1 and OL2 which are found downstream of R operators. When cI dimers bind to OL1 and OL2, OR1 and OR2, as octamer DNA loops over; the the repressor binds to Ol3 and OR3 forming a tight cooperative binding. This leads to repressing cI transcription.
When cI is absent cro genes may be transcribed, in the presence only cI genes are transcribed. But when the concentration of cI is high both are repressed.
Barabara Mayer; http://www.wikiwand.com/
http://www.sci.sdsu.edu/
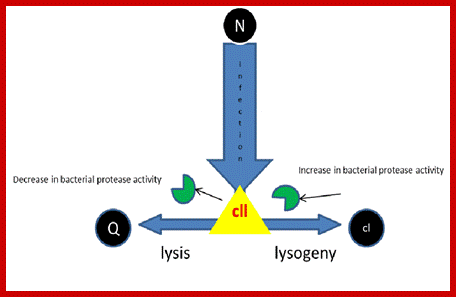
Lysis-lysogeny decision by phage Lambda;
Genetically manipulated with Lac segment in the lambda http://www.sci.sdsu.edu/
http://www.sci.sdsu.edu/
- Uncommitted. Soon after lambda enters a cell:
- RNAP initiates transcription at PR but transcription terminates at tR1, resulting in expression of Cro protein.
- RNAP initiates transcription at PL and the transcription terminates at tL1, resulting in expression of N protein.
- N acts as antiterminator and prevents termination at tR1 and tL1, thus RNAP moves and it allows the expression of cII and cIII proteins.
http://www.sci.sdsu.edu/
- Commitment. The lytic/lysogeny commitment is determined by the accumulation of cII protein.
Lysogeny: If sufficient cII protein produced, cII binds to PRE and activates transcription of cI gene and cII binds to PI and activates transcription of the ‘int’ gene. Because cII protein is rapidly degraded by a protease encoded by the host ‘hfl’ gene, however the accumulation of sufficient cII protein depends upon two factors: growth conditions; if conditions are poor it inhibits Hfl, this leads to more of cII. Multiple infection produces more cIII, which an inhibitor of proteolysis of cII.
http://www.sci.sdsu.edu/
Lysis. If cII protein is degraded, RNAP is unable to initiate transcription from PRE or PI, so transcription continues from PL and PR.
Execution of Lysogeny or Lytic phase: Production cII for either lysogeny or lysis, this result depends upon the accumulation of cII protein during the commitment stage.
Lysogeny:activation of cI expression from PRE results in accumulation of cI protein
- expression of int from PI results in accumulation of integrase protein
- cI protein binds to OL and represses N expression ([N protein] rapidly decreases because N is unstable)
- cI protein binds to OR and represses O, P, and Q expression
- cI protein binds to OR and activates its own expression from PRM
http://www.sci.sdsu.edu/
Lysis:
- decreased [cII protein] cannot activate cI expression from PRE
- transcription from PR continues to express O, P, and Q proteins
- Q protein prevents termination at tR2 allowing expression of PLate operon
- [Cro protein] increases due to transcription from PR
Cro protein binds to OR and OL and represses expression of early gene products. http://www.sci.sdsu.edu/
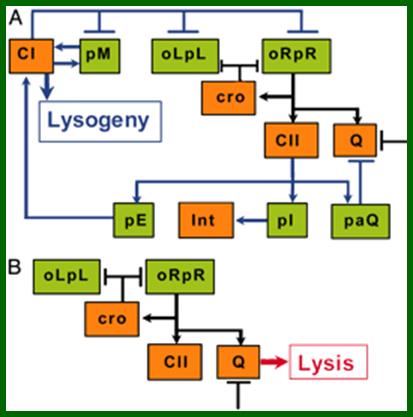
A schematic model for the lysis–lysogeny decision process: (A) Elements of the λ genetic network participating in the lysogenic response. (B) Elements of the λ genetic network active during the lytic pathway. Arrows denote positive actions and bars denote negative actions. A bar also denotes the threshold or cooperativity effect delaying Q activity. Both schemes focus on Genetic model functions studied in this work. (Promoters are shown in green, and proteins are shown in orange.) Black connecting lines represent parts of the network operating in both the lytic and the lysogenic response. (C) Simplified kinetic model suggested by our experiments in which CII (blue line) activates CI synthesis (green line) and represses Q activity (red line) during a lysogenic response. When insufficient levels of CII are accumulated, the lytic response is the default of the genetic network; mol.biol4 masters (borrowed).
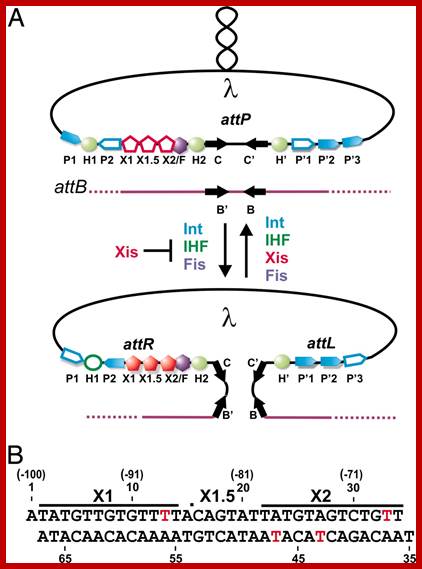
Integration:
Integrase gene product 44.5Kd (356 aa) binds to attachment (att) region and brings the circular DNA on to the host ‘att’ site and by recombination, integrates lambda DNA next to gal gene of the host. Once the lambda genome is integrated, the cI produced, bind to their respective sites and inhibit the transcription of genes required for lytic pathway. cI expression is self regulated. The cI product binding to oR1 and oR2 can recruit RNAP and activate RNAP from pRM through its activator domain located at N-end of the proteins. Thus, more cI is produced and when sufficient amount of cI is produced its own production is blocked
The integrase and the attaching factors produced earlier bind to DNA at ‘att’ site and site specifically integrates the viral DNA into host genome.
Viral DNA ___________p1 att p2___________
X
Host DNA ____________ b1 attb 2___________
I
_________b1_p2________V-DNA____________p1_b2_____
Phage integration into host DNA is site specific and mediated by several factor, among them integrase 44.5Kd and IHF (integration host factor) of 99 and 94 aa heterodimer. Dimeric IHF binds to att site and bends the DNA like ‘U’ shape. The integrase too binds to att sites and bring phage DNA and host DNA together at att site. This integration is also facilitated by two other host factors such as HU and Fis. IHF and Fis are responsible for bending the DNA. Integration uses a 13 BP region of virus and 15bp region of host DNA with perfect matched sequences between viral and host attachment regions. When the integrase brings two DNA together as if it is homologous synapses, it cuts the DNA in both and generates sticky ends, and facilitates the exchange by base pairing. One the homologous sticky ends base pair ligase seals the ends.
Viral DNA att site:
Base pairing at left and right of att:
5’ -----GCTTT TTTATACTAA---
3’------CGAAAAAATAT GATT--
Mohamad A. Abbani
www.pnas.org/content
Dr.Michai Kolot. http://www.tau.ac.il/
The phage DNA integrated into host DNA is called Prophage. The bacterium with prophage is called lysogen. In bacteria with more cI repressors creates a situation called immunity against further viral infection. The immunity region spans from N gene to the end of cII gene. This is because, the entry of another lambda DNA is instantly recognized by cI repressors and prevents transcription that ensures lysogenic condition perpetuated.
Lysoginized lambda DNA can be made to enter into lytic phase by subjecting the bacteria to UV radiations or Mytomycin. The said drug can create DNA with break in single strands, which activate Rec-A complex. In such SOS situations it activates repressor called Lex-A. Cleaving of the LexA activates genes required for repairing damaged DNA. Perhaps the LexA is a regulon.
RecA is also crucial in post replicative repair like the prokaryotic SOS Response. The SOS Response activates multiple genes that allow DNA replication to overlook errors. This response is especially helpful in the survival of Ultra-Violet damaged cells. When a cell is damaged, by UV light for example, the cell initiates its SOS Response (Ptashne). RecA is called in to act as a protease that cleaves the lambda repressor and LexA which is a repressor that normally inhibits proteins such as UvrA and UvrB that aid in the repair of UV damaged DNA (Ptashne). RecA binds with LexA on the DNA and forces it to self-cleave. RecA also cleaves lambda repressor monomers. This decreases the amount of lambda repressor dimers being formed, as the cI production stopped transcription of the repressor from pRM stops. This initiates Cro expression which allows the cell to undergo lytic growth and lysis. (Ptashne).
In lysogenic bacteria activated RecA cleaves a 40 aa connector region of cI at 111-113 amino acids. As the sub domains become free from one another, they no more bind to their respective operator sites and the operator sites with their promoter become free for the host RNAP to activate transcription from their respective promoters. At the same time excisionase binds the borders of lambda DNA and cut and release the DNA. But the released DNA undergoes circularization. Transcriptional activity of different genes required for lytic phase leads to replication of DNA. After several rounds of DNA replication, it switches to rolling circle mode which generates linear concatameric DNA.
Symbols:
pL = promoter for left ward transcription,
oL = operator for promoter pL,
oR = operator for promoter pR,
pR = promoter for right ward transcription-cro onwards,
pRM = promoter for transcription of cI gene-leftwards (from o2).
pRE = promoter for cI. Leftwards from pRE >cro->cI.
pR’ = promoter rightwards of Q across qut and terminates tR’ (for late genes ‘SRR(z)’ and forward).
Pi = Promoter for xis, int, att and b region-leftwards.
P aQ = << anti Q
tL1 = transcriptional termination site for N at the end,
t(i) = second t/L after int site,
tR1 = terminator region for cro (after nutR).
tR2 = terminator region for CII, O ,P (after P),
tR3 = tR’ = terminator site for Q gene.
tR4 =
N = anti terminator factor,
L1 = transcript from pL to tL1, which codes for N protein, if it goes beyond L1 it becomes L2 which includes transcripts for cIII, Xis & int,
R1 – transcript from pR to the end of cro (tR1),
R2 = the transcript from pR to the end of O and P (tR2).
R3 = transcript from pR to the end of Q (tR3).
R4 = transcript from p’R to t’R.
R5 = transcript from p’R to late genes beyond SR.
R4 and R5= Transcripts including R3 and beyond, the R4 does not code for anything, but its extension beyond R4 produces R5. This ensures replication and lytic state.
cI = 27kd, transcriptional inhibitor or Repressor (dimer), induces and maintains lysogeny; binds very strongly to oR1, and binds to oR2, oR3 loosely, but cooperatively
Cro = 7.2Kd regulatory protein a dimer, binds to oR3 tightly, acts as a repressor of cI, if cro succeeds in transcription of early and delayed early genes of the phage, it enters into lytic mode.
Ori = origin of lambda DNA replication is located in O gene. O protein 36.6 kDa binds to this site.
P = 25.6kDa, P-protein binds to O protein, required for initiation of replication,
cII = 10.5kDa another control factor, very unstable, but critical protein, responsible for initiation transcription at pRE leftwards up to the end of cI; involved in lysogeny. It also activates transcription from pi and paQ leftwards.
cIII= 6kDa, it is also involved in lysogeny, assisting cII from degradation.
pRE = promoter for repression establishment,
pRM = promoter for repressor maintenance,
N = 11Kd, an antiterminator substance coded for by the N gene, acts at Nut sites, before tL1, tR1 sites,
Gamma = inhibits host RecBCD.
Int = 44.5Kd, Responsible for the integration of lambda DNA into host DNA at att point.
Xis = 9Kd, Excisionase, responsible for excision of the phage DNA from bacterial DNA.
Q = 22.7Kd, another antiterminator substance, that acts at qut, tR3 and tR4 sites,
NutL and NutR = N utilizing sites, to which the N protein binds and its associated factor bind to RNAP and act as antiterminator,
NusA = N utilizing substances of host, which interact with N, RNAP and other Nus components,
B, C, D, E, W, Nu3, FI and FII = head components and assembly.
G, H, I, J, K, L, M, U, V and Z = Tail components and assembly
A, Nu1 = Cutting and DNA packaging.
R, Rz and S = Involved in the lyses of the cell.
B= an accessory factor, not really required and this region is disposable.
attP = attachment site for phage integration.
attL and attR =borders for phage excision.
Cos = cohesive end sites in the linear DNA, it is the site of ~200bp region where the circular is cut by Terminase, and also contain sequences for DNA packaging.
Transcription termination and antitermination:
In bacterial cells transcription termination at the end of coding region is achieved by two methods; one is called Rho dependent and the other is Rho independent method. Rho independent method uses G-C rich stem loop ending in UUUUs. In addition, lambda factors in association with host factors perform antitermination act at specific Nut sites and qut sites; this helps in transcription to move on.
In the case of Rho dependent process, Rho assembles on to the 5’end of the transcript and moves along the transcript in ATP dependent manner and initiates termination at sequence specific region.
N dependent antitermination:
N (12kd) binds to Nut site at tL1 of N gene and also to NutR. Nut L sequence is located between pL and the proximal portion of N. And the NutR is located between cro and tR1.
Nus-A, bacterial product binds to gpN and RNAP.
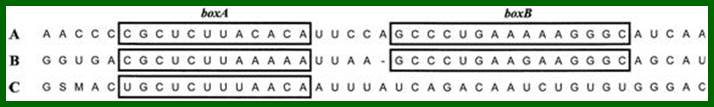
Nus-B, binds to s10 (a ribosomal protein) and to boxA.
Nus-E- it is a ribosomal factor called s10 and it binds to RNAP.
Nus-G- binds to RNAP and it can also interact with RNAP.
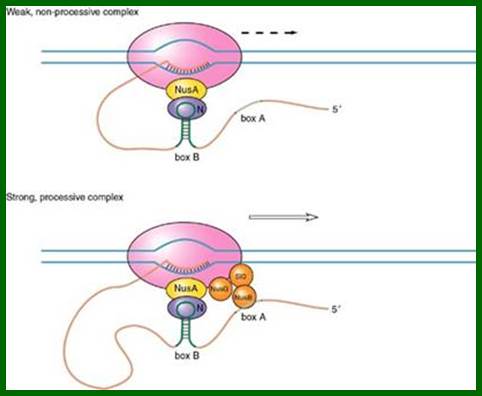
This occurs without the N protein interacting with the DNA; the protein instead binds to the freshly transcribed mRNA. Nut sites contain 3 conserved "boxes," of which only BoxB is essential.
- The boxB RNA sequences are located close to the 5' end of the pL and pR transcripts. When transcribed, each sequence forms a hairpin loop structure that the N protein can bind to.
- N protein binds to boxB in each transcript, and contacts the transcribing RNA polymerase via RNA looping. The N-RNAP complex is stabilized by subsequent binding of several host Nus (N- utilization substance) proteins (which include transcription termination/antitermination factors and, bizarrely, a ribosome subunit).
- The entire complex (including the bound Nut site on the mRNA) continues transcription, and can skip through termination sequences (wikipedia).
Factors- NusA bound to RNAP, N bound to box B (stem loop structure) and intracts with NusA. NusB binds to BoxA (linear) and interact with NusG and s10 thus they form a complex which induce the RNAP to move on. http://www1.nttu.edu.tw/yenlee
3’A--.AAUUAAAA---UUCUCGC…hair-pin-box-B-box-A
Rho dependent antitermination:
In Lambda phage, at transcriptional termination site, whether it is Rho dependent or Rho independent termination process, the transcripts contain two blocks of sequences. One that generates stem loop structure is called B-box and the other is called box-A and it is linear; they are termed as nut regions i.e. N utilizing regions and the Nus is called because the protein is N-utilizing substance.
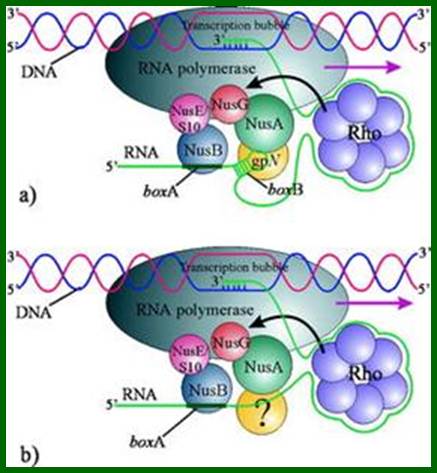
The 5’end of the transcript gets associated with hexamer Rho which moves very close to RNAP where the transcript binds and circularizes. The 5’ end of the transcript produces a stem loop structure called box B to which gpN binds and nusA which binds to RNAP interacts with gpN. NusB binds to box A and interacts with nusE and NusG which are bound to RNAP. All these proteins as complexes prevent rho dependent transcription termination.
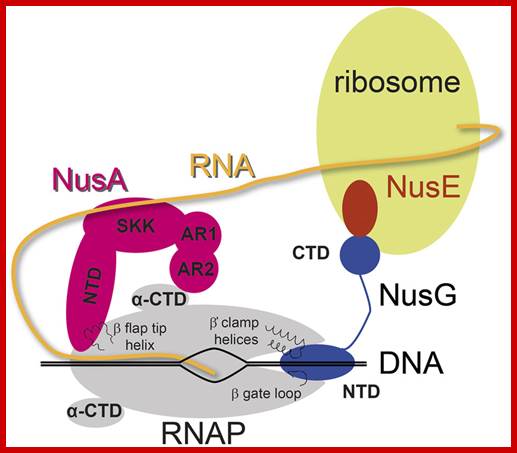
Determination of RNA polymerase binding surfaces of transcription factors by NMR spectroscopy. Johanna Drögemüller et al; In bacteria, RNA polymerase (RNAP), the central enzyme of transcription, is regulated by N-utilization substance (Nus) transcription factors. Several of these factors interact directly, and only transiently, with RNAP to modulate its function. As details of these interactions are largely unknown, we probed the RNAP binding surfaces of Escherichia coli (E. coli) Nus factors by nuclear magnetic resonance (NMR) spectroscopy. Perdeuterated factors with [1H,13C]-labeled methyl groups of Val, Leu, and Ile residues were titrated with protonated RNAP. After verification of this approach with the N-terminal domain (NTD) of NusG and RNAP we determined the RNAP binding site of NusE. It overlaps with the NusE interaction surface for the NusG C-terminal domain, indicating that RNAP and NusG compete for NusE and suggesting possible roles for the NusE: RNAP interaction, e.g. in antitermination and direct transcription: translation coupling. We solved the solution structure of NusA-NTD by NMR spectroscopy, identified its RNAP binding site with the same approach we used for NusG-NTD, and here present a detailed model of the NusA-NTD: RNAP: RNA complex. http://www.nature.com/
The N-protein binds to boxB, nusB binds to box-A interact with other Nus-factors such as nusA, nusG and nusE and s10, which in turn interact with RNAP and the terminal region of the RNA bound by Rho or if the region is free from Rho, then it interacts with upstream region of the mRNA. This reaction actually hastens the activity of RNAP; thus the enzyme doesn’t pause and continues transcription towards their 3’ end.
If the transcription is not terminated it could go all the way to the end of B segment, which produces some accessory factors, without which the replication of viral DNA cannot take place. In the case of Rho independent antitermination, factors that bind weekly to poly (U) and prevent termination and elongation continues. In the case of Rho dependent termination, the complexes prevent Rho overtaking RNAP. Nus-G binds directly to Rho. The N protein recognizes box-B of the transcript and its helix domain binds to major groove and interacts and prevents termination. The ter regions such as tL1 and tR1 and tR2are Rho independent. NutL and nut R are Rho dependent.
Antitermination by gpQ:
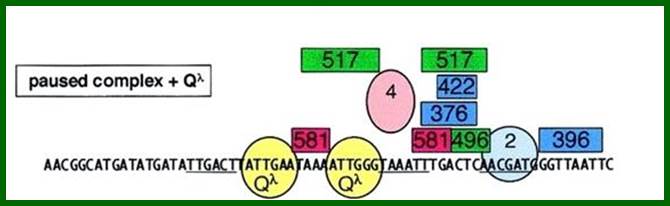
The Q-protein is also an antiterminator factor binds to DNA in sequence specific manner-Qa and Qa ATTGAT and ATTGGT, where the transcript takes stem loop form. In this case qut’ protein directly binds to the DNA in sequence specific manner ATTGAA and ATTGGG, and in association with protein Q and other accessory proteins (7subunts?), react with RNAP and displace sigma 70 and prevent termination.
Q is similar to N in its effect: Q binds to RNA polymerase in Qut sites and the resulting complex can ignore terminators, however the mechanism is very different; the Q protein first associates with a DNA sequence rather than an mRNA sequence.
- The Qut site is very close to the PR’ promoter, close enough that the σ factor has not been released from the RNA polymerase holoenzyme. Part of the Qut site resembles the -10 Pribnow box, causing the holoenzyme to pause.
- Q protein then binds and displaces part of the σ factor and transcription re-initiates WIKIPEDIA).
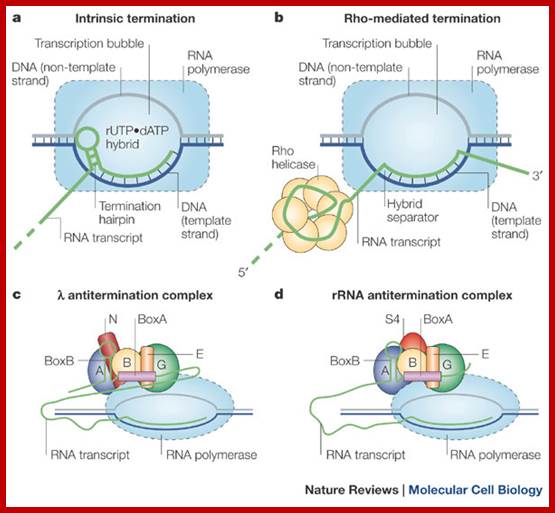
a | The
hairpin stem–loop structure of intrinsic terminators is followed immediately by
a run of rUTP residues in the RNA, which form a particularly weak RNA–DNA
hybrid with the dATP residues of the template, which causes the transcription
complex to pause while the terminator hairpin folds. Transcription termination
occurs when the RNA–DNA hybrid is unwound and the transcription bubble
collapses, thereby causing dissociation of the complex and termination of RNA
transcription. b | Rho is a hexameric helicase that
binds to the nascent RNA and translocates 5'![]() 3' along the transcript until it 'catches
up' with the transcription complex while it is halted at pause sites.
Termination is mediated through hybrid destabilization, potentially through
Rho-mediated conformational changes in RNA polymerase and/or by sequentially
capturing the nascent RNA bases from RNA polymerase and shortening the hybrid
to <8 bp. c | The antitermination complexes of
phage
3' along the transcript until it 'catches
up' with the transcription complex while it is halted at pause sites.
Termination is mediated through hybrid destabilization, potentially through
Rho-mediated conformational changes in RNA polymerase and/or by sequentially
capturing the nascent RNA bases from RNA polymerase and shortening the hybrid
to <8 bp. c | The antitermination complexes of
phage ![]() consist of an RNA element (the nut site) that contains boxA (pink), boxB
(a hairpin loop) and five protein factors — phage
consist of an RNA element (the nut site) that contains boxA (pink), boxB
(a hairpin loop) and five protein factors — phage ![]() N protein (brown) and host proteins NusA (A, purple),
NusB (B, yellow), NusE (E, orange; also known as ribosomal protein S10) and
NusG (G, green) — which bind both to the RNA and to the RNA polymerase within
the transcription complex. This network of interactions stabilizes the
elongation complex to prevent intrinsic termination and results in a twofold
increase in transcription rate that might help to avoid Rho-mediated
termination. d | The ribosomal RNA (rRNA)
antitermination complex functions to abrogate Rho-helicase-mediated termination
(not shown), perhaps by increasing the transcription rate. Antitermination
activity is initiated by the binding of the boxA RNA site by NusB and NusE. Ribosomal
protein S4 (red) and transcription factors NusA and NusG also participate in
the complex that is bound to the RNA polymerase.; Termination and Antitermination; Sandra
J. Grieve & Peter H. von Hippel; http://www.nature.com/
N protein (brown) and host proteins NusA (A, purple),
NusB (B, yellow), NusE (E, orange; also known as ribosomal protein S10) and
NusG (G, green) — which bind both to the RNA and to the RNA polymerase within
the transcription complex. This network of interactions stabilizes the
elongation complex to prevent intrinsic termination and results in a twofold
increase in transcription rate that might help to avoid Rho-mediated
termination. d | The ribosomal RNA (rRNA)
antitermination complex functions to abrogate Rho-helicase-mediated termination
(not shown), perhaps by increasing the transcription rate. Antitermination
activity is initiated by the binding of the boxA RNA site by NusB and NusE. Ribosomal
protein S4 (red) and transcription factors NusA and NusG also participate in
the complex that is bound to the RNA polymerase.; Termination and Antitermination; Sandra
J. Grieve & Peter H. von Hippel; http://www.nature.com/
Replication of lambda DNA:
Replication origin:
The genome replicates in lytic phase provided conditions are favorable.
Replication initiates in a region called origin, which is located within O gene. The origin consists of four 19 bp imperfect palindromes. Next to it there is what is called Dna-A box, which is 40bp A/T rich region, and this is involved in melting of DNA during replication initiation. Next to it is an inverted repeat of 28 bp long. The 19 bp palindrome sequence is given below.
—ACA—[--------AT--------]—[------------] ----
Origin
-CII-----I--------O-------P---------I--------Q-
O-P binding DUE invert segment
-I-><-I-><-I-><-I-><-I-----A/T-----I----<--------I—
4 X 19 bp 40 bp repeats28bp
Lambda DNA replicates only if molecular events lead to the expression O and P genes, otherwise it is not. When conditions are favorable, the following events lead to replication.
The origin for replication is located between cII and Q genes.
For replication to initiate, both O and P gene products are required.
Within the gene O, more towards the promoter of the O gene, a 150 to 172 bp long region contains sequence elements called origin, which is located in the promoter of ‘O’ gene itself. From left to the right of the ‘O’ gene, it has 4 nineteen base pair long; nearly identical palindromic sequences. Next to it, there are about 40 bp long A/T rich sequences. Next to it there is another 28 base pairs of invert repeats.
To function as effective Origin it requires just 4 palindromic sequences and 40 bp AT rich region. A sequence rich in ‘A’ tracts imparts a stable bend to DNA.
Four 19 base palindromic sequences provide the structural motifs for the binding of gpP-O proteins. Each subunit is a 34kd protein, works as homodimer. They bind to each segment as dimers.
The amino terminal of gp-O binds to DNA and the C-terminal region is used for interacting with another protein, a product of P gene, which is next to gpO on the genome.
The gpP protein, 26.5 KD, a dimer, interact with the dimer gpO protein. Binding of each dimer gpO proteins to each of the four 19 base pair regions is like the binding of Dna-A to 9-mer sequences in the origin of E. coli DNA. The protein gpP activity is similar to that of Dna-C in assisting Dna-B. The binding of gpO protein to DNA, induces torsion as the DNA wraps around the cluster of gpO proteins, which leads to the opening of the DNA in A/T rich region.
The factor gp P as dimer interact with C-terminal region of protein gpO and also interacts and binds to Dna-B, a helicase. Dna-B, a helicases and hexamer, (50kd subunits) protein assembles on to the fork in association with gpP protein. Interestingly, the gpP protein by binding to Helicase makes helicase inactive.
Some of the cellular histone like factors by complexing with gpO and gpP prevent DNA from unwinding.
However, another set of host cellular factors like Dna-K, Dna-J and gro-E, (which are chaperone-hsp like proteins) interacts with gpP-Dna-B complex and release inhibition of Dna-B from gpP. Activation requires the input of ATP. With the activation of Dna-B, Dna-k, Dna-j and Dna-E, and gpP are released from the complex.
It is at this juncture, Dna-G, a primase, associates with helicase, forming primosome complexes. One primosome complex each assembles at each fork joints.
DNA Pol-III Holozyme joins as dimer at each of the forks.
The movement of the primosome complex is from the 5’ to 3’ on lagging strand. Other host proteins that are used in replication are ssBs, Gyrase, DNA Pol-I and E. coli DNA ligase.
Transcriptional initiation and progression of the same into gpO-gene is of importance, for transcription through this region which stabilizes the replication bubble. Whether or not the truncated transcripts are used as primers for leading strand replication is not clear.
However, the Dna-G, which is a primase, is responsible for laying sequence specific 9-11 ntds long primers on the lagging strands at an interval of 1000 to 1500 ntds.
DNA-Pol-III Holozyme, which is loaded by gamma complex, synthesizes continuous strand and DNA-Pol-III with gamma complex is responsible for the discontinuous strand synthesis on the lagging strand. The replication of DNA is bidirectional and similar to that of E. coli.
After few rounds of replication by this mode, replication switches to rolling circle mode. What is the protein that generates the nick for rolling circle mode of replication is not yet clear? However, rolling circle mode of replication produces a long double stranded concatameric linear DNA.
Phage Assembly:
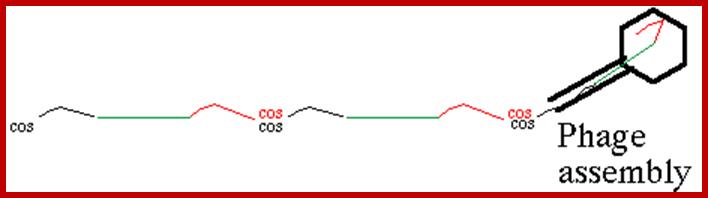
http://www.escience.ws/
Assembly of viral particles:
Robert Edgar and Williams Wood studied the assembly lambda phage from it basic components. Team:SupBiotech-Paris/Concept2:
One can observe in this picture that the initiation and the termination of the encapsulation is mediated by the Cos sequence. This is the fragment between two Cos sequences which is encapsidated.
A model for coupling DNA cutting to DNA packaging. gp17 (blue) is shown as a pentamer, portal (green) as a dodecamer inserted at the special vertex of the icosahedral capsid, and DNA as a grey ribbon. In the presence of gp16, gp17 and ATP, a stimulated holo-terminase complex (tensed state) makes the first cut (A). The terminase-DNA complex then docks onto prohead portal as a pentamer and initiates packaging (B). During active translocation (C), gp17 is incompetent for cutting (relaxed state). After headful packaging, the packaging motor dissociates (D), re-forms holo-terminase complex (tensed state), and makes the headful cut. The terminase-DNA complex once again docks onto the portal of another empty prohead, initiating another round of genome packaging. The stoichiometry of the holo-terminase complexes that makes the first cut or the headful cut is unknown. See ‘Discussion’ section for additional details. https://www.researchgate.net
At a later stage, a complex composed of the viral ATPase and DNA binds to the portal vertex to form a DNA translocating molecular motor. In bacteriophage f 29, the motor can generate a force of up to 57 pN, making it one of the most powerful molecular motors discovered so far (Smith et al , 2001). Such a force is needed to pump DNA against the high internal pressure that increases as the viral DNA is encapsidated. Portal proteins of different phages and herpesviruses show no detectable similarity in the amino-acid sequence and exhibit large variations in their subunit molecular masses, for example, 36 kDa for phage f 29 and 57 kDa for phage SPP1 portals (Valpuesta and Carrascosa, 1994). Nevertheless, available electron microscopy (EM) data show that they all share a common turbine-like shape (Valpuesta and Carrascosa, 1994; Orlova et al , 1999; Trus et al , 2004). Although in all species, the portal protein is a central and essential component of the DNA-translocating machine, the organization of the molecular motor varies. In bacteriophage f 29, the motor consists of three coaxial macromolecular rings: the portal protein, the procapsid RNA (pRNA) and the ATPase (Simpson et al, 2000). There is no evidence for the presence of pRNA in other bacteriophages where an additional protein is normally required for DNA packaging, gp1 in the case of the Bacillus subtilis bacteriophage SPP1 (Chai et al, 1995). In contrast, a circular oligomeric ATPase (Simpson et al, 2000) appears to be a shared feature of the DNA translocating complexes, and such assemblies have been proposed for T3, T4 and lambda phages (Catalano, 2005). In bacteriophage SPP1, the molecular motor, consisting of three proteins (Figure 1)—gp1, gp2 (ATPase) and gp6 (portal protein)—powers translocation of the 45.9 kbp pro-mosome (Camacho et al , 2003; Oliveira et al , 2005). Different components of the DNA-translocating motor possess distinct rotational symmetries, for example, the capsid’s vertex has five-fold symmetry and the B-form DNA has a 10 1 screw axis. In common with the herpesvirus portal protein (Trus et al , 2004), the portal protein of bacteriophage SPP1 can exist as a circular assembly with varying number of subunits: it is found as a 13-subunit assembly in its isolated form and as a 12-subunit assembly when integrated into the functional viral capsid (Orlova et al , 2003). How exactly the molecular motor works has been the subject of much debate. The low-energy barriers to rotation of symmetry mismatching protein rings relative to each other led Hendrix (1978) to propose that DNA translocation is accompanied by rotation of the portal protein inside the capsid vertex. Different models of DNA translocation, all involving the rotation of the portal protein, were put forward following the EM image analysis of the SPP1 portal protein (Dube et al , 1993) and the determination of the X-ray structure of the f 29 portal protein (Simpson et al , 2000; Guasch et al , 2002). However, until the work presented here, the structural data did not characterize the segments defining the most constricted part of the internal tunnel (tunnel loops) that would be in close contact with the DNA during translocation. This issue is addressed by the X-ray structure of the SPP1 portal protein 13-mer and reconstruction of its 12-mer active form. The combination of structural data with geometrical and symmetry constraints imply that DNA translocation is accompanied by sequential conformational changes pro- pagating along the belt of tunnel loops with the portal protein remaining engaged with the DNA. https://www.researchgate.net
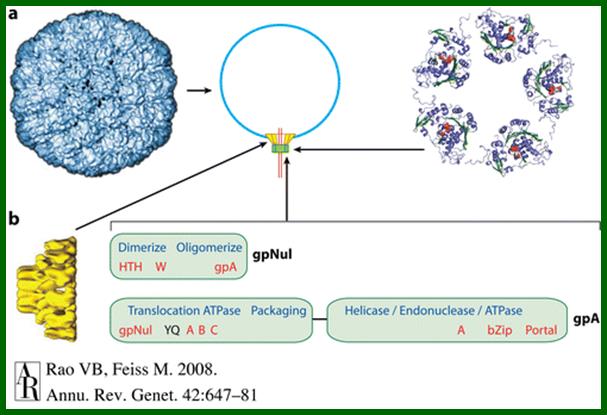
An ATP-powered DNA translocation machine encapsidates the viral genome in the large dsDNA bacteriophages. The essential components include the empty shell, prohead, and the packaging enzyme, terminase. During translocation, terminase is docked on the prohead's portal protein. The translocation ATPase and the concatemer-cutting endonuclease reside in terminase. Remarkably, terminases, portal proteins, and shells of tailed bacteriophages and herpes viruses show conserved features. These DNA viruses may have descended from a common ancestor. Terminase's ATPase consists of a classic nucleotide binding fold, most closely resembling that of monomeric helicases. Intriguing models have been proposed for the mechanism of dsDNA translocation, invoking ATP hydrolysis-driven conformational changes of portal or terminase powering DNA motion. Single-molecule studies show that the packaging motor is fast and powerful. Recent advances permit experiments that can critically test the packaging models. The viral genome translocation mechanism is of general interest, given the parallels between terminases, helicases, and other motor proteins. The Bacteriophage DNA Packaging Motor; Annual Review of Genetics- Venigalla B. Rao1 and Michael Feiss2
Head assembly:
Capsid gpE 38kd and gpD 12Kd protein join in 1:1 ratio; they are the major head proteins; gp D12Kd decorates the capsid. GpE and gpD consists of little less than 420 subunits. They are organized as icasodeltahedron with T-7 pattern geometry. Capsid structure that is assembled in the first stage of assembly is called as prohead. The protein gpNu3, a 19kd protein exist in cytoplasm in about 200 copies and it initiates the organization of prohead (immature) and remains associated with immature prohead as a scaffold, and disassemble from the prohead when mature head is formed. The mature head does not contain gpNu3.
Connector:
Proteins gpB (533aa), gpC, GpFI, gpFII (117aa), gpNu3, gpGroEL, gpGroES (both Gro proteins are chaperones) and gpW also become connector that attaches to the head at one of the vertices and gpNu3 assists in the assembly of head and connector. The gpB (processed) and gpC which form the collar that apparently holds the connector in place. GpB is found in 12 copies organized in the form of a ring with a central orifice. Joining of the connecter makes the head symmetry into asymmetric. Proteins gpU and gpZ facilitate the connector to join at the bottom of the prohead. The connector has a 20 A^o hole.
Initiator: The gpJ, a three-protein subunit structure with gpI, gpL, gpK, gpG, gpH and gpM organizes into an initiator that connects the tail tube at the bottom, where gpJ acts as tail fiber. Tail tube and tail fiber and the connector organization is very elaborate.
Tail tube:
Tail tube consists of 32 stacked hexagonal rings of gpV (31kd), totally made up of 192 subunits. Its assembly is initiated by the initiator protein (gpJ). Actually, initiator acts as the nucleus for the polymerization gpV in the form of 32 stacks of six subunits each. However, the length of the tail is controlled or regulated by gpH (853aa). The tail tube and the terminal fiber are attached through a protein complex of gpG, L, M and H. The fiber protein remains attached to the end of the tail tube and it acts as host binding at selective sites such as maltose receptors.
Tail consisting of tail tube (made up of gpV and initiator made up cup shaped structure to which the fiber binds to its base. The assembly of the tail tube attached to the head via connector is the function of gpU and gpZ.

Bacteriophage
Final assembly:
The above-mentioned individual structures such as head and tail are assembled in hierarchical fashion assisted by a number of host proteins.
Once these are formed the DNA has to be packed into the head and finally the fully formed tail tube with terminal fiber has to be connected to form the complete lambda phage particle. In laboratories, using recombinant-DNA with lambda DNA is used for packaging. The packaging extract contains all the head and tail proteins and also required host factors. What is required is the addition of lambda with inserted clonal DNA. When both are added together lambda particles are fully assemble in 10 to 15 minutes.
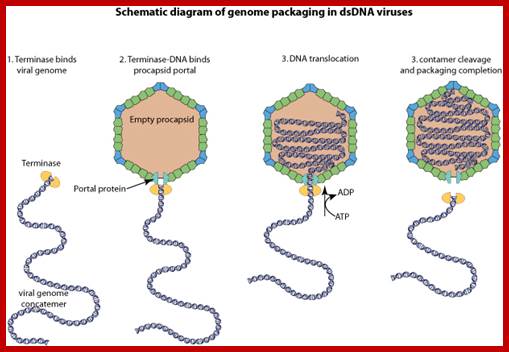
General; Large dsDNA bacteriophages, herpesviruses, adenoviruses and micro viruses encode a powerful DNA-translocating machinery that encapsidates a viral genome into a preassembled capsid or procapsid. The packaging machine is often composed of a portal structure, which provides a gate for DNA entry, and an ATP-driven motor. This motor is composed of the large subunit whose ATPase activity fuels DNA translocation, and most frequently, a small subunit that binds to the viral packaging site. DNA cleavage can be coupled to genome packaging; http://viralzone.expasy.o rg/
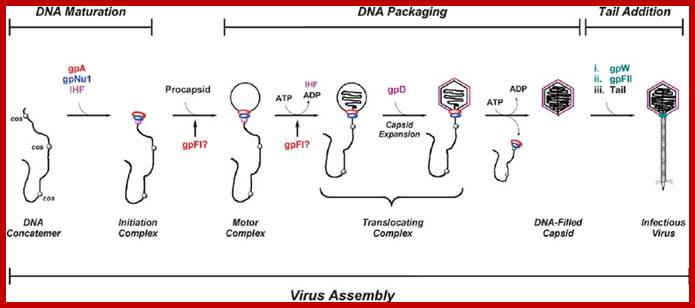
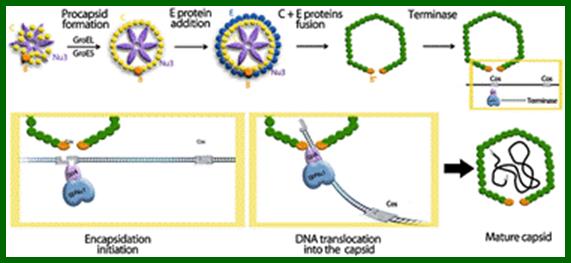
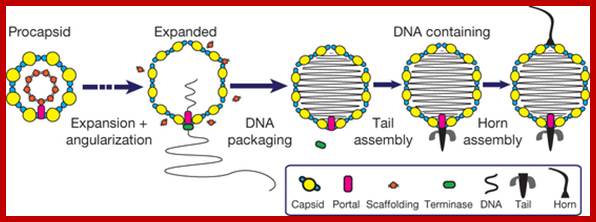
Phage assembly model revealed by ZPC croETLgeneral); The phage’s pathway to maturation starts with the assembly of the precursor procapsid. Initial insertion of DNA into the capsid induces capsid expansion and angularity. The reorganization of the capsid shell culminates before DNA is fully loaded; Wei Dai, Caroline Fu etsl; http://www.nature.com/nature
The Scrunch-worm hypothesis: Transitions between A-DNA and B-DNA provide the driving force for genome packaging in double-stranded DNA bacteriophages:
![]()
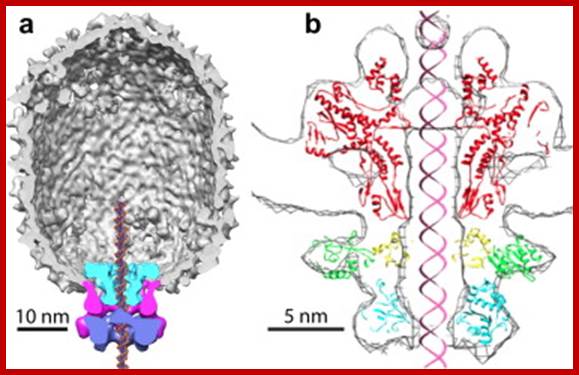
Double-stranded DNA bacteriophages have motors that drive the genome into preformed capsids, using the energy released by hydrolysis of ATP to overcome the forces opposing DNA packaging. Viral packaging motors are the strongest of all biological motors, but it is not known how they generate these forces. Several models for the process of mechanochemical force generation have been put forward, but there is no consensus on which, if any, of these is correct. All the existing models assume that protein-generated forces drive the DNA forward. The scrunch worm hypothesis proposes that the DNA molecule is the active force-generating core of the motor, not simply a substrate on which the motor operates. The protein components of the motor dehydrate a section of the DNA, converting it from the B form to the A form and shortening it by about 23%. The proteins then rehydrate the DNA, which converts back to the B form. Other regions of the motor grip and release the DNA to capture the shortening–lengthening motions of the B → A → B cycle (“scrunching”), so that DNA is pulled into the motor and pushed forward into the capsid. This DNA-centric mechanism provides a quantitative physical explanation for the magnitude of the forces generated by viral packaging motors. It also provides a simple explanation for the fact that each of the steps in the burst cycle advances the DNA by 2.5 base pairs. The scrunch worm hypothesis is consistent with a large body of published data, and it makes four experimentally testable predictions. Stephen C. Harvey http://www.sciencedirect.com/
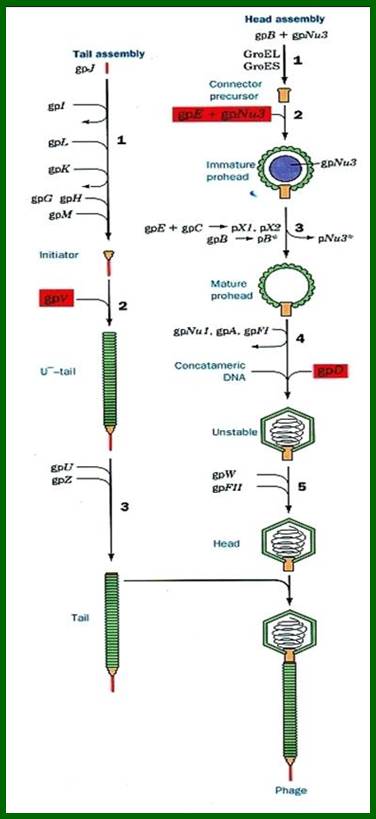
http://mcmanuslab.ucsf.edu/Lambda gene transcription at cI
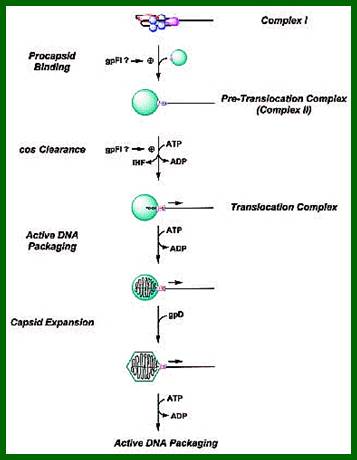
The diagram show how the phage assembled and an infectious viral particle formed.
Under in vivo conditions as and when the said structures assemble the Concatameric DNA formed at a late stage of replication, is used for packaging the DNA into the head. The paradox is the head is of 55nm size and the lambda DNA is 48502 bp x 3.4 = ~ 165000nm. To pack such a long-coiled DNA into such small sized head requires an ingenious method. A solution to this paradox has come from the studies on T7 phage, nearly 39000bp long linear DNA packaging into icosahedral head which has the same size of 55nm. It is now discerned that the DNA from one end is actively pumped into the head through an orifice (20A^o) found in the connector. As the DNA is pumped, it occupies the capsid inner surface and then assembles inwards as coils after coils which appear as nine concentric rings. The center has just folded structures.
In the case of lambda DNA, which is in the form of linear concatamer, first the correct ends have to be recognized for the only the left end of the lambda DNA enters first. This is also true in the case T7 phage. The DNA end that enters first comes last during infection. This holds good for lambda also.
The cos site consists of about 200 bp consisting of at least 3 segments such as cos B, cos N and cos Q. Cos B is subdivided into three sub segments called R1, R2, R3.
The left cos end of the DNA has 12 ntds long sticky tails followed by cos B (R segments) and at the right end cos also contains sticky tail of 12 ntds long.
~~~Bcos.Nu1-A-W-//-J----b--int-xis--N-O-P--Q-S-R-Rz-cosQ~~~
Proteins gpNu1 and gpA bind to cos segment and cleave at cos N segment to generate 12 ntds sticky ends, where they use left end of the cost site. The cos site is 190 to 200 ntds long, but it requires at least 100ntds for efficient packaging.
The packaging is initiated with the binding of Nu1 to R3 region of the B, and terminase binds to R1, both bind cooperatively to each other. Once they bind they take the left end of the DNA to the connector orifice; this process is furthered by the binding IHF, which creates kinks in the DNA ends for the entry into 20A hole. The pumping of DNA into such constrained space enthalpically and entropically unfavorable because opposing intramolecular repulsive charges. Yet they are pumped using ATPase pumps that are located at the connecter orifice. Using phi-29 phage DNA packaging scheme, it is believed, lambda packaging uses connector proteins that are connected to pRNA associated with ATPase. Probably they use six such pumps. This energy driven pumping acts like stator driving the DNA inwards. When the complete 48.502 bp linear DNA is packed, the terminase A again recognizes the cos sites and cuts and only the left is used for packing. In this process gpF1 participates. Unlike some phage the DNA is packed heedful and stops, but here it takes the whole length and little more for the length of DNA is recognized by its cos ends. The lambda DNA has a b-segment which is disposable, so molecular biologists have exploited in removing the b-segment and inserting a foreign DNA in to the phage DNA, thus lambda DNA acts a vector for developing CDNA library and Genomic library.
When one full length DNA is packed into the head, it scans, and with the help of gpF1, it cuts at Cos site, thus releasing concatameric DNA from the packed viral prohead. Again, this protein-DNA complex finds another prohead for loading another full-length DNA into it. The loading of the DNA is not head full as is done in p1 phages.
The gpF1 helps the enzyme Terminase in scanning one full length DNA and finding cos sites for cutting. Once the DNA is packed, the tail tube with terminal tail fiber is added to the connector. This is facilitated by gp Z which activates the tail for binding to the head via the connector. This completes the packaging.

The phage DNA remains as a part and parcel of the host, but its gene expression is totally shut off and lysogeny is established.
This also provides immunity against further infection for any lambda DNA that enters for its gene expression is shut off by the cI factors present in large numbers. Here the cI acts as a guard against any further infectious phages and also maintains its lysogenic state.
From lysogenic to lytic phase:
In lysogenic state, the viral DNA continues to exist as prophage and replicates as a part of host genomic DNA and in this way it can exist as long as the bacterium lives.
But none of its genes are expressed because the cI repressor is constantly produced and it inhibits the expression of other lambda genes. However, it ensures the production of it cI proteins by auto regulation.
Treatment of lysogenic bacteria with Mytomycin or UV, induce lytic phase. Mytomycin or UV causes damage to DNA.
Damage to DNA in the form of any broken ssDNA strands, activates Rec-A protein, by self-proteolysis. Under SOS situations the activated Rec-A partially digests or cleaves the Lex-A at ala-gly bond in the Lex protein. The Lex-A protein is a repressor of a set of genes, which respond to SOS stimuli. The said gene products are involved in repairing damaged DNA in one or the other form.
The activated Rec-A also cleaves the cI repressor at ala 111 and gly 112 position, i.e in the middle of the cI repressor, which is the linker or connector region of between the two domains of the repressor. The digestion or partial cleaving of this protein releases the repressors from the operator sites.
Transcriptional activation of promoter-operators of oLoP and oRpR produces the N an antiterminator, and cro, which inhibits cI expression by binding to oR3 region and at the same time recruits RNA pol to transcribe more cro and other genes on to the right side of cro. This transcriptional activity leading to expression of excisionase, which binds to att site on either side of the inserted lambda and cleaves and releases the lambda DNA, which soon circularizes.
Action of anti terminators at different positions leads to the production of GPO and gpP proteins, which lead to initiation of replication. Several other genes are also expressed required for DNA replication. Initially the DNA goes through several rounds of bidirectional replication, from the replication origin located in O region. After few rounds of replication, the DNA enters into rolling circle mode of replication. How it switches to rolling circle mode of replication is not clear?! This leads to the production of concatameric DNAs.
This also leads to the expression of genes required for viral assembly and maturation (explained above). As viral particles develop in the cell to a density, the gene for punching hole in the membrane, genes R and Rz produce enzymes for hydrolyzing peptidoglycans bonds and peptide bonds between NAG and NAM. The gpS generates enzyme that punches holes in the membrane so the endopeptidases enter into periplasmic space so that they can cleaves cell wall materials for the release of phage particles.
Each of the particles infect bacterial cells in surroundings and kill and lyses the cell in the bacterial lawn, thus it clears the region. The cleared region is called plaque. The number of plaques formed give an estimation of the phage bursts size or titer.
A list of genes and sites in a sequence starting from cos site and their probable functions:
|
Gene
|
Gene product, Mol. Wt (kd) |
Features and functions |
|
Cos-left and cos Right |
|
12bp long cohesive ends, ligation results in circular DNA and cutting linearises the circular DNA |
|
Nu1 |
21 |
Nu1 and terminase bind to the left end of cos |
|
A |
21 |
Terminase? |
|
W |
|
|
|
B B* |
533a.a |
Part of head proteins Part of head proteins |
|
Nu3 |
19 |
Associated with prohead, assists head assembly |
|
C |
439a.a |
C, B, B*, F2, W form a neck between head and the Connector
|
|
D |
12 |
GpD and GpE associate in 1:1, head capsids |
|
E |
38 |
Major capsid proteins |
|
F1 F2 |
117a.a |
Assist Terminase to scan full length concatameric DNA for cutting at cos. . |
|
|
|
|
|
Z |
|
Connector |
|
U |
|
Connector |
|
V |
|
Tail tube, hexagonal components |
|
G and Nu3 |
17Kd 19Kd |
gpG precursor of connector, Nu3 facilitates the prohead formation using gpE |
|
T |
|
|
|
I |
27.7 |
I to M –subunits of connector that binds to terminal fiber J, this connects to tail tube at distal end. |
|
L |
|
initiator |
|
K |
|
initiator |
|
H |
|
initiator |
|
M |
|
initiator |
|
J |
|
Actual terminal fiber |
|
|
|
|
|
B |
|
Accessory factors, can be dispensable |
|
|
|
|
|
Att |
|
site at which integrase binds |
|
Int |
44.5Kd |
Responsible for cutting and joining of viral DNA into host DNA |
|
Xis |
9Kd |
Excision site for viral DNA from the host DNA |
|
P1 |
|
Promoter for integrase and Xis |
|
Exo |
|
Excisionase |
|
Beta |
|
|
|
Gamma Gam |
|
Prevents Rec BCD nuclease activity Inhibits hosts’ exonuclease activity |
|
Kill |
|
|
|
RaL |
|
Alleviates host restriction enzymes B and K |
|
CIII |
|
Stabilizes cII protein |
|
tL1
N |
|
The region at which N transcript terminates An antiterminator for Rho dependent and rho independent transcriptional terminators |
|
oL, pL
|
|
Operator-promoter for N gene, it has o1, o2, o3, to which cI can bind prevent transcription |
|
cI O3
pRM O2 O1 |
~27.7 |
A repressor protein of 236 amino acids long, has two domains connected with linker region of the same polypeptide chain, the N-terminal binds to Major groove of the DNA in sequence specific manner, and the C-terminal region is used for homo-dimrization; it has high affinity in decreasing order, high affinity to o1, then to o2 and the least affinity is to o3, and represses transcription from oRpR, it is a dumbbell shaped protein, binds to DNA and dimerizes, it also binds to N operator region and inhibits transcription, but the protein having acidic domains interact with RNAP and activates it own transcription from o3 pRM, but binding to o1,o2 inhibits its own synthesis. It is the promoter at which cI activates transcription of its own gene. The cI has acidic acid regions, which interact with RNAP, and activates the enzyme. When cI is produced in excess, it also binds to o1 and o2 and shuts of its transcription. CI has greatest affinity to o1 |
|
oRpR
|
|
Is the generalized promoter-operator used for transcriptional initiation for the gene cro onwards? |
|
Cro
pRE
|
7.2 |
It is dumbbell shaped, binds to o3 and inhibits transcription of cI gene Promoter for establishing repression, located at the end of cro. The cII binds to this region and initiates transcription in opposite direction to cro transcription, and proceeds up to oRpR and initiates transcription of cI gene |
|
tR1,2,3, t’R |
|
It is the regions at which transcript terminates. It’s the region at which it has Nut site or qut sites, at which antiterminator factors act to continue transcription to the next cistron |
|
CII |
|
It is an activator of cI and p1 genes. This protein is highly unstable and has a half-life of one minute; hfl-A and B proteins under favorable conditions degrade these proteins. Whether the phage to goes into lytic or lysogenic phase this proteins’ stability determines the fate of the pathway |
|
O-ori |
|
This is gpO protein producing gene, and it also contain a region of 172 bp region, at which replication can be initiated. The gpO protein dimerize and binds to each of the four O binding regions. Binding initiates the melting of DNA at A/T rich region |
|
P |
|
Produces gpP protein, which as a dimer interacts with not only gpO and also binds to Dna-B 9 a helicase). This complex actually inhibits the activity of helicase. |
|
|
|
|
|
Q |
|
It is another antiterminator factor; it is used as an antiterminator factor for the transcription of genes beyond Q to towards S, R and R2 genes, which virtually ensures lytic phase. |
|
|
|
|
|
p’R |
|
Transcription initiation site for the late genes |
|
Qut |
|
Is the region where Q product binds and uses the Q antiterminator factors, for RNAP to continue the transcription |
|
t’R |
|
Another Ter site at which Q acts as antiterminator and the transcript is called R4 and it extends to R5, which virtually ensures DNA replication as well as lytic phase |
|
S |
|
A protein creates or punches holes in the membrane for the transport of few factors |
|
R1 |
|
I is a trans-glycosidase, cleaves glycosidic bonds between NAM and NAG of the host cell wall |
|
R2 |
|
It is an endopeptidase, which hydrolyses peptide bonds of peptidoglycans |
|
Cos-right |
|
The right side of the genome |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Viral Gene Products and Host Factors in Lambda DNA Replication:
|
Viral Genes, Gene Product |
Mol.wt In kds |
Functional features |
|
N |
|
Anti terminator at tL1, tR1, tR2 sites |
|
Q |
|
An anti terminator at tR3, t’R |
|
O P |
34 26.5 |
Bind to Origin, initiate melting of the DNA at A/T rich region |
|
Gamma and Gam |
|
Inhibits host Rec-BCD nuclease and host exonuclease activity |
|
RaL |
|
Alleviates host restriction enzymes B and K |
|
A, Nu1, F1 |
21 |
Bind to Cos sequences at the left 200bp region, scan a full length DNA and cut, and help in packing |
|
S, |
|
Punches holes in the bacterial membrane |
|
R |
|
Trans-glycosidase, cleaves bond between NAM and NAG |
|
R2 |
|
Endopeptidase |
|
Cro |
7.2 |
As dimer binds to o# inhibits transcription of cI gene |
|
CI |
28.3 |
Binds to promoters for N and also pR and inhibits transcription of N genes and cro genes. But it can activates its own gene and can also shut off its own gene |
|
CII |
|
Activates cI at pRE (promoter for repression establishment, it also activate p1 for induction of integrase gene. This protein is very unstable; its half-life is one minute. It concentration is critical for lytic or lysogenic pathway |
|
CIII |
|
It protects cII from protease activity |
|
|
|
|
|
|
|
|
|
Host factors |
Mol.wt |
Functional features |
|
Lam B |
|
Acts as receptor, it is a maltose transport protein |
|
Dna-B |
50x6 |
Helicase, opening the fork |
|
Dna-G |
60 |
Primase |
|
ssBs |
19x4 |
Coats single stranded DNAs |
|
Gyrase |
|
Unwinds or relaxes the super coiled DNA |
|
Ligase |
|
Seals the gaps during replication |
|
Dna-K, Dna-J, Gro-E Gro-EL, Gro-ES |
|
The first three interact with gpP-Dna-B complex and relieve from inhibition, so helicase can active |
|
NUS: A, B, E, G |
|
In the order, A-binds to gpN and RNAP, B-binds to s10, E&G both bind to RNAP, they together interact with RNAP and Rho if found and hastens RNAP to progress. |
|
IHF THF |
|
Host, integration factor Hot, terminator factor |
|
DNAP Holozyme |
|
Replication of both leading and lagging strands |
|
DNAP-I |
|
Removes RNA primers and fills the gap |
|
|
|
|
|
|
|
|
Some of the gene functions:
|
O |
DNA replication |
|
N
|
early gene regulator exclusion |
The regulation region includes the immunity region as well as the genes that are responsible for controlling the switch between lysogenic and lytic growth. The Q antiterminator protein, as well as the anti-Q RNA and PR' constitute a second regulation region.
|
orf28 |
|
The recombination protein genes code for Int and Xis, which are required for integration of the bacteriophages into the bacterial host chromosome during lysogenic growth and excision from the bacterial host chromosome during induction, as well as a number of other genes.
|
Nu1
|
DNA packaging |
|
orf-64 |
|