Amino-acyl tRNA Synthetases:
- Identification of individual distinct tRNA and specific amino acid is performed by specific enzymes.
- Aminoacyl tRNA synthetase is the enzyme that catalyses the addition of amino acids to the 3� end of tRNAs, but amino acids added to each tRNAs is specific.
- Aminoacyl tRNA synthetases are capable of distinguishing individual species of tRNAs and individual species of amino acids.� This absolute specificity of the enzyme stems from individual protein structure, specific binding sites for amino acid and tRNA and catalytic site endowed within the enzyme.�
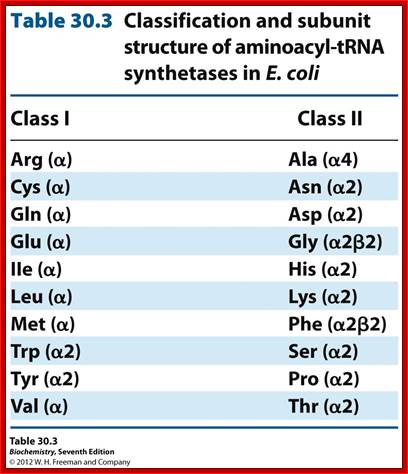
- Basically, aminoacyl synthases are classified into two classes, based on to what 2� or 3� OH group of ribose the amino acid is added.
http://oregonstate.edu/
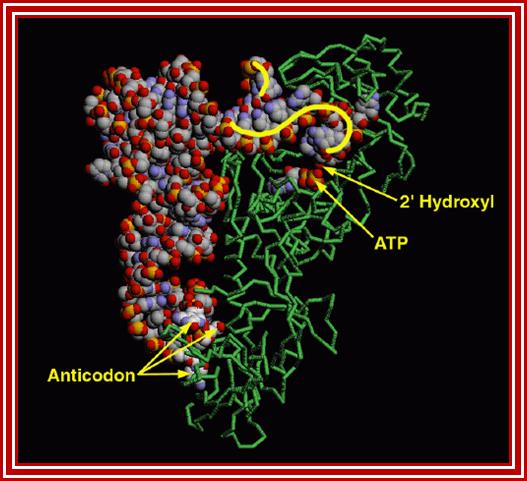
The figure shows the 3-D view of the amino acyl-tRNA synthetase; http://www.rcsb.org/pdb
Important Features of Aminoacyl tRNA Synthases:
http://oregonstate.edu/
|
Class-I, add a.a to 2�OH group of ribose |
Subunits |
Class-II, add a.a to 3�OH group of ribose
|
Subunits |
|
Tryptophan |
a 2? |
--- |
--- |
|
Glutamate |
a 1 |
Glycine |
a2 b2 |
|
Glutamine |
a 1 |
Alanine |
a 4 |
|
Arginine |
a 1 |
Proline |
a 2 |
|
Valine |
a 1 |
Serine |
a 2 |
|
Isoleucine |
a 1 |
Aspartate |
a 2? |
|
Leucine |
a 1 |
Asparagine |
a 2 |
|
Methionine |
a 2 |
Histidine |
a 2 |
|
Tyrosine |
a 2? |
Lysine |
a 2 |
|
|
|
|
|
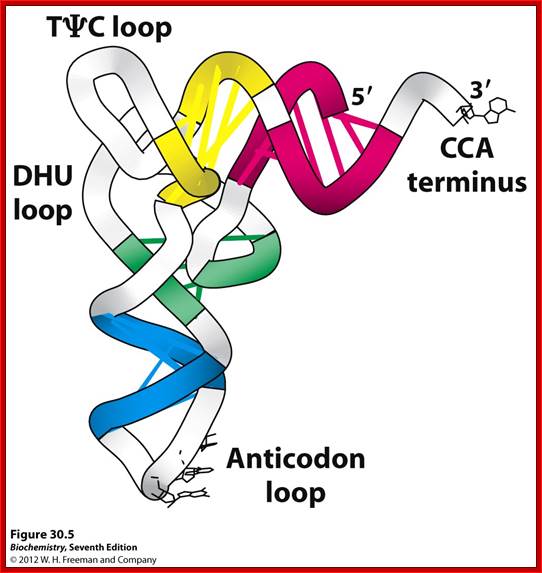
Basically, all tRNAs have distinct 3D structure; amino acids are covalently linked at 5� end to 3�OH of ribose of Adenine, 1-2 nucleotides behind CCA and the last nucleotide at 3� end is A to which amino acids are covalently linked. �All tRNAs have the same ends i.e CCA, but the structure of the tRNA and the aminoacyl-transferase differentiate and determine which amino acid to added to which tRNA.
- It is this property that differentiates the general genetic code from the second genetic code.
- The enzyme has three sites, one for the binding of ATP to activate the enzyme; second has the site for a specific amino acid and the third site is for specific tRNA, it is the two last features are deemed as second half or second genetic code.
- The enzyme, endowed with these features, is said to perform the function of second half of the genetic code.
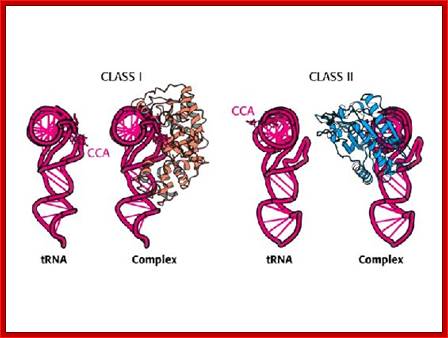
The above diagram shows two types of a.a-tRNA synthetases. BiochemVII edition; http://oregonstate.edu/
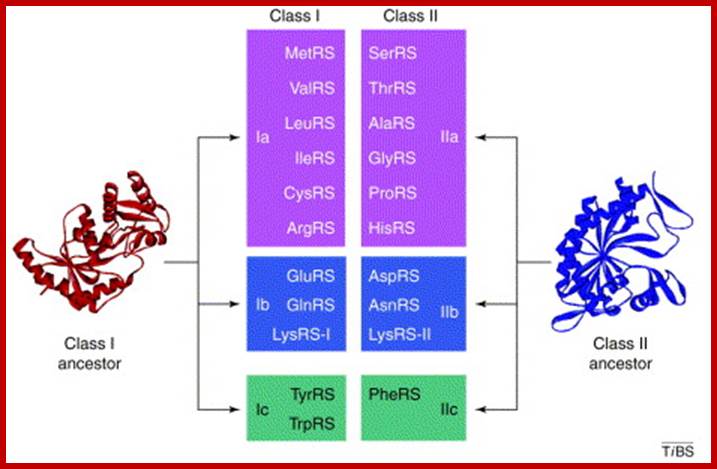
Aminoacylation of tRNAs, each of them are catalyzed by specific aminoacyl-tRNA synthetases, �The enzymes are divided into two classes on the basis of the architectures of their active sites. Members of the two classes also differ in that they bind opposite sides of the tRNA acceptor stem. Importantly, specific pairs of synthetases � one from each class � can be docked simultaneously onto the acceptor stem. This article relates these specific pairings to the organization of the table of codons that defines the universal genetic code. Lluı́s Ribas de Pouplana,Paul Schimmel; ;http://www.cell.com
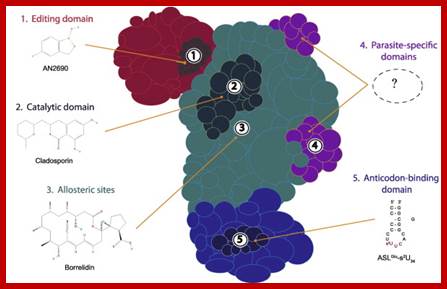
Schematic representation of an aminoacyl-tRNA synthetase. Various aaRS domains are illustrated: the editing domain (red); catalytic domain (cyan); anticodon-binding domain (indigo); and parasite-specific domains (purple). Possible sites of interaction between aaRS and compound (with existing examples) are indicated by numbers: editing site (1); active site (2); allosteric sites (3); parasite-specific domains (4); and anticodon-binding site (5). https://www.researchgate.net
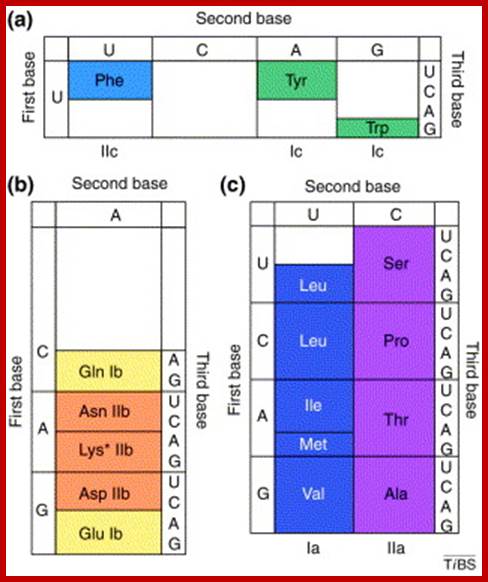
Aminoacyl-tRNA synthetases: potential markers of genetic code development; Aminoacylation of tRNAs, catalyzed by 20 aminoacyl-tRNA synthetases, is responsible for establishing the genetic code. The enzymes are divided into two classes on the basis of the architectures of their active sites. Members of the two classes also differ in that they bind opposite sides of the tRNA acceptor stem. Importantly, specific pairs of synthetases � one from each class � can be docked simultaneously onto the acceptor stem. This article relates these specific pairings to the organization of the table of codons that defines the universal genetic code. Lluı́s Ribas de Pouplana and Paul Schimmel; http://www.cell.com
Mechanism of Amino-Acylation:
The tRNA-aminoacyl synthetase performs admittance, scrutiny and proof reading at post binding stage (kinetic proof reading).�
Binding of amino acid to a.a acyl synthetase is determined by their specific complementary sites. Binding of a specific tRNA to a.a acyl tRNA is also based on their complementary surfaces. While selecting an amino acid hardly it makes mistakes, probably one in ten thousand to hundred thousand, while selecting tRNA, if a mistake has occurred it is corrected by cognate tRNA binding, it also performs what is called chemical proof reading i.e. after charging; if found wrong, it is hydrolyzed and removed.
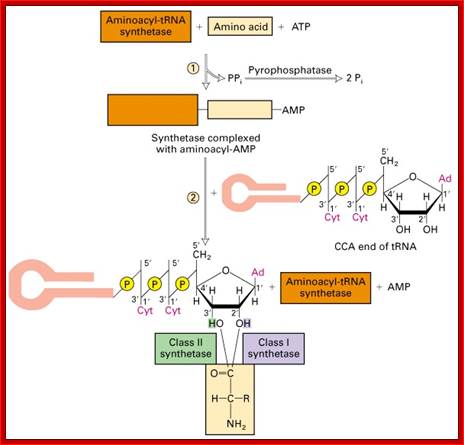
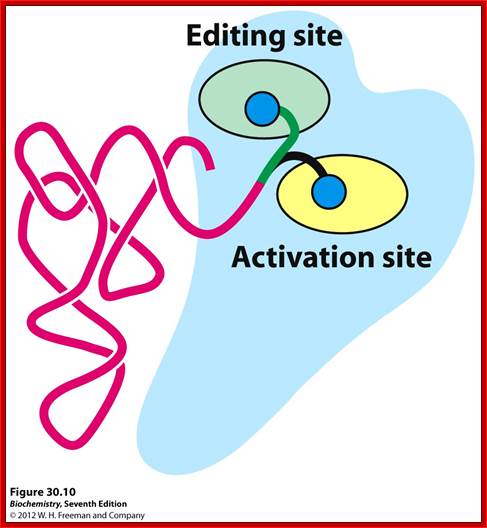
a.a-acylsynthetase-Threonyl-tRNAsynthetase (editing/activation sites); http://oregonstate.edu.
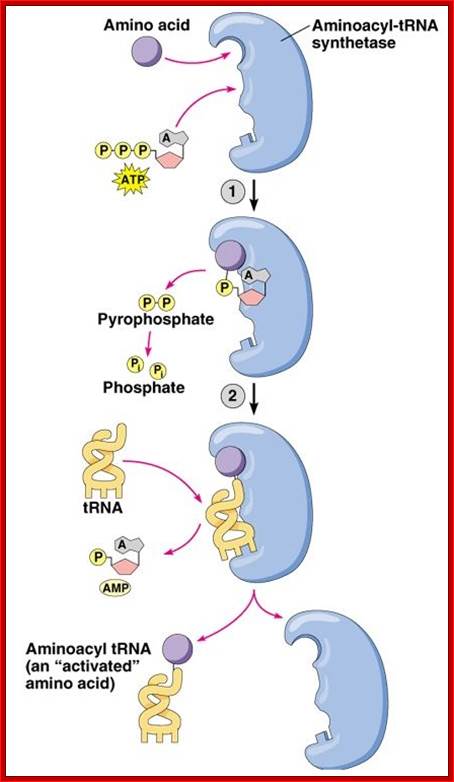
The aminoacylation reaction is shown here as a two-step process, both parts of which occur on the synthetase. All tRNAs have the same sequence CCA at their 3' ends, and this is where the amino acid is added. The amino acid must be 'activated' and ATP provides the activation energy. A complex of AMP-aa forms as an intermediate and the released PPi (pyrophosphate) is hydrolyzed by the enzyme pyrophosphatase with the release of energy to help make the overall reaction pathway more thermodynamically favorable. The aa is then transferred from its AMP carrier to the 3' terminal adenosine. Synthetases are divided into two classes depending on whether they add the amino acid to the 2' hydroxyl group (Class I) or the 3' hydroxyl group (Class II) of the ribose moiety of adenosine.
Three steps in amino acylation of tRNA:
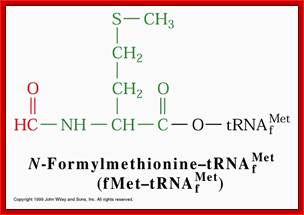
http://www.slideshare.net/ and ; http://web.uconn.edu/
http://urei.bio.uci.edu
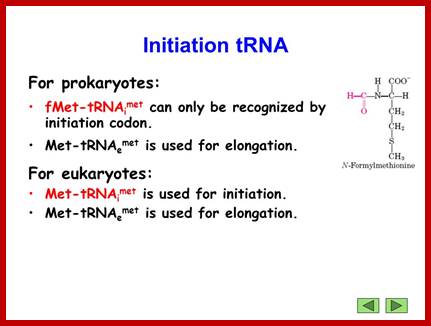
http://slideplayer.com
- First, the enzyme is activated by ATP, to form of enzyme-AMP complex. �The energy of pyrophosphate released is used for activation of the enzyme and induces conformational change for complementary but correct binding sites (active site formation); one for specific amino acid and the other for specific tRNA.�
The binding of amino acid directs its COO- group towards the tRNA site in such a way the COO^- group of the amino acid is positioned close to the 3� or 2� end of �Adenine� of tRNA.� Binding of both substrates induces the enzyme to perform catalytic activity, where carboxyl group of amino acid is covalently added to 2� or 3� OH group of ribose moiety of the terminal Adenine nucleotide, as carbonyl bond which is a high energy bond.
- It is at the time of binding of its substrates the enzymes perform proof reading function; for that matter all enzymes are endowed with such abilities. One class of tRNAs have amino acids linked to 2�OH and the second class of tRNA have amino acids bonded to 3� OH group; such binding does not affect the transfer of amino acid at the time of peptide bond formation and their positions can always exchange. The carbonyl bond thus formed conserves energy; hence it is an energy rich bond (-O=C~O).
The sites or the domains of tRNA that bind to enzymes sites are not yet fully understood.� Based on cross linking studies it is discerned that either acceptor end or the anti-codon region with its geometric dispositions of their nucleotides or both regions can bind to enzyme�s active site.� The DHU loop with species variation in sequence may have an important role in recognition and binding to enzyme surface.
- Aminoacyl tRNA synthases have varying molecular mass ranging from 40kd to 100 KD or more.� Some of them are monomers, made up of only one subunit; some are dimers.� Others may be tetramers with 4 subunits.
From a variety of sources 100 or more of these enzymes have been studied.� The over all size of enzymes is very large in comparison to that of substrates. The enzyme endowed with second genetic code for it has to distinguish which amino acid to pick and to which tRNA it has to add.�� The tRNA are distinguished for specific amino acids and specific anti codons.