Alternative Splicing of Pre mRNAs:
Pre-rRNA, pre-tRNA and pre-mRNAs show a variety of splicing features to generate functional forms of RNAs.�� The features of splicing of pre-mRNAs involve a variety of splicing RNAs and protein factors.� It doesn�t matter which exons and introns are spliced early and which are spliced late, though splicing sequence has some preferences for few Exons and they are joined in linear to produce a reading frame.� This type of splicing is normal and general; still other forms of splicing were discovered with time.
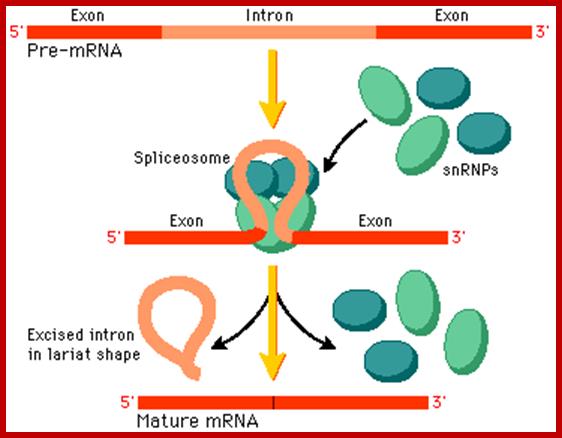
Cis-splicing, with the binding of snRNPs intron segment loops out, which facilitates the binding spliceosome; the spliceosome executes the process of splicing to remove introns and join exons. proteins; http://www.phschool.com/
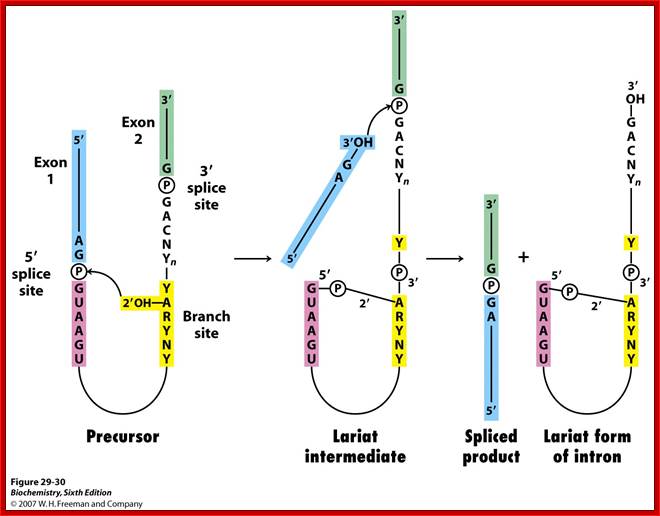
Basic Mechanism of Cis-Splicing. http://pzqin.usc.edu/; http://www.oregonstate.edu
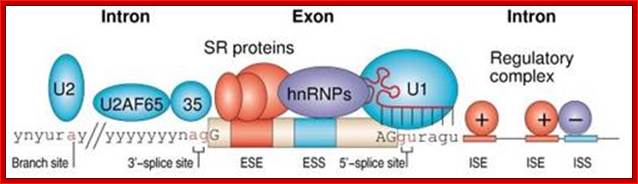
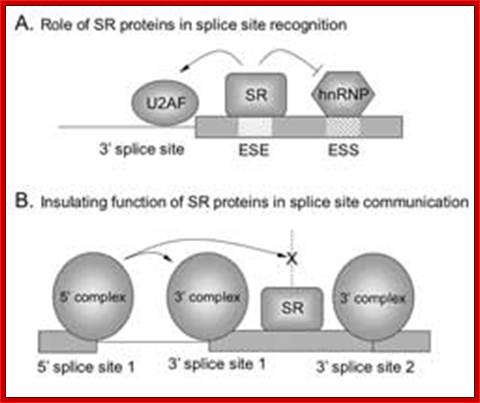
Normal cis-splicing factors� assembly. Assembly of U2snRNA, U2AF65, 35, SR proteins on ESE hnRNPs on ESS U1 at the 5� end of an intron/ 3� 3nd of an exon, functionally if they act they splice an exon and join exons. Elements in pre-mRNA splicing. Five small nuclear ribonucleoproteins (snRNPs) and more than 100 proteins make up the spliceosome. The U1 snRNP binds to the 5'-splice site, and the U2 snRNP binds the branch site through RNA-RNA interactions. Additional enhancer and silencer elements are exon splicing enhancer (ESE) and silencer (ESS) and/or intron splicing enhancer (ISE) and silencer (ISS)). Transacting splicing factors can interact with enhancers and silencers and can accordingly be subdivided into two main groups: members of the serine arginine (SR) family of proteins and of the heterogeneous nuclear ribonucleoprotein particles (hnRNPs).
Matthias Platzer et al;.http://www.sfb604.uni-jena.de/
Pre-mRNA splicing is a process in which intervening sequences (introns) are removed from an mRNA precursor. Splicing consists of two trans-esterification steps, each involving a nucleophilic attack on terminal phosphodiester bonds of the intron. In the first step this is carried out by the 2′ hydroxyl of the branch point (usually adenosine) and in the second step by the 3′ hydroxyl of the upstream (5′) exon1,2. This process is carried out in the spliceosome, a dynamic molecular machine the assembly of which involves sequential binding and release of small nuclear ribonucleoprotein particles (snRNPs) and numerous protein factors as well as the formation and disruption of RNA�RNA, protein�RNA and protein�protein interactions.
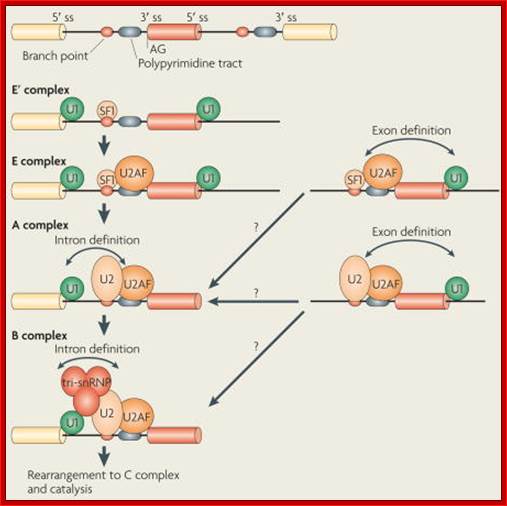
The basic mechanics of spliceosome assembly are well known. Briefly, the process begins with the base pairing of U1 snRNA to the 5′ splice site (ss) and the binding of splicing factor 1 (SF1) to the branch point3 in an ATP-independent manner to form the E′ complex (see the figure; double-headed arrows indicate an interaction). The E′ complex can be converted into the E complex by the recruitment of U2 auxiliary factor (U2AF) heterodimer (comprising U2AF65 and U2AF35) to the polypyrimidine tract and 3′ terminal AG158. The ATP-independent E complex is converted into the ATP-dependent pre-spliceosome A complex by the replacement of SF1 by U2 snRNP at the branch point. Further recruitment of the U4/U6�U5 tri-snRNP leads to the formation of the B complex, which contains all spliceosomal subunits that carry out pre-mRNA splicing. This is followed by extensive conformational changes and remodeling, including the loss of U1 and U4 snRNPs, ultimately resulting in the formation of the C complex, which is the catalytically active spliceosome, Mo Chen and James L. Manley.
The SR (Ser�Arg) proteins are a family of nuclear factors that have many important roles in the splicing of mRNA precursors in metazoan organisms, functions in both constitutive and alternative RNA splicing. They are involved in many steps of splicing regulation, by binding exonic splicing enhancers (ESEs) through their RNA recognition motifs (RRMs) and mediating protein�protein, and perhaps protein�RNA, interactions through their RS (Arg�Ser repeat-containing) domains. All canonical SR proteins have common characteristics (see the table). They have a similar structure, with one or two ribonucleoprotein particle (RNP)-type RNA-binding domains at their amino termini and a variable-length domain enriched in Arg�Ser dipeptides at their carboxyl termini (the RS domain). RS domains are extensively phosphorylated and they function in splicing, usually as activators. Most SR proteins function as pivotal regulators in multiple aspects of mRNA metabolism, such as mRNA nuclear export, nonsense-mediated mRNA decay and translation. Numerous additional RS domain-containing proteins have been identified; proteins known to be involved in alternative splicing are listed in the table.
|
Name |
Domains |
Binding sequence |
Target genes |
|
Canonical SR proteins |
|||
|
SRp20 (SFRS3) |
RRM and RS |
GCUCCUCUUC |
SRP20, CALCA and INSR |
|
SC35 (SFRS2) |
RRM and RS |
UGCUGUU |
ACHE and GRIA1�GRIA4 |
|
ASF/SF2 (SFRS1) |
RRM, RRMH and RS |
RGAAGAAC |
HIPK3, CAMK2D, HIV RNAs and GRIA1�GRIA4 |
|
SRp40 (SFRS5) |
RRM, RRMH and RS |
AGGAGAAGGGA |
HIPK3, PRKCB and FN1 |
|
SRp55 (SFRS6) |
RRM, RRMH and RS |
GGCAGCACCUG |
TNNT2 and CD44 |
|
SRp75 (SFRS4) |
RRM, RRMH and RS |
GAAGGA |
FN1, E1A and CD45 |
|
9G8 (SFRS7) |
RRM, zinc finger and RS |
(GAC)n |
TAU, GNRH and SFRS7 |
|
SRp30c (SFRS9) |
RRM, RRMH and RS |
CUGGAUU |
BCL2L1, TAU and HNRNPA1 |
|
SRp38 (FUSIP1) |
RRM and RS |
AAAGACAAA |
GRIA2 and TRD |
|
Other SR proteins |
|||
|
SRp54 |
RRM and RS |
ND |
TAU |
|
SRp46 (SFRS2B) |
RRM and RS |
ND |
NA |
|
RNPS1 |
RRM and Ser-rich |
ND |
TRA2B |
|
SRrp35 |
RRM and RS |
ND |
NA |
|
SRrp86 (SRrp508 and SFRS12) |
RRM and RS |
ND |
NA |
|
TRA2α |
RRM and two Arg-rich |
GAAARGARR |
dsx |
|
TRA2β |
RRM and two RS |
(GAA)n |
SMN1, CD44 and TAU |
|
RBM5 |
RRM and RS |
ND |
CD95 |
|
CAPER (RBM39) |
RRM and RS |
ND |
VEGF |
*Alternative names are provided in brackets. ACHE, acetylcholine; BCL2L1, BCL-2-like 1; CAMK2D,
Alternative names are provided in brackets. ACHE, acetylcholine; BCL2L1, BCL-2-like 1; CAMK2D, calcium/calmodulin-dependent protein kinase II-δ; CALCA, calcitonin-related polypeptide-α; CAPER, coactivator of activating protein 1 and oestrogen receptors; FN1, fibronectin 1; FUSIP, FUS-interacting serine-arginine-rich protein 1; GNRH, gonadotropin-releasing hormone; GRIA, glutamate receptor, ionotropic, AMPA; HIPK3, homeodomain-interacting protein kinase 3; HNRNPA1, human nuclear RNP A1; INSR, insulin receptor; PRKCB; protein kinase Cβ; RBM, RNA-binding protein; RNPS1, RNA-binding protein with Ser-rich domain 1; RRMH, RRM homology; NA, not applicable; ND, not determined; SFRS, splicing factor, Arg- and Ser-rich; SMN1, survival of motor neuron 1; TNTT2, troponin T type 2; TRA2, transformer 2; TRD, tradin; VEGF, vascular endothelial growth factor, Mo Chen and James L. Manley.
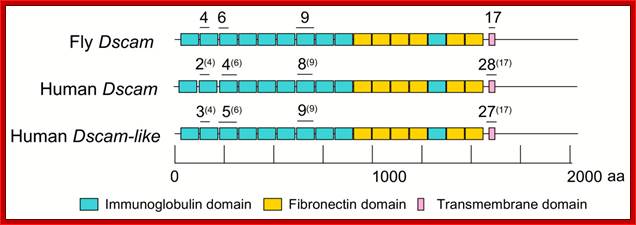
In human beings more than ~90% of mRNAs are spliced in alternative mode.� A single gene by alternative splicing, for example can generate hundreds of alternative forms of mRNAs, so also the products.� Example Slo gene in Rat that encodes Potassium channel protein can generate ~576 alternative forms of proteins from a single pre-mRNA.� A specific Drosophila gene, called Drosophila Cell Adhering Molecule (DSCAM) or it is also called Down�s Syndrome Cell Adhering Molecule can go through alternative splicing resulting in 38016 spliced mRNAs.� The DSCAM gene function is for axon guidance receptor in neurons.� DSCAM gene in Hu is on 21q 22. In D. melanogaster it allows every neuron in the fly to display a unique set of D�scam proteins on its cell surface. It is responsible for the growth of cones to their proper target.� The �DSCAM� gene is 61.2Kbp long, it generates ~7.8kb long transcript (size varies from one mRNA to the other).� It has 95 alternative splice sites and 25 constitutive splicing sites and it has the potential to generate 38 016 different mRNAs and different proteins; this is an incredible achievement by neuronal cells.� If 50% of the 21000 human genes by alternative splicing, they can generate 12,000,000 i.e. 1.2 million proteins. Even 12.5% of such alternative splicing can generate 3 000,000 proteins. Recent genome analysis for Alternative Splicing suggests, among human (Hu) 21000 or 22,000 genes annotated, 90% of them go through Alternative splicing. In fact at least 95% of multiple exonic human transcripts go through alternative splicing. Humans, brain contains approximately Ten billion (10^9) or more neuronal cells and each neuron is connected to ~10,000 synapses; so the network connection for example, must have at least 10^12 i.e 12 trillion or more neuronal network connections and they require such enormous number of specific proteins to perform such network functions; it is amazing and difficult to comprehend. Neurons use parallel processing and the speed of processing is 100Hz; Alternative splicing in most of the cases is tissue specific and the required factors are produced in tissue specific manner; woody Allen and MO Chen and James L. Manley.
Alternative splicing is a very important and powerful tool. To understand the benefit alternative splicing gives the cell we need to understand something about proteins. Proteins can be understood as containing modularized functional units. These functional units can be active sites on enzymes, large structural motifs such as beta-sheets or alpha-helices, or motifs that direct the eventual destination of expressed proteins. A good example of an alternatively spliced pre-mRNA transcript is the mouse IgM immuoglobulin transcript. IgM exists in two forms: excreted and membrane bound. These two forms of the protein differ in the only in the C-terminus: the secreted protein has a secreted terminus motif while the membrane-bound protein has a C-terminal membrane anchor region. Both products come from the same pre-mRNA, but alternative splicing includes either the terminal exon that creates the excreted form of IgM or the membrane-bound form of IgM: http://cnx.org/
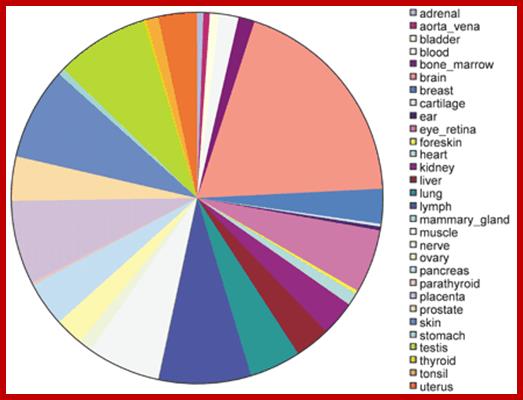
Tissue distribution of human tissue‐specific alternative splicing. Areas on the pie chart are proportional to the total number of alternative splices with high confidence tissue specificity for a particular tissue. http://nar.oxfordjournals.org/
� The �Alternative Splicing�, generally occurs in tissue and cell type specific manner.� The same mRNAs, expressed in many different tissue types, having the same number of Exons and Introns, are spliced differently in different tissues (cell specific manner) or in response to signals, to generate different forms of proteins with certain regions altered. Exons of 150-200ntds long can generate amino acid sequences that can produce a motif or a domain. It is well recorded that proteins have different domains; a domain is a discrete unit of protein structurally it can fold by itself and functionally distinct. Permutation combination of domains has resulted in variety of proteins with different 3-D structures and different functions ex. Zinc finger domain. A short-conserved region of a domain is often called motifs ex. Helix turn-helix turn motif. A domain may contain more than one motifs. . If a sequence dependent exon can generate a protein domain, combination of several hundred of distinct exons can generate several hundreds and thousands of protein domains. The enigma and the paradox of humans having just ~21000 protein coding genes can be explained by mRNA exons and protein domains. Perhaps this is the basis of molecular evolution. Evolution has not stopped; it is still in its infancy and time and scope for another 3.8 billion years at the end of which our solar system condenses and explodes.
� Alternative splicing is executed by alternative splicing factors (ASF), they can be activators or repressors. They are produced in cell specific manner; this leads to change in cell morphology or function or both.
� ESE, ISE, ESS and ISS sequence and their binding factor involved in alternative splicing.� It is to be noted; exons not only code for amino acids but also have information for splicing.
� Even the splice sites (change in the sequence) provide an opportunity to skip the splicing site or use of the splicing site.
� This is also a mechanism which can generates more than one polypeptide from a single transcript without invoking different genes.
Recent genome analysis for Alternative Splicing suggests, among human (Hu) 21000 or 24,000 genes annotated, 90% of them go through Alternative splicing.
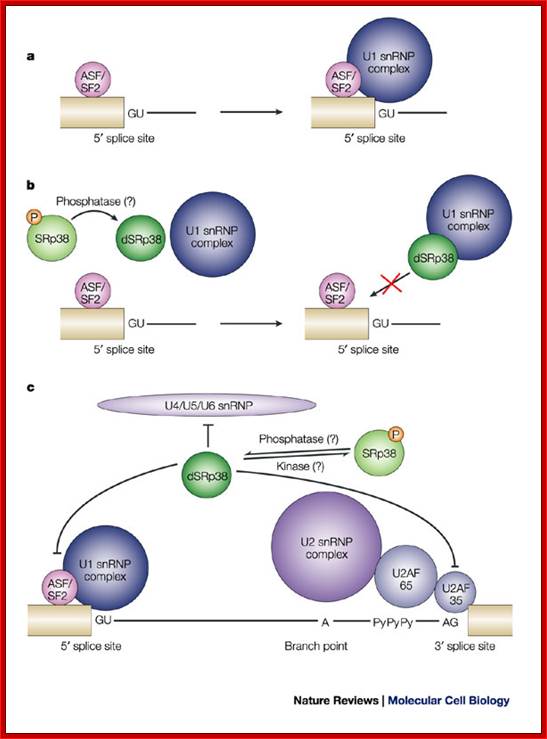
Note: In the case of alternative splicing dephosphorylation of SR38 prevent the binding of p70 and U1 snRNP� thus 5� spicing donor site is incapacitated.
The dSRP38 also prevents the binding of U2SnRNA and U2AF factors at branching site and 3� splicing site of the intron which means the intron cannot spiced out. Dephosphorylation also prevents the binding of and U4/U5/U6 snRNPs associated proteins to 3� splicing site.� Thus block the use of the said introns.� This is one of the mechanisms to skip a specific splicing site of an exon and use another exon that provides favorable sequence for the binding of alternate splicing factors to operate.
�a. Normally, Alternative-Splicing Factor/Splicing Factor-2 (ASF/SF2) or other serine-arginine rich (SR) proteins facilitate the binding of U1 small nuclear ribonucleoprotein particles (U1 snRNPs) to the 5' splice site by interacting with the U1 70K subunit of the U1 snRNP complex. The 5' splice site consists of an invariant GU dinucleotide, which is surrounded by conserved nucleotides (AG/GURAGU, where it denotes the exon�intron boundary, and GU denotes the invariant bases).
�b. �For example, during M phase, or in response to heat shock, SRp38 is dephosphorylated and represses splicing by interacting tightly with the U1RNA-RNPs/70K proteins, thereby interfering with the interaction of ASF/SF2 with the U1snRNP at 5' site for Alternative splicing.
�
c. Dephosphorylated SRp38 (dSRp38) might also repress splicing by
interfering with the recruitment of the U4/U5/U6 snRNP to the spliceosome
through interaction(s) with snRNP-associated RS-domain-containing proteins,
although there are currently no data to support this scenario. The branch point
A, the
2' OH of which serves as the nucleophile in the first step of splicing, the
polypyrimidine stretch (PyPyPy) and the conserved AG at the 3' splice site are indicated.
P= phosphate; U2AF= U2 snRNP auxiliary factor.
Exonic splicing silencer (ESS) and Intronic silencer elements (ISS), in particular, appear to be very prevalent, and may be present in most, if not all cell types where mRNAs are subjected to alternative splicing. The mRNAs subjected to alternative splicing contain inbuilt sequences for splicing.� Alternative splicing in a given cell endowed with specific ASF for specific mRNAs.
Models for alternative splicing regulation; (Lopez, 1998). (Cooper and Mattox, 1997). (Gontarek and Derse, 1996; Kanopka et al., 1996; Lavigueur et al., 1993; Nagel et al., 1998; Ramchatesingh et al., 1995; Staknis and Reed, 1994; Sun et al., 1993), (Caputi et al., 1999; Zhu et al., 2001).
From research over the past 20 years, some general themes have emerged for alternative splicing regulation, although the exact mechanisms still need to be determined. Alternatively spliced exons often have weak consensus sequences at the 5' and 3' ends of the introns, suggesting that additional signals are required for recognition of the exon by the splicing machinery Cis-acting pre-mRNA sequences responsible for regulation of splicing have been identified for many genes. These regions are found in exons or in introns and can be enhancers or silencers of splice site usage. These sequence motifs serve as binding sites for protein factors that can enhance or inhibit the ability of the spliceosome to recognize the exons. The exonic elements not only encode amino acids but also regulate their own ability to be spliced into the mature message. Trans-acting splicing factors that interact with splicing regulatory elements in exons have been identified. Subsets of the SR proteins bind with regulatory sequences important for splicing control. Heterogeneous nuclear ribonucleoprotein (hnRNP) A/B family members can bind to high-affinity sequences in exons and inhibit splicing through blocking SR proteins from binding to the exon.
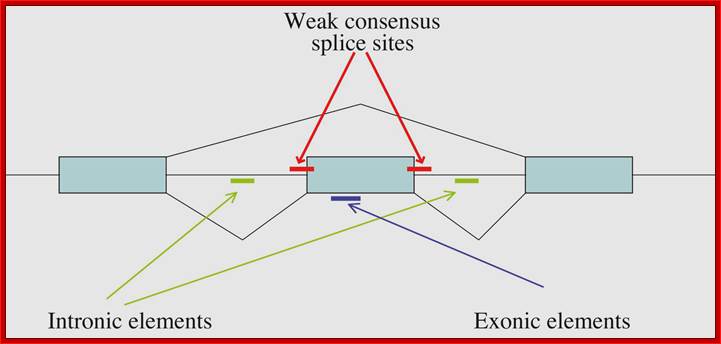
Locations of regions on the pre-mRNA that can affect alternative splicing. http://www.wormbook.org
Locations of regions on the pre-mRNA that can affect alternative splicing; Some combination of these regulatory regions can usually be found. Weaker consensus splice sites surrounding the alternative exon, exonic regulatory regions and intronic regulatory regions are indicated. Alan M. Zahler; http://www.wormbook.org/
Splicing factors important for tissue-specific regulation of vertebrate splicing often assemble into multicomponent complexes on intronic splicing regulatory elements. The downstream control sequence (DCS), found in the intron downstream of the human neural-specific c-src N1 exon, and is one such example. Factors that bind to the DCS regulate N1 splicing; these include hnRNP H, hnRNP F, KH-type splicing regulatory protein (KSRP) and a neural-specific homolog of PTB (nPTB); Comparison of the factors that assemble onto this element from neuronal cell nuclear extract vs. epithelial cell nuclear extract indicate that a subset of these proteins bind from both extracts. Factors in the neuronal extract promote assembly of a different complex that is required to activate splicing of the neural-specific exon. PTB was identified by its ability to bind to polypyrimidine tracts.
Alternative splicing factors
|
|
Genes |
altA |
altD |
altP |
Exons |
IntronR |
Overall |
Ratio |
|
snRNps |
91 |
6 |
3 |
1 |
3 |
11 |
22 |
23.2 |
|
Splicing factors |
109 |
14 |
5 |
0 |
11 |
21 |
38 |
34.9 |
|
Splicing regulators |
60 |
8 |
4 |
2 |
1 |
12 |
20 |
33.3 |
|
Total |
260 |
28 |
12 |
3 |
15 |
44 |
80 |
30.8 |
|
|
|
|
|
|
|
|
|
|
Columns show the number of genes in which alternative splicing occurs,; AltA, alternative acceptor site, AltD, alternative donor site; AltP, alternative intron position; ExonS, exon skipping; IntronR, intron retention; ratio column the number and fraction of genes with any type of alternative splicing. (Genome biology2004)
Cardiac troponin T, alpha-actinin, fibroblast growth factor receptor R2, Calcitonin/CGRP, and alpha tropomyosin and other pre-mRNAs regulate the alternative splicing of their own messages. In these systems, PTB binding to both introns flanking an exon promotes exon skipping. CUG binding protein and related family members known as CELF proteins interact with CUG repeats in introns to regulate splicing. An antagonistic interaction between one of the CELF proteins, ETR-3, and PTB regulates troponin�t� alternative splicing. The nova-1 protein is important for regulation of alternative splicing in the nervous system. This neuronal protein binds the sequence UCAY. A balance between nova activity and PTB is important in regulation of alternative splicing in neurons. The mechanism by which binding of these factors to intronic elements regulates exon inclusion is not clear. Genomic analysis indicates that homologs of all of the major vertebrate splicing factors discussed above can also be found in C. elegans (AMZ, unpublished observations). Chou et al., 1999; Markovtsov et al., 2000; Min et al., 1995; Min et al., 1997). (Wagner and Garcia-Blanco, 2001; Wollerton et al., 2001), (Wagner and Garcia-Blanco, 2001), (Ladd et al., 2001; Philips et al., 1998), (Charlet-B et al., 2002, (Jensen, Dredge, et al., 2000; Jensen, Musunuru, et al., 2000), (Polydorides et al., 2000).
Alternative splicing factors:�
In quoted alternative splicing factors are tissue specific and stage specific.�� They deviate from normal course of splicing, yet they generate linear spliced structures.� But in tissue specific alternative splicing, certain Exons are eliminated and certain Exons are retained.� This is due to the production of one or more alternate splicing factors (ASFs) in that cell type are not same but different.�
- An example for ASF is SF2 in SV40 infected cell, this factor is produced in large amounts, this invokes alternate splicing different from the normal splicing, resulting in the production of small �t� antigen.
- The Jena bioscience provides some Alternative splice factors.� They are ASF2/SF2 (SFRsp30a), ASF/SF2 (SFRS1 or SRp30a, SC35 (SFRS2 or SRp30b), WT-1 (+ KTS), WT-1 (- KTS), B52, SRP55, and HRP48 and many more.� These are available in the market.� They act in combinatorial fashion.
The components involved are exonic sequence enhancer elements and intronic sequence elements.� Binding of specific SR and their related proteins bring about assembly of factors to 5� and 3� splicing joints. ��Similarly, the silence sequences found in introns and exons have the same functions.� The most important splice regulation points are the splice joints at which either promoting or preventing factors bind.� Example sxl binds to 3�splice joint that prevents splicing using the 3� splice joint.� Similarly, the 5� splice joint can be used either to prevent or favor the splicing to specific positions.
SR Proteins and Related Factors in Alternative Splicing: Shengrong Lin and Xiang-Dong Fu.
SR proteins and related factors in Alterative splicing-Shengrong Lin and Xiang-Dong Fu; http://www.landesbioscience.com/
SR proteins are a family of RNA binding proteins that contain a signature RS domain enriched with serine/arginine repeats. The RS domain is also found in many other proteins, which are collectively referred to as SR‑related proteins. Several prototypical SR proteins are essential splicing factors, but the majority of RS domain‑containing factors are characterized by their ability to alter splice site selection in vitro or in transfected cells. SR proteins and SR‑related proteins are generally believed to modulate splice site selection via RNA recognition motif‑mediated binding to exonic splicing enhancers and RS domain‑mediated protein‑protein and protein‑RNA interactions during spliceosome assembly. However, the biological function of individual RS domain‑containing splicing regulators is complex because of redundant as well as competitive functions, context‑dependent effects and regulation by co transcriptional and post‑translational events. This chapter will focus on our current mechanistic understanding of alternative splicing regulation by SR proteins and SR‑related proteins and will discuss some of the questions that remain to be addressed in future research.
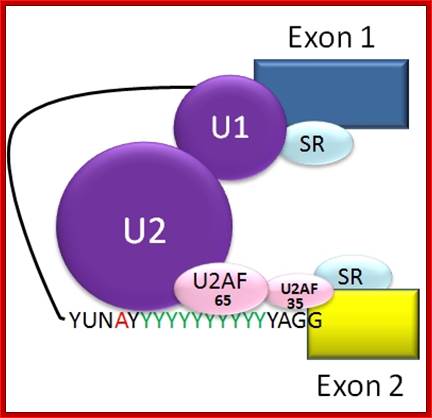
Spliceosome A complex defines the 5� and 3� end of the intron before its removal; http://www.answers.com/topic/alternative-splicing; http://en.wikipedia.org/
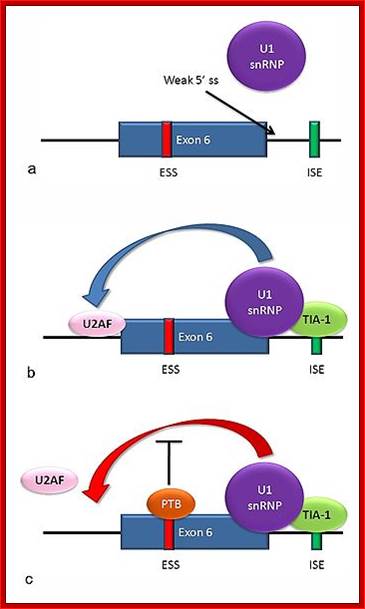
For example, Fas mRNA a) the 5' splice site downstream from exon 6 in the fas pre-mRNA has a weak agreement with the consensus sequence, and is not bound usually by the U1 snRNP. b) Binding of TIA-1 protein to an intronic splicing enhancer site stabilizes binding of the U1 snRNP. The 5' donor site complex assists in binding of the splicing factor U2AF to the 3' splice site upstream of the exon. c) Binding of polypyrimidine tract binding protein (PTB) to the ure6 exonic splicing silencer in exon 6 prevents the 5' complex from assisting in U2AF binding.In situations a and c, exon 6 is skipped, giving an mRNA encoding a soluble protein product. In situation b, exon 6 is included, and the resulting mRNA encodes the membrane-bound isoform of fas protein, which stimulates programmed cell death (apoptosis); http://www.answers.com/topic; http://www.answers.com/topic/alternative-splicing#ixzz31sQlLj64; http://en.wikipedia.org/
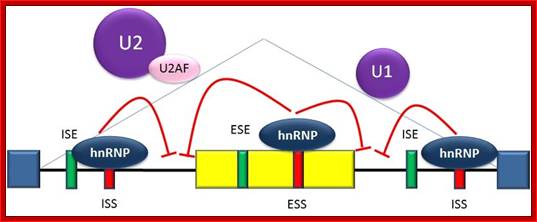
Splicing repression:
Splicing
is regulated by trans-acting proteins (repressors and activators), corresponding cis-acting regulatory
sites (silencers and enhancers) on the RNA, and other RNA features that
influence how splicing will occur, such as secondary structures.� They vary in
sequence, as well as in the types of proteins that bind to them. The majority
of splicing repressors are heterogeneous nuclear ribonucleoproteins (hnRNPs) such as hnRNPA1 and polypyrimidine tract
binding protein (PTB); http://www.answers.com/topic/alternative-splicing#ixzz31sQlLj64
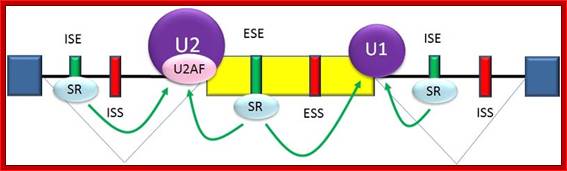
Splicing enhancers are
sites to which splicing activator proteins bind, increasing the probability
that a nearby site will be used as a splice junction. These also may occur in the
intron (intronic splicing enhancers, ISE) or exon (exonic splicing enhancers, ESE). Most of the activator proteins that bind to
ISEs and ESEs are members of the SR
protein family. Such proteins
contain RNA recognition motifs and arginine and serine-rich (RS) domains; http://www.answers.com/topic/alternative-splicing#ixzz31sQlLj64
Alternative Splicing and its Significance:
The vast majority of human genes are broken up into segments (exons) that must be spliced together following transcription of the gene into a precursor mRNA (pre-mRNA). Up to seventy-six to eighty percent of human genes express multiple mRNAs by alternative splicing of their pre-mRNAs (Johnson et al. 2003) in which exons are joined in different patterns. As a result, individual genes express multiple mRNAs that are identical except for discrete regions of variability. Some genes contain multiple alternatively spliced regions and express hundreds or even thousands of different mRNAs. Eighty percent of the time the variability is within the coding region resulting in the expression of different protein isoforms. For many genes, alternative splicing directs expression of functionally divergent protein isoforms according to cell-specific regulation (based on differentiated cell type, developmental stage, gender, or in response to an external signal) (Black 2003; Faustino and Cooper 2003). Alternative splicing can alter the function of proteins by removing or adding specific domains (nuclear localization signals, transcription activation domains, DNA or RNA binding domains, trans-membrane domains), post-translation modification sites, or by causing substantial changes in protein structure by altering even just a few residues (Davletov and Jimenez 2004), (Graveley 2001). Modrek and Lee 2002)..
Alternative splicing Modes:
� This is another versatile mode of differential gene expression, one gene many proteins.
� Alternative form of splicing; is not the only other form of splicing; there are other types such as trans-splicing, self-splicing.� RNAs found in Viroidal RNAs, some ribosomal RNAs and some organelle pre-mRNAs exhibit self-splicing.
� Alternate form of splicing is exemplified in some tissue type expressions such as src-proto-oncogenes, Fibronectin, Troponins, CTF factors, Immunoglobulins, even in tissues involved in sex determination.� A different form of alternative splicing is plausibility.
5�-exonA-I-intron--I-exonB-I-intron-I-exonC-I-intron-I-exonD-3�
� Assume a transcript contains four Exons, A, B and C and D with 3 introns.� They can be spliced differently to generate different sequences such as A-B-C-D (normal), A-B-D-C, A-C-B-D, A-D-B-C, A-C-D, and A-B-D etc.
Exon and Intron Shuffling:
� Exon represents a sequence of amino acids determining a motif or a domain. An exon can code for ~15 aa to ~50 aa (or more in rare cases).� In proteins, exon represents a block of amino acids, so protein is made up of many exons coding for different blocks of amino acids that organize into different motifs.� A single protein may contain one type of motif many times repeated tandemly or in different positions or may contain different motifs or domains in different positions.
� For example, a single exon codes for 10 amino acids and there are 100 different exons coding for different sets of amino acids (means different kinds of motifs and different kinds of functions).� Assume a polypeptide chain of 200 amino acids long, if ten different exons are randomly arranged, what would be the number of different polypeptide chains it can generate. Calculate, if ten different exons are organized by permutation combinations in a 200 ntds long RNA, then what is the number of different combinations it can generate! Alternative splicing causes exon shuffling.� Exon shuffling is one of the many ways to generate new gene products.
Possible Alternative Splicing Modes:
|
Possible form of splicing |
Explanation of splicing process |
|
Optional Exon: |
�Any one of the Exons is spliced out. |
|
Optional introns: |
In place of an Exon an intron can be been retained during splicing |
|
Mutual exclusion of Exons: |
Of the four Exons, any one is excluded to the other.� Ex. In the above-described hypothetical mRNA, out of four Exons, either B is retained instead of C or C is retained in preference to B, and they are mutually exclusive |
|
Internal splicing: |
A part of one intron is included. |
|
Negative control: |
Blocking the splicing of one of the introns by alternative splicing factors. |
|
Positive control: |
Where the splicing is activated for splicing of a specific intron. |
|
|
There can many other alternative forms |
|
|
|
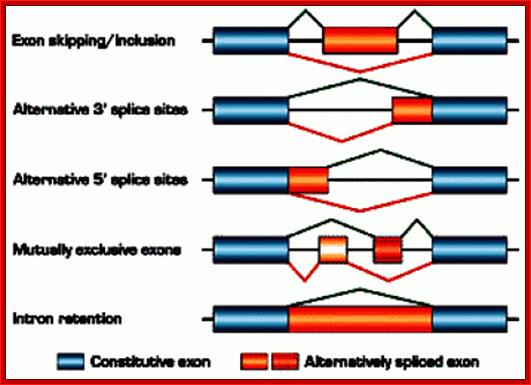
Fig: schematic representation of alternative splicing: The figure illustrates different types of alternative splicing: exon inclusion or skipping, alternative splice-site selection, mutually exclusive exons, and intron retention. For an individual pre-mRNA, different alternative exons often show different types of alternative-splicing patterns. � 2002 Nature Publishing Group Cartegni, L., Chew, S. L., & Krainer, A. R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nature Reviews Genetics 3, 285�298 (2002) doi:10.1038/nrg775.
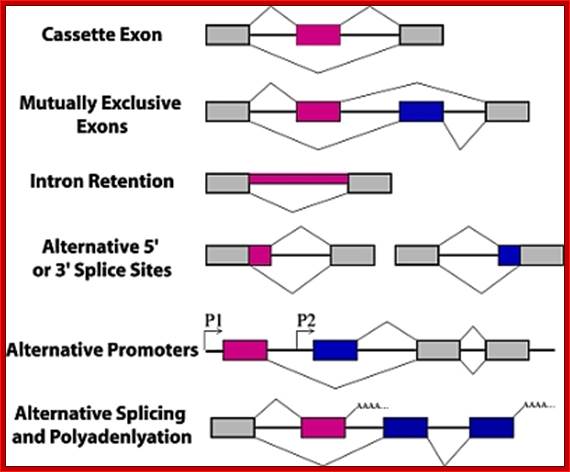
Types of alternative splicing. In these graphics, exons are represented by boxes and introns by lines. Exon regions included in the messages by alternative splicing are colored while constitutive exons are shown in gray. Promoters are indicated with arrows and polyadenylation sites with AAAA.; Alan M. Zahler; http://www.wormbook.org/
There are 4-6 common types of alternative splicing, they are as follows:
- Exon cassette: Certain exons can be skipped in the middle of the protein, resulting in a different transcript.
- Exon cassette: One exonscan be skipped in the middle of the protein, resulting in a different transcript.
- Intron retaining: Introns are used as coding regions. A sequence that is normally considered an Intron is retained in the final transcript that serves as a template for translation.
- Alternative promoter selection: A different promoter is used for different splice variants. This results in a different start of the mRNA transcript.
- Alternative selection of cleavage/polyadenylation sites: Different exons are spliced based on recognition of different cleavage or polyadenylation sites; entire exons can be skipped. Results in a different exon at the 3� end of the transcript.
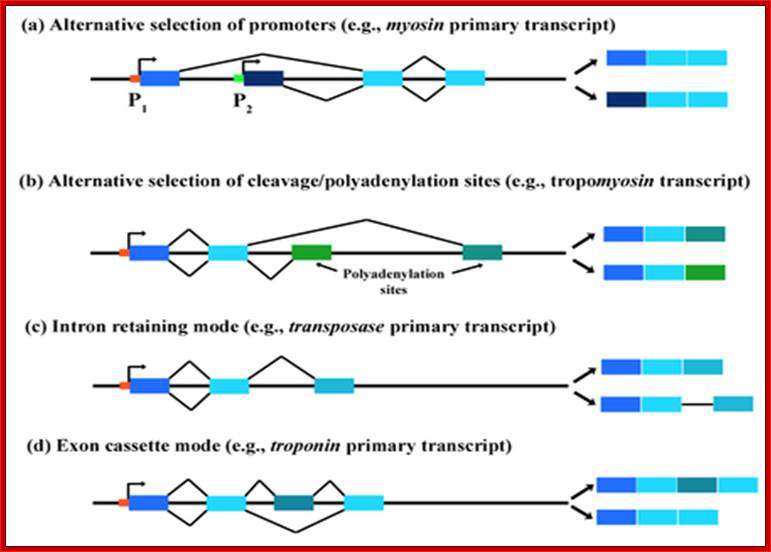 �Four Types of Alternative splicing and their products, using
Alternative promoters. http://en.wikibooks.org/
�Four Types of Alternative splicing and their products, using
Alternative promoters. http://en.wikibooks.org/
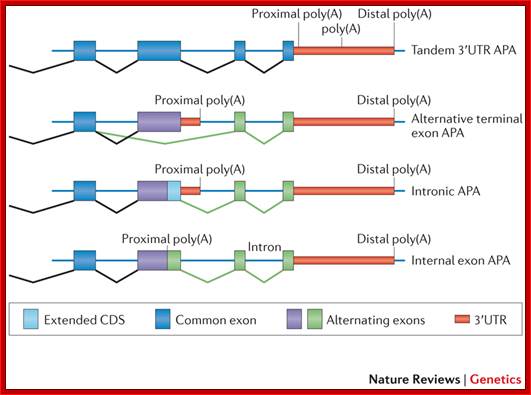
Alternative cleavage and Polyadenylation; The simplest alternative polyadenylation (APA) type, which is termed tandem 3′ untranslated region (UTR) APA, involves the occurrence of alternative poly(A) sites within the same terminal exon and hence generates multiple isoforms that differ in their 3′UTR length without affecting the protein encoded by the gene. The other three types involve APA events, which potentially affect the coding sequences in addition to the 3′UTRs. These types are: alternative terminal exon APA, in which alternative splicing generates isoforms that differ in their last exon; intronic APA, which involves cleaving at the cryptic intronic poly(A) signal (PAS), extending an internal exon and making it the terminal one; and internal exon APA, which involves premature polyadenylation within the coding region. Ran Elkon, Alejandro P. Ugalde & Reuven Agami; ttp://www.nature.com/
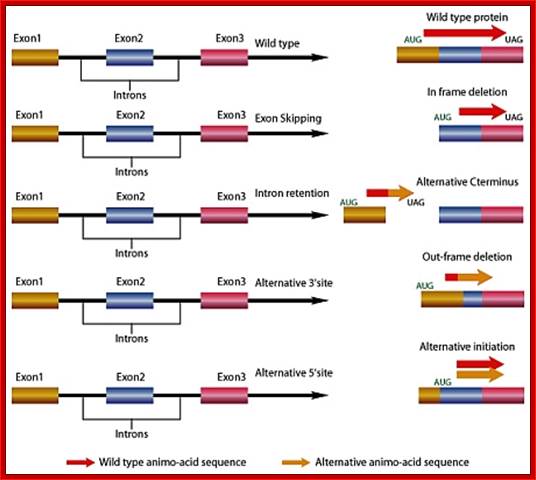
Gene Splicing by Exon skipping, Intron retention and alternative 3�splice site and 5� splice site: http://www.premierbiosoft.com/
A proto-Oncogene called Src, a muscle kinase; occasional mutation in the gene can lead to cancer.� This is a tyrosine kinase gene.� The primary transcript contains, 14 exons including A and B Exons in specific.� This is spliced alternatively in somatic and neuronal tissue to the needs of specific tissues.� Exons A and B provide additional sites for phosphorylation.� In somatic tissues A and B Exons excluded and on the other hand in neuronal tissues A and B are included.� Each form of proteins has distinct functions.
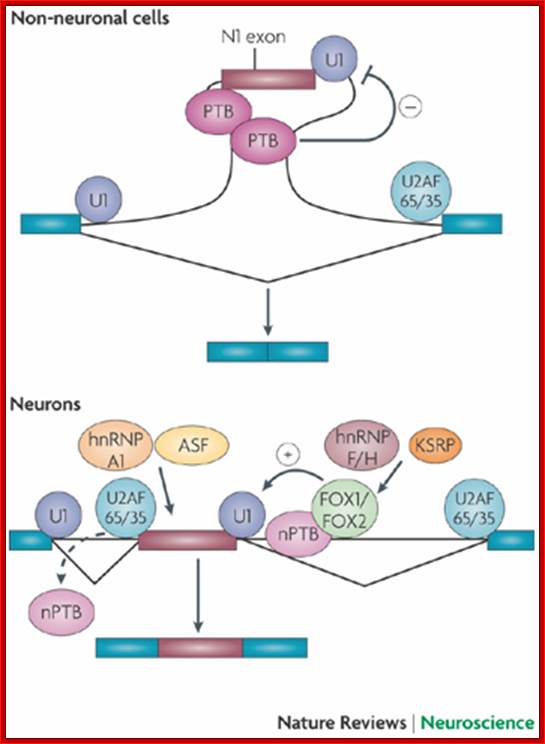
This figure illustrates the regulation of the splicing of the N1 exon of the SRC gene. N1 is repressed in non-neuronal cells by polypyrimidine tract-binding protein (PTB), which binds to elements in the N1 3' splice site and in the downstream intron. This binding block the assembly of an pre-spliceosomal E complex between the N1 exon's 5' splice site and the downstream exon's 3' splice site. In neurons, PTB is replaced by neural PTB (nPTB), which binds to the PTB repressor elements but does not prevent splicing. Neurons also express splicing activators that are members of the Fox family; these bind to enhancer elements downstream of the exon to stimulate its splicing. nPTB must be displaced from the N1 3' splice site (shown as a dashed arrow) to allow its splicing, and it may or may not be displaced from its downstream binding site by the adjacent Fox protein. Other RNA-binding proteins that affect N1 exon splicing include alternate splicing factor (ASF; also known as splicing factor 2 (SF2)), which binds to the exon, and heterogeneous nuclear ribonucleoprotein H (hnRNP H), hnRNP F and KH-type splicing regulatory protein (KSRP), which bind to the downstream intron. These proteins might modulate the function of the key regulators, or might allow regulation in additional cell types (Qin Li, Ji-Ann Lee & Douglas L. Black)
Ovalbumin mRNA Splicing- normal cis splicing:
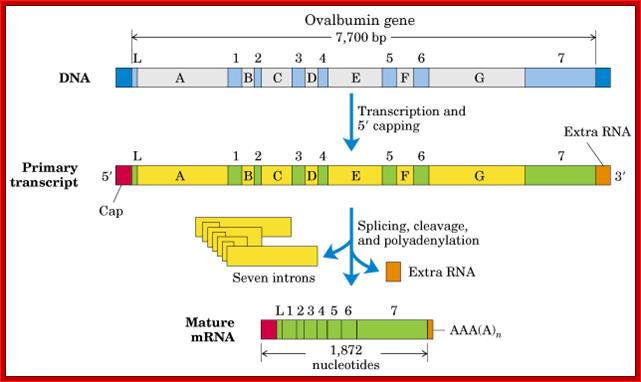
Ovalbumin mRNA cis-splicing; process of splicing was first discovered (I was a student in IISc, and the article on cis splicing of Ovalbumin mRNA published in PNAS); http://www.cs.uni.edu/
Collagen:
Collagen is an important protein in any vertebrates.� It is a large protein and variable in structure and functions. It is synthesized most of the tissues.� The pre-mRNAs contain more than 50 exons and they are spliced in different ways depending on the kind tissue where it is synthesized.� Col1A1 contains 52 exons. There three major forms and they further vary; this is because of alternative splicing.�� Col1A1 gene is 18kb, the two RNA 5.8 and 4.8 generated give rise to 14kDa proteins. Actually because of alternative splicing it can generate 28 different protein types.
Collagen, in the form of elongated fibrils, is mostly found in fibrous tissues such as tendon, ligament and skin, and is also abundant in cornea, cartilage, bone, blood vessels, the gut, and intervertebral disc; Each chain is 1400a.a-long. G.N.Ramachandran (IISc), called this collagens helix as Madras helix.; Elizabeth Jane Kelly; http://www.ebi.ac.uk/
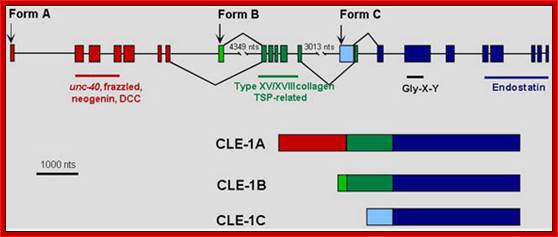
Structure of the cle-1 and protein isoforms. The starts of the three CLE-1 forms, which are transcribed from separate promoters, are shown by arrows above the gene structure. Alternative splicing is indicated by lines above and below the structure connecting between exons. The resulting protein products are illustrated below the gene structure.
Collagen proteins-Alternately spliced, shown only few forms; it consists of 23 exons and by alternative splicing it generate a large number of isoforms; James M.Krammer; http://www.wormbook.org/
Dystrophin:
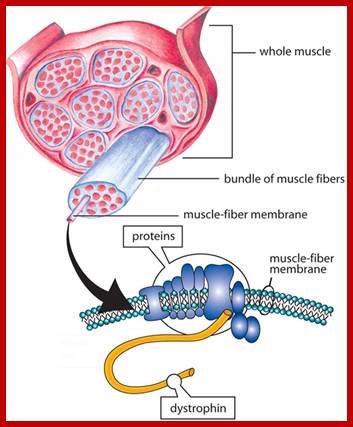
DMD occurs because of mutation in b DMS gene that produces functional Dystrophin protein, it is membrane associated multiplex protein. DMD is inherited in X-linked mode. Most of the time mother carries such genes and sons inherit and sons suffer, not always; https://www.mda.org
There are different types of DMDs, such as Myotonic, Fasciosculohumeral, Congenital, Limb-girdle, and few others.� DMD is a progressive muscle weakness; the disease typically affects male children.� Diseased persons, generally children fall very often, trouble in running, walking on toes, suffer from large calf muscle pain and stiffness and learning disabilities.
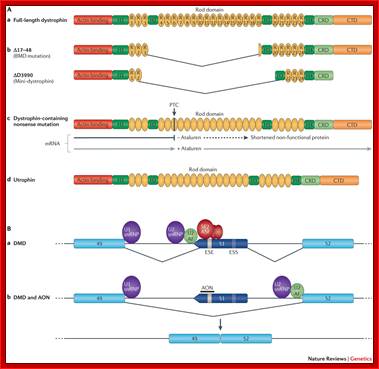
- Aa | Wild-type dystrophin. Full-length dystrophin comprises an amino- terminal actin-binding domain, four hinge domains (H1�H4) and a rod domain consisting of 24 spectrin-like repeats (R1�R24), within which lie a second actin-binding domain, a cysteine-rich domain (CRD) and a carboxy-terminal domain (CTD). Ab | Mini-genes used in gene therapy. A deletion of dystrophin exons 17�48 was identified in a mildly affected patient with Becker's muscular dystrophy (BMD) and was shown to correct 95% of the dystrophic pathology in the Mdx mouse. A clinical trial has been carried out using mini-dystrophin ΔD3990, which consists of a truncated protein that encodes the N-terminal domain, the CRD and the rod domain, but with a reduced number of spectrin repeats (namely, R1, R2, R22, R23 and R24) and three hinges (namely, H1, H3 and H4). Omitting the CTD was not found to be crucial for function. Ac | Nonsense suppression. Ataluren allows read-through of premature termination codons (PTC) to restore production of a full-length functional dystrophin protein. Ad | Utrophin lacks sequence corresponding to spectrin-like repeats 15 and 19 of dystrophin and binds actin only through the N-terminal domain. Utrophin and some of the dystrophin mini-genes will not localize neuronal nitric oxide synthase (nNOS) to the sarcolemma, as is the case in some patients with BMD. B | The principles of antisense oligonucleotide (AON)-mediated skipping of dystrophin exon 51. Ba | The Duchenne muscular dystrophy (DMD) locus in a patient with a deletion of exon 50. As a result of the deletion, exons 49 and 51 are out-of-frame, which leads to an unstable precursor mRNA (pre-mRNA) transcript and a lack of dystrophin protein. Also shown in this region of the locus are some of the key cis- and trans-acting elements that regulate the splicing of the dystrophin pre-mRNA, including U1 and U2 small nuclear RNAs (snRNAs), which define exon�intron boundaries and also exon internal sequences, such as exonic splicing enhancers (ESEs) and exonic splicing silencers (ESSs) that promote and inhibit exon inclusion during pre-mRNA splicing, respectively. Bb | An AON-targeting exon internal sequences within exon 51 adjacent to putative ESE sites may inhibit the association of splicing regulatory proteins (for example, of the SF2 and ASFprotein families) at this recognition site and therefore promote the skipping of this exon during pre-mRNA splicing. Skipping of exon 51 is able to restore a viable mRNA reading frame as axons 49 and 52 are in-frame exons, and therefore a truncated but semi-functional dystrophin isoform is generated. snRNP, small nuclear ribonucleoprotein. Rebecca J. Fairclough, Matthew J. Wood ;& Kay E. Davies; http://www.nature.com/
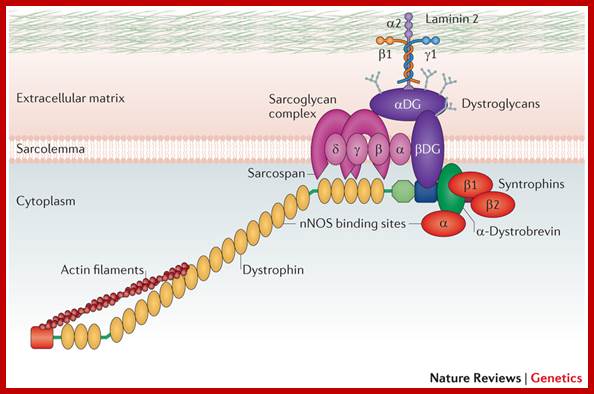
- Dystrophin acts as an important link between the internal cytoskeleton and the extracellular matrix. Neuronal nitric oxide synthase (nNOS) binds to α-syntrophin but also has a binding site in repeat 17 of the rod domain of dystrophin (see Fig. 2A for details of dystrophin domains). αDG, α-dystroglycan; βDG, β-dystroglycan. Rebecca J. Fairclough et al, ��http://www.nature.com/
Fibronectin:
�����������������������������������������������������������������������
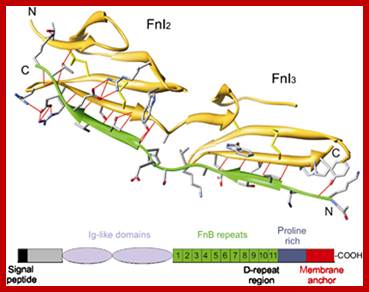
http://femsre.oxfordjournals.org/
Fibronectin is a very important protein that is secreted out of the cell; actually, it is an exoplasmic cellular protein.� In one form it is free from the cell surface and in another form, it is anchored on to the cell surface at the exoplasmic surface, actually Fibronectin can exist in different forms in different tissue type and it is generated by alternative splicing in different tissue types.� This protein has many important functions. It is a multidomain host adhesion protein. It consists of two 250kDa monomers.� The Fibronectin gene is 75 kbp long and the pre-mRNA has 32 Exons.�� Some of the Exons have many repeats located in different positions of the gene in different copy numbers.� In this case there are six types of Exons; they are A, B, C, U, EIII-B, E III-A and two undefined.� They are organized in the following fashion, which shows the kind of Exon and how many repeats of each and the location in the gene.� This is an excellent example to show how one-form Exons can amplify and spread in the same gene or they can be transposed to different genes.� Note each exon codes for a protein motif or domain.� Permutation and combination of exons provide versatility to proteins in terms of structure and function.
---[A]6--[U] 2--[A]3--[B]7--[E IIIB]1--[B]4--[E III-A]1--[B]3--[C]1-[B]1-[A]3---
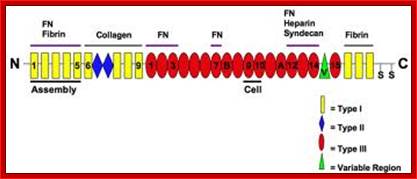
The modular structure of fibronectin and its binding domains: http://en.wikipedia.org/
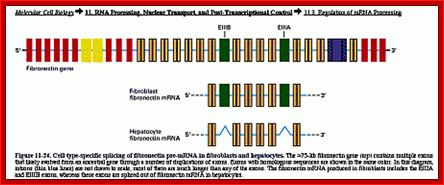
Cell type-specific splicing of fibronectin pre-mRNA in fibroblasts and hepatocytes. The 75-kb FN gene (top) contains multiple exons. Introns are shown in the diagram as thin lines and are not drawn to scale. Most of the introns are much longer than any of the exons. The FN mRNA produced in fibroblasts includes the EIIIA and EIIIB exons, whereas these exons are spliced out of FN mRNA in hepatocytes (from Lodish et al. 2000. Available at http://www.ncbi.nlm.nih.gov/PubMed/). ;http://www.library.utoronto.ca/
--[A]6--[U]2--[A]3--[B]7--[E IIIB]1--[B]4--[E III-A]1--[B]3--[C]1-[B]1-[A]3---
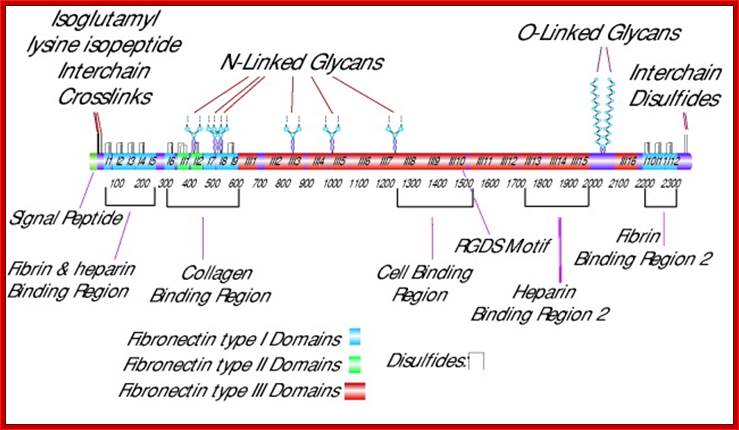
Cellular fibronectin is an adhesion glycoprotein of the extracellular matrix, which exists as a dimer with a molecular mass of ~550 kDa. It contains two heterodimers, the A chain and the B chain containing the type III connecting segment (IIIcs) region. Cellular fibronectin differs from plasma fibronectin, a 200�250 kDa monomer, by the presence of additional polypeptide segments and in altering morphology of transformed cells and hemagglutination. Different forms of fibronectin appear to be generated from tissue specific splicing of fibronectin mRNA, transcribed from a single gene. Multiple domains of fibronectin show binding affinities to collagen, fibrin, heparin, and membrane receptors. The most notable domain, Arg-Gly-Asp (RGD), is recognized by integrins and mediates cell adhesion. Fibronectin is involved in widespread interactions and functions, such as the attachment and migration of many cell types, cytoskeletal assembly, tyrosine phosphorylation, and metastasis, Sigma.http://www.library.utoranto.ca
In Fibroblasts, which is found ubiquitously, processing of Fibronectin pre-mRNA involves in removing E III-B and E III-A.� This protein is secreted.� But in other tissues its E III B and E III-A are included, so the proteins generated remain anchored to the membrane.� Thus these proteins are involved in adhesion and cell-cell interaction with other cells.� In hapatocytes, splicing excludes E III-A and E III-B, when secreted it is left free for circulation in the blood stream and this form of Fibronectin has different kind of functions. �Thus, one can discern that blocks IIIA and IIIB have membrane anchoring features.
A similar type of alternative splicing is found in Intermediate filaments, Troponins, Tropomyosin, SV 40 T and t antigen, Calcitonin, Immunoglobulin, CD4 and their relative lymphocytes and many other structural and transcriptional factors like CAT binding factors (CTFs) and many others.� The above said proteins are produced tissue specifically.
IgM- Alternative Splicing (with different poly-A signals):
- Primary B-lymphocytes individually produce primary Immunoglobulins, which bind to cell surface membranes and act as receptors for specific antigens.� IgM is a pentamer linked by disulfide bonds (970kDa).� Once the cells are activated, cells express the same gene but processing of the pre mRNA changes in such a way the protein produced is released into blood stream.
- but processing of the pre mRNA changes in such a way the protein produced is released into blood stream.
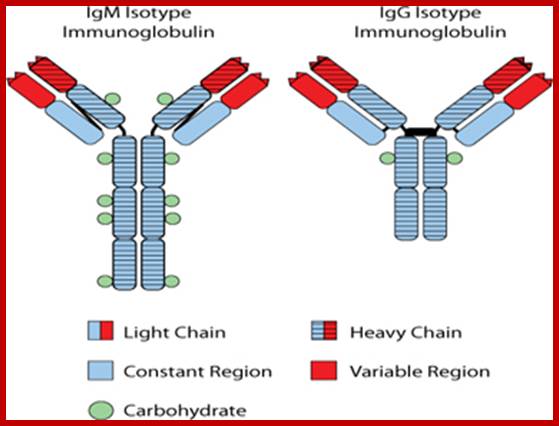
The Id
protein expressed on the surface of FL cells is an immunoglobulin protein
characteristic of the single B-cell from which the tumor arose. The
immunoglobulin protein contains a region known as the �heavy chain� and a
region known as the �light chain�. Almost always in FL, the heavy chain region
is characterized as either an IgM-isotype or an IgG-isotype. The figure
illustrates the dramatic differences in the structure of immunoglobulin protein
characterized as an IgM-isotype as opposed that characterized as an
IgG-isotype.
Read more: http://getfilings.com/sec-filings
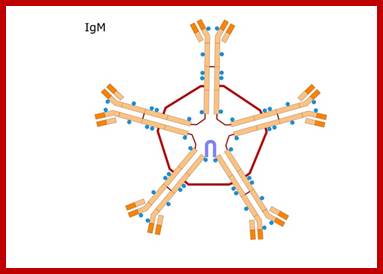
IgM scheme: Heavy chains are blue; light chains are yellow; http://en.wikipedia.org/
- The pre mRNA has a splice donor site and a splice receiver site in the coding region.� In the intron it has Ter (translational termination signal and one RNA cleavage signal and one poly-A signal).� At the end of the codon segments, it has another stop signal and a poly-A signal and cleavage site.
Immunoglobulin M is primarily produced by B lymphocytes.� It is the first antibody to appear in response to an antigen. Both the membrane-associated and secreted Ig proteins are encoded by a single gene whose primary transcript is alternatively processed at its 3' end. The relative use of the alternative processing pathways is regulated during B cell maturation.
�
- The pre mRNA has a splice donor site and a splice receiver site in the coding region.� In the intron it has Ter (translational termination signal and one RNA cleavage signal and one poly-A signal).� At the end of the codon segment, it has another stop signal and a poly-A signal and a cleavage site.
This alternative RNA processing involves two competing reactions, splicing from the last constant region exon to the membrane exon(s) and cleavage-polyadenylation at the secretory-specific poly (A) site. Studies with the IgM-encoding mu gene have shown that cell-specific regulation requires that the efficiencies of these two reactions be balanced; any gene modifications that substantially improve or reduce the efficiency of either reaction also abrogate the regulatory shift in alternative processing pathways. All of the Ig isotypes that undergo a membrane-to-secreted switch during B cell maturation have a similar gene structure, thus suggesting that they might all be regulated by the same mechanism. It has been established previously that cleavage-polyadenylation activity is higher in plasma cells, which secrete IgM, than in B cells, which produce membrane-associated IgM. To determine whether RNA-splicing activity varies during B-lymphocyte development to contribute to micro RNA-processing regulation, we first demonstrate that micro pre-mRNA processing is sensitive to artificial changes in the splice environment by expressing SR proteins with the micro gene, Bruce SR, Dingle RW, Peterson ML. Source-Department of Microbiology and Immunology,. University of Kentucky College of Medicine, Lexington 40536, USA.
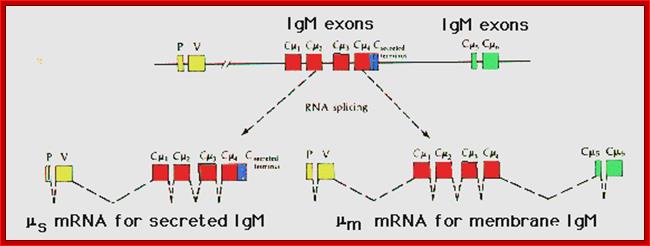
Figure: Proposed alternative splicing patterns for secreted m and membrane mu mRNA. P includes the leader sequence; V encodes the variable (VDJ) region; the possible constant region exons of the mu (IgM) constant region follow downstream. The spliced RNAs (introns) are shown as dashed lines (After Early et al., 1980).
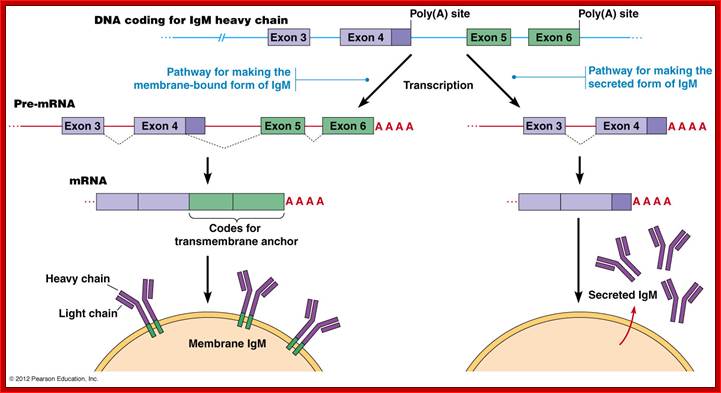
The IgM gene has two possible poly(A) addition
(termination) sites and a number of exons that can produce two alternative
forms. The plasma membrane-bound form contains a transmembrane anchor which is
encoded by exons 5 and 6. If a splice junction within exon 4 is used, exons 5
and 6 (carrying the anchor) are added to generate the IgM heavy chain. The
secreted product is produced when the exon 4 not spliced made and these
transcripts are terminated just after exon 4.
(Principles of Cell Biology (BIOL2060), Department of Biology
Memorial University of Newfoundland.
The secreted IgM contains regions encoded by the VDJ genes and exons Cmu1, Cmu2, Cmu3, and Cmu4. It also contains a terminal portion that allows it to be secreted. The membrane-bound IgM contains the same arrangement, except that instead of the "secretion" exon, it has added a portion encoded by two more exons, Cmu5 and Cmu6, which gives it a hydrophobic tail that can integrate into the lymphocyte membrane. Thus, cleavage and polyadenylation of the Cmu4 exon yield the secretory form of IgM, while cleavage and polyadenylation of the Cmu6 exon (and the differential splicing of Cmu4) yield membrane-bound IgM (Figure 3). Peterson and Perry (1989) have suggested that there is competition between the two sites for the cleavage and polyadenylation factors and that the biases in this competition change as the B cell matures. This has recently been shown to be the case. Takagaki and colleagues (1996) have shown that these two sites compete for limiting amounts of a factor (CstF) that directs the cutting enzymes to the 3′ splice site. The site for the membrane-bound form of the mu-heavy chain is more efficient at binding. Hence, at the low amounts of CstF present in the B-cell, only the membrane bound form of IgM is made. However, when the B cell differentiates into a plasma cell, more CstF is produced, and both sites are utilized. The developing plasma cell makes both the membrane-bound and secreted forms of IgM.
In the primary B lymphocytes, the central intron in the pre-mRNA gets spliced out and poly �A is added at its end, and the translational product has c-terminal end having membrane anchor sequences, so it is anchored on to the cell surface.
- But in activated B-lymphocyte, the poly (A) signal and cleavage sites found in the intron are used to generate a truncated mRNA containing a stop codon in the intron itself; here a part of intron is used to generate a protein.� The protein produced from this mRNA doesn�t contain any C-terminal anchor sequences, so the protein is released into blood stream.
Adenoviral E1 RNA splicing-Alternative types:
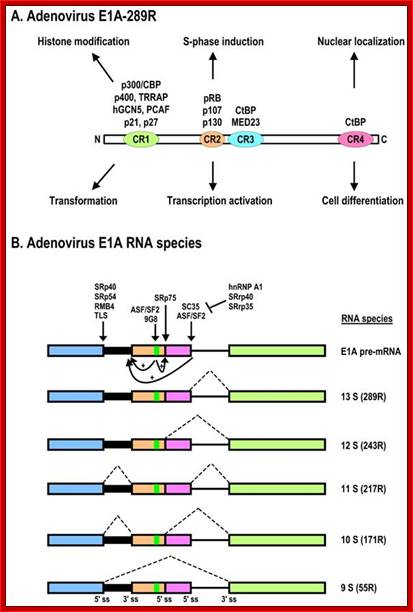
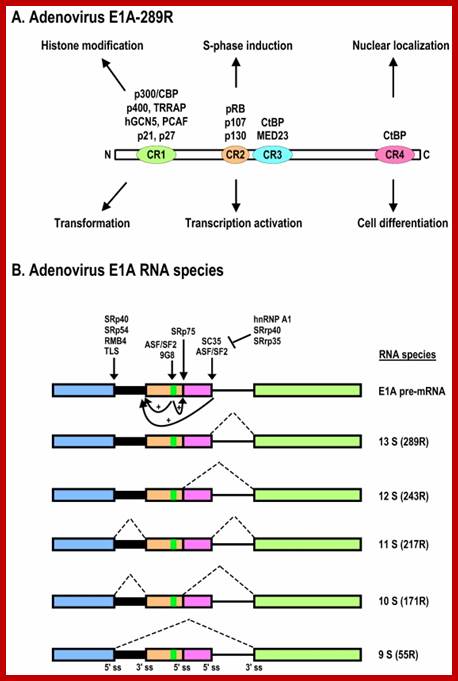
Schematic structures of adenovirus E1A protein and mRNAs; �A. Structure of E1A protein (full-length 289R variant) and its biological functions. Four conserved regions (CR1-CR4) in E1A and mapped domains in E1A are diagramed. �B. RNA structure and alternative spliced species of E1A pre-mRNA. A bidirectional splicing enhancer (BSE) is shown in exon 2 in green, and cellular splicing factors or regulators that control selection of each splice site are indicated by arrows. The panel is modified from reference, with permission. Dotted lines indicate splicing directions. Zhi-Ming Zheng; http://www.ijbs.com/
Adenoviral late gene transcripts:
The transcript of late genes is a massive one of 28 KB length.� This pre mRNA contains three leader sequences L1, L2 and L3 at 5�end and several coding sequences on the right side of the transcript like C1, C2, C3, C4 and so on.�� Alternate splicing leads to the production different transcripts coding for different proteins but with the same common leader sequence.� Common leader sequence is derived from another transcript and spliced to spliced to mRNA by what is called trans-splicing.
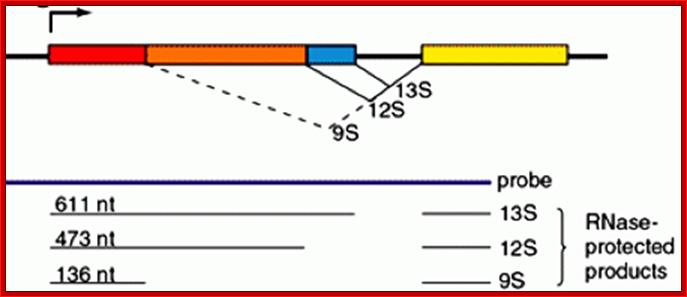
The adenovirus genome encodes two viral oncogenes, E1A and E1B, positioned side-by-side in the left 11.2% of the genome. After an adenovirus infects a human cell, the first viral proteins that are synthesized are products of the E1A region. The full-length E1A protein (289R) is a nuclear protein consisting of 289 aa residues and has four conserved regions: CR1 at the N-terminus, CR2 and CR3 in the middle, and CR4 at the C-terminus; https://www.researchgate.net/
5�-I-L1-II-L2-II-L3-I I-C1--(A*) nII-C2-(A*)nI-I-C3-(A*)nI-I-C4-(A*)nI-3�
5�-L1-I-L2-I-L3-I-�����
5� L1-L2-L3-I---C1�(A) n
5� L1-L2-l-3-I�C2--- (A) n
5� L1-l2--I3-I----C3�(A) n
���� Calcitonin:
Calcitonin (also known as thyrocalcitonin) is a 32-amino acid linear polypeptide hormone that is produced in humans primarily by the Para follicular cells (also known as C-cells) of the thyroid, and in many other animals in the ulti-mobranchial body. Calcitonin the primary gene transcript consists of 6 Exons with two poly-A signals, one at the end of 4th Exon and another at the end of 6 th Exon.
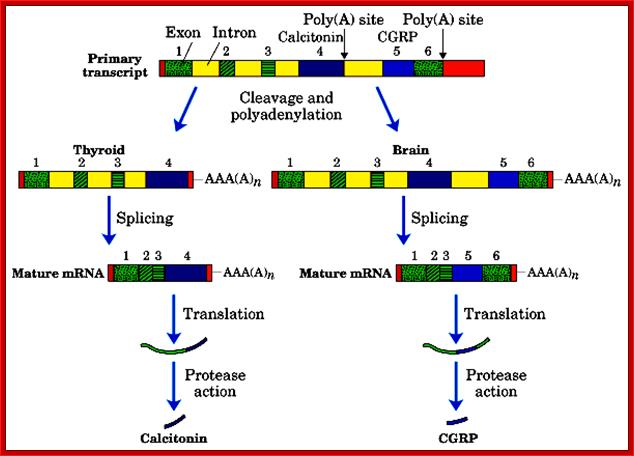
Alternative processing using alternative poly adenylation at specific sites, CGRP = Calcitonin Growth Receptor Protein; http://quizlet.com/
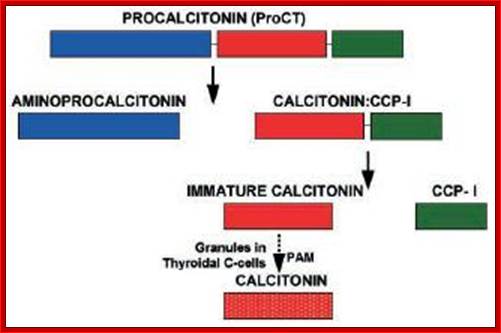
http://www.ufrgs.br/
- Alternative splicing and poly-A addition at the end of 4th exon generate one protein in thyroid cells, which is further subjected to protease to produce the final protein called Calcitonin.
- In another construct it generates a protein with the exclusion of 4th exon, which is also subjected to protease action, and this product is produced in brain cells and it is called Calcitonin growth receptor protein.
Thyroid cells: 1-2--3-4-(A)
Neuronal cells: 1-2- 3-5-6 (A)
This phenomenon obviates the requirement of an additional gene(s) meant for a specific but additional function.�
HIV-mRNA- Alternative splicing:
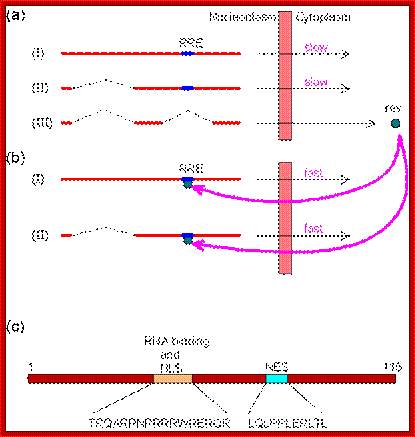
The HIV mRNAs are produced from the primary transcript by three different splicings: unspliced, singly spliced and doubly spliced. Although unspliced and singly sliced mRNAs are made before doubly spliced mRNAs, the protein products of the doubly spliced mRNAs are the first synthesized in the cytoplasm because they are smaller and exported faster. As a matter of fact, expression of unspliced and singly sliced mRNAs would be negligible without the rev protein which is a product of the doubly spliced mRNA. A rev protein consists of 116 amino acids, including a nuclear localization sequence (NLS) and a nuclear export sequence (NES); http://www.web-books.com/MoBio/Free/
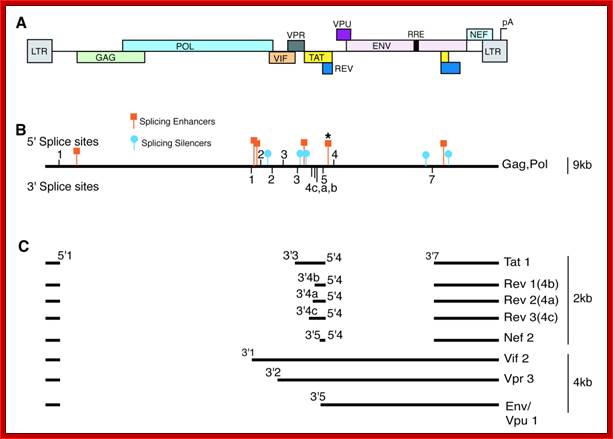
A) The map shows the HIV-1 open reading frames. B) A single pre-mRNA of 9.2 kb is transcribed by the virus. 5� and 3� splice sites are indicated The, unspliced viral mRNA codes for the Gag/Pol gene products. 5� and 3� splice sites are indicated. Splicing silencers (intronic and exonic) and splicing enhancers (intronic and exonic) are indicated. (*) marks the location of the GAR splicing enhancer (see Fig 5). C) Prevalent spliced viral mRNas. Over 40 alternatively spliced mRNAs are originated by the alternative usage of the multiple 5� and 3� splice sites, the most abundant mRNA isoforms are indicated with their approximate size and the splice site utilized to generate them. http://www.intechopen.com/
http://www.pinwallpaper xyz
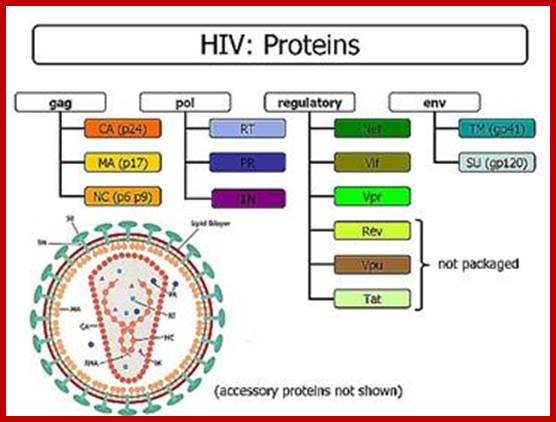
Alternative splicing generates six proteins; Translation terminator in between gag- pol and env generates one full length transcript.� In this, the 5�terminal exon remains constant; can we call this trans splicing combined with alternative splicing- �Trans-Alternative Splicing (TALTVs). Nearly six smaller exons are alternatively spliced; http://www.web-books.com/MoBio/Free
Alpha Tropomyosin:
- In alpha Tropomyosin gene, from rat, the pre-mRNAs have ~12 Exons and they are spliced alternately in different tissues to generate 40 or more different isoforms distinctly different in striated muscles, smooth muscles, brain tissues, Myeloblast, non-muscle fibroblasts and Hepatoma. �Tropomyosin are a large family of integral components of actin filaments which play a critical role in regulating the function of actin filaments in both muscle and nonmuscle cells. These proteins consist of rod-shaped coiled-coil hetero- or homo-dimers that lie along the α-helical groove of most actin filaments. Interaction occurs along the length of the actin filament with dimers aligning in a head-to-tail fashion (wiki).
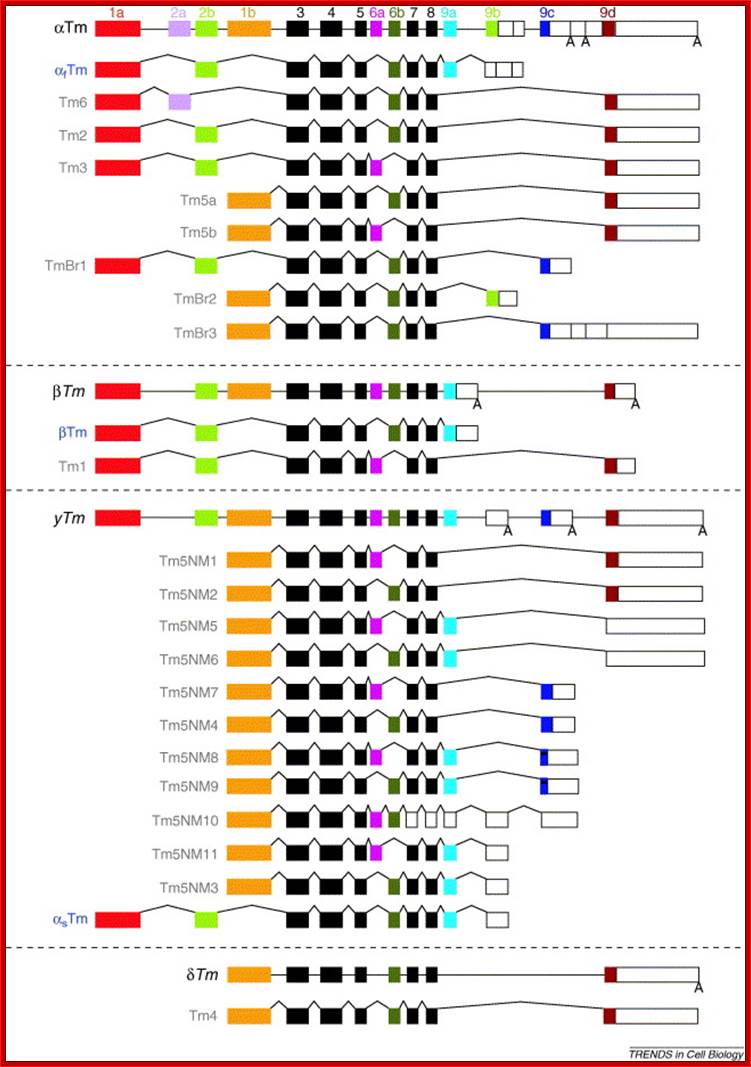
Alternative splicing generates multiple products through the use of alternative promoters, resulting in different amino termini, mutually exclusive internal splicing of 6a versus 6b and alternative carboxyl termini. Colour coding is used to indicate that the 1a exon, for example, from the α-Tropomyosin gene is more similar to the 1a exon from the β-Tropomyosin and γ-Tropomyosin genes than it is to the alternative N-terminal 1b exon from the α-Tropomyosin gene. Not all isoforms generated from these genes are shown, although the existence of those shown has been confirmed by northern blots. In most cases, the isoforms arising from alternative splicing do not contain an exon unique to just one isoform. Rather, the isoforms gain their individuality from a unique combination of exons; http://en.wikipedia.org/wiki (wiki).
SV 40 T and t antigens:
- An animal virus with ds circular DNA of 5300 bp long codes for two types of genes from one region.� Transcription of early genes produce T antigen 90kd, which is required for transformation of cells, later a truncated protein is produced from the same transcript, it is called small t.� This is required for the maintenance of the transformed cells.�
- The production of �t� antigen is by alternate splicing.� Similarly, the late transcript, which codes for capsid proteins are also, produced as single transcript; by alternate splicing it generates three capsid proteins called VP1, VP2 and VP3.
- �But Polyoma virus produces the third middle T protein, again by alternate splicing.
�������������������� Exon 1�������������� ����� ���exon2������������������������� exon3
��� ������� 5�---I------------I - Intron- -I---------ter--*---I - intron -I------------terI----*-3����������������������� * Poly-A signal
����������� �������
������� ��� ����������� ����������� Exon1���������� �������exon3
��������� �5�--------I--------------I-----------------------I--------3�= T antigen
��������������������������������� � Exon1��� exon2
���������� 5�--------I--------------I-------------*������ = t antigen
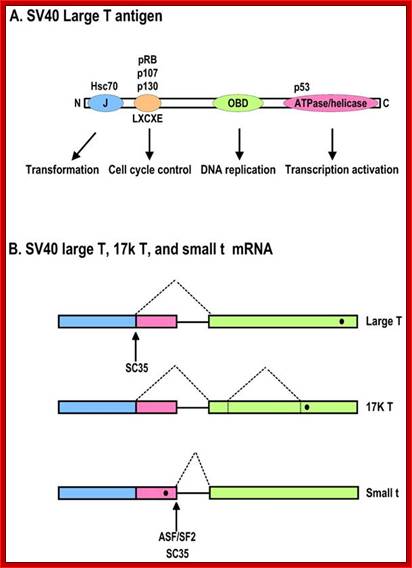
Diagrammatic representation of the SV40 large T antigen and its RNA splicing. A. Schematic protein structure of SV40 large T antigen. J, DnaJ domain; OBD, origin DNA-binding domain. B. Alternative splicing of SV40 T antigen pre-mRNA leads to production of Large T, 17K T, and small t mRNAs. Black dots indicate stop codon locations on spliced RNAs; Zhi-Ming Zheng http://www.ijbs.com/
�����������������������
- The pre-mRNA consists of three Exons namely Exon 1, 2 and 3.� In the exon-2, it has Terminator codon.
- The primary transcript, when spliced normally, it splices out exon-2 and the protein produced from this transcript is T-antigen.
- In the transformed cells splicing includes exon-2.� When the mRNA is translated it generates truncated protein called �t�-antigen.
Chick light Myosin chain splicing:
� In this case, alternate form of splicing produces proteins in tissue specific manner.
� Chick myosin gene produces Cardiac type light myosin chain and in gizzard it produces light chain LC3, but in skeletal muscles it generates both types.
� This is achieved by using two promoters located 10 kbp apart.� By combining two promoters and alternate spicing it produces two different forms of transcripts and proteins, but production of other type of myosin proteins in the same tissue is really fascinating.� The said proteins differ only in the N-terminal ends but at 3�terminal ends they are same.� Myosin heavy chain contains 30 exons.
An intron in one tissue can be an Exon in another tissue: Ex. amylase
����������� Mice:�
In mice, introns of Amylase in one tissue, and it can an exon in another tissue:
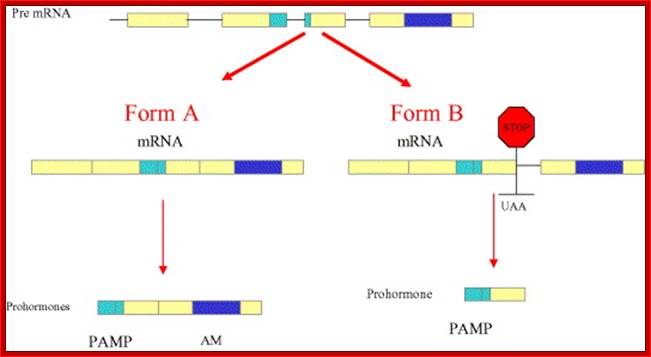
Schematic drawing representing the genomic structure of the AM gene and the alternative splicing mechanism governing the differential expression of AM and PAMP. Removal of the three introns yields form A mRNA, which carries information for both peptides (left hand side). Retention of the third intron results in the introduction of a premature stop codon that prevents AM transcription. Reprinted with permission from Mart�nez et al.
Salivary glands �Amylase:
������������
In salivary glands, promoter and initiator sites lead Exon L partly derived from the intron of amylase transcript.
-IPI�exon-S-I--I-PI-Exon-L-I-intron-I-Exon2-I-intron-I-exon3-I-(A) n
Processed mRNA:��������������������� �
5�capI---L-----I---2----I---3----I--- (A) n
Liver tissue:
--I-p-I--Exon-S-I----I-P-I---Exon-L-I---I--exon-2-I--I---exon-3-I--- (A) n
Processed mRNA:�������������������������
5�capI�S---I---2----I---3-----I--- (A) n
� In this Exon-1 has been spliced out and retained Exon 2 and 3.
Thus, two different forms of amylases are produced in tissue specific ����manner.
Troponin Gene T of Rat muscle:
The Troponin-T pre-mRNA has some 5 Exons that differ from tissues to tissue, but not the 5� proximal Exons and they remain the same.� One form of Troponin is Troponin-T alpha and the other form is Troponin-T beta form.
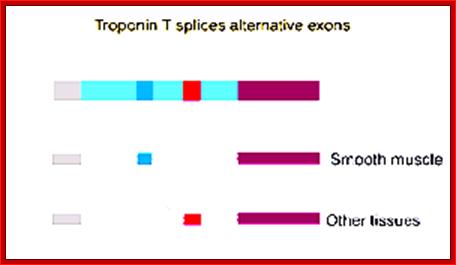
5�G(CH3) -----III---I-w-I- - - I-x-I- - -I---beta-----I-----I-alpha-I - -- -I-Z-I (A) n; main transcript.
�����������������
�During processing the Exon alpha and beta are spliced in mutually exclusive manner in different tissues. The alternative forms of alpha Troponin and beta Troponins are as follows;
Alpha-Troponin form:� 5�cap ---//---Iw-I-X-I Alpha-I-Z-I- (A) n,
Beta-Troponin form: 5�cap ----//--I-W-I-X-I-beta-I-Z-I- (A) n,
- Alternate splicing of Bcl gene transcripts can lead to apoptosis or block apoptosis that can lead to cancer. If the Exon II and III are spliced together the protein produced from the transcript is called Bcl-xl that blocks apoptosis.� On the contrary if a part of Exon II (shaded) is excluded it generates transcript and the protein called Bcl-Xs induces apoptosis
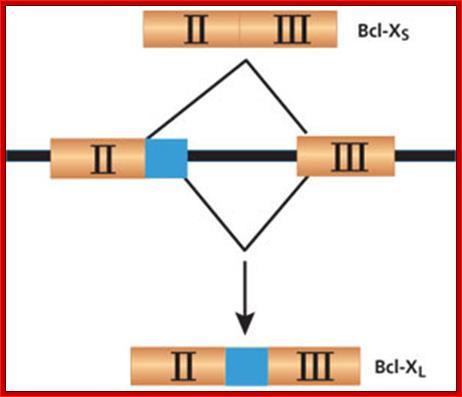
;http://www.the-scientist.com/
The BCL-X gene is a good example of how alternative splicing can make a big difference. If the gene is spliced to include all of Exon II, it will produce Bcl-XLmRNA, which results in a protein that inhibits apoptosis. If the gene is spliced without the non-shaded portion of Exon II, it produces Bcl-XS mRNA, which in turn creates an apoptosis-inducing protein. Many cancers have a high incidence of Bcl-XL, while successful chemotherapy results in a higher proportion of Bcl-XS. ;http://www.the-scientist.com/
�
- Mutations in splicing sites, mutation in splicing factors can lead to abnormal alternative splicing for cis splicing or alternative splicing is pre programmed.��
- So the cells where specific genes are expressed are pre-programmed for the event.� Each cell type, depending upon its needs, produces specific alternate splicing factor or factors in addition to general factors; together they perform alternate splicing. In one tissue the same species of pre-mRNA may be spliced normally, but in another cell type it is spliced in alternative fashion to generate slightly different polypeptide chain.� The exons determine certain motifs or domains. The two different polypeptides generated in this way contain substantial homology, but differ in some segments, that make the difference.
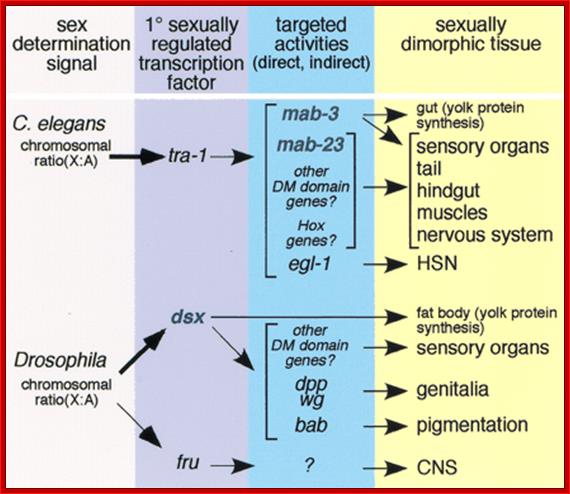
Alternative Splicing and Sex Determination in Drosophila:
Alternate splicing is powerful mechanism by which organisms can generate a greater number of proteins with less number of genes, economy �par excellence�.� Activators and Repressors often regulate alternate splicing.� They use exon and intron enhancer sites.� Regulator proteins bind to Exonic or Intronic splicing enhancer sites (ESE and ISE).� Alternatively, silencers bind to Exon or Intron splice suppressors (ESS or ISS).� Proteins called SR forms are produced constitutively, tissue specifically and produced in response to signals and they are not only responsible for normal splicing but also involved in alternative splicing process.� A specific SR protein performs a specific alternative splicing.� SR proteins have its own specific domains called RNA recognition motif (RRM) and also RS motif (Arginine and Serine) at the Carboxyl end.� The RS motifs interact with SR proteins and splicing proteins and recruit splicing components to nearby splicing sites.� Alternative splicing generates proteins called Isoforms.� They have slightly different structural features and functional properties.
Genetics of Development; http://www.bio.miami.edu/
Chromosomal basis of Sex Determination in Drosophila; XX and XY: http://biology200.gsu.edu/
Ratios of X chromosomes to autosomes in different sexual phenotypes in Drosophila melanogaster
|
X chromosomes |
Autosome sets |
(A)X:A ratio |
Sex |
|
3 |
2 |
1.50 |
Metafemale |
|
4 |
3 |
1.33 |
Metafemale |
|
4 |
4 |
1.00 |
Normal female |
|
3 |
3 |
1.00 |
Normal female |
|
2 |
2 |
1.00 |
Normal female |
|
2 |
3 |
0.66 |
Intersex |
|
1 |
2 |
0.50 |
Normal male |
|
1 |
3 |
0.33 |
Metamale |
Source: After Strickberger 1968.
Phenotypic Consequences of Different Ratios of X Chromosomes to Autosomes
In Drosophila n=4,
where there is 1 sex chromosome and 3 autosomes.
Call each autosome set A, therefore there are 3 autosmes in a diploid fly (A=2)
Sex determination is due to the ratio of X chromosomes to the sets of autosomes
(A)
Therefore:
|
XXAA (diploid) |
X:A ratio = 1 |
Female |
|
XYAA (diploid) |
X:A ratio � 0.5 |
Male |
|
XOAA (diploid) |
X:A ratio = 0.5 |
STERILE Male |
|
XXXAAA (aneuploid) |
X:A ratio = 1 |
Female |
|
XYYAAA (aneuploid) |
X:A ratio = 0.33 |
Male |
|
XXYAAA (aneuploid) |
X:A ratio = 0.67 |
Intersex |
Gynandromorphs:
![]()
Edited by Arjun K Nanda; http://sgugenetics.pbworks.com/
(A) Gynandromorph of D. melanogaster in which the left side is female (XX) and the right side is male (XO). The male side has lost an X chromosome bearing the wild-type alleles of eye color and wing shape, thereby allowing the expression of the recessive alleles, eosin eye and miniature wing on the remaining X chromosome. (B) Photograph of a gynandromorphic Io moth, divided bilaterally into a rose-brown female half and a smaller, yellow male half. ((rom Morgan and Bridges 1919, drawn by Edith Wallace. B; photograph by T. R. Manley, courtesy of The Journal of Heredity.)
(B) The X:A ratio is evaluated through the interaction of numerator and denominator monomeric protein subunits that interact to produce an active complex referred to as NUM�NUM transcription factor. The level of active numerator transcription factor determines whether Sxl (Sex lethal) is to be permanently turned on or is to remain off. If Sxl is on, then the female sex differentiation pathway is turned on, ultimately causing splicing of a form of the dsx mRNA that produces a transcription factor that represses male-specific genes. If Sxl is off, then the sex differentiation pathway is not activated and the default dsx (doublesex) splicing pattern creates an mRNA encoding a female-repressing transcription factor, hence male sex develops in the insect.
(C) The first phase of Drosophila sex determination involves reading the X:A ratio. What elements on the X chromosome are �counted,� and how is this information used? It appears that high values of the X:A ratio are responsible for activating the feminizing switch gene Sex-lethal (Sxl). In XY cells, Sxl remains inactive during the early stages of development. In XX Drosophila, Sxl is activated during the first 2 hours after fertilization, and this gene transcribes a particular embryonic type of Sxl mRNA that is found for only about 2 hours more (Salz et al. 1989). Once activated, the Sxl gene remains active because its protein product is able to bind to and activate its own promoter (S�nchez and N�thiger 1983), (Cline 1983; Salz et al. 1987 and Van Doren et al. 1991); Younger-Shepherd et al. 1992).
D. This female-specific activation of Sxl is thought to be stimulated by �numerator proteins� encoded by the X chromosome. These constitute the X part of the X:A ratio, Cline (1988) has demonstrated that these numerator proteins include Sister less-a and Sister less-b. These proteins bind to the �early� promoter of the Sxl gene to promote its transcription shortly after fertilization.
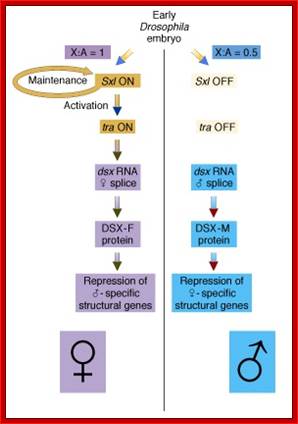
In Drosophila sex is determined via a complex series of genetically programmed developmental choices.� This is in turn determined by the genic balance. ; http://www.bio.miami.edu/
The �denominator proteins� are autosomally encoded proteins such as Deadpan and Extra-macrochaetae. These proteins block the binding or activity of the numerator proteins the denominator proteins may actually be able to form inactive heterodimers with the numerator proteins. It appears, then, that the X:A ratio is measured by competition between X-encoded activators and autosomally encoded repressors of the promoter of the Sxl gene.
Numerator protein: X chromosomes produce what is called numerator proteins; these bind to early promoter of sxl genes and activate transcription.
Denominator proteins: Autosomes produce the said proteins, and they block the activity of numerator proteins.� There is a balance between them.
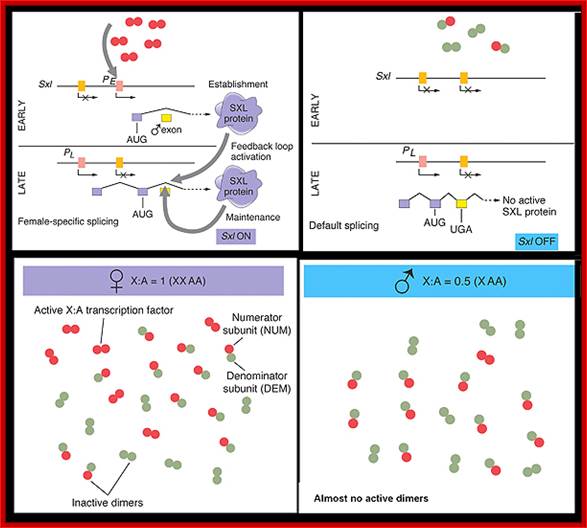
Figure-The initiation and maintenance of the Sxl switch: http://www.bio.miami.edu/dana
�(a) The Sxl switch. High levels of the NUM�NUM transcription-factor dimers activate transcription from the early promoter (PE) of Sxl. (Activated promoters are represented by green rectangles, inactivate promoters by red rectangles. By mid-embryogenesis, PE is turned off, and Sxl is transcribed in all cells of the animal from the constitutively active late promoter (PL). Through binding to the primary PL transcript and regulating its splicing pattern (preventing the male exon from being included in the final mRNA), preexisting active SXL protein ensures the further production of active SXL protein, which continues the splicing pattern leading to its own formation, thus creating an autoregulatory loop. On the other hand, when the X:A ratio is 0.5, PE is not activated. Thus, no SXL protein is present in the early embryo, and the RNA splicing pattern of the PL transcript generates an mRNA that includes the male exon. This exon contains a stop codon (UGA), terminating the Sxl polypeptide prematurely. The short SXL protein made in males is completely nonfunctional. AUG denotes the location of the translation initiation codon for the SXL polypeptide. (b) A plausible mechanism for the molecular basis of the X:A ratio. During early embryogenesis, the numerator and denominator genes are expressed. NUM subunits (red circles) encoded by X chromosome genes and DEM polypeptide subunits (green circles) encoded by autosomal genes form dimers at random. Only NUM�NUM dimers form active transcription factor. If the X:A ratio is 1.0, high levels of these NUM�NUM dimers form, bind to the PE enhancer, and activate transcription from PE. If, by chance, the X:A ratio is 0.5, most NUM subunits are part of NUM�DEM heterodimers and do not function as active transcription factors.
Maintaining the switch in a stable position
The Sxl gene has two promoters. The early promoter is the only one that is activated by the NUM�NUM transcription factors. The early promoter (PE) is active only early in embryogenesis. Later in embryogenesis and for the remainder of the life cycle, the Sxl gene is transcribed from the late promoter (PL) regardless of the X:A ratio or any other condition. This late promoter is active in every cell in the animal, beginning with mid-embryogenesis and persisting for the lifetime of the organism. The primary transcript produced by Sxl transcription from the late promoter is much larger than the primary transcript from the early promoter and is subject to alternative mRNA splicing, depending on the presence or absence of preexisting active SXL protein in the cell.
The SXL protein is an RNA-binding protein that alters the splicing of the nascent Sxl transcript coming from this late promoter. When mRNA splicing occurs in the presence of bound SXL protein, splicing of Sxl produces an mRNA that encodes more active SXL RNA-binding protein. This SXL protein in turn binds to more Sxl primary transcript from the late promoter, creating the spliced form of the mRNA that encodes functional SXL protein, and so forth. Thus a feedback, or autoregulatory, loop, controlled at the level of RNA splicing, maintains SXL activity throughout development in flies with an X:A ratio of 1.0.
Message:
The autoregulatory loop exemplifies how an early developmental decision can be �remembered� for the rest of development, even after the initial signals that established the decisions have long disappeared.
In contrast, when the X:A ratio is 0.5, the Sxl switch is set in the �off� state. The early promoter is not activated early in embryogenesis and hence the early X: A = 0.5 embryo has no SXL protein. As a consequence, in the absence of any active SXL protein, the primary SXL transcript of the late, constitutive Sxl promoter is processed in the default mRNA splicing pattern. This default Sxl mRNA is nonfunctional, in the sense that it encodes a stop codon shortly after the translation-initiation codon of its protein-coding region. The small protein produced from this male-specific spliced mRNA has no biological activity (dysfunctional) . Thus, in Drosophila with a low level of active NUM�NUM transcription factor, the absence of active SXL protein early in development predestines that there will be no SXL activity throughout the remainder of development.
Propagating the decision:
Not only does SXL have to have an autoregulatory maintenance function, but it must be capable of activating the shunt pathway that will lead to female-specific gene expression. It accomplishes this through the same RNA-binding activity. Only in the presence of SXL protein is the primary tra (transformer) transcript spliced to produce an mRNA-encoding active TRA protein (Figure on the following page). In turn, TRA protein is an RNA-binding protein that produces female-specific splicing of the dsx (doublesex) nascent RNA. The mRNA produced by this splicing pattern encodes a DSX-F protein, a transcription factor that globally represses male-specific gene expression.
Regulatory Pathway:
The phenotype is carried out by the �master regulatory switch� and downstream specific sex genes. Sxl gene (Sex lethal gene): choice of pathway is initiated by differential transcription of Sxl gene as well as tra (transformer) gene. The direction of the switch is maintained by an auto feedback loop. �The decision is propagated along the developmental pathway by differential RNA splicing of dsx (doublesex) gene. The default is a male phenotype.
Regulatory Switch (Sxl gene);
Mechanism:
If X:A ratio = 1, then SXL protein is synthesized and a phenotypically, female is the result. If X:A ratio = 0.5, then a non-functional SXL protein is synthesized and a phenotypically male fly is the result.
The X:A ratio set in motion the sex determination pathway by interaction between numerator genes ( X-chromosomal and zygotically expressed) and denominator genes (autosomal, maternally and zygotically expressed). These genes encode transcription factors that are a-Helix�loop-Helix structure NUM, the X-encoded b-HLH and DEM, the autosomal encoded b-HLH is a transcription factor.
The transcription factors have a very short time window, 2-3 hours of fertilization, to determine the sex of the individual via the Sxl gene regulatory switch.
If the gene is turned on, then the level of active X: A the product- NUM transcription factor must be high. They bind to enhancers of Sxl gene, activate, protein transcription from the early promoter (PE) resulting in the production of fully functional SXL protein.
If the gene is turned off, the level of NUM is lower than the threshold, and it is insufficient to promote transcription.
The A:X ratio is measured by NUM proteins competing for dimmer formation with DEM proteins. The transcription factor is only active if NUM forms a dimeric protein complex. NUM monomers have sequence specific DNA binding sites, DEM does not.
NUM monomers have sequence specific DNA binding sites, DEM does not.
Binding site recognition enhancer sequences regulates transcript from promoter of Sxl regulator gene switch.
NUM and DEM polypeptides synthesized at level proportionate to number of copies of each bHLH encoding numerator or denominator gene.
All possible combinations of dimmers can form, but to be active both subunits have to have sequence specific DNA binding sites.
Maintenance of conditions:
Sxl gene has 2 promoters, PE and PL; the PE is activated by NUM:NUM dimer and it is active only in early embryogenesis.
PL promoter is used for transcription the gene regardless of X:A ratio and it is active from mid-embryogenesis to adulthood.
The transcript from PL is larger and it is subject to mRNA splicing. The splicing depends on the presence or absence of the pre-existing SXL proteins. The SXL protein is a RNA binding protein and alters the splicing of the mRNA. If the SXL protein is present, the mRNA is spliced to encode for more SXL protein (female), and if not, a protein is produced but non-functional, results in male.
Propagation of the design:
SXL protein has autoregulatory maintenance function and is capable of activating the shunt pathway leading to the female via mRNA splicing.
In the presence of SXL, the primary tra (transformer) gene transcript is spliced to result in the TRA protein, which like SXL is an RNA binding protein. It binds to mRNA to produce female specific splicing dsx (doublesex) mRNA resulting in the female DSX protein. The DSX protein is a global repressor of male specific gene expression
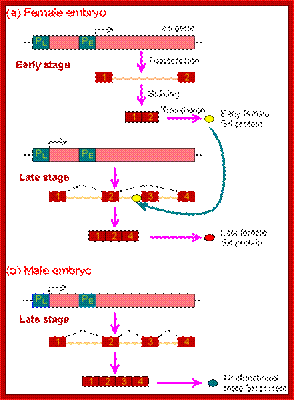
Expression of the Drosophila sex-lethal (sxl) protein. (a) In the early stage of embryogenesis, the sxl protein is expressed in female embryo, but not in the male embryo. (b) In the late female embryo, the sxl protein produced in the early stage may mask the splicing signal for the second intron, resulting in a different protein than in the male embryo. www.webbooks.com .
Alternative splicing events during early development of an embryo leads to sex determination in Drosophila; In this diagram the total number of exons and introns has been shown; the role of sxl, tra1 and tra2 and dsx is depicted and the mode of alternative splicing at different stages has been shown.
Early expression in 2A: XX egg:��
Starts at ~2hrs of fertilization ----> sxl ----> more sxl-� sxl + Tra-1 -------> dsx ->Dsx+Tra-1&2- > sex determination
In the early stage of embryo development, within 2hrs after fertilization, especially during early divisions, differential gene expression leads to the determination of sex. The early sxl transcript remain active for 2hrs, by which time another long lasting sxl product is produced by initiating transcription from the late promoter in embryo that contains 2A:XX chromosome.
First phase: Sxl 1 protein:
During early development, the sex lethal gene (sxl) is expressed in females and not in males.� The gene has two promoters PL and PE (L for late and E for early.� In males the early or late promoter fails to initiate transcription, but in females, gene is expressed from the early promoter. The pre-mRNA has 3 Exons, and normal splicing leads the production sxl protein, which is functional.� Note- sxl lethal means that a mutation in this gene is lethal, other wise this gene is involved in sex determination.
In males:��� -----PL-I--PE-I ex 1-I - -I-ex 2---I- -- - I-ex 3-- -I
Not expressed from PL nor from PE, so no mRNA for sxl.
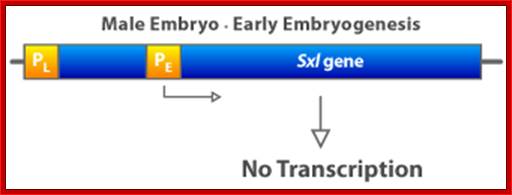
In females: ----PLI- PE-I-ex 1-I- - -I-ex 2-I- - - I- ex 3-------I
Expressed from PE- translation produces sxl.
ProcessedmRNA: 5�--PE---I---ex1---I---2----I----3----I (A) n
The gene expressed from PE and spliced produces normal functional sxl product.
Expressed Sxl in females is functional.
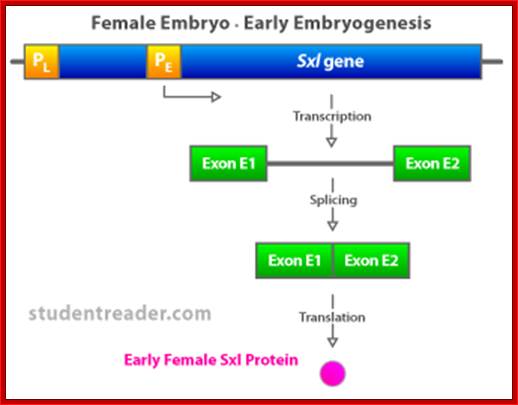
Second phase: Sxl-2 protein:
As the development proceeds, the Sxl genes, both in males and females are expressed from the late promoters.� The transcripts have four Exons.�� In the third Exon a Ter codon is present.�
� In males the splicing is normal, and the transcript contains 8 exons but the transcript has a Ter codon in the 3rd exon; on translation it terminates at Ter codon and the protein produced is not functional.
� In females, the sxl product produced in the first stage (i.e in first round) binds to 3� joint and prevents splicing between 2 and 3 and it favors splicing between 2 and 4 eliminating 3rd exon (exon skipping), (which contains TER codon) and resultant sxl is functional.� Thus sxl, produced at the second level continues to produce more sxl proteins.�
� Note here the sxl, produced first acts as the alternative splicing factor (ASF) that acts at 3� intron splicing site.
Male:� 5�--PL--I�ex1--I----I---ex2---I----I---ex3-ter*-I---I�ex4---I//ex 8�(A) n
Processed:� 5�---I--ex1--I--ex2---I---ex3----ter*--I--ex4---//ex8-- (A) n (no splicing and the protein produced not functional, translation is terminated at the end of Exon3)
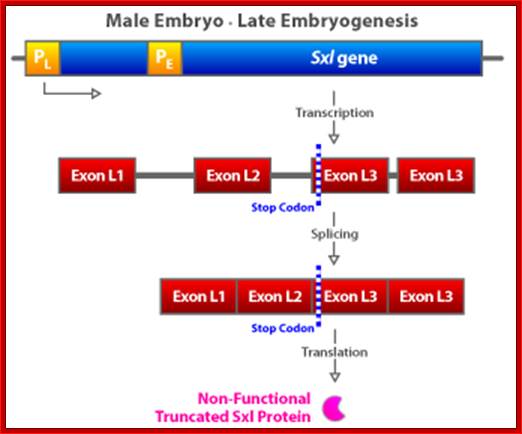
Female:� 5�--PL--I�ex1--I-----I---ex2---I-----I---ex3-----ter*---I�ex4�I//Ex8�(A) n
Processed:� 5�----I---ex1�I---ex2�I---ex4�I//ex8�(A) n ( the exon 3 is spliced out;
�sxl. Protein produced is functional.
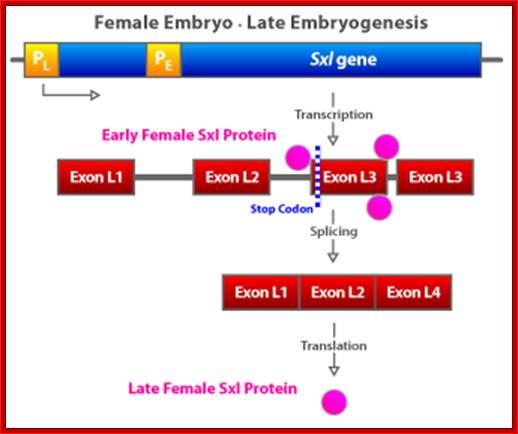
Early female Sxl binds near Exon L3, thus block U2AF to bind to 3�site, thus exon 3 is skipped in females. The late female sxl is transcribed from pL1, consists of exons L1, L2, L3 and L4.� Early sxl binds to 3�splice joint exon L3. Exon L3 is eliminated and the transcript consists of L1, L2 and L4. Late Sxl feeds back on itself and it is expressed through out in female fly.
Third phase:� Tra protein
After few more rounds of cell divisions and differentiation (still at the early development stage), the transformer (Tra) gene is expressed as well as Sxl, however the transcripts from late promoter, the sxl is not functional in males, but both Sxls and Tra are functional in female cells.
�
- The transformer Tra-1 gene transcript has three Exons.� In the second Exon it has a TER sequence.��
- In males there are three exons, splicing is normal and on translation it gets terminated at TER region found in third exon, so the tra-1 protein is rendered nonfunctional.� �
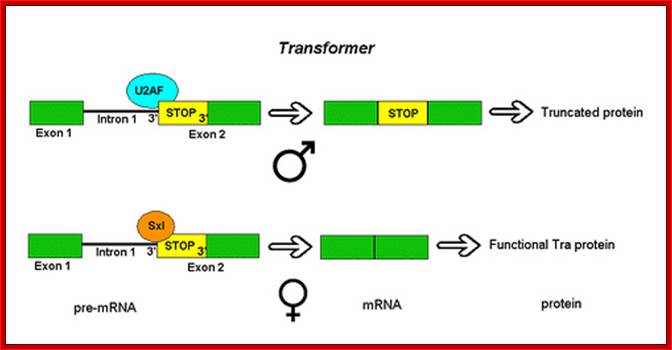
Alternative splicing of the drosophila Transformer gene product; http://en.wikipedia.org/
- But in female cells the sxl produced from the late promoter binds to the intron found between first and second Exon at 3� end of intron-exon splice joint.� It binds to 3� poly-pyrimidine tract and blocks the binding of U2AF and U2-snRNA & snRNPs.� This prevents splicing between first and second Exon, and favor splicing between the first and third Exon. Thus it produces a functional Tra-1 protein.
In Males:
Tra-1:� 5�------I---ex1---I-in---I----ex2--ter*--I--in--I---ex3---I
mRNA:5�-----I---ex--1----I-ex2----ter*--I---ex3�I-(A)n.(Tra-1not functional)
In Females:
Tra-1:5�---I�ex1--I--in--(ppy)n--I�ex2�ter*---I--in--I�ex3---I
mRNA:5�-----I----ex1--I---ex3---I-(A)n, (Tra-1 functional).
Fourth phase: Dsx protein; Alternative splicing of Drosophila dsx gene.
- As the development progresses further, at the stage of male female sex organs development, another gene dsx (double sex), is expressed.� This is the stage of cell determination and differentiation. However, sxl and tra-1 are also expressed in female embryos.� In addition to dsx another transformation gene, tra-2 is also expressed elsewhere.
- The pre-mRNA of dsx gene has 6 Exons and it has a poly-A signal at the end of the 4th Exon.�
- In male determining cells the 4th exon is skipped because of the binding of Tra2, which binds to 3� splice joint between the 3rd and 4th exon, thus the transcript contains 1 to 3 and 5 exons and functional; the dsx produced is functional to determine male.
- In female determining cells 3rd and 4th exons are spliced which contains PolyA signal at the end of the 4th exon and the protein produced is functional in its own way to determine female sex.
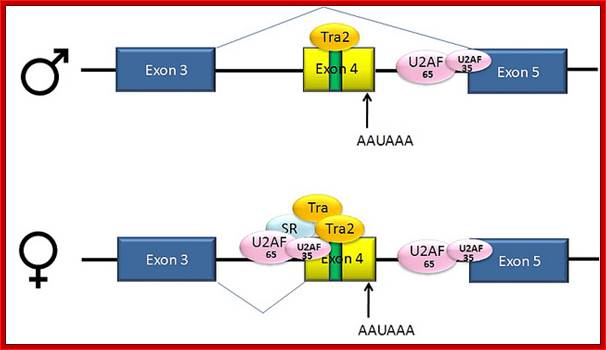
Zhi-Ming Zheng/wiki/File:Dsx_splicing
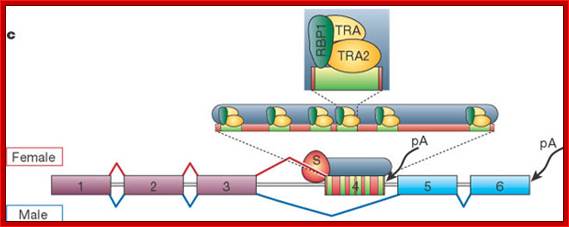
Pre-mRNAs from the D. melanogaster gene dsx contain 6 exons. In males, exons 1, 2, 3, 5 and 6 are joined to form the mRNA, which encodes a transcriptional regulatory protein required for male development. The exon 4 is eliminated in females, Tra1 and Tra2 in splicing between 3 and 4 th exons, thus exons 1,2,3, and 4 are joined, and a polyadenylation signal in exon 4 causes cleavage of the mRNA at that point. The resulting mRNA is a transcriptional regulatory protein required for female development.
This is an example of exon skipping-alternative splicing. The intron upstream from exon 4 has a weak-consensus polypyrimidine tract, to which U2AF proteins bind poorly without assistance from splicing activators. This 3' splice acceptor site is therefore not used in males. Females, however, produce the splicing activator Transformer (Tra). The SR protein Tra2 is produced in both sexes and binds to an ESE in exon 4; if Tra is present; it binds to Tra2 and, along with another SR protein, forms a complex that assists U2AF proteins in binding to the weak polypyrimidine tract. U2 is recruited to the associated branch point, and this leads to inclusion of exon 4 in the mRNA.
The Dsx protein in female prevents male sex organ differentiation and development and the Dsx protein in females blocks male sex organ development.
Maintenance of sxl function
Shortly after Sxl transcription has taken place, a second, �late� promoter on the Sex-lethal gene is activated, and the gene is now transcribed in both males and females. However, analysis of the cDNA from Sxl mRNA shows that the Sxl mRNA of males differs from sxl mRNA of females (Bell et al. 1988). This difference is the result of differential RNA processing. �Moreover, the Sxl protein appears to bind to its own mRNA precursor to splice it in the female manner. Since males do not have any available Sxl protein when the late promoter is activated, their new Sxl transcripts are processed in the male manner (Keyes et al. 1992). The male Sxl mRNA is nonfunctional. While the female-specific Sxl message encodes a protein of 354 amino acids, the male-specific Sxl transcript contains a translation termination codon (UGA) after amino acid 48. The differential RNA processing that puts this termination codon into the male-specific mRNA is shown. In males, the nuclear transcript is spliced in a manner that yields eight exons, and the termination codon is within exon 3. In females, RNA processing yields only seven exons, and the male-specific exon 3 is now spliced out as a large intron. Thus, the female-specific mRNA lacks the termination codon. ( confusing-read carefully).
Figure: Stereogram showing binding of tra pre-mRNA by the cleft of the Sxl protein. The bound 12-nucleotide RNA (GUUGUUUUUUUU) is shown in yellow. The strongly positive regions are shown in blue, while the scattered negative regions are in red. It is worth crossing your eyes to get the three-dimensional effect. (From Handa et al. 1999; stereogram courtesy of S. Yokoyama.).
Tra gene and its role:
- Alternative selection of 3' splice sites preceding exon 2 of tra pre-mRNA is regulated by the SXL protein. In males, the splicing factor U2AF binds to the proximal 3' splice site, leading to an mRNA containing a premature translational stop codon (UAG). In females, SXL binds to the proximal 3' splice site, thus preventing the binding of U2AF. Instead, U2AF binds to the distal 3' splice site, leading to an mRNA that encodes functional TRA protein. In all panels, the exons are indicated by colored rectangles, while introns are shown as pale grey lines.
b, Alternative inclusion of exon 3 of sxl pre-mRNA is regulated by SXL protein. In both males and females, the first step of the splicing reaction results in lariat formation at the branch point sequence upstream from the 3' splice site preceding exon 3. Subsequently, the second-step splicing factor SPF45 binds to the AG dinucleotide of this splice site. In males, SPF45 promotes the second step of the splicing reaction, leading to the inclusion of exon 3. In females, SXL binds to a sequence upstream of the AG dinucleotide, interacts with SPF45 and inhibits its activity. This prevents the second step of the splicing reaction, leading to the exclusion of exon 3 and splicing of exon 2 to exon 4. Seven constitutively spliced exons are not shown.
c, Alternative splicing of dsx pre-mRNA is regulated by the assembly of heterotrimeric protein complexes on female-specific ESEs. The first three exons are constitutively spliced in both sexes. In males, the 3' splice site preceding exon 4 is not recognized by the splicing machinery, resulting in the exclusion of this exon, and splicing of exon 3 to exon 5. In females, the female-specific TRA protein promotes the binding of the SR protein RBP1, and the SR-like protein TRA2 to six copies of an ESE (indicated by green rectangles). These splicing enhancer complexes then recruit the splicing machinery to the 3' splice site preceding exon 4, leading to its inclusion in the mRNA. In females, polyadenylation (pA) occurs downstream of exon 4, whereas in males it occurs downstream of exon 6. 'S' designates the splicing machinery.
The Transformer genes:
The Sxl gene regulates somatic sex determination by controlling the processing of the transformer (tra) gene transcript. The tra message is alternatively spliced to create a female-specific mRNA as well as a nonspecific mRNA that is found in both females and males. Like the male sxl message, the nonspecific tra mRNA contains a termination codon early in the message, making the protein nonfunctional In tra, the second exon of the nonspecific mRNA has the termination codon. This exon is not utilized in the female-specific message (see .How is it that the females make a different transcript than the males? The female-specific protein from the Sxl gene activates a female-specific 3� splice site in the transformer pre-mRNA, causing it to be processed in a way that splices out the second exon. To do this, the Sxl protein blocks the binding of splicing factor U2AF to the nonspecific splice site by specifically binding to the polypyrimidine tract adjacent to it. This causes U2AF to bind to the lower-affinity female-specific 3� splice site and generate a female-specific mRNA. The female-specific tra product acts in concert with the product of the transformer-2 (tra2) gene to help generate the female phenotype. Boggs et al. 1987, Figure 17.19; Handa et al. 1999)., (Valc�rcel et al. 1993)
Alternative splicing of the Transformer gene transcript in Drosophila melanogaster:
In males, the splicing factor U2AF binds to the 3' splice junction of intron 1, allowing the transcript to be spliced at this point. The mRNA then contains the entire exon 2, including an early stop codon. The protein encoded is truncated and non-functional. In females, the master sex determination factor Sex Lethal (Sxl) binds at the 3' splice junction, competing with U2AF. Splicing then occurs at an alternative 3' splice junction within exon 2. The mRNA that results encodes a functional Tra protein.
Doublesex: The switch gene of sex determination
The doublesex (dsx) gene is active in both males and females, but its primary transcript is processed in a sex-specific manner). This alternative RNA processing appears to be the result of the action of the transformer gene products on the dsx gene If the Tra2 and female-specific Tra proteins are both present, the dsx transcript is processed in a female-specific manner. The female splicing pattern produces a female-specific protein that activates female-specific genes (such as those of the yolk proteins and inhibits male development. As discussed in Chapter 5, if functional Tra is not produced, a male-specific transcript of dsx is made. This transcript encodes an active protein that inhibits female traits and promotes male traits. (Baker et al. 1987), (Ryner and Baker 1991)
The functions of the Doublesex proteins can be seen in the formation of the Drosophila genitalia. Male and female genitalia in Drosophila are derived from separate cell populations. In male (XY) flies, the female primordium is repressed, and the male primordium differentiates into the adult genital structures. In female (XX) flies, the male primordium is repressed, and the female primordium differentiates. If the dsx gene is absent (and thus neither transcript is made), both the male and the female primordia develop, and intersexual genitalia are produced. Similarly, in the fat body of Drosophila, activation of the genes for egg yolk production is stimulated by the female Dsx protein and is inhibited by the male Dsx protein (Sch�pbach et al. 1978; Coschigano and Wensink 1993; Jursnich and Burtis 1993), (Baker 1989.
Alternative splicing of dsx pre-mRNA; Pre-mRNAs from the D. melanogaster gene dsx contain 6 exons. In males, exons 1,2,3,5,and 6 are joined to form the mRNA, which encodes a transcriptional regulatory protein required for male development. In females, exons 1,2,3, and 4 are joined, and a polyadenylation signal in exon 4 causes cleavage of the mRNA at that point. The resulting mRNA is a transcriptional regulatory protein required for female development-. Exon skipping; http://en.wikipedia.org/
This is an example of exon skipping alternative splicing. The intron upstream from exon 4 has a weak-consensus polypyrimidine tract, to which U2AF proteins bind poorly without assistance from splicing activators. This 3' splice acceptor site is therefore not used in males. Females, however, produce the splicing activator Transformer (Tra). The SR protein Tra2 is produced in both sexes and binds to an ESE in exon 4; if Tra is present, it binds to Tra2 and, along with another SR protein, forms a complex that assists U2AF proteins in binding to the weak polypyrimidine tract. U2 is recruited to the associated branch point, and this leads to inclusion of exon 4 in the mRNA
According to this model), the result of the sex determination cascade comes down to what type of mRNA is going to be processed from the dsx transcript. If the X:A ratio is 1, then Sxl makes a female-specific splicing factor that causes the tra gene transcript to be spliced in a female-specific manner. This female-specific protein interacts with the Tra2 splicing factor to cause the doublesex pre-mRNA to be spliced in a female-specific manner. If the doublesex transcript is not acted on in this way, it will be processed in a �default� manner to make the male-specific message.
Comparative account of sex differentiation events in C.elegans and Drosophila:
Hierarchy of sexual differentiation genes in C. elegans and Drosophila.
In both C. elegans and Drosophila, the primary sex determining signal, the X chromosome to autosome ratio (X : A), sets the activity of the globally acting sex determination pathway. DM domain genes (blue) in C. elegans and Drosophila differ with respect to their relationship to this pathway. In C. elegans, the primary sexually regulated transcription factor is a Gli-related Zn finger protein, TRA-1A, encoded by the terminal gene of the sex determination pathway, tra-1. tra-1 activity affects the development of most sexually dimorphic tissues, acting through non-sex-specific developmental gene targets (e.g., cell death geneegl-1), as well as dedicated sexual regulators (mab-3). mab-3 (in tissues other than the gut) andmab-23 are not directly targeted by TRA-1A. The sex specificity of their activity derives from the cellular context in which they are expressed. In Drosophila, the primary sexually regulated transcription factor is the DM domain gene dsx. In contrast to C. elegans DM domain genes, dsx is a direct target of the sex-determination pathway in most somatic tissues. Like tra-1, dsx is broadly acting and functions early in sexual differentiation to sex-specifically regulate multiple developmental genes. In the CNS, fru appears to be the primary sexually regulated transcription factor.
A simple view of the human sex differentiation based on X and Y chromosomal factors:
 �
�
All mammals undergo relatively similar sex determination.
The Genetics of Sex Determination
A model for sex initiation was first proposed; by Eva Eicher and Linda Washburn in 1986.
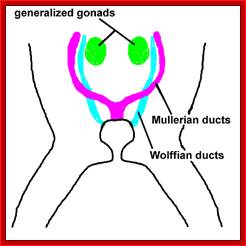
In humans, at the eighth week of development, if the sry gene is present; it turns on.
- Generalized gonads begin their transformation into testes prior to 8 weeks.
- The new testes begin to secrete androgens (testosterone and dihydrotestosterone (DHT) )
- The androgens start a cascade of genetic developmental events that culminate in the embryo becoming male.
- Part of this cascade involves suppression of the expression of the Od ("ovary determining") gene.
- In the presence of androgens, the Mullerian ducts atrophy
- Wolffian ducts develop into the internal male genital tract.
In the absence of androgens, the Od gene turns on at about week 13. Once this happens...
- The OD product triggers a cascade of developmental events that culminate in the embryo's becoming female.
- The Wolffian ducts atrophy
- Mullerian ducts develop into the internal female reproductive organs and tracts.
- Alternative splicing, though produces different proteins most of such proteins have some common Exon motifs some Exons in different positions. Many of the proteins produced by alternate splicing are transcriptional factors, hormone receptors, ion channels; Transposase-P element (in Drosophila), Fibronectin, PDGFA and many others show alternate forms of splicing.�� In ADV early transcripts, SV 40 and ultra bithorax transcripts, due to the availability of 5� donor sites and 3�acceptor sites, certain introns are spliced differently.��
In some cases genes have what is called �cassette Exons�.� This, as a cassette, consists of a group of Exons, can be included or excluded independent of other Exons.� Presence of cassette Exons provides high degree of diversity, ex. Troponin- T has 64 isoforms.
Mechanism of cell-specific alternative splicing:
�Mechanisms of coordinated splicing regulation: " Research scholars have investigated the mechanisms that regulate alternative splicing at several levels. One is the molecular details of how regulatory factors that directly bind the pre-mRNA communicate to the basal splicing machinery and promote inclusion or skipping of a variably spliced region. Another area, described in the section below, is to determine how the activities of splicing regulators are modulated during development (using mouse models) and during cellular differentiation (using cell culture models), to investigate the regulatory networks that coordinate subsets of alternative splicing events during development�.
Alternative splicing decisions are regulated by combinatorial control involving multiple factors that bind to the pre-mRNA. Evidence also indicates that, as in regulation of cell-specific transcription, recruitment (or repression) of the basal machinery requires assembly of a multi-component complex. �The final outcome of a regulated splicing event results from dominance of activation or repression activities.
(Singh et al. 2004 , Han et al. 2005),
To investigate the molecular details of regulation, (work has been by authors (Singh and Han) conducted on two families for splicing regulators: CELF and MBNL. Each of the proteins examined have been shown to have positive or negative effects on splicing of different alternative exons. Interestingly, CELF and MBNL proteins often regulate splicing of the same alternative exons and in all cases act antagonistically.
�Authors primary goal is to understand how splicing regulators activate alternative splicing. The specific questions being addressed are, how does binding of a splicing activator recruit or stabilize binding of the basal splicing machinery; what are the required components of the activation complex; what components bind directly to the splicing activator; and what protein-protein interactions link the bound splicing activator to the basal splicing machinery?�
CELF interacting proteins have identified by yeast two-hybrid analysis, co-immunoprecipitation, and Tandem Affinity Purification (TAP). Recombinant CELF proteins switch splicing from exon skipping to exon inclusion in a cell-free splicing assay. This provides an excellent experimental system to determine the mechanism for activation. This analysis has identified a direct interaction between the CELF protein, CUGBP2 and a basal splicing component, U2 snRNP. Additional CELF-binding proteins with relevance to splicing as well as transcription were identified and will be investigated.
We have identified CELF protein domains required for activation and repression of different pre-mRNA targets. An ongoing analysis of how the same CELF protein activates different pre-mRNA targets and how different CELF paralogues activate the same pre-mRNA has revealed an unexpected diversity of activation mechanisms. This analysis plus comparisons between activation mechanisms of CELF and MBNL proteins will reveal a broad view of the mechanism of splicing activation.
Alternative splicing is the process in which the primary transcript of a gene is reorganized to produce a different protein than the primary transcript. By manipulating of the exons, the sequence of the amino acids produced from the mRNA is affected, resulting in a different protein sequence, and protein structure. This can have a drastic effect on the protein that is produced. Alternative splicing has been observed as a mechanism to produce tissue specific proteins from a gene. Depending on the tissue different proteins can be produced in different tissues from a single gene. This process can be thought of a multiplication process that increases the possible proteins that are produced from a single gene�.
�Alternative Splicing is a major source of protein diversity in living organisms. It has been estimated that at least ~80% of all genes in the human genome are alternatively spliced and this number continues to expand. It was originally thought that the number of alternatively spliced genes accounted for only 5% of proteins in humans. With the unveiling of the human genome, it was revealed that the human genome contains less than 30,000 genes (~ 21000-~22000)). This could potentially account for the huge gap between the number of genes Genome) and the number of proteins (Proteome).
It has been suggested that alternative splicing is the source of higher-level complexity in eukaryotes. This idea is based on the thought that more complex organisms will alternatively splice their genes more often to obtain more possible mRNA sequences. However, evidence shows that the level of alternative splicing between different complexities of organisms is not significant; the evidence is contrary. This study was done using ESTs (expressed sequence tags *link to EST page*). As more EST studies are done, it has become apparent that there is a greater number of alternatively spliced genes than previously thought. ESTs were compared to mRNA sequences using BLAST.�
Examples:
Alternative splicing has been implicated in several diseases. An example of a disease that plays a role in alternative splicing is Rett syndrome. This disease is found primarily in girls and is characterized by problems in forming connections between neurons, or synapses. It is believed that the gene MECP2 (methyltransferase to C base) produces a protein that regulates alternative splicing of some proteins. When this gene is disrupted, transcripts of other genes that would normally be spliced are not spliced, leading to Rett the phenotype of the syndrome.
SLO pre mRNA in chicks: The Regulation of Ion Channel mRNA Alternative- Splicing by Cell Stimulation:
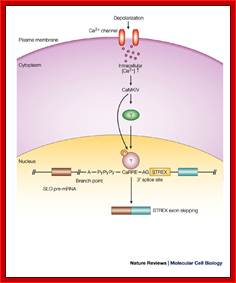
Activated Ca2+ ion channel leads to activation of CaMK-IV and it phosphorylates of an unknown protein that binds to CaRRE element poly-pyrimidine tract of Slo pre-mRNA which is located between the polypyrimidine tract (PyPyPy) and the 3' splice site; thus excludes the STREX region of the mRNA. Gretchen Edwalds-Gilbert; http://www.nature.com/
After depolarization of GH3 pituitary cells in response to neuronal activity, Ca2+/calmodulin-dependent kinase (CaMK)IV is activated. Once activated, CaMKIV phosphorylates an unidentified protein, which binds specifically to the CaMKIV-responsive RNA element (CaRRE) on the SLO pre-mRNA, which is located between the polypyrimidine tract (PyPyPy) and the 3' splice site. The 3' splice site of the stress-axis-regulated (STREX) exon is spliced out which results in skipping of the STREX exon in the mature mRNA. Only the portion of the SLO pre-mRNA that contains the STREX exon is shown. SLO gene in rat codes for potassium channel proteins, which can generate 500 or more alternative forms proteins.�
Slowpoke channels control quantal content of neurotransmitter release at the neuromuscular junction;
Six mutants of SLO-1, a large-conductance and Ca(2+)-activated K(+) channel of C. elegans, were obtained in a genetic screen for regulators of neurotransmitter release. Mutants were isolated by their ability to suppress lethargy of an unc-64 syntaxin mutant that restricts neurotransmitter release. Evoked postsynaptic currents were measured at the neuromuscular junction in both wild-type and mutants; the removal of SLO-1 greatly increases quantal content primarily by increasing duration of release. The selective isolation of slo-1 as the only ion channel mutant derived from a whole genomic screen to detect regulators of neurotransmitter release suggests that SLO-1 plays an important, if not unique, role in regulating neurotransmitter release (Wang, 2001).
The most important compartments of the inner ear, including the cochlea, vestibular system, and endolymphatic sac [Modified from Ref. 83a.].
Cochlea-Internal structural features
Different compartments of the inner ear serve the transduction of specific stimuli. The cochlea transduces mechanical stimuli associated with sound and provides the basis for hearing. The utricle, saccule, and ampullae of the semicircular canals belong to the vestibular labyrinth, which transduces mechanical stimuli associated with head position and head motion. Vestibular sensory transduction provides input to the vestibular system that controls balance, posture, and eye movements. Sensory transduction in the cochlea and vestibular labyrinth has different electrochemical requirements, although all depend on the cycling of K+ between the endolymph and perilymph. In addition to the fluid compartments that house sensory hair cells, the vestibular labyrinth contains another fluid compartment, the endolymphatic sac, which is devoid of sensory hair cells. The function of the endolymphatic sac is poorly understood, although evidence suggests that it controls endolymph fluid volume.
Chick�s inner ear Cochlea showing a layer of cells containing slo protein is partially anchored; each of these cells contain different slo protein to receive the sound wave length from 50Hz to 5000Hz..� This protein is alternatively spliced.
Hair cells, magnified 21,000 times, line the inside of the shell-like cochlea of the inner ear. Damage to these cells, which can�t regenerate, is a major cause of irreversible hearing loss in many people. Photo Researchers; http://online.wsj.com/
In chick inner ear cochlea tube is 5mm long.� This is lined with a layer of epithelial auditory hair cells. These hair cells are tuned to receive and respond to vibration frequency of sound from 50Hz on the left end of the cochlea to 5000Hz at the right.� The hair cells contain ca^+ dependent/calcium gated K-channels. The channel proteins are made up of S0 to S6 transmembrane domains, which organize to form K+ channel and S7 to S10 form cytosolic domains which regulate the opening and closing of the channel.� The channel proteins open and close in response to intracellular concentration of calcium ions and they are regulated by Ca dependent kinases. The slo mRNA contains 27 exons. �In the hair cells the transcript generated is same but it is spliced differently, alternatively in each of the hair cells from the left end to the right of cochlea, thus it can generate 576 isoforms of potassium channels in a series so as to respond to vibration frequency of the sound. The gene is capable of producing 576 alternate forma of proteins lined up in chick cochlear hair cells.� The pre mRNA is� 11kb long and the processed one can be approximately ~4kb long.
The fraction of Slo mRNAs �figure below� containing an exon called STREX (stress axis-regulated exon, shown in yellow) is regulated by neuronal activity. Depolarization of the plasma membrane increases the intracellular Ca2+ concentration, leading to the activation of CaMK IV. This, in turn, is thought to result in the binding of repressor proteins to silencer sequences, one located upstream and the other within STREX. These silencers are shown as red rectangles with hypothetical repressors bound to them. In the presence of the repressors, the STREX exon is excluded from Slo mRNA. The alternatively spliced mRNAs encode two protein isoforms (shown in blue). The channel encoded by mRNA containing the STREX exon has an additional (yellow) domain, and is more sensitive to intracellular Ca2+concentration. For clarity, only the portion of Slo pre-mRNA containing the STREX exon is shown.
Inducible alternative splicing of rat Slo pre-mRNA. Tom Maniatis & Bosiljka Tasic; http://www.nature.com/
 �
�
The top image shows a cross section of cochlear sensory epithelium as shown above, with inner and outer hair cells and the surrounding supporting structures; The slo protein anchored and free are shown; Middle-Tiny hairs in the ear which detect sounds have been regenerated to reverse deafness for the first time, say US researchers in the journal Neuron; James Gallagher ; the bottom figure-http://www.nature.com/;Wanda Layman& Jian Zuo
The pre-mRNA contains 22 exons; with alternate processing it can generate 576 alternate forms of proteins in cell specific manner; so the cells are competent to receive the specific wave length of the incoming sound.� Inclusion of STREX (Stress regulated Axis) exon or the exclusion of STREX exon plays a very important role in K (+) channel opening and closing thus regulate voltage gated channels.� The STREX has calcium binding domain.
The Regulation of Ion Channel; Alternative Splicing by Cell Stimulation:
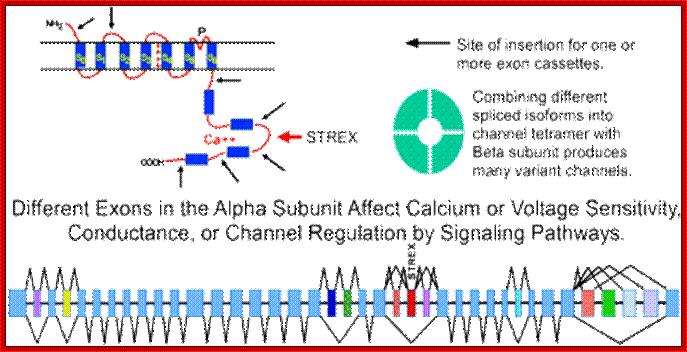
The electrical properties of Calcium Activated K+ Channels (Slo, BK or Maxi-K Channels are tuned through extensive alternative splicing; this figure shows 39exons- STREX is found at 23-25; Douglas Black, Ph.D; http://www.bioeng.ucla.edu/
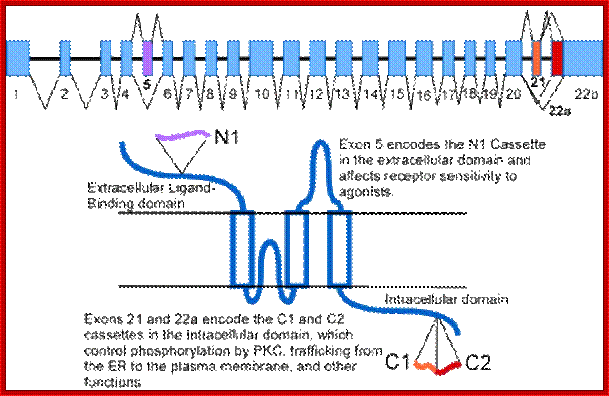
http://www.bioeng.ucla.edu/
The fraction of Slo mRNAs containing an exon called stress axis-regulated exon (STREX;) is regulated by neuronal activity. Depolarization of the plasma membrane increases the intracellular Ca2+ concentration, leading to the activation of the calcium-regulated kinase CaMKIV. This, in turn, is thought to result in the binding of repressor proteins to silencer sequences, one located upstream, and the other within, STREX. These silencers are shown as pink rectangles with hypothetical repressors bound to them. In the presence of the repressors, the STREX exon is excluded from Slo mRNA. The alternatively spliced mRNAs encode two protein isoforms (shown in blue). The channel encoded by mRNA containing the STREX exon has an additional (yellow) domain, and is more sensitive to intracellular Ca2+ concentration than the transporter lacking STREX exon. For clarity, only the portion of Slo pre-mRNA containing the STREX exon is shown. Reproduced from Maniatis, T. & Tasic, B. Alternative pre-mRNA splicing and proteome expansion in metazoans. [Nature 418, 236-243 (2002) � Macmillan Magazines Ltd.]
A poorly understood aspect of splicing regulation is how extracellular stimuli and cell signaling pathways direct changes in splicing. This is especially interesting in the nervous system, where many proteins that determine neuronal excitation are modulated by alternative splicing. To study how specific signaling cascades alter splicing patterns, authors examined the regulation of the STREX exon in the calcium-activated potassium channel transcript (BK channel). BK channels are important in shaping the action potential kinetics of a number of excitable cells and are highly regulated at the level of splicing. The STREX exon adds a peptide insert into the calcium-binding domain of the channel that both makes it more sensitive to intracellular calcium and voltage and allows its modulation by Protein Kinase A. It was found that culturing excitable cells in depolarizing media containing high potassium led to the repression of STREX exon inclusion. This reduction in STREX splicing is blocked by the CaM Kinase inhibitor KN93 and by the L-type calcium channel blocker nifedipine, implicating calcium signaling in this splicing repression. As a model for this regulation, we expressed the STREX exon from a transfected minigene and found that STREX splicing is repressed by a cotransfected constitutive CaMK IV. This approach allowed us to map specific RNA regulatory elements in the BK channel transcript needed to respond to the CaMK signal. We identified a calcium-responsive RNA element (CaRRE-1) that can confer CaMK-dependent repression on a heterologous exon. We are very interested in the mechanism of this repression and how it responds to culture conditions.
The CaMK effect on ion channel splicing has implications for the control of neuronal excitability. Alternative splicing alters the activity of many other proteins that mediate the induction and propagation of action potentials. If splicing is controlled by cell excitation, then these splicing events are likely one component of the plastic changes in cell activity that underlie many aspects of neurophysiology. Additional CaRRE elements were also found in two regulated exons of the NMDA Receptor 1, which are also regulated by CaMK IV. NR1 exon 21 contains a CaRRE element within the exon itself, instead of the 3� splice site, and encodes a peptide that controls the trafficking and assembly of the NR1 subunit. Exon 21 also contains an additional Calcium responsive element, different from the CaRRE-1, which we are currently characterizing. These elements in ion channel exons provide a unique entry point for the dissection of signaling pathways that impinge on the splicing apparatus, and we are working to identify the proteins that bind to them. In an alternative approach, we are studying whether known splicing regulators can be altered in expression or activity by specific signaling pathways. For example, we have found that the nucleo/cytoplasmic distribution of PTB is regulated by phosphorylation by Protein Kinase A. Our goal in these projects is to understand how the information of an extracellular stimulus can be transmitted to the cell nucleus to alter splicing.
DSCAM pre mRNA: Down�s Syndrome Cell Adherence Molecule (or Drosophila Cell Adherence Molecule):
: Structural Biology of Cell Surface Receptors; Meijers Group
A neuron operates by receiving signals from other neurons through connections, called synapses. The combination of these signals, in excess of a certain threshold or activation level, will result in the neuron firing that is sending a signal on to other neurons connected to it. Some signals act as excitations and others as inhibitions to a neuron firing. What we call thinking is believed to be the collective effect of the presence or absence of firings in the pattern of synaptic connections between neurons, excerpt from the NeuralystTM User's Guide.
This sounds very simplistic until we recognize that there are approximately one hundred or 200 billion (1-2 x 10^9, (200,000,000,000) neurons each connected to as many as one thousand trillion in the human brain. The massive number of neurons and the complexity of their interconnections results in a "thinking machine", your brain. �There are about 10^14 synapses (trillion).
One synapse, by itself, is more like a microprocessor�with both memory-storage and information-processing elements�than a mere on/off switch. In fact, one synapse may contain on the order of 1,000 molecular-scale switches. A single human brain has more switches than all the computers and routers and Internet connections on Earth, John Petrie�s life Life-Blag, The Fall of Hyperion; this was based on the premise that human cortex has 22 billion neurions and 220 trillion synapses?
Intercellular interactions occur through supramolecular clusters that form asymmetric synapses when a cell is scanning the environment and symmetric synapses when cell�cell recognition is established. This ancient recognition process has been observed in such diverse systems as predatory amoeba, lymphocytes scanning for antigens and nerve cells seeking connections to form a brain map. At the centre of these interactions are very specific molecular recognition events that trigger a reorganization of the cluster on the cell surface. This in turn amplifies the recognition event, resulting in the activation of a signalling cascade within the cell that leads to physiological changes within the cell. We study the molecular basis of cell surface receptor recognition in the context of the dynamics of the supramolecular cluster as a whole.
A gene that can produce 38016 different proteins (?):
When Brenton Gravely read about a Drosophila melanogaster gene with almost 100 splicing sites and 38,016 putative protein products, he immediately knew what he would be doing over the next few years, if not the rest of his working career. Graveley, a biochemist by trade, taught himself fly genetics so he could work on the provocative Dscam gene. "That gene was the most amazing thing I'd ever seen," says Gravely, an associate professor at the University of Connecticut Health Center, Farmington. "I started working on an experiment on it that day�.
Dscam Proteins Control Self-Non-self-Recognition;
In
the 1980s, Andrew Kramer and Gunter Stent proposed that neurites, the axonal
and dendritic extensions of neurons, have the ability to distinguish between
self and non-self. Self-recognition was proposed to repel processes away from each
other. This would allow neurites of one cell to elaborate uniform coverage of
receptive fields while simultaneously permitting the neurites of different
cells to share the same receptive field. Genetic and biochemical studies over
the past 7 years provide strong evidence that a large family of immunoglobulin
domain-containing cell surface proteins encoded by the Dscam1 locus play a
crucial role in promoting self-avoidance.
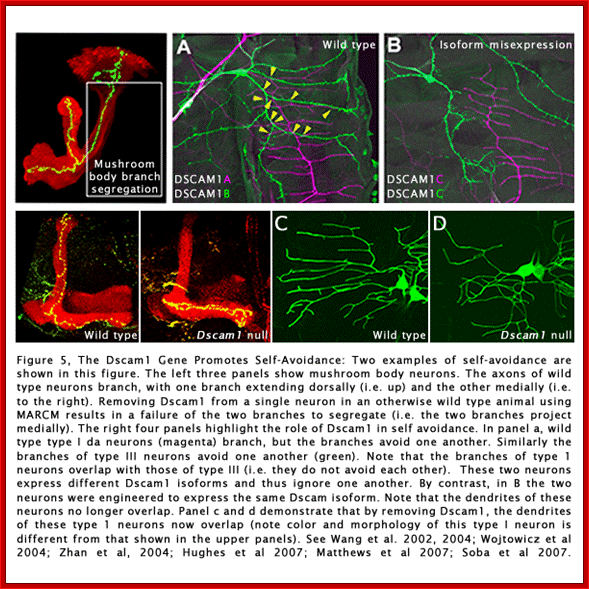
The Dscam gene:
The D�scam gene is 61.2 kb.� One of the drosophila gene transcript processed is ~ 7.8 kb long, the pre mRNA can be spliced in more than 38016 ways.� Term Dscam means �Down Syndrome Cell-Adhering Membrane protein�. The gene in retinal neuron of Drosophila produces axon guidance receptor and it is responsible for guiding the growth cones to their proper target neurons in the brain.� The transcript is ~7.8 kb long; it has 115 exons of which 95 exons that can be splice alternately and other 20 exons are constitutively spliced; overall it has 115 exons.� Exon group4 has 12 alternatives, Exon group6 has 48 alternatives, Exon group9 has 33 alternatives and Exon group17 has 2 alternatives, that can be spliced to generate 38016 combinations; this is an incredible number of transcripts and protein products that they can generate.� Neuronal receptors required for the net work is approximately 250,000, and it is amazingly complex in number and structure and function.� Here Dscam provides that kind of facility to generate required number of proteins in site specific manner in tissues or cells. On the contrary human DSCAM gene is 836140 bp and the mRNA produced is ~6413 ntds long.� In humans the alternate splicing produces only few alternative forms.
In Drosophila this single gene can generate over 38,016 different isoforms by virtue of extensive alternative splicing. In fact, the number of proteins generated by this gene is two to three times the number of genes in the entire Drosophila genome! It is thought that the diversity of Dscam isoforms contributes to the specificity of neuronal wiring. We have found that the alternative splicing of Dscam transcripts is regulated throughout development and in a tissue-specific manner. Moreover, this regulated alternative splicing is evolutionarily conserved. We are now using RNAi, genetics, evolutionary, and biochemical approaches to identify trans-acting factors and cis-acting RNA sequences that participate in controlling this extraordinarily complex alternative splicing event.
�
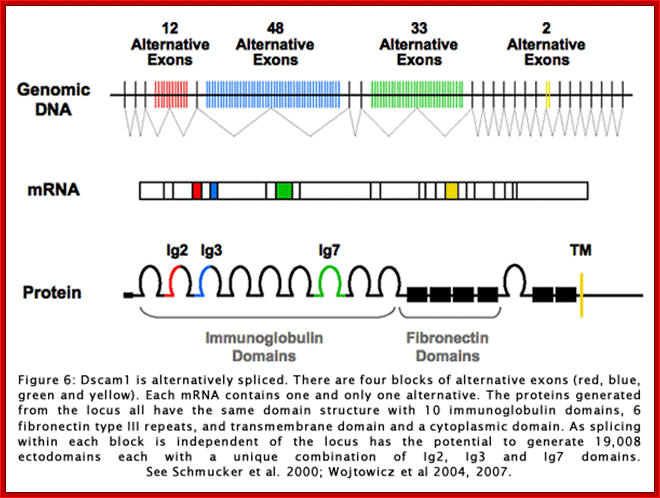
The Drosophila Dscam gene, involved in adhesion between neurons, contains 4 clusters of exons, each with array of possible exons. These are spliced into the mRNA in an exclusive fashion, so that only one of each of the possible exons is represented. If all combinations of these exons are used in alternative splicing, the Dscam gene can produce 38,016 different proteins; http://bio3400.nicerweb.com/
Comparative protein domain structure of Human Dscam, Dscam-like and fly Dscam. Based on results from Ensembl, UniProt, and InterProScan, the four exons in fly that undergo mutually-exclusive alternative splicing are marked above the protein subdomains (exons 4 and 6) or domains (exons 9 and 17) that they encode. The homologous exons in human Dscam and Dscam-L are marked above their corresponding domains in the encoded proteins, with the homologous fly exon for each in parentheses. The vertebrate homologs to fly exon 6 were located for the figure by their position in the global alignment, though the overall similarity was low for these compared to the other homologous exon pairs. BMC Evolutionary Biology 2006
Homologs have very little functional similarities. Human DSCAM is found on chromosome 21 and its products may be the cause of neurodevelopmental defects associated with the disease from which it gets its name.2 But the human gene has few known alternatively spliced products. Other insects appear to splice and dice the gene. The mosquito, for instance, can produce 32,000 proteins. And, Drosophila virilis may make even more proteins than D. melanogaster, almost 40,000, according to Graveley; http://www.biomedcentral.com/
A series of genetic and molecular studies support a model in which binding between isoforms on two opposing membranes triggers a repulsive response: disruption of the cell contacts and extension away from one another. Molecular analyses revealed that each neuron acquires a unique Dscam1 identity via stochastic expression of a set of isoforms. The large number of isoforms encoded at the locus ensures that neurites of different cells are sufficiently different from one another to prevent inappropriate repulsive interactions between them. By contrast, as neurites of the same cell share the same isoforms, they will repel each other. Dscam1-mediated self-avoidance plays an important role in regulating axon and dendrite branching. Studies in progress are directed towards assessing whether Dscam1 may regulate other aspects of patterning neural circuits, including synaptic specificity.
Driving Self-Recognition
A
single gene with the capacity to generate more than 30,000 different proteins
on the surface of a growing neuron reveals a cell-recognition mechanism that
regulates the wiring of the brain in an entirely unexpected fashion.
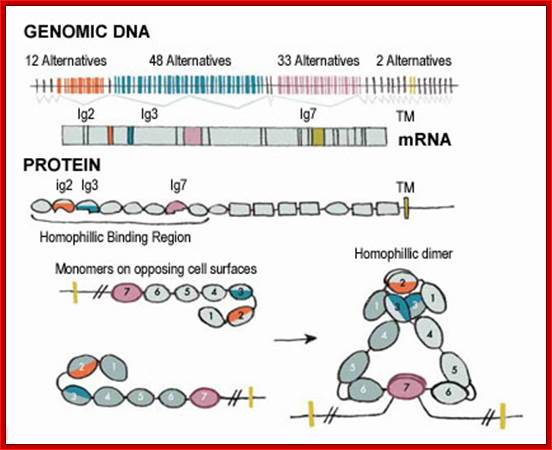
Read more: Driving Self-Recognition; http://www.the- Scientist.com/article/display/57775/#ixzz1C9EkcvZr;;
Vast diversity of recognition molecules.
The parts of the Dscam molecule that give it its
unique shape come from the mixing and matching of four exons that code for the
first halves of the Ig2 and Ig3 domains, all of the Ig7 domain and the entire
transmembrane domain (TM ) of the molecule (top). The Ig7 domain of the
protein, for example, can be chosen randomly from 33 alternative exons, and
mixed with the other Ig domains to produce a total of 38,016 unique Dscam
molecules. The pairing of matched Ig domains on opposing membranes changes the
shape of the molecules (bottom). After Dscam binding, we think that the
branches� adhesion molecules are released and the cytoskeleton remodeled,
causing processes growing way from each other.
�Dscam Pre-mRNA Alternative Splicing�
�A major project in the laboratory is to determine the mechanisms involved in controlling alternative splicing of the Drosophila Down Syndrome Cell Adhesion Molecule (Dscam) gene. The Dscam gene, which was discovered in Larry Zipursky�s lab, encodes an axon guidance receptor that is most similar to one of the human genes implicated in causing Down Syndrome. Perhaps the most interesting aspect of this gene is that it is the most extensively alternatively spliced gene that we know of in any organism. This single gene can generate over 38,016 different isoforms by virtue of extensive alternative splicing. In fact, the number of proteins generated by this gene is two to three times the number of genes in the entire Drosophila genome! It is thought that the diversity of Dscam isoforms contributes to the specificity of neuronal wiring. We have found that the alternative splicing of Dscam transcripts is regulated throughout development and in a tissue-specific manner. Moreover, this regulated alternative splicing is evolutionarily conserved. We are now using RNAi, genetics, evolutionary, and biochemical approaches to identify trans-acting factors and cis-acting RNA sequences that participate in controlling this extraordinary complex alternative splicing events.
By performing an RNAi screen in which hundreds of RNA binding proteins in the Drosophila genome, authors identified about 40 different proteins that regulate the splicing of various Dscam exons. We have also identified several splicing regulatory elements that are required for the inclusion of any alternative exons and we are currently working to identify the mechanisms by which these elements act. In addition, we are using a variety of techniques to determine the role of the different Dscam isoforms in specifying axon guidance in the fly. Although there are differences in the properties of the human and Drosophila Dscam proteins, these studies may lead to insights regarding the role of this gene in the development of Down syndrome in humans�.
DSCAM uses homophilic and heterophilic interactions to initiate signaling underlying multiple functions. Isoform-specific homophilic binding of Drosophila Dscam (red, blue, and green Ig domains) enables an intrinsic self-recognition for repulsion of same-cell axon;) or dendrite; branches.
Although mammalian DSCAM (monochrome receptors) lack diversity of isoforms, homophilic recognition is also important in mammalian receptors. Similar to other guidance receptors, in some biological contexts Dscam or DSCAM signaling might be switched to attraction rather than repulsion. For example, homophilic recognition of vertebrate DSCAMs supports attraction/adhesion of neurites during synaptic targeting Vertebrate DSCAM also binds netrin (i.e., heterophilic ligand) for axon guidance functions .� Dscam's requirement in innate immune responses to pathogens suggests heterophilic binding to different pathogen molecules. By S. Lawrence Zipursky; (Wang et al. 2002; Zhan et al. 2004, Hattori et al. 2007, (Hughes et al. 2007, (Zhan et al. 2004; Yamagata and Sanes 2008), Matthews et al. 2007; Soba et al. 2007), (Yamagata and Sanes 2008). Ly et al. 2008).
1.Netrin proteins and their receptors. (A) Netrins are members of the laminin superfamily. N-terminal netrin sequences encode domains VI and V (green), which are homologous to the N-terminal domains VI and V of laminins (brown). These domains in netrin 1, 2 and 3 are most similar to the laminin γ chain, whereas those in netrin 4, G1 and G2 are most similar to the laminin β chain. The C-terminal C domains (C) of netrins 1-4, G1 and G2 are not homologous to laminin, nor are the C domains of netrins 1-4 homologous to the C domains of the netrin G proteins. (B) The netrin receptors illustrated are all single-pass transmembrane proteins and members of the Ig superfamily. They include deleted in colorectal cancer (DCC), the DCC paralogue neogenin found in vertebrates, members of the UNC5 homologue family, DSCAM and the netrin G ligands (NGLs). CT, C-terminal cysteine-rich capping structure; DB, DCC-binding domain; DD, death domain; FNIII, fibronectin type III domain; Ig, immunoglobulin domain; LRR, leucine-rich repeat; NT, N-terminal cysteine-rich capping structure; P1, P2 and P3, conserved regions in the cytoplasmic domain of DCC; TSP, thrombospondin type 1 (TSP-1) domain; ZU5, zona occludens 5 (ZU-5) domain, with homology to zona occludens 1;. Karen Lai Wing Sun,;http://dev.biologists.org/
The organisation of the C. elegans cadherin-catenin complex. HMR-1A is depicted as forming a dimer, with the N-terminal extracellular cadherin domains of the dimer interacting with those of HMR-1A dimers on opposing cell membranes. This is based on models of the cadherin adhesion interface derived from structural studies of vertebrate classic cadherins. It is not clear if these models hold for the type III classic cadherins. This diagram is by necessity a simplified view; there are many more proteins associated with the CCC, but the details of how they associate with the complex are not fully understood. The domains are identified using the key in (Figure 1). The PCCD is drawn to indicate the putative proteolytic cleavage site based upon sequence similarity to Drosophila E-cadherin.;C Adherins of �C.elegans; �Jonathan Pettitthttp://www.wormbook.org/

Dscam Pride (Biochemistry Joke) ('08)
Regulators of Alternate Splicing:
Exons and Introns have their own Enhancer Splicing Elements and Intron Splicing Elements called ESE and ISE respectively.� They also contain Exon Splice Suppressor and Intron Splice Suppressor elements called ESS and ISS respectively.� Certain activators and repressors regulate alternate splicing.� Activators promote alternate splicing and repressors prevent normal splicing.� It can be said otherwise also.� Activators and repressors bind to sequence elements and recruit or prevent the assembly splicing factors.� Among the nuclear proteins SR proteins are very diverse and produced in response to stimulus or produced in tissue specific manner.� They regulate normal splicing but also alternate splicing events.� Specific SR proteins perform specific alternate splicing.� They bind either to ESEs or silencer sequences.
Bacterial Systems:
- Bacteria are considered as gene-rich organisms.�
- They have no tolerance for not so useful DNA sequences unlike higher organisms.�
- They contain less amount of DNA than higher organisms.�
- They replicate their genome faster unlike eukaryotic genomes.
- They replicate more number of times in a given time yet regulated.�
- The genomic DNA is subjected selective pressure.
Eukaryotic Systems:
- Eukaryotic organisms contain large amount of DNA per chromosomes and contain more than one chromosome.�
- They are not that gene-rich.�
- The genome replication takes longer time and it is highly regulated.�
- The genomic DNA has large amount of which does not code for any proteins.�
- This noncoding DNA absorbs mistakes in chromosomal changes.
However Introns are introduced late in evolution.� Multiple Introns have the potential to generate multiple alternate products. And act as buffer against mutations and mistakes in chromosomal changes
While Introns represent silent DNA sequences, exons represent a domain or a motif. Present proteins have arisen via exon duplication and divergence.� Proteins often have repeat units of amino acid blocks (representative of an exon).� Related exons are found in unrelated genes that have different structure and functions. For example, some of the exons found in LDL receptor gene are also found in EGF gene and some of the exons found in EGF are also found in complement gene C-9.�� Exons can different motifs and they can perform different functions; example, a protein with one motif for DNA binding and the other motif for protein-protein interaction or one motif for DNA binding and the other for activation of transcriptional apparatus.
The present proteins have arisen by exon duplication and divergence.� Proteins are known to have repeating units.� Related exons are found in different proteins, which are totally unrelated in their structure and function.
�In eukaryotes, the removal of introns by splicing is a crucial step in gene expression. For some genes, splicing results in only one single type of mature mRNA, but recent studies (Mironov et al., 1999; Brett et al., 2000, Modrek et al., 2001) revealed that for many genes (up to 60% in human!) alternative splicing (AS) results in two to several types of mRNA isoforms. In extreme cases like the Drosophila Dscam gene (Schmucker et al.,2000) over 38,000 potential transcripts might be produced. This increases essentially the diversity of the transcriptome and has important implications for physiology, development and the genesis of diseases. One approach to investigate alternative splicing is to assemble expressed sequence tags (ESTs) into a list of consensus sequences (putative transcripts) and to analyze them for alternative splicing variants. Unfortunately, in the presence of AS the EST assembly problem usually has multiple solutions. Conventional EST assembly approaches do not explore these solutions exhaustively. This might easily result in truncated, misassembled or even missing transcripts.�
How to compute splicing graphs?
If the genomic template sequence of an EST cluster is known we map each EST to this sequence and build the splicing graph as described above. In the more challenging case where we do not know the genomic sequence we follow the ideas of (Pevzner, 1989; Pevzner et al., 1998) and construct the splicing graph based on the set of all k-words and their reverse complements contained in the EST cluster. In analogy to above we connect to k-words if and only if they occur consecutively in a transcript.�
|
|
|
Idealized splicing graph construction of the adenylosuccinate lyase (ADSL) mRNA.We identify bases of different strands scripts if they are derived from the same genomic position and connect them by an edge, if they occur consecutively in at least one transcript. (Right) Splicing graph of the ADSL gene with/without genomic sequence. Splice sites are marked in red. http://www.ncbi.nlm.nih.gov/ |
|
Results
The splicing graph integrates all cDNAs/ESTs originating from the same gene in a unique data structure. It combines reoccurring sequence parts into single paths, yielding a compact, unambiguous, and biologically meaningful way to represent the huge EST data set. Every transcript can be viewed as a path in the splicing graph and it is easy to generate all possible paths thus exploring all possible transcripts. In the absence of repeats, alternative splicing is depicted by bifurcations in the splicing graph. Since all mRNA isoforms are included, splicing graphs represent all different splicing variants and their relationships simultaneously, an important prerequisite for further explorative data analysis. In difference from many other EST assembly approaches, splicing graphs can be build solely from EST data without knowledge of the genomic sequence - although genome sequencing advances rapidly, still about 40% of the available EST data cannot be mapped onto genomic sequences. To demonstrate the performance of our approach we applied it in a preliminary study to the known genes of chromosome 21. We mapped 153 Uni-Gene EST clusters to chromosome 21. In this set we detected 472 different putative candidates for alternative splicing. The results are shown at the splicing graph gallery of chromosome 21�
�
�The Importance of alternative splicing�;
International Human Genome Sequencing Consortium, 2004; Modrek et al., 2001, Croft et al., 2000; Hide et al., 2001; Kan et al., 2001; Modrek et al., 2001, (Blumenthal and Steward, 1997).
�Advances in animal genome sequencing have led to many questions concerning the very nature and number of genes and how they help to promote the diversity of cellular and organismal functions. One of the interesting disappointments of the analysis of the human genome sequence is that we may possess less than 25,000 genes, only one third more than the number of predicted genes in C. elegans. How can we explain all the complexity and wonder of human biology with such a limited gene set? One way to explain this paradox is to point out that the number of possible proteins from the genome can far exceed the possible number of genes if a large percentage of the genes have the ability to encode multiple proteins. This expansion of the proteome can be accomplished through alternative precursor messenger RNA (pre-mRNA) splicing, which can allow one gene to encode multiple proteins. Comparison of cDNAs with genomic sequence has provided evidence for extensive alternative splicing of human genes. Many different studies predict that from nearly 90% of human genes are alternatively spliced.� �
Diseases due to faulty Alternative splicing:
Alternative splicing is an important mode of generation specific proteins in tissue specific and stage specific and even signal specific manner.� More than 60% of the human genome transcripts exhibit alternative splicing.� But occasionally alternative splicing goes wrong; due to this many diseases develop; only few examples are given below ex. Rett syndrome is due mistakes in alternative splicing of MECP2 gene and its transcript.
Alternative splicing in disease and therapy. Garcia-Blanco MA, Baraniak AP, Lasda EL.
Department of Molecular Genetics and Microbiology, Center for RNA Biology, Box 3053, Research Drive, Duke University Medical Center, Durham, North Carolina 27710, USA. garci001@mc.duke.edu
Alternative splicing is the major source of proteome diversity in humans and thus is highly relevant to disease and therapy. For example, recent work suggests that the long-sought-after target of the analgesic acetaminophen is a neural-specific, alternatively spliced isoform of cyclooxygenase 1 (COX-1). Several important diseases, such as cystic fibrosis, have been linked with mutations or variations in either cis-acting elements or trans-acting factors that lead to aberrant splicing and abnormal protein production. Correction of erroneous splicing is thus an important goal of molecular therapies. Recent experiments have used modified oligonucleotides to inhibit cryptic exons or to activate exons weakened by mutations, suggesting that these reagents could eventually lead to effective therapies.
Alternative splicing in disease and therapy. Garcia-Blanco MA, Baraniak AP, Lasda EL. Department of Molecular Genetics and Microbiology, Center for RNA Biology,
Alternative splicing is the major source of proteome diversity in humans and thus is highly relevant to disease and therapy. For example, recent work suggests that the long-sought-after target of the analgesic acetaminophen is a neural-specific, alternatively spliced isoform of cyclooxygenase 1 (COX-1). Several important diseases, such as cystic fibrosis, have been linked with mutations or variations in either cis-acting elements or trans-acting factors that lead to aberrant splicing and abnormal protein production. Correction of erroneous splicing is thus an important goal of molecular therapies. Recent experiments have used modified oligonucleotides to inhibit cryptic exons or to activate exons weakened by mutations, suggesting that these reagents could eventually lead to effective therapies.
ScienceDaily (Dec. 26, 2008) � An international research team led by Tim Nilsen, Ph.D., a professor of medicine and biochemistry and the director of the School of Medicine's Center for RNA Molecular Biology, has discovered an unexpected mechanism governing alternative splicing, the process by which single genes produce different proteins in different situations. The new mechanism suggests that curing the more than half of genetic diseases that are caused by mutations in the genetic code that in turn create mistakes in alternative splicing may be considerably more complicated than biomedical researchers have previously assumed. Those diseases include a large number of cancers and many neurodegenerative diseases.
The research, titled "Dynamic regulation of alternative splicing by silencers that modulate 5' splice site competition" is published in the December 24 issue of Cell. Nilsen led an international team of researchers from Case Western Reserve University, Columbia University, Memorial Sloan-Kettering Cancer Institute in New York City, and the Max Planck Institute for Biophysical Chemistry in, Germany. Case post-doctoral fellow Yang Yu, Ph.D. was the lead author.
"Regular" splicing is the process by which long strings of nucleotides in a gene's pre-messenger RNA (pre-mRNA) are discarded, and the remaining strings of nucleotides are spliced together into one continuous strand of messenger RNA (mRNA) that produces one unique protein.
Although it creates needed proteins, alternative splicing can also lead to problems. Mistakes in alternative splicing caused by changes (mutations) in DNA sequences create more than half of all genetic diseases. For instance, sometimes the resulting mRNA includes nucleotide sequences that should have been deleted.
If one considers the metaphor that pre-mRNA is a long sentence, Nilsen says, then nucleotides and splice sites are the words of the sentence. "Adding or deleting one word," he says, "can radically change the meaning of the sentence."
Pre-mRNA splicing and human disease; Nuno Andr� Faustino1, and Thomas A. Cooper, Berget et al. 1977; Chow et al. 1977. (Burge et al. 1999.). Hastings and Krainer 2001; Black 2003), (Nissim-Rafinia and Kerem 2002)
The precision and complexity of intron removal during pre-mRNA splicing still amazes even 26 years after the discovery that the coding information of metazoan genes is interrupted by introns (). Adding to this amazement is the recent realization that most human genes express more than one mRNA by alternative splicing, a process by which functionally diverse protein isoforms can be expressed according to different regulatory programs. Given that the vast majority of human genes contain introns and that most pre-mRNAs undergo alternative splicing, it is not surprising that disruption of normal splicing patterns can cause or modify human disease. The purpose of this review is to highlight the different mechanisms by which disruption of pre-mRNA splicing play a role in human disease. Several excellent reviews provide detailed information on splicing and the regulation of splicing;. The potential role of splicing as a modifier of human disease has also recently been reviewed.
Detecting tissue-specific alternative splicing and disease-associated aberrant splicing of the PTCH gene with exon junction microarrays
Kazuaki Nagao, �Naoyuki Togawa, �Katsunori Fujii , �Hideki Uchikawa1, �Yoichi Kohno, �Masao Yamada and �Toshiyuki Miyashita,*
Mutations in the human ortholog of Drosophila patched (PTCH) have been identified in patients with autosomal dominant nevoid basal cell carcinoma syndrome (NBCCS), characterized by minor developmental anomalies and an increased incidence of cancers such as medullo-blastoma and basal cell carcinoma. We identified many isoforms of PTCH mRNA involving exons 1�5, exon 10 and a novel exon, 12b, generated by alternative splicing (AS), most of which have not been deposited in �Gene Bank� nor discussed earlier. To monitor splicing events of the PTCH gene, we designed oligonucleotide arrays on which exon probes and exon�exon junction probes as well as a couple of intron probes for the PTCH gene were placed in duplicate. Probe intensities were normalized on the basis of the total expression of PTCH and probe sensitivity. Tissue-specific regulation of AS identified with the microarrays closely correlated with the results obtained by RT�PCR. Of note, the novel exon, exon 12b, was specifically expressed in the brain and heart, especially in the cerebellum. Additionally, using these microarrays, we were able to detect disease-associated aberrant splicing�s of the PTCH gene in two patients with NBCCS. In both cases, cryptic splice donor sites located either in an exon or in an intron were activated because of the partial disruption of the consensus sequence for the authentic splice donor sites due to point mutations. Taken together, oligonucleotide microarrays containing exon junction probes are demonstrated to be a powerful tool to investigate tissue-specific regulation of AS and aberrant splicing taking place in genetic disorders.
Bringing Genomics and Systems Biology to Pathways and Disease
Within the last two decades, we have witnessed unprecedented progress in genomics and genetics research. The availability of complete genome sequences and systematized catalogs of genetic variation in humans, human pathogens, and other model organisms have had a major impact on biology and have begun to transform clinical practice. The advances in whole genome amplification, genotyping platforms, next generation sequencing, and the coming advent $1000 genome will continue to drive innovation and discovery.
Integration of genomic information with interactomes and evolutionary maps will be leveraged strongly by us to isolate key pathways malfunctioning in human disease for a systems-level approach to diagnostic and treatment.
System level analysis; Integration of genomic information with interactomes and evolutionary maps will be leveraged strongly by us to isolate key pathways malfunctioning in human disease for a systems-level approach to diagnostic and treatment.;XIGM , Institute of Genomic Medicine; http://webcache.googleusercontent.com/
Pathways:
RNA Processing:
Members of the IGM have demonstrated excellence and experience in key areas of RNA processing such as nonsense-mediated decay, alternative splicing, microRNA processing and targeting, non-coding RNAs and translational control. We will capitalize on our expertise to illuminate RNA defects in disease areas.
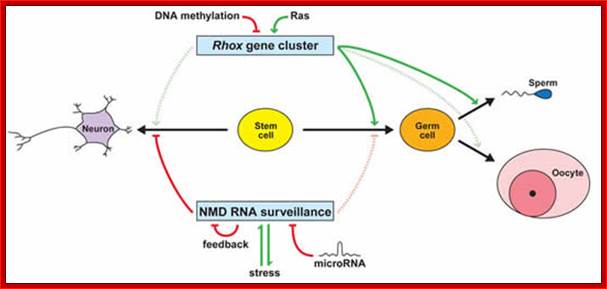
http://webcache.googleusercontent.com/
Epigenetics
IGM comprises of experts in the area of epigenetics such as DNA methylation and histone modification, which are growing areas of importance in disease modeling.
Diseases
Medical scientists have an excellent cadre of leaders applying genetics and genomics approaches to areas neurological and mental health disorders such as epilepsy, ALS, autism, schizophrenia, bipolar disorder cancer and DNA-Repair and �Ataxias.
Neurological and Mental Health Disorder: We have an excellent cadre of leaders applying genetics and genomics approaches to areas neurological and mental health disorders such as epilepsy, ALS, autism, schizophrenia, bipolar disorder and ataxias. IGM boasts prominent pioneers in the area of DNA repair and epigenetics as it applies to cancer biology. http://webcache.googleusercontent.com/
List of diseases caused by missplicing:
Splicing mutations associated with variable phenotypes
- Becker muscular dystrophy.
- Cystic fibrosis.
- Deficiency of the MCAD enzyme,
- Familial Dysautonomia,
- Menke disease.
- Occipital horn syndrome.
- Pyruvate dehydrogenase deficiency.
- Retinitis pigmentosa.
- Sandhoff disease.
- Hutchinson-Gilford Progeria Syndrome.
- Spinal muscular atrophy
(SMA).
Human diseases associated with changes in the relative levels of alternative spliced isoforms
- Breast cancer
- Fragile X syndrome
- Frasier syndrome
- Frontotemporal dementia with Parkinsonism linked to Chromosome 17
- Facioscapulohumeral Muscular Dystrophy (FSHD)
- Gastric cancer
- Giant cell tumors of bones
- Growth hormone deficiency type II
- Head and neck squamos cell carcinoma
- Lung cancer
Export of processed mRNAs:
Almost all mRNAs once spliced they get associated with specific proteins, such exon-exon joint complexes, Cap binding proteins, mRNPs, poly-A binding proteins and others.� Once they are decorated (specifically), they are transported out into the cytoplasm.� Each of the mRNA contains signal sequences for export using Ran-GTP complexes.� For example, HIV mRNA has to be exported without splicing and it requires Rev Proteins which bind to Rev response elements (RRE), then transported.