Trans-Splicing of mRNAs:
As mentioned in the earlier chapter, that transcription and processing including splicing processes are linked.�� The transcripts are subjected to a variety of modifications, which differ from one member of the transcript to the other and also vary from one species to the other.�
� Trypanosomes and C. elegans do show a different type of pre-mRNA splicing called Tran-splicing.� Trans-splicing involves cutting and joining of two different transcripts into a functional RNAs; it is like cut and paste together.
�
� This can be cis form of trans form of splicing and it can be of true to trans-splicing type.
� Even chloroplasts show trans-splicing of their RNA products.� Ex. Rps12 gene in chloroplasts shows trans-splicing.� Chlamydomonas reinhardtii mitochondrial gene for NAD-I and NAD-5 produces functional mRNA by trans-splicing.� In plants, mitochondrial coding regions of few genes are scattered among the other genes.� Each coding segments are flanked by an Intron like sequences. They are transpliced to generate functional RNAs.
� Trans-splicing involves both snRNA and snRnps and few more proteins.
Spliced leader RNA in trans-splicing of metazoan:
Overview and Background:
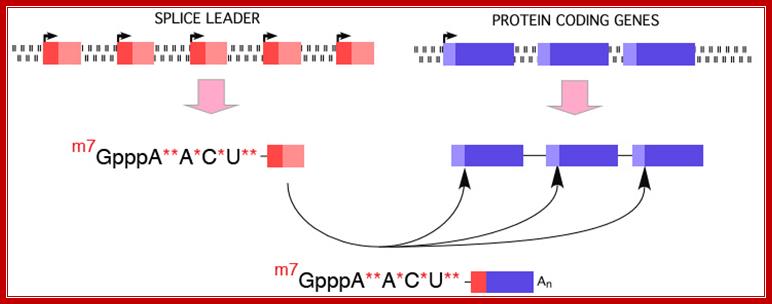
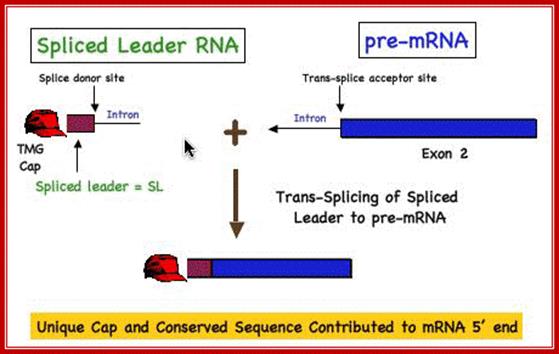
Use of �Splice leader RNA� in trans-splicing is a mechanism of gene expression and it is a form of RNA processing in which a splice leader sequence (SL) is joined to a 5� �leader-less� pre-mRNA to form the mature 5' end of the mRNA. This process adds a conserved sequence to the 5' end of mRNAs (the spliced leader, ranging in size from 15-50 nt) and brings a 5� cap to the mRNA. (see the figure below). This form of RNA maturation was first described in trypanosomes and it has been subsequently demonstrated in other Kinetoplastida single celled animals and flagellated protozoa (Euglena), nematodes, flatworms, and more recently in Cnidarians (Hydra) and in tunicates (Ciona) and some primitive chordates. The identification of trans-splicing in a number of major invertebrate phyla suggests that this particular form of RNA processing may have been common among invertebrates (and perhaps early chordates) and likely represents an evolutionarily important form of gene expression.
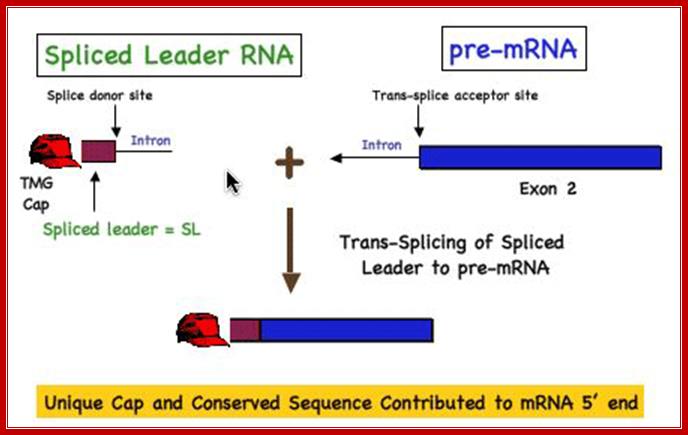
Figure. Illustration of Spliced Leader Trans-Splicing; DAVIS lab. Davis_lab/�Research_Overvie�w.�html
����������������������� The new cap added to the mRNAs as a result of metazoan trans-splicing is a trimethyl guanosine cap (m2, 2,7GpppN), TMG). This cap differs from the typical cap on eukaryotic mRNAs with the addition of two methyl groups at N2. Importantly, not all mRNAs are matured by this processing. Thus, two populations of mRNAs are present in metazoan cells; trans-spliced and not trans-spliced. In nematodes, trans-spliced mRNAs constitute > 60%, whereas in flatworms they constitute ~10% of all mRNAs.
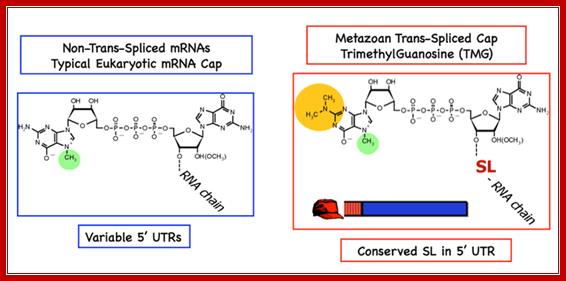
Figure: Illustration of the monomethyl cap on eukaryotic mRNAs compared with the tri-methyl cap on trans-spliced mRNAs; Davis Lab Research
�The mRNA cap and 5' UTR of mRNAs play important roles in mRNA metabolism including mRNA processing, export, translation, and stability. Cap-interacting proteins specifically recognize the mRNA cap and facilitate these aspects of mRNA metabolism. Notably, most eukaryotic cap-interacting proteins do not recognize or have very low affinity for the TMG cap. Therefore, among the questions -1) what function trans-splicing serves in these organisms, 2) how mRNA metabolism is affected or effected? and 3) how the cellular machinery has adapted to recognize and deal with the atypical TMG cap.
As the cap and 5' UTR of mRNAs play important roles in mRNA processing, investigating the contribution of trans-splicing is important. In addition, investigating the adaptation of proteins that interact with the cap to the presence of the novel TMG cap on mRNAs is of importance. To address these questions, scientists have developed and are using in vitro biochemical and in vivo transfection systems in nematode embryos to explore in depth the contribution of the novel cap and SL sequence to mRNA metabolism. They also identified and characterized a number of cap-interacting proteins that have adapted to recognize the atypical TMG cap including the translation initiation factor eIF4E (several isoforms) and decapping proteins (DcpS and Dcp1/2).
The model used for the majority of these studies is the nematode Ascaris (see Ascaris as a Model Nematode). While Ascaris is not amenable to genetic analysis, it enables to obtain large amounts of developmentally synchronized nematode embryos (from fertilization through L3 larvae) and provides a number of very powerful biochemical and molecular tools for studies. These tools include robust cell-free systems for transcription, splicing, translation, and decay as well as biolistic transfection�.
Trypanosome, belong to Euglenoid members, few lower protozoans, even C. elegans and few other nematodes like Ciona (Tunicates) and Euglena, Flatworms, Hydra (Cnidarians), Kinetoplastida and several plant chloroplast and mitochondrial system show trans- splicing, why even Adenoviral transcripts show similar process (not exactly trans slicing).
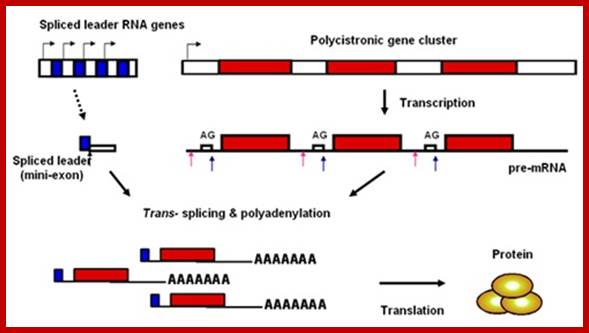
� In Trypanosomes, most of the mRNAs (at least 70%) produced are polycistronic, an exception to eukaryotic rule, where eukaryotic genes are generally monocistronic and prokaryotes have polycistronic mRNAs.� Even C. elegans, a nematode shows substantial number of genes with polycistronic characters, and substantial number of them or spliced by trans-slicing process.
� All functional mRNAs (processed ones) have one common 39-100 or more nucleotides long leader sequence at 5�UTR region.�
� In the above said organisms, the leader sequences, in the genomic DNA, are organized into tandem repeat clusters of 140 ntds long.�
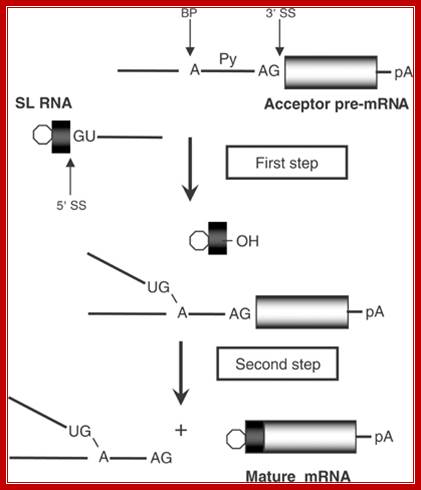
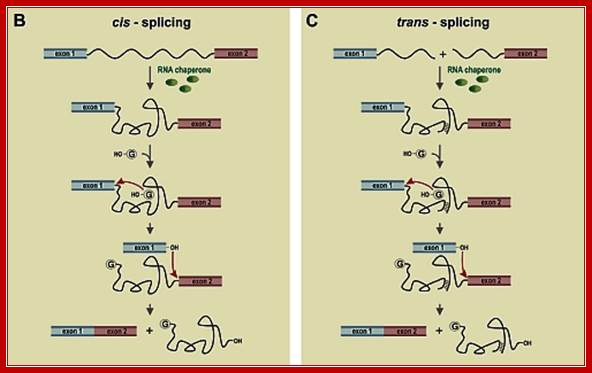
Spliced leader (SL) trans-splicing. In trypanosomes, virtually all mature messages acquire the same 5' terminal sequence called a spliced leader (SL) RNA. This reaction requires most components of the spliceosome and parallels the two steps of both cis-splicing and spliceosome-mediated RNA trans-splicing. The 5' splice site (SS) on the SL RNA and 3' SS on the acceptor pre-mRNA are indicated. BP, branch point; Py, polypyrimidine tract; pA, polyadenylation site; open hexagon, 5' cap structure, L G Mitchell and G J McGarrity
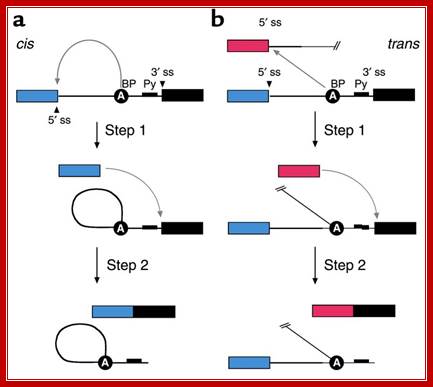
Figure: Messenger RNA reprogramming by spliceosome-mediated RNA trans-splicing; Both cis-splicing and trans-splicing reactions proceed via two phosphoryl transfer reactions. (a) The cis reaction. A schematic of the two phosphoryl transfer reactions required for intron removal. The exons are indicated as boxes (the first or 5′ exon is gray and the second or 3′ exon is black), and the intron is depicted as a line. In the first reaction (step 1), the 2′ OH group of a bulged adenosine at the branch point attacks the 5′ phosphate of the first residue of the intron forming the lariat intermediate and the �free� 5′ exon. The lariat contains the branched adenosine, so called because it is connected via conventional 5′ and 3′ links, but also contains a 2′-5′ linkage to the first residue of the intron (see ref. 16 for a more extensive discussion of the splicing reactions). In the second step, the 3′ OH group of the last residue of the �free� 5′ exon attacks the 5′ phosphate of the first residue of the second exon, forming a product with the two exons ligated and releasing the intron as a lariat. (b) The trans reaction. A schematic of the two phosphoryl transfer reactions required for SMaRT. Icons are as described above, except that the trans-splicing or invading exon is shown as a red box. In step 1, the 2′ OH group of the bulged or branch point adenosine is again the nucleophile and is attacking the 5′ phosphate of the first residue of an intron in a second RNA molecule (this molecule could be identical in sequence to the first or it could be a completely different RNA). Because the reaction proceeds in trans the branched molecule is now a Y-shaped molecule, not a lariat. Step 2 proceeds as described for step 2 in the cis reaction; however, the exon product includes sequences from two RNAs. Mariono A.Garcia-Blanco; http://www.jci.org/
Cis splicing and Trans-splicing
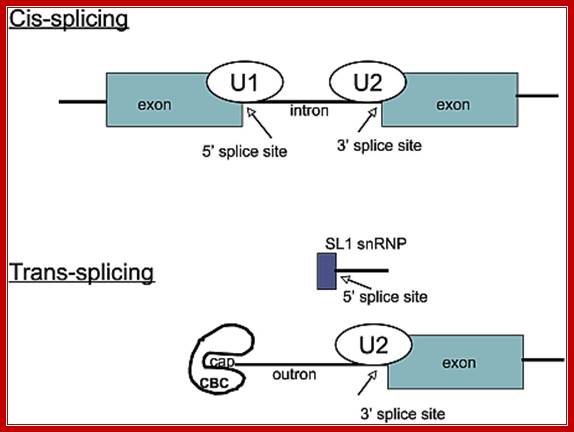
Cis splicing involves two exons are joined by eliminating the intron; while Trans splicing involves using a leader segment from one source and joining to the 5�end of another pre-mRNA which lacks 5� leader segment. In cis-splicing, the U1 snRNP base pairs with the 5' splice site, and U2 snRNP base pairs with the branchpoint near the 3' splice site. The intron is excised and the two exons are spliced together. In trans-splicing there is no 5' splice site on the pre-mRNA for U1 snRNP binding. Instead, the 5' splice site is provided by the donor SL snRNP, which interacts with the U2 snRNP at the 3' splice site, and the SL exon is spliced to the exon on the pre-mRNA. The region between the 5' cap and the trans-splice site is called the outron. CBC: nuclear cap binding complex; Thomas Blumenthal, http://www.wormbook.org/
� In trans splicing, the splice leader RNA is transcribed as 140 ntds long pieces, only a 39-nucleotide sequence at its 5� end is used as leader segment and the rest is used as the Intron segment, for its splice junction has sequences similar to eukaryotic pre-mRNA 5� intron junction sequences. ��The 5� region of the premRNA contains sequence elements similar to that of normal RNA sequences, such as branching site and poly-pyrimidine sequences.� They also contain splice joint sequences such as 5�GU- and AG 3�.� The 5� end of the intron sequences found in this type of pre mRNAs are called �outron� elements.
The basic splicing process operates by using pre-SL region and the 5end of premRNA which have characteristics of regular intronic characters and use the similar snRNAs and snRNPs and bring about slice and joining.� The SL leader shows characterists of snRNA with SM proteins� binding site at the base of stem loop structure
The structurally poly-cistronic mRNAs are separated by introns, each of them having a branching site and AG as the sequence at 3� end of splice joint.
�
� The last part of each cistron contains a poly-A signal (UUUAUU) for cleaving and poly-A addition.�
� The leader sequence is called SL RNA or splice leader RNA.
� Trypanosome contains Sn RNAs like U2, U4 and U5 but not U1 and U6 RNAs. They are associated with sm-proteins and other sn RNA specific proteins. The SL-RNAs perse have structural similarities to that of U1 RNAs. SL-RNAs exist as members of Sm-snRNP complexes.� They can even function without their associated proteins.
�
� The SL-RNAs and other U Sn RNAs and SnRNPs assemble on to their respective targets and bring about splicing, thus one type of leader sequence is provided to all of its mRNAs.
�
� Intron in this kind of polycistronic RNAs, contain a site and sequences for cleaving and releasing them as individual mono cistronic mRNAs with a cap at its 5�end and Poly(A) tail at its 3�end.
� In C.elegans, the pre-leader piece is about 100 ntds long and only a 22-ntd piece from the 5� end of the transcript is spliced as the leader in all the three Actin mRNA transcripts. The leader providing RNA piece is called pre SL RNA and it assumes 3-stem loop structure before it engages in trans-splicing.� The SL RNAs associated with proteins act as SL-RNA-RNP complexes.
� The SL RNA can be capped with 7�CH3G- or 7�CH3-2�CH3-2�CH3 (tri-methylated caps i.e 2�2�7�methylated Gmp).
�
� In trypanosomes, the VSG-221 gene produces surface glycoproteins. Transcription of the said gene initiates, at 5kbp upstream of the VSG 221.� The total length of the transcript is about 57kb.� It is polycistronic and contains information for several mRNAs, rRNAs and tRNA.� It has open reading frames from internal segments.
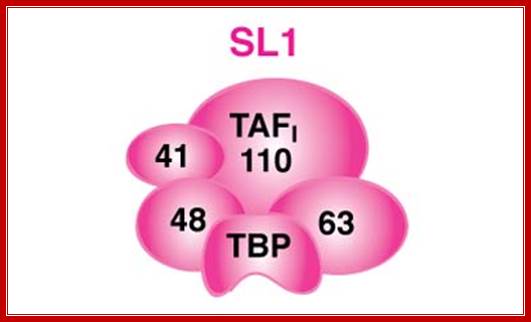
� Based on cap structure SLRNAs are classified as SL1 and SL2.� SL1 contains 7�monomethylated Cap, but the SL2 contains 2�2�7-tri methylated Cap.� SL1 performs splicing at the 5�end of the polycistronic mRNA and SL2 performs trans-splicing at internal cistrons.
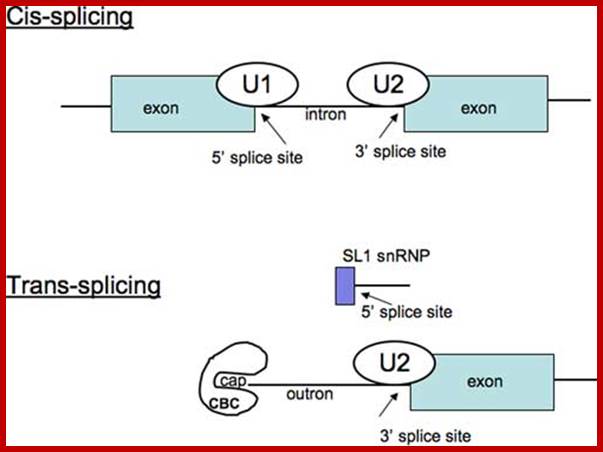
Difference between cis and trans splicing; Comparison of cis- and trans-splicing. In cis-splicing, the U1 snRNP base pairs with the 5� splice site, and U2 snRNP base pairs with the branchpoint near the 3� splice site. The intron is excised and the two exons are spliced together. In trans-splicing there is no 5� splice site on the pre-mRNA for U1 snRNP binding. Instead, the 5� splice site is provided by the donor SL snRNP, which may base pair with a sequence in the outron, the region of the pre-mRNA between the 5� cap and the trans-splice site. The SL exon is then spliced to the first exon on the pre-mRNA. CBC: nuclear cap binding complex. Thomas Blumenthal; http://www.wormbook.org
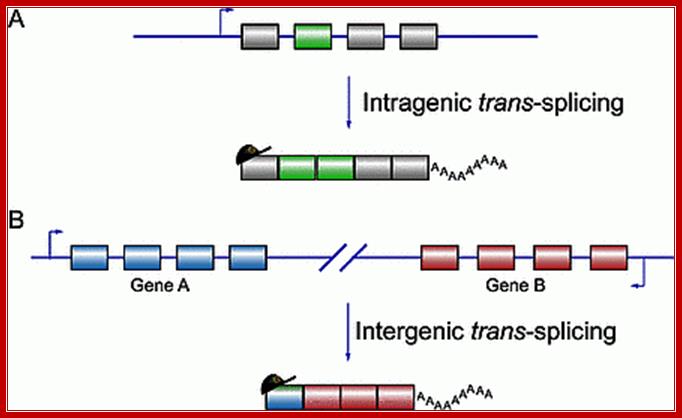
Alternative Trans splicing in Mammals; Trans splicing can be intergenic or intragenic; i.e between two similar mRNA or between two different pre-mRNAs;� Takayuki Horiuchi and Toshiro Aigaki http://submit.biolcell.org/
 �
� 
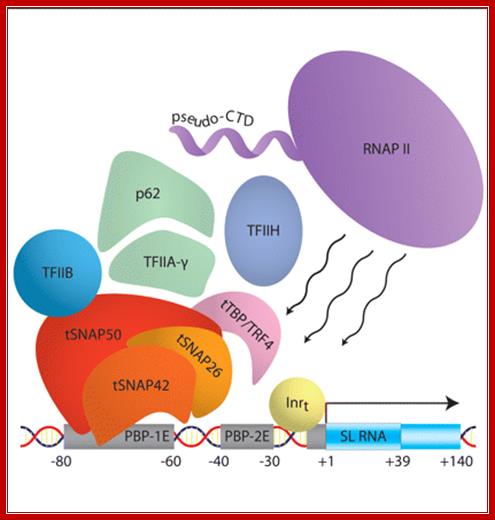
SL1 RNA gene with assembled RNA polymerase complex
�Schematic of the protein components that assemble to form the preinitiation complex at the spliced leader (SL) RNA gene promoter in Trypanosoma brucii: The DNA promoter elements and the transcribed region are shown. The trypanosome SNAP complex consists of the tSNAP50, −42, ~50 and −26 subunits. The pseudo-CTD is the carboxy-terminal ∼300 amino acids of the largest subunit of RNA polymerase II and deviates from the canonical heptapeptide repeats present within the yeast and mammalian enzymes. The tail that is responsible SLRNA processing lacks the 7 (TSPRySP)n� and but contain different set of proteins and the tail is called pseudo tail.� It does not perform� poly-A addition but involved in processing the pre SL RNA.� The SL RNA contains 5�ppp and 3� poly Us because the terminal region of the SLRNA genes ends in T TTTT residues.� SL RNA gets 7� methylated cap.� Then using SLNA- SL associated RNA and proteins base pairs with SL RNA and converts certain Uridines to pseudouridines. More methylation takes place at the 5� end. Then the SM proteins bind; this leads to excision of U from the 3� end.; RNA polymerase I transcription complex�; Dr. joost Zomerdijk; http://www.lifesci.dundee.ac.uk/
Pre-SLRNA Processing:
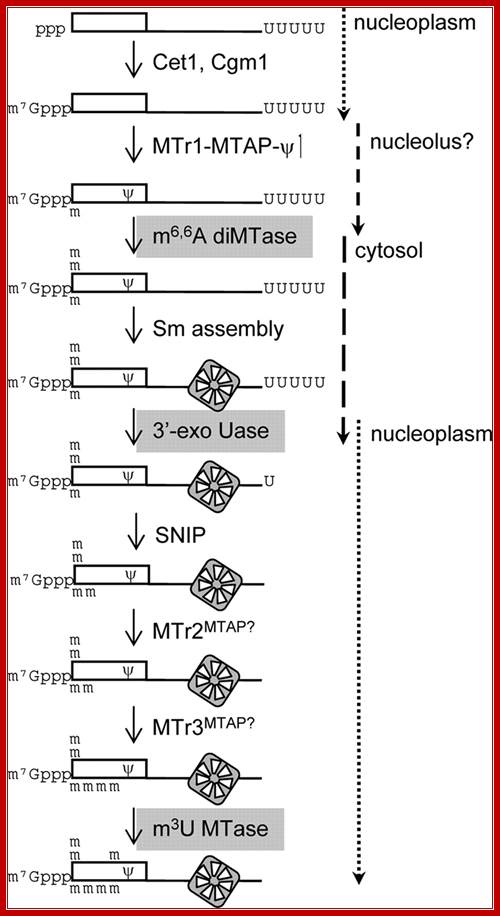
FIG. Working model for SL RNA biogenesis. The enzymatic reactions required for maturation of the SL RNA are shown. Gray backgrounds indicate enzymes that have not been assigned. Assembly of the heptameric Sm ring defines early and late steps in the process. RNA synthesis and cap formation occur at a discrete site in the nucleoplasm. Cap 1 modifications and formation are early modifications; however, their relative order is not known. TbNhp2 and TbNop10 localize to the nucleolus; thus, TbMTr1 complex modifications are placed in that location. The two-step removals of the poly(U) tail and the remaining four cap methylations are late Sm-dependent events. The relative timing of 5' and 3' processing is not known, (Williams et al. 1999).
Nuclear mRNAs in Trypanosomatids are generated by trans-splicing. Although trans-splicing resembles cis-splicing in many ways and most of the U RNA participants have been characterized, relatively few involved proteins have been identified. Herein, authors of this transcript, employed a yeast three-hybrid system to identify a protein, XB1, which binds to the Trypanosoma cruzi SL RNA. �XB1 is a approximately 45 kDa protein which is homologous to the essential pre-mRNA-splicing factor PRP31p from Saccharomyces cerevisiae. Gel shift assays and UV cross-linking experiments with recombinant XB1 confirmed that this T. cruzi protein binds the SL RNA in vitro. The binding site of XB1 on the SL RNA was mapped to stem-loop II by deletion of the SL RNA 'bait' in the three-hybrid system. Finally, UV cross-linking SL RNA with S100 extract indicated native XB1 protein and SL RNA interaction in T. cruzi extract.
Euglena, spliced-leader RNA (SL-RNA) and 5S rRNA
genes are tandemly repeated:
M.Keller, L.H.Tessier, R.L.Chan, J.H.Weil and P.lmbault*
In Euglena gracillis, a 26 nucleotide leader sequence (spliced leader sequence = SL)� is transferred by trans splicing to the 5' end of a vast majority of cytoplasmic mRNAs. The SL originates from the 5' extremity of a family of closely related snRNAs (SL-RNAs) which are about 100 nucleotides long. In this note we present the nucleotide sequences of two SL-RNA genes, confirming the sequences previously established by sequencing purified SL-RNAs. Although some SL-RNA genes are dispersed throughout the genome, we show that the majority of SL-RNA genes are located on 0.6 kb repeated units which also encode the cytoplasmic 5S rRNA. We estimate that the copy number of these repeated units is about 300 per haploid genome. The association of SL-RNA and 5S rRNA genes in tandemly repeated units is also found. �In nematodes but paradoxically does not exist in trypanosomes which are phylogenically much closer to Euglena. We also show that a high number of sequences analogous to the 26 nucleotide SL are dispersed throughout the genome and are not associated with SL-RNAs.
SL RNA Genes of the Ascidian Tunicates Ciona intestinalis and Ciona savignyi: Brendan Yeats1, Jun Matsumoto1, Sandra I. Mortimer1, Eiichi Shoguchi2, Nori Satoh2 and Kenneth E. M. Hastings1*
We (above authors) characterized
by bioinformatics the trans-spliced leader donor RNA (SL RNA) genes of two
ascidians, Ciona intestinalis and Ciona savignyi. The Ciona
intestinalis genome contains ![]() 670 copies of
the SL RNA gene, principally on a 264-bp tandemly repeated element. Fluorescent
in-situ hybridization mapped most of the repeats to a single site on the short
arm of chromosome 8. The Ciona intestinalis genome also contains
670 copies of
the SL RNA gene, principally on a 264-bp tandemly repeated element. Fluorescent
in-situ hybridization mapped most of the repeats to a single site on the short
arm of chromosome 8. The Ciona intestinalis genome also contains ![]() 100 copies of
a >3.6-kb element that carries 1) an SL RNA-related sequence (possible a
pseudogene) and 2) genes for the U6 snRNA and a histone-like protein. The Ciona
savignyi genome contains two SL RNA gene classes having the same SL
sequence as Ciona intestinalis but differing in the intron-like
segments. These reside in similar but distinct repeat units of 575 bp (
100 copies of
a >3.6-kb element that carries 1) an SL RNA-related sequence (possible a
pseudogene) and 2) genes for the U6 snRNA and a histone-like protein. The Ciona
savignyi genome contains two SL RNA gene classes having the same SL
sequence as Ciona intestinalis but differing in the intron-like
segments. These reside in similar but distinct repeat units of 575 bp (![]() 410 copies)
and 552 bp (
410 copies)
and 552 bp (![]() 250 copies)
that are arranged as separate tandem repeats. In neither Ciona species
is the 5S RNA gene present within the SL RNA gene repeat unit. Although the
number of SL RNA genes is similar, there is little sequence similarity between
the intestinalis and savignyi repeat units, apart from the region
encoding the SL RNA itself. This suggests that cis-regulatory elements involved
in transcription and 3′-end processing are likely to be present within
the transcribed region. The genomes of both Ciona species also include
> 100 dispersed short elements containing the 16-nt SL sequence and up to 6
additional nucleotides of the SL RNA sequence.
250 copies)
that are arranged as separate tandem repeats. In neither Ciona species
is the 5S RNA gene present within the SL RNA gene repeat unit. Although the
number of SL RNA genes is similar, there is little sequence similarity between
the intestinalis and savignyi repeat units, apart from the region
encoding the SL RNA itself. This suggests that cis-regulatory elements involved
in transcription and 3′-end processing are likely to be present within
the transcribed region. The genomes of both Ciona species also include
> 100 dispersed short elements containing the 16-nt SL sequence and up to 6
additional nucleotides of the SL RNA sequence.
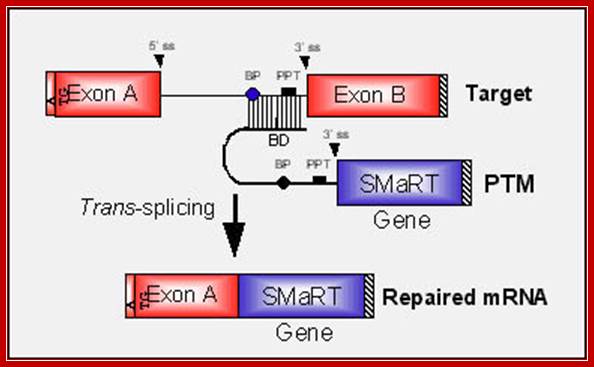
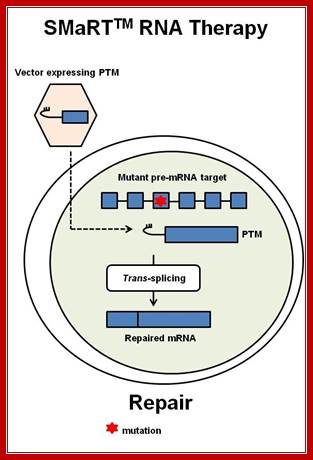
SMaRT technology, which stands forSpliceosome Mediated RNA Trans-Splicing, utilizes pre-therapeutic RNA molecules (PTMs) to promote trans-splicing of specific target precursor mRNA (pre-mRNA) molecules. Previously, Intronn researchers had demonstrated the effectiveness of PTM's when directed against 3'- or 5'-splice sites (references 1 and 2). In the present study, SMaRT was used to precisely target and replace a single internal exon utilizing pre-trans-splicing molecules (PTMs) containing both the 5' and 3' splice sites. Such PTMs promote two trans-splicing reactions with the intended target gene mediated by the spliceosome.;http://www.bioresearchonline.com/
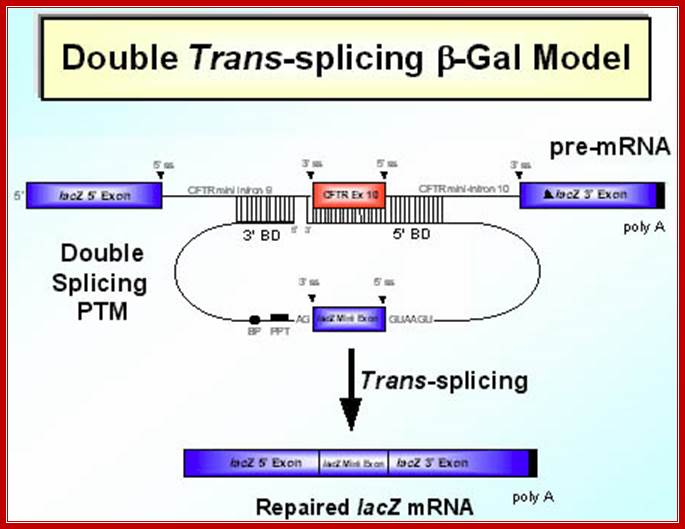
SMaRT mediated gene reprogramming offers several advantages over conventional gene replacement therapy. It's possible with a single PTM to correct all the mutations in a large section of a gene, e.g. cystic fibrosis. In addition, by converting defective message, SMaRT not only repairs but also reduces the ratio of defective to wild type mRNA. And lastly and most importantly, a PTM needs to bring in only a fraction of the target gene, along with relatively short trans-splicing domain. Hence SMaRT technology can be used to develop small, easily deliverable RNA molecular therapeutics for correcting genetic defects or specifically killing tumor cells by expressing toxic genes; SMaRT mediated gene reprogramming offers several advantages over conventional gene replacement therapy. It's possible with a single PTM to correct all the mutations in a large section of a gene, e.g. cystic fibrosis. In addition, by converting defective message, SMaRT not only repairs but also reduces the ratio of defective to wild type mRNA. And lastly and most importantly, a PTM needs to bring in only a fraction of the target gene, along with relatively short trans-splicing domain. Hence SMaRT technology can be used to develop small, easily deliverable RNA molecular therapeutics for correcting genetic defects or specifically killing tumor cells by expressing toxic genes; http://www.bioresearchonline.com/
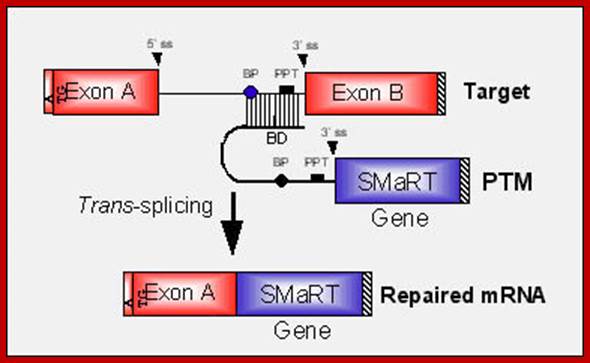
In the above diagram, in-between exons there are introns like sequences with splice joints and branching sequences similar to higher systems. Even Lieshmania contains such long intervening segments between the genes with intron like characteristic features. In this SMaRT Trans splicing, a segment, is chosen from specific pre mRNA, is placed at the intron between B and C and spliced to generate an mRNA with AB and X segments.� The pre trans splicing region of an RNA has features similar to an intron with complementary sequence at its 5� end, which is used for base pairing with later part of the intron and splicing is performed.� Proteins used in their though similar to sn RNPss, �but, some additional proteins are used.
Messenger RNA reprogramming by spliceosome-mediated RNA trans-splicing;
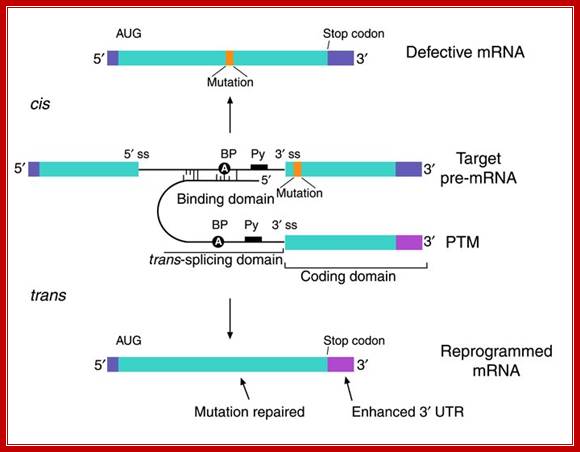
The anatomy of a PTM. The figure depicts a mutant target pre-mRNA (see the LacZ model system in ref. 12) that is incapable of coding for functional protein because it contains a mutation in the second exon (indicated as orange segment). The product of the conventional cis-splicing reaction for this pre-mRNA is a defective mRNA. The PTM shown contains several domains. The binding domain (also referred to as the targeting domain) binds to the pre-mRNA target, thus localizing the PTM near the site of the desired trans-splicing reaction. The binding domain can also be designed to occlude important elements within the 3′ splice site of the target and thus reduce the cis-splicing reaction. Between the binding domain and the trans-splicing domain there is usually a spacer region for flexibility. The trans-splicing domain, which in the case shown includes the elements required to make a potent 3′ splice site, is responsible for the reactivity of the PTM. Finally, the coding domain contains the necessary genetic information that will be imparted into the reprogrammed RNA. This can include protein-coding instructions as well as instructions for the effective processing, transport, and localization of the reprogrammed mRNA. In the example shown, the open reading frame has been changed to repair the mutation in the gene, and the 3′ untranslated region (3′ UTR) has been enhanced to increase mRNA stability and translation. It should be noted that the cis- and trans-splicing reactions are in competition. This implies that as the level of reprogrammed mRNA increases, the level of defective mRNA decreases. A decrease in the level of defective mRNA may be very useful in the case of dominant mutations; Mariano A. Garcia-Blanco; http://www.jci.org/
�Figure: �Messenger RNA reprogramming by spliceosome-mediated RNA trans-splicing;
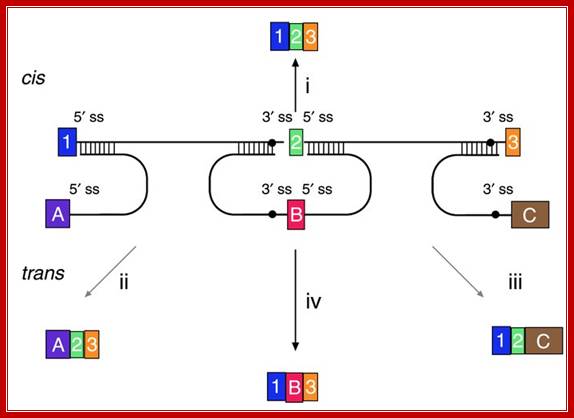
Versatility of mRNA reprogramming by targeted SMaRT. The figure shows a schematic of splicing reactions involving a three-exon pre-mRNA and PTMs. A conventional constitutive cis-splicing reaction leading to the production of the expected 1�2�3 mRNA is shown (i). Three targeted SMaRT reactions are shown. PTM[A] contains a functional 5′ splice site that can trans-splice to the 3′ splice site adjacent to exon 2 in the pre-mRNA target (ii). This trans-splicing produces a chimeric A�2�3 mRNA. PTM[A] is targeted to occlude the naturally occurring 5′ splice site at exon 1 to reduce the use of this site. PTM[C] contains a functional 3′ splice site that can trans-splice to the 5′ splice site adjacent to exon 2 in the pre-mRNA target (iii). This trans-splicing produces a chimeric 1�2�C mRNA. Finally, PTM[B] contains both 3′ and 5′ splice sites bordering an exon, and these splice sites can trans-splice with the 5′ splice site adjacent to exon 1 and the 5′ splice site adjacent to exon 3, respectively (iv). These two trans-splicing reactions lead to internal exon replacement and produce the chimeric 1�B�3 mRNA�. Mariano A. Garcia-Blanco; �http://www.jci.org/
 �
�
Biochemistry lecture notes.;http://www.nsm.buffalo.edu
Davis lab research. Html
www.darwins.blogspot.com
Discovery of operons:
The gpd-3 gene and several other genes whose mRNAs receive SL2 were found to occur at downstream positions in closely-spaced clusters of identically oriented genes. Although these genes in some clusters are trans-spliced to SL1, others are not trans-spliced. The presence of a gene in a downstream location in a closely-spaced cluster signals that its product should be SL2 trans-spliced. A microarray analysis of the entire genome demonstrated how truly robust is this correlation. This analysis identified more than 1000 such clusters in which the downstream mRNAs are trans-spliced to SL2; these clusters contain more than 2600 genes. These operons range from two to eight genes in length and can cover more than 50 kb of the genome. They are found on all chromosomes, but they are rare on the X chromosome. (Blumenthal, 2004; Spieth et al., 1993; Zorio et al., 1994); (Blumenthal et al., 2002)
SL1 operons: SL1-type operon: Trypanosoma transcription factors: Waiting for Godot.�
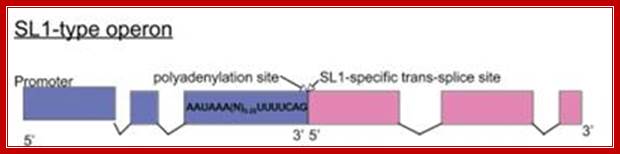
A few C. elegans operons are of the SL1-type, where the site at which 3' cleavage and polyadenylation occurs is also the trans-splice site for the downstream gene, so the intercistronic distance is effectively zero. These operons are always trans-spliced by the SL1 snRNP; Williams et al., 1999, (T. Blumenthal, unpublished); �http://www.wormbook.org/
The C. elegans genome also contains a second type of operon different from those described above in two significant ways. The mRNA of the downstream gene is trans-spliced to SL1, rather than SL2, and there is no intercistronic sequence. The site of polyadenylation of the upstream gene and the trans-splice site are at adjacent nucleotides. Only twenty operons of this type have been identified, so it is quite uncommon. This kind of operon is distinguished from the major type by the fact that 3' end formation of the upstream gene eliminates the trans-splice site of the downstream gene, so that any given pre-mRNA gives rise to either the upstream or the downstream mRNA, but not both.
mRNA maturation by two-step trans-splicing/polyadenylation processing in trypanosomes:
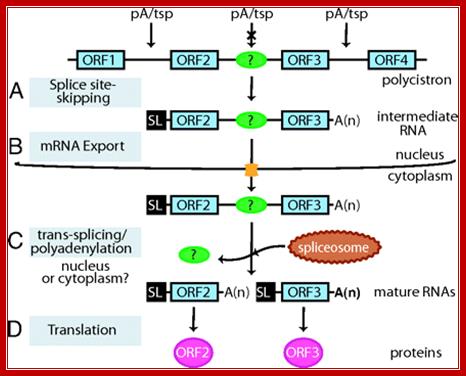
Model of intermediate RNA maturation in T. cruzi. Two steps of trans-splicing and polyadenylation processing generate functional monocistronic mRNAs (see Discussion). In some cases, the second monocistron might not be translated into protein, as is the case for SL-mTS-3′ small RNA. ORF1, ORF2, ORF3, and ORF4, ORFs; pA, polyadenylation site; tsp, trans-splicing site.http://www.pnas.org/
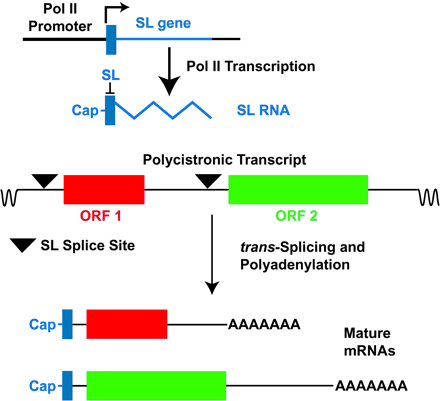
Figure: Transcription of the SL gene (blue) generates SL RNA and trans-splicing of the 39 nucleotide SL (solid blue rectangle) onto polycistronic primary transcripts resolves individual mRNAs. Each SL gene, arranged in a tandem repeat, is transcribed by Pol II from a promoter located immediately upstream of the gene (L-shaped arrow indicates transcriptional start site) and is cotranscriptionally capped. The resulting SL RNA is then trans-spliced onto consensus splice sites (triangles) located on the polycistronic RNAs to generate the 5′ end of each mRNA, and the 3′ end of the mRNA is polyadenylated in a step that is probably associated with trans-splicing at the downstream splice site. ORF 1 (red) and ORF 2 (green) indicate ORFs within the polycistronic RNA that will generate two distinct mRNAs. Intervening sequences that are removed by cis-splicing are exceedingly rare among kinetoplastida species; http://www.pnas.org/
SL2 operons:
(Huang et al., 2001; Kuersten et al., 1997; (Evans and Blumenthal, 2000; Evans et al., 2001). Liu et al., 2001; Liu et al., 2003); (Liu et al., 2003), (Evans and Blumenthal, 2000; Evans et al., 2001).:
In 1989, a second SL was discovered in C. elegans. Huang and Hirsh (1989) reported that the SL at the 5′end of the gpd-3 gene, although it is the same length as the SL found at the 5′ends of other mRNAs, has a different sequence, which they called SL2. Using this sequence, they identified a different SL RNA (SL2 RNA) with a potential secondary structure similar to that of the original SL (now called SL1). SL2 RNA is also present as snRNP; it is bound to Sm antigen and has a TMG cap. Like SL1, SL2 is trans-spliced to a variety of mRNAs. Initial evidence suggested that gpd-3 mRNA receives SL2, but no SL1, whereas other mRNAs receive only SL1. How is this specificity achieved? If SL1 is specific for trans-splicing at the 3′splice sites following outrons, and outrons are simply A + U-rich RNA of any sequence, where is the information to specify SL2?
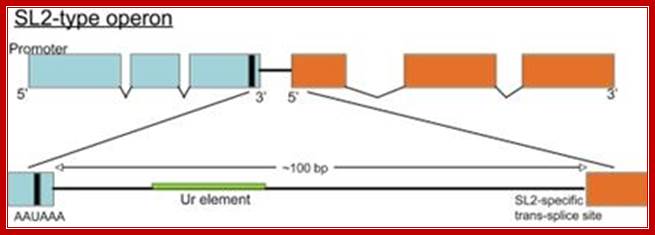
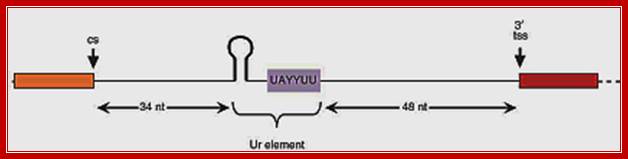
Almost all C. elegans operons are of the SL2-type, shown above. The site of 3' end cleavage and polyadenylation of the upstream gene is determined by the AAUAAA signal, indicated by the black bar. The Ur element serves to protect the downstream RNA from degradation following 3' end cleavage, as well as to attract the SL2 snRNP. Most operons of this type have about 100 bp between the cleavage and polyadenylation site of the upstream gene and the trans-splice site to which SL2 is spliced. Thomas Blumenthal,http://www.wormbook.org/
Signals on the polycistronic pre-mRNA for SL2 type trans-splicing
In eukaryotes one does not find poly cistronic mRNA but some organisms contain such mRNAs, and often they called operons for they have features similar to bacterial operons.� Besides the splice site itself, only two sequences on the pre-mRNA play an important role in SL2-specific trans-splicing: the two presumptive signals for 3' end formation of the gene just upstream. The AAUAAA just 5' of the cleavage site, which is absolutely required for 3' end formation, binds the cleavage and polyadenylation specificity factor (CPSF), and a U-rich sequence just 3' of the cleavage site binds the cleavage stimulatory factor (CStF). CPSF and CStF bind cooperatively to these two sites and together position the site of cleavage. When the AAUAAA is mutated, 3' end cleavage fails to occur, and trans-splicing just downstream becomes less efficient and less specific for SL2. Nevertheless, AAUAAA is not required for SL2 trans-splicing since SL2 trans-splicing downstream still occurs in its absence. CPSF bound to AAUAAA may act by facilitating binding of CstF to the U-rich sequence or by catalyzing 3' end formation itself, which in turn may play a role in SL2 trans-splicing. The protein that binds to the U-rich sequence appears to play the major role in SL2 trans-splicing. When this sequence is mutated, 3' end formation can still occur, albeit somewhat less efficiently, but downstream trans-spliced product fails to accumulate. Thus, either transcription terminates or the downstream RNA is degraded from the site of 3' end formation. When 3' end formation is prevented by mutating both the AAUAAA and the U-rich sequence, downstream product is restored, but all of it is trans-spliced to SL1. The protein that binds to the U-rich element, presumably CstF, performs two functions: it blocks exonucleolytic degradation of the downstream RNA beginning at the site of 3' end cleavage and it recruits the SL2 snRNP. The MS2 phage coat protein tethered to this region in place of the U-rich sequence can substitute for the first function, but not the second.
How does the protein bound to the U-rich element attract the SL2 snRNP? Presuming the protein is CstF, it appears to do so by direct interaction. Antibodies to CstF have been found to immunoprecipitate the SL2 snRNP from C. elegans embryo extracts, and the region of SL2 RNA required for SL2 identity is also required for the CstF interaction, although it is not required for snRNP function. The current model for polycistronic pre-mRNA processing involves first 3' end cleavage at the upstream gene by conventional mechanisms. This sets in motion the chain of events that leads to SL2 trans-splicing: the free 5' phosphate at the cleavage site is attacked by an exonuclease that is then stopped at the U-rich element by the CstF that had bound there for 3' end formation. CstF attracts, or is already bound to, the SL2 snRNP, which splices to the trans-splice site just downstream.
Chloroplast biogenesis and PsaA Trans splicing:
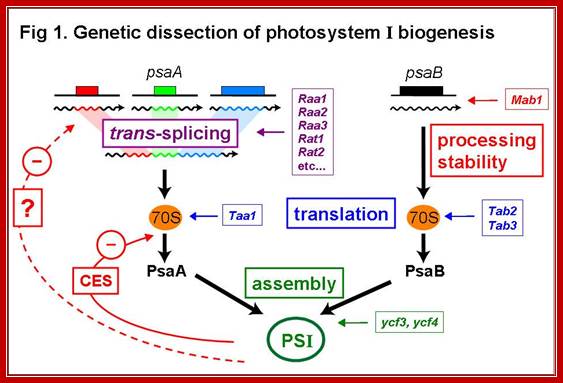
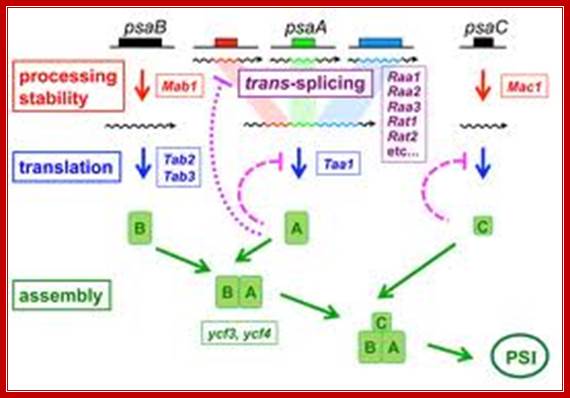
Genetic dissection of Photosystem I Biogenesis; Analysis of psaA and Psab; http://www.molbio.unige.ch/
Description and explanation of the above figures:
The mRNA of PsaA is assembled from three separate transcripts encoded at different loci in the chloroplast genome. The maturation of PsaA mRNA requires two steps of splicing in trans which depend on at least fourteen nuclear genes. We have identified some of these genes (Raa1, Raa2, Raa3) and shown that the corresponding splicing factors belong to large ribonucleoprotein complexes. In mutant�s deficient for PsaA trans-splicing, the precursor of psaA-exon1 is over expressed. This over accumulation is also apparent with chimeric reporter genes under the control of the promoter and 5�untranslated region of psaA-exon1. We have begun to study the mechanism of the underlying negative regulatory feedback loop.
Authors are also studying other proteins that are imported into the chloroplast and are necessary for the maturation of psaB mRNA (Mab1) or for its translation (Tab2, Tab3) and for the translation of psaA (Taa1)
Long-distance splicing:
Long distance trans splicing; http://www.pnas.org/
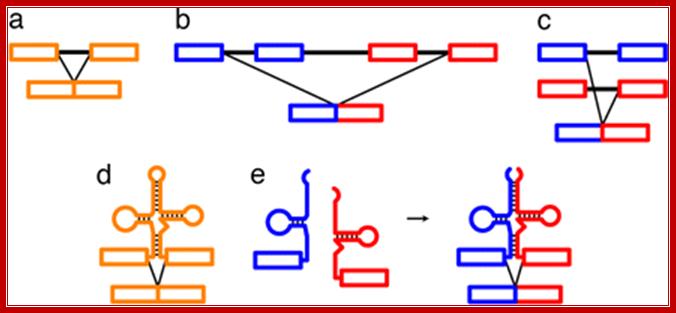
RNA splicing pathways observed in eukaryotes (a�c) and engineered in yeast by Di Segni et al. �(d and e). (a) Canonical pre-mRNA cis-splicing yielding mRNA from pre-mRNA composed of two exons (orange boxes) and an intron (black line). (b) A transcription-induced chimera generated by read-through transcription from one gene (blue) into a downstream gene (red), followed by cis-splicing. (c) Trans-splicing of exons from two distinct pre-mRNAs (red and blue boxes). (d) Cis-splicing of a pre-mRNA embedded with a permuted tRNA. (e) Trans-splicing of two hybrid RNAs each composed of an mRNA fragment and a half tRNA. Note that in the tRNAs the aminoacyl acceptor stem is at the top and the anticodon loop is at the bottom (d and e)
The products of such trans-splicing:
Chimeric mRNAs can be generated by trans-splicing of pretRNA sequences joined to two different pre mRNAs. (A) Scheme of the trans-splicing reaction. Splicing sites are those determined by the tRNA domain formed by the two complementary tRNA halves joined to pre mRNAs 1 and 2. (B) Trans-splicing of STE3 and STE2 pre mRNAs. The STE3 mRNA 5′-half, joined to the SUP4 tRNA 3′-half, associates with the STE2 mRNA 3′-half, joined to the SUP4 tRNA 5′-half, and reconstitutes the mature tRNA domain, including the BHB motif. Each component is encoded on a different PYX vector. Upon splicing (long arrow) at the tRNA-splice sites, a chimeric mRNA (5′-STE3-STE2-3′) is produced, containing a stem�loop insertion. Short arrows indicate splice sites or junction. (C) RNA of yeast cells containing both plasmids was reverse-transcribed and amplified with appropriate primers. The DNA was electrophoresed on an agarose gel (lane 1) together with size markers (M); the DNA band indicated by an arrow was excised and purified. (D) The chromatogram of the sequence of the DNA purified from the gel is shown. The two NcoI sites (CCATGG), corresponding to the sequence at the bottom of the inserted stem, are underlined; the sequence between the two sites corresponds to the inserted stem�loop. The left sequence of the chromatogram matches STE3, and that on the right matches STE2. The short arrow indicates the splice junction.
Yeast tRNA splicing involves cutting and rejoining from two transcripts of Genes VII
Trans splicing; http://genes.atspace.org/
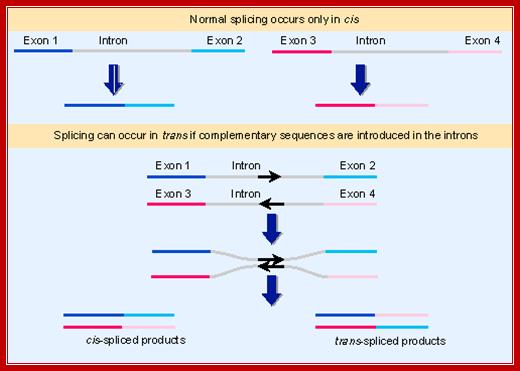
In both mechanistic and evolutionary terms, splicing has been viewed as an intramolecular reaction, amounting essentially to a controlled deletion of the intron sequences at the level of RNA. In genetic terms, splicing occurs only in cis. This means that only sequences on the same molecule of RNA can be spliced together. The upper part of Figure shows the normal situation. The introns can be removed from each RNA molecule, allowing the exons of that RNA molecule to be spliced together, but there is no intermolecular splicing of exons between different RNA molecules. We cannot say that trans splicing never occurs between pre-mRNA transcripts of the same gene, but we know that it must be exceedingly rare, because if it were prevalent the exons of a gene would be able to complement one another genetically instead of belonging to a single complementation group.
Some manipulations can generate trans-splicing. In the example illustrated in the lower part of the above Fig, complementary sequences were introduced into the introns of two RNAs. Base pairing between the complements should create an H-shaped molecule. This molecule could be spliced in cis, to connect exons that are covalently connected by an intron, or it could be spliced in trans, to connect exons of the juxtaposed RNA molecules. Both reactions occur in vitro.
Although trans-splicing is rare, it occurs in vivo in some special situations. One is revealed by the presence of a common 35 base leader sequence at the end of numerous mRNAs in the trypanosome. But the leader sequence is not coded upstream of the individual transcription units. Instead it is transcribed into an independent RNA, carrying additional sequences at its 3′ end, from a repetitive unit located elsewhere in the genome. The figure shows that this RNA carries the 35 base leader sequence followed by a 5′ splice site sequence. The sequences coding for the mRNAs carry a 3′ splice site just preceding the sequence found in the mature mRNA
If the leader and the mRNA are connected by a trans-splicing reaction,
the 3′ region of the leader RNA and the 5′ region of the mRNA will
in effect comprise the 5′ and 3′ halves of an intron. If splicing
occurs by the usual nuclear mechanism, a 5′�V2′
link should form by a reaction between the GU of the 5′ intron
and a branch sequence near the AG of the 3′ intron. Because the two parts
of the intron are not covalently linked, this generates a Y-shaped molecule
instead of a lariat (Sutton and Boothroyd, 1986,Murphy et al., 1986).
Evolution and Role of Trans-Splicing:
(Blumenthal and Steward, 1997); (Blumenthal and Steward, 1997;. Lall et al., 2004). (Lall et al., 2004; Maroney et al., 1995, (Keiper et al., 2000), (Hannon et al., 1990, (Ferguson et al., 1996), (Krause and Hirsh, 1987), (Stein et al., 2003, (Ross et al., 1995) (Stein et al., 2003). (T. Blumenthal, unpublished):
Trans-splicing occurs throughout the nematodes, and there is striking conservation of the SL sequence, whereas the portions of the SL RNAs downstream of the splice site have diverged. The role that SL plays in the cell, however, is not known. In C. elegans the SL tends to be spliced very close to the initiating methionine codon (often immediately adjacent), so it seems likely to play a role in translation initiation; The TMG cap present at the 5' end of the SL becomes the 5' end of trans-spliced mRNAs. A TMG cap stimulates translation in nematodes, at least when it is present at the 5' end of the SL sequence. �In C. elegans, variants of the cap binding translation initiation factor, eIF4E can recognize the TMG cap.
In Ascaris, the SL sequence in the DNA is needed for transcription of the SL RNA gene, which may be one reason why it has been so highly conserved. �Although the roles the SL sequence itself may perform are unknown, trans-splicing is in fact required for viability. Its required role could be a positive effect such as providing a sequence that can facilitate translation initiation, mRNA stability or localization, or it could be required for suppression of a negative effect such as inhibition of translation initiation by AUG codons in the outron.
The C. elegans genome contains 110 SL1 RNA genes on the 1 kb tandem repeat that also contains the genes for 5S rRNA. In contrast, the genome contains only18 dispersed SL2 RNA genes, which specify a variety of variant SL2 RNAs. Some of these have different SL2 sequences, and these have been given different names, such as SL3, SL4 etc. Nonetheless, they are all variants of SL2 and they are used randomly at SL2-accepting trans-splice sites). The C. briggsae genome also contains 18 SL2 RNA genes, and all 36 genes from the two species descended from four primordial SL2 RNA genes present in their last common ancestor.
SMaRT� RNA Applications:
VIRxSYS�s SMaRT� technology provides an effective means to repair and reprogram specific pre-mRNAs and has broad applications (therapeutic and diagnostic), each governed by the nature of the coding sequences included in a specifically engineered PTM. PTMs that encode correct, functional genetic sequences or toxins have been used in RNA therapies to repair mutant mRNAs from genes associated with human disease or to kill diseased cells (e.g. cancer cells).
Because the coding domain can consist of one or more exons, a single PTM can be used to correct all the mutations in the region of the target RNA that is replaced.