E.coli DNA R�plication:
Components of Replication:
An excellent example for delineating structures and the processes involved replication is E.coli and other similar microbes.�� In bacterial cell the DNA of 4.6x10^6 bp, is double stranded, circular and super coiled. �It is associated with histone like basic proteins, and with linker proteins it exhibits looped structure.� Besides the said proteins, there are many proteins bound to various regions of the DNA in sequence specific manner and it is more or less is like a nucleo proteinaceous thread (thin nucleosomal thread).� And in resting stage it is suspended in cytoplasm, but it is attached to mesosomal membranes at a particular point called attachment site.� It is at the time of Replication initiation the DNA gets freed from the mesosome; it is only then replication starts.
E.coli chromosomal DNA is divided into circular map units, which provide information on the length of the DNA that, is transferred to its female counterpart during conjugation in a given unit of time.� The length of DNA transferred in given unit of time is called mpu.� E.coli DNA consists of 100 mpu.
Structural Organization of Origin-C:
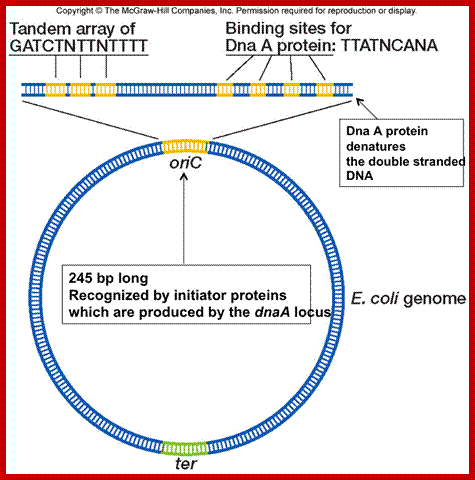
It is the site at which DNA initiates replication.� For that matter all DNAs, whatever may be the source, contain at least one origin locus. In E.coli, it is located at 84.5-mpu position. And it is called Ori-C.� The Ori-C stands for Col E. Chromosome (strain 427).� Origins of other DNA are also named similarly, ex; Ori-lambda, Ori- T7, Ori.Col-E1 etc.� Each of these origins has their own structural features. Almost opposite to Origin one finds replication termination site (Ter).
�
Ori-C site is about ~250 base pairs long and located between gid 'A' (8.5 KB) gene on the left side of Origin and mioC (2.2kb) on the right of origin.� The dna �A� gene is also on its left side.� Actually, this origin is located in a spacer region between the said genes.
https://www.studyblue.com/
�������������������
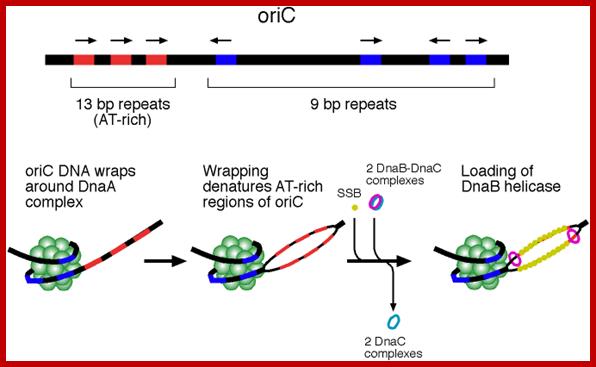
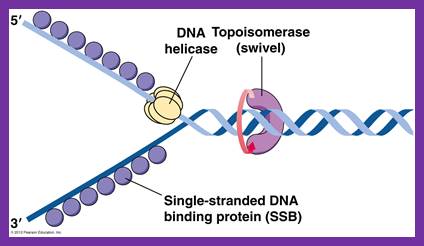
oriC is 245 bp long and contains several conserved sequences: four 9-bp repeats (blue) and three 13-bp repeats (red). The orientation, spacing, and sequences of the 9-bp repeats are critical for function of oriC. The AT-richness of the 13-bp repeats is also crucial for function of oriC. DNA replication is initiated when oriC DNA wraps around a complex of 10-20 DnaA proteins. The wrapping denatures the AT-rich repeats. The denatured region of DNA is held open by the binding of SSB (single strand binding protein). DnaB helicase is loaded onto each strand in an ATP-dependent step requiring DnaA and DnaC. When DnaB is finally loaded onto DNA, DnaC is no longer complexed with DnaB.; http://oregonstate.edu/
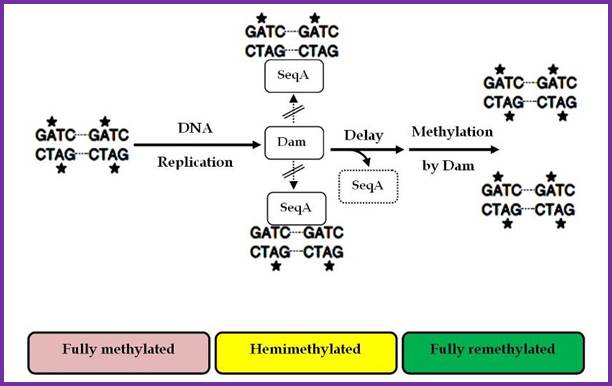
Two or more helically phased GATC sites can be bound by SeqA when they are in hemi methylated state. Binding of SeqA inhibits Dam methylation, maintaining the hemimethylated state for a portion of the cell cycle. Dissociation of SeqA allows Dam to methylate the hemimethylated DNAs, generating fully methylated DNA.; http://www.intechopen.com/
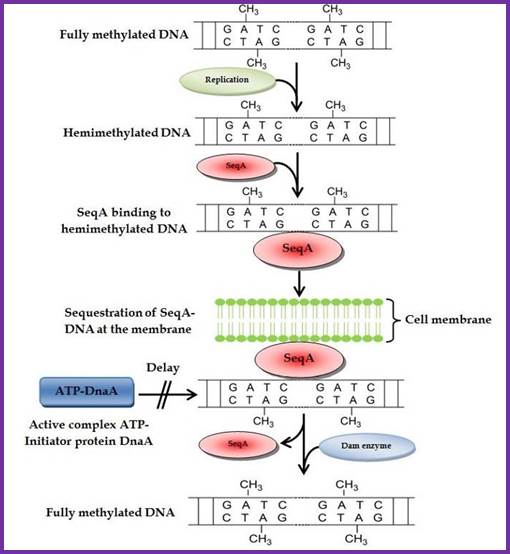
Membrane sequestration of recently replicated origins; http://www.intechopen.com/
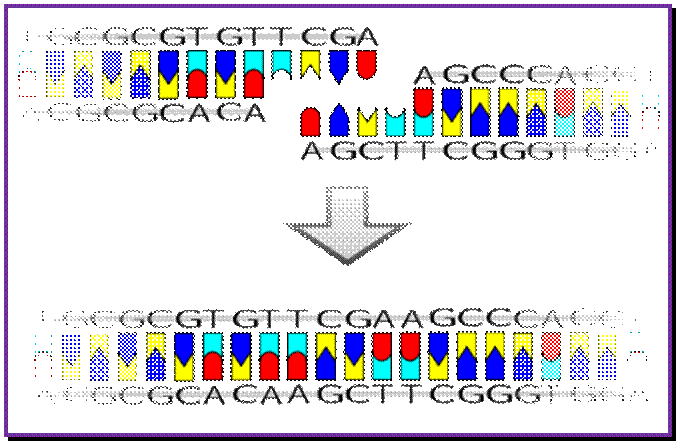
The origin-C consists of two sets of AT rich sequences; one contains 9-mer sequence DAR-TGTGAATAA repeated four times and found on the right part of the Ori.� The13-mer is repeated three times, located on to the left of ninemer sequence DUE, GATCTNTTTATTT. In between the repeats there are spacers and their length is important than their sequences.�� A sequence GATC is dispersed in the origin at least at 14 sites; GATC is also located in the promoter region of dna-�A� gene.� At the beginning of these 9-mer and 13-mer boxes one finds GATC sequence. They are also found in �data� site which acts as a titrator of DnaA proteins. These sites are important for methylation of �A� of GATC by DAM methylases (source of methyl group is S-Adenosyl nucleotide).
Another kind of sequence found is 3� GTC, which is found in the Ori region and also dispersed throughout the length of lagging strand.� These sequences are recognized by Primases, which produce RNA primers.�
The nine-mer sequence is important for it is recognized by a set of proteins, which bind to them, and this leads to conformational changes in the DNA topology.� Similarly, GATC, sequence is also important, because the �A� of the GATC is methylated by DAM-methylases.� The methylation facilitates DNA melting. �Complete methylation of the said sequences in the promoter region of dna-A gene and in the region of origin is sine quo non, without which initiation of replication fails.
�
The protein called Dna-A binds to 9-mer regions co operatively in a sequence specific manner and forms cluster, which makes the DNA wrap over, or coil around the protein cluster. The binding of proteins is aided by ATPs.� This type of wrapping around the protein cluster in left-handed direction induces the DNA to unwind in 13-mer regions into ss DNA.� This region is about 60-80 base pair long, with both strands of DNA unwinded, it looks like an eye or bubble called replication eye, replication bubble.
�����������
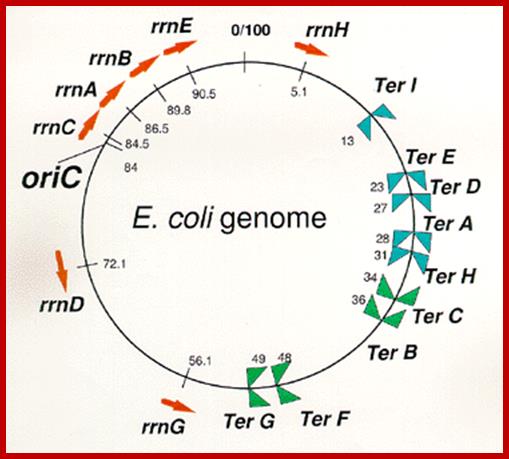
Arrangement
of seven rrn operons and nine Ter sites on the E. coli
genome.
Red arrows indicate locations and transcriptional direction of rrn
operons. It means DNA replication terminus (Ter) site which can block
replication fork approaching from the right, Professor: Takashi
Horiuchi
Exactly opposite to the site of origin, a termination site is located, where replication ends.�� In this region there are six 23 bp long identical, non-palindromic sequences are found, where three each of them are oriented in opposite direction.� They are Ter-I, E, D, A and H on one side and Ter-G, F, B and C are on the other side.� The said sequences are recognized by a protein called Tus (37.08 kd) (Terminator Utilizing proteins), which binds to the said sequences and prevent the movement of replication fork into newly formed DNA for another round of replication cycle, otherwise the replication progresses in to newly made strands and the DNA is subjected to another round of replication.� TER sequences act as a sort of trap, where replication fork can enter but cannot exit.�� However, the TER sequences are not obligatory; for even in the absence of them, or even if they are deleted, replication completes without any hitch.
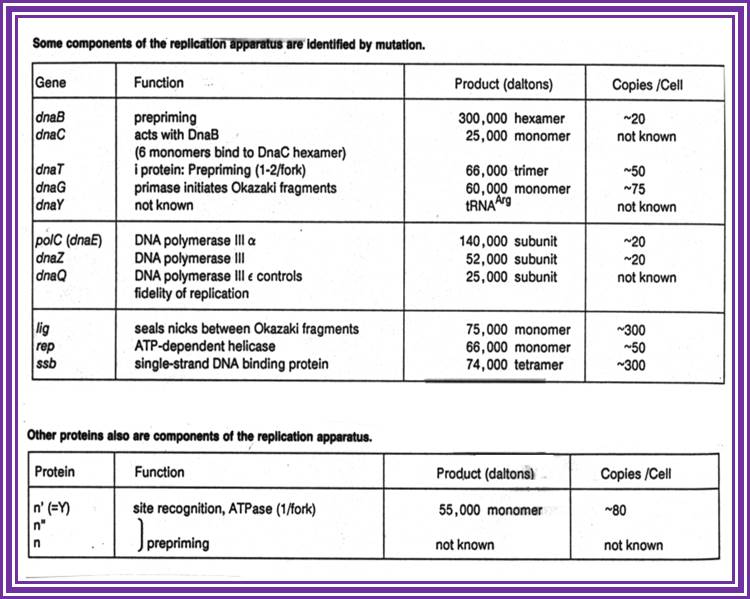
Replication Factors and Replication Enzymes-Required: necat.chem.cornell.edu.
|
Protein |
Gene |
Mol.wt (kd) |
Subunits
|
Dna-A |
dna-A |
50, AAA+ protein |
Monomer, DNA melting |
|
Dna-B |
dna-B (Helicase) |
50 x 6 |
Hexamer, motor protein, helicase |
|
Dna-C |
dna-C |
29 |
Monomer, assists DnaB |
|
SSB |
ssBs |
19 (?) |
Tetramer |
|
Dna-G |
dna-G (primase) |
60 |
Monomer, primase |
|
Rpo enzymes |
Rpo genes |
Mol.wt (kd) |
Subunits
|
|
DNA-pol-I |
Pol-I |
103 |
Monomer, Repair enzyme |
|
DNA pol II |
pOL II |
79-88 |
Repair enzyme, mono to multi- mers |
|
DNA-pol-III |
Many |
>791 |
Multimer, Replicase |
|
Rep-A |
rep-A |
65-76 |
Monomer, helicase3>5� |
|
Hup-A |
hup-A |
9.5 |
Monomer |
|
Hup-B |
hup-B |
9.5 |
Monomer |
|
Top-I |
top-I |
100 |
Topoisomerase |
|
Top-II-A |
top-II-A |
97x2 |
Topoisomerase, dimer |
|
Top-II-B |
top-II-B |
97x2 |
topoisomerase Dimer |
Tus |
Vnu |
37 |
Ter utilizing protein |
|
Top-III/IV |
top-III/IV |
100 |
Remove catenation |
|
DAM |
DAdenine-methylase |
32 |
Monomer |
|
pol IV-(Din B), |
|
|
SoS Repair enzymes |
|
-Pol V-� (� u muc, u muD2 & C) |
|
|
Sos DNA repair enzymes |
|
IHF- |
|
alpha-11.35 beta-10.65 |
Integration host factor, Bends DNA |
|
Fis-x |
|
10.7-dimer |
Binds and DNA-Factor for inversion |
|
Seq A |
|
~20 (181aa), tetramer |
Binds to GATC in origin, repressor |
|
Rob |
|
|
Right arm origin binding protein |
|
SpoOJ |
|
|
Binds DNA to membrane |
|
datA |
|
74 |
Titration� of DnaA protein |
|
DNA ligase |
|
|
|
|
IciA |
htrA |
33>27 |
Binds to GATC and repress initiation (inhibitor) |
|
Protein |
Gene |
Mol.wt (kd) |
Subunits
|
Dna-A |
dna-A |
50, AAA+ protein |
Monomer, DNA melting |
|
Dna-B |
dna-B (Helicase) |
50 x 6 |
Hexamer, motor protein, helicase |
|
Dna-C |
dna-C |
29 |
Monomer, assists DnaB |
|
SSB |
ssBs |
19 (?) |
Tetramer |
|
Dna-G |
dna-G (primase) |
60 |
Monomer, primase |
|
Rpo enzymes |
Rpo genes |
Mol.wt (kd) |
Subunits
|
|
DNA-pol-I |
Pol-I |
103 |
Monomer, Repair enzyme |
|
DNA pol II |
pOL II |
79-88 |
Repair enzyme, mono to multi- mers |
|
DNA-pol-III |
Many |
>791 |
Multimer, Replicase |
|
Rep-A |
rep-A |
65-76 |
Monomer, helicase3>5� |
|
Hup-A |
hup-A |
9.5 |
Monomer |
|
Hup-B |
hup-B |
9.5 |
Monomer |
|
Top-I |
top-I |
100 |
Topoisomerase |
|
Top-II-A |
top-II-A |
97x2 |
Topoisomerase, dimer |
|
Top-II-B |
top-II-B |
97x2 |
topoisomerase Dimer |
Tus |
Vnu |
37 |
Ter utilizing protein |
|
Top-III/IV |
top-III/IV |
100 |
Remove catenation |
|
DAM |
DAdenine-methylase |
32 |
Monomer |
|
pol IV-(Din B), |
|
|
SoS Repair enzymes |
|
-Pol V-� (� u muc, u muD2 & C) |
|
|
Sos DNA repair enzymes |
|
IHF- |
|
alpha-11.35 beta-10.65 |
Integration host factor, Bends DNA |
|
Fis-x |
|
10.7-dimer |
Binds and DNA-Factor for inversion |
|
Seq A |
|
~20 (181aa), tetramer |
Binds to GATC in origin, repressor |
|
Rob |
|
|
Right arm origin binding protein |
|
SpoOJ |
|
|
Binds DNA to membrane |
|
data
|
|
74 |
Titration� of DnaA protein |
|
DNA ligase |
|
|
|
|
IciA |
htrA |
33>27 |
Binds to GATC and repress initiation (inhibitor) |
Dna-A:
�Initiation of replication requires the expression of dna-A gene, which is transcribed earlier to replication and translated to produce DNA-A protein.� It�s gene expression requires full methylation of GATC at its promoter region.� The protein is a monomer, has motifs to bind to unique monomer sites, also they have motifs for protein-protein interaction, thus they can form clusters.�� Proteins don�t have helix turn helix motif nor they have zinc finger structure, but they have hydrophobic regions for helical coiling and protein�protein interactions, yet the characteristic features of protein binding to DNA is still to be understood.� Binding of the monomers to DnaA-A boxes, in ATP dependent manner (proteins have ATPase activity), leads to cooperative binding of more proteins.� This clustering of proteins on DNA makes the DNA to wrap around the proteins, which induces torsional twist and it is this left-handed twist that makes DNA to melt at 13-mer region and AT rich region; perhaps the negative super helical topology in this region may further facilitate the melting of the DNA. Opening or unwinding of dsDNA into single stranded region is an important event in initiation.
�
SS-B:
These proteins, though small (19.5-20kd), as they are translated and released, they bind to negative charged phosphate (P^2-) groups of P-S-P backbone of ssDNA as tetramers, hence they are called single strand DNA binding proteins. The ssDNA wraps around the tetramer SSBs. Binding of the said proteins stabilizes the DNA and makes it as a rigid template and also prevents reannealing of the strands.� When they are bound, they cover approximately 40-65 ntds. The DNA polymerase, while it is generating complementary strands SSBs are displaced. They bind DNA only when the single strand is free from polymerases. In addition, they provide the DNA strand as a straight and unbent template, and stable structures for the enzyme to perform complementary strand synthesis.� They are also required for fork movement.
Dna-C:
It is a monomer, binds to Dna-B in one-to-one manner (1:1).� It helps or facilitates the helicase to be loaded onto ssDNA at replication fork in ATP dependent manner. The DnaC-ATP binds to helicase hexamer and induces the opening of the helicase hexamer ring so that it can load on to single strand and encircle the strand at fork joint.� Once helicase loads on, Dna-C dissociates from the helicase subunits and helicase hexamer in association with ATP acts like a motor protein that moves into the fork and unwinds the DNA ahead of the fork.
Dna-B:
The protein, to begin with it is a monomer, but with the binding of Dna-C in one-to-one manner, subunits start assembling into hexamers to form of a ring. Then with the assistance of Dna-C, helicase ring opens and loads on to the strand, at the joint region of the fork. This assembles only on lagging strand, which is identified by the strand orientation from 5� towards 3� direction.� Two such helicases load, one at each fork joints.
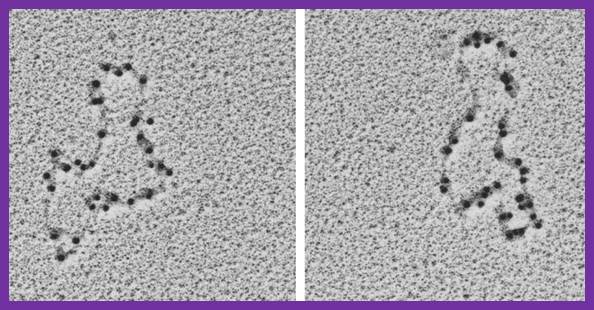
SSBs bound to single replicating DNA strands; http://www.mun.ca/
E.coli- DNA-B subunits in the form a Hexameric ring
Once the Dna-B ring is formed Dna-C dissociates.� The Dna-B complex is a motor protein and acts as DNA dependent ATPase and using the energy it drives into the fork and unwinds the DNA progressively like unzipping the helical DNA.� Its direction of movement is from 5� to 3� on the lagging strand.� There is another protein called Rep A, which as monomer also binds to fork region but to the leading strand and moves in 3� to 5� direction.� However, involvement of this protein in E. coli DNA replication is not substantiated.
Bacterial 3� to 5� DNA helicase; http://ww2.d155.org/
Prokaryotic Helicases:
|
Helicase II (Uvr-D) |
82 |
3�� 5� |
DNA repair |
|
Uvr AB complex |
180 |
5�� 3� |
DNA repair |
|
Helicase-IV |
75 |
3�� 5� |
DNA repair |
|
Rec-BCD |
330 |
3�� 5� |
Recombination |
|
T4 Dda? |
49 |
5�� 3� |
Displaces SsBs |
|
Helicase-I (encoded by F-plasmids) |
180 |
5�� 3� |
Involved in DNA transfer during conjugation |
|
Helicase-III |
20 |
5�� 3� |
Unknown function |
|
Rho
|
50 x 6 |
5�� 3� |
On RNA transcript, performs transcriptional termination |
|
Helicase DnaB |
hexamer |
5�>-3� |
Fork opening |
|
RepA |
Monomer |
3�-5>� |
Fork opening |
|
|
|
|
|
� Note �there are positive and negative helicases; In passive helicases, a significant activation barrier exists (defined as B > kBT, where kB is Boltzmann's constant and T is temperature of the system).� In active helicases, B < kBT, where the system lacks a significant barrier, as the helicase is able to destabilize the nucleic acids, unwinding the double-helix at a constant rate, regardless of the nucleic acid sequence. In active helicases, Vun is approximately equal to Vtrans.� 1976 � Discovery and isolation of E. coli-based DNA helicase.
� 1978 � Discovery of the first eukaryotic DNA helicases, isolated from the lily plant.
� 1982 � "T4 gene 41 protein" is the first reported bacteriophage DNA helicase[13]
� 1985 � First mammalian DNA helicases isolated from calf thymus
� 1986 � SV40 large tumor antigen reported as a viral helicase (1st reported viral protein that was determined to serve as a DNA helicase)
� 1986 � ATPaseIII, a yeast protein, determined to be a DNA helicase
� 1988 � Discovery of seven conserved amino acid domains determined to be helicase motifs
� 1989 � Designation of DNA helicase Superfamily I and Superfamily II
� 1989 - Identification of the DEAD box helicase family
� 1990 - Isolation of a human DNA helicase
� 1992 � Isolation of the first reported mitochondrial DNA helicase (from bovine brain)
� 1996 � Report of the discovery of the first purified chloroplast DNA helicase from the pea[]
- 2002 � Isolation and characterization of the first biochemically active malarial parasite DNA helicase - Plasmodium cynomolgus. https://en.wikipedia.org/wiki/Helicase
The common function of helicases accounts for the fact that they display a certain degree of amino acid sequence homology; they all possess sequence motifs located in the interior of their primary structure, involved in ATP binding, ATP hydrolysis and translocation along the nucleic acid substrate. The variable portion of the amino acid sequence is related to the specific features of each helicase.
Helicases by their structure and functions they classified into super families- Super family (SF1), SF2, SF3, SF4, SF5 and� SF 6.
�Disorder of non-functioning of Helicases can result in mutations.
Human Dead Box RNA helicase; https://en.wikipedia.org
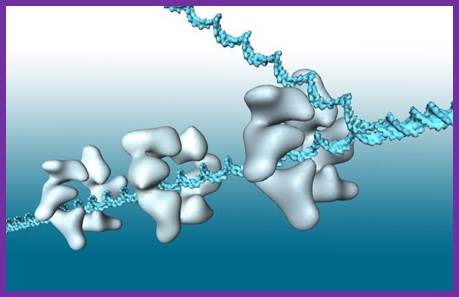
Helicase in motion unwinding ds DNA; http://iespoetaclaudio.centros.educa.jcyl.es/
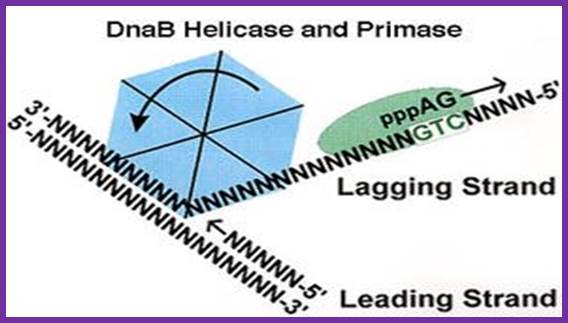 DNA
DnB- Helicase and Primase-polymerases
DNA
DnB- Helicase and Primase-polymerases
������������� The unwinding of double stranded DNA using Dna B helicase. Dna G is the primase. This picture was gathered from� http://chem-mgriep2.unl.edu/replic/Helicase.html� .� The primase DnaD (G) uses GTC sequences in the lagging strand and produces an eleven ntd lng RA as a primer; which provides 3�OH group for the DNA pol to use it for extension of the RNA into complementary DNA; http://www.palaeos.com
Dna-G:
�
It is a monomer, activated by helicase, binds to single strands in the replication bubble.� It specifically recognizes 3�GTC sequences on lagging strands and polymerizes RNA nucleotides on it and generates a short RNA of 11ntds long, and it is called RNA primer, which is used by DNA polymerases to extend the chain by incorporating dNTPs.� It is very active when it is bound to Dna-B.� Whether Dna-G is also responsible for laying primers on leading strand is not clear.� There are few views that RNA primers are laid on leading strand by cellular RNA polymerase, which is actually responsible for all transcriptional activity.� It is known that during initiation, the promoters on either side of the origin, back-to-back, are also active in transcription.� One set of primers is laid on leading strand for continuous strand synthesis.� Another set of primers are laid on lagging strand by Dna G, once in every 1000 or more nucleotides, the primers are laid by primase i.e Dna G.
����������� �����������������������������������������������
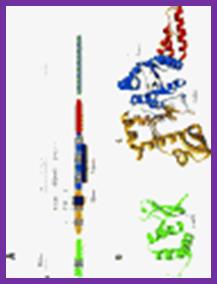 �
�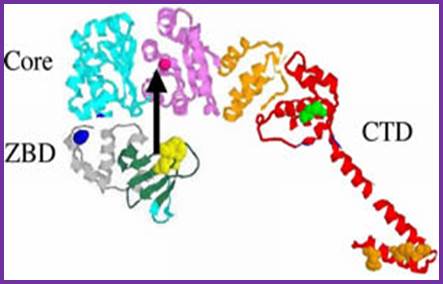
DNA primase- two views. http;//www.chem.unl.edu
The Primase is distinctly different from the regular RNA polymerase, which is multi subunit enzyme of 400kd or more.� This Primase is resistant to Rifamycin, while the Holozyme RNA polymerase is sensitive to Rifamycin.� The primase is a monomer and the RNAP is a pentamer and works as a complex.� The primase uses GTC sequences for laying primers on template strands, once in the leading strand and many times on the lagging strands at an interval of 1500 to 2000 ntds, but RNA polymerase holozyme uses Pribnow box and START nucleotide for initiation of transcription.
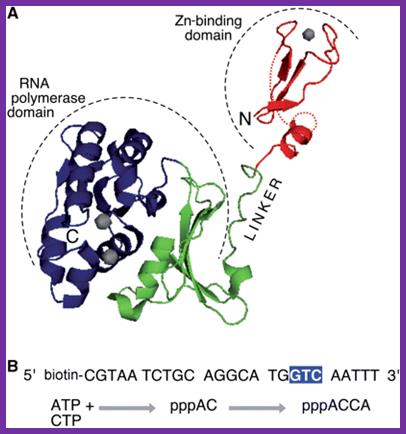
Figure: Organization of T7 DNA primase and template DNA used in this study. (A) The T7 DNA primase domain of gene 4 protein (residues 1�245) is composed of a ZBD (residues 1�54) and an RPD (residues 71�245) connected by a flexible linker region of 16 amino acid residues (6). The primase domain used in this study contains an additional 26 amino acid residues that, in the full-length gene 4 protein, connect the C-terminus of the primase to the helicase domain. (B) Sequence recognition and primer synthesis by T7 DNA primase. T7 primase recognizes a basic trinucleotide sequence (5′-GTC-3′ shown in box) at which the primase catalyzes the synthesis of the dinucleotide pppAC. If the appropriate nucleotides are present in the template, predominately G and T, then the primase will extend the dinucleotide to the functional tetra nucleotide, in this case, pppACCA. Note that the �cryptic� cytosine is not copied into the primer but it is essential for recognition; http://www.pnas.org/;https://richardson.med.harvard.edu
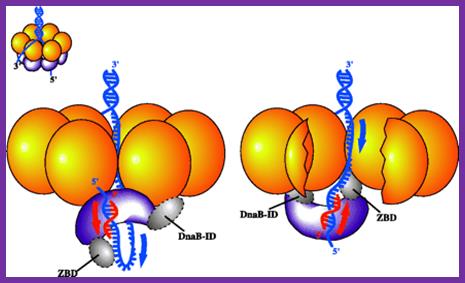
DnaG-primase and DnaB-helicase
Inhibition of this enzymatic activity abrogates microbial propagation and can lead to the development of potent drugs for treating and curing infection. Recently, helicase-primase inhibitors were reported for the treatment of herpes simplex disease. Almost 25 years after the launch of the milestone anti-herpes drug acyclovir (Zovirax�). The helicase-primase enzyme complex appears to be the Achilles heel of herpes simplex viruses and is a prime target of anti-infective drug discovery.
�
Hup and IHF proteins:
They are histone like proteins and found to stimulate initiation of DNA replication, but the exact nature of action of these proteins is not clear.� Similarly, there are few more proteins named as Integration Host Factors (IHF), as dimers, bind to DNA in the region of origin and stimulate replication. They are the products of genes him-A and him-B genes.� The binding of IHF to the major groove induces the DNA bending more than 40�.
Topoisomerases:
There are two types of Topoisomerases in E.coli, one is Topoisomerase I and the other is Topoisomerase II. When replication fork moves during replication, the helically coiled DNA ahead of the fork is rendered positively super coiled and further movement of the fork is blocked.� To facilitate the movement, Topoisomerase-II, also called Gyrase removes positive super coils and introduces negative super coils, which are again relaxed by the same topoisomerase, so the DNA relaxes ahead of the replication fork movement and facilitates uncoiling.

Type1 Topoisomerase; en.wikipedia.org
TP1A Topoisomerase; www.PNAS.org
Topoisomerase I- mechanism of unwinding super coil by cutting one strand and rewinding and ligating the cut strands; www.progchem.ac.cn
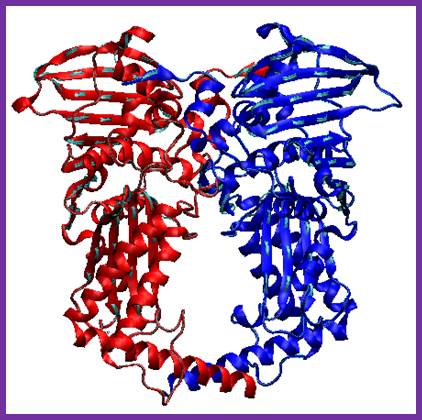
Type II Topoisomerase; Binds to DNA in sequence specific manner, cuts both the strands and swivals brings back the ends and ligate them.www.en.wikipedia.org
�����������������������������������������������������������
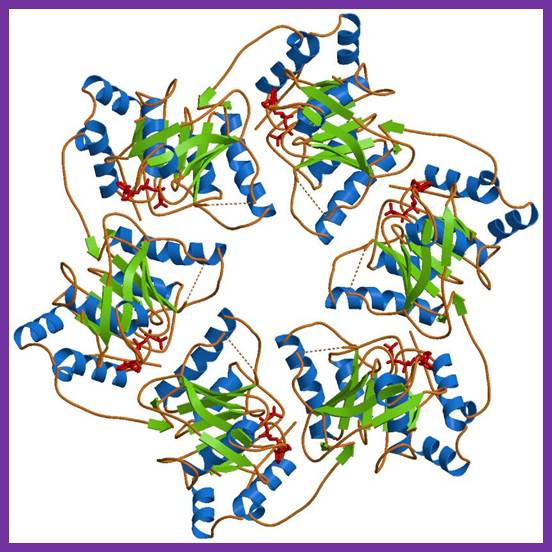
Tus protein:
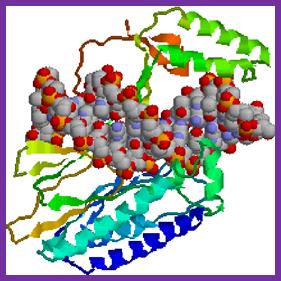
The figure shows 3-D sketch of TER binding proteins called TUS.
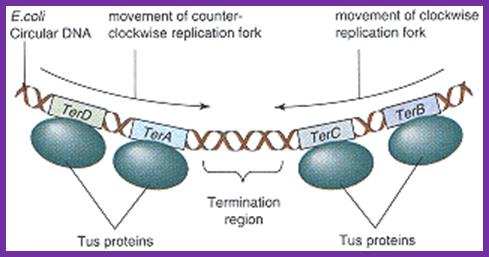
Termination of DNA replication in E. coli.
Termination region (ter) of E. coli incorporates five
asymmetric ter sites. Each ter site can interact with Tus
protein. TerB and terC are oriented in the same direction
and the remaining three ter sites are oriented in the
opposite direction. Because of the orientation of
Tus-bound ter sites, each replisome that reaches the ter
region must cross all the Tuster sites that are
oriented the opposite way before arriving at a site that
causes termination. A replisome moving in the direction
shown by the arrow must first cross terE, terD, and terA
before terminating replication at either the terC or terB
site. This arrangement ensures that each replisome
continues to synthesize DNA until it collides with a
replisome entering the ter region from the opposite
direction, leading to the dissociation of both replisomes
from DNA.
Adapted from Hidaka, M., Kobayashi, T., and
Horiuchi, T. J. Bacteriol. 173:381, 1991.; http://lion.freeoda.com/
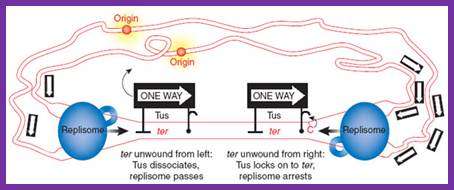
The region in which the two replisomes terminate is defined by multiple ter sites on the DNA, to which the Tus protein (one-way sign) binds. Mulcair et al.2 show that Tus-ter interactions are substantially strengthened upon ter unwinding from the right; this effect is independent of Tus-replisome interactions and is attributed to a binding site on Tus for a conserved cytosine in the ter sequence that is flipped out of its DNA duplex orientation when unwound (see complex on the right). In the cartoon, the replisome on the right has reached its final destination and is arrested, whereas the one on the left still needs to pass two Tus�ter complexes. http://www.nature.com/
Termination sequence utilizing proteins are called Tus proteins. They bind to 23 bp non-palindromic sequences organized in TER site; out of six-seven such sequences three each are oriented in opposite direction.� Binding of Tus proteins stop further movement of helicase and DNA polymerase into newly synthesized DNA strands and also causes disassembly of the polymerase complex?
As the replication forks form, SSBs bind to single stranded regions, helicases assemble on lagging strand at the junction of Y of the fork, primase assemble at the fork and bind to Helicase and lay primers, one each on leading strand complementary to 3�GTC, where the first ribonucleotide that assembles is 5�pppA, then C.� Primers, though on opposite DNA template they are in opposite orientation. Similarly short RNA primers are added on lagging strands using 3�GTC sequences.� Once the primosome complex (Dna-G and Dna-B) is formed, DNA polymerase (a huge complex of proteins) joins the forks, two complexes at each fork, one complex for the leading strand and the other for lagging strand.� For every 1000 to 1500 nucleotides assembled on lagging strand DNA polymerase disassembles and as the primers are produced DNA polymerase is loaded at the 3� end of the primer and replication continues.
DNA Polymerases:
DNA polymerases in prokaryotes, responsible for the replication, have multiple subunits and very complex in organization.� It is not one type of polymerase that works in replication; there are more than four such polymerases that conduct the process.� Mutational studies showed that there are five kinds of DNA polymerases that operate in E.coli, two during replication and others during recombination and DNA repair.
Pol I: Is implicated in DNA repair and has both 5'->3'(Nick translation) and 3�->5' (Proofreading) exonuclease activity. �Pol II: Pol II is involved in replication of damaged DNA and has both 5'->3'chain extension ability and 3' ->5' exonuclease activity. �Pol III: is the main polymerase in bacteria (elongates in DNA replication), as such it has 3'->5' exonuclease proofreading ability. Pol IV: is a Y-family DNA polymerase, Pol V: is a Y-family DNA polymerase and participates in bypassing DNA damage
Among the polymerases, DNA polymerase I, DNA polymerase II and DNA polymerase III are known to be involved in DNA replication and repair.�� Arthur Kornberg and Malcolm Gefter (1970), for the first time showed DNA synthesis in vitro, using a DNA polymerase-I, for which Arthur Kornberg was awarded Nobel Prize, but later Kornberg�s enzyme turned out to be a repair enzyme and DNA polymerase III complex was found to be the real replicating enzyme.� Besides polymerases, another important enzyme required is DNA ligase.� A complex of proteins, enzymes and few uncharacterized factors operate during replication in stepwise and sequence wise mode.
Important Features of DNA polymerases:
|
|
Pol-I |
Pol-II |
Pol-III |
Genes |
Pol-A |
Pol-B |
Pol-C or E |
|
Mol.wt (kd) |
103? |
88-90 |
>830 |
|
Subunits |
One |
1 x4 |
>Twelve |
|
Enzyme no�s per cell |
400 |
Produced when required |
10 to 15 |
|
Functions: |
|
|
|
|
5��3� polymerase |
Yes |
Yes |
Yes |
|
5��3� exonuclease |
Yes |
No |
No |
|
3��5� exonuclease |
Yes |
yes |
yes |
|
|
|
|
|
|
Polymerase |
activity |
template/primer: |
|
|
|
|
|
|
|
Intact duplex |
No |
No |
No |
|
Primed ssDNA |
Yes |
Yes |
Yes |
|
Nicked duplex |
Yes |
No |
No |
|
Duplex with gap (gap filling activity) |
Yes |
Yes |
Yes |
|
Effect of SSBs |
No |
Stimulates |
Stimulates |
|
Synthesis de novo |
Yes |
No |
No |
|
Activity with: 20mM KCl. 50mM KCl. 100mM KCl |
60% 80% 100% |
60% 100% 70% |
100% 50% 10% |
|
Km for nucleotides |
Low |
Low |
High |
|
Beta �subunit |
No stimulation |
Stimulates |
Stimulates |
|
Effect of SH-blocking agents |
No effect |
Blocks |
Blocks |
|
Turnover per second |
16-20/sec |
~40/sec |
250-1000/sec |
|
Rate of synthesis (incorporation of ntds per min) |
100-200/min |
>10 000/min |
>50000/min |
|
Nick translation activity |
Yes |
No |
No |
|
Random priming activity |
Yes |
- |
No |
|
5��3� gap filling activity |
Yes |
- |
No |
|
Can be used in C-DNA preparation? |
Yes, in second strand synthesis |
- |
No |
|
Processivity |
~3-100 ntds |
~10 000 ntds |
2.3 x 10^6 ntds |
|
Fidelity |
Poor |
SoS-repair |
Very high |
|
Overall activity |
Removal of RNA primers, Gap filling, DNA-Repair, |
SOS-repair |
Replication |
|
Primer removal |
yes |
? |
No |
|
DNA mis-match Repair |
Yes |
yes |
yes |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
DNA polymerase I:
This was the first polymerase enzyme discovered and this was the first polymerase that was studied in detail.� Dr. A. Korenberg was awarded Nobel Prize for this discovery; it was thought this enzyme is responsible for DNA replication, but later it turned out, it to be the DNA polymerase involved in Replication.� Using mutants, its functions have been delineated. This gene has been genetically manipulated and shortened as DNA-polymerase large or Klenow fragment, now it is available as recombinant gene and its product is sold all over the world.
�
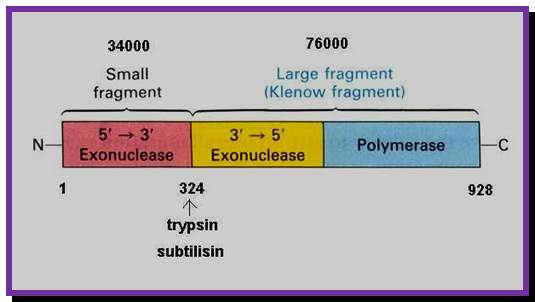
The picture shows different domains of DNA pol I protein.
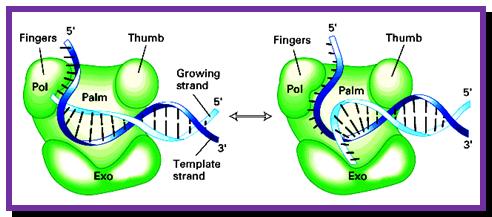
DNA Polymerse-I 3D Model: Palm, Thumb and Fingers each having specific functions; https://www.slideshare.net
DNA polymerase1
The protein is a single polypeptide chain, it is folded into 3-D structure that resembles a palm, where the fingers can be closed and opened. The enzyme 928aa long can dimerize in the presence of Hg^2+, and the dimerized protein exhibits very high enzyme activity.�
- The NH2-terminal region, nearly at 1/3 rd of the polypeptide chain possesses 5� to 3� exonuclease activity; it accounts to about 35 KD.� This property is unique among known polymerases including both DNA and RNA polymerases.�
- The rest of the fragment (68 KD) is called large fragment or Klenow fragment.� The NH2 terminal part of the Klenow has 3� to 5� exonuclease activity and the rest has 5�>3�polymerase activity.
- The palm like part of the enzyme, is folded in such a way, it can hold ds DNA in its palm which contains two sites; one for the binding of the template and the other for the binding to primers 3� OH group.� The thumb part has exonuclease activity.�� Perhaps this is the only enzyme that has versatile activities; 5�.3� exonuclease, 3�>5� exonuclease and 5�>3� polymerase activities.��
- The enzyme, during polymerization activity, shows contractility.� The folding of the protein is in such a way, it generates a groove, so it can accommodate the ssDNA chain as the template and the primer at polymerization point or extension point, which is also called catalytic site.� The intact bound DNA provides template and sequence specificity, while other domain that recognizes the 3� OH group provides active site for catalyzing phosphodiester bond formation using the arriving dNTPs.
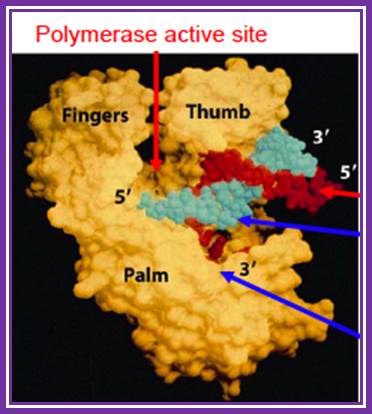 �
�
DNA-Polymerse active http://www.personal.psu.edu/;study.blue.com
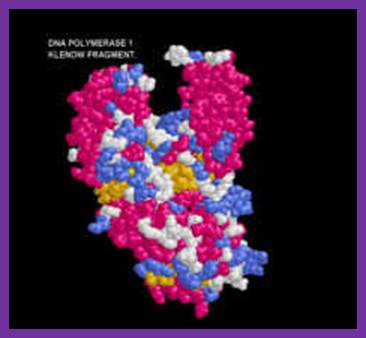
Klenow fragment ; http;//www.users.csbsju.edu
����
����������������������������
- The nucleotide triphosphates actually diffuse on to the active site which is geometrically compatible to accept the correct nucleotide and allows to base pair strictly according to Watson-Crick base pairing rule.� If the diffused nucleotide is not correct, if the pairing is not proper, they are rejected. If the pairing is correct and satisfies both thermodynamic requirements and geometric complementarity, then the enzyme performs catalytic activity by breaking phosphate bond at end of the alpha phosphate position of the tri-phosphate nucleotide and forms the covalent bond with alpha phosphate group with that of 3�OH group of the RNA primer.�
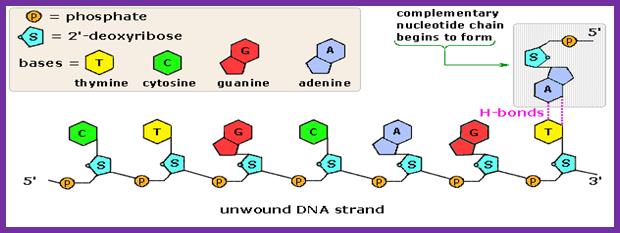
Complementary chain formation; https://www2.chemistry.msu.edu/faculty
- Pyro phosphotase hydrolyses the pyrophosphate (ppi) and the released energy is used for the polymerase to move on the template, one such function is to generate conformational changes in the protein to accommodate the arriving dNTP, screen and activate covalent bond formation in accordance with perfect Watson-Crick base pairing.
- While it performs polymerization it requires optimal number of dNTPs, otherwise the exonuclease domain gets activated.�
- Ions like Mn2+ and Zn2+ are required for its exonuclease activity.� In the larger domain it has preferred site for Mg2+.� Activated nucleotides are the preferred substrates.� Activated dNTPs means, they are bound by bivalent cat ion Mg2+ where the divalent cat ion is bound to both beta and gamma phosphate groups of nucleotides with negative charges. This is the preferred form of nucleotide that is required for the polymerases.�
- Activity wise, it can remove RNA primers from 5� ends and displace the strand by 5 to 10 nucleotides long without cutting each of the phosphodiester bonds individually. It can also remove short DNA fragments from 5� positions on nicked positions.
- The enzyme has a good 3�-5� exonuclease activity by which it removes any mismatches or wrong nucleotides or if there is any wrong base pairing while the enzyme performs polymerization on the template.�
- During polymerization, if it finds any mismatches in base pairing, it stops polymerization and moves backwards, while moving backwards it chips out nucleotide by nucleotide.� In it 3�-5� exonuclease activity it not only removes the incorrect ntds but also removes few more correctly base paired nucleotides, this is an act of precaution than required.�� This ensures high fidelity. The exonuclease activity is very helpful not only during replication but also in DNA damage repair and recombination processes.�
- The 5� �3� exonuclease activity plays an important role in removing errors at 5� end and also in removing Thymidine dimers (cross-linked).�� The 5� exonuclease domain can also perform endonuclease activity by nicking dsDNA to generate a 3�OH group from which it can displace the nicked strand in 5��3� direction up to 10 or more nucleotides long fragment and at the same time it can extend chain from the nicked point by polymerization in 5�-3� direction.� Thus one can consider this enzyme is one of the most utility enzyme and versatile.
- �Perhaps this versatility stems from the fact, during evolution, different genes or parts of genes are brought together as one enzyme.� E.coli 5��3� exonuclease domain of DNA-polymerase-I and its 5� displacement characters are shared with T5 phage and O29 phage DNA polymerases.
����������� ������ -I------38 KD---I-------------I--------68 KD-----I
����������� NH2I -----------------I-------------I-------------------I.COO^-
������� ��������������� 5�>3�exo��������������������� 3�-5� exo. �5�� 3� polymerase
|
Activity |
35kd Fragment |
68 kd Fragment |
|
5��3� pol activity |
No |
Yes |
|
5��3� Exonuclease activity |
Yes |
No |
|
3�� 5� Exonuclease activity |
No |
Yes |
DNA polymerase II:
In 1971, Arthur Korenberg isolated a mutant from E. coli strain; this has led to the discovery of two more DNA polymerases, called DNA POL II and DNA pol III.� There are few conflicting reports about the Mol.wt and the number of subunits found in Pol II.�� Yet the overall structure shows similarities with eukaryotic DNA polymerase� alpha subunit.
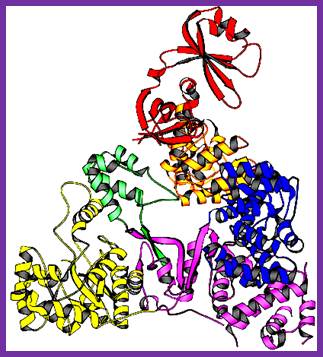
������������� Thermus aquaticus: DNAP II; www.necat.chem.cornell.edu
Its absence doesn�t affect the growth of the bacteria. It has greater affinity towards single strands with primers; it is strongly stimulated by SSB proteins.� It has no 5� �3�exonuclease activity, but it has 5��3� polymerase and 3��5� exonuclease activity.� It is induced during DNA damage and it is involved in repair events and also active during genetic recombination.
DNA polymerase III:
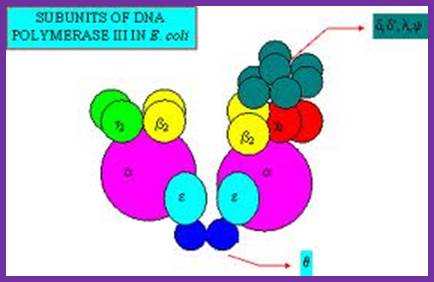
Among the polymerases found in E. coli, DNA pol III is the enzyme that is responsible for actual replication.�� This was discovered by Roger Korenberg, son of Arthur Korenberg (Nobel laureate). The said polymerase is not one polypeptide chain, but made up of a complex of 4 subunits plus beta clamp 2 subunits and 5 subunits beta clamp loading Gamma complex.� Each of the subunits and their genes has been identified and their characters and functions have been delineated to certain extent.� All subunits together are called Holoenzyme.� Basically, it consists of catalytic core complex and an accessory complex.� Assembly of the Holozyme is hierarchical and the Holozyme dissociates and associates very rapidly and exhibits dynamic features.
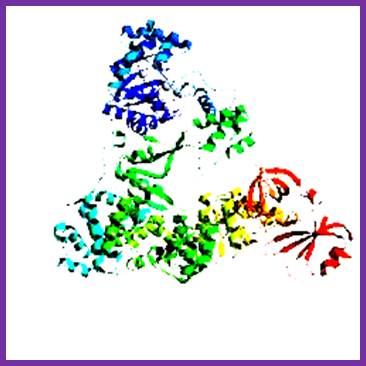
DNAP III core; File: DNS Polymerase III Aufbau.png: From Wikimedia Commons, the free media repository.
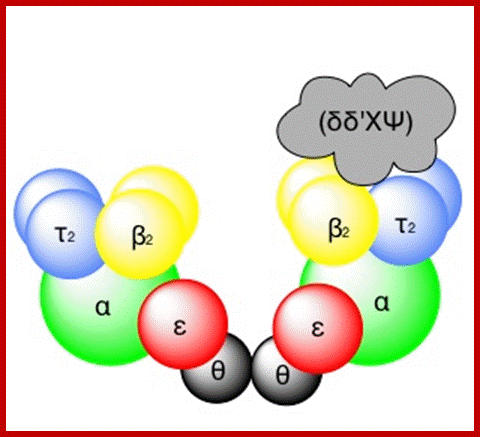
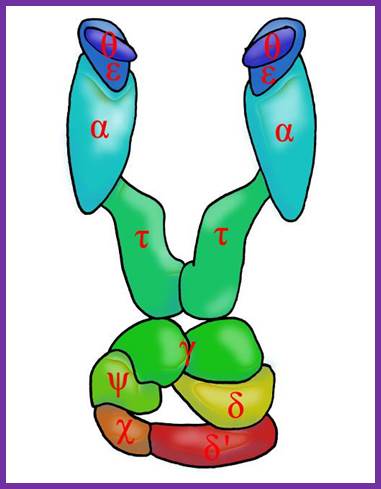
DNA pol III Holozyme complex assembled- overview: 2Theta, 2epsilon, 2alpha 2Tau and 2 beta clamps associated with clamp loader δδ�ΧΨ; https://commons.wikimedia.org
The DNA-pol-III is differentiated from the other two because of its multiple subunits, high molecular mass, high fidelity and super processivity and the rate of synthesis is 50 to 60 kb per minute, fastest among any known multisubunit DNA polymerases.�� Error rate is minimum, possibly one in every 10^10 bp.�� It doesn�t show 5�->3� exonuclease, but it has inherent 3�-5� exo and strand displacement or nick translation activity.� This Holozyme not only performs replication of bacterial DNA but also involved in replication of some parasitic phage DNA such as PhiX174 and M13.� It can work on both linear as well as circular DNAs.�� It can use 3�OH groups of RNA or DNA nicks and use them as primers.
The core part of the enzyme consists of tightly bound subunits- Alpha, Epsilon, Theta and Tau.� Accessory complex is made up of several subunits like gamma, delta, delta�, chi, psi and beta.
Components of DNA Pol-III Holozyme Complex:
|
Protein����������� ����������� ����������� |
Genes |
Mol.wt (kd) |
Subunits |
Activity |
|
Core complex: |
|
|
|
|
Alpha |
Dna-C or E |
132(129) |
1 |
5�>3� pol |
|
Epsilon |
Dna Q |
27.5 |
1 |
3�>5� exonuclease |
|
Theta |
Hol-E |
10 |
1 |
Helps in core assembly |
|
|
|
|
|
|
Tau |
Dna-X |
71 |
1 |
Helps in dimerization of the core and stimulates epsilon�s 3��5� exonuclease activity ? |
|
DnaB |
DnaB |
50 |
6 |
Hexamer; helicase-5->3� |
|
Accessory complex- gamma-delta complex; |
|
|
|
|
Gamma |
Dna-X |
52 (47.5) |
2 |
Binds ATP, Clamp loading, stator |
|
Delta |
Hol-a |
35 (38.7) |
1 |
Binds to Beta, Clamp loading on lagging strand |
|
Delta prime |
Hol-B |
33(36.9) |
1 |
Bind to gamma and beta, Clamp loading |
|
Chi |
Hol-C |
15 (16.6) |
1 |
Binds to ssb,Clamp loading |
|
Psi |
Hol-D |
12 (15.2) |
1 |
Binds to chi and gamma,Clamp loading |
|
Beta subunit |
Dna-N |
37 (40.60 |
homodimer |
Act as DNA clamps |
|
Total Mol.wt and total number of sub units |
|
512 (��� ) |
12 |
|
����������������������������������� �
Alpha subunit:� This is the largest and the important subunit; it has template binding and primer binding sites.� It also contains an active site for free nucleotide binding next to the primer 3�end. The same site acts as a catalytic site for polymerization of properly base paired nucleotides from 5� to 3� direction.� It has no 3�-->5� exonuclease activity.� The rate of DNA synthesis depends upon this subunit.
�
Epsilon subunit:� This is always complexed with alpha subunit in 1:1 ratio.� This has 3��5� exonuclease activity, thus it performs proof reading function.� Association of this increases the polymerase activity of alpha unit by two fold and 50 to 100-fold 3��5� exonuclease activity.� Mutants in this gene, called mut-D, lacks 3��5�exonuclease activity and increases the incorporation of Thymidine nucleotides.� During SOS situations this subunit is synthesized in large amounts and enables DNA repair.
Theta subunit:� Not much is known about this subunit.� However, it is discerned that this favors the association of alpha and epsilon subunit by binding to both and perhaps facilitate in building up of core complex.
Tau subunit:� This is a full-length product of the gene dna-X, which also gives raise to gamma subunit during translation by frame-shift in the reading frame.� This subunit has ssDNA dependent ATPase activity.� It helps in dimerization of the core complexes and enhances processivity of the core enzyme.� The gamma subunit is generated by recoding of the Tau mRNA.
������ �--------------------Tau------------------------�
������������������������� I---------------------------------------------------------I = dna-X
������������������������� I<-----gamma--�I�
Beta subunits:
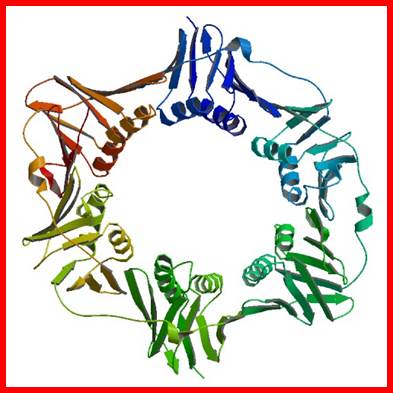
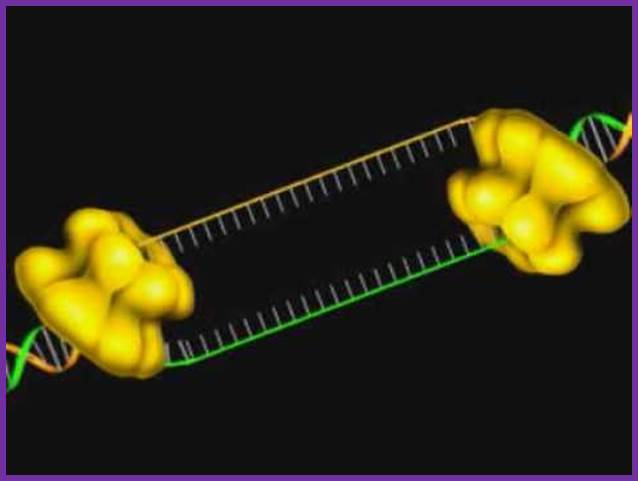
The diagram is a dimmer of DNA pol b, which act as a brace and the same is loaded by the g on to Replicating strand with a primer; Crystal structure; http://www.rcsb.org/
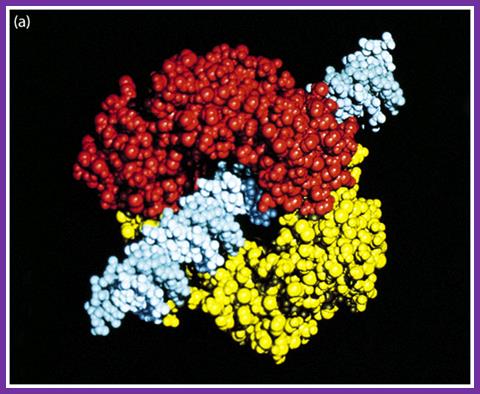
The figure is the model of beta-clamp protein encircling the DNA; http://www.cs.stedwards.edu/
This is a homodimer consists of two clamp shaped or bracket shaped structures.� On dimerization its size is about 80^A thick with a halo of 35 A� to hold ssDNA with a primer. The inner space is filled with water and shows six-fold symmetry and when loaded it encircles the DNA.�
The inner surface of the beta complex has high affinity to DNA with a primer.� This clamp subunits are loaded on to both leading strand and lagging strands behind Holozyme.�� The loading of the clamp is performed by gamma-delta complex in ATP dependent manner.�� Binding of the Beta clamps hold the enzyme and the template together tightly at template primer site, hence it increases the processivity of the enzyme.� Recycling of the Beta clamp takes place at each and every primer position on the lagging strand.� There are about 300 beta dimers (clamps) per cell; this number takes care of replication.� This gene is located in between dna A and rec F genes.� Expression of this gene depends upon the promoter located within dna-A gene.
Gamma complex:
Gamma subunit is a truncated translational product of the full-length mRNA coded for by dna-X gene (codes for tau protein).� It corresponds to 2/3 rd of the N-side of the tau protein.� This segment is generated during translation by frame shift.
Each gamma complex consists of subunits of Gamma, delta, delta�, chi and psi.� In ATP dependent manner the gamma and delta complex can bind the template at initiation complex.� All the components of the complex assemble in cooperative manner, and ATP is required for it.� The Gamma complex in the presence of ATP binds to beta clamp and loads the same on to the primer site where the Holozyme also binds. On the leading strand once the beta clamp is loaded, it need not be there in the complex.� However, on the lagging strand, the beta clamp has to be loaded at intervals for the primers are laid at frequent intervals of length.
Assembly of the Holozyme:
The Holozyme is formed in sequence and stepwise manner.� First alpha and epsilon interact with theta to form of complex, which is a core complex. And their processivity is 10 ntds long.� Then tau interacts with core complexes and dimerizes, this has a processivity of 60 ntds long.� These assemble then onto templates at replication fork where the primosome complex is found; this assembly or loading is assisted by gamma complex.� The same gamma complex also loads the beta clamps onto the template earlier to the core enzymes.� So there will be one gamma complex at each of the forks.� This whole complex, called Holozyme has a processivity of 10^5.� For its activity it requires SsBs.
 ���
���
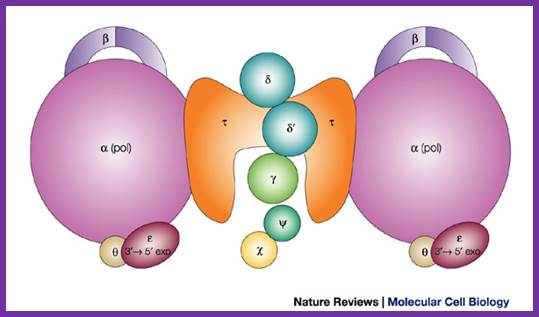
The DNA polymerase III holoenzyme is a multisubunit complex,
which consists of 17 polypeptides. It contains four subassemblies. First, the
core polymerase consists of three subunits: ![]() (the polymerase);
(the polymerase); ![]() (the 3'�5' exonuclease); and
(the 3'�5' exonuclease); and ![]() (the stimulator of the 3'�5' exonuclease). Second, the
(the stimulator of the 3'�5' exonuclease). Second, the ![]() subunit is responsible for dimerization of the core DNA
polymerase. Third, the sliding clamp comprises two homodimers of the
subunit is responsible for dimerization of the core DNA
polymerase. Third, the sliding clamp comprises two homodimers of the ![]() subunit, which provides the ring structure that is needed for
processivity. Fourth, five subunits have clamp-loader functions �
subunit, which provides the ring structure that is needed for
processivity. Fourth, five subunits have clamp-loader functions � ![]() ,
, ![]() ,
, ![]() ',
', ![]() and
and ![]() .--
.--
Architecture of the replicative DNA polymerase III holoenzyme from E. coli. Igor V. Shevelev & Ulrich H�bscher; ;http://www.nature.com/ Schematic picture of DNA polymerase III* (with subunits) and clamp loader; http://en.wikipedia.org/
DNA replication: http://bio.sunyorange.edu
![]()
Replicating DNA with Extraordinary Fidelity: Meet DNA Polymerase:
How does DNA polymerase add new nucleotides to the elongating strand? The polymerase's active site, found in the β-sheet that makes up the palm subdomain catalyes a phosphoryl transfer reaction. It forms a phosphodiester bond by linking the 3' hydroxyl group at the end of the template strand to the nucleotide's 5' phosphoryl group. The first step in the process is a nucleophilic attack on the α-phosphate of the incoming nucleoside triphosphate by the 3' OH of the growing chain. This reaction releases pyrophosphate (PPi). Within the active site, there are two conserved aspartate residues. The magnesium ions on the carboxylate groups of those aspartates is critical to the reaction. These carboxylate groups co-ordinate the magnesium ions and facilitate their participation in the catalysis by holding them in the right orientation. One of the two magnesium ions activates the 3' OH group of the terminal nucleotide. The other is responsible for stabilizing a developing negative charge on the leaving oxygen on the incoming nucleoside triphosphate. Side chains on an alpha helix in the finger domain interact with the incoming triphosphate to also stabilize it. Hydrolysis of the pyrophosphate released in this process generates the energy required for driving the reaction forward. For a more detailed review of the mechanisms involved, I refer readers to Rothwell and Waksman (2005). Jonathan M. http://www.evolutionnews.org/
Gamma + delta = gamma-delta;
Gamma-delta + delta� + chi + psi = gamma-delta complex;
Gamma complex + ATP + beta clamp = gamma-delta-beta complex.
Alpha + epsilon + theta = Core Complex;
Core complex + tau = Dimer Core Complex;
Dimer core complex can assemble on the DNA template, provided replication bubble with the replication fork exists. The gamma-delta complex can load the dimer beta clamps on to the DNA templates with primers.
With primers the beta clamps hold the DNA like a pair of half bracelets with a firm grip. As the beta clamps are in association with core complex and, enzymes won�t fall off easily, this feature enhances the processivity thousand-fold.� Association and dissociation of gamma complex with template DNA and the primer, especially on lagging strand and recycling of gamma-delta and beta clamps, occur frequently.
DNA ligase:� E. coli DNA ligase is a monomer with Mol.wt 74-75Kd.� It is NAD dependent unlike T4 ligase which is ATP dependent.� They bind to 5� end of the DNA strands.� The activated ligase is bound by NAD. A DNA adenylate intermediate is formed in the second step where the bound AMP is transferred to the 5′ end of DNA. The respective enzymes then catalyze the joining of the 3′ nicked DNA to the intermediate and release AMP in the final step. �When the enzyme binds to the 5�of the DNA strand it generates 5�P-P-NAD.� This is an energy dependent process ligates the 3�OH and 5�P and generate �ribose O-P-O ribose.
����������������������������������� DNA ligase; http://www.biochem.umed.edu
NAD dependent Ligase; Tys115; http://www.uea.ac.uk/
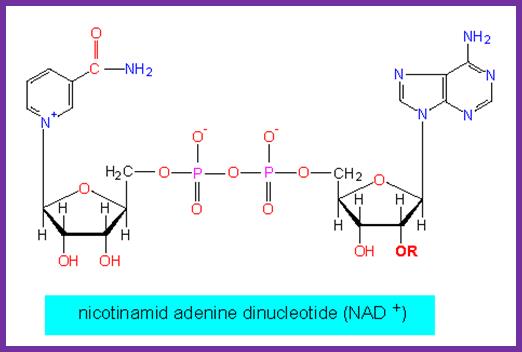
http://www.biologie.uni-hamburg.de/
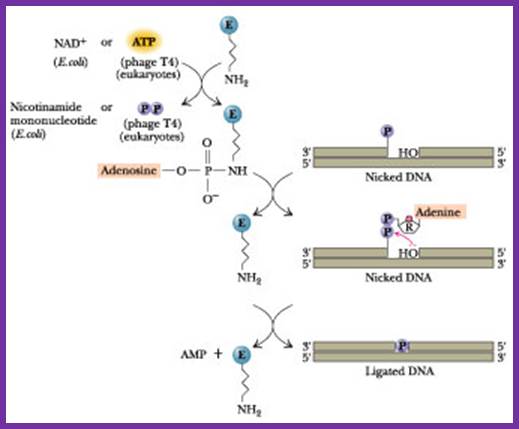
The diagram shown above is ATP dependent T4 ligase in action; http://www.:jwc.neau.edu.cn
Ligated DNA strands; http://en.wikipedia.org/