Self-Splicing Viroidal RNAs:
Discovery:
Viroids were identified as part of the effort to isolate the putative virus that caused potato spindle tuber disease. This disease leads to a second-year harvest of spindly, deformed potatoes. In the early 1960s, William B. Raymer, a plant pathologist at the US Department of Agriculture and Agricultural Research Service (ARS), initiated the project that would lead to the discovery of the viroid. At the time, he was working in the ARS Potato Diseases Investigation Laboratory in Beltsville, Maryland. Raymer and another ARS pathologist, Muriel O'Brien, came up with the first bioassay for the infectious agent that caused potato spindle tuber disease. They were able to transmit the pathogen to tomatoes, which exhibited stunted growth within two weeks of infection (ARS Timeline). With this ready source of diseased leaves, Raymer and O'Brien sought to isolate the virus using standard centrifugation techniques. However, Raymer found that he could not isolate the infectious fraction even at centrifugal forces high enough to sediment all previously identified viruses (Diener 2003), (the mcmanus lab). Flores R, Gas ME, Molina-Serrano D, Nohales MÁ, Carbonell A, Gago S, De la Peña M, Daròs JA
http://people.csail.mit.edu/
Nomenclature:
The term viroid (meaning "virus-like") was coined by Theodor Diener, the discoverer of these small circular plant pathogens (Diener 2003). Viroids are divided into two families named for the prototypical members of each family: Pospiviroidae (from potato spindle tuber viroid, PSTVd) and Avsunviroidae (from avocado sun blotch viroid, ASBVd).
Viroids are characterized by their naked ssRNA genome in circular module, but the genome is base paired extensively with few unpaired open regions (bulges) and it gives an appearance of a rod. There are no capsid protein to encapsidate them; they don’t have open reading frame to generate any protein or proteins. They are present in cells and transmitted either through insect vectors or through mechanical means. The viroids and virusoids cause great damage to a variety of commercially important crop plants and cause economic disaster in many areas of the world. Their mechanism of disease manifestation is more or less similar to interferon-stimulated activation of a specific kind of kinase by a ds RNA.
The Avsunviroidae family contains two genera: Avsunviroids (from ASBVd) and Pelamoviroids (from peach latent mosaic viroid, PLMVd) (Flores et al 1998)
· These Viroidal RNAs, by themselves, don’t code for any proteins for its replication, but replicate using host factors and enzymes.
· Replication of Viroidal RNAs is through what is called rolling circle module. First, the (+) RNA strand generates (-) RNA strand to form, what is called, in molecular parlance, replicative form of DNA (RF form). Then the (–) strand is used for the (+) strand synthesis, which is actually generated as concatameric form, where a long (+) RNA strand is produced where each genomic segment is found as a string of segments.
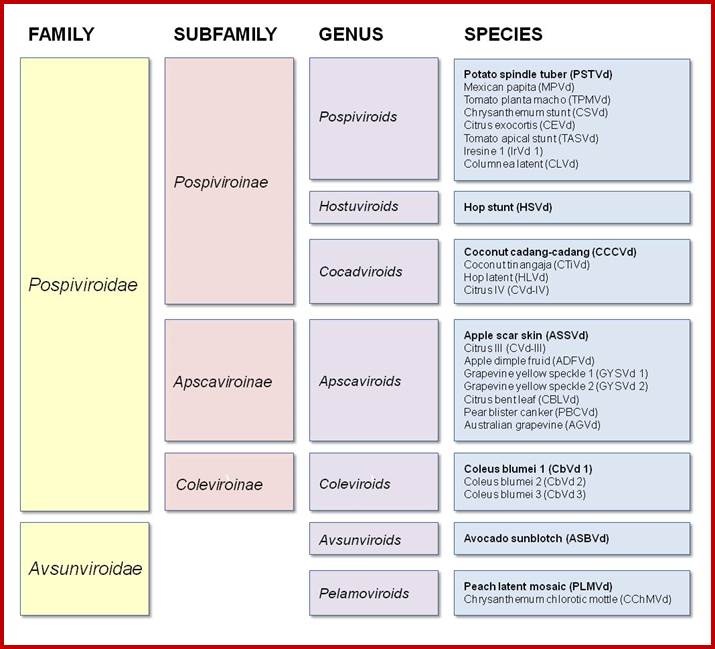
Viroidal RNA; http://mcmanuslab.ucsf.edu/
· These Viroidal RNAs, by themselves, don’t code for any proteins for their replication, but replicate using host factors and enzymes.
· Replication of Viroidal RNAs is through what is called rolling circle module. First, the (+) RNA strand generates (-) RNA strand to form, what is called, in molecular parlance, replicative form of DNA (RF form). Then the (–) strand is used for the (+) strand synthesis, which is actually generated as concatameric form, where a long (+) RNA strand is produced where each genomic segment is found as a string of segments.
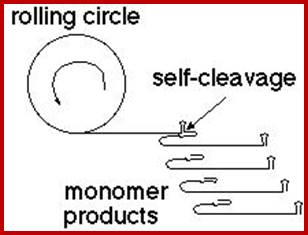
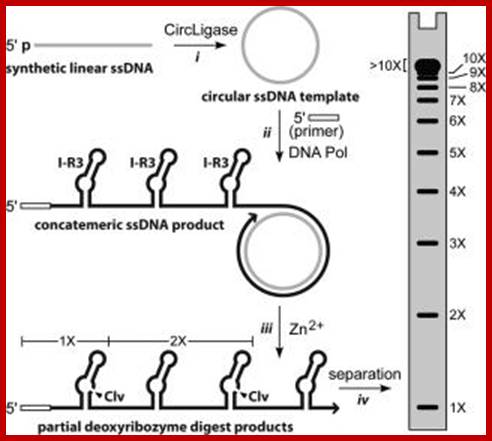
Production of single-stranded DNAs by self-cleavage of rolling-circle amplification products; Production of single-stranded DNAs by self-cleavage of rolling-circle amplification products; Upon incubation of this circular DNA template with Phi 29 DNA polymerase and a complementary DNA primer, a single-stranded concatemer consisting of multiple linear copies of the sequence complementary to the circular template is produced (Figure 2, ii). At the junction of each DNA repeat resides the sequence corresponding to the I-R3 class I self-hydrolyzing deoxyribozyme. However, the deoxyribozyme does not cleave until it is exposed to the conditions needed for robust self-processing (50 mM HEPES, pH 7.0 at 23°C; 100 mM NaCl; 2 mM ZnCl2) (Figure 2, iii). By halting the reaction before all of the deoxyribozymes have cleaved, a mixture of products representing a range of unit-length DNAs is generated. This mixture of deoxyribozyme cleavage products can serve as an ssDNA ladder when separated by gel electrophoresis (Figure 2, iv) or by other methods that can separate large ssDNAs. Hongzhou Gu1,2 and Ronald R. Breaker; http://www.biotechniques.com; Rolling circle mode of viroidal RNA replication
![]()
· The cleaving of such concatameric RNA, at exact positions to generate individual genomes and ligating the ends to produce circular, yet double stranded rod like genome, is performed by its genomic RNA itself.

Viroids: http://people.csail.mit.edu/
· This kind of molecular structure is absolutely safe for no other RNase enzymes capable of digesting from any end or from any internal sites.
· Self-cleavage and ligation requires first cleavage and then ligation, which requires ends such as 5’ OH group and 3’ cyclic phosphate group on the ribose. In order to execute this marvelous catalytic activity, the + form of Viroidal RNA at the region of cleavage and ligation should generate a structure that facilitates self-catalysis. It is an extraordinary event indeed for any system.
· The region of cleavage in the (+) RNA has structural motif called hammer headed structure. The hammer-headed structure emanates from a certain but highly conserved sequence, which has three short stems and a loop; it is this complementary sequence that generates hammer-headed structural configuration.
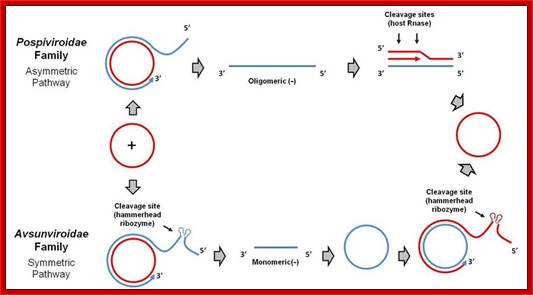
Viroids of the Pospiviroidae family are localized to the nucleus, while viroids of the Avsunviroidae family localize to chloroplasts. The nuclear viroids are dependent upon host DNA-dependent RNA-polymerase II for replication (Rackwitz et al 1981), and the chloroplastic viroids are dependent on either nuclear-encoded but chloroplast-localized DNA-dependent RNA-polymerase (NEP) (Navarro et al 2000) or plastid-encoded DNA-depedent RNA polymerase (PEP) (Motard et al 2008); http://mcmanuslab.ucsf.edu/
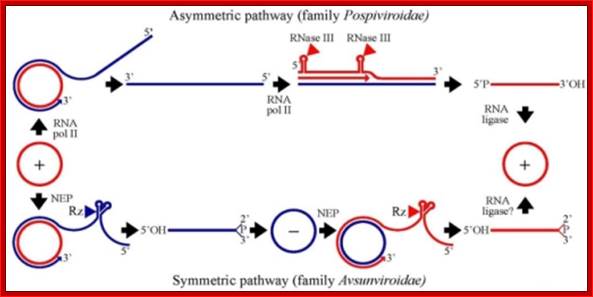
Viroid replication proceeds through an RNA-based rolling-circle mechanism with three steps that, with some variations, operate in both polarity strands: i) synthesis of longer-than-unit strands catalyzed by either the nuclear RNA polymerase II or a nuclear-encoded chloroplastic RNA polymerase, in both instances redirected to transcribe RNA templates, ii) cleavage to unit-length, which in the family Avsunviroidae is mediated by hammerhead ribozymes embedded in both polarity strands, while in the family Pospiviroidae the oligomeric RNAs provide the proper conformation but not the catalytic activity, and iii) circularization. The host RNA polymerases, most likely assisted by additional host proteins, start transcription from specific sites, thus implying the existence of viroid promoters. Cleavage and ligation in the family Pospiviroidae is probably catalyzed by an RNase III-like enzyme and an RNA ligase able to circularize the resulting 5' and 3' termini. Whether a chloroplastic RNA ligase mediates circularization in the family Avsunviroidae, or this reaction is autocatalytic, remains an open issue. Flores R, Gas ME, Molina-Serrano D, Nohales MÁ, Carbonell A, Gago S, De la Peña M, Daròs JA - Viruses (2009), ;http://openi.nlm.nih.gov/
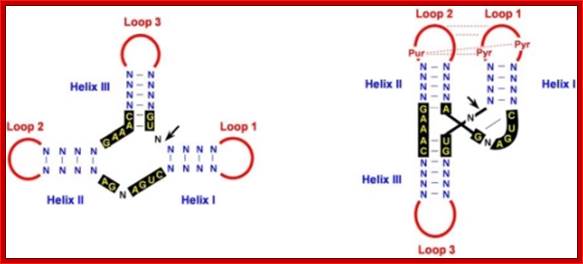
Hammer headed structures; http://openi.nlm.nih.gov/
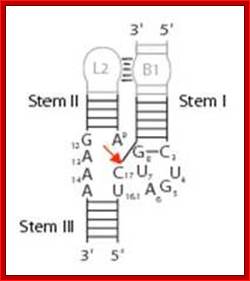
Self cleavage
Hammerhead Ribozyme:
Cleavage to unit-length, which in the family Avsunviroidae is mediated by hammerhead ribozymes embedded in both polarity strands, while in the family Pospiviroidae the oligomeric RNAs provide the proper conformation but not the catalytic activity, and iii) circularization. The host RNA polymerases, most likely assisted by additional host proteins, start transcription from specific sites, thus implying the existence of viroid promoters. Cleavage and ligation in the family Pospiviroidae is probably catalyzed by an RNase III-like enzyme and an RNA ligase able to circularize the resulting 5' and 3' termini. Whether a chloroplastic RNA ligase mediates circularization in the family Avsunviroidae, or this reaction is autocatalytic, remains an open issue; Flores R, Gas ME, Molina-Serrano D, Nohales MÁ, Carbonell A, Gago S, De la Peña M, Daròs JA ; http://openi.nlm.nih.gov/
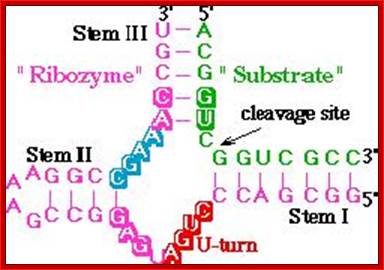
Hammer headed ribozyme sequence and cleavage site. The green colored RNA sequences are the substrate for cutting,\; and the red blue colored RNA is the ribozyme. www.2014.igem.org
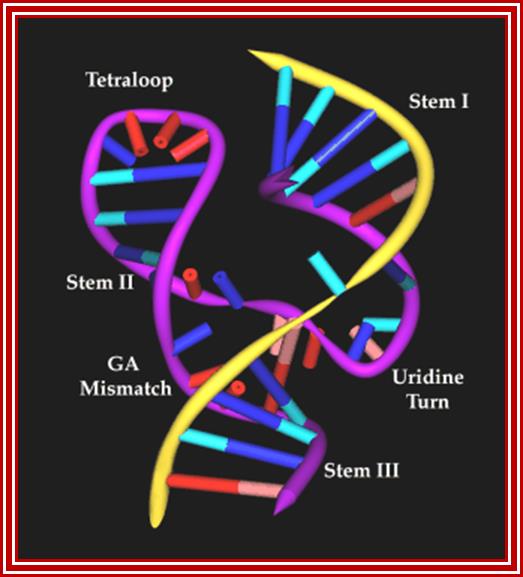
The hammerhead ribozyme (RNA/DNA) structure by Pley, Flaherty and Mckay(1994), purple ribbon shows the RNA ribozyme where as the yellow ribbon represents DNA strand; .http://academic.brooklyn.cuny.edu/
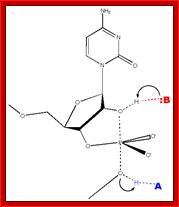
Chemistry of self cleavage
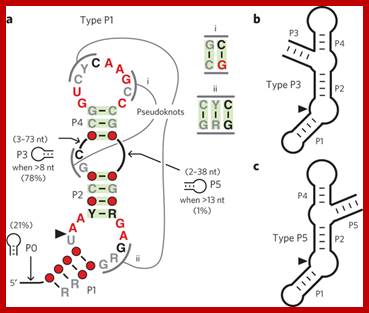
A widespread self-cleaving ribozyme class is revealed by bioinformatics; Ribozymes are noncoding RNAs that promote chemical transformations with rate enhancements approaching those of protein enzymes. Although ribozymes are likely to have been abundant during the RNA world era, only ten classes are known to exist among contemporary organisms. We report the discovery and analysis of an additional self-cleaving ribozyme class, called twister, which is present in many species of bacteria and eukarya. Nearly 2,700 twister ribozymes were identified that conform to a secondary structure consensus that is small yet complex, with three stems conjoined by internal and terminal loops. Two pseudoknots provide tertiary structure contacts that are critical for catalytic activity. The twister ribozyme motif provides another example of a natural RNA catalyst and calls attention to the potentially varied biological roles of this and other classes of widely distributed self-cleaving RNAs.; Adam Roth etal; http://www.nature.com
· Under in vitro conditions, if a synthetic RNA with the said conserved sequence is provided with RNA of suitable sequence as a substrate, the synthetic RNA cleaves the substrate RNA exactly in the position which generates 5’OH and 2’-3’cyclic phosphate groups on ribose moiety.
· Whenever or wherever such sequences produce a hammer headed structure, the RNA itself performs cleavage and ligation reactions simultaneously. In this process a hammer headed structure is a must. For its formation it requires 3 stem loops in which 13 ntds are conserved and located in the center of the structure.
· Self cleavage is not unique to viroidal RNAs, even eukaryotic mRNA go through such self splicing of introns, of course aided by the binding of of spliceosomes.
· The cat-ions required for this process are Mg^2+. Magnesium ions actually in such structural conformations cross-link the lower strand C with the substrate C in its 3-D configuration or conformation. Magnesium confers C and C;
--GUC GGUC---- =upper strand in hammer headed conformation.
---AGCUCCA---- = the lower strand as an enzyme.
· Magnesium, with its divalent cat ionic state, binds to both of the substrate C and enzyme C, that is, it links Cytidine to Cytidine. It may also link to Uridine. In this state, it extracts proton from the 2’OH of target Cytidine (i.e. from its ribose group), then it attacks directly to the labile phosphodiester bond and cleaves it to generate a cyclic 5’OH group and a 2’ to 3’ cyclic phosphate at its ribose moiety. The released segment can easily be circularized by reacting 2’-O-P-O-3’cyclic phosphate end with that of 5’ OH group of the same molecule.
· In Newts, Ntopthalamus viridescens, which is a salamander, presence of self-cleaving plant Viroidal RNA segments, have been discovered. The newt genome has many repetitious 330 bp blocks organized in tandem repeats in head to tail mode.
-------------->I-------------->I--------------- >I-------------->I--------------->
Concatameric RNA
Some of these bocks are transcribed into large precursor RNAs and the same are exported into cytoplasm. In cytoplasm they are self cleaved by the mechanism that Viroidal RNAs have used Hammer headed conformation or folding, to produce exactly 330 ntds long fragment.