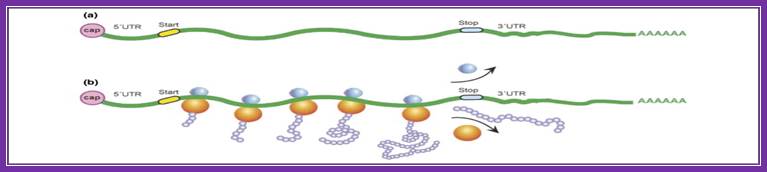
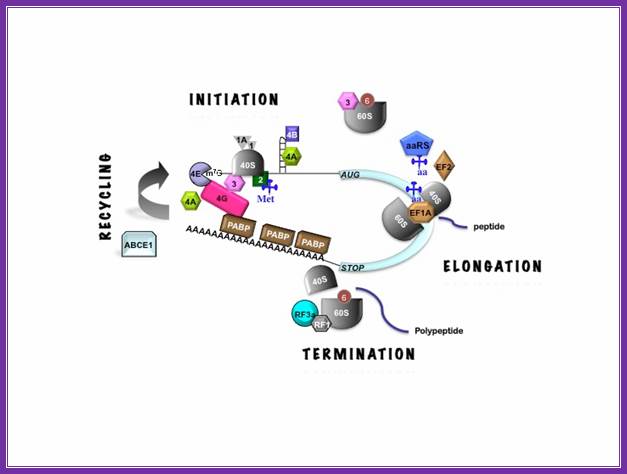
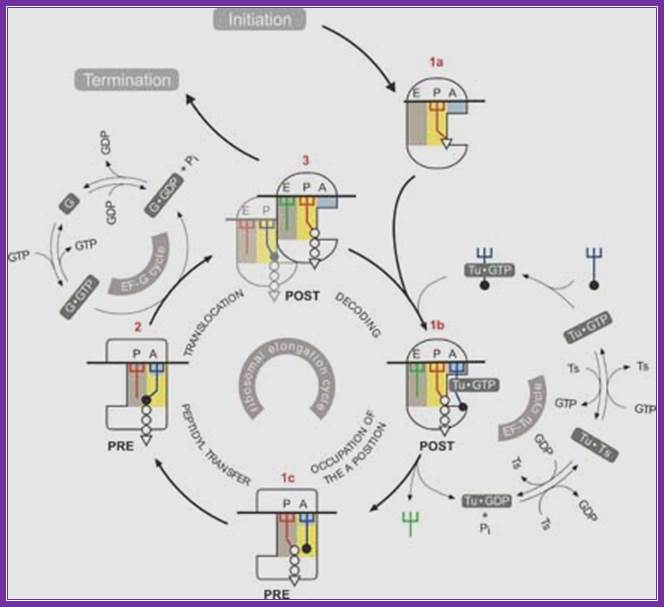
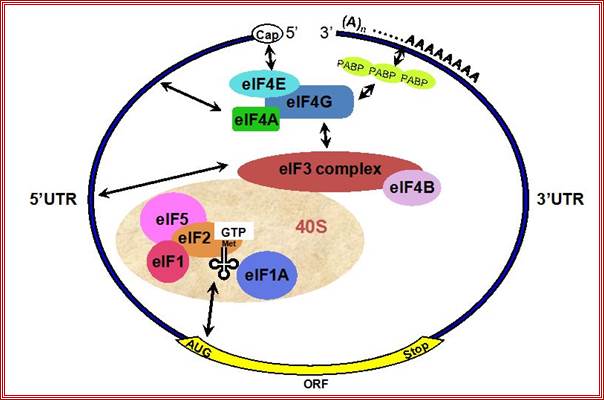
Protein_synthesis_eukaryotic_mechanism_of_synthesis:
All mrnas are the encoded informational copies of a gene.� Processed mrnas in eukaryotes are monocistronic, while bacterial mrnas (most) are polycistronic.� Though all mrnas have open reading frame; the 5�utr and 3�utr regions vary, for they have regulatory structures.� Only few organisms contain polycistronic mrna, which are processed into monocistronic mrna.
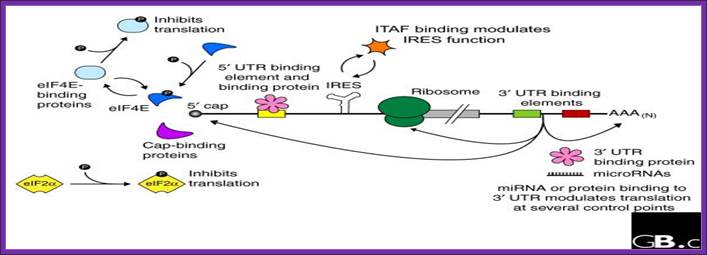
A schematic drawing of a eukaryotic mrna, illustrating some post-transcriptional regulatory elements that affect initiation of protein synthesis.;www.intechopen.com
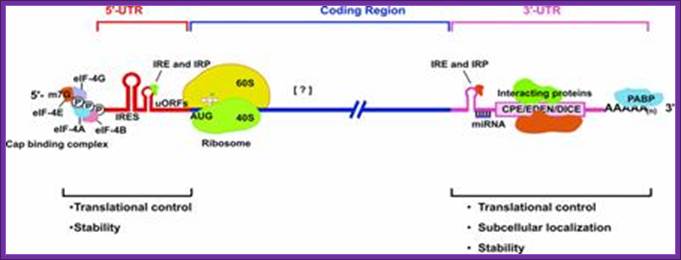
Diagram above and diagram below show few structural features of processed mrna, several features found at 5�cap,� 5�utr (ire-irp, ire and kozak) and 3�utr (ire, are, cpe, antisense mirna binding site, zip code, embryonic deadenylation element (eden) and dice play important roles in the stability and translation.
structural feature of processed m.RNAs
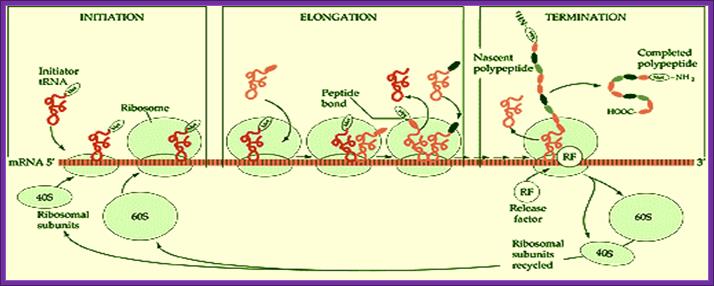
Basic view of translation steps.
Only the processed mRNAs with certain mRNPSs are transported out of the nucleus facilitated by ran gtp associated exportins into cytoplasm.� It is in cytoplasm many chain initiation factors bind.� The poly-A binding factor pab-ii is replaced by pab-i in the cytoplasm and pab-ii returns to the nucleus via nls signal sequences and ran-gdp associated importins.�
Mrnas with 5uorf and uaugs:
Among the 5� UTRs some cis-acting elements play a role in translation regulation; they are called upstream open reading frames (uORFs) and upstream uAUGs located in the 5'utr of mRNAs. Upstream open reading frames uORFs are found in some transcripts of Arabidopsis, yeast, bacteria and many others. We, lacono m migone f and pesole g 2005, present here a genome-wide analysis of uAUGs and uORFs in a curated set of human and rodent mRNAs. Functions of predicted morf proteins could be inferred for 16 homology groups and many of these proteins appear to have a regulatory function, including 6 transcription factors, 5 signal transduction factors, 3 developmental signal molecules, a homolog of translation initiation factor eif5, and a ring finger protein, celine a hayden and richard a jorgensen. Authors (above) study shows that the occurrence of uAUGs is suppressed more strongly than that of uORFs and that in-frame uAUGs are more strongly suppressed than out-of-frame uAUGs. A very similar pattern of uAUGs/ uORFs frequency was also observed in mouse m. RNAs. The analysis of orthologous 5'UTR sequences revealed a remarkable degree of evolutionary conservation only of those uORFs which acquired some functional activity. Our data suggest that besides leaky scanning and reinitiation, which likely occur with variable and gene-specific efficiency, the ribosome-shunt mechanism, eventually coupled to reinitiation after uORFs translation, may be a widespread mode of translation regulation in eukaryotes, in 5�utr uAUGs are conspicuously common in certain classes of genes, including two-thirds of oncogenes and many other genes involved in the control of cellular growth and differentiation.
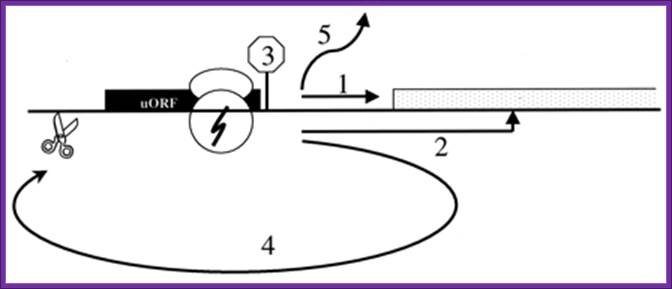
Upstream long 5�utr translation is based on the number and position uAUG and uORF, if the uORF is combined with proper kozak sequence the initiation continues till the transcript ends with its attenuator or termination sequences. Celine a hayden and richard a jorgensen.
Regulation of uorfs small open reading frames in 5�utr:
Regulation of such uORFs are found in gene transcripts of polyamine�s production, amino acid�s production, and sucrose response, and there are several such �5�utr with uORFs. �Interestingly 20-30% of yeast and mammalian and surprisingly plant transcripts contain such elements.� In yeast the transcript produced for gcn4 gene encoding a transcriptional activator (under amino acid starvation and stress contains four uORFs and only the fourth works.� Another example is cpa1 which codes for arginine attenuator peptide (AAP), which reduces the synthesis of synthase under high arginine concentration also contain such uORFs. Significantly uORFs reduce protein expression of the downstream ORF.� This is based on analysis of 11,649 matched mRNAs and protein measurements (sarah e. Calvoa et al).
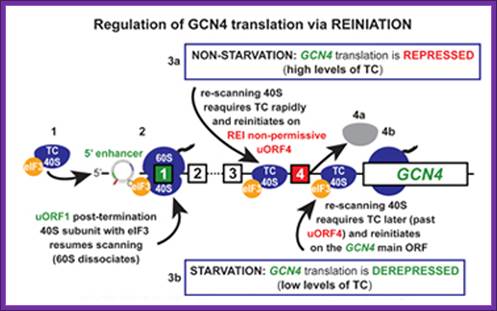
translation-born gcn- phenotypes can arise from a variety of reasons such as for example: 1) reduced rate of mrna recruitment; 2) reduced initiation at uorf1 (leaky scanning phenotype; pic complexes miss the correct start codon with increased frequency and continue scanning downstream); 3) failure of the 40s ribosome to resume scanning after terminating at uorf1; 4) reduced rate of scanning (slow scanning phenotype); 5) instability of re-scanning ribosomes on the mrna (defect in processivity of scanning).http://www.biomed.cas.cz/
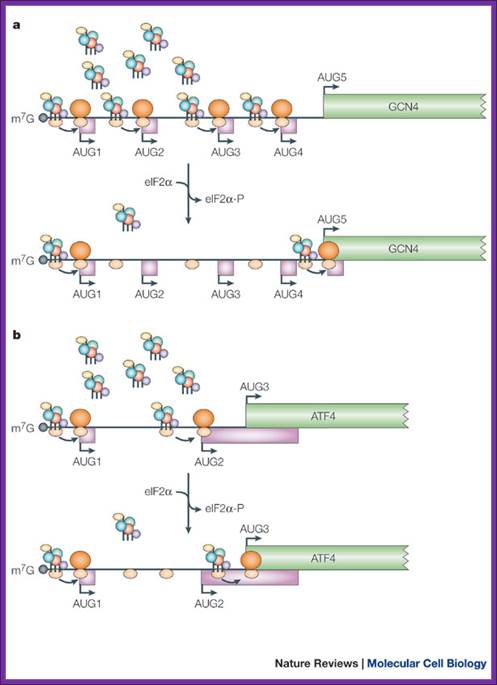
A |
translation of the yeast transcriptional activator gcn4 is regulated by four
upstream open reading frames (uorfs). At low levels of eukaryotic initiation
factor-2![]() (eif2
(eif2![]() ) phosphorylation, when the
ternary complex is abundant, ribosomes initiate at uorf1 and resume scanning to
reinitiate at uorf2, uorf3 or uorf4. However, ribosomes that terminate at these
latter uorfs cannot resume scanning, thereby decreasing the probability of
initiation at the gcn4 orf. By contrast, during amino-acid starvation,
increased levels of eif2
) phosphorylation, when the
ternary complex is abundant, ribosomes initiate at uorf1 and resume scanning to
reinitiate at uorf2, uorf3 or uorf4. However, ribosomes that terminate at these
latter uorfs cannot resume scanning, thereby decreasing the probability of
initiation at the gcn4 orf. By contrast, during amino-acid starvation,
increased levels of eif2![]() phosphorylation lower the abundance of the
ternary complex and reinitiation at uorf2�4 becomes less frequent, which allows
scanning ribosomes to reach the gcn4 orf. B |
translation of the mammalian activating transcription factor-4 (atf4) is
regulated by two uorfs. Similar to the regulation of gcn4, when the ternary
complex is abundant (in the presence of low levels of eif2
phosphorylation lower the abundance of the
ternary complex and reinitiation at uorf2�4 becomes less frequent, which allows
scanning ribosomes to reach the gcn4 orf. B |
translation of the mammalian activating transcription factor-4 (atf4) is
regulated by two uorfs. Similar to the regulation of gcn4, when the ternary
complex is abundant (in the presence of low levels of eif2![]() phosphorylation), the ribosomes initiate at uorf1
and frequently reinitiate at uorf2. As uorf2 overlaps with the atf4 orf, the
translation of uorf2 suppresses the translation of atf4. During endoplasmic
reticulum (er) stress, when the level of the ternary complex is reduced, the
ribosome scans through uorf2 and initiates at the atf4 initiation codon. Coding
regions are shown as green rectangles, uorfs are shown as pink rectangles, 5'
untranslated regions (utrs) are shown as thin lines, initiation codons (aug)
are indicated with arrows and ribosomes are shown in orange (60s subunit, dark
orange; 40s subunit, light orange). The ternary complex is represented as in fig. 2. Gcn4, general control non-derepressible; m7g,
cap structure. Martin holcik & nahum sonenberg; nature reviews-2005
phosphorylation), the ribosomes initiate at uorf1
and frequently reinitiate at uorf2. As uorf2 overlaps with the atf4 orf, the
translation of uorf2 suppresses the translation of atf4. During endoplasmic
reticulum (er) stress, when the level of the ternary complex is reduced, the
ribosome scans through uorf2 and initiates at the atf4 initiation codon. Coding
regions are shown as green rectangles, uorfs are shown as pink rectangles, 5'
untranslated regions (utrs) are shown as thin lines, initiation codons (aug)
are indicated with arrows and ribosomes are shown in orange (60s subunit, dark
orange; 40s subunit, light orange). The ternary complex is represented as in fig. 2. Gcn4, general control non-derepressible; m7g,
cap structure. Martin holcik & nahum sonenberg; nature reviews-2005
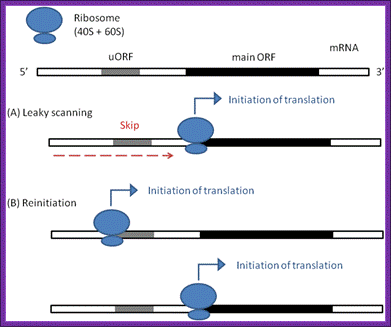
The irregular models of ribosome scanning on eukaryotic mRNAs. (a) leaky scanning and (b) reinitiation. Gray regions indicate uORFs on the mRNA, whereas black ones represent the main ORFs.
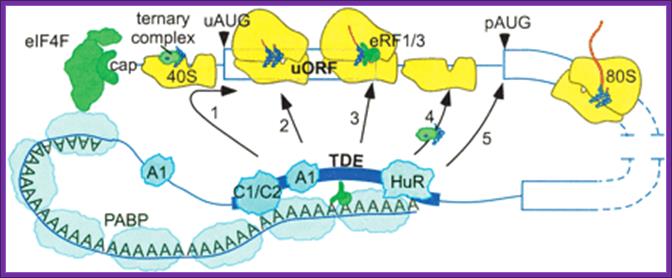
Potential mechanisms of 3′utr sequence regulation of uorf inhibitory activity. A 3′utr sequence that alleviates the inhibitory effects of uorfs could, in principle, act at one or more of several steps. It could promote leaky scanning past the uaug codon (arrow 1) or eliminate an elongation block during uorf translation (arrow 2). It also could alter the efficiency of termination (arrow 3), of recharging the 40s with eif2�gtp�met-trnaimet (ternary complex) or other initiation factors (arrow 4), or of paug codon recognition (arrow 5). Proteins such as hnrnpc1/c2 (c1/c2) and hur that bind specifically to the 3′ tde in the her-2 3′utr may mediate the derepression mechanism through their direct or indirect interactions with other 3′utr-binding factors such as pabp, hnrnpa1 (a1), and eukaryotic release factors 1 and 3 (erf1/3). The illustration is not to scale; for example, the entire her-2 uorf coding region could be covered by only one ribosome, matthew s. Sachs and adam p. Geballe.
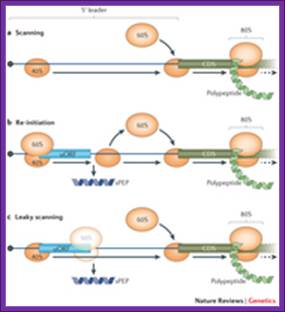
Leaky scanning and reinitiation; a | in the standard scanning model of eukaryotic translation, the 40s ribosomal subunit, together with pre-initiation factors (not shown), binds to the 5′ cap of the mrna and scans along the transcript until the first initiation codon�shea j. Andrews,& joseph a. Roth Nagel, nature review article 2014
Chain initiation:
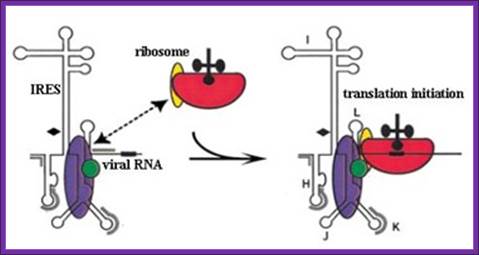
Ires mediated initiation:
Chain initiation takes place by cap mediated or internal ribosome entry sites Ires mediated mechanisms that depend upon the kind and structure of mRNAs. Many eukaryotic plus RNA strands have Ires. But Ires mediated initiation may skip eif2a and even eif-g.��� But there are mRNAs without such cap structures; in such cases the initiation complex enters using 5� Ires sequences.� Often one finds bicistronic mrnas where the intercistronic spacer has to provide Ires structure element for the initiation at the second cistron.
It is interesting to note eukaryotic ribosomes can initiate translation on circular RNAs (+), but require Ires elements and they can translate multiple consecutive rounds of translation; chen cy, sarnow p.
|
|
|
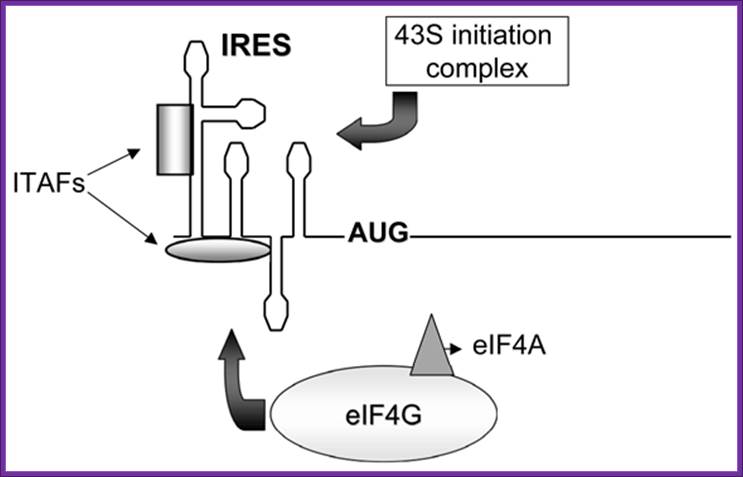
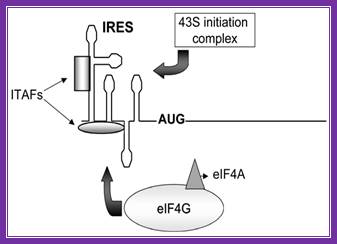
Figure. Schematic representation of internal ribosome entry site (ires)-mediated translation initiation. Internal initiation is an alternative mechanism to cap-dependent translation initiation which allows loading of the 40s ribosomal subunit on the mrna in a 5' end- and cap- independent fashion. Among the different iress canonical initiation factor requirements are variable. However, most iress require specific cellular proteins, ires trans-acting factors (itafs), to be functional. See the text for details. For diagram simplicity, other proteins, as well as a second eif4a molecule known to interact with eif4g, have been omitted. Chen cy, sarnow p |
|
|
|
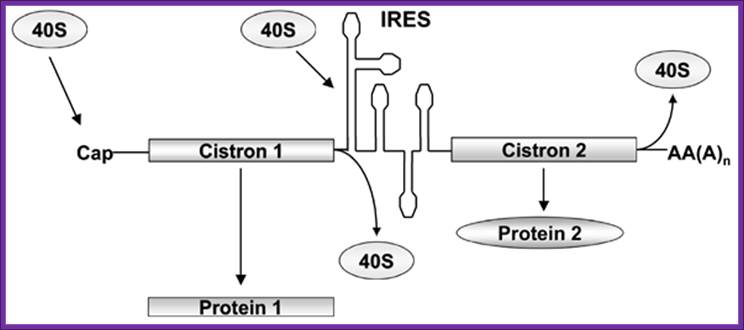
Bicistronic mRNAs-discistro-viridae- in bicistronic constructs, the first cistron is cap-dependent while the second cistron will be translated only if the intercistronic sequences can function as an ires since ribosome recruitment to the intercistronic spacer is independent from the 5'cap structure. For diagram simplicity, eif and other proteins known to participate in these processes have been omitted; biol. Res. V.38 n.2-3 santiago 2005; marcelo l�pez-lastra1,2,*, andrea rivas1 and mar�a in�s barr�a1 Bicistronic mrnas. In bicistronic constructs, the first cistron is cap-dependent while the second cistron will be translated; biol. Res. V.38 n.2-3 santiago 2005, marcelo l�pez-lastra et al
Ires mediated initiation; the encephalomyocarditis virus Ires binds ribosomes which enable the translation initiation to occur; (a) schematic diagram of human eif4g1 protein. Shown are the binding sites for pabp, eif4e, eif4a, and eif3 and the target site for picornaviral proteinase 2apro. The minimum eif4g1 fragment that binds specifically to the emcv Ires and supports 48s complex formation corresponds to amino acid residues 697�949. (b) model for assembly of 48s complexes on emcv-like Iress. Structural domains of the Ires and regions of contact with the following factors as determined by foot printing are shown: eif4g/4a complex (blue/green), itaf 45 (diamonds), ptb (gray). Ptb contains four rrm domains and binds multiple sites on emcv-like Ires; such binding (indicated by a dotted line) therefore may stabilize a specific conformation of the Ires. The recruitment of a 40s ribosomal subunit (red) carrying initiator tRNA and eif3 (yellow) is shown. �(adapted from u.t. hellen & peter sarnow); (note, there are some mistakes in typography) |
Only if the intercistronic sequences can function as an ires since ribosome recruitment to the intercistronic spacer is independent from the 5'cap structure.
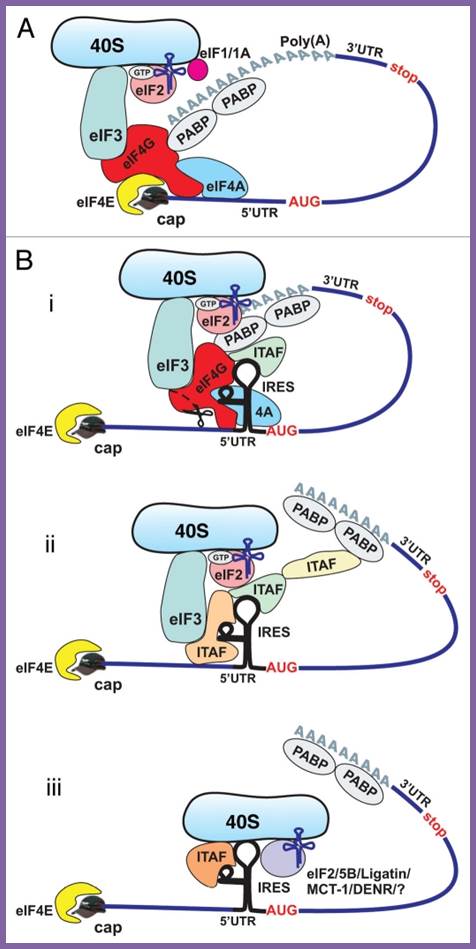
Cap-dependent and ires-mediated mechanisms of translation initiation in eukaryotic cells. (a) cap-dependent initiation is believed to require all canonical initiation factors and involve circularization of the mrna via interaction of pabp with eif4g. B) cellular ires-mediated translation does not require the cap- binding proteins eif4e and ? Or intact eif4g, but may involve circularization of the mrna. The requirement for canonical initiation factors and itafs can vary between different ires-containing mrnas. Potential mechanisms of cellular ires-mediated translation: 1; most ,if not all canonical factors and many itafs are required; 2. A limited number of canonical factors and itafs are required middle, 3.canaonical factors are dispensible , but some itafs may be required (bottom).� Delivery of met-trnaimet to 40s ribosomal subunit may be performed by eif%b/ligatin/mct-1/denr and perhaps some others, yet unidentified proteins, acting in place of eif2; Anton a Komar and Maria Hatzoglou
Chain initiation takes place simultaneously using different chain initiation factors. In most of the mrnas 5� cap is used.� In some viruses vpg and in some others ires are used for chain initiation at the 5� end.
� In the case of picorna viral rnas and encephalo myocarditis viral rnas, both are (+) rnas, they don�t have cap structures but they are bound by vpg protein, yet on translation the transcript produces proteins of which protein 2a, which acts on cap binding proteins of host mrnas especially eif-4g and cleaves a part of 5�utr; so none of the host mrnas are translated, but only viral rnas are translated because of the ribosomal entry site is internal (identified by stem loop structures) ires.� If the mrna does not contain cap, it usually possesses ires through which the small subunit 40s enters and initiates protein synthesis.� Even some capped mrnas or non-capped mrna have ires, in such cases ribosomes initiates translation at ires. Quite a number of viral rnas contain ires for initiation protein synthesis by their positive rna genomes ex. Polio, rhino, foot and mouth virus, hepatitis a virus maloney murine leukemia virus etc. Even many cellular mrnas have ires ex. Some growth factor mrnas,, nf-kb, eif4ga, apaf-1 and many others.
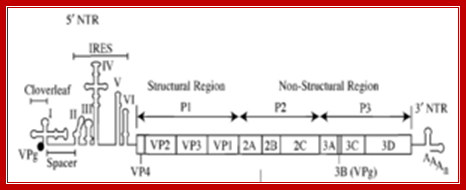
poliovirus genome, including an ires https://commons.wikimedia.org
Cap mediated initiation:
Eif-3 interacts with 80s ribosome and binds to 40s subunit and induces dissociation of 80s into 40s and 60s units. Factors like eif1 assist in eif3 activation.� Binding of eif3, also activates the 40s ribosomal subunit.
Eif-6 then binds to 60s subunit and it further prevents 60s to associate with 40s. They are anti dimeric ribosomes.
Ribosomal wholesome model; http://www.biochem.umd.edu
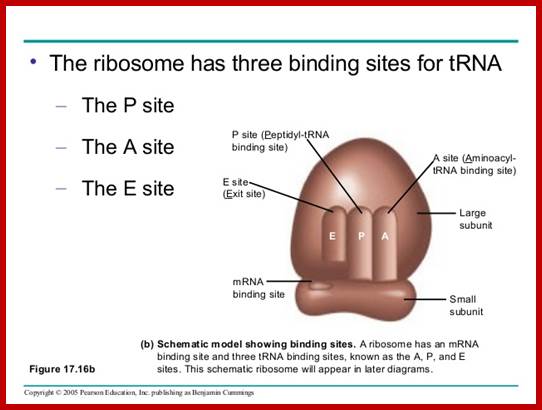
Https://www.slideshare.net
� Chain initiation factor eif-2-αβγ is activated by the binding of gtp, which induces conformational change in the protein so as to accept initiator amino-acyl met.tRNA only. In eukaryotes, initiator met.tRNA has characteristic structural features (see tRNA chapter) to act as initiator tRNA.� In yeast cells, the 64th nucleotide of met i.tRNAa is phosphorylated at r2�oh position. It is the only trna that can bind to p-site during initiation of translation. The first and the 72nd nucleotides of i.tRNA are a and u respectively. The tRNAs are loaded with methionine and remain unformylated that is the difference between eukaryotic and prokaryotic initiator tRNA. The initiator met.tRNA is structurally and functionally different from the rest of a.a tRNAs.
Activation of eif2a and its regulation is very important protein chain initiation and protein synthesis. Eif2 plays a central role in the maintenance of what is generally considered a rate-limiting step in mrna translation. Phosphorylation of eif2 on its smallest, or α-, subunit converts eif2 from a substrate of eif2b into a competitive inhibitor. Thus, phosphorylation of eif2α effectively prevents formation of the eif2�gtp�met-trnai complex and inhibits global protein synthesis. Phosphorylation of eif2α occurs under a variety of conditions including viral infection, apoptosis, nutrient deprivation, heme-deprivation, and certain stresses, scot r kimball
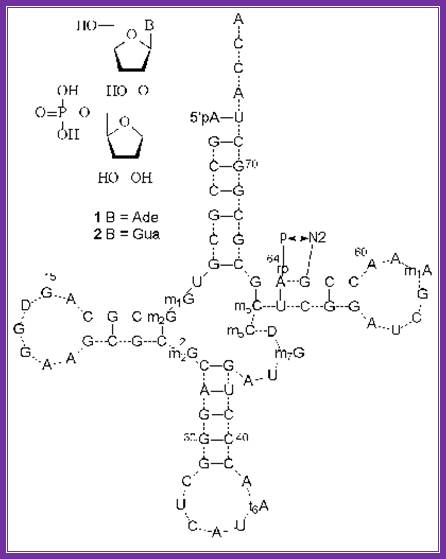
the structures of minor nucleosides 1 and 2 ] and their positions in yeast trnaimet . A hydrogen bond between the 5��-phosphate residue and guanine 2-amino group is shown. Http://www.protein.bio.msu.ru/
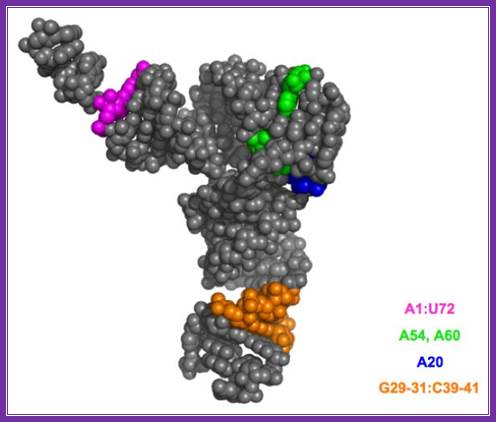
Eukaryotic initiator met.tRNA:� the 1st and 72nd nucleotides are base paired in initiator tRNA; there are 3 GC pairs in anticodon stem region. Unique CCu sequence is found in tUCG loop. Http://www.angelfire.com
�the individual components of eif-4f complex assemble at the 5� end of mrna one by one.� The first factor that binds directly to the cap is eif-4e, then eif-4a, eif-4b and eif-4g.� The critical feature in this assembly is the binding of phosphorylated eif-4e to the cap structure. It is cap-binding protein.� Phosphorylation of eif4e increases the efficiency of translation.�
The binding of pab1 bound the eif-g binds to the cap binding protein eif-4e; this leads to the binding of pab1 and eif-g and circularization of the mRNA. The binding of eif-4e leads to a chain of events in recruiting other components.� Circularization of mRNA with factor g enhances the efficiency and rate of translation.
As eif-g binds, the eif-4a assisted by eif-4b, has helicase activity, scans the leader and removes any stem-loop structures in the leader.�� The components act like a mobile complex move from 5� towards 3� end of the leader.
�
� Eif3 assisted by eif1a bound to 40s ribosomal subunit, recruits eif2-gtp-initiator.met i.tRNA on to its surface which form 43s complex.
The eif4g (for it interacts and binds to eif3), recruits activated 43s if-2a-gtp-initiator.met.i. trna-40s ribosome complexes on to 5�end of the leader sequence of mRNA.� The binding of ribosome-initiator eif-2(abg)-gtp-trna- complex strongly enhances scanning. The mRNA is threaded through the cleft of the 40s ribosome subunit.� The association of small ribosomal subunit and met-i.tRNA complex with 5�ends of the mRNA and its associated complex forms 48s complex.
��
�this mobile complex not only removes any hurdles like stem-loops in the leader, but also scans for correct initiator-AUG codon (of the orf) in the mRNA, that is present in correct sequence context called Kozak sequence (g/accaugg).� If there are any other AUGs, in the leader sequence called uAUGs, if they are not in proper context, they are skipped, but when it finds one, it halts and positions the 40s ribosome-eif2a-gtp-met i.tRNAin such a way the initiator met.tRNA is placed against the first AUG codon of the uORF in mrna, at the p site of 40s ribosome. Normally initiation and elongation at uorf or uAUG is aborted.� Here P and A sites are found in both small and large ribosome subunits. In these reactions eif-4g and eif-1 play very important roles.� The eif4g also binds to pab-i as mentioned earlier, thus the poly (a) end of the mRNA is also recruited on to the initiation complex, where the 5� end and the 3� end of the mRNA are in closed loop.� This configuration enhances initiation and also the rate of protein synthesis.
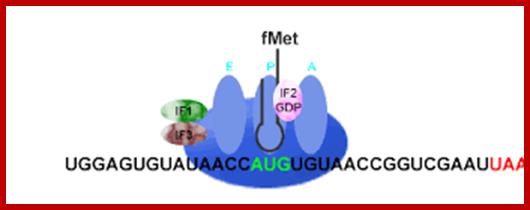
When the eif2-met i.trna-gtp complex is placed in proper position called p-site and base pairs with the first aug codon, provided kozak sequence embeds aug codon.� Then eif2-gtp hydrolyses and all factors bound to 40s are released; the eif-g remains still bound to 4e of mrna-5�end and pabi of the 3�end.� Hydrolysis of gtp to gdp bound to eif-2αβγ, induces conformational change in the ribosome that assures further correctional changes in positioning and base pairing of initiator trna with the initiator codon. Then gdp dissociate from the eif2 initiation complex and recycled. At this juncture eif5 associates with this complex, ribosome undergo conformational changes and now it is in active sate with its p site bound to met i.trna and free a site fully formed.�
� The 60s subunit, which was inactive hitherto with the binding of eif-6, is activated by eif-4c. The factor eif5b enables 60s subunit to join 40s.
The 80s complex formation is also assisted by eif-4d. Sparsomycin blocks translational initiation.
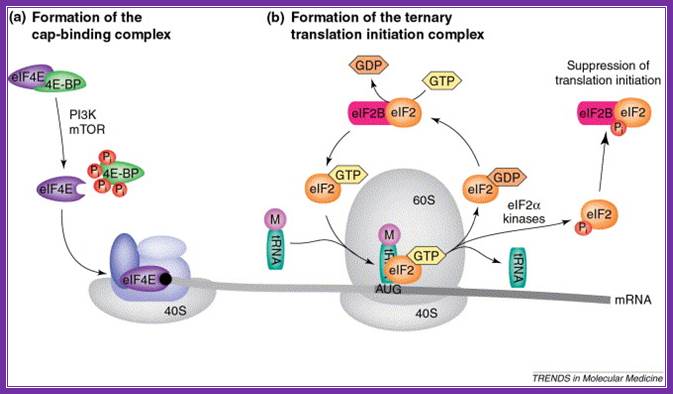
Rate-limiting steps in the initiation of translation. (a) the eukaryotic translation initiation factor 4e (eif4e) is retained in an inactive form by 4e-binding proteins (4e-bps), but is released after phosphorylation of the 4e-bps by phosphoinositol 3-kinase (pi3k) and mtor (mammalian target of rapamycin). Free eif4e then binds to the mrna cap (black circle) as part of the eif4f complex that recruits the small 40s ribosomal subunit. (b) a ternary complex, consisting of eif2, met�trnaimet and gtp, facilitates aug-codon recognition and initiation of protein synthesis. Eif2 is recycled through the exchange of gdp for gtp by the associated guanine-nucleotide-exchange factor, eif2b. However, phosphorylation of the eif2α subunit by an eif2α kinase prevents dissociation of eif2 from eif2b, leading to inhibition of translation initiation. Abbreviations: m, methionine; pi, phosphate; cornelis f calkhoven, christine m�ller, achim leutz
Kozak sequence:
�initiation of protein synthesis takes place at the 5�end of the mRNA with AUG codon as the first codon in the open reading frame.��� The initiator codon determines the first amino acid position of the n-terminal end of the protein chain.��� If there are more than one AUG codons called uAUG s in the leader sequence, correct initiation takes place only from that AUG, which has specific Kozak (marilyn kozak) sequence in the right context.�
the Kozak sequence is, 5�---CCrCC AUG -, where r can be a or g.� Positionally the a of AUG is +1; the u, g and g are in +2, +3, and +4th position.� In the upstream, which are referred to as �1, -2 and �3 are c, c and r (a or g).
����������������������������������������������� � -4 �3 �2 �1-+1 +2 +3 +4
����������������������������������������������� 5� c� g� c� c� a �u� g g 3�,[ r =a or g]
�sv-40 mRNAs recombinants constructs with AUGs in different positions and different sequence context, when translated in monkey�s cos-cells, chain initiation was at its best when g or a at (�) 3 position and g at +4th position.�
If two as of two AUG s, which are, placed eight ntds apart and if the downstream aug is in correct sequence context, initiation takes place from the second AUG.� If the u instead of g at +4 th position is placed, initiation, though, takes place from the second aug, but not as efficient as in the first case.
�
Ires:� in the absence of cap structure, if the 5� leader region contains Ires, 43s directly binds to� Ires and initiates translation.
In eukaryotic systems, if there are two such AUGs with few nucleotides between them, generally one finds a terminator codon in between them, thus the second AUG represents reinitiation.�
Schematic representation of internal ribosome entry site (ires)-mediated translation initiation: internal initiation is an alternative mechanism to cap-dependent translation initiation which allows loading of the 40s ribosomal subunit on the mrna in a 5' end- and cap- independent fashion. Among the different iress canonical initiation factor requirements are variable. However, most iress require specific cellular proteins, ires trans-acting factors (itafs), to be functional. See the text for details. For diagram simplicity, other proteins, as well as a second eif4a molecule known to interact with eif4g, have been omitted. Http://www.scielo.cl/
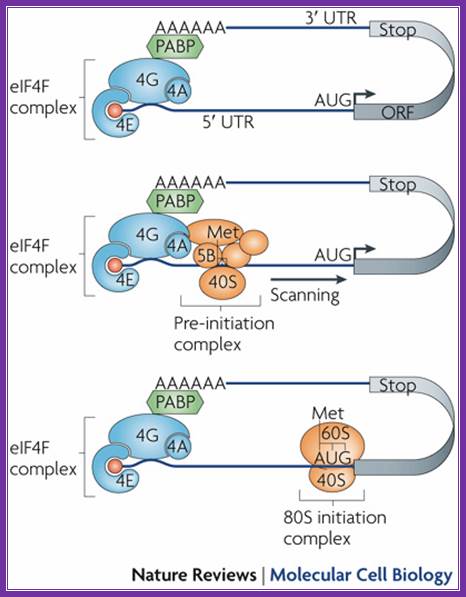
Binding of eif4e to the 5� cap is important for chain initiation.� The binding of 4e is possible only when 4ebp are released from 4e.� Phosphorylation 4e accelerates chain initiation. Once 4e binds met-eif2-gtp -40s ribosome complex joins the leader sequence.� The multifaceted 4g, a scaffold, binding brings the 5� end and poly-a tail bound pabi in circular mode.� The 40s fmet-eif2gtp complex threads through the leader sequences assisted by 4a (atp dependent helicase) -4b.� The helicase removes any stem loop structures that act as hindrance.� When this 40s complex reach and recognizes the aug codon, if it is in right context determined by the kozak sequence, the large ribosomal subunit activated by eif2b and eif5b joins the pre-initiation complex. www.sfsoo.com
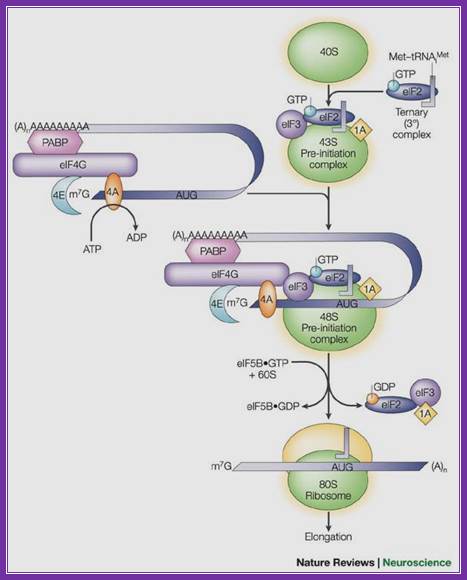
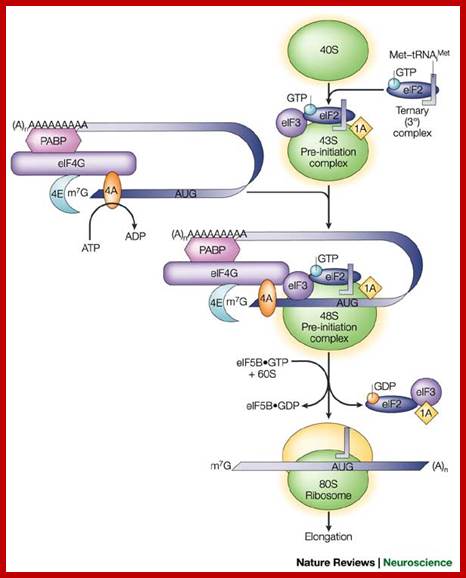
A binary complex of eukaryotic translation initiation factor 2 (eif2) and gtp binds to methionyl�transfer rna (met�trnaimet), and the ternary complex associates with the 40s ribosomal subunit. The association of additional factors, such as eif3 and eif1a (1a), with the 40s subunit promotes ternary complex binding and generates a 43s pre-initiation complex. The cap-binding complex, which consists of eif4e (4e), eif4g and eif4a (4a), binds to the 7-methyl-gtp (m7gtp) cap structure at the 5' end of a messenger rna (mrna). Eif4g also binds to the poly(a)-binding protein (pabp), thereby bridging the 5' and 3' ends of the mrna. This mrna circularization and the atp-dependent helicase activity of eif4a are thought to promote the binding of the 43s pre-initiation complex to the mrna, which produces a 48s pre-initiation complex. Following scanning of the ribosome to the aug start codon, gtp is hydrolysed by eif2, which triggers the dissociation of factors from the 48s complex and allows the eif5b- and gtp-dependent binding of the large, 60s ribosomal subunit. Although the precise timing and requirements for the release of factors from the pre-initiation complexes are not clear, the 80s product of the pathway is competent for translation elongation and protein synthesis. Biochemical mechanisms for translational regulation in synaptic plasticity; eric klann & thomas e. Dever.
Pathway of translation initiation in eukaryotes; a binary complex of eukaryotic translation initiation factor 2 (eif2) and gtp binds to methionyl�transfer rna (met�trnaimet), and the ternary complex associates with the 40s ribosomal subunit. The association of additional factors, such as eif3 and eif1a (1a), with the 40s subunit promotes ternary complex binding and generates a 43s pre-initiation complex. The cap-binding complex, which consists of eif4e (4e), eif4g and eif4a (4a), binds to the 7-methyl-gtp (m7gtp) cap structure at the 5' end of a messenger rna (mrna). Eif4g also binds to the poly(a)-binding protein (pabp), thereby bridging the 5' and 3' ends of the mrna. This mrna circularization and the atp-dependent helicase activity of eif4a are thought to promote the binding of the 43s pre-initiation complex to the mrna, which produces a 48s pre-initiation complex. Following scanning of the ribosome to the aug start codon, gtp is hydrolysed by eif2, which triggers the dissociation of factors from the 48s complex and allows the eif5b- and gtp-dependent binding of the large, 60s ribosomal subunit. Although the precise timing and requirements for the release of factors from the pre-initiation complexes are not clear, the 80s product of the pathway is competent for translation elongation and protein synthesis.
;biochemical mechanisms for translational regulation in synaptic plasticity;
Eric klann & thomas e. Dever;http://www.nature.com
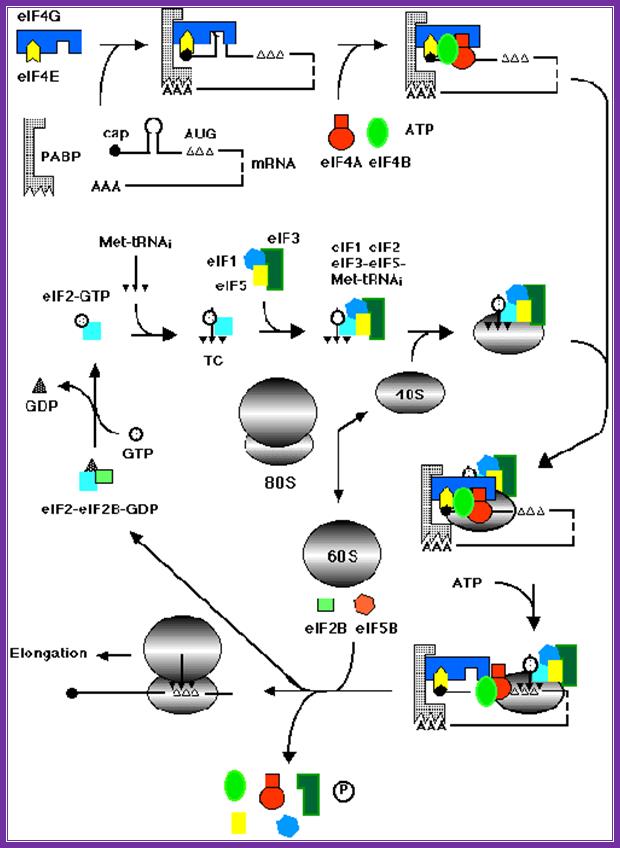
Assembly of various factors and mRNA and ribosomal subunits during chain initiation. www.theses.ulaval.ca
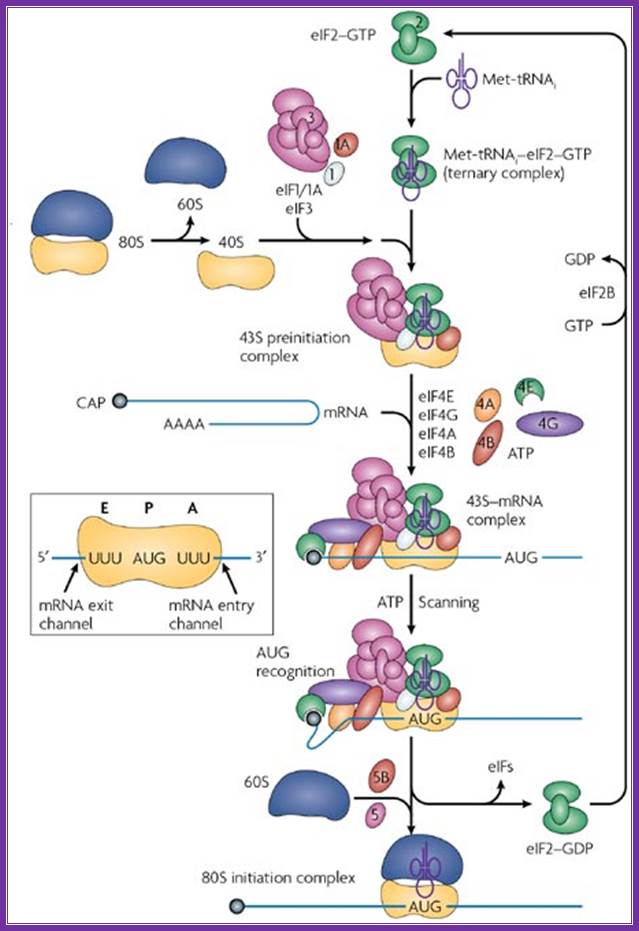
In most cellular mrnas, recognition of the start site for protein synthesis occurs by ribosome scanning, a proposed model of which is shown in the figure (reviewed in refs.� This mechanism requires all of the canonical initiation factors that have been characterized so far and, in contrast to the hcv ires, also requires the 5'-cap structure found on all cellular mrnas. Translation initiation begins on the 40s (small) ribosomal subunit, which is composed of an mrna binding cleft, an aminoacyl (a) site, peptidyl (p) site, and exit (e) site. The mrna enters the 40s ribosomal subunit through the mrna entry channel, passes through the a, p and e sites and leaves through the mrna exit channel. To initiate translation, a free pool of 40s ribosomal subunits, stabilized by association with the large multisubunit initiation factor eif3, binds to met-trnai and mrna (reviewed in. The met-trnai is brought to the 40s ribosomal subunit as part of an eif2�gtp complex, and together with two small initiation factors, eif1 and eif1a, forms the 43s preinitiation complex. Assembly of the 43s preinitiation complex on an mrna by this pathway requires the 5'end of the mrna, as ribosomes are unable to bind an mrna that has been covalently circularized92. 43s preinitiation complex formation is enhanced by the presence of the cap-binding protein eif4e, which in turn associates with the scaffold protein eif4g. The interaction between eif4g and eif3 stabilizes the 43s preinitiation complex, leading to its migration (scanning) to the aug start codon by the unwinding of any rna structure present in the 5'-untranslated region (utr). On pairing between the met-trnai anticodon and the aug start codon, eif2 hydrolyses its gtp with the help of the gtpase activating protein eif5. The eif2�gdp complex dissociates from the 40s ribosomal complex and is recycled by a guanine nucleotide exchange factor, eif2b, so that it can associate with a new met-trnai and take part in another round of initiation. The exchange of gdp for gtp by eif2b is highly regulated in eukaryotic cells, by a mechanism involving the phosphorylation of eif2 by one of at least four distinct kinases, preventing the efficient recycling of this initiation factor90, 93, 94. Finally, a second gtpase, eif5b, promotes the joining of the 60s ribosomal subunit to form an 80s initiation complex and the removal of the remaining initiation factors. Structural and mechanistic insights into hepatitis c viral translation initiation ;christopher s. Fraser and jennifer a. Doudna
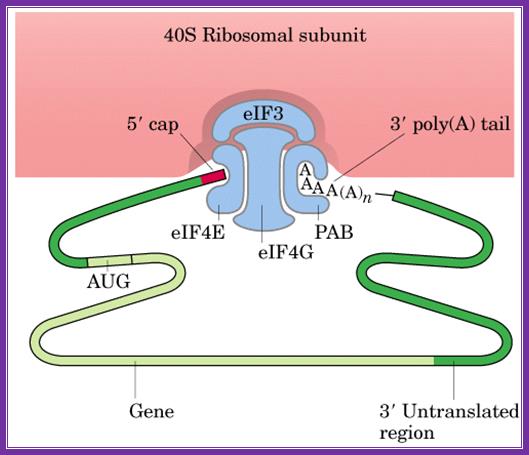
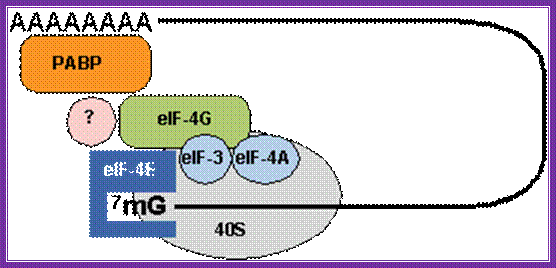
The diagrams above and below show the 5�end and the 3�end of the m. RNAs come together because of the association of various initiation factors that assemble at that time chain initiation; important one is eif4e-eif4g and pabii bring the RNA in circular form. Www.biochem4.okstate.edu
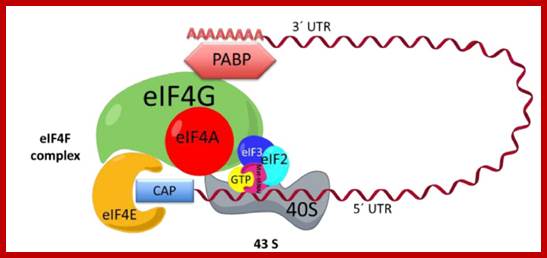
Initiation complex. The interaction between the eif4f complex, 43s, and mrna is shown. Eif4f is formed by eif4a, eif4g, and eif4e. The complex 43s is formed by eif3, the small ribosomal subunit, and eif2, which in turn is formed by methionine-trna-initiator (met-trnai) and gtp. The mrna is recruited to the eif4f complex across the interaction of the 3′ end and poly -a- binding protein (pabp) and the 5′ cap and eif4e. Utr: untranslated region. ;initiation complex with all factors except large ribosomal subunit. What is missing in this initiation complex? Https://www.researchgate.net
Regulation at cap biding elements:
Eukaryotic mRNAs contain 5�cap 7�methylated guanine linked to first nucleotide by 5�~5� triple phosphate bonds. The initiation factor 4e binds to cap.� Phosphorylation of 4e at ser209 by mnk1 and mnk2 enhances the activation of translation.� Another factor called 4ebp, when binds to 4e blocks chain initiation.� But phosphorylation of 4ebp at specific ser makes it release from the 4e.
Open reading frame (ORF) defines the coding region of mRNA with an initiator codon in correct context and subsequent sequences are read in three nucleotides as units, which end up in three-nucleotide terminator codon.��
If there are two ORFs one after the other i.e. one in the upstream and the other in the downstream, invariably the upstream uORFs is translated provided the Kozak sequence in proper context.
In the leader sequence, if there are any strong stem-loop sequences at the upstream of aug, it can hinder chain initiation for it hinders ribosomal scanning.� In general stem loop structures are removed by eif4a and eif4b (helicase).� On the other hand if the aug is present within the stem-loop, initiation is not inhibited and chain initiation takes place.
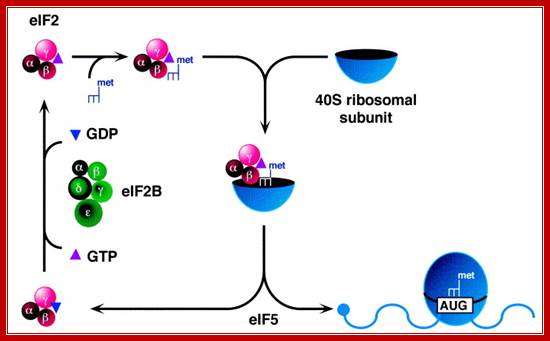
Recycling of eif2-αβγ:
The ubiquitous eif2-αβγ-GP-t.RNA complex binds to the ribosomal surface, places the initiator met-t.RNA against iAUG codon, with the association of 60s ribosomal subunit, GTP gets hydrolyzed and released from the ribosomal surface as eif-2-αβγ-GDP + pi.
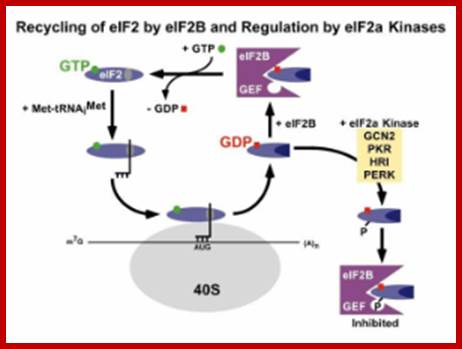
Https://science.nichd.nih.gov
The eif-2αβγ-gdp is recycled by eif-2b; first by interacting with eif-2αβγ gdp.� This interaction releases gdp from eif-2αβγ and forms a complex with eif2b.� Now this complex interacts with gtp, loads the gtp on to generate eif-αβγ-gtp and eif-2b with gdp is released.� This factor is regulated by various kinase factors such as heme-hemin induced kinase, gcn2, viral-ifn induced kinase and many others.
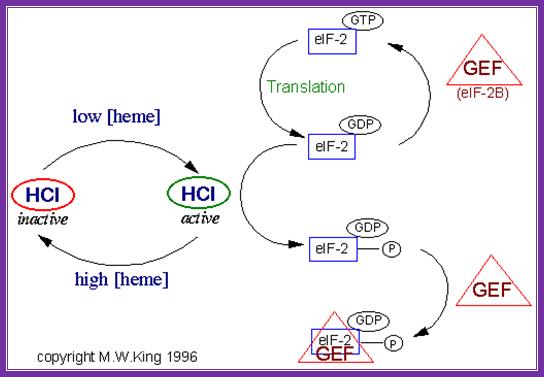
In response to the levels of iron mediated heme the activity of eFABG is controlled.� When the heme content is low the heme dependent repressor is active it binds gef and renders recycling of eIFABG inactive.� But if the concentration of heme is high HCL becomes inactive, it does not bind to eIFABG and the initiation is normal.
The eif-2a cycle involves the regeneration of gtp-bound eif-2a following the hydrolysis of gtp during translational initiation. When the 40s preinitiation complex is engaged with the 60s ribosome to form the 80s initiation complex, the gtp bound to eif-2a is hydrolyzed providing energy for the process. In order for additional rounds of translational initiation to occur, the gdp bound to eif-2a must be exchanged with gtp. This exchange is performed by eif-2b which is also called guanine nucleotide exchange factor (gef).
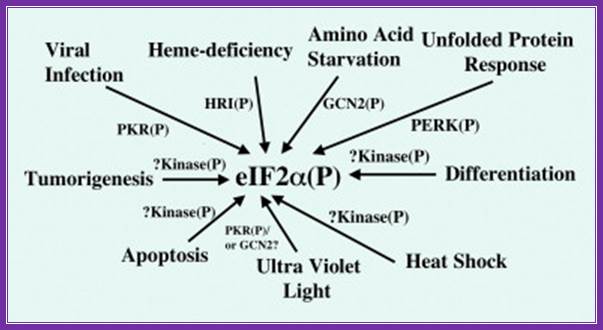
Regulation of protein chain initiation is executed by several factors acting on protein chain initiation factor eif2alpha (serine 51); in response to environmental stresses, a family of protein kinases phosphorylate eif2 (eukaryotic initiation factor 2) to alleviate cellular injury or alternatively induce apoptosis. Phosphorylation of eif2 reduces global translation, allowing cells to conserve resources and to initiate a reconfiguration of gene expression to effectively manage stress conditions. Accompanying this general protein synthesis control, eif2 phosphorylation induces translation of specific m. RNAs, such as that encoding the bzip (basic leucine zipper) transcriptional regulator atf4 (activating transcription factor 4). Atf4 also enhances the expression of additional transcription factors, atf3 and chop (ccaat/enhancer-binding protein homologous protein)/gadd153 (growth arrest and Dna-damage-inducible protein), that assist in the regulation of genes involved in metabolism, the redox status of the cells and apoptosis. Reduced translation by eif2 phosphorylation can also lead to activation of stress-related transcription factors, such as nf-κb (nuclear factor κb), by lowering the steady-state levels of short-lived regulatory proteins such as iκb (inhibitor of nf-κb). While many of the genes induced by eif2 phosphorylation are shared between different environmental stresses, eif2 kinases function in conjunction with other stress-response pathways, such as those regulated by mitogen-activated protein kinases, to elicit gene expression programmes that are tailored for the specific stress condition. Loss of eif2 kinase pathways can have important health consequences. Mice devoid of the eif2 kinase gcn2 [general control non-derepressible-2 or eif2ak4 (eif2α kinase 4)] show sensitivity to nutritional deficiencies and aberrant eating behaviors, and deletion of pek [pancreatic eif2α kinase or perk (rna-dependent protein kinase-like endoplasmic reticulum kinase) or eif2ak3] leads to neonatal insulin-dependent diabetes, epiphyseal dysplasia and hepatic and renal complications;
Coping with stress: eif2 kinases and translational control r.c. wek, h.-y. Jiang, t.g. anthony \ http://www.biochemsoctrans.org
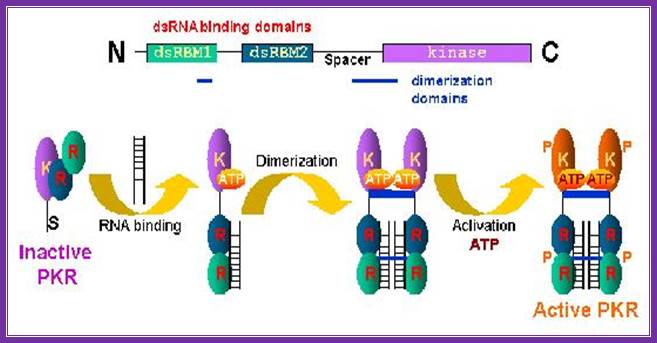
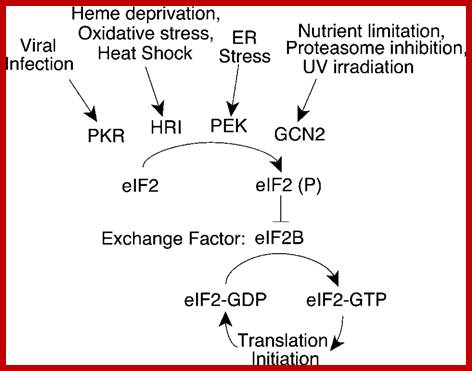
Rna dependent-pkr=pkr is a major mediator of antiviral and inflammatory responses. This interferon- and tnf-inducible stress signal kinase depends for its activation strictly on dsrna, a hallmark of viral infection. Once pkr is activated, it potently inhibits protein synthesis in the cell by phosphorylating the α chain of eukaryotic translation initiation factor eif2, the protein that binds trnaimet in dependence on gtp, blocking gdp-gtp exchange needed for the recycling of eif2 between successive rounds of translation and thereby rendering eif2 inactive. Https://medicine.ekmd.huji.ac.il
Infection of cell with dna or rna viruses leads to the synthesis of dsrna by transcription. The double-stranded rna (dsrna)-activates protein kinase (pkr) provides a fundamental control step in the regulation of protein synthesis initiation through phosphorylation of the alpha subunit of eukaryotic translation initiation factor 2 (eif-2α), this leads to blocking of eifb as gef, a process that prevents polypeptide chain initiation, kotio u.kumar etal.
Four stress-responsive protein kinases, including gcn2 and pkr, phosphorylate eukaryotic translation initiation factor 2alpha (eif2alpha) on ser51 to regulate general and gene-specific protein synthesis. Phosphorylated eif2a is an inhibitor of its guanine nucleotide exchange factor, eif2b, dey mtrieselmann etal. The rna-regulated protein kinase (pkr) is an interferon-inducible enzyme of widespread occurrence in eukaryotic organisms. This serine/threonine-specific protein kinase is activated by double-stranded rna by a mechanism involving autophosphorylation. Under stress gcn2 is active and it phosphorylates eif2a and makes it inactive with binding of exchange factor eif2b
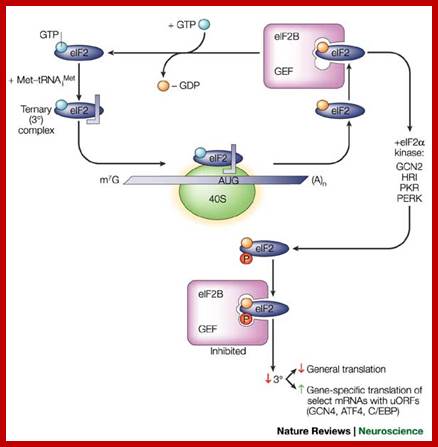
this recycling reaction is a key control point to
regulate translation initiation and it is the target of the eif2
protein kinases. Four protein kinases have been identified that specifically
phosphorylate the subunit of eif2 alfa on the residue ser-51.
Activated gcn2,pkr, prk and hri when activated they phosphorylate serine or
threonine at 51 amino acid. When the phosphorylated alpha subunit binds to
eif2b-they don�r dissociate and remain bound, thus eif2abg remain inactive.
The eukaryotic translation initiation factor 2
(eif2)�gtp binary complex binds to methionyl�transfer rna (met�trnaimet) and forms a ternary complex that then associates with
the 40s ribosomal subunit. After start-codon recognition, gtp is hydrolysed by
eif2 and the binary eif2�gdp complex is then released. The guanine
nucleotide-exchange factor (gef) eif2b converts inactive eif2�gdp to active eif2�gtp,
a process that is inhibited by phosphorylation (p) of the ![]() -subunit of eif2 on serine 51 by one of
the four known eif2
-subunit of eif2 on serine 51 by one of
the four known eif2![]() kinases. Phosphorylation of eif2
kinases. Phosphorylation of eif2![]() converts eif2 to a competitive inhibitor of eif2b, and
inhibition of eif2b results in lowered levels of ternary complexes, which
reduces general translation but increases translation of a specific class of
messenger rnas (mrnas) with upstream open reading frames (uorfs)22.
Atf4, activating transcription factor 4; c/ebp, ccaat/enhancer-binding protein;
gcn, general control non-derepressible; hri, haem-regulated initiation factor 2
converts eif2 to a competitive inhibitor of eif2b, and
inhibition of eif2b results in lowered levels of ternary complexes, which
reduces general translation but increases translation of a specific class of
messenger rnas (mrnas) with upstream open reading frames (uorfs)22.
Atf4, activating transcription factor 4; c/ebp, ccaat/enhancer-binding protein;
gcn, general control non-derepressible; hri, haem-regulated initiation factor 2![]() kinase; m7g,
7-methyl-gtp; perk, eif2
kinase; m7g,
7-methyl-gtp; perk, eif2![]() kinase 3; pkr, protein kinase-rna regulated, interferon-inducible
double-stranded rna dependent.� Eric klann & thomas e. Dever;http://www.nature.com
kinase 3; pkr, protein kinase-rna regulated, interferon-inducible
double-stranded rna dependent.� Eric klann & thomas e. Dever;http://www.nature.com
When activated by their cognate upstream stress signals, the mammalian eif2 kinase perk and gcn2 repress translation of most mrnas but selectively increase translation of activating transcription factor 4 (atf4), resulting in the induction of the downstream gene chop (gadd153), harding h.p etal.
Chain elongation:
Chain initiation complex positions the initiator met-i.trna on p site in proper perspective and in correct base pairing geometry. Binding of eif2a-met i.trna to p� site opens up �a� site into active form.� Each of the ribosomal sites should be fully formed for the binding of incoming a.a trnas.
Elongation factor called eef1a-alpha gets activated with the binding of gtp, which is performed by eef1- (beta-gamma), it is recycling factor.
The activated eef1a-alpha-gtp has binding site for amino acyl-trnas. This is an active complex.� Unlike eif2a, these factors accept any a.acyl trnas.� It does not accept initiator trna, because of initiator trnas have their own structural conformation and structural constraint.
� This activated complex, now binds to a� site on the ribosomal surface in correct base pair geometry.�� If the base pairing is proper, it triggers hydrolysis of gtp and the eef1alpha is released from the surface as eef1alpha-gdp complex.� The energy released during hydrolysis induces conformational change in the active site for peptide bond formation.� If the base pairing does not match, the trna complex is ejected out, where it uses gtp hydrolysis. Eef1alpha-gdp gets recycled by eef2b (a gef).
Release of eefa-GDP makes way to the binding of eef-g.GTP also binds to large ribosomal subunit at what is called factor binding site (fbs).� It is located at the right side of a� site( overlapping). Eefg-GTP is translocation factor.
Peptide bond formation:
Which of the components of ribosome is responsible for peptide bond formation?� Is it one of the riboproteins or rRNA (23/28s or 18s/28s RNA)?� Present understanding is that the 23srna (prokaryote) and 28srna in eukaryotes located between p and a sites (?) Responsible for peptide bond formation.� It has some sequences that actuate peptide bond formation; especially 2541 ntd �a� is believed to be responsible for peptide bond formation.�
To prove this concept scientist have treated ribosomes with proteinase-k it removes most of the proteins from the ribosome, yet with few proteins still remain bound to the rrna, under in vitro conditions (proteinase-k treated) ribosomes perform peptide bond formation.� On the other hand, if ribosomes are treated with rnase, riboproteins alone cannot form peptide bonds. So it seems RNA nucleotides at particular position require few proteins for stabilization of the conformation; so RNA can execute bond formation. Free 2�oh group of RNA nucleotides have the ability to perform catalytic functions.
The factor involved in making the bond between two amino acids on the ribosomes is one of the nucleotides in the rRNA.� The complex is called PTC (peptide transferase complex).� In prokaryotes specific adenine with 2�oh is considered to be involved in bond formation.� What was believed, previously that one of the riboproteins is responsible for peptide bond formation, does not hold good now.
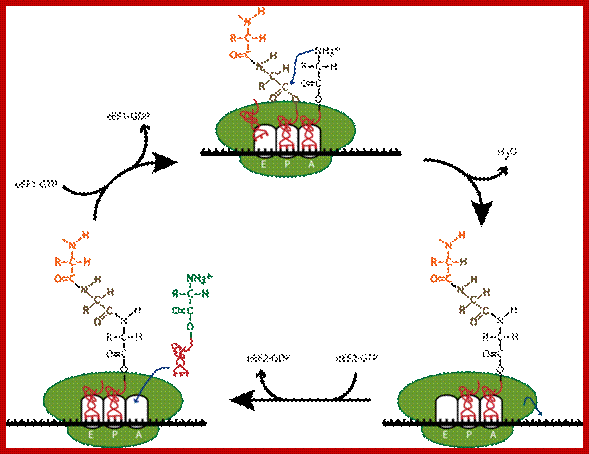
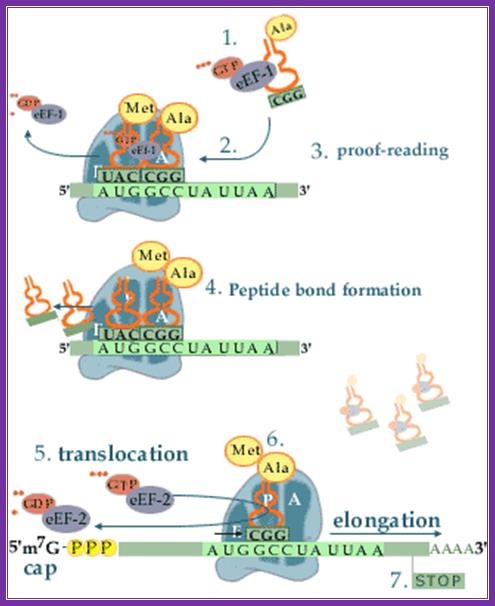
Top fig; amino
acid-containing tRNA molecules (aminoacyl-trnas, aa-trna) are picked up by
elongation factor eef-1 in the presence of GTP.
2. The formed complex enters the empty a-site on a ribosome carrying an
initiator met-trnai or a peptidyl-trna.
3. On the ribosome, the anticodon of the incoming aminoacyl-trna is matched
against the mrna codon positioned in the a-site. As the three bases in the codon can be
arranged in 64 different combinations, the translational machinery must be able
to select the aminoacyl-trna carrying the matching anticodon. During this
proof-reading, aminoacyl-tRNAs with non-cognate anticodons are thrown out of
the ribosome and replaced by new aa-tRNAs that are to be checked.
4. When the right aminoacyl-tRNA enters the a-site the growing polypeptide in
the p-site is almost immediately linked to the new
amino acid in the a-site via a peptide bond. Formation of the peptide bond is
catalysed by the ribosome itself. The reaction leaves an empty tRNA in the
ribosomal p-site and the new peptidyl-tRNA in the a-site.
5. In the next step the ribosome moves one codon forward on the mRNA.
Simultaneously, the empty tRNA is displaced from the p-site to the e-site as the peptidyl trna is translocated
from the a-site to the p-site. The process is facilitated by elongation factor
eef-2 and gtp.
6. After the translocation, the peptidyl-tRNA is positioned in the p-site and
the next codon on the mRNA is made available for interaction with a new aminao-acyl-tRNA
in the a-site.
7. These reaction steps are repeated until the ribosome encounters an in-frame
stop-codon. At this point the translation is terminated. Www.nobel prize.org
Translation elongation in eukaryotes. During translation elongation, the incoming aminoacyl-trna enters the ribosome a site, where it binds if the trna anticodon is complementary to the a site mrna codon. The elongation factor eef1 assists in loading the aminoacyl-trna, powering the process through the hydrolysis of gtp. The growing polypeptide chain is attached to the trna in the ribosome p site. The ribosome's peptidyl transferase catalyses the transfer of the growing polypeptide chain from the p site trna to the amino group of the a site amino acid. This creates a peptide bond between the c terminus of the growing polypeptide chain and the a site amino acid. After the peptide bond is created, the growing polypeptide chain is attached to the a site trna, and the trna in the p site is empty. The ribosome translocates once codon on the mrna. The elongation factor eef2 assists in the translocation, powering the process through the hydrolysis of gtp. During translocation, the two trnas remain basepaired to their mrna codons, so the ribosome moves over them, putting the empty trna in the e site (where it will be expelled from the ribosome) and the trna with the growing polypeptide chain in the p site. The a site moves over an empty codon, and the process repeats itself until a stop codon is reached. ���http://www.boundless.com
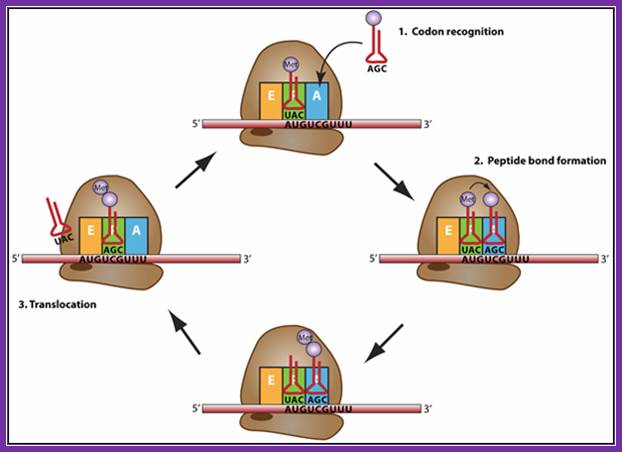
Elongation: a trna bound to its amino acid (known as an aminoacyl-trna) that is able to base pair with the next codon on the mrna arrives at the a site. The preceding amino acid (met at the start of translation) is covalently linked to the incoming amino acid with a peptide bond. The initiator trna moves to the e site and the ribosome moves one codon downstream. This shifts the more most recent trna from the a site to the p site, opening up the a site for the arrival of a new aminoacyl-trna. Polypeptide synthesis repeats, the trna residing in the e site is released from the complex, the trnas in the p site and a site shift over and the next amino acid is added to the growing polypeptide chain. This cycle repeats until a stop codon is reached.
Positioning of the a.a- t.RNA at a site, in correct base pairing geometry, triggers peptidyl transferase, whatever may be the source of the enzyme (may it be from one of the ribo-protein? Or may be one of the rRNA nucleotide), to bring about the transfer of energy rich carbonyl bond from the p site bound met-t.RNA to the amino group of the a.a t.RNA that is located on a site.� With this transfer, the t.RNA in p site is deacylated and free.� And the t.RNA at a site loaded with peptide bond (peptide chain), so it is called peptidyl tmRNA. Dissociation of eef1-α GDP makes way for the binding of eef2-gtp (= ef-g) E.coli), a 100kd factor, which is considered as translocation factor.� It binds to factor binding site next to a� site.
With peptide bond formation GTP is hydrolyzed by eef-2 that releases eef2-gdp complex, for recycling and recharging.
The energy released in this process is used for the translocation of ribosome complex by 3 nucleotides on m. RNAs, from 5� towards 3� end.� Whether m.RNAs is pulled or ribosome translocate on m.RNAs is yet to be discerned.� The process of chain elongation repeated.
As the elongation process progresses in processive manner, amino acid-by-amino acid, and codon-by-codon; in the process the 5�end of m.RNAs is made free.� The free end is again used for initiating the binding of I. Met t.RNA gtp-40s ribosomal subunits as another chain initiating complex.
In this way, a single m.RNA can be associated with more than 10 or more ribosomes (depending upon the length of the m. RNAs) to form polysome complex.� Visualize the titin m. RNAs, perhaps the longest m. RNAs. How many ribosomes it can hold? Polysomal complex enhances the rate of protein synthesis.� In some case, where the cell is stimulated for cell division or stimulated for cellular activation, inactive m.RNAs get activated and they can be loaded with more than 15 to 20 or more ribosomes per m.RNA.
�as the process of chain initiation, chain elongation and termination goes on simultaneously, at any given time the m.RNA is always loaded with 15-20 or more ribosomes.� The rate of protein synthesis can be 15a.a to 20a.a per second. This decoding process takes care of proof-reading during decoding by anticodons.
�the efficiency of translation is furthered by the circularized m.RNA�� 5�end cap and 3�polya tail by cap binding protein, pab1 and eief4g.� How exactly it enhances the rate of protein synthesis is yet to be clearly defined. Perhaps as the ribosome comes to the end of m.RNA the small ribosomal subunit slides on to the 5� end of the m.RNA directly and start translation. It means translation termination should have to be completed and polypeptide chain should be released at this point of time. But the problem is if the 5� of m.RNA is not free, new ribosomes cannot enter? (So, what is the problem?)
When the m.RNA is translated, the n-terminal end of the polypeptide chain is threaded through the exit tunnel in the large ribosomal subunit. Depending upon the sequence of n-terminal region, the ribosomal-m.RNA-polypeptide complex can be loaded onto endoplasmic reticulum.� Otherwise, the ribosomal�m.RNA complex can be localized in a specific position within the cell enmeshed in a network of actin filaments or free in cytoplasm.
Recycling of eef1alpha:
The eef1a-GTP-a.a. tRNA after loading on to the �A� site of ribosomal surface; i.e. the GTP-m.RNA on to P site and a.a tRNA to the a site properly, GTP hydrolysis, leads to the release of eef-1alpha-GDP.� This factor is found in large numbers and sequesters all met.RNAs.� Regeneration of this factor as eef1a alpha is very important. If it is prevented the whole process of translation comes to stand still. The eef1a-alpha-GDP interacts with eef1b-gtp and binds to it; this releases GDP from its surface.� The GTP is loaded on to eef1a aided by eef1b.� Thus, regenerated eef1a-alpha-gtp binds to a.at-RNA to form a complex that is required for elongation process.� This recycling process can be subjected regulation.
Chain termination:
Eukaryotes have only one chain termination factor, in comparison to prokaryotes, which have 3 chain releasing factors.� The erf-1 is a GTP dependent protein.
When the ribosomal machinery, with it�s a site reaches any one of the ter codons, ribosome halts or stalls).� Erf1 protein mimics t.RNA in its structure. Then rf1 binds to a site and executes the release of polypeptide chain by peptidyl transferase (peptidyl hydrolysis).
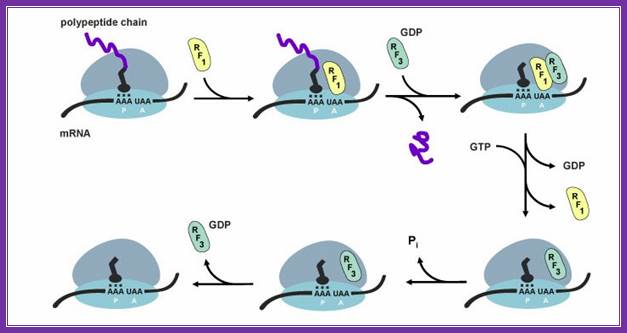
Important differences found in the interaction with class ii release factors, Rf3 and eRf3.� Erf1 has a specific region in its c domain which allows formation of a stable complex with erf3.� E. coli Rf1/2 does not contain such regions and no complex formation was seen in between rf1/2 and rf3. The third difference is in stop codon recognition. While in eukaryotes erf1 recognizes all three stop codons, prokaryotic rf1 and rf2 recognize UAA plus either UAG (rf1) or UGA (rf2).
Even more significant differences are found on comparison of Rf3 and eRf3 (table). First, erR3 is encoded by an essential gene, while this is not true for E.coli Rf3. Second, though both Rf3 and eRf3 are GTP-binding proteins, their structural differences are obvious: E. coli Rf3 is similar to ef-G (elongation factor, translocase), while eRf3 is similar to eef1alpha (elongation factor, analog of prokaryotic eF-tu). Third, the c domain of eRF3 contains an erf1-binding site, while no similar site was found in RF3. Fourth, RF3 activates GTPase in the presence of ribosomes and in the absence of RF1/2, while eRF3 requires not only ribosomes, but also eRF1 for its GTPase activity.
During termination erf1 activates eRF3-gtpase.� With the hydrolysis of GTP, eRF1 is released but RF3 is still bound to ribosome.� Ribosome recycling is executed by abce complex. The action of this complex in chain release and ribosome recycling is not yet clarified well.
Peptide bond formation leads to shifting the peptidyl tRNA to P site.� At this point, the erf-1 binds to a site to ter codon.� It transfers a water molecule to c- terminal end of the polypeptide chain still covalently bound to the tRNA; GTP hydrolysis leads to the release the protein chain. To remove erf1 from the surface of ribosome, it requires erf3 (which acts like pk rf3).� It requires eEF-GTP to perform the removal of tRNA from the P-site and E-site.� Both bind to factor binding site and GTP hydrolysis removes them. The release of ribosomal subunits, what is now called ribosomal recycling. The rrF of prokaryotic is absent in ek.� eF-G performs the process using abce GTPase complex and terminates the process, but eif3-ef-GTP is required for dissociation ribosomal subunits.
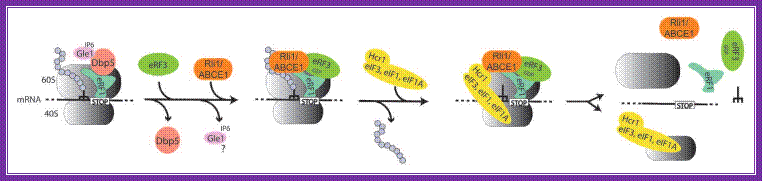
Model for translation termination in eukaryotes; when a translating ribosome has reached a stop codon, it arrests at this position: the trna mimicking eukaryotic release factor 1 (erf1) recognizes the stop codon and its binding is supported by the dead-box rna helicase dbp5, possibly by rearranging the surrounding rna/protein structures to fit erf1 into its correct position. For this conversion the atp ase activity of dbp5 is required, which is stimulated by the cofactors gle1 and inositol hexakisphosphate (ip 6). After this remodeling process dbp5 dissociates, which in turn allows the entrance of gtp -bound erf3 into the ribosome complex. It is currently unclear if gle1 dissociates or remains bound to the ribosome. The entry point of rli1/abce1 is currently unknown, but it is likely that it supports the termination accuracy in pre-termination complexes (termination complexes before polypeptide chain release). Erf3, which is bound to erf1 subsequently hydrolyzes gtp , resulting in the polypeptide chain release. For recycling of the ribosomal subunits, eif 3 associates with its stimulating factors eif 1 and eif 1a and dissociate supported by ntp hydrolysis through rli1/abce1,
The rna-bound ribosome into free 60s, 40s, trna and mrna. If the initiation factors and rli1 and/or gle1 remain bound to ribosomal subunits for initiation is unclear and remains to be shown.
The process of translation. (a) a typical eukaryotic mRNA is shown, with start (initiation) and stop (termination) codons indicated. Protein synthesis occurs in three steps: initiation, elongation and termination. Initiation involves recognition of the 5′-cap by proteins called initiation factors, which recruit the small ribosomal subunit. After this subunit identifies the start codon, the large ribosomal subunit associates, and translation begins. (b) an mRNA occupied by several actively elongating ribosomes (which together constitute a polysome) is shown. As translation proceeds, newly synthesized proteins (nascent polypeptides) emerge from the large ribosomal subunit. Termination occurs when an elongating ribosome encounters the stop codon; the ribosome dissociates from the mRNA, and the completed protein is released. (science direct)
Comparison between rf3 and erf3 of prokaryotes and eukaryotes:�
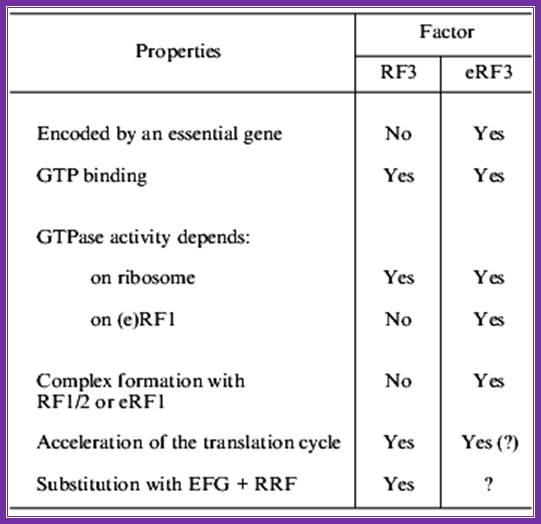
Some differences between prokaryotic and eukaryotic rf3s
����������������������������������������������� Three stages of translation
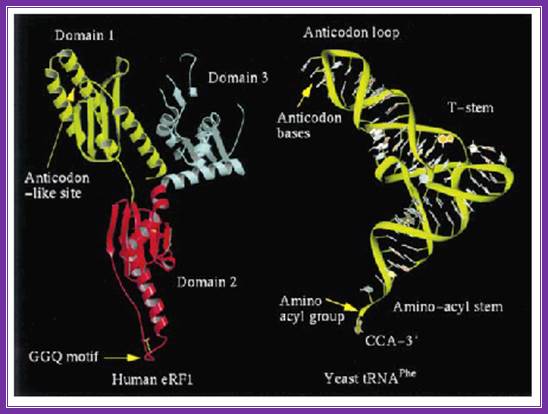
The picture shows the shape of erf1 with that of an trna; both occupy the same site so they have similar shape.
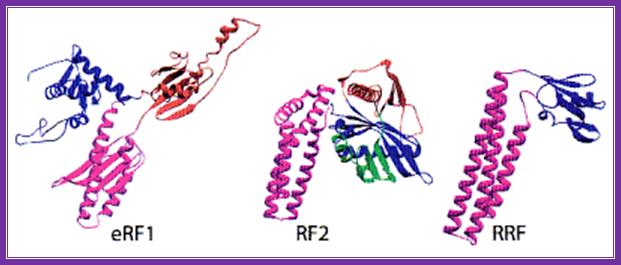
The above three ribbon models of prokaryotes show the shapes of the rf1, rf2 and rrf; they almost look similar and mimic tRNA structure.
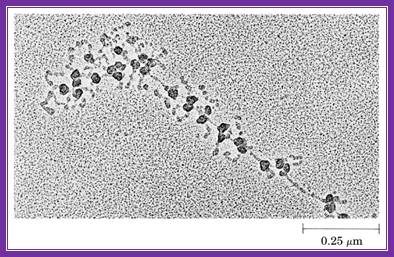
������������� ������������������������������������������ This electron microscopic figure shows a polysome
The role of abce1 in eukaryotic post-termination ribosomal recycling;
Andrey v. Pisarev, maxim a. Skabkin, vera p. Pisareva,1 olga v. Skabkina, aur�lie m. Rakotondrafara, matthias w. Hentze,2 christopher u. T. Hellen, and tatyana v. Pestova*
After termination, eukaryotic 80s ribosomes remain associated with mrna, p-site deacylated trna and release factor erf1. They must be recycled by dissociating these ligands and separating ribosomes into subunits. Although recycling of eukaryotic post-termination complexes (post-tcs) can be mediated by initiation factors eif3, eif1 and eif1a (pisarev et al., 2007), this energy-free mechanism can function only in a narrow range of low mg2+ concentrations. Here we report that abce1, a conserved and essential member of the atp-binding cassette (abc) family of proteins, promotes eukaryotic ribosomal recycling over a wide range of mg2+ concentrations. Abce1 dissociates post-tcs into free 60s subunits and mrna- and trna-bound 40s subunits. It can hydrolyze atp, gtp, utp and ctp.� Nucleotide triphosphate (ntp) hydrolysis by abce1 (which is stimulated by post-tcs) is required for its recycling activity. Importantly, abce1 dissociates only post-tcs obtained with erf1/erf3 (or erf1 alone), but not post-tcs obtained with puromycin in erf1's absence.
Reader-Students; Note, Note,Note; the following problems- Short form vs exapanded forms;
trna=tRNA, mrna=mRNA, abc= Atp Binding Cassette. pcs= post termination complex. Atp= ATP,gtp=GTP,utp=UTP,ctp=CTP, ntp=Nucleotide terminal complex, ntp=Nucleotide triphosphate, post tcs= Post terminl complex, atp, ,gtp, utp, ctp==Aenosine, Guanosine, Uracil, cytosine-tri-phospho ntds,
Recycling of eukaryotic post termination ribosomal complexes:
Andrey v. Pisarev1, christopher u.t. hellen1 and tatyana v. Pestova
After translational termination, mrna and p site deacylated trna remain associated with ribosomes in post-termination complexes (post-tcs), which must therefore be recycled by releasing mrna and deacylated trna and by dissociating ribosomes into subunits. Recycling of bacterial post-tcs requires elongation factor ef-g and a ribosome recycling factor rrf. Eukaryotes do not encode a rrf homolog, and their mechanism of ribosomal recycling is unknown. We investigated eukaryotic recycling using post-tcs assembled on a model mrna encoding a tetra peptide followed by a uaa stop codon and report that initiation factors eif3, eif1, eif1a, and eif3j, a loosely associated subunit of eif3, can promote recycling of eukaryotic post-tcs. Eif3 is the principal factor that promotes splitting of post-termination ribosomes into 60s subunits and trna- and mrna-bound 40s subunits. Its activity is enhanced by eifs 3j, 1, and 1a. Eif1 also mediates release of p site trna, whereas eif3j ensures subsequent dissociation of mrna.
Translation termination in eukaryotes: polypeptide release factor erf1 is composed of functionally and structurally distinct domains.
l y frolova, t i merkulova, and l l kisselev
Abstract
Class-1 polypeptide chain release factors (rfs) trigger hydrolysis of peptidyl-trna at the ribosomal peptidyl transferase center mediated by one of the three termination codons. In eukaryotes, apart from catalyzing the translation termination reaction, erf1 binds to and activates another factor, erf3, which is a ribosome-dependent and erf1-dependent gtpase. Because peptidyl-trna hydrolysis and gtp hydrolysis could be uncoupled in vitro, we suggest that the two main functions of erf1 are associated with different domains of the erf1 protein. We show here by deletion analysis that human erf1 is composed of two physically separated and functionally distinct domains. The "core" domain is fully competent in ribosome binding and termination-codon-dependent peptidyl-trna hydrolysis, and encompasses the n-terminal and middle parts of the polypeptide chain. The c-terminal one-third of erf1 binds to erf3 in vivo in the absence of the core domain, but both domains are required to activate erf3 gtpase in the ribosome. The calculated isoelectric points of the core and c domains are 9.74 and 4.23, respectively. This highly uneven charge distribution between the two domains implies that electrostatic interdomain interaction may affect the erf1 binding to the ribosome and erf3, its activity in the termination reaction and activation of erf3 gtpase. The positively charged core of erf1 may interact with negatively charged rrna and peptidyl-trna phosphate backbones at the ribosomal erf1 binding site and exhibit rna-binding ability. The structural and functional dissimilarity of the core and erf3-binding domains implies that evolutionarily erf1 originated as a product of gene fusion.
Nmd:
In mammalian systems, if a stop codon is generated, during transcription, either due to mutation or transcriptional error; before the normal ter position, the mrna is degraded by what is called nonsense mediated mRNA decay (nmd).� When such stop codons are found, bound ribosomes are released from the template, but a complex of proteins found at the upstream region of exon-exon junction (ejc), get activated and recruits rnase complexes and destroy the mrna from 5�cap onwards, which is called 5�exonuclease degradation.� In some cases mrnas without stop codons are also degraded by process called no-stop codon mediated degradation. In the case of mrnas not having any stop codon translation continues into poly (a) region where it generates poly-lysines.� This leads to stalling of ribosomes.� At this juncture a protein called ski3 (related to erf3) binds to ribosomes, stimulates ribosomal release and recruit 3�>>5�exonuclease for degradation.
���������������������������� ribosomes are targets for several antibiotics
Energy
cost for synthesis of a protein with n amino acids:
|
2n |
|
|
|
1 |
|
|
|
N-1 |
|
|
|
N-1 |
|
|
|
1 |
|
|
|
|
|
|
|
|
|
|
Each atp or gtp cleavage generates ~40 kj/mol.� Each peptide bond costs ~160 kj/mol in the cell, yet an uncatalyzed chemical reaction to form a peptide bond costs only ~20 kj/mol.
Translation in the archaea:
Most archaea live in environments such as hot sulfur springs where they experience temperatures as high as 80 degrees c and a ph of 2. These are called thermoacidophiles. Others live in methane-containing (methanogens) or high salt (extreme halophiles) environments. Archaea are biochemically more like eukaryotes than they are the eubacteria. Example rna pol in archaea is complex like ek. Archaeal ribosomes have a size and composition similar to those of their bacterial counterparts: they contain three ribonucleic acid (rna) molecules, 16s, 23s and 5s rna and 50�70 proteins depending on the species. Gene promoters more or less similar to that of ek.� But archaea like eubacteria contain operons with polycistronic mrnas. In view of the increasing number of similarities between the archaea and the eukaryotes, the term archaea-bacteria is no longer used. All other cellular forms of life (including plants, animals, and fungi) are referred to as eukaryotes.
Similarities and dissimilarities between eubacteria, archaea �������������������� ����������and� eukaryotes
|
Eubacteria |
Archaea |
Eukaryotes |
|
|
Nucleus |
No |
No |
Yes: membrane-bound |
|
Nucleosomes/histones |
No |
Yes |
Yes |
|
Operons/polycistronic mrnas |
Yes |
Yes |
No |
|
Introns |
No |
No |
Yes |
|
Tata box binding protein |
No |
Yes |
Yes |
|
Organelles |
No |
No |
Yes: mitochondria, lysosomes, endoplasmic reticulum etc. |
|
Chromosomes |
One circular |
One circular |
More than one |
|
Rna polymerase |
One (simple) |
More than one (complex) |
More than one (complex) |
|
Protein initiator amino acid |
N-formyl methionine |
Methionine |
Methionine |
|
Protein synthesis sensitivity to diphtheria toxin |
Insensitive |
Sensitive |
Sensitive |
|
Peptidoglycan |
Yes |
No |
No |
|
Protein synthesis |
Of archaea are more similar to those of eukaryotes than eubacteria |
||
In most respects, translation in the archaea more closely resembles the equivalent events in the eukaryotic cytoplasm rather than in bacteria. The one apparent exception is that the archaeal ribosome, at 70s, is comparable in size to the bacterial ribosome and, like bacterial ribosomes, contains 23s, 16s and 5s rrnas. This apparent similarity is illusory because the archaeal rrnas form base-paired secondary structures that are significantly different from the equivalent bacterial structures. The archaeal structures are also different from the eukaryotic versions, but the ribosomal proteins that attach to the rrnas are homologs of the eukaryotic proteins. Archaeal mrnas are capped and polyadenylated, and translation initiation is thought to involve a scanning process similar to that described for eukaryotic mrnas. Archaeal trnas display a few unique features, including the absence of thymine in the so-called tψc arm of the cloverleaf, and the presence at various positions of modified nucleotides not seen in either bacteria or eukaryotes. The methionine carried by the initiator trna is not n-formylated and the initiation and elongation factors resemble the eukaryotic molecules.
The three major divisions of life, archaea, bacteria, and eukaryota (used with permission from the website of the university of california museum of paleontology: http://www.ucmp.berkeley.edu/alllife/threedomains.html).