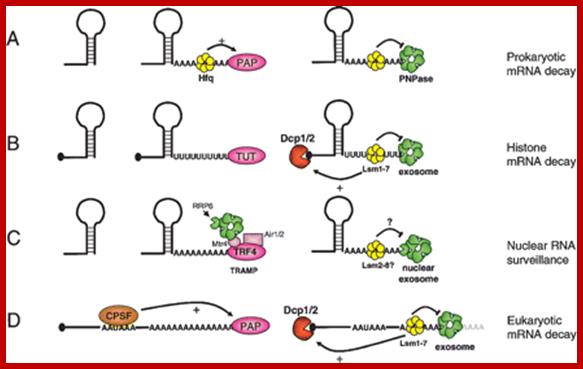
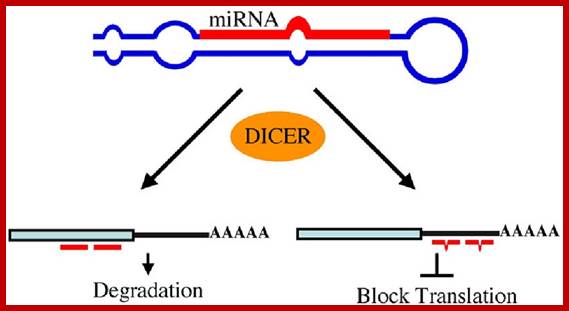
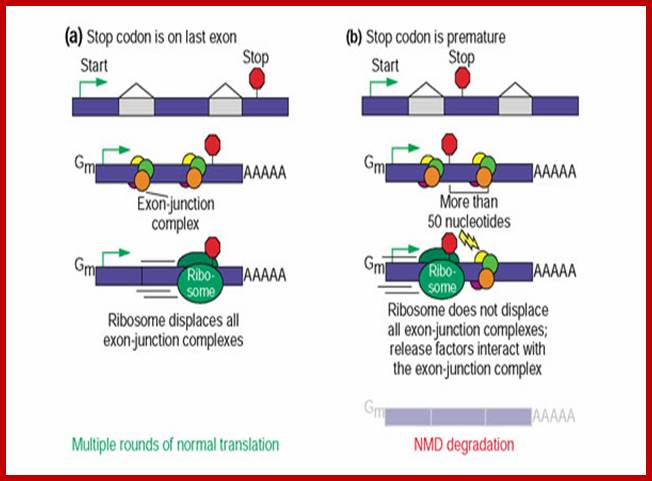
Stability of mRNAs and its Regulation:
�The translation of messenger RNA (mRNA) transcripts, a string of codons into a string of amino acids called polypeptide is a vital part of the central dogma in the �DOMAIN� of molecular biology�. Messenger RNA molecules are, however, prone to a host of fidelity errors which can cause errors in translation into quality of proteins. RNA surveillance mechanisms are methods cells use to assure the quality and fidelity of the mRNA molecules. This is generally achieved through marking aberrant mRNA molecule for degradation by various endogenous and exonucleases.
Surveillance of mRNAs has been well documented in bacteria. In eukaryotes, these mechanisms are known to function in both the nucleus and cytoplasm. �Fidelity checks of mRNA molecules in the nucleus results in the degradation of improperly processed transcripts before export into the cytoplasm. Transcripts are subject to further surveillance again in cytoplasm. Cytoplasmic surveillance mechanisms assess mRNA transcripts for the absence of stop codons or presence of premature stop codons or other insurmountable knots or problems.
Life span of cellular mRNAs is very critical for cellular developmental stages or phases or for a cell to be stable and long lasting in a particular tissue.�
Often the stability of mRNA is measured in terms of chemical half-life and functional half-life. Chemical half-life was determined for each RNA by the following �twofold� algorithm: 1. The earliest time point at which the transcript was detected was used as the baseline abundance. 2. The earliest successive time point for which a twofold decrease was detected was used as the experimental abundance, and the half-life was calculated assuming exponential decay; the chemical half-life of total hoxP mRNA as an example was 8 min. �There is a clear correlation between the functional mRNA half-life and the ribosome spacing in the mRNA region approximately between codon 20 and codon 45. From this finding, we (authors mentioned below) predicted that inserts of slowly translated codons before codon 20 or after codon 45 should shorten or prolong the functional mRNA half-life by altering the ribosome density in the important regions. These predictions were tested on eight new lacZ variants. �It is suggested that that translation-rate-mediated differences in the spacing between ribosomes in this early coding region is a parameter that determines the mRNAs functional half-life (Pedersen M, Nissen S, Mitarai N, Lo Svenningsen S, Sneppen K, Pedersen S.). U Oelmuller et al. , Douglas W. Selinger, Rini Mukherjee Saxena. �
The concentration of any species of RNA in the cell is proportional to its rate of turnover. In general prokaryotic mRNAs have very short half-life of 2 to 5 minutes, for they don�t have protective features that are found in Eukaryote (EK) mRNAs, such as cap at 5�end and poly-A at 3� end.�
In bacterial cells, as they are synthesized they are either subjected to translation. �If the 5� end fails to initiate translation, exonuclease acts upon them. Most of the mRNAs are destined to be degraded, so the half-life is very short, three to five minutes. In bacteria if the first two 5� PPs of the three 5�PPP are removed and then it is acted upon by an endonuclease and then by nucleases from the 3� and 5� ends or and by endonucleases.
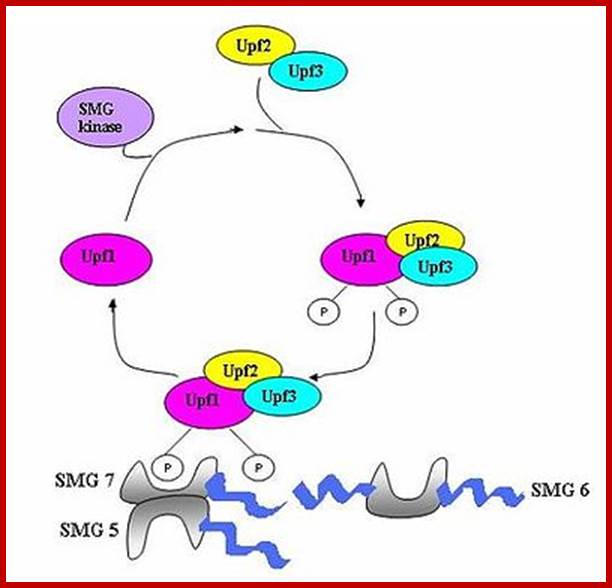
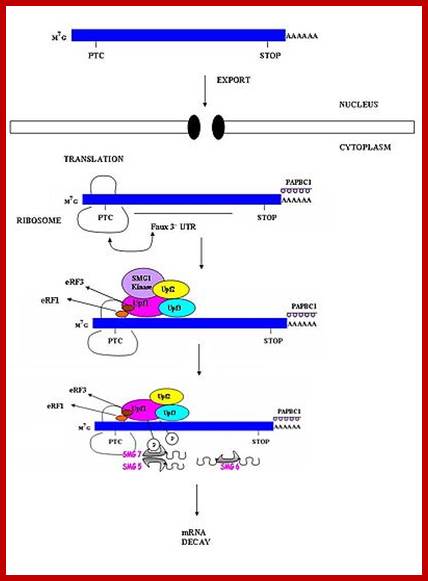
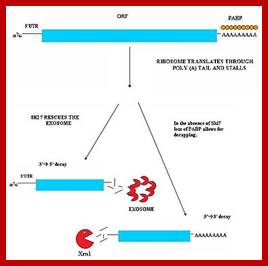
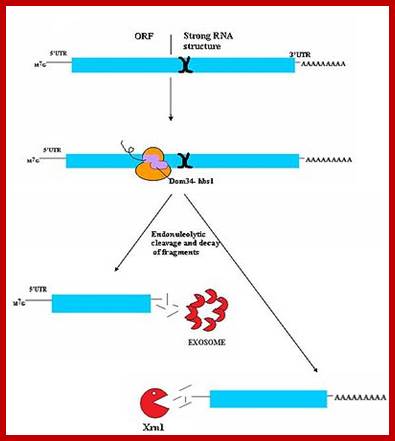
Three surveillance and decay mechanisms are currently known to function within cells: the Nonsense Mediated mRNA decay (NMD); the Nonstop Mediated mRNA decay (NSD); and the No-go mediated mRNA decay (NGD).�
Rel E, REL B and Rel A: Toxin � antitoxin (TA) genes of prokaryotic cells; Prof Kenneth Keiler: Pennsylvania State University.
The figure below shows the components of bacterial toxin�antitoxin gene loci, exemplified by the model system relBE of Escherichia coli. The relBE genes encode RelE toxin and RelB antitoxin. RelB and RelE proteins combine very efficiently and form a non-toxic RelBE complex. Thus, the cells can grow and survive even though the genes express and they form RelBE complex. The RelBE complex binds to DNA in the relBE promoter region (prelB; symbolized by a broken arrow pointing left-ward) and negatively regulates transcription of the relBE genes. Lon is an intracellular protease that degrades RelB antitoxin in response to environmental cues (such as nutrient limitation). Degradation of RelB leads to activation of the RelE toxin. RelE is an enzyme (endonuclease) that cleaves mRNA positioned at the ribosomal A-site. Thus, during nutrient limitations, RelE inhibits translation. In turn, such metabolic regulation of translation increases cell survival during stressful conditions.
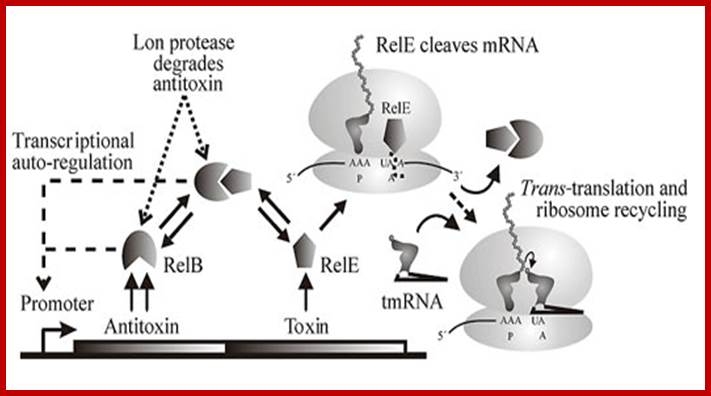
Genetic organization and components of the relBE locus from E. coli: During nutritional stress, Lon protease is activated by an unknown signal and cleaves RelB. In turn, this activates RelE that cleaves mRNAs positioned at the ribosomal A-site. Since RelE acts catalytically, translation can be very efficiently inhibited by RelE; Genetic organization and regulatory components of the relBE Toxin-Antitoxin operon(left), the site of mRNA cleavage by relE(middle), and ribosome rescue by tmRNA after mRNA truncation by relE(right). From The Bacterial Toxin RELE Displays Codon-Specific Cleavage of mRNAs in the Ribosomal A site. http://2009.igem.org/
On the other hand, cut mRNAs has to be rescued and it is done by the way of tmRNA in dormant cells. It does accelerate the rescue efficiency of rel-B. �Overproduction of tmRNA efficiently counteracts rel-E toxicity.
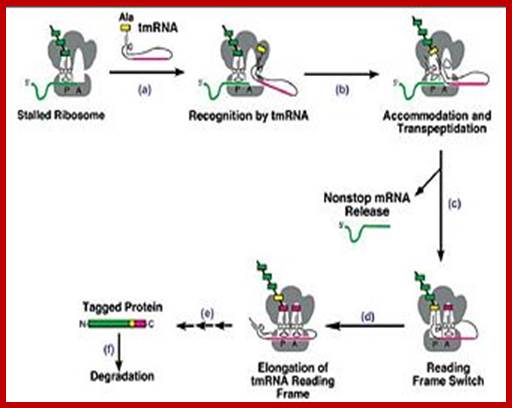
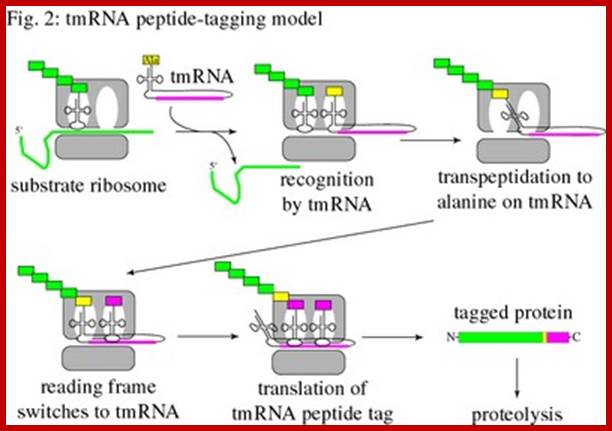
Trans-translation model for tmRNA-mediated protein tagging and ribosome rescue from Trans-Translation: The tmRNA-Mediated Surveillance Mechanism for Ribosome Rescue, Directed Protein Degradation, and Nonstop mRNA Decay. http://2009.igem.org/
The tmRNA structure and the trans-translation pathway: �
The 5� and 3� ends of tmRNA fold into a structure resembling alanyl-tRNA, and the 3� end is charged with alanine by alanyl-tRNA synthetase. However, tmRNA is much larger than a tRNA, and does not contain an anticodon stem-loop structure. Instead, there are several pseudo knots, (depending on the species) and a specialized open reading frame encoding the peptide tag. The tag reading frame has 8-35 codons and ends with a stop codon, but does not have a translation initiation sequence or a typical start codon. Like many RNAs, tmRNA requires protein cofactors for its activity. SmpB is a small protein that binds tightly and specifically to tmRNA, and it is important for tmRNA structure, stability, and activity. The translation elongation factor EF-Tu also binds to tmRNA and promotes productive interaction with the ribosome.
In Caulobacter, a deletion of tmRNA results in a delay in the initiation of DNA replication during the swarmer-to-stalked cell transition. By identifying the substrates of tmRNA and determining how these substrates are responsible for the cell-cycle phenotype, we aim to understand the role of tmRNA in cell-cycle regulation and bacterial differentiation. We have also found that the levels of tmRNA are regulated with respect to the cell cycle by transcription and RNA degradation. By studying the mechanism of regulation of tmRNA activity, we will identify new cell-cycle regulatory factors, and understand how tmRNA is integrated into the Caulobacter regulatory network. �Kenneth Keiler; http://bmb.psu.edu/
The dual functions of tmRNA (Public domain from Wikipedia: tmRNA):
In 1996, Keiler et al. described the ability of 10Sa RNA bind to both 1) free ribosomes from messenger RNA sequences lacking stop codons and 2) add a tag to the aberrant peptide, tagging it for degradation. In 1999, Karzai et al. pinpointed SmpB as a key binding partner to tmRNA in Salmonella enterica mutants that phenocopied tmRNA deletion strains of E. coli.
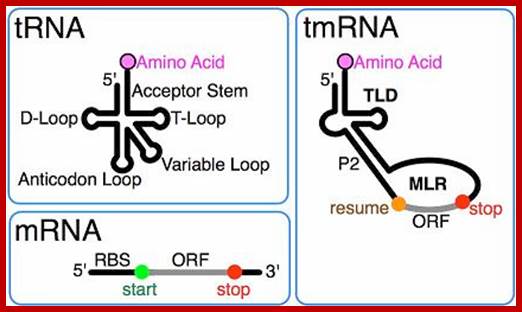
tmRNA combines features of tRNA and mRNA.; http://en.wikipedia.org/
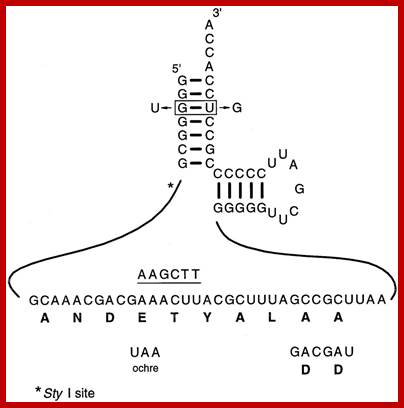
The Tm RNA sequence that is translated is ANDETYALLA; Variants used in determining the properties of gonococcal tmRNA required for N.gonorrhoeae growth. The DD mutation was generated by replacing the two terminal alanine codons with two aspartic acid codons, while the ochre mutation was generated by changing the fourth codon from GAA to the sequence encoding the ochre translation terminator UAA. Both mutants also carry a single base substitution (from AAACTT to AAGCTT) that provides an identifying HindIII site, as indicated by the underlined sequence above the tag sequence. Two changes were made in the sequences encoding the acceptor stem: these result in the change of the G:U base pair required for recognition of alanyl-tRNA synthetase to a U:G base pair. This mutant also carries a single base substitution (from CCTTGG to CGTTGG) that provides an identifying marker by eliminating a StyI site that is only five nucleotides away from the G3 position, indicated by the asterisk. Canhui Huang, et al ; http://emboj.embopress.org/;
Two types of tmRNAs are known: Single-chain tmRNAs and two-piece tmRNAs. In most eubacteria, a single tmRNA molecule is encoded by the ssrA gene. However, in alpha-proteobacteria, beta-proteobacteria and cyanobacteria, the ssrA gene has undergone a circular permutation, leaving the mature 3' end of the RNA upstream of the mature 5' end. As a result, in these bacterial lineages tmRNA is actually formed from two distinct RNA molecules.
Conservation of structure
The tRNA-like domain of tmRNA contains a D-loop, a T-arm and an acceptor stem. As with all tRNA, the 3� end of the acceptor stem of this domain terminates in CCA. In order to be recognized by alanyl-tRNA synthetase, the acceptor stem also contains a G: U wobble base pair. However, the anticodon stem is absent, with a �connecting� structure linking the tRNA-like domain to the rest of the tmRNA in its place. The rest of the tmRNA is comprised of the ORF of the mRNA-like domain, and two to four pseudo knots. Although all known tmRNAs contain pseudo knots, reason for their conservation remains a mystery, as tmRNA mutants where pseudo knots are replaced by simple hairpin structures do not show a significant loss in tmRNA function.
Conservation of function
Though all eubacteria have been found to possess the tmRNA/SmpB system, it appears to play a role of varying importance throughout the kingdom. In some organisms, such as Neisseria gonorrhoeae, if tmRNA is disrupted, the mutants are unable to survive. Yet if the same mutation is present in E. coli, the population only sees a mild decrease in rate of growth. It is likely that organisms that are less severely affected by loss of function in tmRNA/SmpB have developed an alternative though less efficient mechanism for releasing stalled ribosomes.
Biogenesis:
As with pre-tRNAs, pre-tmRNAs are processed by cellular ribonucleases which remove 5� and 3� nucleotides. This results in the formation of a mature tmRNA where 5� and 3� ends fold together, and the 3� hydroxyl group at the terminal CCA of the acceptor arm can then be charged with alanine. In some bacteria, e.g. Bacillus subtilis, the 3� CCA region is added on by tRNA nucleotidyl transferase. In bacterial lineages that have undergone a circular permutation in the tmRNA sequence, an excision event must also occur to form a mature two-piece tmRNA.
Rates of tmRNA biogenesis and degradation are highly influenced by intracellular signals. In E. coli, each cell usually has about 700 tmRNA molecules, and about one per 10-20 ribosomes. However, when its 1:1 binding partner SmpB reaches sub-stoichiometric levels, tmRNA is degraded more quickly. Levels of tmRNA can be dependent on normal cellular processes such as cell cycle, as in Caulobacter cresecentus. Concentrations can also change drastically in response to cellular stress, as in. subtilis where tmRNA can reach 10-fold its normal levels in conditions of heat shock.
The ssrA RNA is transcribed as a 457-nt precursor which is processed at both 5' and 3' ends to generate the mature tmRNA. Processing at the 5' end requires RNase P; processing at the 3' end requires RNase E, which cuts the precursor at three sites immediately adjacent to the CCA-3'end.
Cellular functions:
The tmRNA system has two main functions: 1) rescue of stalled ribosomes and 2) mRNA/protein quality control. This is accomplished in a process called trans-translation.
The tmRNA system is able to recognize, bind and release stalled ribosomes that would otherwise be unusable and decrease the overall efficiency of translation. It also enables degradation of the defective mRNA so that it cannot re-engage active ribosomes. Lastly, it adds a tag to the aberrant protein product, targeting it for proteolysis.
Functions in bacteria
How commonly is the tmRNA system used in bacteria? In E. coli, tmRNA rescue of ribosomes occurs in 1 out of every 250 translational events. Even in this species, where the tmRNA system is inessential, tmRNA deletion mutants constitutively activate heat shock response as in a state of stress. For many other bacteria, the tmRNA system is crucial for viability (see Evolution and Conservation: conservation of function). The tmRNA system is also necessary for virulence in pathogenic species such as Salmonella enterica and Yersinia pseudotuberculosis where it increases survival in macrophages. There is also evidence that tmRNA is involved in motility, cell cycle, type III secretion systems and flagella synthesis.
Recently, tmRNA has also been shown to help maintain low levels of Lac repressor in E. coli. When the Lac repressor tetramer binds its own operator, transcription of the gene is interrupted, forming a nonstop mRNA. In the absence of the tmRNA system, as in the deletion mutants, the resulting Lac repressor fragments are still able to bind operators and repressor activity is increased, only to be amplified with further translation. However, when intact, the tmRNA system degrades the aberrant mRNA and Lac repressor fragments, limiting repressor levels for efficient Lac induction when lactose metabolism may be required later on.
Functions in phages
Phages may use tmRNA activity to gauge the translational abilities of the host. For example, when ribosomes in E. coli stall during translation of the hybrid λ-P22 phage Mu lysogen repressor, tmRNA-mediated release mediates the induction of the lysogenic cycle.
Mechanism of action:
In the current model, the process of trans-translation occurs as follows: 1) the alanine-charged tmRNA system recognizes a stalled ribosome. 2) the tmRNA manifests its tRNA-like properties in binding the A-site of the ribosome and donating its alanine to the polypeptide chain. 3) The mRNA-like properties of tmRNA allow it to code for proteolysis tag on the aberrant protein, as well as a stop codon for release of the ribosome. 4) The defective mRNA is degraded. Recognizing and binding a stalled ribosome. For a tRNA, EF-Tu and GTP mediate delivery to the ribosome in an active process where GTP is hydrolyzed to form GDP. GDP is then released along with EF-Tu after the correct codon-anticodon interactions to stabilize binding.
For tmRNA a similar process presumably occurs, although the tRNA-like region lacks an anticodon. Instead, SmpB, a roughly 160 residue protein which binds specifically to tmRNA, is necessary for association with EF-Tu, GTP and the A- site of the ribosome. The actual stoichiometric proportions of SmpB, ribosomal subunit and tmRNA binding are still unknown.
It has recently been proposed that the tmRNA complex recognizes stalled ribosomes via steric restrictions. In ribosomes undergoing active translation, both the P and A sites are filled and the 3� mRNA is associated tightly with ribosomes and RNA polymerase, preventing the bulky tmRNA complex from binding. However, the 3� ends of nonstop mRNAs remain caught at the P site, leaving the A site open to the tmRNA complex.
tRNA-like functions:
When the tmRNA complex is bound to the ribosomal A-site, transpeptidation occurs as in normal peptide synthesis, where the nascent polypeptide is transferred to the tmRNA alanine and shifted to the ribosomal P-site.
mRNA-like functions:
Amazingly, translation is then switched from the nonstop mRNA to the tmRNA ORF without a classic AUG start codon, Shine-Dalgarno sequence, or ribosomal reassembly. Though the mechanism by which this occurs is still unknown, through mutagenesis studies, it has been shown that a GCA resume codon and a conserved UAG sequence two nucleotides upstream are necessary for establishing the correct peptide reading frame.
Upon translation of the tmRNA ORF terminating in a proper stop codon, a tag is added to the nascent polypeptide, directing it for targeted proteolysis. The mRNA is then released, and SmpB and tmRNA mediate its degradation by RNase R.
Significant partner RNAs or proteins for tmRNA:
As mentioned previously, alanyl-tRNA synthetase is required for tmRNA activation, EF-Tu is necessary for ribosome binding, RNase R is needed for degradation of nonstop RNAs, and SmpB is crucial for almost all tmRNA processes.
Degradation of proteins tagged by tmRNAs is accomplished by three energy-dependent proteases, ClpXP, ClpAP, FtsH as well as Tsp, a periplasmic energy-independent protease. The majority of degradation is performed by ClpXP in vitro, while FtsH degrades only a small subset of proteins with low thermodynamic stability. Various adaptor proteins are also involved in mediating protein degradation.
Potential as a tool:
Specific bacteria can be identified in a mixture by creating a fluorescent probe to tmRNA, which is present at high copy numbers in the cell. In the past, probes have been made against 16S rRNA, but it has been difficult to distinguish between closely related bacteria because an rRNA sequence is highly conserved across species. In 2001, Sch�nhuber et al. found that by designing probes after sequence alignment analysis, individual species such as Lactococcus lactus, or general classes such as gram-positive bacteria could be pinpointed using FISH (fluorescent in situ hybridization).
Role in human disease:
The tmRNA system is vital for growth of many pathogenic bacteria species including Mycobacterium pneumoniae, of the same genus as the causative agent of tuberculosis, and Neisseria gonorrheae, the causative agent of the sexually transmitted infection, gonorrhea. Many more bacteria that do not require the tmRNA system to grow under optimal conditions in vitro will exhibit defects without it when under environmental stress. For example, Salmonella typhimurium, an enteric pathogen loses viability in macrophages in the absence of a functional tmRNA system. Yersinia pseudotuberculosis, a relative of the causative agent of plague, is unable to use its type III secretion system within a host without the tmRNA system. Thus unable to deliver effector molecules to target cells it is rendered avirulent, and does not cause disease in mouse models.
Importance of 5'- end of mRNA in E.coli:
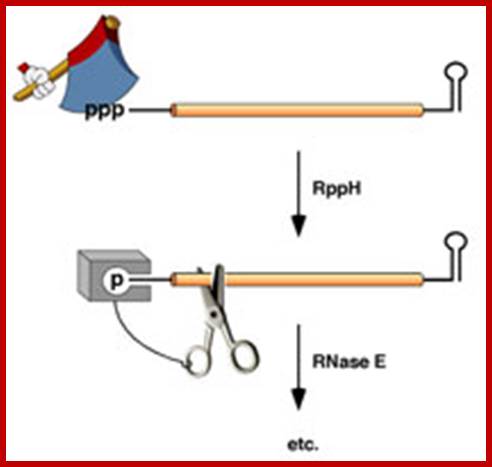
�In bacteria, the lifetimes of mRNAs can differ by more than an order of magnitude, with profound consequences for gene expression. For many years it had been assumed that mRNA degradation in E. coli begins with endonucleolytic cleavage at internal sites. However, recent findings have challenged that view by showing that mRNA decay can instead be triggered by a prior non-nucleolytic event that marks transcripts for rapid turnover: the rate-determining conversion of the 5' terminus from a triphosphate to monophosphates. This modification creates better substrates for the endonuclease RNase E, whose cleavage activity is greatly enhanced when the RNA-5' end decay to mono phosphorylated 5� end. The pyrophosphate-removing hydrolase responsible for that 5'-terminal event, the first such bacterial enzyme ever characterized. That the action of the pyrophosphohydrolase is impeded when the 5' end is structurally sequestered by a stem-loop helps to explain the stabilizing influence of 5'-terminal base pairing on mRNA lifetimes in vivo. Interestingly, this master regulator of 5'-end-dependent mRNA degradation in E. coli not only catalyzes a process functionally reminiscent of eukaryotic mRNA decapping but also bears an evolutionary relationship to the eukaryotic decapping enzyme Dcp2.
RNase E is an essential ribonuclease with a key role in the degradation and/or processing of RNA. In E. coli, RNase E is present as a membrane bound multicomponent complex called a "degradosome". The other major components are polynucleotide phosphorylase (PNPase), the RhlB RNA helicase, and enolase while the DnaK chaperonin protein, GroEL, and polynucleotide phosphate kinase (PPK) are minor components. The ratios of the major components may vary during different stages of cell growth though the RhlB helicase remains at close to a 1:1 ratio.
Post Transcriptional processes; Mechanism of 5'-end-dependent RNA degradation in E. coli; In bacteria, the lifetimes of mRNAs can differ by more than an order of magnitude, with profound consequences for gene expression. bacterial mRNA degradation begins with endonucleolytic cleavage at internal sites. However, our recent findings have challenged that view by showing that mRNA decay can instead be triggered by a prior non-nucleolytic event that marks transcripts for rapid turnover: the rate-determining conversion of the 5' terminus from a triphosphate to a monophosphate. In Escherichia coli, this modification creates better substrates for the endonuclease RNase E, whose cleavage activity is greatly enhanced when the RNA 5' end is monophosphorylated, whereas in Bacillus subtilis it triggers 5'-exonucleolytic degradation by RNase J; BELASC LAB; http://www.med.nyu.edu/
The C-terminal part of RNaseE appears to be responsible for binding these other components. However, these other components are not essential for RNaseE function since a truncated RNase E protein lacking the C-terminal half is sufficient for cell viability and for RNA degradation and processing in vivo in E. coli.
The
degradosome has also been shown to contain RNA molecules such as RNAi and
fragmented rRNAs which are known to be substrates for RNase E. �The RNaseE can
cleave RNAi at three or four internal sites. More surprising was an observation
that degradosomes could also contain the10Sa/ssrA RNA/tmRNA.
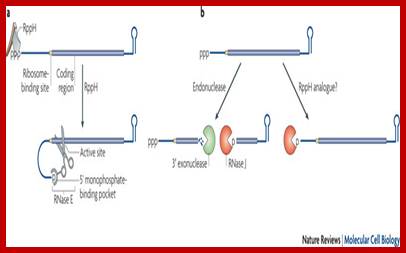
Pathways for 5′ end-dependent mRNA degradation in bacteria.; Fig 2a. RNA decay in bacteria that contain the endonuclease RNase E or a homologue. Pyrophosphate removal by RppH generates a 5′-terminal monophosphate that binds to a discrete pocket on the surface of RNase E, thereby facilitating mRNA cleavage at a downstream location by the active site of that enzyme. InEscherichia coli, RNase E cleavage of primary transcripts can also occur by an alternative, 5′ end-independent mechanism that does not require prior pyrophosphate removal (Fig. 2a). b RNA decay in bacteria that contain the 5′ exonuclease RNase J. Internal cleavage by an endonuclease generates a monophosphorylated intermediate that is susceptible to 5′-to-3′ digestion by RNase J, the exonucleolytic activity of which is impeded by a 5′ triphosphate. Alternatively, it is possible that 5′ exonucleolytic digestion by RNase J may be triggered by pyrophosphate removal from primary transcripts by an as yet unidentified RppH analogue. Ribosomes have been omitted from this figure for simplicity. Joel G. Belasco; http://www.nature.com/
Role of Poly-Adenylation:
The role of polyadenylation in regulating copy number control of ColEI plasmids was discovered when a new mutation affecting plasmid copy number (pcnB) was discovered and shown to be the same gene as that coding for a bacterial polyA polymerase. More recently, Sarkar, Cao and Jain (Mol Genet Genomics (2002) carried out a study to identify additional polyA polymerases and, instead, discovered two unexpected regulatory molecules. (pcnB mutants display residual polyadenylation activity)
They started with a pcnB strain of E. coli carrying a ColEI based plasmid which contained the lac operon. Expression of beta-Galactosidase is then an approximate measure of plasmid copy number. Colonies containing this plasmid were less blue than wild-type strains of E. coli carrying the same plasmid.
A genomic library of E. coli DNA was constructed in a p15 based plasmid. This plasmid has a replicon which is compatible with that of ColEI plasmids. The plasmid library was transformed into the pcnB strain of E. coli carrying the lac operon plasmid.
A small number of colonies appeared to have a darker blue color than the others. This indicated that the p15 plasmid was expressing some function which was interfering with the copy number control of the ColEI plasmid. The isolated and measure the beta-Galactosidase activity in 14 colonies and found it to be 1.5 to 3.6 times higher than in the 'control' colonies. Analysis of mRNAs showed the presence of Poly (A) at the end of 3� end.� Polyadenylated segment acts as the target for polysedenylase enzyme which starts the degradation of mRNAs in bacteria (generalized).
Poly-A additional mode of degradation:
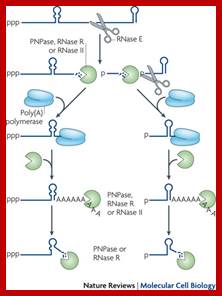
Facilitation of 3� exonuclelytic degradation of bacterial mRNA decay intermediates by polyadenylation; Endonucleolytic cleavage of mRNA by RNase E generates multiple fragments, one of which ends with the original 3′-terminal stem-loop. The others undergo 3′ exonucleolytic attack by polynucleotide phosphorylase (PNPase), RNase R and/or RNase II until an upstream stem-loop is encountered, which interrupts further degradation owing to the preference of these ribonucleases for unpaired 3′ ends. The resulting decay intermediates are then polyadenylated by poly(A) polymerase, thereby enabling the exonucleases to re-engage. The repeated addition of single-stranded poly(A) tails to the 3′ ends of these intermediates provides many opportunities for PNPase and RNase R to overcome structural impediments to exonucleolytic degradation, and eventually they succeed. The ability of PNPase to digest base-paired RNA is enhanced by its association with the RNA helicase RhlB, whereas RNase R requires no such assistance. By contrast, RNase II can degrade poly(A) and other types of unstructured RNA but not structured RNA. Ribosomes and coding regions have been omitted from this figure for simplicity. Joel G. Belasco; http://www.nature.com/
Polyadenylation in bacteria helps polynucleotide phosphorylase (PNPs) degrade past secondary structure:
In many bacteria, both mRNAs and non-coding RNAs can be polyadenylated. This poly(A) tail promotes degradation by the degradosomes, which contains two RNA-degrading enzymes: polynucleotide phosphorylase and RNase E. Polynucleotide phosphorylase binds to the 3' end of RNAs and the 3' extension provided by the poly(A) tail allows it to bind to the RNAs whose secondary structure would otherwise block the 3' end. Successive rounds of polyadenylation and degradation of the 3' end by polynucleotide phosphorylase allows the degradosomes to overcome these secondary structures. The poly (A) tail can also recruit RNase that cut the RNA into two. These bacterial poly (A) tails are about 30 nucleotides long.
In as different groups as animals and trypanosomes, the mitochondria contain both stabilizing and destabilizing poly (A) tails. Destabilizing polyadenylation targets both mRNA and noncoding RNAs. The poly (A) tails is 43 nucleotides long on average. The stabilizing ones start at the stop codon and without them the stop codon (UAA) is not complete as the genome only encodes the U or UA part. Plant mitochondria only have destabilizing polyadenylation, and yeast mitochondria have no polyadenylation at all.
While many bacteria and mitochondria have polyadenylate polymerases, they also have another type of polyadenylation, performed by polynucleotide phosphorylase itself. This enzyme is found in bacteria, mitochondria, plastids and as a constituent of the archaeal exosome (in those archaea that have an exosome). They can synthesize a 3' extension where the vast majority of the bases are adenines. Like in bacteria, polyadenylation by polynucleotide phosphorylase promotes degradation of the RNA in plastids and likely also in Archaea.
The first indication that poly(A) tails destabilize RNA came from the discovery that PAP I controls the stability of the small RNA (RNA I) which regulates the replication of ColE1 plasmids (see above). �Further study led to the conclusion that PNPase does not bind RNA I, whose 3' end is sequestered in a secondary structure and that poly(A) tails provide sites where PNPase can bind and initiate the exonucleolytic degradation of RNA I. Polyadenylation has since been shown to be involved in the degradation of other RNA species and of RNA in general and it is admitted that the mechanism of degradation of RNA I can be extended to the poly(A)-dependent degradation of any RNA with a 3' secondary structure.
The current idea is that PNPase can carry out the complete processive degradation of RNAs containing weak secondary structures but is blocked when it encounters stable hairpins which cause dissociation of the ribonuclease from its substrate. It has been proposed that an Exonucleolytic Impeding Factor, referred to as EIF, might provoke PNPase stalling at secondary structures. When blocked at secondary structures, PNPase releases RNAs devoid of a 3' single-stranded stretch of nucleotides that cannot be bound by exoribonucleases. The current model of RNA decay postulates that these tailless RNAs are readenylated thus allowing PNPase to reinitiate exonucleolytic decay. Again, PNPase can generate tail-less RNA or, possibly, continue to degrade the RNA upstream of the tail and remove few nucleotides at the bottom of the hairpin before to dissociate from the RNA.
Polyadenylation in Escherichia coli and other prokaryotes:
The
focus of this (by authors) is to delineate the molecular mechanism of
polyadenylation in E. coli. Previous experiments in our laboratory have
shown that poly (A) tails target mRNAs for rapid decay, but also increase the
stability of the mRNAs encoding both RNase E and polynucleotide phosphorylase
(PNPase), two ribonucleases that are involved in degrading all types of RNA
molecules in the cell. �We have also shown that poly(A) polymerase I is part of
a multiprotein complex that also includes PNPase and Hfq, a small RNA binding
protein. In addition, several lines of experimentation indicate that
rho-independent transcription terminators serve as polyadenylation signals in
exponentially growing cells). A genomic analysis of the entire E. coli
transcriptome has shown that the vast majority of E. coli ORFs undergo
some degree of polyadenylation, which facilitates RNA degradation.� .��������
However, many questions remain to be answered. For example, why is immature 23S ribosomal RNA a primary substrate for poly (A) polymerase? �In addition, the analysis of polyadenylation is complicated by the fact that while poly(A) polymerase accounts for 90% of the poly(A) tails (Mohanty & Kushner, 1999), PNPase also synthesizes polynucleotide tails in wild-type bacteria. The heteropolymeric tails added by PNPase are generally found near the 5� termini of transcripts and do not appear to perform the same function as poly (A) tails. We are currently trying to determine whether polyadenylation is used primarily to target full-length mRNAs or to help promote the degradation of mRNA fragments generated by endonucleolytic digestion by enzymes such as RNase E, RNase G and RNase III. We are also working to determine if the polyadenylation complex contains additional proteins. Furthermore, we are trying to establish what makes a rho-independent transcription terminator an effective polyadenylation signal. Our current working model of polyadenylation is shown in figures.
A model for polyadenylation of mRNAs in E. coli: mRNAs with Rho-independent transcription terminators will contain a stem-loop structure at their 3' ends. This structure inhibits the activity of both PNPase and RNase II, the two major 5� -> 3� exonucleases in the cell, because of its very short single-stranded region at the 3� terminus. In the presence of the riboregulator protein Hfq, a complex containing Hfq, PNPase and poly (A) polymerase I (PAP I) binds to the terminus. PAP I then initiates the addition of A residues to form a poly (A) tail. It is not clear whether Hfq remains associated with PAP I after polyadenylation commences. At some point, PAP I is displaced by PNPase and degradation starts in the 3� -> 5� direction, proceeding through the stem-loop structure, releasing the Hfq protein. As PNPase approaches the 5' end of the mRNA, its rate of degradation slows as the localized concentration of inorganic Pi declines. Eventually, the enzyme begins to synthesize a polynucleotide tail on the transcript. A cycling process then ensues until the mRNA is either degraded into a small oligonucleotide (4-7 nt) that is no longer a substrate for PNPase or the enzyme dissociates from the substrate.
Poly-A addition and mRNA degradation is prevalent in chloroplasts also, for plastids were derived from ancestral Cyanophytes.
Endonucleolytic Degradation of mRNA in Bacteria:
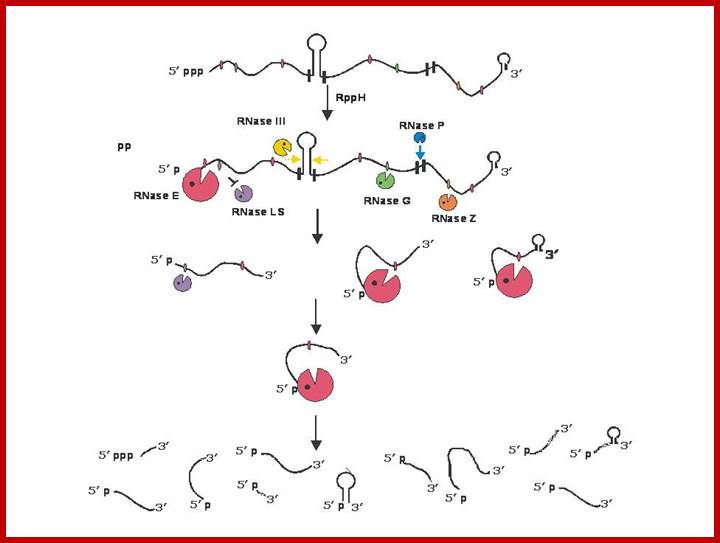
Endonucleolytic initiation of mRNA decay for a polycistronic transcript. Intercistronic regions are marked by small black vertical bars. Since RNase E, the most abundant endoribonuclease is inhibited by the presence of a 5� triphosphate; the first step in the decay of many mRNAs is the action of RppH to convert the 5� triphosphate to a 5� phosphomonoester. Subsequently, RNase E, will bind to the 5� phosphomonoester terminus to initiate decay. Its binding sterically prevents the binding of RNase LS at a contiguous site. However, the ability of RNase III to cleave the stem-loop structure in the intercistronic region is independent of RNase E action. Similarly, RNase P cleavage within the downstream intercistronic region is also independent of the initial RNase E cleavage. In addition, the downstream RNase G and RNase Z cleavage sites may be recognized, independent of RNase E binding at the 5� terminus, if there are sufficient amounts of each enzyme present. Thus the first round of endonucleolytic cleavage events could yield from between 5-7 decay intermediates. Subsequent cleavages by RNase E, RNase LS, RNase G and RNase Z could lead to a total of 11 decay intermediates if all of the sites are cleaved. It is possible, that some cleavages will not take place, if exonucleolytic degradation of the initial decay intermediates proceeds so rapidly that some endonucleolytic cleavage sites are actually degraded before they are recognized by their respective enzymes. In addition, it should also be noted that Baker and Mackie (2003) have shown that under certain circumstances RNase E can cleave at internal sites without binding to a 5� terminus. Sizes of the various endonucleases reflect an estimate of their relative participation in mRNA decay. For the sake of simplicity, RNase E is shown without the other components of the degradosome. The products of the endonucleolytic cleavages will subsequently be degraded by a combination of PNPase, RNase II, RNase R and poly(A) polymerase. http://www.genetics.uga.edu/
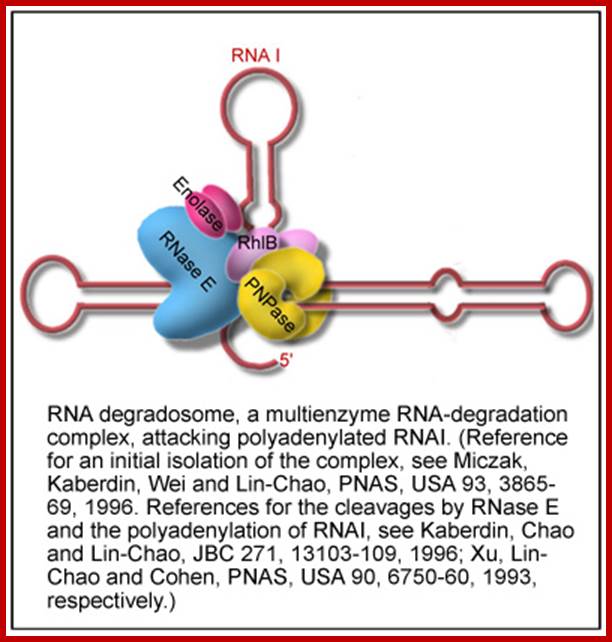
A view of degradosome consisting of RNaseE, PPK, DnaK, PNPase, Enolase RhlB helicase and RNAi; Dr.Sue-Chao http://iao.sinica.edu.tw/
In E. coli, all of the ribo-exonucleases that have been identified degrade RNA molecules from the 3� terminus. These 3� > 5� exonucleases fall into two classes depending on their mechanism of catalysis. Hydrolytic enzymes such as RNase II and RNase R release nucleoside monophosphates, while enzymes such as polynucleotide phosphorylase and RNase PH use a phosphorolytic mechanism that requires the presence of inorganic phosphate and releases nucleoside diphosphates. Polynucleotide phosphorylase is a reversible enzyme. It is already known that PNPase is the primary enzyme involved in the degradation of poly(A) tails associated with mRNAs and that it functions both degradatively and biosynthetically in E. coli Genome-wide analysis using RNase II and PNPase deletion mutants has demonstrated that PNPase plays a greater role in mRNA decay than RNase II. An unexpected observation from this work was the large percentage of E. coli mRNAs that are destabilized in the absence of RNase II. The recent observation that RNase R is required for the degradation of mRNAs contained large stem-loop structures called repetitive extragenic elements has led us to construct a series of mutants containing a combination of mutations in PNPase, RNase II and RNase R so that we can gain a better understanding of how these three exonucleases function in mRNA decay.
RNase E encoded by r ne, was first identified in the late 1970s based on its involvement in the processing of a 9S rRNA precursor into a p5S species). Subsequently, it was shown to be involved in mRNA decay and the maturation of tRNAs. Since this enzyme is essential for cell viability, it was assumed that either the defect in mRNA decay or 9S rRNA processing was responsible for the loss of cell viability. However, experiments carried out with a series of RNase E deletion mutations demonstrated that these assumptions were not correct. In fact, experiments employing a series of truncated RNase E proteins led us to hypothesize that the ability of RNase E to initiate the maturation of tRNAs was its essential function. Of further interest is the fact that RNase E serves as the scaffold for a multiprotein complex called the "degradosome". Since this complex contains endo and exonucleases as well as an RNA helicase activity, it was assumed that it accounted for the bulk of mRNA decay. However, analysis of RNase E deletion mutants has demonstrated that assembly of the degradosome is not required for normal mRNA decay In contrast; degradosome assembly is required for the cell to be able to detect alteration at the level of poly adenylation.
Additional experiments have shown that the RNase-E �gene is controlled by a complex regulatory systems that involves three distinct promoters. With the completion of the crystal structure of the catalytic region of RNase E, it is now possible to examine the interaction of the various domains in the activity of RNase E on various mRNA, tRNA and rRNA substrates. For example, we have recently identified second-site intragenic suppressor mutations that complement the growth defects associated with the rne-1 and rne-3071 alleles at 44oC. �Interestingly, these suppressor mutations restore normal activity on tRNA precursors but mRNA decay remains defective. We are now employing a variety of genetic and biochemical techniques to help understand how RNase E distinguishes among mRNA, rRNA and tRNA substrates, (Apirion & Lasser, 1978; (Mohanty & Kushner, 2002), (Ow et al., 2000). (Arraiano et al., 1988), (Ow & Kushner, 2002). (Mohanty & Kushner, 2003). (Ow et al., 2002). Ow & Kushner, 2002). (Ow et al., 2000). (Callaghan et al., 2005),
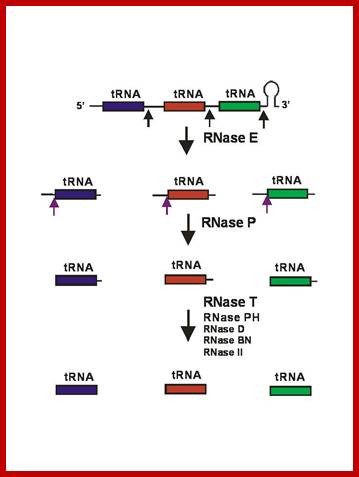
tRNA processing RNases: (Mohanty & Kushner, 2007). (Mohanty & Kushner, 2008)
While this model explains the initial processing of many tRNA precursors, we have recently shown that there are some polycistronic operons that are processed without any involvement of RNase E. In particular, the valV valW and leuQ, leuP and leuV operons are separated into pre-tRNAs by RNase P. In this pathway, the initial cleavages by RNase P generate pre-tRNAs with mature 5� termini after the rho-dependent terminator is removed by a combination of RNase II and PNPase. The processing of the 3� termini is similar to that described in Fig. Subsequently, we have shown that the secG, leuU and metT operons also employ RNase P to separate the tRNAs. In the case of the metT operon, which contains seven tRNAs, the processing pathway actually involves both RNase P and RNase E. We are currently studying the maturation of additional tRNAs that use a different mechanism for removing terminator sequences.
General model for tRNA processing in E. coli. In the first step the endoribonuclease RNase E cleaves in the intercistronic regions of polycistronic tRNA precursors to generate pre-tRNAs that have a small number of extra nucleotides at both their 5� and 3� ends. The mature 5� termini are generated by cleavage with the ribozyme, RNase P, while the mature 3� end arises from the action of a series of 3�-> 5� exonucleases. The most important of these enzymes are RNase T and RNase PH. http://www.genetics.uga.edu/
Bacterial mRNAs are degraded in the 3′>5′ direction also:
Studies of mutant bacteria whose mRNAs have extended half-lives have identified a range of ribonucleases and other RNA-degrading enzymes that are thought to be involved in mRNA degradation. These include (Carpousis et al., 1999):-
� RNase E and RNase III, which are endonucleases that make internal cuts in RNA molecules;
� RNase II, which is an exonuclease that removes nucleotides in the 3>>5� direction;
� Polynucleotide phosphorylase (PNPase), which also removes nucleotides sequentially from the 3′ end of an mRNA but, unlike true nucleases, requires inorganic phosphate as a co-substrate.
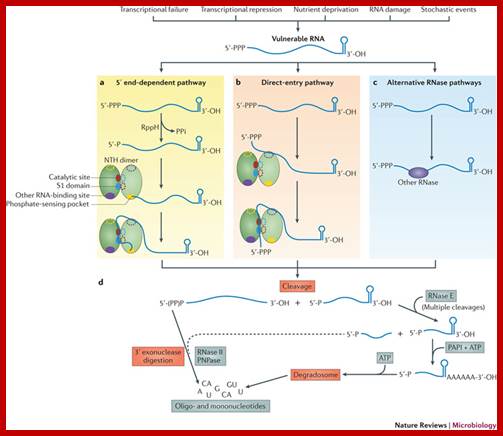
A variety of events can influence the fate of an mRNA before the first phosphodiester bond is actually cleaved. Internal and external cues, including temperature and supply of nutrients, signal to the transcriptional and translational machineries, which in turn influence the accumulation and utilization of each mRNA. The presence of translating ribosomes is generally considered to be protective. Conversely, a naked mRNA (one that is free from ribosomes) becomes vulnerable to endonuclease attack. Independently, the 5′ and 3′ termini of mRNAs are processed by pyrophosphohydrolases and by either 3′ exonucleases or poly(A) polymerase (PAPI), respectively (not shown). PAPI activity is stimulated by the RNA chaperone Hfq68. Depending on the status of the 5′ terminus and the density of ribosomes in the cell, a vulnerable mRNA can enter one of at least three pathways. a | The 5′ end-dependent pathway relies on the phosphate-sensing pocket of RNase E (or, less frequently, RNase G) to facilitate substrate binding. Following the initial recognition of the 5′ end, the enzyme�substrate complex rearranges to thread the RNA through the RNA channel (S1 domain) and the cleavage site (catalytic site) of RNase E. In principle, each dimer in an RNase E tetramer could engage a separate RNA substrate. b | Direct entry by RNase E by-passes the 5′ end and recognizes the substrate directly, possibly using contacts in the carboxy-terminal half of an Rne monomer (purple). This complex also rearranges to thread the RNA through the RNA channel and the cleavage site. c | In alternative pathways, other endonucleases such as RNase III bind to those RNAs that contain their specific recognition features. d | All three pathways converge at the initial endonucleolytic cleavage step, which generates two products. The 3′ hydroxylated product containing the 5′ end of the substrate is vulnerable to 3′ exonucleases such as RNase II and polynucleotide phosphorylase (PNPase). RNase II is believed to be responsible for the bulk of exonucleolytic RNA degradation in Escherichia coli. Both of these enzymes processively degrade RNAs, releasing 5′ mononucleotides (RNase II) or 5′ dinucleotides (PNPase). The 5′ monophosphorylated product of the initial cleavage is preferentially recognized by RNase E (and more rarely by RNase G) owing to the 5′ sensor in these enzymes. This sensor confers a 25�30-fold enhancement on the rate of degradation in vitro. �Thus, following the initial cleavage, there is a wave of additional endonucleolytic cleavages by RNase E, moving in the 5′�3′ direction, each generating a 3′-OH upstream product (which is vulnerable to 3′ exonucleases) and a nested set of progressively shortened distal products, all retaining a 5′ monophosphate. RNase E does not readily cleave RNAs containing stable secondary structures. Thus, terminal fragments containing Rho-independent terminators, REP (repetitive extragenic palindromic) sequences or other structures require oligo(A) addition, which is catalysed by PAPI. This modification enables PNPase and RhlB in the RNA degradosome to attack structured RNAs coupled to ATP hydrolysis. �Short (5 nucleotide) oligonucleotides are substrates for oligo-RNase, a 3′ exonucleases; George A. Mackie� http://www.nature.com
Coordinate gene expression system is similar in prokaryotes and Eukaryotes:
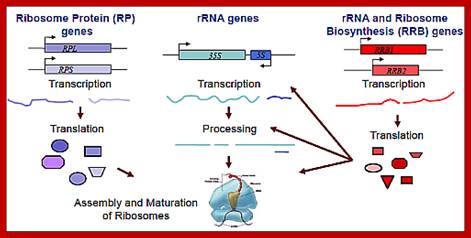
Ribosome biogenesis requires the coordinated regulation of three extensive gene networks, including 137 cytoplasmic RP genes, 150 rRNA genes and some 200 RRB genes.;This figure holds good for both prokaryotes and eukaryotes with small differences i.e tRNA synthesis is missing in the diagram. Anand Soorneedi et al http://mmcalear.faculty.wesleyan.edu
Eukaryotic processed mRNA-fine features:
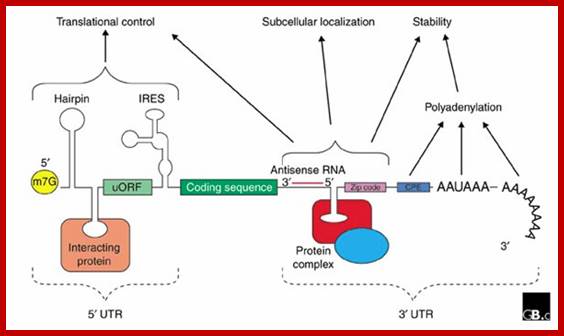
The generic structure of a eukaryotic mRNA;: Illustrating some post-transcriptional regulatory elements that affect gene expression. Abbreviations (from 5' to 3'): UTR, untranslated region; m7G, 7-methyl-guanosine cap; hairpin, hairpin-like secondary structures; uORF, upstream open reading frame; IRES, internal ribosome entry site; CPE, cytoplasmic polyadenylation element, RNA transport signals RTS, antisense RNA binding region zip code for mi/si RNAs? And AAUAAA, polyadenylation signal. Mignone et al. Genome Biology 2002 3:reviews0004.1 doi: 10.1186/gb-2002-3-3-reviews0004
Structural organization of eukaryotic mRNA and the different points of possible regulation of translation through various trans-acting factors
Structural organization of eukaryotic mRNA and the different points of possible regulation of translation through various trans-acting factors; m7G, cap structure; eIF, eukaryotic initiation factor; CPE, cytoplasm polyadenylation element; EDEN, embryonic deadenylation signal; DICE, differential control element; PABP, poly(A)-binding protein. [?], possible sites of interaction of transacting factors (yet unknown) in the coding sequence. Regions of mRNA involved in subcellular localization and stability are also indicated.; http://www.biolcell.org/
5′-m7G, cap structure; eIF-4G eukaryotic initiation factor; IRE- Iron response elements, Ire- Internal ribosome entry elements, CPE, cytoplasm polyadenylation element; EDEN, embryonic deadenylation signal; DICE, differential control element; mRNA binding elements, mRNA localization signal elements, PABP, poly(A)-binding protein [?], possible sites of interaction of transacting factors (yet unknown) in the coding sequence. Regions called Zip code of mRNA involved in subcellular localization and stability are also indicated.
mRNA regulatory elements:
Baker and Coller; http://genomebiology.com/
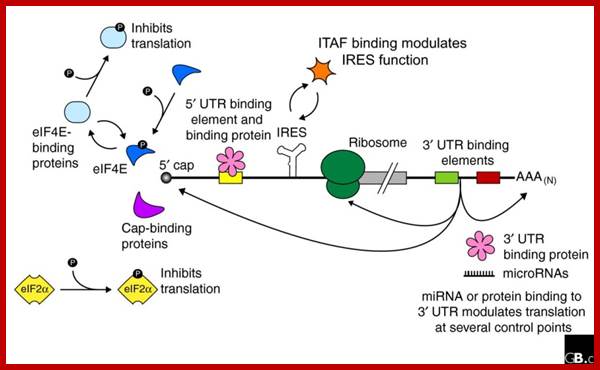
Regulation of eukaryotic mRNA translation occurs at numerous control points. Recognition of 3' UTR sequence or structural elements (green and red boxes) by RNA-binding proteins leads to either activation or repression of translation, often through alteration of the 3' poly(A) tail or through interactions with proteins that bind at the 5' terminal cap structure (that is, the initiation factor eIF4E or cap-binding proteins). Repression of translation by miRNAs can occur through inhibition of translation initiation or elongation, and may also lead to changes in the status of the mRNA 3' poly(A) tail. Elements found within the mRNA 5' UTR (yellow box) can bind regulatory proteins that repress translation by inhibiting 48S ribosome scanning. Global regulation of mRNA translation is commonly achieved through modification of the translational apparatus (that is, by phosphorylation of the translation initiation factors eIF2α and eIF4E) and the ribosome itself, or modulation of protein partner binding affinities (such as the phosphorylation of the eIF4E-binding proteins). Translation can be initiated independent of the mRNA 5' cap through a structured internal ribosome entry site (IRES) in the 5' UTR whose efficiency in initiating translation is, in turn, modulated by trans-acting factors (ITAFs).
Half-life of mRNA with or without Cap and Poly-A tail:
Luciferase mRNAs have been used to determine its half-life and translation efficiency with or without cap and poly- (A) tail.�
Half-life of Luciferase mRNA without cap and without poly (A) is just 31 minutes, and translational activity is 2900 (as measured in terms of light emitted by ug of radioactive protein).
But mRNAs without cap but with poly (A) tail shows half-life of 44 minutes. And its activity is 4480.� The capped mRNA without poly (A) has half-life of 53 minutes and translation activity is 62000 a virtual 50% increase in its half-life and translational efficiency.�
The capped mRNA with poly- (A) tail, has a half-life of 100 minutes and its translational activity is 1,333 000; the relative effect of cap on its activity 200 fold.
In eukaryotes, mRNAs have half-life ranging from 10 minutes to hours and in some cases it can be as long as 120 days in RBCs or so.� Plant Sieve tube cells contain mRNAs that can live for six months are so.
- In RBC cells, after the breakdown of nuclear membrane, Globin mRNAs, found in massive amount, remain very stable in cytoplasm up to 120 days or so.��
A similar situation is also found in sieve cells of vascular plants, where the unicellular sieve cells at the early stage of differentiation, the nucleus disintegrates; transcription is massive and the mRNAs produced are specific to the cell type and they remain active as long as one year. How?
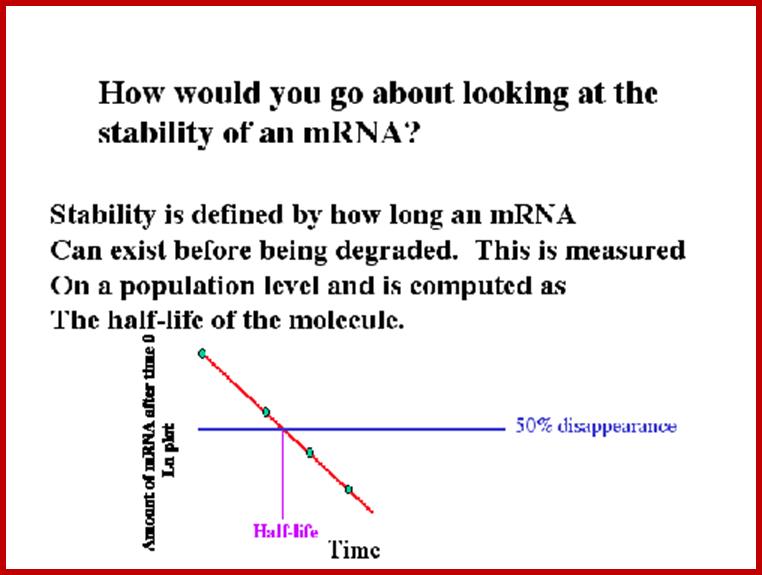
Stabilty Meausurement
Fate of Transient mRNAs:
- During certain stages of development, the cells are going through transient phases of differentiation; in such cases the mRNAs have very short half-life.� Some such transcripts are produced only once in the life span of an organism and they are never to be produced again.
The mRNAs produced in such stage have to be transient and they have to be degraded, otherwise stage specific differentiation won�t be executed and results in wrong programming.
- Many signal mediated induction of transcripts, once their function is over they have to be degraded immediately.
In other situations mRNAs produced in response to certain stimulus, whose effect is transient and in such conditions the half-life of mRNA should be as short as possible.
These are some of the needs or demands of the cells; it is indeed programmed during speciation and evolution. Half-life of proteins is determined by N terminal sequences, and other sequence tags.
- Many of the mRNAs are made transiently during development stages, where cell cycle operates in multiplication of cells.� The expression and the stability of mRNAs is cell cycle specific, mRNAs are transient, and once their function is complete they are immediately degraded. In some cases, mRNAs are made and degraded as quickly as possible.� Some mRNAs are stabilized and long lasting.�
�
Another example of degradation of transiently expressed mRNAs in response to stimuli is those of GMCSF (Granulocyte Macrophage Colony Stimulating Factor), TNF (Tumor Necrosis Factor), ILs (Interleukins), INFs (Interferon - alpha, beta and gamma), C-fos, and C-myc and others.� The above said factors are synthesized in response to stimuli and the product remains for a short duration and this is made possible by their short half-life of their respective mRNA.�
- Lymphokines are required for short duration only, and if they persist longer it leads to harmful effects.�
Most of the mRNAs with very short half-life contain certain sequences called ARE (AU rich elements) such as AUUUA, AUUUAUUUA or multiple copies of such sequences at 3�UTR (untranslated region) regions of their mRNAs.�� These sequences are either located in a stem-loop structure or outside the stem-loop structures at 3�ends. These sequences are recognized by specific endonucleases and degrade mRNAs in quick time.�
Structural motifs of mRNAs with specific sequences are important for regulating the stability of mRNAs.
|
Name of the mRNA |
3� sequence motifs |
|
GM-CSF |
-AUUUA----AUUUA---AUUUAn---- |
|
ILs |
-AUUUUAUUUUA------ |
|
TNF |
-AUUUA-------AUUUAAUUUA---AUUUAn--- |
|
C-fos |
-AUUUA-----------AUUUA------ (n) |
|
C-myc |
----------------------AUUUAn |
GMSF = granulocyte/macrophage stimulating factor,
TNF = Tumor necrosis factor
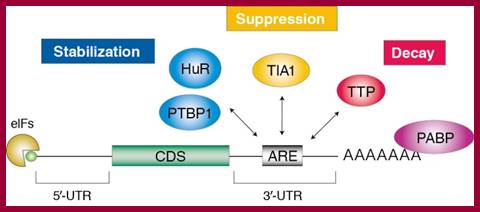
Adenine-Uracil-rich element (ARE)-mediated mRNA decay (AMD): Adenine rich elements found in mRNAs regulates the concentration of a class of mRNAs that contain AU-rich sequences within their 3′ untranslated regions (3′UTRs). The AMD-regulated genes are involved in cellular proliferation, immune response and cardiovascular toning. Impaired AMD results in cancer insurgence and progression and is linked to inflammatory diseases such as Crohn-like inflammatory bowel disease and inflammatory arthritis.
ARE-binding proteins (ABPs) recruit the cytoplasmic mRNA degradation machinery to the target mRNAs leading to their 3′-to-5′ degradation. ABP recognition of specific AREs provides therefore the selectivity of the degradation mechanism. The K-homology splicing regulator protein (KSRP) is a multi-domain protein that has been used as a functional model for AMD1. Investigation is going on, how KSRP uses different combinations of its four RNA-binding domains to recognize selectively its targets and how protein-protein interactions modulate KSRP-RNA recognition and KSRP function2,3. ARE mediated mRNA degradation. KSRP recruits the exosome and PARN to the target mRNAs leading to 3′-to-5′ degradation.
Recent studies suggest that certain disease- or function-related mRNAs or microRNAs (miRNAs) bind to RNA-binding proteins (RBPs), forming clusters which are termed here as RiboClusters. RBPs play an important role in post-transcriptional regulation of gene expression at various steps such as splicing, nuclear export, subcellular localization, mRNA stability and translation. RiboCluster Profiler� is an optimized unique tool which enables customers to extensively analyze the certain disease- or function-related genes. http://ruo.mbl.co.jp
Adenine-uracil-rich element (ARE)-mediated mRNA decay (AMD) regulates the concentration of a class of mRNAs that contain AU-rich sequences within their 3′ untranslated regions (3′UTRs).;ARE-binding proteins (ABPs) recruit the cytoplasmic mRNA degradation machinery to the target mRNAs leading to their 3′-to-5′ degradation (See figure).;�mRNA decay mediated by the AU-rich element (ARE)�; KSRP recruits the exosome and PARN to the target mRNAs leading to 3′-to-5′ degradation; Andre Ramos Group; http://www.nimr.mrc.ac.uk/
Controlling the rate at which an mRNA is degraded is an important mechanism that allows cells to regulate gene expression at the posttranscriptional level. The most abundant group of mRNAs subjected to this mode of regulation are those containing an AU-rich element (ARE) in the 3' untranslated region (UTR). The presence of an ARE in a particular transcript inhibits translation of the mRNA and induces ARE-mediated mRNA decay (AMD). Potent AREs have been identified in the 3'UTRs of growth factors (e.g. GM-CSF, IL-3), cytokines (e.g. TNF-alpha, IL-2, IL-6), pro-inflammatory proteins (e.g. COX-2, MMP-13), and proto-oncogenes (e.g. c-myc, c-fos). AMD is a highly conserved regulatory mechanism that functions in yeast, trypanosomes, drosophila cells and virtually all mammalian cell lines. Thus, AMD is a general mechanism that regulates gene expression in most, if not all eukaryotic cells. Based on the frequency of AU-rich sequences in the human genome database, it has been estimated that 5-10% of all mRNAs may contain AREs. We use functional approaches such as RNA-Immunoprecipitation followed by microarray analysis to explore the spectrum of mRNAs regulated by AREs.
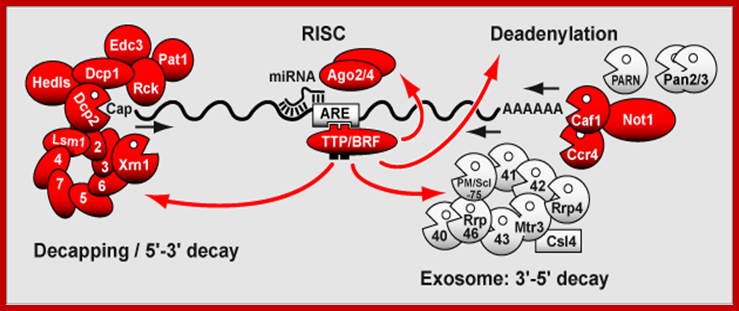
The rapid degradation of mRNAs containing AREs requires RNA binding proteins such as TTP/BRF1. Upon binding to the ARE, TTP (Tri Tetra Proline)/BRF activate deadenylation of the mRNA. TTP/BRF further interact with the exosome responsible for 3'-5' decay, and with the decapping/Xrn1 complex responsible for 5'-3' decay. Interaction with the RISC complex may contribute to AMD. Proteins associated with P-bodies are depicted in red.�; http://www.cellnetworks.uni-hd.de/
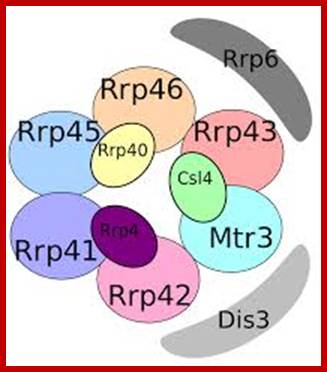
Yeast Exosome composition; -additional-Exonuclease RRP6 and DISC3;
The rapid degradation of mRNAs containing AREs requires RNA binding proteins such as TTP/BRF1. Upon binding to the ARE, TTP (Tri Tetra Proline)/BRF activate deadenylation of the mRNA. TTP/BRF further interact with the exosome responsible for 3'-5' decay, and with the decapping/Xrn1 complex responsible for 5'-3' decay. Interaction with the RISC complex may contribute to AMD. Proteins associated with P-bodies are depicted in red.� http://www.genesilico.pl/
Half-life of certain mRNAs depends upon the presences of certain hormones; for ex. Prolactin hormone has an effect on the longevity of Casein mRNA; this has been established in cultured mammalian cells.
mRNA with (+) Prolactin- increases the production casein mRNA (casein is a phospho protein) two fold and the mRNA half-life is increased by 48hrs.
mRNA without (-) Prolactin- casein mRNA s are degraded very fast.
- Most of the above mentioned mRNAs produce proteins of short-term duration and activity.�
- If such A/T sequences are transferred to the 3�ends of some stable mRNAs genes such as goblins and introduced into cells, the Globin mRNAs are degraded as fast as the short-term mRNAs.
- On the contrary if mRNA containing AU rich sequences are removed by recombinant methods; the mRNAs will have longer life, ex. c Fos.
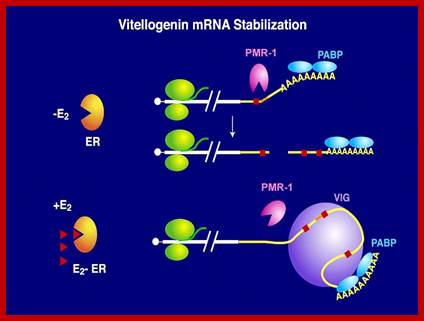
- Vitellogenin mRNA half-life: The half-life of vitellogenin mRNA is approximately 3 weeks in the presence of estrogen and 16 hr after estrogen is withdrawn from the culture medium.
www.life.illinois.edu
The half-life of vitellogenin mRNA is approximately 3 weeks in the presence of estrogen and 16 hr after estrogen is withdrawn from the culture medium. Total poly-(A) mRNA exhibits the same half-life (16 hr) in the presence or absence of estrogen. The rapid cytoplasmic degradation of vitellogenin mRNA in the absence of estrogen is fully reversible upon restimulation with estrogen, indicating that nuclear modification of vitellogenin mRNA transcripts is not responsible for their stability. Intermediate levels of vitellogenin mRNA stability and changes in the relative rate of vitellogenin gene transcription are not observed late in estrogen induction, when vitellogenin mRNA levels plateau. Instead, Xenopus liver cells achieve fine control over the level of vitellogenin mRNA through down-regulation of the overall rate of total nuclear RNA synthesis.
- Certain stage specific transcriptional factors are produced once in the lifetime of the organism, and such transcripts should have structural feature for degradation in quick time.
- Experimentally, it has been proved that any mRNAs with methylated cap at 5�end and poly-A tail at 3� end are stable because of their ability to synthesize their proteins for a long time and they are more active than the others.�� Such mRNAs don�t have any destabilizing 3� end sequences.� Stability to such mRNAs is due to the binding of proteins to 5�cap and the binding of PAB-1 to poly-A tail, which actually prevent degradation from exonuclease by circularization of the mRNA by the binding of cap, bound eIFG with poly-A tail bound PAB1 proteins.
- Many a time�s the stability or the longevity of mRNAs is coupled to its translation activity.� For example histone mRNAs, which don�t have ploy-A tail are very stable as long as they are not translated.�
Regulation of ARE-mediated mRNA decay (AMD)
Activation of the immune system involves a rapid and transient production of cytokines. Many cytokine mRNAs contain AREs, and their expression is under control of the RNA-binding protein Tris tetra proline (TTP). In unstipulated cells, these mRNAs are rapidly degraded. When the immune system is stimulated, transcription of cytokine mRNAs is activated in the nucleus, and the cytokine mRNAs are stabilized in the cytoplasm. The signal for stabilizing these mRNAs is mediated via the p38-MAPK � MK2 kinase pathway. Previous work has shown that TTP is an important target of MK2 (Figure 2). MK2 phosphorylates TTP at serine 52 and 178, which leads to binding of 14-3-3 adaptor proteins. As a consequence, the activity of TTP is reduced and cytokine mRNAs are stabilized. The phosphatase PP2A acts as an antagonist of MK2 by dephosphorylation and thereby activating TTP. We are currently investigating how the phosphorylation of TTP regulates its activity.
![]()
ARE-mediated decay of mRNA. In the cytoplasm, AUBPs (like TTP) bind to ARE (AUUUA) in
the mRNA. The binding of AUBPs recruits either Dcp, which promotes decapping of
mRNA, or deadenylase, which removes the poly(A) tail. After exposure of
terminal mRNA, exonucleases act to degrade the mRNA in either the
5′�3′ direction using Xrn1 or in the 3′�5′ direction
using the exosome. ARE, AU-rich element;
AUBPs, AU-binding proteins. Firoz Ahmed, et al;
http://webcache.googleusercontent.com/
Histone mRNA turnover:
- Histone mRNAs though don�t have poly-A tail; they have a small stem loop structure at their 3� end, but contain 7�CH3-G�p-p-p A�cap at 5� end. However stored histone mRNA are added with short oligo (A)s which are bound by SLBP2 on 3� side of stem loop structure.
- Activation of oocytes lead to destruction of Slbp2 and removal of Oligo (A) and translation is initiated.�
- During DNA replication at �S� stage histone mRNAs are stable, once replication is over, histone mRNAs get degraded.� This is because during DNA replication, translation activity is mostly shut off of histone mRNAs, so no translation and no degradation.� As replication ends, translation of histone is renewed, which is absolutely required for the next replication and histone mRNA are used vigorously for translation.�
� �It is believed that during translation ribosomes as they pass through mRNA ends, ribosome bound nucleases degrade mRNAs. The histone mRNAs contain a small stem loop structure at the 3�UTR. �The 3� end small loop binding protein SLBP plays an important role. The release of SLBp2 triggers the decay. At the end of DNA replication i.e at the end of S-phase histone mRNAs are degraded by addition several Us to the 3� end of mRNAs. �Histone mRNAs undergo oligouridinylation by a cytoplasmic terminal uridinyl transferase at the end of S phase. This degradation of Poly (Us) is by 3�exonucleases. This leads to association of Lsm11�7, and recruitment of the decapping and 5′�3′ decay machinery. How Lsm1�7 association influences exosome activity is unclear, although there is evidence for an inhibitory role This leads to the degradation of histone mRNA from 5� end also, first by removing 7�CH3.Gpp by decapsase (Dcp1 and Dcp2). Then the 5�end with P� groups is degraded by Xrn�s by 5� exonuclease activity. Decay also occurs 3′�5′ by the exosome.
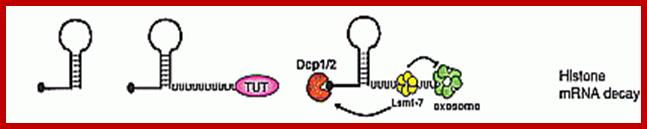
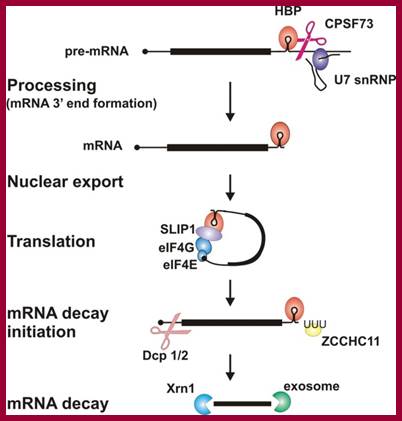
Histone proteins are essential for the packaging of DNA into chromosomes. Histone gene expression is cell-cycle-regulated and coupled to DNA replication. Control of histone gene expression occurs at the transcriptional and post-transcriptional level and ensures that a fine balance between histone abundance and DNA replication is maintained for the correct packaging of newly replicated DNA into chromosomes. In the present paper, we review histone gene expression, highlighting the control mechanisms and key molecules involved in this process.; The control of histone gene expression; Alexander M.J. Rattray Berndt M�ller; http://www.biochemsoctrans.org
|
|
|
� |
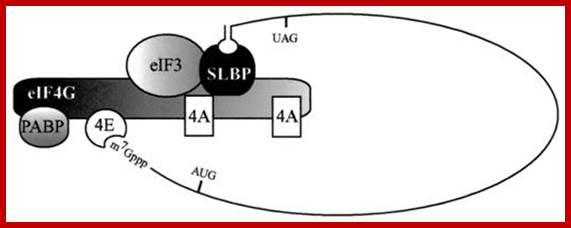
Circularization of mRNA with binding of Translation Initiation Factors; Proposed physical and function model of the association of SLBP with the eIF4F/eIF3/PABP complex. SLBP is shown bound to the 3'-terminal stem-loop of a cell cycle-regulated histone mRNA. Association of SLBP with the eIF4F/eIF3/PABP complex requires eIF4G and perhaps eIF3. eIF4E is not required for the physical association of SLBP with the complex but is required for SLBP function in that it is necessary for the binding of the complex to the 5' cap; Biochemistry Faculty; http://webcache.googleusercontent.com/
Proposed physical and function model of the association of SLBP with the eIF4F/eIF3/PABP complex:
�
SLBP (stem loop binding protein) is shown to be bound to the 3'-terminal stem-loop of a cell cycle-regulated histone mRNA. Association of SLBP with the eIF4F/eIF3/PABP complex requires eIF4G and perhaps eIF3. But eIF4E is not required for the physical association of SLBP with the complex but it is required for SLBP function in that it is necessary for the binding of the complex to the 5' cap.
Many mRNAs, having either long or short half life span, have stem loop structure with certain sequences either at 5�UTR or at 3�UTR, along with their consensus caps and poly-A-tails. Stability of mRNAs is confirmed by their 5�end and 3�end structures in corresponding to the cellular requirements.
Relationship between mRNA degradation and translational repression of mRNAs:
A fundamental aspect of control of mRNA degradation is the interface between mRNA translation in polysomes and mRNA degradation in P-bodies. What controls the transit of mRNAs between these particles? Recent exciting results have shown that reporter miRNAs (micro-RNAs) that are targeted for translational repression by mRNAs become concentrated in P-bodies in a miRNA-dependent manner. �Argonaute proteins which are a core component of the RISC are found in P-bodies, suggesting that Argonaute binds to translationally repressed RNAs and delivers them to P-bodies. Alternatively, Argonaute, bound to the translationally repressed RNA, may nucleate the assembly of a P-body, which would be consistent with the dynamic nature of these structures. Localization of the translationally repressed RNAs to P-bodies would prevent protein synthesis, as the translation machinery is excluded from this structure. Since Argonaute binds to the Dcp1�Dcp2 complex, it might then promote decapping and subsequent degradation of the mRNA targets. These data therefore suggest that the sequestering of targeted mRNAs in P-bodies is crucial for translational repression.
The concept of de-adenylated mRNAs being stored in a repressed state is common in many biological contexts including early development and mRNA transport. For example, during Drosophila oogenesis, oskar mRNA is translationally repressed and is transported in particles to the posterior end of the oocyte before it is translationally activated. In axons and dendrites of human neurons, repressed RNAs are transported in large particles from the nucleus to the tip of the axon or dendrite where they are then translated. In some cases, similar proteins are involved in both translational repression and mRNA degradation. For example, the Drosophila homologue of the helicase Dhh1p, Me31b, which, in yeast, is required for efficient decapping, is necessary for the translational repression of bicoid and oskar mRNAs and is located in cytoplasmic particles in egg chambers. The Xenopus homologue of Dhh1p, Xp54, appears to repress mRNA translation directly and also is a major constituent of maternal storage particles. For C. elegans, the Dhh1p homologue cgh-1 is expressed specifically in the germ line and early embryo, and is localized to P-granules and other putative mRNA�protein particles. Therefore transport of RNAs to P-bodies may not be one-way; as with other RNA granules, the mRNAs may re-emerge to be activated and translated.
U1-A Auto regulation on and off:
- One of the small Mol.wt SnU1 RNA is involved in processing of pre-mRNAs.�
- U1 Sn RNAs are associated with SnRNPs.�
- One of the SnRNPs is U1A protein.�
- Its synthesis is controlled by U1A snRNP gene.�
- The pre-mRNA of U1A protein contains two sequences at its 3� end called U1A binding boxes.� These are located upstream of poly-A signal site.�
- It is well known that poly-A provides stability to mRNA.�
- When U1A proteins, present in the cells more than required, they bind to the U1A boxes of its mRNA.�
- In the presence of these factors at U1A boxes does not prevent the assembly of other cleavage specific protein factor (CPSFs), CF1 and CF2 (cleavage factors) and PAP (poly A polymerase) which are required for cleaving and 3�end poly-adenylation.�
- The assemblies of these factors in association with U1A, perform cleavage at specific site, but prevent poly-adenylation to the free 3� end.� The mRNAs, with free 3�end without long poly-A and its associated PAB-II proteins, are immediately subjected to 3�exonuclease degradation. �Thus U1A brings down the level of nU1 RNA level.
On the contrary if the concentration of these factors is less than required, they don�t bind to U1A boxes and the U1A pre-mRNA is processed and the mRNA produced translates to generate the functional U1A factor.
Mechanism of Translational Control Involving the Poly-A tail:
Viruses employ a variety of unconventional strategies to usurp the cellular translation machinery. eIF4G and its interaction partners are preferred targets for viruses. During infection by certain picorna viruses, the N-terminal third of eIF4G that links the protein to cap structure and poly (A) tail of cellular mRNAs is cleaved from the remainder of the protein by proteolytic attack. Similarly, the PABC domain is removed from PABP. These manipulations lead to an inhibition of translation of cellular mRNA in favor of selective translation of the viral mRNA. Another example is the case of the above-mentioned, rotaviruses, which employ a translational strategy that directly targets the PABP-eIF4G interaction.
Similarly, the PABC domain is removed from PABP. �These manipulations lead to an inhibition of translation of cellular mRNA in favor of selective translation of the viral mRNA. Another example is the case of the above-mentioned rotaviruses, which employ a translational strategy that directly targets the PABP-eIF4G interaction.
- Cells are also endowed with enzymes that can decap mRNAs with an enzyme called Decappase coded for by dcp-1addna2 genes.�
- When the concentration of PAB-I goes down below certain level, poly-A is removed very fast by poly-A nuclease (PAN).�
- This is followed by rapid degradation of 5� ends by an exonuclease enzyme called Xrn1 gene product.��
- Degraded of mRNA takes place in two modes; one deadenylation dependent manner and the other are deadenylation independent manner.� In this process, if the translation of the mRNA is terminated before its normal TER codons, and an exonuclease degrades mRNA from 5�end
- Increase in the level of translatable mRNA can be achieved either by excessive synthesis of mRNA or by preventing the degradation of synthesized mRNA. Either way translatable mRNA levels remain high.
�
mRNA degradation:
RNA abundance is regulated by balancing transcription and degradation, processes that control the temporal and spatial distribution of cellular RNA. In eukaryotic cells, mRNA decay is catalyzed by two major pathways, and both can be initiated by deadenylation of the polyadenylated (poly-A) tail. After decapping, 5' to 3' RNA degradation is accomplished by Xrn1, a 5' to 3' exoribonuclease. �In the 3' to 5' pathway, RNA degradation is catalyzed by a multi-subunit 3' to 5' exoribonuclease complex, the RNA exosome. The cap structure and the poly (A) tail also play important roles in mRNA degradation. It is thus not surprising that there are numerous indications for an intimate connection between translation and mRNA turnover. Four distinct pathways of mRNA decay are known in eukaryotes and have been primarily studied in Saccharomyces cerevisiae. Two pathways start with deadenylation as the initial step and are thought to occur on many if not all mRNA species. In the predominant pathway first start of de adenylation a 3�end; this is followed by a removal of the cap structure, termed �decapping�, and 5' to 3' exonucleolytic degradation of the body of the mRNA. Alternatively, mRNAs can be degraded in 3' to 5' direction following deadenylation. Nonsense-mediated decay (NMD) is a more specific mechanism, which ensures that aberrant mRNA molecules are rapidly decapped and degraded 5' to 3' independently of Deadenylation. Finally, some mRNAs are known to contain cleavage sites for specific endonucleases that can trigger their degradation.
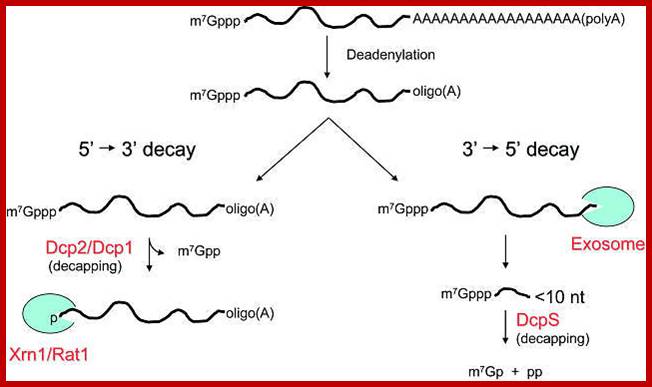
Decapping and Deadenylation dependent mRNA decay; http://www.mskcc.org/
�There are good indications that the predominant mRNA turnover pathway is regulated by the status of the translation initiation machinery on the mRNA. Analyses of yeast strains containing mutations in the translation initiation factors eIF4E, eIF4G, eIF4A or a subunit of eIF3 show increased rates of deadenylation and subsequent decapping of both unstable MFA2 and stable PGK1 mRNAs. �By contrast, inhibiting the elongation step of translation slows down deadenylation and decapping, perhaps indicating a window of opportunity for mRNA degradation in the translation cycle. �An attractive model proposes that deadenylation rates reflect the degree of accessibility of the poly(A) tail for the deadenylase. Likewise, decapping rates may reflect a competition between eIF4E and the decapping enzyme Dcp1p for the cap structure. Several observations provide initial molecular explanations for these models. eIF4E has a much lower intrinsic affinity for cap than Dcp1p. During active translation initiation, this is probably compensated for by an enhancement of eIF4E-binding to the cap structure, through the interaction with eIF4G. �Dcp1p also binds directly to eIF4G and Pab1p, either as individual proteins or as part of the eIF4F-Pab1p complex.� �Pab1p can modulate the interactions of eIF4G and eIF4E with the mRNA24 while eIF4G can stimulate Dcp1p activity. The latter effect is blocked by eIF4E. �It has also been shown that Pab1p is required for the poly(A) tail to function as an inhibitor of decapping. Thus, the emerging picture is one of a dynamic complex that bridges between the mRNA ends and is involved in the switch from translation initiation to mRNA degradation. Loss of Pab1p-binding to the deadenylated 3' end of the mRNA could trigger a rearrangement at the 5' end that allows Dcp1 access to the cap structure�
Multifunctional control of mRNA decay and translation.
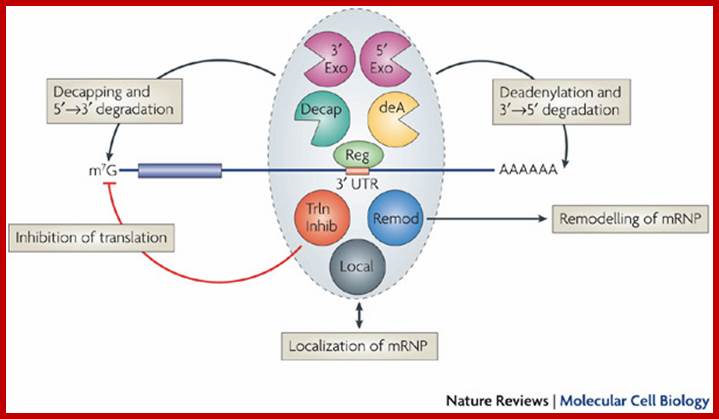
Aaron C. Goldstrohm & Marvin Wickens; http://www.nature.com/
Regulators (Reg) can control mRNA deadenylation, decay and translation by recruiting multifunctional complexes to mRNAs. The composition, assembly and enzymatic activities of the complex dictate the regulatory outcome. These complexes (shown in the grey oval) can include enzymes that deadenylate (deadenylases; deA), decap and degrade mRNAs (for example, 5' exonucleases such as XRN1 or 3' exonucleases such as the exosome). Complexes can also consist of proteins that inhibit translation (5'-cap- or eIF4E-binding proteins, or helicases such as Dhh1 orthologues; shown as Trln Inhib), remodel protein�mRNA complexes (for example, helicases; shown as Remodeller or control mRNA localization (shown as Local). The remodeling of the mRNA�protein complex (mRNP) could affect one or multiple regulatory steps. The recruitment of a multifunctional regulatory complex can occur through a single molecular contact with a single subunit of that complex. Likewise, the regulator can nucleate the assembly of a multifunctional complex on the mRNA through separate contacts with multiple regulatory factors. In other cases, the recruitment can occur through combinatorial interactions between regulators that are bound to the 3' untranslated region and several subunits of the complexes. Aaron C. Goldstrohm & Marvin Wickens; http://www.nature.com/
Translational control by changes in poly(A) tail length: recycling mRNAs:
Examples of Cytoplasmic Deadenylation:
Beyond the well-known function of poly(A) tail length in mRNA stability, recent years have witnessed an explosion of information about how changes in tail length and the selection of alternative polyadenylation sites contribute to the translational regulation of a large portion of the genome. The mechanisms and factors mediating nuclear and cytoplasmic changes in poly(A) tail length have been studied in great detail, the targets of these mechanisms have been identified�in some cases by genome-wide screenings�and changes in poly(A) tail length are now implicated in a number of physiological and pathological processes. However, in very few cases have all three levels�mechanisms, targets and functions�been studied together.
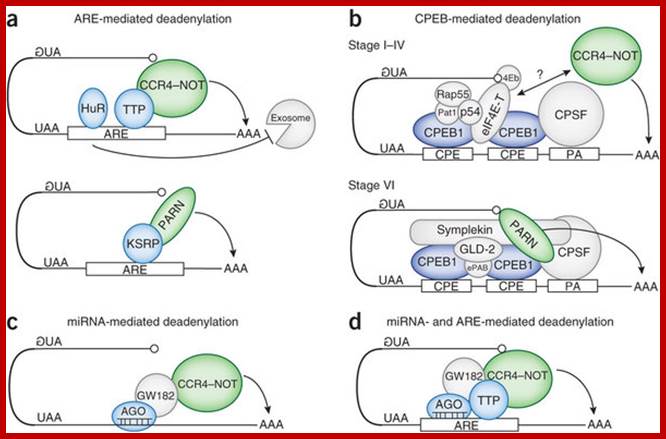
(a) AREs drive mRNA deadenylation. The binding of the ARE-BPs (in blue) TTP and KSRP on AREs induces rapid mRNA deadenylation by the recruitment of the deadenylases CCR4�NOT complex and PARN (in green), respectively. Laure Well et al.; http://www.nature.com/
Components of the mRNA degradation:
�The most complete set of factors involved in mRNA decay has been identified in yeast and many of them have recognized homologs in other eukaryotes. The DCP1 gene encodes the decapping enzyme. Dcp2p is another factor required for decapping, probably by regulating Dcp1p activity. The related proteins Edc1p and Edc2p also stimulate decapping. �Xrn1p is the exo ribonuclease responsible for 5' to 3' degradation of the body of the mRNA. �Degradation 3' to 5' is carried out by the exosome complex and the accessory proteins Ski2p. Recently, Ccr4p and Caf1p/Pop2p have been identified as components of the elusive major yeast cytoplasmic deadenylases. Both proteins have nuclease domains, Ccr4p being a member of a magnesium-dependent endonuclease-related family while Caf1p/Pop2p belongs to the RNase D family of 3' to 5' exonuclease. While Caf1p/Pop2p displays significant similarity to the human deadenylase PARN, it does not appear to represent a PARN homologue in yeast. Both Ccr4p and Caf1p/Pop2p are highly conserved in eukaryotic cells.
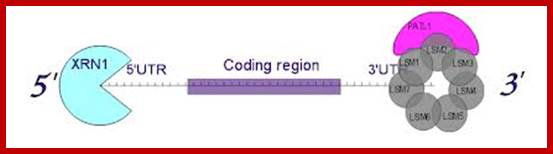
Decapped mRNA: LSM1-7: XRN1 complex in Hoosapiens; http://www.genesilico.pl
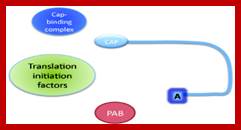
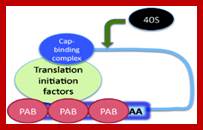
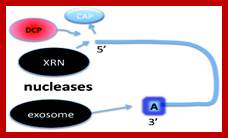
The mRNA with its Cap at the 5� end and Poly-A tail at the 3� end, bind to many factors. Translation initiation factors associate at the 5�end with CAP structure and PABII bind to Poly-A tail.� This leads to circularization and translation is initiated. With time Poly-A tail gradually shortened, with shortening of Poly-A tail and another enzyme Xrn bind to Cap and removes the cap. With PAB absent and Cap absent the mRNA is subjected to both 5� exonucleases-Decapsase and 3� exonucleases (Exosome). DA.Magnus et al; http://aghunt.wordpress.com
Additionally, a group of seven Sm-like proteins (Lsm1p-Lsm7p) and the associated factor Pat1p/ Mrt1p activate decapping in the major mRNA degradation pathway. �Sm and Sm-like proteins can form heptameric complexes in the shape of a ring. �Consistent with their role in mRNA degradation, several Lsm proteins (Lsm1, 2, 3, 5, 6, 7) co-purified biochemically with the Xrn1p exonuclease. Lsm1-7 proteins co-immunoprecipitate with Dcp1, Pat1/Mrt1 and mRNA. Two-hybrid screens suggest interactions between various Lsm proteins (Lsm1p-Lsm8p), and components of the mRNA degradation pathway: Pat1p/Mrt1p, Dcp1, Dcp2p, and Xrn1p.145 While the above results may not all represent bona fide protein-protein interactions, together they make a strong case for a role of the Lsm1-7 complex in mRNA degradation.�
�New ways to meet your (3′) end-oligouridylation (B) as one of the steps on the path to destruction�:
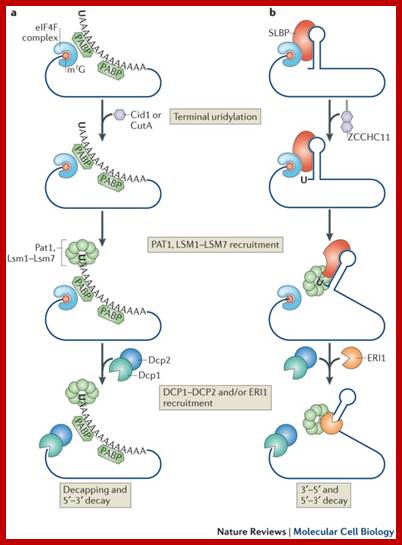
a | A polyadenylated cytoplasmic mRNA in Schizosaccharomyces pombe or Aspergillus nidulans is subject to 3′ uridylation by Cid1 or CutA, respectively. The uridylated mRNA is a preferred binding target of the Pat1 and Lsm1�Lsm7 complex, which in turn recruits the decapping enzyme Dcp1�Dcp2. This leads to translational silencing of the mRNA and allows its degradation by 5′�3′ exonucleolysis. b | A mammalian replication-dependent histone mRNA, which possesses a 3′ stem�loop rather than a poly(A) tail, is uridylated by the Cid1 orthologue zinc-finger CCHC domain-containing protein 11 (ZCCHC11). In this case, PAT1 and LSM1�LSM7 are thought to recruit the ERI1 3′�5′ exonuclease, which degrades the 3′ stem�loop to the extent that the stem�loop binding protein (SLBP) is displaced. The demonstration that such mRNAs are often degraded from both ends suggests that DCP1�DCP2 is also recruited. The single 3′ uridylyl (U) residue on the mRNA represents either monouridylation or oligouridylation.; Chris J. Norbury, http://www.nature.com/
������������� Various modes of mRNA decay is depicted
3′ Single-stranded extensions lead to similar degradation pathways in prokaryotes and eukaryotes. (A) Prokaryotic mRNAs degrade from the 3′ end through polyadenylation by PAP1 and subsequent 3′�5′ exonucleolytic decay mediated either by PNPase as shown here, or other 3′�5′ exonucleases. �Hfq, an Sm-like protein, associates with the 3′ poly (A) tract and modulates PAP and PNPase activity. (B) Histone mRNAs undergo oligouridylation by a cytoplasmic terminal uridyl transferase at the end of S phase. This leads to association of Lsm11�7, and recruitment of the decapping and 5′�3′ decay machinery. Decay also occurs 3′�5′ by the exosome. How Lsm1�7 association influences exosome activity is unclear, although there is evidence for an inhibitory role (see the text). (C) In yeast, aberrant nuclear RNAs undergo polyadenylation by the TRAMP complex, which contains the Trf4 noncanonical PAP as well as a helicase (Mtr4) and an RNA-binding protein (Air1 or Air2). TRAMP recruits the nuclear exosome (containing Rrp6) to the polyadenylated RNA leading to rapid degradation. It is not clear whether the nuclear Lsm complex (Lsm2�8) influences this process. (D) Eukaryotic mRNAs undergo polyadenylation in the nucleus in a co transcriptional process involving the CPSF and nuclear PAP, among other factors. In the cytoplasm, these mRNAs undergo degradation that is initiated by removal of the majority of the poly (A) tail (not shown). Lsm1�7 is thought to associate with the remaining 3′ oligo (A) tract and recruit the decapping machinery to induce 5′�3′ decay. In addition, the exosome degrades the transcript 3′�5′. The interplay between the exosome and Lsm1�7 is not clear, although evidence suggests that Lsm inhibits exosome activity.� http://genesdev.cshlp.org/
�All organisms require a reliable mechanism to turn genes on and off. This regulation of gene expression underlies cellular processes ranging from the response to environmental signals to the development of multi-cellular organisms and cell-cell communication. Understandably, the cell tightly controls gene expression at every step from DNA to protein. Recent work has given new insights into these control mechanisms and revealed dedicated pathways that target mRNA for degradation, thereby efficiently turning genes off.
�After export to the cytoplasm, mRNA is protected from degradation by a 5� cap
structure and PAB1 at 3� poly adenine tail. In the deadenylation dependent mRNA
decay pathway, the polyA tail is gradually shortened by exonucleases. This
ultimately attracts the degradation machinery that rapidly degrades the mRNA in
both in the 5� to 3� direction and in the 3� to 5� direction. Additional
mechanisms, including the nonsense mediated decay pathway, bypass the need for
deadenylation and can remove the mRNA from the transcriptional pool
independently. Interestingly, the same enzymes are responsible for the actual
degradation of the mRNA independent of the pathway taken (see figure).
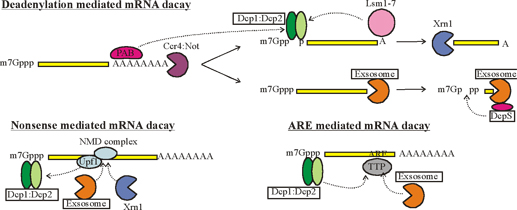
Figure: Simplified mechanism of mRNA decay. Dotted arrows indicate (indirect) interactions. Details of the pathways vary between different eukaryotes. PAB: poly-A binding protein; Max Planck Institute for developmental Biology; http://www.eb.tuebingen.mpg.de/
�Crystal structures for some of the complexes that play a role in mRNA decay have been solved, including the DCP1:DCP2 decapping complex, the DCPS scavenger decapping complex and the multi-component exosome complex. Understanding of enzyme function is, however, limited to a static 3 dimensional fold of one of the many conformations that these proteins can adopt. To fully understand how molecular motions lead to catalytic activity a complete picture of the protein dynamics is required. In addition, the catalytic activity of these enzymes must be tightly regulated to prevent premature degradation of mRNA and to ensure maximum activity as soon as an mRNA has been identified as a substrate. As such, intermolecular interactions that modulate catalytic activity, e.g. by restricting molecular motions are of foremost interest.�
![]()
Composition of a degradosome with previously shown Degadosomes: http://www.artheritis-research.com
Roles of PABP in mRNA translation and stability. This model depicts different stages of a cytoplasmic mRNA 'life cycle', in which distinct roles can be ascribed to PABP. (a) Association of PABP with the mRNA poly(A) tail. (b) Interaction of PABP with elongation initiation factor eIF4G to promote formation of the 'closed loop', thus (c) initiating translation and antagonizing decapping. (d) Interaction of PABP with the termination factor eRF3 and recycling of the ribosome from the 5' to the 3' end of the same mRNA. (e) Poly(A) shortening by the Ccr4p-Pop2p-Notp deadenylases complex. (f) Loss of the poly(A) tail and PABP, facilitating (g) dissociation of the proteins of the mRNP, binding of the Lsm1-7p-Pat1p complex, and decapping by the decapping proteins Dcp1p and Dcp2p, and subsequent (h) 5'-to-3' degradation of the mRNA by the exonuclease Xrn1p or (i) 3'-to-5' degradation by the Exosome; Magnus et al.; http://genomebiology.com/
Schematic representation of the human exosome complex:
The associations between individual components of the human exosome are hypothetical, since no structural data have been presented to date. All human exosome components analyzed so far (PM/Scl-100, PM/Scl-75, hRrp4p, hRrp40p, hRrp41p, hRrp42p, hRrp46p, and hCsl4p) are recognized by auto antibodies present in IIM sera, although some (PM/Scl-100, PM/Scl-75, and hRrp4p) are preferentially recognized. Brouwer et al. Arthritis Res 2001 3:102 doi:10.1186/ar147
This model depicts different stages of a cytoplasmic mRNA 'life cycle', in which distinct roles can be ascribed to PABP. (a) Association of PABP with the mRNA poly (A) tail. (b) Interaction of PABP with elongation initiation factor eIF4G to promote formation of the 'closed loop', thus (c) initiating translation and antagonizing decapping. (d) Interaction of PABP with the termination factor eRF3 and recycling of the ribosome from the 5' to the 3' end of the same mRNA. (e) Poly (A) shortening by the Ccr4p-Pop2p-Notp deadenylase complex. (f) Loss of the poly(A) tail and PABP, facilitating (g) dissociation of the proteins of the mRNP, binding of the Lsm1-7p-Pat1p complex, and decapping by the decapping proteins Dcp1p and Dcp2p, and subsequent (h) 5'-to-3' degradation of the mRNA by the exonuclease Xrn1p or (i) 3'-to-5' degradation by the exosome.
Micro RNA function:
�Human cells contain thousands of different micro RNAs, short RNA molecules that function as negative genetic regulators. In animal cells, micro RNAs act by annealing to mRNAs to which they are imperfectly complementary.� Several studies have shown that micro RNAs inhibit gene expression not only by repressing translation but also by directing rapid poly (A) tail removal, thereby hastening mRNA degradation. The ability of micro RNAs to expedite deadenylation does not result from decreased translation; nor does translational repression by micro RNAs require a poly (A) tail. These findings suggest that micro RNAs utilize two distinct post-transcriptional mechanisms to down regulate gene expression�.
Joel G. Belasco; http://microbiology-parasitology.med.nyu.edu/
Human cells contain hundreds of different microRNAs, short RNA molecules that function as negative genetic regulators. In animal cells, microRNAs act by annealing to mRNAs to which they are partially complementary. Our studies have shown that microRNAs inhibit gene expression not only by repressing translation but also by directing rapid poly(A) tail removal, thereby hastening mRNA degradation. Analogously, small interfering RNAs (siRNAs), the mediators of RNA interference, inhibit the function of fully complementary mRNAs both by guiding endonucleolytic cleavage of those messages and by repressing their translation. Human cells contain hundreds of different microRNAs, short RNA molecules that function as negative genetic regulators. In animal cells, microRNAs act by annealing to mRNAs to which they are partially complementary. Our studies have shown that microRNAs inhibit gene expression not only by repressing translation but also by directing rapid poly(A) tail removal, thereby hastening mRNA degradation. Analogously, small interfering RNAs (siRNAs), the mediators of RNA interference, inhibit the function of fully complementary mRNAs both by guiding endonucleolytic cleavage of those messages and by repressing their translation. Joel G. Belasco.
![]()
Mechanisms of gene repression by micro RNAs. http://mirmap.ezlab.org/
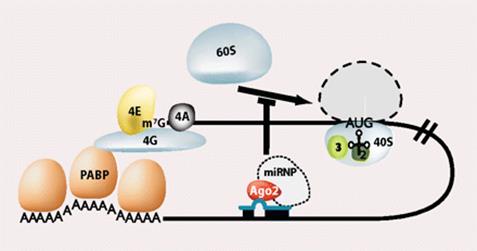
A model of miRNA-directed repression of translation initiation. Several translation initiation factors may interact with a recruited Ago protein to repress translation including the cap-binding factor, eIF4E; the protein associated with the polyA tail, PABP; the bridging protein between cap structures and the polyA tails, eIF4G; the RNA helicase that unwinds local mRNA secondary structure, eIF4A; and the multicomponent proteins associated with the 40S ribosome, eIF3 and eIF2. CrossMark:http://www.pnas.org/
Small interfering RNAs (si RNAs), the mediators of RNA interference, are closely related to micro RNAs. Although si RNAs were originally thought to inhibit the function of fully complementary messages solely by guiding endonucleolytic cleavage, recent data indicate that they can also repress translation of those messages that contain sequences not perfectly matched. Interference by miRNAs is more or less similar.� In addition, we have found that the specificity of RNA interference by si or mi RNAs is cell-type-dependent due to disparities in the tissue distribution and activity of the four Ago proteins that deliver si or mi RNAs to their mRNA targets in human cells.
Still Confused about siRNA vs. miRNA?; Here is an illustration from a recent PLoS Biology paper:
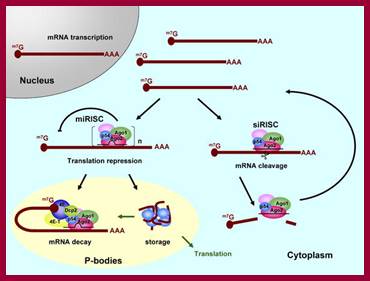
Two complexes: 1-miRNA-imperfect base pairing between the small RNA and the target leads this complex sorts the RNA to p-bodies (processing bodies) where other proteins join in. The mRNA, it is either destroyed or stored. 2-siRNA, perfect base pairing or imperfect the mRNA is destroyed and that's it.

A role for eIF4II in miRNA mediated mRNA silencing; Elisa Izaurraide; http://www.nature.com/
Because of its ability to turn off individual gene expression, RNA interference provides a remarkably precise tool for studying the effects of individual genes. There are several ways to deliver dsRNA to cells. It can be injected into a single cell or placed in a viral chromosome that infects the cells being studied. Roundworms will absorb dsRNA if they are soaked in a solution containing it, or if they eat bacteria that contain it. Unlike gene targeting, administration of dsRNA does not require long and laborious breeding of the target organism carrying the knockout.
�New Developments in DsRNA:
Recent research has also shown that a class of similar dsRNA fragments, called small temporal RNAs, plays important roles in development in the roundworm, fruit fly, and other animals. Although little is so far known about them, these fragments are made by dicer from the cell's own RNA as a normal part of the developmental process and appear to help control gene expression. This is an exception to the statement that the presence of dsRNA signals a threat to the cell; how these are distinguished from threatening dsRNA is not yet known.� In humans approximately 50% of known miRNAs are found in clusters, and they are transcribed as polycistronic primary transcripts. There are usually two or three gene miRNAs per cluster and the largest cluster at 13q31 is composed of seven genes transcripts.(Calin et al., 2004a) (He et al., 2005). (Lagos-Quintana et al., 2001; Lau et al., 2001, (Lee et al., 2002).
Micro RNA coding genes or regions;
It was initially thought that most miRNA genes are located in intergenic regions. A recent analysis of miRNA gene locations and transcription units� which are involved in combining genome assemblies and expressed sequence tag databases demonstrated more than 70% of mammalian miRNA genes are located in defined transcription units. Moreover, two thirds of miRNA genes are found in the introns in the sense orientation. About 80% of these 117 intronic miRNAs (out of 232 studied miRNAs) were in introns of protein-coding genes and just a small part of 117 miRNAs were in the introns of noncoding RNAs (ncRNAs). Interestingly miRNAs can be also present in either an exon or an intron depending on the alternative splicing pattern (14 miRNAs). This indicates that bioinformatic searches focused on intergenic regions might have missed some miRNA genes. Bioinformatic searches for miRNA-specific promoter elements upstream of miRNA sequences have not yet been successful.
Interestingly, some miRNA genes are located in the Hox clusters which play role in development. In the mammalian Hox clusters are the miR-10 and miR-196 families of miRNA genes (Lagos-Quintana et al., 2003; Lim et al., 2003b). (Rodriguez et al., 2004). Lagos-Quintana et al., 2001).
Transcription of miRNA Genes:
Scientists are searching for Pri-miRNAs transcripts and discussing their length. In case of lin-4 all the elements required for the regulation and initiation of transcription are located in a 693 bp genomic fragment. The pri-miRNA precursor for human miRNAs cluster miR-23a-miR-27a-miR-24-2 and for isolated miR-21 are unspliced ~2,2 and ~3, 4 kilobase long RNAs. Both are capped, polyadenylated non-coding RNAs. In contrast, human pri-miRNA for miR-155 contains two introns, two poly-A sites and can give two alternatively spliced pri-miRNA precursors of ~0,6 and ~1,4 kb.
There is a key question which RNA polymerase transcribes a miRNAs gene. The two candidate RNA polymerases for pri-miRNA transcription are pol II and pol III. Recently, it has been suggested that pri-miRNAs with their own promoters are Pol II products, because:
1. Pri-miRNAs are sometimes several kilobases long - longer than typical pol III transcripts.
2. Many miRNAs are differentially expressed during development - typical for pol II but not pol III products.
3. Fully functional miRNAs can be generated from a plasmid containing a pri-miRNA sequence under the control of pol II promoter.
4. Pri-miRNAs were shown to contain both CAP STRUCTURES (m7GpppN located at the 5�end) and Poly(A) tails (25�200 adenine nucleotides at the 3�end).
5. MicroRNA transcription activity is sensitive to alpha-amanitin at a concentration that specifically inhibits pol II, but not pol III.
6. Association of pol II with the promoter of miR-23a~27a~24-2 and other miRNAs was demonstrated by chromatin immunoprecipitation analyses; (Lee et al., 2004).
(Cai et al., 2004; Lee et al., 2004; see Fig 4). (Lee et al., 2002). (Lagos-Quintana et al., 2002; Krichevsky et al., 2003; Calin et al., 2004b; Miska et al., 2004). (Cai et al., 2004). (Cai et al., 2004; Lee et al., 2004), (Lee et al., 2004), (Lee et al., 1993).
�Biogenesis of RNAi�s:
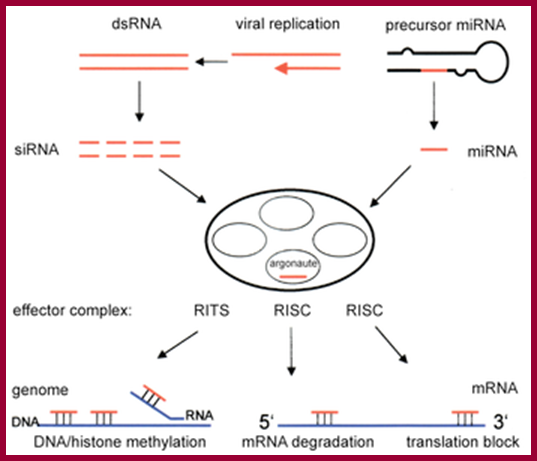
The enzyme dicer trims double stranded RNA, to form small interfering RNA or microRNA. These processed RNAs are incorporated into the RNA-induced silencing complex (RISC), which targets messenger RNA to prevent translation; http://en.wikipedia.org/
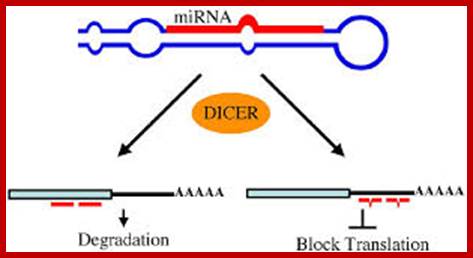
C.elegans miRNA, miRNAs use two mechanisms to exert gene regulation. Some animal miRNAs can bind to mRNA targets with exact complementarity and induce the RNAi pathway. miRNAs also bind to targets with imperfect complementarity and block translation. There is no evidence that C. elegans miRNAs use the former pathway; http://www.wormbook.org/
Biogenesis and function of intronic miRNAs: (A) The native intronic miRNA is co-transcribed with a precursor messenger RNA (pre-mRNA) by Pol-II and cleaved out of the pre-mRNA by RNA splicing machinery, spliceosome. The spliced intron with hairpin-like secondary structures is further processed into mature miRNAs capable of triggering RNAi effects, while the ligated exons become a mature messenger RNA (mRNA) for protein synthesis. (B) We, the above mentioned authors, designed an artificial intron containing pre-miRNA, namely SpRNAi, mimicking the biogenesis processes of the native intronic miRNAs. (C) When a designed miR-EGFP (280-302)-stem loop RNA construct was tested in the EGFP-expressing Tg (UAS: gfp) in zebra fishes, they are detected a strong RNAi effect only on the target EGFP. �No detectable gene silencing effect was observed in other lanes from left to right: 1, blank vector control (Ctl); 2, miRNA-stem-loop targeting HIV-p24 (mock); 3, miRNA without stem loop (anti); and 5, stem loop-miRNA* complementary to the miR-EGFP (280-302) sequence (miR*). The off-target genes such as vector RGFP and fish actin were not affected, indicating the high target specificity of miRNA-mediated gene silencing. (D) Three different miR-EGFP(280-302) expression systems were tested for miRNA biogenesis from left to right: 1, vector expressing intron-free RGFP, no pre-miRNA insert; 2, vector expressing RGFP with an intronic 5'-miRNA-stemloop-miRNA*-3' insert; and 3, vector similar to the 2 construct but with a defected 5'-splice site in the intron. In Northern bolt analysis probing the miR-EGFP (280-302) sequence, the mature miRNA was released only from the spliced intron resulted from the vector 2 construct in the cell cytoplasm.
MicroRNA (miRNA) genomic organization, biogenesis and function:
Genomic distribution of miRNA genes: The sequence encoding miRNA is shown in red. TF: transcription factor. (A) Clusters throughout the genome transcribed as polycistronic primary transcripts and subsequently cleaved into multiple miRNAs; (B) intergenic regions transcribed as independent transcriptional units; (C) intronic sequences (in grey) of protein-coding or -non-coding transcription units or exonic sequences (black cylinders) of non-coding genes. Primary miRNAs (pri-miRNAs) are transcribed and transiently receive a 7-methylguanosine (7mGpppG) cap and a poly(A) tail. The pri-miRNA is processed into a precursor miRNA (pre-miRNA) stem-loop of ∼60 nucleotides (nt) in length by the nuclear RNase III enzyme Drosha and its partner DiGeorge syndrome critical region gene 8 (DGCR8). Exportin-5 actively transports pre-miRNA into the cytosol, where it is processed by the Dicer RNaseIII enzyme, together with its partner TAR (HIV) RNA binding protein (TRBP), into mature, 22 nt-long double strand miRNAs. The RNA strand (in red) is recruited as a single-stranded molecule into the RNA-induced silencing (RISC) effector complex and assembled through processes that are dependent on Dicer and other double strand RNA binding domain proteins, as well as on members of the Argonaute family. Mature miRNAs then guide the RISC complex to the 3′ untranslated regions (3′-UTR) of the complementary messenger RNA (mRNA) targets and repress their expression by several mechanisms: repression of mRNA translation, destabilization of mRNA transcripts through cleavage, de-adenylation, and localization in the processing body (P-body), where the miRNA-targeted mRNA can be sequestered from the translational machinery and degraded or stored for subsequent use. Nuclear localization of mature miRNAs has been described as a novel mechanism of action for miRNAs. Scissors indicate the cleavage on pri-miRNA or mRNA.
Biogenesis of miRNAs and assembly into RISC complex:
�RNA pol II generates capped and polyadenylated pri-miRNAs which are processed by Drosha in the nucleus to generate pre-miRNAs. After translocation into the cytoplasm by exportins-5, pre-miRNAs are processed by Dicer to form the mature miRNA/miRNA* duplex. Following processing, miRNAs are assembled into the RISC complex. Only one strand of the duplex is stably associated with the RISC complex. The mature miRNA directs repression of mRNA containing partially complementary miRNA binding sites within the 3'UTR.
Pri-miRNA to Pre - miRNA:
MicroRNAs are first expressed as part of transcripts termed primary microRNAs and then processed enzymatically to generate pre-microRNA molecules. After transport into the cytoplasm a second enzymatic process produces active microRNA molecules that can interfere with gene expression by inducing targeted mRNA degradation or by active repression of the translation machinery of the cell.
MicroRNAs (miRNAs) are small, non-coding regulatory RNAs found in many phyla that control such diverse events as development, metabolism, cell fate and cell death. They have also been implicated in human cancers. The C. elegans genome encodes hundreds of miRNAs, including the founding members of the miRNA family lin-4 and let-7. Despite the abundance of C. elegans miRNAs, few miRNA targets are known and little is known about the mechanism by which they function. However, C. elegans research continues to push the boundaries of discovery in this area.
lin-4 and let-7 are the best understood miRNAs. They control the timing of adult cell fate determination in hypodermal cells by binding to partially complementary sites in the mRNA of key developmental regulators to repress protein expression. For example, lin-4 is predicted to bind to seven sites in the lin-14 3' untranslated region (UTR) to repress LIN-14, while let-7 is predicted to bind two let-7complementary sites in the lin-41 3' UTR to down-regulate LIN-41. Two other miRNAs, lsy-6 and mir-273, control left-right asymmetry in neural development, and also target key developmental regulators for repression. Approximately one third of the C. elegans miRNAs are differentially expressed during development indicating a major role for miRNAs in C. elegans development. Given the remarkable conservation of developmental mechanism across phylogeny, many of the principles of miRNAs discovered in C. elegans are likely to be applicable to higher animals. Biogenesis of miRNA and its action on mRNAs; www.wormbook.org
RNAi and the P-body connection
John J. Rossi; Nature Cell Biology 7, 643 - 644 (2005)
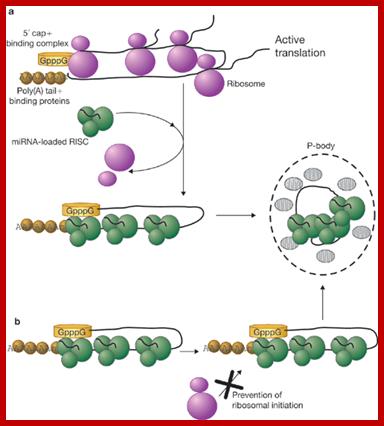
This model depicts a typical message with a 5' cap sequence bound by a cap-binding complex, and a 3' poly(A) tract bound by poly(A)-binding proteins. Two possible models could be envisaged for miRNA-loaded RISC, blocking translation and forming P-bodies. (a) First, RISC may bind to the 3' UTR of a message that is undergoing translation and block entry of ribosomal subunits at the 5' cap. (b) Alternatively, RISC may bind to the mRNA before initiation of translation, possibly as the RNA leaves the nucleus, and prevent ribosomal initiation. In both models, binding of RISC to the message is followed by formation of a P-body. Once in the P-body, the message is either stored or degraded; John J. Rossi; http://www.nature.com/
Main factors involved in nuclear degradation and mRNP surveillance:
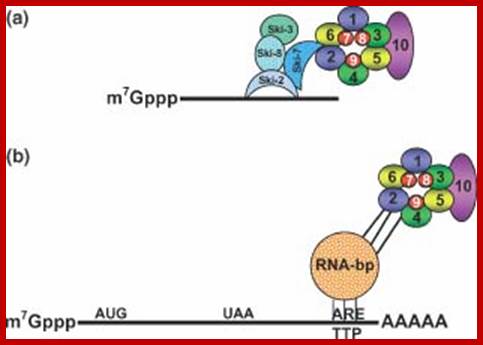
Trf4/Air2/Mtr4p Polyadenylation complex-TRAMP; it is a is a multi-protein complex consisting of the RNA helicase Mtr4, a poly(A) polymerase (PAP) (either Trf4 or Trf5) and a zinc knuckle protein (either Air1 or Air2). The TRAMP complex interacts with the exosome complex in the nucleus of eukaryotic cells and is involved in the 3' end processing of ribosomal RNA and snoRNAs; http://www.onlinelibraray.com
�(A) Structure of the RNA exosome and the TRAMP complex. The exosome core is composed of a hexameric ring with a Cap structure containing three polypeptides. In eukaryotes, the catalytic activity of the complex depends on two additional subunits, Rrp6p and Rrp44p that interact with the core exosome on the opposite site of the Cap. Rrp6p has a mainly nuclear localization, whereas Rrp44 is present both in the nucleus and in the cytoplasm. TRAMP is an exosome cofactor composed by either one of two RNA-binding proteins (Air1p and Air2p) of one helicase (Mtr4p) and either one of two poly (A) polymerases (Trf4p and Trf5p). (B) TRAMP is believed to stimulate the RNA degradation activity of the exosome by at least two mechanisms: (1) by adding an unstructured poly(A) tail to the RNA to favor the progression of the exosome through secondary structures (left-hand pathway) and (2) by helping substrate recognition through the RNA-binding activities of Air1p and Air2p (right-hand pathway).
PNPase in mRNA degradation:
Polynucleotide phosphorylase PNPase is an important enzyme responsible for mRNA turnover from 3�- to 5�-end in bacteria. It shares a similar structural organization to eukaryotic exosome core complexes. Our structural and biochemical study on E. coli PNPase shows that the trimeric structure of a KH/S1-truncated PNPase is more expanded, containing a slightly wider central RNA-binding channel than that of the wild-type PNPase. This result suggests that the KH/S1 domain is involved not only in RNA binding but it also helps PNPase to assemble into a more compact trimer. This finding is likely a general phenomenon since only bacterial PNPase and archaeal exosome with constricted channels are efficient enzymes in RNA degradation. We also study the structures and biochemical properties of human exosome component proteins and mitochondrial PNPase to further characterize the structure-and-function relationship of PNPase in mRNA binding and degradation.
mRNA quality control: Background - the central dogma of genetics:
Maintenance of life requires a myriad of biochemical processes, which turn carbohydrates, fats and proteins into energy and building blocks used by the cells of the body. These processes are carried out by molecular machines, mainly comprising proteins, which are produced and assembled based on the genetic information stored in our genome - the DNA. However, translation of DNA into protein does not occur directly, but rather via so-called messenger RNAs (mRNAs), which act as the physical moieties transferring genetic information from DNA to the molecular factories, which decode the messages of mRNAs into proteins.
Mutated messengers are eliminated by cellular quality control systems:
One type of mutation, called a "nonsense codon", often appears in mRNAs regardless of which genes they originate from. Nonsense mutations are dangerous to the cell as they often result in mRNAs, which produce truncated and toxic proteins. Fortunately, cells exercise "molecular surveillance" to detect and eliminate nonsense mRNAs before they cause too much trouble. This quality control system is called "nonsense-mediated decay" (or short "NMD"), and research by the Danish/Swiss team has now unraveled the details behind how NMD works to destroy mutated messengers.
Details revealed - human nonsense mRNAs are degraded "inside out":
The Danish/Swiss researchers have demonstrated that a particular enzyme (called SMG6), which is best described as a "molecular scissor", associates with nonsense mRNAs in the vicinity of the mutated region to simply cut the messenger in two halves (see illustration below). This cleavage exposes fragile inner parts of the mRNA, which are subject to rapid elimination by efficient cellular degradation machines. The discovery is a break-through because it goes against the prevailing view of the research field by suggesting that the NMD process is conserved in all multi-cellular organisms.
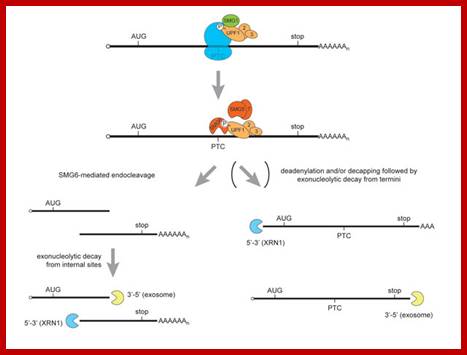
Schematic drawing illustrating how the mutated area (denoted PTC) of the nonsense mRNA (black horizontal line) is recognized by specific NMD factors (in color). After recognition the mRNA is cleaved into two halves. The published study discovered the pathway shown to the left, whereas earlier studies suggested the pathway to the right to be prevalent (the illustration is from the published work.
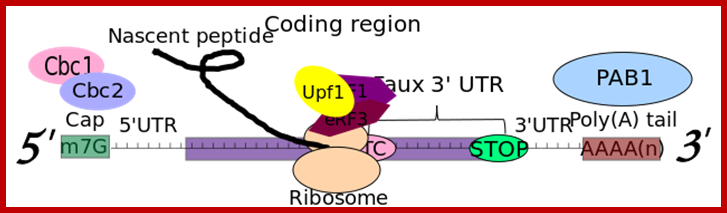
NMD pathway: mRNA transcript with PTC , ribosome bound and Upf3 bound in S.cerevisiae; http://www.genesilico.pl/http://www.genesilico.pl/
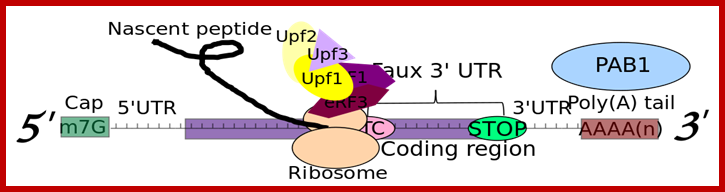
NMD pathway-Upf2, Upf3 bound to mRNA- it is marked for degradation in S.cerevisiae, http://www.genesilico.pl/; Proteins involved in the above reactions- PAB cytoplasmic and nuclear); RF1/2-GTP; ATP dependent helicase NAM7, NMD decay protein2 and 3 (all S.cerevisiae)
Removal of nonsense mRNA is crucial for the well-being of human cells. Nevertheless, nonsense mRNAs are involved in the disease progression of up to 1/3 of all described disease-causing mutations. A detailed understanding of the molecular mechanisms underlying the NMD process is imperative for the future development of targeted medicine. The published work represents a major step in this direction.
The Cap-to-Tail guide to mRNA turnover: Carol J. Wilusz, Michael Wormington and Stuart W. Peltz :
Authors observed that there are a few messenger RNAs that degrade by a minor pathway known as endoribonucleolytic decay. Endo ribonucleases recognize specific sequence elements within the transcript and cleave the mRNA internally. The cleavage event generates free 3' and 5' ends that are easily accessible to exonucleases and the products of the cleavage reaction are therefore rapidly degraded.
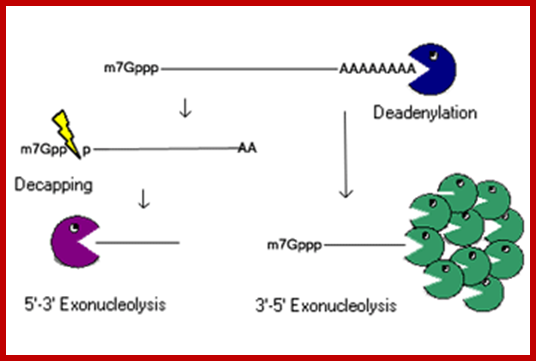
mRNA degradation in mammalian cells; Jeens Lykke-Anderson; http://labs.biology.ucsd.edu/
Interestingly,
several mRNAs that are degraded by this pathway also interact with proteins
that block access of the endo ribonuclease to its cleavage site. For example,
an endonuclease from Xenopus laevis hepatocytes, PMR1, can cleave the vitellogenin mRNA but its action is
prevented by binding of the vigilin protein to a site that overlaps the PMR1 cleavage site. Similarly, the ![]() -globin
mRNA is cleaved at a site in its 3' UTR by an erythroid-enriched endonuclease.
In this case, cleavage is inhibited by binding of the
-globin
mRNA is cleaved at a site in its 3' UTR by an erythroid-enriched endonuclease.
In this case, cleavage is inhibited by binding of the ![]() -CP
complex of proteins to an overlapping sequence.
-CP
complex of proteins to an overlapping sequence.
Different Modes of mRNA: NMD mode of Decay:
Recognition of premature termination codons in humans is splicing dependent. (a) During pre-mRNA processing, introns are removed and a set of proteins called the exon-junction complex is deposited. According to the current model for mammalian NMD, these complexes serve to facilitate transport from the nucleus and to remember the gene structure. During the first, pioneering, round of translation, the ribosome will displace all exon-junction complexes in its path until it reaches a stop codon. If the termination codon is on or near the final exon, as is the case for most genes, the ribosome will have displaced all exon-junction complexes. The mRNA will then undergo multiple rounds of translation. (b) If the termination codon is sufficiently far upstream of the final intron position, exon-junction complexes will remain. mRNA by NMD.
mRNA surveillance; UPF1 is a conserved helicase which is phosphorylated in the process of NMD. This phosphorylation is catalyzed by SMG1 kinase. This process requires UPF2 and UPF3. Dephosphorylation of UPF1 is catalyzed by SMG5, SMG6 and SMG7 proteins; UPF1 is a conserved helicase which is phosphorylated in the process of NMD.� This phoshorylation is catalyzed by SMG1 kinase which requires UPF2 and Upf3. http://en.wikipedia.org/
Nonsense Mediated decay is involved in detection and decay of mRNA transcripts which contain premature termination codons (PTCs). PTCs can arise in cells through various mechanisms: germ line mutations in DNA; somatic mutations in DNA; errors in transcription; or errors in post transcriptional mRNA processing. Failure to recognize and decay these mRNA transcripts can result in the production of truncated proteins which may be harmful to the organism. By causing decay of C-terminally truncated polypeptides, the NMD mechanism can protect cells against deleterious dominant-negative, and gain of function effects. �PTCs have been implicated in approximately 30% of all inherited diseases; as such, the NMD pathway plays a vital role in assuring overall survival and fitness of an organism.
Nonsense mediated decay in mammals is mediated by the exon-exon junction. This junction is marked by a group of proteins which constitute the exon junction complex (EJC). The EJC recruits UPF1/SMG by transcription factors eRF1/eRF3. Interactions of these proteins lead to the assembly of the surveillance complex. This complex is ultimately responsible for the degradation of the nonsense mRNA; http://en.wikipedia.org/
A surveillance complex consisting of various proteins (eRF1, eRF3, Upf1, Upf2 and Upf3) is assembled and scans the mRNA for premature stop codons. The assembly of this complex is triggered by premature translation termination. If a premature stop codon is detected then the mRNA transcript is signaled for degradation � the coupling of detection with degradation occurs.
Nonsense mediated decay in mammals is mediated by the exon-exon junction. This junction is identified by a group of proteins which constitute, the exon junction complex (EJC). The EJC recruits UPF1/SMG by transcription factors eRF1/eRF3. Interactions of these proteins lead to the assembly of the surveillance complex. This complex is ultimately responsible for the degradation of the nonsense mRNA.
A premature stop codon must be recognized as different from a normal stop codon so that only the former triggers a NMD response. It has been observed that the ability of a nonsense codon to cause mRNA degradation depends on its relative location to the downstream sequence element and associated proteins [1]. Studies have demonstrated that nucleotides more than 50-54 nucleotides upstream of the last exon-exon junction can target mRNA for decay. Those downstream from this region are unable to do so. Thus, nonsense codons lie more than 50-54 nucleotides upstream from the last exon boundary whereas natural stop codons are located within terminal exons. Exon Junction Complexes (EJCs) mark the exon-exon boundaries. EJCs are multiprotein complexes that assemble during splicing at a position about 20-24 nucleotides upstream from the splice junction. It is this EJC that provides position information needed to discriminate premature stop codons from natural stop codons. Recognition of PTCs appears to be dependent on the definitions of the exon-exon junctions. This suggests involvement of the spliceosome in mammalian NMD. Research has investigated the possibility of spliceosome involvement in mammalian NMD and has determined this is a likely possibility. Furthermore, it has been observed that NMD mechanisms are not activated by nonsense transcripts that are generated from genes that naturally do not contain introns (i.e. Histone H4, Hsp70, melanocortin-4-receptor).
When the ribosome reaches a PTC, the translation factors eRF1 and eRF3 interact with retained EJC complexes though a multiprotein bridge. The interactions of UPF1 with the terminating complex and with UPF2/UPF3 of the retained EJCs are critical. It is these interactions which target the mRNA for rapid decay by endogenous nucleases.
Nonsense mediated mRNA decay in invertebrates is postulated to be mediated by the presence of a faux 3' untranslated region (UTR). These faux 3'UTRs are distinguished from natural 3'UTRs which follow natural stop codons. This is due to the lack of binding proteins which are normally present in natural 3'UTR. These binding proteins include the poly(A)-binding protein (PABP). http://en.wikipedia.org/
NMD Avoidance:
mRNAs with nonsense mutations are generally thought to be targeted for decay via the NMD pathways. The presence of this premature stop codon about 50-54 nts 5� to the exon junction appears to be the trigger for rapid decay; however, it has been observed that some mRNA molecules with a premature stop codon are able to avoid detection and decay. �In general, these mRNA molecules possess the stop codon very early in the reading frame (i.e. the PTC is AUG-proximal). This appears to be a contradiction to the current accepted model of NMD as this position is significantly 5� of the exon-exon junction.
This has been demonstrated in β-globulin. β-globulin mRNAs containing a nonsense mutation early in the first exon of the gene are more stable than NMD sensitive mRNA molecules. The exact mechanism of detection avoidance is currently not known. It has been suggested that the poly-A binding protein (PABP) appears to play a role in this stability. It has been demonstrated in other studies that the presence of this protein near AUG-proximal PTCs appears to promote the stability of these otherwise NMD sensitive mRNAs. It has been observed that this protective effect is not limited only to the β-globulin promoter. This suggests that this NMD avoidance mechanism may be prevalent in other tissue types for a variety of genes. The current model of NMD may need to be revisited upon further studies.
Degradation of aberrant mRNAs with nonsense codons: Ana Eulalio, Isabelle Behm-Ansmant and Elisa Izaurralde.
mRNAs
with premature translation-termination codons PTCs, also known as nonsense
codons) are degraded by a conserved quality-control mechanism, known as the
nonsense-mediated mRNA decay (NMD) pathway. Current models indicate that
prematurely terminating ribosomes signal the presence of a nonsense codon, and
this leads to the recruitment of NMD factors that assemble onto faulty mRNAs to
form a surveillance complex. The surveillance complex then recruits the general
decay enzymes. In Saccharomyces cerevisiae and mammals
(see the figure, part a), PTC-containing mRNAs are
degraded by decapping (which is catalyzed by the decapping enzyme DCP2 and the
co-activator DCP1) and 5'![]() 3'-exonucleolytic degradation by XRN1 (without undergoing
deadenylation), or by deadenylation and 3'
3'-exonucleolytic degradation by XRN1 (without undergoing
deadenylation), or by deadenylation and 3'![]() 5' degradation by the exosome and the SKI complex. In Drosophila melanogaster (see the figure, part b),
degradation is initiated by endonucleolytic cleavage, the resulting RNA
fragments are degraded from the newly generated 5' ends by XRN1 and from the 3'
ends by the exosome and the SKI complex. This implies a link between NMD
factors and P-body components. In agreement with this, human SMG7, which is an
NMD factor, localizes to P bodies and recruits the surveillance-complex protein
UPF1 and another NMD factor, SMG5. Furthermore, UPF1 triggers P-body formation
and the accumulation of the other surveillance-complex components UPF2 and UPF3
and NMD substrates in these bodies in S. cerevisiae cells
that lack DCP1.
5' degradation by the exosome and the SKI complex. In Drosophila melanogaster (see the figure, part b),
degradation is initiated by endonucleolytic cleavage, the resulting RNA
fragments are degraded from the newly generated 5' ends by XRN1 and from the 3'
ends by the exosome and the SKI complex. This implies a link between NMD
factors and P-body components. In agreement with this, human SMG7, which is an
NMD factor, localizes to P bodies and recruits the surveillance-complex protein
UPF1 and another NMD factor, SMG5. Furthermore, UPF1 triggers P-body formation
and the accumulation of the other surveillance-complex components UPF2 and UPF3
and NMD substrates in these bodies in S. cerevisiae cells
that lack DCP1.
m7G, 7-methylguanosine; PTC, premature translation-termination codon:
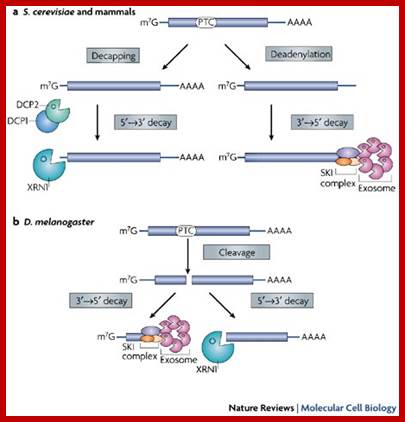
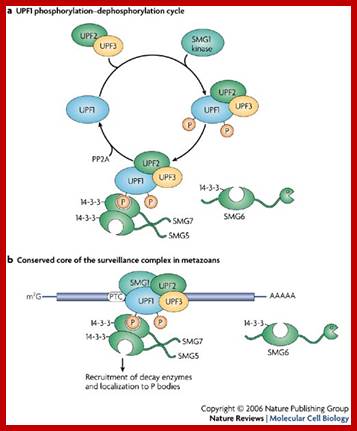 |
|
http://www.naturereviwes.com
a | UPF1 is a conserved RNA helicase that is essential for nonsense-mediated mRNA decay (NMD). UPF1 has N- and C-terminal extensions with multiple serine residues that are targets for phoshorylation. Phoshorylation of UPF1 is catalyzed by SMG1, a phosphoinositide-3-kinase-related protein kinase, and requires UPF2 and UPF3. Dephosphorylation of UPF1 is mediated by SMG5, SMG6 and SMG7, which are three similar, but not redundant, proteins. These proteins are characterized by the presence of a 14-3-3-like domain that binds phosphorylated UPF1, and they trigger UPF1 dephosphorylation by recruiting protein phosphatase-2A (PP2A). b | The precise mechanism by which NMD effectors assemble onto mRNAs that terminate translation prematurely is not completely understood and differs among species. It is generally accepted that UPF1 is recruited by terminating ribosomes and then interacts with UPF2 and UPF3. Formation of the UPF1�UPF2�UPF3 complex on the mRNA triggers UPF1 phoshorylation by SMG1. Phoshorylation of UPF1 leads to the recruitment of SMG7 (most likely in association with SMG5) through specific interactions with the 14-3-3-like domains. SMG7 then targets the bound mRNA for decay. Decay might occur in the cytoplasm or in P bodies. Indeed, SMG7 localizes to P bodies and promotes the accumulation of UPF1 and SMG5 in P-bodies. Additionally, SMG7 and SMG5 recruit PP2A, resulting in UPF1 dephosphorylation and dissociation from the 14-3-3-like binding sites (not shown). This probably has a role in the recycling of NMD effectors for a new round of NMD. The role of SMG6 in NMD remains unclear. This protein does not localize to P bodies, but has a C-terminal domain with nuclease activity.
Nonstop Mediated mRNA Decay: Overview;
mRNA surveillance; http://en.wikipedia.org/
Nonstop mediated mRNA. Translation of a mRNA without a stop codon results in the translation of the ribosome into the 3' poly-A tail region. This results in a stalled ribosome. The ribosome is rescued by two distinct pathways. The mechanisms are dependent of the absence or presence of the Ski7 protein. Ski proteins form a complex); http://en.wikipedia.org/
Nonstop Mediated decay (NMD) is involved in the detection and decay of mRNA transcripts which lack a stop codon. These mRNA transcripts can arise from many different mechanisms such as premature 3� adenylation or cryptic polyadenylation signals within the coding region of a gene. This lack of a stop codon results a significant issue for cells. Ribosomes translating the mRNA eventually translate into the 3�poly-A tail region of transcripts and stalls. As a result it cannot eject the mRNA. Ribosomes thus may become sequestered associated with the nonstop mRNA and would not be available to translate other mRNA molecules into proteins. Nonstop mediated decay mediates this problem by both freeing the stalled ribosomes and marking the nonstop mRNA for degradation in the cell by nucleases. Nonstop mediated decay consists of two distinct pathways which likely act in concert to decay nonstop mRNA.
NMD mediated Decay:
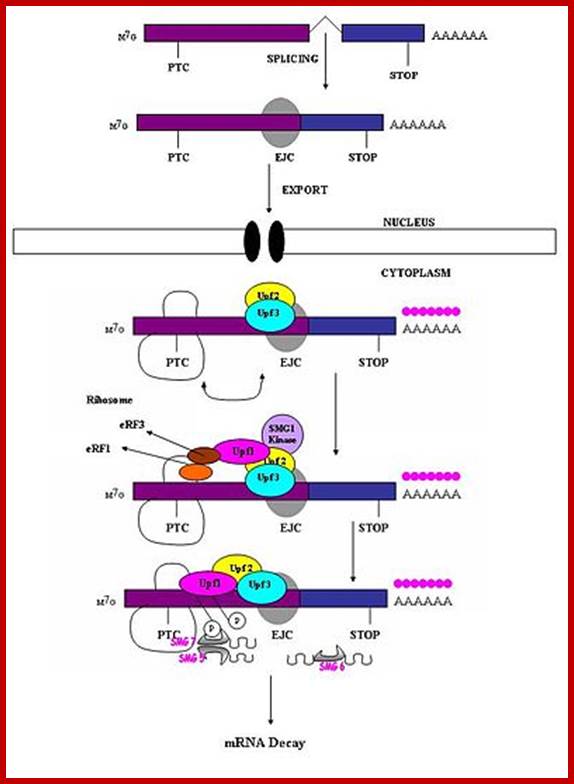
Nonsense mediated decay in mammals is mediated by the exon-exon junction. This junction is marked by a group of proteins which constitute the exon junction complex (EJC). The EJC recruits UPF1/SMG by transcription factors eRF1/eRF3. Interactions of these proteins lead to the assembly of the surveillance complex. This complex is ultimately responsible for the degradation of the nonsense mRNA; http://en.wikipedia.org/
Ski7 Pathway
This pathway is active when Ski7 protein is available in the cell. The Ski7 protein is thought to bind to the empty A site of the ribosome. This binding allows the ribosome to eject the stuck nonstop mRNA molecule � this even frees the ribosome and allows it to translate other transcripts. The Ski7 is now associated with the nonstop mRNA and it is this association which targets the nonstop mRNA for recognition by the exosome. The Ski7-exosome complex rapidly deadenylates the mRNA molecule which allows the exosome to decay the transcript in a 5� to 3� fashion.
Non Ski7 Pathway:
A second type of non-stop mediated decay has been observed in yeast. In this mechanism, the absence of Ski7 results in the loss of poly-A tail binding PABP proteins by the action of the translation ribosome. The removal of these PABP proteins then results in the loss of the protective 5�m7G cap. The loss of the cap results in rapid degradation of the transcript by an endogenous 5�-3� exonuclease such as XrnI
No-Go Mediated mRNA Decay:
No-Go mediated mRNA decay-ribosomes are stalled due various factors and even the secondary structure of mRNA; this leads to the binding of Dom34/Hbs1 at A� site of the ribosome. This leads to the cleaving of the mRNA at ribosome stalled site and the fragments are degraded using exosomes from 3� to 5� and Xrn degrade from 5� to 3�. http://en.wikipedia.org/
Stalled ribosomes due to strong secondary structure can be rescued by the Dom34-Hbs1 proteins. These proteins bind near the stall site and cleave the mRNA in an endonucleolytic fashion. The resulting mRNA fragments are decayed by the exosome and Xrn1.� As such, it is not currently well understood. This mechanism targets mRNA transcripts which have caused the ribosome to stall part way down the mRNA during translation. This stall may be caused by such factors as strong secondary structures which may physically block the translational machinery from moving down the transcript. �NGD recognizes these stall sites and prevents further translation factors from associating. The transcript is then cleaved in an endonucleolytic fashion at the stall site by Dom34-Hbs1. This process both releases the stalled ribosome and partially degrades the no-go mRNA molecule. The fragmented mRNA molecules are then fully degraded by the exosome in a 3� to 5� fashion and by Xrn1 in a 5� to 3� fashion.
It is not currently known how this process releases the mRNA from the ribosomes, however, Hbs1 is closely related to the Ski7 protein which plays a clear role in ribosome release in Ski7 mediated NSD. It is postulated that Hbs1 may play a similar role in NGD
Cellular Consequences of Impaired Translation Termination:
Efficient translation termination is essential for the production of all proteins. However, because different stop codons have different termination efficiencies depending on their sequence context, reductions in translation termination efficiency can affect the production of one protein much more than the production of another. Relatively small reductions in translation termination efficiency will therefore have significant impact on the composition of the proteome. Moreover, because of the close connection between translation termination and mRNA turnover, this may be exacerbated by additional differential effects on mRNA stability.
The exosome and RNA quality control in the nucleus: Stepanka Vanacova and Richard Stefl.
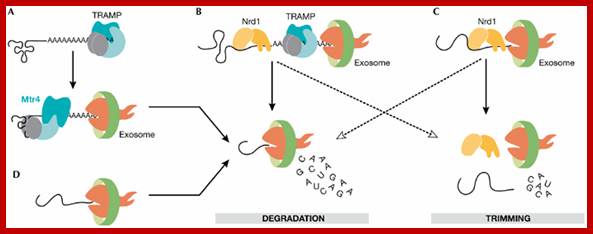
The exosome and RNA quality control in the nucleus; Stepanka Vanacova, Richard Stefl; http://embor.embopress.org/
Exonucleolytic activity of the exosome is stimulated by accessory protein complexes in vitro and in vivo. (A) The Trf4�Air2�Mtr4 polyadenylation (TRAMP) complex tags aberrant RNAs with short stretches of oligo(A)s, which initiates RNA digestion by the exosome (LaCava et al, 2005; Vanacova et al, 2005). (B) Mtr4 helicase of the TRAMP complex unwinds the structured parts of RNAs. The TRAMP complex associates with the Nrd1 complex that binds to short sequence elements on a subset of nuclear RNAs (Vasiljeva & Buratowski, 2006). The interaction between the specific RNA recognition mediated by the Nrd1 complex and the polyadenylation activity mediated by the TRAMP complex acts as the initiation step for RNA degradation by the exosome. (C)The Nrd1 complex can stimulate exosome activity on RNAs with the Nrd1 complex-specific binding sites (Vasiljeva & Buratowski, 2006). This often leads to partial digestion of the RNA (trimming), but can also cause RNA degradation. (D) The exosome destroys the leftovers of RNA processing, such as the products of endonucleolytic cleavage, apparently by itself. Air- Arginine methyltransferase-Interacting RING finger protein; Mtr- mRNA transport; Nrd-nuclear pre-mRNA down-regulation; Trf-Topoisomerase one-related function.
Transcription Termination and RNA Degradation Contribute to Silencing of RNA Polymerase II Transcription within Heterochromatin:
Lidia Vasiljeva1, Minkyu Kim1, Nihal Terzi1, Luis M. Soares1 and Stephen Buratowski1,
Within
the heterochromatin of budding yeast, RNA polymerase II (RNAPII) transcription
is repressed by the Sir2 deacetylase. Although heterochromatic silencing is
generally thought to be due to limited accessibility of the underlying DNA,
there are several reports of RNAPII and basal transcription factors within
silenced regions. Analysis of the rDNA array revealed cryptic RNAPII
transcription within the ![]() nontranscribed
nontranscribed![]() spacer region. These transcripts are terminated by the Nrd1/Sen1 complex and
degraded by the exosome. Mutations in this pathway lead to decreased silencing
and dramatic chromatin changes in the rDNA locus. Interestingly, Nrd1
mutants also show higher levels of rDNA recombination, suggesting that the
cryptic RNAPII transcription might have a physiological role in regulating rDNA
copy number. The Nrd1/Sen1/exosome pathway also contributes to silencing at
telomeric loci. These results suggest that silencing of heterochromatic genes
in Saccharomyces cerevisiae occurs at both transcriptional and
posttranscriptional levels.
spacer region. These transcripts are terminated by the Nrd1/Sen1 complex and
degraded by the exosome. Mutations in this pathway lead to decreased silencing
and dramatic chromatin changes in the rDNA locus. Interestingly, Nrd1
mutants also show higher levels of rDNA recombination, suggesting that the
cryptic RNAPII transcription might have a physiological role in regulating rDNA
copy number. The Nrd1/Sen1/exosome pathway also contributes to silencing at
telomeric loci. These results suggest that silencing of heterochromatic genes
in Saccharomyces cerevisiae occurs at both transcriptional and
posttranscriptional levels.
Mechanisms of RNA Degradation by the Eukaryotic Exosome: by Rafal Tomecki, Karolina Drazkowska, Andrzej Dziembowski
�
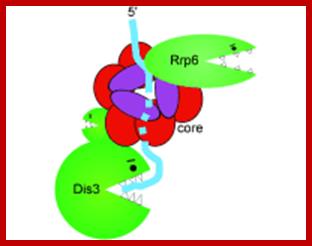
Mechanisms of RNA Degradation by the Eukaryotic Exosome; The exosome is a multi-subunit protein complex involved in essentially all phenomena associated with RNA metabolism in eukaryotic cells. This review discusses recent discoveries in the fields of biochemistry and structural biology that have shed new light on the mechanisms of RNA recruitment to the catalytic subunits of the Exosome; http://onlinelibrary.wiley.com; http://eprints.ibb.waw.pl/
RNA on its way to destruction: The exosome is a multi-subunit protein complex involved in essentially all phenomena associated with RNA metabolism in eukaryotic cells. This review discusses recent discoveries in the fields of biochemistry and structural biology that have shed new light on the mechanisms of RNA recruitment to the catalytic subunits of the exosome.
E. coli mRNA and Eukaryotic mRNAs� decay: The end defines the means in bacterial mRNA decay;
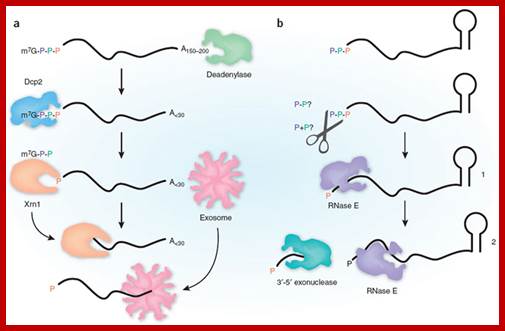
Eukaryotic mRNA decay involves poly-A deadenylation and decapping, then 5�exonuclease action by Xrn.� Prokaryotic mRNA degradation starts with dephosphorylation at 5�end then endonuclease action by RNase E and RNase II.; Daniel R Schoenberg.
E.coli decay is activated by the conversion of the mRNA 5' end from triphosphate to monophosphate and in Eukaryotes removal of the cap initiates decay
�(a) Eukaryotic mRNAs begin with an m7GpppX cap structure on the 5' end and end with a 150�200-residue poly (A) tail. Decay generally begins with shortening of the poly (A) tail to <30 residues by one or more deadenylases. The m7GpppX cap is cleaved by Dcp2 to generate RNA with a 5'-monophosphate end, which is subsequently degraded with 5'-3' polarity by Xrn1 and with 3'-5' polarity by the exosome. (b) Bacterial mRNAs begin with a 5'-triphosphate and end with a stem-loop structure, and decay involves endonuclease cleavage by RNase E. The preferred substrate for RNase E is RNA with a 5'-monophosphate, a property that is determined by the presence of a monophosphate-binding pocket within the catalytic domain of the enzyme. Celesnik et al. describe a previously unknown step in mRNA decay in which pyrophosphate is removed from the 5' end by an unknown enzyme(s) (scissors), in a manner analogous to decapping in eukaryotes, to generate a 5'-monophosphate substrate for cleavage by RNase E. This is the rate-limiting step in decay, and subsequent cleavage by RNase E generates an upstream product with a 3'-hydroxyl that is degraded by 3'-5' exonucleases and a downstream product with a 5'-monophosphate. This cycle is then repeated to complete degradation of the mRNA.
Processing bodies (PBs) and Stress granules (SGs):
Sites of translational suppression and mRNA decay
Processing (P)-bodies are small cytoplasmic foci that contain many proteins of the RNA decay machinery including deadenylases, the decapping enzymes Dcp1/Dcp2 and the 5'-3' exonuclease Xrn1. Since mRNA decay intermediates accumulate in P-bodies after inhibition of the RNA decay machinery, P-bodies are believed to be the actual sites where deadenylation, decapping and 5'-3' mRNA decay occurs. We have found that the ARE-binding proteins TTP, BRF1 and BRF2 also colocalize with P-bodies (Figure 3A), indicating that P-bodies are important for AMD. In addition, mRNAs translationally silenced by micro-RNAs are also recruited to P-bodies. These points to a dual role of P-bodies as sites of translational suppression and mRNA decay. A goal in our lab is to identify novel P-body proteins and characterize their function.
Polysomal RNA is circularized by interactions between poly(A)-binding protein 1 (PABP1) and eukaryotic translation initiation factor 4G (eIF4G), which are stabilized by eIF3.
Linearized messenger ribonucleoproteins (mRNPs) seem to be destined for processing bodies (PBs), whereas circularized mRNPs are directed to stress granules (SGs).
It is possible that mRNPs in PBs or SGs can be remodeled to nucleate the assembly of other types of RNA granules. Alternatively, selected mRNPs might move from one type of granule to another, thus creating transient tethers between different granules.
Outlines the regulatory influence of intracellular RNA-protein complex, such as processing bodies.� P-bodies and stess granules ,SGs; the RNA binding proteins RBPs and the A-to-I RNA editing that influence the efficiency of target site recognition and miRNA function and are likely to influence RNAi and antiviral defence. http://www.biomedcentral.com
Outlines the regulatory influence of intracellular RNA-Protein complexes, such as processing bodies, P-Bodies and stress granules, SGs; the RNA-binding proteins RBPs and the A-to-I RNA-editing that influence the efficiency of target site recognition and miRNA function, and are likely to influence RNAi and antiviral defense.
����������������������������������� RNA processing Bodies: mRNA processing bodies, known as P-bodies, are cytoplasmic ribonucleoprotein granules in which proteins involved in mRNA decay, silencing and translational repression accumulate with mRNAs that are destined for degradation or translational repression50. P-body components include decapping protein 2 (DCP2), decapping activators such as DCP1, enhancer of decapping 4 (EDC4) and DEAD box protein 6 (DDX6; also known as RCK), 5′-to-3′ exoribonuclease 1 (XRN1) and Argonaute and GW182 proteins. The role of P-bodies is not completely understood but it has been shown that silencing and mRNA degradation can occur in their absence22, 50, 51. Furthermore, a Drosophila melanogaster GW182 mutant that does not localize to P-bodies can rescue silencing in cells depleted of endogenous GW182 (Ref. 20). This, together with the observation that the silencing domains of GW182 proteins do not localize to P-bodies, indicates that the silencing activity of these proteins is independent of P-body localization20, 26. The image shows P-bodies (yellow) in HeLa cells expressing green fluorescent protein (GFP) fused to DCP1. Cells were stained with antibodies cross-reacting with EDC4 and a human nuclear antigen26 (red). Image courtesy of D. Lazzareti, Max Planck Institute for Developmental Biology, T�bingen, Germany.RNA processing bodies;
Felix Tritschler, Eric Huntzinger & Elisa Izaurraldehttp://www.nature.com/
Protective P bodies (green), which shelter maternal mRNAs, are just one type of P body shown in the above fig.
Silent mRNA in P-Bodies and Stress granules:
One of the most exciting recent discoveries in the
field of mRNA turnover is that mRNA degradation
in the cytoplasm seems to take place in specific particles known as P-bodies.
In both yeast and human cells, P-bodies have been shown to harbor many enzymes
of the basic decay machinery, including deadenylating enzymes, the decapping
complex and the 5'�3' exoribonucleases Xrn1. Recent studies have shown that
mRNA itself can be visualized in P-bodies when mRNA degradation is inhibited
suggesting that P-bodies are actual sites of mRNA decay. P-bodies are dynamic
structures that increase dramatically both in size and in number in response to
different forms of stress such as energy deprivation or heat shock.
In his keynote lecture, R. Parker (Tucson, AZ, USA) presented
evidence that the helicase Dhh1 and the Pat1 protein have important roles as
inhibitors of translation. These data also indicate
that P-bodies in yeast might be where mRNA is not only degraded, but relocated
after translational arrest.
In mammalian cells, translationally silenced mRNAs accumulate in stress granules, which are cytoplasmic aggregates distinct from P-bodies. Stress granules are not present under normal conditions, but form as a result of environmental stress such as heat shock, oxidative stress or energy deprivation. �Messenger RNA in stress granules remains polyadenylated; by storing translation initiation factors, small 40S ribosomal subunits and mRNAs together in one location, the cell is prepared for a rapid return to active translation as soon as the stress is removed. However, stress granules also seem to be sites where mRNAs are selected for degradation, (Cougot et al, 2004; Sheth & Parker, 2003),, (Brengues et al, 2005, (Kedersha & Anderson, 2002).
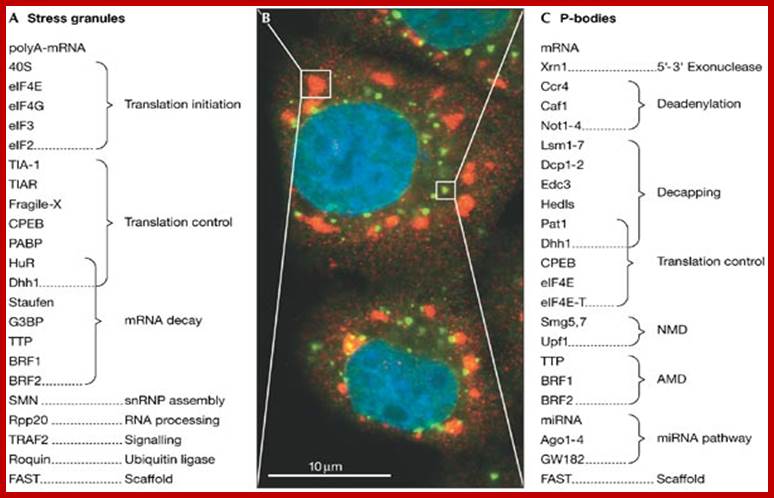
http://www..embor.emborpress.org
Stress granules and processing bodies have a central role in controlling messenger RNA translation and stability. (A) Stress granules contain aggregates of translationally stalled poly(A)-mRNA, pre-initiation complexes including the small 40S ribosomal subunit, as well as various RNA-binding proteins involved in regulating the translation and decay of specific mRNAs. Note that stress granules have not been observed in Saccharomyces cerevisiae. (B) Immunofluorescence micrograph of human HeLa cells that were subject to oxidative stress by treatment with arsenite. Fixed cells were stained in red with a polyclonal eIF4E antibody (and in green with a human auto-antiserum that recognizes GW182. Nuclei were stained in blue with Hoechst dye. Areas delineated by boxes show a stress granule (red) and a processing (P)-body (green, with partial yellow overlap). (C) P-bodies contain components of the basic mRNA degradation machinery and are thought to be the actual sites of mRNA decay. P-body-associated proteins suppress translation, induce mRNA deadenylation, decapping and 5'�3' exonucleolytic degradation. P-bodies also contain proteins that are required for specific mRNA degradation pathways such as nonsense-mediated mRNA decay (NMD), ARE-mediated mRNA decay (AMD) and microRNA (miRNA)-directed gene silencing. P-bodies are found in both yeast and mammalian cells. Cell Signaling, Beverly, MA, USA;. M.J. Fritzler, University of Calgary, AB, Canada.
Stress granules and processing bodies are related mRNA-containing granules implicated in controlling mRNA translation and decay. A genomic screen identifies numerous factors affecting granule formation, including proteins involved in O-GlcNAc modifications. These results highlight the importance of post-translational modifications in translational control and mRNP granule formation.
Localization of ribonuclease in cytoplasmic particles:
Recent work has shown that Xrn1p forms a multicomponent complex with the decapping proteins Dcp1 and Dcp2, the Lsm proteins (which are likely to form a heptameric ring encircling the RNA), the mRNA-degradation factor Pat1p and the DEAD (Asp-Glu-Ala-Asp)-box-containing helicase protein Dhh1p. These proteins are located in cytoplasmic processing bodies known as P-bodies. P-bodies have also been observed in human, mouse cells and Drosophila, and M.V. Zabolotskaya and S.F. Newbury, unpublished work. In both yeast and human cells, it has been shown that these P-bodies are the sites where mRNAs are specifically decapped and degraded.
P-bodies are dynamic structures which can change in size and number under different conditions. In yeast and human cells, the P-bodies have been shown to be affected dynamically by the availability of untranslated mRNA. For example, if the cells are treated with translation elongation inhibitors, which trap mRNAs in polysomes, P-bodies disappear within 5 min. In contrast, when mRNAs are driven off polysomes by conditions that decrease translation initiation, such as glucose deprivation, P-bodies rapidly increase in number and size . In Drosophila and human cells, the de-adenylase Ccr4 also accumulates in P-bodies, suggesting a link between de-adenylation and decapping in these particles. These results are consistent with the polysome pool and the P-body pool of RNAs being spatially distinct.
Stress granules (SGs): a historical perspective
The TIA proteins exhibit a remarkably altered sub cellular distribution in stressed cells, moving from the nucleus into the cytoplasm where they accumulate at phase-dense sub cellular sites, which were termed stress granules (SGs). Analogous structures were first described in heat shocked tomato cells, and shown to contain small heat-shock proteins (HSPs); similar structures were also noted in mammalian cells. Plant SGs were later shown to contain RNA in addition to HSPs. Remarkably, the RNA content of plant heat-shock SGs was selective; they contained mRNAs encoding housekeeping genes, but excluded newly synthesized heat-shock mRNAs. s were sites in which mRNAs were stored during stress, for later translation. The SG RNAs were translationally inactive, but could be translated in vitro, and could also be translated after the cells had recovered from the stress.
Dynamic behavior:
Mammalian SGs exhibit dynamic behavior in vivo. Arsenite treatment causes both TIA proteins to move from the nucleus to the cytoplasm and coalesce to form SGs; this process takes approx. 15 min. If the stress is removed, the SGs continue to increase in size, but decrease in number (owing to fusion of the smaller SGs), for an additional 1�2 h, depending on the severity of the stress. As each cell recovers, its SGs disperse suddenly and concurrently, again taking 6�10 min. It shows the co-ordinate recruitment of TIA-1 (green) and the RNA-stabilizing protein HuR (red) to SGs during this process. This very dynamic behavior is incompatible with the plant model of SGs as storage depots of untranslated mRNA. Alternatively, it appears that mammalian SGs at least are dynamically maintained as a result of altered mRNA metabolism, e.g. they are domains through which mRNAs pass, but whose contents are in continual flux. Many such nuclear structures exist [e.g. nucleoli, GEMS (gemini of coiled bodies), speckles or PML (promyelocytic leukemia) bodies, and some nuclear bodies are stress-induced, but SGs appear uniquely cytoplasmic.
Stress granules in chloroplasts
Stress granules: Accumulation of translationally repressed mRNAs:
Stress granules represent a different cytoplasmic structure that only appears in cells exposed to environmental stress. Stress granules contain large amounts of untranslated mRNA that accumulates during stress-induced shut-down of protein synthesis. Although stress granules are distinct from P-bodies, they often appear juxtaposed (Figure 3B). We found that the over expression of TTP, BRF1 and BRF2 further enhances the close physical association between stress granules and P-bodies (Figure 3C), which indicates a functional link between the two compartments. We have also shown that the recruitment of TTP to stress granules is regulated by the same phosphorylation events that inhibit TTP activity: phosphorylation at serine 52 and 178 prevents TTP from associating with stress granules. We are further exploring the connection between P-bodies and stress granules with the aim of better understanding how the translation and degradation of mRNAs is coordinately regulated.� The common SG inducing stimuli is the phosphorylation of alpha subunit of eIF2 (eIF2 alpha-GTP-a.atRNA complex), translation initiator factor.
�(A) In transfected COS7 cells, HA-tagged TTP (red) co localizes with P-bodies (PB, marked by Dcp1a in blue). (B) Stress granules (SG, marked by TIA1 in green) are induced after treatment with the mitochondrial inhibitor FCCP, and appear juxtaposed to P-bodies. (C) In cells transfected with HA-TTP, P-bodies are tightly associated with stress granules.
Stress granules: Cytoplasmic mRNA is routed to "stress granules" (blue) after translational silencing, and delivered to "processing bodies" (yellow) for degradation. http://www.zmbh.uni-heidelberg.de/
Protein composition of SGs:
The TIA proteins and PABP accumulate quantitatively at SGs in stressed cells, yet sedimentation experiments indicated that only a minor fraction of these proteins co-sediment in sucrose gradients, in the region corresponding to 80s monosomes. Moreover, the great preponderance of TIA-1 from stressed cells migrates as soluble mRNPs, despite its localization to seemingly large (diameters in the order of micrometers) SGs by microscopy. In light of the dynamic nature of SGs revealed by FRAP, we questioned their stability on sucrose gradients, and assessed SG composition using immunofluorescence, rather than the more obvious (but more ambiguous) biochemical purification.
Fluorescence
microscopy reveals that a subset of small ribosomal subunits is components of
SGs. Of the eIFs tested, eIF3, eIF4E and eIF4G were recruited to SGs that were
induced both by stress and by enforced expression of the eIF2 Ser51Asp
phospho-mimetic mutant, establishing them as genuine SG components. However,
eIF5 and eIF2 are missing from SGs. As eIF5 normally serves to link
eIF2�GTP�tRNA F-met to eIF3, we propose a model in which eIF5�eIF2�GTP�tRNA ![]() and
the TIA proteins compete for the same site(s) on the preinitiation complex.
Thus, the relative amounts of the active ternary complex and cytoplasmic TIA
would compete to determine whether a given transcript is initiated productively
or routed to an SG.
and
the TIA proteins compete for the same site(s) on the preinitiation complex.
Thus, the relative amounts of the active ternary complex and cytoplasmic TIA
would compete to determine whether a given transcript is initiated productively
or routed to an SG.
Other protein components of SGs include the RNA-binding protein HuR, an ARE-binding protein that stabilizes RNA, and tristetraprolin (TTP) (N. Kedersha and P. Anderson, unpublished work), another ARE-binding protein that acts to destabilize target mRNAs. Other ARE-binding proteins, such as hnRNPA1 (heteronuclear RNP-A1) and hnRNPD (heteronuclear RNP-D)/AUF-1 (AU-rich RNA binding factor), however, are not recruited to SGs. The selective recruitment of both stabilizing and destabilizing proteins to the SG supports a model in which these dynamic micro domains are sites of mRNA triage during stress.������������������������������������������������������������
Translational initiation in the absence or presence of stress:
In the absence of stress, the eIF2�GTP�tRNAMet ternary complex (green) is available to form a canonical 48 S preinitiation complex at the 5� end of capped transcripts (green arrow: normal) and scanning begins. Upon recognition of the initiation codon by the anticodon of tRNAMet, eIF5 promotes GTP hydrolysis and early initiation factors are displaced by the 60 S ribosomal subunit. As additional ribosomes are added to the transcript, the mRNA is converted into a polysome (bottom left). In stressed cells, phosphorylation of eIF2 depletes the stores of eIF2�GTP�tRNAMet, and the cytoplasmic amount of TIA-1 (yellow) increases. Under these conditions, TIA-1 is included in a non-canonical, eIF2/eIF5-deficient preinitiation complex that is translationally silent. TIA-1 auto-aggregation then promotes the accumulation of these complexes at discrete cytoplasmic foci known as SGs (composed of all components of the 48 S pre-initiation complexes except eIF2 and eIF5). Translational components that are present in SGs are shown in red; non-translation proteins present in SGs are yellow, those absent from SGs are shown in green.
RNA content and the role of SGs:
In mammalian cells, in situ staining reveals that approx. 50% of all poly(A)+ mRNA is recruited to SGs, indicating that a significant fraction of total mRNA is actively recruited to SGs. In heat- shocked HeLa cells, digoxygenin-labelled probe specific for HSP70 and double immuno-staining was used to examine the localization of HSP70 mRNA relative to the localization of TIAR protein. As described unstimulated cells exhibit the normal, predominantly nuclear distribution of TIAR (green). HSP70 mRNA (red) is not detectable in unstimulated cells, left panel. Following a 90 min treatment at 45 �C, the reorganization of TIAR into cytoplasmic SGs is apparent, middle panel, green). Newly-synthesized HSP70 mRNA (red) is apparent both in the cytoplasm as well as in a few nuclear foci, representing sites of new mRNA synthesis. It is apparent from the region shown enlarged (inset, right panels) that HSP70 mRNA is excluded from TIA-positive SGs. Thus, mammalian SGs, like their plant counterparts, specifically exclude HSP70. Whether they exclude other stress-induced transcripts that are preferentially translated during stress, such as ATF4 (activating transcription factor-4), GADD34 (growth arrest and DNA damage-34), and BiP (binding immunoglobulin protein), remains to be determined. Of the so-called 'housekeeping' transcripts, we have examined glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA as an example of a non-stress-induced transcript and found it to be quantitatively routed to SGs upon arsenite-induced stress
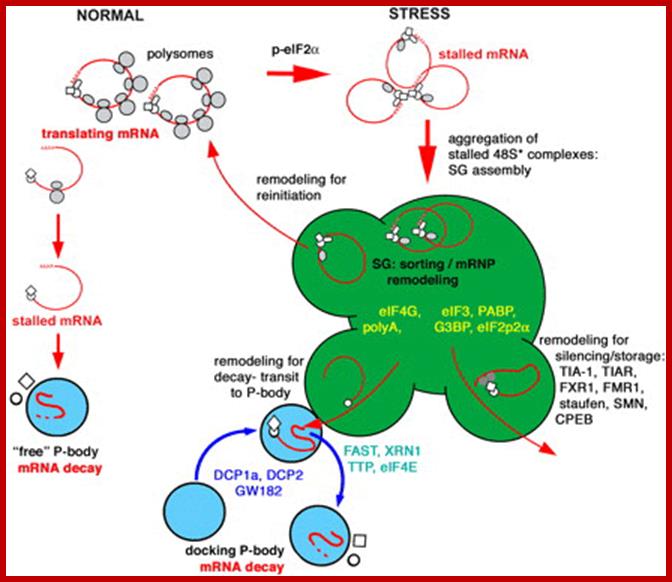
Figure. Hypothetical model of the relationship between SGs (stress granules) andPBs (processing bodies). Proteins found exclusively in SGs are shown in yellow; proteins found in both SGs and PBs which are distinct cytoplasmic sites of mRNA degradation are depicted in green; and proteins restricted to PBs are shown in blue type; Dr.Paul J.Anderson; http://bric.postech.ac.kr/
o-o