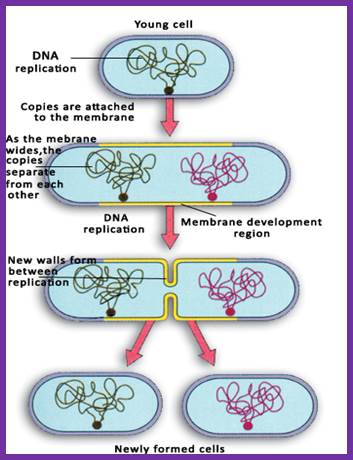
Mechanism of Replication:
Initiation:�
A unit of DNA that undergoes replication id called Replicon.� The position of replication Origin varies and structural features of Origin also varies. But the origin is recognized by specific transacting factors that initiates replication.� The concept of replicon model has been under review.
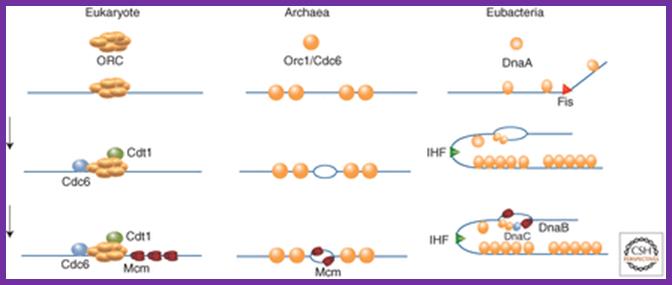
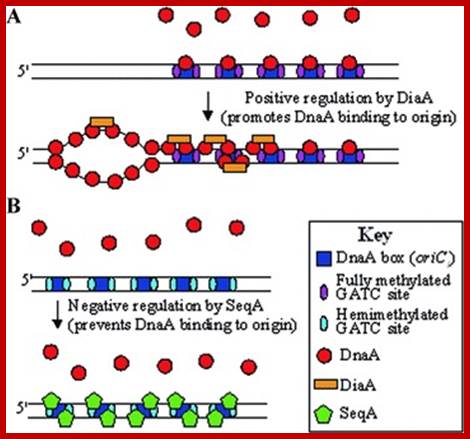
Revised versions of the replicon model for all domains of life. For cells of each domain type, trans-acting initiators recognize replication origins to assemble prereplicative complexes required to unwind the DNA and load DNA helicase. Eukaryotic initiators are preassembled into hexameric origin recognition complexes (ORCs) before interacting with DNA. In prokaryotes, single initiators (archaeal Orc1/Cdc6 or bacterial DnaA) bind to recognition sites and assemble into complexes on DNA. In all cases, the DNA helicases (MCMs or DnaB) are recruited to the origin and loaded onto single DNA strands. In bacteria, DNA-bending proteins, such as Fis or IHF, may modulate the assembly of pre-RC by bending the origin DNA. Two activities of DnaA are described in the figure. The larger version binds to recognition sites, and the smaller version represents DnaA required to assist DnaC in loading DnaB helicase on single-stranded DNA. Alan C. Leonard1 and Marcel M�chali;
aleonard@fit.edu; mechali@igh.cnrs.fr; �http://cshperspectives.cshlp.org/
Under favorable conditions, as the cell-mass: cell volume ratio gains, the cell enters into cell division mode.
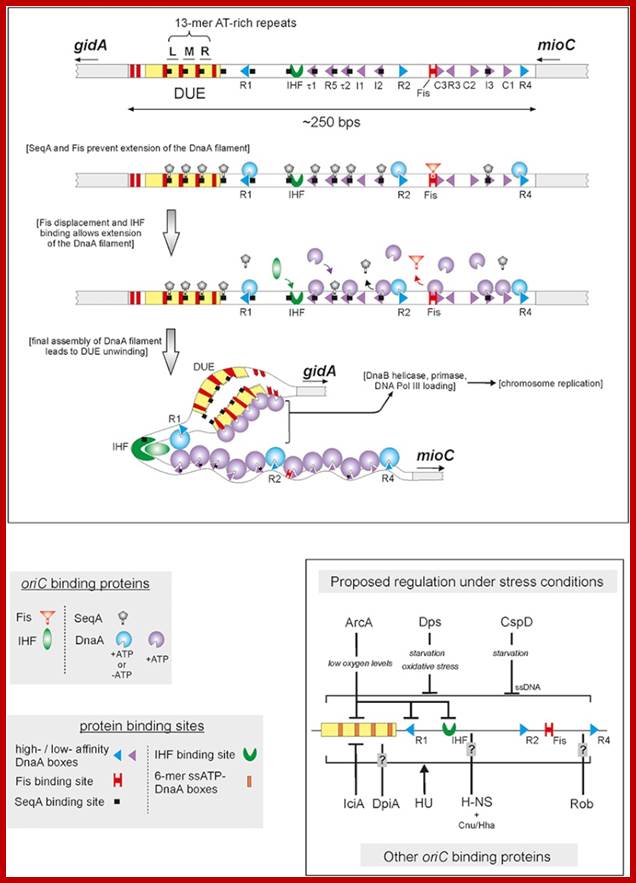
Replication origin called Ori-C is located at 84.3 mpu position of the genome.� The oriC locus is about ~250-~300 bp long and located in between gidA and mio C gene.
�
- To start with, besides activation of required genes in triggering cell cycle events (several of them mentioned in a table in the beginning of this chapter), activated Dam methyl transferases start methylating Adenine moiety in 5�GATC3� sequences, which are spread all over the origin-C (at least they exist in 14 blocks, five in M13mer region and others in 9mer sequences) and also in the promoter region of the dna A gene. Activation of Dam Methylases is of critical importance.
- The replication is bidirectional; interestingly one of the strands is called Watson�s strand and the other complementary is called Crick�s strand.
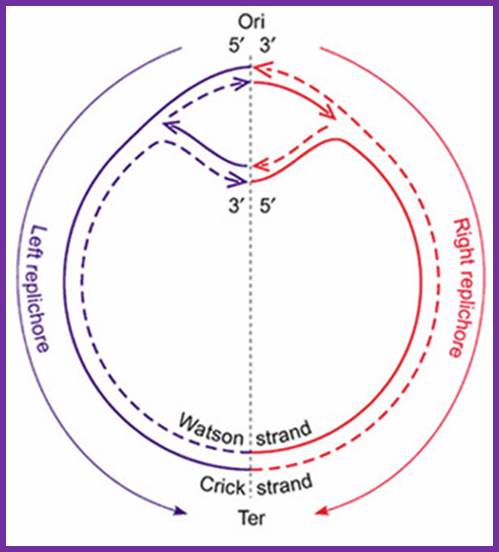
The topology of bi-directional replication of a circular prokaryotic chromosome: The continuous line is the DNA strand replicated as the leading strand; the dashed line is the DNA strand replicated as the lagging strand; Ori, the origin of replication; Ter, the terminus of replication. Ori and Ter divide the chromosome into two replichores, arbitrarily called left and right. Mickiewicz et al. Genome Biology 2001 2: interactions1004.1 doi:10.1186/gb-2001-2-12-interactions1004; http://www.uam.es/
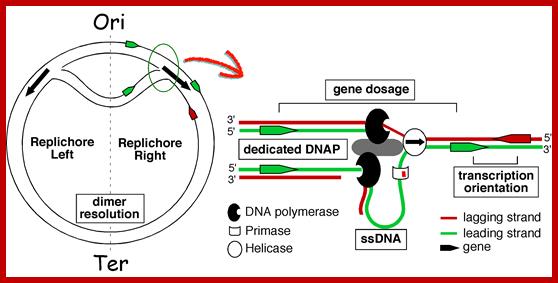
Replication of the bacterial chromosome proceeds bi-directionally from the origin to the terminus of replication. At each replication fork, two DNA polymerases replicate the leading and the lagging strand. Boxes indicate elements of asymmetry. See the text for details.
�Wduardo p. C Rocha; http://wwwabi.snv.jussieu.fr
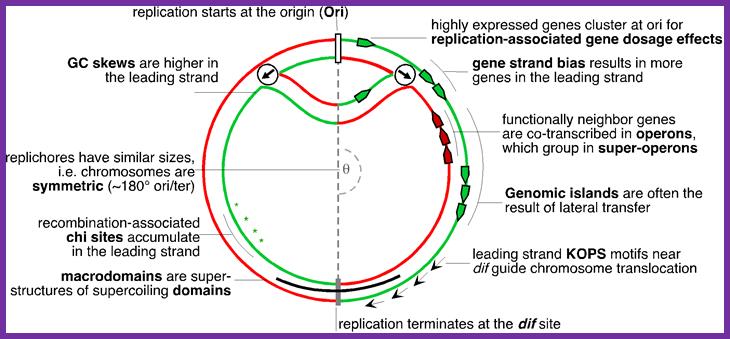
Elements associated with the organization of the bacterial chromosome. Green and Red distinguish between the leading and the lagging strands. Ori and Ter identify the origin and terminus of replication, encircled arrows indicate the direction of replication fork progression. Besides these elements, several mutational biases related with replication have been described in bacterial genomes (reviewed in Rocha, Annual Rev Genetics, 08). https://www.pasteur.fr; Eduardo P.C Rocha:�http://wwwabi.snv.jussieu.fr/; during replication
Gene transcription around Ori site in leading strand is more than the lagging strand, 75% in B subtilis and 55% in E.coli
In pre4vious round replicated daughter DNA molecules not only get supercoiled but also compacted by bacterial SMC proteins. Newly replicated DNA exists in hemi-methylated state and �the newly formed circular DNA remains bound to mesosomal membranes at a site called attachment site.� This attachment is facilitated by certain proteins that are bound to DNA, which in turn bind to membrane phospholipid head groups; the suspected proteins are SPoOJs which are bound to Origin region and they prevent DNA release from membrane and initiation of replication is inhibited by the inhibitor present at hemi-methylated sites at the origin of DNA. They are also involved in chromosomal separation.� Once it was believed that DNA binds to mesosomal membrane at specific DNA attachment sites.� However, it is not clear, but it is assumed that the DNA attachment site (there are twenty sites) to membrane is very close to replication origin. The site is recognized as OCB1 to which oriC binds. �This site is perhaps the adhesion region of outer and inner membranes. This region is located to the left of OriC where 10 out of 11 Dam methylation sites are concentrated. The probable protein binding site of DNA sequence can be� �a/gCCa/tGGg. The inhibitor is called IciA (inhibitor of chromosome initiation). �On full methylation, Dna-A proteins release DNA from the inhibitors and from the membrane.
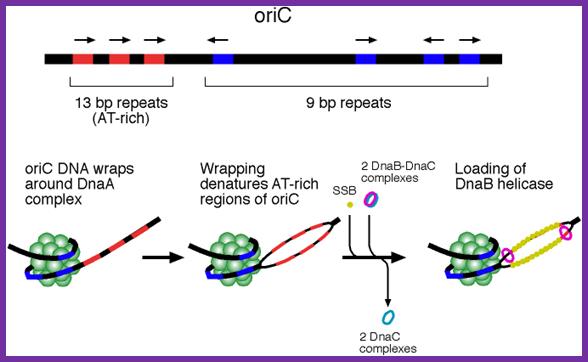
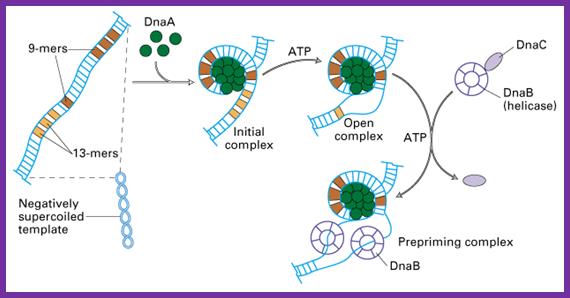
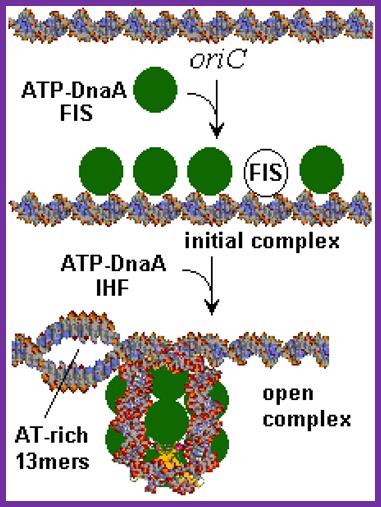
This picture depicts positions of 9-mer and 13-mers sequences called AT rich boxes; Fully methylated sites in 9-mer region are bound by Dna-A proteins cooperatively; it this binding leads to the DNA to wrap around the clusters that leads to torsion in opposite direction and brings about the unwinding of the DNA at 13-mer regions. http://oregonstate.edu/
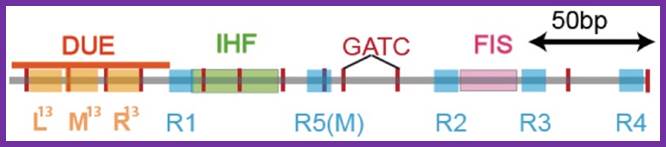
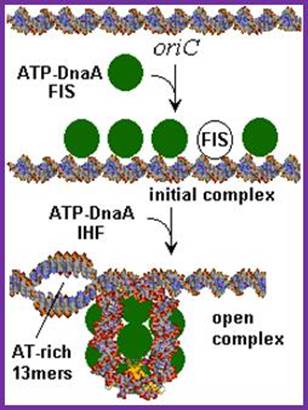
The E. coli origin of replication bears five 9-mer DnaA-binding sites (R1, R2, R3, R4 and R5) as well as three 13-mer binding sites included in an A+T-rich DNA unwinding element (DUE). Although the 9-mers show no differential specificity between DnaA-ATP and DnaA-ADP, the 13-mers specifically recruit DnaA-ATP. In addition to DnaA-binding sites, oriC hosts-specific binding sites for IHF-Integration Host factor, SeqA and FIS-Factor for inversion, three proteins that regulate the activity of DnaA; Sylvain Zorman,1 H. Seitz,2 B. Sclavi,3 and T. R. Strick1,*
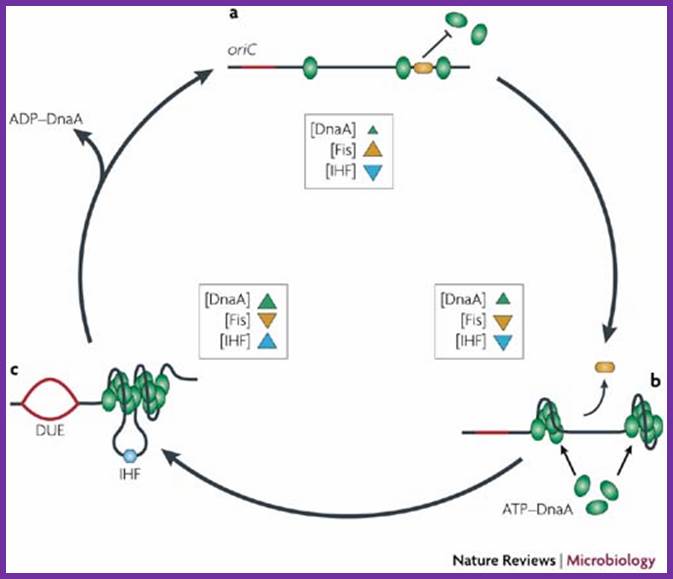
A model for the dynamic interplay between DNA architectural proteins and DnaA during replication initiation.
The relative increases and decreases in the concentrations of factors at oriC as a function of progression through the cell cycle, as derived from experimental observations10, 98, are shown. a | Before the start of initiation, DnaA (green) is bound to several high-affinity DnaA boxes at oriC. Fis (factor for inversion stimulation, orange) is also associated with its primary binding site, whereas IHF (integration host factor, blue) is absent. b | At the start of initiation, the local concentration of ATP�DnaA at oriC increases, while at the same time Fis occupancy decreases. c | As initiation progresses, IHF binds oriC. The severe bending of DNA by IHF probably assists the interaction of DnaA with its weaker affinity binding sites during the formation of the final nucleoprotein complex and unwinding of the DNA-unwinding element (DUE). Melissa L. Mott & James M. Berger; http://www.nature.com/
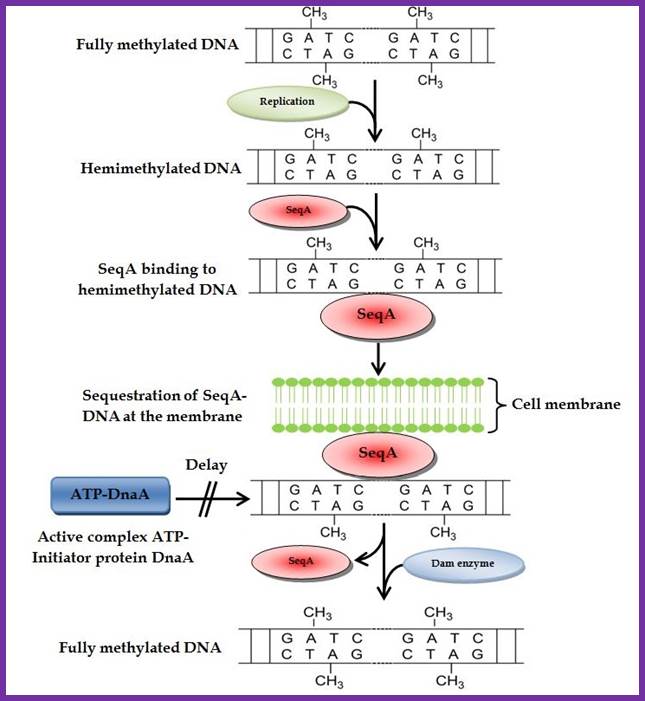
Methylation and sequestration- the mechanisms- DNA replication; Amine Auloi et al: www.intechopen.com/. http://nurseps.com/
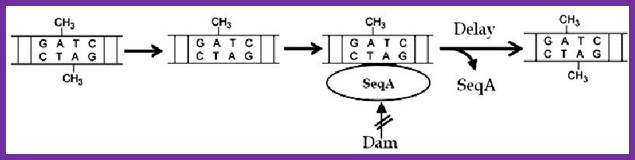
In previous round of replication, fully methylated DNA in the mentioned GATC regions becomes hemi-methylated. In this state initiation of replication is stalled for the lack of fully methylated strands (methylation in both strands) in OriC region. �This OriC region many GATC sequences in hemi-methylated state are bound by �SeqA (called �A� boxes) and masks the sequences from methylation. The SeqA is a tetramer protein has greater preference to hemi-methylated GATC sequences in newly replicated E. coli DNAs. This time period is called eclipse period.�
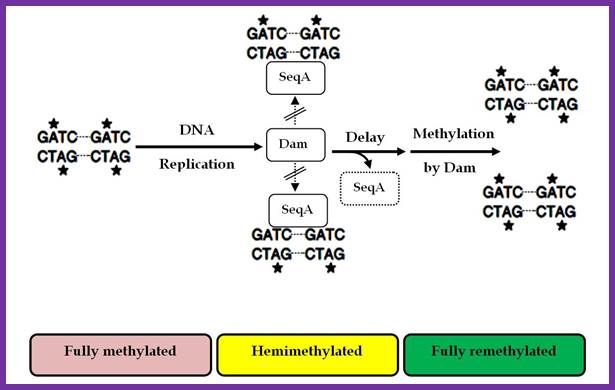
The vast majority of chromosomal GATC sites are fully methylated until DNA replication generates two hemi methylated species, one methylated on the top strand and one methylated on the bottom strand. Within a short time after replication (less than 5 minutes), Dam methylates the nonmethylated GATC site, regenerating a fully methylated GATC site. http://www.intechopen.com/
http://www.intechopen.com
Helically phased GATC sites can be bound by SeqA when they are in the hemi-methylated state. Binding of SeqA inhibits Dam methylation, maintaining the hemi-methylated state for a part of the cell cycle. Dissociation of SeqA allows Dam to methylate the hemi-methylated DNA, thus generating fully methylated DNA. But SeqA spontaneously dissociate and reassociate.� During this period more DnaAs are produced due to activation of dnA gene.� The activation of dnaA gene is due to fully methylated state of the gene promoters. The dnaA gene is located at 82 min site (mpu).� Its promoter is ~945 bp long.� And the gene codes for 54kDa protein.� It is located left of OriC.� It contains multiple promoters with abundance of GATC sequences (at least 9 of them, while OriC contains 11 of them.� The DnaA protein is involved in activation of its own gene by the binding of DnaA proteins to their promoter sequences or what is called DnaA box.� The gene is auto regulated by its own product DnaA at transcription level.� The promoter consists of several sites, of which one is strong binding site to DnaA, two are week binding sites and other two in flanking regions contain DnaA-ATP binding sites.� Once the hemi methylated sites are fully methylated, proteins bind and dnaA gene is expressed.
Two or more helically phased GATC sites can be bound by SeqA when they are in the hemimethylated state. Binding of SeqA inhibits Dam methylation, maintaining the hemimethylated state for a portion of the cell cycle. Dissociation of SeqA allows Dam to methylate the hemimethylated DNAs, generating fully methylated DNA.� Amine Aloui, Alya El May, Saloua Kouass Sahbani and Ahmed Landoulsi http://www.intechopen.com
Signals for activation of cell division leads to methylation of GATC at gene promoter dnaA as well as at other regions.� This leads to the activation of dna-A gene to produce DnaA proteins. �Expression of dnaA gene is regulated by its own product DnaA-ATP (it has two promoter regions).� The DnaA is AAA+ protein and it is activated by the binding of ATP.� The DnaA protein contains four domains; domain III contains ATP binding, others have protein-protein interaction and DNA binding domains. Proteins organize into right handed oligomeric forms around which the DNA wraps around in left handed form so as to render the DNA into negative supercoil (Bramhill and Kornberg). This has an effect on 13-mer region called DUE (DNA Unwinding Element) region opens up into single stranded templates.
Activation of cell division process at cytoplasmic level involves many signal transduction pathways similar to that of Arc two component systems.� In this process, the dnaA gene is activated; accumulation of DnaA protein to certain concentration is very important, till then the DnaA proteins are sequestered at dat-A region. Factor titration is done sequestering of DnaA proteins till adequate number of DnaA proteins is generated.
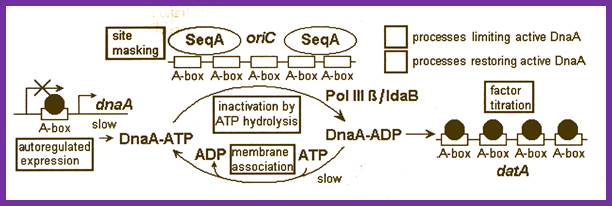
GATC hemi methylation and A box masking: Newly synthesized DNA lack methylation, however OriC and GATC remain unmethylated or hemimethylated for extended period.� These sites are bound by SeqA proteins near DNA-A binding sites.� The hemimethylated state remains for about 10 minutes. DnaA titration by data:� Multiple binding sites for DNA at data (Dna A titration) at 94.7mpu of E.coli gene map.� As this site is the origin, it is replicated early causing the doubling of the sites thus DnaA protein level is kept low until the copy number builds by expression of weak constitutive promoter.� Initiation of second round replication is kept in check till DnaA-ATP levels are high enough that weak R3 site is occupied (Katayama et al2001)
Model outlining the proposed functions of the DiaA & SeqA proteins in the initiation of replication of chromosome 1 in P. profundum SS9 and in piezophilic growth. (A) DiaA is a positive regulator of the initiation of chromosome 1 replication and removal of DiaA results in piezo-sensitive growth. (B) SeqA is a negative regulator of the initiation of chromosome I replication and removal of SeqA results in piezo-enhanced growth. Model based on the functions of the E. coli DnaA, DiaA, and SeqA proteins. https://www.researchgate.net
- Protein Seq-A delays initiation after one round of replication.� This protein binds to hemi methylated DNA specifically at or near Dna-A boxes in the origin region.� Mutation in Seq-A gene causes initiation early. And produce many opened origins too soon. Full methylation of GATC sequences makes SeqA nonfunctional and the DNA is ready for another round of replication.
- SeqA binding to hemimethylated GATC sequence at OriC, may help in association of DNA, Ori region to bacterial membrane.
---- Abox- SeqA-Abox-ori C-Abox-seqA-Abox--
- There is another site called datA region (DnaA titration), which also contain four A-boxes to which DnaA-ADP proteins bind; the binding DnaA proteins to these sites acts as factor titration process.� When the concentration of DnaA-ADP proteins is low they are all bound to data sites.� When sufficient amount is produced, they used for initiation of replication by activating DnaA protein into DnaA-ATP.
http://humbio.ru
The diagram depicts how the wrapping of the DNA around Dna-A proteins leads to opening of the DNA at 13-mer regions.� ATP mediates the loading of Dna-A on to methylated Dna-Boxes.� The opening also facilitates the loading of the Dna-B hexamer in ATP dependent manner, Dna-B is an Helicase-a ATP dependent motor protein.
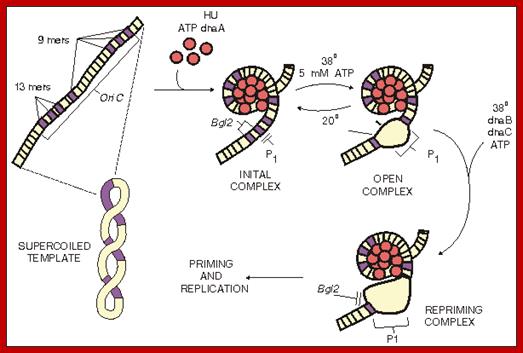
Bottom Fig. Scheme of replication initiation in oriC E.coli. dnaA
protein binds with
fragmetami 4 to 9 bp in length, organizing in oriC DNA region around the protein core and creating initiation
complex. Thereafter 3
fragments of DNA each of length 13 bp successively melted by dnaA
and create an open complex. Now
complex dnaB-dnaC proteins can
connect to a fragment of 13 bp and expand disclosure duplex, generating "prepriming
complex". Details are given
in the article Bramha and Korenberg �� http://humbio.ru
Hemi methylation prevents initiation of replication, at the same time hemimethylation in promoter elements of dna-A gene also leads to repression of it.� It is only on full methylation, the gene becomes active.� Protein such as regulatory inhibitor of DnaA called RIDA and few others are involved in interacting with one another in binding and releasing the DNA from the membrane.
The release of the DNA is also assisted by full methylation of GATC sites in the origin region, it at this time SeqA are released.� SeqA binding provides time for the synthesis and accumulation DnaA proteins; this happens in between two cycles of replication events. SeqA overlaps A-boxes thus mask the GATC sites from methylation.
Methylation and sequestration he mechanism of DNA replication;
Moeka Kurihara; http://nurseps.com/
---- Abox- SeqA-Abox-ori C-Abox-seqA-Abox--
- During initiation of cell division, especially the said hemi-methylated segments become fully methylated. This triggers transcriptional activation of dna-A gene.
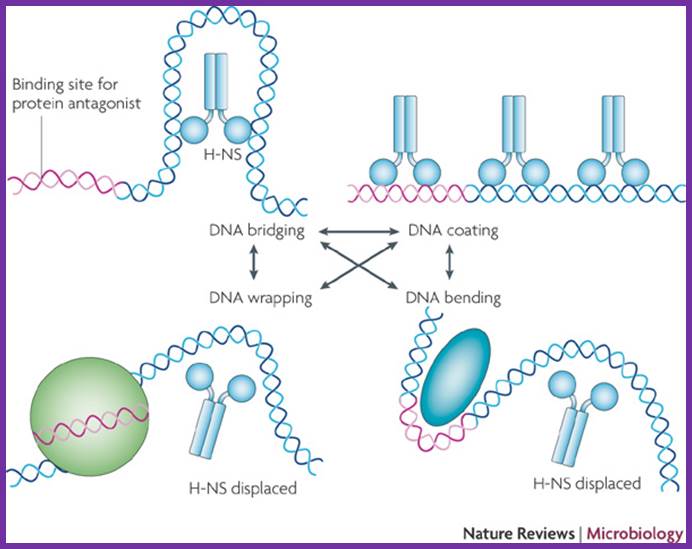
The circular DNA 4.6 x 10^bp long is approximately 1mm long exceeds the size of the cell.� Thus the long DNA is compacted into supercoiled structured regions bound by proteins called nucleoids. They are like pearls on necklace. The bacterial SMC proteins (structural maintenance of chromosomes) act as molecular clamps. �Each of the nucleoids contains highly compacted supercoiled DNA (60%), nucleoid proteins NAPs (different from histones), topoisomerases and transcriptional factors and mRNAs.� These nucleoids are transcriptionally active. These regulatory NAPs are: Fis (factor for inversion stimulation), HU (histone-like protein), H-NS (histone-like nucleoid structuring protein), and IHF (integration host factor). The concentrations of these proteins vary in different growth phases, from 10,000 to 60,000 monomers per cell.� The said number of nucleoids can be more than 2063. The size of DNA can be approximately 1 to 1.5kbp.
����� ����������������������� Nucleoids; a model; www.keywordhut.com
�Sam iksha; http://www.yourarticlelibrary.com
50-100 kbp long loops studded with specific proteins; these organized structures are called Nucleoids.� Such loops are attached at the base by high salt insoluble nuclear matrix/scaffold like proteins.
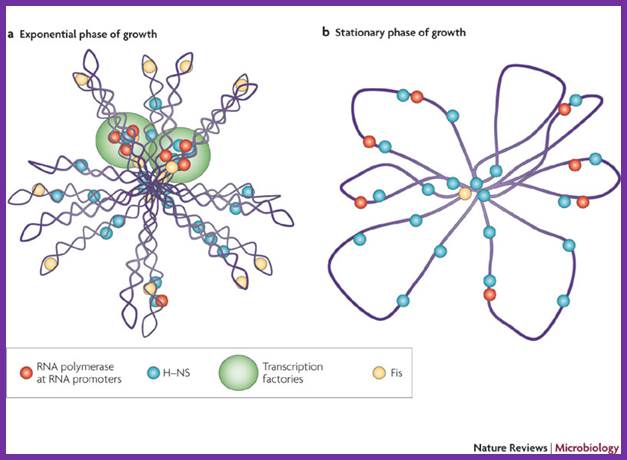
The folded chromosome is organized into looped domains that are negatively supercoiled during the exponential phase of growth. In this phase, the abundant nucleoid-associated proteins histone-like nucleoid-structuring protein (H-NS) and factor for inversion stimulation (Fis) bind throughout the nucleoid and are associated with the seven ribosomal RNA operons. As shown here in two cases, these are organized into superstructures called transcription factories. b | In stationary phase the rRNA operons are quiescent and Fis is almost undetectable. The chromosome has fewer looped domains, and those that are visible consist of relaxed DNA, Shane C. Dillon & Charles J. Dorman
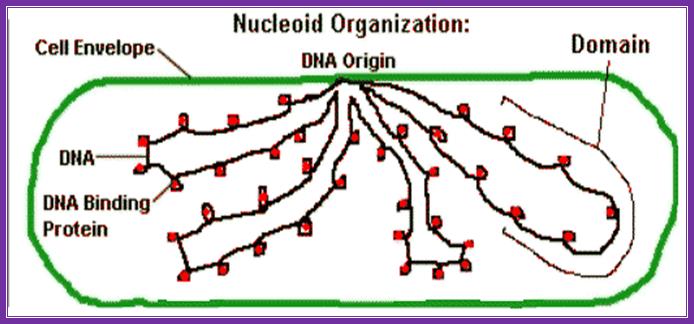
E.coli DNA is coiled loops with histone like proteins form nucleoids and one can see OriC region bound to plasma membrane; Kenyon Biology. www.nptel.ac.in
www.cell.com
Shane C. Dillon & Charles J. Dorman;http://www.nature.com
Nucleoid-associated proteins (NAPs) to assist in chromosome condensation and organization. By bending or bridging DNA, NAPs also facilitate chromosome segregation and regulate gene expression. Dan Song,Joseph J.Loparo; http://www.cell.com
E.coli DNA replicative origin binds to outer membrane with high affinity at a 463bp region (-46 and +417).� This region is rich in GATC sequences. It binds to membranes when it is hemi methylated. This also facilitates the binding of SeqA and SeqB. Seq proteins help the DNA to bind to membrane. This hemi methylated state remains for 8-10 minutes after replication.� The methylation of A at �GATC to the promoter of dnaA gene facilitates transcription of it. Once GATC sites are fully methylated at the region of Ori and E.coli DNA membrane binding site, the circular compacted DNA detaches from the membrane. These two events are very important in initiating DNA replication cycle. The binding of hemi methylated ori region to membrane is implicated in preventing premature reinitiation of the newly replicated DNA. Methylation of promoter region of dna-A gene leads to transcription and translation. The Dna-A proteins build up in cytoplasm of the cell.
- Dna-A, are monomeric proteins, have hydrophobic features, as well as DNA binding motifs.� As the number of DnaA proteins build up they start binding to four nine-mer sequences, also called DnaA boxes.� Binding of the said proteins is ATP dependent. Once they start binding in sequence specific manner, proteins also start protein-protein hydrophobic coiled- coil interactions. This leads to clustering of proteins, which makes strongly DnaA bound DNA to wrap around the protein clusters in left-handed manner; the number of proteins bound can be anywhere 20-30.
- The wrapping of DNA around the cluster is also facilitated by the binding of IHF and Fis proteins, which bind at a site in between R1 and MM and R2 and R3 respectively; these proteins on binding cause bending of DNA to �U� form. �Right arm binding protein also contributes to DNA bending.�
Wrapping of DNA around the proteins lead to negative twisting of ds DNA on its axis, which transmits as an unwinding force to the neighboring region, and the DNA sequence at 13mer segments (DUE) unwinds (all the 3 segments including in between spacers) and open into single stranded bubble of 60-68 base pair length.
- As the ss DNA bubble is formed, single strand binding proteins, the SSBs bind to negatively charged sugar-phosphate-sugar backbone, as tetramers.� This association makes strands rigid, straight and prevents reannealing with each other.�� Limited opening of the DNA in the ORI-C region, it is about 60 bp long, is further stabilized by the transcriptional activity at the adjacent promoters (back-to-back oriented).� This structural feature also presents an opportunity for several proteins to bind.
Ref ?
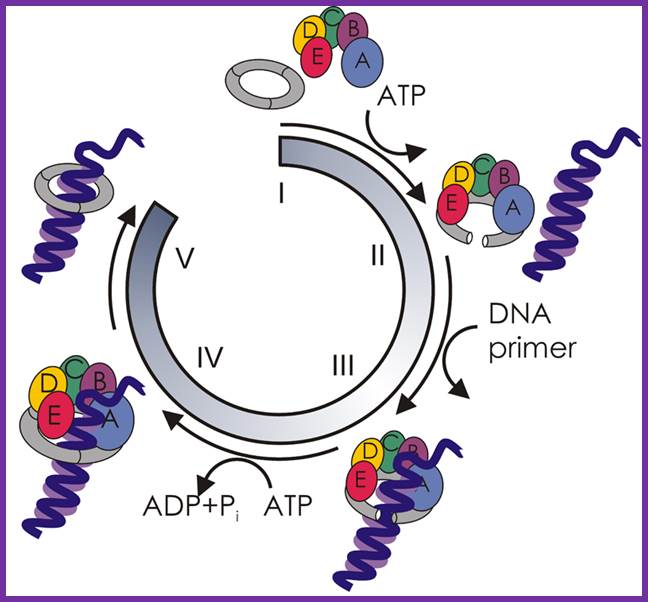
Dna-B (hexamer ring) assisted by Dna-C in 1:1 ratio is loaded onto the fork in ATP dependent manner.� During loading the circularly oriented helicase ring opens and clamps on to ss DNA at the joint of replication fork.� One of the domains has ATPase activity. Dna-B, is an helicase, encircle the lagging stand at the joint of the fork in 5��3� orientation.� Helicase loading is assisted by Dna-C in ATP dependent manner.� This motor protein is responsible for driving the fork opening further like unzipping. Dna-G, called primase or priming RNA polymerase, which is distinct from the pentameric regular RNA polymerase, associates with Dna-B on the ssDNA template. This is also known as �sliding clamp� increase the rate of DNA synthesis by 1000 fold(WIKI). This type of sliding clamp is also found in Archaea (trimer) and eukaryotes (trimer) and viruses as gp43/gp45 as trimmers.
|
Sliding clamp protein |
Aggregation state |
Associated polymerase |
|
|
beta subunit of pol III |
dimer |
||
|
archaeal PCNA |
trimer |
pol ε |
|
|
trimer |
|||
|
gp43 / gp45 |
trimer |
RB69 Pol / T4 Pol |
www.wikipedia.org-
During replication once the clamp is loaded on, it moves along the DNA pol III complex on both lagging and leading strands in ATP dependent manner- like a motor protein. This provides excellent processivity to DNA pol. Ivanov, Jaguar, Petascale; https://www.olcf.ornl.gov
sandwalk.blogspot.com/.../dna-replication-in-e-coli-problem...
�The replication bubble at the origin region shows direction of new strand synthesis. http://oregonstate.edu
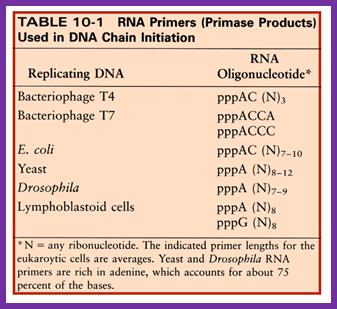
- Using 3� GTC sequences DnaG the primase produces a short stretch of RNA strand complementary to the template strand, invariably starting with 5� pppA G�ends at 3�OH of eleven ntds long RNA primer.� Sequencing of the DNA in this region to find where the primer initiates and where it terminates, for DNA extension shows GTC at �11 and at -1, it has C nucleotide.� The RNA and DNA joint sequence always starts with 5�CGG-.� The primer RNA -DNA junction shows the following sequence in several species, such as G4 phage, alpha 3 phages, and R 100 plasmid DNA, Ff6 DNA and R6k ori alpha.� This region is 93% rich in purines.
The primase is a monomeric protein, resistant to Rifamycin, while the regular RNA-pol-III is sensitive to Rifamycin.� The primase is responsible for laying short, ~11 ntds long primers on leading strand once at the beginning and lay primers on lagging strand at frequent intervals of 1000 to 1500 nucleotides all along the length.
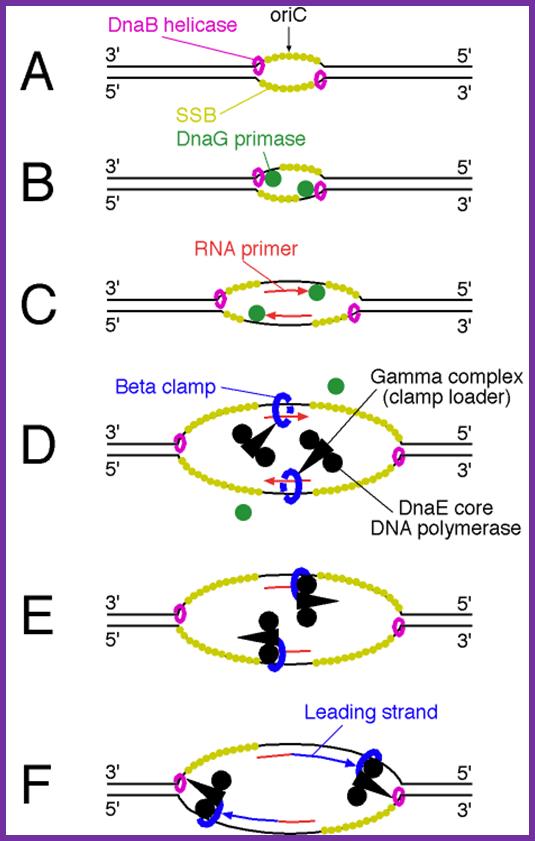
Binding of DnaA to to sequence specific sequences leads to opening and loading of SSB stabilizes the replication bubble.� The Helicase is loaded� and moves from 3�to 5� direction. At the same time Primase lays a short RNA primer; the DNA pol complex is loaded with the assistance of Clamp loading complex; DNA pol II now copies DNA on the leading strand and on the other strand called lagging strand the DNA is copied in short fragments called Okazaki fragments;� http://oregonstate.edu/
DNA is threaded through helicase complex as if the dsDNA is unzipped.
� A huge protein complex catalyzes the reactions of DNA replication.
� This replication complex recognizes an origin of replication on a chromosome.
� DNA replicates in both directions from the origin, forming two replication forks.
� In DNA replication, both parental strands of DNA act as templates.
� Until recently, it was believed that the replication complex perse moves along the strands of DNA.
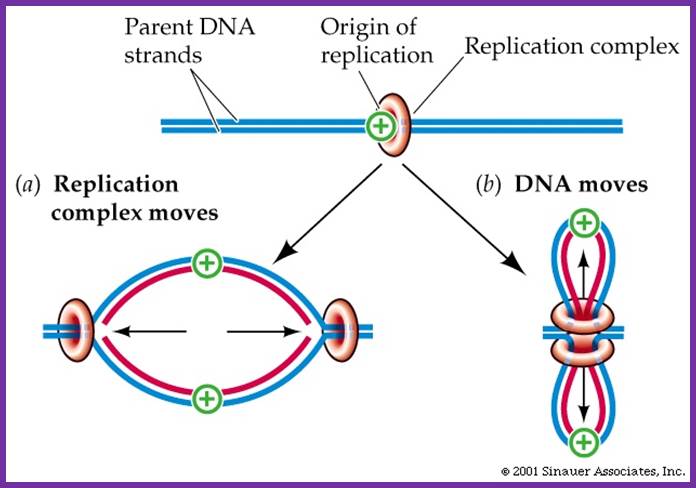
Once the replication complex loads on to their trmplates Replication complex moves along with new strand synthesized; but another view id the Replication complex is stationary and it pulls the DNA inwards and newly synthesized move out of it. So Pol complex remains in the same position. Sinauser associates.inc; http://bioserv.fiu.edu/
Recent evidence suggests that the replication complex is stationary, and DNA strand is pulled and threads through it.
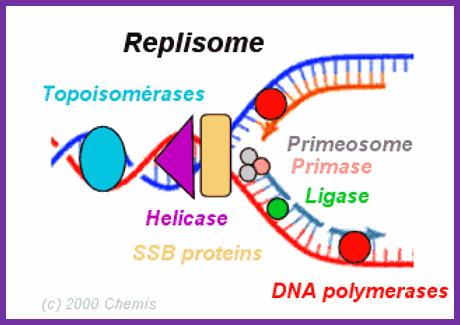
���� � Replication complexes consist of several proteins with different roles.
� DNA helicase denatures the double helix.
� Single-strand binding proteins keep the two strands separate.
� RNA primase makes a primer strand that serves as a starting point for replication.
� DNA polymerase uses the 3�OH of the RNA primer and adds complementary deoxy ribo nucleotides to the growing strand, proofreads the DNA, and repairs it.
� � DNA ligase seals up if any breaks in the sugar�phosphate backbone.
- Primase is always tagged on to the helicase.�
This diagram shows how DNA-B helicase progresses the replication fork
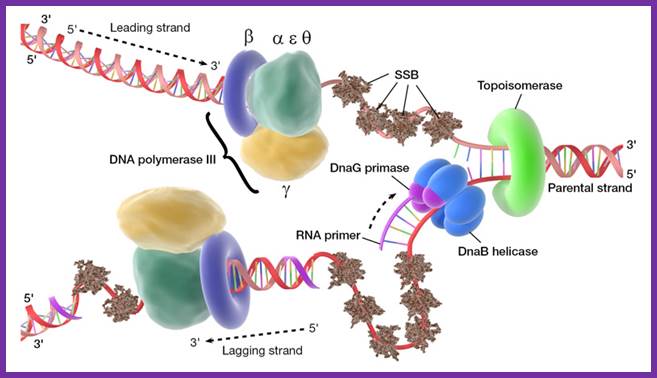
At this juncture Gamma complex loads beta-clamps not only on leading strand but also on lagging strand. The gamma complex binds to ATP and uses the energy to open the dimeric beta clamps and close on the DNA strand.� The beta clamps are positioned on the template at the base of DNA pol III provides firm binding to the DNA strand and helps processivity.� The gamma complex also facilitates the assembly of DNA-Pol III as dimers on to their respective beta clamps on to both strands.� DNA pol III, as a dimer complex, joins template at the joint of the fork.
�
Assembled components at one end of the replication fork.; http://pixgood.com/
So, one finds one sets of two DNA-Pol dimers, i.e. at one end of the fork and the other set at the other end of replication fork.� Perhaps in the dimeric Holozyme configuration one that is bound to leading strand is oriented toward fork end and the other in opposite direction.�� Beta clamps bind to complex assembles on to the template by displacing SSBs.�� The new strands always grow in 5� > 3� direction.�� One strand grows on leading -strand till the end of the DNA.� The other called lagging strand the DNA polymerase assembles nucleotides up to 1000 to 1500 ntds and dissociates. Then RNA primase adds primers on to it beta clamp with DNA pol is loaded with the help of Gamma complex.
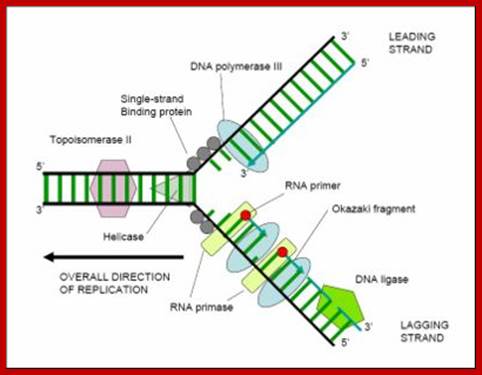
DNA
synthesis at a replication fork (in case of E. coli):
Two new ds DNAs are created by using the two parent ssDNAs as templates near
the replication fork.
STEP1: DnaB helicase(blue) separates a dsDNA into two ssDNAs, as cutting the hydrogen bands between base pairs. It seems to as if unzip. Topoisomerase (lime green) has a role in rewinding the twist of double helices which was generated by the dsDNA separation.
STEP2: The separated ssDNA has a tendency of annealing. For preventing annealing, Single-Stranded DNA Binding Protein: SSB (brown) bind to separated ssDNA.
STEP3: DnaG primase(purple) is activated by binding to DnaB helicase, and synthesizes a short RNA primer approximately 10 nucleotides long using a ssDNA as a template.
�STEP4:
DNA polymerase III elongates a new ssDNA strand by adding a deoxyribonucleotide
at a time in the 5'-3' direction to the RNA primer, using a ssDNA as a
template.
The E.coli polymerase III is a complex consisting of multiple different protein
subunits. The core part(green) consists of three subunits (alpha, epsilon,
delta). clamp part is beta subunit (navy blue), and clamp-loader part is gamma
complex(yellow). https://www.cs.duke.edu
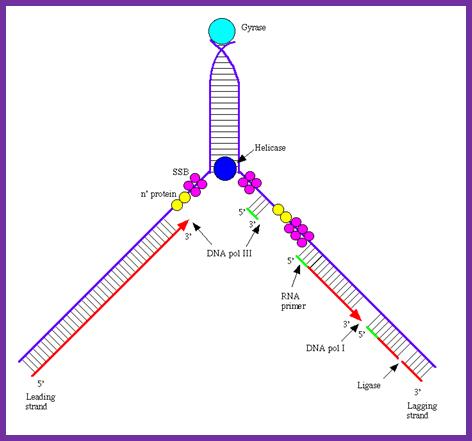
A simple representation of the components at replication fork
is figure represents the assembled components in action. http://en.citizendium.org
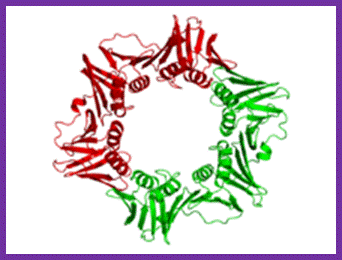
The structure represents two subunits (homodimer); acts as clamp around the ssDNA at the base of DNA pol III Holozyme; http://en.wikipedia.org/
�
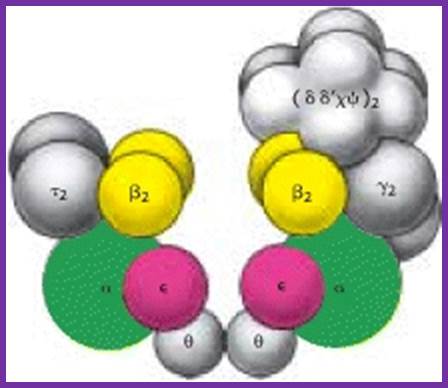
Components of DNAPol and gamma complex with beta clamps; http://sci.bio.lmu.de; http://de.academic.ru/
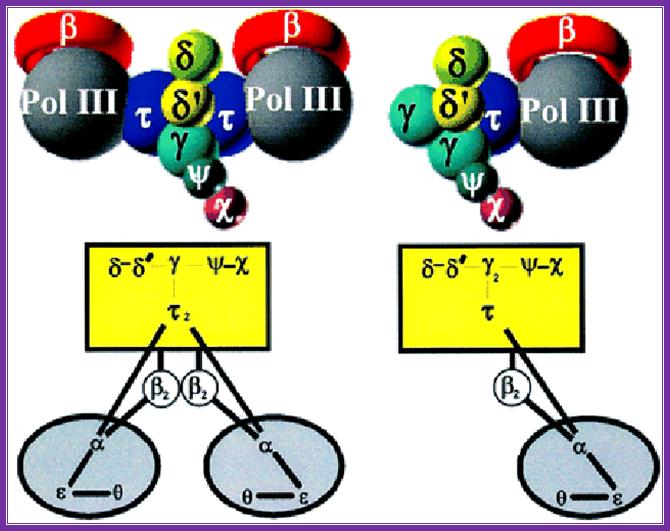
An illustration of the dimeric holozyme with all its components and a single holozyme; Arthur E. Pritchard, et al ;http://emboj.embopress.org
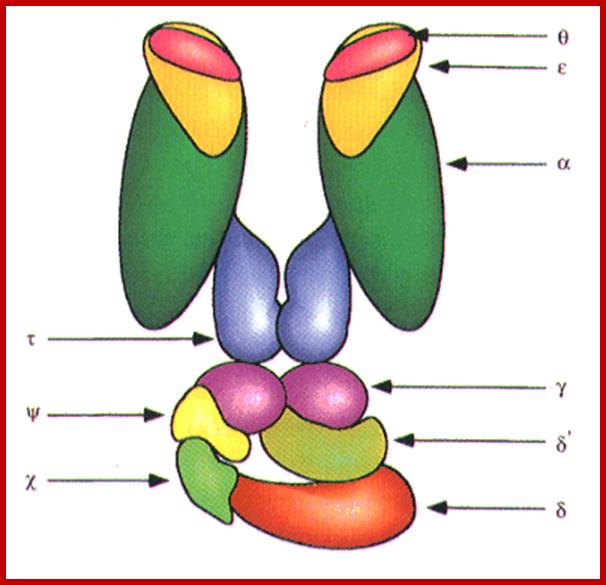
http://oregonstate.edu
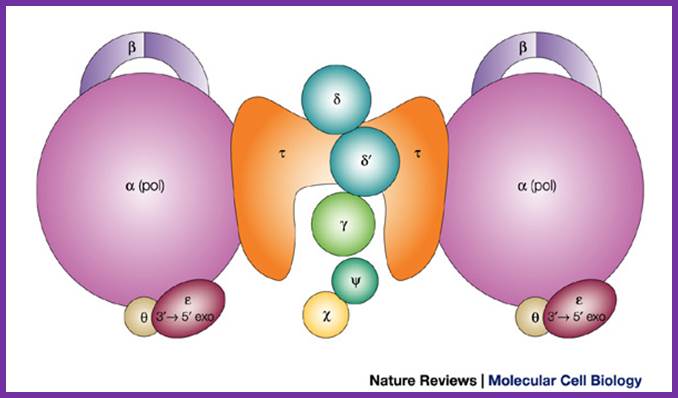
Assembled Holozyme components; http://weloveteaching.com/ and http://trdocs.org/; http://cytology-cytogenetics.blogspot.in/
Elongation:
Let us clarify certain points. At the replication fork, which can accommodate just two pairs of enzyme complexes, one pair at each fork joint, they are bound in such a way; they are positioned back-to-back.
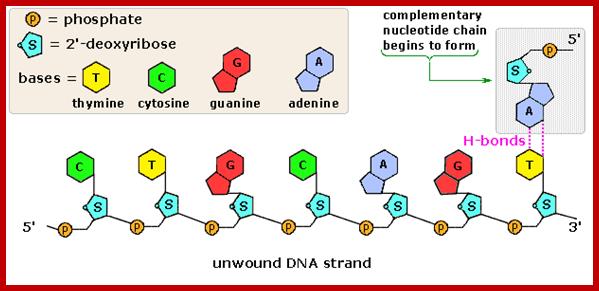
A ssDNA template with a primer http://intranet.tdmu.edu.ua
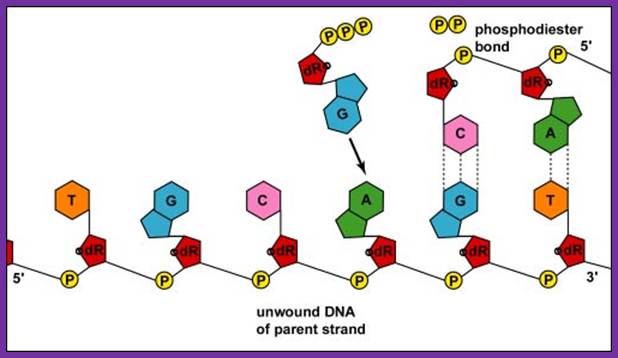
The diagram shows the addition of Nucleotides on the template strand; https://pt.slideshare.net/
- Among the two core complexes (at one fork), one is bound to leading strand positioned to extend the primer laid by RNA pol G (?) and the other to the lagging strand at the primer�s 3� end. The growth of the new strand is always from 5� towards 3� end.
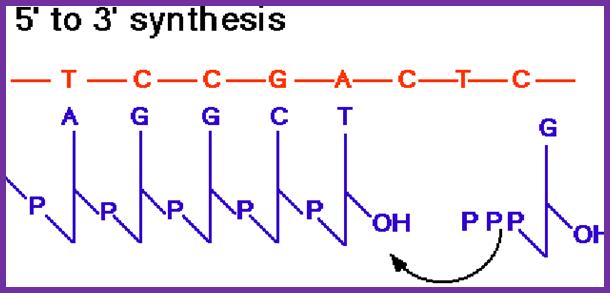
This figure shows a new strand formation on a template >>>.
- At each fork, the leading strand (as a template) having a polarity 3��5�, at initiating position contains primers for complementary strand synthesis in 5�-> 3� direction.� On the leading strand, the primer is laid at the start only once and it has 5��3� direction.��
- The partner parental strand to the leading strand is called lagging strand, whose template polarity is 5��3�.� The primer is laid on lagging strand at specific sequences in opposite direction yet the primer is in 5��3� direction.� On lagging strand, primers are laid at frequent intervals as the Helicase, Primase and DNA-pol-III together move along replicating strand.
- Synthesis of a new strand on leading strand is called continuous strand and the strand synthesized on lagging strand is termed as discontinuous strand, for this strand is synthesized in short segments of 1000 to 1500 nucleotides long.
- Complementary strands synthesize on both templates in 5��3� direction, this is because both core enzyme complexes that associate at each of the fork point move in only one direction on the parental strands.
The core enzyme bound on leading strand moves in 3�� 5�direction and the other core enzyme bound to lagging strand moves in 5�� 3� direction.� Perhaps this movement is propelled and facilitated by the Helicase complex that moves on the lagging strand in 5��3� direction. ��The Helicase is known as the motor protein, it uses ATP energy as driving force for movement.� The Helicase protein is endowed with DNA dependent ATPase activity.�
�
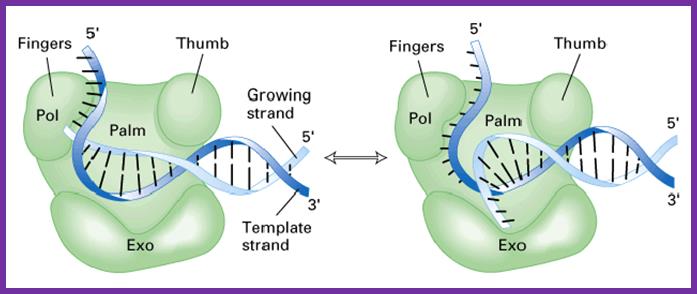
The DNA polymerase performing synthesis and editing; mol.biol
DNA Polymerase I
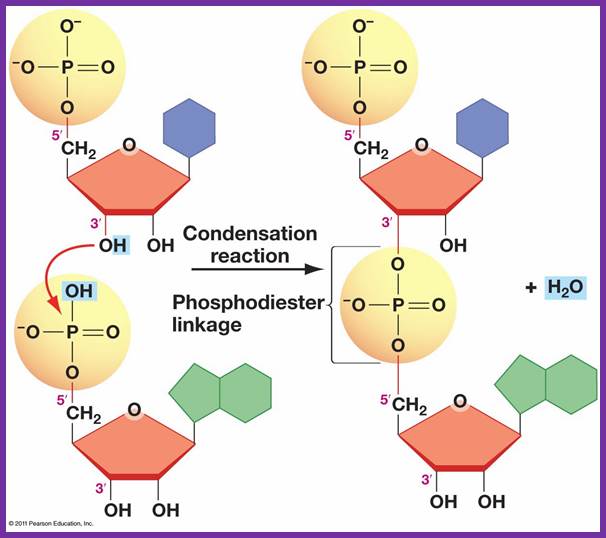
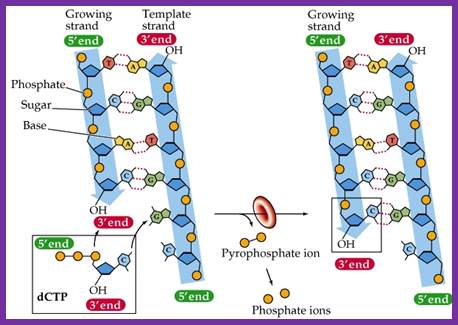
The diagram shows the formation of Phosphodiester bond formation between the incoming triphospho nucleotide and primer nucleotide between alpha phosphate with 2�OH group; http://www.uic.edu/
- The movement of core complexes and the direction of the synthesis of new strands pose a paradoxical situation.� Leading strand (3�-->5' direction) 5�) is primed with an RNA with 5��3� orientation.� On the contrary the lagging strand is in opposite direction i.e. 5� to 3�. The primer is laid in 5�-3�, but the dimer enzymes move in the direction of replication fork movement.
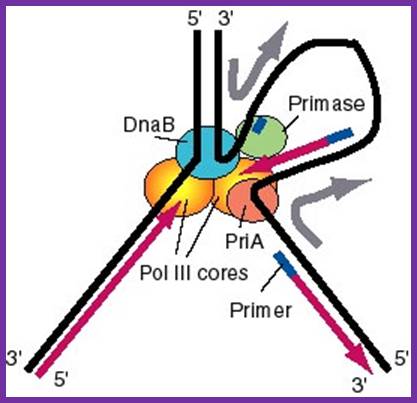
Replisome: A model for the activities of helicases DnaB and PriA at replication fork. In this model, the replication machinery in a large replisome (polymerase and primosomes) are postulated to be stationary, with the DNA strands moving through it like fabric through a sewing machine. After unwinding the duplex DNA at the replication fork, the DnaB helicase can also track along single stranded DNA in a 5' to 3' direction. If the helicase is stationary, then the template strand for lagging strand synthesis moves in the direction of the upper gray arrow in the diagram, away from the replication fork. This template strand is also bound to the PriA helicase at the "back" end of the replisome. The 3' to 5' tracking activity of PriA will also pull the template strand, now in the direction of the bottom gray arrow. The result of the DnaB and PriA tracking activities is to pull the template for lagging strand synthesis into a loop, which is tethered to the replisome at both ends. Primase is also in the replisome, and can synthesize primers along this strand at appropriate intervals (thick blue lines), and one of the core polymerases of the DNA polymerase III holoenzyme can synthesize an Okazaki fragment. Newly synthesized DNA is a thick violet line, and parental DNA strands are thick black lines. http://biowiki.ucdavis.edu/
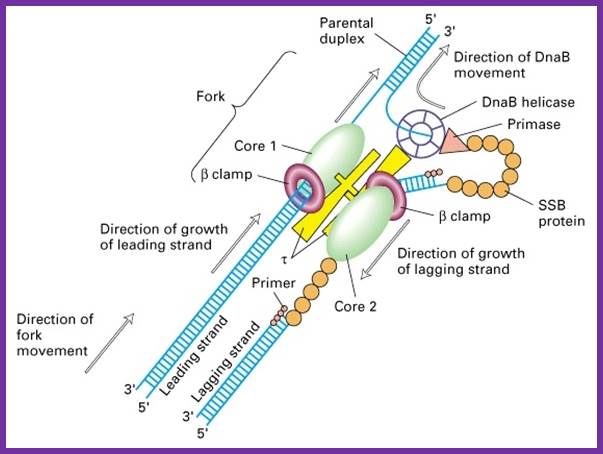
�
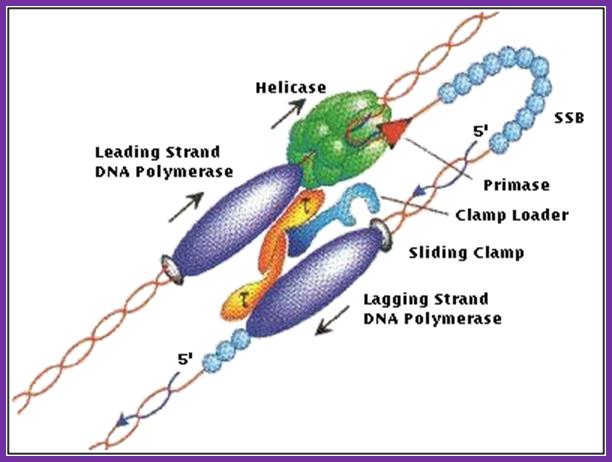
Molecular machines: Mechanochemistry of DNA duplication is highly complex and dynamic. The diagram shows a different model for the synthesis of� a new DNA strand on leading and lagging strand as well. http://www.nat.vu.nl/
- Orientation.� The extension of the primer as DNA in 5��3� direction is absolutely perfect for the enzyme for the enzyme is traversing the leading strand in 3�� 5� direction.� On the contrary the core enzyme, which is moving on, the lagging strand, is in 5��3 direction.� The primers are laid in the opposite direction.� In order to synthesize the new strand on lagging strand one of the two core enzyme has to move in the opposite direction to obey the direction of new strand synthesis. This is what is called a paradox.
- It is more or less established, by electron microscopy that the core enzymes at each fork move as pairs.� Extension of primers on leading strand is perfectly OK.� While the core complex that is bound to lagging strand has to achieve new strand synthesis in 5��3� direction by wrapping the lagging strand around the core complex and positions the strand on the core enzyme in reverse orientation, the enzyme extends the primer as new DNA strand and the movement of the core complex is in the same direction.
- In this process whatever may the orientation of the template strands, the new strand is synthesized in 5�-3� direction.
- The new strand on lagging template is synthesized till it meets another primer. This is because the helicase-primase complexes are moving ahead of the DNA-pol complexes, the primase activated by the helicase, lays primers at intervals of 100 to 1500 nucleotides, where they find their 3�GTC-5� sequences.� Using these sequences the primase lays 11 ntds long primers ahead of the core enzyme, thus the paradox is explained.
- The shape of the catalytic site that accepts the incoming nucleotide triphosphate is determined by the nucleotide found in the template strand.� The nucleotide-binding site decides correct complementary base pairing.�� Once it scrutinizes, it can reject the inappropriate nucleotides and accepts correct nucleotide, which form perfect complementarity as dictated by their respective geometric shapes. If the pairing is perfect, the enzyme catalyses the phosphodiester bond formation between the 3�OH group of the nucleotide found on the strand and 5�PPP of the incoming nucleotide. The pyrophosphate released is hydrolysed to release the energy, which is used by the enzyme for forward movement.
- If the base pairing is not perfect, it is rejected instaneously on the spot.� By chance if the accepted base is wrong, the enzyme, by the virtue of its 3��5� exonuclease activity moves backwards and removes not only the incorrect nucleotide but also few correct base paired nucleotides too, then moves forward.�
- A perfect catalytic site formation with proper geometrical shape is an important conformational event and such conformational changes are induced by the nucleotide geometry of the template.
- The enzyme is shaped in the form of a human right hand palm, the base, the thumb and fingers.� The palm base is made of beta sheets, and possesses a catalytic site, two metal ions binding sites (Mg or Zn).� Fingers and thumb are made up of alpha helices.
- At catalytic site where the arriving nucleotide sits, contains a pocket, where the gamma and beta phosphate chain of triphosphate is drawn into and the alpha phosphate group is positioned against the 3�OH group of the primer nucleotide or any other nucleotide that already exists. Thus, it provides electronegative rich surrounding for the catalysis, to break a bond and make a bond.
- Then the core complex moves forward to continue to synthesize the complementary strand.�
- It assembles nucleotides one after another up to 7 to 8 ntds in one stretch with out leaving the hold on the template strand, which results in contraction of the enzyme, then it releases the hold and moves to the next position; its movement is like an inchworm.�
- The movement of replication fork ahead and the movement of enzyme, while polymerization ensuing is well coordinated events.�
- Continuous strand synthesis on leading strand conforms to 5��3� direction of the chain synthesis. But the other enzyme unit on the opposite template, to be complaint with the direction of polymerization, without dissociating from the template, yet to retain the direction, the enzyme holds the lagging strand and wraps around its surface in such a way, it extends the primer by incorporating dNTPs in 5��3� polarity and progresses till encounters another primer.� At which time the core enzyme leaves free of the segment that is replicated, and now holds on to the next primer on the template.�
- This is achieved by the enzyme, which is still bound to its partner enzyme, by synthesizing the complementary strand in fragments for; the primase, which is associated with the helicase, produces RNA primers at the interval of 1000 or more nucleotides.�
- Energy required for the bond formation and movement of the enzyme is provided by pyro phophorolysis.��
- The helicase motor protein drives ahead of the fork using ATP energy; the positive super coils produced ahead of the fork in response to helicase movement are relaxed by the activity of Gyrase or Topoisomerase II, again in ATP dependent manner.�
- Discontinuous synthesis of DNA on lagging strand produces fragments of DNA, called OKAZAKI fragments named after the discoverer.�
- As the enzymes move along the strands, they displace SsBs�, which reassociate on the single strand regions made free by helicase movement.�
- Association of Ssb�s with helicase on one side and an enzyme on the other side activates both the enzymes. �
- Replication process is processive, more or less free from errors.� The high processivity is due to the association of enzyme to beta clamps. The rate of synthesis is of very high order i.e. 50000 to 60000 ntds per minute, which is amazingly fastest when compared to any other polymerizing enzymes, yet error rate is extremely low i.e. about one in every 10 8 nucleotides incorporated. This exhibits high fidelity of the enzyme activity even though it exhibits fastest rate of synthesis with least number of errors.� This is the quintessence of replication of genetic material.
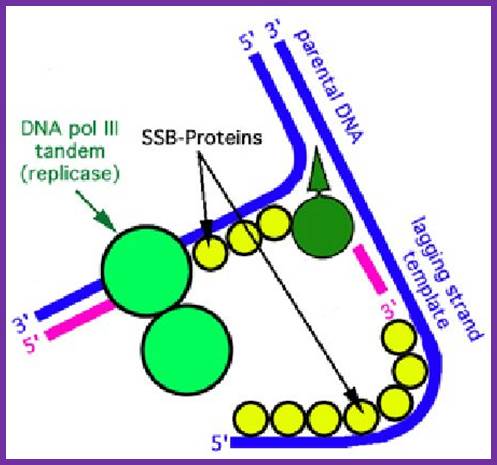
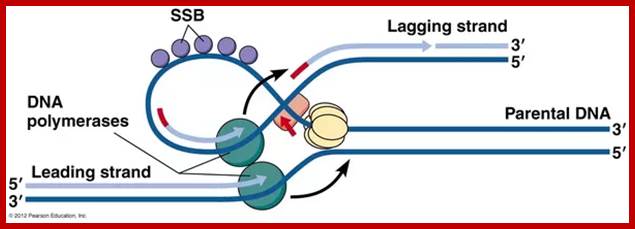
The above diagram and the diagram below show how leading and lagging strand are copied simultaneously. https://www.quora.com
This diagram� is self-explanatory; http://bioserv.fiu.edu/
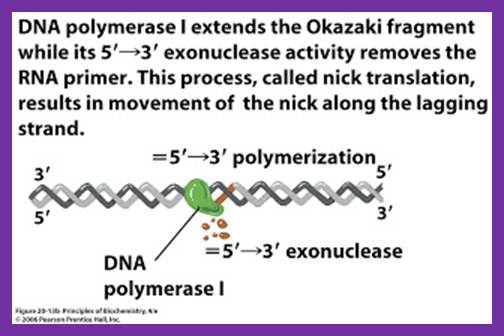
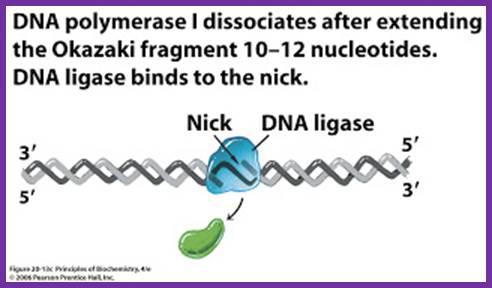
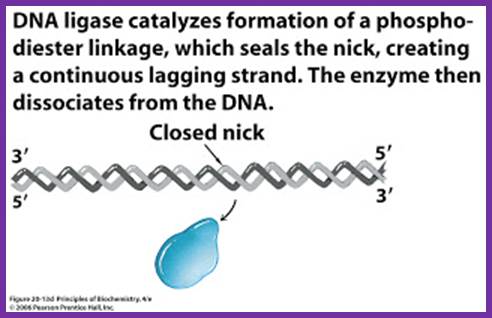
While the replication is progressing with a zing, the RNA primers are removed and gaps are filled, and the ends are ligated by DNA pol -I and DNA ligase respectively.� As DNA pol-I has both 3��5� and 5�>3� exonuclease and 5��3� polymerase activity, the enzyme can recognizes the 3�OH end of primer RNA and removes RNA nucleotides by strand displacement till the entire RNA is removed.� While the enzyme is engaged in progressive removal of RNA primer, it can also use the 3�OH of the DNA and extends till it reaches 5�end of next Okazaki fragment, at which it stops and dissociates from the dsDNA. DNA pol-Is� processivity is not great and also its fidelity, yet its activity is very important for it is also involved damage repair.
- If the polymerase makes any mistakes in incorporating wrong nucleotides or wrong base pairing, or if there are any existing thymidine dimers, the DNA repairing enzymes swing into action and repair errors.� Beta clamps are used during DNA repair process.
- Removal of primers and filling the gap with DNA nucleotides leave the two ends of the DNA segments free, without a covalent bond, i.e. 3�OH of the preceding strand and 5� phosphate end of the next segment. A covalent phosphodiester bond seals these two ends by an E.coli enzyme called NAD dependent DNA ligase.

- Thus the elongation process is series of successive processes, where new strand formation is determined by the template sequence, the process is mostly error free; this is in spite of tremendous speed with which it polymerizes the new strand.� It is a molecular marvel and a par excellent molecular process.
�
Termination:
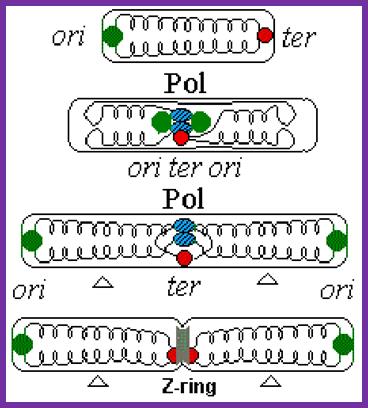
- As in the case of replication initiation, replication is also terminated at a particular site.� In E.coli, the termination site is located exactly opposite to the site of origin.�
- The TER region is quite extended, but it consists of six 23 bp non-palindromic sequences, organized into two groups of three and four each. TER-A, B and D are on one side i.e. towards the zero mpu and the other three TER-C, F, G are on the opposite side, where each of the groups, in their sequence they are oriented in opposite direction.
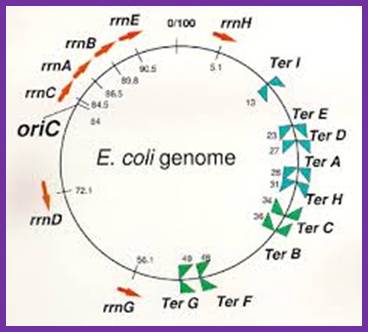
Arrangement
of rrn operons and nine Ter sites on the E. coli genome.
Reds arrows indicate locations and transcriptional direction of rrn operons. ![]() means
DNA replication terminus (Ter) site
which can block replication fork approaching from the right, Professor: Takashi Horiuchi
means
DNA replication terminus (Ter) site
which can block replication fork approaching from the right, Professor: Takashi Horiuchi
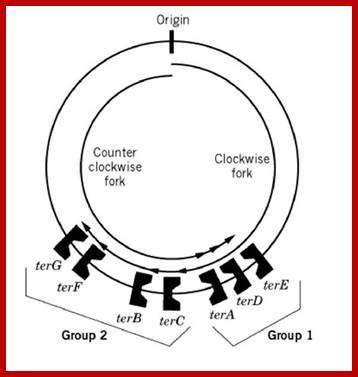
���������������������������� Location TER region; http://what-when-how.com
- When replication forks from the initiation point move in bi-directional manner, they are likely to meet each other from opposite direction and actually meet in the TER region.�
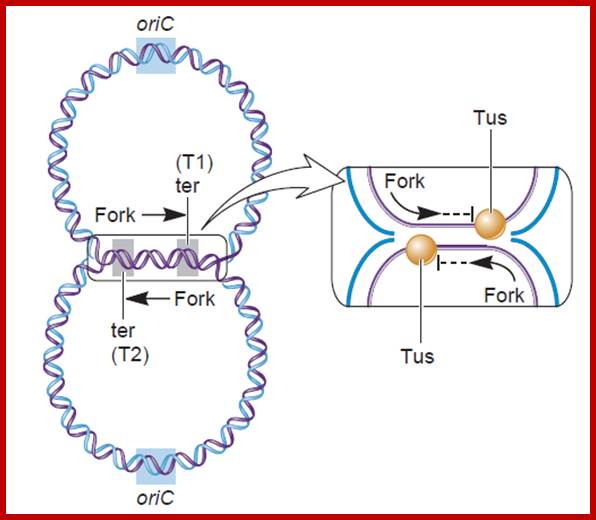
- If the forks move unhindered, they are destined to move into the newly synthesized daughter DNA, for the DNA in E. coli is in circular form.� If they do so, a new cycle of replication is initiated even before the termination of the replication of the first strand.� To prevent this from happening, proteins called Termination utilizing substances (Tus) bind to these sequences and prevent helicase to move into the newly formed DNA molecules.
- The Tus protein, which is also called replication terminator protein (RTP), consists of two domains connected by two anti parallel B-sheets, thus create a central cleft into which DNA fits in.� Tus protein grips the DNA and blocks helicase movement towards B-end.� But the proteins allow Helicase movement into P-end, thus Helicase moves to completion of replication.
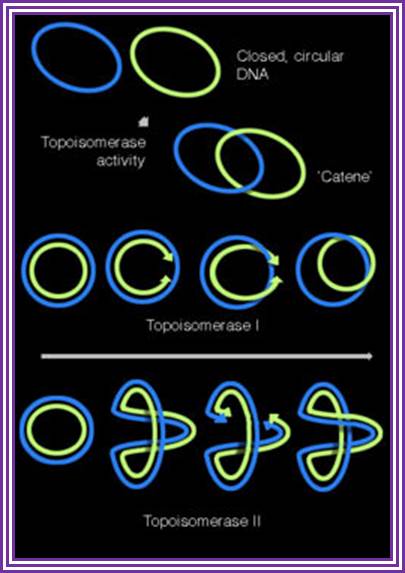
- Though the exact mechanism is not clear, it is possible that newly synthesized daughter molecules are likely to get entangled in catenation.
Replication of the DNA separating the opposing replication forks, leaves the completed chromosomes joined as �catenanes� or topologically interlinked circles. The circles are not covalently linked, but cannot be separated because they are interwound and each is covalently closed. The catenated circles require the action of topoisomerases to separate the circles [decatanation]. In E.coli, DNA topoisomerase IV plays the major role in the separation of the catenated chromosomes, transiently breaking both DNA strands of one chromosome and allowing the other chromosome to pass through the break. Confusion arises when some scientific literature state that DNA gyrase is the sole enzyme responsible for decatanation. In an experiment conducted by Zechiedrich, Khodursky and Cozzarelli in 1997, it was found that topoisomerase IV is the only important decatenase of DNA replication intermediates in bacteria. �In this particular experiment, when DNA gyrase alone were inhibited, most of the catenanes were unlinked. However, when Topoisomerase IV alone was inhibited, decatenation was almost completely blocked. The results obtained suggest that Topoisomerase IV is the primary decatenase in vivo, and although DNA gyrase does play a role in decatenation, its function is not as essential as topoisomerase IV in the decatenation of interlinked chromosomes.
(A) The positions of the six terminator sequences on the E. coli genome are shown, with the arrowheads indicating the direction that each terminator sequence can be passed by a replication fork. (B) Bound Tus proteins allow a replication fork to pass when the fork approaches from one direction but not when it approaches from the other direction. The diagram shows a replication fork passing by the left-hand Tus, because the DnaB helicase that is moving the fork forwards can disrupt the Tus when it approaches it from this direction. The fork is then blocked by the second Tus, because this one has its impenetrable wall of β-strands facing towards the fork.
Role of topoisomerases in replication termination. Replication of the DNA separating opposing replication
forks leaves the completed chromosomes joined as catenanes, or topologically
interlinked circles. The circles are not covalently linked, but because they
are interwound and each is covalently closed, they cannot be separated�
except by the action of topoisomerases. In E.
coli, a type II topoisomerase known as DNA topoisomerase IV plays the primary role in the
separation of catenated chromosomes, transiently breaking both DNA strands of
one chromosome and allowing the other chromosome to pass through the break.
�http://reasonandscience.heavenforum.org/http://reasonandscience.heavenforum.org/
�
- The TER sequences, however, not all together required for termination of replication.� There are many plasmid DNAs that complete replication without TER sequence; such DNA are too many and too far to understand. If a plasmid containing TER sequences are deleted replication is not affected.
- The daughter DNA molecules thus produced in the region of origin, the GATC sequences are hemi-methylated and it is in this state they get attached to mesosomal membrane.� How?� What is the mechanism, what are the enzymes involved in this process?�
- The daughter molecules are segregated through mesosomal membranes and cytoplasm divides by fission process.� Genetic studies suggest the role of many gene products are involved in segregation of DNA, membrane invagination in the middle of the cell, all-round the cytoplasm and peptido-glycan are laid in between the membrane space.� Once the cell wall is fully formed, it splits and cells two daughter cells are released. Cells then grow to full size to initiate another round cell cycle.
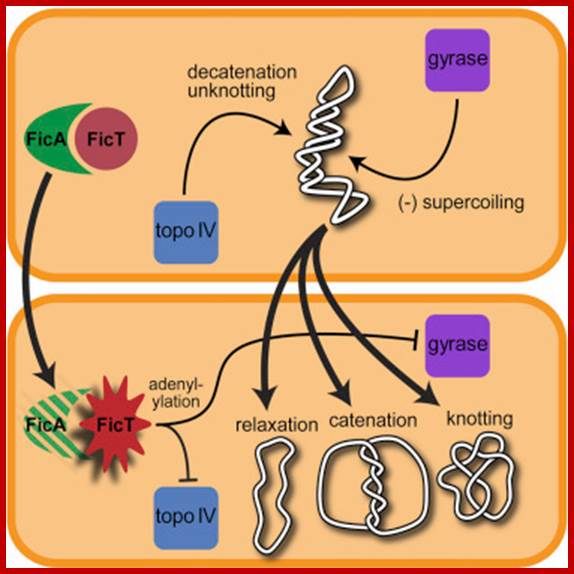
Adenylylation of Gyrase and Topo IV by FicT Toxins Disrupts Bacterial DNA Topology; Toxin-antitoxin (TA) modules are ubiquitous molecular switches controlling bacterial growth via the release of toxins that inhibit cell proliferation. Most of these toxins interfere with protein translation, but a growing variety of other mechanisms hints at a diversity that is not yet fully appreciated. Here, we characterize a group of FIC domain proteins as toxins of the conserved and abundant FicTA family of TA modules, and we reveal that they act by suspending control of cellular DNA topology. We show that FicTs are enzymes that adenylylate DNA gyrase and topoisomerase IV, the essential bacterial type IIA topoisomerases, at their ATP-binding site. This modification inactivates both targets by blocking their ATPase activity, and, consequently, causes reversible growth arrest due to the knotting, catenation, and relaxation of cellular DNA. Our results give insight into the regulation of DNA topology and highlight the remarkable plasticity of FIC domain proteins.; et al; http://www.sciencedirect.com
Topoisomerase activities illustrated with on covalently closed circular DNA. Topoisomerase enzymes are able to form supercoils in DNA, and interconvert covalently closed circular DNA and their catenated forms.
Second round of DNA replication has to wait till the cell mass increases with cell volume (size).
As the two daughter DNA molecules are segregated, they are sequestered to mesosomal membrane at attachment point.� At this point the cytoplasm divides to generate two daughter cells which grow in time.
Immediate replication is prevented for one of the newly produced strands at ori site and the promoter region of dnaA gene is yet hemi methylated at GATC sites. Hemi methylated GATC sites in ori c region makes the DNA bind to membrane.� The region of origin has several GATC sequences; in hemi methylated state is covered by proteins called SeqA, which prevents methylation. At the same time the promoter region of dnaA gene, as it is hemi ethylated fails to initiate transcription.� The promoter has sequences which contains strong and weak sites for the binding of DnaA binding and DnaA-ATP binding respectively. Complete methylation of hemi ethylated sites at promoter region makes the gene active. Complete methylation at attachment site releases the DNA from the membrane.
With time methylation at other GATC sites make SeqA to dissociate as more and more DnaA protein are formed and active Dna-ATP subunits are produced.� This leads to the binding of DnaA-ATP to 9-mer sites initiates another round of replication.� Between the events, completion and initiation there is a time lapse called eclipse period.� It is at this point of time cell acquires the competency for the second-round replication.
Segregation, Partioning of Chromosomes and Cytokinesis:
Daughter DNA molecules produced in general move away from the central region, but at the end of replication the daughter DNA molecules also have chances of recombination before partitioning or segregation. If they undergo recombination by Xer-C and Xer-D, which are recombinases, they generate Holliday junctions.� This leads to problems in segregation.� Xer-C and Xer-D dependent recombination takes place in a region determined by �Dif� site, which is about 30kbp long near Ter region.� However resolution of Holliday junction by the enzyme Fts-K releases circular DNA from catenated condition into independent circles; this function is essential for segregation.� Fts-K is a membrane protein; its Carboxyl end causes Xer to resolve this.� The Fts-K has ATPase activity and it can traverse along the DNA.
Mutation that affects partioning of DNA is also found in Topoisomerases. These enzymes are required for separation of daughter DNA molecules; otherwise they will be tangled at TER regions.� Such mutations can result in cell containing 2n DNA in some cells and some cells without DNA.� Mutations that are cis-acting and trans-acting have been identified, but gene products that are transacting have been isolated.� If protein synthesis is blocked before termination of replicated DNA molecules fail to segregate, but if protein synthesis is allowed to resume chromosomes segregate and move away from the middle region.� It is now known that segregation requires Muk-genes.� Mutations affect this function.� Muk-A is identical to an envelope protein called Tol-C that is involved in chromosomal attachment.� Gene Muk-B produces a protein, which is 180KD; it works like structure maintenance chromosome (SMC) proteins.� Muk-B has some sequence relation to that of Dynamin a motor protein.� Muk-BEF proteins act on chromosomal DNA in condensing into individual nucleoids which helps in proper segregation, higher super coiled density helps in proper segregation.
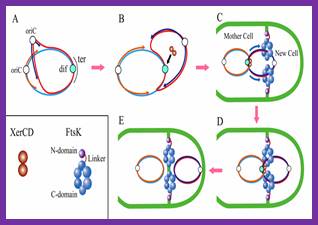
The replication of E. coli circular DNA begins from the region called oriC. Replication forks run to both directions and they are stopped by the Tus protein at the region of ter which is opposite to oriC. The dimer of double-stranded DNA (dsDNA) which finished replication is recomposed in the region� dif �and separates into two monomers. The enzyme recognizing the dif sequence and making recombination is a complex of XerC and XerD. Furthermore, it is noted that XerCD divides the dimer into two monomers with the help of the FtsK protein. FtsK is a membrane-bound DNA translocase found in many eubacteria, such as E. coli.
The DNA translocase is an adenosine triphosphate (ATP)-dependent molecular motor which move DNA rapidly in the case of chromosome division, DNA recombination, and DNA transport. T7gp4, DnaB, SV40 SpoIIIE, etc. belong to this type of protein. The structure of FtsK may be divided into three regions (N, linker, C). The N domain is locates on cell membrane which invaginates in cell division. The length and structure of the linker region varies depending on bacterial species. The C domain transposes DNA to the direction of a daughter cell at the rate of more than 6.7 kbp/s. At the same time, the C domain proceeds to the dif region and separates dsDNA by activating XerCD complex. FtsK expresses the function by forming a hexamer-ring. A large channel is formed in the center of each subunit and dsDNA passes through this channel.
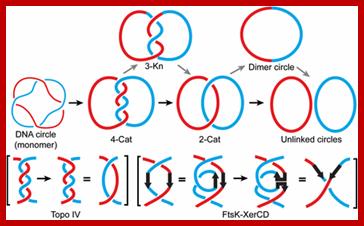
1. Replication of a circular DNA; Resolution of a (4)-torus catenane by Topo IV or by FtsK�XerCD. A (4)-torus catenane formed by the replication of an imaginary circular DNA with two helical turns is shown. The thin red and blue lines indicate the two strands of the double helix. The thick red and blue lines represent double helical daughter DNA molecules. The catenane is unlinked by Topo IV in two steps (the straight path) or by FtsK�XerCD in four steps (the zig-zag path). The reduction of crossing number, by two for each step of Topo IV action and by one for each step of FtsK�XerCD action, is shown in parentheses. Mathematical validation of a biological model for unlinking replication catenae�s by recombination; Makkuni ; Jayaram1; http://www.pnas.org
Step A:
The replication of circular DNA proceeds with two replication forks from the
region of oriC
to the region of ter
in opposite directions.
Step B: When replication has finished at ter,
XerCD complex bind to dif
site located in the ter.
Step C: One replicated circular DNA is dragged into the daughter cell through a
channel in the center of the FtsK(C-domain). At the same time, this means dif move toward the
FtsK(C-domain).
Step D: When dif
approaches FtsK(C-domain), XerCD is activated by the FtsK.
Step E : The circular DNA dimer is completely resolved by the activated XerCD
site-specific recombination.
(*) Although only two FtsK is shown in the Fig.1, many FtsK exist on invaginated region of cell membrane.�]
Bacterial DNA has been found to be in association with membrane fraction, the membrane fraction have been found to be enriched with genetic markers associated with Origin, replication fork� and Ter regions.� Protein in this region is involved in initiating replication at origin, but mutations affect this process.� Association of origins to membranes is the key for segregation of daughter chromosomes in condensed state.
A similar situation is also found in the case of single copy plasmids, when the single copy plasmid replicates it has to be segregated equally otherwise the plasmid is missing in one of the daughter cells.� But there are genes involved in segregation of single copy plasmids, they are called Par- genes; par-A and par-B whose products are transacting components.
The plasmids also contain one cis acting site called �par-S�.� Par-A is an ATPase binds to Par-B protein and forms dimmers.� Meanwhile a dimer protein called Integration Host Factor (IHF) binds to �S� , a 24 bp long site, which is located in between par box-A and par box-B.� These boxes are the sites to which Par-A and par-B proteins bind as dimmers.� Binding of IHF to �S� bends DNA in such a way the par boxes are brought very close to one another. �This facilitates the binding of Par-A and Par-B proteins as dimmers to these boxes.� This, complex proteins is called partitioning complexes which assemble on each plasmid DNA.� These Complexes some how are associated with partioning membranes, which are segregated by septal membranes.�
A list of Genes Involved in Cytoplasmic Division:
|
Event
|
Gene |
Gene product n function |
|
Maintains rod shape |
Rod-A and pBP2
|
Rod �A is a member of SEDS family proteins; involved in periseptal ring formation |
|
Septation |
Fts-A = Transpeptidase Fts-L Fts-F Fts-E Fts-I Fts-K Fts-Q Fts-N Fts-W= transmembrane protein Fts-Z= like tubulin proteins Zip A |
Fts-Z is Involved in mid septal ring formation not polar septa, it is temperature sensitive, mutation produces long filamentous cells. |
|
|
|
|
|
Cell separation or splitting the septal structure |
Env-A |
|
|
Inactivation of septation site |
min B, min-C, min- D, and min-E |
Mutation in min-B leads to septation even at poles leading to produce mini cells; mutation in C and D leads to filamentous morphology.
|
|
Regulation of septation |
Sfi-A Lon |
|
|
|
|
|
|
|
Muk-A, Muk-B, Muk-E, Muk-F |
Involved in segregation of chromosomes; they are like �smc� proteins |
|
|
Par-A, Par-B Par-C |
They are involved in portioning of chromosomes |
Cytoplasmic Division:
Bacterial cell division involves cleavage of the cytoplasm in the middle and cell separation.� A set of genes is involved in septal formation that leads to cell division; they are many such genes, such as min genes, PBP2, PBP3 and Rod-A, Fts-genes, Xer and Muk and other genes.� Information on the details, though scanty, conditional mutants have provided some significant information.
- Process of cytoplasmic division starts in about 1-2 minutes of DNA replication and it will be completed in about 15 to 20 minutes. �
- Development of middle wall is initiated at predefined position where membrane components from the earlier cell division act as the initiating centers.� It is at this site the annulus is formed where the inner and outer membranes are connected, this is periseptal ring.� At this point invagination of the envelope develops into septal ring or Z-ring.�
- This membrane structure has the same composition as that of the envelope.� The rigid structure of peptido-glycan is found in between outer and inner membranes.� Peptidoglycans are synthesized and transported in to the middle space by trans peptidase and trans glycosylation activity.
http://www.eva.university.edu.uy
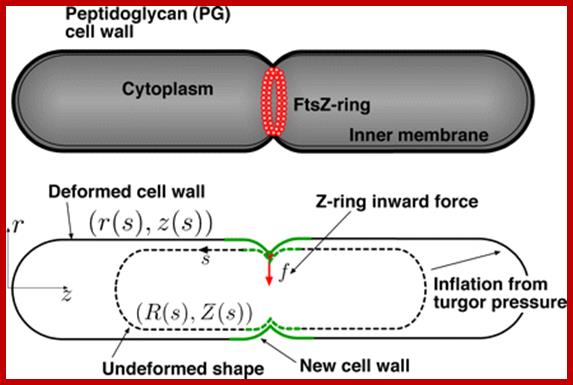
Cell division in bacteria is dependent on the precise placement of components of the division machinery at the cell center, a process initiated by assembly of a medial ring of the tubulin analog FtsZ.
In E. coli, movement of the growing fork is about 1000 bp per second. It takes about 42 minutes to duplicate the entire genomic DNA. In eukaryotic DNA, the fork movement is only about 100 bp per second. This is probably due to the association of DNA with histones, which may hinder the fork movement. In humans, replication of the entire genome requires about 8 hours. In fruit flies, it takes only 3 - 4 minutes.�
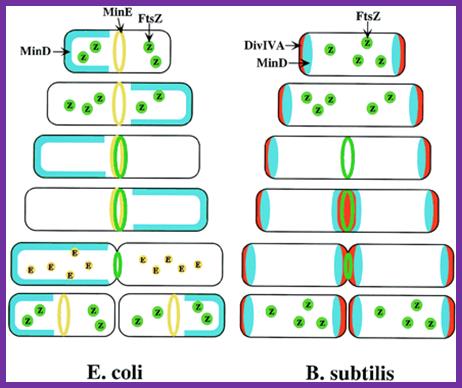
Models for division-site selection in E. coli (Left) and B. subtilis (Right). MinD is in blue, MinE in yellow, FtsZ in green and DivIV in red. Shown are different stages of the cell cycle, beginning with a newborn cell and finishing with cell division that produces two daughter cells. (Left) In E. coli, MinE localizes to a ring-like structure at or near the middle of the cell early in the division cycle. MinD accumulates alternately at the membrane periphery on either side of the MinE ring (3). The alternation of MinD localization from one pole to the other occurs at a frequency of the order of tens of seconds. The rapid relocation of MinD ensures that no FtsZ ring is assembled at either the � or � sites in the cell halves. The presence of MinE at midcell prevents the MinD inhibitory activity at this site, allowing assembly of the FtsZ ring at this site. The MinE ring disassembles before completion of constriction. (Right) In B. subtilis, DivIVA and MinD are localized to the cell poles in a newborn cell, and therefore the presence of the MinD inhibitor prevents the formation of the FtsZ ring at these sites. Later, presumably after completion of DNA replication, a new potential division site is created at midcell. The sequestration of the MinD inhibitor to the poles allows assembly of the FtsZ ring at midcell and recruitment of other cell division proteins. At this point, the division machinery presumably becomes resistant to the MinD inhibition. DivIVA and MinD proteins then are recruited to the mid- cell. Constriction then is initiated. When constriction, is completed, the FtsZ ring disassembles, but DivIVA and MinD remain at the newly formed poles, preventing further divisions from taking place in these polar sites.;http://www.pnas.org/
A simple illustration of Bacterial cell division using various components.
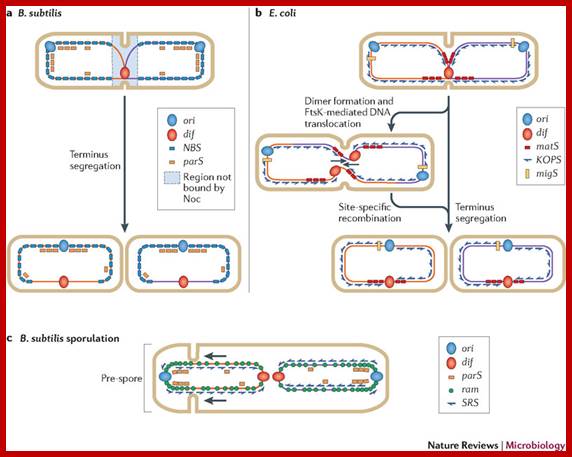
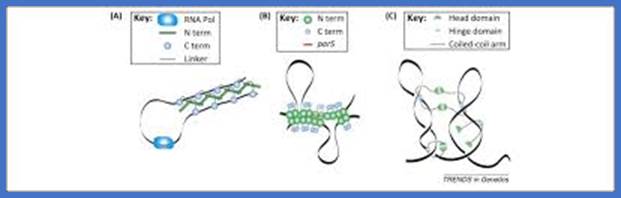
Motifs implicated in chromosome segregation and cell division. a | In Bacillus subtilis, most of the parS motifs are in the origin (ori) domain: when bound by SpoOJ, they stimulate segregation of the replichores. The distribution of the NBS (Noc-binding sites) motifs away from the terminus ensures that nucleoid occlusion protein (Noc), which binds NBS motifs, does not bind this region, and this allows assembly of the cell division machinery at the middle of the cell while replication is terminating. b | In Escherichia coli, the migS motif, situated ~200 kb from ori, is implicated in segregation of the sister ori regions. The matS (macrodomain TER site) motifs that are present in TER macrodomains (which overlap with the terminus region) are bound by MatP, which results in compaction and late segregation of this region. When a chromosome dimer is formed, KOPS (FtsK-orienting polar sequences; also known as FRS) motif-dependent DNA translocation mediated by FtsK brings the two dif sites together (indicated by arrows), which stimulates dimer resolution into two monomers by site-specific recombination. After the DNA molecules are separated, cell division can occur. c | The ori domain of B. subtilis is enriched in ram (RacA-binding motif) sites and parS motifs that are bound by RacA and SpoOJ, respectively, during sporulation. This stimulates proper segregation of ori to the pre-spore compartment. SRS (SpoIIIE recognition sequences) motifs also have a role in translocating DNA that is trapped at a late step of septum formation and during asymmetrical cell division in sporulation.;Fabrice Touzain, Marie-Agn�s Petit, Sophie Schbath & Meriem El Karoui;www.nature.com
Mark Gonzalez;http://beckwith.med.harvard.edu/
![]()
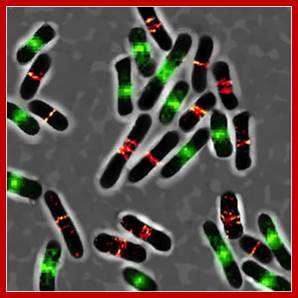
The Z-ring is made up of FtsZ proteins which are constantly polymerizing and depolymerizing through a GTP-dependent mechanism. During this dynamic polymerization, the ring becomes smaller until the daughter cells are finally split. Although FtsZ is the star of the show, many other Fts* (filamentation-forming temperature sensitive) proteins are involved in binary fission. In addition, there is a thick layer of regulatory genes that changes the time interval between assemblies of the Z-ring. (Such as my personal favorite example, sdiA, which in Escherichia coli can modulate the interval between divisions in response to the presence of bacteria other than Escherichia coli!); Nathan Brown; https://www.quora.com
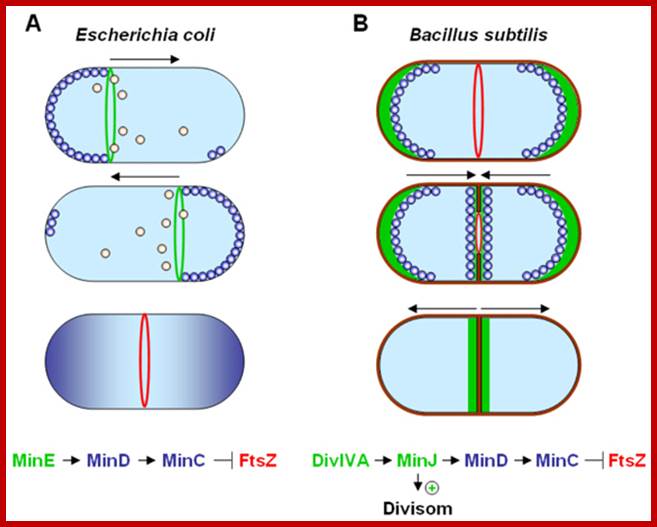
Dynamic control of cytokinesis. (A) In E. coli an oscillating Min System spatially organizes divisome (red) placement. MinCD (blue) complexes are released from the membrane by MinE (green) triggered ATP hydrolysis (orange). (B) In B. subtilis MinCD is recruited to the cell poles and division sites by a DivIVA/MinJ (green) complex. Protein dynamics are indicated by arrows; http://www.bacteriology.bio.lmu.de/
- PBP2 and Rod-A gene products are involved in peptidoglycans formation.� Rod-A protein belongs to a family of proteins called SEDS (Shape, Elongation, Division and Sporulation). PBP2 and Rod-A are coded from the same operon.� Each member of SEDS functions together leading to synthesis and cross-linking peptidoglycans.� PBP2 is transpeptidase interacts with Rod-A.� Mutation in either of the genes results in the loss of rod shape and cell become spherical.� The gene product of Fts-Z is very essential for the peptidoglycans layer formation.
- Mutations that fail to divide, also fail to segregate nucleoids because cells fail to produce septum and such cells exhibit filamentous morphology. They are called Fts mutants for they are obtained as temperature sensitive mutants producing filamentous morphology.
- Some mutants instead producing normal cells generate mini-cells because of frequent septal formation resulting in some cells with 2n nuclei and some cells without a nucleus; there are partition mutants (par mutants).
- Fts-Z mutants produce filamentous morphology and over expression of it leads to increased septal formation per unit mass of cells.� Fts-Z is required at pre existing periseptal sites.� Fts-Z does not affect periseptal annuli or their location.� As the constriction begins Fts-Z proteins move from cytoplasm into septal membrane and organize into Z-ring.� The Z-rings develops in about 1-2 minutes after DNA replication and after Z-ring formation within another 15-20 minutes constriction leads to division of cytoplasm into two compartments.� Fts-Z proteins are like Tubulins, and have GTPase activity, so involved in polymerization of Fts-Z proteins into Microtubule like filaments and assemble into Z-ring.� Hence it is dynamic structure and exchanges subunits between the Z-ring and cytoplasmic pool.�
- Association and assembly of Fts-Z are facilitated by another protein called Zip-A protein, found in inner membranes of the septal ring.
- Fts-A is also associated with Zip-A in the membrane.� If both are absent, ring formation fails, at least one of them is required for the formation of the septal ring.� After Fts-A association, other members of Fts family start assembling one after another and the last to associate is Fts-W, which is expressed as a part of Fts-I operon.� The Fts-I is a transpeptidase called PBP3 (penicillin binding protein), it is located in the membrane, but its catalytic site is at periplasmic side.� This protein is responsible for the growth of peptidoglycans in the septal ring, which causes the membranes, both outer and inner membranes of the ring to grow inwards.
- Fts-Z1 and Fts-Z2 are found in the chloroplasts of Arabidopsis thaliana; even plant mitochondria though don�t contain Fts-Z proteins, they have dynamin like proteins for mitochondrial membrane constriction.� Dynamin is a motor protein involved in contractile and motor activity.
- Another set of gene products involved in septal formation are min genes.� Mutation or deletion min-B genes results in mini cells due to formation septa not only at mid region but also at polar region.� Wild min-B suppresses septal formation at Polar Regions but not at middle region.�
- Polar sites are found as remnant from previous cell division and they act as nucleating centers for septal formation.� Min-B locus contains genes like min-C, min-D and min-E.� Min-C activated min-D and prevents Fts-Z protein assembly into Z-ring.� Very interesting feature is that expression of C and D in the absence of Min-E or over expression min-c and D even in the presence of Min-E inhibit cell division and produce filamentous form.� Experiments indicate equal level expression of min-E and Min-C and D produce proper mid septum.� Accumulation of min-E in the septal region prevents and suppresses the ring formation by utilizing Fts-Z and Zip-A.� Paradoxically min-D is required for min-E for ring formation.
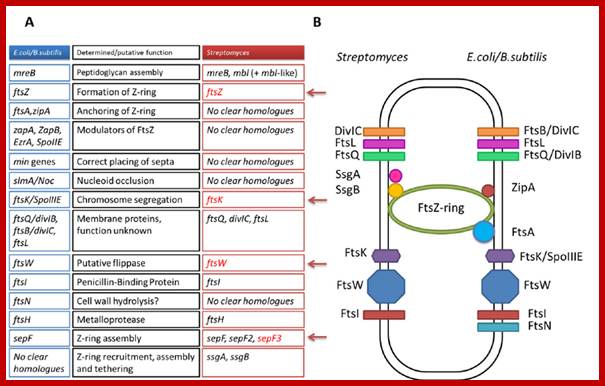
Comparison of the cell division machinery of E. coli/B. subtilis and Streptomyces� (A) Genes encoding key components of the divisome in E. coli and Streptomyces
https://www.researchgate.net.
�
Fidelity of replication:
- Fidelity of is the measure of faithfulness in function and the efficiency of it is measured by its activity in not making mistakes (errors).��
- Replication process is a mechanistic process and a complex process hence it is error prone.� The rate of replication is so fast enzymes are bound to make errors.�
- However, cells are empowered with mechanisms to prevent such errors, as well as repair the errors during replication or in post replication stages.�
- Any error less than 10^-10 to 10^-12 is tolerable.
- Fidelity is mostly ensured by balanced level of dNTPs made available.� Synthesis of dNTPs is itself highly regulated.� High levels of dNTPs ensure high fidelity. Low concentration leads for error-prone, especially in the incorporating wrong nucleotides.�
- Incorporation of Uracils is minimized by the activity of UTPase and UTP.N� glycosylases.
- Complementary base pairing by Watson- Crick rule further ensures specifity and virtual fidelity.�
- DNA polymerases have induced fit conformation change, as in any other enzymes, where the active site adopts to the correct base pairing using geometrical complementarity, which prevents wrong base pairing.
- DNA-pol III Enzyme complexes have 3��5� exonuclease activity further corrects the wrong incorporation of nucleotides or mismatched bases.�
- DNA pol-I is another enzyme that involves in removing mismatched bases by and incorporating correct bases.�
- DNA Ligases don�t ligate if the ends are not correctly base paired or if the gaps are more than one nucleotide long.
- Cells are provided with a battery of enzymes, which readily respond to any damage or any error, and perform repair with promptness and accuracy.