Cis Splicing of Pre-mRNAs:
Synthesis of different kinds of RNAs and their processing takes place in the nucleus. Processing sites within the nucleus has been discerned using labeled fluorescent dyes. Nucleolar rRNAs are processed in specific bodies in the nucleus called Cajal bodies. Similarly the pre-mRNAs are processed within the nuclear sap but at specific sites, called nuclear speckles; these can be discerned by using labeled dyes.
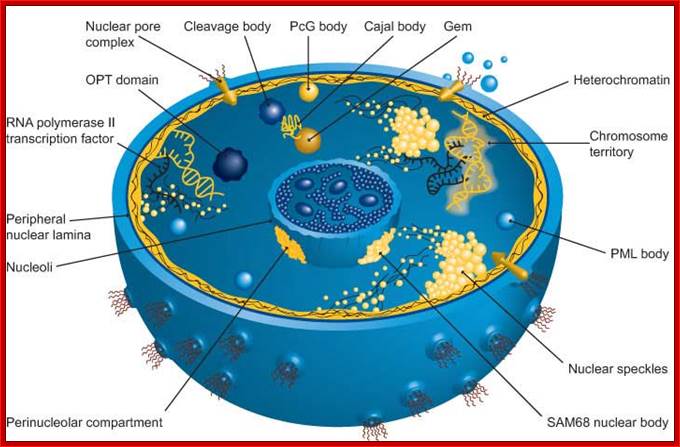
The nucleus is separated from the cytoplasm by a double membrane. The outer nuclear membrane is continuous with the endoplasmic reticulum (Spector 2001; Lamond and Sleeman 2003). Exchange of proteins and mRNA between the cytoplasm and the nucleus occur through multi-protein structures situated in the nuclear envelope known as nuclear pores. The nucleus is compartmentalized and contains numerous sub-nuclear bodies, including nucleoli, splicing speckles, Cajal bodies (CB), gems, and promyelocytic leukemia (PML) bodies in addition to chromosomes. In contrast to cytoplasmic compartments, the sub-nuclear bodies lack a membrane separating them from the nucleoplasm. The buildup of factors in these distinct sub-nuclear bodies may serve to enhance the efficiency of specific nuclear processes (ABCAM). http://www.abcam.com/
Nuclear Speckles: Speckles are irregular shaped structures of varied size and the nucleus typically contains 25-50 of such sub-nuclear bodies (Spector 2001; Lamond and Sleeman 2003). Nuclear speckles are rich in splicing factors including small nuclear ribonucleoprotein particles (snRNPs) and non-snRNP protein splicing factors such as the SC35. Speckles are often found close to actively transcribed genes and are thought to act as a reservoir for the splicing of nascent pre-mRNA at nearby genes. They found associated with active chromatin sites. The DNA free of chromatin proteins loops out at the time of transcription. Many such transcription clusters in a given location of chromatin is often called ‘transcription factory’.
Cajal bodies: Numerous coiled bodies called
Cajal bodies are found in many cell types and are typically 0.2-1 um in
diameter (Matera 1999). These structures appear as a tangle of coiled threads
and are characterized by the presence of the p80 Coilin protein. Cajal bodies are
thought to play a role in snRNP biogenesis and in the
trafficking of snRNPs and small
nucleolar RNPs (snoRNPs). Cajal bodies are rich
in spliceosomal U1, U2, U4/U6 and U5
snRNAs and snRNPs as well as U7 snRNA/snRNPs involved in histone 3’-end
processing (most histone transcripts are not polyadenylated rather their 3’
ends are produced by an endonucleolytic cleavage and U3 and U8 snoRNPs involved
in processing of pre-rRNA. It is believed that snRNPs and snoRNPs move through Cajal bodies then on
to nuclear speckles or nucleoli respectively. Gems or Gemini of Cajal bodies
are found adjacent to Cajal bodies characterized by
the presence of the survival of motor neuronal gene products (SMN) and Gemin 2. PML
bodies are characterized by the presence of PML protein. The
primary role of PML
bodies remains unclear; but they may play a role in transcriptional
regulation and anti-viral responses.
|
Splicing Diversity |
||
|
Eukaryotes |
Prokaryotes |
|
|
Spliceosomal |
+ |
- |
|
Self-splicing |
+ |
+ |
|
tRNA |
+ |
+ |
From WIKIPEDIA
PcG–Polycomb proteins of Polycomb repressive complex (PRC1), located near nuclear interchromatin compartment.
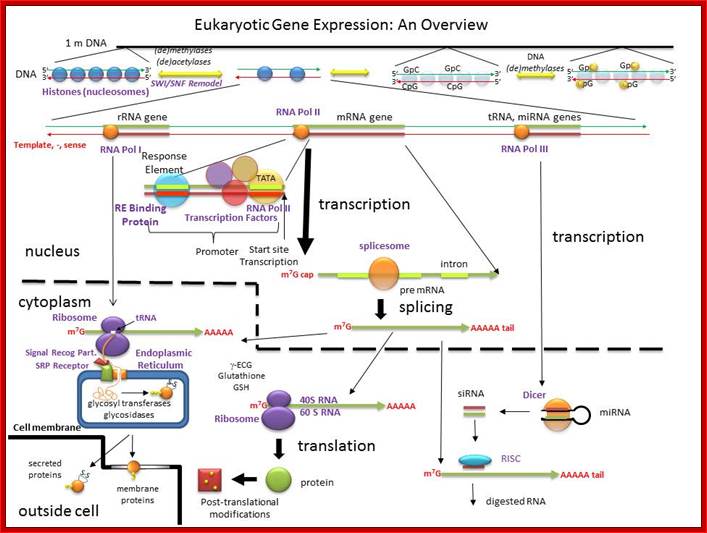
An Overview of the Regulation of Gene Expression in Eukaroytes; hjakubowski@cssju.edu; Prof. Henry Jakubowski; http://biowiki.ucdavis.edu/
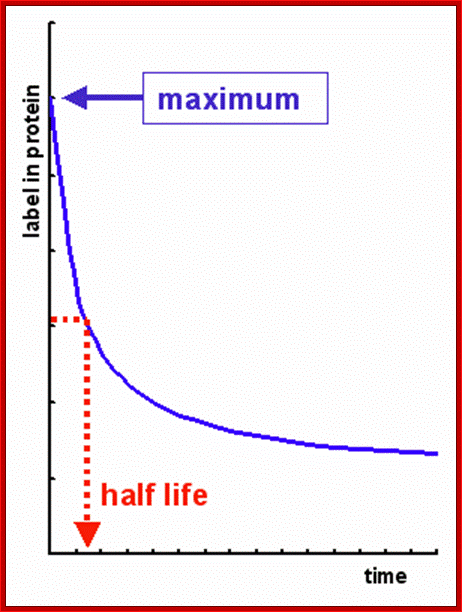
Time taken for the degradation of half of proteins; http://david-bender.co.uk/
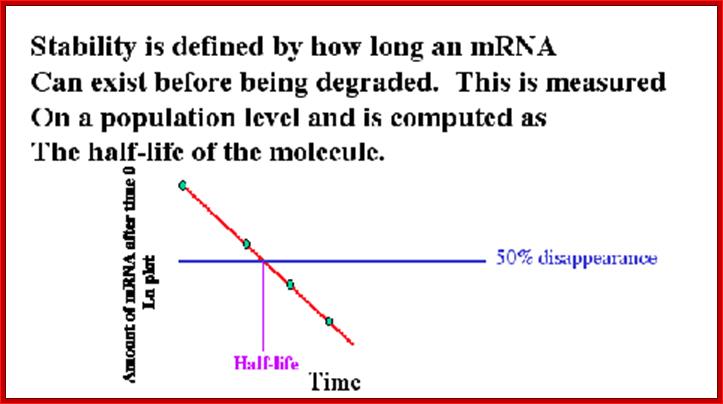
Half life of mRNAs; Ribose Nucleic Acid: http://david-bender.co.uk/
Introduction:
Ribose nucleic acids, when researchers realized that life originated on this planet as RNA world nearly 3.8 billion years ago. It is now considered as primeval molecule of life, and the life originated with RNA as the genetic material. Once DNA was deemed as the sole genetic materials of all living organisms, till the discovery of Tobacco Mosaic Virus (TMV) (Russian scientist), which contained RNA, now RNA can act as the genetic material. Once RNA was considered as an accessory form of nucleic acids sub-serving, but soon people realized, that some RNAs have coded information and some were capable of performing catalytic activity, where proteins were once considered as the sole macromolecules capable performing enzymatic catalytic functions. When life originated around 3.1-3.2 billion years ago, it was the RNA world. Everyone now trying to understand, how RNA world gave rise to DNA world. DNA world as genetic material is dominant, yet RNA has a hold on DNA’s function. Ever since Feulgan demonstrated, by chromogenic reactive agents, that there are two types of nucleic acids, one Feulgan positive called DNA and the other Feulgan’s negative called RNA, understanding of the chemistry, structure and functions in detail has been the goal of a large number of molecular biologists, yet there are many aspects of RNA remain shrouded with mystery.
Quantity of RNA in a cell:
More than 90% of the total RNA of the cell is found in cytoplasm, the rest of it is in the nucleus especially in nucleolus region.
- Total concentration of RNA changes depending upon the kind of cell and its state.
Active cells contain more RNA than inactive or resting cells. Perhaps highest concentration RNA is found in brain cells, as they are the most active cells in human body.
- In comparison to DNA, certain types of RNAs are very unstable; their half-life is just few minutes, but some may remain longer than 120 days or more.
Shorter half-life of some types of RNAs is of importance to cellular development and stability.
- Some RNA species are common to all cells; some are specific to cell types.
Chemistry:
Chemical analysis of RNA shows greater similarities with DNA with respect to the components it contains. It is basically made-up of a ribose sugar (where as DNA contains deoxyribose sugar), a phosphate group (DNA also contain the similar phosphate groups), and nitrogenous bases such as adenine, guanine, cytosine (all the three are also present in DNA) and uracil (DNA contains Thymidine in the place of Uracil).
- These individual nucleotides are synthesized in a series of enzymatic reactions. They are named after their respective bases, such as rATP, rGTP, rCTP and rUTP.
- Among them rATP is the most important energy provider for a variety of cellular functions, whereas others like rGTP or rATP derived- cyclic AMP rGTP-cyclic GMP act as a secondary messenger. rGTP also plays an important role in protein synthesis, polymerization of Tubulins (~55kDa) into microtubules, protein synthesis, enzyme regulation, glycosylation, and carbohydrate metabolism, to say they are the few functions of rGTP. Similarly, rCTP, rUTPs are also involved in glycosylation and carbohydrate synthesis. rATPs are involved in polymerization of G-Actins to F-actins. Many of the nucleotides are substantially, in a specific manner, modified after RNA synthesis.
Composition of Nucleotides in RNA varies from one species to the other, and depends on what template on which it is formed and at what time.
- There is no equivalence of purines and pyrimidines in terms of quantity unlike DNA, where total pyrimidines is equal total purines, In most of the cases DNA is double stranded, in most cases RNA is single stranded with occasional secondary structures with helical or hair pin structures.
Structure:
All RNAs, whatever may be the type, structure and functions they perform exist as polynucleotide chains with 5’ to 3’ polarity.
- Each of the polynucleotide chains is synthesized on DNA or RNA templates.
- Pyro-phosphorylase called (Polynucleotide Phosphorylase) is the enzyme found in cells capable of producing polynucleotide chains by random polymerization and without templates.
Size and molecular weight of RNAs molecules vary from one species to the other, which range from 20-22 ntds (60x365= 21900 Daltons) to 10 000 or more (365 0000 Daltons).
- Basically, RNAs are synthesized as polynucleotide chains (a primary structure), but they immediately or while they are in the process of synthesis, they are processed and fold, because of complementary base pairing into secondary structures and even possibly into tertiary structures. Binding of specific protein provides stability of 3-D structures of RNAs.
Even higher order of organization is possible with association of more than one RNA chains and a variety of proteins into a complex and compact 3-D structure ex. RNA+RNPs-Ribosomes.
Based on structural organization and functions, RNAs have been classified, into Ribosomal RNA (rRNA more than 90% of cellular RNA), Transfer RNA (tRNA) and Messenger RNA (mRNA). These are the major class of RNAs involved in decoding the information into polypeptides. But there is another category of small Mol.wt RNAs called non coding RNAs (ncRNAs). Recent estimation of them is approximately 10, 000 or more. They are now named as ncRNAs most of them are functional RNAs ex. Sc RNA, Sn RNA, Antisense RNAs such as Si RNAs or Mi micro RNAs, Primer Pri-RNAs, Guide RNAs, Efference eRNAs, Xist RNAs, piRNAs, pRNAs (p1RNase), Tm RNAs (transfer-mRNA like), SnoRNAs, , Telomeric RNA, 7sRNA, 7sK RNA, B2 RNA, Srp RNA, and many more (Refer to the chapter ‘small molecular wt. RNAs’). According to ENCODE information the HUGO people have given ~3000 specific names for the small ncRNAs. With the exception of mRNA, tRNA and Genetic RNAs all others can be clubbed into Non-Coding RNAs (Nc small RNAs). Most of these ncRNA are transcribed by non-coding DNA and many of them derived from regular transcripts but most of them are regulatory in nature.
Ribose Nucleic Acid:
Introduction:
Ribose nucleic acids, when researchers realized that life originated on this planet as RNA world nearly 3.8 billion years ago. It is now considered as primeval molecule of life, and the life originated with RNA as the genetic material. Once DNA was deemed as the sole genetic materials of all living organisms, till the discovery of Tobacco Mosaic Virus (TMV) (Russian scientist), which contained RNA, now RNA can act as the genetic material. Once RNA was considered as an accessory form of nucleic acids sub-serving, but soon people realized, that some RNAs have coded (Genetic) information and some were capable of performing catalytic activity, where proteins were once considered as the sole macromolecules capable performing enzymatic catalytic functions. When life originated around 3.1-3.2 billion years ago, it was the RNA world. Everyone now trying to understand, how RNA world gave rise to DNA world. Now, DNA world as genetic material is dominant, yet RNA has a hold on DNA’s function. Ever since Feulgan demonstrated, by chromogenic reactive agents, that there are two types of nucleic acids, one Feulgan positive called DNA and the other Feulgen’s negative called RNA, understanding of the chemistry, structure and functions in detail has been the goal of a large number of molecular biologists, yet there are many aspects of RNA remain shrouded with mystery.
Quantity of RNA in a cell:
More than 90% of the total RNA of the cell is found in cytoplasm, the rest of it is in the nucleus especially in nucleolus region.
- Total concentration of RNA changes depending upon the kind of cell and its state.
Active cells contain more RNA than inactive or resting cells. Perhaps highest concentration RNA is found in brain cells, as they are the most active cells in human body.
- In comparison to DNA, certain types of RNAs are very unstable; their half-life is just few minutes, but some may remain longer than 120 days or more.
Shorter half-life of some types of RNAs is of importance to cellular development and stability.
- Some RNA species are common to all cells; some are specific to cell types.
Chemistry:
Chemical analysis of RNA shows greater similarities with DNA with respect to the components it contains. It is basically made-up of a Ribose sugar (where as DNA contains deoxyribose sugar), a phosphate group (DNA also contain the similar phosphate groups), and nitrogenous bases such as adenine, guanine, cytosine (all the three are also present in DNA) and uracil (DNA contains Thymidine in the place of Uracil).
- These individual nucleotides are synthesized in a series of enzymatic reactions. They are named after their respective bases, such as rATP, rGTP, rCTP and rUTP.
- Among them rATP is the most important energy provider for a variety of cellular functions, whereas others like rGTP or rATP derived- cyclic AMP, rGTP-cyclic GMP, act as a secondary messenger. rGTP also plays an important role in protein synthesis, polymerization of Tubulins (~55kDa) into microtubules, protein synthesis, enzyme regulation, glycosylation, and carbohydrate metabolism, to say they are the few functions of rGTP. Similarly, rCTP, rUTPs are also involved in glycosylation and carbohydrate synthesis. rATPs are involved in polymerization of G-Actins to F-actins. Many of the nucleotides are substantially, in a specific manner, modified after RNA synthesis.
Composition of Nucleotides in RNA varies from one species to the other, and depends on what template on which it is formed and at what time.
- There is no equivalence of purines and pyrimidines in terms of quantity unlike DNA, where total pyrimidines is equal total purines, In most of the cases DNA is double stranded, in most cases RNA is single stranded with occasional secondary structures with helical or hair pin structures.
Structure:
All RNAs, whatever may be the type, structure and functions they perform exist as polynucleotide chains with 5’ to 3’ polarity.
- Each of the polynucleotide chains is synthesized on DNA or RNA templates.
- Pyro-phosphorylase called (Polynucleotide Phosphorylase) is the enzyme found in cells capable of producing polynucleotide chains by random polymerization and without templates.
Size and molecular weight of RNAs molecules vary from one species to the other, which range from 20-22 ntds (60x365= 21900 Daltons) to 10 000 or more (365 0000 Daltons).
- Basically, RNAs are synthesized as polynucleotide chains (a primary structure), but they immediately or while they are in the process of synthesis, they are processed and fold, because of complementary base pairing into secondary structures and even possibly into tertiary structures. Binding of specific protein provides stability of 3-D structures of RNAs.
Even higher order of organization is possible with association of more than one RNA chains and a variety of proteins into a complex and compact 3-D structure ex. RNA+RNPs-Ribosomes.
Based on structural organization and functions, RNAs have been classified, into Ribosomal RNA (rRNA more than 90% of cellular RNA), Transfer RNA (tRNA) and Messenger RNA (mRNA). These are the major class of RNAs involved in decoding the information into polypeptides. But there is another category of small Mol.wt RNAs called non coding RNAs (ncRNAs). Recent estimation of them is approximately 10, 000 or more. They are now named as ncRNAs most of them are functional RNAs ex. Sc RNA, Sn RNA, Antisense RNAs such as Si RNAs or Mi micro RNAs, Primer PriRNAs, Guide RNAs, Efference eRNAs, Xist RNAs, piRNAs, pRNAs (p1RNase), Tm RNAs (transfer-mRNA like), SnoRNAs, , Telomeric RNA, 7sRNA, 7sK RNA, B2 RNA, Srp RNA, and many more (Refer to the chapter ‘small molecular wt. RNAs’). According to ENCODE information the HUGO people have given ~3000 specific names for the small ncRNAs. With the exception of mRNA, tRNA and Genetic RNAs all others can be clubbed into Non-Coding RNAs (Nc small RNAs). Most of these ncRNA are transcribed by non-coding DNA and many of them derived from regular transcripts but most of them are regulatory in nature.
Ribose Nucleic Acid:
Introduction:
Ribose nucleic acids, when researchers realized that life originated on this planet as RNA world nearly ~~3.8 billion years ago. It is now considered as primeval molecule of life, and the life originated with RNA as the genetic material. Once DNA was deemed as the sole genetic materials of all living organisms, till the discovery of Tobacco Mosaic Virus (TMV) (Russian scientist), which contained RNA, now RNA can act as the genetic material. Once RNA was considered as an accessory form of nucleic acids sub-serving, but soon people realized, that some RNAs have coded information and some were capable of performing catalytic activity, where proteins were once considered as the sole macromolecules capable performing enzymatic catalytic functions. When life originated around 3.6-3.8 billion years ago, it was the RNA world. Everyone now trying to understand, how RNA world gave rise to DNA world. DNA world as genetic material is dominant now, yet RNA has a hold on DNA’s function. Ever since Feulgan demonstrated, by chromogenic reactive agents, that there are two types of nucleic acids, one Feulgan positive called DNA and the other Feulgen’s negative called RNA, understanding of the chemistry, structure and functions in detail has been the goal of a large number of molecular biologists, yet there are many aspects of RNA remain shrouded with mystery.
Quantity of RNA in a cell:
More than 90% of the total RNA, of the Present-day cell, is found in cytoplasm, the rest of it is in the nucleus especially in nucleolus region.
- Total concentration of RNA changes depending upon the kind of cell and its state.
Active cells contain more RNA than inactive or resting cells. Perhaps highest concentration RNA is found in brain cells, as they are the most active cells in human body.
- In comparison to DNA, certain types of RNAs are very unstable; their half-life is just few minutes, but some may remain longer than 120 days or more.
Shorter half-life of some types of RNAs is of importance to cellular development and stability.
- Some RNA species are common to all cells; some are specific to cell types.
Chemistry:
Chemical analysis of RNA shows greater similarities with DNA with respect to the components it contains. It is basically made-up of a ribose sugar (where as DNA contains deoxyribose sugar), a phosphate group (DNA also contain the similar phosphate groups), and nitrogenous bases such as adenine, guanine, cytosine (all the three are also present in DNA) and uracil (DNA contains Thymidine in the place of Uracil).
- These individual nucleotides are synthesized in a series of enzymatic reactions. They are named after their respective bases, such as rATP, rGTP, rCTP and rUTP.
- Among them rATP is the most important energy provider for a variety of cellular functions, whereas others like rGTP or rATP derived- cyclic AMP, rGTP, cyclic-GMP act as a secondary messenger. The rGTP also plays an important role in protein synthesis, polymerization of Tubulins (~55kDa) into microtubules, protein synthesis, enzyme regulation, glycosylation, and carbohydrate metabolism, to say they are the few functions of rGTP. Similarly, rCTP, rUTPs are also involved in glycosylation and carbohydrate synthesis. rATPs are involved in polymerization of G-Actins to F-actins. Many of the nucleotides are substantially, in a specific manner, modified after RNA synthesis.
Composition of Nucleotides in RNA varies from one species to the other, and depends on what template on which it is formed and at what time.
- There is no equivalence of purines and pyrimidines in terms of quantity unlike DNA, where total pyrimidines is equal total purines, In most of the cases DNA is double stranded, in most cases RNA is single stranded with occasional secondary structures with helical or hair pin structures.
Structure:
All RNAs, whatever may be the type, structure and functions they perform exist as polynucleotide chains with 5’ to 3’ polarity.
- Each of the polynucleotide chains is synthesized on DNA or RNA templates.
- Pyro-phosphorylase called (Polynucleotide Phosphorylase) is the enzyme found in cells capable of producing polynucleotide chains by random polymerization and without templates.
Size and molecular weight of RNAs molecules vary from one species to the other, which range from 20-22 ntds (60x365= 21900 Daltons) to 10 000 or more (365 0000 Daltons).
- Basically, RNAs are synthesized as polynucleotide chains (a primary structure), but they immediately or while they are in the process of synthesis, they are processed and fold, because of complementary base pairing into secondary structures and even possibly into tertiary structures. Binding of specific protein provides stability of 3-D structures of RNAs.
Even higher order of organization is possible with association of more than one RNA chains and a variety of proteins into a complex and compact 3-D structure ex. RNA+RNPs-Ribosomes.
Based on structural organization and functions, RNAs have been classified, into Ribosomal RNA (rRNA more than 90% of cellular RNA), Transfer RNA (tRNA) and Messenger RNA (mRNA). These are the major class of RNAs involved in decoding the information into polypeptides. But there is another category of small Mol.wt RNAs called non coding RNAs (ncRNAs). Recent estimation of them is approximately 10, 000 or more. They are now named as ncRNAs most of them are functional RNAs ex. Sc RNA, Sn RNA, Antisense RNAs such as Si RNAs or Mi micro RNAs, Primer PriRNAs, Guide-G RNAs, Efference eRNAs, Xist RNAs, piRNAs, pRNAs (p1RNase), Tm RNAs (transfer-mRNA like), SnoRNAs, Telomeric RNA, 7sRNA, 7sK RNA, B2 RNA, Srp RNA, and many more (Refer to the chapter ‘small molecular wt. RNAs’). According to ENCODE information the HUGO people have given ~3000 specific names for the small ncRNAs. With the exception of mRNA, tRNA and Genetic RNAs all others can be clubbed into Non-Coding RNAs (Nc small RNAs). Most of these ncRNA are transcribed by non-coding DNA and many of them derived from regular transcripts but most of them are regulatory in nature.
Ribose Nucleic Acid:
Introduction:
Ribose nucleic acids, when researchers realized that life originated on this planet as RNA world nearly 3.8 billion years ago. It is now considered as primeval molecule of life, and the life originated with RNA as the genetic material. Once DNA was deemed as the sole genetic materials of all living organisms, till the discovery of Tobacco Mosaic Virus (TMV) (Russian scientist), which contained RNA, now RNA can act as the genetic material. Once RNA was considered as an accessory form of nucleic acids sub-serving, but soon people realized, that some RNAs have coded information and some were capable of performing catalytic activity, where proteins were once considered as the sole macromolecules capable performing enzymatic catalytic functions. When life originated around 3.1-3.2 billion years ago, it was the RNA world. Everyone now trying to understand, how RNA world gave rise to DNA world. DNA world as genetic material is dominant, yet RNA has a hold on DNA’s function. Ever since Feulgan demonstrated, by chromogenic reactive agents, that there are two types of nucleic acids, one Feulgan positive called DNA and the other Feulgen’s negative called RNA, understanding of the chemistry, structure and functions in detail has been the goal of a large number of molecular biologists, yet there are many aspects of RNA remain shrouded with mystery.
Quantity of RNA in a cell:
More than 90% of the total RNA of the cell is found in cytoplasm, the rest of it is in the nucleus especially in nucleolus region.
- Total concentration of RNA changes depending upon the kind of cell and its state.
Active cells contain more RNA than inactive or resting cells. Perhaps highest concentration RNA is found in brain cells, as they are the most active cells in human body.
- In comparison to DNA, certain types of RNAs are very unstable; their half-life is just few minutes, but some may remain longer than 120 days or more.
Shorter half-life of some types of RNAs is of importance to cellular development and stability.
- Some RNA species are common to all cells; some are specific to cell types.
Chemistry:
Chemical analysis of RNA shows greater similarities with DNA with respect to the components it contains. It is basically made-up of a ribose sugar (where as DNA contains deoxyribose sugar), a phosphate group (DNA also contain the similar phosphate groups), and nitrogenous bases such as adenine, guanine, cytosine (all the three are also present in DNA) and uracil (DNA contains Thymidine in the place of Uracil).
- These individual nucleotides are synthesized in a series of enzymatic reactions. They are named after their respective bases, such as rATP, rGTP, rCTP and rUTP.
- Among them rATP is the most important energy provider for a variety of cellular functions, whereas others like rGTP or rATP derived- cyclic-AMP rGTP-cyclic GMP act as a secondary messenger. rGTP also plays an important role in protein synthesis, polymerization of Tubulins (~55kDa) into microtubules, protein synthesis, enzyme regulation, glycosylation, and carbohydrate metabolism, to say they are the few functions of rGTP. Similarly, rCTP, rUTPs are also involved in glycosylation and carbohydrate synthesis. rATPs are involved in polymerization of G-Actins to F-actins. Many of the nucleotides are substantially, in a specific manner, modified after RNA synthesis.
Composition of Nucleotides in RNA varies from one species to the other, and depends on what template on which it is formed and at what time.
- There is no equivalence of purines and pyrimidines in terms of quantity unlike DNA, where total pyrimidines are equal total purines. In most of the cases DNA is double stranded, in most cases RNA is single stranded with occasional secondary structures with helical or hair-pin structures.
Structure:
All RNAs, whatever may be the type, structure and functions they perform exist as polynucleotide chains with 5’ to 3’ polarity.
- Each of the polynucleotide chains is synthesized on DNA or RNA templates.
- Pyro-phosphorylase called (Polynucleotide Phosphorylase) is the enzyme found in cells capable of producing polynucleotide chains by random polymerization and without templates.
Size and molecular weight of RNAs molecules vary from one species to the other, which range from 20-22 ntds (60x365= 21900 Daltons) to 10 000 or more (365 0000 Daltons).
- Basically, RNAs are synthesized as polynucleotide chains (a primary structure), but they immediately or while they are in the process of synthesis, they are processed and fold, because of complementary base pairing into secondary structures and even possibly into tertiary structures. Binding of specific protein provides stability of 3-D structures of RNAs.
Even higher order of organization is possible with association of more than one RNA chains and a variety of proteins into a complex and compact 3-D structure ex. RNA+RNPs-Ribosomes.
Based on structural organization and functions, RNAs have been classified, into Ribosomal RNA (rRNA more than 90% of cellular RNA), Transfer RNA (tRNA) and Messenger RNA (mRNA). These are the major class of RNAs involved in decoding the information into polypeptides. But there is another category of small Mol.wt RNAs called non coding RNAs (ncRNAs). Recent estimation of them is approximately 10, 000 or more. They are now named as ncRNAs most of them are functional RNAs ex. Sc RNA, Sn RNA, Antisense RNAs such as Si RNAs or Mi micro RNAs, Primer PriRNAs, Guide G-RNAs, Efference eRNAs, Xist RNAs, piRNAs, pRNAs (p1RNase), Tm RNAs (transfer-mRNA like), SnoRNAs, , Telomeric RNA, 7sRNA, 7sK RNA, B2 RNA, Srp RNA, and many more (Refer to the chapter ‘small molecular wt. RNAs’). According to ENCODE information the HUGO people have given ~3000 specific names for the small ncRNAs. With the exception of mRNA, tRNA and Genetic RNAs all others can be clubbed into Non-Coding RNAs (Nc small RNAs). Most of these ncRNA are transcribed by non-coding DNA and many of them derived from regular transcripts but most of them are regulatory in nature.
Ribose Nucleic Acid:
Introduction:
Ribose nucleic acids, when researchers realized that life originated on this planet as RNA world nearly 3.8 billion years ago. It is now considered as primeval molecule of life, and the life originated with RNA as the genetic material. Once DNA was deemed as the sole genetic materials of all living organisms, till the discovery of Tobacco Mosaic Virus (TMV) (Russian scientist), which contained RNA, now RNA can act as the genetic material. Once RNA was considered as an accessory form of nucleic acids sub-serving, but soon people realized, that some RNAs have coded information and some were capable of performing catalytic activity, where proteins were once considered as the sole macromolecules capable performing enzymatic catalytic functions. When life originated around 3.1-3.2 billion years ago, it was the RNA world. Everyone now trying to understand, how RNA world gave rise to DNA world. DNA world as genetic material is dominant, yet RNA has a hold on DNA’s function. Ever since Feulgan demonstrated, by chromogenic reactive agents, that there are two types of nucleic acids, one Feulgan positive called DNA and the other Feulgen’s negative called RNA, understanding of the chemistry, structure and functions in detail has been the goal of a large number of molecular biologists, yet there are many aspects of RNA remain shrouded with mystery.
Quantity of RNA in a cell:
More than 90% of the total RNA of the cell is found in cytoplasm, the rest of it is in the nucleus especially in nucleolus region.
- Total concentration of RNA changes depending upon the kind of cell and its state.
Active cells contain more RNA than inactive or resting cells. Perhaps highest concentration RNA is found in brain cells, as they are the most active cells in human body.
- In comparison to DNA, certain types of RNAs are very unstable; their half-life is just few minutes, but some may remain longer than 120 days or more.
Shorter half-life of some types of RNAs is of importance to cellular development and stability.
- Some RNA species are common to all cells; some are specific to cell types.
Chemistry:
Chemical analysis of RNA shows greater similarities with DNA with respect to the components it contains. It is basically made-up of a ribose sugar (where as DNA contains deoxyribose sugar), a phosphate group (DNA also contain the similar phosphate groups), and nitrogenous bases such as adenine, guanine, cytosine (all the three are also present in DNA) and uracil (DNA contains Thymidine in the place of Uracil).
- These individual nucleotides are synthesized in a series of enzymatic reactions. They are named after their respective bases, such as rATP, rGTP, rCTP and rUTP.
- Among them rATP is the most important energy provider for a variety of cellular functions, whereas others like rGTP or rATP derived- cyclic AMP rGTP-cyclic GMP act as a secondary messenger. rGTP also plays an important role in protein synthesis, polymerization of Tubulins (~55kDa) into microtubules, protein synthesis, enzyme regulation, glycosylation, and carbohydrate metabolism, to say they are the few functions of rGTP. Similarly, rCTP, rUTPs are also involved in glycosylation and carbohydrate synthesis. rATPs are involved in polymerization of G-Actins to F-actins. Many of the nucleotides are substantially, in a specific manner, modified after RNA synthesis.
Composition of RNA varies from one species to the other, and depends on what template on which it is formed and at what time.
- There is no equivalence of purines and pyrimidines in terms of quantity unlike DNA, where total pyrimidines are equal total purines, in most of the cases DNA is double stranded, in most cases RNA is single stranded with occasional secondary structures with helical or hair pin structures.
Structure:
All RNAs, whatever may be the type, structure and functions they perform exist as polynucleotide chains with 5’ to 3’ polarity.
- Each of the polynucleotide chains is synthesized on DNA or RNA templates.
- Pyro-phosphorylase called (Polynucleotide Phosphorylase) is the enzyme found in cells capable of producing polynucleotide chains by random polymerization and without templates.
Size and molecular weight of RNAs molecules vary from one species to the other, which range from 20-22 ntds (60x365= 21900 Daltons) to 10 000 or more (365 0000 Daltons).
- Basically RNAs are synthesized as polynucleotide chains (a primary structure), but they immediately or while they are in the process of synthesis, they are processed and fold, because of complementary base pairing into secondary structures and even possibly into tertiary structures. Binding of specific protein provides stability of 3-D structures of RNAs.
Even higher order of organization is possible with association of more than one RNA chains and a variety of proteins into a complex and compact 3-D structure ex. RNA+RNPs-Ribosomes.
Based on structural organization and functions, RNAs have been classified, into Ribosomal RNA (rRNA more than 90% of cellular RNA), Transfer RNA (tRNA) and Messenger RNA (mRNA). These are the major class of RNAs involved in decoding the information into polypeptides. But there is another category of small Mol.wt RNAs called non coding RNAs (ncRNAs). Recent estimation of them is approximately 10, 000 or more. They are now named as ncRNAs most of them are functional RNAs ex. Sc RNA, Sn RNA, Antisense RNAs such as Si RNAs or Mi micro RNAs, Primer PriRNAs, Guide-G RNAs, Efference eRNAs, Xist RNAs, piRNAs, pRNAs (p1RNase), Tm RNAs (transfer-mRNA like), SnoRNAs, Telomeric RNA, 7sRNA, 7sK RNA, B2 RNA, Srp RNA and many more (Refer to the chapter ‘small molecular wt. RNAs’). According to ENCODE information the HUGO people have given ~3000 specific names for the small ncRNAs. With the exception of mRNA, tRNA and Genetic RNAs all others can be clubbed into Non-Coding RNAs (Nc small RNAs). Most of these ncRNA are transcribed by non-coding DNA and many of them derived from regular transcripts but most of them are regulatory in nature.
Ribose Nucleic Acid:
Introduction:
Ribose nucleic acids, when researchers realized that life originated on this planet as RNA world nearly ~3.8 billion years ago. It is now considered as primeval molecule of life, and the life originated with RNA as the genetic material. Once DNA was deemed as the sole genetic materials of all living organisms, till the discovery of Tobacco Mosaic Virus (TMV) (Russian scientist), which contained RNA, now RNA can act as the genetic material. Once RNA was considered as an accessory form of nucleic acids sub-serving, but soon people realized, that some RNAs have coded information and some were capable of performing catalytic activity, where proteins were once considered as the sole macromolecules capable performing enzymatic catalytic functions. When life originated around 3.1-3.8 billion years ago, it was the RNA world. Everyone now trying to understand, how RNA world gave rise to DNA world. DNA world as genetic material is dominant, yet RNA has a hold on DNA’s functions. Ever since Feulgan demonstrated, by chromogenic reactive agents, that there are two types of nucleic acids, one Feulgan positive called DNA and the other Feulgen’s negative called RNA, understanding of the chemistry, structure and functions in detail has been the goal of a large number of molecular biologists, yet there are many aspects of RNA remain shrouded with mystery.
Quantity of RNA in a cell:
More than 90% of the total RNA of the cell is found in EK cellular cytoplasm, the rest of it is in the nucleus especially in nucleolus region.
- Total concentration of RNA changes depending upon the kind of cell and its state.
Active cells contain more RNA than inactive or resting cells. Perhaps highest concentration RNA is found in brain cells, as they are the most active cells in human body.
- In comparison to DNA, certain types of RNAs are very unstable; their half-life is just few minutes, but some may remain longer than 120 days or more.
Shorter half-life of some types of RNAs is of importance to cellular development and stability.
- Some RNA species are common to all cells; some are specific to cell types.
Chemistry:
Chemical analysis of RNA shows greater similarities with DNA with respect to the components it contains. It is basically made-up of a ribose sugar (where as DNA contains deoxyribose sugar), a phosphate group (DNA also contain the similar phosphate groups), and nitrogenous bases such as adenine, guanine, cytosine (all the three are also present in DNA) and uracil (DNA contains Thymidine in the place of Uracil).
- These individual nucleotides are synthesized in a series of enzymatic reactions. They are named after their respective bases, such as rATP, rGTP, rCTP and rUTP.
- Among them, rATP is the most important energy provider for a variety of cellular functions, whereas others like rGTP or rATP derived- cyclic AMP, rGTP, cyclic GMP act as a secondary messenger. rGTP also plays an important role in protein synthesis, polymerization of Tubulins (~55kDa) into microtubules, protein synthesis, enzyme regulation, glycosylation, and carbohydrate metabolism, to say they are the few functions of rGTP. Similarly, rCTP, rUTPs are also involved in glycosylation and carbohydrate synthesis. rATPs are involved in polymerization of G-Actins to F-actins. Many of the nucleotides are substantially, in a specific manner, modified after RNA synthesis.
Composition of Nucleotides in RNA varies from one species to the other, and depends on what template on which it is formed and at what time.
- There is no equivalence of purines and pyrimidines in terms of quantity unlike DNA, where total pyrimidines is equal total purines, In most of the cases DNA is double stranded, in most cases RNA is single stranded with occasional secondary structures with helical or hair pin structures.
Structure:
All RNAs, whatever may be the type, structure and functions they perform exist as polynucleotide chains with 5’ to 3’ polarity.
- Each of the polynucleotide chains is synthesized on DNA or RNA templates.
- Pyro-phosphorylase called (Polynucleotide Phosphorylase) is the enzyme found in cells capable of producing polynucleotide chains by random polymerization and without templates.
Size and molecular weight of RNAs molecules vary from one species to the other, which range from 20-22 ntds (60x365= 21900 Daltons) to 10 000 or more (365 0000 Daltons).
- Basically, RNAs are synthesized as polynucleotide chains (a primary structure), but they immediately or while they are in the process of synthesis, they are processed and fold, because of complementary base pairing into secondary structures and even possibly into tertiary structures. Binding of specific protein provides stability of 3-D structures of RNAs.
Even higher order of organization is possible with association of more than one RNA chains and a variety of proteins into a complex and compact 3-D structure ex. RNA+RNPs-Ribosomes.
Based on structural organization and functions, RNAs have been classified, into Ribosomal RNA (rRNA more than 90% of cellular RNA), Transfer RNA (tRNA) and Messenger RNA (mRNA). These are the major class of RNAs involved in decoding the information into polypeptides. But there is another category of small Mol.wt RNAs called non coding RNAs (ncRNAs). Recent estimation of them is approximately 10, 000 or more. They are now named as ncRNAs most of them are functional RNAs ex. Sc RNA, Sn RNA, Antisense RNAs such as Si RNAs or Mi micro RNAs, Primer PriRNAs, Guide G-RNAs, Efference eRNAs, Xist RNAs, piRNAs, pRNAs (p1RNase), Tm RNAs (transfer-mRNA like), SnoRNAs, , Telomeric RNA, 7sRNA, 7sK RNA, B2 RNA, Srp RNA, and many more (Refer to the chapter ‘small molecular wt. RNAs’). According to ENCODE information the HUGO people have given ~3000 specific names for the small ncRNAs. With the exception of mRNA, tRNA and Genetic RNAs all others can be clubbed into Non-Coding RNAs (Nc small RNAs). Most of these ncRNA are transcribed by non-coding DNA and many of them derived from regular transcripts but most of them are regulatory in nature.
Ribose Nucleic Acid:
Introduction:
Ribose nucleic acids, when researchers’ realized that life originated on this planet as RNA world nearly 3.8 billion years ago. It is now considered as primeval molecule of life, and the life originated with RNA as the genetic material. Once DNA was deemed as the sole genetic materials of all living organisms, till the discovery of Tobacco Mosaic Virus (TMV) (Russian scientist), which contained RNA, now RNA can act as the genetic material. Once RNA was considered as an accessory form of nucleic acids sub-serving, but soon people realized, that some RNAs have coded information and some were capable of performing catalytic activity, where proteins were once considered as the sole macromolecules capable performing enzymatic catalytic functions. When life originated around 3.1-3.2 billion years ago, it was the RNA world. Everyone now trying to understand, how RNA world gave rise to DNA world. DNA world as genetic material is dominant, yet RNA has a hold on DNA’s function. Ever since Feulgan demonstrated, by chromogenic reactive agents, that there are two types of nucleic acids, one Feulgan positive called DNA and the other Feulgen’s negative called RNA, understanding of the chemistry, structure and functions in detail has been the goal of a large number of molecular biologists, yet there are many aspects of RNA remain shrouded with mystery.
Quantity of RNA in a cell:
More than 90% of the total RNA of the cell is found in cytoplasm, the rest of it is in the nucleus especially in nucleolus region.
- Total concentration of RNA changes depending upon the kind of cell and its state.
Active cells contain more RNA than inactive or resting cells. Perhaps highest concentration RNA is found in brain cells, as they are the most active cells in human body.
- In comparison to DNA, certain types of RNAs are very unstable; their half-life is just few minutes, but some may remain longer than 120 days or more.
Shorter half-life of some types of RNAs is of importance to cellular development and stability.
- Some RNA species are common to all cells; some are specific to cell types.
Chemistry:
Chemical analysis of RNA shows greater similarities with DNA with respect to the components it contains. It is basically made-up of a ribose sugar (where as DNA contains deoxyribose sugar), a phosphate group (DNA also contain the similar phosphate groups), and nitrogenous bases such as adenine, guanine, cytosine (all the three are also present in DNA) and uracil (DNA contains Thymidine in the place of Uracil).
- These individual nucleotides are synthesized in a series of enzymatic reactions. They are named after their respective bases, such as rATP, rGTP, rCTP and rUTP.
- Among them rATP is the most important energy provider for a variety of cellular functions, whereas others like rGTP or rATP derived- cyclic-AMP rGTP-cyclic-GMP act as a secondary messengers. The rGTP also plays an important role in protein synthesis, polymerization of Tubulins (~55kDa) into microtubules, protein synthesis, enzyme regulation, glycosylation, and carbohydrate metabolism, to say they are the few functions of rGTP. Similarly, rCTP, rUTPs are also involved in glycosylation and carbohydrate synthesis. rATPs are involved in polymerization of G-Actins to F-actins. Many of the nucleotides are substantially, in a specific manner, modified after RNA synthesis.
Composition of Nucleotides in RNA varies from one species to the other, and depends on what template on which it is formed and at what time.
- There is no equivalence of purines and pyrimidines in terms of quantity unlike DNA, where total pyrimidines is equal total purines, In most of the cases DNA is double stranded, in most cases RNA is single stranded with occasional secondary structures with helical or hair pin structures.
Structure:
All RNAs, whatever may be the type, structure and functions they perform exist as polynucleotide chains with 5’ to 3’ polarity.
- Each of the polynucleotide chains is synthesized on DNA or RNA templates.
- Pyro-phosphorylase called (Polynucleotide Phosphorylase) is the enzyme found in cells capable of producing polynucleotide chains by random polymerization and without templates.
Size and molecular weight of RNAs molecules vary from one species to the other, which range from 20-22 ntds (60x365= 21900 Daltons) to 10 000 or more (365 0000 Daltons).
- Basically, RNAs are synthesized as polynucleotide chains (a primary structure), but they immediately or while they are in the process of synthesis, they are processed and fold, because of complementary base pairing into secondary structures and even possibly into tertiary structures. Binding of specific protein provides stability of 3-D structures of RNAs.
Even higher order of organization is possible with association of more than one RNA chains and a variety of proteins into a complex and compact 3-D structure ex. RNA+RNPs-Ribosomes.
Based on structural organization and functions, RNAs have been classified, into Ribosomal RNA (rRNA more than 90% of cellular RNA), Transfer RNA (tRNA) and Messenger RNA (mRNA). These are the major class of RNAs involved in decoding the information into polypeptides. But there is another category of small Mol.wt RNAs called non coding RNAs (ncRNAs). Recent estimation of them is approximately 10, 000 or more. They are now named as ncRNAs most of them are functional RNAs ex. Sc RNA, Sn RNA, Antisense RNAs such as Si RNAs or Mi micro RNAs, Primer PriRNAs, Guide-G RNAs, Efference eRNAs, Xist RNAs, piRNAs, pRNAs (p1RNase), Tm RNAs (transfer-mRNA like), SnoRNAs, Telomeric RNA, 7sRNA, 7sK RNA, B2 RNA, Srp RNA, and many more (Refer to the chapter ‘small molecular wt. RNAs’). According to ENCODE information the HUGO people have given ~3000 specific names for the small ncRNAs. With the exception of mRNA, tRNA and Genetic RNAs all others can be clubbed into Non-Coding RNAs (Nc small RNAs). Most of these ncRNA are transcribed by non-coding DNA and many of them derived from regular transcripts but most of them are regulatory in nature.
Introduction: Ribose Nucleic Acid:
Ribose nucleic acids, when researchers realized that life originated on this planet as RNA world nearly 3.8 billion years ago. It is now considered as primeval molecule of life, and the life originated with RNA as the genetic material. Once DNA was deemed as the sole genetic materials of all living organisms, till the discovery of Tobacco Mosaic Virus (TMV) (Russian scientist), which contained RNA, now RNA can act as the genetic material. Once RNA was considered as an accessory form of nucleic acids sub-serving, but soon people realized, that some RNAs have coded information and some were capable of performing catalytic activity, where proteins were once considered as the sole macromolecules capable performing enzymatic catalytic functions. When life originated around 3.1-3.2 billion years ago, it was the RNA world. Everyone now trying to understand, how RNA world gave rise to DNA world. DNA world as genetic material is dominant, yet RNA has a hold on DNA’s function. Ever since Feulgan demonstrated, by chromogenic reactive agents, that there are two types of nucleic acids, one Feulgan positive called DNA and the other Feulgen’s negative called RNA, understanding of the chemistry, structure and functions in detail has been the goal of a large number of molecular biologists, yet there are many aspects of RNA remain shrouded with mystery.
Quantity of RNA in a cell:
More than 90% of the total RNA of the cell is found in cytoplasm, the rest of it is in the nucleus especially in nucleolus region.
- Total concentration of RNA changes depending upon the kind of cell and its state.
Active cells contain more RNA than inactive or resting cells. Perhaps highest concentration RNA is found in brain cells, as they are the most active cells in human body.
- In comparison to DNA, certain types of RNAs are very unstable; their half-life is just few minutes, but some may remain longer than 120 days or more.
Shorter half-life of some types of RNAs is of importance to cellular development and stability.
- Some RNA species are common to all cells; some are specific to cell types.
Chemistry:
Chemical analysis of RNA shows greater similarities with DNA with respect to the components it contains. It is basically made-up of a ribose sugar (where as DNA contains deoxyribose sugar), a phosphate group (DNA also contain the similar phosphate groups), and nitrogenous bases such as adenine, guanine, cytosine (all the three are also present in DNA) and uracil (DNA contains Thymidine in the place of Uracil).
- These individual nucleotides are synthesized in a series of enzymatic reactions. They are named after their respective bases, such as rATP, rGTP, rCTP and rUTP.
- Among them rATP is the most important energy provider for a variety of cellular functions, whereas others like rGTP or rATP derived- cyclic AMP rGTP-cyclic GMP act as a secondary messengers. rGTP also plays an important role in protein synthesis, polymerization of Tubulins (~55kDa) into microtubules, protein synthesis, enzyme regulation, glycosylation, and carbohydrate metabolism, to say they are the few functions of rGTP. Similarly, rCTP, rUTPs are also involved in glycosylation and carbohydrate synthesis. rATPs are involved in polymerization of G-Actins to F-actins. Many of the nucleotides are substantially, in a specific manner, modified after RNA synthesis.
Composition of Nucleotides in RNA varies from one species to the other, and depends on what template on which it is formed and at what time.
- There is no equivalence of purines and pyrimidines in terms of quantity unlike DNA, where total pyrimidines is equal total purines, In most of the cases DNA is double stranded, in most cases RNA is single stranded with occasional secondary structures with helical or hair pin structures.
Structure:
All RNAs, whatever may be the type, structure and functions they perform exist as polynucleotide chains with 5’ to 3’ polarity.
- Each of the polynucleotide chains is synthesized on DNA or RNA templates.
- Pyro-phosphorylase called (Polynucleotide Phosphorylase) is the enzyme found in cells capable of producing polynucleotide chains by random polymerization and without templates.
Size and molecular weight of RNAs molecules vary from one species to the other, which range from 20-22 ntds (60x365= 21900 Daltons) to 10 000 or more (365 0000 Daltons).
- Basically, RNAs are synthesized as polynucleotide chains (a primary structure), but they immediately or while they are in the process of synthesis, they are processed and fold, because of complementary base pairing into secondary structures and even possibly into tertiary structures. Binding of specific protein provides stability of 3-D structures of RNAs.
Even higher order of organization is possible with association of more than one RNA chains and a variety of proteins into a complex and compact 3-D structure ex. RNA+RNPs-Ribosomes.
Based on structural organization and functions, RNAs have been classified, into Ribosomal RNA (rRNA more than 90% of cellular RNA), Transfer RNA (tRNA) and Messenger RNA (mRNA). These are the major class of RNAs involved in decoding the information into polypeptides. But there is another category of small Mol.wt RNAs called non coding RNAs (ncRNAs). Recent estimation of them is approximately 10, 000 or more. They are now named as ncRNAs most of them are functional RNAs ex. Sc RNA, Sn RNA, Antisense RNAs such as Si RNAs or Mi micro RNAs, Primer PriRNAs, Guide G-RNAs, Efference eRNAs, Xist RNAs, piRNAs, pRNAs (p1RNase), Tm RNAs (transfer-mRNA like), SnoRNAs, , Telomeric RNA, 7sRNA, 7sK RNA, B2 RNA, Srp RNA, and many more (Refer to the chapter ‘small molecular wt. RNAs’). According to ENCODE information the HUGO people have given ~3000 specific names for the small ncRNAs. With the exception of mRNA, tRNA and Genetic RNAs all others can be clubbed into non-coding RNAs (Nc small RNAs). Most of these ncRNA are transcribed by non-coding DNA and many of them derived from regular transcripts but most of them are regulatory in nature.
Ribose Nucleic Acid:
Introduction:
Ribose nucleic acids, when researchers’ realized that life originated on this planet as RNA world nearly 3.8 billion years ago. It is now considered as primeval molecule of life, and the life originated with RNA as the genetic material. Once DNA was deemed as the sole genetic materials of all living organisms, till the discovery of Tobacco Mosaic Virus (TMV) (Russian scientist), which contained RNA, now RNA can act as the genetic material. Once RNA was considered as an accessory form of nucleic acids sub-serving, but soon people realized, that some RNAs have coded information and some were capable of performing catalytic activity, where proteins were once considered as the sole macromolecules capable performing enzymatic catalytic functions. When life originated around 3.1-3.2 billion years ago, it was the RNA world. Everyone now trying to understand, how RNA world gave rise to DNA world. DNA world as genetic material is dominant, yet RNA has a hold on DNA’s function. Ever since Feulgan demonstrated, by chromogenic reactive agents, that there are two types of nucleic acids, one Feulgan positive called DNA and the other Feulgen’s negative called RNA, understanding of the chemistry, structure and functions in detail has been the goal of a large number of molecular biologists, yet there are many aspects of RNA remain shrouded with mystery.
Quantity of RNA in a cell:
More than 90% of the total RNA of the cell is found in cytoplasm, the rest of it is in the nucleus especially in nucleolus region.
- Total concentration of RNA changes depending upon the kind of cell and its state.
Active cells contain more RNA than inactive or resting cells. Perhaps highest concentration RNA is found in brain cells, as they are the most active cells in human body.
- In comparison to DNA, certain types of RNAs are very unstable; their half-life is just few minutes, but some may remain longer than 120 days or more.
Shorter half-life of some types of RNAs is of importance to cellular development and stability.
- Some RNA species are common to all cells; some are specific to cell types.
Chemistry:
Chemical analysis of RNA shows greater similarities with DNA with respect to the components it contains. It is basically made-up of a ribose sugar (where as DNA contains deoxyribose sugar), a phosphate group (DNA also contain the similar phosphate groups), and nitrogenous bases such as adenine, guanine, cytosine (all the three are also present in DNA) and uracil (DNA contains Thymidine in the place of Uracil).
- These individual nucleotides are synthesized in a series of enzymatic reactions. They are named after their respective bases, such as rATP, rGTP, rCTP and rUTP.
- Among them rATP is the most important energy provider for a variety of cellular functions, whereas others like rGTP or rATP derived- cyclic AMP rGTP-cyclic GMP act as a secondary messenger. rGTP also plays an important role in protein synthesis, polymerization of Tubulins (~55kDa) into microtubules, protein synthesis, enzyme regulation, glycosylation, and carbohydrate metabolism, to say they are the few functions of rGTP. Similarly, rCTP, rUTPs are also involved in glycosylation and carbohydrate synthesis. rATPs are involved in polymerization of G-Actins to F-actins. Many of the nucleotides are substantially, in a specific manner, modified after RNA synthesis.
Composition of Nucleotides in RNA varies from one species to the other, and depends on what template on which it is formed and at what time.
- There is no equivalence of purines and pyrimidines in terms of quantity unlike DNA, where total pyrimidines is equal total purines, In most of the cases DNA is double stranded, in most cases RNA is single stranded with occasional secondary structures with helical or hair pin structures.
Structure:
All RNAs, whatever may be the type, structure and functions they perform exist as polynucleotide chains with 5’ to 3’ polarity.
- Each of the polynucleotide chains is synthesized on DNA or RNA templates.
- Pyro-phosphorylase called (Polynucleotide Phosphorylase) is the enzyme found in cells capable of producing polynucleotide chains by random polymerization and without templates.
Size and molecular weight of RNAs molecules vary from one species to the other, which range from 20-22 ntds (60x365= 21900 Daltons) to 10 000 or more (365 0000 Daltons).
- Basically, RNAs are synthesized as polynucleotide chains (a primary structure), but they immediately or while they are in the process of synthesis, they are processed and fold, because of complementary base pairing into secondary structures and even possibly into tertiary structures. Binding of specific protein provides stability of 3-D structures of RNAs.
Even higher order of organization is possible with association of more than one RNA chains and a variety of proteins into a complex and compact 3-D structure ex. RNA+RNPs-Ribosomes.
Based on structural organization and functions, RNAs have been classified, into Ribosomal RNA (rRNA more than 90% of cellular RNA), Transfer RNA (tRNA) and Messenger RNA (mRNA). These are the major class of RNAs involved in decoding the information into polypeptides. But there is another category of small Mol.wt RNAs called non coding RNAs (ncRNAs). Recent estimation of them is approximately 10, 000 or more. They are now named as ncRNAs most of them are functional RNAs ex. Sc RNA, Sn RNA, Antisense RNAs such as Si RNAs or Mi micro RNAs, Primer PriRNAs, Guide G-RNAs, Efference eRNAs, Xist RNAs, piRNAs, pRNAs (p1RNase), Tm RNAs (transfer-mRNA like), SnoRNAs, , Telomeric RNA, 7sRNA, 7sK RNA, B2 RNA, Srp RNA, and many more (Refer to the chapter ‘small molecular wt. RNAs’). According to ENCODE information the HUGO people have given ~3000 specific names for the small ncRNAs. With the exception of mRNA, tRNA and Genetic RNAs all others can be clubbed into Non-Coding RNAs (Nc small RNAs). Most of these ncRNA are transcribed by non-coding DNA and many of them derived from regular transcripts but most of them are regulatory in nature.
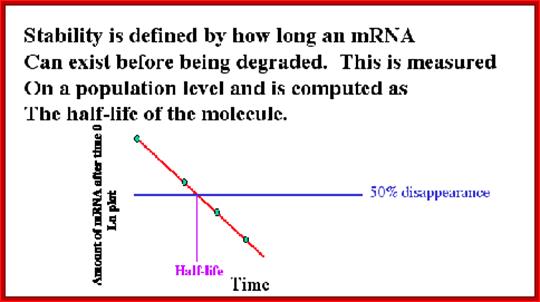
http://david-bender.co.uk/;http://slideplayer.com/
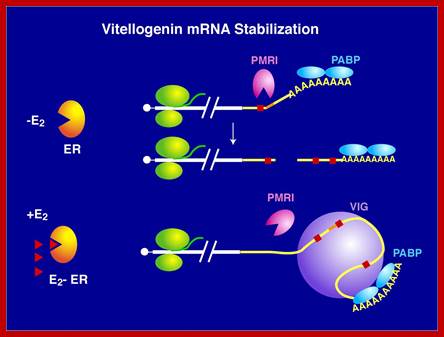
The level of an mRNA in the cytoplasm represents a balance between the rate at which the mRNA precursor is synthesized in the nucleus, and the rates of nuclear RNA processing and export, and cytoplasmic degradation. Long ago, we showed that estrogen induction of the mRNA encoding the egg yolk precursor protein vitellogenin in liver cells of Xenopus laevis increased the half-life of vitellogenin mRNA from 16 hours to 500 hours (about 3 weeks). This work helped establish the regulation of mRNA stability as a major regulatory site in vertebrate cells. We identified the estrogen-inducible mRNA binding protein vigilin as playing a key role in this process. The role of vigilin in regulation of vitellogenin mRNA stability is shown in schematic form in the figure.; http://www.life.illinois.edu/
Daniel R Schoenberg; http://www.nature.com/
Half-life of mRNAs can be measured by labeling RNA and look for time at which half of the labeled mRNA available as functional ones. Different species of mRNAs in a population show different half lives. Some can as short as few minutes and some stay put for a period of 120 days or so; perhaps the longest half life of mRNAs are from plant Phloem sieve tubes (3 moths to 6 months or more?).
When pre mRNAs are transcribed, they are of different sizes, so they are called heterogeneous nuclear RNAs (hnRNAs). They get associated with variety of proteins (hnRNPs), some are specific and some are general which serve splicing of pre mRNAs.
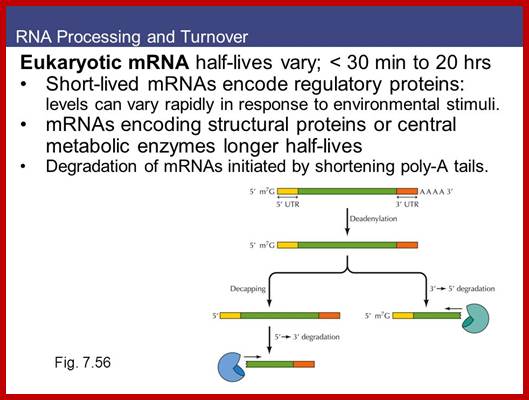
The pre mRNAs contain coding and intervening non-coding regions; the coding regions are called Exons and non-coding regions are called Introns or intervening sequences. In 1938 Walter Gilbert from Harvard coined the terms exons and introns. Note that 5’ upstream region from Initiator codon is called 5’UTR and downstream beyond Terminator codon is 3’UTR, in some cases these may contain coding sequences but they are required for initiation of protein synthesis or/and regulation of mRNA transcripts. There are different kinds of introns; among them some are involved in normal cis splicing processes, some are spliced alternative, some are trans-spliced and some are self-splicing called Group I , Group-II and group III introns. Surprisingly yeast cells have very few introns. Though eukaryotic mRNAs are monocistronic, usually one finds intervening sequences (non-coding) between coding segments. Majority of eukaryotic mRNAs are monocistronic, only few in lower organisms contain polycistronic mRNAs.
In general, eukaryotic transcripts of >2000-2500 ntds long have 6-8 exons. Exon size ranges from 100 to 200ntds, but the intron size can vary from 500 to 15000 ntds long or more. Dystrophin mRNA is 14,000 nts long with 75 introns. In general, after splicing, the normal size of mRNAs (average) is about ~1800 to ~2500 nucleotides; Titin has 178-312 exons. The largest exon is 17106 ntds long and the largest eukaryote gene transcript is Dystrophin (apoDp40) -2.2 ntds long with 76-78 introns (427kDa protein). This gene is located on x chromosome atp 21.2. Its transcription requires 14-16hrs.
The 26,564 (21,746) annotated genes in the human genome (October, 2003) show a total of 233,785 exons and 207,344 introns. On average, there are 8.8 exons and 7.8 introns per gene. About 80% of the exons in each chromosome are <200 bp in length. Nearly 0.01% of the introns are <20 bp in length and <10% of introns are more than 11,000 bp in length. Average size of the introns in Hu is 5419 ntds. The first exon and first intron in genes are always longer. There are genes with single introns or single exon. Longest intron is from Heparan Sp46.0 sulfotransferase, it is 740,920 ntds long. Many eukaryotic genes such as Histone, GPCR, HSPB3, INF-alfa and INF-beta genes are 'without intron'. Human genome consists of 1760 intron-less genes. Human angiogenesis factor genes are intron less. Human serotonin 1D receptor variant is an intron-less gene. Largest exon in humans from PRDM11 gene 8034 ntds (=2678 a.a). Most of the introns contain Sno RNA segments, some even contain miRNA segments. Some introns are copied and translated to generate a protein.
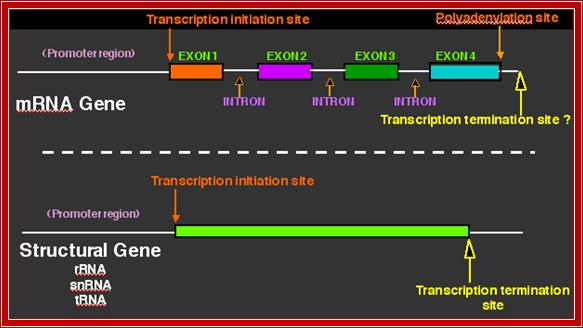
Eukaryotic Gene structural elements:
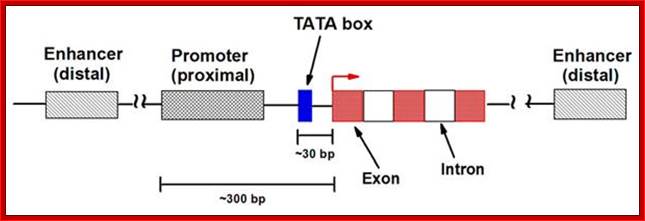
The gene structural features.
http://nitro.biosci.arizona.edu/
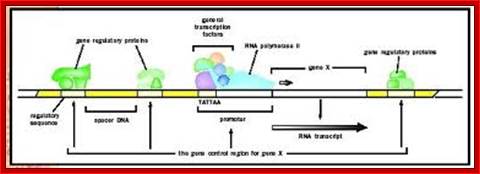
Gene promoter elements bound by enzymes and factors; http://www.biologyreference.com/
Diagrams of mRNAs of eukaryotic upper and prokaryotic lower; topsy.fr/hashtag?
Processed eukaryotic mRNA; http://dnaofbioscience.blogspot.in/
As soon as the 5’ end mRNA emerges out of RNAP complex, they get coated with core proteins, which are in the form of beads consisting of 20 or so subunits and they have a size of 30-40KD (40s and 200Å) and contain proteins by name A1, A2, B1, B2, C1 and C2 etc. Proteins are found in 108 copies per nucleus while the number of hnRNAs is about one million. Proteins found in the nuclei have many functions such as chromosomal replication, transcription, DNA repair, rRNA processing, tRNA modifications, mRNA splicing, transport and degradation.
As already mentioned, pre-mRNAs of different sizes and heterogeneity is very characteristic; it is the name of the ‘Game’, hence they are called hnRNAs. Understanding of the structure and function of cis acting and trans-acting factors that perform splicing is important. For example an eukaryotic intron (self-splicing) can be introduced into bacterial beta galactosidase gene and the same can be transfected into a bacterial cell by transformation protocols. Bacteria don’t have self-splicing facility. But the transfected beta galactosidase mRNA undergoes splicing. Today we know that bacteria do have self-splicing introns. The splicing apparatus is generic and not tissue specific, but alternative and trans-splicing components are tissue specific and generic. Nonetheless, if a self splicing intron is introduced and transfected, such introns get self-spliced.
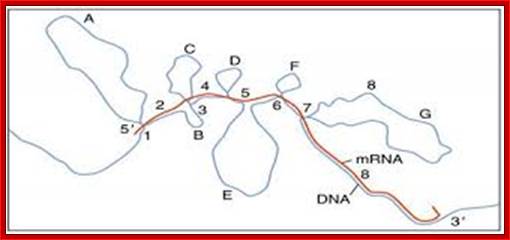
The pre mRNA having exons and introns was identified in Adenoviral pre-mRNAs. This is the diagram of ovalbumin processed mRNA hybridized to its DNA of the gene. The spliced regions of mRNA are hybridized and unhybridized regions loop out. Ovalbumin mRNA consists of 7 introns and 8 exons. http://genome.cbs.dtu.dk/
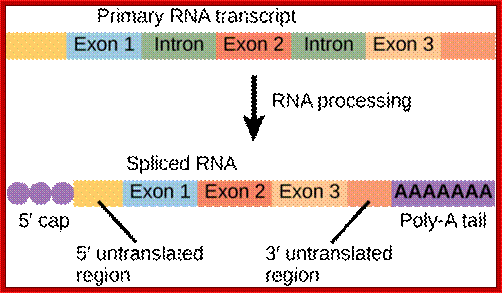
mRNA central coding region is divided into coding and non coding segments called Exons and Introns respectively; Cnx.org
.
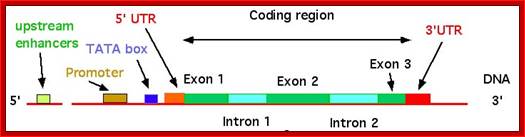
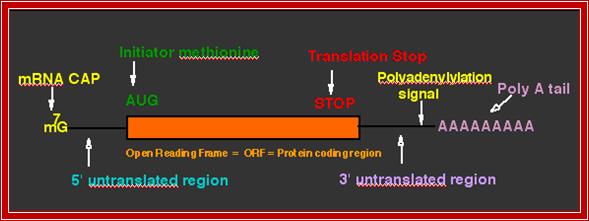
The structure of a mature eukaryotic mRNA. A fully processed mRNA includes a 5' cap, 5' UTR, coding region, 3' UTR, and poly(A) Processed mRNA; http://en.wikipedia.org/
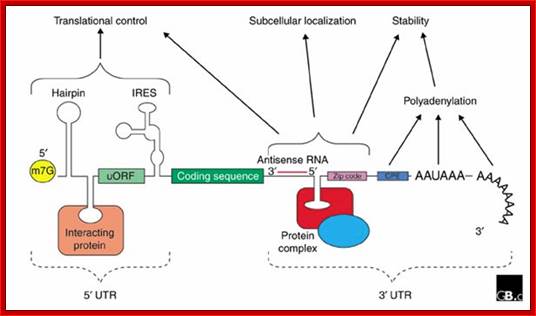
The generic structure of a eukaryotic mRNA, illustrating some post-transcriptional regulatory elements that affect gene expression. Abbreviations (from 5' to 3'): UTR, untranslated region; m7G, 7-methyl-guanosine cap; hairpin, hairpin-like secondary structures; uORF, upstream open reading frame; IRES, internal ribosome entry site; CPE, cytoplasmic polyadenylation element; AAUAAA, polyadenylation signal;The above diagram is the processed mRNA showing various structural and functional features; http://genomebiology.com/
mRNA with several protein binding elements:
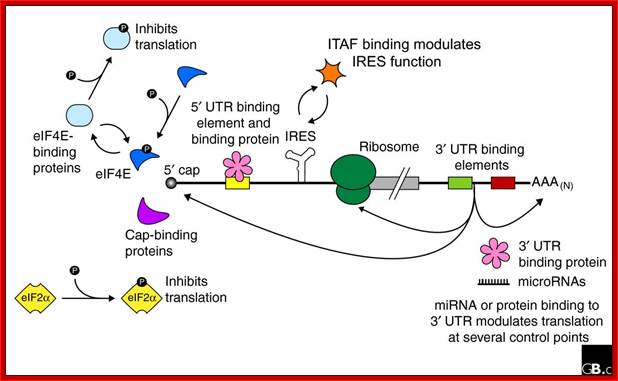
Regulation of eukaryotic mRNA translation occurs at numerous control points. Recognition of 3' UTR sequence or structural elements (green and red boxes) by RNA-binding proteins leads to either activation or repression of translation, often through alteration of the 3' poly(A) tail or through interactions with proteins that bind at the 5' terminal cap structure (that is, the initiation factor eIF4E or cap-binding proteins). Repression of translation by miRNAs can occur through inhibition of translation initiation or elongation, and may also lead to changes in the status of the mRNA 3' poly-(A) tail. Elements found within the mRNA 5' UTR (yellow box) can bind regulatory proteins that repress translation by inhibiting 48S ribosome scanning. Global regulation of mRNA translation is commonly achieved through modification of the translational apparatus (that is, by phosphorylation of the translation initiation factors eIF2α and eIF4E) and the ribosome itself, or modulation of protein partner binding affinities (such as the phosphorylation of the eIF4E-binding proteins). Phosphorylation of eF4E prevent the binding of eIF4E. Translation can be initiated independent of the mRNA 5' cap through a structured internal ribosome entry site (IRES) in the 5' UTR whose efficiency in initiating translation is, in turn, modulated by trans-acting factors (ITAFs), often leaderless mRNA can be initiated by ribosome binding to the first AUG sequence of the mRNA; http://genomebiology.com/
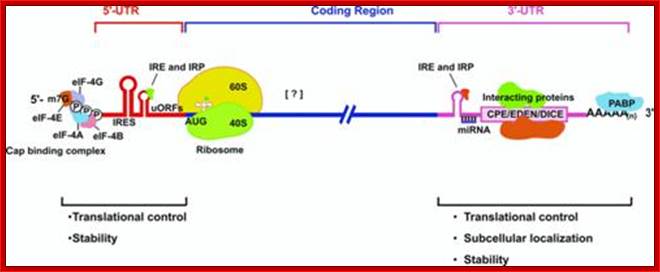
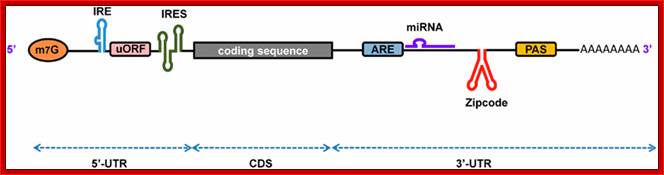
Processed eukaryotic mRNA containing various structural features such 7’CH3GpppA-cap, IRE-internal ribosomal entry site, regulatory loops IREbp iron bound protein binding site, Kozak at ORF; 3’ IRE–Bp binding loops, mi/si RNA binding sites, zip code-mRNA localizing sequence, CPE cytoplasmic polyadenylation sequence, AUUUA sequence ARE regulatory, and poly-A tail. He ARE elements are characterized as AREi, II and III (II--IV). Domain I that contains sites for trans-acting factors exhibiting single stranded RNA binding specificity is mainly unstructured. By contrast, each core domains (II-V) is highly organized and folds into helices interrupted by bulges and interior loops and closed by very exposed apical loops. These elements mostly built specific determinants for trans-acting factors. Besides, these findings provide a valuable database for structure/function studies. Bottom figure A schematic representation of eukaryotic mRNA with functional elements. UTR, untranslated region; CDS, coding sequence; m7G, 7-methyl-guanosine cap; IRE, iron-responsive element; uORF, upstream open reading frame; IRES, internal ribosome entry site; ARE, AU-rich element; PAS, poly(A) signal; Firoz Ahmed http://journal.frontiersin.org/
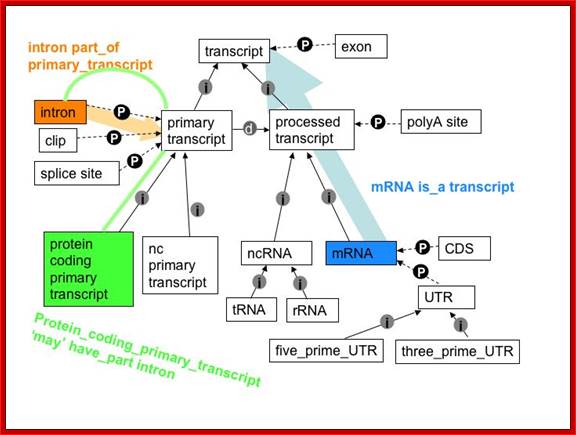
Primary transcript and its processed products; it is a repeat figure to emphasize: http://www.sequenceontology.org/
The 5’ UTR region in many mRNAs contains few regulatory loops, uORF, Internal ribosome entry loops and Iron Response Elements IRES; the 3’UTR contains a variety of sequence elements such as TCS (targeting cis sequences, which includes RTS (RNA transport sequences; (CTTGCTT, CGCAGAGATC, CATTTCTTTGTC)- RNAi binding elements, CPE-Cytoplasmic Polyadenylation Elements (CPE 4–6U1–2A1–2), Stem loop ARE (AUUUUA) elements, poly(A) addition signal sequences (AAUAAA) and EDEN–Deadenylation signal sequence).
If there are 10 introns, the number exons will be 11, but the first 5’ end and the last 3’ end exons are mostly noncoding (UTRs) but they may contain Initiator codon and Terminator codons in 5’UTR and 3’UTR respectively.
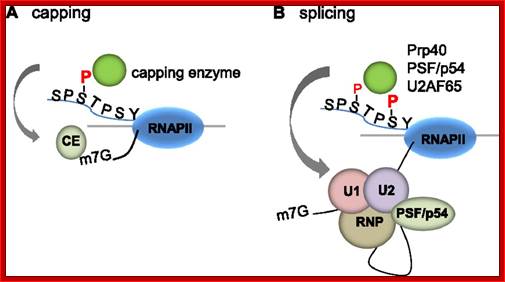
The processing starts as the pre mRNA emerges out RNAPII complex. The CTD tail of RNAPII complex has the required components for capping, splicing and Poly-A addition to pre mRNAs. The CTD tails contain seven ntds sequences repeats from 22-52. Tyr-Ser-Pro-Thr-Ser-Pro-Ser are the sequences. It is rich in hydroxyl aminoa cids and some of them get phosphorylated. The 5’ end of the emerging pre-mRNA is captured by set of protein subunits which are found associated with phosphorylated CTD tail of the RNAPII’s major subunit. There is specific rule for splicing of introns, if there are seven introns; normally intron 3 is removed last. The order can be 1, 2, 5/6, 7/4, and 3, but it need not be a hard and fast rule.
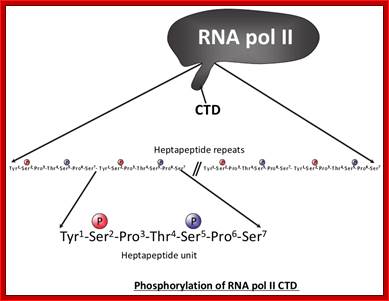
Structure and composition of the RNA pol II large subunit. The C-Terminal Domain (CTD) contains up to 52 repeats of the heptapeptide unit YSPTSPS. Note that the consensus sequence of the heptapeptide repeats 26-52 is often degenerated. Post-translational modifications of heptade residues affect the RNA pol II functions. Phosphorylation of Serine 5 (by TFIIH and cyclin-dependent kinase 7) is important for promoter clearance during initiation and elongation. Phosphorylation of Serine 2 (by P-TEFb and cyclin-dependent kinase 9) is associated with elongation and transcriptional termination. Adam Hall and Philippe T. Georgel; http://www.intechopen.com/
The CTD facilitates capping and splicing by recruitment of RNA processing factors. (A) Capping enzyme (CE) is recruited to the vicinity of nascent mRNA by the CTD phosphorylated on Ser5. (B) During transcription, the CTD is phosphorylated on Ser2, while the Ser5-P is dephosphorylated and is involved in recruiting the indicated splicing factors, which defines splice sites and facilitates assembly of the spliceosome. In this and subsequent figures, green spheres above the CTD represent relevant CTD-binding proteins, while assembled functional complexes are indicated; J.Adam Hall and Philippe T.Gweorgel; http://genesdev.cshlp.org/

Processed mRNA shows to contain all its basic components
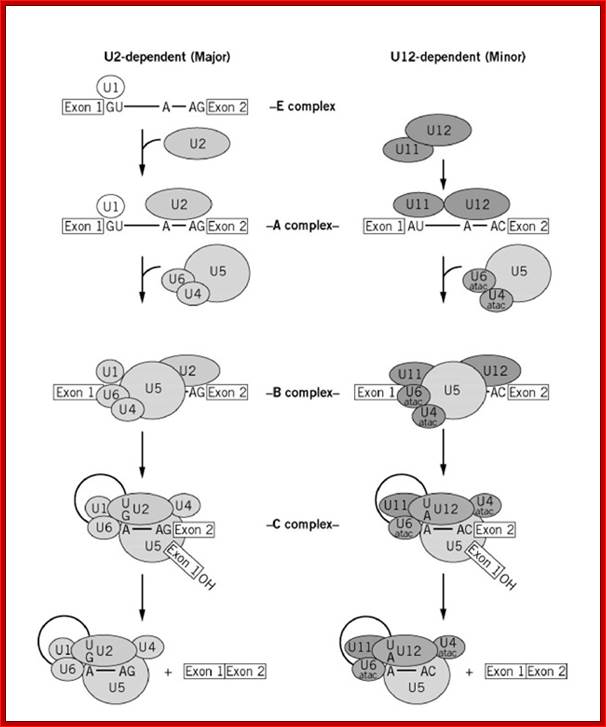
- A group of small molecular weight RNAs and their associated proteins play a role in organizing the splicing material into a structural form, which is employed for cutting and joining i.e. splicing of mRNA. The complex is called spliceosome of ~12MD (mega-Daltons). Each of them consists of 5 different snRNAs/ (3.3Mda), 41 to 45 snRNPs (2.2Mda), 70 splicing factors (4.7Mda) and an additional 30 factors (2 Mda); the spliceosome is almost as large as a ribosome. The total number of spliceosomal snRNAs found in a eukaryotic cell is about one million or more.
- The Intron, a region of the RNA a non coding segment, that is to be cut and removed from the bordering exons, themselves undergo certain structural conformation, required for interaction with other components in splicing processes.
The nuclear sap is loaded with small Mol.wt RNAs called Sn RNAs (small molecular weight nuclear RNAs) and snRNPs.
- There are other small nucleolar RNA called sno RNAs. Sno RNAs (small molecular weight nucleolar RNA) are different for they are involved in modification of rRNA precursor in the nucleolus. There are other small molecular weights cytoplasmic RNAs such as ScRNAs and many coding and non-coding ncRNAs, which have different roles; they are involved in regulatory processes.
- Usn RNAs are named for they are rich in Us; here several Usn RNAs participate in splicing. Usn RNAs are associated with their specific proteins and also some common proteins found in all SnRNA species. The proteins are called SnRNPS. These SnRNAs and SnRNPS were discovered from patients suffering from Systemic Lupus Erythematosus (SLE), where patients develop autoimmunity to his or her own SnRNPs; they are called SNURPs.
- SCYRPS is the name for cytoplasmic ScRNA/ScRNPS complexes, and SNURPS is the name for SnRNA/SnRNPs complex.
- Concentration of snRNA and their associated proteins is about 10^6 and10^8 per cell respectively.
A list of Sn, Sc and Sno RNAs:
|
Size + 5’end |
Location + RNAP transcribed |
Function |
|
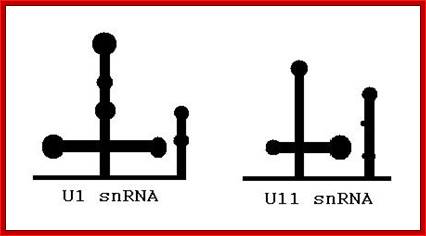
U1 sn RNA |
165ntds, 2,2,7capped |
Nucleus; RNAPII |
Pre-mRNA splicing |
|
U2 sn RNA
|
188, 2,2,7-CH3- cap |
Nucleus; RNAP-II |
Pre-mRNA splicing; ten 2'-O-methylated residues and 13 pseudouridines |
|
U3 Sn RNA |
210ntds, 2,2,7-Trimethyl cap |
Nucleolus, Cajal bodies, RNAPII |
Pre-rRNA processing |
|
U4 sn RNA
|
142,(145)ntds, 2,2,7 cap |
Nucleus; RNAP-II |
Extensively base paired with U6 |
|
U5 sn RNA |
116 ntds, Tri methyl cap 2,2,7capped |
Nucleus; RNAP-II |
Base pairs with the last two ntds of the first exon and the first two ntds of the second exon |
|
U6 sn RNA |
107,(106)ntds 5’CH3-O-ppp;cap 5’CH3-O-ppp cap
|
Nucleus; RNAP-II activated ? RNAPIII |
Base pairs with 5’end of Intron, base pairs with 5’ of U2 RNA |
|
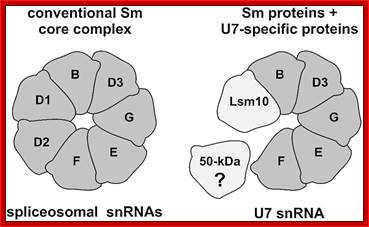
U7 sn RNA |
56ntds, 2’2’-7cap –Droso, Xenopus |
Nucleus; RNAP-II |
Processing histone mRNAs |
|
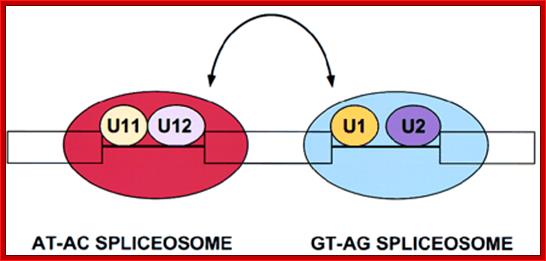
U10,U11, U12, U14 U4-atac, U6-atac |
5’2,2,7-capped |
Nucleus |
Some animals and Some plant mRNAs; rRNA processing |
|
Other small RNAs |
|
|
|
|
7SL sc RNA
7s RNA Archaea 4.5SRNA- Bacteria |
~300, no cap, SRP RNA |
Cytoplasm; RNAP-III |
Involved in docking ribosome-mRNA complex on to ER |
|
SnoRNA/snoRNPs |
20-200 ntds C/D and H/ACA snoRNAs, also capped; RNAPII transcripts; |
Nucleolus, Intron derived or independent gene coded |
rRNA modification and even sn RNA and mRNA modifications |
|
7SK RNA |
330ntd, Capped by methylation at 5’ P? |
RNAP III |
Inhibits p-TEFb CTD p-lation[KG1] |
Note-Three nucleolar RNAs are, 207, 154, and 135 nucleotides long, and are named El, E2, and E3, respectively, and their unique nucleotide sequences suggest that they may belong to an additional family of small nucleolar RNAs. The 5' ends of these three RNAs do not appear to have a trimethy-lguanosine cap or another type of cap. Apparent homologs of these three RNAs were detected in mouse, rabbit, and frog cells, suggesting their universal importance. They are housekeeping RNA species.
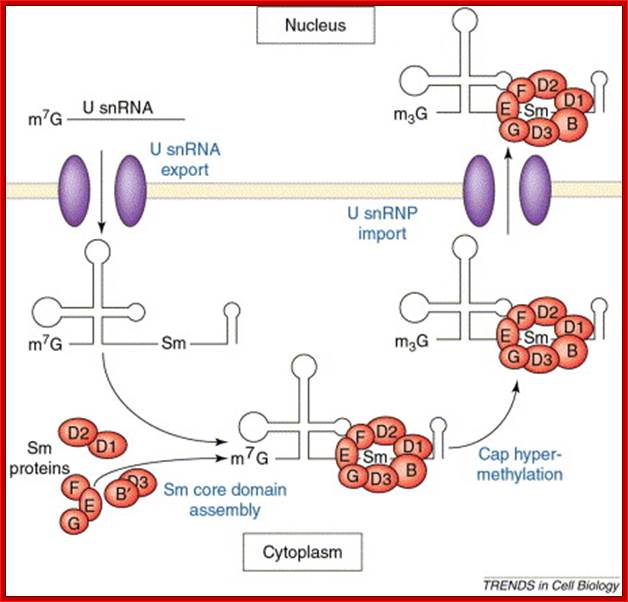
Sm proteins
|
Sm proteins |
Mol.wt |
Sm proteins |
Mol.wt |
|
A |
34 |
|
|
|
B’ |
29 |
D3 |
18 |
|
B |
28 |
E |
13 |
|
C |
22 |
|
|
|
D1 |
16 |
F |
12 |
|
D2 |
16.5 |
G |
11 |
|
|
|
|
|
B/B’ (alternative splicing products), A, D1, D2, D3, E, F and G are smProteins that bind to 5’stem loop structures of U-snRNAs. The common protein is called SM protein; it consists of seven subunits and binds to the base of a stem loop of snRNAs. In sn7 RNA two of the seven protein subunits are different., they are called LSM Proteins.
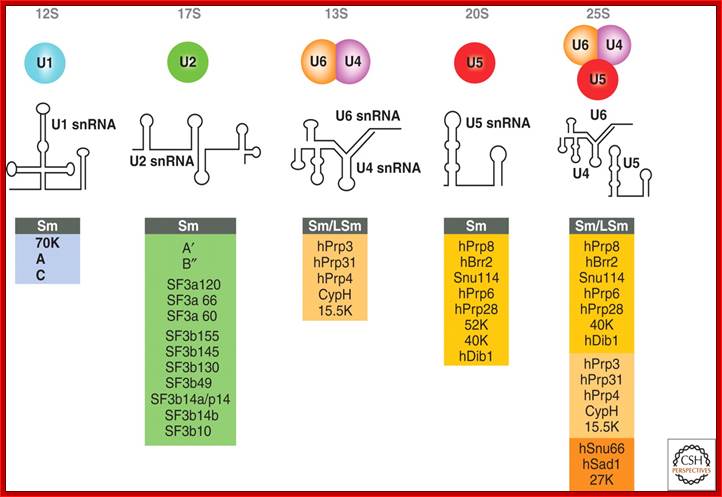
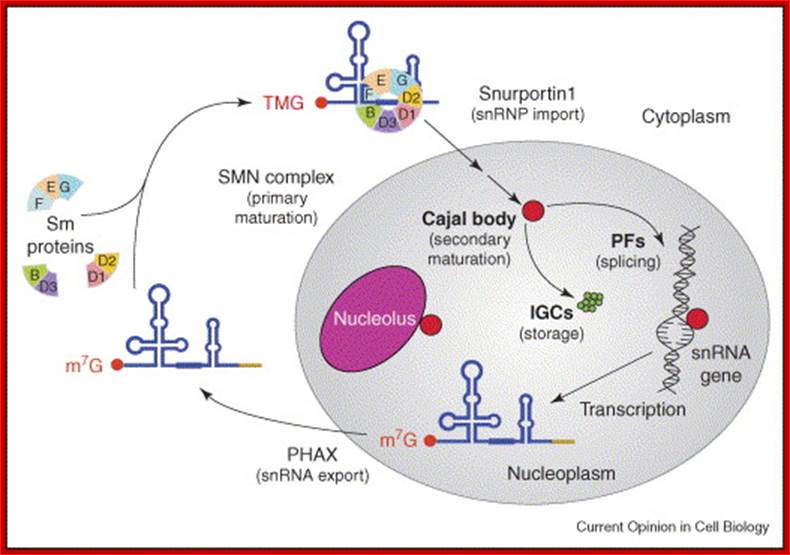
Splicesome atructure and functions; Protein composition and snRNA secondary structures of the major human spliceosomal snRNPs. All seven Sm proteins (B/B’, D3, D2, D1, E, F, and G) or LSm proteins (Lsm2-8) are indicated by “Sm” or “LSm” at the top of the boxes showing the proteins associated with each snRNP. The U4/U6./U5 tri-snRNP contains two sets of Sm proteins and one set of LSm proteins. Cindy L. Will and Reinhard Lührmann; http://www.cshperspectives.com/
Cis Splicing Intron Structure:
Intron, a kind of spacer within the coding region of mRNAs, has some structural features common to all cis splicing pre mRNAs. The spacers found in polycistronic bacterial mRNA are different from eukaryotic introns. The size of introns varies. The DNA coding for introns can be more than 20-30% of the protein coding region of the genome, whereas the exons that code for amino acids is just or less than 1.2-2% of the coding region of the genomic DNA.
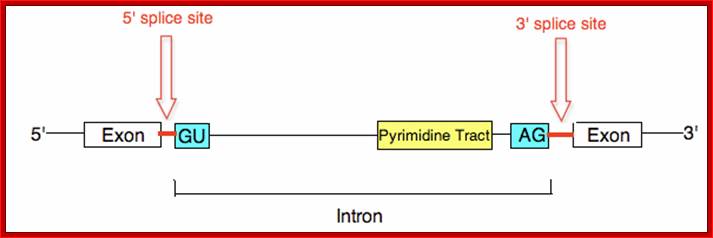
The 5’ splice joint of the introns has GU sequence and the 3’ splice joint sequence has AG sequence. From the 1/3rd of the 3’ splicing site one finds a branching site, CUA/G A* C/G, where the penultimate nucleotide is A which provides 2’OH group for nucleophilic reactions; it is involved in the first splicing reaction.
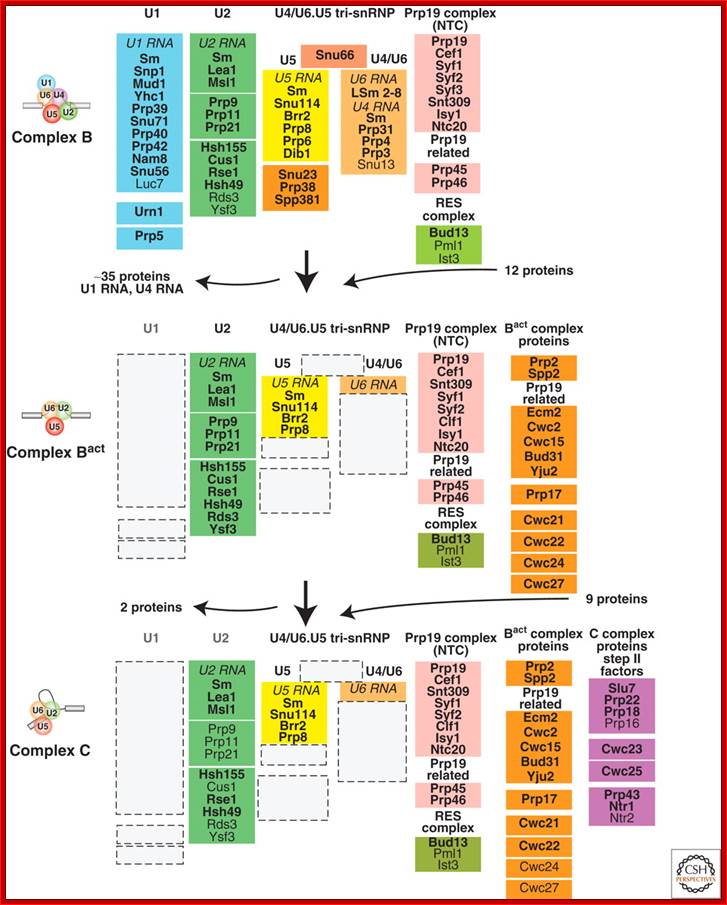
Compositional dynamics of the yeast spliceosome. Proteins identified by mass spectrometry in S. cerevisiae B, Bact, and C spliceosomal complexes are shown. Proteins are grouped according to their function or association with an snRNP, protein complex or spliceosomal complex. The relative abundance of the indicated proteins is indicated by light (sub stoichiometric) or dark (stoichiometric) lettering. (Reprinted, with permission,
Fabrizio [© Elsevier].); Cindy L. Will and Reinhard Lührmann; http://www.cshperspectives.com/
Generalized Intron Structure with Reference to Exon Blocks:
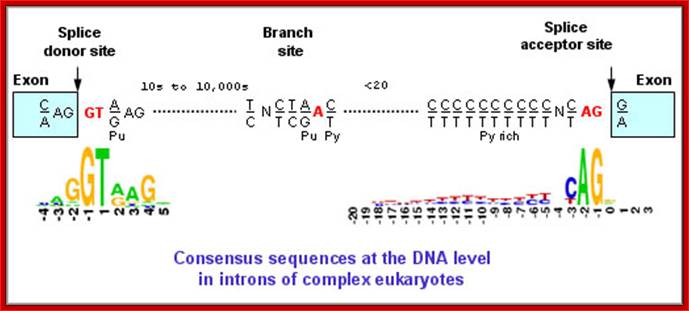
5’and 3’ splicing sites, Branch sequences, Py rich sequence they act as core elements for cis-splicing; http://www.geneinfinity.org/
Exon1GU A//GA [5’GU----Branch Site *A---(Ppy)n---AG3’]-I 5’exon 2
Branching site (B. Site): CUA/G A* C/G (mammalian), UACUA A* C (yeast), and poly-pyrimidine tract (Ppyn), the nucleotide A* has 2’OH group
Mammalian system:
-Exon1—G [ GURAGU---CUA/G A* C/G---(Ppy) 12-15 –YAG] Exon-2 (GU-AG)
Yeast: Exon1---G[GUAUGU---UACUA A* C-- (Ppy) 12-15---AG] Exon2 (GU-AG)
Plants: Exon G[ AUAUCCU---UCCUUA* A---YCCAC ] (AU-AC)
Group I Intron: G [CU-----XXXXXXXX U ],
Group II Introns: G [ GUYG—nnn-A*Y---y—AG(Y) ]—
Group III: -G [ GU--//GU—nnA* Y—AG//--XXXA*X—AG ]—
ORFs, Splicing and coding sequences;
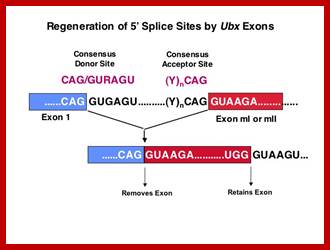
Regeneration of splicing sites by ubx exon
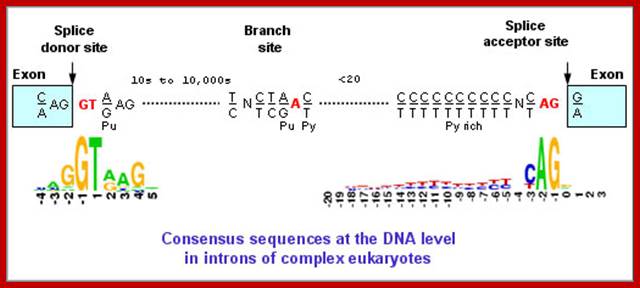
Top Fig; http://orchid.bio.cmu.edu; Bottom Fig; http://www.geneinfinity.org
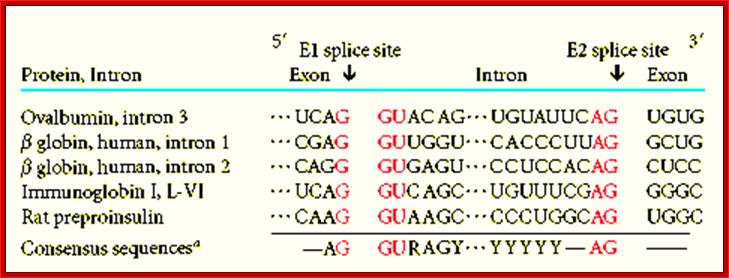
Intron-Exon- splicing sequence elements: http://www.geneinfinity.org
Each of the intron and exon junctions at 5’ and 3’ ends has defined sequences. The introns mammalian systems have GU at 5’ splice sites and AG at 3’ splice site. Within the intron there is branching site little nearer to 3’ splicing site. The branching site has a sequence of which A* at penultimate position and it is very important for it is this nucleotide with its 2’OH group nucleophilically acts to cleave at 5’joint and covalently joins with the 5’ end of the intron to 2’OH of its ribose. In between branching and at 3’ splice site there is another sequence called poly pyrimidine tract.
- Introns by themselves can undergo various secondary structural organizations, based on their complementary nucleotide sequences, but such self determined secondary structural formation is somehow precluded by the binding of rather base pairing of splicing snRNA/snRNP structures. As the pre-mRNA is synthesized, and as and when the 5’end is made available, the said splicing snRNAs assemble on to them. This prevents intron’s self-generating secondary structures. If any folding takes place, it is more or less guided by one or the other spliceosome elements.
The components required for Splicing (the list is incomplete):
- Five Sn RNAs are U1, U2, U4, U5 and U6. There are other snRNAs like U11 and U12 similar to U1 and U2 respectively in few plants.
- The snRNAs are associated with proteins called SR proteins, (20, 35, 40, 55 and 75kDa), for they are rich in Serine (S) and Arginine (R) residues at C-terminal regions and the N-terminal regions contain domains for RNA binding, termed as RRMs (RNA Recognition Motif, RRM). They also contain SR protein-protein interacting domains. Another class of proteins called SM proteins and they bind to certain conserved sequences such as 5’AUAAUUUGUGGUAGU-3’.
- SnRNPs and SnRNAs commit pre-mRNA for splicing for they bind to splicing sites.
- Their associated RNPs are about 70 or more in number.
- Some Sno RNAs modify mRNA at specific sites in the form of methylation of 2’OH of ribose.
The snRNP life cycle. The snRNP life cycle consists of cytoplasmic and nuclear phases, culminating in delivery of mature snRNPs to sites of active splicing. Following nuclear (pol II) transcription, 7-methylguanosine (m7G) capped pre-snRNAs are exported to the cytoplasm via the transport adaptor, PHAX. Upon reaching the cytoplasm, the SMN complex assembles them into a complex with seven Sm proteins, followed by hypermethylation of the 5′ cap and trimming of the 3′ end. The trimethyl guanosine (TMG) cap is bound by snurportin1, which, together with the SMN complex and importin-β, mediates nuclear import. Upon nuclear re-entry, the Sm snRNPs first target to Cajal bodies for secondary maturation, before they either participate in splicing at peri chromatin fibrils (PFs) or are stored within inter-chromatin granule clusters (IGCs) for later use; A Gregory Matera,Karl B Shpargel
Transcripts of U1, U2, U4, U5 and U7 sn RNA genes after processing they are transported into cytoplasm where their monomethyl cap is trimethylated. Then seven sm proteins are added to the base of their stem loop structure in the form of a ring. Then they are transported back to the nucleus and reach Cajal bodies, where they are further get associated with other snRNPs. It is these snRNAs with their snRNPs perform splicing reactions. Nuclear speckles are the sites considered to be splicing regions.
Among the common smRNPs, seven of them associate with snRNAs in the form of a ring at consensus RNA sequences, except U6 snRNAs. But U7snRNA consists of five common sm proteins and two other specific to U7snRNA, called Lsm.
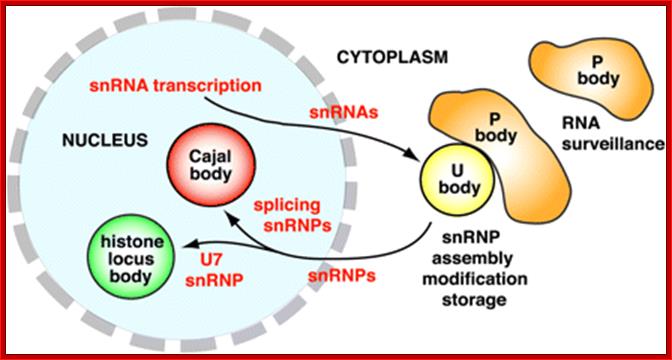
Nuclear and cytoplasmic bodies involved in snRNP assembly. U snRNAs are transcribed in the nucleus and exported to the cytoplasm, where they form snRNP complexes with Sm and Lsm proteins. U bodies may be sites for assembly, modification, or storage of cytoplasmic snRNPs. On return to the nucleus, snRNPs target to Cajal bodies (splicing snRNPs) or the histone locus body (U7 snRNP). Cytoplasmic U bodies invariably associate with P bodies, which function in RNA surveillance and decay, P bodies are aggregates of translationally repressed mRNPs associated with the translation repression and mRNA decay machinery. U bodies- Uridine-rich small nuclear ribonucleoproteins (U snRNPs) are involved in key steps of pre-mRNA processing in the nucleus of eukaryotic cells,
Ji-Long Liu
Pre mRNA splicing involved proteins:
Polypurine tract binding proteins: U2AF1 (U2AF 65 and 35; U2 Auxiliary Factors) and SF upstream of U2AF; and PTP and PSF. U2AF65 of mammalian system is equivalent to yeast’s Mud 2 protein. They act like positioning factors and guide branch site binding proteins.
5’splice joint binding protein: p70: p70 along with U1 RNA/RNPs binds to 5’ splice joint region; provides positioning of U1 RNA; U1 RNA base pairs with 5’ proximal region of the intron.
Branch site binding proteins: BBPs (branch site binding proteins 65p and 35pU2AF) in mammalian system bind to branching point. Several hnRNP proteins like hnRNPs A1 and hnRNPs C1 are also involved.
U2 Sn RNA associated non-snRNP proteins: They are called splicing factors named as SF1 and SF2; SF2 is also called alternative splicing factor ASF. SF4 performs catalytic steps.
SR and RS proteins: Few other splicing factors like Sc35 (RS), which is a member of SRs, which contain Serine and Arginine protein-protein interacting motifs. The other SRPs may be SRP 55 and SRP20.
PPSF and ATPs; they are required for disassembly of spliceosome.
Enhancers/Suppressors: They are required for augmenting the rate of splicing. They bind to ESE (exon splicing enhancer) and ESS (exon splicing suppressor). Some enhancer proteins are 20. kDa. They have N-terminal RRM motifs and the C-end as SR domains for protein-protein interaction.
Yeast SRPs protein: In yeast cells SRPs are not produced, but they have different SnRNPs and also some other proteins called Prps like prp40p, which are found associated with U1 snRNP and Prp8p are associated with U5 snRNP. They also have MUD proteins (mutant-u-die). One of the Muds called Mud2 binds to both 5’ and 3’ splice sites.
Branch point and (Y) n binding protein (BBP): Mud2p and BBP resemble mammalian U2AF65, 35 and SF1 (SF5). Yeast also produces Prp16; it is an ATP dependent helicase, binds to spliceosome and participates in the second step. Prp 22 is again ATP dependent helicase but functions in the release of mature mRNA from the spliceosome.
Sm Ribonucleoproteins, RNA Processors;
Sm proteins in the cytoplasm:
The Sm proteins are synthesized in the cytoplasm stored in the cytoplasm in the form of partially assembled ring complexes, all associated with the pICln protein. The complex consists of SmD1, SmD2, SmF, SmE and SmG with pICln, a 2-4S complex of B, SmD3, SmB and SmD1; they undergo post-translational modification in the methylosome. The three Sm proteins have repeated arginine-glycine motifs in the C-terminal ends and the arginine side chains are symmetrically dimethylated to ω-NG, NG'-dimethyl-arginine. It has been suggested that pICln, which occurs in all three precursor complexes but is absent in the mature snRNPs, acts as a specialized chaperone, preventing premature assembly of Sm proteins.
The Sm core proteins of U1, U2, U4, U5 and U7 snRNPs consists of seven proteins: B/B’ (alternative splicing products), D1, D2, D3, E, F and G. These proteins gained their name from their reaction with Sm serotype antibodies from patients with the autoimmune disease systemic lupus erythematoses. The Sm proteins form a seven-member ring core structure that encircles the RNA. All the Sm proteins share a conserved Sm motif, consisting of two sets of conserved sequences (Sm1 and Sm2) separated by a large loop region, which appear to be the sites of protein-protein interactions necessary for the core structure to form. The Sm core is essential for the biogenesis, transport and function of the snRNP particles. In sn7 RNA two of the seven substituted with others. A different sm core proteins assemble with Usn7 RNA called LSM proteins, (Birkhäuser Verlag, Basel, Switzerland) Oct 61(19-20), D. Schumperli and R. S. Pillai).
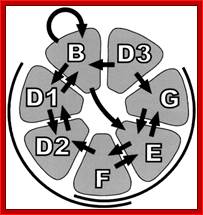
snRNP core complex; strong interactions are shown; heterotrimer complexes F/D2/D1 and F/E/G are indicated; http://www.pnas.org/(Trypanosome
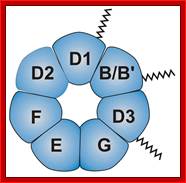
A 7-member ring Sm core proteins; They bind as a ring of seven sm proteins ( B, D1–D3, E, F and G) , but one of the seven is different in U7 sn RNA binding, at the base of stem loop structure of snRNAs and they bind to a sequence RAUUu/GUUGR; snRNPs for U11,U12 and U4atac/U6atacproteins;) Ramesh S. Pillai, et al; http://emboj.embopress.org/
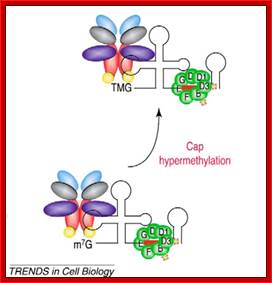
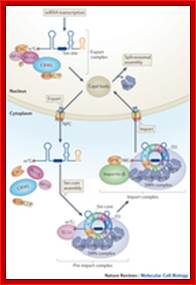
The snRNA move out of the nucleus, where they are loaded with Smn RNAPs and trimethylation of cap takes place; then the snRNA-RNPs s move into the nucleus and reach cajal bodies. Maturation of snRNA;;Small nuclear RNAs (snRNA) pre-export complex consists of the heterodimeric cap binding complex (CBC), arsenic resistance protein2 (ARS2) with export adaptor; http://www.nature.com/
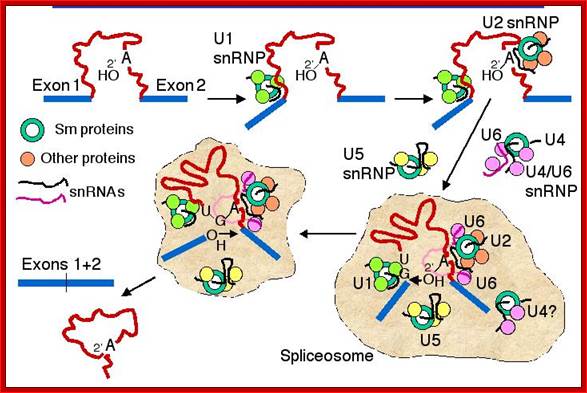
Spliceosome assembly and catalysis; BMP400, part 3;
http://www.personal.psu.edu/
The
Sm proteins are methylated by the PRMT5-complex (1). Hereafter, the
PRMT5-complex docks onto the SMN-complex and the transfer of Sm proteins is
permitted. The SMN-complex then hands over the Sm proteins onto the U snRNAs
and enables RNP formation.
- Any of the proteins found associated with pre mRNAs are often called hnRNPs denoting heterogeneity of the proteins and heterogeneity of mRNA sizes, which may bind to hnRNAs.
- Analyses of them show a variety of proteins, at least 70 or more, having Mol.wt from as low as 40kd to as high as 120kd.
- They are named A1, A2 (30 to 40kd) to U (116kd).
For their binding ability to poly-Us, poly-As, poly-Cs and poly-G nucleotides they are characterized as U RNPs and so on; there are many of them. Using specific polynucleotide columns, it is possible to purify the proteins.
- Those proteins that bind to RNAs are called RNPs, for they have RNA binding motifs and such motifs are named as RRN motifs. They are derived from many genes and alternate splicing of many of these gene transcripts compounds its heterogeneity.
- Some of the said proteins have a conserved 90 amino acid stretch with sequences of Serine and Arginine dipeptides at C-terminal end called SR proteins. The N-terminal hydrophobic sequence has four beta sheets and 2 alpha helices. The beta motifs bind to certain RNA sequences.
- RNPs-C end has such 4 beta and 2 alpha helices.
- Many of them, such as hnRNP-Us have five RGG boxes (Arginine, Glycine, Glycine) in a 26aa domain. In few other HnRNP proteins, in a 45 amino acid domain, contain KH motifs (Lysine and Histidine) motifs, mixed with KGG and LIG motifs.
- Some of the hnRNPs like As, Bs and Cs form core proteins, which associate with the newly synthesized hnRNAs give a bead like appearance to mRNAs.
- Each of the core proteins are of 40s size and 20nm thick.
- Many of the hnRNPs move out of the nucleus and some return and some don’t. Many of them are involved in the transportation of tRNA, ribosomes, mRNAs and even sn RNAs
- Splicing components when they associate at splicing site, consisting of 5 sn-RNAs, several SnRNPs, HnRNPs, non-sn RNPs and other splicing factors, as a complex, are as big as 60s almost the size of a large ribosomal subunit called spliceosome.
Eukaryotic pre-mRNAs have Introns and Exons of different sizes and of different numbers. Whatever may be that, all pre-mRNAs have, with out exceptions, a 5’ noncoding region, called leader sequence, it runs from the first cap binding nucleotide to the first initiator codon or beyond that. The size of the leader varies from one species to the other. In Schizosaccharomyces one of the mRNA (steII) has 5’UTR 2,273 ntds long. The leader sequence has important functions.
Some mRNAs produced by some protozoan parasites, C.elegans and Adenoviral late gene products have a common leader sequence with a 5’cap. This invariant noncoding segment is a must for the function of mRNA. Similarly at 3’ end, from the last terminator codon, mRNAs have another non-coding region whose sequence and structures vary from one species to the other. They play an important role in the stability of mRNAs. The size and structure of mRNAs are individualistic. In many mRNAs the structure determines the stability of it.
The coding region, whatever may be the size or sequence, individual species have specific number of codons intervened by a noncoding sequence (in most of the cases, but there are exceptions to this rule) called Introns. Cutting out introns and joining of Exons exactly in a particular sequence is not without certain common structural elements identifying the borders between Exons and introns. In cis splicing process, most of the introns, whatever may be the size, have specific sequence at 5’ splicing site and 3’ splicing site. These sequences are invariant (with certain exceptions), that is how they differentiate between Introns and exons, there need not be any consensus sequences at the ends of Exons.
- The overall process is more or less is generalized. As there are no perfect evidence(s), it is agreed tacitly to update the information as and when they come to fore.
- The presence of cap has some influence on the splicing of first or the last Exon. It is more or less an observed fact, that the Cap in pre-mRNA facilitates and augments the splicing of introns preferentially from the 5’ end of the pre-mRNA, and poly-A influences the splicing from the last Intron.
- As and when the 5’end of the nascent pre-mRNA emerges out of the RNAP-II complex, the phosphorylated (at specific sites) CTD tail bound by many mRNA processing complexes, such as cap complex, splicing machinery and polyadenylation proteins, perform the said functions.
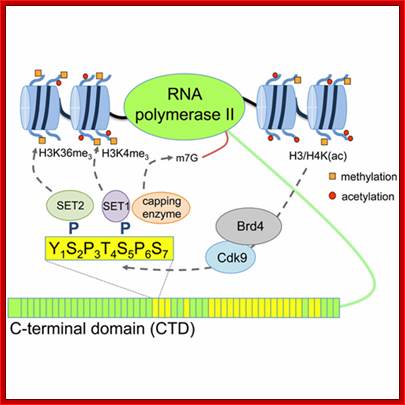
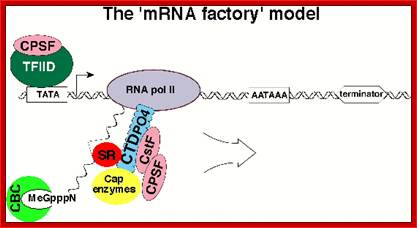
This molecular structure is placed at the carboxy-terminus of the large subunit (Rpb1) of RNA polymerase II (RNAPII) and will be termed the CTD (Cterminal domain) ; Andreas W. Thomae et al: http://www.cipsm.de/;
The CTD tail is long enough to get associated with a variety of factors bound to several aminoacid repeats; they perform functions like cappin mRNA at 5’ end, Splicing process and Polyadenylation; with Polymerase the complex acts like a factory; David Bently; http://www.ucdenver.edu/
This is expected, as the 5’end of the newly formed pre-mRNA gets capped, a protein complex called CBC (Cap Binding Complex), consisting of 80kd and 20 KD subunits bind to the cap.
- CBC also facilitates the splicing of introns near to it first. HOW? Perhaps the protein complexes may help in the binding of spliceosome elements to it.
- When an mRNA containing A, B and C Exons from the left or 5’ end, one with the cap and the other with out cap, was subjected to processing with Hela cell nuclear extract, capped ones spliced out both introns, but mRNA without cap showed splicing of the last Intron.
- Even, if the Intron positions are changed like B, A and C, from A, B, and C, but with cap, splicing favored between B and A first.
- Similarly, presence of poly-A favors the splicing from last introns.
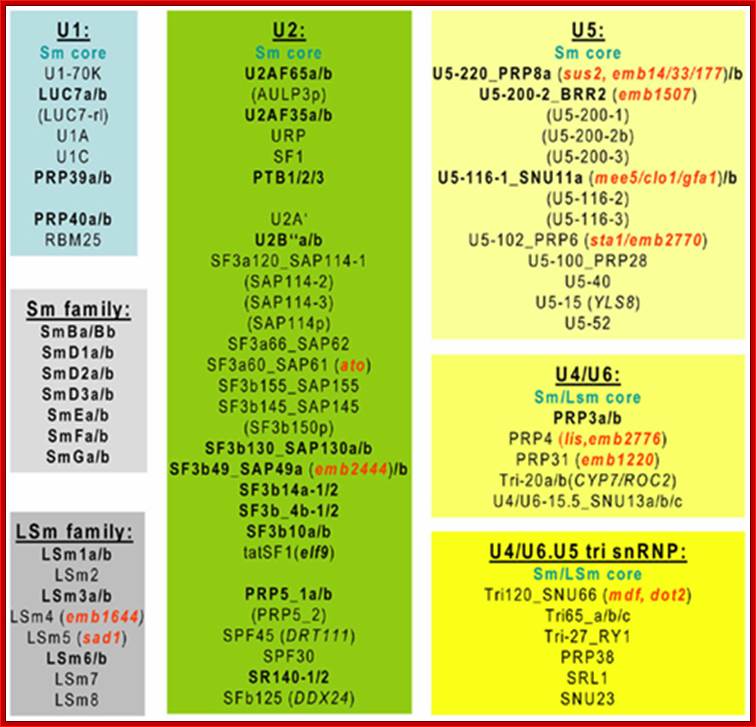
Spliceosomal U snRNP components conserved in Arabidopsis. Subunits encoded by two or more genes are highlighted in bold and characterized gene mutations are indicated in red. http://journal.frontiersin.org/
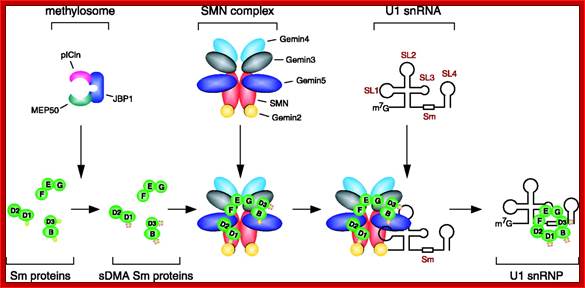
SMN proteins 6-7 play an important role in splicing; .http;//www.scincemag,org
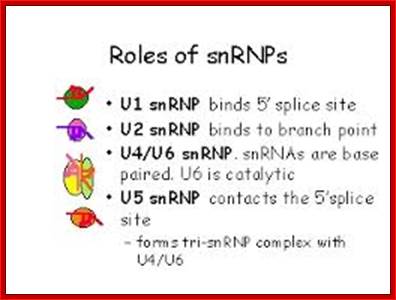
Mechanism of Splicing:
- During splicing, as pre-mRNA’s 5’ end is emerging out of DNA bound transcriptional complex, it is immediately gets covered with core hnRNP complexes.
- The long CTD tail (often phosphorylated at specific positions) of large subunit of RNAP-II recruits or gets associated with spliceosome components; this facilitates what is called co-transcriptional splicing, which ensures correct splicing.
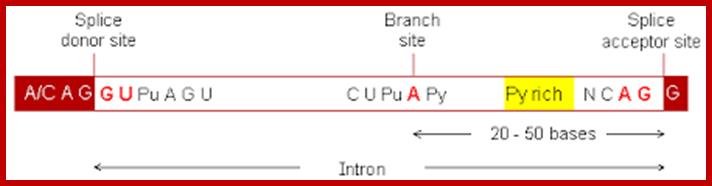
www.zaiza.net; http://dnaofbioscience.blogspot.com
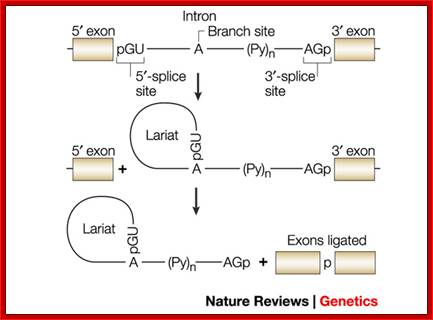
There are several conserved motifs in the nucleotide sequences near the intron–exon boundaries that act as essential splicing signals: GU and AG dinucleotides at the exon–intron and intron–exon junctions, respectively (5'- and 3'-splice sites), a polypyrimidine tract (Py)n and an A nucleotide at the branch site. Splicing takes places in two TRANSESTERIFICATION steps. In the first step, the 2'-hydroxyl group of the A residue at the branch site attacks the phosphate at the GU 5'-splice site. This leads to cleavage of the 5' exon from the intron and the formation of a lariat intermediate. In the following step, a second transesterification reaction, which involves the phosphate (p) at the 3' end of the intron and the 3'-hydroxyl group of the detached exon, ligates the two exons. This reaction releases the intron, still in the form of a lariat.; Splicing reactions and essential splicing signals; Franco Pagani & Francisco E. Baralle; http://www.nature.com/
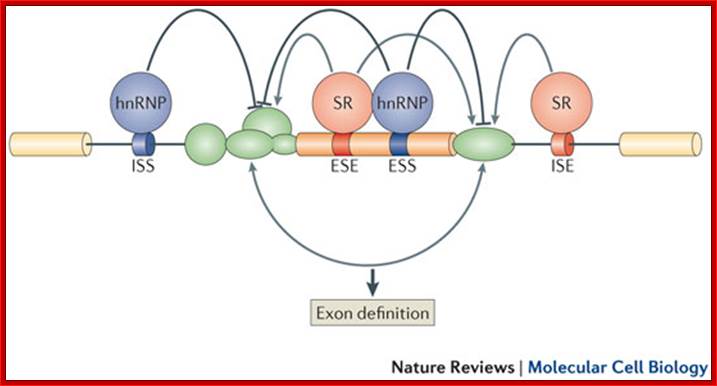
Splicing is governed by cis-regulatory sequences in the pre-mRNA (that is, exonic splicing enhancers (ESEs), exonic splicing silencers (ESSs), intronic splicing enhancers (ISEs) and intronic splicing silencers (ISSs)) and two main families of alternative splicing regulatory proteins, Ser/Arg-rich proteins (SRs) and heterogeneous nuclear ribonucleoproteins (hnRNPs). These regulatory proteins target components of the spliceosome (shown in green) that associate with both the 5′ and the 3′ splice sites flanking the alternative exon and can have either activating or inhibitory effects on the recognition and use of that site. In addition, interactions among components of the spliceosome that are recruited to the 3′ and 5′ splice sites can mediate exon definition, by, for example, ensuring that the recognition of one of these sites by a Ser/Arg-rich protein may indirectly stimulate recognition of the other one, Alberto R. Kornblihtt, Ignacio E. Schor,
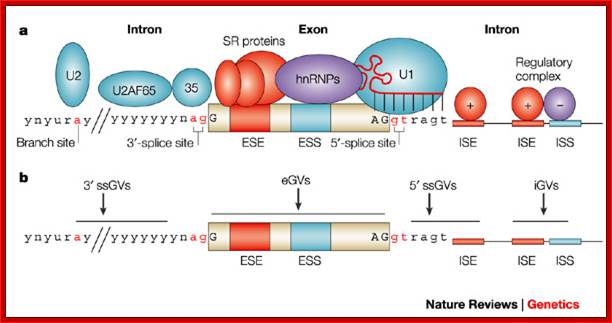
Regulatory elements in pre-mRNA splicing and GVs that can affect them; a | The essential splicing signals that define the exon boundaries are relatively short and poorly conserved sequences. Only the GU and the AG dinucleotides that directly flank the exon (at the 3' and 5' ends, respectively) and the branch-point adenosine (all in red) are always conserved. In most cases, there is also a polypyrimidine tract of variable length (the consensus symbol 'y' represents a pyrimidine base — cytosine or thymine) upstream of the 3'-splice site. The branch point is typically located 18–40 nucleotides upstream from the polypyrimidine tract. Components of the basal splicing machinery bind to the consensus sequences and promote assembly of the splicing complex. This multiprotein complex, known as a spliceosome, performs the correct identification of the splicing signals and catalysis of the cut-and-paste reactions (Fig. 1). Five small nuclear ribonucleoproteins (snRNPs) and more than 100 proteins make up the spliceosome. The U1 snRNP binds to the 5'-splice site, and the U2 snRNP binds the branch site through RNA–RNA interactions. Additional enhancer and silencer elements in the exons (EXON SPLICING ENHANCER (ESE); EXON SPLICING SILENCER (ESS)) and/or introns (INTRON SPLICING ENHANCER (ISE); INTRON SPLICING SILENCER (ISS)) allow the correct splice sites to be distinguished from the many cryptic splice sites that have identical signal sequences. Trans-acting splicing factors can interact with enhancers and silencers and can accordingly be subdivided into two main groups: members of the serine arginine (SR) family of proteins and of the HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN PARTICLES (hnRNPs). In general, SR protein binding at ESE facilitates exon recognition whereas hnRNPs are inhibitory. Protein–protein interactions in the spliceosome that modulate the recognition of the splice sites are the probable cause of splicing inhibition or activation. b | Genomic variants (GVs) can affect different splicing regulatory elements, leading to aberrant splicing. Exonic GVs (eGVs) can either change the amino acid, result in synonymous GVs in exons (sGVs) or introduce a nonsense codon. Intronic GVs might be located within approximately 50 bp from the splice sites (that is, 3'-splice site GVs (ssGVs) and 5' ssGVs) or deep in the introns (intronic GVs (iGVs)). Franco Pagani & Francisco E. Baralle; http://www.nature.com/
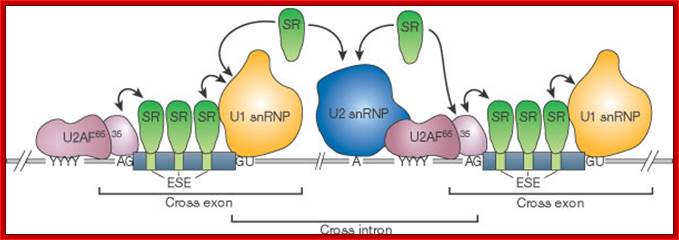
The correct 5' (GU) and 3' (AG) splice sites are recognized by the splicing machinery on the basis of their proximity to exons. The exons contain exonic splicing enhancers (ESEs) that are binding sites for SR proteins. When bound to an ESE, the SR proteins recruit U1 snRNP to the downstream 5' splice site, and the splicing factor U2AF (65 and 35 kDa subunits) to the pyrimidine tract (YYYY) and the AG dinucleotide of the upstream 3' splice site, respectively. In turn, U2AF recruits U2 snRNP to the branchpoint sequence (A). Thus, the bound SR proteins recruit splicing factors to form a 'cross-exon' recognition complex. SR proteins also function in 'cross-intron' recognition by facilitating the interactions between U1 snRNP bound to the upstream 5' splice site and U2 snRNP bound to the branchpoint sequence. Tom Maniatis & Bosiljka Tasic; http://www.nature.com/
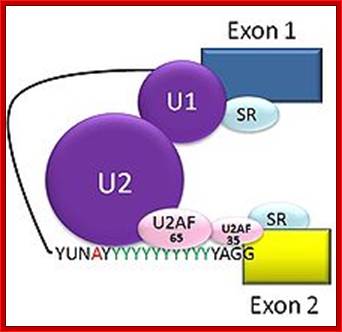
Spliceosome A complex defines the 5' and 3' ends of the intron before removal; WIKIPEDIA
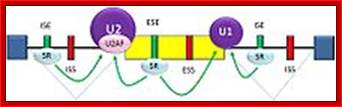
Cis splicing activation; http://en.wikipedia.org/
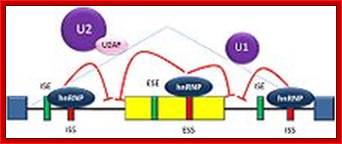
Splicing repression; http://en.wikipedia.org/
Splicing is regulated by trans-acting proteins (repressors and activators) and corresponding cis-acting regulatory sites (silencers and enhancers) on the pre-mRNA. There are two major types of cis-acting RNA sequence elements present in pre-mRNAs and they have corresponding trans-acting RNA-binding proteins.
Splicing silencers are sites to which splicing repressor proteins bind, reducing the probability that a nearby site will be used as a splice junction. These can be located in the intron itself (intronic splicing silencers, ISS) or in a neighboring exon (exonic splicing silencers, ESS). The majority of splicing repressors are heterogeneous nuclear ribonucleoproteins (hnRNPs) such as hnRNPA1 and polypyrimidine tract binding protein (PTB). Splicing enhancers are sites to which splicing activator proteins bind, increasing the probability that a nearby site will be used as a splice junction. These also may occur in the intron (intronic splicing enhancers, ISE) or exon (exonic splicing enhancers, ESE). Most of the activator proteins that bind to ISEs and ESEs are members of the SR protein family. Such proteins contain RNA recognition motifs and arginine and serine-rich (RS) domains
- First exons should be recognized, this is achieved by the binding of SR proteins to exonic enhancer sequences.
- When SRs are bound at 3’end of the exon, recruit U1snRNA-snRNPs (70kd) to 5’ exon-intron splicing site (5’ splicing site); it is assisted by base pairing between the snRNA at a consensus sequence in Intron and Exon joint 5’GUAU: GU-
- U1-SnRNA with its 70kd RNPs binds to 5’splicing site The SR proteins bound at 5’end of exons recruits splicing factors such as p65 and p35 to the pyrimidine tract and the AG region at 3’splicing site.
U2AFs recruit U2 snRNA/snRnps to branching site. SR proteins also function in cross-intron' recognition by facilitating the interactions between U1 snRNP bound to the upstream 5' splice site and U2 snRNP bound to the branch point sequence.
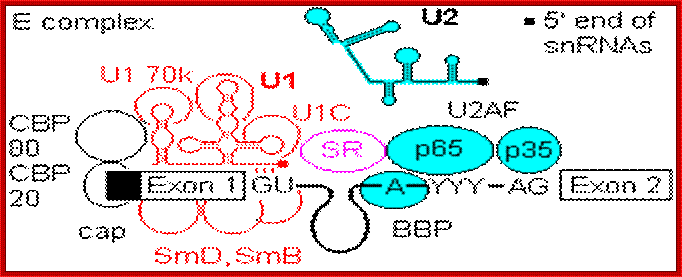
Formation of the commitment or E complex involves binding of factor U1 snRNP (complex of U1 RNA, U1A RRM protein, U1C and U1 70K protein) to the 5'-intron GU site. Recognition is by base pairing of the 3' end of U1 with the consensus sequence AG|GUAGGU (vertical bar is the exon-intron junction), and ATP is consumed in the base pairing process. SR accessory factors associate with the exon towards the 5' direction, and facilitate binding of U1. 1a). Pre-mRNA processing; http://www.hixonparvo.info/
The bindings of SR proteins assure accuracy and efficiency of splicing. Many of the SR proteins that bind to ESE are produced constitutively, but some are produced in tissue specific manner and induced in response to certain signals.
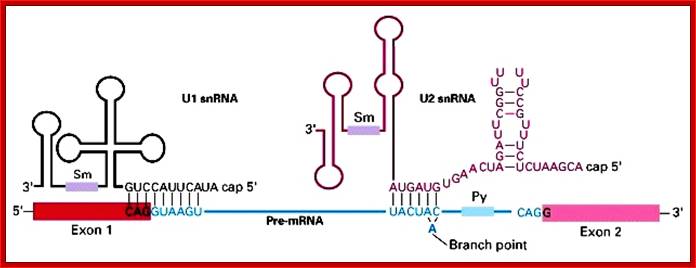
U1 and U2 sn RNA-sm proteins base pair with exon-intron junction at 5’ site and U2 RNA and RNPs base pair with branch sites of the intron; this facilitates the intron to be modulated for bringing the introns 5’ site very near to branch point ‘A’2’OH for splice reaction,
;http://regulatorygenomics.upf.edu/
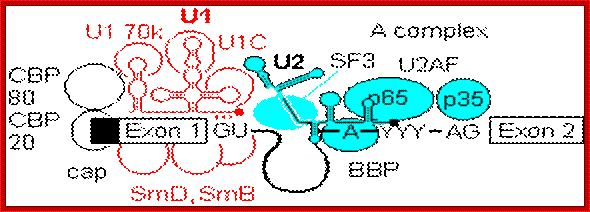
U1 with p70base pairs with 5’intron sequence. This is generally followed by binding of U2 auxiliary factor U2AF p65 and p35 (Mud2 in yeast) to the pyrimidine rich tract between the branch site and the 3'- end of the intron. U2, plus the associated SF3a/b can then base pair with the metazoan branch point sequence YNYURAY to give the A complex. An additional protein factor, BBP (branch binding protein) binds in the region of the branch point A. In yeast, the branch point is more conserved, UACUAAC.; Pre-mRNA processing; http://www.hixonparvo.info/
U2 base pairs with branch point sequences; note the ‘A’ loops out for it has no complementary base to pair. These snRNA base pairing are facilitated by specific proteins bound at 5’ end of the intron and proteins bound at branch and pyrimidine tract. Note the Sm protein binding sites in each of the U1, U2 and U4; these bound snRNAs undergo further conformational changes (aided by their proteins) in to structural form shown below.
U2 interaction with U4/U5/U6 complex leads to the branch point unpaired ‘A’ very close to the intron 5’ splice joint loops out. The nucleophilic 2’OH of A group of the branch site hydrolyses the phosphate bond between 3’ end of exon and 5’end of intron G. (Birkhäuser Verlag, Basel, Switzerland) Oct 61(19-20), D. Schumperli and R. S. Pillai,.
---Exon---G-G---intron-----------A*------AG-Exon 2—
---Exon 1---G3’OH 5’pG---intron------A*---AG-Exon2
With the binding of U4/5/6 complex, positions are changed and the U1 is released. The branch site A2’O (-) interacts with the 5’ splice site and cuts the at the site and at same time the G nucleotide covalently links with the A forming a lariat. This leaves the 3’ end of the first exon 3’OH free. After lariat formation and the rearrangement and the binding releases U4.
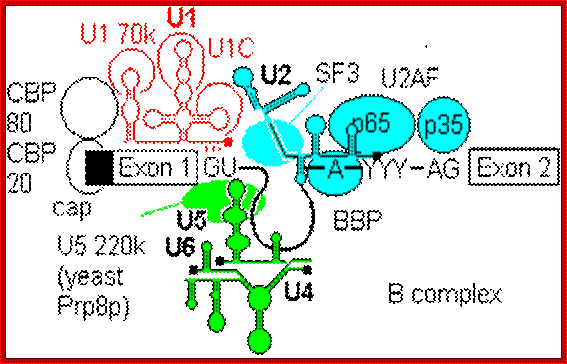
The U4-U6, U5 tri snRNP is then recruited to give the B complex. There is some evidence that U4/U6 recruitment to the 5'-splice site can precede U2 assembly at the branch point. Finally, some radical ATP-dependent base pair rearrangements occur to organize the catalytically competent C complex. Two tri-snRNP factors, U5 100p and U5200p have been shown to contain DExD/H box domains.; Pre-mRNA processing; http://www.hixonparvo.info/
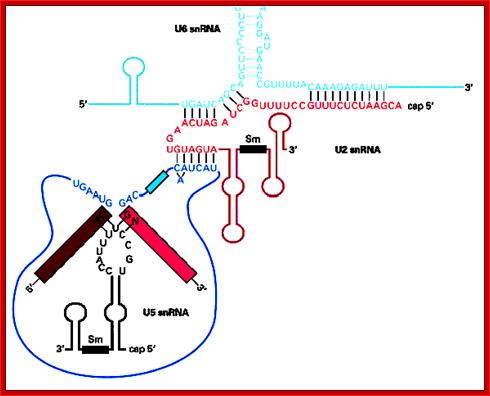
The diagram shows the branch site ‘A2OH’ protruded from U2 mediated base pairing. It is the A2’OH cuts the 5’ splice joint and forms covalent bond between first exon 3’OH and 2’OH of the ‘A’ and U5 RNA base pairs with the last two ntds of the first exon and first two ntds of the second exon, thus the two ends are brought close to each other for splicing reactions.
U2 also base pairs with branch site on one side and the other side it base pairs with U6. The U5 base pairs with 3’ last two nucleotides of first exon and first two nucleotides of second exon; this brings 3’end of the first exon very close to 5’ end of the second exon. The lariat is still looped out. This rearrangement leads to the final configuration for the excision of the lariat intron and joining of the exons.
The looped out lariat intron gets debranched and degraded. But some of these introns are used as Sno RNAs.
The G3’OH group of the first exon hydrolyses the phosphodiester bond between the last nucleotides of the 3’end of the intron- exon splice site and also produces a phosphodiester bond between the two exons. So far no enzyme was found to slice and splice in joining two exons; it is the RNA molecules that perform such catalytic functions.
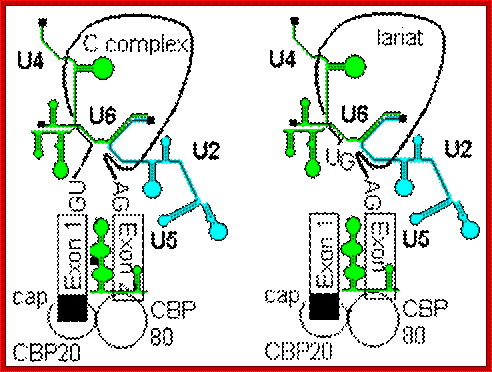
Pre-mRNA processing; http://www.hixonparvo.info/
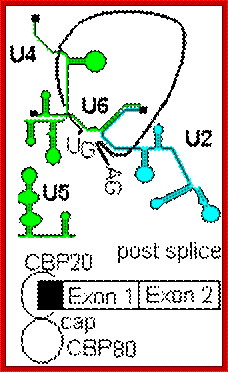
Pre-mRNA processing; http://www.hixonparvo.info/
Pre-mRNA processing;
U5 first base pairs to the upstream exon and the
5'-splice site, a process that requires RNA unwindase activity to displace U1
from the exon.
U6 base-pairs to U2, resulting in displacement of U4. Finally, U5 base pairs to exon 2 near the 3'- splice site on the same stem loop that already holds Exon1, bringing the 3'-OH of Exon 1 into close proximity to 5'-p of exon 2. http://www.hixonparvo.info/
Exons contain elements called exonic enhancers which are targets for binding SR and related RRM containing proteins. The organization that lays out the splicing pattern starts with the Cap binding complex of CBP20 and CBP80, and possibly even with the CTD of the RNA Pol II (Zeng and Berget, 2000). An array of protein factors, e.g. SC35 bind in a cooperative manner between cap and first splice site to define its location. Other SR proteins bridge the intron gap from U1 70k to facilitate U2AF binding, and establish branch point and 3' splice site. Once the U2 complex is in place, SR proteins link up to the next 5' splice site, to continue the process. Thus the pattern of splice sites is established progressively from the 5' cap towards the 3' end, and the spliceosome does not select intron targets for splicing at random.
Process of splicing is cryptic for the U sn RNAs with their specific proteins interact with exons and introns and seek complementary sequences for base pairing. Base pairing with introns and exons and base pairing between different U sn RNAs, this in addition to SR proteins binding to Exonic and intronic sequences leads to secondary structural looping and adjustment in such a way the branch site A’2OH group is brought close to 5’ intronic splice site. The A’2OH highly negative charge brings about the hydrolysis of phosphodiester bond, at the same time A gets covalently linked to the 5’ end of the intron, this creates a lariat. The base pairing of U5 sn RNA to ends of Exons bring the 3’OH (-) group of the exon to the 3’intron-exon splicing site leading to hydrolysis and covalent bond formation to release the intron in the form of lariat and Exon-p-Exon joined.
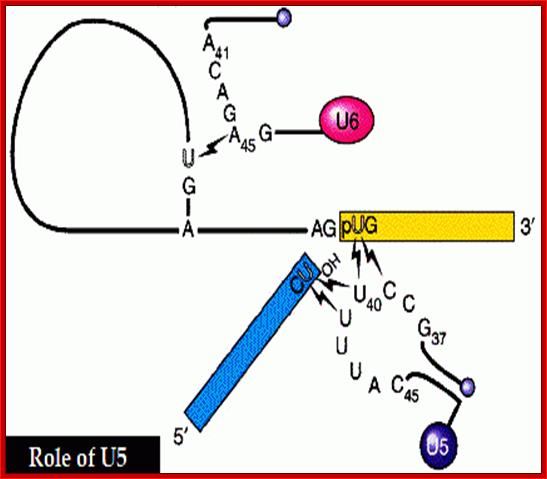
Binding of U5 to the last two nucleotides of the first exon and first two nucleotides of the second exon bring the two exons very close to each other for splicing reaction.
In metazoans, certain members of the hnRNP (heterogeneous nuclear ribonucleoprotein) class bind to sites in particular in the introns. These include hnRNP A1, which binds indiscriminately to pre-mRNA and has a negative effect on spliceosome assembly. The function of the enhancers and SR proteins seems to be to exclude hnRNAs from the exons, and a gap in the chain of enhancers and SR proteins allows hnRNP A1 to act as a splicing repressor.
Splice site specificity is reasonably conserved across species, allowing expression of transgenes. Occasionally splice sites may be misread, for example when wild type Green Fluorescent Protein is expressed in higher plants, the polypeptide may be disrupted by misinterpretation of a coding sequence as a plant specific splicing site.
[GU-U1-70---U2-U2AF-65-35-AG] -SRExon1SR-[GU-U1-70--U2-U2AF-65-35-AG] exon2--GU-SR--SR---AG[--SR Exon-SR- ]GU-SR--SR---AG-
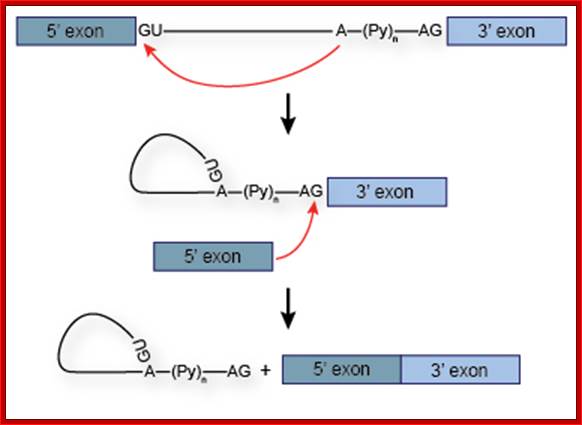
The above illustration depicts first and the second steps of trans-esterification reactions. http://mutagenetix.utsouthwestern.edu/
Steps one and two nucleophilic attack of the phosphodiester bond completes the splicing process, releasing the intron as a lariat carrying the various splicing factors.
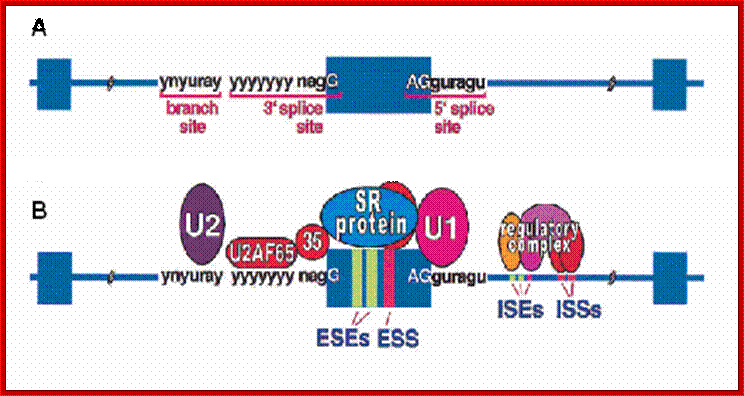
The diagram illustrates the assembly of various splicing components in sequence specific manner. SR proteins are evolutionarily conserved phosphoproteins. They contain one or two N-terminal RNA-recognition motifs (RRMs) and a C-terminal domain rich in repeating arginines and serines (the RS domain) that can be phosphorylated at multiple positions. SR proteins shuttle continuously between the nucleus and the cytoplasm. Both exons and intrns contain certain protein binding sequences that act as exon enhancer or exon supressors. The same holds good for ISE and ISS. http://edoc.hu-berlin.de/
Both exonic splicing enhancer (hESE [5′ GAAGAAGA 3′]) and a human exonic splicing silencer (hESS [5′ CAAGG 3′] sequences are found in exons. The spliceosome is a very complex RNA-protein aggregate that has been estimated to contain approximately 145 different proteins in addition to the five spliceosomal snRNAs.
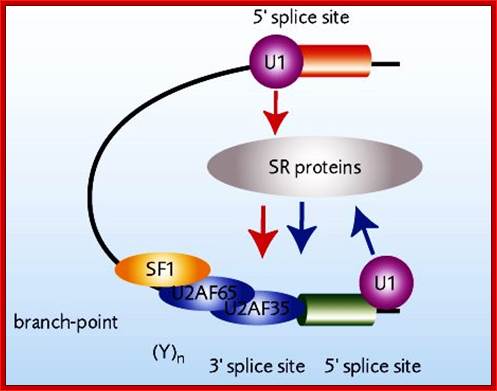
The assembly of the required components (not all are shown) makes the splicing components as commitment complex.
In this assembly, protein-protein interaction and base pairing interaction play an important role in positioning of each of the components at their respective positions and in an order.
This leads to further readjustments in the complex involving the positions of RNA sequences.
- The U5 sn RNA has sequences, which is very critical, involve in base pairing with the last two nucleotides of the first Exon and the first two nucleotides of the second Exon, which virtually brings 3’ end of the exon-1 with the 5’ end of exon-2 absolutely next each other.
- With the close proximity of the 3’OH group of the exon-1 base and the 3’ joint leads to another trans-esterification reaction similar to the first; this is driven by 3’OH group of the first exon.
- The Intron that is released with a lariat is still associated with whatever spliceosome complex left behind in place. Later the lariat Intron is debranched and degraded by nucleases.
- Some of the introns act as rRNA processing sno RNA complexes
- In these two trans-esterification reactions, whether RNA bases are directly involved in covalent bond formation or RNA associated protein perform bond formation, is still an enigma to scientists. However, there are strong indications that SF 4 factor (splicing factor 4) is involved in catalytic steps.
- Spliceosome associated with released intron is disassembled assisted by PSF factors, which is aided by ATP.
Splicing of Minor Class of AT-AC Introns:
- Invariably, a majority of metazoans, animals and plants including, have GU-AG (GT-AG) rule for introns, but in substantial number of plants as well as in few animals, some of the genes have AU-AC (AT-AC) rule.
- The gene associated with nuclear proliferation in mammals has this AU-AC rule. Splicing in these require Sn RNPs such as snRNA U11 and U12 and also other 3 Sn RNAs i.e. U5, U4 and, U6 SnRNA. Note U11 and U12 with their associated snRNPs are similar to U1 and U2 snRNAs in structure and function.
- The branching site is UCCA ‘A’ C. U11 interacts with 5’ splice site and U12 interacts with branching site. The rest of the mechanism is similar to the normal cis-splicing process.
- The U4/U6 RNAs and their protein components are similar to the normal spliceosome components but distinct. But the U5 snRNA component is same as the normal spliceosome complex.
- These are called ATàAC (AU-AC) spliceosomes.
- It is believed that group-II Introns gave rise to AT—AC forms, then AT—AC s have give rise to GU—AG form of splicing components.
- Plants: --IAUAUCCU---UCCUUA*A------------YCCACI
Exon 1-[-------------Intron----------] Exon2
Mammals: ---I GURAGU------UNYURA*Y—-py—----YAG I---
Yeast: ---I GUAUGU—---UAYUAA*Y----py-------CAG I—
AT-AC: ---I AUAUCUU—UCYUUAA*Y--py—-YCCAC I—
Trans: ---I GUAA-----------UAGUAA*Y—py—UUYAC I—
Plants : --IAUAUCCU---UCCUUA*A------------YCCACI
Group I: ----ICU ------------------XXXXXXX-----------------------UI----
Group II: ---IGUYG--------nnnnn--A*Y----py-------AG(Y) I—
Group III: ----IGU--// GU--nnA*Y---AG//---XXXA*X—AG I---
Structure of U12-type introns and minor spliceosome:
Although originally referred to as ATAC introns, U2-type introns have GT-AG 5’ and 3’ splice sites while U12-type introns have AT-AC at their 5’ and 3’ ends.
Minor spliceosomal RNAs; U1 and U11 can be folded similarly; http://en.wikipedia.org/
The main determinants for distinguishing U2- and U12-type introns are 5’ splice site and branch site sequences.
Minor spliceosome consists of U11, U12, U4atac, and U6atac, together with U5 and an unknown number of non-snRNP proteins. The U11, U12 and U4atac/U6atac snRNPs are functional analogs of the U1, U2 and U4/U6 snRNPs in major spliceosome. Although the minor U4atac and U6atac snRNAs are functional analogs of U4 and U6, respectively, they share only limited sequence homology (ca. 40%). Furthermore, the sequence of U11 in comparison with U1, as well as U12 compared with U2, are completely unrelated. Despite this fact, the minor U11, U12, U4atac and U6atac snRNAs can be folded into structures similar to U1, U2, U4 and U6, respectively.
U12 intron-specific spliceosomes contain U11 and U12 small nuclear ribonucleoproteins and mediate the removal of U12 introns from precursor-mRNAs. Among the several proteins unique to the U12-type spliceosomes, an Arabidopsis thaliana AtU11/U12-31K protein has been shown to be indispensible for proper U12 intron splicing and for normal growth and development of Arabidopsis plants. The Minor Spliceosomal Protein U11/U12-31K Is an RNA Chaperone Crucial for U12 Intron Splicing and the Development of Dicot and Monocot Plants, Kyung Jin Kwang et al.
Stepwise assembly of the U2- and U12-dependent spliceosomes: Only those steps that can be resolved by biochemical methods (eg, native gel electrophoresis or gel filtration) under normal conditions with mammalian splicing extracts are shown. For the sake of simplicity, the ordered interactions of the snRNPs (indicated by ellipses), but not those of non-snRNP proteins, are shown. The various spliceosomal complexes are named according to the metazoan nomenclature. Although not yet identified, a post-splicing complex (containing only the excised intron) similar to that formed upon dissociation of the U2-dependent spliceosome is depicted for the U12-dependent spliceosome. Exon and intron sequences are indicated by boxes and lines, respectively. The first two and last two intron nucleotides, as well as the branch site adenosine, are also shown. http://what-when-how.com/
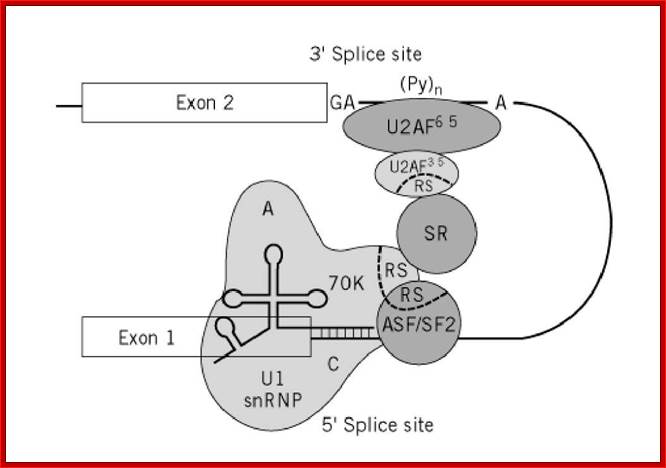
Network of protein-protein and protein-RNA interactions that occur during spliceosomal E complex formation. The U1 snRNP consists of the U1 snRNA (thick solid line), whose 5′ end base-pairs with the pre-mRNA’s 5 ‘splice site, the snRNP core proteins (not indicated), and the U1-specific 70K, A, and C proteins. The U1 snRNA/5 ‘splice site interaction is facilitated by the interaction of the RS domain of the SR protein ASF/SF2 with that of the U1-70K protein. SR proteins are thought to bridge the 5′ and 3′ splice sites by interacting simultaneously with U1-70K and the 35 kDa subunit of U2AF, which also contains an RS domain. Exon and intron sequences are indicated by boxes and thin solid lines, respectively. RS domains are delineated by a dotted line. The branch point adenosine (A), the polypyrimidine tract (Py)n, and the conserved dinucleotide at the 3-splice site (GA) are indicated. What-When-How-in depth Tutorial and Information-Spliceosome Biology;
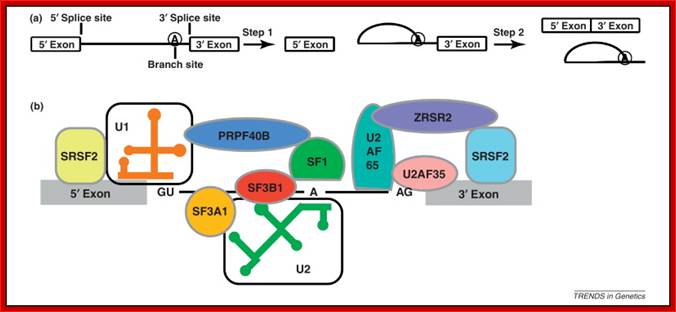
The splicing reaction and interactions in the early phase of intron recognition and spliceosome formation.: (a) The splicing reaction catalyzed by the spliceosome occurs in two steps. In the first step, the phosphodiester bond at the 5′ splice site is broken and the 5′ end of the intron is joined to the 2′ hydroxyl group of the branch-site adenosine residue. In the second step, the phosphodiester bond at the 3′ splice site is broken and joined to the free 3′ hydroxyl group of the 5′ exon. The products are the ligated exons and the intron in the form of a lariat RNA. (b) During the initial steps in splicing, the splice sites and adjacent RNA sequences are bound by a network of interacting factors. A subset of these factors is shown here, of which several have been implicated in myeloid malignancies as discussed in the text. Some of the initial interactions include the binding of SR family proteins such as SRSF2 to splicing enhancer elements located within exons which recruit the U2AF65/U2AF35 heterodimer to the 3′ splice site either directly or through additional factors such as ZRSR2. The U2AF65 subunit binds to the pyrimidine-rich region of the 3′ splice site whereas the U2AF35 subunit binds to the AG dinucleotide at the splice junction. SF1 binds to the branch site A residue whereas U1 snRNP binds to the 5′ splice site through base-pairing interactions. The 5′ and 3′ splice-site complexes are joined together by protein–protein interactions mediated by factors such as PRPF40B. Subsequent to these steps, the U2 snRNP is recruited by U2AF to the branch site where it base-pairs to the intron RNA. U2 snRNP binding is also stabilized by binding of the SF3b complex (which includes the SF3B1 protein) to the RNA upstream of the branch site. Richard A. Padgett
U2 dependent-Splice sites Exon1-5’GT A*---AG3’-Exon2
U12 dependent splice sites. Exon1 5’AT or GT----A*---AG or AC3’-Exon2
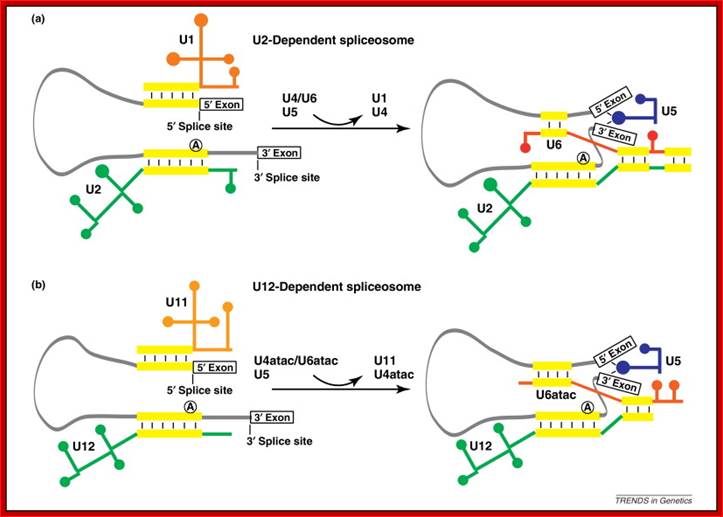
Formation of the spliceosomes: (a) Splicing of U2-dependent introns. (b) Splicing of U12-dependent introns. The early steps of spliceosome formation culminate in the base-pairing of U1 or U11 and U2 or U12 to the 5′ and 3′ splice sites of U2-dependent or U12-dependent introns respectively (base-pairings are indicated by the yellow bars). In the next phase of assembly, tri-snRNP complexes composed of U4, U5 and U6 or U4atac, U5 and U6atac are joined to the forming spliceosome. The base-pairs connecting U4 and U6 or U4atac and U6atac are unwound and new pairings are made between U6 and U2 or U6atac and U12 leading to the release of U4 or U4atac. U6 or U6atac also form base-pairs to the 5′ splice site, displacing U1 or U11 from the complex. U5 interacts with the exons to hold the RNAs in place during the splicing reactions.Richard A. Padgett
AT-AC introns:
Shortly after the discovery of split genes in 1977, a conserved sequence feature at both ends of cellular and viral introns was recognized, i.e., the presence of GT at the 5’ splice site and AG at the 3’ splice site, giving rise to the so-called GT-AG rule (8). This rule holds in most cases, but exceptions have been found. For example, GC is occasionally found at the 5’ end of certain introns, GC-AG introns are processed by the same splicing pathway as conventional GT-AG introns (3). It had long been assumed that removal of all introns from eukaryotic pre-mRNAs took place by the same splicing pathway, until recent developments demonstrated the existence of a second pre-mRNA splicing pathway.
AT-AC introns have unique and highly conserved 5’-splice-site and branch site elements, which are recognized by a unique set of minor snRNAs, U11, U12, U4atac, and U6atac. These snRNAs lack extensive sequence homology to the major snRNAs, but they appear to have related secondary structures, and more importantly, they play analogous roles in splice site recognition and perhaps in splicing catalysis. Despite the remarkable parallels, there are some significant differences between major and minor snRNAs. For example, U11 and U12 form a stable di-snRNP particle that probably enters the AT-AC spliceosome as a single entity, whereas U1 and U2 are discrete snRNP mono-particles. Although the major U5 snRNA appears to be involved in both splicing pathways, this snRNA assembles onto the major spliceosome as part of a U4/U6 · U5 tri-snRNP particle, whereas an analogous U4atac/U6atac · U5 tri-snRNP particle has not yet been described. Notwithstanding the fact that the AT-AC and major spliceosomes have different snRNA constituents, the catalytic core of the AT-AC spliceosome is thought to resemble that of the major spliceosome.
AT-AC Pre-mRNA Splicing Mechanisms and Conservation of Minor Introns in Voltage-Gated Ion Channel Genes. Exon definition interactions between consecutive minor and major introns. Components of the minor and major spliceosomes interact across the intervening exon. The spliceosomes are denoted by the ellipses. U11 and U12 are shown bound to the AT-AC 5′ splice site and branch site, respectively, and U1 and U2 are shown bound to the conventional 5′ splice site and branch site, respectively. The interaction across the exon requires U1 snRNA base pairing at the conventional 5′ splice site. Bound U1 snRNP probably interacts indirectly with U12 snRNP components, perhaps through bridging by SR proteins. U11 and U12 form a di-snRNP particle, although it is not known if their interactions are maintained in the spliceosome. http://mcb.asm.org/
Exon definition interactions between consecutive minor and major introns:
Components of the minor and major spliceosomes interact across the intervening exon. The spliceosomes are denoted by the ellipses. U11 and U12 are shown bound to the AT-AC 5’ splice site and branch site, respectively, and U1 and U2 are shown bound to the conventional 5’ splice site and branch site, respectively. The interaction across the exon requires U1 snRNA base pairing at the conventional 5’ splice site. Bound U1 snRNP probably interacts indirectly with U12 snRNP components, perhaps through bridging by SR proteins. U11 and U12 form a di-snRNP particle, although it is not known if their interactions are maintained in the spliceosome.
U11 and U12 snRNAs. Human U11 and U12 are rare snRNAs that have Sm antigen binding sites but exhibit no sequence homology to other snRNAs. U11 and U12 snRNP particles presumably contain all the Sm core proteins. U12 interacts with a fraction of the more abundant U11 to form a di-snRNP complex. A 65-kDa protein of the U11/U12 complex, identified by virtue of its reactivity with a scleroderma patient antiserum has been described previously, although its sequence is not known. The predicted secondary structures of U11 and U12 snRNAs are similar to those of U1 and U2, respectively. U11 and U12 localize in the nucleoplasm and are concentrated in coiled bodies and nuclear speckles, but they are excluded from nucleoli. This distribution is very similar to that of the major spliceosomal snRNAs U1 and U2. U12 orthologues have been cloned from mouse, chicken, and frog, species in which AT-AC introns are known to exist (reviewed in references) .
The role of U11 and U12 snRNAs in AT-AC splicing has been firmly established. U11 snRNA is present in in vitro-assembled P120 AT-AC spliceosomes and can be cross-linked to the P120 AT-AC 5’ splice site. U11 also interacts with the P120 AT-AC 5’ splice site in vivo through base pairing. U12 snRNA is essential for AT-AC splicing in vitro and in vivo, U12 functions in the AT-AC splicing pathway by base pairing with the highly conserved branch site sequences. Thus, U11 is analogous to U1 snRNA in the major pathway, whereas U12 is analogous to U2.
U11 snRNA interacts in vivo with the 5' splice site of U12-dependent (AU-AC) pre-mRNA introns. Kolossova and R A Padgett.
A notable feature of the newly described U12 snRNA-dependent class of eukaryotic nuclear pre-mRNA introns is the highly conserved 8-nt 5' splice site sequence. This sequence is virtually invariant in all known members of this class from plants to mammals. Based on sequence complementarity between this sequence and the 5' end of the U11 snRNA, we proposed that U11 snRNP may play a role in identifying and/or activating the 5' splice site for splicing. Here we show that mutations of the conserved 5' splice site sequence of a U12-dependent intron severely reduce correct splicing in vivo and that compensatory mutations in U11 snRNA can suppress the effects of the 5' splice site mutations to varying extents. This provides evidence for a required interaction between U11 snRNA and the 5' splice site sequence involving Watson-Crick base pairing. This data, in addition to a report that U11 snRNP is bound transiently to the U12-dependent spliceosome, suggests that U11 snRNP is the analogue of U1 snRNP in splicing this rare class of introns.
Splicing of Plant’ UA rich introns:
In plants introns are rich in UA sequences spread all along the length of an Intron. These help in recognizing 5’ and 3’ splicing sites. Potential 5’ splicing sites have complementarity with U1 SnRNAs, which bind to AG rich Exon and AC rich intronic elements. At 3’ splicing region it has UA transition point and a 3’ consensus sequence having U rich elements followed by GCAG.
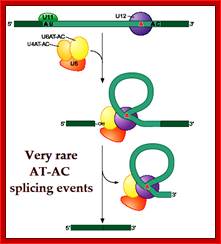
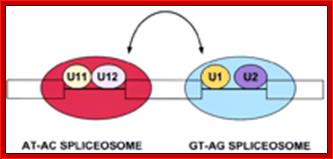
The AT_AC (AU- AC) intron based splicing is rare but it is prevalent in Plants and chloroplasts; AT-AC splicing process; Mol.Cell.Biol; http://mcb.asm.org/
- U11 binds at 5’ splice site.
- U12 tract binding factor binds to U rich sequence preceding 3’ splice site.
- UA island binding factors associate with internal UA islands.
- Exon sequence elements binding factors associate with AG elements in the adjacent Exons.
- In plants basically splice selection is primarily defined by UA rich sequence within an Intron structure.
- However splicing in monocots varies a little bit.
- The U4 and U6 are termed U4AT-AC and U6 as U6AT-AC
Assembly and processing Sequence:
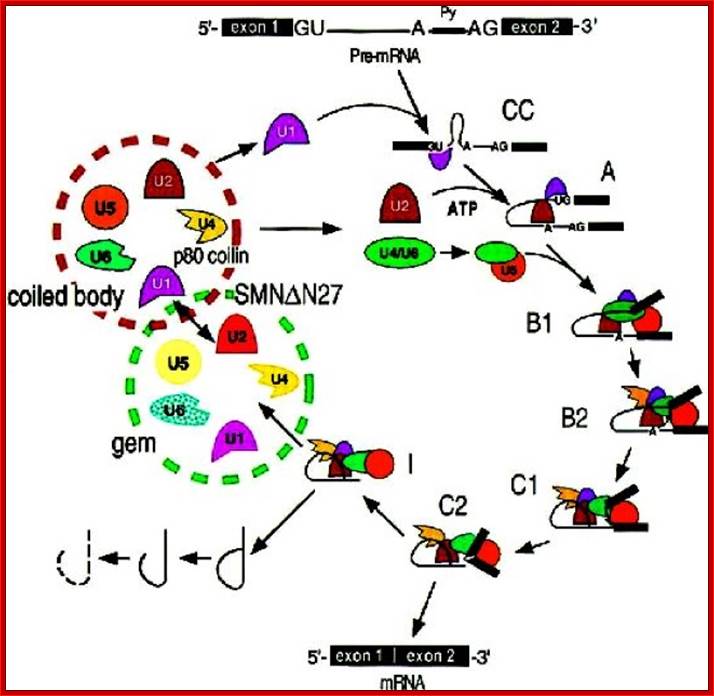
The above figure illustrates the involvement of various splicing components and their action and cycling events; Eukaryotic mRNA maturation; http://bricker.tcnj.edu/
Capping complexes, splicing complexes and Polyadenylation complexes assemble on CTD tail.
As the 5’End of the pre-mRNA emerges, the 5’ end is added with 7’methyl Guanosine in 5’ppp5’ Adenosine.
![]() I
I
SR proteins bind to exon located ESE-(ESS) loci
![]()
Spliceosomes assemble one after another at splicing sites.
![]()
U1/URNPAs and 70kd protein assembles at 5’ splice site of Intron.
![]()
U2AF65/35 assemble at (py)n tract.
![]()
SF1/BBP assembles at branch site.
![]()
U2/U2 snRNPs assemble and base pair at branch site. BBP is released.
![]()
Meanwhile SR proteins bind to ESE of Exon 1 and Exon2 found on either sides of the intron.
![]()
They interact with spliceosome factors for proper assembly.
![]()
U6/U4/U5 RNA and RNPs assemble on to U1/snRNP and U2/snRNP complex, interact and bring 5’splice site closer to branch site. U5 binds to 5’site, U6 binds to U2.
![]()
U2AF65/35 released readjustments among them.
![]()
U1/snRNPs are released.
![]()
Readjustment among the snRNA and snRNPs among themselves, where. U5 shifts to Intron, U6 binds to 5’ site.
![]()
U4/snRNps are released.
![]()
U6 and U2 catalyze the reaction of cleaving at 5’splice site and covalent bond formation between Intron’s 5’ –P with 2’OH group of A of the branch site.
![]()
U5 brings the 3’ end of the exon1 and 5’ end of the exon 2 close to each other by base paring of the last two bases of Exon1 and first 2bases of exon2 respectively.
![]()
The free 3’OH group of the exon1 attacks the 3’splice site, and cleaves releasing the intron in lariat form; simultaneously a covalent bond is formed between the 3’end of the Exon1 and the 5’end of the exo2. Thus exon1 and exon2 are spliced.
![]()
Exon1-o-P-o-Exon2.
![]()
The lariat associated introns in splicing is debranched and degraded with specific RNases or they are processed to act as snoRNA for modification of rRNAs. Some of the introns after debranching they are modified into rRNA processing snoRNAs
Splicing and Export Link:
Pre-mRNAs are prevented from exporting from the nucleus to cytoplasm. But the processed mRNAs are transported for they are associated with specific proteins. This is due to presence of splicing components at splicing sites. However there are a set of proteins called Exon Junction complex (EJC). They are made up of 8 protein subunits; they bind to spliceosome elements when they are still bound to splicing sites. The EJC complex includes REF components which interact with Export proteins such as TAP and Mex. Once the pre-mRNA is spliced, mRNA is exported and TAP/Mex proteins are released.
However, there are many mRNAs without introns (~5%) are also transported out, by mechanism called facilitated transport. Polyadenylation signal, TREXmRNA export components and the mRNA export receptor TAP are required for the intron less mRNA transport into cytoplasm. Such mRNAs are found to be stable. mRNA lacking introns contain a portion of their coding sequence, known as a cytoplasmic accumulation region (CAR), which is essential for stable accumulation of the intron-less mRNAs in the cytoplasm. The CAR in each mRNA is unexpectedly large, ranging in size from ∼160 to 285 nts.
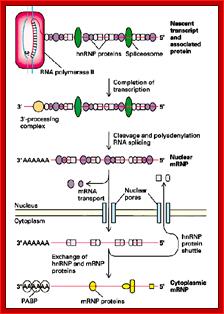
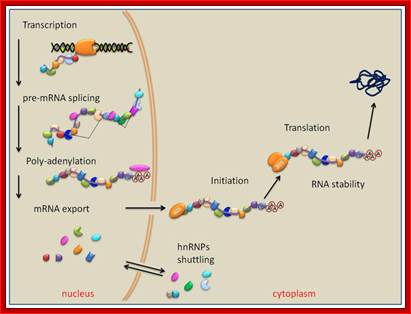
This diagram shows most of the pre and post splicing events that happen till the processed mRNA bound by nuclear mRNPs transported out of the nucleus, which soon get associated with cytoplasmic mRNPs and the nuclear mRNPs are transported back into the nucleus. Exon-exon joint complexes are bound to spliced mRNAs.
Many specific RNA binding proteins are involved in mRNA processing then transport of the same with utilization of Ran GTP/GDP;hnRNP plays avery important role. http://www.mdpi.com/