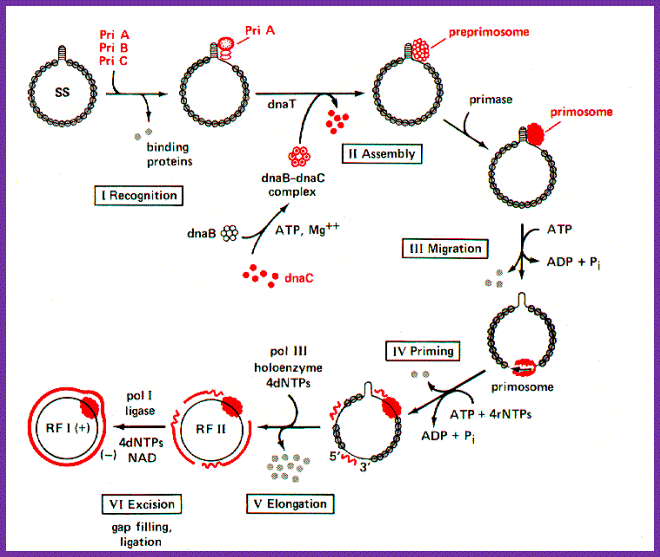
Primosomes:
- A group of proteins, that bind to replication origin site and synthesize primers to initiate replication, is called Primosomes.
- Constituents of Primosomes differ from one system to the other.�
- In E. coli assembly Dna-A binding leads to Dna-B and Primase G binding-all together form the primosome complex.
- In the case of phiX174 phage DNA, the primosome complex is made up of Pri-A, Pri-B, Pri-C, Dna-T, Dna-B, Dna-C and Dna-G.�
- The said components assemble at a particular site called �pas�; means primosome assembling site.�
- Such assemblies are found in Origin-C of E. coli, Col E-1 Ori, phiX174 phage origin (phi ori), m13 phage DNA and few other plasmid DNAs for that matter all chromosomal DNAs have their own specific origins and their own specific primosomes.�
- Based on the sequence, structures and the components assembled to form Primosomes; they are grouped in to Dna-A dependent and Pri-A dependent primosomes.
DnaA- and PriA-dependent primosomes: two distinct replication complexes for replication of Escherichia coli chromosome. Masai H, Arai K. Department of Molecular and Developmental Biology, Institute of Medical Science, University of Tokyo, 4-6-1, Shirokanedai, Minato-ku, Tokyo, 108, Japan. hisao@ims.u-tokyo.ac.jp
Primosomes are complex of proteins that produce RNA primer at the time of DNA replication. Enzymatic analyses of primosome assembly at chromosomal and plasmid origins as well as that at single-stranded DNA replication origins revealed that the presence of two distinct primosomes in Escherichia coli for primer RNA synthesis and duplex unwinding.
�The DnaA-dependent primosome is assembled at oriC, the chromosomal origin of Escherichia coli, as well as at the A site, a single-stranded DNA hairpin containing a DnaA box sequence within its stem.
�In contrast, PriA protein recognizes a small stem-loop, called n'-pas (primosome assembly site), and initiates assembly of the phiX174-type PriA- primosome in conjunction with other prepriming proteins. Genetic analyses of the prepriming proteins required specifically for the latter primosome strongly suggested that it is responsible for RecA-dependent, DnaA/oriC-independent replication. �
Furthermore, primosome assembly in replication of various plasmids may also be classified into either DnaA-dependent or PriA-dependent types. Escherichia coli possess two distinct, mutually exclusive primosomes which are differentially utilized by the chromosome as well as by the plasmids. PriA protein appears to be conserved in a wide range of prokaryotic species, and the same will be discussed and its possible biological functions ex. PriA-dependent primosome in responses to DNA damages.
Kinds of Primosomes:
|
Dna-A Dependent Primosomes |
Pri-A dependent Primosomes |
||||
|
|
|
||||
|
Chromosomes: E.coli-DNA |
Ori-C |
Present; operates |
In damaged E.coli DNA |
Ori-Ms (isdr) |
Yes, operates |
|
Plasmids: |
|
|
|
|
|
|
F- |
Ori |
Yes |
- |
- |
- |
|
PSc 101 |
Ori |
Yes |
Col-E1 |
Ori Col E-1 |
Yes, operates |
|
R6K |
Ori |
Yes |
pBR322 |
Ori pBR |
Yes |
|
RK2 |
Ori |
Yes |
RSF 1030 |
Ori |
Yes |
|
Rts 1 |
Ori |
Yes |
Col e2 ori |
Ori cool E 2 |
Yes |
|
SS DNA: |
Ss DNA Ori |
Nil |
Phi X 174 |
Ori (-), and Ori (+) |
Yes |
|
Requirements for primosome formation: |
|
|
|
|
|
|
Dna binding proteins |
Dna-A |
|
Pri-A? |
|
|
|
|
Hu |
|
Pri-B |
|
|
|
|
|
|
Pri-C |
|
|
|
|
|
|
|
|
|
|
Helicase |
Dna-B |
|
Dna-B, Dna-T |
|
|
|
|
Dna-C |
Assists DnaB |
Dna-C |
|
|
|
Priming |
Dna-G |
|
Dna-G |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
- Replicons depending upon Dna-A proteins produce what is called�� Dna-A dependent primosomes, consisting Dna-A, Dna-B, Dna C and Dna-G components.� They are celled ABC dependent primosomes.
- On the other hand, Pri-A dependent primosomes constitute Pri-A, Pri-B, Pri-C, Dna-T, Dna-B/C complex and Dna-G for initiation; they are called Pri-A dependent primosomes; they are well characterized in phiX174.
Pri-A dependent Primosome Component:
|
Pri-A Primosome component |
Gene |
Mol.wt (KD) |
Subunit |
�Function |
Pri-A |
|
76 |
Monomer |
|
|
Pri-B |
|
11.5 |
Dimer |
|
|
Pri-C |
|
33 |
Monomer |
|
|
Dna-T |
|
22 |
Trimer |
|
|
Dna-B |
|
50 |
Hexamer |
|
|
Dna-C |
|
29 |
Monomer |
|
|
Dna-G |
|
60 |
Monomer |
|
|
RNA-pol |
|
|
Pentamer (Holozyme) |
Lays primer on leading strand |
|
RNase-H |
|
Dimer |
|
By exonuclease activity produces a primer |
|
|
|
|
|
|
Note: In Col E 1, on lagging strand, in addition to the above, requires Rom protein (7.5kd), a monomer.
Components of primosome in E. coli:
|
Protein |
Gene |
Activities and Functions |
|
PriA |
priA |
helicase, 3' to 5' movement, site recognition |
|
PriB |
priB |
|
|
PriC |
priC |
|
|
DnaT |
dnaT |
needed to add DnaB‑DnaC complex to preprimosome |
|
DnaC |
dnaC |
forms complex with DnaB |
|
DnaB |
dnaB |
helicase, 5' to 3' movement. DNA dependent ATPase. |
|
DnaG |
dnaG |
synthesize primer |
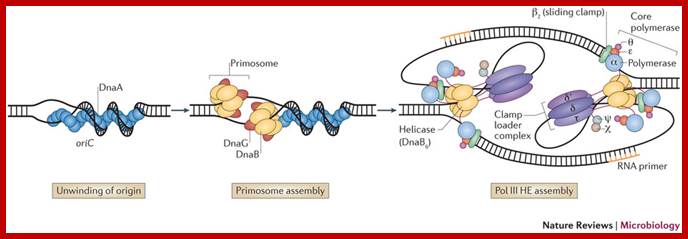
Five different proteins are found in a prepriming complex, PriA, PriB, PriC, DnaT, and DnaB (Table above), Dna A and DnaC are also needed for the assembly of this complex. In the case of X174 viral DNA template coated with SSB, PriA recognizes a primer assembly site. The proteins PriB and PriC are then added to form a complex. The hexameric protein DnaB is in a complex with six molecules of DnaC when it is not on the DNA. In an ATP-dependent process, and with help from DnaT, DnaB is transferred to the template and DnaC is released.
In contrast to the other DNA polymerases discussed, terminal deoxynucleotidyl transferase does not require a template. It adds dNTPs (as dNMP) to the 3' end of DNA, using that 3' hydroxyl as a primer. It is found in differentiating lymphocytes, and appears to be used physiologically to introduce somatic mutations in immunoglobulin genes. In the laboratory, it is used to add "homopolymeric tails" to the ends of DNA molecules by incubating a linear DNA with one particular dNTP and terminal deoxynucleotidyl transferase.
The pre-priming complex and DnaG bind and make the active primosome. Although the role of each of the proteins in the primosome is not yet clear, information is available on some of the steps in primosome action. The preferred binding site on the template for primase is CTG (in E.coli), with the T being used as the template for the first nucleotide of the primers. A high affinity complex between DnaB and ATP forms or stabilizes a secondary structure in the single-stranded template DNA that is used by primase; this is thought to be how DnaB "activates" the primase to begin synthesis. After ATP hydrolysis by DnaB, the low affinity ADP-DnaB dissociates from the template. The primosome can now move to the next site for primer synthesis.
Another depiction PriA-primosomes involved PhiX174 DNA replication; multiple primers are produced. http://shomusbiology.weebly.com/
Figure: Assembly and migration of the primosome. After assembly of the pre-priming complex, DnaG joins the complex to complete the primosome. After synthesis of the primer (dark black line), the primosome moves to the next site for synthesis. This tracking along the SSB-coated single stranded DNA requires ATP hydrolysis and causes dissociation of some of the SSB. In the diagram, the primosome is shown moving in a 5' to 3' direction along the template strand (clockwise), which is the opposite of the direction of primer synthesis. This would be the direction of movement catalyzed by DnaB. The primosome also contains PriA, which catalyzes movement along single-stranded DNA in the opposite direction. Once primers have been synthesized, DNA polymerase III can synthesize Okazaki fragments from them, the primers are excised and gaps repaired by DNA polymerase I, and then the fragments are ligated together.
Components required for Dna-A dependent Primosomes:
|
Types |
Proteins |
Mol.wt (KD) & subunits |
|
Functions |
|
Dna binding proteins |
Dna-A |
50, monomers |
|
Bind to Dna-A boxes induce opening at 13 mer regions |
|
|
Hu |
|
|
Binds to Ori-C, induces DNA bending |
|
|
|
|
|
|
|
|
|
|
|
|
|
Helicase |
Dna-B |
50, hexamer |
|
Binds to lagging strand at fork region and acts as the motor protein in unwinding DNA helix, in 5�. 3� direction |
|
|
Dna-C |
, Monomer (1:1) |
|
Assists Dna-B in 1:1 manner |
Priming |
Dna-G |
60, monomer |
|
Lays RNA primer of lagging strand |
Primers:
�
- They are short segments of RNA or DNA or a nicked end of DNA on the replication strands, whichever that provides 3�OH group for the DNA polymerase to extend the chain.� Such segments are called primer for they prime the extension or synthesis.�
- The RNA polymerases don�t require primers for transcriptional initiation, but require sequences to which they bind and initiate RNA synthesis at a specific nucleotide called START site.�
- When Life originated on this planet, its genetic material was discerned as RNA (an RNA world). With evolution, the RNA, which was acting not only as the genetic material but also as a catalyst, replaced RNA with DNA as the genetic material.�
- There are many evidences to show that RNA in association with few proteins or with out them is capable of performing a variety of enzymatic activities, such as splicing, processing and copying the RNA.
- RNA is known to act as genetic material in a large number of animal, plant and bacterial viruses including viroids and virusoids.
- Though DNA is now predominant form of genetic material, yet it requires RNA as a primer for its replication, perhaps a molecular memory.�
- The RNA primers are not produced by the regular RNA-Pols (multisubunit complexes) but by specialized RNAps called Primases (Dna-G).
- Primers are produced by either by A, B, C dependent primosome or Pri-A dependent primosome complex mode.�
- RNA-pol as a holozyme can initiate transcription and produce RNA, which is trimmed by RNase-H and provide RNA as the primer.
- A nick in the DNA an act as the primer for it provides free 3�OH end for the DNA polymerase to extend it. Such DNA primers are found in a variety of systems ex. rolling circle mode of replication in several viral genomes, bacteria and even in higher organisms.�
- In some systems tRNA paired to RNA or DNA templates acts as the primer for DNA synthesis.
- Apart from the primers mentioned above there are few novel forms of primers.� In some viral genomes, ex. Adeno virus and Picorna virus and in few other viral genomes, proteins bound to certain nucleotides act as primers by pairing with the first nucleotide at the 3�end of the replicating strand.
- In influenza and other Orthomyxo viruses, leader sequences from the host mRNAs are cut and placed on viral RNAs at their 3� ends, which act as primers.
- Very often replication is stopped (stalled) due to many reasons, but break spots act as the site of re-replication site at which a set of proteins assemble the restart replication, perhaps these are secondary primosomes.
Models of primosomes and their activity during primer synthesis.
From the following article-
Coupling-DNA unwinding activity with primer synthesis in the bacteriophage T4 primosome; Maria Manosas, Michelle M Spiering, Zhihao Zhuang, Stephen J Benkovic & Vincent Croquette.
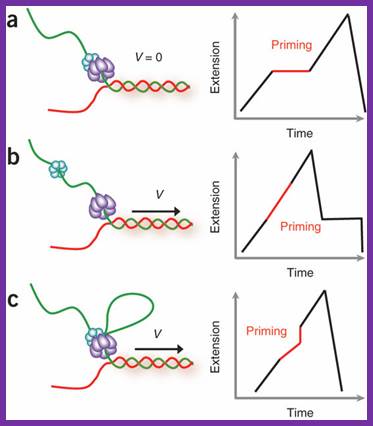
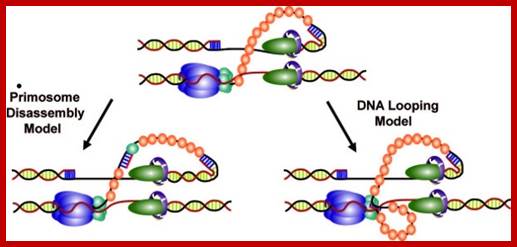
Schematic representation of three possible models for helicase and primase interaction during primer synthesis (left) and the real-time DNA extension traces expected for each model (right). (a) In the pausing model, the helicase and primase temporarily stop translocating during priming. (b) In the disassembly model, the primase dissociates from the helicase to synthesize a primer while the helicase continues unwinding DNA. (c) In the DNA looping model, the primosome remains intact, and DNA unwound during priming forms a loop.
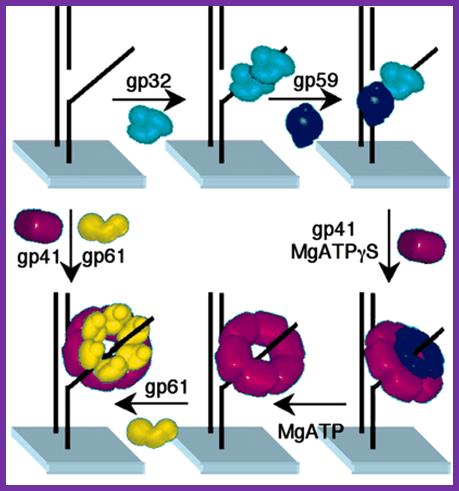
Assembly of the bacteriophage T4 primosome: Single-molecule and ensemble studies
1. Zhiquan Zhang * , �, Michelle M. Spiering � , �, Michael A. Trakselis � , �,
2. Faoud T. Ishmael � , �, Jun Xi �, Stephen J. Benkovic �, and Gordon G. Hammes * , ∥
Assembly mechanism of the T4 lagging-strand primosome on fDNA. The gp32 protein binds to fDNA with either subsequent or concurrent binding of gp59. Subsequently, gp41 binds to gp59 and is loaded onto the fDNA in the presence of nucleotide. ATP hydrolysis is required for gp41 to displace gp32 and gp59, either directly or by translocation. The gp61 protein then binds and interacts closely with gp41 on fDNA. In the absence of gp32 and gp59, both gp41 and gp61 bind to DNA.
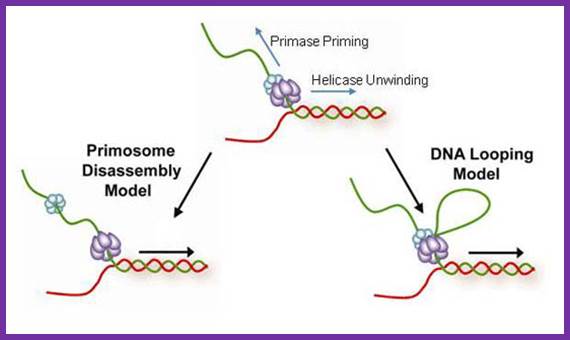
The coordination between the continuous leading strand DNA synthesis and the discontinuous lagging strand counterpart is regulated by a subassembly of the replisome called the primosome. In the T4 system the primosome is formed by a hexameric helicase (gp41) that unwinds dsDNA and an oligomeric primase (gp61) that synthesizes RNA primers to initiate repetitive Okazaki fragment synthesis. Three possible models have been suggested to explain how helicase is able to unwind dsDNA translocating 5′ to 3′ on the lagging strand while primase travels in the opposite direction (3′ to 5′) in order to synthesize an RNA primer. In the first model (pausing), the helicase temporarily pauses or stops translocating to allow for primer synthesis and then resumes unwinding the DNA; helicase pausing would necessitate the pausing of the entire replisome while a primer is being synthesized. In the second model (disassembly), one or more primase subunits dissociate from the helicase and remain behind to synthesize a primer while the helicase and any remaining primase subunits continue to translocate along the lagging strand. In this model, leading strand synthesis would continue uninterrupted; however, new primase subunits might need to be recruited for each cycle of Okazaki fragment synthesis. In the third model (DNA looping), the primosome remains intact and the DNA that is continuously unwound by the helicase during primer synthesis forms a loop which is released once the primer is transferred to the lagging strand polymerase.
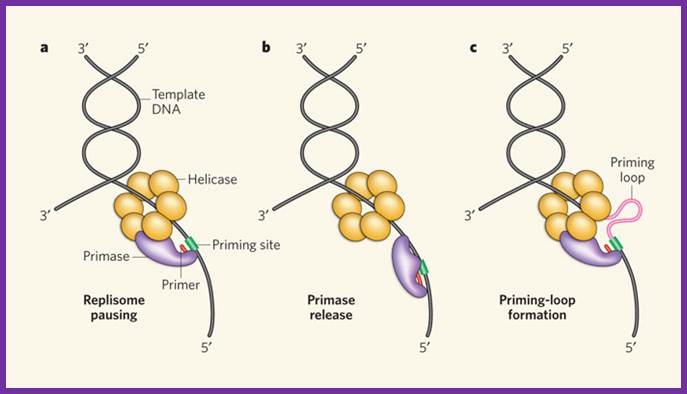
Three priming mechanisms;
Interaction of the primase with the helicase is necessary for primer synthesis at a lagging-strand priming site. The primase makes primers in the opposite direction to helicase movement, leaving three ways by which the replisome might resolve this directional problem. a, The whole replisome can pause for primer synthesis; b, it can promptly release the primase; or c, as described for the first time by Pandey et al.1 and Manosas et al., the replisome can continue to move forward while the primase�helicase interaction persists. This produces a priming loop that eventually collapses into the lagging-strand trombone loop, probably on transfer of the primer to the lagging-strand polymerase.Three priming mechanisms; Nicholas E. Dixon; http://www.nature.com/
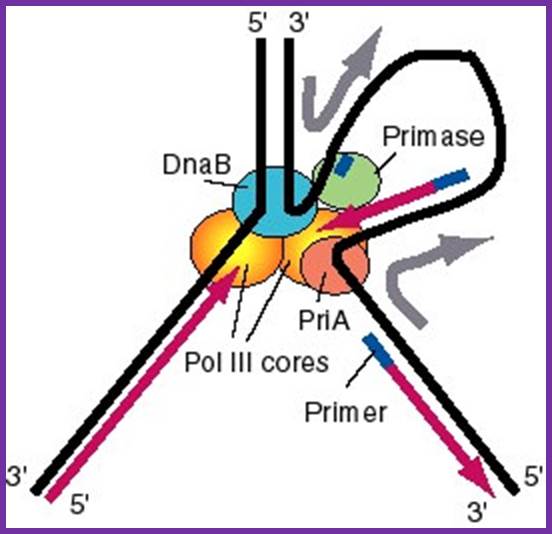
http://www.bx.psu.edu; http://slideshare.net
Figure: A model for the activities of helicases DnaB and PriA at replication fork: In this model, the replication machinery in a large Replisome (polymerase and primosome) is postulated to be stationary, with the DNA strands moving through it like fabric through a sewing machine. After unwinding the duplex DNA at the replication fork, the DnaB helicase can also track along single stranded DNA in a 5' to 3' direction. If the helicase is stationary, then the template strand for lagging strand synthesis moves in the direction of the upper gray arrow in the diagram, away from the replication fork. This template strand is also bound to the PriA helicase at the "back" end of the replisome. The 3' to 5' tracking activity of PriA will also pull the template strand, now in the direction of the bottom gray arrow. The result of the DnaB and PriA tracking activities is to pull the template for lagging strand synthesis into a loop, which is tethered to the replisome at both ends. Primase is also in the replisome, and can synthesize primers along this strand at appropriate intervals (thick blue lines), and one of the core polymerases of the DNA polymerase III holoenzyme can synthesize an Okazaki fragment. Newly synthesized DNA is a thick violet line, and parental DNA strands are thick black lines.
Decades of research have resulted in a remarkably detailed understanding of the molecular mechanisms of bacterial DNA replication, transcription and translation. Our understanding of the kinetics and physical mechanisms that drive these processes forward has been expanded by the ability of single-molecule in vitro techniques, such as force spectroscopy and single-molecule F�rster (fluorescence) resonance energy transfer (smFRET), to capture short-lived intermediate states in complex pathways. Furthermore, these technologies have revealed novel mechanisms that support enzyme processivity and govern the assembly of large multicomponent complexes. Here, we summarize the application of in vitro single-molecule studies to the analysis of fundamental bacterial processes, with a focus on the most recent functional insights that have been gained from fluorescence-based methods.www.nature.com.
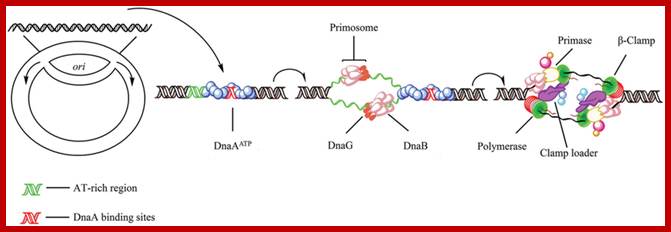
Initiation of replication in bacteria. In Escherichia coli, replication initiation requires binding of the DNA-binding protein DnaA to DnaA-boxes at the chromosome origin oriC which is regulated by SeqA (Dame et al., 2011). Then, with the activation of ATP, two DnaB hexamers and the helicase loader DnaC, one double hexamer for each replication direction, are positioned by DnaA into the loop (Wahle et al., 1989; Skarstad and Katayama, 2013). Primase (DnaG) which can enter the complex and synthesize two leading strand primers, stimulates release of the regulatory protein DnaC from DnaB after transiently binding to the DnaB replicative helicase (Arias-Palomo et al., 2013). Also, DnaB binds to the sliding clamp loader, a ring-shaped dimer of the β-subunit which in turn binds the DNA polymerases III (Kelman and O�Donnell, 1995; O�Donnell et al., 2013). https://www.researchgate.net
Schematic of replication process; coupling Primase and Helicase-primosome and replisomes activity; Priming activities of the bacteriophage T4 primosome; Is primosome and replisome same or different; http://pimprenelle.lps.ens.fr
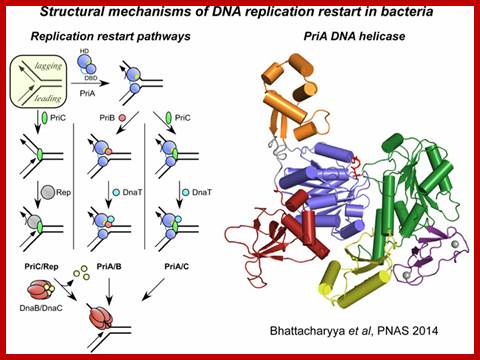
Mechanisms of DNA replication restart at Replication stalled sites;
Nearly all replication complexes (replisomes) formed at bacterial replication origins are thought to stall during the course of DNA replication. This stalling may occur at sites of DNA damage and can lead to dramatic rearrangement of the replication fork DNA and disassembly of the replisome. Reassembly of the DNA replication machinery on abandoned replication forks is therefore often required for complete DNA replication. "DNA replication restart" is mediated several proteins (PriA, PriB, PriC, and DnaT in E. coli) that function in a coordinated manner to reload the replisome. Currently, little is known about the structural mechanisms that support this essential cellular function.
In this model, PriA acts as a first-responder protein, binding directly to the abandoned replication fork DNA. Subsequent stepwise assembly of PriB and DnaT or PriC onto the PriA/DNA complex creates a platform for recruiting the replisome on the replication fork.
Coordination of human DNA repair complexes. Eukaryotic cells have an astonishing number of DNA repair pathways that are regulated and coordinated, at least in part, by dynamic physical interactions. We have recently begun a study of a key protein interaction interface that links the Fanconi Anemia Core Complex and Bloom Dissolvasome, two DNA repair complexes. Our lab has determined the high-resolution crystal structure of the minimal protein interface that links these complexes, which is comprised of a short peptide from the FANCM protein from the Fanconi Anemia Core Complex binding to the RMI heterodimer from the Bloom Dissolvasome. Using our structure as a guide, we have shown that mutations that block interaction between these complexes induces genomic instability in cells-a hallmark of nearly all cancers. We are continuing to investigate this and other repair interfaces as examples of regulation by dynamic protein assembly and as potential chemotherapeutic targets. https://bmolchem.wisc.edu
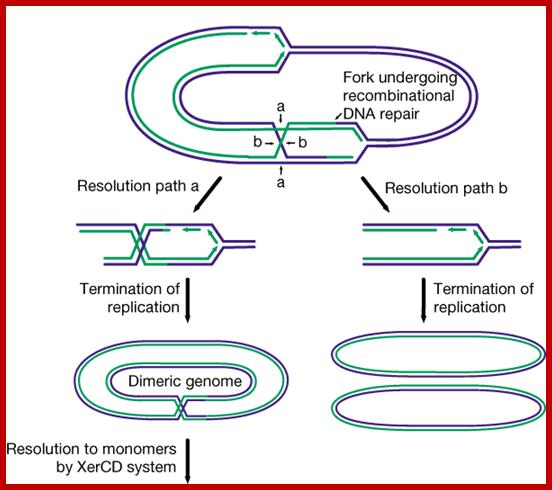
David J. Sherratt4, Steven J. Sandler & Kenneth J. Marians; www.nature.com
The bacterial SOS response to unusual levels of DNA damage has been recognized and studied for several decades. Pathways for re-establishing inactivated replication forks under normal growth conditions have received far less attention. In bacteria growing aerobically in the absence of SOS-inducing conditions, many replication forks encounter DNA damage, leading to inactivation. The pathways for fork reactivation involve the homologous recombination systems, are non-mutagenic, and integrate almost every aspect of DNA metabolism. On a frequency-of-use basis, these pathways represent the main function of bacterial DNA recombination systems, as well as the main function of a number of other enzymatic systems that are associated with replication and site-specific recombination. https://www.nature.com
Note- there are multiple pathways to restart replication in the regions of of stalled replication- refer to http://www.pnas.org content/101/35/12783.figures-only
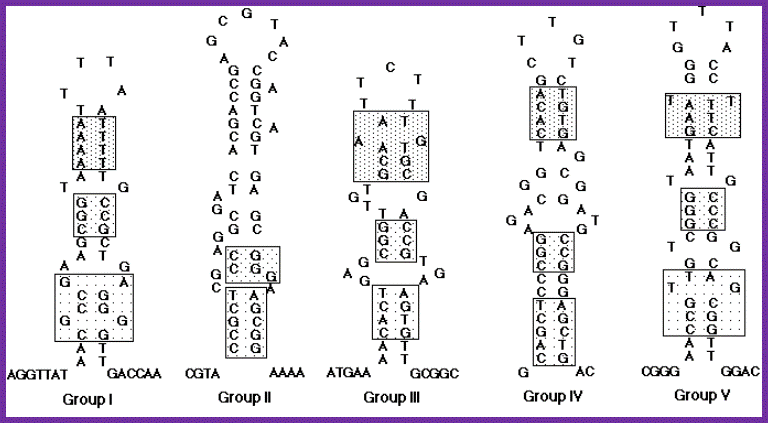
Nucleotide sequences and possible stem-loop structures of n'-pas isolated from various plasmid DNAs. Eight newly isolated n'-pas in addition to previously reported ones were classified into five groups on the basis of their primary structures. n'-pas of X174 phage, R100 plasmid and F�f2b belong to group I, n'-pas on the lagging strand of ColE1-type plasmids (ssiA) and n' sites of F�f5, F�f7 and ColE2 plasmid to group II, n'-pas of the F�f2a to group III, n'-pas on the leading strand of ColE1-type plasmids (ssiB) to group IV, and n'-pas from Rts1 plasmid to group V. Each group is represented by the sequences of the first listing described above. The sequences indicated by boxes show some sequence similarities, although none of them are conserved among all the groups. The group I and group II were described before. Hisao Masai and Ken-ichi Arai. http://www.bioscience.org/
A Specific Docking Site for DNA Polymerase α-Primase on the SV40 Helicase Is Required for Viral Primosome Activity, but Helicase Activity Is Dispensable*
1. Hao Huang,� Kun Zhao,� Diana R. Arnett and� Ellen Fanning1
1. From the Department of Biological Sciences, Vanderbilt University, Nashville, Tennessee 37235-1634; 1 To whom correspondence should be addressed. Tel.: 615-343-5677; Fax: 615-343-6707; E-mail: Ellen.fanning@vanderbilt.edu.Replication of simian virus 40 (SV40) DNA, a model for eukaryotic chromosomal replication, can be reconstituted in vitro using the viral helicase (large tumor antigen, or Tag) and purified human proteins.
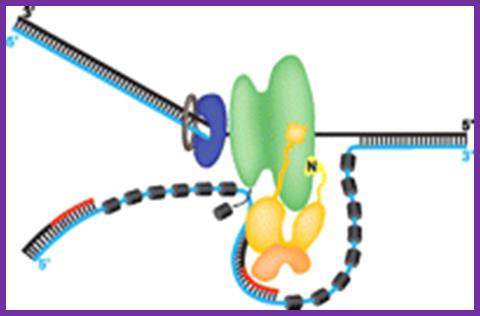
DNA polymerase α-primase (pol-prim), constituting the viral primosome: an example for an eukaryote: http://www.jbc.org
Like the well characterized primosomes of phages T7 and T4, this trio of proteins coordinates parental DNA unwinding with primer synthesis to initiate the leading strand at the viral origin and each Okazaki fragment on the lagging strand template. We recently determined the structure of a previously unrecognized pol-prim domain (p68N) that docks on Tag, identified the p68N surface that contacts Tag, and demonstrated its vital role in primosome function. Here, we identify the p68N-docking site on Tag by using structure-guided mutagenesis of the Tag helicase surface. A charge reverse substitution in Tag disrupted both p68N-binding and primosome activity but did not affect docking with other pol-prim subunits. Unexpectedly, the substitution also disrupted Tag ATPase and helicase activity, suggesting a potential link between p68N docking and ATPase activity. To assess this possibility, we examined the primosome activity of Tag with a single residue substitution in the Walker B motif. Although this substitution abolished ATPase and helicase activity as expected, it did not reduce pol-prim docking on Tag or primosome activity on single-stranded DNA, indicating that Tag ATPase is dispensable for primosome activity in vitro.