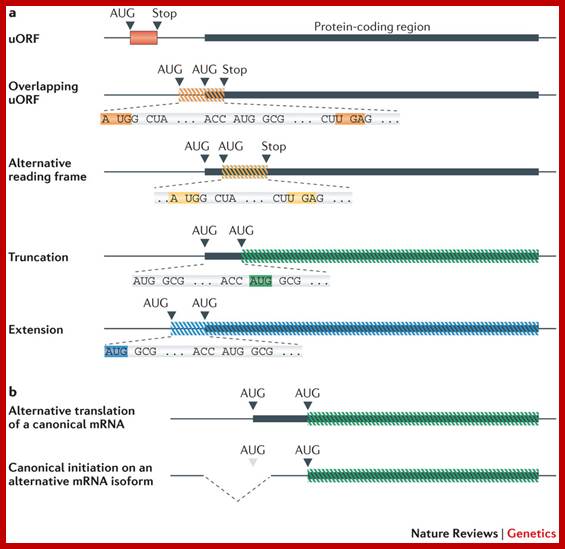
Anticodons and Wobble Base Pairing:
- The total number of codons of universal codon directory is 64, of which UGA, UAG and UAA are terminator or nonsense codons and AUG is an initiator codon, rarely GUG can act as the initiator codon.
- The actual number of sense codons for amino acids is 61 and the other 3 are terminator codons. One or two of the sense codons, AUG and GUG perform initiator function.
- Some of the codons are redundant or degenerate or two or more different codons code for the same amino acid. This is due to third base degeneracy and it is an acquired genetic character during the course of evolution.
- The tRNAs have anticodons to read or decipher the codons.
- To match nucleotide-by-nucleotide, employing complementary base pairing, there should be 61 different tRNA with different anti codon nucleotide sequences, but the total number of tRNAs found in any system is less than 61 and in most cases, it is 22 to 31 maximum. If that is the case how on earth the tRNAs can match 61 codons.
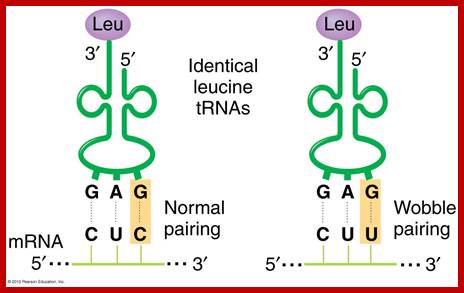
- This paradox is solved by what is famously called wobble base pairing proposed by F.C. Crick. In this, the anticodon 5’ end base in tRNA has the ability to pair with more than one base found at the 3rd base at 3’ end of the codon (mRNA).
- Orientation of codons and anti codons is anti parallel. The third base in the codon base pairs with the first base of the anti codon.
- The wobble base pairing is an innovative mechanism acquired during the course of evolution and it enhances the efficiency of translation, where a smaller number of tRNAs, 20-30 match 61 codons. This mode of base pairing does not impair or violate the 64 codons’ directory. The efficiency is furthered by many codons having 3rd base redundancy. Inosine in the first base can recognize U and C of the codon in the third position.
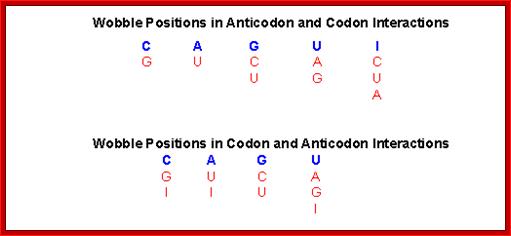
Anti codon 3’ <----5’:
3’ XXC XXA XXU XXU XXG XXG XXI XXI XXI
IIIIII IIIIII IIIIII IIIIII IIIIII IIIII IIIII IIIII IIIII
5’ XXG, XXU, XXA, XXG, XXC, XXU, XXU, XXC, XXA
Codon 5’---->3’:
Wobble Base pairing Between Anti Codon 5’ Position: Codons’3’ Position:
AntiCodon 5’ = 3’ Codon
C = G
A = U
U = A
U = G
G = C
G = U
I = U
I = C
In addition to wobble base pairing, the third base in the codon (in most of the cases) shows degeneracy. What is the advantage of such degeneracy? - Point mutations are absorbed.
- In normal situations anti codons match codons base by base. Occasionally point mutations change the bases, if the bases change the meaning of a codon for an amino acid also changes, then such mutations is called missense mutations.
- If the base changes make amino acid coding combinations into nonsense codons where the codons don’t recognize any anti codons of aa.tRNA. There are no aa-tRNAs with anticodons, which can recognize such codons. Such mutations are called nonsense mutations.
Missense and nonsense mutations are often suppressed by mutations in respective tRNAs; such mutations are called suppressor mutations or intergenic suppressors.
- Most of the times only 3 bases are used as anti codons, but under certain cases four bases are read by tRNAs, which also suppresses point mutations.
- In many cases tRNAs are also involved in frame shift reading frames where tRNA instead of reading just three nucleotides it reads four nucleotides either by retracting one base backwards or one base forward.
- In certain viruses, few tRNAs act as primers for the replication of the viral genomes.
Seleno-Cysteine:
- Seleno-cysteine is newest amino acid added to the existing list of 20 amino acid codon directory. Selenium is a trace element found in several enzymes, such as Formate dehydrogenase and several other enzymes including, seleno-Glutaredoxin, and glutathione peroxidase.. Seven such enzymes that require Sec are identified (Artemy Beiaminov et al)
- Sel-cys-tRNA is unique for it contains 3’ACU5’ as anticodon sequence.
- First the serine is added to serine tRNA by serine-aa-tRNA synthetase; -R=CH2-OH. Then selenium is incorporated into side chain as R containing R= CH2-Se-H, which looks more like a cysteine, so it is called Seleno-cysteine.
- In bacteria, during translation of N-formyl dehydrogenase mRNA fused to upstream of beta-Galactosidase transcript, it requires Sel-cys tRNA to read through UGA (terminator codon) to produce a functional protein. But a change in sequence of UGA to UGU or UCA abolishes selenium requirement.
--------A C U---=---5’ sel-cys
--U G A----=--3’ mRNA
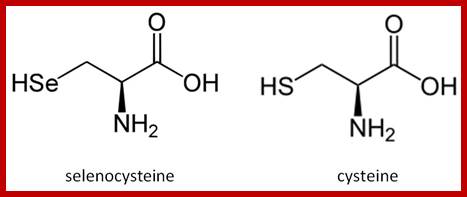
Addition of selenium to the SH group of the R-chain; http://bioinformatica.upf.edu/
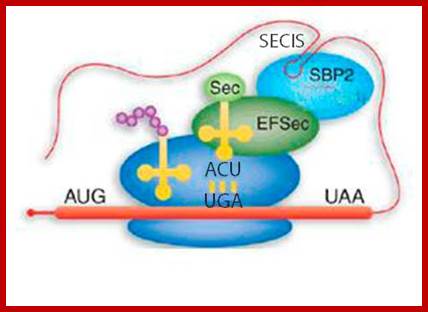
Selenocysteine (Sec) uses the UGA codon and it is read as sel-cysteine; http://bioinformatica.upf.edu/
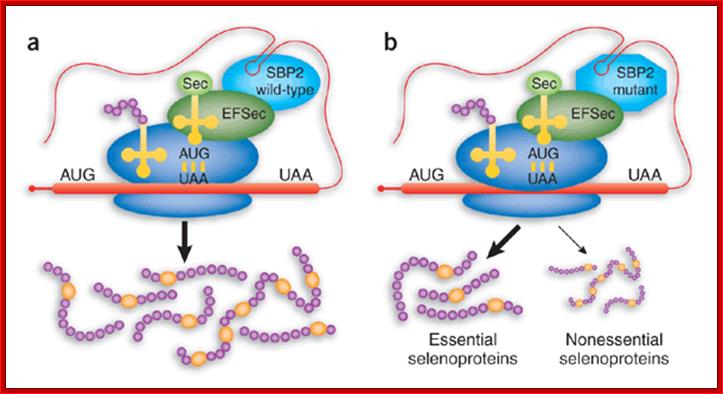
a) The SECIS element in the 3' untranslated region of the mRNA (stem loop in light blue) recruits SBP2 (red), which in turn recruits EFSec (blue) and tRNASec (yellow). The complex interacts at the ribosome to decode UGA as selenocysteine. (b) Mutation of SBP2 results in preservation of the synthesis of essential selenoproteins but a decrease in the synthesis of nonessential ones. Essential proteins include thioredoxin reductase, plasma glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase; nonessential proteins include type 2 iodothyronine deiodinase, selenoprotein P and glutathione peroxidase 1. Sec, selenocysteine. Marla J.Berry; http://www.nature.com/
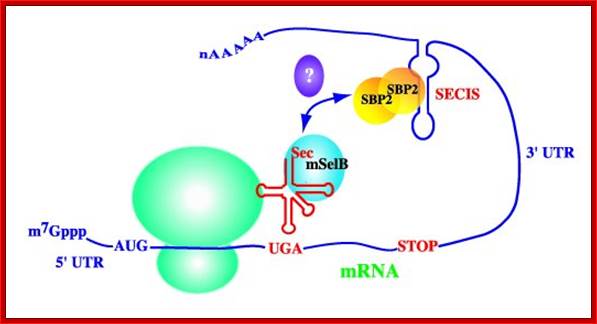
Artemy Beniaminov , Akiko Takeuchi , Laurence Wurth , Christine Allmang; http://www-ibmc.u-strasbg.fr/
The tRNA Sec and a stem-loop structure in the 3'UTR of the mRNA of selenoproteins (the SECIS) play a key role in reprogramming the codon UGA Sec. Molecular biologists have previously proposed structural models for tRNA SecSECIS pattern and isolated and functionally characterized the elongation factor of the mammalian EFSEC specialized dictionaries and the SBP2 protein binding to SECIS element. To better understand the principles of interaction SBP2-SECIS RNA and the function of SBP2, which are at the heart of the mechanism of synthesis of selenoproteins, the resolution of the crystal structure of this complex was undertaken in collaboration with the team Philippe Dumas. In addition, they devoted great efforts to research and functional characterization of proteins interacting with SBP2.
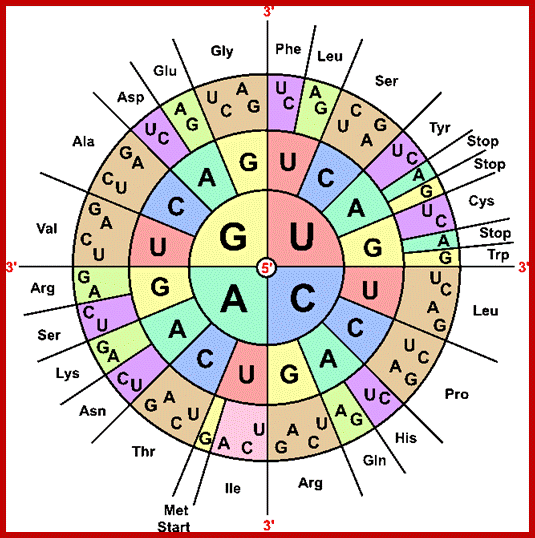
Alejandro Arroyo saved to Genetic: Allen Crowley saved to SciFi;
Codon table; codon: three adjacent nucleotides (triplet) in mRNA that base-pair with the corresponding anticodon of tRNA molecule that carries a particular amino acid, hence, specifying the type and sequence of amino acids for protein synthesis; there are 64 codons but only 20 amino acids so it is highly redundant; How many bp's for 120 amino acids? 3 x 120 = 360; https://iaincarstairs.files.wordpress.com
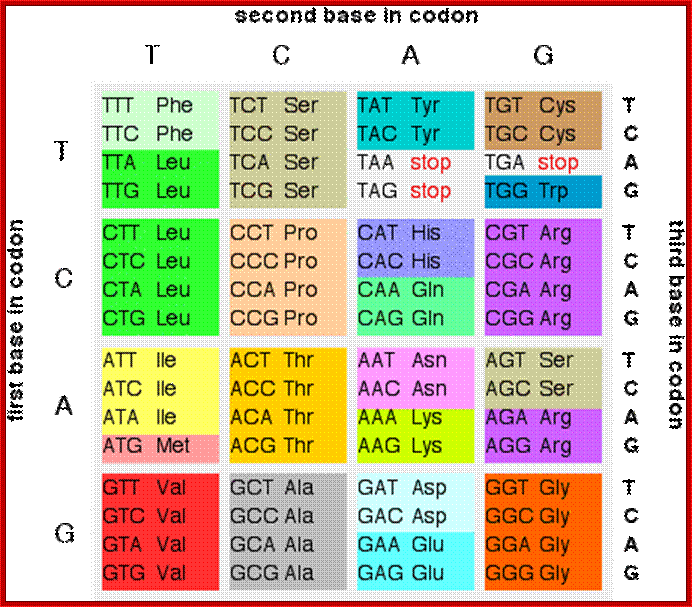
http://www.chemguide.co.uk/
Codon-Reverse Codon Symmetry
A reverse codon of any codon XYZ is defined as ZYX, where X,Y,Z can be any base. The arrows in table represent pairs of “codon - reverse codons”. For instance, the reverse codon of CCU (Pro) is UCC (Ser). There exist 15 different amino acids in the rows 000, 101, 010 and 111 where the codon is reverse to itself, e.g. Lys (AAA), Tyr (UAU). Considering codons and their reverse codons we report three observations.
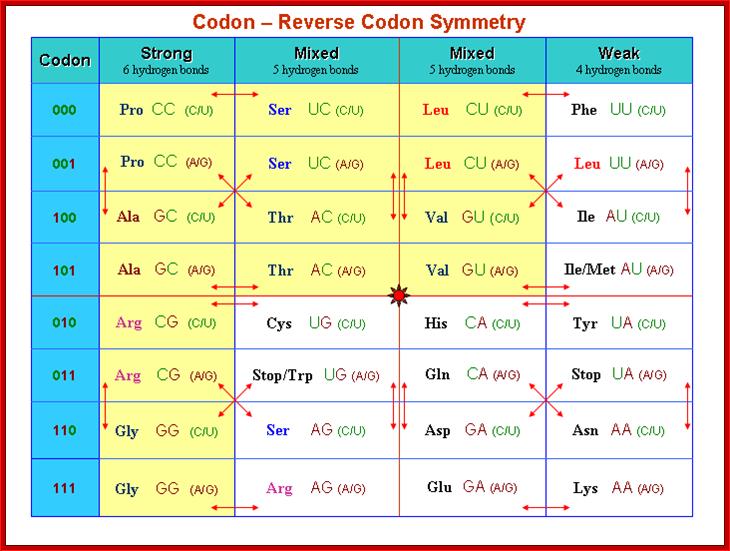
http://www.keyword-suggestions.com/;http://www.imb-jena.de/
Codon-Reverse Codon Symmetry:
1. Table can be divided into four blocks (codon - reverse codon groups) of the same size, for instance the upper left block with Pro (P), Ser (S), Ala(A) and Thr (T). Each block shows the same arrow pattern. All strongly evolutionary conserved groups of amino acids (Thompson et al., 1994) are subsets of exactly one codon - reverse codon group, e.g. the MILV amino acids belong to the upper right block in the table. The other conserved strong groups belonging to one block are STA, NEQK, NHQK, NDEQ, QHRK, MILF, HY. The only exception is FYW.
2. Genetists studied all known tRNA genes of 104 different organisms (Sprinzl et al., 1999). It is now known that the STOP codons do not have any tRNA. They have found that there are also no tRNA genes containing anticodons reverse to the STOP anticodons (ACT, ATC and ATT in Table 3). This is true for archaea (16), bacteria (81) and most eukaryotes (7). The only exception is H. sapiens, possessing one tRNAAsn gene with the anticodon ATT, but humans also have three different possible suppressor tRNA genes (Lowe and Eddy, 1997).
3. There are some codon - reverse codon pairs where there only exist tRNAs for one codon, but no tRNA for the reverse codon, e.g. all 104 studied organisms have at least one tRNA for Tyr with anticodon GTA (some organisms, for instance H. sapiens, have different tRNAs with the same anticodon, altogether there are 189 different tRNAs with the anticodon GTA in the 104 species, but no organism has a tRNA for His with anticodon ATG. Table 3 lists different codon - reverse codon pairs. It is interesting to note that in the whole super-kingdom bacteria the number of tRNA genes for the reverse part of table 3 is always zero, the few exceptions are only in eukaryotes and archaea. Another unique property of all bacteria is that there is no tRNA with anticodons for the self-reverse codons UUU (Phe), UCU (Ser), UGU (Cys) and UAU (Tyr). Generally, table 3 shows that anticodons with an A at the third position are strongly preferred, whereas A** anticodons are significantly suppressed, http://www.imb-jena.de/
Codon-Anticodon-Wobble hypothesis’
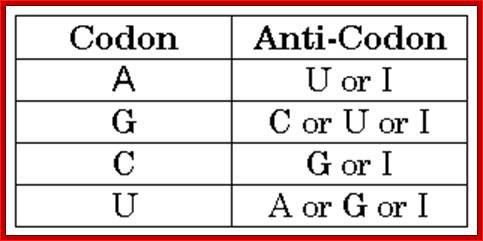
http://www.codefun.com/
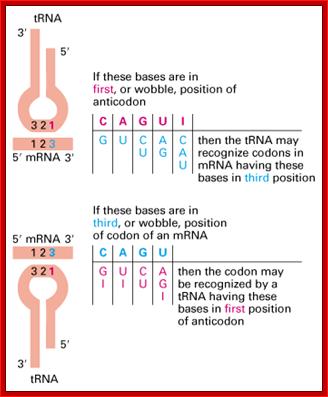
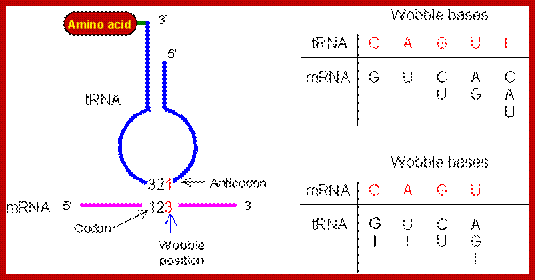
Wobble between the last alphabet of Codon (mRNA) and the first alphabet of the anticodon (tRNA) http://www.web-books.com/MoBio/Free; www.keyword-suggestions.com
The medical biochemistrypage.org; Translation of Proteins; http://themedicalbiochemistrypage.org/
Biochemistry: http://biology.stackexchange.com/
Extended anti-codon: frame shift reading frame:
This was proposed by S.H Kim (1973). In this model the efficiency of anticodon in the recognition of codon is determined by the relationship between anticodon loop and the proximal anticodon stem sequence.
Cardinal Nucleotides
|
|
A |
U |
G |
C |
|
VII |
------- |
-------- |
Py/pu |
--------- |
|
VI |
GC / GC |
--------- |
PY / PU |
GC / CG |
|
V |
PU / PY |
---- |
-------- |
---- |
|
IV |
GC |
GC / CG |
--- |
CG |
|
III |
AU /UA |
AU / CG |
--- |
CG |
|
II |
A |
A |
A / U |
A / C |
|
I |
Me2I6A |
I6A |
M2A/ G |
Am2A/ m6A |
|
|
|
|
|
|
Extended reading frame:
If we take ubiquitin protein when it’s mRNA is translated, the protein produced is longer than the Ubiquitin itself. The human message encodes 80 amino acids beyond ubiquitin’s carboxyl terminus. The additional sequence contains several cysteine residues, which are absent from ubiquitin, and it is extremely basic with 30% of the residues being lysine and arginine. The extended region 10,000 Da may act as binding sites for DNA or RNA.
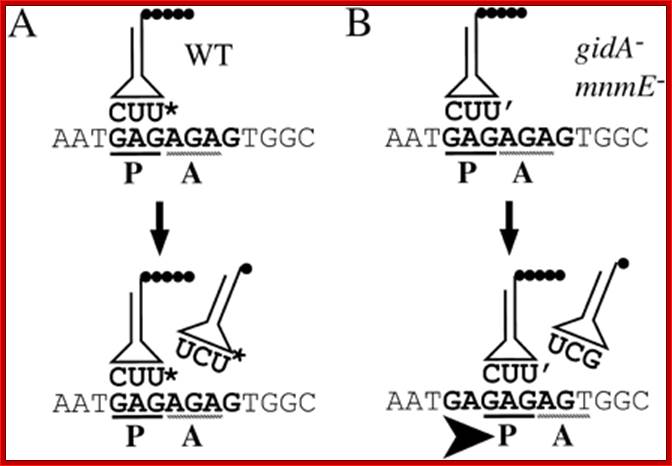
Figure :
Model for the observed +2 frameshift. (A) The normal process of
translation in NECB1 lacZ(+GA)begins with the
peptidyl-tRNA ![]() (U* is the modified uridine at the wobble position, which could be
either mnm5-s2-U or mnm5-U) bound to the mRNA in the P-site of the ribosome.
The empty A-site is normally filled with a tRNA
(U* is the modified uridine at the wobble position, which could be
either mnm5-s2-U or mnm5-U) bound to the mRNA in the P-site of the ribosome.
The empty A-site is normally filled with a tRNA ![]() and the translational complex can proceed to the next step of
elongation by transferring the peptide and translocating the accepted tRNA in
the P-site. (B) In mnmE mutants, and
probably in gidA mutants, the uridine
at the wobble position of the tRNAGlu is hypomodified
(U‘: s2-U or some other hypomodified form of the mnm5-s2-U). This hypomodified
nucleotide is less efficient at pairing with a guanine in the wobble position.
The presence of a rare AGA codon at the A-site results in a pause during the
translation process. Therefore, the conjunction of a less efficient pairing
between the tRNA and the mRNA at the P-site and the presence of a rare codon at
the A-site renders the ribosome prone to shifting. This translation complex
moves to (or possibly scans for) the next GAG cognate codon of the tRNA in the
P-site, which is easy on this repetitive sequence and then resumes translation,
but in a different frame (Damien
Brégeon1,4, Vincent
Colot2,3, Miroslav
Radman1, and François
Taddei1; http://genesdev.cshlp.org/
and the translational complex can proceed to the next step of
elongation by transferring the peptide and translocating the accepted tRNA in
the P-site. (B) In mnmE mutants, and
probably in gidA mutants, the uridine
at the wobble position of the tRNAGlu is hypomodified
(U‘: s2-U or some other hypomodified form of the mnm5-s2-U). This hypomodified
nucleotide is less efficient at pairing with a guanine in the wobble position.
The presence of a rare AGA codon at the A-site results in a pause during the
translation process. Therefore, the conjunction of a less efficient pairing
between the tRNA and the mRNA at the P-site and the presence of a rare codon at
the A-site renders the ribosome prone to shifting. This translation complex
moves to (or possibly scans for) the next GAG cognate codon of the tRNA in the
P-site, which is easy on this repetitive sequence and then resumes translation,
but in a different frame (Damien
Brégeon1,4, Vincent
Colot2,3, Miroslav
Radman1, and François
Taddei1; http://genesdev.cshlp.org/