Replication-Factors and Enzymes:
|
Protein |
Gene |
Mol.wt (kd) |
Subunits/molecules per cell/turn over
|
Dna-A |
dna-A |
50 |
Monomer |
|
Dna-B |
dna-B (Helicase) |
50x6 |
Hexamer |
|
Dna-C |
dna-C |
29 |
Monomer |
|
SSB |
ssBs |
19x4 |
Tetramer |
|
Dna-G |
dna-G (primase) |
60 |
Monomer |
|
Rpo |
Rpo genes (many) |
~400 Kd |
Pentamer |
|
DNA-pol-I
|
Pol-I |
103 ? |
Monomer, a major repair enzyme/400/600 |
|
DNA-Pol-II |
Pol II |
~90 |
Major repair enzyme/30 |
|
DNA-pol-III |
3 genes |
>600 |
Multimer/40/1200 |
|
DNA pol alpha |
dna D |
129.9 |
Dimer |
|
DNA pol epsilon |
dna Q |
27.5 |
monomer |
|
DNA pol theta |
hol E |
8.6 |
monomer |
|
Gamma complex |
|
|
|
|
Beta clamp |
dnaN |
40.6x2 |
dimer |
|
Rep-A |
rep-A |
65-76 |
Monomer, helicase 3�>5 |
|
Hup-A |
hup-A |
9.5 |
Monomer |
|
Hup-B |
hup-B |
9.5 |
Monomer |
|
Top-I |
top-I |
100 |
Monomer, breaks single strand |
|
Top-II-A |
top-II-A |
97 |
Dimer, together breaks both strands |
|
Top-II-B |
top-II-B |
97 |
Dimer |
|
Tus |
Vnu |
37 |
Monomer |
|
Top-III/IV |
top-III/IV |
100 |
Monomer |
|
DAM methylase |
dam-methylase |
32 |
Monomer |
|
Pol IV-Din B |
|
|
SOS repair enzyme |
|
Pol-V-Umuc and UmudD2C |
|
|
SOS repair enzymes |
|
IHF |
Integration host factor-like histones |
|
Bends DNA and stabilizes |
|
Fis |
|
|
Factor for inversion |
|
Rob |
|
|
Right arm ori binding protein |
|
Seq A |
|
|
Binds to GATC� at Dna-A boxes |
|
Dna ligase |
|
|
|
|
|
|
|
|
Dna-A:
�Initiation of replication requires the expression of dna-A gene, which is transcribed and translated to produce Dna-A protein.� Its gene expression requires full methylation in its promoter region at GATC sites.� The protein is a monomer, has motifs to bind to unique monomer sites, also they have motifs for protein-protein interaction, thus they can form clusters.�� Proteins don�t have helix turn helix motif nor they have zinc finger structure, but they have hydrophobic regions for helical coiling and protein �protein interactions, yet the characteristic features of protein binding to DNA is still to be understood.� Binding of the monomers to Dna-A boxes, in ATP dependent manner (proteins have very slow ATPase activity), leads to cooperative binding of more proteins.� This clustering on DNA makes the DNA to wrap around the proteins, which induces torsional twist and it is this twist that makes DNA to melt at 13-mer facilitate the melting of the DNA. Opening or unwinding of dsDNA into single stranded region is an important event in initiation.
�
SS-B:
These proteins, though small, as they are translated, they bind to phosphate (P^2-) group of P-S-P backbone of DNA as tetramers, hence they are called single strand DNA binding proteins. Binding of the said proteins stabilizes the DNA and makes it as a rigid template and also prevents reannealing of the strands.� When they are bound, they cover approximately 40-60 ntds. The DNA polymerase, while it is producing complementary strand SsBs are displaced and they bind DNA only when the single strand is free from polymerases. In addition, they provide the DNA strand as a straight rod like template structures for the enzyme to perform complementary strand synthesis.� They are also required for fork movement.
Dna-C:
It is a monomer, binds to Dna-B in one to one manner.� It helps or facilitates the helicase to load onto ssDNA at replication fork.� Once helicase assembles, Dna-C dissociates from the helicase subunits.
Dna-B:
The protein, to begin with it is a monomer, but with the binding of Dna-C in one-to-one manner, they start assembling on ssDNA as a hexamer in the form of a ring around the strand, at the joint region of the fork. This assembles only on lagging strand, which is identified by the strand orientation from 5� towards 3� direction.� Once the Dna-B ring is formed Dna-C dissociates.� The Dna-B complex is a motor protein and acts as DNA dependent ATPase and using the energy it drives into the fork and unwinds the DNA progressively like an unzipped.� Its direction of movement is from 5� to 3� on the lagging strand.� There is another protein called Rep A, which as monomer also binds to fork region but to the leading strand and moves in 3� to 5� direction.� However involvement of this protein in E.coli DNA replication is not substantiated.
Prokaryotic Helicases:
|
Helicase II (Uvr-D) |
82 |
3�� 5� |
DNA repair |
|
Uvr AB complex |
180 |
5�� 3� |
DNA repair |
|
Helicase-IV |
75 |
3�� 5� |
DNA repair |
|
Rec-BCD |
330 |
3�� 5� |
Recombination |
|
T4 Dda? |
49 |
5�� 3� |
Displaces SsBs |
|
Helicase-I (encoded by F-plasmids) |
180 |
5�� 3� |
Involved in DNA transfer during conjugation |
|
Helicase-III |
20 |
5�� 3� |
Unknown function |
|
Rho |
50 x 6 |
5�� 3� |
On RNA transcript, performs transcriptional termination |
Dna-G:
�
It is a monomer, binds to single strands in the replication bubble.� It specifically recognizes 3�GTC sequences on lagging strands and polymerizes RNA nucleotides on it and generates a short RNA strand called RNA primer, which is used by DNA polymerases to extend the chain by incorporating dNTPs.� It is very active when it is bound to Dna-B.� Whether Dna-G is also responsible for laying primers on leading strand is not clear.� There are few views that RNA primers are laid on leading strand by cellular RNA polymerase, which is actually responsible for all transcriptional activity.� It is known that during initiation, the promoters on either side of the origin, back-to-back, are also active in transcription.� One set of primers is laid on leading strand for continuous strand synthesis.� Another set of primers are laid on lagging strand, once in every 1000 or more nucleotides, the primers are laid by primase. The Primase is distinctly different from the regular RNA polymerase, which is multiple subunit enzyme 0f 400kd or more. Primase is resistant to Rifamycin, while the Holozyme RNA polymerase is sensitive to Rifamycin.� The primase is a monomer and the RNAP is a pentamer and works as a complex.
�
Hup proteins:
They are histone like proteins and found to stimulate initiation of DNA replication, but the exact nature of action of these proteins is not clear.� Similarly, there are few more proteins named as IHF (integration host factors, bind to DNA in the region of origin and stimulate replication. They are the products of genes him-A and hup- genes.
Topoisomerases:
There are two types of Topoisomerases in E.coli, one is Topoisomerase I and the other is Topoisomerase II. When replication fork moves during replication, the helically coiled DNA ahead of the fork is rendered positively super coiled and further movement of the fork is blocked.� To facilitate the movement, Topoisomerase-II, also called Gyrase removes positive super coils and introduces negative super coils, which are again relaxed by the same topoisomerase, so the DNA relaxes and facilitates uncoiling.
Tus protein:
It is 36kd protein; It binds to 23 bp non-palindromic sequences organized in TER site; out of six such sequences three each are oriented in opposite direction.� Binding of Tus proteins stops further movement of helicase and enzymes into newly synthesized dsDNA.
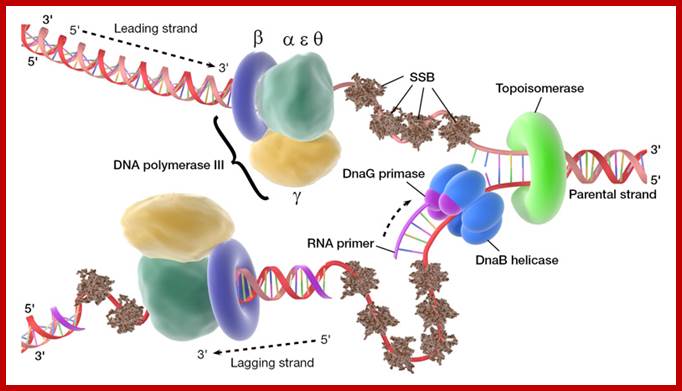
Dna B hexamer protein is in ATP dependent manner acts as a motor protein.� Loading of helicase on to ssDNA at fork requires the binding of Dna-C to DnaB one to one and it loads on to DNA in ATP dependent manner, where the ring like hexamer opens and threads on to ssDNA and closes. As the replication fork forms, SSBs bind to single stranded regions, helicases assemble on lagging strand at the junction of Y of the fork, Primases assemble at the fork and bind to Helicase and lay primers, one each on leading strand complementary to 3�GTC, where the first ribonucleotide assembled is 5�A, then G.� Primers, though on opposite DNA template they are in opposite orientation. Similarly short RNA primers are added on lagging strands using 3�GTC sequences.� Once the primosome complex (Dna-G and Dna-B) is formed, DNA polymerase (a huge complex of proteins) joins the forks, two complexes at each fork, one complex for the leading strand and the other for lagging strand.
DNA Polymerases:
DNA polymerases, responsible for the replication in prokaryotes, have multiple subunits and very complex in organization.� It is not one type of polymerase that works in replication; there are more than two such polymerases that conduct the process.� Mutational studies showed that there are three kinds of DNA polymerases that operate in E.coli, two during replication and another during recombination and DNA repair.
They are DNA polymerase I, DNA polymerase II and DNA polymerase III.�� Aurther Korenberg, for the first time showed in vitro DNA synthesis, using a DNA polymerase-I, for which he was awarded Nobel Prize, but later Kornberg�s enzyme turned out to be a repair enzyme and DNA polymerase III complex was found to be the real replicating enzyme.� Besides polymerases, another important enzyme required is DNA ligase.� A complex of proteins, enzymes and few uncharacterized factors operate during replication in stepwise and sequence wise mode.
DNAP III was discovered by Thomas Kornberg (son of Arthur Kornberg) and Malcolm Gefter in 1970. It has high processivity. �It works in conjunction with PolIV and PolV.
Important Features of DNA polymerases:
|
|
Pol-I |
Pol-II |
Pol-III |
Genes |
Pol-A |
Pol-B |
Pol-C or E |
|
Mol.wt (kd) |
103? |
88-90 |
>830 |
|
Subunits |
One |
Four? |
>Fifteen |
|
Enzyme no�s per cell |
400 |
? |
10 to 40 |
|
Functions: |
|
|
|
|
5��3� polymerase |
Yes |
Yes |
Yes |
|
5��3� exonuclease |
Yes |
No |
No |
|
3��5� exonuclease |
Yes |
No |
No |
|
Polymerase activity with template/primer: |
|
|
|
|
Intact duplex |
No |
No |
No |
|
Primed ssDNA |
Yes |
Yes |
Yes |
|
Nicked duplex |
Yes |
No |
No |
|
Duplex with gap (gap filling activity) |
Yes |
Yes |
Yes |
|
Effect of SSBs |
No |
Stimulates |
Stimulates |
|
Synthesis de novo |
Yes |
No |
No |
|
Activity with: 20mM KCl. 50mM KCl. 100mM KCl |
60% 80% 100% |
60% 100% 70% |
100% 50% 10% |
|
Km for nucleotides |
Low |
Low |
High |
|
Beta �subunit |
No stimulation |
Stimulates |
Stimulates |
|
Effect of SH-blocking agents |
No effect |
Blocks |
Blocks |
|
Turnover per second |
16-20 |
~7 |
250-1200 |
|
Rate of synthesis (incorporation of ntds per min) |
600-800ntds/min, (1000ntds per sec)? |
>10 000/min |
>50000/min |
|
Nick translation activity |
Yes |
No |
No |
|
Random priming activity |
Yes |
- |
No |
|
5��3� gap filling activity |
Yes |
- |
No |
|
C-DNA preparation |
Yes, in second strand synthesis |
- |
No |
|
Processivity |
~3-100 ntds |
~10 000 ntds |
2.3 x 10^6 ntds |
|
Fidelity |
Poor |
SoS-repair |
Very high |
|
Overall activity |
Removal of RNA primers, Gap filling, DNA-Repair, |
SOS-repair |
Replication |
|
|
A major repair enzyme |
A minor repair enzyme |
Replicase |
|
Rate/sec |
16-20/sec |
40/sec |
250-1000/sec |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Note that din B and Um U D2 C are SOS repair enzymes.� They are so good the error rate is one in one billion base pair incorporation.
DNA polymerase I:
This was the first polymerase enzyme discovered and this was the first polymerase that was studied in detail.� Using mutants, its functions have been delineated. This gene has been genetically manipulated and shortened as DNA-polymerase large or Klenow fragment, now it is available as recombinant gene and its product is sold all over the world.
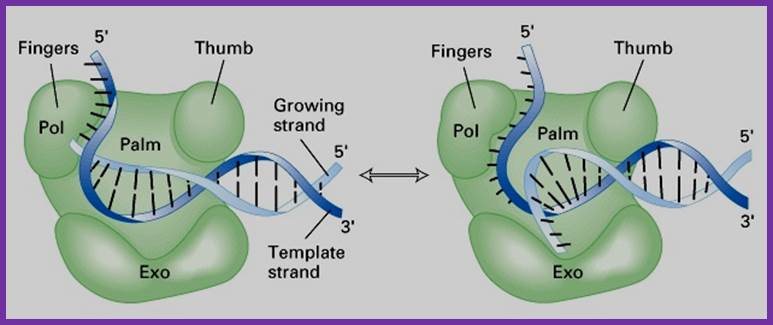
- The protein is a single polypeptide chain, it is folded into 3-D structure that resembles a palm, where the fingers can close and open on to the palm.�
The enzyme 928aa long can dimerize in the presence of Hg^2+, and dimerized protein shows very high enzyme activity.
�
- The NH2-terminal region, nearly at 1/3 rd of the polypeptide chain possesses 5� to 3� exonuclease activity; it accounts to about 35 KD.� This property is unique among known polymerases including both DNA and RNA polymerases.�
The rest of the fragment (68 KD) is called large fragment or Klenow fragment.� The NH2 terminal part of the Klenow has 3� to 5� exonuclease activity and the rest has 5�->3� polymerase activity.
- The palm like part of the enzyme folded in such a way it can hold ds DNA and contains two sites, one for the binding of the template and the other has the binding site of primer3� OH group.� The thumb part has exonuclease activity.�� Perhaps this is the only enzyme that has versatile activities. ��
The enzyme, during polymerization activity, shows contractility.� The folding of the protein is in such a way it has a groove, so it can accommodate the ssDNA chain as the template and the primer at polymerization point or extension point, which is also called catalytic site.� The intact bound DNA provides template and sequence specificity, while other domain that recognizes the 3� OH group provides active site for catalyzing phosphodiester bond formation using the arriving dNTPs.
�
- The nucleotide triphosphates actually diffuse on to the site which is formed geometrically to accept the correct nucleotide and allows to base pair strictly according to Watson-Crick base pairing rule.� If the diffused nucleotide is not correct, if the pairing is not proper, they are rejected. If the pairing is correct and satisfies both thermodynamic requirements and geometric complementarity, then the enzyme performs catalytic activity by breaking phosphate bond at alpha position of the tri-phosphate nucleotide and forms the covalent bond with alpha phosphate group to that of 3�OH group of the RNA primer.�
Pyro phosphotase hydrolyses the pyrophosphate (ppi) and the released energy is used for a variety of purposes, one such function is the movement of the enzyme on the template, produce conformational changes to accommodate the arriving dNTP, screen and form a covalent bond in accordance with perfect Watson-Crick base pairing.
While it performs polymerization it require optimal amount of dNTPs, otherwise the exonuclease domain gets activated.�
�
- Ions like Mn2+ and Zn2+ are required for its exonuclease activity.� In the larger domain it has preferred site for Mg2+.� Activated nucleotides are the preferred substrates.� Activated dNTPs means, they are bound by bivalent cat ion Mg2+ where the divalent cat ion is bound to both beta and gamma phosphate groups of nucleotides with negative charges. This is the preferred form of nucleotide that is required for the polymerases.�
Activity wise, it can remove RNA primers from 5� ends and displace the strand by 5 to 10 nucleotides long without cutting each of the phosphodiester bonds individually.�
It can also remove short DNA fragments from 5� positions on nicked positions.
The enzyme has a good 3�-5� exonuclease activity by which it removes any mismatches or wrong nucleotides or if there is any wrong base pairing while the enzyme performs polymerization on the template.�
- During polymerization, if it finds any mismatches in base pairing, it stops polymerization and moves backwards, while moving backwards it chips out nucleotide by nucleotide.� In it 3�-5� exonuclease activity it not only removes the incorrect ntds but also removes few more correctly base paired nucleotides, this is an act of precaution than required.�� This ensures high fidelity. The exonuclease activity is very helpful not only during replication but also in DNA damage repair and recombination processes.�
The 5� �3� exonuclease activity plays an important role in removing errors at 5� end and also in removing Thymidine dimers (cross-linked).�� The 5� exonuclease domain can also perform endonuclease activity by nicking dsDNA to generate a 3�OH group from which it can displace the nicked strand in 5��3� direction up to 10 or more nucleotides long fragment and at the same time it can extend chain from the nicked point by polymerization in 5�-3� direction.� Thus one can consider this enzyme is one of the most utility enzyme and versatile.
�
- Perhaps this versatility stems from the fact during evolution, different genes or parts of genes are brought together as one enzyme.� E.coli�s 5��3� exonuclease domain of DNA-polymerase-I and its 5� displacement characters are shared with T5 phage and O29 phage DNA polymerases.
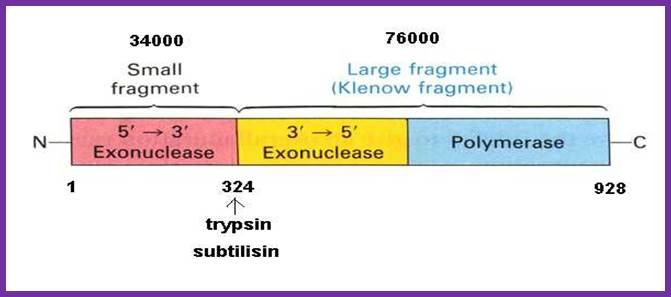
http://cpctas-lcmb.pmf.kg.ac.rs/
I-------(34) 38 KD---I-------------------------68 (76) KD-------------I
NH2 ------------------I-----------------I----------------------------I.COO^-
5�>3�exo��������� 3�-5� exo. ��5�� 3� polymerase
|
Activity |
35kd Fragment |
68 kd Fragment |
|
5��3� pol activity |
No |
Yes |
|
5��3� Exonuclease activity |
Yes |
No |
|
3�� 5� exonuclease activity |
No |
Yes |
DNA polymerase II:
In 1971 T. Korenberg isolated a mutant from E.coli strain; this has led to the discovery of two more DNA polymerases, called DNA Pol II and DNA pol III.� There are few conflicting reports about the Mol.wt and the number of subunits found in Pol II.�� Yet the overall structure shows similarities with eukaryotic DNA polymerase� alpha subunit.
Its absence doesn�t affect the growth of the bacteria.� Its activity increases when DNA is damaged.� It has greater affinity towards single strands with primers; it is strongly stimulated by SSB proteins.� It has no 5� �3�exonuclease activity, but it has 5��3� polymerase and 3��5� exonuclease activity.� It is induced during DNA damage and repair events as well as during genetic recombination.
DNA polymerase III:
Among the polymerases found in E.coli, DNA pol III is the enzyme that is responsible for actual replication.�� The said polymerase is not one polypeptide chain, but made up of more than 12 subunits having a molecular mass more than 512 KD.� Each of the subunits and their genes has been identified and their characters and functions have been delineated to certain extent.� All subunits together as a complex is called Holozyme.� Basically it consists of catalytic core complex and an accessory complex.� Assembly of the Holozyme is hierarchical and the holozyme dissociates and associates very rapidly and exhibits dynamic features.
The DNA-Pol-III differs from the other two because of its multiple subunits, high molecular mass, high fidelity and super processivity and the rate of synthesis is ~1000bp per sec, i.e 50 to 60 kb per minute, fastest among any known multisubunit DNA polymerases, while RNAP performs 80bp/sec (?).� Error rate is minimum, possibly one in 10^-8 bp.�� It doesn�t show 5�->3� exonuclease and strand displacement or nick translation activity.� This Holozyme not only performs replication of bacterial DNA but also involved in replication of some parasitic phage DNA such as phi X174 and M13.� It can work on both linear as well as circular DNAs.�� It can use 3�OH groups of RNA or DNA nick and use them as primers.
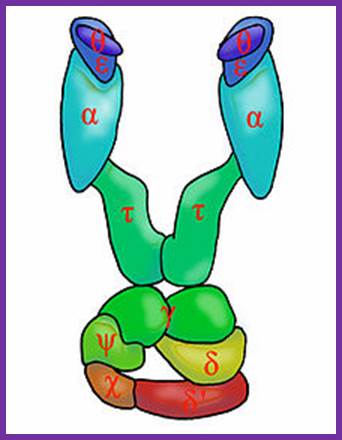
The core part of the enzyme consists of tightly bound subunits- alpha, epsilon. Theta and Tau.� The accessory complex is made up of several subunits like gamma, delta, delta�, chi, psi and beta subunits.
Components of DNA pol-III Complex:
|
Protein����������� ����������� ����������� |
Genes |
Mol.wt (kd) |
Subunits |
Activity |
|
Core complex: |
|
|
|
|
Alpha |
dna-C or D |
132(129) |
1 |
5�>3� pol |
|
Epsilon |
dna Q |
27.5 |
1 |
3�>5� exonuclease |
|
Theta |
hol-E |
10(8.6) |
1 |
Helps in core assembly |
|
Tau |
dna-X |
71 |
1 |
Helps in dimrization of the core |
|
Accessory complex; |
|
|
|
|
Gamma |
dna-X |
52(47.5) |
2 |
Clamp loading |
|
Delta |
hol-a |
35(38.7) |
1 |
Clamp loading on lagging strand |
|
Delta prime |
hol-B |
33(36.9) |
1 |
Clamp loading |
|
Chi |
hol-C |
15(16.6) |
1 |
Clamp loading |
|
Psi |
hol-D |
12(15.2) |
1 |
Clamp loading |
Beta subunit
|
dna-N |
37 (40.6) |
2 (4) |
Act as DNA clamps |
|
|
|
|
|
|
|
Total Mol.wt and total number of sub units |
|
512 |
12 |
|
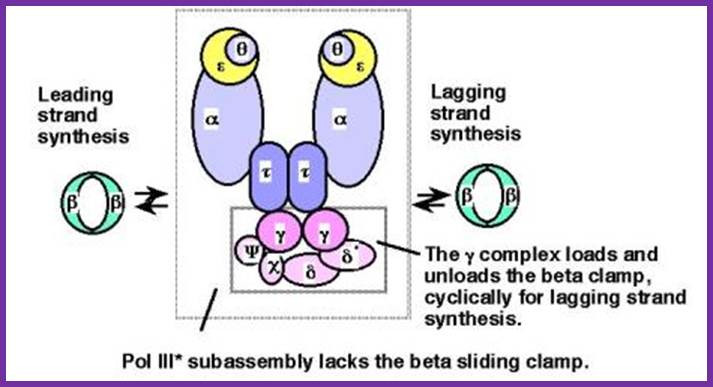
Schematic picture of DNA polymerase III* (with subunits including clamp loading complexes); http://www.personal.psu.edu/
![The 3|[prime]||[ndash]|5|[prime]| exonucleases](Prokaryotic_DNA_Replication4-Factors_and_Enzymes_files/image005.jpg)
Architecture
of the replicative DNA polymerase III holoenzyme from E. coli; The
DNA polymerase III holoenzyme is a multisubunit complex, which consists of 17
polypeptides. It contains four subassemblies. First, the core polymerase
consists of three subunits:  (the polymerase);
(the polymerase); 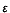 (the 3'�5'
exonuclease); and
(the 3'�5'
exonuclease); and  (the stimulator of the
3'�5' exonuclease). Second, the
(the stimulator of the
3'�5' exonuclease). Second, the  subunit is responsible
for dimerization of the core DNA polymerase. Third, the sliding clamp comprises
two homodimers of the
subunit is responsible
for dimerization of the core DNA polymerase. Third, the sliding clamp comprises
two homodimers of the 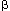 subunit, which provides
the ring structure that is needed for processivity. Fourth, five subunits have
clamp-loader functions �
subunit, which provides
the ring structure that is needed for processivity. Fourth, five subunits have
clamp-loader functions � 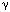 ,
,  ,
,  ',
', 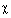 and
and  . For details, Igor V. Shevelev &
Ulrich H�bscher; http://www.nature.com/nrm/journal
. For details, Igor V. Shevelev &
Ulrich H�bscher; http://www.nature.com/nrm/journal
Alpha subunit:�
This is the largest and the important subunit; it has template binding and primer binding sites.� It also contains an active site for free nucleotide binding next to the primer 3�end. The same site acts as a catalytic site for polymerization of properly base paired nucleotides from 5� to 3� direction.� It has no 3�-->5� exonuclease activity.� The rate of DNA synthesis depends upon this subunit.
�
Epsilon subunit:�
This is always complexed with alpha subunit in 1:1 ratio.� This has 3��5� exonuclease activity, thus it performs poof reading function.� Association of this increases the polymerase activity of alpha unit by two fold and 50 to 100-fold 3��5� exonuclease activity.
Mutants in this gene, called mut-D, lacks 3��5�exonuclease activity and increases the incorporation of Thymidine nucleotides.� During SOS situations this subunit is synthesized in large amounts and enables DNA repair.
Theta subunit:� Not much is known about this subunit.� However it is discerned that this favors the association of alpha and epsilon subunit by binding to both and perhaps facilitate in building up of larger complex.
Tau subunit:� This is a full-length product of the gene dna-X, which also gives raise to gamma subunit during translation by frame-shift in the reading frame.� This subunit has ssDNA dependent ATPase activity.� It helps in dimrization of the core complexes and enhances processivity of the core enzyme.
������ �--------------------Tau----------------------�
������������������������ ����I--------------------------------------------------I = dna-X gene.
������������������������ ����I<-----gamma--�I�
Beta subunit:
This is a homodimer consists of two clamp shaped or bracket shaped structures.� On dimerization it encloses a halo of 20 �-35� to hold ssDNA with a primer. It has six-fold symmetry contain 12 α helices.� The hole is filled with water, when it holds DNA it surrounds the DNA.� This clamp can be a dimer but in some it can be trimer.� The clamps are loaded on to both leading strand and lagging strands behind holozyme.�� The loading of the clamp is performed by gamma-delta complex in ATP dependent manner.�� Binding of the Beta clamps hold the enzyme and the template together tightly at template primer site, hence it increases the processivity of the enzyme.� Recycling of the Beta clamp takes place at each and every primer position on the lagging strand. A single gamma complex, positioned in the center of the fork, loads beta clamp on to both strands There are about 300 beta dimers (clamps) per cell; this number takes care of replication.� This gene is located in between dna A and rec F genes.� Expression of this gene depends upon the promoter located within dna-A gene.
�������������
Gamma complex:
Gamma subunit perse is a truncated translational product of the full length mRNA coded for by dna-χ gene (codes for tau protein).� It corresponds to 2/3 rd of the N-side of the tau (τ) protein.�
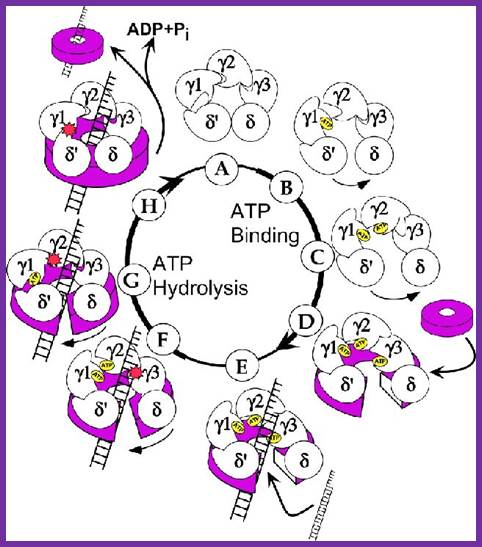
����������������������������������� Gamma complex the clamp loader; http://www.jbc.org/
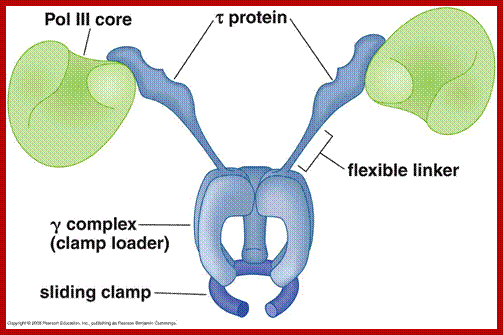

Beta-subunit
is a homodimer. The diameter of it's center channel is ~35Å.
This is large enough to let dsDNA(diameter : ~25Å) through; http://ipr.pdbj.org
This segment is generated during translation by frame shift in the reading frame.
Gamma complex consists of Gamma (γ), delta (δ), delta� (δ�), chi χ and psi (ψ) subunit proteins.� In ATP dependent manner the gamma and delta complex can bind the template at initiation as complex.� All the components of the complex assemble in cooperative manner, and ATP dependent.� The Gamma complex is considered as clamp loader.
The Gamma complex in the presence of ATP binds to beta clamp and loads the same on to the primer site where the holozyme also bind (for the core enzyme has high affinity to the dimeric clamp). On the leading strand once the beta clamp is loaded, it need not be there in the complex.� Positioning in between the two strands the Gamma complex loads the beta clamps on to each of the strands. However on the lagging strand, the beta clamp has to be loaded at intervals for the primers are laid at frequent intervals of length.�
Assembly of the Holozyme:
The Holozyme is formed in sequence and stepwise manner.� First alpha and epsilon interact with theta to form of complex, which is a core complex. And their processivity is 10 ntds long.� Then tau interacts with core complexes and dimerizes, this has a processivity of 60 ntds long.� These assemble then onto templates at replication fork where the primosome complex is found.� This region is recognized by the gamma complex and loads the beta clamps onto the template with the core enzymes.� This complex, called Holozyme has a processivity of 10^5.� For its activity it requires SsBs.
Gamma + delta = gamma-delta;
Gamma-delta + delta� + chi + psi = Gamma-delta complex;
Gamma complex + ATP + beta clamp = gamma-delta-beta complex.
Alpha + epsilon + theta = Core Complex;
Core complex + tau = Dimer Core Complex;
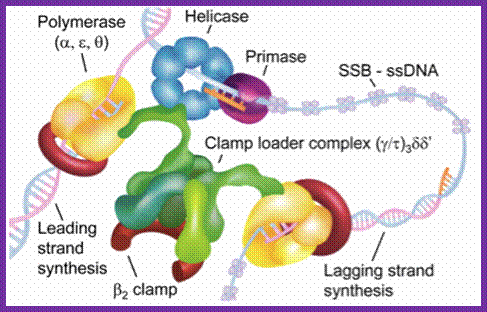
The above diagram shows position of each component assembled on the DNA at replication fork. http://www.umich.edu/
Dimer core complex can assemble on the DNA template, provided replication bubble with the replication fork exists. A single gamma-delta complex can load the dimer beta clamps on to the DNA templates with primers.� The beta clamps hold the DNA with the primer like a pair of half bracelets holding the DNA with a firm grip. The gamma complex also helps in loading the DNA Pol complex next to Beta clamp.� As the beta clamps are in association with core complex enzymes won�t dissociate easily, this feature enhances the processivity thousand fold.� Association and dissociation of gamma complex with template DNA and the primer, and recycling of gamma-delta and beta clamps, especially on lagging strand occur frequently.