Protein Synthesis; e � Ribosome-_as_Translation_Machine.doc:
Ribosomal chemistry determines the structure of ribosomes.� Both prokaryotic and eukaryotic ribosomal structure is built more or less on the same molecular architecture.� Though the number of rRNAs and the number of proteins found in each of the subunits may slightly vary, yet their function is more or less same.� Ribosome can be considered as an assemblage of different types of rRNA and variety of proteins; together the complex acts as a dynamic molecular machine, which churns out protein after protein.�� It is well known that RNAs can perform a variety of enzymatic functions.� The structural organization is designed for interaction with variety components, especially for translation.� Ribosomes are dynamic structures, which can undergo subtle conformational changes when factors and other components bind.� It has a highly flexible structural form, with many domains of rRNA exposed at various regions and sites of ribosome and plays important functions along with riboproteins.� Both rRNA and riboproteins have a role not only in organization of the ribosome, but also have a role in performing variety of functions.� Mutation and RNase digestions of intact ribosomes support the above said view.� It has sites for the following binding or interactive functions:
- For the binding of initiator tRNA and aminoacyl tRNAs at �P� and �A� sites respectively.
- For the binding of mRNA to 30s or 40s ribosome subunit.
- For the binding of a variety of chain initiating factors.
- For the binding of different chain elongation factors.
- For the binding of terminating factors.
- For interacting smaller and larger ribosomal subunits.
- There is tunnel like structure for the exit of nascently synthesized polypeptide chain with N-terminal end in front.
- It has a site for the binding of ribosomes to endoplasmic reticulum to facilitate the transport of protein as it is synthesized.
- Large ribosomal surface has enzymes for peptide bond formation.
- Large ribosomal surface also contains discharged tRNA exit site.
- It has binding site for EFG-GTP/ eFG-GTP,
- It has a binding site for ribosome remodeling factor.
- Small ribosomal subunit has an additional site for the entry of aminoacyl tRNA, before it enters into �A� site.
- Some ribosomal proteins are involved in activating stringent responsive reactions.
A variety of techniques and methods have been employed to locate sites and the regions which perform certain functions, the techniques are affinity labeling, kethoxal modification of guanines in single stranded RNAs, RNase digestion, protease digestion and neutron scattering techniques.� Here prokaryotic ribosomal components are taken for description.
� The S1 protein, which is also called as split protein and it is one of the largest riboproteins; structurally exhibits elongated form. It is required for the binding of mRNA and it is also required for the mRNA to be in single stranded state.
� S1 is associated with s18 and s21; this complex is involved in initiator tRNA binding to the AUG of mRNA.� This complex is located in the vicinity of the cleft of the small ribosomal subunit.� The 3� end of 16s rRNA is located at this region.
� The 3� end of the 16s rRNA is involved in the binding of mRNA through a sequence called Shine-Dalgarno located in the leader region of the mRNA.
� IF3 binds to this site in the cleft region.
� IF3 cross-links to the 3� end of 16s rRNA.
� IF3 stabilizes mRNA and 30s ribosome binding, which is slightly displaced when 50 s ribosomal subunit binds.
� IF3 activates 30s ribosomes and prevents the binding of 50; it has similar effect on large ribosomal subunits.
� The 16s rRNA at 791 ntd region has a loop, which facilitates the binding of 50s ribosome, where 16s rRNA and 23s rRNA interact and base pair with each other to form 30s and 50s subunit complex of 70s.� The binding of IF3 factor prevents this interaction.
� The 16s rRNA at 1392-1407 region, which region is also called 1400 region, is actually the part of �A� site, which interacts with aminoacyl tRNA complexed with EF-Tu.GTP.
� The 5s rRNA associated with L5, L8, L25, also bind to 23s rRNA.� This complex is located near �A� and �P� sites.
� The peptidyl transferase is located in the central protuberance region, to which site Puromycin binds.
� Polypeptide tunnel is located at 150^o from peptidyl transferase region.
� The L7 and L12 complex is located in the stalk region of the large subunit and possess GTPase activity or function.
� EF-G site is located in 50s subunit and it is very close to s12, also EF-G will be placed at the interface between subunits.
� EF-Tu is located at 30s at 2660 loop of 16s rRNA.
� EF-G and EF-Tu are inter-related, but the binding of them is mutually exclusive.� This location has a counterpart in eukaryotic ribosome, which is the target for plant lectins which inhibit elongation, and recin depurinates rRNA.
� The distance between peptidyl tRNA and aa-tRNA is less than 10 A^o.��
� Closer positioning of the reacting amino acids is very essential for any covalent bond formation.� Though the diameter of tRNAs is 20A^o, the positioning of adjacent tRNA is about 3.4 A^o.
� When the L-shaped tRNAs with their respective amino acids are placed in their respective positions, conformational changes in the ribosomes bring in amino acids within the bonding distances.
� While translocating, ribosomes are guided along mRNA in a stepwise fashion and GTP hydrolysis provides the energy for their movements.
Which of them is involved in translocation as a motor protein?
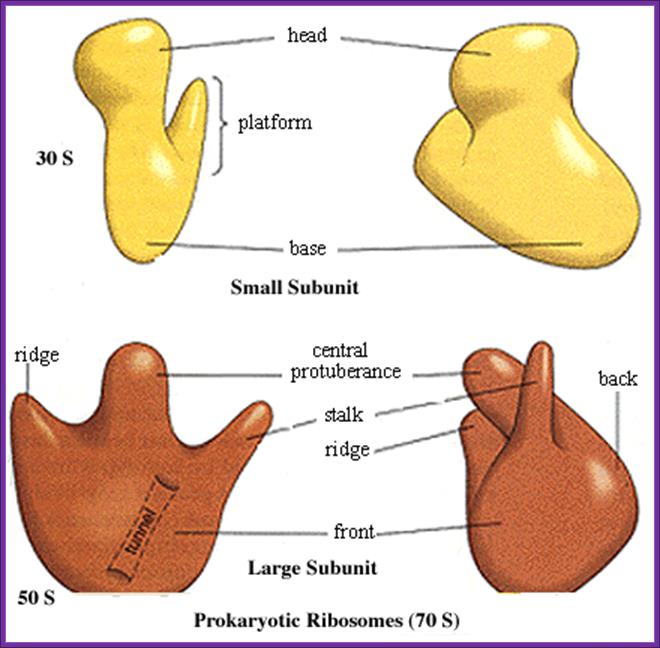
A general view of small and large subunits; http://ec.asm.org/
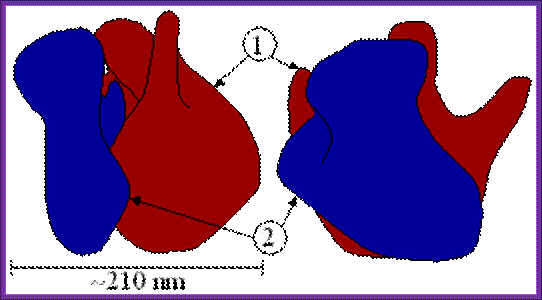
Another view One Bind to the other;
Ribosome
structure indicating small subunit (A) and large subunit (B). Side and front
view.
(1) Head. (2) Platform. (3) Base. (4) Ridge.
(5) Central protuberance. (6) Back. (7) Stalk. (8) Front. Large (1) and small (2) subunit fit
together; http://www.newworldencyclopedia.org/
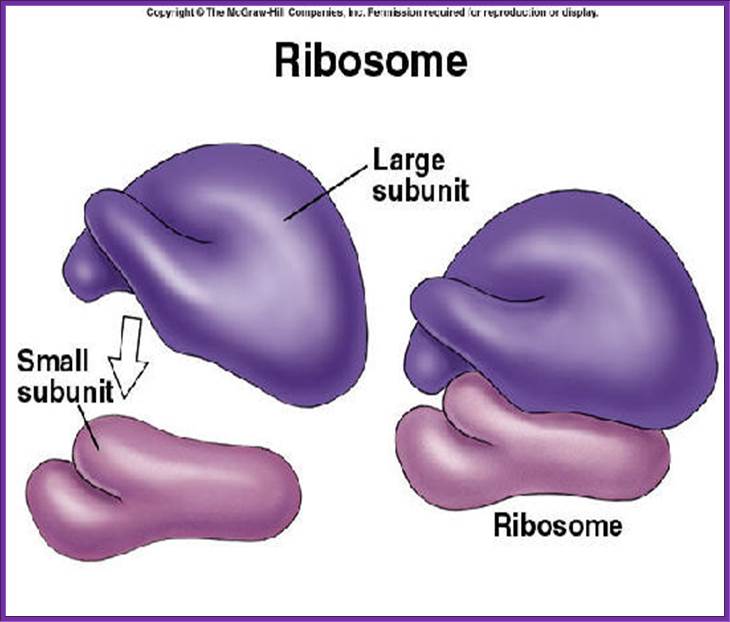
Large subunit on the top of the small subunit; look at the finger like projections; https://socratic.org
Ribosome at the best; https://ajweinmann.wordpress.com
Another 3-D view of crystalkine structure; http://www.brighthub.com/science
�����������������������������������������������
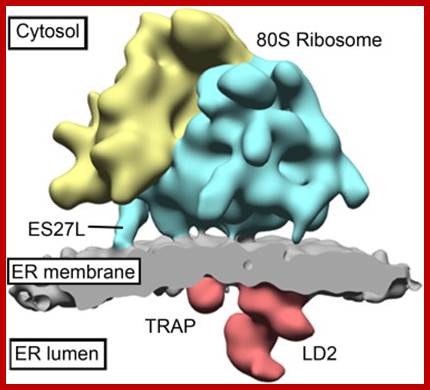
Structure and 3D Arrangement of Endoplasmic Reticulum Membrane-Associated Ribosomes;� Subtomogram average of the canine ER-associated ribosome in situ at 31 � resolution; Large subunit rRNA ES27L is in direct contact with the ER membrane; Sec61, TRAP, and potentially OST and the SP complex are resolved; ER-associated ribosomes adopt a preferred arrangement, likely polyribosome specific�; In eukaryotic cells, cotranslational protein translocation across the endoplasmic reticulum (ER) membrane requires an elaborate macromolecular machinery. While structural details of ribosomes bound to purified and solubilized constituents of the translocon have been elucidated in recent years, little structural knowledge of ribosomes bound to the complete ER protein translocation machinery in a native membrane environment exists. Here, we used cryoelectron tomography to provide a three-dimensional reconstruction of 80S ribosomes attached to functional canine pancreatic ER microsomes in situ. In the resulting subtomogram average at 31 � resolution, we observe direct contact of ribosomal expansion segment ES27L and the membrane and distinguish several membrane-embedded and lumenal complexes, including Sec61, the TRAP complex and another large complex protruding 90 � into the lumen. Membrane-associated ribosomes adopt a preferred three-dimensional arrangement that is likely specific for ER-associated polyribosomes and may explain the high translation efficiency of ER-associated ribosomes compared to their cytosolic counterparts; Stefan Pfeffer et al; http://www.cell.com
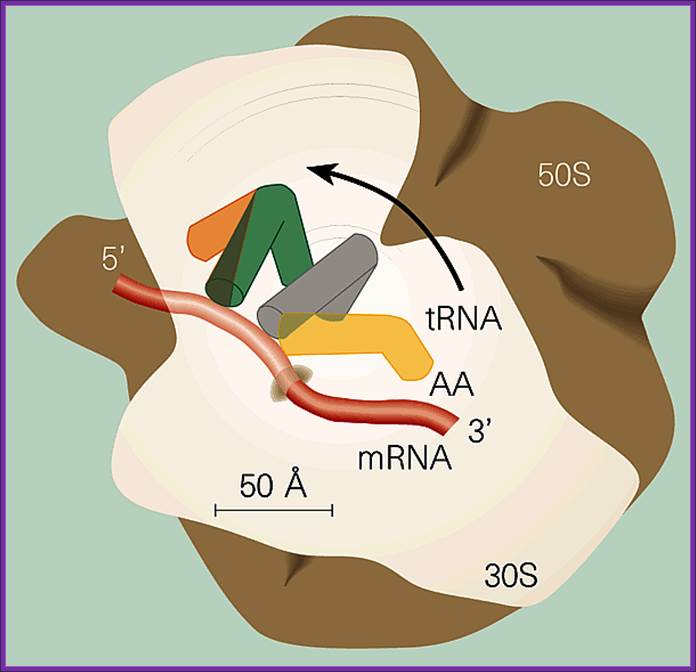
A general view small and large subunits bound to each other with mRNA and tRNAs; Figure 1: The ribosome, showing some of the states through which the tRNA-mRNA complex moves between the ribosomal subunits.Mechanics of the ribosome; Electron-density maps of a cell's protein-producing machinery � the ribosome � have been partially solved. This breakthrough shows that high-resolution structures of the whole ribosome will soon be available. This provides electron-density maps of the two ribosomal subunits, obtained by X-ray diffraction at resolutions of 5.5 � and 5 �, respectively. Roger Garrett;� https://www.nature.com
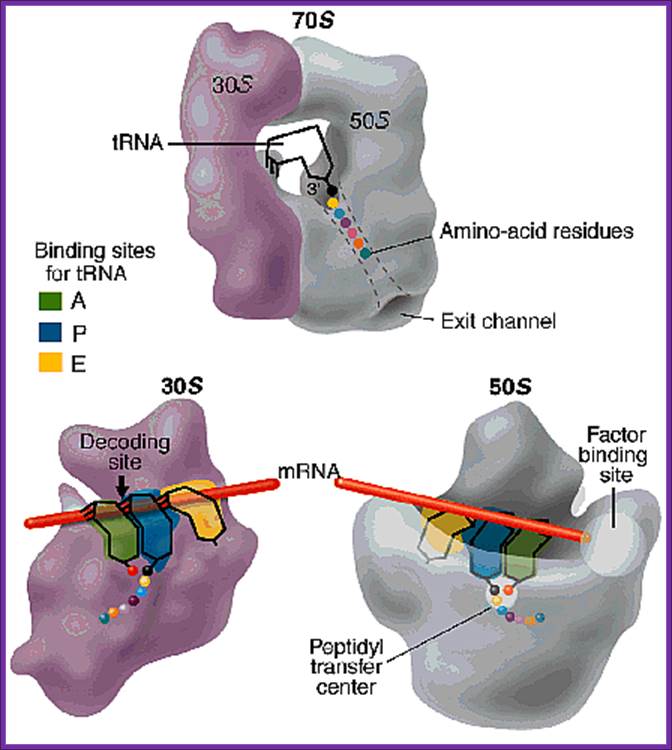
Positioning of the mRNA in terms of tRNAs from A site to P site to E site; www.,sule-gratis.blogspot.com
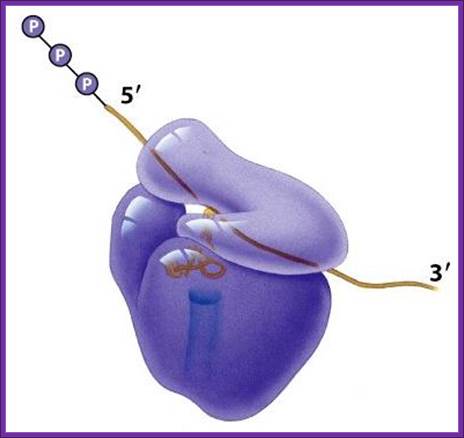
Translation of mRNA is from 5� end of mRNA to 3� end; mRNA is threaded though the groove found in small ribosomal subunit.
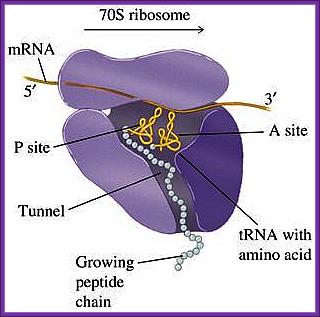
www.blueprintsforliving.com
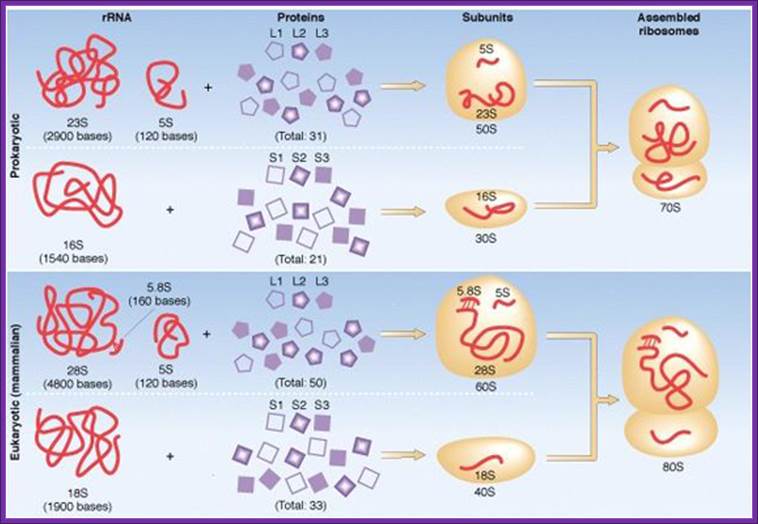
Composition of ribosomal components; http://www.tutorvista.com/
----o---