Protein Synthesis:
Prokaryotes Introduction:
The genetic material, in bacteria and eukaryotes, is double stranded DNA; In viruses it can be DNA or RNA. The mRNA is another polynucleotide chain with a specific sequence with a polarity. Protein is a polypeptide chain of amino acids with sequence specificity and it has a polarity. All of them exhibit linearity, where the information flow is from DNA to RNA and from RNA to protein, all are linear chains, which is the basis for Crick’s Central Dogma. Francis Crick proposed this par excellent dogma before the discovery of Reverse Transcriptase by Baltimore and Timmins. But the revised Central Dogma, forced to revise the central dogma, on the basis of findings that RNA can be transcribed into DNA, so DNA to RNA and RNA to DNA is now an accepted fact, however RNA to protein is the final step in flow of information.
Protein is a functional molecule, without which no life is possible. Protein is built with amino acids; there are only 20 amino acids, proteins are countless. For protein to be synthesized DNA/RNA are required.
Codon Directory:
|
|
U |
|
C |
|
A |
|
G |
|
|
|
U |
UUU UUCphe UUA UUGleu |
F F L L |
UCU UCC UCA UCGser |
S S S S |
UAU UACtyr UAAstop UAGstop |
Y Y - - |
UGU UGCcys UGAstopUGGtrp |
C C - W |
U C A G |
|
C |
CUU CUC CUA CUGleu |
L L L L |
CCU CCC CCA CCGpro |
P P P P |
CAU CAChis CAA CAGgln |
H H Q Q |
CGU CGC CGA CGGarg |
R R R R |
U C A G
|
|
A |
AUU AUC AUAileAUGmet |
I I I M |
ACU ACC ACA ACGthr |
T T T T
|
AAU AACasn AAA AAGlys |
N N K K |
AGU AGCser AGA AGGarg |
S S R R |
U C A G |
G |
GUU GUC GUA GUGval |
V V V V |
GCU GCC GCA GCGala |
A A A A |
GAU GACasp GAA GAGglu |
D D E E |
GGU GGC GGA GGGgly |
G G G G |
U C A G
|
Anti Codon Directory ?:
|
|
A |
|
G |
|
U |
|
C |
|
|
|
|
AAA AAGphe AAU AACleu |
|
AGA AGG AGU AGCser |
|
AUA AUGtyr - - |
|
ACA ACGcys - ACCtrp |
|
A G U C |
|
G |
GAA GAG GAU GACleu |
|
GGA GGG GGU GGCpro |
|
GUA GUGhis GUU GUCgln |
|
GCA GCG GCU GCCarg |
|
A G U C |
|
U |
UAA UAG UAUilu UACmet |
|
UGA UGG UGU UGCthr |
|
UUA UUGasn UUU UUClys |
|
UCA UCGser UCU UCCarg |
|
A G U C |
|
C |
CAA CAG CAU CACval |
|
CGA CGG CGU CGCval |
|
CUA CUGasp CUU CUCglu |
|
CCA CCG CCU CCCgly |
|
A G U C |
Expected anticodon sequences but one does not find all the expected anticodons in tRNAs, but tRNAs with isoforms decode all the 63 codons; no anticodons for Terminator codons.
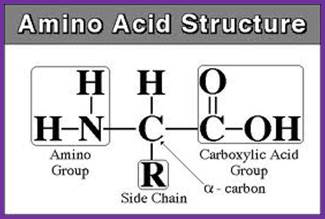
Amino acid basic structure
|
Amino acid specificity. Acronyms |
|
|
|
|
|
Alanine A |
|
|
Arginine R |
Lysine K |
|
Phenylalanine |
|
|
Cysteine C |
Proline P |
|
Serine S |
|
|
|
|
|
Glycine G |
Threonine |
|
Tyrosine Y |
|
|
Leucine L |
Valine V
|
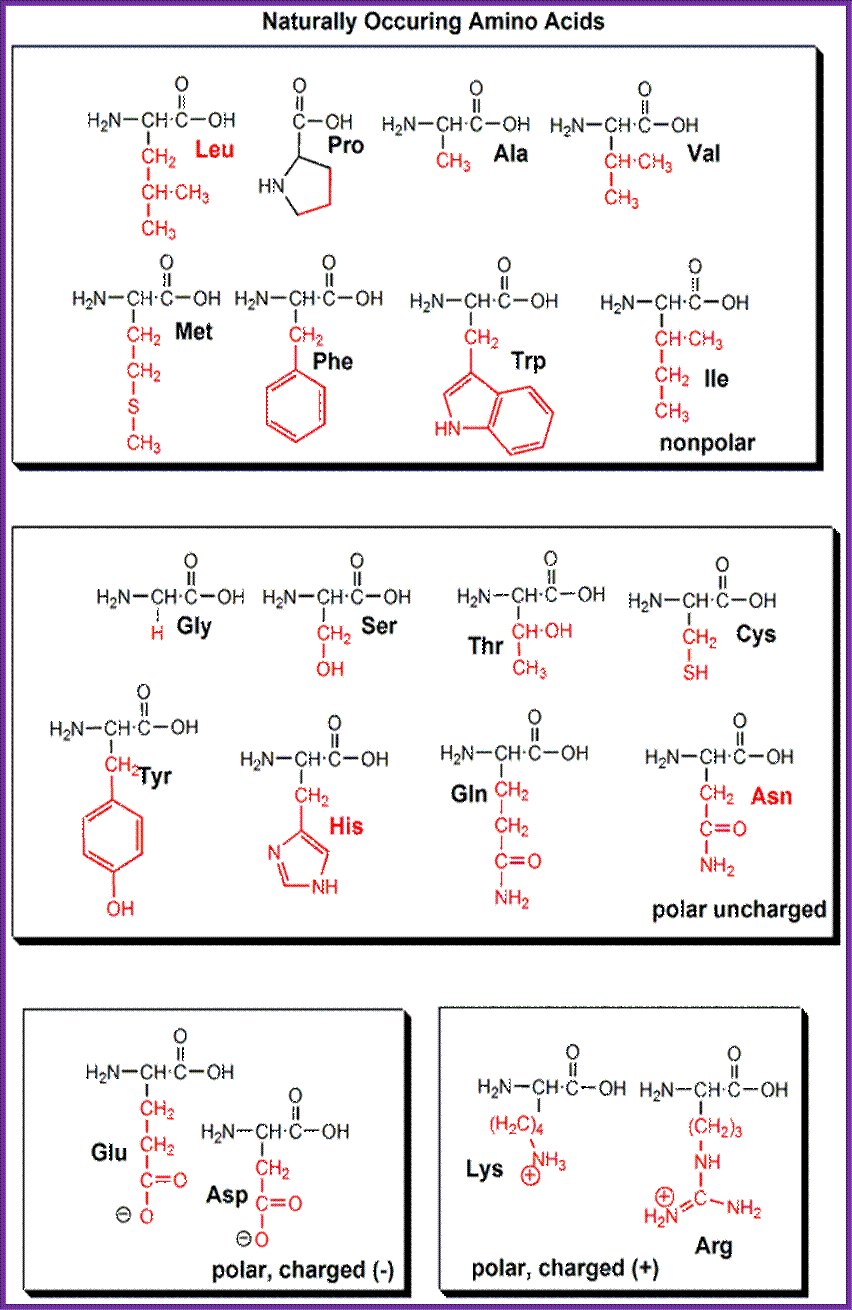
Each amino acid contains an "amine" group (NH3) and a "carboxy" group (COOH) (shown in black in the diagram). Amino acids vary in their side chains (indicated in red in the diagram).The eight amino acids in the red side chains are nonpolar and hydrophobic. The other amino acids are polar and hydrophilic ("water loving"). The two amino acids in the magenta box are acidic ("carboxy" group in the side chain).
The three amino acids in the light blue box are basic ("amine" group in the side chain). http://employees.csbsju.edu/
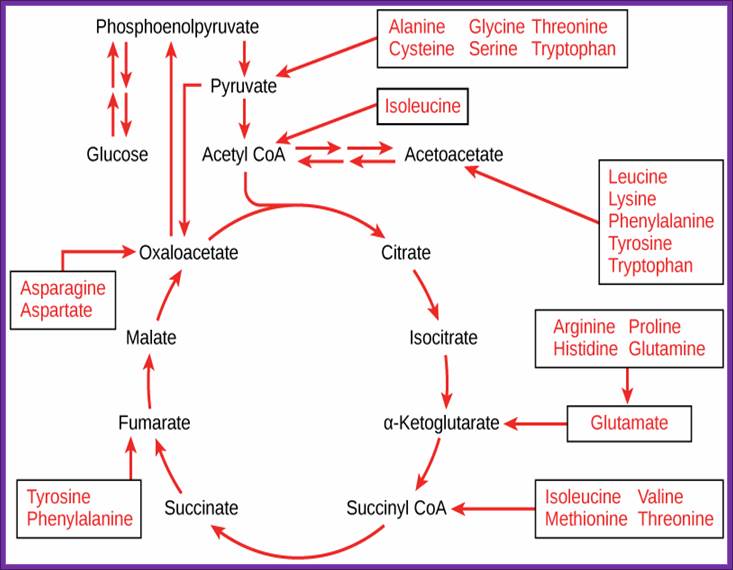
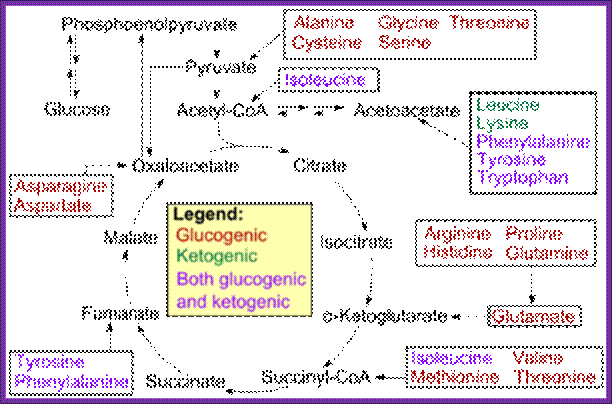
Glycolytic and Krebs’s cycle reactions are used in generating most of the amino acids. https://www.boundless.com
The genetic code:
• Degeneracy of the genetic code
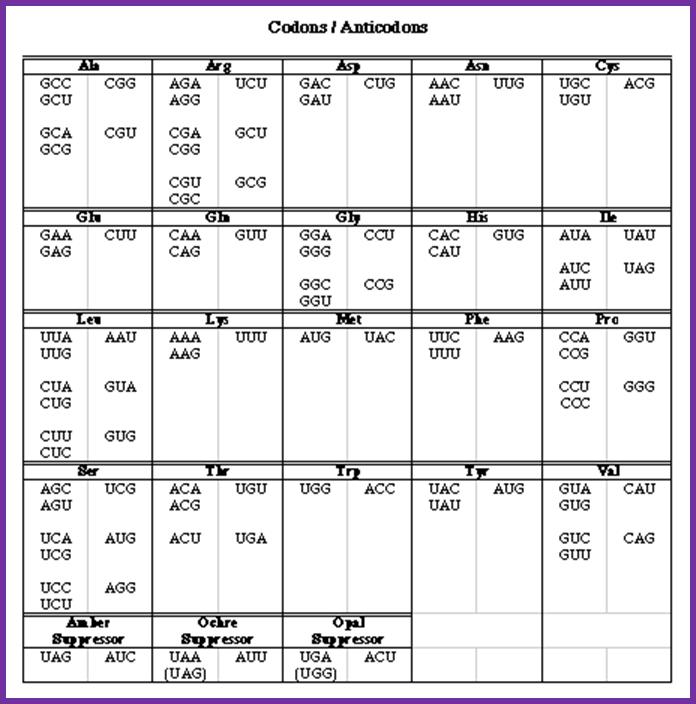
To encode 20 (21) amino acids, combination of two nucleotides is not enough (4X4=16). Three nucleotides are too many (4x4x4=64), 44 excess. This excess number is compensated for having isoforms of tRNA and degenerate codons where the last nucleotide wobbles. The same amino acid can be recognized by more than one codon and the same codon can be recognized by more than one of the isoforms of anticodon. Room for innovation. Moreover, amino acids with similar properties can be substituted for each other without changing the structure and function of a protein.
• Six possible translation frames for every nucleotide stretch
– GCU.UGU.UUA.CGA.AUU.A -Ala – Cys – Leu – Arg – Ile -
– G.CUU.GUU.UAC.GAA.UUA - Leu – Val – Tyr – Glu – Leu
Most of the tRNAs carry three nucleotides as anticodons, but the anticodons need not be the same number as of codons; they have evolved what we call as wobble mechanism, where the last nucleotide in tRNA that is from 5’end can base pair with different degenerate nucleotides found in mRNA codons. There are no tRNAs that can decode terminator codons. The tRNAs are essential reading codons in the mRNAs.
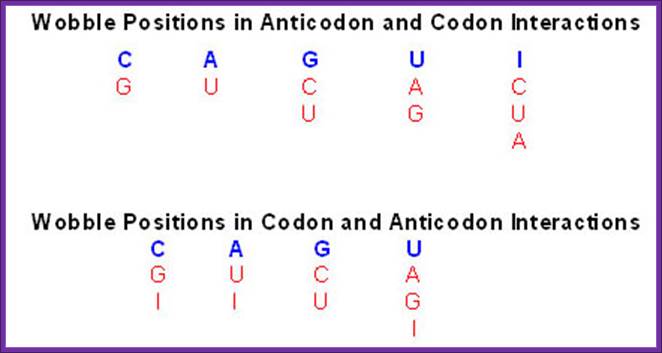
Diagram showing the various modified nucleotides of tRNAs that are found in the wobble position in the anticodon. The top half shows the wobble nucleotides of the anticodon in blue and the various nucleotides (in red) of the wobble position of the codon that can be found in non-Watson-Crick base-pairs. The lower panel illustrates the opposite showing the wobble nucleotides of the codon in blue and the associated wobble nucleotides of the anticodon in red; http://themedicalbiochemistrypage.org/
Revised wobble base pairing;
|
tRNA5’ anticodon |
mRNA 3’codon |
|
G |
U, C |
|
C |
G
|
|
K2C |
A |
|
A |
U, C (A), G |
|
U unmodified |
U, C,’ A’, G |
|
Xm^5S@, Xm^5Um, Um, Xm^5U
|
A, (G) |
|
Xo^5U |
U, A ,G |
|
I |
A, A, G |
|
|
|
Revised codon and anticodon showing tRNA 5’nucleotide can base pair with more than one non complementary last nucleotide of the codon.
tRNAs and Anticodon Wobble;;http://www.sci.sdsu.edu/
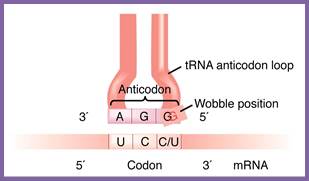
Wobble; https://www.mun.ca/biology
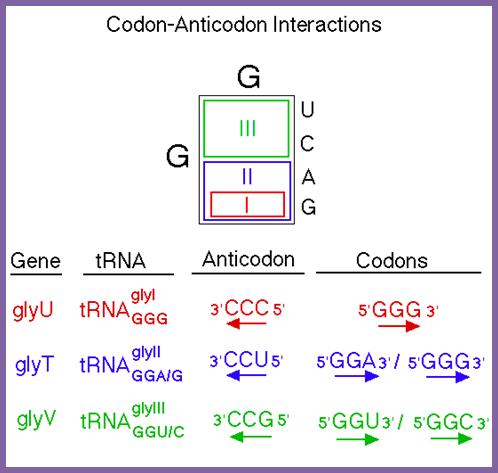
There are 3 stop codons for every 64 combination of triplet nucleotides; long ORFs are unlikely; probably genes in some viruses contain as many as four overlapping frames and they are functional.
In bacterial systems, transcription and translation events are coupled and in eukaryotes the events are uncoupled. Bacterial cells don’t have intracellular compartmentation. In eukaryotes cytoplasm is compartmentalized where a large number of the ribosomes translate in free-state and equal numbers of proteins, which are secretary and membrane components, are translated on endoplasmic membranes. In bacteria, coupled transcription and translation has advantages, for the 5’end, as bound by ribosomes, is protected from exonuclease activity. In EK cells, RNAs are transcribed and processed in the nucleus, and then transported into the cytoplasm. In cytoplasm ribosomes engage mRNAs in protein synthesis at different loci, one at the so-called Free State and another bound on endoplasmic reticulum. These are designed to the needs of cells, where certain proteins are secreted out or destined to plasma membranes and endoplasmic reticulum and other cellular milieu. Many proteins are destined to different cell organelles and intracellular structures. Localization of protein synthesis is of a significant feature.
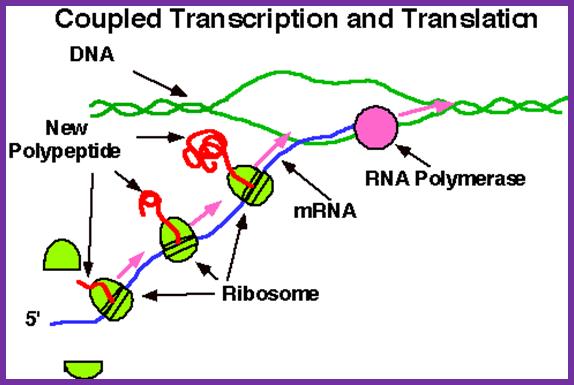
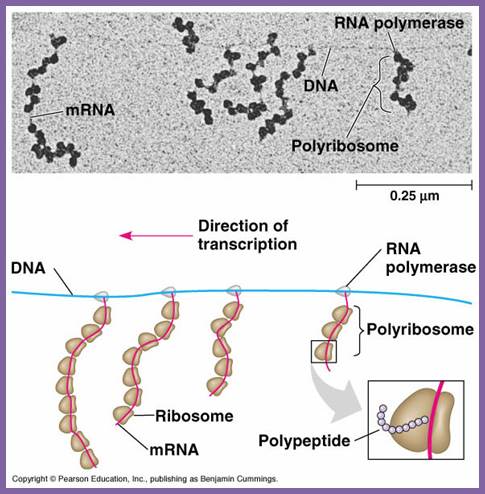
This simple diagram shows the bacterial transcription and translation goes hand in hand; this coupled process is absolutely required for the 5’ end of the emerging mRNA has no protection from 5’exonucleases. http://nitro.biosci.arizona.edu/ and http://www.bio.utexas.edu/
The rate of protein synthesis varies, from 15 to 20 amino acids per second ?. A single mRNA can be associated with more than one ribosome, ten to thirty or more; this depends on the length of mRNA. Polysome formation further enhances the rate of protein synthesis. How ribosomes, molecular machines accelerate or decelerate the rate of synthesis is not very clear.
Process of translation goes through in stepwise but progressive mode. Chain initiation, chain elongation and chain termination are three important steps and each step can be regulated. Translation is also regulated.
Factors and Other Components Required for Protein Synthesis:
|
Events |
Factor |
Mol.wt (KD) |
Number per cell |
Function |
||
|
Chain Initiation |
|
|
|
|
||
|
|
IF-1 |
8.1(7.8) |
1 per7 ribosomes |
Assists IF3 |
||
|
|
IF-2 |
97.9 (97.9) |
1 per/7 ribosomes |
Bind to i tRNA |
||
|
|
IF-3 |
20.7 (19.8) |
1 per 7 ribosomes |
aids 30S dissociation , binds mRNA |
||
|
Chain elongation |
|
|
|
|
||
|
|
EF-Tu |
43.225 |
10:1(~100,000) |
binds to a.a tRNA |
||
|
|
EF-Ts |
31.0 |
1:1 (~10000) |
GEF |
||
|
|
EF-G |
77.444 |
1:1 (~20000) |
translocation |
||
|
Chain Termination |
- |
|
|
|
||
|
|
RF-1 |
39.6 UAA,UAG |
1 per 20 ribs |
Release protein chain |
||
|
|
RF-2 |
UAA.UGA |
1 per 20 ribs |
|
||
|
|
RF-3 |
58 |
|
Release of RF1 and 2 |
||
|
|
RRF |
20.3 (~184a.a) |
|
Ribosome recycling factor |
||
|
Initiator tRNAs |
~6000 per cell |
|
|
|
||
|
Amino acyl tRNAs |
|
|
~375,000/cell |
|
||
|
Ribosomal subunits |
|
|
~20000-65000 each per cell |
|
||
|
mRNAs |
|
|
~50000/cell |
|
||
|
GTP and ATPs |
|
|
???? |
|
||
|
Ribosome Remodeling factor-RMF |
50a.a |
|
|
Binds to 50s, Dimerize it; Stalls under stress |
||
|
Hibernation factor-Hib |
|
|
|
|
||
Bacteria:
rrn rRNA operons: -8-11 operons
Ribosomal protein coding genes: -6 operons,
~20,000 --70000 ribosomes per cell, (550000-70000),
tRNA: Genes per genome-55.6; ~375,000 tRNAs per cell, mRNA: 50-60K per cell, RNAPs-4600?